Abstract
miR-101–3p may play a therapeutic role in various tumours. However, its anti-tumour mechanism remains unclear, and a definitive strategy to treat tumour cells in vivo is lacking. The objective of this study was to investigate the inhibitory mechanism of miR-101–3p on tumour cells and to develop relevant nanomedicines for in vivo therapy. The expression levels of miR-101–3p and its target protein TBLR1 in tumour tissues and cells were detected, and their relationship with ferroptosis was clarified. Furthermore, the efficacy of nanocarriers in achieving in vivo therapeutic gene delivery was evaluated. Nanomedicine was further developed, with the anti-proliferative in vivo therapeutic effect validated using a subcutaneous xenograft cancer model. The expression level of miR-101–3p negatively correlated with clinical tumour size and TNM stage. miR-101–3p restores ferroptosis in tumour cells by directly targeting TBLR1, which in turn promotes apoptosis and inhibits proliferation. We developed nanomedicine that can deliver miR-101–3p to tumour cells in vivo to achieve ferroptosis recovery, as well as to inhibit in vivo tumour proliferation. The miR-101–3p/TBLR1 axis plays an important role in tumour ferroptosis. Nanopharmaceuticals that increase miR-101–3p levels may be effective therapies to inhibit tumour proliferation.
Keywords: Ferroptosis, Nanomedicine, ROS, Proliferation, Apoptosis, TBLR1
1. Introduction
Nanopharmaceutical carriers have been known to effectively transport nucleic acid-based drugs, as well as small-molecule drugs, targeting tumours in vivo [11]. To achieve better in vivo anti-tumour effects, more effective therapeutic targets need to be selected for nanocarriers [13,21]. Ferroptosis, an important mode of programmed death in normal cells, is significantly suppressed in most malignancies, resulting in uncontrolled proliferation and metastasis of tumour cells. Furthermore, there is mounting evidence that ferroptosis is a reliable target for the treatment of malignant tumours [1]. However, specific regulatory genes need to be carefully selected based on the molecular biology of different tumour ferroptosis regulators, to produce precise therapeutic effects. MicroRNAs (miRNA/miR) are small non-coding RNAs that directly control the protein expression of numerous genes by binding to the 3′-untranslated regions (3′UTRs) of target genes. Additionally, it has been demonstrated that some microRNAs (miRNAs) are regulatory agents that target cellular ferroptosis. Therefore, a better therapeutic effect could be achieved by exploring potential miRNAs that inhibit the proliferation of cancer [19]. miR-101–3p is closely associated with the cell death processes, including autophagy and apoptosis [17,37], and may also be associated with the regulation of ferroptotic processes. Our previous study regarding miR-101–3p has reached similar conclusions [3]. Therefore, the cancer therapeutic role and molecular biological mechanisms of miR-101–3p need to be further investigated, determining its potential application as a target for nanomedicine. In this study, the relationship between miR-101–3p and pathological features in clinical tumour specimens was used as a clue to further analyse its effect and mechanism for achieving tumour therapy through the regulation of ferroptosis. Furthermore, it was used as a therapeutic target to develop a nanomedicine that could achieve in vivo therapeutic efficacy.
2. Materials and methods
2.1. Patients and clinical samples
Samples of tumour tissue and adjacent non-tumour tissue were collected from 32 patients who underwent surgery resection between February 2013 and February 2015 at the First Affiliated Hospital of Sun Yat-sen University. All patients were pathologically diagnosed as pulmonary cancer after surgery by two independent pathologists. All tissue samples were pathologically characterised, and clinical information concerning the tumour samples was summarised. In this study, no patient received preoperative radiation or chemotherapy. The use of the clinical specimens was approved by the local institutional review board and the ethics committees of the First Affiliated Hospital of Sun Yat-sen University and conformed with the ethical guidelines of the Helsinki Declaration. All clinical tissue samples were obtained for research purposes after receiving written informed consent from respective patients. Research protocols were approved by the Clinical Research Ethics Committees of the First Affiliated Hospital of Sun Yat-sen University. The protocol was approved by the Clinical Research Ethics Committee and conformed with the ethical guidelines of the Helsinki Declaration.
2.2. Cell lines, cell culture, and in vitro cell transfection
The immortal human cell line, BEAS-2B, and the tumour cell lines A549, L78, NCI–H460, GLC-82, SPC-A1, and PC9 were provided by the Stem Cell Bank of the Chinese Academy of Sciences. The cell culture was performed in the same way as we reported previously [40]. In vitro transfection was typically performed on cells at approximately 70% density. To regulate the intracellular expression of miR-101–3p, miR-101–3p mimic, miR-101–3p inhibitor, and negative control miR were designed and synthesised (Sagan Corporation, Shanghai). miRNA sequences used in the study were listed in Supplementary Table 1. The oligonucleotide plasmid vector was designed and synthesised following the sequence in our previous study [15]. Cells were transfected with the miR-101–3p mimic, miR-101–3p inhibitor, negative control miR, or TBLR1 plasmid in vitro using Lipofectamine® 3000 (Thermo Fisher Scientific) according to the manufacturer's instructions [9]. Successfully transfected cells were used for further in vitro experiments. All constructs were verified by sequencing.
