Abstract
While significant insights have been gained concerning COVID-19, superspreading of coronaviruses remains a mystery. The vast majority of cases have been linked to a relatively small portion of infected individuals. Yet, the genetic sequence of the virus, severity of disease, and underlying host parameters, such as age, sex, and health conditions, are not clearly driving the superspreading phenomenon. In this commentary we discuss what is known and what is not known about coronavirus superspreader transmission and explore whether characteristics of the virion, the donor, or the environment contribute to this phenomenon.
Keywords: COVID-19, SARS-CoV-2, Coronavirus, superspreading, transmission
Introduction
The emergence of severe acute respiratory syndrome coronavirus (SARS-CoV) marked a new era in zoonotic transmission of coronaviruses. Nearly 20 years later, a litany of new viruses (influenza H5N1, H1N1, MERS-CoV) and known viruses (Ebola, Nipah, Zika) have produced significant outbreaks in humans. While major efforts have been made to improve recognition of zoonotic infections, limit their damage, and identify their sources in nature, the most important factor in the initiation of an epidemic may be the ability of the pathogen to transmit. Numerous deadly viruses, such as avian influenza, Nipah virus, and others, have been limited by their relatively poor human-to-human transmission. In contrast, major epidemics associated with coronaviruses (CoV), flaviviruses, and Ebola virus have been driven by more efficient transmission. Efficient transmission is critical to the epidemiological success of zoonotic viruses, yet little is understood about how coronaviruses transmit.
During the current COVID-19 pandemic, SARS-CoV-2 transmission is marked by cluster transmission phenotypes in which 80% of new infections are driven by <20% of infected individuals [1., 2., 3., 4.]. In contrast, influenza virus transmission is likely less cluster driven, with a more uniform transmission trajectory. Detailed contact tracing of influenza infections is not routinely conducted, complicating similar analysis between SARS-CoV-2 and influenza virus. However, retrospective analysis of influenza seasons has indicated that epidemics are driven by frequent short-distance local transmission, which may be consistent with coronavirus as well [5,6]. Cluster transmission was also observed during previous coronavirus outbreaks – such as SARS and the outbreak caused by Middle East respiratory syndrome coronavirus (MERS-CoV) – and is the result of superspreader events. While consistent in epidemic strains, the superspreading phenomenon is not limited to coronaviruses. Reports from the West African outbreak of Ebola indicated that just 3% of infected patients accounted for more than 60% of all infections [7]. Similarly, several superspreaders have been identified with measles outbreaks in the last few years [8]. For bacteria that cause tuberculosis and typhoid, human superspreaders have had a long, and complex history [9].
In addition to social connectivity and susceptibility of hosts, we contend that transmission of respiratory pathogens can be influenced by four parameters: (i) donor-specific modifications of the virion, (ii) the donor microbiome, (iii) physical constraints, and (iv) environmental conditions. Modulation of these parameters can impact transmission fitness and superspreading. In this Opinion article, we discuss what is known and what is not known about coronavirus superspreader transmission and explore whether features of the virus, the donor, or the environment contribute to this phenomenon.
Defining Superspreading
The reproduction number (R) describes the average number of infections spread from an individual case; it will often decide the success or limitations of a pathogen [10,11]. The basic reproductive number of a virus, R0, is defined as the initial spread of a virus through a completely susceptible population, while Rt describes the effective reproductive value at a given point in time. For viruses with a high R0/Rt, infection can spread exponentially and requires major efforts to quarantine. Measles has an R0 >10, indicating that the average infected patient passes the virus to 10 or more people [11,12]. In contrast, a virus with a low R0 is expected to transmit to fewer people. Importantly, virulence and transmissibility are often independent, meaning that many deadly viruses may not spread efficiently. For example, both highly pathogenic avian influenza (R0 <2) and Ebola (R0 <2) viruses have relatively low transmissibility despite their high mortality [10,13]. Similarly, for most patients, SARS-CoV, SARS-CoV-2, and MERS-CoV have an R0 <1, limiting the number of infections from an individual host.
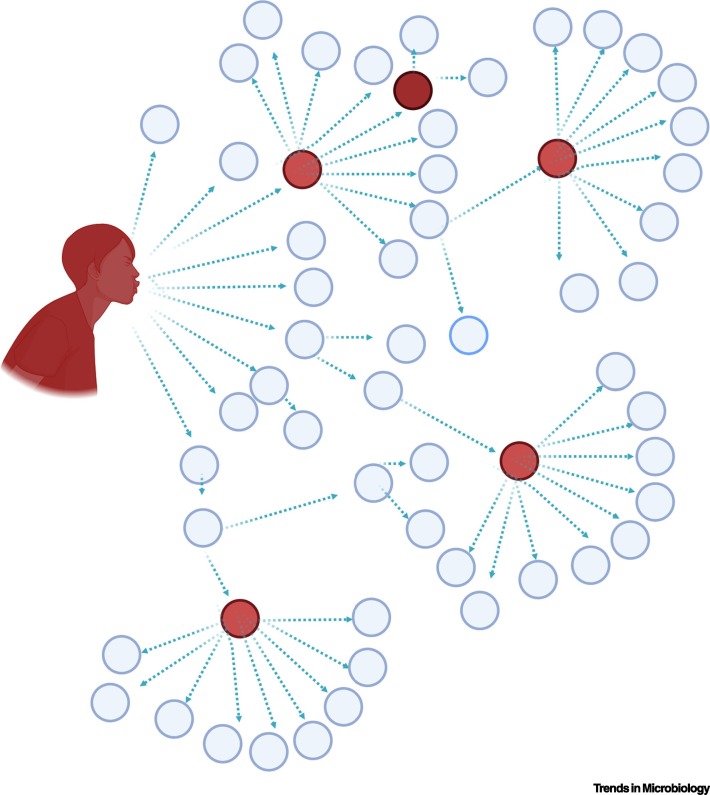
Notably, R0 is not a static value for most respiratory viruses, including influenza and measles [11]; similar variations can be observed with secondary infections of SARS-CoV and SARS-CoV-2. The majority of SARS-CoV patients seed <1 secondary infection (Figure 1 ). However, on average, one in ten SARS-CoV patients were found to be superspreaders; these superspreaders result in >10 secondary infections and seeded a significant portion of the cases around the world [13]. A breakdown of selected superspreading events of the three recent emerging coronaviruses is shown in Table 1 . These events demonstrate the impact of a single source infecting tens to hundreds of other individuals. During the MERS-CoV outbreak, one traveler seeded the infection of 29 others in South Korea, and one of these latter patients subsequently infected additional people; this pattern differed significantly from the vast majority of MERS-CoV patients in South Korea who infected <1 other person [14]. For SARS-CoV-2, a similar trend has been observed, with the vast majority of cases (80%) seeded by <20% of COVID-19 patients [1,2]. Therefore, the relative number of secondary infections is heterogeneous and can vary between individuals.
