ABSTRACT
Intracellular bacteria have evolved various strategies to evade host defense mechanisms. Remarkably, the obligately intracellular bacterium, Ehrlichia chaffeensis, hijacks host cell processes of the mononuclear phagocyte to evade host defenses through mechanisms executed in part by tandem repeat protein (TRP) effectors secreted by the type 1 secretion system. In the past decade, TRP120 has emerged as a model moonlighting effector, acting as a ligand mimetic, nucleomodulin and ubiquitin ligase. These defined functions illuminate the diverse roles TRP120 plays in exploiting and manipulating host cell processes, including cytoskeletal organization, vesicle trafficking, cell signaling, transcriptional regulation, post-translational modifications, autophagy and apoptosis. This review will focus on TRP effectors and their expanding roles in infection and provide perspective on Ehrlichia chaffeensis as an invaluable model organism for understanding infection strategies of obligately intracellular bacteria.
Keywords: Ehrlichia chaffeensis, tandem repeat proteins, moonlighting, effector proteins, intracellular bacteria, effector–pathogen interactions
This review covers the current status of Ehrlichia effector proteins and the complex network of molecular effector–pathogen interactions that they exploit to cause infection and persist intracellularly.
INTRODUCTION
Nearly 30 years ago, Ehrlichia chaffeensis (E.ch.) was identified as an emerging tick-borne pathogen responsible for the life-threatening zoonosis and human monocytic ehrlichiosis (HME) (Anderson et al. 1992). E.ch. is a Gram-negative, obligatory intracellular bacterium capable of surviving and replicating within mononuclear phagocytes (Paddock and Childs 2003). Previous reviews have provided broad overviews of E.ch. pathogenesis. This review will focus on more recent findings regarding the continually expanding roles of various E.ch. effector proteins and illuminate the versatile and important roles of E.ch. tandem repeat proteins (TRPs) in reprogramming the host cell to promote intracellular infection.
E.ch. exhibits tropism for mononuclear phagocytes which are crucial for innate host defenses, thus it is important to understand the sophisticated immunoevasion strategies that have evolved within the confines of a small genome (∼1.3 Mb). The E.ch. genome encodes just 882 proteins, containing low GC content (∼30%), various tandem repeats (TRs) and long non-coding regions (Andersson and Andersson 1999). The creation and deletion of TRs occur through an unknown mechanism that is compatible with DNA slippage (Dunning Hotopp et al. 2006). DNA TRs are small (12 bp) or large (100–300 bp) and are known to play a critical role in gene expression and phase variation. Further, TRs found in proteins encoded by the genome allow E.ch. adaptation to the host and are phylogenetically distinct from TRs found in other Ehrlichia species (Frutos et al. 2006).
TRs are found in proteins throughout all kingdoms but have been best recognized in multicellular eukaryotes where they have evolved to create functional diversity. The diversity of TRs gives rise to structures capable of interacting with a variety of binding partners (Lin, Hsu and Chang 2012). Well recognized examples in eukaryotes include the C2H2-type zinc finger, WD40 repeats and leucine rich repeat (Björklund, Ekman and Elofsson 2006). They interact with various proteins, small molecules, DNA and RNA to mediate an array of biological outcomes, including cell adhesion, protein folding, signal transduction, immune response, transcription, RNA processing and apoptosis (Lin, Hsu and Chang 2012). The E.ch. genome reveals an array of genes involved in host–pathogen interactions, including genes that encode tandem and ankyrin repeat containing proteins (Wakeel et al. 2011). Although extensive research on TRs has been focused on eukaryotic organisms, there has been a rapid increase in information regarding pathogen utilization of TRs to interface with the host cell. Many pathogenic bacteria are known to possess TRs, and the variation of the repeat domains generates functional and antigenic diversity (Lin, Hsu and Chang 2012). Streptococcus is a well characterized group of pathogens that employ TR surface-associated proteins and secreted effectors for infection. One study identified 52 streptococcal genomes in which 3748 proteins were identified to contain TRs. Highly repeated sequences within bacterial proteins have been associated with increased virulence, implying that Streptococcus tandem domain-containing proteins may be involved in the pathogenesis of streptococci (Scholze and Boch 2011; Lin, Hsu and Chang 2012).
TRP effectors and the type 1 secretion system
During infection, E.ch. utilizes both a type 1 secretion system (T1SS) and a type 4 secretion system (T4SS). E.ch. contains genes which code for VirB and VirD proteins associated with the inner membrane channel and ATPase components of T4SS. Many hypothetical T4SS substrates exist as well as confirmed substrates including Etf-1 and Etf-2, which are involved in apoptosis and endosomal maturation respectively (Rikihisa 2017; Yan et al. 2018). However, perhaps less studied and underappreciated, particularly with respect to intracellular bacteria, is the role of the T1SS during infection. Notably, E.ch. secretes many effector proteins by the type 1 secretion system (T1SS) as recently predicted by bioinformatic analysis (Luo et al. 2020). TRPs contain type 1 secretion signal sequences located in the C-terminal domain and were the first ehrlichial proteins to be identified as T1SS substrates using an E. coli complemented with the hemolysin secretion system (Fig. 1; Wakeel et al. 2011). The T1SS is widely utilized by Gram-negative bacteria and is employed to secrete various exotoxins, adhesins and enzymes (Green and Mecsas 2016). TRPs have many similarities with the repeats-in-toxins (RTX) family comprised of exotoxins, lipases, S-layer proteins and adhesins. The features consistent between TRPs and RTX members are glycine and aspartate rich tandem repeats, a non-cleavable C-terminal T1SS signal, homology with ATP-transporters and acidic pls (Wakeel et al. 2011). Both TRP and RTX family members utilize the T1SS to employ various proteins, which identifies the emerging importance and role of T1SS effectors in promoting intracellular infection.
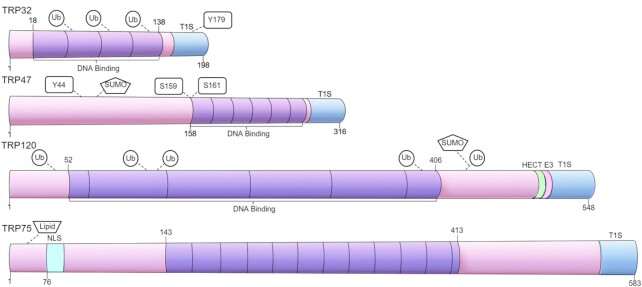
Figure 1.
E. chaffeensis TRP effectors. Schematic of E. ch. TRPs illustrating TR domains and other important features. TRPs contain molecularly distinct TR domains that vary in sequence, length and number. TRPs are secreted by the T1SS and have predicted type 1 secretion signals located in the terminal amino acids of the C-terminal domain. The TRP effectors are nucleomodulins that bind host cell DNA via TR DNA binding domains to modulate host gene transcription. TRPs are modified by PTMs and sites of ubiquitination, SUMOylation and phosphorylation have been identified. TRP120 is a HECT E3 ubiquitin ligase and contains a conserved catalytic site in the C-terminal domain, allowing it to ubiquitinate and target host proteins for degradation.
The T1SS is an ATP-binding cassette (ABC) transporter that forms a channel for one-step secretion of effector proteins from the bacterial cytoplasm to the extracellular environment. It consists of an ATP-binding cassette protein (ECH0383), a TolC outer membrane protein (ECH1020) and a membrane fusion protein (ECH0970), all of which are located within the cell envelope and are essential components of the secretion nanomachine (Delepelaire 2004). The membrane fusion protein is an adaptor that contains a cytoplasmic domain at the N-terminus, a membrane anchor and a periplasmic domain to connect the outer and inner membrane components of the T1SS, in response to the substrate binding the cytoplasmic side. Additionally, the TolC outer membrane protein is a trimeric protein that is responsible for channel formation throughout the outer membrane and periplasm. Remarkably, the ATP-binding cassette protein is fused to a transmembrane domain and recognizes the substrate's secretion signal, assuring that only specific substrates are recognized. The secretion signal is located at the C-terminal, and although the exact region is not defined, the T1SS substrates typically contain repeat sequences that are enriched in [LDAVTSIF] amino acids and occasionally comprised of [KHPMWC] amino acids within the 50 amino acid C-terminal region of the protein (Delepelaire 2004).
Localization of TRPs has been demonstrated on the surface of ehrlichiae as well as extracellularly. Although the T1SS is traditionally recognized as a one step process, recent investigations describe a two-step process or intermediate step that stalls protein secretion resulting in surface localization (Spitz et al. 2019). A primary example of the intermediate step has been demonstrated in E. coli. The discovery of a retention module (RM) at the N terminus was found to anchor the adhesin to the cell surface to stall further translocation, leaving a stalled plug in the periplasm, most likely wedged in TolC and the translocated E. coli adhesin in the extracellular space. During unfavorable conditions where biofilm formation cannot occur, the RM is removed by proteolysis and LapA is secreted. Therefore, the adhesin–TolC–RM complex occurs as a pseudoperiplasmic intermediate. The RTX adhesin model identifies the secretion of an unfolded substrate with its C terminus facing the OM protein. Ca2+ ions bind the GG repeats to induce folding of the substrate into a β-roll (Spitz et al. 2019). In the case of E. coli LapA and IBA substrates, the N-terminal domain folds before secretion completes to plug the translocon, demonstrating a two-step process (Spitz et al. 2019). The two-step process describes a mechanism in which secreted proteins may act as both a surface protein and an effector protein. The documented surface localization and secreted forms of E.ch. TRP120 suggests it uses this type of multi-mechanistic protein secretion via the T1SS.
TRPs and the pathogen–host interface
E.ch. has emerged as a model organism for understanding the pathobiology of intracellular bacteria, and for investigating the role of TRP effectors (Rogan et al. 2019). TRPs are major immunoreactive proteins that elicit strong host antibody responses and are known to interact with many host cell proteins during infection (Tables 1–3). Using Y2H analyses, an array of pathogen–host interactions involved in diverse cellular processes have been identified. Notably, TRP32, TRP47, TRP75 and TRP120 interact with host proteins associated with cell signaling, cytoskeleton organization, vesicle trafficking and intracellular transport, transcriptional regulation, PTMs and apoptosis (Luo, Dunphy and McBride 2017; Luo, Mitra and McBride 2018).
Table 1.
