Version Changes
Revised. Amendments from Version 1
We have revised the manuscript with specific suggestions in the same line, as suggested by the reviewers. Some of the recommendations were also re-written into more realistic phrases. We have re-edited the text and added additional explanations in the introduction highlighting recently published data showing that NNRTI resistance are also linked to poor response to first-line dolutegravir based regimen. In the revision, we have revised the statement “sequence detection system”by replacing it with “real-time PCR system”. We have modified table 4 to clearly state the costing comparison. In the new version of our manuscript, we have included more parameters and clearly indicated the cost inputs for cost comparison in table 4. We have modified the Discussion to clarify that although most patients are currently initiating on DTG based regimen in Botswana, enfavirenz is also currently being used in some patients especially women of child bearing age as DTG was at some point associated with neural tube defects when taken during prenancy. We have added a new limitation addressing the small samples with HIVDR. We expanded the last sentence in the discussion that future work will build on the findings of this study. We are thankful to the reviewers for their suggestions.
Abstract
Background: HIV-1 drug resistance poses a major threat to the success of antiretroviral therapy. The high costs of available HIV drug resistance assays prohibit their routine usage in resource-limited settings. Pan-degenerate amplification and adaptation (PANDAA), a focused genotyping approach based on quantitative PCR (qPCR), promises a fast and cost-effective way to detect HIV drug resistance mutations (HIVDRMs). Given the high cost of current genotyping methods, we sought to use PANDAA for screening key HIVDRMs in antiretroviral-naïve individuals at codons 103, 106 and 184 of the HIV-1 reverse transcriptase gene. Mutations selected at these positions have been shown to be the most common driver mutations in treatment failure.
Methods: A total of 103 samples from antiretroviral-naïve individuals previously genotyped by Sanger population sequencing were used to assess and verify the performance of PANDAA. PANDAA samples were run on the ABI 7500 Sequence Detection System to genotype the K103N, V106M and M184V HIVDRMs. In addition, the cost per sample and reaction times were compared.
Results: Sanger population sequencing and PANDAA detected K103N mutation in three (2.9%) out of 103 participants. There was no evidence of baseline V106M and M184V mutations observed in our study. To genotype the six HIVDRMs it costs approximately 40 USD using PANDAA, while the reagents cost per test for Sanger population sequencing is approximately 100 USD per sample. PANDAA was performed quicker compared to Sanger sequencing, 2 hours for PANDAA versus 15 hours for Sanger sequencing.
Conclusion: The performance of PANDAA and Sanger population sequencing demonstrated complete concordance. PANDAA could improve patient management by providing quick and relatively cheap access to drug-resistance information.
Keywords: HIV-1 drug resistance testing, Assay performance, Pan-degenerate amplification and adaptation
Introduction
HIV remains a major global health problem; currently, 37.9 million adults and children are estimated to be living with HIV with sub-Saharan Africa being the most severely affected region 1. In Botswana, 380 000 people are estimated to be living with HIV of which 310 713 are on treatment 2. In 2016, Botswana introduced universal HIV treatment to all HIV positive individuals regardless of their immune status 3. Combination antiretroviral therapy (cART) has been successful in reducing morbidity and mortality in individuals infected with HIV as well as in prevention of mother-to-child transmission (PMTCT) of HIV 4. Despite the availability of antiretroviral drugs, which inhibits HIV replication and reducing mortality, one public health concern about the wide scale rollout of cART is the increase in emergence and transmission of HIV drug resistance 5– 7, which has the potential to reduce the efficacy and compromise the success of ART programmes 8, 9.
Although first generation NNRTIs, Nevirapine(NVP) and Efavirenz(EFV) have been replaced by DTG as part of the first line cART regimen, presence of baseline NNRTI resistance mutations has been linked to poor response to first line DTG based regimen 10, therefore it is still important to analyze NNRTI mutations that would affect the efficacy of DTG based regimen. HIV-1 reverse transcriptase, protease and integrase mutations introduced into the viral genome contribute to the development of resistance to antiretroviral drugs. Major non-nucleoside reverse transcriptase inhibitor (NNRTI) mutations, such as K103N and V106M, are selected when HIV is exposed to nevirapine (NVP) and efavirenz, which is still used in both low and high resource settings as part of patient management. Also, resistance mutations that develop in patients exposed to the first generation NNRTIs, NVP and EFV have been shown to confer some cross-resistance to second generation NNRTIs like etravirine and rilpivirine 11. M184V is a major NRTI mutation selected for under tenofovir and lamivudine. HIV drug resistance testing is routinely used for clinical care in high-income countries; however, routine HIV drug resistance testing is not available to majority of patients in resource-limited settings due to the high costs of implementation and limited trained manpower. While Sanger sequencing-based methodologies remain the gold standard for mutation detection, the assays are costly and resource-intensive. Thus, it is urgent to use a simple and cheaper detection method for HIV drug resistance. Detecting known specific mutations provides important information that guides patient treatment options. Moreover, utilising point mutation assays could provide a faster crucial information regarding the mutations present in the patient.
