Abstract
We report 3 cases of severe COVID-19 due to the SARS-CoV-2 P.1 lineage in a familial cluster detected in Salvador, Bahia–Brazil. All cases were linked to travel by family members from the state of Amazonas to Bahia in late December 2020. This report indicates the cryptic transmission of the SARS-CoV-2 P.1 lineage across Brazil and highlights the importance of genomic surveillance to track the emergence of new SARS-CoV-2 variants of concern.
Keywords: SARS-CoV-2 P.1 lineage, COVID-19, SARS-CoV-2 variants of concern
Introduction
The SARS-CoV-2 virus, which causes COVID-19, reportedly emerged in December 2019 in Wuhan, China (Zhu et al., 2020). SARS-CoV-2 rapidly spread around the world, and on March 11, 2020, the World Health Organization (WHO) declared the outbreak as a pandemic (WHO/Europe, 2020). More than one year since its emergence, with more than 153 954 491 reported cases and 3 221 052 deaths (WHO, 2021), the COVID-19 pandemic continues to impact countries worldwide. In addition, in late 2020, new SARS-CoV-2 Variants of Concern (VOC) were identified, including the 501Y.V1 (B.1.1.7 lineage) reported in the United Kingdom (Public Health England, 2020) and 501Y.V2 (B.1.351 lineage) first detected in South Africa (Tegally et al., 2021).
In early January 2021, another SARS-CoV-2 VOC, termed P.1 (or 20J/501Y.V3), was first identified (Fujino et al., 2021). This variant is believed to have emerged in early December in Manaus in northern Brazil and has been associated with increased transmissibility and high viral load (Faria et al., 2021, Naveca et al., 2021a).
Herein, we report and characterize 3 cases of severe COVID-19 linked to the SARS-CoV-2 P.1 lineage detected in Salvador, Bahia, northeast Brazil, part of a familial cluster of infections resulting from travel between the states of Amazonas and Bahia in late December 2020.
Case series
Three members of the same family, residents of Manaus (Amazonas, Brazil), traveled to Salvador (Bahia, Brazil) on December 19, 2020, to visit 2 relatives for the holidays. Table 1 lists the clinical characteristics and laboratory results from all 5 cases of infection.
Table 1.
Demographic and clinical characteristics of 5 cases of COVID-19 linked to the P.1 variant of SARS-CoV-2.
| Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | |
|---|---|---|---|---|---|
| Age (years) | 40 | 19 | 69 | 41 | 71 |
| Sex | Female | Male | Male | Male | Female |
| Place of residence | Manaus | Manaus | Salvador | Manaus | Salvador |
| RT-PCR for SARS-CoV-2 | Detectable | Detectable | Detectable | Detectable | Detectable |
| SARS-CoV-2 sequencing | ND | ND | P.1 lineage | P.1 lineage | P.1 lineage |
| Onset of symptoms | 23/12/2020 | 27/12/2020 | 25/12/2020 | 27/12/2020 | 25/12/2020 |
| Comorbidities | None | None | SAH | None | COPD |
| Hospital admission | No | No | Yes | Yes | Yes |
| SpO2 at admission | ND | ND | 92% | 90% | 80% |
| % lung involvement on Thorax HRCT | ND | ND | 50%–75% | 50% | 50%–75% |
| d-dimer at admission (ng/mL) | ND | ND | 1054 | 1038 | 755 |
| C reactive protein at admission (mg/L) | ND | ND | 17.8 | 5.3 | 14.7 |
| ICU admission | No | No | Yes | Yes | Yes |
| Respiratory support | No | No | HFNC | HFNC | Mechanical ventilation |
| Days in ICU | 0 | 0 | 12 | 11 | 65 |
| Days in hospital | 0 | 0 | 19 | 21 | 65 |
| Outcome | Recovery | Recovery | Recovery | Recovery | Death |
ND = not done; SpO2 = oxygen saturation (breathing room air); HRCT = high resolution computed tomography; SAH = systemic arterial hypertension; COPD = chronic obstructive pulmonary disease; HFNC = high-flow nasal cannula; RT-PCR = real-time polymerase chain reaction; ICU = intensive care unit.
Case 1
On December 23, 2020, a 40-year-old female with no comorbidities began to experience myalgia and nasal obstruction lasting for 3 days and then promptly recovered.
Case 2
On December 27, 2020, a previously healthy 19-year-old male reported fever and myalgia and recovered after 3 days of mild symptoms.