TRP–host protein interactions that influence transcriptional regulation and PTMs.
| Host protein | Function | ||
|---|---|---|---|
| TRP | Symbol | Full name | Transcriptional regulation and PTMs |
| TRP32 | DAZAP2 | DAZ-associated protein 2 | Binds various proteins to regulate transcription |
| HHEX | Hematopoietically-expressed homeobox protein | Transcription factor | |
| TRP47 | ARID2 | AT-rich interactive domain-containing protein 2 | SWI/SNF chromatin remodeling complex |
| HDAC2 | Histone deacetylase 2 | Deacetylation of lysine residues | |
| PIWIL4 | Piwi-like protein 4 | piRNA metabolic process | |
| STAT5A | Signal transducer and activator of transcription 5A | Signal transduction and activation of transcription | |
| STAT6 | Signal transducer and activator of transcription 6 | Signal transduction and activation of transcription | |
| STT3B | Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit | Catalytic subunit of the oligosaccharyl transferase complex | |
| TFEC | Transcription factor EC | Transcriptional regulator: repressor, activator | |
| TRP75 | AFF1 | AF4/FMR2 family member 1 | Gene expression regulation |
| CSDE1 | Cold shock domain containing E1 | RNA-binding protein | |
| FAM208A | Protein TASOR | Epigenetic repression regulation | |
| GMEB1 | Glucocorticoid modulatory element-binding protein 1 | Trans-acting factor | |
| HERC2 | E3 ubiquitin-protein ligase | Ubiquitin-dependent retention regulator | |
| KDM3B | Lysine-specific demethylase 3B | Histone demethylase | |
| PIAS1 | E3 SUMO-protein ligase PIAS1 | E3-type small ubiquitin-like modifier (SUMO) ligase | |
| PPP1R11 | E3 ubiquitin-protein ligase PPP1R11 | E3 ubiquitin-protein ligase | |
| STAT3 | Signal transducer and activator of transcription 3.1 | Transcription factor | |
| UBE2I | SUMO-conjugating enzyme | SUMO activity | |
| USP15 | Ubiquitin carboxyl-terminal hydrolase 15 | Hydrolase, removes conjugated ubiquitin from target proteins | |
| USP3 | Ubiquitin carboxyl-terminal hydrolase 3 | Hydrolase, deubiquitinates monoubiquitinated target proteins | |
| USP8 | Ubiquitin carboxyl-terminal hydrolase 8 | Hydrolase, removes conjugated ubiquitin from target proteins | |
| TRP120 | ARID1B | AT-rich interactive domain-containing protein 1B | SWI/SNF chromatin remodeling complex, represses Wnt |
| ATAD2B | ATPase family AAA domain-containing protein 2B | Chromatin/histone binding | |
| BAHCC1 | BAH and coiled-coil domain-containing protein 1 | Chromatin binding | |
| BMP2K | BMP-2-inducible protein kinase | Phosphatase regulator activity | |
| BTBD6 | BTB/POZ domain-containing protein 6 | Adapter protein for the cul3 E3 ubiquitin-protein ligase complex | |
| CAND1 | Cullin-associated NEDD8-dissociated protein 1 | Assembly factor of SKP1-CUL1-F-box E3 ubiquitin ligase complexes | |
| CDK12 | Cyclin-dependent kinase 12 | Transcription elongation | |
| CLK1 | Dual specificity protein kinase CLK1 | Phosphorylates serine/arginine-rich proteins of spliceosomal complex | |
| DDX5 | Probable ATP-dependent RNA helicase | Alternative regulation of pre-mRNA splicing | |
| FUS | RNA-binding protein FUS | Transcription regulation, RNA splicing, RNA transport, DNA repair | |
| HNRNPA2B1 | Heterogeneous nuclear ribonucleoproteins A2/B1 | Packaging pre-mRNAs into hnRNP particles | |
| ILF3 | Interleukin enhancer-binding factor 3 | RNA-binding protein | |
| KDM6B | Lysine-specific demethylase 6B | Histone demethylase | |
| KLHL12 | Kelch-like protein 12 | Substrate-specific adapter of BTB-CUL3-RBX1 E3 ubiquitin ligase | |
| MBNL1 | Muscleblind-like protein 1 | Pre-mRNA alternative splicing regulation | |
| NSD1 | Histone–lysine N-methyltransferase, H3 lysine-36 specific | Histone methyltransferase | |
| OTUB1 | Ubiquitin thioesterase OTUB1 | Hydrolase with regulatory role in protein turnover | |
| PPP6R1 | Serine/threonine-protein phosphatase 6 regulatory subunit 1 | Regulatory subunit of protein phosphatase 6 | |
| SFRS2 | Serine/arginine-rich splicing factor 2 | Splicing of pre-mRNA | |
| TRIM24 | Transcription intermediary factor 1-alpha | Transcriptional coactivator | |
| UBC | UBC core domain-containing protein | Ubiquitin-protein transferase activity, ATP binding, DNA binding | |
| TRP120/TRP47 | PCGF5 | Polycomb group RING finger protein 5 | Component of Polycomb group multiprotein PRC1-like complex |
| UBB | Polyubiquitin-B | Conjugates to target proteins for various functions | |
| TRP120/TRP75 | IRF2BP2 | Interferon regulatory factor 2-binding protein 2 | Transcription corepressor |
Table 3.
TRP–host protein interactions to influence apoptosis.
| Host protein | Function | ||
|---|---|---|---|
| TRP | Symbol | Full name | Apoptosis |
| TRP32 | CD14 | Carbonic anhydrase 1 | Cell signaling |
| GLCCI1 | Glucocorticoid-induced transcript 1 protein | Apoptotic function | |
| TP53I11 | Tumor protein p53-inducible protein 11 | Negative regulation of cell population proliferation | |
| TRP47 | CDK1 | Cyclin dependent kinase 1 | Cell cycle modulator |
| CAP1 | Adenylate cyclase associated protein 1 | Homeostasis | |
| GNB1 | G protein subunit beta 1 | Apoptotic function and cell proliferation | |
| HDAC2 | Histone deacetylase 2 | Negative regulator of apoptosis and cell differentiation | |
| PTPN2 | Protein tyrosine posphatase, non-receptor type 2 | Homeostasis and positive apoptosis regulation | |
| STAT5A | Signal transducer and activator of transcription 5A | Cell proliferation regulator | |
| STAT6 | Signal transducer and activator of transcription 6 | Cell proliferation regulator | |
| TRP75 | PRDX3 | Thioredoxin-dependent peroxide reductase, mitochondrial | Homeostasis and cell proliferation |
| TPT1 | Translationally-controlled tumor protein | Negative regulation of apoptotic process | |
| TRP120 | ADAM17 | ADAM metallopeptidase domain 17 | Cell signaling and regulator of apoptosis and cell proliferation |
| CAT | Catalase | Metabolism | |
| CXCL12 | C-C-C motif chemokine ligand 12 | Negative regulation of apoptosis | |
| DDX5 | DEAD-box helicase 5 | Transcriptional regulation | |
| ERAL1 | Era-like 12S mitachondrial rRNA chaperone 1 | Mitochondrial protection | |
| FBXW7 | F-box and WD repeat domain containing 7 | Cell signaling, apoptosis regulator and PTM | |
| ICAM3 | Intercellular adhesion molecule 3 | Vesicle trafficking and PTM | |
| IRF2BP | Interferon regulatory factor 2 binding protein 2 | Transcriptional corepressor and negative regulator | |
| KDM6B | Lysine demethylase 6B | Cell fate commitment | |
| KRAS | KRAS proto-oncogene, GTPase | Homeostasis and negative regulator of apoptosis | |
| LGALS1 | Galectin 1 | Regulator of apoptosis, cell proliferation and cell differentiation | |
| ORAOV1 | Oral cancer overexpressed 1 | Biogenesis | |
| PDE1B | Phosphodiesterase 1B | Apoptosis regulation | |
| PPP3R1 | Protein phosphatase 3 regulatory subunit B, alpha | Apoptotic signaling pathway and Wnt signaling | |
| SEPX1 | Selenoprotein X, 1 | Innate immune response | |
| SPTA1 | Spectrin alpha and erythrocytic 1 | Cell proliferation and cell shape regulator | |
| TRIM24 | Tripartite motif containing 24 | Apoptosis regulation. Negative regulation of cell proliferation | |
| TRP120/TRP32 | EEF1A1 | Eukaryotic translation elongation factor 1 alpha 1 | Autophagy and GTPase activity |
| IGHA1 | Immunoglobulin heavy constant alpha 1 | Innate immune response | |
| IGLL5 | Immunoglobulin lambda like polypeptide 5 | Innate immune response and phagocytosis | |
| TRP120/TRP47 | CLC | Charcot-Leyden crystal galectin | Protein aggregate and cytotoxic |
| IGKC | Immunoglobulin kappa constant | Innate immune response |
Table 2.
TRP–host protein interactions to influence cell signaling.
