Abstract
Background
Precision medicine, space exploration, drug discovery to characterization of dark chemical space of habitats and organisms, metabolomics takes a centre stage in providing answers to diverse biological, biomedical, and environmental questions. With technological advances in mass-spectrometry and spectroscopy platforms that aid in generation of information rich datasets that are complex big-data, data analytics tend to co-evolve to match the pace of analytical instrumentation. Software tools, resources, databases, and solutions help in harnessing the concealed information in the generated data for eventual translational success.
Aim of the review
In this review, ~ 85 metabolomics software resources, packages, tools, databases, and other utilities that appeared in 2020 are introduced to the research community.
Key scientific concepts of review
In Table 1 the computational dependencies and downloadable links of the tools are provided, and the resources are categorized based on their utility. The review aims to keep the community of metabolomics researchers updated with all the resources developed in 2020 at a collated avenue, in line with efforts form 2015 onwards to help them find these at one place for further referencing and use.
Keywords: Metabolomics, Tool, Database, Software, Annotation, Metabolite, In silico, Recourse, Program
Introduction
The year 2020 has seen an enormous rise in applications of ion mobility mass-spectrometry (IMS), and data-independent acquisition (DIA) methods of analyses in both metabolomics and lipidomics. In terms of application, mass spectrometry as a technology promises advance care for cancer patients in clinical and intraoperative use (J. Zhang, Ge, et al., 2020; Zhang, Sans, et al., 2020), imaging mass spectrometry (MSI) based natural products (NPs) discovery (Spraker et al. 2020), nanoscale secondary ion mass spectrometry (nanoSIMS) usage in subcellular MS imaging and quantitative analysis in organelles (Thomen et al. 2020), capturing urban sources of contamination from high resolution mass spectrometry (HRMS) (Bowen et al., 2020) to detection of COVID-19 disease signatures (Mahmud & Garrett, 2020).
From an analytical method development stand point, interesting developments such as plasma pseudotargeted metabolomics method using ultra-high-performance liquid chromatography–mass spectrometry (UHPLC-MS) (Zheng et al. 2020) and the need for combined use of nuclear magnetic resonance spectroscopy and mass spectrometry approaches in metabolomics (Letertre et al. 2020) are notable. For volume-limited samples, solutions such as sub-nanoliter metabolomics via LC–MS/MS such as pulsed MS ion generation method known as triboelectric nanogenerator inductive nanoelectrospray ionization (TENGi nanoESI) MS (Li et al. 2020) was introduced. Flow-injection Orbitrap mass spectrometry (FI-MS) enabled reproducible detection of ~ 9,000 and ~ 10,000 m/z features in metabolomics and lipidomics analysis of serum samples, respectively, with a sample scan time of ~ 15 s and duty time of ~ 30 s; a ~ 50% increase versus current spectral-stitching FI-MS methods (Sarvin et al. 2020). A spatial metabolomics pipeline (metaFISH) that combined fluorescence in situ hybridization (FISH) microscopy and high-resolution atmospheric-pressure matrix-assisted laser desorption/ionization mass spectrometry to image host–microbe symbioses and their metabolic interactions (Geier et al. 2020) was also reported. Another study that compared the full-scan, data-dependent acquisition (DDA), and data-independent acquisition (DIA) methods in HR LC–MS/MS based metabolomics to reveal that spectra quality is better in DDA with average dot product score 83.1% higher than DIA and the number of MS2 spectra (spectra quantity) is larger in DIA (Guo & Huan, 2020a). Furthermore, it was shown that DDA mode consistently generated fewer uniquely found significant features than full-scan and DIA modes (Guo & Huan, 2020b).
Using with Raman spectroscopy, followed by stimulated Raman scattering (SRS) microscopy and Raman-guided subcellular pharmaco-metabolomics in metastatic melanoma cells revealed intracellular lipid droplets that helped identify a previously unknown susceptibility of lipid mono-unsaturation within de-differentiated mesenchymal cells with innate resistance to BRAF inhibition (Du et al. 2020). Application of 31P NMR was shown to hold potential of expanding the coverage of the metabolome by detecting phosphorus-containing metabolites (Bhinderwala et al. 2020).
The effectiveness of the flow injection analysis-continuous accumulation of selected ions Fourier transform ion cyclotron resonance mass spectrometry (FIA-CASI-FTMS) workflow utilizing isotopic fine structure (IFS) for molecular formula assignment was realized for metabolomics applications (Thompson et al. 2020). A buffer modification workflow (BMW) in which the same sample is run by LC–MS in both liquid chromatography solvent with 14NH3–acetate buffer and in solvent with the buffer modified with 15NH3–formate, resulted in characteristic mass and signal intensity changes for adduct peaks, facilitating their annotation (Lu et al. 2020). Towards reference materials standardization, quantitative measures of approximately 200 metabolites for each of three pooled reference materials (220 metabolites for Qstd3, 211 metabolites for CHEAR, 204 metabolites for NIST1950) were obtained and supported harmonization of metabolomics data collected from 3677 human samples in 17 separate studies analyzed by two complementary HRMS methods (K. H. Liu, Mrzic, et al., 2020; Liu, Nellis, et al., 2020). Another review highlighted the recent progresses (since 2016) in the field of chemical derivatization LC–MS for both targeted and untargeted metabolome analysis (Zhao & Li, 2020). The characterization of compounds by the number of labile hydrogen and oxygen atoms in the molecule, which can be measured using hydrogen/deuterium and 16O/18O-exchange approaches allows reduction of the search space by a factor of 10 and considerably increases the reliability of the compound identification (Kostyukevich et al. 2020). Preference for monophasic methods that are quicker and simpler than biphasic methods for their amenability and integration into future automation for hydrophilic interaction chromatography (HILIC) ultrahigh-performance liquid chromatography–mass spectrometry (UHPLC–MS) and nonpolar extracts by C18 reversed-phase UHPLC–MS based metabolomics in animal tissues and biofluids (Southam et al. 2020) was also demonstrated. In other innovative applications, use of short columns and direct solvent switches allowed for fast screening (3 min per polarity), where a total of 50 commonly reported diagnostic or explorative biomarkers were validated with a limit of quantification that was comparable with conventional LC–MS/MS (van der Laan et al. 2020).
From the stand point of data analysis, metabolomics as a field is starting to benefit by applying machine learning (ML) (Liebal et al. 2020) and deep learning (DL) (Pomyen et al. 2020; Sen et al. 2020) approaches to address diverse challenges from data preprocessing to biological interpretation. In the context of systems and personalized medicine LIONESS (Linear Interpolation to Obtain Network Estimates for Single Samples) and ssPCC (single sample network based on Pearson correlation) were evaluated and compared in the context of metabolite–metabolite association networks (Jahagirdar & Saccenti, 2020). In annotation domains for low resolution GC–MS data, usage of DL ranking for small molecules identification, a deep learning ranking model outperformed other approaches and enabled reducing a fraction of wrong answers (at rank-1) by 9–23% depending on the used data set (Matyushin et al. 2020). In the age of artificial intelligence, spatial metabolomics and IMS promise to revolutionize biology and healthcare (Alexandrov, 2020). Approaches such as an integrated strategy of fusing features and removing redundancy based on graph density (FRRGD) were proposed that greatly enhanced the metabolome detection coverage with low abundance (Ju et al. 2020).
For a software survey of other mass-spectrometry derived omics tools, packages, resources, softwares and databases, readers can consult other treatise for metaproteomics (Sajulga et al. 2020), data‐independent acquisition mass spectrometry‐based proteomics (F. Zhang, Ge, et al., 2020; Zhang, Sans, et al., 2020), single cell and single cell-type metabolomics (B. B. Misra, 2020a) among others.
Diverse online resources such as OMICtools (http://omictools.com/) (Henry et al. 2014), Fiehn laboratory pages (http://fiehnlab.ucdavis.edu/ and http://metabolomics.ucdavis.edu/Downloads), the International Metabolomics Society’s resource pages, software repositories such as Comprehensive R Archive Network (CRAN) (https://cran.r-project.org/web/packages/available_packages_by_name.html), Bioconductor (https://www.bioconductor.org/), the Python Package Index (PyPI) (https://www.pypi.org), GitLab (https://www.gitlab.com), and GitHub (https://www.github.com/) are excellent resources to obtain software tools, databases and resources for metabolomics research. Metabolomics Tools Wiki claimed to be an updated resource for metabolomics tools, databases and software resources has ceased to be updated since 2017 (Spicer et al. 2017). Whilst there exists a plethora of programming languages, modern interpreted scripting languages such as R, Python, Raku, Ruby, and MATLAB are evidently popular in metabolomics.
Building on the previously established review structure this overview of major tools and resources in metabolomics, spanning 2015–2019 (B. Misra & van der Hooft, 2015; O’Shea & Misra, 2020) is organized into the following sections: (1) Platform-specific tools, (2) Preprocessing and QC tools, (3) Annotation tools, (4) Multifunctional tools, (5) Tools for statistical analysis and visualization, (6) Databases, and (7) Other specialized tools.
Table 1 provides a summary of all reviewed resources and their availability. Furthermore, in Table 2, highlighted are unpublished tools that can be found in the CRAN and PyPI software repositories that are deemed useful for the metabolomics research community, but are not associated with a scholarly article that is published.
Table 1.
The entire list of reviewed tools is organized by important analytical steps in metabolomics data analysis and includes details regarding their platform dependency, and implementation, e.g., programming language (R, Python, Java, C/C ++, etc.) or web browser based and their availability
| Name of the Software Tool | Category | Platform dependency | Implementation/ use dependency | Software availability | References |
|---|---|---|---|---|---|
| AlpsNMR | Platform | NMR | R | https://github.com/sipss/AlpsNMR | (Madrid-Gambin et al. 2020) |
| SigMa | Platform | NMR | MATLAB, Standalone | https://github.com/BEKZODKHAKIMOV/SigMa_Ver1 | (Khakimov et al. 2020) |
| NMRfilter | Platform | NMR | NA | https://github.com/stefhk3/nmrfilterprojects | (Kuhn et al. 2020) |
| MSHub + EI-GNPS | Platform | GC–MS | GNPS, Web | https://bitbucket.org/iAnalytica/mshub_process/src/master/; https://github.com/CCMS-UCSD/GNPS_Workflows/tree/master/mshub-gc/tools/mshub-gc/proc | (Aksenov et al. 2020) |
| RGCxGC toolbox | Platform | GCXGC-MS | R, TeX | https://github.com/DanielQuiroz97/RGCxGC | (Quiroz-Moreno et al. 2020) |
| CROP | Preprocessing | LC–MS/MS | R | https://github.com/rendju/CROP | (Kouřil et al. 2020) |
| ncGTW | Preprocessing | LC–MS/MS | R, C ++ | https://github.com/ChiungTingWu/ncGTW | (Wu et al. 2020) |
| TidyMS | Preprocessing | LC–MS/MS | Python | https://github.com/griquelme/tidyms | (Riquelme et al. 2020) |
| AutoTuner | Preprocessing | LC–MS/MS | R | https://github.com/crmclean/Autotuner | (McLean & Kujawinski, 2020) |
| hRUV | Preprocessing | LC–MS/MS | R | https://shiny.maths.usyd.edu.au/hRUV/ | (Kim et al. 2020) |
| MetumpX | Preprocessing | Any | R | https://github.com/hasaniqbal777/MetumpX-bin | (Wajid et al. 2020) |
| MetaQuac | QC | Targeted LC–MS | R | https://github.com/bihealth/metaquac | (Kuhring et al. 2020) |
| dbnorm | QC | Any | R | https://github.com/NBDZ/dbnorm | (Bararpour et al. 2020) |
| MetaClean | QC | LC–MS/MS | R | https://cran.r-project.org/web/packages/MetaClean/index.html | (Chetnik et al. 2020) |
| NeatMS | QC | LC–MS/MS | Python | https://github.com/bihealth/NeatMS | (Gloaguen et al. 2020) |
| MESSAR | Annotation | LC–MS/MS | Web | https://messar.biodatamining.be/ | (Liu, Mrzic, et al., 2020; Liu, Nellis, et al., 2020) |
| SMART 2.0 | Annotation | 2D NMR | Web | https://smart.ucsd.edu/classic | (Reher et al. 2020) |
| MetFID | Annotation | MS/MS data | NA | NA | (Fan et al. 2020) |
