Abstract
In eukaryotic cells, pre-mRNA splicing is catalyzed by the spliceosome, a highly dynamic molecular machinery that undergoes dramatic conformational and compositional rearrangements throughout the splicing cycle. These crucial rearrangements are largely driven by eight DExD/H-box RNA helicases. Interestingly, the four helicases participating in the late stages of splicing are all DEAH-box helicases that share structural similarities. This review aims to provide an overview of the structure and function of these DEAH-box helicases, including new information provided by recent cryo-electron microscopy structures of the spliceosomal complexes.
RNA Helicases in Splicing
The multiple conformational and compositional changes of the spliceosome are largely driven by eight superfamily 2 (SF2) helicases, the largest and most diverse helicase superfamily (for reviews, see [1,2]). SF2 helicases are generally made up of a helicase core consisting of two RecA domains. The core is flanked by more variable N- and C-terminal extensions. The two RecA domains are characterized by 13 conserved motifs (Figure 1A). Four motifs (Q, I, II, and VI) are responsible for nucleoside triphosphate (NTP) binding and hydrolysis, seven motifs (Ia, Ib, Ic, IV, IVa, V, and Vb) bind the nucleic acid, and two motifs (III and Va) coordinate the nucleic acid-binding site and the NTP-binding site [3]. SF2 can be further divided into subfamilies including the DEAD-box, DEAH-box, and ski2-like subfamilies based on differences in their sequence motifs as well as structural and mechanistic features [3]. Of the four spliceosomal helicases important for early steps of spliceosome assembly and activation, Prp5, Sub2 (UAP56 in human), and Prp28 are members of the DEAD-box subfamily and Brr2 belongs to the Ski2-like subfamily. Four additional helicases (Prp2, Prp16, Prp22, and Prp43) act during the catalysis and disassembly stages of the splicing cycle and belong to the DEAH-box subfamily. In metazoans, five additional helicases, Aquarius, SF3b125, elFAIII, DDX35, and Abstrakt, are involved in splicing [4-7].
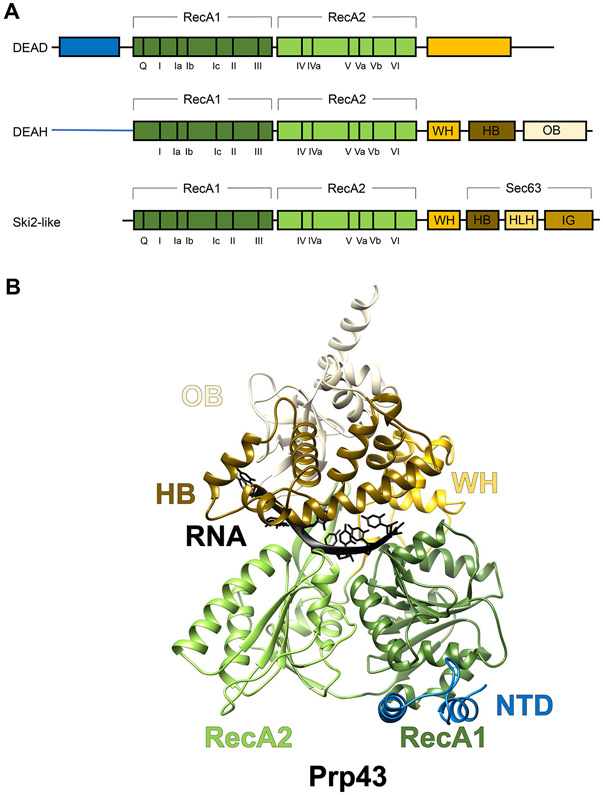
Figure 1.
Overall Domain Organization and Structure of the Splicing Helicases.
(A) Domain organizations and conserved motifs of the three subfamilies of superfamily 2 (SF2) helicases that the splicing helicases belong to. (B) The structure of Chaetomium thermophilum DEAH-box helicase Prp43 (PDB ID 5LTA) illustrating the domain architecture of DEAH-box helicases using the same color scheme as in A. Abbreviations: HB, helix bundle; HLH, helix–loop–helix; IG, immunoglobulin-like; NTD, N-terminal domain; OB, oligosaccharide-binding fold; WH, winged helix.
In this review, we will provide a summary of what is known about the structure, function, mechanism, and regulation of the four DEAH-box helicases in splicing as well as new insights provided by recent cryo-electron microscopy (cryo-EM) structures of the spliceosome
Overall Structure of DEAH-Box Spliceosomal Helicases
The X-ray crystallographic structures (FL or domains) of spliceosomal DEAH-box helicases of various species, other than Prp16, have been determined. Their N-terminal domain (NTD) varies in length (84–475 residues) and is the least conserved region in DEAH-box helicases [2]. The NTDs are predicted to be disordered and are either truncated or not visible in all DEAH-box crystal structures except for yeast Prp43 which has the shortest NTD (84 residues). In Prp43, residues 10–36 of NTD form a helix that interacts with the winged helix (WH) and helix bundle (HB) subdomains and the rest of the NTD extends across the RecA1 domain [8].
All DEAH-box helicase structures contain the helicase core made of two RecA domains packed against each other (Figure 1B). The tandem RecA domains bind RNA with the conserved helicase motifs. The relative movement of the two RecA domains to each other driven by ATP hydrolysis is crucial for RNA translocation.
Unlike the DEAD-box helicases which have more variable C-terminal extensions, the C-terminal domain (CTD) of DEAH-box helicases shares a common organization including a WH, an HB, and an oligosaccharide-binding fold (OB) subdomain (Figure 1A) [9]. The CTD and the helicase core together create an RNA-binding tunnel that involves residues from the RecA domains to the OB subdomain (Figure 1B). This RNA-binding tunnel provides the helicase a stronger ‘grip’ on the RNA, consistent with the role of DEAH-box helicases in translocating on long RNA segments. By contrast, DEAD-box helicases, lacking the conserved domain organization in the CTD as seen in DEAH-box helicases, bind and unwind short duplexes through local destabilization of the RNA [10]. The CTD of DEAH-box helicases interacts strongly with the RecA1 domain through its WH subdomain and has a weaker association with the RecA2 domain. The close interactions between the CTD and the RecA domains provide an opportunity for additional proteins to regulate the helicase activity through their binding to the CTD (such as G-patch proteins that bind to the DEAH-specific OB domain).
Functions of DEAH-Box Spliceosomal Helicases
An obvious function of helicases in splicing is to drive the conformational rearrangements of the spliceosome as it goes from one stage to the next in the splicing cycle. For example, Prp2 (together with G-patch protein Spp2) mainly promotes the transition from the Bact complex to the catalytically active B* complex [11-14], presumably by destabilizing the SF3a and SF3b subcomplexes of U2 small nuclear ribonucleoprotein (snRNP) bound near the branch site, thus freeing the branch point (BP) for nucleophilic attack of the 5′ splice site (ss) [15,16,17] (Box 1). Of the other three DEAH-box spliceosomal helicases, Prp16 is necessary for the transition between the first and second step of splicing, likely by facilitating the release of first-step splicing factors Yju2 and Cwc25 [18]. Subsequently, Prp22 acts to release the ligated exons and convert the P to intron lariat spliceosome (ILS) complex [19]. Finally, Prp43 together with the Ntr1–Ntr2 dimer disassembles the ILS complex [20-22].
