Abstract
Metallothionein (MT1M) belongs to a family of cysteine-rich cytosolic protein and has been reported to be a tumor suppressor gene in multiple cancers. However, its role in esophageal carcinoma carcinogenesis remains unclear. In this study, MT1M expression was correlated with tumor type, stage, drinking and smoking history, as well as patient survival. We also studied the regulation and biological function of MT1M in esophageal squamous cell carcinoma (ESCC). We have found that MT1M is significantly downregulated in ESCC tissues compared with adjacent non-cancer tissues. Furthermore, restoration of expression by treatment with the demethylation agent A + T showed that MT1M downregulation might be closely related to hypermethylation in its promoter region. Over-expression of MT1M in ESCC cells significantly altered cell morphology, induced apoptosis, and reduced colony formation, cell viability, migration and epithelial-mesenchymal transition. Moreover, based on reactive oxygen species (ROS) levels, a superoxide dismutase 1 (SOD1) activity assay and protein analysis, we verified that the tumor-suppressive function of MT1M was at least partially caused by its upregulation of ROS levels, downregulation of SOD1 activity and phosphorylation of the SOD1 downstream pathway PI3K/AKT. In conclusion, our results demonstrated that MT1M was a novel tumor-suppressor in ESCC and may be disrupted by promoter CpG methylation during esophageal carcinogenesis.
Keywords: esophageal squamous cell carcinoma, MT1M, PI3K/AKT, SOD1, tumor suppressor
INTRODUCTION
China has the highest incidence of esophageal cancer worldwide. This type of cancer consists of two main histological types: esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC) (Siegel et al., 2020). In Asian countries, especially China, 90% of esophageal cancer cases are identified as ESCC. In low-risk areas, EAC is the most common histologic type (Zeng et al., 2016). Therefore, understanding the mechanisms of ESCC development is of great interest in China. The molecular mechanisms of esophageal carcinogenesis are not clear, but multiple step processes and very complex signaling pathways are involved. Silencing of tumor suppressor genes (TSGs) by genetic and epigenetic pathways has recently been revealed as an important step in esophageal tumorigenesis (Cheng et al., 2012; Ye et al., 2019). Understanding the function and mechanisms of these TSGs in esophageal cancer will help us to find molecular biomarkers to predict the occurrence and outcome of esophageal cancer earlier. Biomarkers for EAC are currently a hotspot for research. However, ESCC is not as well studied as EAC in terms of TSGs. The molecular biomarkers used clinically in ESCC diagnosis are rare. So, study of the function and regulation of silencing TSGs in ESCC is urgently needed.
Metallothioneins (MTs) are a family of cysteine-rich cytosolic proteins with a very low molecular weight (ranging from 6 to 7 kDa). The MT family plays a vital role in metal ion homeostasis and detoxification. MTs are involved in metalloregulatory processes by binding to heavy metals. Specifically, through binding to the heavy metal zinc, MTs participate in regulating cell growth and proliferation (Kumari et al., 1998). Many studies have shown that some members of the MT family, such as MT2, 3 and 4, are highly expressed in different tumors, suggesting that MTs may play a vital role as an oncogene in carcinogenesis (Arriaga et al., 2012; Ferrario et al., 2008; Zheng et al., 2017). However, recent studies have shown that MT1M, a member of the MT1 family, possesses tumor suppressive function in hepatocellular carcinoma (Mao et al., 2012), Papillary thyroid cancer (Chen et al., 2019) and breast cancer (Jadhav et al., 2015). Additionally, the downregulation of MT1M in HCC is an indicator of unfavorable patient prognosis. These recent reports suggested that MT1M, unlike most of its family members, might serve as TSG in some tumor types.
Recently, our group found that in the UALCAN database MT1M is downregulated in ESCC and that low expression of MT1M is associated with poor patient survival, suggesting that MT1M may be a novel tumor suppressor gene in ESCC. Therefore, in the present study, we have investigated the expression and promoter methylation of MT1M in ESCC and explored its tumor suppressive mechanisms in relation to the SOD1/PI3K signaling pathway. Most importantly, we analyzed the correlation between MT1M expression, the clinicopathological characteristics and patient out-come. We have provided experimental evidence demonstrating that MT1M may function as a TSGs and possess potential clinical significance in ESCC. In the future, it could be used as a molecular biomarker for diagnosis and prognosis of ESCC.
MATERIALS AND METHODS
Cell lines and tissue samples
Esophageal cancer cell lines KYSE150, KYSE510, KYSE960 (cell lines were gifts from Chongqing Key Laboratory of Molecular Oncology and Epigenetics, The First Affiliated Hospital of Chongqing Medical University, cells were originally from the Japanese Cancer Research Resources Bank, Japan) were used in this study. Cell lines were maintained in a 1:1 combination of Ham’s F12 and RPMI 1640 media (Gibco-BRL, Germany) with 10% fetal bovine serum (FBS; PAA Laboratories, Austria) and 1% penicillin, streptomycin. Cells were incubated at 37°C in a humidified atmosphere containing 5% CO2. 293T human epithelial cells and BEAS-2B cells were cultured in DMEM with 10% FBS. 293T cell and BEAS-2B cells are used as a positive control.
Fresh NSCLC tissues and tumor-adjacent tissues were obtained from patients who underwent esophagectomy at the Department of Cardiothoracic Surgery in the First Affiliated Hospital of Chongqing Medical University (Patient clinical features were listed in Table 1). This research was approved by the Institutional Ethics Committees of the First Affiliated Hospital of Chongqing Medical University (No. 2019-11) and followed the principles of the Declaration of Helsinki. Patient consent forms were signed by each patient who participated in this study.
Table 1.
Clinicopathological features of 15 ESCC patients
| Clinicopathological features | No. of patients (n = 15) |
|---|---|
| Sex | |
| Male | 11 |
| Female | 4 |
| Age | |
| ≤ 50 y | 3 |
| 51-59 y | 8 |
| ≥ 60 y | 4 |
| Phase | |
| I | 5 |
| II | 7 |
| III | 3 |
| IV | 0 |
| Tumor size | |
| < 3.0 cm | 4 |
| 3.0 to < 5.0 cm | 10 |
| 5.0 to < 7.0 cm | 1 |
| ≥ 7.0 cm | 0 |
| Lymph node metastasis | |
| Present | 12 |
| Absent | 3 |
| Distant metastasis | |
| Present | 2 |
| Absent | 13 |
| Smoking history | |
| Smoker | 11 |
| Non-smoker | 4 |
| Drinking history | |
| Drinking | 9 |
| Never-drinking | 6 |
| Tumor histology | |
| Adenocarcinoma | 0 |
| Squamous cell carcinoma | 15 |
Analyses using online databases
The UALCAN database (http://ualcan.path.uab.edu/) was used to analyze the correlation between MT1M expression and patient survival, the relationships between MT1M expression and ESCC patient clinical signatures and MT1M promotor methylation status in ESCC. The STRING database (http://string-db.org/) was used to explore the possible related genes of MT1M. The threshold se/arch value used for this study was a P value < 0.05.