| Host protein | Function | ||
|---|---|---|---|
| TRP | Symbol | Full name | Cell signaling |
| TRP32 | CD14 | CD14 molecule | MAPK, TLR, IKK/NFκB and LPS |
| CD63 | CD63 molecule | Integrin and VEGF | |
| IGHA1 | Immunoglobulin heavy constant alpha 1 | B cell receptor | |
| IGHV | Immunoglobulin heavy chain variable region | Notch and B cell receptor | |
| IGHLL5 | Immunoglobulin lambda like polypeptide 5 | B cell receptor | |
| RC3H1 | Ring finger and CCCH-type domain 1 | T cell receptor and NFκB | |
| TRP47 | BPI | Bactericidal/permeability-increasing protein | IL, TNF and TLR |
| CAB39 | Calcium binding protein 39 | PI3K/Akt/mTOR and IGF1R, | |
| CDK1 | Cyclin dependent kinase 1 | p53, MAPK and Hedgehog | |
| CDK10 | Cyclin dependent kinase 10 | MAPK | |
| FYN | FYN proto-oncogene, Src family tyrosine kinase | Fcγ receptor, T cell receptor, MAPK, IKK/NFκB, PI3 and C-type lectin receptor | |
| GNB1 | G protein subunit beta 1 | GPCR, Ras, Wnt, PI3K/Akt, Hedgehog, CXCR3/4 and cytokine | |
| IGLL1 | Immunoglobulin lambda like polypeptide 1 | B cell receptor | |
| PRTN3 | Proteinase 3 | Cytokine and IL | |
| PTPN2 | Protein tyrosine phosphatase, non-receptor type 2 | ERK1/2, EGF receptor, IFNγ, IL, TNF, IFN and T cell receptor | |
| TRP75 | ANXA5 | Annexin A5 | NFκB, PI3K/Akt/mTOR, ERK and p38 MAPK |
| CD84 | Cluster of differentiation 84 | Cell survival | |
| CSF1 | Colony stimulating factor 1 | PI3K/AKT/mTOR | |
| IFNLR1 | Interferon lambda receptor 1 | Cytokine ligands IFNL2 and IFNL3 receptor | |
| ITGB1 | Integrin beta-1 | Collagen receptor | |
| ITGB2 | Integrin beta-2 | ICAMs and ubiquitin-like proteins receptor | |
| MMP9 | Matrix metalloproteinase-9 | Cytokine-mediated signaling and peptidase activity | |
| NPTN | Neuroplastin | FGFR1 signaling | |
| PI4KA | Phosphatidylinositol 4-kinase alpha | Signal transduction | |
| PRKAA1 | 5'-AMP-activated protein kinase catalytic subunit alpha-1 | Catalytic subunit of AMP-activated protein kinase (AMPK) | |
| RAB3GAP1 | Rab3 GTPase-activating protein catalytic subunit | GTPase activity | |
| RAD50 | DNA repair protein RAD50 | Cellular response to DNA damage | |
| RB1CC1 | RB1-inducible coiled-coil protein 1 | Autophagy | |
| SEPW1 | Selenoprotein W | Glutathione (GSH)-dependent antioxidant | |
| SGSM3 | Small G protein signaling modulator 3 | GTPase activity | |
| SH3BP5 | SH3 domain-binding protein 5 | BTK-related cytoplasmic signaling in B-cells | |
| SPP1 | Secreted Phosphoprotein 1 | Hedgehog, PTH and Integrin | |
| TRP120 | ADAM17 | ADAM metallopeptidase domain 17 | Hedgehog, EGFR, TGFβ, GPCR, Notch, TNF and cytokine/chemokine |
| AKAP2 | A kinase anchor protein 2 | GPCR | |
| ANXA2 | Annexin A2 | NFκB, IL, EGFR, STAT3, Calcium and Wnt | |
| CXCL12 | C-X-C motif chemokine ligand 12 | NFκB, GPCR and chemokine | |
| GCSAM | Germinal center associated signaling and motility | B cell receptor | |
| GNAI2 | G protein subunit alpha i2 | MAPK, GPCR and chemokine | |
| GPS1 | G protein pathway suppressor 1 | MAPK, JNK and GPCR | |
| IFNGR2 | Interferon gamma receptor 2 | JAK-STAT and IFNγ | |
| IL2RG | Interleukin 2 receptor subunit gamma | MAPK, PI3K/Akt, IL and FGFR | |
| KRAS | KRAS proto-oncogene, GTPase | MAPK, NFκB, EGFR, Ras and Rac | |
| LGALS1 | Galectin 1 | IKK/NFκB | |
| PDE1B | Phosphodiesterase 1B | GPCR, PLC, EGFR and FGFR | |
| PPP3R1 | Protein phosphatase 3 regulatory subunit B, alpha | Wnt, MAPK | |
| TLE4 | Transducin like enhancer of split 4 | Wnt, Notch | |
| TRP120/TRP32 | IGHA1 | Immunoglobulin heavy constant alpha 1 | B cell receptor |
| IGLL5 | Immunoglobulin lambda like polypeptide 5 | B cell receptor | |
| TRP120/TRP47 | IGKC | Immunoglobulin kappa constant | B cell receptor signaling pathway |
TRP32
Studies indicate that TRP32 has many functional roles in reprogramming host cellular processes through host protein interactions, and that nearly all TRP32 interacting partners promote infection (Luo et al. 2017). Y2H analysis determined that TRP32 has many binding partners with varying functions, including elongation factor 1 alpha (EF1A), immunoglobulin heavy constant alpha 1 (IGHA1), DAZ-associated protein 2 (DAZAP2), p53 inducible protein 11 (TP53I11) and hematopoietically expressed homeobox (HHEX; Tables 1–3; Luo et al. 2011; Luo and McBride 2012; Farris et al. 2016). EF1A1 is one of the most abundant proteins in eukaryotes and has many functional roles including cytoskeletal remodeling, apoptosis, translation and enzyme regulation. Others include IGHA1, involved in antigen binding, transcription factor DAZAP2 that functions during canonical Wnt signaling, TP53I11 which plays a role in inducing apoptosis, and HHEX a homeobox protein involved in hematopoietic cell differentiation (Maruyama et al. 2007; Lukas et al. 2009; Goodings et al. 2015).
TRP47
E.ch. effector TRP47 was the first ehrlichial TRP examined using the Y2H approach to identify pathogen–host interactions (Wakeel et al. 2010). TRP47 was found to interact with an array of host proteins that positively influence infection (Tables 1–3). TRP47 interacting partners included host proteins with functional roles in cell signaling, vesicle trafficking, intracellular transport, metabolism, PTMs and transcription (Luo et al. 2017). The effector also interacts with both actin binding protein (CAP1) and the Src family tyrosine kinase, Fyn (Wakeel et al. 2010). TRP47 interactions with CAP1 at the morula membrane interface modify CAP1 distribution, which may heavily influence host cell homeostasis. Additionally, TRP47 binds cofilin, actin, SH3 domain, adenylyl cyclase and profilin to potentially guide receptor-mediated endocytosis and vesicle trafficking (Kibler et al. 2018).
TRP75
E.ch. TRP75 which has been shown to interact with a variety of host cell targets that regulate cell signaling, vesicle trafficking, intracellular transport, cytoskeleton organization, metabolism, PTMs and cellular homeostasis (Tables 1–3). TRP75 is tyrosine phosphorylated and predicted to be a lipoprotein, based on its lipobox sequence in the N-terminal region (Luo, Mitra and McBride 2018). Y2H analysis identified TRP75 interaction with 13 human proteins, including solute carrier family 4 member 7 (SLC4A7), actin binding and actin related proteins; actin-related protein 2/3 complex subunit 5 (ARPC5), lymphocyte cystolic protein 1 (LCP1), pleckstrin (PLEK), tropomyosin 4 (TPM4) and apoptosis regulators; eukaryotic translation elongation factor 1 alpha 1 (EEF1A1), integrin subunit beta 2 (ITGB2), peroxiredoxin 3 (PRDX3), protein kinase AMP-activated catalytic subunit alpha 1 (PRKAA1), proteasome 26S subunit, ATPase 5 (PSMC5), RB1-inducible coiled-coil 1 (RB1CC1), selenoprotein W,1 (SEPW1) and signal transducer and activator of transcription 3 (STAT3; Luo, Mitra and McBride 2018). Although the role of TRP75 has not been fully elucidated, like other TRPs, interactions with various host targets suggests an important role in regulating host cellular processes to promote infection.
TRP120
Y2H studies of E.ch. TRP120 have identified a multitude of molecular interactions between TRP120 and host proteins including a diverse group of eukaryotic proteins involved in multiple cellular processes, including cell signaling, vesicle trafficking, PTMs, transcriptional regulation, apoptosis and homeostasis (Tables 1–3; Luo et al. 2011). Many of these interactions have been investigated in detail confirming the role of TRP120 in modulating host cell processes through interactions with host cell proteins. TRP120 modulates Notch signaling through direct interactions with ADAM17 and FBXW7, exploits host PTM machinery through interactions with NEDD4L and UBC9, and highjacks chromatin remodeling via its interaction with PCGF5 to regulate transcription (Dunphy, Luo and McBride 2014; Mitra et al. 2018; Wang et al. 2020). These novel interactions described in more detail in this review indicate that TRP120 has complex and diverse functions, and further research will elucidate the unique mechanisms underlying these host–pathogen molecular interactions and their importance in ehrlichial pathobiology.
TRP posttranslational modifications
TRP120 effector activity is due in part to its ability to exploit host cell machinery to acquire PTMs such as ubiquitin (Ub) and SUMO (Dunphy, Luo and McBride 2014; Zhu et al. 2017). Notably, studies have revealed that TRP120 is both intrinsically ubiquitinated and ubiquitinated by host HECT ligase activity to enhance interactions between TRP120 and host cell targets to promote infection. TRP120 utilizes intrinsically and extrinsically generated PTMs to promote effector–host interactions (Zhu et al. 2017). Studies have demonstrated that human HECT E3 Ub ligase NEDD4L interacts with and facilitates modification of TRP120 with ubiquitin (Fig. 2; Wang et al. 2020). In addition, TRP120 is selectively conjugated with SUMO2/3 isoforms and ehrlichial inclusions co-localize with SUMO1 and UBC9 (Fig. 2; Dunphy, Luo and McBride 2014). Identified TRP120 interacting host proteins contain SUMO interacting motifs (SIMs) that are short hydrophobic domains decorated with acidic residues essential for SUMO-mediated protein interactions. TRP120 exhibits various interactions with SIM-containing proteins including cytoskeleton component Myo10 (unconventional myosin) and GGA1 (Golgi-localizing, γ-adaptin ear domain homology and Arf-binding protein) recruitment and trafficking regulator, indicating that SUMOylation contributes to the numerous molecular interactions between TRP120 and host proteins (Dunphy, Luo and McBride 2014; Zhu et al. 2017). Initial studies demonstrated that TRP120 is conjugated to SUMO at a carboxyl-terminal canonical consensus SUMO conjugation motif and is specifically SUMOylated at Lys 432 to facilitate interactions with PCGF5 and other host proteins. Inhibition of the SUMO pathway negatively impacts E.ch. infection and prevents PCGF5 interaction, indicating that SUMOylation is critical in this regard (Dunphy, Luo and McBride 2014). Exploitation of the SUMO pathway to mediate the effector–host interactions demonstrates the importance of acquiring such PTMs for pathogen–host interactions that promote intracellular infection.
Figure 2.