| CPVA | Annotation | Any | Web | https://github.com/13479776/cpvamscpva.med.sustech.edu.cn | (Luan et al. 2020) |
| NRPro | Annotation | LC–MS/MS | Java, Web | https://bioinfo.cristal.univ-lille.fr/nrpro/ | (Ricart et al. 2020) |
| MetENP/MetENPWeb | Annotation | LC–MS/MS | R, Web | https://www.metabolomicsworkbench.org/data/analyze.php | (Choudhary et al. 2020) |
| CANOPUS | Annotation | LC–MS/MS | Standalone | https://bio.informatik.uni-jena.de/software/canopus/ | (Dührkop et al. 2020) |
| MolDiscovery | Annotation | LC–MS/MS | Python | https://github.com/mohimanilab/molDiscovery | (Cao et al. n.d.) |
| MetIDfyR | Annotation | LC–MS/MS | R | https://github.com/agnesblch/MetIDfyR | (Delcourt et al. 2020) |
| Qemistree | Annotation | LC–MS/MS | Python | https://github.com/biocore/q2-qemistree | (Tripathi et al. 2020) |
| IIMN | Annotation | LC–MS/MS | GNPS, Web | https://ccms-ucsd.github.io/GNPSDocumentation/fbmn-iin/ | (Robin Schmid, Daniel Petras, Louis-Félix Nothias, Mingxun Wang, Allegra T. Aron, Annika Jagels, Hiroshi Tsugawa, Johannes Rainer, Mar Garcia-Aloy, Kai Dührkop, Ansgar Korf, Tomáš Pluskal, Zdeněk Kameník, Alan K. Jarmusch, Andrés Mauricio Caraballo-Rodrígu 2020) |
| FOBI | Annotation | Any | R, Web | https://github.com/pcastellanoescuder/FOBI_Visualization_Tool | (Castellano-Escuder et al. 2020) |
| Biodendro | Annotation | LC–MS/MS | Python | https://github.com/ccdmb/BioDendro | (Rawlinson et al. 2020) |
| AllCCS atlas | Annotation | IM-MS | Web | https://github.com/ZhuMetLab/AllCCS; http://allccs.zhulab.cn/ | (Zhou et al. 2020) |
| Binner | Annotation | LC–MS/MS | Java | https://binner.med.umich.edu/ | (Kachman et al. 2019) |
| MS-CleanR | Annotation | LC–MS/MS | R | https://github.com/eMetaboHUB/MS-CleanR | (Fraisier-Vannier et al. 2020) |
| Retip | Annotation | LC–MS/MS | R | https://www.retip.app/ | (Bonini et al. 2020) |
| QSRR Automator | Annotation | LC–MS/MS | Python | https://github.com/UofUMetabolomicsCore/QSRR_Automator/releases/tag/v1_exe | (Naylor et al. 2020) |
| MFAssignR | Annotation | LC–MS/MS | R, HTML | https://github.com/skschum/MFAssignR | (Schum et al. 2020) |
| McSearch | Annotation | LC–MS/MS | R | https://github.com/HuanLab/McSearch; http://cloudmetabolomics.ca/mcsearch | (Xing et al. 2020) |
| REDU | Annotation | LC–MS/MS | GNPS, Web | https://redu.ucsd.edu/ | (Jarmusch et al. 2020) |
| MASST | Annotation | LC–MS/MS | GNPS, Web | https://masst.ucsd.edu/ | (Wang, Jarmusch, et al., 2020; Wang, Leber, et al., 2020) |
| NPClassifier | Annotation | Any | Web | https://npclassifier.ucsd.edu/ | (kim et al. 2020) |
| patRoon | Annotation | HR MS/MS | R | https://github.com/rickhelmus/patRoon | (Helmus et al. 2021) |
| LipidLynxX | Annotation | LC–MS/MS | Python, Standalone | http://lipidmaps.org/lipidlynxx/ | (Ni & Fedorova, 2020) |
| Skyline | Multifunctional | Any | Standalone | https://skyline.ms/project/home/software/Skyline/begin.view | (Adams et al. 2020) |
| NoTaMe | Multifunctional | LC–MS/MS | R, Web | https://github.com/antonvsdata/notame | (Klåvus et al. 2020) |
| BALSAM | Multifunctional | IMS, GC–MS, LC–MS | Web, Python, HTML, Java | https://exbio.wzw.tum.de/balsam/, https://github.com/philmaweb/balsam_django | (Weber et al. 2020) |
| MRMkit | Multifunctional | Targeted LC–MS | Python, R | https://github.com/cssblab/MRMkit | (Teo et al. 2020) |
| MetaboShiny | Multifunctional | Any | R | https://github.com/joannawolthuis/MetaboShiny | (Wolthuis et al. 2020) |
| SmartPeak | Multifunctional | Many | C#, Python | https://github.com/AutoFlowResearch/SmartPeak | (Kutuzova et al. 2020) |
| MS-DIAL 4.0 | Multifunctional | LC–MS/MS, GC–MS, IMS | Standalone | http://prime.psc.riken.jp/compms/msdial/main.html | (Tsugawa et al. 2020) |
| IP4M | Multifunctional | LC–MS/MS | Java, Perl, R, Standalone | https://IP4M.cn | (Liang et al. 2020) |
| DropMS | Multifunctional | HR MS | Web | http://www.dropms.online/ | (Rosa et al. 2020) |
| Epimetal | Statistics, visualization | Any | JavaScript, Web | https://github.com/amergin/epimetal; http://epimetal.computationalmedicine.fi/ | (Ekholm et al. 2020) |
| Metabolite AutoPlotter | Statistics, visualization | Quantitative metabolomics data, any | R, Web | https://mpietzke.shinyapps.io/AutoPlotter/ | (Pietzke & Vazquez, 2020) |
| Metabolite-Investigator | Statistics, visualization | LC–MS | R, Web | https://github.com/cfbeuchel/Metabolite-Investigator; https://apps.health-atlas.de/metabolite-investigator/ | (Beuchel et al. 2020) |
| VIIME | Statistics, visualization | Any | Web | https://viime.org/#/ | (Choudhury et al. 2020) |
| struct | Statistics, visualization | Any | R | http://bioconductor.org/packages/struct | (Lloyd et al. 2020) |
| lipidr | Statistics, visualization | LC–MS/MS | R | https://www.lipidr.org/ | (Mohamed et al. 2020) |
| NOREVA | Statistics | Any | Web, R, Standalone | http://idrblab.cn/noreva/ | (Yang et al. 2020) |
| %polynova_2way | Statistics | Processed data | SAS | https://doi.org/10.1371/journal.pone.0244013.s002 | (Manjarin et al. 2020) |
| rawR | Visualization | LC–MS | R, C ++ | https://github.com/fgcz/rawR | (Kockmann & Panse, 2020)R |
| Metaboverse | Visualization | Any | Java, HTML, Standalone | https://github.com/Metaboverse | (Jordan A. Berg, Youjia Zhou, T. Cameron Waller, Yeyun Ouyang, Sara M. Nowinski, Tyler Van Ry, Ian George, James E. Cox, Bei Wang 2020) |
| JS-MS 2.0 | Visualization | LC–MS/MS | Java, JavaScript, HTML | https://github.com/optimusmoose/jsms | (Henning & Smith, 2020) |
| COCONUT | Database | Any | Web | https://coconut.naturalproducts.net/ | (Sorokina et al. n.d.) |
| METLIN MS2 molecular standards database | Database | LC–MS/MS | Web | http://metlin.scripps.edu/ | (Xue et al. 2020) |
| CSMDB | Database | NMR | MATLAB | https://github.com/cibionnmrlab/CSMDB-with-ConQuer-ABC | (Charris-Molina et al. 2020) |
| EMBL-MCF | Database | LC–MS | NA | https://curatr.mcf.embl.de/ | |
| MIAMI | Isotopic | GC–MS | C ++ | http://miami.tu-bs.de/ | (Dudek et al. 2020) |
| isoSCAN | Isotopic | GC–MS | R | https://github.com/jcapelladesto/isoSCAN | (Capellades et al. 2020) |
| LiPydomics | Lipidomics | Ion Mobility, Lipidomics | Python, HTML | https://github.com/dylanhross/lipydomics | (Ross et al. 2020) |
| LipidCreator | Lipidomics | LC–MS | C#, HTML, Skyline plugin | https://github.com/lifs-tools/lipidcreator | (Peng et al. 2020) |
| Lipid Annotator | Lipidomics | LC–MS/MS | NA | NA | (Koelmel et al. 2020) |
| Raman2imzML | MSI | Raman | C ++, R | https://github.com/LlucSF/Raman2imzML | (Iakab et al. 2020) |
| SUMMER | Multiomics | Any | R, Web | http://igc1.salk.edu:3838/summer/ and https://bitbucket.org/salkigc/summer/src/master/ | Huang et al. (2020) |
| metpropagate | Analysis, visualization | Untargeted LC–MS/MS | R, Python | https://github.com/emmagraham/metPropagate | Graham Linck et al. (2020) |
The tools generally follow their order of appearance in the manuscript text
Table 2.
List of useful R/ Bioconductor packages that surfaced/ were improved in 2020
| CRAN package name | Title | Description | Link |
|---|---|---|---|
| lilikoi | Metabolomics personalized pathway analysis tool | Helps map metabolites data into pathways and calculates pathway deregulation scores, and enables perform exploratory analysis, classification and prognosis analysis on both metabolites and pathways | https://cran.r-project.org/web/packages/lilikoi/index.html |
| omu | A metabolomics analysis tool for intuitive figures and convenient metadata collection | Helps generate intuitive figures for metabolomics data by using Kyoto Encyclopaedia of Genes and Genomes (KEGG) hierarchy data, and gathers functional orthology and gene data using the package 'KEGGREST' to access the 'KEGG' API | https://cran.r-project.org/web/packages/omu/index.html |
| eRah | Automated spectral deconvolution, alignment, and metabolite identification in GC/MS-based untargeted metabolomics | Updated to 2016 published tool eRah, that aids in automated compound deconvolution, alignment across samples, and identification of metabolites by spectral library matching in untargeted GC–MS metabolomics workflows | https://cran.r-project.org/web/packages/erah/index.html |
| MetaDBparse | Annotate mass over charge values with databases and formula prediction | Useful for parsing functionality for over 30 metabolomics databases, and calculates given adducts and isotope patterns and inserts into one big database which can be used to annotate unknown m/z values | https://cran.r-project.org/web/packages/MetaDBparse/index.html |
| MetaClean | Detection of low-quality peaks in untargeted metabolomics data | Uses 11 peak quality metrics and eight diverse machine learning algorithms to build a classifier for the automatic assessment of peak integration quality of peaks from untargeted metabolomics analyses | https://cran.r-project.org/web/packages/MetaClean/index.html |
| tmod | Feature set enrichment analysis for metabolomics and transcriptomics | Feature or gene set enrichment analysis in transcriptomics and metabolomics data and the allows enrichment based on ranked list of features, visualization and multivariate data analysis | https://cran.r-project.org/web/packages/tmod/index.html |
| ccmn | CCMN and other normalization methods for metabolomics data | Allows implementation of Cross-contribution Compensating Multiple standard Normalization (CCMN) method | https://cran.r-project.org/web/packages/crmn/index.html |
| LipidMS | Lipid annotation for LC–MS/MS DIA data | Aids in annotation of lipids in untargeted LC-DIA-MS lipidomics data based on fragmentation rules | https://cran.r-project.org/web/packages/LipidMS/index.html |
| enviGCMS | GC/LC–ms data analysis for environmental science | For environmental mass spectrometry (GC/LC-MS) data analysis for molecular isotope ratio, matrix effects and short-chain chlorinated paraffins analysis etc | https://cran.r-project.org/web/packages/enviGCMS/index.html |
| nontarget | Detecting isotope, adduct and homologue relations in LC–MS data | Allows screening of HRMS data set for peaks related by (1) isotope patterns, (2) different adducts of the same molecule and/or (3) homologue series; thus yielding isotopic pattern and adduct groups called 'components' with homologue series information. Further plotting, filtering of MS data for mass defects etc. are facilitated | https://cran.r-project.org/web/packages/nontarget/index.html |
| mosaic.find | Finding rhythmic and non-rhythmic trends in multi-omics data (MOSAIC) | MOSAIC (Multi-Omics Selection with Amplitude Independent Criteria) provides a function (mosaic_find()) designed to find rhythmic and non-rhythmic trends in multi-omics time course data using model selection and joint modelling | https://cran.r-project.org/web/packages/mosaic.find/index.html |
| ActivePathways | Integrative pathway enrichment analysis of multivariate omics data | A framework for analysing multiple omics datasets in the context of molecular pathways, biological processes and other types of gene sets. The tool uses p-value merging to combine gene- or protein-level signals, followed by ranked hypergeometric tests to determine enriched pathways and processes | https://cran.r-project.org/web/packages/ActivePathways/index.html |
| wilson | Web-based interactive omics visualization | Provides modules for creating web-based applications that use plot-based strategies to visualize and analyse multi-omics data | https://cran.r-project.org/web/packages/wilson/index.html |
| mixKernel | Omics data integration using kernel methods | The package aims at providing methods to combine kernel for unsupervised exploratory analysis, that can help integration of heterogenous types of data | https://cran.r-project.org/web/packages/mixKernel/index.html |
Platform-specific tools
Metabolomics as a discipline depends on mass spectrometry and spectroscopy analytical platforms to generate high through put omics scale data. These include, and are not limited to liquid chromatography-mass spectrometry (LC–MS), gas chromatography-mass spectrometry (GC–MS), capillary electrophoresis-mass spectrometry (CE-MS), and spectroscopic methods such as 1H-NMR, 13C-NMR, Raman, and Fourier transform infrared (FTIR) among others. In this section, I discuss all the tools that appeared in 2020 for analyses of datasets that are specific to a metabolomics platform or technology, i.e., LC–MS, GC–MS, and NMR.
Automated spectraL processing system for NMR (AlpsNMR), is an R-package that provides automated signal processing for untargeted NMR metabolomics datasets by performing region exclusion, spectra loading, metadata handling, automated outlier detection, spectra alignment and peak-picking, integration and normalization (Madrid-Gambin et al. 2020). The tool can load Bruker and JDX samples and can preprocess them for downstream statistical analysis.