RNA extraction
Cell lines and tissue samples treated with DNase I was used for RNA was extraction by TRIzol reagent (Molecular Research Center, USA). The RNA concentrations were measured by spectrophotometry and the store temperature was –80°C. By Promega Reverse Transcription System (Promega, USA) RNA was reversely transcribed. Semi-quantitative polymerase chain reaction (PCR) was carried out using Go-Taq DNA polymerase (Promega) and reaction conditions were as we reported before (Ye et al., 2018), GAPDH were used as internal control. Real-time PCR used ABI SYBR green on an ABI 7500 real-time PCR detection system (Applied Biosystems, USA) and conditions were as reported (Ye et al., 2018). GAPDH was used as a loading control.
The primers used were as follows; MT1M: forward primer, 5’-AATAGAACAAGCTGCACAAC-3’; reverse primer, 5’-TGGCTCAGTATCGTATTGAA-3’. The primer for GAPDH was as follows; forward primer, 5’-GGAGTCAACGGATTTGGT-3’; reverse primer, 5’-GTGATGGGATTTCCATTGAT-3’.
Demethylation treatment
KYSE 150 and KYSE960 cell lines (1 × 105 cells/ml) were seeded in 100 mm dishes. As soon as cells settling down, DNA demethylating agent 5-Aza (Sigma-Aldrich, USA) and histone deacetylase inhibitor trichostatin A (TSA; Cayman Chemical, USA) were applied as previously described (Ye et al., 2019). Cells were collected 24 h after TSA treatment and MT1M mRNA were analyzed by reverse transcription PCR (RT-PCR).
Plasmid and generation of stable cell lines
pcDNA3.1(+)-Flag-MT1M was generated as previously described (Li et al., 2014), and the sequence was verified. To establish cells pools stably expressing MT1M, full-length MT1M expression plasmid was transfected into KYSE150and KYSE960 cells by Lipofectamine 2000 system (Invitrogen, USA). The cells were maintained in 350 μg/ml of G418 for 14 days to establish stable cell lines and the ectopic expressing of MT1M were confirmed by RT-PCR and western blot.
Colony-formation assay
KYSE150 and KYSE960 cells (800 cells/well) stably transfected with pcDNA3.1-MT1M or empty vector were plated in a 6-well plates and incubated for 14-21 days, cells were fixed with 4% paraformaldehyde solution (PFA) and stained with crystal violet solution. Images were captured by a camera. Colonies with > 50 cells/colony were counted. All counting was done by three different people, separately.
Cell proliferation assay
Cell proliferation was carried out by MTS assay. Stable MT1M-expressing and empty vector cells were seeded in 96-well plates (3,000 cells/well) with 100 μl of medium. Cells were incubated for 24, 48, or 72 h. Following, 20 μl MTS diluted in 100 μl/well serum free media was applied to each well. Subsequently, after 2 h incubated additionally at 37°C. Absorbance was measured at 490 nm through a microplate reader (Multiskan MK3; Thermo Fisher Scientific, USA). For each group, data from 5 different wells were pooled. All the experiments were performed in triplicate.
Cell migration and invasion assay
Cell migration ability was also demonstrated using Transwell chambers (8-μm pore size; Corning, USA). For migration assay, KYSE150 and KYSE960 cells stably transfected with pcDNA3.1-MT1M or empty vector were harvested and washed twice with phosphate-buffered saline (PBS) by centrifuging 6 min at 800 rpm. Aliquots of 3 × 104 cells/well were diluted in 100 μl serum-free medium and plated directly onto the inserts surface in 24-well plates. The lower chambers added 800 μl culture medium and 10% FBS. The plates were maintained at 37°C. After 24 h, cells passed through the inserts membrane were fixed with and dyed with 4% PFA and 0.1% crystal violet respectively. At last, pictures were taken and quantified under microscope in six random fields, the assay was done 3 times separately.
Flow cytometry analysis of apoptosis
For apoptosis, KYSE150 and KYSE960 cells were transiently transfected with 4 μg pcDNA3.1-MT1M or empty vector as described before, then analyzed using Annexin V-FITC/PI staining. Data were analyzed using CellQuest™ Pro (BD Biosciences, USA).
Three-dimensional culture
Stable MT1M expressing KYSE150 and control cell three-dimensional (3D) culture were carried out as described by Bissell’s protocol (Lee et al., 2007). Briefly, 120 μl/well matrigel (BD MatrigelTM; BD Biosciences) was coated on pre-cooled 24 well plates, and kept at 37°C for at least 30 min. Cells (4 × 104/well) were diluted in culture media with 10% matrigel. Cell mixtures were seed onto the stiffed gel surface. Incubated for 24 h, whole well cells were fixed with 4% PFA and images were taken.
Western blot
Cell lines stable expressing vector or pcDNA3.1-MT1M were washed with ice-cold PBS for 3 times and treated with lysis buffer. Protein sample (40 μg) was separated by SDS-PAGE, transferred to polyvinylidene difluoride membrane, and immune-stained as the manufactory instruction. Bands were detected through ECL detection system. The primary antibodies were used as follows: anti-MT1M antibody (#ab158927; Abcam, USA), PCNA(#ab92592; Abcam), E-cadherin (#ab231303; Abcam), N-cadherin (#ab76011; Abcam), Vimentin (#ab8069; Abcam), SOD1 (#ab51254; Abcam), SOD2 (#13194; Cell Signaling Technology, USA), SOD3 (sc-271170; Santa Cruz Biotechnology, USA), Erk (#4372; Cell Signaling Technology), p-Erk (sc-7383; Santa Cruz Biotechnology), Akt (#4685; Cell Signaling Technology), Nrf2 (sc-365949; Santa Cruz Biotechnology), GPx2 (ab137431; Abcam), p-Akt (sc-4060; Santa Cruz Biotechnology), rabbit polyclonal anti-PI3 kinase p85 alpha (phosphor-Y607, #ab182651; Abcam), cleaved caspase-3 (#9661; Cell Signaling Technology), bcl-2 (#ab32124; Abcam),BAX (#9942; Cell Signaling Technology), and GAPDH (bsm-51010M; BIOSS, China) as a control. The dilution of primary and secondary antibodies was used according to the instructions. The signals were detected with an enhanced chemiluminescence kit (Amersham Pharmacia Biotech, USA) and the band intensities were quantified by Image-Pro Plus 6.0 (Bio-Rad, USA).
Immunofluorescence staining
Cells were cultured on coverslips and transfected with MT1M or empty vector. Forty-eight hours later, cells were fixed by 4% PFA and permeabilized with 0.1% Triton X-100. Cells were blocked for 1 h with 1% bovine serum albumin and incubated with primary antibodies 4°C overnight, the antibody used were: antibody against Flag (#F9291-.2MG; Sigma-Aldrich), SOD1 (#ab51254; Abcam). Nuclei were stained with DAPI. Images were recorded using a fluorescence microscope (Leica DM IRB; Leica, Germany).
SOD1 inhibitor treatment
Cell lines stable expressing vector or pcDNA3.1-MT1M were treated with SOD1 inhibitor LD100 at 100 μM for 24 h. Protein was then extracted and western blotting was examined as described.
Zinc supplementation
To detect the effect of ZnSO4 supplementation on SOD1 signaling, KYSE150 cells were treated with 5 μM ZnSO4 for 24 h, protein were extracted and western blot was applied.
