Abstract
Hypoxia-inducible factors (HIFs) mediate metabolic reprogramming in response to hypoxia. However, the role of HIFs in branched-chain amino acid (BCAA) metabolism remains unknown. Here we show that hypoxia upregulates mRNA and protein levels of the BCAA transporter LAT1 and the BCAA metabolic enzyme BCAT1, but not their paralogs LAT2-4 and BCAT2, in human glioblastoma (GBM) cell lines as well as primary GBM cells. Hypoxia-induced LAT1 protein upregulation is mediated by both HIF-1 and HIF-2 in GBM cells. Although both HIF-1α and HIF-2α directly bind to the hypoxia response element at the first intron of the human BCAT1 gene, HIF-1α is exclusively responsible for hypoxia-induced BCAT1 expression in GBM cells. Knockout of HIF-1α and HIF-2α significantly reduces glutamate labeling from BCAAs in GBM cells under hypoxia, which provides functional evidence for HIF-mediated reprogramming of BCAA metabolism. Genetic or pharmacological inhibition of BCAT1 inhibits GBM cell growth under hypoxia. Together, these findings uncover a previously unrecognized HIF-dependent metabolic pathway that increases GBM cell growth under conditions of hypoxic stress.
Electronic supplementary material
The online version of this article (10.1007/s00018-020-03483-1) contains supplementary material, which is available to authorized users.
Keywords: Hypoxia, Metabolism, Hypoxia-inducible factor, Branched-chain amino acid, Gene regulation, Glioblastoma
Introduction
Metabolism is frequently reprogrammed in cancer cells to meet rapid cell growth under stress conditions. Aerobic glycolysis (also named as the Warburg effect) is the best known altered metabolic phenotype in cancer cells, which is crucial for high demands of macromolecular building blocks (nucleotides, amino acids, and acetyl-CoA) for cancer cell proliferation [1]. Glutamine is also an important carbon and nitrogen source for cancer cell growth by breakdown through glutaminolysis to glutamate [2], and the latter contributes to energy metabolism, nucleotide and protein synthesis, and redox homeostasis in cancer cells [3]. It has been shown that glutamate is released from glioma and has an autocrine effect on glioma growth [4]. In the brain, about one-third of glutamate is de novo synthesized from the branched-chain amino acids (BCAAs: leucine, isoleucine, valine) [5]. BCAAs are transported into the cytosol via a family of L-type amino acid transporters (LAT1-4), and catabolized to branched-chain α-ketoacids (BCKAs) by cytosolic branched-chain aminotransferase (BCAT) 1 and mitochondrial BCAT2 with a transfer of an α-amino group from BCAAs to α-ketoglutarate to synthesize glutamate [5]. BCKAs are further metabolized by the branched-chain α-ketoacid dehydrogenase complex into isovaleryl-CoA, isobutyryl-CoA, and α-methylbutyryl-CoA, followed by multi-step biochemical reactions to generate their end products acetyl-CoA, succinyl-CoA, and acetoacetate [5]. While BCAT2 is ubiquitously expressed, BCAT1 expression is limited to certain tissues, including brain. Recent studies showed that BCAT1 is exclusively expressed in glioblastoma (GBM) with wild-type (WT) isocitrate dehydrogenase (IDH) 1 and promotes GBM cell proliferation by increasing glutamate production [6]. High levels of LAT1 are also found in GBM tissues, and pharmacological inhibition of LAT1 decreases rat glioma growth [7]. However, the molecular mechanism of reprogramming of BCAA metabolism in GBM remains elusive.
The tumor microenvironment of GBM is often hypoxic and hypoxia is a key driver of tumor malignancy. Hypoxia-inducible factor (HIF) is a family of transcription factors that primarily mediate cellular responses to hypoxia to drive GBM progression. Three family members (HIF-1–3) have been identified in mammals [8–10]. Under hypoxia, HIF is formed by dimerization of α(HIF-1α, HIF-2α, or HIF-3α) and β(HIF-1β or ARNT2) subunits and the heterodimeric HIF binds to the hypoxia response element (HRE, 5′-A/GCGTG-3′) to enhance transcription of the downstream target genes [11]. It is well known that HIF-1 and HIF-2 upregulate many metabolic enzymes to reprogram cellular metabolism in cancer cells. They induce the expression of glucose transporters, glycolytic enzymes, and pyruvate dehydrogenase kinases to enhance glucose metabolism and cause pyruvate shunt away from the oxidative phosphorylation pathway towards lactate production in cancer cells [11–13]. HIF-1 and HIF-2 also control reductive glutamine metabolism for proliferation of GBM cells and renal cancer cells [14, 15]. A recent report showed that HIF-2 binds to the HRE of SLC7A5 gene (encoding LAT1) to upregulate LAT1 protein, leading to increased leucine uptake and subsequent mTORC1 activation in renal carcinoma cells [16]. Yet, whether or not HIF regulates BCAA metabolism is unknown.
In the present study, we discovered a new HIF-dependent metabolic pathway in GBM cells. HIF-1 and HIF-2 upregulated LAT1 protein in GBM cells, whereas HIF-1 bound to the HRE of the BCAT1 gene to induce its gene transcription, leading to reprogramming of BCAA metabolism and GBM cell growth under hypoxia.
Materials and methods
Plasmid constructs
DNA oligonucleotides containing WT or mutant BCAT1 HRE were annealed and ligated into MluI/BglII-linearized pGL2-promoter vector (Promega). DNA oligonucleotides of the single guide RNA (sgRNA) targeting human BCAT1 (Supplementary Table 1) were annealed and ligated into BsmBI-linearized lentiCRISPRv2 vector (Addgene, #52961). Other constructs have been described previously [17–20]. All recombinant plasmids were verified by Sanger sequencing.
Cell culture, transfection, and hypoxia
HeLa, HEK293T, U251MG, and U87MG cells were cultured in DMEM supplemented with 10% heat-inactivated fetal bovine serum (FBS) at 37 °C in a 5% CO2/95% air incubator. Cells were transfected using PolyJet (SignaGen) or FuGENE6 (Promega) transfection reagent according to the manufacturer’s protocol. Hypoxia was conducted by placing cells in a modular incubator chamber (Billups-Rothenberg) flushed with a gas mixture of 1% O2, 5% CO2, and balanced N2. Lentivirus was produced by transfection of HEK293T cells with sgRNA vector and packaging vectors (psPAX2 and pMD2.G). All cell lines are mycoplasma-free and have been authenticated by STR DNA profiling analysis.
Primary GBM culture
The study was approved by the Institutional Review Board at UT Southwestern Medical Center with informed consent. The deidentified fresh human GBM specimens were dissociated into single cells using collagenase D (1 mg/ml, Sigma) and DNase I (0.1 mg/ml, Sigma) and filtered with a sterile 70-μm cell strainer. A single-cell suspension was plated on a 6-well ultra-low attachment culture plate and cultured in DMEM/F-12 supplemented with B27 supplements (Thermo Fisher), basic fibroblast growth factor (10 ng/ml, Sigma), epidermal growth factor (20 ng/ml, Sigma), and penicillin (50 U/ml)/streptomycin (50 μg/ml)/neomycin (100 μg/ml, Sigma).
