Abstract
Even though mesenchymal stem cells (MSCs) are known for cartilage regeneration, their therapeutic efficacy needs to be enhanced. In the present study, we produced genome-edited silent information regulator 2 type 1 (Sirt1)-overexpressing MSCs, and evaluated their therapeutic potential in a damaged cartilage mouse liver fibrosis model. The Sirt1 gene was successfully inserted into a ‘safe harbor’ genomic locus in amniotic mesenchymal stem cells (AMMs), and the chondrogenic properties of the Sirt1 gene overexpressing AMMs (AMM/S) were characterized using quantitative PCR and histology. Therapeutic potentials were investigated in a collagen-induced arthritis (CIA) mouse model. Chondrocyte-differentiated AMM/S expressed cartilage-specific genes and were positive for Safranin O staining. Transplantation of AMM/S attenuated CIA progression and suppressed T helper (Th)-17 cell activation while increasing the Treg cell population in CIA mice. Pro-inflammatory factors, such as interleukin (IL)-1β, IL-6, monocyte chemoattractant protein (MCP)-1, and tumor necrosis factor (TNF)-α were significantly decreased in AMM/S-injected joint tissues. In conclusion, genome-edited AMM/S may represent a safe and alternative therapeutic option for the treatment and repair of damaged cartilage, or in inflammatory joint arthritis.
Keywords: anti-inflammation, cell therapy, genome editing, mesenchymal stem cells, osteoarthritis
INTRODUCTION
Osteoarthritis (OA) is a chronic joint disease in the elderly population due to the destruction of cartilage and other joint tissues. The pathogenesis of OA is known to be related to pro-inflammatory cytokines such as interleukin-1 (IL-1), IL-6, IL-17, and tumor necrosis factor (TNF) (Hedbom and Hauselmann, 2002). The prevalence of OA increases with age, and patients require long-term treatment including pain-control drugs, physical therapy, and other surgical procedures.
Tissue engineering using stem cells has been of interest in the treatment of OA. However, obstacles remain regarding the therapeutic potential or control of stem cell dysfunction in the environment of host tissues. Thus, more sophisticated or advanced technologies are required to improve therapeutic efficacy. Recently, genome editing technology has attracted attention for its highly specific cellular genome engineering capability. It is possible to precisely modify the genomes of mammalian cells. Guide RNA directs an endonuclease to a specific genomic target and editing cuts the chromosomal DNA in living cells (Jinek et al., 2012). This process enables the activation of endogenous cellular DNA repair pathways and genome editing, such as the addition or disruption of genes.
Strategies for enhancing the therapeutic effects of stem cells in OA have been applied using various growth factors, chemicals, and scaffolding applications (Qasim et al., 2020). Silent information regulator 2 type 1 (Sirt1) plays a role in cartilage extracellular matrix synthesis and promotes cell survival, even under proinflammatory stress (Dvir-Ginzberg and Steinmeyer, 2013). Sirt1 is an epigenetic regulator of particular relevance to OA and is associated with the modulation of aging and caloric intake (Dvir-Ginzberg and Steinmeyer, 2013). In fact, Sirt1-deficient mice exhibit altered cartilage phenotypes (Gabay et al., 2013). In this study, we investigated the therapeutic properties of Sirt1-overexpressing amniotic mesenchymal stem cells (AMM/S) generated using gene editing in a damaged cartilage during inflammatory process.
MATERIALS AND METHODS
Cell culture and mice
Human amniotic mesenchymal stem cells (AMMs) were purchased from Thermo Fisher Scientific (USA). The AMMs were cultured in low-glucose Dulbecco’s modified Eagle’s medium (DMEM; Gibco, USA) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 µg/ml streptomycin (Gibco). Six-week-old male DBA/1 mice were purchased from Orientbio (Korea).
Donor vector construction
Sirt1 was synthesized and inserted into the adeno-associated virus integration site 1 (AAVS1) safe harbor site, targeting the donor vector (System Biosciences, USA) at the NdeI and SalI restriction sites.
Transfection and selection, fluorescence-activated cell sorting (FACS)
AMMs were maintained in DMEM supplemented with 10% FBS. For electroporation, human AMMs were harvested, counted, and 1 × 105 cells were resuspended with 0.6 μg of AAVS1 left Transcription activator-like effector nuclease (TALEN) vector (System Biosciences), AAVS1 right TALE-Nuclease vector (System Biosciences), and AAVS1 HR Donor (System Biosciences) in 10 μl of electroporation buffer. The cells were electroporated using the Neon Transfection System (Thermo Fisher Scientific). Five days after transfection, Sirt1 knock-in cells were selected by incubation with 5 μg/ml puromycin for 7 days. Puromycin-selected cells were resuspended in FACS buffer and sorted as previously described (Choi et al., 2019).
Genomic DNA extraction and junction polymerase chain reaction (PCR)
Genomic DNA was extracted from the cultured cells using a G-spinTM Total DNA Extraction Mini Kit (iNtRON Biotechnology, Korea) according to the manufacturer’s instructions. Next, 120 ng of genomic DNA was amplified by touch-down PCR (36 cycles) and a second-round PCR, as previously described.
Reverse transcription PCR (RT-PCR)
To confirm gene expression, RNA isolation and cDNA synthesis were conducted as previously described (Han et al., 2020). RT-PCR primers were designed and synthesized by Bioneer (Korea) targeting (Table 1).