Case 3
On December 25, 2020, a 69-year-old male with systemic arterial hypertension and dyslipidemia began to experience fever, chills, headache, and myalgia. After 10 days of symptoms and the onset of dyspnea, he was admitted to the emergency service of a local hospital. His pulse oxygen saturation (SpO2) level was 92% when breathing room air, and high-resolution computed tomography (HRCT) of the thorax revealed 50%–75% of bilateral ground-glass pulmonary opacities. The day after admission, the patient was transferred to the hospital’s intensive care unit (ICU) for oxygen support by high-flow nasal cannula (HFNC). He remained in the ICU for 12 days and was discharged after 19 days. Methylprednisolone was administered during his hospital stay.
Case 4
On December 27, 2020, a 41-year-old male reported myalgia and fever. After 9 days of persistent fever and the onset of dyspnea, he was admitted to the same hospital for oxygen support. His SpO2 was 90% when breathing room air, with an arterial oxygen pressure (pO2) of 60.5 mmHg. Thorax HRCT revealed 50% of bilateral ground-glass pulmonary opacities. After 3 days, he was transferred to the ICU due to worsening respiratory symptoms. During 11 days in the ICU, the patient required oxygen delivery by HFNC. Methylprednisolone was administered, and the patient was discharged after 21 days.
Case 5
On December 25, 2020, a 71-year-old female with chronic obstructive pulmonary disease and dyslipidemia experienced fever, odynophagia and diarrhea. After 7 days, worsening dyspnea prompted her to seek medical attention. Upon arrival at the same hospital, she was admitted to the ICU due to severe respiratory distress. Her SpO2 was 80% when breathing room air, and thorax HRCT revealed 50%–75% of bilateral ground-glass pulmonary opacities. The patient initially received non-invasive oxygen support with HFNC, but due to worsening respiratory distress, endotracheal intubation and invasive mechanical ventilation support for moderate acute respiratory distress syndrome were required. Methylprednisolone was administered upon admission. Her course became complicated by nosocomial infection, persistent respiratory failure, and multiple organ failure, culminating in death after 65 days in the ICU.
SARS-CoV-2 diagnosis, sequencing, and lineage designation
All 5 cases reported herein tested positive for SARS-CoV-2 by real-time polymerase chain reaction (RT-PCR). Nasopharyngeal swab samples were tested by multiplex RT-PCR using the Allplex SARS-CoV-2 assay (Seegene Inc., Seoul, Korea) on an ABI7500 Fast real-time PCR instrument (Thermo Fisher Scientific). SARS-CoV-2 viral genome sequencing was run on a PGM Ion System; reads were analyzed using Torrent Suite Software v.5.12.1 (Thermo Fisher Scientific).
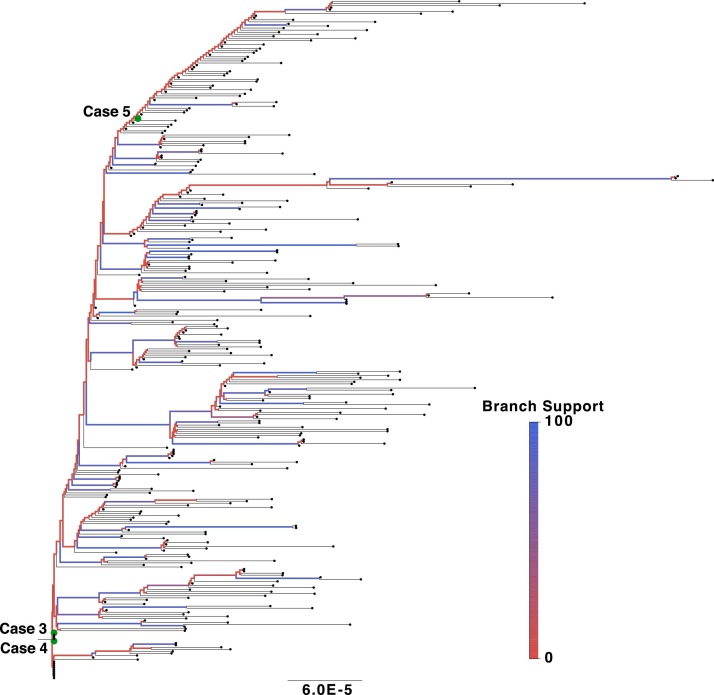
High-quality SARS-CoV-2 genome sequences were generated for Cases 3–5 and submitted to the online pangolin tool for lineage classification (Rambaut et al., 2020). All 3 sequences were classified as the P.1 lineage and are available at the EpiCov database, maintained by the GISAID initiative, with accession codes EPI_ISL_1443197, EPI_ISL_1443196 and EPI_ISL_1443198. A phylogenetic tree was constructed considering the available P.1 diversity. All high-quality Brazilian P.1 sequences (>29,000 bp and <1% Ns) were retrieved from GISAID on March 25, 2021. Alignment was performed using the MAFFT sequence alignment program, and the phylogenetic tree was inferred using the IQ-TREE software package with Maximum Likelihood (ML) analysis, as previously described (Naveca et al., 2021a). This approach resulted in a tree trunk with very low branch support, reflecting the rapid spread of P.1 and low genetic genome diversity in this early phase of dissemination (Figure 1 ). The majority of the obtained sequences branching out from the tree trunk had been isolated in the Amazonas region or were cases of individuals with a history of travel to the region. Cases 3–5 clustered within the obtained sequences, indicating that these originated from the early diversity of P.1 prior to the further diversification that can be observed in more recently sampled genomes.