Generation of CRISPR/Cas9-based knockout (KO) cell lines
BCAT1 KO, HIF-1α KO, HIF-2α KO, and HIF-1/2α double KO (DKO) cell lines were generated using the CRISPR/Cas9 technique. Cells were transiently transfected with sgRNA vector using PolyJet (SignaGen). A single KO cell was selected after treatment with puromycin. KO cells were verified by genotyping and/or immunoblot assays.
Colony formation assay
Cells were seeded on a 48-well plate and cultured for 7 days. Gabapentin (5 mM) or vehicle (H2O) was added into media immediately after seeding. Colonies were washed with PBS, fixed with 4% paraformaldehyde, and stained with 0.01% crystal violet. The crystal violet dye was dissolved in 10% acetic acid and measured by a plate reader at OD 570 nm.
Quantitative reverse transcription-polymerase chain reaction (RT-qPCR) assay
Total RNA was isolated from cultured cells using Trizol reagent (Thermo Fisher), treated with DNase I (Ambion), and reverse transcribed using an iScript cDNA Synthesis Kit (Bio-Rad) as described previously [17, 19]. Real-time qPCR was performed with primers listed in Supplementary Table 2 in a CFX96 cycler (Bio-Rad) using iTaq Universal SYBR Green Supermix (Bio-Rad). Target mRNA expression was normalized to 18S rRNA and its fold change was calculated based on the threshold cycle as , where and.
Immunoblot assay
Cells were lysed in modified lysis buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM β-mercaptoethanol, 1% Igepal, and protease inhibitor cocktail) for 30 min on ice, followed by sonication and centrifugation at 13,000g for 15 min. Equal amounts of lysates were fractionated by SDS-PAGE and subject to immunoblot assay with the following antibodies: LAT1 (Proteintech, Cat.#: 13752-1-AP), BCAT1 (Proteintech, Cat.#: 13640-1-AP), BCAT2 (Proteintech, Cat.#: 16417-1-AP), HIF-1α (BD Biosciences, Cat.#: 610959), HIF-2α (Bethyl Laboratories, Cat.#: A700-003), or actin (Proteintech, Cat.#: 66009-1-Ig).
Luciferase reporter assay
HeLa, HIF-1α or HIF-2α KO HeLa, or HEK293T cells were plated on 48-well plates or poly-l-lysine-coated 48-well plates (for HEK293T cells), and transiently transfected with WT or mutant BCAT1 HRE reporter plasmid; control reporter plasmid pSV40-Renilla; and p3XFLAG-HIF-1α, p3XFLAG-HIF-2α, or empty vector. Twenty-four hours later, cells were exposed to 20% or 1% O2 for 24 h. Firefly and Renilla luciferase activities were measured using the Dual Luciferase Reporter Assay System (Promega) as described previously [17, 19].
Chromatin immunoprecipitation (ChIP)-qPCR assay
U251MG cells were exposed to 20% or 1% O2 for 24 h, crosslinked with 1% formaldehyde for 20 min at room temperature, and quenched in 0.125 M glycine. Cells were lysed in lysis buffer (50 mM Tris–HCl, 10 mM EDTA, 1% SDS, protease inhibitor cocktail), sonicated, and subjected to immunoprecipitation in the presence of salmon sperm DNA/protein A beads with antibodies against HIF-1α, HIF-2 α, HIF-1β, or IgG overnight at 4 °C. Precipitated chromatin DNA was extensively washed, eluted with freshly prepared elution buffer (0.1 M NaHCO3, 1% SDS), decrosslinked at 65 °C for 4 h, incubated with proteinase K at 45 °C for 45 min, purified using phenol/chloroform/isoamyl alcohol (25:24:1, v/v), and quantified by real-time qPCR as described previously [17, 19]. The primers used for ChIP-qPCR are listed in Supplementary Table 3.
Isotope tracing assay
Cells were seeded on 6-cm dishes and exposed to 20% or 1% O2 for 48 h. After rinsing with warm PBS, cells were incubated for 1 h with freshly prepared DMEM containing 10% FBS, 0.105 g/L L[15N]leucine, 0.105 g/L L[15N]isoleucine, and 0.094 g/L L[15N]valine, which achieved about 50% of BCAAs labeling in the total pool. For hypoxia group, labeled media were preequilibrated with 1% O2 for 30–60 min. At the end of incubation, cells were rinsed with ice-cold saline. To measure metabolites in media, cells were replaced with fresh DMEM and media were collected after 8 h-culture. To measure intracellular metabolites, 0.5 ml of cold methanol/water (1:1, v/v) was added to the dishes. Cells were scraped off the dishes and lysed by repeated freeze–thaw cycles, followed by centrifugation at 13,000g for 10 min. The cell pellets were collected and dissolved in 0.1 N NaOH. Proteins were quantified by Bradford assay. The supernatant containing the intracellular metabolites was analyzed by gas chromatography-tandem mass spectrometry (GC–MS). To prepare samples for GC–MS analysis, myristic acid was added to the supernatant as an internal standard. Samples were then evaporated and derivatized by tert-butyldimethylsilylation. The methodology of GC–MS analysis was described previously [21]. The total pool size of each isotopomer was calculated by summing up area under their curve and then normalized to both protein concentration and internal standard. The relative pool size of each isotopomer was further calculated by their total pool size x their enrichment.
Statistical analysis
Data were repeated for at least three times and expressed as mean ± SEM. Statistical analysis was performed by Student’s t test between two groups, and one-way or two-way ANOVA within multiple groups. p < 0.05 is considered significant.