Table 1.
RT-PCR primer sequences
| Gene | Sequence (5’-3’) | Size (bp) | |
|---|---|---|---|
| GAPDH | Forward | TCTTCACCACCATGGAGAAG | 224 |
| Reverse | CATGAGTCCTTCCACGATAC | ||
| SOX9 | Forward | GAGGAAGTCGGTGAAGAACG | 362 |
| Reverse | GCAGGTACTGGTCAAACTCG | ||
| COMP | Forward | GGAGATCGTGCAGACAATGA | 424 |
| Reverse | GAATCGCACCCTGATGTAGC | ||
| COL10A1 | Forward | CACTACCCAACACCAAGACAC | 495 |
| Reverse | GACGACCAGGAGCACCATA |
Quantitative real-time (qRT)-PCR
qRT-PCR assays were conducted according to previous studies (Choi et al., 2012; Kim et al., 2010). Briefly, total RNA was isolated from cells using RNA-stat (Iso-Tex Diagnostics, USA). The genomic DNA contamination was removed using DNAse (Thermo Fisher Scientific). Extracted RNA was reverse-transcribed using TaqMan reagents (Applied Biosystems, USA) according to the manufacturer’s specifications. The synthesized cDNA was subjected to qRT-PCR using specific primers and probes. RNA levels were quantitatively measured using an ABI PRISM 7000 instrument (Applied Biosystems). Relative mRNA expression was normalized to that of GAPDH expression. The qRT-PCR primers used were as follows: human Sirt1 (Hs01009006_m1), GAPDH (Hs99999905_m1), and mouse IL-1β (Mm00434228_m1), IL-6 (Mm00446190_m1), MCP-1 (Mm00441242_m1), TNF-α (Mm00443258_m1), and GAPDH (Mm99999915_g1). All primers and probe were purchased from Applied Biosystems.
Safranin O staining
After 3 weeks of culture in chondrocyte differentiation medium (Lonza, USA), which consisting of chondrocyte differentiation basal medium, insulin growth factor, transforming growth factor (TGF)-β, Insulin, transferrin and 10% of FBS, the cells were fixed with 4% paraformaldehyde for 10 min and stained using a Safranin O staining kit (ScienCell Research Laboratories, USA) following the manufacturer’s instructions.
Splenocyte co-culture and enzyme-linked immunosorbent assay (ELISA)
Splenocyte co-culture assays have previously been reported (Wu et al., 2016). Briefly, spleens from healthy male DBA/1 mice were harvested, and tissues were minced in phosphate-buffered saline (PBS). Splenocytes were isolated using Ficoll-Hypaque density-gradient centrifugation and suspended in RPMI 1640 medium. To determine the effects of AMM/S on T cells, 1 × 105 AMMs or AMM/S were treated with or without 10 ng/ml TNFα for 1 day and then co-cultured with 1 × 106 splenocytes in RPMI 1640 containing 10% FBS. After 2 days, supernatants from co-cultures were collected and cytokine levels were measured. The cytokine concentration levels in the supernatant or serum were examined using murine IL-10 or IL-17A ELISA kits (R&D Systems, USA) according to the manufacturer’s specifications.
Induction of collagen-induced arthritis (CIA) model and treatment
All experimental protocols were approved by the Institutional Animal Care and Use Committee of the Catholic Kwandong University (CKU-01-2020-013). Bovine type II collagen (Chondrex, USA) was emulsified at a ratio 1:1 with complete Freund’s adjuvant (Chondrex) containing 2 mg/ml heat-killed Mycobacterium tuberculosis. Six-week-old male DBA/1 mice (OrientBio) received a primary immunization, followed by booster immunization on day 21 using the same concentration of bovine type II collagen and incomplete Freund’s adjuvant (Chondrex). Injection was conducted intradermally at the base of the tail. The severity of arthritis was observed for 28 days after the first injection. The severity of arthritis was monitored and scored as determined by hind paw swelling and clinical scoring: 0 = normal, 1 = slight swelling, 2 = moderate swelling, 3 = severe swelling and reversible joint immobility, and 4 = severe swelling and irreversible joint immobility (Delgado et al., 2001). To evaluate therapeutic efficacy, 1 × 106 AMMs and AMM-S were injected intraperitoneally twice a week when the arthritis score reached 3 or more.
Flow cytometric analysis
Th17 and Treg cell populations were examined using flow cytometry. The antibodies used were phycoerythrin-conjugated rat anti-mouse CD4 (eBioscience, USA), fluorescein isothiocyanate (FITC)-conjugated rat anti-mouse IL-17A (eBioscience), and FITC-conjugated rat anti-mouse Foxp3 (eBioscience). Analyses were conducted using CellQuest software (BD, USA).
Measurement of cytokines
The concentrations of cytokines were examined using Platinum ELISA kits (eBioscience) and murine IL-17A ELISA kits (R&D Systems). IL-17A from serum was quantified according to the kit manufacturer’s instructions.