In this study, we compare an HIV genotyping method, pan-degenerate amplification and adaptation (PANDAA), a focused point mutation genotyping assay, with Sanger population based sequencing 12. It is anticipated that PANDAA could serve as an alternative method to rapidly detect HIV-1 drug resistance mutations in HIV patients in Botswana.
Methods
Study population
This was a retrospective study utilizing existing data and stored PCR products from 103 specimens previously genotyped by Sanger based population sequencing from a previous completed study conducted at Botswana Harvard AIDS Institute Partnership, Gaborone, Botswana: Novel strategy for HIV drug resistance monitoring in developing countries (BHP063 study) 13. Briefly, this study enrolled 234 pregnant women diagnosed with HIV and 188 pre-ART from Infectious Diseases Care Clinics (IDCC) between 2012 and 2015. These participants were enrolled to determine the prevalence of HIV transmission at three different geographical locations in Botswana (Gaborone, Molepolole, Mochudi). In samples collected between 2014 and 2015, the following mutations were detected in the main cohort; K103N, G190A and L90M 13.
For the current study, a convenience sampling method was employed to maximize the number of samples available for analysis and the current study used baseline samples collected between 2014 and 2015 from the main cohort, provided that the stored sample(s) were still available with sufficient remaining volume for PCR products. At the time of the current study, the first-line ART regimen in Botswana consisted of tenofovir + emtricitabine (or lamivudine) + efavirenz (or NVP).
Ethical considerations
Ethical clearance for the BHP063 study was obtained from the Human Research Development Committee (HRDC) at the Botswana Ministry of Health (Ethics permit number: HPDME 13/18/1 Vol (366). All participants consented prior to participation in the study.
The current study was approved by the University of Botswana Institute Review Board (IRB) and the Human Research Development Committee at the Botswana Ministry of Health (Ethics permit number: HPDME 13/18/1 Vol (833)) and the need for informed consent was waived since remnant plasma samples were used for this study.
RNA extraction, reverse transcription and PCR amplification
RNA extraction using EZ1 Advanced XL (Qiagen) automated instrument and PCR were performed in the main cohort as described previously 13. The primers used were CWF1-LNA2 and CWR1-LNA3 for first round, whereas second-round primers were CWF1-LNA2 and RT20C13 ( Table 1).
Table 1. Detailed sequences of the primers used for PCR and sequencing 13.
| Primer Name | Primer Sequence | HXB2 position |
|---|---|---|
| CWF1-LNA2 | 5′+GAA+G+GACACCAAATGAAAGAYTG-3′ | 2044-2066 |
| CWR1-LNA3 | 5′-G+CA+TAC+TTYCCTGTTTTCAG-3′ | 3613-3594 |
| CWF1 | 5′ -GAAGGACACCAAATGAAAGAYTG-3′ | 2044-2066 |
| CWCS2 | 5′ -AGAACTCAAGA CTTTTGGG-3′ | 2044-2066 |
| CWCS3 | 5′ -TGCTGGGTGCGGTATTC-3′ | 3145-3129 |
| CWCS5 | 5′ -TGGTAAA TTTGATATGTCCAT-3′ | 3577-3557 |
| Seq6 | 5′ -CCATCCCTGTGGAAGCACATTA-3′ | 3008-2987 |
| Seq2.1-F2 | 5′ -GGCCAGGGAATTTTCTTCAGAGC-3′ | 2120-2142 |
| RT20C | 5′ -CTGCCAATTCTAATTCTG CTTC-3′ | 3462-3441 |
The primers used for Sanger sequencing are those shown in bold.
Drug resistance genotyping by population sequencing
Direct population sequencing of the pol gene was previously performed on an ABI 3130xl genetic analyser (Applied Biosystems, Foster City, CA, USA) using BigDye Terminator cycle sequencing kit (Life Technologies, Carlsbad, CA, USA) 13.