Determination of intracellular ROS level
Intracellular ROS levels were measured using 2’,7’-dichlorofluorescin diacetate (H2DCFDA; Invitrogen) as described in the instructions. Generally, cells stably expressing vector and MT1M were seeded on 12-well plates. Then cells were incubated with medium with 2 mM H2DCFDA for 15 min at 37°C in the dark. The DCF fluorescence intensity was observed and quantified under a fluorescence microscope. Images were captured using Axio Imager Z1 (Zeiss, Germany). The fluorescence intensity were measured using Image J, the quantification was done three separated times.
Statistical analysis
All data are representative of three independent experiments and presented as mean ± SD. SPSS software (ver. 16.0; SPSS, USA) was used for statistical analyses. Student’s t-test was used to evaluate the statistical significance. For all tests, P < 0.05 was considered statistically significant.
RESULTS
MT1M is a downregulated and hypermethylated gene in ESCC
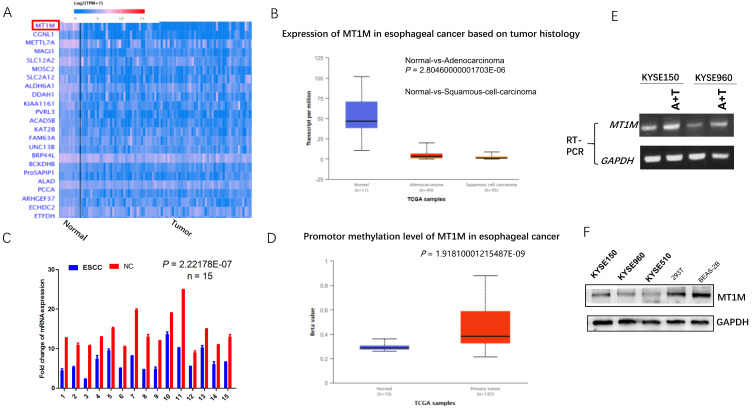
To understand the function of MT1M, we first examined this gene in the UALCAN (TCGA) database and found that MT1M was downregulated in ESCC tissues compared to normal adjacent tissues (Fig. 1A). Additionally, MT1M was downregulated in ESCC compared to EAC (Fig. 1B). Moreover, the MT1M promoter CpG island was highly methylated compared to normal tissue (Fig. 1D). Oka et al. (2009) also reported that MT1M is highly methylated in ESCC and its methylation levels had significant correlations with smoking duration. These findings all suggested that this gene might be a potential tumor suppressor gene in ESCC. To verify this hypothesis, we examined the expression of MT1M mRNA through semi-quantitative RT-PCR in 15 paired samples of human esophageal cancer tissues and the surgical margin. MT1M expression was downregulated in 86.67% (13/15) of primary esophageal tumor tissues compared with their corresponding adjacent tissues (P = 0.017; Fig. 1C). By western blot, we have also demonstrated that MT1M was downregulated in three ESCC cell lines KYSE150, KYSE510, KYSE960 compared to 293T human epithelium cell and human lung epithelial BEAS-2B cells (Fig. 1F). Thus, we determined that MT1M was a gene that was frequently downregulated in esophageal cancer.
Fig. 1. MT1M is downregulated in esophageal cancer.
(A) MT1M is downregulated in ESCC tissue compared to normal adjacent tissue (http://ualcan.path.uab.edu/). (B) MT1M is especially low expression in ESCC (http://ualcan.path.uab.edu/). (C) The expression of MT1M mRNA through semi-quantitative RT-PCR in 15 paired samples of human esophageal cancer tissues and surgical margin. MT1M expression was downregulated in 86.67% (13/15) of primary esophageal tumor tissues compared with their corresponding adjacent tissues (P = 0.017). NC, non-cancerous tissue. (D) MT1M promotor CpG island is hypermethylated compared to normal tissue (http://ualcan.path.uab.edu/). (E) Pharmacological demethylation reagent (A + T) restores the expression of MT1M in KYSE150 and KYSE960 cell lines. (F) MT1M is downregulated in ESCC cell lines compared to 293T human epithelium cell and Human lung epithelial BEAS-2B cells.
To further analyze whether its downregulation was correlated with MT1M promoter methylation status as reported by the Ushijima group and UALCAN database, KYSE150 and KYSE960 cell lines were treated with demethylating agents 5-Aza-2'-deoxycytidine (5-Aza) and trichostatin A (TSA), and restored expression of MT1M was observed by means of RT-PCR (Fig. 1E). Taken together, these data indicated that the MT1M gene was downregulated in ESCC, and that hypermethylation of the promoter’s CpG island was at least part of the underlying mechanism of its down regulation in ESCC.
MT1M expression is correlated with patient clinicopathological features and survival
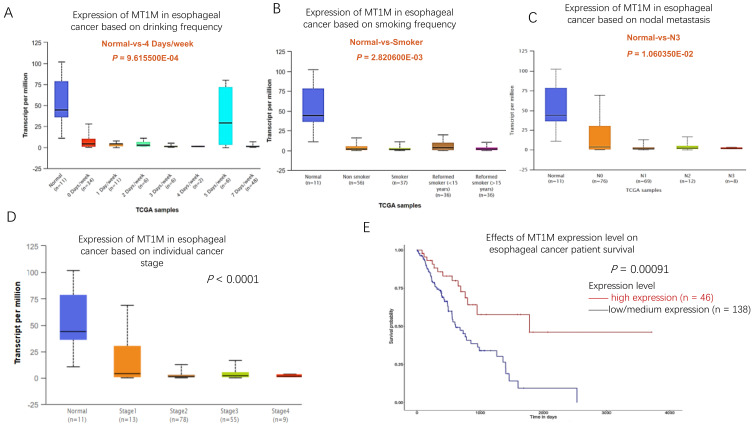
To assess whether there was any correlation between MT1M gene expression and patient clinicopathological features in ESCC samples, the UALCAN database was explored. The results showed that in esophageal cancer, low MT1M expression was associated with high frequency of alcohol use (Fig. 2A). In addition, its low expression was related to patient smoking status, with MT1M being expressed at lower levels in smokers and former smokers than in non-smokers (Fig. 2B). Moreover, low MT1M levels were inversely correlated with lymph node metastasis (Fig. 2C) and tumor stage (Fig. 2D). We further analyzed the relationship between MT1M expression and esophageal cancer patient survival using the UALCAN database and confirmed that in esophageal cancer, including both squamous cell cancer and adenocarcinoma, low MT1M expression was associated with poor patient survival (P = 0.00091; Fig. 2E). These results indicated that MT1M expression might serve as a biomarker to predict the prognosis of esophageal cancer patients.
Fig. 2. MT1M expression level is correlated with patients’ clinical features.
(A) MT1M level is inversely correlated with patients’ drinking frequency (Normal-vs-4 Days/week: P = 9.615500E-04). (B) MT1M level is inversely correlated with patients’ smoking frequency (Normal- vs-Smoker: P = 2.820600E-03). (C) MT1M expression in esophageal cancer is associated with patients’ lymph metastasis status (Normal- vs-N3: P = 1.060350E-02). (D) The expression of MT1M in esophageal cancer is reversely coordinated with patients’ tumor stage (Normal- vs-Stage4: P = 1.060350E-02). (E) Low MT1M expression was associated with poor patient survival in esophageal cancer (P = 0.00091).
Together, these findings indicated that the downregulation of MT1M expression was associated with advanced tumor stage, lymph node metastasis status, former smoking habit and frequent alcohol usage. Thus, low-MT1M expression was a risk factor for poor patient outcome in esophageal cancer. MT1M may serve as a biomarker for esophageal cancer outcome and prognosis.