Figure 1.
Maximum likelihood phylogenetic tree detailing the P.1 lineage in Brazil.
Discussion
The emergence of SARS-CoV-2 VOCs in different parts of the world presents new challenges to COVID-19 pandemic control. Increased transmissibility (Tegally et al., 2021) and mortality (Davies et al., 2021) have been associated with VOCs, and cases of reinfection have also been reported (Naveca et al., 2021b). The capacity to decrease the efficacy of currently available vaccines (Madhi et al., 2021) represents an additional threat.
With COVID-19 incidence of 7104 cases/100 000 individuals, corresponding to a mortality rate of 197 cases/100 000 individuals, Brazil is currently facing a critical phase of the pandemic, with alarming increases in cases and deaths notified across all regions of the country (https://covid.saude.gov.br/, accessed on May 5, 2021). One underlying cause behind the surge in case numbers is the emergence of SARS-CoV-2 lineage P.1, which has already been detected in all regions of Brazil (http://www.genomahcov.fiocruz.br, accessed on April 21, 2021).
The 3 cases of P.1 lineage identified herein exemplify the cryptic transmission of this VOC throughout Brazil before it had been identified. P.1 could have been introduced into other Brazilian states due to holiday-associated travel at the end of 2020 and early 2021. The cases reported here predate 11 other cases identified in the state of Bahia in mid-January, all of which were associated with travel from/to Manaus (Tosta et al., 2021). For instance, in the Southern Brazilian region, community transmission of P.1 was detected in January 2021 (Martins et al., 2021), and the first local P.1 case was identified in a popular Brazilian tourist destination (Salvato et al., 2021).
In conclusion, the present report details a familial cluster of COVID-19 linked to the SARS-CoV-2 P.1 lineage. Notably, 3/5 reported cases evolved as severe COVID-19, entailing long ICU stays with 1 associated death. One patient, a 41-year-old male, had no prior risk factors that would predispose him to severe disease. On the basis of the alarming surge in COVID-19 deaths recently reported in Brazil, it has been assumed that the P.1 lineage could imply an increased risk of severe infection or higher mortality. This association is speculative; further studies are urgently needed to assess this assumption more comprehensively.
The present report further supports the cryptic transmission of the SARS-CoV-2 P.1 lineage across Brazil, highlighting the importance of genomic surveillance to track the emergence of new viral variants. Coordinated implementation of non-pharmaceutical interventions, including social distance, airport screening and quarantine for travelers, are pivotal tools to slow down viral transmission and diminish the burden on the national public health system.
Three newly obtained genomic sequences were analyzed together with 327 P.1 background genomes retrieved from GISAID. Internal branches are colored according to support (SH-like aLRT), and sequences from the reported cases are highlighted with green circles. The tree was rooted in the branch connecting to the earliest sampled genome.
Ethical approval
This study was approved by National Commission for Ethics and Research (3.980.128/2020). All participants provided written informed consent.
Funding source
B.S.F.S. was supported by the Conselho Nacional de Desenvolvimento Cientifico e Tecnologico (CNPq-310874/2019-0), Serrapilheira Institute (IDOR/Covid-19) and the Inova Fiocruz grant (48112178997324). A.A.C. and V.C.A. were supported by the Fundação Maria Emília Freire de Carvalho.
Conflict of interest
A.A.C. works as a part-time Internal Expert Consultant at GSK-Brazil, and has no conflicts of interest with regard to the present publication.