PANDAA qPCR
The stored pol-derived PCR products were diluted prior to PANDAA focused genotyping.
PANDAA qPCR reactions for detecting drug-resistant point mutations K103N, M184V, V106M were performed on an ABI 7500 real-time PCR System (Applied Biosystems).
PANDAA is provided as a 10x mix of primers and probes that are specific for each DRM in three triplex qPCR reactions 14. A single target codon is amplified by the PANDAA primers (proprietary properties of Aldatu Biosciences) and the wild-type variants in each patient is detected using a VIC-labelled TaqMan MGB probe, which is differentiated from the resistant variant, which is detected by a FAM-labelled probe (Life Technologies, MA, USA). Components of the PANDAA reaction contained 5 µL buffer (kappa Probe Fast, kappa Biosystems), 1 µL PANDAA probes (VIC labelled wild-type and DRM-specific FAM-labelled) and primer mix (forward and reverse primers), 4 µL template to a final volume of 10 µl. Each sample was performed in triplicate under the following thermal cycling conditions: 98°C for 3 minutes followed by 40 cycles of 95°C for 5 seconds then 60°C for 90 seconds during which fluorescence data were acquired. Each sample was run in triplicate for each DRM. PANDAA primers include locked nucleic acids (LNAs) which increase affinity for their target sequences and contain an adaptor region (ADR) that is matched to the probe-binding site and a pan-degenerate region (PDR) that incorporates degenerate bases in the targeted primer-binding site upstream of the ADR. The principle of PANDAA is shown in Figure 1 12.
Figure 1. Overview of PANDAA.
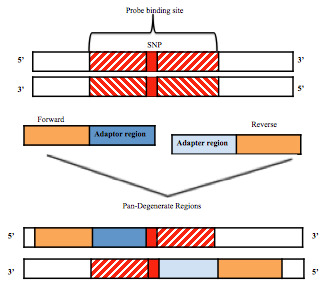
Adaptor regions of PANDAA primers that is matched to the probe-binding site and a pan-degenerate region. This figure has been reproduced with permission from MacLeod et al. 12.
The different protocols (K103N, V106M and M184V) were performed separately, each with a corresponding set of standards.
PANDAA data analysis
The threshold was set at 0.02 and using the ABI 7500 software, raw qPCR fluorescence data were exported from Applied Biosystems SDS software to excel and Cq values were corrected for differences in probe-binding efficiencies. All reactions were performed in triplicate, and the mean of the three values was used for calculation.
Cycle quantification (Cq) values were recorded for each sample. Samples were considered positive when the amplification of the mutant was statistically significant with respect to control sample. The percent abundance of the DRM was calculated using E^ΔCq, whereby E is the efficiency of probe-binding, and ΔCq is the Cq difference between the wild-type and DRM probes, after correcting for variations in probe-binding efficiency.
Reagent cost comparison
The costs of reagents were estimated according to updated prices. Cost of equipment such as ABI 3130XL and ABI 7500 real-time PCR system were not considered as these items of equipment were already available in the laboratory.
Reaction time
To establish the total time to perform each method, we considered the total time to perform genotyping method and interpretation of results.
Concordance statistics
Agreements between PANDAA and Sanger population sequencing were calculated using Cohen’s kappa coefficient. The Mann-Whitney U-test was used to test for differences in CD4 counts and viral loads between the groups with drug resistance mutations and those without drug resistance mutations. Two-sided tests were used and a p-value less than 0.05 implied statistically significant differences. All statistical analysis was carried out using R version 3.5.1 15, other than R 2, which was calculated using the linear regression function in Microsoft Excel.
Results
Characteristics of participants
All participants were female. The median age was 28 (Q1; Q3: 24; 32) years ( Table 2).