Figure 1.
Superspreading in the Context of Coronaviruses.
While coronaviruses such as severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) have a low R0 for most patients (<1), about 10% of patients have been found to be superspreaders (red circles), accounting for >80% of infections associated with coronavirus outbreaks. Factors that contribute to superspreading are unknown but are not due to changes in viral RNA sequence or severity of disease in the host. Created with BioRender.com.
Table 1.
Select Superspreading Events from SARS-CoV, MERS-CoV, and SARS-CoV-2
| Coronavirus | Event date | Event location | Venue | Number of seeded infections | Sex of donor | Age of donor | Refs |
|---|---|---|---|---|---|---|---|
| SARS-CoV | February 21, 2003 | Hong Kong | Hotel | 13 | Male | ND | [74] |
| March 15, 2003 | Hong Kong to Beijing | Airplane | ~20 | Male | 72 | [75] | |
| March 1–May 31, 2003 | Singapore | Hospital | >10 | Female | 22 | [76] | |
| March 1–May 31, 2003 | Singapore | Hospital | >10 | Female | 27 | [76] | |
| March 1–May 31, 2003 | Singapore | Hospital | >10 | Female | 53 | [76] | |
| March 1–May 31, 2003 | Singapore | Hospital | >10 | Male | 60 | [76] | |
| March 1–May 31, 2003 | Singapore | Hospital | >10 | Male | 64 | [76] | |
| MERS | May 11, 2015 | Multiple | Hospital and clinics | 26 | Male | 68 | [77] |
| May 15–17, 2015 | Seoul, South Korea | Hospital | 6 | Male | 35 | [78] | |
| May 15–17, 2015 | Seoul, South Korea | Hospital | 23 | Male | 41 | [78] | |
| May 15–17, 2015 | Seoul, South Korea | Hospital | 11 | Female | 75 | [78] | |
| May 27–29, 2015 | Seoul, South Korea | Emergency room | 82 | Male | 35 | [79] | |
| SARS-CoV-2 | January 19, 2020 | Ningbo, China | Bus | 30 | Female | 64 | www.nytimes.com/2020/09/01/health/coronavirus-bus-china.html |
| January 24–28, 2020 | France | Resort | 11 | Male | 53 | www.theguardian.com/world/2020/feb/10/super-spreader-brought-coronavirus-from-singapore-to-sussex-via-france | |
| February 10, 2020 | Daegu, South Korea | Church | 38 | Female | 61 | www.theguardian.com/world/2020/feb/20/south-korean-city-daegu-lockdown-coronavirus-outbreak-cases-soar-at-church-cult-cluster | |
| Feb 26–27, 2020 | Boston, MA | International conference | 97 | ND | ~100 | [4] | |
| Monday, March 2, 2020 | New York, USA | Hospital (multiple) | 90 | Male | 50 | www.nytimes.com/2020/03/10/nyregion/coronavirus-new-rochelle-pneumonia.html | |
| Tuesday, March 10, 2020 | Washington, USA | Choir practice | 52 | ND | ND | [80] | |
| June 17-20 2020 | Georgia, USA | Summer camp | 260a | ND | Teenage | [81] | |
| Monday, August 17, 2020 | Paju, South Korea | Coffee shop | 56 | Female | Mid 30s | www.businessinsider.com/56-got-coronavirus-south-korea-starbucks-mask-wearers-did-not-2020-8 | |
| May 27-29, 2020 | Utah, USA | Daycare facility | 5 | ND | Adult | [71] |
ND, not disclosed.
Limitation: may include infections resulting from before or after camp exposure and not based on a single transmission event.
From the examination of limited data from known superspreaders, no common host traits or viral mutations have been observed. Initial studies of superspreaders of SARS-CoV and MERS tended to identify males, suggesting a potential sex bias (Table 1). However, during the ongoing COVID-19 pandemic, infections occurred in equal proportions within males and females, but male patients presented with more severe disease and higher viral loads [15]. Yet, examination of many of the documented COVID-19 superspreading events revealed both female and male spreaders across a wide age range (Table 1). Importantly, for both SARS-CoV and MERS-CoV, superspreading events were not associated with mutations in the virus sequences that drive increased transmission [16]. Similarly, for SARS-CoV-2, no genetic mutants to date have been linked to superspreading events. Based on these observations, it is unlikely that changes to the viral genome are driving coronavirus superspreading. Instead, we propose that cluster transmission is driven by a combination of underlying host factors, nongenetic variations within the virus, or environmental constraints.