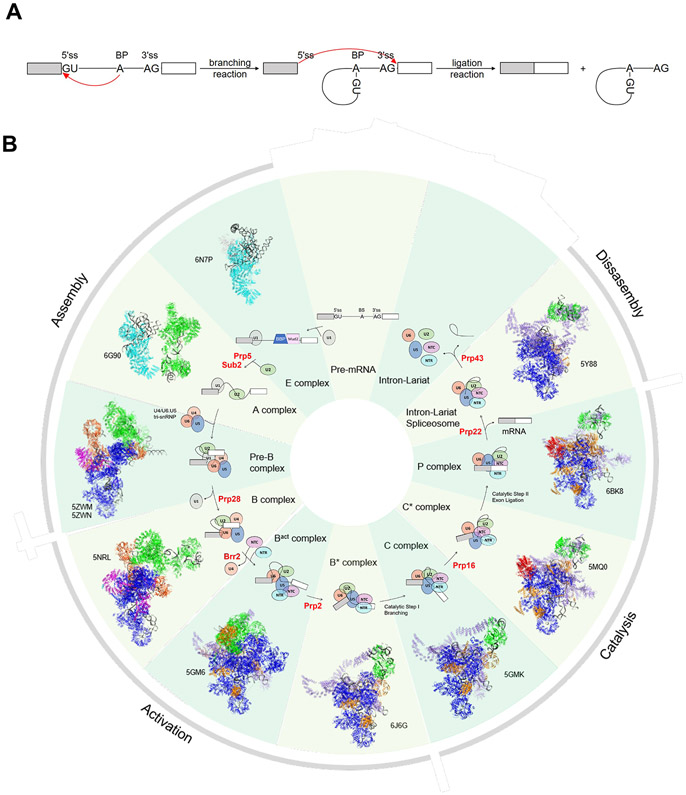
Text box 1: Pre-mRNA Splicing.
In eukaryotes, introns are removed through two transesterification reactions. In the first (branching) reaction, the 2′ OH group of the BP adenosine attacks the 5′ ss to form a lariat intermediate. In the second (ligation) reaction, the newly freed 3′ OH group of the 5′ exon attacks the 3′ ss to ligate the two exons and release an intron lariat (Figure IA) [80-82]. The splicing reaction is catalyzed by the spliceosome, a multi-megadalton protein–RNA complex that is assembled on each pre-mRNA substrate anew in a stepwise manner (Figure IB) [83]. Initial ss recognition is carried out by the U1 snRNP through base pairing of the U1 snRNA with the 5′ ss in both yeast and human. The BP is recognized by the BBP–Mud2 heterodimer in yeast and by SF1 in human. The polypyrimidine tract and 3′ ss are recognized by the U2AF65–U2AF35 heterodimer in human. These initial components form the spliceosomal E complex. In the second step, BBP/SF1 and Mud2/U2AF are displaced, allowing the U2 snRNP to bind the BPS through the U2 snRNA, leading to formation of the A complex. The U4/U6.U5 tri-snRNP is recruited to the A complex to form the pre-B complex. At this point, all five snRNPs necessary for splicing are present but must undergo major compositional and conformational rearrangements before the spliceosome becomes catalytically active. The U1 and U4 snRNPs are dissociated from the spliceosome during this activation process to form the Bact complex, and the spliceosome is further rearranged to generate the activated spliceosome B* complex. In the B* complex, the U6 snRNA substitutes the U1 snRNA to base pair with the 5′ ss and the U2 and U6 snRNAs are extensively base paired to bring the 5′ ss and BP close together for the first transesterification reaction. The resulting C complex, after the branching reaction, is remodeled to form the C* complex that is ready for the second catalytic step. After this second step, the postcatalytic P complex is formed containing the ligated exons and the lariat intermediate. Finally, the ligated exons are released, the resulting intron lariat spliceosome (ILS) complex is disassembled, and the ILS components are recycled to take part in a new splicing cycle.
Pre-mRNA Splicing. (A) Schematic representation of the two transesterification reactions in pre-mRNA splicing. Boxes and solid lines represent the exons and introns, respectively. The red arrows show the nucleophilic attacks at the phosphodiester bond at the 5′ and 3′ ss during splicing. (B) A schematic representation of the splicing cycle in yeast is shown in the inside ring. Only the snRNPs (ovals) but not non-snRNP proteins are shown for simplicity. The spliceosomal helicases are indicated in red. The outside ring shows the cryo-EM structure of each corresponding spliceosomal complex and its PDB ID.
As a consequence of their role in driving these essential rearrangements, many of spliceosomal helicases become gatekeepers guarding against the use of suboptimal substrates. This is accomplished through the kinetic proofreading mechanism first introduced by Hopfield and Ninio [23,24]. In this mechanism, a catalytic reaction branches into a second pathway that kinetically competes with the main productive pathway and antagonizes suboptimal substrates [23-25]. At least five spliceosomal helicases, Prp5, Prp28, Prp16, Prp22, and Prp43, have been shown to reject suboptimal substrates and promote optimal substrates, thus increasing splicing fidelity through two possible models of kinetic proofreading [26]. In the timer model, the helicase allots a limited window of opportunity for a particular event in the splicing cycle. If the substrate (e.g., an optimal substrate) passes the proofreading step faster than the respective helicase can act, the substrate proceeds to the product. If the substrate (e.g. a suboptimal substrate) goes through proofreading more slowly, the helicase acts and rejects the substrate [25]. Alternatively, in the sensor model, the helicase senses and rejects a suboptimal substrate more quickly than an optimal substrate, achieved through differences in the stability of the substrate-containing spliceosome complex or by regulation of the ATPase activity of the helicase [25]. The kinetic proofreading mechanism has also been observed as a mechanism for GTPases to ensure high fidelity in protein translocation [27], suggesting a general mechanism that allows NTPases to take part in quality control in addition to performing their dedicated rearrangement function.
Mechanism of Action
DEAH-box helicases bind and translocate along a single-stranded 3′ overhang in a 3′ to 5′ direction to unwind the RNA duplex (reviewed in [28]). Comparison of several spliceosomal DEAH-box helicase structures in various nucleotide and RNA-binding states suggests a model by which these helicases translocate along the RNA [29]. When ATP is bound in this model, the two RecA domains are in a closed conformation that binds four nucleotides (Figure 2A). Upon ATP hydrolysis and the transition to an ADP-bound state, the RecA2 domain is reoriented, resulting in weakened interaction with the RNA. The release of ADP leads to the open conformation of the two RecA domains that now bind five nucleotides of the RNA (Figure 2B). Cycling through the open and closed states driven by ATP hydrolysis therefore potentially allows translocation of the helicase along the RNA with a rate of one nucleotide per hydrolyzed ATP [29].