Signature mapping (SigMa), developed as a standalone tool using MATLAB dependencies, for processing raw urine 1H-NMR spectra into a metabolite table (Khakimov et al. 2020). SigMa relies on the division of the urine NMR spectra into Signature Signals (SS), Signals of Unknown spin Systems (SUS) and bins of complex unresolved regions (BINS), thus allowing simultaneous detection of urinary metabolites in large-scale NMR metabolomics studies using a SigMa chemical shift library and a new automatic peak picking algorithm.
NMR filter, is a stand-alone interactive software for high-confidence NMR compound identification that runs NMR chemical shift predictions and matches them with the experimental data, where it defines the identity of compounds using a list of matching rates and correlating parameters of accuracy together with figures for visual validation (Kuhn et al. 2020).
MSHub/ electron ionisation (EI)-Global Natural Product Social (GNPS) Molecular Networking analysis, as a platform enables users to store, process, share, annotate, compare and perform molecular networking of both unit/low resolution and GC–HRMS data (Aksenov et al. 2020). GNPS-MassIVE is a public data repository for untargeted MS2 data, EI-MS data, with sample information (metadata) and annotated MS2 spectra (Aron et al. 2020). MSHub performs the auto-deconvolution of compound fragmentation patterns via unsupervised non-negative matrix factorization and quantifies the reproducibility of fragmentation patterns across samples, followed by GNPS molecular networking analyses.
RGCxGC toolbox, is an R-package that aids in analysis of two dimensional gas chromatography-mass spectrometry (2D GC–MS) data by offering pre-processing algorithms for signal enhancement, such as baseline correction based on asymmetric least squares, smoothing based on the Whittaker smoother, and peak alignment 2D Correlation Optimized Warping and multiway principal component analysis (Quiroz-Moreno et al. 2020).
Preprocessing and quality control (QC) tools
In untargeted metabolomics workflows that use either LC–MS/MS, GC–MS or NMR, depend a lot on pre-processing of the acquired raw datasets prior to statistical analyses and interpretation. Preprocessing typically involves tools that aid in the detection of masses (as m/z’s) from mass spectra (i.e., feature detection), construct and display extracted ion chromatograms, detect chromatographic peaks, deconvolution, peak alignment, data matrix curation steps such as batch and blank corrections to filtration and normalization steps, and quality assessments. Though, there are decade old popular preprocessing tools available to the community in the form of xcms (Tautenhahn et al. 2008), MZmine 2 (MZmine Development Team 2015), MS-DIAL (Tsugawa et al. 2015) there is a consistent effort to improve the workflows- from reducing computational time, to developing graphical user interfaces (GUIs) for users to render them user friendly to addressing challenges associated with interpretation of data from advanced platforms such as HRMS data or those from IMS, MSI etc. In fact, a recent comparative effort (among software tools such as software packages MZmine 2, enviMass, Compound Discoverer™, and XCMS Online) demonstrated a low coherence between the four processing tools, as overlap of features between all four programs was only about 10%, and for each software between 40 and 55% of features did not match with any other program (Hohrenk et al. 2020). Moreover, quality control (QC) tools are important to take care of systematic and random variations/ errors induced during experimental and analytical workflows. Batch effects can pose a lot of challenges, i.e., introduction of experimental artifacts that can interfere with the measurement of phenotype‐related metabolome changes in metabolomics data (Han & Li, 2020), and data normalization strategies, tools, and software solutions available are reviewed to circumvent some of these challenges (B. B. Misra, 2020b). In this section, I cover the preprocessing and the QC tools that appeared in 2020.
Correlation-based removal Of multiPlicities (CROP), implemented as an R-package is a visual post-processing tool that removes redundant features from LC–MS/MS based untargeted metabolomic data sets (Kouřil et al. 2020), where it groups highly correlated features within a defined retention time (RT) window avoiding the condition of specific m/z difference making it a second-tier strategy for multiplicities reduction. The output is a graphical representation of correlation network allowing a good understanding of the clusters composition that can aid in further parameter tuning.
neighbor-wise compound-specific Graphical Time Warping (ncGTW), is an integrated reference-free profile alignment method, implemented as an R-package and is available as a plugin for xcms that aids in detecting and fixing the bad alignments (misaligned feature groups) in the LC–MS data to render accurate grouping and peak-filling (Wu et al. 2020).
TidyMS, is a Python package for preprocessing of untargeted LC–MS/MS derived metabolomics data that reads raw data fro-m a .mzML file format, generates spectra and total ion chromatograms (TICs), allows peak picking, feature detection, reads processed data from xcms, MZmine 2 among others, offers functionalities for data matrix curation, normalization, imputation, scaling, quality metrics, QC-based batch corrections and interactive visualization of results (Riquelme et al. 2020).
AutoTuner, available as an R-package, is a parameter optimization algorithm that obtains parameter estimates from raw data in a single step as opposed to many iterations in a data-specific manner to generate robust features from untargeted LC–MS/MS runs (McLean & Kujawinski, 2020). For input, AutoTuner requires at least 3 samples of raw data converted from proprietary instrument formats (e.g. .mzML, .mzXML, or .CDF).
remove unwanted variation in a hierarchical structure (hRUV), is an R-package (also available as Shiny app) that aids in removal of unwanted variation from large scale LC–MS metabolomics studies which it accomplishes by progressively merging the adjustments in neighboring batches (Taiyun Kim, Owen Tang, Stephen T Vernon, Katharine A Kott, Yen Chin KoaTaiyun Kim, Owen Tang, Stephen T Vernon, Katharine A Kott, Yen Chin Koay, John Park, David James, Terence P Speed, Pengyi Yang, John F. O’Sullivan, Gemma A Figtree, Jean Yee Hwa Yangy, 2020). The package uses sample replicates to integrate data from several batches for removal of intra-batch signal drift and inter-batch unwanted variation and outperforms existing tools while retaining biological variation. For assessment of the results, a user can visualize results as three kinds of diagnostic plots, i.e., principal component analysis (PCA) plots, relative log expression (RLE) plots, and metabolite run plots.
MetumpX, is a Ubuntu-based R- package that facilitate easy download and installation of 103 tools spread across the standard untargeted MS- based metabolomics pipeline (Wajid et al. 2020). The package can aid in automatically installation of software pipelines truly speeding up the learning curve to build software workstations.
MeTaQuaC, is an R- package and aids in implementation of concepts and methods for Biocrates kits and its application in targeted LC–MS metabolomics workflows and creates a QC report containing visualization and informative scores, and provides summary statistics, and unsupervised multivariate analysis methods among others (Kuhring et al. 2020).
Dbnorm, is an R-package that allows visualization and removal of technical heterogeneity from large scale metabolomics dataset, after allowing inspection at both in macroscopic and microscopic scales at both sample batch and metabolic feature levels, respectively (Bararpour et al., 2020). dbnorm includes several statistical models such as, ComBat (parametric and non-parametric)-model from sva package that are already in use for metabolomics data normalization, and ber function.
MetaClean, available as an R-package, uses 11 peak quality metrics and 8 diverse ML algorithms to build a classifier for the automatic assessment of peak integration quality of peaks from untargeted metabolomics datasets (Chetnik et al. 2020). It was shown that AdaBoost algorithm and a set of 11 peak quality metrics were best performing classifiers, and applying this framework to peaks retained after filtering by 30% relative standard deviation (RSD) across pooled QC samples was able to further distinguish poorly integrated peaks that were not removed from filtering alone.
NeatMS, is a Python package that is available for untargeted LC–MS signal labelling and filtering, which enables automated filtering out of false positive MS1 peaks reported by routine LC–MS data processing pipelines. It relies on neural networking-based classification, and can process outputs from MZMine 2 and xcms analysis.
Annotation tools
Metabolite annotation remains a critical step that defines the success or failure of untargeted metabolomics efforts. With newer technologies such as collision cross section (CCS) data for ion mobility, high resolution mass spectra from Orbitrap, direct injection data, data independent acquisition (DIA)/ all ion fragmentation (AIF), imaging MS and multi-dimensional chromatography the annotation results have gained additional impetus in compound identification, but these methods have offered newer challenges in themselves for tool development. False discovery rates (FDRs) of annotations indicate that low FDRs yield low number yet reliable annotations, whereas higher FDR report high number of annotations by those of poor-quality annotations. Though metabolite annotation efforts can benefit from RT as an orthogonal information, efforts for combining RT predictions with MS/MS data is currently lacking (Witting & Böcker, 2020). Clearly reference spectra and spectral DBs/ libraries are not enough to annotate roughly 5–30% of the total features captured (depending on the environmental/ biological matrices in question) in a given mass spectrometry-based metabolomics dataset. Though experimentally obtained MS/MS data and NMR data on pure standards are precious, and aid in development of computational solutions for compound identification, they do not suffice at their current numbers, accessibility, and availability. Moreover, in 2020, the Metabolite Identification Task Group of the International Metabolomics Society assessed and proposed a set of revised reporting standards for metabolite annotation/ identification and requested community feedback for levels from A-G, from defining an enantiomer or a chiral metabolite (level A) (to unknown molecular formula with specific spectral features (G). Once formalized, these would positively affect and improve reporting standards in studies and the publication landscape in metabolomics research. In Fig. 1, 2, 3, shown are the software interfaces and analysis outputs for some of the annotation tools discussed in the following sections.
Fig. 1.
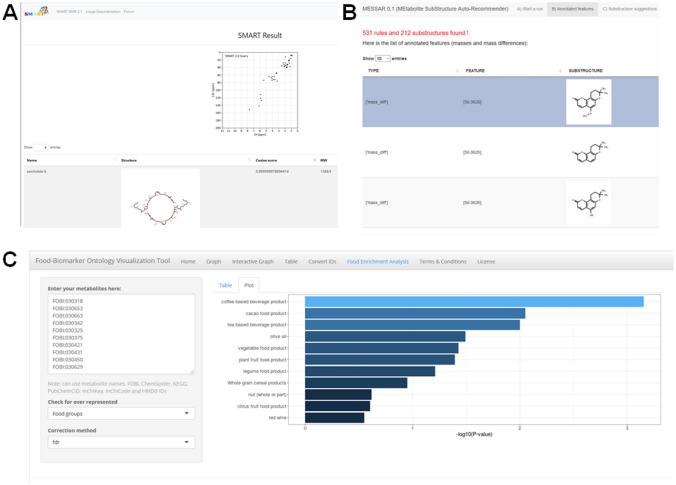
Snapshots of a subset of tools and resources discussed in this review. a SMART 2.0 outputs for a demo metabolite swinholide A, b Outputs from MESSAR for corosollic acid, c Demo analysis on FOBI
Fig. 2.
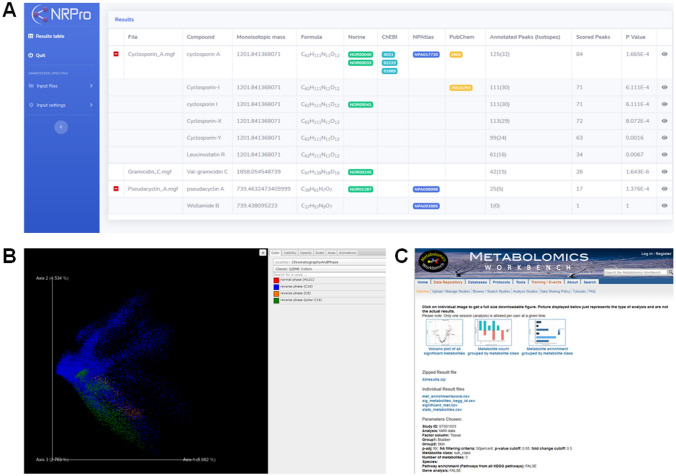
Snapshots of a subset of tools and resources discussed in this review. a Outputs on demo data on NRPro, b REDU analysis results for a demo data, c MetENPWeb analysis results
Fig. 3.
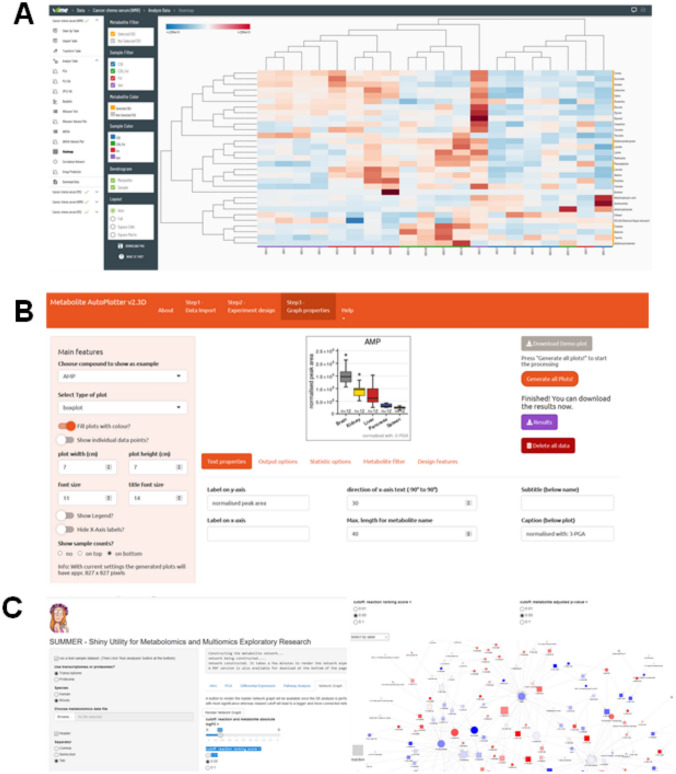
Snapshots of a subset of tools and resources discussed in this review. Web interfaces and snapshot of outputs for demo data on a VIIME, b MetaboliteAutoPlotter, and c SUMMER
MEtabolite SubStructure Auto-Recommender (MESSAR), is a web-based tool that provides an automated method for substructure recommendation guided by association rule mining, captures potential relationships between spectral features and substructures as learned from public spectral libraries for suggesting substructures for any unknown mass spectrum (Y. Liu, Mrzic, et al., 2020; Liu, Nellis, et al., 2020). Though the interface does not perform batch processing currently, it provides an open-source approach to annotate substructures.