Potential Mechanisms Driving Cluster Transmission
With no evidence for host sex/age biases or changes in the viral genome, we considered other factors that could influence coronavirus superspreading. A successful transmission event requires the virus to maintain infectivity to infect a recipient host. However, it is clear from the infection data that other factors impact the ability of a person to become infected. For example, virus expelled from a superspreader may be more stable in the environment or have modifications that improve attachment or entry. Specific environmental or physical conditions may influence superspreading capacity and may be governed by permanent or transient host factors. Together, these possibilities, both individually and in combination, are key drivers of superspreading in coronavirus infection.
Modification to the Coronavirus Virion
While analysis of the viral RNA genome has not found mutations, the virions from superspreaders may still be distinct. One possibility is that post-transcriptional modification of proteins on the virion may alter transmissibility and infection. For example, the coronavirus spike protein is cleaved by host proteases, which has implications for infection and spread [17]. Host-specific differences in protease expression or distribution may alter virion processing and infectivity. Similarly, the lipid composition of the coronavirus envelope may be distinct among different donors, thus impacting the spread of the virus [18]. Other post-translational modifications of the coronavirus spike, including phosphorylation or ubiquitination, may also vary between hosts and should be studied further for their role in superspreading.
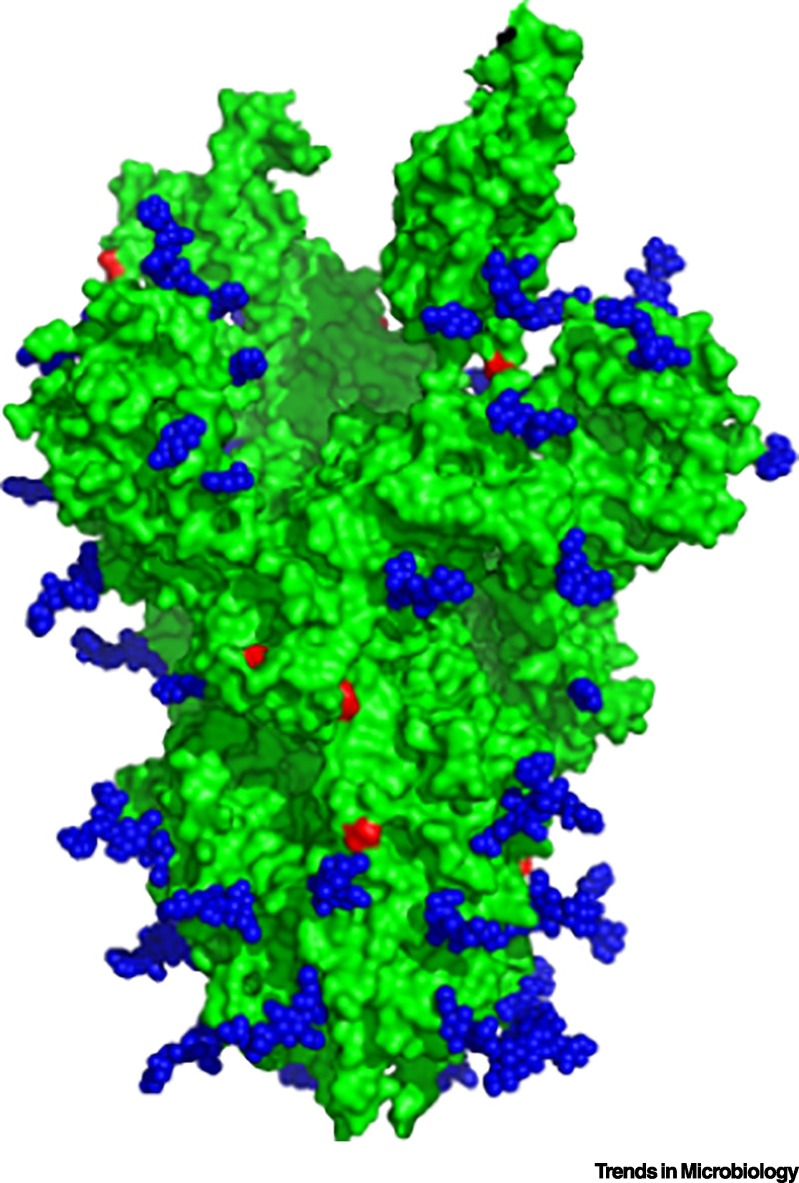
Notably, glycosylation is a key post-translational modification known to play a critical role in attaching carbohydrates to proteins. Glycosylated proteins are abundant within the mucus layer of the respiratory tract [19] and can impact the infectivity and spread of viruses. Work by several structural biology groups has indicated the formation of a glycan shield on the surface of the coronavirus spikes [20,21]. N-linked glycosylation has been identified throughout the spike (Figure 2 ), but strikingly, regions adjacent to the receptor-binding domain and the S1/S2 cleavage site, needed for entry and fusion, lack glycan motifs. Other groups have predicted sites for O-linked glycosylation, but the lack of data from purified virions creates a gap in this analysis [22]. Additional analysis of variations in spike glycosylation between hosts would provide strong evidence that this feature could contribute to transmission heterogeneity.
Figure 2.
Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) Spike with Glycan Shield.
SARS-CoV trimer (green, PDB: 6NB6) [20] displaying N-linked glycan distribution (blue spheres) and the predicted sites for potential O-linked glycosylation based on Net-O-Gly server 4.0 (red residues) [22].