Ectopic expression of MT1M suppressed ESCC cell growth
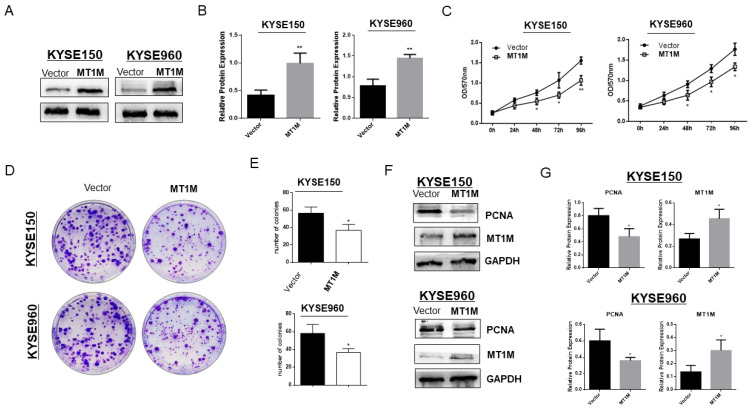
In order to verify whether MT1M possesses tumor suppressive functions as our data and database have predicted, we transfected a pcDNA3.1-Flag-MT1M expressing plasmid into the esophageal cancer cell lines KYSE150 and KYSE960 with low expression levels. Restoration of MT1M after stable transfection was verified using western blot (Figs. 3A and 3B). The tumor growth suppressive effects of MT1M on both KYSE150 (P < 0.01) and KYSE960 (P < 0.05) were demonstrated using a MTS assay (Fig. 3C). The inhibition of tumor growth was further confirmed using a colony formation assay (P < 0.05; Fig. 3D). The colonies in MT1M-expressing cells were significantly reduced in both size and quantity in KYSE150 and KYSE960 compared to control cells (Fig. 3E). Next, using western blotting, we determined that PCNA, a marker reported to be related to tumor growth, was reduced upon MT1M re-expression (Figs. 3F and 3G). These results all suggested that MT1M could inhibit ESCC cell growth.
Fig. 3. Ectopic expression of MT1M suppressed ESCC cell growth.
(A) Western blot confirmed the stable re-expression of MT1M in MT1M-transfected KYSE150 and KYSE960 cell lines. (B) Quantitative analysis of western blots. Asterisks indicate a significant level of proliferation compared with controls (**P < 0.01). (C) MTS assay for cell proliferation on vector- and MT1M-transfected stable cell lines. Asterisks indicate a significant level of proliferation compared with controls (*P < 0.05, **P < 0.01). (D) Ectopic expression of MT1M inhibited colony formation in KYSE150 and KYSE960 cell lines. (E) Quantitative analysis of colony formation. Values are expressed as the mean ± SD from three independent experiments, and the asterisk indicates the statistical significance compared to the controls (*P < 0.05). (F) Western blot indicated proliferation marker PCNA expression was inhibited by MT1M expression. (G) Quantitative analysis of western blots.
MT1M induces apoptosis of ESCC cells
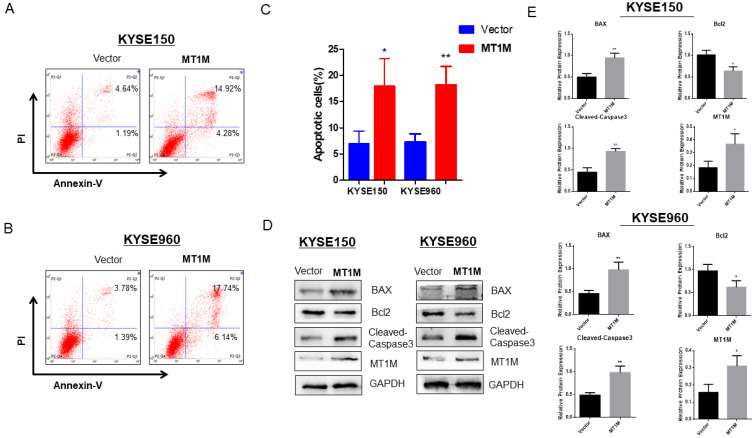
To investigate the impact of MT1M on ESCC cell apoptosis, a flow cytometry assay was used. The results showed that the ectopic expression of MT1M significantly induced apoptosis in KYSE150 (18.21% ± 3.48% vs 7.31% ± 1.57%, P < 0.05) and KYSE960 cell lines (18.04% ± 5.13% vs 6.97% ± 2.43%, P < 0.01) compared to empty vector (Figs. 4A-4C). To further verify the mechanisms of MT1M inducing apoptosis, Western blotting was used. Results suggested that, in both cell lines, MT1M induced caspase-3 cleavage and BAX expression, but inhibited the anti-apoptotic protein Bcl-2 level (Figs. 4D and 4E).
Fig. 4. MT1M induces cell apoptosis in KYSE150 and KYSE960 cell lines.
(A) MT1M Induction of apoptosis detected by flow cytometric analysis with Annexin V-FITC and PI-staining in KYSE150 cells. (B) MT1M Induction of apoptosis detected by flow cytometric analysis with Annexin V-FITC and PI-staining in KYSE960 cells. (C) Quantitative analysis of apoptosis (*P < 0.05, **P < 0.01). (D) Western blot indicated MT1M expression induced caspase-3 cleavage, BAX expression but inhibited anti-apoptotic protein bcl-2 level. (E) Quantitative analysis of western blots (*P < 0.05, **P < 0.01).
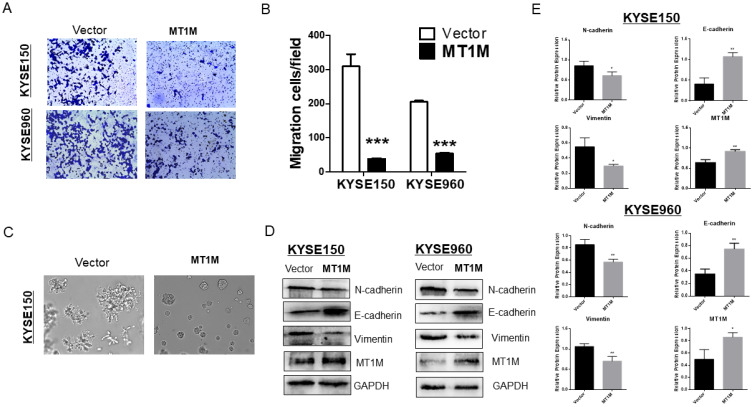
Overexpression of MT1M suppresses ESCC cell migration and epithelial-mesenchymal transition
In order to further explore the tumor suppressive effects of MT1M, a Transwell assay system was used. As shown in Fig. 5A, MT1M significantly suppressed migration through the insert membrane to the lower side of the chamber in 10% FBS media (P˂ 0.001; Fig. 5B). Since MT1M could inhibit cell migration, we next investigated whether MT1M had the potential to regulate the epithelial-mesenchymal transition (EMT). First, in 3D cell culture, we demonstrated that KYSE150 cells transfected with an empty vector were bigger in size, with more tubules and a more invasion phenotype (Fig. 5C, left panel) compared to MT1M expressing cells, which were smaller in size and sphere (Fig. 5C, right panel). These 3D morphology changes suggested that MT1M could inhibit ESCC cell EMT. Further, by Western blot (Fig. 5D) and its quantification results (Fig. 5E), EMT markers vimentin and N-cadherin were downregulated. E-cadherin was elevated after the restoration of MT1M expression. Taken together, these findings demonstrated that MT1M suppressed ESCC cell EMT through cell morphological changes and protein expression mechanisms.