Acknowledgments
The authors are grateful to the physicians and nurses involved in the patients’ clinical care, the GISAID team, and all researchers who submitted sequences to the EpiCoV database. The GISAID table containing detailed data on the sequences used in this study and the associated researchers is included as Appendix Table 1. The authors also thank Andris K. Walter for English language revision and manuscript copy editing assistance.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ijid.2021.05.010.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Davies N.G., Jarvis C.I., CMMID COVID-19 Working Group, Edmunds W.J., Jewell N.P., Diaz-Ordaz K., et al. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021;593(May (7858)):270–274. doi: 10.1038/s41586-021-03426-1. Epub 2021 March 15. PMID: 33723411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.D.S., Mishra S., et al. Genomics and epidemiology of a novel SARS-CoV-2 lineage in Manaus, Brazil. medRxiv. 2021 doi: 10.1101/2021.02.26.21252554. 2021.02.26.21252554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino T., Nomoto H., Kutsuna S., Ujiie M., Suzuki T., Sato R., et al. Novel SARS-CoV-2 variant identified in travelers from Brazil to Japan. Emerg Infect Dis. 2021;27(April (4)):1243–1245. doi: 10.3201/eid2704.210138. Epub 2021 February 10. PMID: 33567247; PMCID: PMC8007308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhi S.A., Baillie V., Cutland C.L., Voysey M., Koen A.L., Fairlie L., et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B.1.351 variant. N Engl J Med. 2021;384(May (20)):1885–1898. doi: 10.1056/NEJMoa2102214. Epub 2021 March 16. PMID: 33725432; PMCID: PMC7993410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins A.F., Zavascki A.P., Wink P.L., Volpato F.C.Z., Monteiro F.L., Rosset C., et al. Detection of SARS-CoV-2 lineage P.1 in patients from a region with exponentially increasing hospitalisation rate, February 2021, Rio Grande do Sul, Southern Brazil. Euro Surveill. 2021;26(March (12)) doi: 10.2807/1560-7917.ES.2021.26.12.2100276. PMID: 33769251; PMCID: PMC7995561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveca F.G., Nascimento V., de Souza V.C., Corado A.L., Nascimento F., Silva G., et al. COVID-19 in Amazonas, Brazil, was driven by the persistence of endemic lineages and P .1 emergence. Nat Med. 2021;(May (25)) doi: 10.1038/s41591-021-01378-7. Epub ahead of print. PMID: 34035535. [DOI] [PubMed] [Google Scholar]
- Naveca F., da Costa C., Nascimento V., Souza V., Corado A., Nascimento F., et al. 2021. Three SARS-CoV-2 reinfection cases by the new Variant of Concern (VOC) P.1/501Y.V3. [DOI] [Google Scholar]
- Public Health England . 2020. Investigation of novel SARS-COV-2 variant: variant of concern 202012/01.www.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variant-variant-of-concern-20201201 [Google Scholar]
- Rambaut A., Holmes E.C., O’Toole Á., Hill V., McCrone J.T., Ruis C., et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5(November (11)):1403–1407. doi: 10.1038/s41564-020-0770-5. Epub 2020 July 15. PMID: 32669681; PMCID: PMC7610519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvato R.S., Gregianini T.S., Campos A.A.S., Crescente L.V., Vallandro M.J., Ranieri T.M.S., et al. 2021. Epidemiological investigation reveals local transmission of SARS-CoV-2 lineage P.1 in Southern Brazil. March 2 [cited 2021 Apr 21]; Available from: [DOI] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592(April (7854)):438–443. doi: 10.1038/s41586-021-03402-9. Epub 2021 March 9. PMID: 33690265. [DOI] [PubMed] [Google Scholar]
- Tosta S., Giovanetti M., Nardy V.B., da Silva L.R.O., Gómez M.K.A., Lima J.G., et al. Early genomic detection of SARS-CoV-2 P.1 variant in Northeast Brazil. medRxiv. 2021 doi: 10.1101/2021.02.25.21252490. 2021.02.25.21252490. [DOI] [Google Scholar]
- WHO/Europe . 2020. Coronavirus disease (COVID-19) outbreak — WHO announces COVID-19 outbreak a pandemic.https://www.euro.who.int/en/health-topics/health-emergencies/coronavirus-covid-19/news/news/2020/3/who-announces-covid-19-outbreak-a-pandemic [13 March 2021] [Google Scholar]
- World Health Organization COVID-19 dashboard. https://covid19.who.int [Accessed 5 May 2021]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(February (8)):727–733. doi: 10.1056/NEJMoa2001017. Epub 2020 January 24. PMID: 31978945; PMCID: PMC7092803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.