Small Molecule Accurate Recognition Technology (SMART 2.0), is an artificial intelligence (AI) -based ML tool for mixture analysis in NMR data analysis workflow that aid in subsequent accelerated discovery and characterization of new NPs. SMART 2.0 generates structure hypotheses from two dimensional NMR data [1H-13C- Hetero‐nuclear Single Quantum Coherence (HSQC) spectra], then compares with a query HSQC spectrum against a library of > 100,000 NPs to provide outputs as simplified molecular-input line-entry system (SMILES), structures, cosine similarity, and molecular weights for a given compound of interest.
MetFID, is a tool that uses an artificial neural network (ANN) trained for predicting molecular fingerprints based on experimental MS/MS data (Fan et al. 2020). MetFID retrieves candidates from metabolite databases using molecular formula or m/z value of the precursor ion of the analyte and the candidate whose fingerprint is most analogous to the predicted fingerprint which is used for metabolite annotation. However, no codes or accessible tools/ repositories are provided with the published scholarly article.
CPVA, is a web-based tool that is aimed at the analyses of untargeted LC–MS/MS generated metabolomics data for visualization and annotation of LC peaks, where the tool performs functions such as annotation of adducts, isotopes and contaminants, and allows visualization of peak morphology metrics (Luan et al. 2020). Further, the tool aids in capturing potential noises and contaminants encountered in chromatographic peak lists generated from LC–MS/MS data, thus resulting in a reduced false positive peak calling in order to help data quality and downstream data processing.
NRPro, is a web-based application dedicated for dereplication and characterization of peptidic natural products (PNPs) from LC–MS/MS datasets that performs automatic peak annotation through a statistically validated scoring system (Ricart et al. 2020). An example NRPro dereplication effort revealed that the software was able to identify 169 PNPs in a dataset of 352 spectra with an FDR of 3.55.
MetENP/MetENPWeb, is available as an R-package on the Metabolomics Workbench repository, also deployed as a web-based application that allows extending the metabolomics data enrichment analysis to include Kyoto Encyclopedia of Genes and Genomes (KEGG)-based species-specific pathway analysis, pathway enrichment scores, gene-enzyme data, and enzymatic activities of the significantly altered metabolites on any Metabolomics Workbench submitted studies/ datasets (Choudhary et al. 2020). Various plots and visualizations such as volcano plots and bar graphs are available to the user of the tool after the analyses.
Class Assignment aNd Ontology Prediction Using mass Spectrometry (CANOPUS), available as a part of SIRIUS (Dührkop et al. 2019) suite of software, is a computational tool for systematic compound class annotation from fragmentation spectra (Dührkop et al. 2020). CANOPUS uses a deep neural network to predict 2,497 compound classes from fragmentation spectra, including all biologically relevant classes, and explicitly targets compounds for which neither spectral nor structural reference data are available in addition to predicting compound classes lacking MS/MS training data. Recently, CANOPUS was made available for analysis of MS/MS spectra obtained from both positive and negative mode ionization datasets.
molDiscovery, is a mass spectral database search method that improves both efficiency and accuracy of small molecule identification by (i) using an efficient algorithm to generate mass spectrometry fragmentations, and (ii) learning a probabilistic model to match small molecules with their mass spectra (Mohimani et al. 2020). A search of over 8 million spectra from the GNPS molecular networking infrastructure demonstrated that this probabilistic model can correctly identify nearly six times more unique compounds than other previously reported methods.
MetIDfyR, developed as an R-package that aids in in silico drug phase I/II biotransformation prediction and mass-spectrometric data mining from untargeted LC-HRMS/MS datasets (Delcourt et al. 2020) to help with feature annotation. With the ability to predict drug metabolism products from in vitro and in vivo studies, this tool holds potential in annotation workflows in drug discovery programs.
Qemistree, is a cheminformatics tool available as an advanced analysis workflow on GNPS infrastructure that allows mass spectrometry data to be represented in the context of sample metadata and chemical ontologies (Tripathi et al. 2020). This tree-guided data exploration tool allows comparison of metabolomics samples across different experimental conditions such as chromatographic shifts. The Qemistree software pipeline is freely available to the microbiome and metabolomics communities in the form of a QIIME2 plugin as well.
Ion identity molecular networking (IIMN), a workflow available within the GNPS ecosystem that complements the feature based molecular networking (FBMN) by aiding in annotating and connecting related ion species in feature-based molecular networks (Schmid et al. 2020). Though, MS1-based ion identity networks (IIN), are well-known, IIMN helps to integrate IIN into MS2-based molecular networks in the GNPS environment, thus adding MS/MS information on top of MS1 characteristics of ions.
Food-Biomarker Ontology (FOBI), is a tool developed in R language, is a web-based analysis and visualization package that is focused on interactive visualization of the FOBI structure (Castellano-Escuder et al. 2020). FOBI (Food-Biomarker Ontology) is a new ontology that describes food and their associated metabolite entities and is composed of two interconnected sub-ontologies, the ‘Food Ontology’ consisting of raw foods and ‘multi-component foods’ and a second: ‘Biomarker Ontology’ containing food intake biomarkers classified by their chemical classes. These two sub-ontologies are conceptually independent but interconnected by different properties. Functionalities of the tool include static and dynamic network visualization, downloadable tables, compound ID conversions, classical and food enrichment analyses.
BioDendro, is a Python package, for feature analysis of LC–MS/MS metabolomics data as a workflow that enables users to flexibly cluster and interrogate thousands of MS/MS spectra and quickly identify the core fragment patterns causing groupings leading to identification of core chemical backbones of a larger class, even when the individual metabolite of interest is not found in public databases (Rawlinson et al. 2020).
AllCCS, is a freely accessible database/ CCS atlas that covers vast chemical structures with > 5000 experimental CCS records and ~ 12 million calculated CCS values for > 1.6 million small molecules, with medium relative errors of 0.5–2% for a broad spectrum of small molecules (Zhou et al. 2020) for annotation of both known and unknown structures. Further, the tool facilitates a strategy for metabolite annotation using known or unknown chemical structures in IMS metabolomics workflows.
Binner, implemented as a standalone Java executable software package eliminates degenerate feature signals present in untargeted ESI-LC–MS/MS metabolomic datasets (Kachman et al. 2019). When a user provides an aligned feature table, with unique compound (feature) identifier, m/z, RT, and feature intensities, the Binner annotation file specifies information on annotation, mass, mode, charge, and tier information when annotating the final set of features.
MS-CleanR, is an R-package that provides functions for feature filtering and annotation of LC–MS data, that depends on the outputs of an need MS-DIAL (v4.00 or higher) and MS-FINDER (3.30 or higher) (Fraisier-Vannier et al. 2020). It uses MS-DIAL peak list processed in DDA or DIA obtained using either positive ionization mode (PI) or negative ionization mode (NI) or both as its input. MS-CleanR applies generic filters encompassing blank injection signal subtraction, background ions drift removal, unusual mass defect filtering, RSD threshold based on sample class and relative mass defect (RMD) window filtering. Furthermore, all selected features are exported to MS-FINDER program for in silico-based annotation using hydrogen rearrangement rules (HRR) scoring system and it was shown that implementation of MS-CleanR reduced the number of signals by nearly 80% while retaining 95% of unique metabolite features.
Retention time prediction for metabolomics (Retip), is an R-package, for predicting RT for small molecules in high pressure liquid chromatography (HPLC) MS data analysis workflows (Bonini et al. 2020). In order to help annotate unknowns and removing false positive annotations, it uses five different machine learning algorithms [i.e., random forest (RF), Bayesian regularized neural network, XGBoost, lightGBM, and Keras] to build a stable, accurate and fast RT prediction model. It also includes useful biochemical (structural) databases like: BMDB, ChEBI, DrugBank, ECMDB, FooDB, HMDB, KNApSAcK, PlantCyc, SMPDB, T3DB, UNPD, YMDB and STOFF.
QSRR Automator, is a software package that helps automate RT prediction model creation that’s been tested with metabolomics and lipidomics data from multiple chromatography columns from published literature and in-house work from the authors (Naylor et al. 2020).
MFAssignR, is an R-package that performs noise estimation, 13C and 34S polyisotopic mass filtering, mass measurement recalibration, and molecular formula assignment for UHPLC-MS data analysis in environmental complex mixtures (Schum et al. 2020). The function of this tool includes determination of noise, S/N threshold, identification of isotopes, potential recalibrant series, mass list recalibration, assignment of molecular formula to the recalibrated mass list, and output plots to evaluate the quality of the assignments.
Metabolite core structure-based Search (McSearch), is a program available both as an R-package and a web-based tool for automated metabolite annotation for LC–MS/MS data. It utilizes a Core Structure-based Search (CSS) algorithm, hypothetical neutral loss (HNL) library and biotransformation database to achieve metabolite annotation using the structural analogs of query compounds (Xing et al. 2020). The tool is available both as single search mode (.csv files) and batch search mode (.mgf or .mzXML formats). The input for single search mode is a Core Structure-based Search (see input_single_search.csv as a template). For batch mode, we currently accept raw data of .mgf or .mzXML format as input.
ReDU, is a GNPS based system for metadata capture of public deposited MS-based metabolomics data, with validated controlled vocabularies that captures knowledge by enabling reanalysis of public data and/or co-analysis of one’s own data for finding chemicals and associated metadata for a repository-scale molecular networking (Jarmusch et al. 2020). Currently, 38,305 files in GNPS (19.6% of GNPS) are ReDU compatible which includes data collected from natural and human-built environments, human and animal tissues, biofluids and food from all over the world.
Mass Spectrometry Search Tool (MASST), is a web-based MS search engine avaialble within the GNPS infrastructure that enables searches of all small-molecule MS/MS data in public metabolomics repositories (Wang, Jarmusch, et al., 2020; Wang, Leber, et al., 2020). MASST comprises a web-based system to search the public data repository part of the GNPS/MassIVE knowledge base and an analysis infrastructure for a single MS/MS spectrum. All public data submitted to/ available in GNPS/MassIVE becomes MASST-searchable. MASST searches yield results according to user-defined search parameters.
NPClassifier, is a DL tool for automated structural classification of NPs (Wang, Leber, et al. 2020). Currently available as a web-based tool for a simple search effort. The tool aims to accelerate NP discovery by facilitating and enabling large-scale genome and metabolome mining efforts and linking NP structures to their bioactivity.
LipidLynxX, developed in Python and available both as a standalone tool and web interface, is a software to convert diverse lipid annotations to unified identifiers and cross-ID matching (Ni & Fedorova, 2020). It primarily offers three models, the Converter, that allows conversion of different abbreviations to uniformed LipidLynxX IDs, an Equalizer that allows cross comparison of different levels of IDs on selected levels, and a Linker module that allows linking abbreviations to available resources.
patRoon, is a R-package that aids in non-target HR MS data analysis workflows (Helmus et al. 2021). The tool offers various functionality and strategies to simplify and perform automated processing of complex (environmental) data effectively using well-tested algorithms by harmonizing various open-source software tools and with reduced computational times.
Multifunctional tools
Multifunctional tools, in this review are defined as tools that allow an user to start with raw mass spectrometric or spectroscopic data and go through pre-processing steps, QCs, statistical analyses, data visualization and interpretation. In this section I cover software solutions that surfaced in 2020.
Skyline, originally developed for SWATH (and DIA) and targeted proteomics workflows, has now expanded to data analysis for small molecule analysis, including selected reaction monitoring (SRM), HRMS datasets, and calibrated quantification, for data visualization and interrogation features already available in Skyline, such as peak picking, chromatographic alignment, and transition selection among others (Adams et al. 2020).
notame, available as an R-package and a Wranglr Shiny web application for automation of worklist files, is a multifunctional tool for untargeted LC–MS/MS metabolomics data analyses that aids in reading outputs from MS-DIAL, allows drift correction, flagging and removal of low-quality features, imputation of missing values, batch effect correction, offers novel clustering methods, statistical analyses and visualization for QC, explorative analyses aiding in interpretation of statistical tests (Klåvus et al. 2020).