Fig. 5. Ectopic MT1M expression inhibits cell migration and EMT.
(A) Cell migration assay in vector control and MT1M expressing KYSE150 and KYSE960 cell lines by transwell assay. Original magnification, ×10. (B) Quantitative analysis of cell migrated in vector and MT1M expressing KYSE150 and KYSE960 cell line by a 24-transwell system (***P < 0.001). (C) MT1M induces KYSE150 cell line morphology changes in three-dimensional cell culture model. Fixed cells were photographed using phase-contrast microscope at 10× magnification. (D) MT1M expression regulated EMT markers N-cadherin, E-cadherin, Vimentin. (E) Quantitative analysis of western blots (*P < 0.05, **P < 0.01).
Ectopic expression of MT1M inhibits SOD1/PI3K signaling pathway
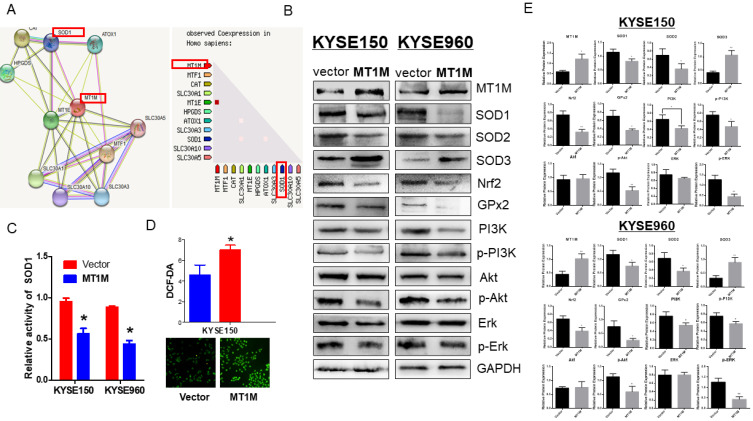
Taken together, through the study of cellular function, we have shown that MT1M may serve as a tumor suppressor gene in ESCC and regulate proliferation, apoptosis and EMT. To clarify the mechanisms underlying the function of MT1M, we first predicted the possible MT1M-interacting genes using the STRING database (https://string-db.org/). Interestingly, we found that SOD1 was a gene with direct interaction with MT1M (Fig. 6A, left panel), and that the expression of these two genes was inversely correlated with each other (Fig. 6A, right panel). SOD1 is an essential antioxidant enzyme that is widely distributed in a cell and provides 80% of total SOD activity. Recently, increasing evidence has shown that SOD1 is a hub gene in reactive oxygen species (ROS) signaling (Carter et al., 2009). Furthermore, phosphatidylinositol 3-kinase-Akt (PI3K-Akt) is one of the main pathways regulated by ROS signaling (Shadel and Horvath, 2015). Additionally, through intracellular ROS level measurement (Fig. 6D), ectopic expression of MT1M significantly evaluated the ROS level, which may, in turn, cause the apoptosis of ESCC cells (Gao et al., 2020). Because SOD1 is the hub gene in the ROS signaling pathway, we next examined the SOD1 activity through a SOD1 activity assay and verified that SOD1 enzymatic activity was suppressed by MT1M re-expression (P < 0.05; Fig. 6C). SOD1 expression and phosphorylation of the downstream targets in the PI3K-Akt signaling pathway such as PI3K, AKT and ERK were also observed to be downregulated by the restored expression of MT1M. Nrf2 and GPx2 are also involved in ROS signaling and both were inhibited by MT1M, confirming that MT1M regulates the ROS pathway (Fig. 6B). Further, Western blotting demonstrated that a SOD1 family member, SOD2, was also downregulated by MT1M expression, however SOD3 was induced by MT1M (Fig. 6B), the quantitative analysis of the Western blot is shown in Fig. 6E. These findings clarified the inhibitory role of MT1M in the ROS-SOD1 signaling pathway.
Fig. 6. Ectopic expression of MT1M inhibits SOD1/PI3K signaling pathway.
(A) SOD1 is a directly interacting gene of MT1M. MT1M expression is inversely correlated with SOD1 level (https://string-db.org/). (B) Expression levels of SOD1, SOD2, SOD3, Nrf2, GPx2, phosphorylation of SOD1 downstream targets PI3K, Akt, ERK were lowered by western blot in vector and MT1M expressing cells. (C) MT1M suppressed SOD1 activity (*P < 0.05). (D) MT1M evaluated intracellular ROS level (*P < 0.05). (E) Quantitative analysis of western blots (*P < 0.05, **P < 0.01).
MT1M is involved in SOD1/PI3K signaling pathway
Moreover, to further demonstrate whether MT1M may be involved in the SOD1/PI3K-AKT axis, the SOD1 inhibitor LD100 and ZnSO4 were used. The results demonstrated that re-expression of MT1M could further enhance the down-regulative effects of LD100 on the SOD1/PI3K-AKT pathway, while zinc supplementation induced MT1M expressing and suppressed SOD1 and its downstream targets (Figs. 7A and 7B). Immunofluorescence results suggested that SOD1 was significantly downregulated by the ectopic expression of MT1M (Fig. 7C). Thus, these findings suggested that MT1M may regulate esophageal cancer development not only by EMT, but also through its involvement in the repression of the ROS-SOD1 pathway (Fig. 7D).
Fig. 7. MT1M is involved in SOD1/PI3K signaling pathway.
(A) SOD1 inhibitor LD100 as well as ZnSO4 were used, re-expressing of MT1M can further enhance the down-regulative effects of inhibitor LD100 on the SOD1/PI3K-AKT pathway, while Zinc supplementation induced MT1M expressing and suppressed SOD1 and its downstream targets. (B) Quantitative analysis of western blots (*P < 0.05, **P < 0.01). (C) MT1M downregulated SOD1 expression by Immuno-fluorescence microscopy. MT1M stained red, which was hardly seen in the vector-transfected cells. SOD1 stained green mainly in the in the cytoplasm and nucleus membrane. (D) Schematic representation of the possible pathogenic mechanism of MT1M/SOD1 axis in ESCC progression.
DISCUSSION
In humans, MTs are encoded by a family of genes located on chromosome 16q13 and include at least 11 functional members (Babula et al., 2012; Krezel and Maret, 2017; Miles et al., 2000).The biological functions of MTs family are well-known as they possess high affinity for heavy metals and protect cells and tissues against heavy metal toxicity. This is vital for cell proliferation and differentiation, and to protect cells against DNA damage, apoptosis (Theocharis et al., 2004). Recently studies have demonstrated that MT expression is associated with the process of carcinogenesis and cancer progression (West et al., 1990). However, the differential expression of MTs depends on the type and differentiation status of the tumor, as well as other environmental stimuli and/or gene mutations (Si and Lang, 2018). For example, some studies have shown that MT expression is upregulated in nasopharyngeal cancer (Du et al., 2006), ovarian cancer (Kawahara et al., 2019), urinary bladder cancer (Tsui et al., 2019), and melanoma (Emri et al., 2013). In contrast, recent studies have shown that the downregulation of MT1M is closely associated with poor prognosis particularly in thyroid cancer (Chen et al., 2019) and hepatocellular carcinoma (Changjun et al., 2018). However, its role in esophageal cancer remains unclear. By searching data in the UALCAN database, we found that family members of MTs are significantly downregulated in esophageal cancer.