Histological analysis
To obtain cartilage and paw samples, mice were euthanized with CO2 gas and tissues were obtained by dissection. The limbs and paws were fixed overnight in 4% paraformaldehyde and decalcified. Cartilage and paw tissues were embedded in optimal cutting temperature compound and cryosectioned to 10 µm. To analyze inflammation, sections were stained with H&E. To confirm cartilage destruction in the CIA model, the specimens were stained using Safranin O (ScienCell Research Laboratories) or Alcian blue (Newcomer Supply, USA) following the manufacturer’s instructions. Cartilage degradation was measured by using a degradation score and the following scale: from 0 to 3 was defined as either no loss or complete loss of staining for proteoglycans (Wu et al., 2016). To analyze inflammation, H&E staining was performed according to previous study (Jang et al., 2020). The degree of inflammation was scored as reported previously (Razawy et al., 2020) using the following scale: 0, no inflammation; 1, minimal inflammation; 2, mild inflammation; 3, moderate inflammation; and 4, severe inflammation.
Statistical analysis
All data are presented as mean ± SD. Statistical analyses were performed using Student’s t-test for comparisons between two groups, and ANOVA with Bonferroni’s test with multiple comparison correction using SPSS (ver. 12.0; SPSS, USA). Data with P < 0.05 were considered to be statistically significant.
RESULTS
Targeted knock-in of Sirt1 in AMMs
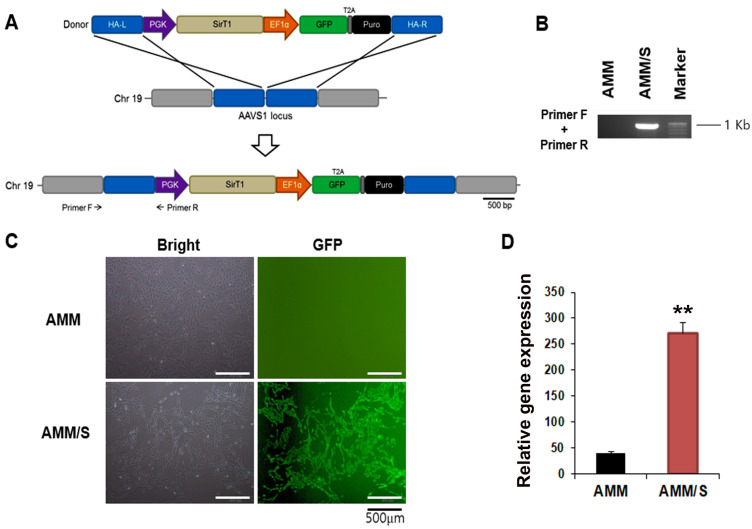
To produce a stem cell line overexpressing Sirt1 using gene editing, we used TALEN-mediated gene integration methodology. The targeting donor plasmid carried the phosphoglycerate kinase (PGK) promoter-driven Sirt1, and elongation factor-1 alpha (EF1α) promoter-driven green fluorescent protein [GFP]-T2A-puromycin, and was designed to be integrated into AAVS1 on chromosome 19 (Fig. 1A). AMMs were transfected with the donor plasmid and a pair of TALENs. The transfected cells (<10% of GFP-positive cells) underwent in vitro selection with puromycin and were additionally isolated using FACS (98.9% GFP-positive cells) (Fig. 1B). To confirm genomic integration of the donor plasmid into the AAVS1 site, we verified the genomic DNA using standard PCR, followed by touch-down PCR (Don et al., 1991; Korbie and Mattick, 2008). The correct insertion of the donor plasmid was detected via 5-junction fragment (960 bp) amplification (Fig. 1C). Finally, Sirt1 expression by AMM/S was confirmed via qPCR, and Sirt1 mRNA levels were found to be significantly increased in AMM/S compared to the control AMMs (Fig. 1D). A successfully gene-edited AMM/S line was subsequently used in all experiments.
Fig. 1. Generation of AMM/S cell line using TALEN gene editing.
(A) Schematic diagram of the donor vector carrying TGF-β1-bearing donor plasmid DNA. An expression cassette containing the PGK promoter-driven TGF-β1 and EF1α promoter-driven GFP-T2A-puromycin was inserted into the AAVS1 site via homology-directed repair. The locations of primers for junction detection are indicated (primers F and R). HA-L, left homology arm; HA-R, right homology arm; PGK, phosphoglycerate kinase promoter; EF1α, elongation factor-1 alpha promoter; Puro, puromycin. (B) Inserted donor plasmid was confirmed using junction PCR. (C) GFP-expressing AMM/S. Transfected cells were selected using puromycin followed by FACS. Scale bars = 500 μm. (D) Expression levels of TGF-β1 were examined using qPCR. **P < 0.01, n = 4.