Breath AnaLysis viSualizAtion Metabolite discovery (BALSAM), is an interactive web-based tool that integrates state-of-the-art preprocessing and analysis techniques for supervised feature extraction and visualization of multi capillary column—ion mobility spectrometry (MCC-IMS) data preprocessing workflows that deals with breath analysis (Weber et al. 2020). In addition, it supports peak detection and peak alignment as well as RT based GC–MS and LC–MS data analysis.
MRMkit, is a software solution for data processing of large scale targeted LC–MS -based metabolomics data that performs automated peak detection, peak integration, normalization, batch effect correction, quality metric calculations, visualizations of chromatograms, and removal of redundant peaks from multimodal classes by RT selection (Teo et al. 2020).
MetaboShiny, available as a MetaboShiny, a R/Shiny-based package featuring data analysis, database- and formula prediction -based annotation and visualization on diverse MS datasets (Wolthuis et al. 2020). MetaboShiny allows a diverse set of customization and global settings to an user, in addition to adding databases, data normalization and filtering, statistical functions ranging from dimension reduction [from PCA, partial least-squares-discriminant analysis (PLS-DA) to t-distributed stochastic neighbor embedding (t-SNE)], univariate analyses on sets of features, range of visualizations from volcano plots to heatmaps, Venn diagrams, power calculations, and metabolite enrichment analyses.
SmartPeak, is a programmable software application that offers novel algorithms for RT alignment, calibration curve fitting, and peak interrogation for facilitating reproducibility by reducing operator bias to ensure high QC/quality assurance (QA) for automated processing of CE-, GC- and LC–MS(/MS) data, and HPLC data for targeted and semi-targeted metabolomics, lipidomics, and fluxomics experiments (Kutuzova et al. 2020).
MS-DIAL 4, is a standalone DIA software tool that provides a comprehensive lipidome atlas with RT, CCS, and MS/MS information encapsulating mass spectral fragmentations of lipids across 117 lipid subclasses and includes analysis of ion mobility MS/MS data (Tsugawa et al. 2020). Using lipidomics data from diverse samples the study reported semi quantified 8,051 lipids using MS-DIAL 4 with a 1–2% estimated FDR.
Integrated mass spectrometry-based untargeted metabolomics data mining (IP4M), is a multifunctional tool for untargeted MS-based metabolomics data processing and analysis that has 62 functions categorized into 8 modules (Liang et al. 2020). The modules cover all majority of the steps of metabolomics data mining, including raw data preprocessing (alignment, peak de-convolution, peak picking, and isotope filtering), peak annotation, peak table preprocessing, basic statistical description, classification and biomarker detection, correlation analysis, cluster and sub-cluster analysis, regression analysis, receiver operating characteristic (ROC) analysis, pathway and enrichment analysis, and sample size and power analysis.
DropMS, is a online tool with a user-friendly and browser-based interface to facilitate the processing of high resolution and precision oil mass spectrometry data for petroleomics applications (Rosa et al. 2020). Uploaded mass spectra to the server are processed using various algorithms reported in the literature, such as S/N ratio filters, recalibrations, chemical formula assimilations and data visualization using graphs and diagrams popularly known in mass spectrometry such as Van Krevelen and Kendrick diagrams among other visualizations.
Tools for statistical analysis and visualization
In this section, described are tools dedicated for statistical analyses and visualization of metabolomics data visualization.
EpiMetal, is a web-based application that allows statistical analyses and visualization of large datasets for epidemiological analyses and self-organizing maps (SOMs) for metabolomics (Ekholm et al. 2020). A pilot data with > 500 quantitative molecular measurements for each sample and two large-scale epidemiological cohorts (N > 10,000) are available to users on the interface as well.
Metabolite AutoPlotter, is an R-package and wrapped into a Shiny web application that can be run online in a web browser, which uses pre-processed metabolite-intensity tables as inputs and accepts different experimental designs, with respect to the number of metabolites, conditions and replicates and process and plots metabolite data sets (different types), converts and cleans-up the data, allows data normalization for sample descriptions and metabolite names as well as sorting experimental conditions (Pietzke & Vazquez, 2020).
Metabolite-Investigator, is a free and open web-based tool and stand-alone Shiny application, that provides a scalable analysis workflow for quantitative metabolomics data from multiple studies by performing data integration, cleaning, transformation, batch analysis and multiple statistical analysis methods including uni- and multivariable factor-metabolite associations, network analysis, and factor prioritization in one or more cohorts (Beuchel et al. 2020).
VIIME (VIsualization and Integration of Metabolomics Experiments), available as a web server, provides a workflow for metabolomics research by offering state-of-the-art integration algorithms and visualizations (Choudhury et al. 2020). A user starts with an uploaded spreadsheet of quantitative metabolomics data and runs a semi-automated process which informs about low-variance and high-missingness data, allows arbitrary sample and metabolite exclusion, and performs adjustable missing data imputation, informs about data pretreatment, runs PCA and block PCA, statistical analyses such as Wilcoxon and analysis of variance (ANOVA), and finally provides interactive tables, charts, heatmaps and networks diagrams as outputs on a given metabolomics data.
struct (Statistics in R using Class-based Templates), is an R/ Bioconductor package that defines a suite of class-based templates to allow users to develop and implement standardized and readable statistical analysis workflows for metabolomics and other omics technologies (Lloyd et al. 2020). Struct integrates with the STATistics Ontology to ensure consistent reporting and maximizes semantic interoperability. A related package, the structToolbox, which includes an extensive set of commonly used data analysis methods using the templates provided in the struct package. struct includes a suite of S4 class-based templates (i.e., model, sequence, iterator, chart and metric classes) to facilitate the standardization of R-based workflows for statistics and ML. The toolbox includes pre-processing methods (e.g. signal drift and batch correction, normalisation, missing value imputation and scaling), univariate (e.g. t-test, various forms of ANOVA, Kruskal–Wallis test and more) and multivariate statistical methods [e.g. PCA and partial least squares (PLS), including cross-validation and permutation testing] as well as machine learning methods (e.g. support vector machines).
lipidr, is an R/Bioconductor-package for data mining and analysis of lipidomics datasets that implements a lipidomic-focused analysis workflow for targeted and untargeted lipidomics (Mohamed et al. 2020). lipidr imports numerical matrices, Skyline exports, and Metabolomics Workbench files directly into R interface, and allows thorough data inspection, normalization, and uni- and multivariate analyses, resulting in interactive visualizations as well as a novel lipid set enrichment analysis.
NORmalization and EVAluation (NOREVA 2.0), is a web-server (also available as a standalone R-package) for normalization of metabolomics data, with the latest version’s capabilities to deal with time-course and multi-class metabolomics datasets (Yang et al. 2020). In addition, NOREVA 2.0 integrates a total of 168 normalization methods and combinations thereof leading to removal of unwanted variations, correction of signal drifts based on QCs, performance evaluation of the datasets, thus, pointing to the best normalization methods for a given dataset.
%polynova_2way, is a Macro written for the statistical software Statistical Analysis System (SAS) to help identify metabolites differentially expressed in study designs with a two-way factorial treatment and hierarchical design structure (Manjarin et al. 2020). The Macro calculates the least squares means using a linear mixed model with fixed and random effects, runs a 2-way ANOVA, corrects the P-values for the number of metabolites using the FDR or Bonferroni procedure, and calculates the P-value for the least squares mean differences for each metabolite.
rawR, available as an R-package that provides operating system (OS) independent access to all spectral data and chromatograms logged in the mass spectrometry vendor, Thermo Fisher Scientific’s .RAW files obtained from MS runs (Kockmann & Panse, 2020).
Metaboverse, is an interactive standalone software tool for the exploration and automated extraction of potential regulatory events, patterns, and trends from multi-omic data within the context of a metabolic network and other global reaction networks (Berg et al. 2020). The tool aids in analysis of Reactome knowledgebase derived networks for 90 + model organisms, helps integrate multi-condition and time course data, in addition to facilitating exploration of super-pathway specific reaction perturbation networks among others.
JavaScript mass spectrometry (JS-MS) 2.0, is a standalone visualization GUI software suite that provides a dependency-free, browser-based, one click, cross-platform solution for creating MS1 ground truth set of features (i.e., defined as raw data points manually curated into features, whether extracted ion chromatograms or isotopic envelopes) (Henning & Smith, 2020). The tool enables loading, viewing, and navigating MS1 data in 2- and 3-dimensions, and adds tools for capturing, editing, saving, and viewing isotopic envelope and extracted isotopic chromatogram features. It further interfaces via Hypertext Transfer Protocol (HTTP) to the MsDataServer application programming interface (API) for access to the MS data stored in the MzTree format.
Databases
In this section, I discuss the databases (both spectral and structural) that have appeared or updated in 2020.
COlleCtion of Open Natural prodUcTs (COCONUT), is available as a webserver (with downloadable structural data on NPs) an aggregated dataset of NPs from different open resources and offers a subsequent web interface to browse, search and easily and quickly download NPs (Sorokina & Steinbeck, 2020). The DB contains structures and sparse annotations for over 400,000 non-redundant NPs.
METLIN MS2, is chemical standards spectral DB that is well annotated and structurally diverse database consisting of over 850,000 chemical standards with MS/MS data generated in both positive and negative ionization modes at multiple collision energies (CEs), collectively containing over 4,000,000 curated HR MS/MS data that covers almost 1% of PubChem’s 93 million compounds (Xue et al. 2020).
EMBL-MCF, is an open LC–MS/MS spectral library that currently contains over 1600 fragmentation spectra obtained from 435 authentic standards of endogenous metabolites and lipids (Phapale et al. 2021). The EMBL-MCF spectral library is created and shared using an in-house developed web-application.
The Wake Forest CPM GC–MS spectral and RT libraries consist of HR EI-MS and HR chemical ionization (CI)-MS/MS spectra obtained from silylated chemical standards obtained from the Mass Spectrometry Metabolite Library of Standards (MSMLS Kit™) (B. B. Misra & Olivier, 2020).
Chemical Shift Multiplet Database (CSMDB), is a database that uses JRES spectra obtained from the Birmingham Metabolite Library (BML), to provide scores by accounting for both matched and unmatched peaks from a query list and the database hits (Charris-Molina et al. 2020). This input list is generated from a projection of a 2D statistical correlation analysis on the J-RESolved (JRES) spectra, p-[JRES- Statistical TOtal Correlation SpectroscopY (STOCSY)], being able to compare the multiplets for the matched peaks. The CSMDB is complemented with “consecutive queries to assess biological correlation” (ConQuer ABC), a simple inspection of peaks left unmatched from the query list and consecutive queries to assign all (or most) peaks in the original query list.
Other specialized tools
This section covers numerous tools that did not quite fall into the six categories listed above, and are developed with a purpose to address a specialized application to facilitate metabolomics data analysis. These tools include the ones developed for isotopic data analysis in stable isotope labelling experiments, softwares for analysis of lipidomics data, mass spectrometry imaging data, and multiomics/ integrated omics analysis.
Mass isotopolome analysis for mode of action identification (MIAMI), is a tool that uses MetaboliteDetector (https://md.tu-bs.de/) and non-targeted tracer fate detection (NTFD) libraries (http://ntfd.mit.edu/), combines the strengths of targeted and non-targeted efforts for estimation of metabolic flux changes in GC–MS datasets (Dudek et al. 2020). In stable isotope labeling experimental data, MIAMI determines a mass isotopomer distribution-based (MID) similarity network and incorporates the data into metabolic reference networks and aids in the identification of MID variations of all labeled metabolites across conditions, targets of metabolic changes are detected.
isoSCAN, is an R-package that automatically quantifies all isotopologues of intermediate metabolites of glycolysis, tricarboxylic acid (TCA) cycle, amino acids, pentose phosphate pathway, and urea cycle, from low resolution (LR)MS and HRMS data (i.e., GC-chemical ionization -MS) in stable isotope labeling experiments (Capellades et al. 2020).
LiPydomics, is available as a Python package which performs statistical and multivariate analyses (“stats” module), generates informative plots (“plotting” module), identifies lipid species at different confidence levels (“identification” module), and performs a text-based interface (“interactive” module) aiding in further interpretation (Ross et al. 2020).
LipidCreator, is available both as a Skyline plugin and a standalone/command-line operation, is a lipid building block-based workbench and knowledgebase for semi-automatic generation of targeted lipidomics MS assays and in silico spectral libraries (Peng et al. 2020). It can support diverse lipid categories, allows SRM/ parallel reaction monitoring (PRM) assay generation for both labeled and unlabeled lipid species and their derived fragment ions, allows in silico spectral library generation and CEs optimization and the entire workflow can be integrated into Konstanz Information Miner (KNIME™) and Galaxy workflows as a native node.
Lipid Annotator, is a standalone software for lipidomic analysis of data collected by HR LC–MS/MS (Koelmel et al. 2020). Lipid Annotator algorithm, intended for lipid annotation based on in-silico libraries, consists of five general steps: feature finding, association of MS/MS scans with features, annotation of possible lipids for each feature, calculation of the percent abundance of each fatty acyl constituent under a single chromatographic peak in the case of mixed spectra, and filtration of final annotated features. Lipid Annotator can be used on large datasets for rapid annotation, relative quantification, and statistics (using a downstream workflow with commercial tools such as MassHunter Profinder (Agilent Technologies) and MassHunter Mass Profiler Professional softwares (Agilent Technologies).