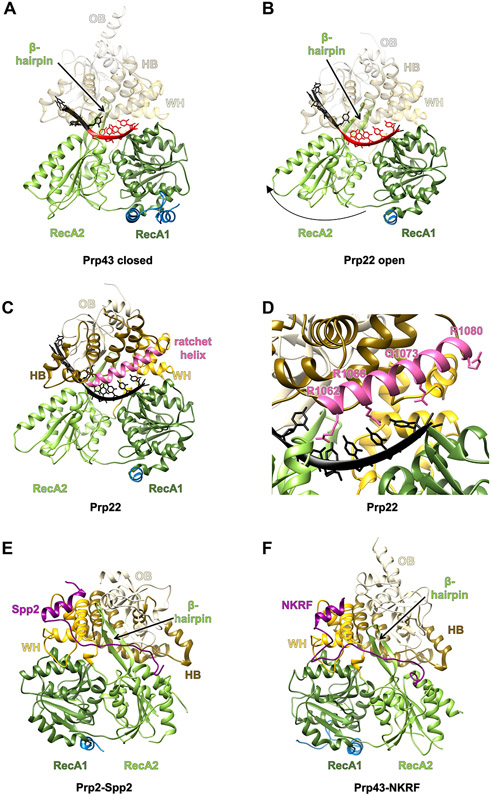
Figure 2.
Structural Basis for the DEAH-Box Mechanism of Action and Regulation by G-Patch Domains.
The domains are colored as in Figure 1A. (A) The closed form of Chaetomium thermophilum DEAH-box helicase Prp43 bound to ADP and RNA (PDB ID 5LTA). (B) The open form of C. thermophilum DEAH-box helicase Prp22 bound to RNA (PDB ID 6I3P). Both proteins are shown in the same orientation after aligning their RecA1 domains. The RNA stacked in the binding tunnel is highlighted in red. (C) Location of the ratchet helix in C. thermophilum Prp22 (PDB ID 6I3P). The ratchet helix is highlighted in pink. (D) A zoomed in view of the ratchet helix and its surroundings. Residues on the ratchet helix that interact, or potentially interact, with a longer RNA are shown as sticks. (E) The structure of C. thermophilum Prp2 in complex with Spp2 (purple) (PDB ID 6RM9). (F) The structure of Homo sapiens Prp43 with NKRF (purple) bound (PDB ID 6SH7). Abbreviations: HB, helix bundle; OB, oligosaccharide-binding fold; WH, winged helix.
An alternative mechanism of translocation involves a ratchet helix in the HB subdomain of the CTD (Figure 2C). Ratchet helices have been described for several SF2 family helicases, including the DNA helicase Hel308 [30], Ski2-like helicase Brr2 [31], and DEAH-box helicase Prp22 [32]. The ratchet helix interacts with single-stranded DNA or RNA (often through base stacking with aromatic residues as well as electrostatic or h-bond interactions with positively charged or polar residues), and the N terminus of the ratchet helix contacts the RecA2 domain (Figure 2D). Movement of RecA2, driven by ATP binding and hydrolysis, likely shifts the position of the ratchet helix toward RecA2, resulting in strand translocation of the RNA bound by the ratchet helix. It is possible that both the open-closed conformation cycling, and the ratchet helix contribute to strand translocation to a certain extent.
DEAH-box helicases can clearly unwind double-stranded RNA (dsRNA) in vitro and the unwinding mechanism may involve a β-hairpin that extends from the RecA2 domain to contact the HB subdomain of the CTD [33-37]. In the structure of DNA helicase Hel308 bound to double-stranded DNA (dsDNA), a similar β-hairpin disrupts 2 base pairs of the dsDNA [30], leading to the speculation that the β-hairpin in DEAH-box helicases also participates in strand separation [8]. However, the structure of Prp22 in complex with single-stranded RNA suggests that this β-hairpin is likely located outside of the potential RNA duplex, mainly facilitating translocation instead of strand separation [29]. Therefore, it remains to be determined whether the β-hairpin is required for unwinding in vitro or what other mechanisms may be responsible for this activity.
Intriguingly, spliceosomal DEAH-box helicases may not directly unwind their dsRNA substrate in vivo. Biochemical data, including single-molecule fluorescence resonance energy transfer and UV crosslinking on yeast Prp16 and Prp22, demonstrated that these helicases never get close to the dsRNA region they are supposed to unwind [38]. Instead, the DEAH-box helicase is thought to be loaded onto a 3′ overhang and begin translocation, resulting in pulling of the single-stranded RNA and disruption of base pairs by a winching mechanism [38].
Regulation of Substrate Specificity and Helicase Activity
An important question in the study of DExD/H-box helicases is how their specificity and activity are regulated, so that they unwind the right substrate at the right time. DEAH-box helicases typically do not show substrate specificity in vitro and are able to unwind any RNA duplexes with 3′ overhang [39,40]. One possible mechanism of achieving substrate specificity in vivo is by specifically recruiting a helicase to the right substrate, possibly mediated by protein–protein interactions. Given that the NTDs are the least conserved between the otherwise similar spliceosomal DEAH-box helicases [2], whether the NTD is responsible for recruiting them to the spliceosome will be an interesting topic of future investigation.
Another potential mechanism to convey specificity is by activating DEAH-box helicases with additional protein factors such as G-patch proteins [13,41] at the right time and place. G-patch domains are characterized by a 40–45-amino acid glycine-rich stretch [42]. In yeast, one of the five known G-patch proteins interacts with Prp2 during splicing and the other four with Prp43 in several RNA-processing pathways [43]. In humans, identification of more than 20 G-patch proteins indicates that the regulation of DEAH-box helicases may be more widespread and complex [43].
Two recent studies reveal the structural basis for G-patch-mediated activation of DEAH-box helicases. Chaetomium thermophilum Prp2 was crystallized with the G-patch domain of Spp2 in five different crystal forms [44] and human Prp43 was crystallized with the G-patch domain of ribosome biogenesis factor NKRF in its apo and ADP-bound form (Figure 2E,F) [45]. In both cases the N terminus of the G-patch motif forms an amphipathic α-helix that interacts with the WH subdomain of its helicase partner. This is followed by a linker region that runs across the β-hairpin in the RecA2 domain of its helicase partner. Downstream of the linker, the C terminus of the G-patch domain binds to the region of the helicase RecA2 domain adjacent to the C-terminal HB subdomain (Figure 2E,F). The authors suggest that binding of the G-patch motif stabilizes the helicase in an intermediate conformation. It is mostly closed with high RNA-binding affinity but can still accommodate the necessary movement induced by ATP hydrolysis [46]. The G-patch protein would thus allow the helicase core to translocate without losing its overall grip on the RNA, therefore stimulating the activity of its helicase partner.