Results
Hypoxia upregulates LAT1 and BCAT1 in GBM cells
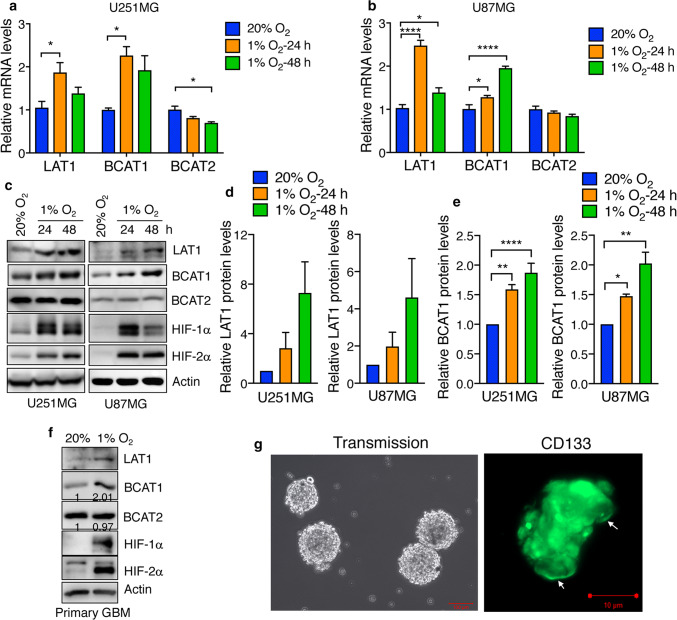
To determine whether hypoxia regulates the expression of the metabolic proteins that control BCAA metabolism in GBM cells, human GBM U251MG cells were exposed to 20% or 1% O2 for 24 or 48 h. RT-qPCR assay showed that hypoxia significantly increased LAT1 mRNA levels in U251MG cells (Fig. 1a), which is consistent with a previous study in renal carcinoma cells [16]. LAT2 mRNA expression was not affected by hypoxia in U251MG cells (Supplementary Fig. 1), while LAT3 and LAT4 mRNAs were downregulated by hypoxia in U251MG cells (Supplementary Fig. 1). BCAT1 mRNA was significantly induced at 24 or 48 h after hypoxia exposure in U251MG cells (Fig. 1a). In contrast, BCAT2 mRNA was modestly downregulated by hypoxia in U251MG cells (Fig. 1a). Hypoxia-induced LAT1 and BCAT1 mRNA upregulation was similarly observed in another human GBM U87MG cell line (Fig. 1b). In line with their mRNA expression, the protein levels of LAT1 and BCAT1, but not BCAT2, were increased in U251MG and U87MG cells under hypoxia in a time-dependent manner (Fig. 1c–e). We further confirmed hypoxia-induced expression of LAT1 and BCAT1 in primary human GBM cells (Fig. 1f). Primary GBM cells were validated by CD133 immunostaining (Fig. 1g). These data indicate that hypoxia specifically induces the expression of LAT1 and BCAT1, but not LAT2-4 and BCAT2, in human GBM cells.
Fig. 1.
Hypoxia upregulates LAT1 and BCAT1, but not BCAT2, in human GBM cells. a, b RT-qPCR analysis of LAT1, BCAT1, and BCAT2 mRNAs in U251MG (a) and U87MG (b) cells exposed to 20% or 1% O2 for 24 or 48 h. Data are shown in mean ± SEM, n = 3–6. *p < 0.05; ****p < 0.0001 (One-way ANOVA). c–e Immunoblot analysis of indicated protein levels in U251MG (left) and U87MG (right) exposed to 20% or 1% O2 for 24 or 48 h. Representative blots from 3–6 independent experiments are shown in c. LAT1 (d), BCAT1 (e) and actin western blot bands were quantified by densitometry and the ratio of LAT1 and BCAT1 to actin was normalized to 20% O2 (mean ± SEM, n = 3–6). *p < 0.05; **p < 0.01; ****p < 0.0001 (One-way ANOVA). f Immunoblot analysis of indicated protein levels in primary human GBM cells exposed to 20% or 1% O2 for 48 h. Representative blots from two independent experiments are shown. Intensity of BCAT1 and BCAT2 western blots is quantified and normalized to 20% O2. g Primary GBM cells were stained with a mouse monoclonal anti-CD133 antibody followed by a donkey anti-mouse Cy2 antibody, fixed with 4% paraformaldehyde, and visualized by a Zeiss microscopy. Scale bar, 100 μm (left) and 10 μm (right)
Distinct role of HIFs in hypoxia-induced LAT1 and BCAT1 expression in GBM cells
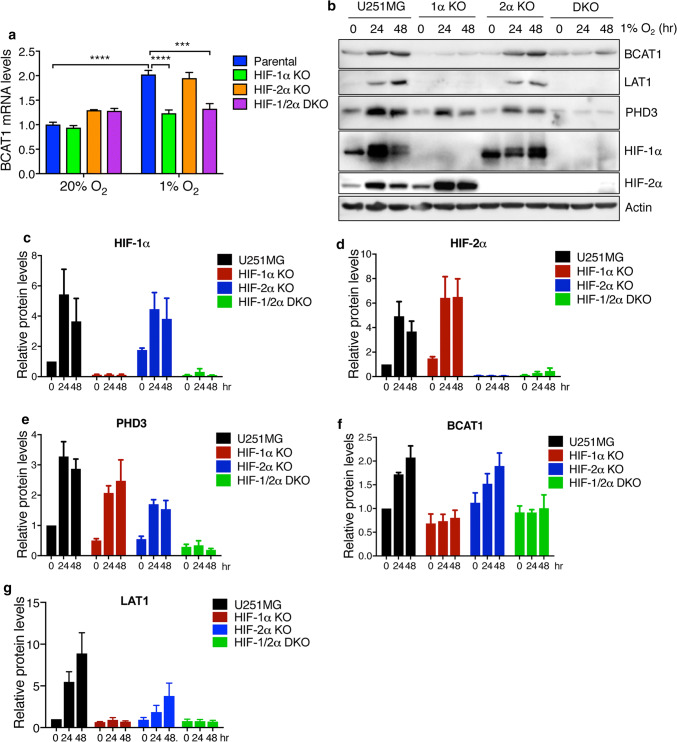
To study whether HIFs are required for hypoxia-induced BCAT1 and LAT1 expression in GBM cells, we generated HIF-1α KO, HIF-2α KO, and HIF-1/2α DKO U251MG cells by the CRISPR/Cas9 technique [20]. KO of HIF-1α and HIF-2α proteins was confirmed in these cells (Fig. 2b–d). Expression of PHD3, a known HIF-1 and HIF-2 target protein [22], was downregulated in both HIF-1α KO and HIF-2α KO cells, and further reduced in HIF-1/2α DKO cells under hypoxia (Fig. 2b, e), functionally validating HIF-1α KO, HIF-2α KO, and HIF-1/2α DKO U251MG cells. Both HIF-1α KO and HIF-1/2α DKO abolished hypoxia-induced BCAT1 mRNA expression in U251MG cells (Fig. 2a). However, HIF-2α KO failed to affect BCAT1 mRNA expression in U251MG cells (Fig. 2a). Consistently, hypoxia-induced BCAT1 protein levels were inhibited in HIF-1α KO or HIF-1/2α DKO, but not HIF-2α KO, U251MG cells (Fig. 2b, f). It was reported that LAT1 is induced by HIF-2 in kidney cancer cells [16]. As expected, HIF-2α KO inhibited hypoxia-induced LAT1 protein expression in U251MG cells (Fig. 2b, g). In addition, we found that HIF-1α KO or HIF-1/2α DKO completely abolished hypoxia-induced LAT1 protein upregulation in U251MG cells (Fig. 2b, g). Together, these data indicate that both HIF-1 and HIF-2 are required for LAT1 expression in human GBM cells, and that hypoxia upregulates BCAT1 expression in human GBM cells in a HIF-1-dependent manner.