In vitro chondrogenic differentiation potential of AMM/S
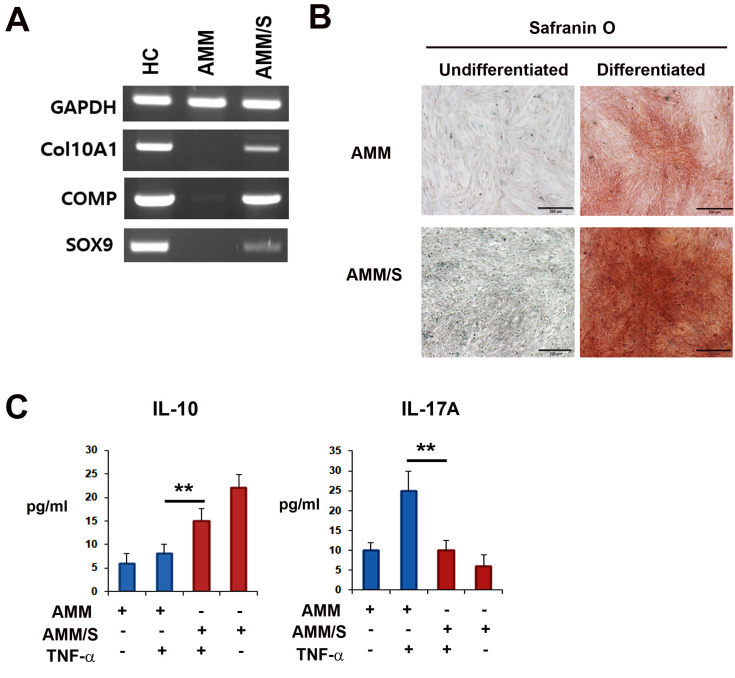
To investigate their chondrogenic potential, we performed qPCR analysis. Intriguingly, chondrogenically-differentiated AMM/S expressed chondrogenic-specific markers, such as type A1, 10 collagen (COL10A1), cartilage oligomeric matrix protein (COMP), and SRY-box transcription factor (SOX) 9 in AMM/S (Fig. 2A). However, undifferentiated AMM did not express these chondrogenic-specific markers. Additionally, we examined chondrogenic potential using histological experiments. Safranin O staining results revealed that AMM/S had a greater degree of staining than AMM (Fig. 2B).
Fig. 2. Chondrogenic and immunomodulatory potential of AMM/S.
(A) The expression of chondrogenic-specific genes was measured using RT-PCR. (B) Histological analysis was performed using Safranin O staining. Scale bars = 200 μm. (C) AMMs or AMM/S were treated or not with TNF-α, and then co-cultured with splenocytes. The supernatants were collected and the concentrations of IL-10 and IL-17A were measured by ELISA, n = 4 each, **P < 0.01.
Next, to evaluate the in vitro immunomodulatory effects of AMM/S on T cells, AMMs or AMM/S were treated with or without TNF-α, and then co-cultured with splenocytes. Supernatants from co-cultures were evaluated for cytokine levels after 2 days. Interestingly, ELISA results revealed that co-culture with AMM/S revealed significantly higher IL-10 and lower IL-17A levels in the culture supernatant compared with AMMs (Fig. 2C).
Therapeutic properties of AMM/S in murine CIA model
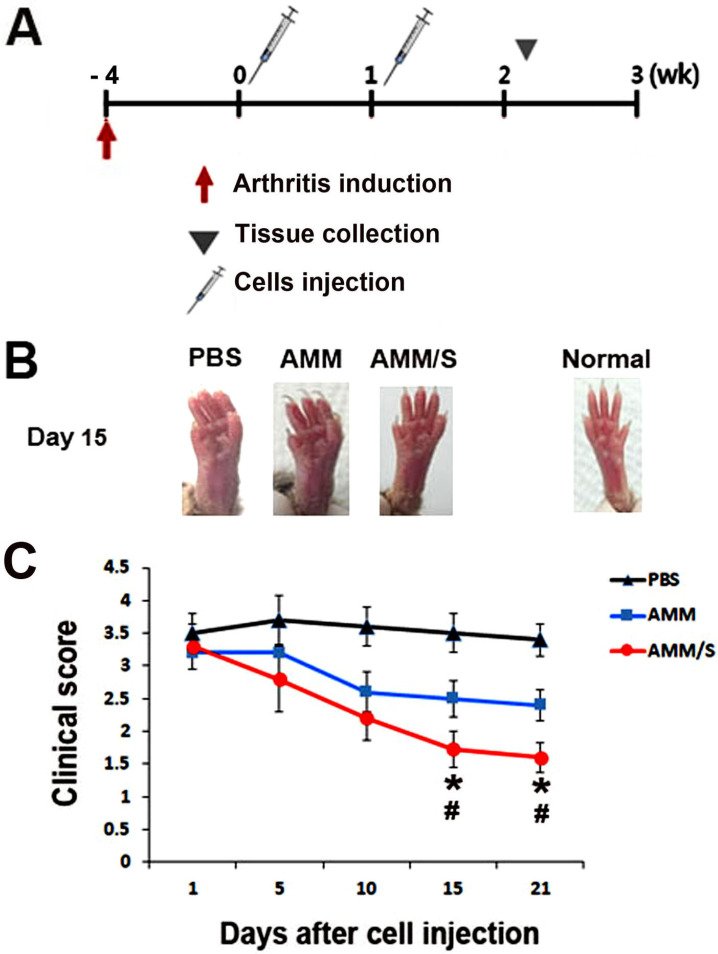
To investigate the therapeutic potential of AMM/S for restoring damaged cartilage in vivo, we induced a CIA mouse model using bovine type II collagen. The arthritis clinical score was evaluated after cell injection (Fig. 3A). Interestingly, there were significantly lower arthritis clinical scores at 15 and 21 days in the AMM/S-injected group compared with the control PBS- or AMM-injected groups (Figs. 3B and 3C).
Fig. 3. AMM/S transplantation results in protection against disease progression.
(A) Schematic representation of the procedure for the induction of arthritis, cell injection, and the collection of specimens. (B) Representative images of paws after cell injection. (C) Quantification of arthritis scores. Arthritis scores were measured using severely swelled paws. *P < 0.05 AMM/S vs AMM, # P < 0.01 AMM/S vs PBS, n = 5 each.