Insights from Cryo-EM Structures
The recent cryo-EM structures of the spliceosomal complexes in the yeast and human splicing cycle [32,47-70] provided a direct visualization of the physiological context of these helicases. We can now ‘see’ the likely substrates of these helicases, their possible mechanism of action, and how they can be specifically recruited or regulated (by what factors) so they can perform their dedicated function in splicing. We will briefly summarize the information gleaned from these cryo-EM structures in the following text, with the aforementioned questions in mind.
Prp2
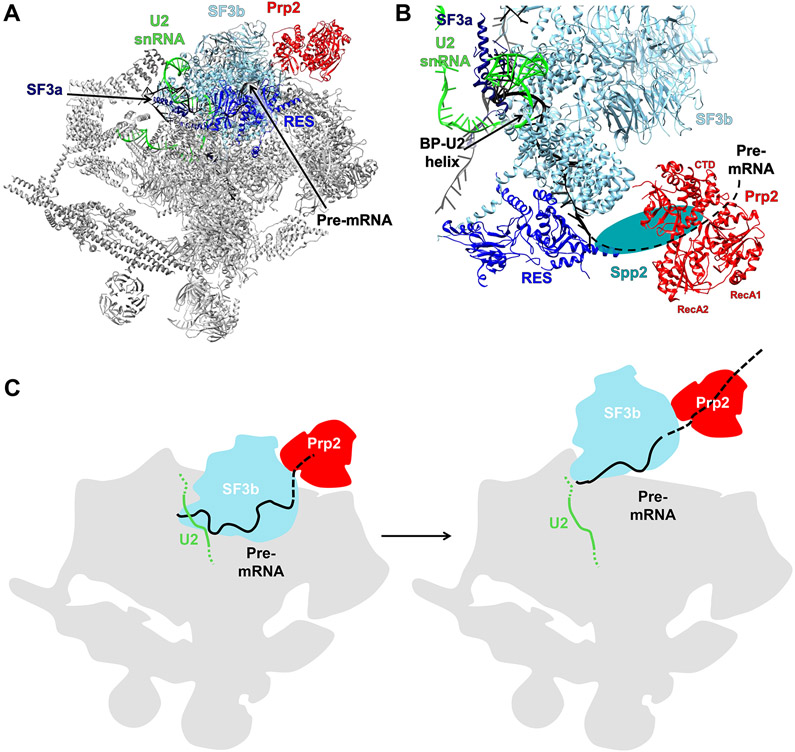
Cryo-EM structures of the yeast Bact complex determined by both the Shi and Lührmann laboratories contain Prp2 [48,50]. In these structures, the C terminus of Prp2 contacts the HEAT (Huntingtin, elongation factor 3, protein phosphatase 2A, and TOR1) repeats of Hsh155 in SF3b and is close to the RES complex important for the efficient formation of the Bact complex [48,71] (Figure 3A,B). Crosslinking analyses suggest that some additional density between Prp2 and the RES complex can be modeled as G-patch protein Spp2. The N terminus of Spp2 interacts with Prp2, Brr2, and Rse1 (a component of the SF3b complex) and its C terminus interacts with the OB and RecA2 domains of Prp2. These crosslinking data are largely consistent with the recent crystal structure of the helicase core of Prp2 in complex with the G-patch domain of Spp2 [44,48]. Thus, Spp2 may not only activate the helicases activity at the right time but also recruit Prp2 to the appropriate location through Spp2’s contacts with both the RES complex and Prp2. Through the action of Prp2, the SF3b complex is displaced from U2-branch point sequence (BPS; it remains loosely associated with the U2 snRNP until the spliceosome is disassembled [72]), which may contribute to the release of Prp2 from the spliceosome.
Figure 3.
Prp2 in the Spliceosome Bact Complex.
(A) Overall structure of the yeast Bact complex (PDB ID 5GM6). (B) A zoomed in view of the region containing Prp2. The black dotted line is the hypothetical path of the pre-mRNA and the teal oval represents the likely location of Spp2. (C) A model depicting SF3b displacement and freeing of the branch point (BP) by Prp2. Abbreviation: CTD, C-terminal domain.
In the Bact structures, no RNA binding of Prp2 is visible. Previous biochemical data [73,74] suggest that Prp2 binds the pre-mRNA 25–30 nt downstream of the branch site (approximately 60 Å away) [48,50], thus enabling movement along the pre-mRNA in a 3′ to 5′ direction and displacement of the SF3a and SF3b complexes from the branch site region [15,16,17]. The location of Prp2 in the periphery of the spliceosome (Figure 3A) suggests that instead of Prp2 translocating along the pre-mRNA, the pre-mRNA is actually pulled into Prp2 through a winching model [38]. This action destabilizes the SF3a and SF3b subcomplexes (Figure 3C). A common theme among spliceosomal structures is the localization of DEAH-box helicases in the periphery of the complex. The helices are therefore far away from the duplexes they are supposed to unwind which lie buried within the spliceosome. These observations provide a structural basis for the winching mechanism [38] discussed earlier. The peripheral location of these helicases enables them to join and leave the spliceosome easily which likely facilitates the regulation of their action. The human Bact structure shows that Prp2 likely also binds the 3′ end of pre-mRNA and contacts the RES complex and SF3 in a similar manner as the yeast structure, suggesting a similar mechanism of action [61,65].
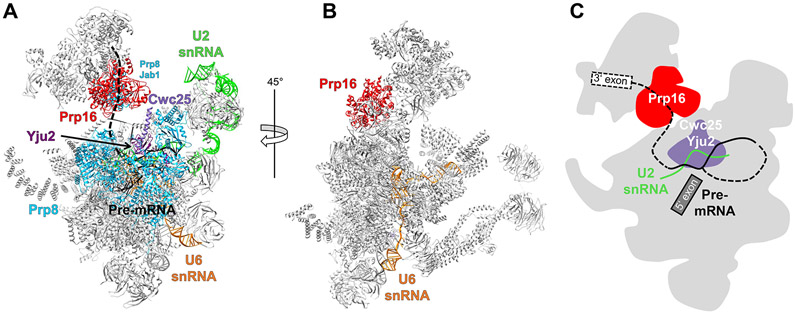
Prp16
Prp16 is present in the cryo-EM structures of the yeast and human C complexes [47,58,64]. In the yeast C complex structure, a large region of density contacting Cwc25 is located near the intron exit channel by the branch site [47]. The density allows unambiguous fitting of two RecA domains which has been interpreted as Prp16 based on its contact with Cwc25 (Figure 4A). Prp16 is located in the space formerly occupied by the SF3b factor Hsh155, suggesting that Prp16 cannot be recruited to the spliceosome until Prp2 displaces the SF3b complex from the U2–BPS. Prp16 also contacts the Prp8 Jab1 domain in the C complex (Figure 4A), which (together with the N-terminal cassette of Brr2) undergoes dramatic positional shift from the Bact to C complex [65]. This stage-specific rearrangement of the otherwise omnipresent Brr2 and Prp8 may contribute to the specific recruitment of Prp16.