Fig. 2.
HIFs are required for hypoxia-induced LAT1 and BCAT1 expression in GBM cells. a RT-qPCR analysis of BCAT1 mRNA in parental, HIF-1α KO, HIF-2α KO, and HIF-1/2α DKO U251MG cells exposed to 20% or 1% O2 for 24 h. Data are shown in mean ± SEM, n = 3. ***p < 0.001; ****p < 0.0001 (Two-way ANOVA). b–g Immunoblot analysis of indicated protein levels in parental, HIF-1α KO, HIF-2α KO, and HIF-1/2α DKO U251MG cells exposed to 20% or 1% O2 for 24 or 48 h. Representative blots from three independent experiments are shown in b. Intensity of HIF-1α (c), HIF-2α (d), PHD3 (e), BCAT1 (f), LAT1 (g) and actin western blot bands was quantified by densitometry and their ratio to actin was normalized to U251MG cells at 0 h (mean ± SEM, n = 3)
BCAT1 is a direct HIF-1 target gene
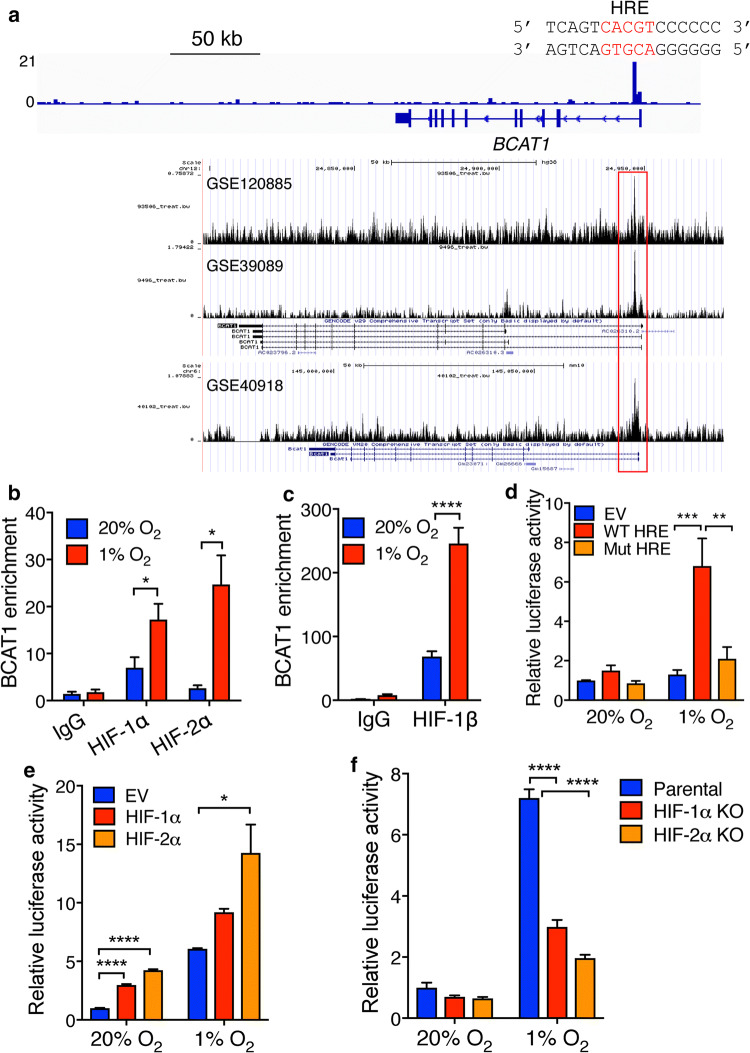
The HRE has been found at the promoter of the SLC7A5 gene [16]. Thus, we decided to identify the HRE on the BCAT1 gene to determine whether BCAT1 is a direct HIF-1 target gene. To this end, we analyzed HIF-1α ChIP-sequencing data [19] and found a strong HIF-1α ChIP peak in the first intron of BCAT1 gene (Fig. 3a). This HIF-1α binding peak was conserved in multiple types of human and mouse cells (Fig. 3a). The putative core HIF-1 binding sequence 5′-ACGTG-3′ (on the antisense strand) was found at this ChIP peak (Fig. 3a). To validate, we performed ChIP-qPCR assay in U251MG cells exposed to 20% or 1% O2 for 24 h. HIF-1α was enriched at the BCAT1 HRE in nonhypoxic U251MG cells, which was significantly increased > twofold by hypoxia (Fig. 3b). Conversely, hypoxia did not affect IgG enrichment at this HRE, suggesting the specificity of the assay (Fig. 3b). Interestingly, although HIF-2α KO did not alter BCAT1 mRNA and protein levels (Fig. 2), HIF-2α was also enriched at this HRE in U251MG cells under hypoxia (Fig. 3b). As expected, increased HIF-1β occupancy at this HRE was similarly detected in hypoxic U251MG cells (Fig. 3c). These data indicate that both HIF-1 and HIF-2 directly bind to the HRE of the BCAT1 gene in GBM cells.
Fig. 3.