Glycosylation of the spike protein could have functional consequences for coronavirus infection. It has been suggested that the coronavirus glycan shield helps to mediate antibody escape [23], similar to reports for other respiratory viruses [24., 25., 26., 27.]. Similarly, glycosylation may also play a role in binding and attachment, as seen in a subset of coronaviruses [28]. In addition, glycosylation of viral proteins may influence interactions with specific glycans in the respiratory tract. Recent data have demonstrated that human respiratory mucus from differentiated airway epithelial cells protects enveloped viruses (influenza and bacteriophage Phi6) from decay in aerosols and droplets [29]. Therefore, it is feasible that glycosylation of the spike protein will enhance interaction with airway components. Importantly, the efficiency of glycosylation and branching may be host-specific and independent of viral sequence changes. Taken together, the glycosylation profile of the spike protein by different donors may modulate interactions within the airway and thus contribute to coronavirus infectivity and transmissibility.
Building on this idea, blood-group antigens confer phenotypic differences in glycosylation between individuals which may alter the virion in a host-dependent and viral sequence-independent manner. Human blood types can be divided into four different groups based on the glycan epitopes expressed on their red blood cells. Inherited from parents, the glycan epitopes are encoded by glycosyltransferases expressed from the ABO, FUT1, and FUT2 loci in the human genome [30,31]. The resulting blood groups all form the H antigen on erythrocytes but are further subdivided into A, B, and O blood groups. The A allele encodes N-acetylgalactosamine (N-Gal) as a terminal glycan, and the B allele encodes galactose (gal) as its terminal glycan. O blood group individuals express inactive A/B glycosyltransferases and lack these terminal glycans [32]. Finally, A/B blood group individuals express both A and B glycosyltransferase alleles and thus express both terminal glycans on their red blood cells. Together, the blood group antigens identify key differences in the glycosylation machinery based on individual genetics. Importantly, an individual’s blood group can contribute to differences in the glycosylation profiles of host glycoproteins, lectins, and mucins [33].
Differences in blood-group antigens have been implicated in susceptibility to virus infections. Human challenge studies, examining asymptomatic gastrointestinal infection caused by Norwalk virus, have revealed that a lack of FUT2 in the ABH histo-blood group was sufficient to block infection [34]. Similarly, worldwide distribution of rotavirus infection suggests an evolutionary impact of human blood groups since the VP8 protein in human rotavirus strains can interact with A-type histo-blood group antigen [35,36]. Blood-group status has also been identified as playing a role in susceptibility to HIV [37]. Multiple studies have suggested an increase in COVID-19 infections in non-O blood groups [38., 39., 40., 41.]. A similar observation of individual blood-group status and infectivity was reported during the original SARS-CoV outbreak indicating [42] a potenial role in infection and spread. Recently, two reports have suggested a link between SARS-CoV-2 binding to sugars associated with blood group A [43,44]. These data leave the intriguing possibility that SARS-CoV-2 produced in individuals with distinct blood groups may differ in their glycosylation patterns and subsequent transmission. Future studies combining blood-group status with contact tracing will provide critical insight into the transmissibility and susceptibility of individuals based on blood-group antigen.
Impact of the Host Microbiome on Virus Transmission
Similar to glycosylation, the host microbiome may influence superspreading. Comprising a diverse group of commensal bacteria inhabiting spaces within the body, the host microbiome is unique to each individual host. Research examining enterovirus transmission has revealed a critical need for virus–microbiome interactions to enhance virus stability, virulence, and spread between hosts [45,46]. Interestingly, respiratory viruses, such as coronaviruses and influenza viruses, are maintained within the enteric systems of their reservoir hosts (bats and aquatic fowl, respectively). Therefore, given the gut origins of these viruses, it is possible that bacterial–viral associations may be maintained within the respiratory tract. Supporting this notion, several reports have linked SARS-CoV infection in activating an immune response to bacterial moieties, including Toll-like receptors (TLRs) and complement pathways [47., 48., 49., 50.]. In addition, the coronavirus spike protein has domains known to interact with polysaccharide moieties [28,51,52].
To date, the majority of studies examining the effect of the host microbiome on spread of pathogens have focused on enteric pathogens. However, during SARS and MERS-CoV infections the host respiratory tract is the primary site of infection with limited links to the enteric pathways in humans [53,54]. Recent studies in the ferret model have revealed that treatment with topical antibiotics in the nasal cavity results in reduced airborne transmission of influenza viruses [55], suggesting a link between nasal microbiota composition and transmission fitness of respiratory viruses. Notably, studies in mice and ferrets have suggested that coinfection with a common commensal bacterium, Streptococcus pneumoniae, decreases viral replication of respiratory syncytial virus and influenza viruses and reduces airborne transmission of influenza viruses [56,57]. These observations suggest that nasal microbiome communities can influence efficient airborne transmission of respiratory viruses. Integration of microbiome analysis in animal and human transmission studies may provide critical knowledge on the interplay between microbial communities and the airborne transmission of viruses.