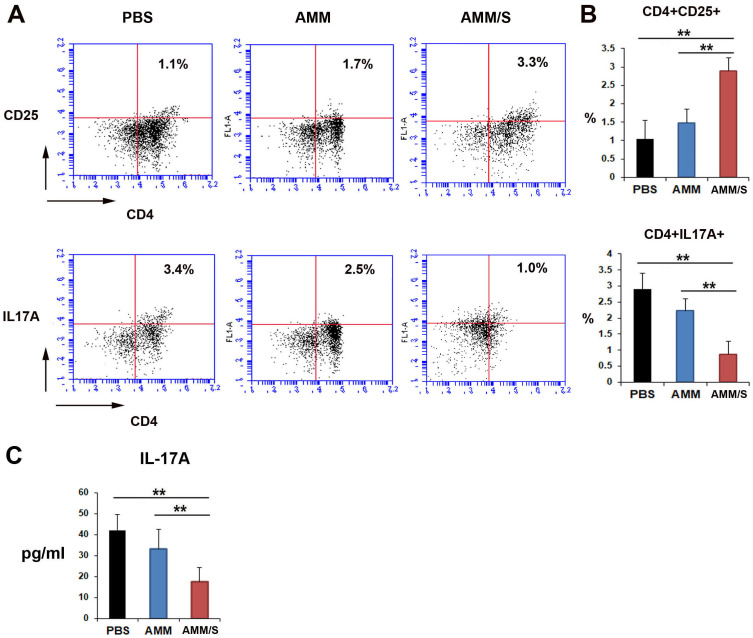
Next, to investigate possible mechanisms underlying the favorable therapeutic effects of AMM/S, we examined the influence of T cells after cell injection. The population of Treg cells increased after AMM/S was introduced into the bloodstream of mice compared to that in the PBS- or AMM-injected cohorts (Figs. 4A and 4B). However, the Th17 cell population was significantly decreased in AMM/S-injected mice compared with PBS control or AMM-injected mouse groups (Figs. 4A and 4B). We also determined the concentration of IL-17A after injection of cells in CIA mice. IL-17A levels were significantly decreased in AMM/S-injected CIA mice (Fig. 4C).
Fig. 4. AMM/S transplantation influences Treg and Th17 cell populations in CIA mice.
(A) Representative figures illustrating flow cytometric data for the identification of Treg and Th17 cells. (B) Quantitative data for Treg and Th17 cells were measured using murine CIA blood samples 2 weeks after injection of cells. n = 5 each; **P < 0.01. (C) Concentration of IL-17A in serum of CIA mice 2 weeks after injection of cells. n = 5 each; **P < 0.01.
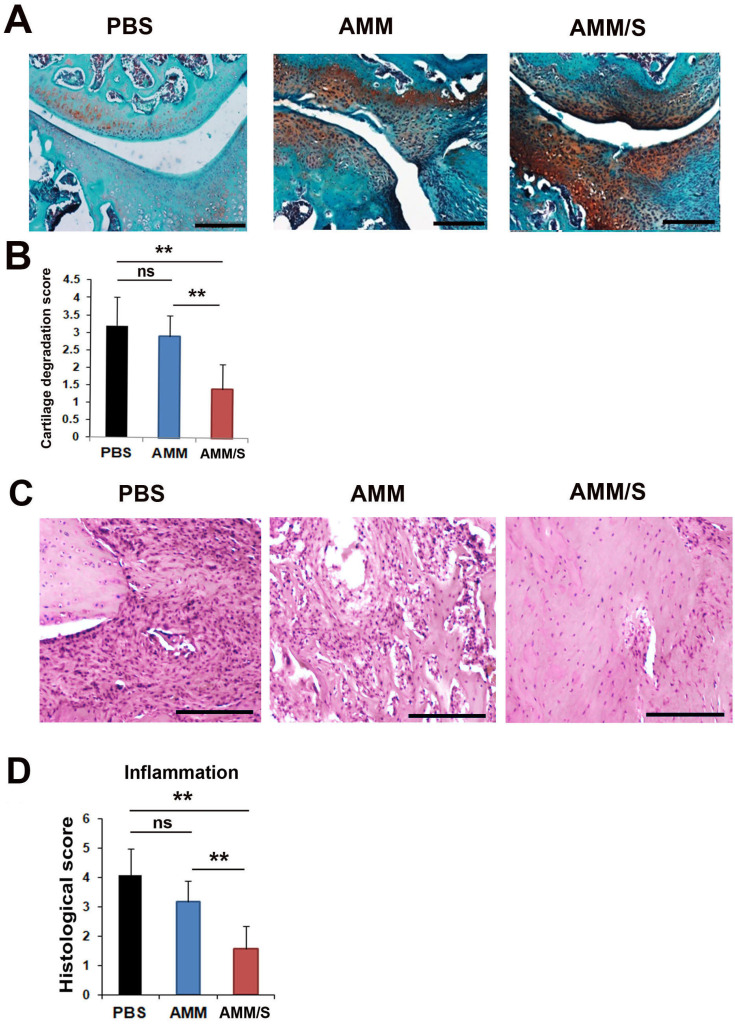
Histological analysis in joints from CIA mice
To investigate the protection of cartilage degradation in vivo, mouse joint tissues were stained with Safranin O. Such staining can detect the expression of proteoglycans in cartilage. The AMM/S-injected group exhibited increased proteoglycan expression in articular cartilage compared with the PBS- or AMM-injected control groups, suggesting protection against cartilage damage (Figs. 5A and 5B). Next, to evaluate inflammatory responses in joint tissues, H&E staining was conducted. Histological analysis showed that AMM/S-injected joint tissues showed significantly lower inflammatory cell infiltration than PBS- or AMM-injected control joint tissues (Figs. 5C and 5D).