BCAT1 is a direct HIF-1 target gene. a The genome browser snapshots of the HIF-1α ChIP peak at the BCAT1 locus in human and mouse cells. The nucleotide sequence of the HRE (shown in red) within the first intron of the human BCAT1 gene is shown. b, c ChIP-qPCR analysis of enrichment of HIF-1α (b), HIF-2α (b), HIF-1β (c), and IgG at the BCAT1 HRE in U251MG cells exposed to 20% or 1% O2 for 24 h. *p < 0.05; ****p < 0.0001 (Student’s t test). d HEK293T cells were cotransfected with empty pGL2p vector (EV), pGL2p-WT BCAT1 HRE, or pGL2p-mutant (Mut) BCAT1 HRE, and pSV40-Renilla, and exposed to 20% or 1% O2 for 24 h. The ratio of firefly/Renilla activities was normalized to EV at 20% O2 (mean ± SEM, n = 3). **p < 0.01; ***p < 0.001 (Two-way ANOVA). e HeLa cells were cotransfected with pGL2p-WT BCAT1 HRE, pSV40-Renilla, and vector encoding FLAG-HIF-1α or FLAG-HIF-2α, or FLAG EV, and exposed to 20% or 1% O2 for 24 h. The ratio of firefly/Renilla activities was normalized to EV at 20% O2 (mean ± SEM, n = 3). *p < 0.05; ****p < 0.0001 (Two-way ANOVA). f Parental, HIF-1α KO, or HIF-2α KO HeLa cells were cotransfected with pGL2p-WT BCAT1 HRE and pSV40-Renilla, and exposed to 20% or 1% O2 for 24 h. The ratio of firefly/Renilla activities was normalized to parental at 20% O2 (mean ± SEM, n = 3). ****p < 0.0001 (Two-way ANOVA)
To further determine whether the BCAT1 HRE is functional, we performed a dual-luciferase reporter assay. HEK293T and HeLa cells were used for this assay because of high transfection efficiency in these cell lines. A 53-bp oligonucleotide sequence containing the BCAT1 HRE was inserted upstream of the firefly luciferase coding sequence in a pGL2-promoter reporter plasmid. Another firefly luciferase reporter plasmid containing mutant BCAT1 HRE, in which 5′-CGT-3′ within the HIF binding site was mutated into 5′-AAA-3′, was similarly generated. HEK293T cells were co-transfected with WT or mutant BCAT1 HRE reporter plasmid or empty vector (EV) and a constitutively expressed Renilla luciferase control reporter pSV40-Renilla, and exposed to 20% or 1% O2 for 24 h. WT BCAT1 HRE significantly increased the expression of the firefly luciferase gene, but not the Renilla luciferase gene, in hypoxic HEK293T cells (Fig. 3d), which was abolished by mutation of the HIF binding site (Fig. 3d). Ectopic expression of FLAG-HIF-1α or FLAG-HIF-2α significantly increased BCAT1 HRE-driven firefly luciferase gene expression in HeLa cells (Fig. 3e). Less BCAT1 HRE activity by FLAG-HIF-1α is possibly due to less FLAG-HIF-1α protein levels as compared to FLAG-HIF-2α (Supplementary Fig. 2a). In contrast, CRISPR/Cas9-based KO of HIF-1α or HIF-2α significantly decreased hypoxia-induced BCAT1 HRE activity in HeLa cells (Fig. 3f and Supplementary Fig. 2b). Together, these data indicate that HIF-1 directly binds to the HRE at the first intron of BCAT1 gene to activate its gene transcription and that HIF-2 has the binding ability to the BCAT1 HRE but is not sufficient to activate BCAT1 mRNA transcription in GBM cells.
HIF-1 and HIF-2 mediate reprogramming of BCAA metabolism in GBM cells
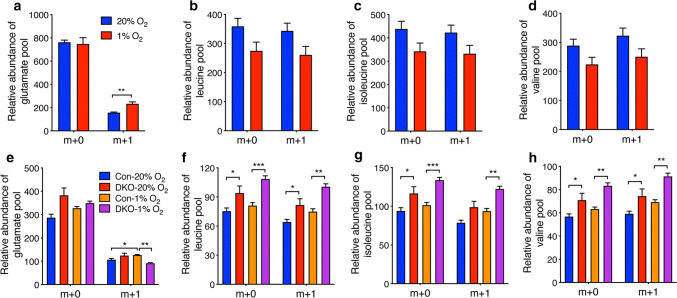
To determine whether hypoxia regulates reprogramming of BCAA metabolism, we performed isotope tracing assays using three BCAA tracers L[15N]leucine, L[15N]isoleucine, and L[15N]valine to measure the relative changes of intracellular levels of BCAAs and transamination of BCAAs in nonhypoxic and hypoxic GBM cells. As shown in Fig. 4a, the fractional amount of intracellular glutamate m + 1 was significantly increased by hypoxia. In contrast, hypoxia had no effect on levels of extracellular glutamate m + 1 in U87MG cells (Supplementary Fig. 3). These data indicate elevated transamination between BCAAs and glutamate in hypoxic U87MG cells. The fractions of leucine, isoleucine, and valine were modestly decreased by hypoxia in U87MG cells (Fig. 4b–d). We further studied whether HIF is required for hypoxia-induced reprogramming of BCAA metabolism. Because HIF-1 and HIF-2 differentially controlled the expression of LAT1 and BCAT1 (Fig. 2), HIF-1/2α DKO U251MG cells were used in this study. Hypoxia also similarly increased BCAAs-derived glutamate labeling in U251MG cells, which was attenuated by HIF-1/2α DKO (Fig. 4e). Conversely, m + 0 and m + 1 fractions of leucine, isoleucine, and valine were enhanced in HIF-1/2α DKO U251MG cells under 20% O2 and 1% O2 (Fig. 4f–h), indicating reduced BCAA metabolism by HIF-1/2α DKO. Of note, U251MG cells expressed HIF-1α and HIF-2α under 20% O2 (Fig. 1c). Together, these data indicate that HIF-1 and HIF-2 enhance reprogramming of BCAA metabolism in GBM cells.
Fig. 4.
HIF-1 and HIF-2 enhance BCAA metabolism in GBM cells. a–d Enrichment of glutamate (a), leucine (b), isoleucine (c), valine (d) fractions in nonhypoxic and hypoxic U87MG cells cultured in L[15N]leucine, L[15N]isoleucine, and L[15N]valine. Data are shown in mean ± SEM, n = 4. **p < 0.01 (Student’s t test). e–h Enrichment of glutamate (e), leucine (f), isoleucine (g), valine (h) fractions in nonhypoxic and hypoxic parental (Con) or HIF-1/2α DKO U251MG cells cultured in L[15N]leucine, L[15N]isoleucine, and L[15N]valine. Data are shown in mean ± SEM, n = 4. *p < 0.05; **p < 0.01; ***p < 0.001 (Two-way ANOVA)
BCAT1 promotes GBM cell growth under hypoxia
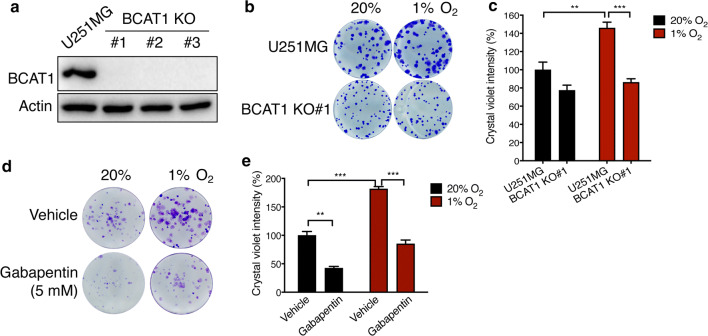
To determine the functional significance of BCAT1 upregulation by hypoxia in GBM cells, we generated three independent BCAT1 KO U251MG cell lines by the CRISPR/Cas9 technique (Fig. 5a) and studied the effect of BCAT1 on cell growth by colony formation assay. Hypoxia significantly increased colony growth of U251MG cells, which was reduced by BCAT1 KO#1 (Fig. 5b, c). Similar results were also observed in BCAT1 KO#2 and KO#3 U251MG cells (Supplementary Fig. 4a–d). To determine if the enzymatic activity of BCAT1 is required for GBM cell growth, we applied a BCAT1 inhibitor Gabapentin in the colony formation assay. Treatment of Gabapentin (5 mM) significantly reduced U251MG colony formation under both 20% and 1% O2 (Fig. 5d, e). These data indicate that BCAT1 increases GBM cell growth under hypoxia and that its enzymatic activity is required for GBM cell growth.