Figure 4.
Prp16 in the Spliceosome C Complex.
(A) Overall structure of the yeast C complex (PDB ID 5LJ5) showing the hypothetical path of the 3′ end of the pre-mRNA that would allow for remodeling by Prp16. (B) C complex rotated by 45° with respect to panel A. The U6 small nuclear RNA (snRNA; orange) does not come close to Prp16 and thus can be ruled out as a target of Prp16. (C) A mechanistic model shows that Prp16 pulls the pre-mRNA, dislodging Cwc25 and Yju2 and leading to destabilization of the U2–BP interaction. Abbreviation: BP, branch point.
In the yeast C complex structure, the intron downstream of the BP binds to the RT and linker domains as well as the α-finger of Prp8 to project in the direction of Prp16, making it a possible target of Prp16 (Figure 4A). By contrast, essentially all of the U6 small nuclear RNAs (snRNAs; 97 out of 112 nt) are modeled which are far from Prp16 (Figure 4B), ruling out the previous hypothesis that U6 is the target of Prp16 based on genetic interactions [75]. Although the RNA-binding site of Prp16 is located approximately 45 Å away from the last ordered nucleotide of the pre-mRNA, ATP-driven translocation of the pre-mRNA in a 3′ to 5′ direction could result in pulling the branch helix out of its pocket. This pulling will likely destabilize the binding of Yju2 and Cwc25 (Figure 4C), consistent with the role of Prp16 in facilitating the release of Yju2 and Cwc25 [9]. The newly vacant space around the branch helix then allows the 3’ exon to enter the active site and bind the U5 snRNA loop 1.
Subsequently, a human C complex structure containing Prp16 was determined [64]. Overall, the core regions of the yeast and human C complexes as well as the spatial arrangement of Prp16 are similar. This suggests a common mechanism of action of Prp16 between the two organisms.
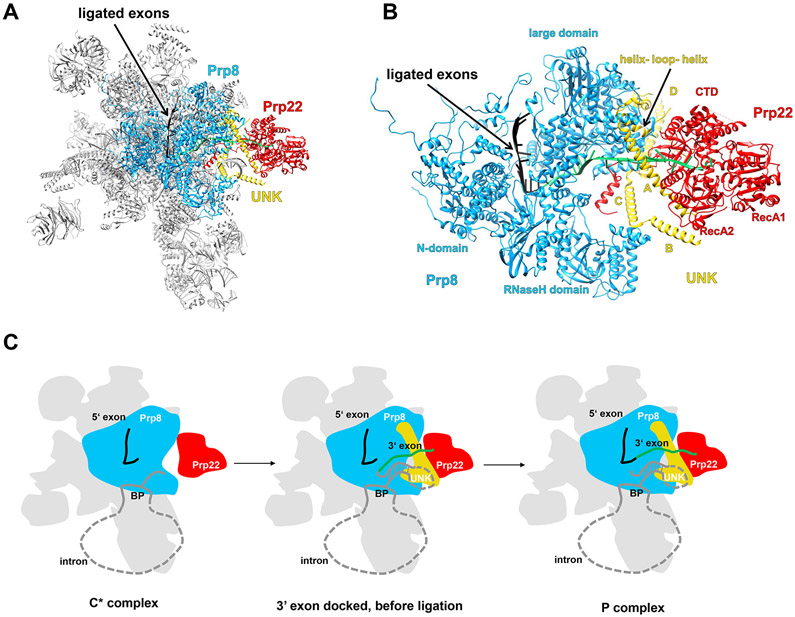
Prp22
Prp22 is present in the cryo-EM structures of the yeast C* and P as well as the human P complexes [32,51,54,57,67,70] in similar positions, suggesting a comparable mechanism of action in both species. Prp22 is loosely associated with the yeast C* complex, resulting in poor density [54]; however, it is more defined in the P-complex. In the C* and P complexes, Prp22 occupies the space formerly held by Prp16 in the C complex. This suggests that Prp22 could play a role in displacement of Prp16 from the spliceosome. Prp22 is located in the periphery of the spliceosome and mainly contacts the Prp8 RNase H and large domain (consisting of residues 885–1824; Figure 5A,B). The interaction with Prp8 alone is likely not enough to ensure specific recruitment, as Prp8 is present in most of the spliceosomal complexes. Specific recruitment of Prp22 may involve multiple protein–protein interactions (potentially mediated through the Prp22 NTD) working in concert [2].
Figure 5.
Prp22 in the Spliceosome P Complex.
(A) Overall structure of the yeast P complex (PDB ID 6BK8) with Prp22 located in the periphery. (B) A zoomed in view of the region containing Prp22. Prp22 attaches to the spliceosome through its interaction with Prp8, protein UNK, and the 3′ exon (green). (C) A schematic representation of the winching and 3′ splice site proofreading mechanisms by which Prp22 pulls on and releases the 3′ exon from the spliceosome (adapted from [32]). Abbreviations: BP, branch point; CTD, C-terminal domain.
Prp22 is the only DEAH-box helicase for which RNA binding could be observed in the cryo-EM structures, which clearly demonstrates that Prp22 binds the 3′ exon through its two RecA domains and the CTD (Figure 5B). Prp22 is positioned to pull the exon out of the spliceosome in a 3′ to 5′ direction to release the ligated exons from the spliceosome using a winching movement, as proposed by earlier biochemical studies [38] (Figure 5C). Since one of the main contacts Prp22 makes with the spliceosome is with the 3′ exon, it is plausible that Prp22 falls off the spliceosome with the ligated exons that are pulled out.
The structure of Prp22 in the context of the C* and P complex also provides a visual illustration of the kinetic proofreading model that operates in the other spliceosomal DEAH-box helicases as well. In the C* complex, Prp22 can pull on the 3′ exon and compete with the ligation reaction by weakening the interaction between the 3′ ss and the spliceosome. For a suboptimal substrate, the pulling by Prp22 wins which would remove the 3′ ss from the spliceosome, and thus prevent splicing. For an optimal substrate, the translocation by Prp22 takes place after successful exon ligation, which will promote splicing by releasing the ligated exons from the spliceosome.