Fig. 5. Histological staining of joints in CIA mice after injection of cells.
(A) Proteoglycan expression was identified using Safranin O staining of the joints of CIA mice after injection of MSCs. Scale bars = 200 μm. (B) Quantification of cartilage degradation scores. Loss of proteoglycans was identified after staining for proteoglycans. n = 5 each; **P < 0.01. ns, not significant. (C) Representative images of H&E-stained sections of joint tissues. Scale bars = 200 μm. (D) Quantification of inflammatory response histological scores. Mononuclear cell infiltration and inflammatory pathological scores were measured after cell transplantation. AMM/S-injected Treg and Th17 cells exhibited low mononuclear cell infiltration and normal cartilage surface morphology. n = 5 each; **P < 0.01.
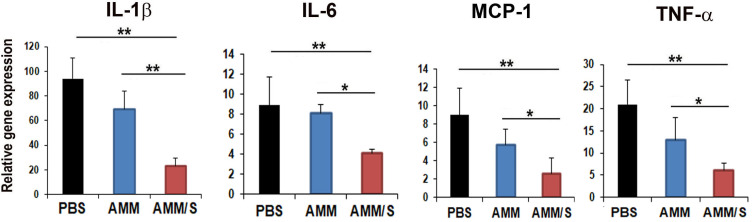
Inflammatory gene expression in joints from CIA mice
To further elucidate the therapeutic mechanisms of AMM/S, we analyzed the expression levels of pro-inflammatory factors in joint tissues after injection of cells. Interestingly, pro-inflammatory factors, such as IL-1β, IL-6, MCP-1, and TNF-α were significantly decreased in AMM/S-injected joint tissues compared with PBS- or AMM-injected joint tissues (Fig. 6).
Fig. 6. AMM/S transplantation suppresses inflammation in mouse joints.
The expression levels of inflammation genes in joint tissues were measured using qPCR. AMM/S transplantation revealed low expression of representative pro-inflammatory factors and high expression of anti-inflammatory factors. n = 5 each; *P < 0.05, **P < 0.01.
DISCUSSION
Using targeted gene editing, we generated Sirt1-overexpressing MSCs for enhanced cartilage protection or regeneration. In this study, we first demonstrated that genome-edited AMM/S tended to protect against arthritis progression through their therapeutic effects on chondrogenesis and T lymphocyte activation. These results indicated that Sirt1-overexpressing MSCs could be an alternative therapeutic option for the treatment of OA.
Genome editing technology is a highly useful tool that can control endogenous gene expression with minimal off-target effects. A recent report showed that CRISPR genome editing of cytokine receptor genes in stem cells promotes cell survival and tissue deposition in inflammatory environments (Farhang et al., 2017). Erythropoietin (EPO) gene-edited MSCs successfully secrete high levels of EPO (Benabdallah et al., 2010) and HGF-overexpressing MSCs generated by gene editing exhibit improved therapeutic properties in an animal model of ischemia (Chang et al., 2016). These data indicate that genome editing can provide an ideal platform for the generation of specific, safe, and novel stem cell lines. Thus, these reports prompted us to investigate the therapeutic potential of genome-edited stem cells.
Over the past decade, our laboratory has sought to identify the best sources of stem cells. Among these, AMMs offer great benefits as a source of allogeneic stem cells, as they can be readily obtained without any ethical concerns, and express low levels of immunological responses (Alviano et al., 2007). In addition, they have high cell proliferative, survival, and trans-differentiation properties (Alviano et al., 2007; Tsuji et al., 2010). Specifically, we found that AMMs are the best MSC source for genome editing because of their high transfection efficiency compared to other stem cell sources.
Even though stem cells have become attractive tools for tissue regenerative applications, controversy regarding their low therapeutic efficacy has become a major obstacle. The aim of this study was to explore genome engineering technologies to address the challenges involved in stem cell therapy. To identify favorable factors driving anti-inflammation and regeneration, we examined one of the important factors, Sirt1, involved in cartilage repair. Recently, it has been reported that Sirt1 promotes chondrogenic differentiation and reduces MSC apoptosis (Ou et al., 2020). In addition, activation of Sirt1 inhibits inflammation and degradative processes in cartilage (Backesjo et al., 2009; Buhrmann et al., 2014). Sirt1 is an enzyme that deacetylates transcription factors that contribute to cellular regulation (Peng et al., 2011). Sirt1-deficient mice exhibit an altered cartilage phenotype (Gabay et al., 2013) and overexpression of Sirt1 inhibits osteoarthritic gene expression in human chondrocytes (Matsushita et al., 2013). In line with these reports, our results also revealed that AMM/S exhibited higher chondrocyte differentiation in vitro and AMM/S transplantation decreased levels of pro-inflammatory factor in joints of CIA mice. These results may indicate that Sirt1 plays important roles in the pathogenesis of OA.