Table 2. Characteristics of participants.
| Characteristics | Value |
|---|---|
| Age, median (Q1, Q3) years | 28 (24, 32) |
| CD4+ T cell count, median (Q1,Q3) (cells/uL) | 331 (207.5, 495.5) |
| HIV-1 RNA copies, median (Q1, Q3), log 10 copies/ml (Q1,Q3) | 4.1 (3.49, 4.55) |
Performance of PANDAA
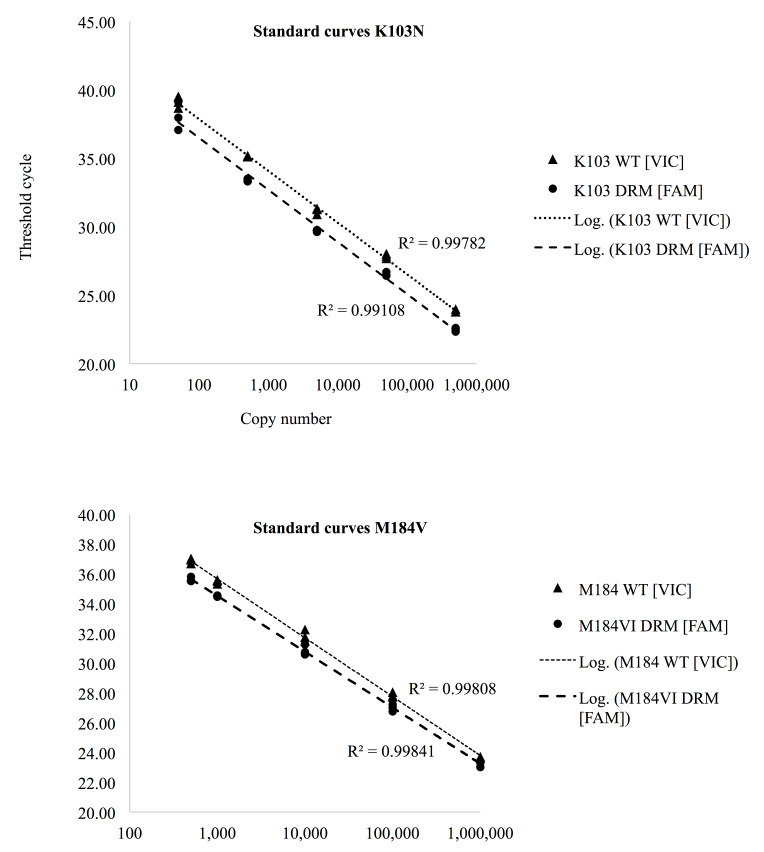
The amplification efficiency was determined by analysing serial dilutions of positive control. A linear standard curve generated from 10-fold dilution was obtained as shown in Figure 2.
Figure 2. Standard curves generated from ten-fold serial dilutions.
Correlation coefficients (r 2) were higher than 99.4%.
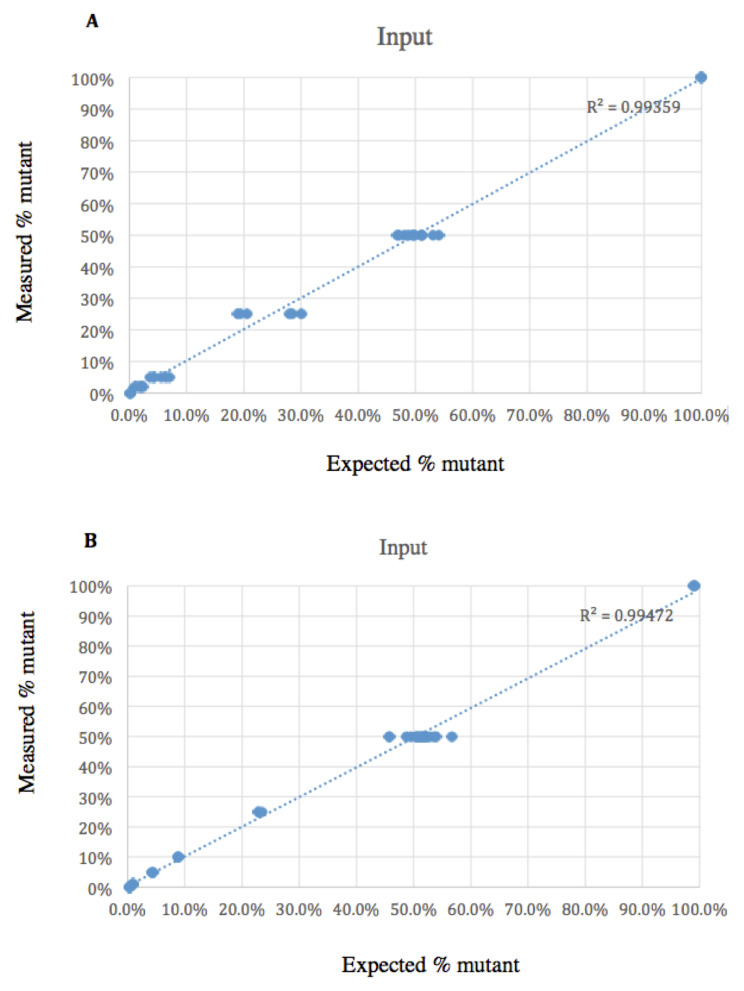
PANDAA showed reproducible results when 1:1 mix of wild-type and DRM templates over a range of copy numbers tested in triplicate. The correlation of each mutant detected by PANDAA correlated with expected mutant as shown in Figure 3.