Fig. 5.
BCAT1 increases GBM cell growth under hypoxia. a Immunoblot analysis of BCAT1 and actin proteins in parental and BCAT1 KO U251MG cells. Representative blots from three independent experiments are shown. b, c Colony growth of parental and BCAT1 KO#1 U251MG cells cultured under 20% or 1% O2 for 7 days. Representative images from three independent experiments are shown in b. The crystal violet dye is measured by a plate reader and data are normalized to U251MG cells at 20% O2 (mean ± SEM, n = 3, c). **p < 0.01; ***p < 0.001 (Two-way ANOVA). d, e Colony growth of U251MG cells exposed to 20% or 1% O2 for 7 days in the presence or absence of Gabapentin (5 mM). Representative images from three independent experiments are shown in d. The crystal violet dye is measured by a plate reader and data are normalized to vehicle at 20% O2 (mean ± SEM, n = 3, e). **p < 0.01; ***p < 0.001 (Two-way ANOVA)
Discussion
In the present study, we for the first time uncover a critical role of HIFs in reprogramming of BCAA metabolism in GBM cells under hypoxia. GBM cells adapt to increased malignant phenotypes under conditions of hypoxic stress through elevating BCAA metabolism. Our biochemical studies of gene regulation clearly reveal that LAT1 protein is upregulated by both HIF-1 and HIF-2 in GBM cells, whereas BCAT1 expression is regulated by HIF-1 only, but not HIF-2, in hypoxic GBM cells. Upregulation of LAT1 expression by hypoxia in GBM cells has been supported by a previous study in renal carcinoma cells [16]. Distinct to an exclusive role of HIF-2 in LAT1 upregulation in renal carcinoma cells, both HIF-1 and HIF-2 are required for hypoxia-induced LAT1 protein expression in GBM cells. Moreover, HIF-1 plays a more dominant role in LAT1 induction than HIF-2, indicating that LAT1 is regulated by HIFs in a cell type-specific manner. LAT1 is a primary transporter for BCAAs and highly expressed in brain as a specific LAT1 inhibitor JPH203 can completely block leucine uptake in cancer cells [23, 24]. LAT3 and LAT4 are downregulated by hypoxia in GBM cells, different from LAT1. Previous studies implicated the role of LAT3 and LAT4 in the efflux of intracellular BCAAs into the bloodstream [25]. Thus, hypoxia not only increases BCAA uptake, but also inhibits BCAA efflux, leading to elevated levels of intracellular BCAAs in GBM cells.
Similar to differential regulation of LATs by hypoxia, HIF-1 upregulates BCAT1 but has no effect on BCAT2 protein expression in GBM cells. Interestingly, our ChIP-qPCR and luciferase reporter assays showed that HIF-2α is able to bind to the BCAT1 HRE to enhance its activity, but HIF-2α KO has no effect on the mRNA and protein levels of BCAT1, suggesting that HIF-2α alone is not sufficient to activate BCAT1 gene transcription in GBM cells. Epigenetic regulators play a crucial role in HIF transactivation [26]. Whether or not an epigenetic regulator differentially controls the transcriptional activity of HIF-1 and HIF-2 to regulate BCAT1 transcription requires further investigation in the future.
A previous report showed that BCAT1, but not BCAT2, is induced by the T cell receptor in CD4+ T cells [27]. These findings and ours suggest that BCAT1 expression is more prone to be altered in response to stimuli, which may compensate its limited distribution in certain tissues. In addition to differential regulation of BCAT1 and BCAT2 by stimuli, redox plays an important role in BCAT activity. The conserved CXXC motif was identified in mammalian BCAT1 and BCAT2 and functions as a redox sensor [28]. Oxidation of this motif by redox inhibits the activity of both BCAT1 and BCAT2, although BCAT2 is more oxidized than BCAT1 [29–32], as well as reduces BCAT2 binding to protein disulfide isomerase in the brain [33]. Nitric oxide also acts on thiol groups of BCAT1 and BCAT2 and inactivates these enzymes [34]. It has been shown that the nitric oxide reactive agent S-nitrosoglutathione inhibits BCAT1 in vitro through S-nitrosation at the Cysteine residue 335, but may exert a different mechanism of BCAT2 inhibition [34]. Chronic hypoxia is known to increase the redox production in the mammalian cells [35], but it remains unknown to what extent BCAT1 and BCAT2 are regulated by redox under chronic hypoxia.
Our GC–MS studies showed that transamination of BCAAs is enhanced by HIFs in GBM cells under hypoxia. Several outstanding questions remain to be answered in the future. It remains unclear whether HIF-1 increases BCAT1 activity to enhance BCAA transamination in GBM cells. The BCAT activity assay is needed in the future to address this question. BCAT1 catalyzes the reversible transamination of BCAAs, and whether or not HIFs increase reductive metabolism of BCAAs in GBM cells remains to be investigated. Although cancer cells express both BCAT1 and BCAT2, BCAT1 is expressed in limited normal tissues. It would be interesting to investigate in the future how hypoxia regulates BCAA metabolism in tissues expressing BCAT2 alone. Nevertheless, our findings provide functional evidence for HIF-mediated reprogramming of BCAA metabolism in GBM cells. Importantly, we showed that BCAT1 is required for GBM cell growth under hypoxia through its transaminase activity. Cancer cells are addicted to altered metabolism for survival. Hypoxia causes a shift from glucose oxidation to lactate production and increases reductive glutamine metabolism in cancer cells [12, 14, 15, 17], indicating a critical role of glutamine → glutamate → the TCA cycle in cell growth under hypoxia. Metabolomics studies showed that the concentration of glutamine is significantly lower in the hypoxic regions than the peripheral nonhypoxic areas of solid tumors [36]. In contrast, the levels of glutamate are not reduced in the hypoxic regions [36]. Likewise, BCAA levels are comparable in both hypoxic and nonhypoxic regions of tumors [36]. Thus, HIF-mediated BCAA reprogramming may be an important mechanism that maintains the level of glutamate in the hypoxic regions of tumors to support reductive metabolism and subsequent cancer cell survival. Recent studies showed a critical role of BCAT1 in malignancy of human cancers, including wild-type IDH1 GBM, lung cancer, breast cancer, leukemia, and ovarian cancer [6, 37–41]. Our findings here suggest that BCAT1, like other HIF-induced metabolic genes [42, 43], may reprogram cellular metabolism and contribute to HIF-mediated malignancy of a wide range of solid tumors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would thank to Jennifer E. Wang for proofreading. This work was supported by grants from the NIH (R00CA168746, R01CA222393), Welch Foundation (I-1903), and the CPRIT (RR140036, RP190358) to W.L., and the NIH (R00NS078049, R01AG066166, and R35GM124693), CPRIT (RP170671), and Welch Foundation (I-1939) to Y.W. W.L. is a CPRIT Scholar in Cancer Research.