The present report aimed to study the role of MT1M in ESCC. Results revealed by semi-quantitative PCR that MT1M was frequently downregulated in 15 cancer tissues compared to adjacent normal tissues at the mRNA level. Western blot also confirmed that in ESCC cell lines MT1M was expressed at low levels compared to 293T human epithelia cells and BEAS-2B normal human bronchial epithelium cells. Data from the UALCAN online database identified MT1M is a frequently methylated gene in ESCC. The loss or downregulation of MT1M was at least partially associated with its CpG methylation, as further verified by UALCAN methylation data and the restored expression of MT1M after treatment with demethylating reagents 5-Aza and TSA. Downregulation and methylation of MT1M in esophageal cancer suggested that it could be a possible candidate TSGs in ESCC. Moreover, exploration of the UALCAN revealed that low MT1M expression was associated with high alcohol usage, smoking habit and advanced tumor stage. Notably, low MT1M expression was associated with poor patient survival. These data suggested the methylation of MT1M in ESCC was tumor specific and could serve as a potential biomarker in esophageal cancer prognosis. Next, we investigated whether MT1M possesses tumor suppressive functions in ESCC cell lines. Ectopic expression of MT1M significantly suppressed cell growth in the esophageal cancer cell lines KYSE150 and KYSE960, verified by colony formation assays and MTS. Flow cytometry analysis and western blots both showed that MT1M induced esophageal cell apoptosis. Transwell assays demonstrated that cell migration was inhibited by MT1M. Cell morphology changes are the initial steps in epithelial-mesenchymal transition (EMT) and play an essential role in cancer migration (Brabletz et al., 2018). 3D cell culture indicated the re-expressing MT1M was able to induce a cell morphological change to a less invasive phenotype compared to empty vector control cells. Western blot demonstrated that the EMT markers Vimentin and N-cadherin were downregulated. E-cadherin was upregulated after restoration of MT1M expression. Taking together, these findings indicated that MT1M possessed a tumor suppressive function in esophageal cancer through inhibition of apoptosis and EMT.
To understand the exact mechanisms of MT1M regulating apoptosis and EMT, we searched for possible signaling pathway involved with MT1M. In the STRING database, we have identified that MT1M directly interacted with SOD1. SOD1 is an essential antioxidant enzyme (Dimayuga et al., 2007). The regulatory role of SOD1 in signaling of the ROS hydrogen peroxide (H2O2) has been increasingly recognized recently (Juarez et al., 2008). ROS plays a vital role as physiological regulator of many intracellular signaling transduction pathways (Cui et al., 2018). However, in cancer cell the role of ROS is a double-edged sword (Schumacker, 2015). In cancer cell, excessive ROS production causes oxidative stress which will in turn increase mitochondrial DNA damage, further activates the oncogenes and tumor progression (Sosa et al., 2013; Trachootham et al., 2009). Paradoxically, as long as intracellular level ROS exceeds the toxicity threshold or the antioxidant system is disturbed, it will induce significant DNA damage and cell apoptosis (Cairns et al., 2011). Moreover, elevated ROS production can also drive programmed cell death in cancer cells (Castaldo et al., 2016). This indicates that cancer cells require a robustly active antioxidant system to prevent cellular damage. Superoxide dismutases (SODs) are enzymes that catalyze the removal of superoxide free radicals (Patel et al., 2017). There are three distinct members of this metalloenzyme family in mammals: SOD1 (Cu/ZnSOD), SOD2 (MnSOD), and SOD3 (ecSOD) (Che et al., 2016). SOD1, SOD2 as well as Nrf2, and GPx2 are antioxidant genes of the ROS signaling pathway (Du et al., 2020; Griess et al., 2017; Rojo de la Vega et al., 2018). These genes can suppress the ROS level and help cancer cell maintain the ROS levels below the toxicity threshold to prevent the cells from undergoing apoptosis or programmed cell death. So, in general, these genes can promote cancer cell proliferation and invasion. However, in contrast to its family members, SOD1 and SOD2, the downregulation of SOD3 has been examined in lung and mammary carcinomas and found to be due to DNA copy number change or hypermethylation in the promoter of methylation (Yamada et al., 2000). Overexpressed SOD3 causes hypoxic accumulation of hypoxia-inducible factor (HIF)-1α in PDA cells and oncogenic VEGF is also suppressed by SOD3 (Sibenaller et al., 2014). SOD3 is considered to possess tumor suppressive function. In accordance with these findings, our results suggested that over-expressing of MT1M suppressed the SOD1 activity and intracellular ROS level. MT1M also inhibited the expression of ROS downstream targets SOD1, SOD2, Nrf2 and GPx2 while SOD3 expression was increased. This confirmed that MT1M regulated ESCC progression through the SOD1-ROS pathway.
Phosphatidylinositol 3-kinase-Akt (PI3K-Akt), mitogen-activated protein kinase (MAPK), AMP-activated protein kinase (AMPK), nuclear factor-κB (NF-κB), and c-Jun N-terminal kinase (JNK) are the downstream signaling pathways regulated by the ROS-SOD1 axis (Bindoli and Rigobello, 2013). All of these signaling pathways promote unfavorable cell growth, proliferation and differentiation of cells, and are hallmarks of tumorigenesis, angiogenesis, and metastasis (D'Autreaux and Toledano, 2007). Among these signaling pathways, PI3K-AKT regulates cell apoptosis and EMT in several cancer types (Hoxhaj and Manning, 2020; Salt et al., 2014). We hypothesized that the underlying mechanisms of the inhibition of cell growth and EMT of MT1M was related to the SOD1/PI3K-Akt axis. Western blotting further demonstrated that MT1M inhibited the SOD1/PI3K-Akt axis and, additionally, MT1M expression strengthened the inhibitory effects of LD100, a SOD1 inhibitor, on the SOD1/PI3K-AKT axis. Zinc supplementation induced MT1M expression and enhanced the inhibitory effects of SOD1/PI3K-AKT signaling. These results indicated that selective inhibition of SOD1 by MT1M promoted apoptosis and inhibited the epithelial-mesenchymal transition (EMT) of ESCC cells via regulation of the ROS-SOD1-PI3K/AKT signaling network. However, to further understand whether MT1M has the ability to regulate other pathways involved with ROS signaling, such as the NF-κB pathway, and to study exactly how MT1M regulates SOD1, the interacting loci and protein structure, is an important future direction to reveal the full biological regulatory function of MT1M in ESCC and other human tumors.
In conclusion, in this study we identified MT1M as a tumor suppressor gene in ESCC that it was inactivated by promoter hypermethylation in esophageal cancer. MT1M contributed to the inhibition of esophageal cancer progression by inhibiting cell proliferation, migration, EMT and inducing tumor cell apoptosis through regulating SOD1/PI3K-AKT signaling pathway. Therefore, the expression of MT1M has the potential to serve as a biomarker for esophageal cancer diagnosis and prognosis.
ACKNOWLEDGMENTS
This study was supported by Natural Science Foundation of Chongqing, Commission of Science and Technology of Chongqing, China (cstc2019jcyj-msxmX0861).