Figure 3. Measured mutant correlated with expected mutant.
( A) K103N: R 2=0.99339. ( B) M184V: R 2=0.99472.
Quantification of drug resistance of patient samples by PANDAA
PANDAA was completed on patient-derived amplicons of 103 ARV naïve individuals for the K103N, V106M and M184V DRMs using PANDAA. PANDAA identified the presence of K103N in three samples. The three samples with K103N were the same samples that Sanger sequencing detected. Only wild-type sequences at codons 106 and 184 of the RT could be identified by both PANDAA and population sequencing. There was a complete concordance between population sequencing and PANDAA assay as PANDAA qPCR confirmed the presence of HIV drug-resistant mutations as identified by population-based sequencing as shown in Table 3.
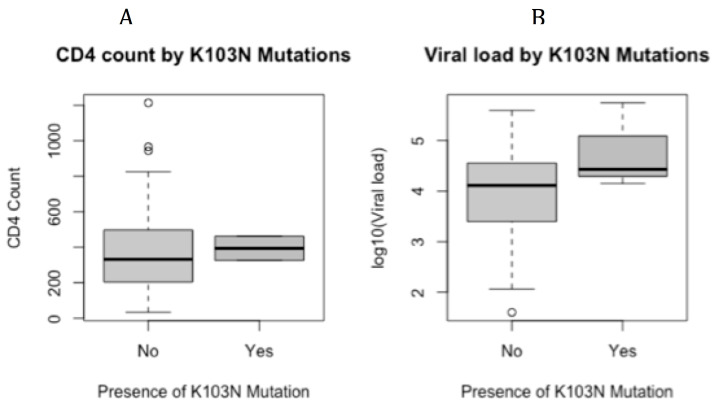
Differences in CD4 counts and viral loads between the groups with drug resistance mutations and those without drug resistance mutations are shown in Figure 4A and Figure 4B.
Figure 4.
Median CD4 ( A) and median viral load ( B) between samples with detected K103N and samples without K103N mutation. Samples without K103N mutation (n=100). Samples with K103N mutation (n=3)
Table 3. Comparison of drug resistance mutations identified by Sanger sequencing and PANDAA.
| PANDAA | |||||
|---|---|---|---|---|---|
| Assay | Yes | No | Total | ||
| K103N | Sanger | Yes | 3 | 0 | 3 |
| No | 0 | 100 | 100 | ||
| M184V | Sanger | Yes | 0 | 0 | 0 |
| No | 0 | 103 | 103 | ||
| V106M | Sanger | Yes | 0 | 0 | 0 |
| No | 0 | 103 | 103 |
Cost and time analysis of each reaction
We calculated the costs for materials and reagents including those associated with the running of samples on the ABI 3130XL sequencer.
The cost of genotyping six drug resistance mutations per patient using PANDAA is 40 USD and Sanger population sequencing is estimated at 100 USD per sample. The turnaround time for PANDAA and Sanger sequencing is approximately 2 hours and 24 hours, respectively ( Table 4).
Table 4. Comparison of sequencing cost and time required for PANDAA and Sanger population sequencing.
| Sequencing method | Laboratory parameter | Time (Minutes) | Cost /sample US$ | |
|---|---|---|---|---|
| Estimated Hands on time | Instrument time | |||
| Sanger Sequencing ** | RNA extraction | 20mins | 43min | 12 |
| RT-PCR | 10mins | 240min | 10.68 | |
| Nested PCR | 10mins | 180min | 2 | |
| Gel electrophoresis | 10mins | 30min | 4.45 | |
| PCR product purification | 20mins | - | 1.45 | |
| Cycle sequencing | 10mins | 106mins | 20.5 | |
| Sequence purification | 15mins | - | 3.00 | |
| Sequence detection | 10mins | 420mins (Overnight) | ||
| Data analysis | 20mins | |||
| Total | 1hr 20mins | 19hrs 12min | $54.08 | |
| PANDAA * | RNA extraction | 10min | 86min | 12 |
| One-step RT-PCR | 10minutes | 120min | 24 | |
| Data analysis | 20min | 20min | ||
| Total | 40min | 4hrs 16mins | $36 | |
These cost do not include costs for gloves, tips and instruments.