For cell-based therapies, another important function of MSCs is immunomodulation. The release of immunomodulatory factors, such as hepatocyte growth factor (HGF), IL-10, and TGF-β1, protect cartilage in the synovium (Kehoe et al., 2014). In addition, it has been reported that T cells regulate arthritic pathogenesis. Immunomodulatory factors induce Treg cells and suppress inflammation by reducing proliferation of Th17 cell (Aggarwal and Pittenger, 2005). However, Th17 cells are involved in inflammatory processes, and Th17/Treg cell imbalances could present problems in arthritis therapy. Interestingly, AMM//S transplantation resulted in a significant suppression of Th17 and protection of cartilage against damage. These data indicated that therapeutic functions of AMM/S might affect reciprocal regulation of Th17/Treg cell imbalances in CIA mice.
In summary, this study revealed that Sirt1 overexpression after AMM gene editing resulted in robust therapeutic effects without changes in MSC properties in injured cartilage. Our observations indicated that transplantation of AMM/S involved in the pathogenesis of OA and Sirt1 overexpression might contribute to the prevention of OA and chondrocyte degradation. Further investigations are required to evaluate the efficacy and safety of AMM/S for treating joint OA in the context of clinical settings.
ACKNOWLEDGMENTS
This work was financially supported through National Research Foundation (NRF) of Korea grants funded by the Korean Government (No. NRF-2016R1A2B4012683) and the research fund of Catholic Kwandong University for Dr. S.-W. Kim; the research fund of Dong-A University for Dr. S. Han; and an NRF of Korea grant funded by the Korean government (No. NRF-2020R1C1C101316611) for Dr. D.-S. Chae.
Footnotes
AUTHOR CONTRIBUTIONS
S.W.K. conceived and designed the experiments. D.S.C. and S.H. performed the experiments. S.W.K. analyzed the data. M.K.L. contributed reagents, materials, and analysis tools. S.W.K. wrote the paper.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Aggarwal S., Pittenger M.F. Human mesenchymal stem cells modulate allogeneic immune cell responses. Blood. 2005;105:1815–1822. doi: 10.1182/blood-2004-04-1559. [DOI] [PubMed] [Google Scholar]
- Alviano F., Fossati V., Marchionni C., Arpinati M., Bonsi L., Franchina M., Lanzoni G., Cantoni S., Cavallini C., Bianchi F., et al. Term Amniotic membrane is a high throughput source for multipotent Mesenchymal Stem Cells with the ability to differentiate into endothelial cells in vitro. BMC Dev. Biol. 2007;7:11. doi: 10.1186/1471-213X-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backesjo C.M., Li Y., Lindgren U., Haldosen L.A. Activation of Sirt1 decreases adipocyte formation during osteoblast differentiation of mesenchymal stem cells. Cells Tissues Organs. 2009;189:93–97. doi: 10.1159/000151744. [DOI] [PubMed] [Google Scholar]
- Benabdallah B.F., Allard E., Yao S., Friedman G., Gregory P.D., Eliopoulos N., Fradette J., Spees J.L., Haddad E., Holmes M.C., et al. Targeted gene addition to human mesenchymal stromal cells as a cell-based plasma-soluble protein delivery platform. Cytotherapy. 2010;12:394–399. doi: 10.3109/14653240903583803. [DOI] [PubMed] [Google Scholar]
- Buhrmann C., Busch F., Shayan P., Shakibaei M. Sirtuin-1 (SIRT1) is required for promoting chondrogenic differentiation of mesenchymal stem cells. J. Biol. Chem. 2014;289:22048–22062. doi: 10.1074/jbc.M114.568790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.K., Kim P.H., Cho H.M., Yum S.Y., Choi Y.J., Son Y., Lee D., Kang I., Kang K.S., Jang G., et al. Inducible HGF-secreting human umbilical cord blood-derived MSCs produced via TALEN-mediated genome editing promoted angiogenesis. Mol. Ther. 2016;24:1644–1654. doi: 10.1038/mt.2016.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B., Chun E., Kim S.Y., Kim M., Lee K.Y., Kim S.J. Notch-induced hIL-6 production facilitates the maintenance of self-renewal of hCD34+ cord blood cells through the activation of Jak-PI3K-STAT3 pathway. Am. J. Pathol. 2012;180:351–364. doi: 10.1016/j.ajpath.2011.09.030. [DOI] [PubMed] [Google Scholar]
- Choi J.S., Jeong I.S., Han J.H., Cheon S.H., Kim S.W. IL-10-secreting human MSCs generated by TALEN gene editing ameliorate liver fibrosis through enhanced anti-fibrotic activity. Biomater. Sci. 2019;7:1078–1087. doi: 10.1039/c8bm01347k. [DOI] [PubMed] [Google Scholar]
- Delgado M., Abad C., Martinez C., Leceta J., Gomariz R.P. Vasoactive intestinal peptide prevents experimental arthritis by downregulating both autoimmune and inflammatory components of the disease. Nat. Med. 2001;7:563–568. doi: 10.1038/87887. [DOI] [PubMed] [Google Scholar]