Footnotes
AUTHOR CONTRIBUTIONS
D.L., W.P., and B.W. performed experiments and analyzed data. H.L. and R.Z. (Ruizhen Zhang) prepared and provided samples and reagents. R.Z. (Ruiqin Zhou) and L.Y. (Lijun Yao) collected patient samples and recorded patient information. L.Y. (Lin Ye) designed and supported the whole study and wrote the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Arriaga J.M., Levy E.M., Bravo A.I., Bayo S.M., Amat M., Aris M., Hannois A., Bruno L., Roberti M.P., Loria F.S., et al. Metallothionein expression in colorectal cancer: relevance of different isoforms for tumor progression and patient survival. Hum. Pathol. 2012;43:197–208. doi: 10.1016/j.humpath.2011.04.015. [DOI] [PubMed] [Google Scholar]
- Babula P., Masarik M., Adam V., Eckschlager T., Stiborova M., Trnkova L., Skutkova H., Provaznik I., Hubalek J., Kizek R. Mammalian metallothioneins: properties and functions. Metallomics. 2012;4:739–750. doi: 10.1039/c2mt20081c. [DOI] [PubMed] [Google Scholar]
- Bindoli A., Rigobello M.P. Principles in redox signaling: from chemistry to functional significance. Antioxid. Redox Signal. 2013;18:1557–1593. doi: 10.1089/ars.2012.4655. [DOI] [PubMed] [Google Scholar]
- Brabletz T., Kalluri R., Nieto M.A., Weinberg R.A. EMT in cancer. Nat. Rev. Cancer. 2018;18:128–134. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]
- Cairns R.A., Harris I.S., Mak T.W. Regulation of cancer cell metabolism. Nat. Rev. Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Carter B.J., Anklesaria P., Choi S., Engelhardt J.F. Redox modifier genes and pathways in amyotrophic lateral sclerosis. Antioxid. Redox Signal. 2009;11:1569–1586. doi: 10.1089/ars.2008.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaldo S.A., Freitas J.R., Conchinha N.V., Madureira P.A. The tumorigenic roles of the cellular REDOX regulatory systems. Oxid. Med. Cell. Longev. 2016;2016:8413032. doi: 10.1155/2016/8413032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changjun L., Feizhou H., Dezhen P., Zhao L., Xianhai M. MiR-545-3p/MT1M axis regulates cell proliferation, invasion and migration in hepatocellular carcinoma. Biomed. Pharmacother. 2018;108:347–354. doi: 10.1016/j.biopha.2018.09.009. [DOI] [PubMed] [Google Scholar]
- Che M., Wang R., Li X., Wang H.Y., Zheng X.F.S. Expanding roles of superoxide dismutases in cell regulation and cancer. Drug Discov. Today. 2016;21:143–149. doi: 10.1016/j.drudis.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Quan R., Bhandari A., Chen Z., Guan Y., Xiang J., You J., Teng L. Low metallothionein 1M (MT1M) is associated with thyroid cancer cell lines progression. Am. J. Transl. Res. 2019;11:1760–1770. [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Liang P., Geng H., Wang Z., Li L., Cheng S.H., Ying J., Su X., Ng K.M., Ng M.H., et al. A novel 19q13 nucleolar zinc finger protein suppresses tumor cell growth through inhibiting ribosome biogenesis and inducing apoptosis but is frequently silenced in multiple carcinomas. Mol. Cancer Res. 2012;10:925–936. doi: 10.1158/1541-7786.MCR-11-0594. [DOI] [PubMed] [Google Scholar]
- Cui Q., Wang J.Q., Assaraf Y.G., Ren L., Gupta P., Wei L., Ashby C.R., Jr., Yang D.H., Jr., Chen Z.S., Jr. Modulating ROS to overcome multidrug resistance in cancer. Drug Resist. Updat. 2018;41:1–25. doi: 10.1016/j.drup.2018.11.001. [DOI] [PubMed] [Google Scholar]
- D'Autreaux B., Toledano M.B. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- Dimayuga F.O., Wang C., Clark J.M., Dimayuga E.R., Dimayuga V.M., Bruce-Keller A.J. SOD1 overexpression alters ROS production and reduces neurotoxic inflammatory signaling in microglial cells. J. Neuroimmunol. 2007;182:89–99. doi: 10.1016/j.jneuroim.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H., Chen B., Jiao N.L., Liu Y.H., Sun S.Y., Zhang Y.W. Elevated glutathione peroxidase 2 expression promotes cisplatin resistance in lung adenocarcinoma. Oxid. Med. Cell. Longev. 2020;2020:7370157. doi: 10.1155/2020/7370157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H.Y., Li Y., Olivo M., Yip G.W., Bay B.H. Differential up-regulation of metallothionein isoforms in well-differentiated nasopharyngeal cancer cells in vitro by photoactivated hypericin. Oncol. Rep. 2006;16:1397–1402. [PubMed] [Google Scholar]
- Emri E., Egervari K., Varvolgyi T., Rozsa D., Miko E., Dezso B., Veres I., Mehes G., Emri G., Remenyik E. Correlation among metallothionein expression, intratumoural macrophage infiltration and the risk of metastasis in human cutaneous malignant melanoma. J. Eur. Acad. Dermatol. Venereol. 2013;27:e320–e327. doi: 10.1111/j.1468-3083.2012.04653.x. [DOI] [PubMed] [Google Scholar]
- Ferrario C., Lavagni P., Gariboldi M., Miranda C., Losa M., Cleris L., Formelli F., Pilotti S., Pierotti M.A., Greco A. Metallothionein 1G acts as an oncosupressor in papillary thyroid carcinoma. Lab. Invest. 2008;88:474–481. doi: 10.1038/labinvest.2008.17. [DOI] [PubMed] [Google Scholar]
- Gao L., Loveless J., Shay C., Teng Y. Targeting ROS-mediated crosstalk between autophagy and apoptosis in cancer. Adv. Exp. Med. Biol. 2020;1260:1–12. doi: 10.1007/978-3-030-42667-5_1. [DOI] [PubMed] [Google Scholar]
- Griess B., Tom E., Domann F., Teoh-Fitzgerald M. Extracellular superoxide dismutase and its role in cancer. Free Radic. Biol. Med. 2017;112:464–479. doi: 10.1016/j.freeradbiomed.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoxhaj G., Manning B.D. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer. 2020;20:74–88. doi: 10.1038/s41568-019-0216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav R.R., Ye Z., Huang R.L., Liu J., Hsu P.Y., Huang Y.W., Rangel L.B., Lai H.C., Roa J.C., Kirma N.B., et al. Genome-wide DNA methylation analysis reveals estrogen-mediated epigenetic repression of metallothionein-1 gene cluster in breast cancer. Clin. Epigenetics. 2015;7:13. doi: 10.1186/s13148-015-0045-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez J.C., Manuia M., Burnett M.E., Betancourt O., Boivin B., Shaw D.E., Tonks N.K., Mazar A.P., Doñate F. Superoxide dismutase 1 (SOD1) is essential for H2O2-mediated oxidation and inactivation of phosphatases in growth factor signaling. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7147–7152. doi: 10.1073/pnas.0709451105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara B., Ramadoss S., Chaudhuri G., Janzen C., Sen S., Mascharak P.K. Carbon monoxide sensitizes cisplatin-resistant ovarian cancer cell lines toward cisplatin via attenuation of levels of glutathione and nuclear metallothionein. J. Inorg. Biochem. 2019;191:29–39. doi: 10.1016/j.jinorgbio.2018.11.003. [DOI] [PubMed] [Google Scholar]