Phyisical Factors Driving Transmission
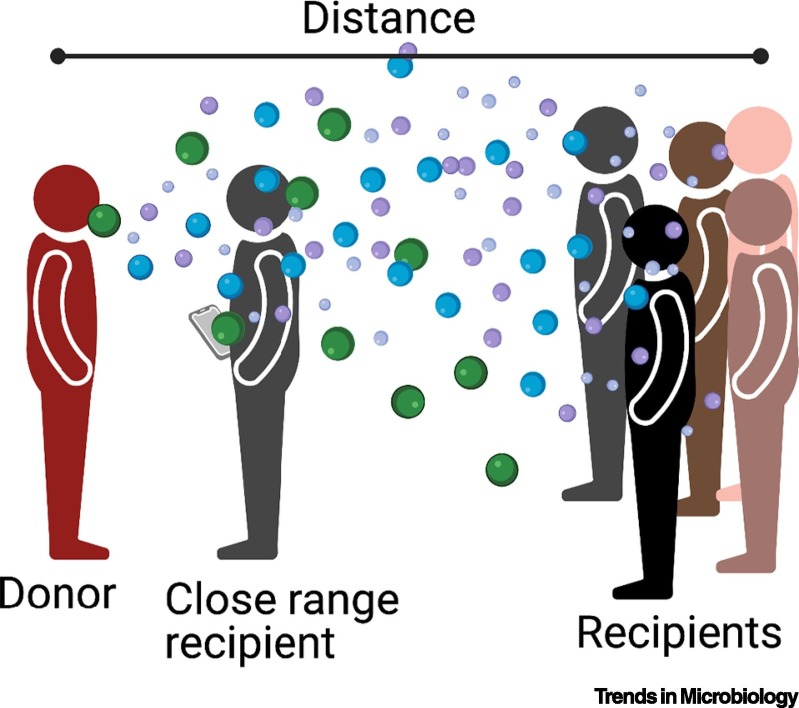
Independent of the host, physical constraints of a given space may play a critical role in superspreading. Respiratory viruses transmit through multiple modes: (i) direct contact, (ii) indirect contact through a contaminated surface, (iii) large droplet spray, or (iv) aerosol transmission (Figure 3 ). The relative efficiency of each mode is still unknown, but all modes are feasible. Release of virus-laden aerosols in a large-size range contributes to both close-up and long-range transmission [11,58]. At close contact all modes of transmission are possible and transmission is highly efficient. Close-contact transmission likely accounts for linear secondary transmission events (Figure 1, gray circles), although not all infections will result in a secondary infection. However, in suprespreader events long-range transmission is likely to mediate a large proportion of the cases. For example, in a recent superspreader event in a South Korean coffee shop, many of the unmasked costumers became infected even with a >6 f. (~2 m) distance between the source and recipients, while the masked employees were not infectedi. Therefore, persistence of SARS-CoV-2 in virus-laden aerosols is critical for long-range transmission in a superspreader event. Aerosol release from COVID-19 patients revealed that ~20% of the patients expel signifcantly more aerosols into the environment, and this was found to be related to the body mass index (BMI) age of the individual [59], suggesting that an obese donor may contribute to superspreading through the release of more virus-laden aerosols. Other factors that can influence aerosol release include singing, speaking in certain dialects, and the volume of one’s voice [60]. Together, the mechanical and physical factors of a space or donor may strongly contribute to superspreading.
Figure 3.
Airborne Transmission of Respiratory Viruses.
An infected donor can expel a range of aerosol sizes that can be virus-laden. These aerosols can fall onto surfaces to create fomites or be inhaled by a donor. At close range a recipient is exposed to all aerosol sizes, increasing infection probability. At a further distance away, recipients will inhale smaller aerosols. Created with BioRender.com
Environmental Contributors to Transmission
The environmental conditions provide another factor that may affect superspreading. Persistence of viruses in the environment is primarily driven by temperature and humidity [61]. However, indoor spaces have a well-controlled temperature with small fluctuations; therefore, it is possible that relative humidity is more likely to alter the stability of viruses in the environment. Previous studies have determined that the SARS-CoV and MERS viruses are stable at low and high relative humidity but have a higher rate of decay at mid-range humidity conditions [62., 63., 64.]. This is similar to other respiratory viruses such as influenza viruses [65,66]. Examination of SARS-CoV-2 stability in virus-laden aerosols, using a rotating Goldberg drum, revealed a half-life of 1.1 h at 65% relative humidity [67]. However, these stability studies were done in the absence of respiratory mucus which has been shown to protect enveloped viruses from humidity-mediated decay in submicron aerosols and droplets [29,68,69]. Therefore, it is feasible that virus persistence in the environment is longer than previously reported, and at short time scales and relative humidity may contribute to cluster transmission of SARS-CoV-2.
Examination of multiple documented superspreader events reveals a variety of location types ranging from hospital settings, churches, and coffee shops, including events at the White Houseii (Table 1). This wide variety in the type of environment means a range of air-exchange rates and ventilation capacities, suggesting that these factors do not directly contribute to superspreader events. However, it is clear that increased ventilation and reduced capacity in indoor spaces will decrease transmission of SARS-CoV-2. Thus, engineering parameters of a space, such as ventilation rates, occupancy, and air exchange, may enhance transmission. Therefore, until the host specifics of a superspreader are known, basic nonpharmaceutical interventions, like increased ventilation and air-exchange rates, may alter the consequences of a superspreading event.
Other Considerations for COVID-19 Superspreading Transmission
While the parameters we outline in the preceding text have the potential to play a role in superspreading, many other factors may also contribute in ways that are not yet clear. Immunocompromised individuals have been shown to lead to enhanced viral burden and persistence viral shedding, which may contribute to increased transmissibility [70]. Thus, variations in host immune status could be factors specific to superspreaders. The infectious dose of SARS-CoV-2 is still unknown and may vary based on age, pre-existing immunity of individual recipients, and the viral population swarm expelled by a donor. Similarly, transmission within pediatric populations has not been as prevalent, but more recent examples from contact tracing in day-care and classroom outbreaks [71,72] indicate a threat for spread despite mild severity of SARS-CoV-2 infection in children. The emergence of SARS-CoV-2 variants with altered transmission fitness may also be driven by superspreading [73]. Mutations within the spike protein can alter tissue tropism, receptor avidity, and have consequences for protein glycosylation, interaction with the host mucus, and overall viral persistence in the environment. While no such data are available for the currently circulating variants, the impact of these variant mutations on factors that influence superspreading events must be considered. Notably, vaccination against SARS-CoV-2 has the potential to mitigate COVID-19 transmission; however, understanding the factors that contribute to superspreading may be crucial to maintaining vaccine efficacy and preventing breakthrough infections.