Raman2imzML, available as an R-package is a converter that transforms Raman imaging data in text format exported from WiRe 5.2 (Renishaw) and FIVE 5.1 (WiTec) into the .imzML data format (Iakab et al. 2020). The .mzML is a standardized common data format created and adopted by the mass spectrometry community and this tool exclusively handles imaging data for further exploratory imaging analysis.
Metabolomics datasets play an indispensable role in multi-omics data integration and analytics workflows as metabolites are the closest to the phenotype and helps connect with the genotype (Fiehn, 2002). Recent efforts in multi-omics domain encompass harmonization of quality metrics and power calculation in multi-omic studies (Tarazona et al. 2020) to standardized data sharing guidelines (Krassowski et al. 2020). A recent review introduced the tools for computational methods and resources in metabolomics and multiomics integration (Eicher et al. 2020). Another review focused on metabolomics-centric integration of data for biomedical research (Wörheide et al. 2021). Integration of omics datasets, such as those of metabolomics and microbiome/ metagenomics present challenges of their own (B. B. Misra, 2020c), and hence, more effective tools are necessary to address the challenges in this area. In this section, I capture a couple of the multi-omics tools developed in 2020.
Shiny Utility for Metabolomics and Multiomics Exploratory Research (SUMMER), is a Shiny-based tool that enables mechanistic interpretation of steady-state metabolomics data by integrating transcriptomics or proteomics data with metabolomics datasets by helping capture enzyme activities estimated from transcriptomics or proteomics data by calculating changes in reaction rate potentials (Huang et al. 2020). The tool offers several modules to perform PCA, differential expression analysis, pathway analysis, and network analysis.
metPropagate, is a network-based approach that uses untargeted metabolomics data from a single patient and a group of controls to prioritize candidate genes in patients with suspected inborn errors of metabolism (IEMs) (Graham Linck et al. 2020). This approach determines whether metabolomic evidence could be used to prioritize the causative gene from this list of candidate genes, where each gene in a patient’s candidate gene list is ranked using a per-gene metabolomic score termed the “metPropagate score”, which represented the likely metabolic relevance of a particular gene to each patient.
Summary of current tools
In this section, I summarize the observed trends for the tools reported in 2020, which are:
Majority of the software tools and packages focus on ‘annotations’, i.e., almost 35% of the total 72 tools reported for the year deal with untargeted metabolomics data annotation.
82% of the total tools reported are concerned with data analysis challenges with “LC–MS/MS”, mostly untargeted LC–HRMS/MS efforts.
Programming languages used for these tools mostly are R language packages (28 tools), Python language packages (11 tools), Java language (5 tools) or are web-servers/ web-based tools (23 tools).
48% of the reported tools are ‘easy to use’ (click to start, web-based, or plug-and-play type tools) from a user stand point for community of biologists and chemists who are not computational savvy.
Of the total tools reported here, 57% of the tools have a GitHub repository associated with them.
Couple of tools are improved versions, suggesting these are active tools that are being developed/maintained.
Lot of tools reported in the year deal with specialized applications: ranging from data integration (i.e., metabolomics data with proteomics/transcriptomics data), epidemiological metabolomics data, lipidomics, MSI data.
Concluding remarks
In summary, one can observe that there are numerous tools that were either developed from scratch or evolved from their previous versions in 2020 alone. Some tools and approaches found new applications, such as GNPS in the domain of GC–MS-based metabolomics (Aksenov et al. 2020), or released as a beta/ advanced version, i.e., MS-DIAL for lipidomics (Tsugawa et al. 2020) workflows. Only the future years will dictate as to which of these 2020 tools live on to see another year in terms of utility/ application, stays maintained and remain available, get improved, and get adopted by the metabolomics research community. Irrespective, all these tools help understanding metabolomics data from diverse stand points and are welcome additions to the community going forward into the big data-driven precision medicine era. In general, the trend is to develop, fast, computationally less intensive, robust, open-source, user-friendly tools that can adhere to findable, accessible, interoperable, and reproducible (FAIR) guidelines. Undoubtedly, the metabolomics research community needs more of these improved tools, and in the coming years the tools, resources, and databases will keep coming and getting better.
Acknowledgements
I acknowledge the efforts of the informatics and computational resource developers who help drive the field forward with their codes, packages, tools, and resources that enable the metabolomists, biologists and analytical chemists to keep pace with the volume and complexity of the metabolomics data generated. I do also apologize to all investigators whose tools and resources might have been missed in this review, inadvertently. I would like to acknowledge the independent reviewers and the editor for their comments to help improve this manuscript.
Abbreviations
- AIF
All ion fragmentation
- ANOVA
Analysis of variance
- ANN
Artificial neural network
- CE
Capillary electrophoresis
- DDA
Data dependent acquisition
- DIA
Data independent acquisition
- DB
Database
- FDR
False discovery rate
- FIA
Flow injection analysis
- GC
Gas chromatography
- GNPS
Global Natural Product Social molecular networking
- GUI
Graphical user interface
- HRMS
High-resolution mass spectrometry
- HR MS/MS
High-resolution tandem mass spectrometry
- Q-ToF
Hybrid quadrupole orthogonal time-of-flight
- IMS
Ion-mobility mass spectrometry
- KEGG
Kyoto encyclopedia of genes and genomes
- LC
Liquid chromatography
- ML
Machine learning
- MSI
Mass spectrometry imaging
- MS
Mass spectrometry
- m/z
Mass-to-charge
- DL
Meep learning
- MSI
Imaging mass spectrometry
- MRM
Multiple reaction monitoring
- NMR
Nuclear magnetic resonance
- PLS-DA
Partial least-squares-discriminant analysis
- PCA
Principal component analysis
- CCS
Collision cross section
- QA
Quality assurance
- QC
Quality control
- RSD
Relative standard deviation
- RT
Retention time
- R
R-Statistical programming
- S/N
Signal to noise ratio
- SRM
Single reaction monitoring
- MS/MS
Tandem mass spectrometry
- QqQ
Triple quadruple
- UPLC-TOF
Ultra-performance liquid chromatography-time-of-flight mass spectrometry
- XCMS
Various forms (X) of chromatography mass spectrometry
Funding
None.
Compliance with ethical standards
Conflict of interest
None.
Ethical approval
This article does not contain any studies with human and/or animal participants performed by the authors.
Research involving human and/or animal participants
None.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Adams KJ, Pratt B, Bose N, Dubois LG, St. John-Williams L, Perrott KM, et al. Skyline for small molecules: A unifying software package for quantitative metabolomics. Journal of Proteome Research. 2020;19(4):1447–1458. doi: 10.1021/acs.jproteome.9b00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksenov AA, Laponogov I, Zhang Z, Doran SLF, Belluomo I, Veselkov D, et al. Auto-deconvolution and molecular networking of gas chromatography–mass spectrometry data. Nature Biotechnology. 2020 doi: 10.1038/s41587-020-0700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov T. Spatial metabolomics and imaging mass spectrometry in the age of artificial intelligence. Annual Review of Biomedical Data Science. 2020;3:1. doi: 10.1146/annurev-biodatasci-011420-031537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron AT, Gentry EC, McPhail KL, Nothias LF, Nothias-Esposito M, Bouslimani A, et al. Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nature Protocols. 2020;15(6):1954–1991. doi: 10.1038/s41596-020-0317-5. [DOI] [PubMed] [Google Scholar]
- Bararpour, N., Gilardi, F., Carmeli, C., Sidibe, J., Ivanisevic, J., Caputo, T., Augsburger, M., Grabherr, S., Desvergne, B., Guex, N., Bochud, M., Thomas, A. (2020). Visualization and normalization of drift effect across batches in metabolome-wide association studies. biorx, 914051. 10.1101/2020.01.22.914051
- Berg, J. A., Zhou, Y., Cameron Waller, T., Ouyang, Y., Nowinski, S. M., Van Ry, T., George, I., Cox, J. E., Wang, B., Rutter, J. (2020). Gazing into the Metaboverse: Automated exploration and contextualization of metabolic data. bioRxiv, 171850.10.1101/2020.06.25.171850
- Beuchel C, Kirsten H, Ceglarek U, Scholz M. Metabolite-Investigator: An integrated user-friendly workflow for metabolomics multi-study analysis. Bioinformatics. 2020 doi: 10.1093/bioinformatics/btaa967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhinderwala F, Evans P, Jones K, Laws BR, Smith TG, Morton M, Powers R. Phosphorus NMR and its application to metabolomics. Analytical Chemistry. 2020;92(14):9536–9545. doi: 10.1021/acs.analchem.0c00591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonini P, Kind T, Tsugawa H, Barupal DK, Fiehn O. Retip: Retention time prediction for compound annotation in untargeted metabolomics. Analytical Chemistry. 2020;92(11):7515–7522. doi: 10.1021/acs.analchem.9b05765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen Du, Tian Z, Peter KT, Kolodziej EP. Developing unique nontarget high-resolution mass spectrometry signatures to track contaminant sources in urban waters. Environmental Science & Technology Letters. 2020;7(12):923–930. doi: 10.1021/acs.estlett.0c00749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capellades, J., Junza, A., Samino, S., Brunner, J. S., Schabbauer, G., Vinaixa, M., & Yanes, O. (2020). Exploring the use of gas chromatography coupled to chemical ionization mass spectrometry (GC-CI-MS) for stable isotope labeling in metabolomics. Analytical Chemistry.10.1021/acs.analchem.0c02998 [DOI] [PubMed]
- Castellano-Escuder P, González-Domínguez R, Wishart DS, Andrés-Lacueva C, Sánchez-Pla A. FOBI: An ontology to represent food intake data and associate it with metabolomic data. Database: The journal of biological databases and curation. 2020;2020:2020. doi: 10.1093/databa/baaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charris-Molina A, Riquelme G, Burdisso P, Hoijemberg PA. Consecutive queries to assess biological correlation in NMR metabolomics: Performance of comprehensive search of multiplets over typical 1D 1H NMR database search. Journal of Proteome Research. 2020;19(8):2977–2988. doi: 10.1021/acs.jproteome.9b00872. [DOI] [PubMed] [Google Scholar]
- Chetnik K, Petrick L, Pandey G. MetaClean: A machine learning-based classifier for reduced false positive peak detection in untargeted LC–MS metabolomics data. Metabolomics. 2020;16(11):117. doi: 10.1007/s11306-020-01738-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary, K. S., Fahy, E., Coakley, K., Sud, M., Maurya, M. R., & Subramaniam, S. (2020). MetENP/MetENPWeb: An R package and web application for metabolomics enrichment and pathway analysis in Metabolomics Workbench. bioRxiv, 2020.11.20.391912.10.1101/2020.11.20.391912
- Choudhury, R., Beezley, J., Davis, B., Tomeck, J., Gratzl, S., Golzarri-arroyo, L., et al. (2020). Viime: Visualization and Integration of Metabolomics Experiments. Journal of Open Source Software 5, 1–13.10.21105/joss.02410 [DOI] [PMC free article] [PubMed]
- Delcourt V, Barnabé A, Loup B, Garcia P, André F, Chabot B, et al. MetIDfyR: An open-source r package to decipher small-molecule drug metabolism through high-resolution mass spectrometry. Analytical Chemistry. 2020 doi: 10.1021/acs.analchem.0c02281. [DOI] [PubMed] [Google Scholar]
- Du J, Su Y, Qian C, Yuan D, Miao K, Lee D, et al. Raman-guided subcellular pharmaco-metabolomics for metastatic melanoma cells. Nature Communications. 2020;11(1):4830. doi: 10.1038/s41467-020-18376-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek C-A, Reuse C, Fuchs R, Hendriks J, Starck V, Hiller K. MIAMI––a tool for non-targeted detection of metabolic flux changes for mode of action identification. Bioinformatics. 2020;36(12):3925–3926. doi: 10.1093/bioinformatics/btaa251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dührkop, K., Fleischauer, M., Ludwig, M., et al. SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nature Methods, 16, 299–302 (2019). 10.1038/s41592-019-0344-8 [DOI] [PubMed]
- Dührkop K, Nothias L-F, Fleischauer M, Reher R, Ludwig M, Hoffmann MA, et al. Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nature Biotechnology. 2020 doi: 10.1038/s41587-020-0740-8. [DOI] [PubMed] [Google Scholar]