- Krezel A., Maret W. The functions of metamorphic metallothioneins in zinc and copper metabolism. Int. J. Mol. Sci. 2017;18:1237. doi: 10.3390/ijms18061237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari M.V., Hiramatsu M., Ebadi M. Free radical scavenging actions of metallothionein isoforms I and II. Free Radic. Res. 1998;29:93–101. doi: 10.1080/10715769800300111. [DOI] [PubMed] [Google Scholar]
- Lee G.Y., Kenny P.A., Lee E.H., Bissell M.J. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat. Methods. 2007;4:359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Ying J., Tong X., Zhong L., Su X., Xiang T., Shu X., Rong R., Xiong L., Li H., et al. Epigenetic identification of receptor tyrosine kinase-like orphan receptor 2 as a functional tumor suppressor inhibiting beta-catenin and AKT signaling but frequently methylated in common carcinomas. Cell. Mol. Life Sci. 2014;71:2179–2192. doi: 10.1007/s00018-013-1485-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J., Yu H., Wang C., Sun L., Jiang W., Zhang P., Xiao Q., Han D., Saiyin H., Zhu J., et al. Metallothionein MT1M is a tumor suppressor of human hepatocellular carcinomas. Carcinogenesis. 2012;33:2568–2577. doi: 10.1093/carcin/bgs287. [DOI] [PubMed] [Google Scholar]
- Miles A.T., Hawksworth G.M., Beattie J.H., Rodilla V. Induction, regulation, degradation, and biological significance of mammalian metallothioneins. Crit. Rev. Biochem. Mol. Biol. 2000;35:35–70. doi: 10.1080/10409230091169168. [DOI] [PubMed] [Google Scholar]
- Oka D., Yamashita S., Tomioka T., Nakanishi Y., Kato H., Kaminishi M., Ushijima T. The presence of aberrant DNA methylation in noncancerous esophageal mucosae in association with smoking history: a target for risk diagnosis and prevention of esophageal cancers. Cancer. 2009;115:3412–3426. doi: 10.1002/cncr.24394. [DOI] [PubMed] [Google Scholar]
- Patel G.K., Khan M.A., Bhardwaj A., Srivastava S.K., Zubair H., Patton M.C., Singh S., Khushman M., Singh A.P. Exosomes confer chemoresistance to pancreatic cancer cells by promoting ROS detoxification and miR-155-mediated suppression of key gemcitabine-metabolising enzyme, DCK. Br. J. Cancer. 2017;116:609–619. doi: 10.1038/bjc.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo de la Vega M., Chapman E., Zhang D.D. NRF2 and the hallmarks of cancer. Cancer Cell. 2018;34:21–43. doi: 10.1016/j.ccell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salt M.B., Bandyopadhyay S., McCormick F. Epithelial-to-mesenchymal transition rewires the molecular path to PI3K-dependent proliferation. Cancer Discov. 2014;4:186–199. doi: 10.1158/2159-8290.CD-13-0520. [DOI] [PubMed] [Google Scholar]
- Schumacker P.T. Reactive oxygen species in cancer: a dance with the devil. Cancer Cell. 2015;27:156–157. doi: 10.1016/j.ccell.2015.01.007. [DOI] [PubMed] [Google Scholar]
- Shadel G.S., Horvath T.L. Mitochondrial ROS signaling in organismal homeostasis. Cell. 2015;163:560–569. doi: 10.1016/j.cell.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si M., Lang J. The roles of metallothioneins in carcinogenesis. J. Hematol. Oncol. 2018;11:107. doi: 10.1186/s13045-018-0645-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibenaller Z.A., Welsh J.L., Du C., Witmer J.R., Schrock H.E., Du J., Buettner G.R., Goswami P.C., Cieslak J.A., 3rd, Cullen J.J., 3rd Extracellular superoxide dismutase suppresses hypoxia-inducible factor-1alpha in pancreatic cancer. Free Radic. Biol. Med. 2014;69:357–366. doi: 10.1016/j.freeradbiomed.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- Sosa V., Moliné T., Somoza R., Paciucci R., Kondoh H., LLeonart M.E. Oxidative stress and cancer: an overview. Ageing Res. Rev. 2013;12:376–390. doi: 10.1016/j.arr.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Theocharis S.E., Margeli A.P., Klijanienko J.T., Kouraklis G.P. Metallothionein expression in human neoplasia. Histopathology. 2004;45:103–118. doi: 10.1111/j.1365-2559.2004.01922.x. [DOI] [PubMed] [Google Scholar]
- Trachootham D., Alexandre J., Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat. Rev. Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- Tsui K.H., Hou C.P., Chang K.S., Lin Y.H., Feng T.H., Chen C.C., Shin Y.S., Juang H.H. Metallothionein 3 is a hypoxia-upregulated oncogene enhancing cell invasion and tumorigenesis in human bladder carcinoma cells. Int. J. Mol. Sci. 2019;20:980. doi: 10.3390/ijms20040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.K., Stallings R., Hildebrand C.E., Chiu R., Karin M., Richards R.I. Human metallothionein genes: structure of the functional locus at 16q13. Genomics. 1990;8:513–518. doi: 10.1016/0888-7543(90)90038-v. [DOI] [PubMed] [Google Scholar]
- Yamada H., Yamada Y., Adachi T., Fukatsu A., Sakuma M., Futenma A., Kakumu S. Protective role of extracellular superoxide dismutase in hemodialysis patients. Nephron. 2000;84:218–223. doi: 10.1159/000045580. [DOI] [PubMed] [Google Scholar]
- Ye L., Xiang T., Fan Y., Zhang D., Li L., Zhang C., He X., Xiang Q., Tao Q., Ren G. The 19q13 KRAB Zinc-finger protein ZFP82 suppresses the growth and invasion of esophageal carcinoma cells through inhibiting NF-kappaB transcription and inducing apoptosis. Epigenomics. 2019;11:65–80. doi: 10.2217/epi-2018-0092. [DOI] [PubMed] [Google Scholar]
- Ye L., Xiang T., Zhu J., Li D., Shao Q., Peng W., Tang J., Li L., Ren G. Interferon consensus sequence-binding protein 8, a tumor suppressor, suppresses tumor growth and invasion of non-small cell lung cancer by interacting with the Wnt/beta-catenin pathway. Cell. Physiol. Biochem. 2018;51:961–978. doi: 10.1159/000495399. [DOI] [PubMed] [Google Scholar]
- Zeng H., Zheng R., Zhang S., Zuo T., Xia C., Zou X., Chen W. Esophageal cancer statistics in China, 2011: estimates based on 177 cancer registries. Thorac. Cancer. 2016;7:232–237. doi: 10.1111/1759-7714.12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Jiang L., Hu Y., Xiao C., Xu N., Zhou J., Zhou X. Metallothionein 1H (MT1H) functions as a tumor suppressor in hepatocellular carcinoma through regulating Wnt/beta-catenin signaling pathway. BMC Cancer. 2017;17:161. doi: 10.1186/s12885-017-3139-2. [DOI] [PMC free article] [PubMed] [Google Scholar]