Concluding Remarks
Many aspects of the ongoing COVID-19 pandemic are still unknown and will take years to understand. Among these, the factors that drive superspreading may be the most unclear. The spread of COVID-19 has been heterogeneous in terms of age, gender, and genetic features. Yet, we know that the vast majority of cases are linked to a small proportion of infected individuals. While transmission of a virus requires that two individuals be within a given space at the same time, other parameters influence the likelihood of getting infected. These may include donor-derived modification of the virion, altered infectivity due to the host microbiome, or physical/environmental conditions that play a role in optimizing transmission. With the continuing spread of SARS-CoV-2, the opportunity exists to explore this question and develop an understanding of how superspreading transmission occurs. These insights will be critical to disrupt the ongoing outbreak and mitigate the spread of future emergent virus strains (see Outstanding Questions).
Outstanding Questions.
Can key features that lead to superspreading be defined?
Once identified, are superspreaders always likely to transmit to high numbers of people?
Can understanding superspreading parameters limit the emergence of future events?
Will findings from coronavirus superspreaders have utility against other infections, including influenza and Ebola?
Alt-text: Outstanding Questions
Acknowledgments
Acknowledgments
We would like to thank Ruth Nwego, a NSURP (National Summer Undergraduate Research Project) student in the Lakdawala laboratory, for compiling the list of superspreader events displayed in Table 1. This work is supported by National Institute of Allergy and Infectious Diseases (1R01AI139063-01A1, JHU CEIRS HHSN272201400007C to S.S.L and 1R01AI153602, R21AI145400 to V.D.M.).
Declaration of Interests
There are no interests to declare.
Resources
iwww.businessinsider.com/56-got-coronavirus-south-korea-starbucks-mask-wearers-did-not-2020-8iiwww.newsweek.com/amy-coney-barrett-rose-garden-event-was-wh-covid-superspreader-new-data-suggests-1537865References
- 1.Yang X., et al. Genetic cluster analysis of SARS-CoV-2 and the identification of those responsible for the major outbreaks in various countries. Emerg. Microbes Infect. 2020;9:1287–1299. doi: 10.1080/22221751.2020.1773745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu X.K., et al. Reconstruction of transmission pairs for novel coronavirus disease 2019 (COVID-19) in mainland China: estimation of super-spreading events, serial interval, and hazard of infection. Clin. Infect. Dis. 2020;71:3163–3167. doi: 10.1093/cid/ciaa790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adam D.C., et al. Clustering and superspreading potential of SARS-CoV-2 infections in Hong Kong. Nat. Med. 2020;26:1714–1719. doi: 10.1038/s41591-020-1092-0. [DOI] [PubMed] [Google Scholar]
- 4.Lemieux J.E., et al. Phylogenetic analysis of SARS-CoV-2 in Boston highlights the impact of superspreading events. Science. 2021;371 doi: 10.1126/science.abe3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charu V., et al. Human mobility and the spatial transmission of influenza in the United States. PLoS Comput. Biol. 2017;13 doi: 10.1371/journal.pcbi.1005382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo D., et al. Multi-scale modeling for the transmission of influenza and the evaluation of interventions toward it. Sci. Rep. 2015;5:8980. doi: 10.1038/srep08980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau M.S., et al. Spatial and temporal dynamics of superspreading events in the 2014–2015 West Africa Ebola epidemic. Proc. Natl. Acad. Sci. U. S. A. 2017;114:2337–2342. doi: 10.1073/pnas.1614595114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seto J., et al. Detection of modified measles and super-spreader using a real-time reverse transcription PCR in the largest measles outbreak, Yamagata, Japan, 2017 in its elimination era. Epidemiol. Infect. 2018;146:1707–1713. doi: 10.1017/S095026881800211X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stein R.A. Super-spreaders in infectious diseases. Int. J. Infect. Dis. 2011;15:e510–e513. doi: 10.1016/j.ijid.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser C., et al. Pandemic potential of a strain of influenza A (H1N1): early findings. Science. 2009;324:1557–1561. doi: 10.1126/science.1176062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leung N.H.L. Transmissibility and transmission of respiratory viruses. Nat. Rev. Microbiol. 2021 doi: 10.1038/s41579-021-00535-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerra F.M., et al. The basic reproduction number (R0) of measles: a systematic review. Lancet Infect. Dis. 2017;17:e420–e428. doi: 10.1016/S1473-3099(17)30307-9. [DOI] [PubMed] [Google Scholar]
- 13.Khan A., et al. Estimating the basic reproductive ratio for the Ebola outbreak in Liberia and Sierra Leone. Infect. Dis. Poverty. 2015;4:13. doi: 10.1186/s40249-015-0043-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong G., et al. MERS, SARS, and Ebola: the role of super-spreaders in infectious disease. Cell Host Microbe. 2015;18:398–401. doi: 10.1016/j.chom.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein S.L., et al. Biological sex impacts COVID-19 outcomes. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park D., et al. Analysis of intrapatient heterogeneity uncovers the microevolution of Middle East respiratory syndrome coronavirus. Cold Spring Harb. Mol. Case Stud. 2016;2 doi: 10.1101/mcs.a001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson B.A., et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature. 2021;591:293–299. doi: 10.1038/s41586-021-03237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson B.A., et al. Peptidoglycan-associated cyclic lipopeptide disrupts viral infectivity. J. Virol. 2019;93 doi: 10.1128/JVI.01282-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ridley C., Thornton D.J. Mucins: the frontline defence of the lung. Biochem. Soc. Trans. 2018;46:1099–1106. doi: 10.1042/BST20170402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walls A.C., et al. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176:1026–1039 e15. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe Y., et al. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shajahan A., et al. Deducing the N- and O- glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology. 2020;30:981–988. doi: 10.1093/glycob/cwaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walls A.C., et al. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2016;23:899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das S.R., et al. Glycosylation focuses sequence variation in the influenza A virus H1 hemagglutinin globular domain. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kosik I., et al. Influenza A virus hemagglutinin glycosylation compensates for antibody escape fitness costs. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medina R.A., et al. Glycosylations in the globular head of the hemagglutinin protein modulate the virulence and antigenic properties of the H1N1 influenza viruses. Sci. Transl. Med. 2013;5 doi: 10.1126/scitranslmed.3005996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zost S.J., et al. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc. Natl. Acad. Sci. U. S. A. 2017;114:12578–12583. doi: 10.1073/pnas.1712377114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tortorici M.A., et al. Structural basis for human coronavirus attachment to sialic acid receptors. Nat. Struct. Mol. Biol. 2019;26:481–489. doi: 10.1038/s41594-019-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kormuth K.A., et al. Influenza virus infectivity is retained in aerosols and droplets independent of relative humidity. J. Infect. Dis. 2018;218:739–747. doi: 10.1093/infdis/jiy221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van den Steen P., et al. Concepts and principles of O-linked glycosylation. Crit. Rev. Biochem. Mol. Biol. 1998;33:151–208. doi: 10.1080/10409239891204198. [DOI] [PubMed] [Google Scholar]
- 31.Varki A., et al., editors. Essentials of Glycobiology [Internet] 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor (NY): 2015–2017. PMID: 27010055. [PubMed] [Google Scholar]
- 32.Hara A., et al. A new chemical approach to human ABO histo-blood group type 2 antigens. Molecules. 2013;19:414–437. doi: 10.3390/molecules19010414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vitiazeva V., et al. The O-Linked glycome and blood group antigens ABO on mucin-type glycoproteins in mucinous and serous epithelial ovarian tumors. PLoS One. 2015;10 doi: 10.1371/journal.pone.0130197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindesmith L., et al. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 2003;9:548–553. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- 35.Hu L., et al. Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature. 2012;485:256–259. doi: 10.1038/nature10996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Midgley S.E., et al. Suspected zoonotic transmission of rotavirus group A in Danish adults. Epidemiol. Infect. 2012;140:1013–1017. doi: 10.1017/S0950268811001981. [DOI] [PubMed] [Google Scholar]
- 37.Cooling L. Blood groups in infection and host susceptibility. Clin. Microbiol. Rev. 2015;28:801–870. doi: 10.1128/CMR.00109-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silva-Filho J.C., et al. The influence of ABO blood groups on COVID-19 susceptibility and severity: A molecular hypothesis based on carbohydrate–carbohydrate interactions. Med. Hypotheses. 2020;144:110155. doi: 10.1016/j.mehy.2020.110155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ellinghaus D., et al. Genomewide association study of severe Covid-19 with respiratory failure. N. Engl. J. Med. 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao J., et al. Relationship between the ABO blood group and the COVID-19 susceptibility. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1150. Published online August 4, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu B.B., et al. Association between ABO blood groups and COVID-19 infection, severity and demise: A systematic review and meta-analysis. Infect. Genet. Evol. 2020;84:104485. doi: 10.1016/j.meegid.2020.104485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng Y., et al. ABO blood group and susceptibility to severe acute respiratory syndrome. JAMA. 2005;293:1450–1451. doi: 10.1001/jama.293.12.1450-c. [DOI] [PubMed] [Google Scholar]
- 43.Ryzhikov A.B., et al. Recombinant SARS-CoV-2 S protein binds to glycans of the lactosamine family in vitro. Biochem. (Moscow) 2021;86:243–247. doi: 10.1134/S0006297921030019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu S.C., et al. The SARS-CoV-2 receptor-binding domain preferentially recognizes blood group A. Blood Adv. 2021;5:1305–1309. doi: 10.1182/bloodadvances.2020003259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuss S.K., et al. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011;334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson C.M., et al. Bacterial lipopolysaccharide binding enhances virion stability and promotes environmental fitness of an enteric virus. Cell Host Microbe. 2014;15:36–46. doi: 10.1016/j.chom.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gralinski L.E., et al. Complement activation contributes to severe acute respiratory syndrome coronavirus pathogenesis. mBio. 2018;9 doi: 10.1128/mBio.01753-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Totura A.L., et al. Toll-like receptor 3 signaling via TRIF contributes to a protective innate immune response to severe acute respiratory syndrome coronavirus infection. mBio. 2015;6 doi: 10.1128/mBio.00638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gralinski L.E., et al. Allelic variation in the Toll-like receptor adaptor protein Ticam2 contributes to SARS-coronavirus pathogenesis in mice. G3 (Bethesda) 2017;7:1653–1663. doi: 10.1534/g3.117.041434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheahan T., et al. MyD88 is required for protection from lethal infection with a mouse-adapted SARS-CoV. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li W., et al. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E8508–E8517. doi: 10.1073/pnas.1712592114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Milewska A., et al. Human coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. J. Virol. 2014;88:13221–13230. doi: 10.1128/JVI.02078-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou J., et al. Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Sci. Adv. 2017;3 doi: 10.1126/sciadv.aao4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gu J., et al. Multiple organ infection and the pathogenesis of SARS. J. Exp. Med. 2005;202:415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rowe H.M., et al. Respiratory bacteria stabilize and promote airborne transmission of influenza A virus. mSystems. 2020;5 doi: 10.1128/mSystems.00762-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown K.M., et al. Coinfection of Streptococcus pneumoniae reduces airborne transmission of influenza virus. bioRxiv. 2020 2020.11.10.376442. [Google Scholar]