- Eicher T, Kinnebrew G, Patt A, Spencer K, Ying K, Ma Q, et al. Metabolomics and multi-omics integration: A survey of computational methods and resources. Metabolites. 2020;10(5):202. doi: 10.3390/metabo10050202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekholm J, Ohukainen P, Kangas AJ, Kettunen J, Wang Q, Karsikas M, et al. EpiMetal: An open-source graphical web browser tool for easy statistical analyses in epidemiology and metabolomics. International Journal of Epidemiology. 2020;49(4):1075–1081. doi: 10.1093/ije/dyz244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z, Alley A, Ghaffari K, Ressom HW. MetFID: artificial neural network-based compound fingerprint prediction for metabolite annotation. Metabolomics. 2020;16(10):104. doi: 10.1007/s11306-020-01726-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O. Metabolomics—The link between genotypes and phenotypes. Plant Molecular Biology. 2002;48(1–2):155–171. doi: 10.1023/A:1013713905833. [DOI] [PubMed] [Google Scholar]
- Fraisier-Vannier O, Chervin J, Cabanac G, Puech V, Fournier S, Durand V, et al. MS-CleanR: A feature-filtering workflow for untargeted LC–MS based metabolomics. Analytical Chemistry. 2020;92(14):9971–9981. doi: 10.1021/acs.analchem.0c01594. [DOI] [PubMed] [Google Scholar]
- Geier B, Sogin EM, Michellod D, Janda M, Kompauer M, Spengler B, et al. Spatial metabolomics of in situ host–microbe interactions at the micrometre scale. Nature Microbiology. 2020;5(3):498–510. doi: 10.1038/s41564-019-0664-6. [DOI] [PubMed] [Google Scholar]
- Graham Linck EJ, Richmond PA, Tarailo-Graovac M, Engelke U, Kluijtmans LAJ, Coene KLM, et al. metPropagate: network-guided propagation of metabolomic information for prioritization of metabolic disease genes. npj Genomic Medicine. 2020;5(1):25. doi: 10.1038/s41525-020-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Huan T. Comparison of full-scan, data-dependent, and data-independent acquisition modes in liquid chromatography-mass spectrometry based untargeted metabolomics. Analytical Chemistry. 2020;92(12):8072–8080. doi: 10.1021/acs.analchem.9b05135. [DOI] [PubMed] [Google Scholar]
- Guo J, Huan T. Evaluation of significant features discovered from different data acquisition modes in mass spectrometry-based untargeted metabolomics. Analytica Chimica Acta. 2020;1137:37–46. doi: 10.1016/j.aca.2020.08.065. [DOI] [PubMed] [Google Scholar]
- Han W, Li L. Evaluating and minimizing batch effects in metabolomics. Mass Spectrometry Reviews. 2020 doi: 10.1002/mas.21672. [DOI] [PubMed] [Google Scholar]
- Helmus R, ter Laak TL, van Wezel AP, de Voogt P, Schymanski EL. patRoon: Open source software platform for environmental mass spectrometry based non-target screening. Journal of Cheminformatics. 2021;13(1):1. doi: 10.1186/s13321-020-00477-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning J, Smith R. A web-based system for creating, viewing, and editing precursor mass spectrometry ground truth data. BMC Bioinformatics. 2020;21(1):418. doi: 10.1186/s12859-020-03752-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, V. J., Bandrowski, A. E., Pepin, A.-S., Gonzalez, B. J., & Desfeux, A. (2014). OMICtools: an informative directory for multi-omic data analysis. Database, 2014, bau069–bau069.10.1093/database/bau069 [DOI] [PMC free article] [PubMed]
- Hohrenk LL, Itzel F, Baetz N, Tuerk J, Vosough M, Schmidt TC. Comparison of software tools for liquid chromatography–high-resolution mass spectrometry data processing in nontarget screening of environmental samples. Analytical Chemistry. 2020;92(2):1898–1907. doi: 10.1021/acs.analchem.9b04095. [DOI] [PubMed] [Google Scholar]
- Mohimani H, Cao L, Guler M, Tagirdzhanov A. MolDiscovery: Learning Mass Spectrometry Fragmentation of Small Molecules. Research Square. 2020 doi: 10.21203/rs.3.rs-71854/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Currais A, Shokhirev MN. SUMMER, a shiny utility for metabolomics and multiomics exploratory research. Metabolomics. 2020;16(12):126. doi: 10.1007/s11306-020-01750-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakab SA, Sementé L, García-Altares M, Correig X, Ràfols P. Raman2imzML converts Raman imaging data into the standard mass spectrometry imaging format. BMC Bioinformatics. 2020;21(1):448. doi: 10.1186/s12859-020-03789-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahagirdar S, Saccenti E. Evaluation of single sample network inference methods for metabolomics-based systems medicine. Journal of Proteome Research. 2020 doi: 10.1021/acs.jproteome.0c00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmusch AK, Wang M, Aceves CM, Advani RS, Aguirre S, Aksenov AA, et al. ReDU: a framework to find and reanalyze public mass spectrometry data. Nature Methods. 2020 doi: 10.1038/s41592-020-0916-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju R, Liu X, Zheng F, Zhao X, Lu X, Lin X, et al. A graph density-based strategy for features fusion from different peak extract software to achieve more metabolites in metabolic profiling from high-resolution mass spectrometry. Analytica Chimica Acta. 2020;1139:8–14. doi: 10.1016/j.aca.2020.09.029. [DOI] [PubMed] [Google Scholar]
- Kachman M, Habra H, Duren W, Wigginton J, Sajjakulnukit P, Michailidis G, et al. Deep annotation of untargeted LC-MS metabolomics data with Binner. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakimov B, Mobaraki N, Trimigno A, Aru V, Engelsen SB. Signature mapping (SigMa): An efficient approach for processing complex human urine 1H NMR metabolomics data. Analytica Chimica Acta. 2020;1108:142–151. doi: 10.1016/j.aca.2020.02.025. [DOI] [PubMed] [Google Scholar]
- Klåvus A, Kokla M, Noerman S, Koistinen VM, Tuomainen M, Zarei I, et al. “Notame”: Workflow for non-targeted LC–MS metabolic profiling. Metabolites. 2020;10(4):135. doi: 10.3390/metabo10040135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kockmann, T., & Panse, C. (2020). rawR - Direct access to raw mass spectrometry data in R. bioRxiv, 2020.10.30.362533. 10.1101/2020.10.30.362533
- Koelmel JP, Li X, Stow SM, Sartain MJ, Murali A, Kemperman R, et al. Lipid annotator: Towards accurate annotation in non-targeted liquid chromatography high-resolution tandem mass spectrometry (LC-HRMS/MS) lipidomics using a rapid and user-friendly software. Metabolites. 2020;10(3):101. doi: 10.3390/metabo10030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyukevich Y, Zherebker A, Orlov A, Kovaleva O, Burykina T, Isotov B, Nikolaev EN. Hydrogen/deuterium and 16 O/ 18 O-exchange mass spectrometry boosting the reliability of compound identification. Analytical Chemistry. 2020;92(10):6877–6885. doi: 10.1021/acs.analchem.9b05379. [DOI] [PubMed] [Google Scholar]
- Kouřil Š, de Sousa J, Václavík J, Friedecký D, Adam T. CROP: correlation-based reduction of feature multiplicities in untargeted metabolomic data. Bioinformatics. 2020;36(9):2941–2942. doi: 10.1093/bioinformatics/btaa012. [DOI] [PubMed] [Google Scholar]
- Krassowski M, Das V, Sahu SK, Misra BB. State of the field in multi-omics research: From computational needs to data mining and sharing. Frontiers in Genetics. 2020 doi: 10.3389/fgene.2020.610798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn S, Colreavy-Donnelly S, de Andrade Silva Quaresma LE, de Andrade Silva Quaresma E, Borges RM. Applying NMR compound identification using NMRfilter to match predicted to experimental data. Metabolomics. 2020;16(12):123. doi: 10.1007/s11306-020-01748-1. [DOI] [PubMed] [Google Scholar]
- Kuhring M, Eisenberger A, Schmidt V, Kränkel N, Leistner DM, Kirwan J, Beule D. Concepts and software package for efficient quality control in targeted metabolomics studies: MeTaQuaC. Analytical Chemistry. 2020;92(15):10241–10245. doi: 10.1021/acs.analchem.0c00136. [DOI] [PubMed] [Google Scholar]
- Kutuzova S, Colaianni P, Röst H, Sachsenberg T, Alka O, Kohlbacher O, et al. SmartPeak automates targeted and quantitative metabolomics data processing. Analytical Chemistry. 2020;92(24):15968–15974. doi: 10.1021/acs.analchem.0c03421. [DOI] [PubMed] [Google Scholar]
- Letertre MPM, Dervilly G, Giraudeau P. Combined nuclear magnetic resonance spectroscopy and mass spectrometry approaches for metabolomics. Analytical Chemistry. 2020 doi: 10.1021/acs.analchem.0c04371. [DOI] [PubMed] [Google Scholar]
- Li Y, Bouza M, Wu C, Guo H, Huang D, Doron G, et al. Sub-nanoliter metabolomics via mass spectrometry to characterize volume-limited samples. Nature Communications. 2020;11(1):5625. doi: 10.1038/s41467-020-19444-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, Liu Q, Zhou K, Jia W, Xie G, Chen T. IP4M: an integrated platform for mass spectrometry-based metabolomics data mining. BMC Bioinformatics. 2020;21(1):444. doi: 10.1186/s12859-020-03786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebal UW, Phan ANT, Sudhakar M, Raman K, Blank LM. Machine learning applications for mass spectrometry-based metabolomics. Metabolites. 2020;10(6):243. doi: 10.3390/metabo10060243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Nellis M, Uppal K, Ma C, Tran V, Liang Y, et al. Reference standardization for quantification and harmonization of large-scale metabolomics. Analytical Chemistry. 2020 doi: 10.1021/acs.analchem.0c00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Mrzic A, Meysman P, De Vijlder T, Romijn EP, Valkenborg D, et al. MESSAR: Automated recommendation of metabolite substructures from tandem mass spectra. PLoS ONE. 2020;15(1):e0226770. doi: 10.1371/journal.pone.0226770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd GR, Jankevics A, Weber RJM. struct: An R/Bioconductor-based framework for standardized metabolomics data analysis and beyond. Bioinformatics. 2020 doi: 10.1093/bioinformatics/btaa1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Xing X, Wang L, Chen L, Zhang S, McReynolds MR, Rabinowitz JD. Improved annotation of untargeted metabolomics data through buffer modifications that shift adduct mass and intensity. Analytical Chemistry. 2020;92(17):11573–11581. doi: 10.1021/acs.analchem.0c00985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan H, Jiang X, Ji F, Lan Z, Cai Z, Zhang W. CPVA: A web-based metabolomic tool for chromatographic peak visualization and annotation. Bioinformatics. 2020;36(12):3913–3915. doi: 10.1093/bioinformatics/btaa200. [DOI] [PubMed] [Google Scholar]
- Madrid-Gambin F, Oller-Moreno S, Fernandez L, Bartova S, Giner MP, Joyce C, et al. AlpsNMR: Asn R package for signal processing of fully untargeted NMR-based metabolomics. Bioinformatics. 2020;36(9):2943–2945. doi: 10.1093/bioinformatics/btaa022. [DOI] [PubMed] [Google Scholar]
- Mahmud I, Garrett TJ. Mass spectrometry techniques in emerging pathogens studies: COVID-19 Perspectives. Journal of the American Society for Mass Spectrometry. 2020;31(10):2013–2024. doi: 10.1021/jasms.0c00238. [DOI] [PubMed] [Google Scholar]
- Manjarin R, Maj MA, La Frano MR, Glanz H. %polynova_2way: A SAS macro for implementation of mixed models for metabolomics data. PLoS ONE. 2020;15(12):e0244013. doi: 10.1371/journal.pone.0244013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyushin DD, Sholokhova AY, Buryak AK. Deep learning driven GC-MS library search and its application for metabolomics. Analytical Chemistry. 2020;92(17):11818–11825. doi: 10.1021/acs.analchem.0c02082. [DOI] [PubMed] [Google Scholar]
- McLean C, Kujawinski EB. AutoTuner: High fidelity and robust parameter selection for metabolomics data processing. Analytical Chemistry. 2020;92(8):5724–5732. doi: 10.1021/acs.analchem.9b04804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra BB. Open-source software tools, databases, and resources for single-cell and single-cell-type metabolomics. In: Shrestha B, editor. Single cell metabolism. Methods in molecular biology. New York: Humana; 2020. pp. 191–217. [DOI] [PubMed] [Google Scholar]
- Misra BB. Data normalization strategies in metabolomics: Current challenges, approaches, and tools. European Journal of Mass Spectrometry. 2020;26:165–174. doi: 10.1177/1469066720918446. [DOI] [PubMed] [Google Scholar]
- Misra BB. The connection and disconnection between microbiome and metabolome: A critical appraisal in clinical research. Biological Research For Nursing. 2020;22:561. doi: 10.1177/1099800420903083. [DOI] [PubMed] [Google Scholar]
- Misra BB, Olivier M. High resolution GC-orbitrap-MS metabolomics using both electron ionization and chemical ionization for analysis of human plasma. Journal of Proteome Research. 2020;19(7):2717–2731. doi: 10.1021/acs.jproteome.9b00774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra B, van der Hooft J. Updates in metabolomics tools and resources: 2014–2015. Electrophoresis. 2015;37(1):86–110. doi: 10.1002/elps.201500417. [DOI] [PubMed] [Google Scholar]
- Mohamed A, Molendijk J, Hill MM. lipidr: A software tool for data mining and analysis of lipidomics datasets. Journal of Proteome Research. 2020;19(7):2890–2897. doi: 10.1021/acs.jproteome.0c00082. [DOI] [PubMed] [Google Scholar]
- MZmine Development Team. (2015). MZmine 2 manual, (c), 14.