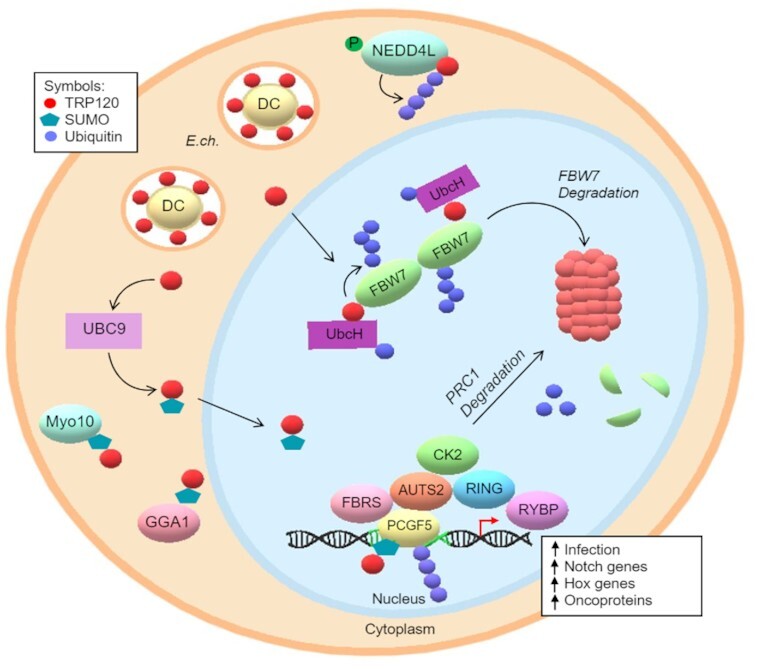
TRP120 exploitation of host PTM pathways and host protein interactions. TRP120 is SUMOylated at canonical SUMO motif by host cell PTM machinery (UBC9), which promotes the direct interaction between Myo10 and GGA1. TRP120 auto-ubiquitinates via intrinsic HECT E3 Ub ligase activity and interacts with host NEDD4L to mediate self-ubiquitination. In the nucleus, TRP120 uses Ub ligase activity to target FBW7and PCGF5 for Ub-mediated degradation. TRP120 binds to FBW7 in a trans conformation and ubiquitinates with K48-Ub chains, resulting in the upregulation of Notch genes and oncoproteins for cell survival. SUMOylated TRP120 binds PCGF5 resulting in PCGFs degradation and the upregulation of HOX genes.
Effectors TRP120 and AmpA of Ehrlichia and Anaplasma were the first reported examples of bacterial proteins post translationally modified with SUMO (Dunphy, Luo and McBride 2014; Beyer et al. 2015). Previous studies had shown that other pathogens utilize PTMs to interface with the host cell. They mimic, inhibit and serve as substrates of the SUMOylation and Ub pathways to modulate host cellular functions. Several Gram-negative bacteria directly target SUMOylation as a survival strategy, including the multi-drug resistant bacteria, K. pneumoniae, which prevents SUMOylation of host target proteins to subvert innate immunity of the host cell. K. pneumoniae increases deSUMOylase SENP2 levels in the cytosol through K48 ubiquitylation and degradation by the ubiquitin proteasome (Sá-Pessoa et al. 2020). The direct SUMOylation of host proteins by intracellular bacteria was recently determined in studies where S. Typhimurium mediated the host endocytic vesicular transport pathway (VTP) through SUMOylation of RAB7, a key component of VTP (Mohapatra et al. 2019).
TRP nucleomodulins
Many E.ch. effectors translocate to the host cell nucleus including tandem and ankyrin (Anks) repeat proteins TRP32, TRP120, TRP47 and Ank200 and thus are considered nucleomodulins (Wakeel et al. 2011; Luo, Dunphy and McBride 2017; Rogan et al. 2019). E.ch. Ank200 was the first ehrlichial effector identified as a nucleomodulin, due to its localization in the nucleus and ability to directly bind genomic Alu elements (AT-rich regions) responsible for controlling ATPase activity, transcriptional regulation and cell fate (Zhu et al. 2009). More recently, TRP32, TRP120 and TRP47 have been identified as nucleomodulins (Table 4). TRPs appear to modulate gene expression using various mechanisms, including direct binding through the protein–DNA complexes, interacting with host proteins to modify epigenetics and degrading nuclear host ubiquitin ligases such as FBW7 to upregulate genes associated with cell survival (Kibler et al. 2018; Klema et al. 2018; Mitra et al. 2018; Wang et al. 2020).
Table 4.
Gene regulation of TRP nucleomodulins.
| Host protein | Function | ||
|---|---|---|---|
| TRP | Symbol | Gene description | |
| TRP32 | AKT3 | AKT Serine/Threonine Kinase 3 | Cell signaling and glycogen synthesis |
| ATF4 | Activating Transcription Factor 4 | Transcriptional regulation | |
| BTK | Bruton Tyrosine Kinase | Cell signaling | |
| CALM2 | Calmodulin 2 | Cell signaling and homeostasis | |
| FOS | Fos Proto-Oncogene, AP-1 Transcription Factor Subunit | Cell proliferation and differentiation and transformation | |
| JUN | Jun Proto-Oncogene, AP-1 Transcription Factor Subunit | Cell signaling | |
| MALAT1 | Homo sapiens metastasis associated lung adenocarcinoma transcript 1 | Controls cell cycle via B-MYB and mRNA processing | |
| MED1 | Mediator Complex Subunit 1 | Transcriptional regulation | |
| MIR142 | Homo sapiens miRNA 142 | Homeopoietic cell development and function | |
| MIR17HG | Homo sapiens miR-17–92 cluster | Cell proliferation and differentiation | |
| MIR21 | Homo sapiens miRNA 21 | Oncomir, anti-apoptotic, targets: PTEN, Bcl2 and TGFBRII | |
| MIR200C | Homo sapiens miRNA 200c | MET progression, TLR, targets: IKBKB, KRAS and MYD88 | |
| MIR505 | Homo sapiens miRNA 505 | Inhibits cell proliferation and induces apoptosis | |
| NRAS | NRAS Proto-Oncogene, GTPase | Vesicle trafficking | |
| RPS23 | Ribosomal Protein S23 | Protein synthesis | |
| TNFAIP | TNF Alpha-induced Protein 3 | Cell signaling | |
| TRP47 | ACTG1 | Actin Gamma 1 | Cell motility and cytoskeleton organization |
| ACTR2 | Actin Related Protein 2 | Cell motility | |
| ARTN | Artemin | Gene regulation | |
| CACNA | Calcium Voltage-Gated Channel Subunit Alpha | Calcium influx | |
| CACNA1G | Calcium Voltage-Gated Channel Subunit Alpha1 G | Calcium influx | |
| CAPZB | Capping Actin Protein of Muscle Z-Line Subunit Beta | Actin filament regulation | |
| CD74 | CD74 Molecule | Immune response | |
| DMTN | Dematin Actin Binding Protein | Cytoskeleton organization | |
| EZR | Ezrin | Cell adhesion, migration and organization | |
| GFRA2 | GDNF Family Receptor Alpha 2 | Cell fate and differentiation | |
| KCNA10 | Potassium Voltage-Gated Channel Subfamily A Member 10 | Potassium transport | |
| KCNA2 | Potassium Voltage-Gated Channel Subfamily A Member 2 | Potassium transport | |
| SLC2A1 | Solute Carrier Family 2 Member 1 | Glucose transporter | |
| SPTB | Spectrin Beta, Erythrocytic | Cytoskeleton organization | |
| TIRAP | TIR Domain Containing Adaptor Protein | Cell signaling and immune response | |
| TNF | Tumor Necrosis Factor | Immune response | |
| TRP120 | ADAM17 | ADAM Metallopeptidase Domain 17 | Notch and homeostasis |
| ADRBK1 | Adrenergic, beta and receptor kinase 1 | Transcriptional regulation and cytoskeletal organization | |
| CARD9 | Caspase recruitment domain family, member 9 | Apoptosis | |
| CD79 | CD79 molecule | Adhesion, leukocyte recruitment and activation | |
| ERN1 | Endoplasmic Reticulum to Nucleus Signaling 1 | Protein folding | |
| FOXA2 | Forkhead Box A2 | Transcriptional regulation | |
| GTF2H1 | General Transcription Factor IIH Subunit 1 | Transcriptional regulation | |
| IKBKB | Inhibitor of kappa light polypeptide gene enhancer in B cells | Cell signaling | |
| Jak2 | Janus kinase 2 | Cell signaling | |
| NOTCH1 | Homo sapiens notch1 | Notch, homeostasis | |
| NPM2 | Nucleoplasmin 2 | Chromatin reprogramming | |
| LRP5 | Low-density lipoprotein receptor-related protein 5 | Wnt and homeostasis | |
| PTK2 | Protein tyrosine kinase 2 | Cell signaling | |
| TLR5 | Toll-like receptor 5 | Innate immune response | |
| TNFRSF14 | Tumor necrosis factor receptor superfamily, member 14 | Inflammatory response | |
| TNFRSF9 | Tumor necrosis factor receptor superfamily, member 9 | Cell proliferation and inflammatory response | |
| ZNF670 | Zinc finger protein 670 | Transcriptional regulation | |
| ZNF250 | Zinc finger protein 250 | Transcriptional regulation | |
| ZNF684 | Zinc finger protein 250 | Transcriptional regulation | |
| ZNF282 | Zinc finger protein 282 | Transcriptional regulation | |
| TRP120/TRP47/TRP32 | BMP8B | Bone Morphogenetic Protein 8b | Cell signaling |
| CAP1 | Cyclase Associated Actin Cytoskeleton Regulatory Protein 1 | Cell signaling and cytoskeleton | |
| CD20 | Membrane Spanning 4-Domains A1 | Cell differentiation | |
| CITED4 | Cbp/P300-Interacting Transactivator 4 | Transcriptional regulation | |
| CLDN19 | Claudin 19 | Cell adhesion | |
| COL9A2 | Collagen Type IX Alpha 2 Chain | Collagen structure | |
| CTPS1 | CTP Synthase 1 | Biosynthesis | |
| FOXJ3 | Forkhead Box J3 | Transcriptional regulation | |
| GUCA2A | Guanylate Cyclase Activator 2A | Cell signaling | |
| IRF2BP2 | Interferon Regulatory Factor 2 Binding Protein 2 | Transcriptional regulation | |
| KCNQ4 | Potassium Voltage-Gated Channel Subfamily Q Member 4 | Potassium channel | |
| LMNA | Lamin A/C | Cell structure | |
| MFSD2A | Major Facilitator Superfamily Domain Containing 2A | Sodium transportation | |
| MYCL | MYCL Proto-Oncogene, BHLH Transcription Factor | Transcriptional regulation | |
| NFYC | Nuclear Transcription Factor Y Subunit Gamma | Transcriptional regulation | |
| PPT1 | Palmitoyl-Protein Thioesterase 1 | Lysosomal degradation | |
| PSMB1 | Proteasome 20S Subunit Beta 1 | Protein degradation | |
| RIMS3 | Regulating Synaptic Membrane Exocytosis 3 | Exocytosis regulation | |
| RLF | RLF Zinc Finger | Transcriptional regulation | |
| TRIT1 | TRNA Isopentenyltransferase 1 | Translation regulation |
TRP32 is considered a nucleomodulin because of its ability to control the host cell through direct interactions with host target genes within the nucleus. TRP32 binds G-rich motifs with GGTGGC-like sequence repeats and targets genes that mediate cell signaling, transcription, cell proliferation/differentiation and apoptosis (Table 4; Farris et al. 2018). Further analysis has demonstrated that TRP32 lysine residues are sites of ubiquitination by host ubiquitin machinery. Although TRP32 lacks a PPxY motif, the effector is modified by host protein NEDD4L, similarly to TRP120. NEDD4L-mediated ubiquitination of TRP32 promotes nuclear localization and transcriptional repressor function (Farris et al. 2016). In addition, TRP32 is phosphorylated at Y179, located in the C-terminal tri-tyrosine motif, which plays a role in directing TRP32 nuclear translocation (Farris et al. 2016).