** Batch of 13 samples and seven primers
*Batch of 32 samples in triplicate.
Discussion
Here, we show that the HIV drug resistance mutations results of PANDAA are comparable to those produced by Sanger population sequencing. Our study provides baseline data of PANDAA performance and has added an insight that monitoring HIV drug resistance mutations is possible using PANDAA. Having protocols in place for detecting HIV drug resistance mutations using fast and low-cost platforms is important for guiding treatment options and patient management, thereby achieving WHO goal of eliminating HIV by 2030.
When the duration of each method was compared, the results showed that PANDAA required the shortest time for genotyping and had the lowest cost, when compared to Sanger sequencing. It is important to note that PANDAA cost 40 USD for six relevant drug resistance mutations, thus making it much more affordable compared to Sanger sequencing which costs. Sanger sequencing is the widely used and validated method and it is still a relevant platform to use; however, using PANDAA to detect key drug resistance mutations will reduce the cost, especially in this test-and-treat era, thereby enabling quicker results to patients.
Our study had small number of positive samples used to compare the results; however, PANDAA was shown to produce concordant results with sanger sequencing. PANDAA can be considered to rapidly detect drug resistance mutations at a cheaper cost. In addition, PANDAA kit is more cost-effective, and after preparation genotyping results can be obtained in less than two hours.
Botswana has recently introduced universal HIV therapy; however, additional patients are likely to develop drug resistance and transmit these drug-resistant HIV strains to their uninfected partners. As more patients will be receiving ART in Botswana, there is a need to consider investing in fast, low-cost assays to detect mutations associated with drug resistance.
Although most patients are currently initiating on DTG based regimen in Botswana, efavirenz-based regimen is still being used for pregnant women 16, 17 and patients on TB treatment 18. Common drug resistance mutations associated with resistance to efavirenz include K103N (AAA/G to AAC/T) and V106M 19. The key M184V (ATG to GTG) mutation in HIV-1 RT is associated with high-level resistance to the lamivudine (3TC) and emtricitabine (FTC) 20; however, M184V has been shown to rapidly decay in the absence of treatment as a result of its impact on viral fitness 21. HIVDR testing is important to clinicians for patient management, however the cost of reagents and equipment maintenance for resistance testing is the biggest obstacles in resource-limited settings.
In this study, we used PANDAA, to screen for NRTI and NNRTI drug-resistant viruses in 103 newly diagnosed HIV-infected pregnant women from the BHP063 cohort and compared the PANDAA results to those obtained by Sanger based population sequencing. Standard curves generated proved PANDAA to accurately differentiated mutants from wild type. In one hundred and three samples included in our study, the use of PANDAA assay enabled detection of K103N in 3 antiretroviral naïve individuals. Both PANDAA and Sanger sequencing did not detect any mutations at codons 106 and 184 in the HIV strains from this cohort. This study provides insights on the performance of PANDAA, a simple method that utilises primers and probes on any available real-time qPCR platform to detect key HIV drug resistance mutations.
The data generated by our study confirm the ability of PANDAA to detect K103N HIV drug resistance mutation as a point mutation assay, and these data correspond to Sanger sequencing data. The results generated from the use of PANDAA provide evidence that this assay represents an alternative strategy for rapid, specific detection of mutations of interest. At the time the samples were collected for this study, 2014–2015, the standard of care for treatment of HIV infection in Botswana was a regimen that included tenofovir, emtricitabine, and efavirenz co-formulated into one pill, Atripla, taken once a day. By using PANDAA, we targeted the most likely mutations to develop to these medications in HIV-1 subtype C, the M184V, K103N and V106M mutations in reverse transcriptase, a targeted and cost-effective approach to genotyping is possible 22.