In the P complex structure, density for a unidentified protein (UNK) consisting of four helices (A–D) is located near Prp22 and Prp8 (Figure 5B). In the C* complex, UNK is present but without obvious helices A and B. A helix–loop–helix region located near helices A and D of UNK contacts the Prp22 CTD (Figure 5B) and potentially acts to promote the helicase activity of Prp22. In the same manner, two G-patch proteins (Spp2 and Ntr1) stimulate the helicase activity of Prp2 and Prp43 by binding to the OB subdomains of these helicases [11,41,74,76,77]. It is conceivable that UNK may regulate the winching activity of Prp22 at different stages in the splicing cycle, which could potentially explain why Prp22 can pull off the ligated exons in the P complex but not the intron–exon in the C* complex. Additionally, density for UNK could only be observed in the C* and P complex, possibly explaining how Prp22 is specifically recruited to the spliceosome.
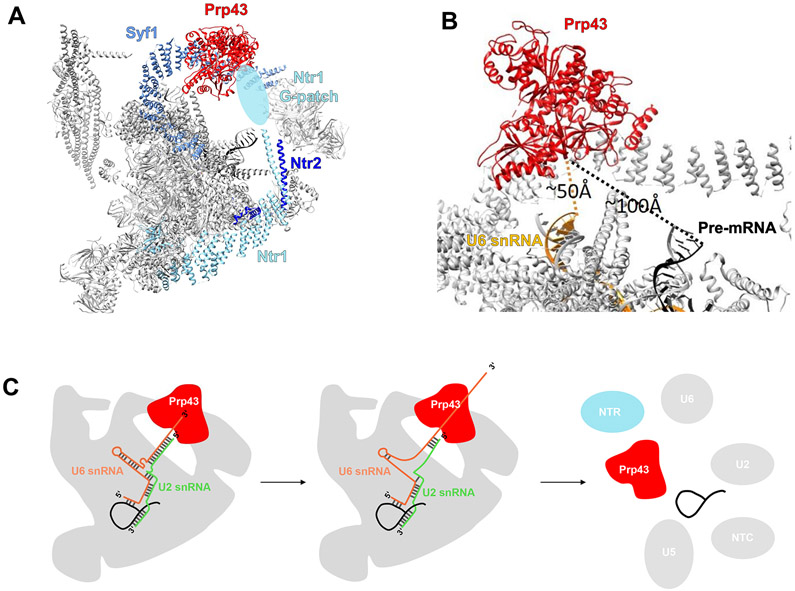
Prp43
The yeast and human ILS structures both contain Prp43 [56,70]. In the yeast ILS structure [56], Prp43 is located near the center of the superhelical protein Syf1, a protein important for Yju2 recruitment to the spliceosome [78] (Figure 6A). The G-patch portion of Ntr1 that is believed to interact with Prp43 could not be modeled due to limited local resolution. However, a recent structure of human Prp43/DHX15 in complex with the G-patch domain of the ribosome biogenesis factor NKRF suggests that the unmodeled lobe of density across the WH and RecA2 domains of Prp43 corresponds to Ntr1 [45,56]. Analogous to Spp2 and Prp2, Ntr1 may convey substrate specificity to Prp43 by recruiting it to the appropriate location in the spliceosome and activating it at the correct time.
Figure 6.
Prp43 in the Spliceosome ILS Complex.
(A) Overall structure of the yeast ILS complex (PDB ID 5Y88). The light blue oval represents the likely location of the Ntr1 G-patch with insufficient density for modeling. (B) A zoomed in view of the Prp43-containing region of the ILS complex. Dotted lines indicate the distance from the U6 small nuclear RNA (snRNA) and the pre-mRNA to the Prp43 RNA-binding groove. (C) A model of how Prp43 translocation along the U6 snRNA could lead to spliceosome disassembly (adapted from [79]). Abbreviations: ILS, intron lariat spliceosome; NTC, NineTeen complex; NTR, NTC related.
The location of Prp43 in yeast ILS suggests two possibilities for how it facilitates spliceosomal disassembly as its RNA target cannot be determined from the structures alone. The first possibility is that Prp43 pulls on the intron lariat sequence between the 5′ ss and the branch site. This pulling results in the unwinding of the intron–U2 snRNA and intron–U6 snRNA duplexes as well as the dissociation of the NineTeen complex (NTC) and NTC-related components. The second scenario relies on Prp43 pulling on the 3′ end of U6 snRNA, resulting in the unwinding of the U2–U6 and the intron–U6 duplex, dissociation of bound proteins, and spliceosome disassembly (Figure 6C). Although the visible 3′ end of the U6 snRNA and intron-lariat are located approximately 50 and 85–100 Å, respectively, away from the RNA-binding site of Prp43, (Figure 6B), both are feasible to reach Prp43 if the unmodeled regions of these RNAs are considered. Recent biochemical data confirmed the second scenario by demonstrating that the 3′ end of U6 snRNA directly crosslinks to Prp43, it is necessary for intron release and spliceosome disassembly, and plays a role in splicing fidelity by discarding suboptimal lariat intermediates [79].
Two different conformations of the human ILS complex can be observed, one without Prp43 bound (ILS1) and one with Prp43 bound (ILS2) [70]. The rigid spliceosomal core remains unchanged upon Prp43 recruitment, but the transition from ILS1 to ILS2 results in conformational shifts of elements surrounding Prp43. For example, the U2 snRNP undergoes translocation and becomes even more flexible in the ILS2 complex compared with the ILS1 complex, likely in preparation for spliceosome disassembly. Comparison of the human and yeast ILS shows that most of the core components, including Prp43, are organized in a nearly identical fashion, suggesting that Prp43 functions via the same mechanism in both species. However, in the human structure two lobes of unidentified density were observed near Prp43, indicating that additional proteins may play a role in the disassembly of the spliceosome in humans, potentially by modulating the activity of Prp43.
Summary
Since the discovery of splicing about 40 years ago, we have learned a great deal about the structure, function, mechanism, and regulation of the four DEAH-box helicases that drive the late stages of the splicing cycle through genetic, biochemical, and structural analyses. The flurry of recent spliceosomal cryo-EM structures has provided direct visualization of how these spliceosomal helicases are associated with the splicing machinery. These structures have often provided direct support for previous models inferred from genetic and biochemical observations, as well as offered new insight into the structure and function of these helicases. However, one major shortcoming of these cryo-EM structures is that the RNA helicases are often located in the periphery of the spliceosome with poor local resolution. Consequently, details on conformational changes or interactions with key protein or RNA components for these helicases often cannot be directly obtained from cryo-EM structures alone. The lack of any RNA density for three of the helicases makes their respective RNA substrate uncertain; furthermore, the missing protein interactions leaves the question of how these helicases are specifically recruited to the spliceosome unanswered (see Outstanding Questions). For example, the missing NTD of these helicases interacting with another flexible component (a protein/domain/structural element that is specific for a particular complex but invisible due to its flexibility) may be an important contributor to the specific recruitment of the helicase. Continued improvement in sample preparation and computational methods may lead to better capture and visualization of dynamic interactions and flexible components. In addition, combining cryo-EM with other biochemical, biophysical, and genetics approaches will likely be the best strategy to answer these questions.
Outstanding Questions.
Does the open-closed conformational cycling or the ratchet helix or both contribute to the translocation activity of DEAH-box helicases?