TRP47 has most recently been identified as a nucleomodulin and the gene encoding TRP47 is the most highly expressed ehrlichial gene during infection in mammalian cells (Kuriakose et al. 2011). TRP47 has a TR domain, consisting of seven 19-mer (ASVSEGDAVVNAVSQETPA) TRs within the C-terminal region of the protein (Luo et al. 2010; Fig. 1). The TR region shows homology with various eukaryotic proteins including DNA polymerase III subunit gamma and tau-conserved domain, ribonuclease E and renin receptor/ATP6AP2/CAPER protein (McBride and Walker 2013). TRP47 translocates to the nucleus at least in part by utilizing a MYND-binding domain-dependent mechanism to bind enhancers of host genes (Kibler et al. 2018). Consistent with other TRPs, the tandem repeat domain of TRP47 binds host DNA and primarily targets genes that influence cell signaling, immune responses, cytoskeleton organization and glucose/potassium transport to promote infection (Table 4).
E. ch. TRP120 was the second ehrlichial nucleomodulin identified and shown to directly bind host DNA. Notably, sequence analysis has revealed that TRP120 does not contain DNA-binding domains typical of eukaryotic transcription factors. Instead, TRP120 binds GC-rich DNA in an ordered structure to form a protein–DNA complex (Klema et al. 2018). Studies demonstrate that TRP120 is highly acidic, which allows its interaction with negatively charged DNA to regulate transcription. In addition, TRP120 may recruit host proteins to stabilize repulsive interactions between TRP120 and varying pH regions of DNA, further promoting transcriptional regulation (Klema et al. 2018). As a nucleomodulin, TRP120 binds DNA to regulate multiple functions, including cell signaling, cytoskeletal organization, transcription, translation and apoptosis (Table 4). Additionally, TRP120 interacts with an array of chromatin-modifying proteins, including proteins of the SWI/SNF chromatin remodeling complex and polycomb comb group (PcG) proteins to positively influence infection (Zhu et al. 2017).
Notably, E.ch. nucleomodulins resemble Xanthomonas transcription activator-like (TAL) effectors, which are unique proteins that contain predictable and modifiable sequences, which makes them a DNA-targeting technology (Zhou, Aertsen and Michiels 2014). TALs are secreted via a T3SS into plant cells, where they translocate to the nucleus and directly bind target promoters to induce gene expression favorable for infection (Scholze and Boch 2011). TALs are used in a variety of DNA-specific applications, including DNA probing, mutation, activation, repression and replacement (Zhou, Aertsen and Michiels 2014; Rinaldi et al. 2017). Further understanding the intricate role of bacterial nucleomodulins, including TALs and TRPs, will potentially impact the medical field through the discovery of novel therapeutics.
TRP120 moonlighting portfolio
E.ch. TRP120 is the most characterized TRP and has numerous interactions with host proteins to promote E.ch. survival (Tables 1–3). In the past decade, the various functions of TRP120 as a moonlighting protein have been well defined and demonstrate that E.ch. relies on this effector to reprogram the host cell (Wang et al. 2020). TRP120 is a model moonlighting protein that utilizes intricate molecular strategies to mediate host cell processes. To date, the well documented roles include ligand mimic, nucleomodulin and E3 ubiquitin ligase activity. Further, TRP120 has several other defined functions, including its roles in cell entry, cytoskeletal organization, vesicle trafficking, cell signaling, transcription regulation and apoptosis. In the section below, we will explore the known TRP120 moonlighting functions during infection in more detail.
TRP120 invasin
Several studies have identified mechanisms involved in E.ch. invasion of monocytes including Ca2+ signaling, actin filamentation and Wnt signaling (McBride and Walker 2013; Rogan et al. 2019). Surface protein DNaseX, and potentially other glycophosphatidylinositol (GPI)-anchored proteins associated with caveolae are involved in infectious dense-core cell (DC) ehrlichiae adherence and entry into host cells (Lin and Rikihisa 2003b; Mohan Kumar et al. 2015). Studies have determined that the C-terminus of EtpE (ECH1038) triggers E.ch. entry through its interactions with DNaseX, CD147, N-Wiskott-Aldrich syndrome protein (N-WASP) and hnRNP-K (Mohan Kumar et al. 2013, 2015). However, TRP120 expressed on infectious DC ehrlichiae has also been shown to play a role in ehrlichial host cell entry (Popov et al. 2000). Studies have demonstrated that E.ch. invasion of the host cell requires TRP120 and the stability of TRP120 is regulated by bacterial second messenger, cyclic di-GMP and ehrlichial surface serine protease, HtrA (Kumagai et al. 2010).
Recent studies have demonstrated that E.ch. internalization is dependent on Wnt signaling and TRP–receptor interactions (Rogan et al. 2019). TRP120 serves as an adhesin and interacts with Wnt FZD receptors on the host cell to activate the canonical and non-canonical Wnt signaling pathways to stimulate phagocytosis and entry (Luo et al. 2011; Rogan et al. 2021). TRP-coated microspheres-induced phagocytic uptake in macrophages; however, in the presence of a Wnt signaling small molecule inhibitor, the TRP coated microspheres failed to stimulate uptake phagocytosis (Luo et al. 2016). This data suggests that TRP-induced phagocytosis occurs via non-canonical Wnt signaling, which is further supported through studies demonstrating that Wnt5-FZD5-PI3K signaling induces the uptake of E. coli without negatively impacting infection (Maiti et al. 2012; Luo et al. 2016). Further evidence suggests that E.ch. hijacks the canonical and non-canonical Wnt signaling pathways via the FZD receptor. FZD knockdowns at 1-day p.i. significantly reduce E.ch. infection (Via et al. 2015).
Genes involved in membrane trafficking during E.ch. infection show decreased expression, including synaptosomal associated protein, 23 kDa (SNAP23), membrane of RAS oncogene family (Rab5A) and syntaxin 16 (STX16). Further, it is likely that TRP120 interacts with actins to promote entry, since inhibition of actin polymerization in E.ch. infected cells disrupts filopodia formation (Thomas et al. 2010) and TRP120 is known to interact with host proteins involved in cytoskeletal organization, including actin gamma 1 (ACTG1), actin related protein 2/3 complex (ARPC2) and unc-13 homolog D (UNC13D; Luo et al. 2011). E.ch. TRPs bind genes involved in cytoskeletal rearrangement and vesicle trafficking, such as syntaxins (SNX14, SNX11 and SNX17), clathrin (CTLA), TSNARE1 and caotomer (COPA). Thus, E.ch. TRP120 and other TRPs potentially modulate genes associated with cytoskeletal organization to facilitate ehrlichial entry, vesicular trafficking and exocytosis to promote infection.
TRP120 cellular signaling ligand mimetic
Recent studies have demonstrated that E.ch. exploits host cellular processes to establish a favorable niche through the activation of conserved cell signaling pathways, including the Notch and Wnt signaling pathways (Fig. 3). Other survival strategies employed by E.ch. include the suppression of tyrosine and mitogen activated protein kinase (MAPK) activity and the regulation of Toll-like receptors and transcription factors in monocytes and macrophages (Rogan et al. 2019). The activation of the Notch and Wnt signaling pathways are thought to occur through specific ligand–receptor interactions (e.g. Frizzled (FZD), Notch) to promote E.ch. survival (Guruharsha, Kankel and Artavanis-Tsakonas 2012; Hori, Sen and Artavanis-Tsakonas 2013; Luo et al. 2016; Rogan et al. 2019; Wang et al. 2020). Studies demonstrate that TRP120 likely serves as a ligand mimic to promote intracellular survival, providing a useful model to investigate intracellular bacterial reprograming of the host cell to create a favorable environment for infection.
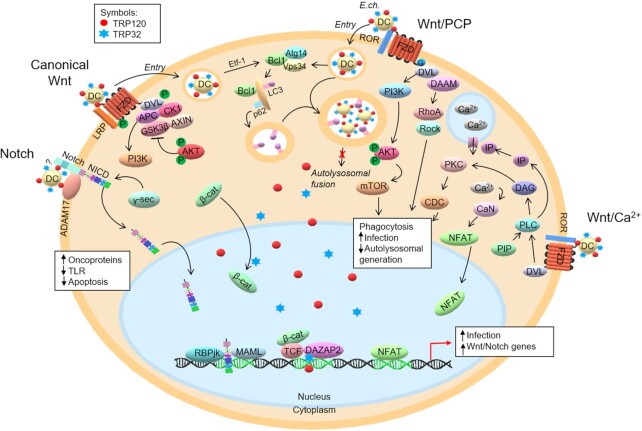
Figure 3.
TRP-mediated activation of conserved signaling pathways and role in infection. TRPs act as ligand mimetics and interact with Notch and Wnt receptors to activate host cell signaling. On the cell surface, E.ch. TRP120 expressed on the surface of dense-cored ehrlichiae interacts with ADAM17 and possibly Notch receptor to activate Notch signaling. Similarly, TRP120 interacts with FZD receptors to activate canonical and non-canonical Wnt signaling to regulate apoptosis and autophagy. Notably, expression of the FZD5 receptor increases during infection. In addition, TRP32 interactions with Wnt transcription factor DAZAP2 to potentially influence Wnt gene transcription.