- Don R., Cox P., Wainwright B., Baker K., Mattick J. 'Touchdown' PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvir-Ginzberg M., Steinmeyer J. Towards elucidating the role of SirT1 in osteoarthritis. Front. Biosci. (Landmark Ed.) 2013;18:343–355. doi: 10.2741/4105. [DOI] [PubMed] [Google Scholar]
- Farhang N., Brunger J.M., Stover J.D., Thakore P.I., Lawrence B., Guilak F., Gersbach C.A., Setton L.A., Bowles R.D. (*) CRISPR-based epigenome editing of cytokine receptors for the promotion of cell survival and tissue deposition in inflammatory environments. Tissue Eng. Part A. 2017;23:738–749. doi: 10.1089/ten.TEA.2016.0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay O., Zaal K.J., Sanchez C., Dvir-Ginzberg M., Gagarina V., Song Y., He X.H., McBurney M.W. Sirt1-deficient mice exhibit an altered cartilage phenotype. Joint Bone Spine. 2013;80:613–620. doi: 10.1016/j.jbspin.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J.H., Han S., Jeong I.S., Cheon S.H., Kim S.W. Minicircle-based GCP-2 ex vivo gene therapy enhanced the reepithelialization and angiogenic capacity. J. Tissue Eng. Regen. Med. 2020;14:829–839. doi: 10.1002/term.3049. [DOI] [PubMed] [Google Scholar]
- Hedbom E., Hauselmann H.J. Molecular aspects of pathogenesis in osteoarthritis: the role of inflammation. Cell. Mol. Life Sci. 2002;59:45–53. doi: 10.1007/s00018-002-8404-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang K.H., Hwang Y., Kim E. PARP1 impedes SIRT1-mediated autophagy during degeneration of the retinal pigment epithelium under oxidative stress. Mol. Cells. 2020;43:632–644. doi: 10.14348/molcells.2020.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehoe O., Cartwright A., Askari A., El Haj A.J., Middleton J. Intra-articular injection of mesenchymal stem cells leads to reduced inflammation and cartilage damage in murine antigen-induced arthritis. J. Transl. Med. 2014;12:157. doi: 10.1186/1479-5876-12-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.W., Kim H., Cho H.J., Lee J.U., Levit R., Yoon Y.S. Human peripheral blood-derived CD31+ cells have robust angiogenic and vasculogenic properties and are effective for treating ischemic vascular disease. J. Am. Coll. Cardiol. 2010;56:593–607. doi: 10.1016/j.jacc.2010.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbie D.J., Mattick J.S. Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nat. Protoc. 2008;3:1452–1456. doi: 10.1038/nprot.2008.133. [DOI] [PubMed] [Google Scholar]
- Matsushita T., Sasaki H., Takayama K., Ishida K., Matsumoto T., Kubo S., Matsuzaki T., Nishida K., Kurosaka M., Kuroda R. The overexpression of SIRT1 inhibited osteoarthritic gene expression changes induced by interleukin-1beta in human chondrocytes. J. Orthop. Res. 2013;31:531–537. doi: 10.1002/jor.22268. [DOI] [PubMed] [Google Scholar]
- Ou X., Ying J., Bai X., Wang C., Ruan D. Activation of SIRT1 promotes cartilage differentiation and reduces apoptosis of nucleus pulposus mesenchymal stem cells via the MCP1/CCR2 axis in subjects with intervertebral disc degeneration. Int. J. Mol. Med. 2020;46:1074–1084. doi: 10.3892/ijmm.2020.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Yuan Z., Ling H., Fukasawa K., Robertson K., Olashaw N., Koomen J., Chen J., Lane W.S., Seto E. SIRT1 deacetylates the DNA methyltransferase 1 (DNMT1) protein and alters its activities. Mol. Cell. Biol. 2011;31:4720–4734. doi: 10.1128/MCB.06147-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qasim M., Le N.X.T., Nguyen T.P.T., Chae D.S., Park S.J., Lee N.Y. Nanohybrid biodegradable scaffolds for TGF-beta3 release for the chondrogenic differentiation of human mesenchymal stem cells. Int. J. Pharm. 2020;581:119248. doi: 10.1016/j.ijpharm.2020.119248. [DOI] [PubMed] [Google Scholar]
- Razawy W., Asmawidjaja P.S., Mus A.M., Salioska N., Davelaar N., Kops N., Oukka M., Alves C.H., Lubberts E. CD4(+) CCR6(+) T cells, but not gammadelta T cells, are important for the IL-23R-dependent progression of antigen-induced inflammatory arthritis in mice. Eur. J. Immunol. 2020;50:245–255. doi: 10.1002/eji.201948112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H., Miyoshi S., Ikegami Y., Hida N., Asada H., Togashi I., Suzuki J., Satake M., Nakamizo H., Tanaka M., et al. Xenografted human amniotic membrane-derived mesenchymal stem cells are immunologically tolerated and transdifferentiated into cardiomyocytes. Circ. Res. 2010;106:1613–1623. doi: 10.1161/CIRCRESAHA.109.205260. [DOI] [PubMed] [Google Scholar]
- Wu C.C., Liu F.L., Sytwu H.K., Tsai C.Y., Chang D.M. CD146+ mesenchymal stem cells display greater therapeutic potential than CD146- cells for treating collagen-induced arthritis in mice. Stem Cell Res. Ther. 2016;7:23. doi: 10.1186/s13287-016-0285-4. [DOI] [PMC free article] [PubMed] [Google Scholar]