Our study had some limitations. Firstly, we only examined the most common relevant resistant mutations, V106M, K103N and M184V of the reverse transcriptase; therefore, there was a limited number of positive mutations available for analysis. There was no clear correlation between viral load and mutations identified due to small sample size of patients with mutations. Secondly, at the time of the study we utilized samples from ART naïve patients and not exposed to ART leading to few cases with drug resistant mutations. Another limitation in this study was the lack of samples with M184V and V106M, making it difficult to draw a conclusion on the performance of PANDAA in detecting V106M and M184V. There is a need for further studies utilising samples with more HIV drug mutations. The applicability of this assay can be demonstrated further by testing a larger number of samples with known mutations. Nevertheless, we have shown that it is possible to genotype HIV drug resistance mutations in HIV naïve individuals using PANDAA and future work will build on the findings of this study.
Conclusion
Our findings proved the potential use of PANDAA assay for testing drug resistance mutations in resource-limited settings. This study demonstrates that applying this cost-effective assay to samples from treatment-naïve individuals where background HIV drug resistance may be increasing can provide valuable insight into baseline resistance and allow for decisions to be made to ensure the best prospect of successful HIV treatment. PANDAA holds the same promise for detection of HIV DRM in patients failing ART, although the current study did not include any participants with known ART exposure. Given the simplicity and cost-effectiveness of PANDAA, it can be performed in any laboratory with real-time PCR capability and its principle could be easily adapted to other clinically relevant point mutations. Overall, the comparative results indicate that PANDAA assay provides similar results with Sanger population sequencing at a much lower cost.
Data availability
Sequence data generated in this study has been deposited with NCBI GenBank under sequential accession numbers MT908833– MT908846 and MT919428– MT919516.
Figshare: Use of a mutation-specific genotyping method to assess for HIV-1 drug resistance in antiretroviral-naïve HIV-1 Subtype C-infected patients in Botswana; https://doi.org/10.6084/m9.figshare.12644930 23.
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
Acknowledgements
The authors would like to acknowledge the study participants, principal investigator, and study coordinator from the Novel strategy completed study. We would like to also extend our acknowledgements to University of Botswana and the Botswana Harvard HIV Reference Laboratory for their support and contribution to the success of the study. We thank Aldatu Biosciences for their support throughout the experiment.
Funding Statement
DM, SM, SG were supported by Sub-Saharan African Network for TB/HIV Research Excellence (SANTHE), a DELTAS Africa Initiative [grant # DEL-15-006]. The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)’s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust [grant # 107752/Z/15/Z] and the UK government. The views expressed in this publication are those of the author(s) and not necessarily those of AAS, NEPAD Agency, Wellcome Trust or the UK government.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 2; peer review: 2 approved]
References
- 1. UNAIDS: UNAIDS, Global H. I. V. AIDS statistics.2019. (accessed 03 March 2020); Reference Source [Google Scholar]
- 2. UNAIDS: Country factsheets.Botswana.2018. (accessed 03 March 2020). Reference Source [Google Scholar]
- 3. WHO. World Health organization: Progress report 2016: prevent HIV test and treat all: WHO support for country impact.World Health Organization.2016. (accessed 27/11/2019): Reference Source [Google Scholar]
- 4. Braitstein P, Brinkhof MW, Dabis F, et al. : Mortality of HIV-1-infected patients in the first year of antiretroviral therapy: comparison between low-income and high-income countries. Lancet. 2006;367(9513):817–824. 10.1016/S0140-6736(06)68337-2 [DOI] [PubMed] [Google Scholar]
- 5. Bennett DE, Myatt M, Bertagnolio S, et al. : Recommendations for surveillance of transmitted HIV drug resistance in countries scaling up antiretroviral treatment. Antivir Ther. 2008;13(Suppl 2):25–36. [PubMed] [Google Scholar]