- 57.Manna S., et al. Synergism and antagonism of bacterial-viral co-infection in the upper respiratory tract. bioRxiv. 2020 doi: 10.1128/msphere.00984-21. 2020.11.11.378794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prather K.A., et al. Airborne transmission of SARS-CoV-2. Science. 2020;370:303–304. doi: 10.1126/science.abf0521. [DOI] [PubMed] [Google Scholar]
- 59.Edwards D.A., et al. Exhaled aerosol increases with COVID-19 infection, age, and obesity. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2021830118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Asadi S., et al. Aerosol emission and superemission during human speech increase with voice loudness. Sci. Rep. 2019;9:2348. doi: 10.1038/s41598-019-38808-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marr L.C., et al. Mechanistic insights into the effect of humidity on airborne influenza virus survival, transmission and incidence. J. R. Soc. Interface. 2019;16:20180298. doi: 10.1098/rsif.2018.0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chan K.H., et al. The effects of temperature and relative humidity on the viability of the SARS Coronavirus. Adv. Virol. 2011;2011:734690. doi: 10.1155/2011/734690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ijaz M.K., et al. Survival characteristics of airborne human coronavirus 229E. J. Gen. Virol. 1985;66:2743–2748. doi: 10.1099/0022-1317-66-12-2743. [DOI] [PubMed] [Google Scholar]
- 64.van Doremalen N., et al. Stability of Middle East respiratory syndrome coronavirus (MERS-CoV) under different environmental conditions. Euro. Surveill. 2013;18:20590. doi: 10.2807/1560-7917.es2013.18.38.20590. [DOI] [PubMed] [Google Scholar]
- 65.Schaffer F.L., et al. Survival of airborne influenza virus: effects of propagating host, relative humidity, and composition of spray fluids. Arch. Virol. 1976;51:263–273. doi: 10.1007/BF01317930. [DOI] [PubMed] [Google Scholar]
- 66.Yang W., et al. Relationship between humidity and influenza A viability in droplets and implications for influenza's seasonality. PLoS One. 2012;7 doi: 10.1371/journal.pone.0046789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Doremalen N., et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kormuth K.A., et al. Environmental persistence of influenza viruses is dependent upon virus type and host origin. mSphere. 2019;4 doi: 10.1128/mSphere.00552-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thomas Y., et al. Survival of influenza virus on banknotes. Appl. Environ. Microbiol. 2008;74:3002–3007. doi: 10.1128/AEM.00076-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baang J.H., et al. Prolonged severe acute respiratory syndrome coronavirus 2 replication in an immunocompromised patient. J. Infect. Dis. 2021;223:23–27. doi: 10.1093/infdis/jiaa666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lopez A.S., et al. Transmission dynamics of COVID-19 outbreaks associated with child care facilities – Salt Lake City, Utah, April–July 2020. MMWR Morb. Mortal. Wkly Rep. 2020;69:1319–1323. doi: 10.15585/mmwr.mm6937e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zimmerman K.O., et al. Incidence and secondary transmission of SARS-CoV-2 infections in schools. Pediatrics. 2021;147 doi: 10.1542/peds.2020-048090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Plante J.A., et al. The variant gambit: COVID’s next move. Cell Host Microbe. 2021;29:508–515. doi: 10.1016/j.chom.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Centers for Disease Contol and Prevention Update: outbreak of severe acute respiratory syndrome – worldwide, 2003. MMWR Morb. Mortal. Wkly Rep. 2003;52:241–246. 248. [PubMed] [Google Scholar]
- 75.Olsen S.J., et al. Transmission of the severe acute respiratory syndrome on aircraft. N. Engl. J. Med. 2003;349:2416–2422. doi: 10.1056/NEJMoa031349. [DOI] [PubMed] [Google Scholar]
- 76.Chen M.I., et al. Understanding the super-spreading events of SARS in Singapore. Ann. Acad. Med. Singap. 2006;35:390–394. [PubMed] [Google Scholar]
- 77.Korea Centers for Disease Control and Prevention Middle East Respiratory Syndrome Coronavirus Outbreak in the Republic of Korea, 2015. Osong. Public Health Res. Perspect. 2015;6:269–278. doi: 10.1016/j.phrp.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Choe S., et al. Exploration of superspreading events in 2015 MERS-CoV outbreak in Korea by branching process models. Int. J. Environ. Res. Public Health. 2020;17:6137. doi: 10.3390/ijerph17176137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cho S.Y., et al. MERS-CoV outbreak following a single patient exposure in an emergency room in South Korea: an epidemiological outbreak study. Lancet. 2016;388:994–1001. doi: 10.1016/S0140-6736(16)30623-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hamner L., et al. High SARS-CoV-2 attack rate following exposure at a choir practice – Skagit County, Washington, March 2020. MMWR Morb. Mortal. Wkly Rep. 2020;69:606–610. doi: 10.15585/mmwr.mm6919e6. [DOI] [PubMed] [Google Scholar]
- 81.Szablewski C.M., et al. SARS-CoV-2 transmission and infection among attendees of an overnight camp – Georgia, June 2020. MMWR Morb. Mortal. Wkly Rep. 2020;69:1023–1025. doi: 10.15585/mmwr.mm6931e1. [DOI] [PMC free article] [PubMed] [Google Scholar]