Author contributions
WL, YW conceived the study, analyzed the data, and wrote the paper; BZ, YC, XS, MZ, BL performed experiments and analyzed the data; KH, TP provided fresh human glioblastoma tissues; RD analyzed the data. All authors read and approved the manuscript.
Compliance with ethical standards
Conflict of interest
R.J.D. is an advisor for Agios Pharmaceuticals. Other authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Bo Zhang and Yan Chen contributed equally to this work.
Contributor Information
Yingfei Wang, Email: Yingfei.Wang@UTSouthwestern.edu.
Weibo Luo, Email: Weibo.Luo@UTSouthwestern.edu.
References
- 1.Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci. 2016;41(3):211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Investig. 2013;123(9):3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coloff JL, Murphy JP, Braun CR, Harris IS, Shelton LM, Kami K, Gygi SP, Selfors LM, Brugge JS. Differential glutamate metabolism in proliferating and quiescent mammary epithelial cells. Cell Metab. 2016;23(5):867–880. doi: 10.1016/j.cmet.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 4.Takano T, Lin JH, Arcuino G, Gao Q, Yang J, Nedergaard M. Glutamate release promotes growth of malignant gliomas. Nat Med. 2001;7(9):1010–1015. doi: 10.1038/nm0901-1010. [DOI] [PubMed] [Google Scholar]
- 5.Yudkoff M. Brain metabolism of branched-chain amino acids. Glia. 1997;21(1):92–98. doi: 10.1002/(SICI)1098-1136(199709)21:1<92::AID-GLIA10>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 6.Tonjes M, Barbus S, Park YJ, Wang W, Schlotter M, Lindroth AM, Pleier SV, Bai AHC, Karra D, Piro RM, Felsberg J, Addington A, Lemke D, Weibrecht I, Hovestadt V, Rolli CG, Campos B, Turcan S, Sturm D, Witt H, Chan TA, Herold-Mende C, Kemkemer R, Konig R, Schmidt K, Hull WE, Pfister SM, Jugold M, Hutson SM, Plass C, Okun JG, Reifenberger G, Lichter P, Radlwimmer B. BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nat Med. 2013;19(7):901–908. doi: 10.1038/nm.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nawashiro H, Otani N, Shinomiya N, Fukui S, Ooigawa H, Shima K, Matsuo H, Kanai Y, Endou H. L-type amino acid transporter 1 as a potential molecular target in human astrocytic tumors. Int J Cancer. 2006;119(3):484–492. doi: 10.1002/ijc.21866. [DOI] [PubMed] [Google Scholar]
- 8.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92(12):5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11(1):72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 10.Gu YZ, Moran SM, Hogenesch JB, Wartman L, Bradfield CA. Molecular characterization and chromosomal localization of a third alpha-class hypoxia inducible factor subunit, HIF3alpha. Gene Expr. 1998;7(3):205–213. [PMC free article] [PubMed] [Google Scholar]
- 11.Semenza GL, Jiang BH, Leung SW, Passantino R, Concordet JP, Maire P, Giallongo A. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem. 1996;271(51):32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- 12.Semenza GL, Roth PH, Fang HM, Wang GL. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J Biol Chem. 1994;269(38):23757–23763. doi: 10.1016/S0021-9258(17)31580-6. [DOI] [PubMed] [Google Scholar]
- 13.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3(3):177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, Kelleher JK, Vander Heiden MG, Iliopoulos O, Stephanopoulos G. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2011;481(7381):380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, DeMatteo RG, Simon MC, Thompson CB. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci USA. 2011;108(49):19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elorza A, Soro-Arnaiz I, Melendez-Rodriguez F, Rodriguez-Vaello V, Marsboom G, de Carcer G, Acosta-Iborra B, Albacete-Albacete L, Ordonez A, Serrano-Oviedo L, Gimenez-Bachs JM, Vara-Vega A, Salinas A, Sanchez-Prieto R, Martin del Rio R, Sanchez-Madrid F, Malumbres M, Landazuri MO, Aragones J. HIF2alpha acts as an mTORC1 activator through the amino acid carrier SLC7A5. Mol Cell. 2012;48(5):681–691. doi: 10.1016/j.molcel.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 17.Luo W, Hu H, Chang R, Zhong J, Knabel M, O'Meally R, Cole RN, Pandey A, Semenza GL. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145(5):732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo W, Chen I, Chen Y, Alkam D, Wang Y, Semenza GL. PRDX2 and PRDX4 are negative regulators of hypoxia-inducible factors under conditions of prolonged hypoxia. Oncotarget. 2016;7(6):6379–6397. doi: 10.18632/oncotarget.7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Zhang B, Bao L, Jin L, Yang M, Peng Y, Kumar A, Wang JE, Wang C, Zou X, Xing C, Wang Y, Luo W. ZMYND8 acetylation mediates HIF-dependent breast cancer progression and metastasis. J Clin Investig. 2018;128(5):1937–1955. doi: 10.1172/JCI95089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bao L, Chen Y, Lai HT, Wu SY, Wang JE, Hatanpaa KJ, Raisanen JM, Fontenot M, Lega B, Chiang CM, Semenza GL, Wang Y, Luo W. Methylation of hypoxia-inducible factor (HIF)-1alpha by G9a/GLP inhibits HIF-1 transcriptional activity and cell migration. Nucleic Acids Res. 2018;46(13):6576–6591. doi: 10.1093/nar/gky449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang C, Ko B, Hensley CT, Jiang L, Wasti AT, Kim J, Sudderth J, Calvaruso MA, Lumata L, Mitsche M, Rutter J, Merritt ME, DeBerardinis RJ. Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol Cell. 2014;56(3):414–424. doi: 10.1016/j.molcel.2014.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schodel J, Oikonomopoulos S, Ragoussis J, Pugh CW, Ratcliffe PJ, Mole DR. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood. 2011;117(23):e207–217. doi: 10.1182/blood-2010-10-314427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boado RJ, Li JY, Nagaya M, Zhang C, Pardridge WM. Selective expression of the large neutral amino acid transporter at the blood–brain barrier. Proc Natl Acad Sci USA. 1999;96(21):12079–12084. doi: 10.1073/pnas.96.21.12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yun DW, Lee SA, Park MG, Kim JS, Yu SK, Park MR, Kim SG, Oh JS, Kim CS, Kim HJ, Kim JS, Chun HS, Kanai Y, Endou H, Wempe MF, Kim DK. JPH203, an L-type amino acid transporter 1-selective compound, induces apoptosis of YD-38 human oral cancer cells. J Pharmacol Sci. 2014;124(2):208–217. doi: 10.1254/jphs.13154FP. [DOI] [PubMed] [Google Scholar]