- Naylor BC, Catrow JL, Maschek JA, Cox JE. QSRR automator: A tool for automating retention time prediction in lipidomics and metabolomics. Metabolites. 2020;10(6):237. doi: 10.3390/metabo10060237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, Z., & Fedorova, M. (2020). LipidLynxX: a data transfer hub to support integration of large scale lipidomics datasets, 33894.10.1101/2020.04.09.033894
- O’Shea K, Misra BB. Software tools, databases and resources in metabolomics: Updates from 2018 to 2019. Metabolomics. 2020;16(3):1–23. doi: 10.1007/s11306-020-01657-3. [DOI] [PubMed] [Google Scholar]
- Peng B, Kopczynski D, Pratt BS, Ejsing CS, Burla B, Hermansson M, et al. LipidCreator workbench to probe the lipidomic landscape. Nature Communications. 2020;11(1):2057. doi: 10.1038/s41467-020-15960-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phapale P, Palmer A, Gathungu RM, Kale D, Brügger B, Alexandrov T. Public LC-orbitrap tandem mass spectral library for metabolite identification. Journal of Proteome Research. 2021 doi: 10.1021/acs.jproteome.0c00930. [DOI] [PubMed] [Google Scholar]
- Pietzke M, Vazquez A. Metabolite AutoPlotter—an application to process and visualise metabolite data in the web browser. Cancer & Metabolism. 2020;8(1):15. doi: 10.1186/s40170-020-00220-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomyen Y, Wanichthanarak K, Poungsombat P, Fahrmann J, Grapov D, Khoomrung S. Deep metabolome: Applications of deep learning in metabolomics. Computational and Structural Biotechnology Journal. 2020;18:2818–2825. doi: 10.1016/j.csbj.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroz-Moreno C, Furlan MF, Belinato JR, Augusto F, Alexandrino GL, Mogollón NGS. RGCxGC toolbox: An R-package for data processing in comprehensive two-dimensional gas chromatography-mass spectrometry. Microchemical Journal. 2020;156:104830. doi: 10.1016/j.microc.2020.104830. [DOI] [Google Scholar]
- Rawlinson C, Jones D, Rakshit S, Meka S, Moffat CS, Moolhuijzen P. Hierarchical clustering of MS/MS spectra from the firefly metabolome identifies new lucibufagin compounds. Scientific Reports. 2020 doi: 10.1038/s41598-020-63036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricart E, Pupin M, Müller M, Lisacek F. Automatic annotation and dereplication of tandem mass spectra of peptidic natural products. Analytical Chemistry. 2020 doi: 10.1021/acs.analchem.0c03208. [DOI] [PubMed] [Google Scholar]
- Riquelme G, Zabalegui N, Marchi P, Jones CM, Monge ME. A python-based pipeline for preprocessing LC–MS data for untargeted metabolomics workflows. Metabolites. 2020;10(10):416. doi: 10.3390/metabo10100416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, R., Petras, D., Nothias, L-F., Wang, M., Aron, A. T., Jagels, A., Tsugawa, H., Rainer, J., Garcia-Aloy, M., Dührkop, K., Korf, A., Pluskal, T., Kameník, Z., Jarmusch, A. K., Andrés Mauricio Caraballo-Rodrígu, P. C. D. (2020). Ion Identity Molecular Networking in the GNPS Environment. bioRxiv, 088948.10.1101/2020.05.11.088948
- Rosa TR, Folli GS, Pacheco WLS, Castro MP, Romão W, Filgueiras PR. DropMS: Petroleomics data treatment based in web server for high-resolution mass spectrometry. Journal of the American Society for Mass Spectrometry. 2020;31(7):1483–1490. doi: 10.1021/jasms.0c00109. [DOI] [PubMed] [Google Scholar]
- Ross DH, Cho JH, Zhang R, Hines KM, Xu L. LiPydomics: A python package for comprehensive prediction of lipid collision cross sections and retention times and analysis of ion mobility-mass spectrometry-based lipidomics data. Analytical Chemistry. 2020 doi: 10.1021/acs.analchem.0c02560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajulga R, Easterly C, Riffle M, Mesuere B, Muth T, Mehta S, et al. Survey of metaproteomics software tools for functional microbiome analysis. PLoS ONE. 2020;15(11):e0241503. doi: 10.1371/journal.pone.0241503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvin B, Lagziel S, Sarvin N, Mukha D, Kumar P, Aizenshtein E, Shlomi T. Fast and sensitive flow-injection mass spectrometry metabolomics by analyzing sample-specific ion distributions. Nature Communications. 2020;11(1):3186. doi: 10.1038/s41467-020-17026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schum SK, Brown LE, Mazzoleni LR. MFAssignR: Molecular formula assignment software for ultrahigh resolution mass spectrometry analysis of environmental complex mixtures. Environmental Research. 2020;191:110114. doi: 10.1016/j.envres.2020.110114. [DOI] [PubMed] [Google Scholar]
- Sen P, Lamichhane S, Mathema VB, Mcglinchey A, Dickens AM, Khoomrung S, Ore M. OUP accepted manuscript. Briefings Bioinformatics. 2020 doi: 10.1093/bib/bbaa204. [DOI] [PubMed] [Google Scholar]
- Sorokina M, Steinbeck C. Review on natural products databases: where to find data in 2020. Journal of Cheminformatics. 2020;12(1):20. doi: 10.1186/s13321-020-00424-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southam AD, Pursell H, Frigerio G, Jankevics A, Weber RJM, Dunn WB. Characterization of monophasic solvent-based tissue extractions for the detection of polar metabolites and lipids applying ultrahigh-performance liquid chromatography-mass spectrometry clinical metabolic phenotyping assays. Journal of Proteome Research. 2020 doi: 10.1021/acs.jproteome.0c00660. [DOI] [PubMed] [Google Scholar]
- Spicer R, Salek RM, Moreno P, Cañueto D, Steinbeck C. Navigating freely-available software tools for metabolomics analysis. Metabolomics. 2017;13(9):106. doi: 10.1007/s11306-017-1242-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spraker JE, Luu GT, Sanchez LM. Imaging mass spectrometry for natural products discovery: A review of ionization methods. Natural Product Reports. 2020;37(2):150–162. doi: 10.1039/C9NP00038K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiyun Kim, Owen Tang, Stephen T Vernon, Katharine A Kott, Yen Chin KoaTaiyun Kim, Owen Tang, Stephen T Vernon, Katharine A Kott, Yen Chin Koay, John Park, David James, Terence P Speed, Pengyi Yang, John F. O’Sullivan, Gemma A Figtree, Jean Yee Hwa Yangy, J. Y. H. Y. (2020). hRUV: Hierarchical approach to removal of unwanted variation for large-scale metabolomics data. bioRxiv, 423723.10.1101/2020.12.21.423723
- Tarazona S, Balzano-Nogueira L, Gómez-Cabrero D, Schmidt A, Imhof A, Hankemeier T, et al. Harmonization of quality metrics and power calculation in multi-omic studies. Nature Communications. 2020;11(1):3092. doi: 10.1038/s41467-020-16937-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautenhahn R, Böttcher C, Neumann S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics. 2008;9(1):504. doi: 10.1186/1471-2105-9-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo G, Chew WS, Burla BJ, Herr D, Tai ES, Wenk MR, et al. MRMkit: Automated data processing for large-scale targeted metabolomics analysis. Analytical Chemistry. 2020 doi: 10.1021/acs.analchem.0c03060. [DOI] [PubMed] [Google Scholar]
- Thomen A, Najafinobar N, Penen F, Kay E, Upadhyay PP, Li X, et al. Subcellular mass spectrometry imaging and absolute quantitative analysis across organelles. ACS Nano. 2020;14(4):4316–4325. doi: 10.1021/acsnano.9b09804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CJ, Witt M, Forcisi S, Moritz F, Kessler N, Laukien FH, Schmitt-Kopplin P. An enhanced isotopic fine structure method for exact mass analysis in discovery metabolomics: FIA-CASI-FTMS. Journal of the American Society for Mass Spectrometry. 2020;31(10):2025–2034. doi: 10.1021/jasms.0c00047. [DOI] [PubMed] [Google Scholar]
- Tripathi A, Vázquez-Baeza Y, Gauglitz JM, Wang M, Dührkop K, Nothias-Esposito M, et al. Chemically informed analyses of metabolomics mass spectrometry data with Qemistree. Nature Chemical Biology. 2020 doi: 10.1038/s41589-020-00677-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugawa H, Cajka T, Kind T, Ma Y, Higgins B, Ikeda K, et al. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nature Methods. 2015;12(6):523–526. doi: 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugawa H, Ikeda K, Takahashi M, Satoh A, Mori Y, Uchino H, et al. A lipidome atlas in MS-DIAL 4. Nature Biotechnology. 2020;38(10):1159–1163. doi: 10.1038/s41587-020-0531-2. [DOI] [PubMed] [Google Scholar]
- van der Laan T, Dubbelman A-C, Duisters K, Kindt A, Harms AC, Hankemeier T. High-throughput fractionation coupled to mass spectrometry for improved quantitation in metabolomics. Analytical Chemistry. 2020;92(21):14330–14338. doi: 10.1021/acs.analchem.0c01375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajid B, Iqbal H, Jamil M, Rafique H, Anwar F. MetumpX—a metabolomics support package for untargeted mass spectrometry. Bioinformatics. 2020;36(5):1647–1648. doi: 10.1093/bioinformatics/btz765. [DOI] [PubMed] [Google Scholar]
- Wang M, Jarmusch AK, Vargas F, Aksenov AA, Gauglitz JM, Weldon K, et al. Mass spectrometry searches using MASST. Nature Biotechnology. 2020;38(1):23–26. doi: 10.1038/s41587-019-0375-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M., Leber, C., Nothias, L., Reher, R., Kang, K. Bin, Hooft, J. J. Van Der, et al. (2020). NPClassifier: A deep neural network-based structural classification tool for natural products, (1).10.26434/chemrxiv.12885494.v1 [DOI] [PMC free article] [PubMed]
- Weber P, Pauling JK, List M, Baumbach J. BALSAM—An interactive online platform for breath analysis visualization and classification. Metabolites. 2020;10(10):393. doi: 10.3390/metabo10100393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witting M, Böcker S. Current status of retention time prediction in metabolite identification. Journal of Separation Science. 2020;43(9–10):1746–1754. doi: 10.1002/jssc.202000060. [DOI] [PubMed] [Google Scholar]
- Wolthuis JC, Magnusdottir S, Pras-Raves M, Moshiri M, Jans JJM, Burgering B, et al. MetaboShiny: Interactive analysis and metabolite annotation of mass spectrometry-based metabolomics data. Metabolomics. 2020;16(9):99. doi: 10.1007/s11306-020-01717-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wörheide MA, Krumsiek J, Kastenmüller G, Arnold M. Multi-omics integration in biomedical research—A metabolomics-centric review. Analytica Chimica Acta. 2021;1141:144–162. doi: 10.1016/j.aca.2020.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-T, Wang Y, Wang Y, Ebbels T, Karaman I, Graça G, et al. Targeted realignment of LC-MS profiles by neighbor-wise compound-specific graphical time warping with misalignment detection. Bioinformatics. 2020;36(9):2862–2871. doi: 10.1093/bioinformatics/btaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, Hu Y, Yin Z, Liu M, Tang X, Fang M, Huan T. Retrieving and utilizing hypothetical neutral losses from tandem mass spectra for spectral similarity analysis and unknown metabolite annotation. Analytical Chemistry. 2020 doi: 10.1021/acs.analchem.0c02521. [DOI] [PubMed] [Google Scholar]
- Xue J, Guijas C, Benton HP, Warth B, Siuzdak G. METLIN MS2 molecular standards database: a broad chemical and biological resource. Nature Methods. 2020 doi: 10.1038/s41592-020-0942-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Wang Y, Zhang Y, Li F, Xia W, Zhou Y, et al. NOREVA: Enhanced normalization and evaluation of time-course and multi-class metabolomic data. Nucleic Acids Research. 2020;48(W1):W436–W448. doi: 10.1093/nar/gkaa258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Ge W, Ruan G, Cai X, Guo T. Data-independent acquisition mass spectrometry-based proteomics and software tools: A glimpse in 2020. Proteomics. 2020;20(17–18):1900276. doi: 10.1002/pmic.201900276. [DOI] [PubMed] [Google Scholar]
- Zhang J, Sans M, Garza KY, Eberlin LS. Mass spectrometry technologies to advance care for cancer patients in clinical and intraoperative use. Mass Spectrometry Reviews. 2020 doi: 10.1002/mas.21664. [DOI] [PubMed] [Google Scholar]
- Zhao S, Li L. Chemical derivatization in LC-MS-based metabolomics study. TrAC Trends in Analytical Chemistry. 2020;131:115988. doi: 10.1016/j.trac.2020.115988. [DOI] [Google Scholar]
- Zheng F, Zhao X, Zeng Z, Wang L, Lv W, Wang Q, Xu G. Development of a plasma pseudotargeted metabolomics method based on ultra-high-performance liquid chromatography–mass spectrometry. Nature Protocols. 2020;15(8):2519–2537. doi: 10.1038/s41596-020-0341-5. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Luo M, Chen X, Yin Y, Xiong X, Wang R, Zhu Z-J. Ion mobility collision cross-section atlas for known and unknown metabolite annotation in untargeted metabolomics. Nature Communications. 2020;11(1):4334. doi: 10.1038/s41467-020-18171-8. [DOI] [PMC free article] [PubMed] [Google Scholar]