Notch signaling
In order to mediate the Notch signaling pathway, TRP120 interacts with a variety of host genes and proteins associated with Notch signaling (Fig. 3). The Notch signaling pathway is a conserved signaling pathway with critical roles in cellular homeostasis, including cell proliferation and differentiation (Hoyne 2003; Palaga et al. 2013; Barth and Köhler 2014; Song et al. 2015). Recent studies have identified Notch signaling as a pathway targeted by numerous intracellular bacteria for infection and survival, including Salmonella, Mycobacterium bovis and Bacillus anthracis, potentially due to Notch activity regulating innate and adaptive immune responses including inflammation, lymphocyte cell development, Toll-like receptor (TLR) expression and apoptosis (Narayana and Balaji 2008; Zhu et al. 2011; Larabee et al. 2013; Larabee and Ballard 2014; Lina et al. 2016). Notably, studies have determined that TRP120 directly activates Notch signaling resulting in the downregulation of innate immune sensing (Lina et al. 2016). Specifically, TRP120 activation of the Notch signaling pathway results in the downregulation of toll-like receptor (TLR) 2/4, which is caused by inhibition of the extracellular signal-regulated kinase 1/2 (ERK1/2) and p38 mitogen-activated protein kinase (MAPK) pathways required for PU.1 expression (Hao et al. 2007; Welcker and Clurman 2008; Matsumoto et al. 2011).
Studies using pharmacological inhibitors and small interfering RNAs (siRNAs) against Notch activating proteins demonstrated that Notch signaling is required for ehrlichial survival (Zhu et al. 2011). In subsequent Y2H studies, TRP120 was found to interact with Notch pathway proteins ADAM17, NEDD4L and the host nuclear tumor suppressor F-BOX and WD domain repeating-containing 7 (FBW7; Wang et al. 2020). Further analysis regarding the interaction between FBW7 and TRP120 demonstrated that TRP120 directly interacts with FBW7 FBOX and WD40 domains and ubiquitinates FBW7 for degradation, which regulates Notch signaling and stabilizes oncoproteins involved in cell survival and apoptosis (Wang et al. 2020).
In addition to influencing PRR expression, TRP120 appears to exploit Notch signaling to prevent apoptosis for E.ch. survival (Wang et al. 2020). TRP120 has been specifically associated with proteins involved in the regulation of apoptosis, including eukaryotic translation elongation factor 1 alpha 1 (EEF1A1), and cytochrome c oxidase subunit II (COX2), which indicated a potential role in influencing cell survival (Tsujimoto 1998; Carrington et al. 2017). Further, E.ch. infection promotes the upregulation of apoptotic inhibitors during infection, including NF-κB, IER3, MCL1, BCL-2 and BirC3, and the downregulation of apoptotic inducers, such as Bik and BNIP3L (Luo et al. 2011). Interestingly, the BCL-2 anti-apoptotic protein MCL1 level is increased during E.ch. infection due to Notch activation and degradation of the MCL-1 negative regulator FBW7 via TRP120 ubiquitination (Lin, Hsu and Chang 2012; Rinaldi et al. 2017). Therefore, E.ch. stabilizes and increases MCL1 levels for ehrlichial survival. Based on this information, E.ch. may be exploiting host cell intrinsic apoptosis to promote survival partially through TRP120 ubiquitination, effector–host protein interactions and modulation of host gene transcription.
Wnt signaling
TRP120 plays a major role in mediating Wnt signaling during infection. Initially, studies using Y2H identified interactions between TRP120 and various components and regulators of the Wnt signaling pathway, including positive regulators (PPP3R1 and VPS29) and negative regulators (ARID1B, CEP164, KLHL12, ILF3 and LMO2; Luo et al.2011; Luo and McBride 2012; Rogan et al. 2019). Recent studies determined that TRP120 utilizes ligand mimicry to bind Wnt FZD receptors and activates the Wnt signaling pathway in human monocytes (Fig. 3; Rogan et al. 2021). The Wnt signaling pathway is evolutionarily conserved and critical to eukaryotic development and cell fate (Luo et al. 2016). Notably, Wnt signaling has emerged as a key player in pathogenesis by several bacteria, including E.ch., S. enterica, M. tuberculosis, C. difficile, P. aeruginosa and E. coli. A TRP120 binding motif within the promoter region of Wnt target genes has been identified, which suggests that TRP120 regulates transcription genes associated with Wnt signaling (Luo et al. 2016; Rogan et al. 2019). Additionally, gene silencing of non-canonical and canonical Wnt signaling components was shown to have detrimental effects on E.ch. infection. Specifically, RNA silencing of Wnt components, including β-catenin, CK1, CAMKII, Wnt5a FZD5, FZD9, LRP6 and NFAT resulted in significantly reduced E.ch. infection (Rogan et al. 2019). In contrast, the silencing of Wnt antagonist DKK3 increased infection (Luo et al. 2016).
Wnt signaling is known to regulate autophagy and other innate immune responses. A recent study demonstrated that E.ch. hijacks the Wnt signaling pathway via TRP120 to inhibit autolysosome generation and autophagic destruction (Fig. 3; Lina et al. 2017). E.ch. utilizes TRP120 to exploit both the Wnt and PI3k/AKT pathways to activate mTOR signaling and regulate TFEB nuclear translocation to inhibit lysosomal biogenesis and autolysosomal fusion with E.ch. containing vacuoles (Lina et al. 2017). Increased levels of GSK3-β, a Wnt antagonist, were detected in E.ch. infected cells and were shown to be stimulated by TRP120 (Lina et al. 2017). These effects were abrogated with the treatment of a Wnt-Dvl inhibitor. Therefore, TRP120 is a key player in activating the PI3K/Akt pathway and inhibiting GSK3 activity via phosphorylation to prevent lysosomal fusion.
Molecular mimicry
TRP120 utilizes molecular mimicry and is known as a Wnt ligand mimetic that initiates Wnt signaling through its direct interactions with the FZD family of receptors (Rogan et al. 2021). Recent studies demonstrate the direct binding between TRP120 and multiple FZD receptors to activate the Wnt signaling pathway via a short linear motif (SLiM) in TRP120 that is homologous to Wnt ligands. This study reveals the first example of bacterial mimicry of Wnt signaling ligands. Further investigation is needed to elucidate E.ch. TRP120 interactions with Notch receptors. However, since TRP120 is known as a Wnt ligand mimic and to independently activate Notch signaling, it is likely that TRP120 directly interacts with Notch receptors to activate the Notch signaling pathway via ligand mimicry.
Molecular mimicry is a powerful mechanism utilized by pathogens to exploit host cell functions to promote replication and dissemination. Studies determined that viral mimicry occurs for roughly 30% of motif classes identified in the Eukaryotic Linear Motif (ELM) database. Pathogenic molecular mimicry of host cell components occurs via protein SLiMs, which mimic host SLiMs (Davey, Cyert and Moses 2015; Via et al. 2015). SLiMs were first identified as conserved sequences in evolving regions and have arisen through convergent and co-evolution to enhance ligand binding, protein stability and cell signaling (Via et al. 2015). SLiMs are short stretches of contiguous amino acids that reside within natively disordered protein regions and can be found in accessible loops of folded domains (Davey, Cyert and Moses 2015). A single protein can contain various SLiMs, creating an intricate network of balance and competition between each motif (Davey, Cyert and Moses 2015; Via et al. 2015). Further, bacterial pathogens, including T. gondii, H. pylori, C. trachomatis, A. phagocytophilum, M. tuberculosis and L. monocytogenes, utilize SLiMs that share both composition and function with host SLiMs for molecular mimicry, which poses a threat on the delicate balance of host SLiMs and protein interaction networks due to their ability to titrate the motif-binding partners (Via et al. 2015). Unlike viruses, bacteria produce toxins and secreted proteins, and have larger genomes allowing numerous opportunities for molecular mimicry of various host proteins involved in internalization, transcription, immune response and cell signaling (Lina et al. 2016). Thus, the function of TRP120 SLiMs that mimic endogenous ligands highlights the pathogen's ability to upregulate developmental signaling pathways and makes E.ch. an important model organism for infectious diseases to further understand the pathobiology of bacterial pathogens.
HECT E3 ubiquitin ligase
TRP120 contains a functional HECT E3 ligase domain located at the C-terminal, allowing the effector to target host proteins for degradation. This unique characteristic allows TRP120 to regulate signaling through the ubiquitination of its host binding partners in addition to activating conserved signaling pathways through ligand mimicry. Recent studies identified the interaction between TRP120 and F-BOX and WD domain repeating-containing 7 (FBW7). FBW7 is the F-box protein subunit of the Skp1-cullin-1-FBOX E3 Ub ligase complex (SCF) and is required for substrate recognition to regulate an array of oncoproteins involved in Notch signaling (Elmore 2007). FBW7 negatively regulates oncoproteins (Notch, MCL1, cJun and cMyc) involved in cell survival. Thus, TRP120 not only activates, but regulates Notch signaling and identifies FBW7 as a substrate of the TRP120 HECT E3 Ub ligase to maintain Notch signaling for E.ch. survival (Fig. 2; Wang et al. 2020).
Further, TRP120 interacts with proteins of the SWI/SNF chromatin remodeling complex and polycomb comb group (PcG) proteins (Zhu et al. 2017). Polycomb repressive complexes (PRCs) are multi-subunit complexes divided into two groups (PRC1 and PRC2), important for the regulation of chromatin conformation and transcriptional regulation in eukaryotic cells. PRC1 results in the monoubiquitination of histone 2A at lysine 119, while PRC2 is involved in the trimethylation of histone 3 at lysine 27. PRC histone modifications result in chromatin conformational and transcriptional changes of eukaryotic genes. TRP120 is shown to directly interact with the RING domain of PCGF5, a unit of PRC1 (Mitra et al. 2018). PCGF5 is essential in epigenetics to maintain the transcriptionally repressive state of many host cell genes. TRP120 targets PCGF5 for Ub-mediated degradation within the nucleus, providing evidence that TRP120 mimics mammalian E3 ubiquitin ligase activity (Fig. 2). The interaction between TRP120 and PCGF5 occurs during early infection at the TR domain while the HECT domain remains active, demonstrating that PCGF5 is a substrate of TRP120 ligase activity (Mitra et al. 2018). At 48 h, PCGF is redistributed from the nucleus to the ehrlichial vacuole and PCGF isoforms are shown to undergo proteasomal degradation facilitated by TRP120. The proteasomal degradation of PCGF isoforms causes chromatin conformational changes and altered transcriptional activity of PRC1-associated Hox genes (Wang et al. 2020). The direct interaction between TRP120 and PCGF5 demonstrates a unique strategy whereby E.ch. employs to exploit host epigenetic machinery to modulate gene expression for infection.