Is the β-hairpin required for the in vitro unwinding activity of DEAH-box helicases? Is it important for the in vivo activity of DEAH-box helicases in the spliceosome?
How are spliceosomal DEAH-box helicases recruited to the right place at the right time? Do their NTDs play a role? What triggers the release of the DEAH-box helicases from the spliceosome?
If the NTDs are not the driving force behind the specific recruitment of DEAH-box helicases, what is their biological function?
How can we improve the local resolution of RNA helicases in spliceosomal structures so we can better visualize their interactions with RNA substrates and protein partners?
Highlights.
Pre-mRNA splicing is catalyzed by the spliceosome, a multi megadalton protein RNA machinery that undergoes dramatic conformational and compositional rearrangements throughout the splicing cycle, largely driven by eight DExD/H-box RNA helicases.
The four helicases participating in the late stages of splicing are all DEAH-box helicases that share structural similarities.
Prior genetic, structural, and biochemical studies have revealed much information on the structure, function, mechanism, and regulation of the four DEAH-box splicing helicases.
The recent cryo-EM structures of spliceosomal complexes provided a direct visualization of the physiological context of these helicases, offering new insights into their substrates, mechanism of action, and regulation.
Acknowledgments
We acknowledge support from NIH grants GM126157 and GM130673 (to R.Z.). S.E. is a Howard Hughes Medical Institute Gilliam Fellow. We thank Dr Florian Heyd for help and support while F.D.B worked on early versions of the manuscript.
References
- 1.Staley JP and Guthrie C (1998) Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92, 315–326 [DOI] [PubMed] [Google Scholar]
- 2.Cordin O and Beggs JD (2013) RNA helicases in splicing. RNA Biol. 10, 83–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fairman-Williams ME et al. (2010) SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol 20, 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Irion U and Leptin M (1999) Developmental and cell biological functions of the Drosophila DEAD-box protein abstrakt. Curr. Biol 9, 1373–1381 [DOI] [PubMed] [Google Scholar]
- 5.Chan CC et al. (2004) eIF4A3 is a novel component of the exon junction complex. RNA 10, 200–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Will CL et al. (2002) Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 21, 4978–4988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sam M et al. (1998) Aquarius, a novel gene isolated by gene trapping with an RNA-dependent RNA polymerase motif. Dev. Dyn 212, 304–317 [DOI] [PubMed] [Google Scholar]
- 8.He Y et al. (2010) Structural basis for the function of DEAH helicases. EMBO Rep. 11, 180–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozgur S et al. (2015) The conformational plasticity of eukaryotic RNA-dependent ATPases. FEBS J. 282, 850–863 [DOI] [PubMed] [Google Scholar]
- 10.Gilman B et al. (2017) Distinct RNA-unwinding mechanisms of DEAD-box and DEAH-box RNA helicase proteins in remodeling structured RNAs and RNPs. Biochem. Soc. Trans 45, 1313–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silverman EJ et al. (2004) Interaction between a G-patch protein and a spliceosomal DExD/H-box ATPase that is critical for splicing. Mol. Cell. Biol 24, 10101–10110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim SH and Lin RJ (1996) Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol. Cell. Biol 16, 6810–6819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roy J et al. (1995) The final stages of spliceosome maturation require Spp2p that can interact with the DEAH box protein Prp2p and promote step 1 of splicing. RNA 1, 375–390 [PMC free article] [PubMed] [Google Scholar]
- 14.King DS and Beggs JD (1990) Interactions of PRP2 protein with pre-mRNA splicing complexes in Saccharomyces cerevisiae. Nucleic Acids Res. 18, 6559–6564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bao P et al. (2017) Yeast Prp2 liberates the 5′ splice site and the branch site adenosine for catalysis of pre-mRNA splicing. RNA 23, 1770–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lardelli RM et al. (2010) Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA 16, 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warkocki Z et al. (2009) Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat. Struct. Mol. Biol 16, 1237–1243 [DOI] [PubMed] [Google Scholar]
- 18.Tseng CK et al. (2011) DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps. RNA 17, 145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwer B and Gross CH (1998) Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 17, 2086–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arenas JE and Abelson JN (1997) Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl. Acad. Sci. U. S. A 94, 11798–11802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai RT et al. (2005) Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 19, 2991–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fourmann JB et al. (2016) The target of the DEAH-box NTP triphosphatase Prp43 in Saccharomyces cerevisiae spliceosomes is the U2 snRNP-intron interaction. Elife 5, e15564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ninio J (1975) Kinetic amplification of enzyme discrimination. Biochimie 57, 587–595 [DOI] [PubMed] [Google Scholar]
- 24.Hopfield JJ (1974) Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl. Acad. Sci. U. S. A 71, 4135–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Semlow DR and Staley JP (2012) Staying on message: ensuring fidelity in pre-mRNA splicing. Trends Biochem. Sci 37, 263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koodathingal P and Staley JP (2013) Splicing fidelity: DEAD/ H-box ATPases as molecular clocks. RNA Biol. 10, 1073–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X et al. (2010) Sequential checkpoints govern substrate selection during cotranslational protein targeting. Science 328, 757–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pyle AM (2008) Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys 37, 317–336 [DOI] [PubMed] [Google Scholar]
- 29.Hamann F et al. (2019) Structural basis for RNA translocation by DEAH-box ATPases. Nucleic Acids Res. 47, 4349–4362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buttner K et al. (2007) Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat. Struct. Mol. Biol 14, 647–652 [DOI] [PubMed] [Google Scholar]
- 31.Pena V et al. (2009) Common design principles in the spliceosomal RNA helicase Brr2 and in the Hel308 DNA helicase. Mol. Cell 35, 454–466 [DOI] [PubMed] [Google Scholar]
- 32.Liu S et al. (2017) Structure of the yeast spliceosomal postcatalytic P complex. Science 358, 1278–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He Y et al. (2017) Structure of the DEAH/RHA ATPase Prp43p bound to RNA implicates a pair of hairpins and motif Va in trans- location along RNA. RNA 23, 1110–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka N and Schwer B (2006) Mutations in PRP43 that uncouple RNA-dependent NTPase activity and pre-mRNA splicing function. Biochemistry 45, 6510–6521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim SH et al. (1992) The purified yeast pre-mRNA splicing factor PRP2 is an RNA-dependent NTPase. EMBO J. 11, 2319–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwer B and Guthrie C (1991) PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature 349, 494–499 [DOI] [PubMed] [Google Scholar]
- 37.Company M et al. (1991) Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature 349, 487–493 [DOI] [PubMed] [Google Scholar]