- 6. Gupta RK, Jordan MR, Sultan BJ, et al. : Global trends in antiretroviral resistance in treatment-naive individuals with HIV after rollout of antiretroviral treatment in resource-limited settings: a global collaborative study and meta-regression analysis. Lancet. 2012;380(9849):1250–1258. 10.1016/S0140-6736(12)61038-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pennings PS: HIV Drug Resistance: Problems and Perspectives. Infect Dis Rep. 2013;5(Suppl 1):e5. 10.4081/idr.2013.s1.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hirsch MS, Brun-Vezinet F, Clotet B, et al. : Antiretroviral drug resistance testing in adults infected with human immunodeficiency virus type 1: 2003 recommendations of an International AIDS Society-USA Panel. Clin Infect Dis. 2003;37(1):113–128. 10.1086/375597 [DOI] [PubMed] [Google Scholar]
- 9. Clavel F, Hance AJ: HIV drug resistance. N Engl J Med. 2004;350(10):1023–35. 10.1056/NEJMra025195 [DOI] [PubMed] [Google Scholar]
- 10. Siedner MJ, Moorhouse MA, Simmons B, et al. : Reduced efficacy of HIV-1 integrase inhibitors in patients with drug resistance mutations in reverse transcriptase. Nat Commun. 2020;11(1):5922. 10.1038/s41467-020-19801-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Diphoko T, Gaseitsiwe S, Kasvosve I, et al. : Prevalence of Rilpivirine and Etravirine Resistance Mutations in HIV-1 Subtype C-Infected Patients Failing Nevirapine or Efavirenz-Based Combination Antiretroviral Therapy in Botswana. AIDS Res Hum Retroviruses. 2018;34(8):667–671. 10.1089/AID.2017.0135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. MacLeod IJ, Rowley CF, Essex M: PANDAA-monium: Intentional violations of conventional qPCR design enables rapid, HIV-1 subtype-independent drug resistance SNP detection. bioRxiv. 2019;795054. 10.1101/795054 [DOI] [Google Scholar]
- 13. Rowley CF, MacLeod IJ, Maruapula D, et al. : Sharp increase in rates of HIV transmitted drug resistance at antenatal clinics in Botswana demonstrates the need for routine surveillance. J Antimicrob Chemother. 2016;71(5):1361–1366. 10.1093/jac/dkv500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Denaro HG M, Long JE, Renzette N, et al. : Validation of PANDAA qDx HIVDR RTI a simple and scalable real-time PCR-based HIV drug resistance genotyping kit for the management of NNRTI-based ART failure. IAS2019 Conference on HIV science, 21-24 July 2019, Mexicocity, Mexico.Abstract number MOPEB141.2019. [Google Scholar]
- 15. Core TR: R: A language and environment for statistical computing.(Accessed 28 November 2019). R foundation for statistical computing. 2019. [Accessed 28/11/2019]. Reference Source [Google Scholar]
- 16. Zash R, Jacobson DL, Diseko M, et al. : Comparative safety of dolutegravir-based or efavirenz-based antiretroviral treatment started during pregnancy in Botswana: an observational study. Lancet Glob Health. 2018;6(7):e804–e810. 10.1016/S2214-109X(18)30218-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zash R, Holmes L, Diseko M, et al. : Neural-tube defects and antiretroviral treatment regimens in Botswana. N Engl J Med. 2019;381(9):827–840. 10.1056/NEJMoa1905230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. MoHW: Handbook of the Botswana 2016 integrated HIV clinical care guidelines.2019; Accessed 27 November 2019. Reference Source [Google Scholar]
- 19. Manasa J, Katzenstein D, Cassol S, et al. : Primary drug resistance in South Africa: data from 10 years of surveys. AIDS Res Hum Retroviruses. 2012;28(6):558–65. 10.1089/aid.2011.0284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Diallo K, Götte M, Wainberg MA: Molecular impact of the M184V mutation in human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 2003;47(11):3377–83. 10.1128/aac.47.11.3377-3383.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cane PA: Stability of transmitted drug-resistant HIV-1 species. Curr Opin Infec Dis. 2005;18(6):537–42. 10.1097/01.qco.0000191506.10363.e1 [DOI] [PubMed] [Google Scholar]
- 22. Nasir IA, Emeribe AU, Ojeamiren I, et al. : Human Immunodeficiency Virus Resistance Testing Technologies and Their Applicability in Resource-Limited Settings of Africa. Infect Dis (Auckl). 2017;10:1178633717749597. 10.1177/1178633717749597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maruapula D, MacLeod IJ, Moyo S, et al. : Use of a mutation-specific genotyping method to assess for HIV-1 drug resistance in antiretroviral-naive HIV-1 Subtype C-infected patients in Botswana. Figshare Dataset. 2020. [DOI] [PMC free article] [PubMed]
- 24. Shafer RW: Human Immunodeficiency Virus Type 1 Drug Resistance Mutations Update. J Infect Dis. 2017;216(suppl_9):S843–S846. 10.1093/infdis/jix398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. http://hivdb.stanford.edu/. [Google Scholar]
- 26. Shafer R: Stanford HIV Drug Resistance Database.(Accessed 02 June 2017). Reference Source [Google Scholar]