Various pathogens have been shown to target host proteins for degradation (Ribet and Cossart 2010). Shigella effector proteins hijack host ubiquitination machinery to promote infection, whereby effectors OspI and OspG interact with host E2 enzymes to disrupt their functions, while NEL family effectors mimic E3 ligases to target host proteins for Ub-mediated degradation (Kim et al. 2005; Sanada et al. 2012; Nishide et al. 2013). Additionally, the Salmonella T3SS substrates SopA and SopB modulate host functions using ubiquitination for SopA E3 ubiquitin activity and SopB relocation from the host cell surface to the Salmonella-containing vacuole (SCV) to recruit Rab5 (Knodler et al. 2009; Herhaus and Dikic Zhang et al. 2006; Patel et al. 2009, 2018).
Temporal interactions
The diverse interactions between TRPs and host proteins is thought to occur in a temporal pattern through different stages of infection. Although the direct temporal interactions have not been fully elucidated, the location of TRPs during infection has been determined at specific timepoints. TRP32 is detected around the morulae at 24 h post-infection (hpi). Within 48 hpi TRP32 is observed in the perinuclear region and by 72 hpi TRP32 is found in the perinuclear region and in the nucleus (Farris et al. 2016). In addition, TRP47 is observed in the morulae at 24 hpi and within the nucleus at 24–72 hpi (Kibler et al. 2018). TRP120 is expressed on the surface of infectious dense-cored ehrlichiae and initiates infection within 3 hpi and is found in the nucleus at 24 hpi.
The diverse functions of TRP120 are thought to be temporally coordinated during infection, beginning within 3 hpi with its interactions with cell surface receptors including the Notch receptor complex and Wnt FZD receptors to activate Notch and Wnt signaling pathways to stimulate phagocytosis and entry, downregulate innate immune recognition receptors and inhibit lysosomal fusion to promote infection (Lina et al. 2016; Wang et al. 2020; Rogan et al. 2021). During bacterial replication, TRP120 interacts with many host proteins involved in an array of cellular functions, including its interactions with NEDD4L and UBC9 to hijack host PTM machinery. Sequentially, TRP120 is found in the nucleus at 24 h interacting with and ubiquitinating FBXW7 and PCGF5 to regulate host cell transcription and cell fate (Dunphy, Luo and McBride 2014; Mitra et al. 2018; Wang et al. 2020). Notably, the elaborate interactions of TRPs throughout infection occur to modulate cell processes in cell signaling, vesicle trafficking and intracellular transport, metabolism, PTMs, transcriptional regulation and apoptosis to avoid host defense systems and permit intracellular replication (Luo et al. 2017).
Summary and perspective
Understanding the strategies that are used by intracellular bacteria to exploit the host cell and survive a hostile environment have been revealed by studying various pathogens. Most notably, E.ch has one of the smallest bacterial genomes, yet has evolved strategies within the confines of its limited genome to reprogram the complex and sophisticated mononuclear phagocyte and circumvent the innate defenses. Pathogen–host interactions that have recently been described using E.ch. are new to science and thus highlighting the value of this organism for understanding how intracellular pathogens cause infection and evade host defense mechanisms. E.ch. secretes TRP effectors via a T1SS to hijacks host cell processes, including cell signaling, cytoskeletal organization, vesicle trafficking, transcriptional regulation, post-translational modifications, autophagy and apoptosis to evade host defenses. Investigations have led to the understanding that many other pathogenic bacteria utilize TR containing proteins for survival. Further studies regarding E.ch. TRP exploitation of host cell processes and the moonlighting mechanisms involved will unravel the role of bacterial effectors during infection and will potentially reveal the breath and complexity of moonlighting effectors in other bacterial species. Understanding the interactions between TRPs and host interacting partners will reveal how pathogens modulate host cell processes for survival and may facilitate the creation of new and innovative broad-based therapeutics for E.ch. and other intracellular bacteria.
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health grants AI149136, AI137779 and AI123610 awarded to JWM, T32AI007526-20 biodefense training fellowship to CDB, and NIH 1F31AI152424-01 fellowship to LLP.
Contributor Information
Caitlan D Byerly, Departments of Pathology, University of Texas Medical Branch, Galveston, TX 77555, USA.
LaNisha L Patterson, Departments of Pathology, University of Texas Medical Branch, Galveston, TX 77555, USA.
Jere W McBride, Departments of Pathology, University of Texas Medical Branch, Galveston, TX 77555, USA; Microbiology and Immunology, University of Texas Medical Branch, Galveston, TX 77555, USA; Center for Biodefense and Emerging Infectious Diseases, University of Texas Medical Branch, Galveston, TX 77555, USA; Sealy Institute for Vaccine Sciences, University of Texas Medical Branch, Galveston, TX 77555, USA; Institute for Human Infections and Immunity, University of Texas Medical Branch, Galveston, TX 77555, USA.
Conflicts of interest
None declared.
REFERENCES
- Anderson BE, Sumner JW, Dawson JEet al. . Detection of the etiologic agent of human ehrlichiosis by polymerase chain reaction. J Clin Microbiol. 1992;30:775–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson JO, Andersson SG. Genome degradation is an ongoing process in Rickettsia. Mol Biol Evol. 1999;16:1178–91. [DOI] [PubMed] [Google Scholar]
- Barth JMI, Köhler K. How to take autophagy and endocytosis up a notch. Biomed Res Int. 2014;2014:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer AR, Truchan HK, May LJet al. . The Anaplasma phagocytophilum effector AmpA hijacks host cell SUMOylation. Cell Microbiol. 2015;17:504–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björklund ÅK, Ekman D, Elofsson A. Expansion of protein domain repeats. PLoS Comput Biol. 2006;2:0959–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington EM, Zhan Y, Brady JLet al. . Anti-apoptotic proteins BCL-2, MCL-1 and A1 summate collectively to maintain survival of immune cell populations both in vitro and in vivo. Cell Death Differ. 2017;24:878–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey NE, Cyert MS, Moses AM. Short linear motifs - ex nihilo evolution of protein regulation short linear motifs - The unexplored frontier of the eukaryotic proteome. Cell Commun Signal. 2015;13:9–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delepelaire P. Type I secretion in Gram-negative bacteria. Biochimica et Biophysica Acta (BBA) Mol Cell Res. 2004;1694:149–61. [DOI] [PubMed] [Google Scholar]
- Dunning Hotopp JC, Lin M, Madupu Ret al. . Comparative genomics of emerging human ehrlichiosis agents. PLos Genet. 2006;2:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy PS, Luo T, McBride JW. Ehrlichia chaffeensis exploits host SUMOylation pathways to mediate effector-host interactions and promote intracellular survival. Infect Immun. 2014;82:4154–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35:495–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris TR, Dunphy PS, Zhu Bet al. . Ehrlichia chaffeensis TRP32 is a nucleomodulin that directly regulates expression of host genes governing differentiation and proliferation. Infect Immun. 2016;84:3182–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris TR, Zhu B, Wang JYet al. . Ehrlichia chaffeensis TRP32 nucleomodulin function and localization is regulated by NEDD4L-mediated ubiquitination. Front Cell Infect Microbiol. 2018;7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frutos R, Viari A, Ferraz Cet al. . Comparative genomic analysis of three strains of Ehrlichia ruminantium reveals an active process of genome size plasticity. J Bacteriol. 2006;188:2533–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodings C, Smith E, Mathias Eet al. . Hhex is required at multiple stages of adult hematopoietic stem and progenitor cell differentiation. Stem Cells. 2015;33:2628–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green ER, Mecsas J. Bacterial secretion systems: an overview. Microbiol Spectr. 2016;4:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruharsha KG, Kankel MW, Artavanis-Tsakonas S. The Notch signaling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet. 2012;13:654–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao B, Oehlmann S, Sowa MEet al. . Structure of a Fbw7-Skp1-Cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26:131–43. [DOI] [PubMed] [Google Scholar]
- Heraus L, Dikic I. Regulation of Salmonella-host cell interactions via the ubiquitin system. Int J Med Microbiol. 2018;308:176–84. [DOI] [PubMed] [Google Scholar]
- Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. J Cell Sci. 2013;126:2135–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyne GF. Notch signaling in the immune system. J Leukoc Biol. 2003;74:971–81. [DOI] [PubMed] [Google Scholar]
- Kibler CE, Milligan SL, Farris TRet al. . Ehrlichia chaffeensis TRP47 enters the nucleus via a MYND-binding domain-dependent mechanism and predominantly binds enhancers of host genes associated with signal transduction, cytoskeletal organization, and immune response. PLoS One. 2018;13:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Lenzen G, Page ALet al. . The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc Natl Acad Sci. 2005;102:14046–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klema VJ, Sepuru KM, Füllbrunn Net al. . Ehrlichia chaffeensis TRP120 nucleomodulin binds DNA with disordered tandem repeat domain. PLoS One. 2018;13:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler LA, Winfree S, Drecktrah Det al. . Ubiquitination of the bacterial inositol phosphatase, SopB, regulates its biological activity at the plasma membrane. Cell Microbiol. 2009;11:1652–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai Y, Matsuo J, Hayakawa Yet al. . Cyclic di-GMP signaling regulates invasion by Ehrlichia chaffeensis of human monocytes. J Bacteriol. 2010;192:4122–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriakose JA, Miyashiro S, Luo Tet al. . Ehrlichia chaffeensis transcriptome in mammalian and arthropod hosts reveals differential gene expression and post transcriptional regulation. PLoS One. 2011;6:e24136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larabee JL, Ballard JD. Modulation of Notch signaling by intracellular bacterial toxins. FASEB J. 2014;28. [Google Scholar]