- 38.Semlow DR et al. (2016) Spliceosomal DEAH-Box ATPases remodel pre-mRNA to activate alternative splice sites. Cell 164, 985–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagner JD et al. (1998) The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J. 17, 2926–2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka N and Schwer B (2005) Characterization of the NTPase, RNA-binding, and RNA helicase activities of the DEAH-box splicing factor Prp22. Biochemistry 44, 9795–9803 [DOI] [PubMed] [Google Scholar]
- 41.Tanaka N et al. (2007) Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev. 21, 2312–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aravind L and Koonin EV (1999) G-patch: a new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem. Sci 24, 342–344 [DOI] [PubMed] [Google Scholar]
- 43.Sloan KE and Bohnsack MT (2018) Unravelling the mechanisms of RNA helicase regulation. Trends Biochem. Sci 43, 237–250 [DOI] [PubMed] [Google Scholar]
- 44.Hamann F et al. (2020) Structural analysis of the intrinsically disordered splicing factor Spp2 and its binding to the DEAH-box ATPase Prp2. Proc. Natl. Acad. Sci. U. S. A 117, 2948–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Studer MK et al. (2020) Structural basis for DEAH-helicase activation by G-patch proteins. Proc. Natl. Acad. Sci. U. S. A 117, 7159–7170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tauchert MJ et al. (2017) Structural insights into the mechanism of the DEAH-box RNA helicase Prp43. Elife 6, e21510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galej WP et al. (2016) Cryo-EM structure of the spliceosome immediately after branching. Nature 537, 197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rauhut R et al. (2016) Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science 353, 1399–1405 [DOI] [PubMed] [Google Scholar]
- 49.Wan R et al. (2016) Structure of a yeast catalytic step I spliceosome at 3.4 A resolution. Science 353, 895–904 [DOI] [PubMed] [Google Scholar]
- 50.Yan C et al. (2016) Structure of a yeast activated spliceosome at 3.5 A resolution. Science 353, 904–911 [DOI] [PubMed] [Google Scholar]
- 51.Bai R et al. (2017) Structure of the post-catalytic spliceosome from Saccharomyces cerevisiae. Cell 171, 1589–1598.e8 [DOI] [PubMed] [Google Scholar]
- 52.Bertram K et al. (2017) Cryo-EM structure of a pre-catalytic human spliceosome primed for activation. Cell 170, 701–713.e11 [DOI] [PubMed] [Google Scholar]
- 53.Bertram K et al. (2017) Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature 542, 318–323 [DOI] [PubMed] [Google Scholar]
- 54.Fica SM et al. (2017) Structure of a spliceosome remodelled for exon ligation. Nature 542, 377–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Plaschka C et al. (2017) Structure of a pre-catalytic spliceosome. Nature 546, 617–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wan R et al. (2017) Structure of an intron lariat spliceosome from Saccharomyces cerevisiae. Cell 171, 120–132.e12 [DOI] [PubMed] [Google Scholar]
- 57.Wilkinson ME et al. (2017) Postcatalytic spliceosome structure reveals mechanism of 3′-splice site selection. Science 358, 1283–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan C et al. (2017) Structure of a yeast step II catalytically activated spliceosome. Science 355, 149–155 [DOI] [PubMed] [Google Scholar]
- 59.Zhang X et al. (2017) An atomic structure of the human spliceosome. Cell 169, 918–929.e14 [DOI] [PubMed] [Google Scholar]
- 60.Bai R et al. (2018) Structures of the fully assembled Saccharomyces cerevisiae spliceosome before activation. Science 360, 1423–1429 [DOI] [PubMed] [Google Scholar]
- 61.Haselbach D et al. (2018) Structure and conformational dynamics of the human spliceosomal B(act) complex. Cell 172, 454–464.e11 [DOI] [PubMed] [Google Scholar]
- 62.Plaschka C et al. (2018) Prespliceosome structure provides insights into spliceosome assembly and regulation. Nature 559, 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhan X et al. (2018) Structures of the human pre-catalytic spliceosome and its precursor spliceosome. Cell Res. 28, 1129–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhan X et al. (2018) Structure of a human catalytic step I spliceosome. Science 359, 537–545 [DOI] [PubMed] [Google Scholar]
- 65.Zhang X et al. (2018) Structure of the human activated spliceosome in three conformational states. Cell Res. 28, 307–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Charenton C et al. (2019) Mechanism of 5′ splice site transfer for human spliceosome activation. Science 364, 362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fica SM et al. (2019) A human postcatalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation. Science 363, 710–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li X et al. (2019) A unified mechanism for intron and exon definition and back-splicing. Nature 573, 375–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wan R et al. (2019) Structures of the catalytically activated yeast spliceosome reveal the mechanism of branching. Cell 177, 339–351.e13 [DOI] [PubMed] [Google Scholar]
- 70.Zhang X et al. (2019) Structures of the human spliceosomes before and after release of the ligated exon. Cell Res. 29, 274–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bao P et al. (2017) The RES complex is required for efficient transformation of the precatalytic B spliceosome into an activated B(act) complex. Genes Dev. 31, 2416–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun C (2020) The SF3b complex: splicing and beyond. Cell. Mol. Life Sci 77, 3583–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu HL and Cheng SC (2012) The interaction of Prp2 with a defined region of the intron is required for the first splicing reaction. Mol. Cell. Biol 32, 5056–5066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Warkocki Z et al. (2015) The G-patch protein Spp2 couples the spliceosome-stimulated ATPase activity of the DEAH-box protein Prp2 to catalytic activation of the spliceosome. Genes Dev. 29, 94–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Madhani HD and Guthrie C (1994) Genetic interactions between the yeast RNA helicase homolog Prp16 and spliceosomal snRNAs identify candidate ligands for the Prp16 RNA-dependent ATPase. Genetics 137, 677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fourmann JB et al. (2017) Regulation of Prp43-mediated disassembly of spliceosomes by its cofactors Ntr1 and Ntr2. Nucleic Acids Res. 45, 4068–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Walbott H et al. (2010) Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J. 29, 2194–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu YC et al. (2007) A novel splicing factor, Yju2, is associated with NTC and acts after Prp2 in promoting the first catalytic reaction of pre-mRNA splicing. Mol. Cell. Biol 27, 5403–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Toroney R et al. (2019) Termination of pre-mRNA splicing requires that the ATPase and RNA unwindase Prp43p acts on the catalytic snRNA U6. Genes Dev. 33, 1555–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brody E and Abelson J (1985) The "spliceosome": yeast premessenger RNA associates with a 40S complex in a splicing-dependent reaction. Science 228 (4702), 963–967 [DOI] [PubMed] [Google Scholar]
- 81.Frendewey D and Keller W (1985) Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell 42 (1), 355–367 [DOI] [PubMed] [Google Scholar]
- 82.Grabowski PJ et al. (1985) A multicomponent complex is involved in the splicing of messenger RNA precursors. Cell 42 (1), 345–353 [DOI] [PubMed] [Google Scholar]
- 83.Wahl MC et al. (2009) The spliceosome: design principles of a dynamic RNP machine. Cell 136 (4), 701–718 [DOI] [PubMed] [Google Scholar]