- 25.Bodoy S, Fotiadis D, Stoeger C, Kanai Y, Palacin M. The small SLC43 family: facilitator system l amino acid transporters and the orphan EEG1. Mol Aspects Med. 2013;34(2–3):638–645. doi: 10.1016/j.mam.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 26.Luo W, Wang Y. Epigenetic regulators: multifunctional proteins modulating hypoxia-inducible factor-alpha protein stability and activity. Cell Mol Life Sci. 2018;75(6):1043–1056. doi: 10.1007/s00018-017-2684-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ananieva EA, Patel CH, Drake CH, Powell JD, Hutson SM. Cytosolic branched chain aminotransferase (BCATc) regulates mTORC1 signaling and glycolytic metabolism in CD4+ T cells. J Biol Chem. 2014;289(27):18793–18804. doi: 10.1074/jbc.M114.554113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conway ME, Yennawar N, Wallin R, Poole LB, Hutson SM. Identification of a peroxide-sensitive redox switch at the CXXC motif in the human mitochondrial branched chain aminotransferase. Biochemistry. 2002;41(29):9070–9078. doi: 10.1021/bi020200i. [DOI] [PubMed] [Google Scholar]
- 29.Coles SJ, Hancock JT, Conway ME. Differential redox potential between the human cytosolic and mitochondrial branched-chain aminotransferase. Acta Biochim Biophys Sin (Shanghai) 2012;44(2):172–176. doi: 10.1093/abbs/gmr103. [DOI] [PubMed] [Google Scholar]
- 30.Conway ME, Coles SJ, Islam MM, Hutson SM. Regulatory control of human cytosolic branched-chain aminotransferase by oxidation and S-glutathionylation and its interactions with redox sensitive neuronal proteins. Biochemistry. 2008;47(19):5465–5479. doi: 10.1021/bi800303h. [DOI] [PubMed] [Google Scholar]
- 31.Conway ME, Poole LB, Hutson SM. Roles for cysteine residues in the regulatory CXXC motif of human mitochondrial branched chain aminotransferase enzyme. Biochemistry. 2004;43(23):7356–7364. doi: 10.1021/bi0498050. [DOI] [PubMed] [Google Scholar]
- 32.Yennawar NH, Islam MM, Conway M, Wallin R, Hutson SM. Human mitochondrial branched chain aminotransferase isozyme: structural role of the CXXC center in catalysis. J Biol Chem. 2006;281(51):39660–39671. doi: 10.1074/jbc.M607552200. [DOI] [PubMed] [Google Scholar]
- 33.El Hindy M, Hezwani M, Corry D, Hull J, El Amraoui F, Harris M, Lee C, Forshaw T, Wilson A, Mansbridge A, Hassler M, Patel VB, Kehoe PG, Love S, Conway ME. The branched-chain aminotransferase proteins: novel redox chaperones for protein disulfide isomerase-implications in Alzheimer's disease. Antioxid Redox Signal. 2014;20(16):2497–2513. doi: 10.1089/ars.2012.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coles SJ, Easton P, Sharrod H, Hutson SM, Hancock J, Patel VB, Conway ME. S-Nitrosoglutathione inactivation of the mitochondrial and cytosolic BCAT proteins: S-nitrosation and S-thiolation. Biochemistry. 2009;48(3):645–656. doi: 10.1021/bi801805h. [DOI] [PubMed] [Google Scholar]
- 35.Luo W, Wang Y. Hypoxia mediates tumor malignancy and therapy resistance. Adv Exp Med Biol. 2019;1136:1–18. doi: 10.1007/978-3-030-12734-3_1. [DOI] [PubMed] [Google Scholar]
- 36.Pan M, Reid MA, Lowman XH, Kulkarni RP, Tran TQ, Liu X, Yang Y, Hernandez-Davies JE, Rosales KK, Li H, Hugo W, Song C, Xu X, Schones DE, Ann DK, Gradinaru V, Lo RS, Locasale JW, Kong M. Regional glutamine deficiency in tumours promotes dedifferentiation through inhibition of histone demethylation. Nat Cell Biol. 2016;18(10):1090–1101. doi: 10.1038/ncb3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang ZQ, Faddaoui A, Bachvarova M, Plante M, Gregoire J, Renaud MC, Sebastianelli A, Guillemette C, Gobeil S, Macdonald E, Vanderhyden B, Bachvarov D. BCAT1 expression associates with ovarian cancer progression: possible implications in altered disease metabolism. Oncotarget. 2015;6(31):31522–31543. doi: 10.18632/oncotarget.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thewes V, Simon R, Hlevnjak M, Schlotter M, Schroeter P, Schmidt K, Wu Y, Anzeneder T, Wang W, Windisch P, Kirchgassner M, Melling N, Kneisel N, Buttner R, Deuschle U, Sinn HP, Schneeweiss A, Heck S, Kaulfuss S, Hess-Stumpp H, Okun JG, Sauter G, Lykkesfeldt AE, Zapatka M, Radlwimmer B, Lichter P, Tonjes M. The branched-chain amino acid transaminase 1 sustains growth of antiestrogen-resistant and ERalpha-negative breast cancer. Oncogene. 2017;36(29):4124–4134. doi: 10.1038/onc.2017.32. [DOI] [PubMed] [Google Scholar]
- 39.Mayers JR, Torrence ME, Danai LV, Papagiannakopoulos T, Davidson SM, Bauer MR, Lau AN, Ji BW, Dixit PD, Hosios AM, Muir A, Chin CR, Freinkman E, Jacks T, Wolpin BM, Vitkup D, Vander Heiden MG. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science. 2016;353(6304):1161–1165. doi: 10.1126/science.aaf5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hattori A, Tsunoda M, Konuma T, Kobayashi M, Nagy T, Glushka J, Tayyari F, McSkimming D, Kannan N, Tojo A, Edison AS, Ito T. Cancer progression by reprogrammed BCAA metabolism in myeloid leukaemia. Nature. 2017;545(7655):500–504. doi: 10.1038/nature22314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raffel S, Falcone M, Kneisel N, Hansson J, Wang W, Lutz C, Bullinger L, Poschet G, Nonnenmacher Y, Barnert A, Bahr C, Zeisberger P, Przybylla A, Sohn M, Tonjes M, Erez A, Adler L, Jensen P, Scholl C, Frohling S, Cocciardi S, Wuchter P, Thiede C, Florcken A, Westermann J, Ehninger G, Lichter P, Hiller K, Hell R, Herrmann C, Ho AD, Krijgsveld J, Radlwimmer B, Trumpp A. BCAT1 restricts alphaKG levels in AML stem cells leading to IDHmut-like DNA hypermethylation. Nature. 2017;551(7680):384–388. doi: 10.1038/nature24294. [DOI] [PubMed] [Google Scholar]
- 42.Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL, Dang CV. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci USA. 2010;107(5):2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolf A, Agnihotri S, Micallef J, Mukherjee J, Sabha N, Cairns R, Hawkins C, Guha A. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J Exp Med. 2011;208(2):313–326. doi: 10.1084/jem.20101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.