- Larabee JL, Shakir SM, Barua Set al. . Increased cAMP in monocytes augments notch signaling mechanisms by elevating RBP-J and transducin-like enhancer of split (TLE). J Biol Chem. 2013;288:21526–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lina T, Dunphy P, Luo Tet al. . Ehrlichia chaffeensis TRP120 activates canonical Notch signaling to downregulate TLR2/4 expression and promote intracellular survival. mBio. 2016;7:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lina TT, Luo T, Velayutham TSet al. . Ehrlichia activation of Wnt-PI3K-mTOR signaling inhibits autolysosome generation and autophagic destruction by the mononuclear phagocyte. Infect Immun. 2017;85:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin IH, Hsu MT, Chang CH. Protein domain repetition is enriched in Streptococcal cell-surface proteins. Genomics. 2012;100:370–9. [DOI] [PubMed] [Google Scholar]
- Lin M, Rikihisa Y. Obligatory intracellular parasitism by Ehrlichia chaffeensis and Anaplasma phagocytophilum involves caveolae and glycosylphosphatidylinositol-anchored proteins. Cell Microbiol. 2003;5:809–20. [DOI] [PubMed] [Google Scholar]
- Lukas J, Mazna P, Valenta Tet al. . Dazap2 modulates transcription driven by the Wnt effector TCF-4. Nucleic Acids Res. 2009;37:3007–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, Dunphy PS, Lina TTet al. . Ehrlichia chaffeensis exploits canonical and noncanonical host Wnt signaling pathways to stimulate phagocytosis and promote intracellular survival. Infect Immun. 2016;84:686–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, Dunphy PS, McBride JW. Ehrlichia chaffeensis tandem repeat effector targets differentially influence infection. Front Cell Infect Microbiol. 2017;7:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, Kuriakose JA, Zhu Bet al. . Ehrlichia chaffeensis TRP120 interacts with a diverse array of eukaryotic proteins involved in transcription, signaling, and cytoskeleton organization. Infect Immun. 2011;79:4382–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, McBride JW. Ehrlichia chaffeensis TRP32 interacts with host cell targets that influence intracellular survival. Infect Immun. 2012;80:2297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, Mitra S, McBride JW. Ehrlichia chaffeensis TRP75 interacts with host cell targets involved in homeostasis, cytoskeleton organization, and apoptosis regulation to promote infection. mSphere. 2018;3:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, Patel JG, Zhang Xet al. . Ehrlichia chaffeensis and E. canis hypothetical protein immunoanalysis reveals small secreted immunodominant proteins and conformation-dependent antibody epitopes. NPJ Vaccines. 2020;5:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, Zhang X, Nicholson WLet al. . Molecular characterization of antibody epitopes of Ehrlichia chaffeensis ankyrin protein 200 and tandem repeat protein 47 and evaluation of synthetic immunodeterminants for serodiagnosis of human monocytotropic ehrlichiosis. Clin Vaccine Immunol. 2010;17:87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti G, Naskar D, Sen M. The Wingless homolog Wnt5a stimulates phagocytosis but not bacterial killing. Proc Natl Acad Sci. 2012;109:16600–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Nara K, Yoshikawa Het al. . Txk, a member of the non-receptor tyrosine kinase of the Tec family, forms a complex with poly(ADP-ribose) polymerase 1 and elongation factor 1 alpha and regulates interferon-gamma gene transcription in Th1 cells. Clin Exp Immunol. 2007;147:164–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A, Onoyama I, Sunabori Tet al. . Fbxw7-dependent degradation of notch is required for control of “Stemness” and neuronal-glial differentiation in neural stem cells. J Biol Chem. 2011;286:13754–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride JW, Walker DH. Molecular and cellular pathobiology of Ehrlichia Infection: targets for new therapeutics and immunomodulation strategies. Nat Inst Health. 2013;13:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S, Dunphy PS, Das Set al. . Ehrlichia chaffeensis TRP120 effector targets and recruits host polycomb group proteins for degradation to promote intracellular infection. Infect Immun. 2018;86:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan Kumar D, Lin M, Xiong Qet al. . EtpE binding to DNase X induces ehrlichial entry via CD147 and hnRNP-K recruitment, followed by mobilization of N-WASP and actin. Mol Biol. 2015;6:e01541–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan Kumar D, Yamaguchi M, Miura Ket al. . Ehrlichia chaffeensis uses its surface protein EtpE to bind GPI-anchored protein DNase X and trigger entry into mammalian cells. PLoS. 2013;9:e1003666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra G, Gaur P, Mujagond Pet al. . A SUMOylation-dependent switch of RAB7 governs intracellular life and pathogenesis of Salmonella typhimurium. J Cell Sci. 2019;132. DOI: 10.1242/jcs.222612. [DOI] [PubMed] [Google Scholar]
- Narayana Y, Balaji KN. NOTCH1 up-regulation and signaling involved in Mycobacterium bovis BCG-induced SOCS3 expression in macrophages. J Biol Chem. 2008;283:12501–11. [DOI] [PubMed] [Google Scholar]
- Nishide A, Kim M, Takagi Ket al. . Structural basis for the recognition of Ubc13 by the Shigella flexneri effector OspI. J Mol Biol. 2013;425:2623–31. [DOI] [PubMed] [Google Scholar]
- Paddock D, Childs E. Ehrlichia chaffeensis: a prototypical emerging pathogen. Clin Microbiol Rev. 2003;16:37–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaga T, Ratanabunyong S, Pattarakankul Tet al. . Notch signaling regulates expression of Mcl-1 and apoptosis in PPD-treated macrophages. Cell Mol Immunol. 2013;10:444–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JC, Hueffer K, Lam TTet al. . Diversification of a Salmonella virulence protein function by ubiquitin-dependent differential localization. Cell. 2009;137:283–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov VL, Xj Yu, Walker DH. The 120 kDa outer membrane protein of Ehrlichia chaffeensis: preferential expression on dense-core cells and gene expression in Escherichia coli associated with attachment and entry. Microb Pathog. 2000;28:71–80. [DOI] [PubMed] [Google Scholar]
- Ribet D, Cossart P. Pathogen-mediated posttranslational modifications: a re-emerging field. Cell. 2010;143:694–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikihisa Y. Role and function of the type IV secretion system in Anaplasma and Ehrlichia species. Curr Top Microbiol Immunol. 2017;413:297–321. [DOI] [PubMed] [Google Scholar]
- Rinaldi FC, Doyle LA, Stoddard BLet al. . The effect of increasing numbers of repeats on TAL effector DNA binding specificity. Nucleic Acids Res. 2017;45:6960–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogan MR, Patterson LL, Byerly CDet al. . Ehrlichia chaffeensis TRP120 is a Wnt ligand mimetic that interacts with Wnt receptors and contains a novel repetitive short linear motif that activates Wnt signaling. mSphere. 2021;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogan MR, Patterson LL, Wang JYet al. . Bacterial manipulation of wnt signaling: a host-pathogen tug-of-wnt. Front Immunol. 2019;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sá-Pessoa J, Przybyszewska K, Vasconcelos FNet al. . Klebsiella pneumoniae reduces SUMOylation to limit host defense responses. mBio. 2020;11:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada T, Kim M, Mimuro Het al. . The Shigella flexneri effector OspI deamidates UBC13 to dampen the inflammatory response. Nature. 2012;483:623–6. [DOI] [PubMed] [Google Scholar]
- Scholze H, Boch J. TAL effectors are remote controls for gene activation. Curr Opin Microbiol. 2011;14:47–53. [DOI] [PubMed] [Google Scholar]
- Song BQ, Chi Y, Li Xet al. . Inhibition of Notch signaling promotes the adipogenic differentiation of mesenchymal stem cells through autophagy activation and PTEN-PI3K/AKT/mTOR pathway. Cell Physiol Biochem. 2015;36:1991–2002. [DOI] [PubMed] [Google Scholar]
- Spitz O, Erenburg IN, Beer Tet al. . Type I secretion systems—One mechanism for all?. Protein Secret Bact. 2019:215–25. DOi: 10.1128/microbiolspec.PSIB-0003-2018. [Google Scholar]
- Thomas S, Popov VL, Walker DH. Exit mechanisms of the intracellular bacterium Ehrlichia. PLoS One. 2010;5:e15775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimoto Y. Role of Bcl-2 family proteins in apoptosis: apoptosomes or mitochondria?. Genes Cells. 1998;3:697–707. [DOI] [PubMed] [Google Scholar]
- Via A, Uyar B, Brun Cet al. . How pathogens use linear motifs to perturb host cell networks. Trends Biochem Sci. 2015;40:36–48. [DOI] [PubMed] [Google Scholar]
- Wakeel A, den Dulk-Ras A, Hooykaas PJJet al. . Ehrlichia chaffeensis tandem repeat proteins and Ank200 are type 1 secretion system substrates related to the repeats-in-toxin exoprotein family. Front Cell Infect Microbiology. 2011;1:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeel A, Zhu B, Yu XJet al. . New insights into molecular Ehrlichia chaffeensis-host interactions. Microbes Infect. 2010;12:337–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY, Zhu B, Patterson LNLet al. . Ehrlichia chaffeensis TRP120-mediated ubiquitination and proteasomal degradation of tumor suppressor FBW7 increases oncoprotein stability and promotes infection. PLoS Pathog. 2020;16:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. [DOI] [PubMed] [Google Scholar]
- Yan Q, Lin M, Huang Wet al. . Ehrlichia type IV secretion system effector Etf-2 binds to active RAB5 and delays endosome maturation. Proc Natl Acad Sci. 2018;115:E8977–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Higashide WM, McCormick BAet al. . The inflammation-associated Salmonella SopA is a HECT-like E3 ubiquitin ligase. Mol Microbiol. 2006;62:786–93. DOi: 10.1111/j.1365-2958.2006.05407.x. [DOI] [PubMed] [Google Scholar]
- Zhou K, Aertsen A, Michiels CW. The role of variable DNA tandem repeats in bacterial adaptation. FEMS Microbiol Rev. 2014;38:119–41. [DOI] [PubMed] [Google Scholar]
- Zhu B, Das S, Mitra Set al. . Ehrlichia chaffeensis TRP120 moonlights as a HECT E3 ligase involved in self and host ubiquitination to influence protein interactions and stability for intracellular survival. Infect Immun. 2017;85:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Kuriakose JA, Luo Tet al. . Ehrlichia chaffeensis TRP120 binds a G+C-rich motif in host cell DNA and exhibits eukaryotic transcriptional activator function. Infect Immun. 2011;79:4370–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Nethery KA, Kuriakose JAet al. . Nuclear translocated Ehrlichia chaffeensis ankyrin protein interacts with a specific adenine-rich motif of host promoter and intronic Alu elements. Infect Immun. 2009;77:4243–55. [DOI] [PMC free article] [PubMed] [Google Scholar]