2.3. Reverse transcription-polymerase chain reaction (RT-PCR) assay and Western blot assay
The reverse transcription polymerase chain reaction (RT-PCR) experiment was carried out according to the method published earlier. In brief, total RNA was extracted at room temperature from tissue lysates or cell samples and reverse transcribed to cDNA. Information of primers were listed in Supplementary Table 1. The relative expression quantities of miR-101–3p and TBLR1were determined by using the 2−ΔΔCq method [10]. The relative amount of mRNA was normalized by using β-actin mRNA levels. Experiments were performed three times.
The protein contents of the tissue or cell lysates were measured according to the method published previously [15]. A suitable amount of total protein from each sample (10–20 μg) was separated by 10% SDS-PAGE. Following vertical electrophoretic separation, proteins were transferred to polyvinylidene fluoride membranes. The membranes were blocked with 5% fat-free milk for 1 h at room temperature and then incubated with rabbit anti-TBLR1, rabbit anti-Ki67, rabbit anti-PCNA, rabbit anti-cleaved-Caspase3, rabbit anti-Bcl-2, rabbit anti-GPX4, rabbit anti-PTGS2 or rabbit anti-β-actin antibodies (all from Abcam, Cambridge, UK) at 4 °C overnight [34]. Information and dilution of antibodies used in this study were listed in Supplementary Table 2. Following washing the membranes three times in Tris-buffered saline with Tween-20 (TBST), the membranes were incubated with an HRP-conjugated secondary antibody (Abcam, Cambridge, UK) at room temperature for 2 h. After washing three times with TBST again, the exposure and development were performed using an ECL Chemiluminescence Detection kit (GE Healthcare Life Sciences, Little Chalfont, UK), and the densities were measured using a GS-900™ Calibrated Densitometer with ImageLab 4.1 software from Bio-Rad Laboratories, Inc. (Hercules, CA, USA) [31]. All of the assays were performed in triplicate.
2.4. CCK-8 cell proliferation assay and colony formation assay
The proliferation ability of A549 cells was measured using a Cell Counting Kit-8 (CCK-8) kit (Dojindo, Japan) according to the manufacturer's instructions [39]. Briefly, transfected A549 cells were seeded into 96-well plates at a density of 5 × 103 cells/well and incubated for 48 h. Subsequently, 10 μL CCK-8 reagent was added to wells, and the cells were cultured for an additional 1 h. Miniature microplate readers were used to determine the optical density (OD) at 450 nm [31]. All experiments were performed in triplicate.
Colony formation assay was carried out according to the method previously reported [40]. Base layers of complete culture medium (2 ml) containing 0.6% Difco Noble agar (BD Biosciences) were set in each well of a 6-well cell culture plate. This was further overlaid with 2 ml of 0.3% agar containing a suspension of 1 × 104 single cells in culture medium plus 20% FBS. After 10 days, the colony images were photographed and quantified. Colonies larger than 0.1 mm in diameter were scored. The reproducibility of our study was guaranteed by repeating all experiments at least three times.
2.5. Annexin V/Propidium iodide (PI) flow cytometry apoptosis assay
Briefly, transfected cells in different groups were digested with pancreatin and subsequently washed with phosphate-buffered saline (PBS). After cells were suspended in 100 μL binding buffer, 5 μL anti-Annexin V‐FITC antibody was added and allowed to stand in the dark for 15 min at room temperature. Then, PI (5 μL) was added immediately before the flow cytometric assay (Merck Group, Germany) [38]. Flow cytometry was used to analyse the results [30]. All experiments were performed in triplicate.
2.6. Target prediction and luciferase reporter assays
The miRNA-targeted proteins were predicted using miRanda, TargetScan, and PicTar databases [25]. A site-directed mutagenesis kit (Stratagene; Agilent Technologies, Inc., Santa Clara, CA, USA) was used to produce the mutant type of the TBLR1 3′UTR luciferase reporter vector (pmirGLO-TBLR1-MUT) according to the manufacturer's protocol. The luciferase reporter contained regions of the TBLR1 3′UTR with the indicated miR-101–3p target sites, or mutant sites to determine whether TBLR1 was a direct target of miR-101–3p. Briefly, cells were seeded in triplicate in 6-well plates and allowed to settle for 12 h. The indicated plasmids and equal amounts of miR-101–3p or negative control (NC) RNA were transfected into cells (Life Technologies). Twenty-four hours after transfection, luciferase activity was measured using a Dual-Luciferase Reporter Assay System (Promega Corporation). All experiments were performed in triplicate.
2.7. Ferroptosis-associated oxidative stress detection
The DCFDA cellular reactive oxygen species (ROS) assay was performed following the manufacturer's instructions (ab113851, Abcam, UK). Briefly, cells were incubated with DCFDA (20 μM, 100 μL/well) for 0.5 h at 37 °C in the dark. DCFDA is a fluorescent probe that can detect ROS. After diffusion into cells, non-fluorescent DCFDA is deacetylated by cellular esterases and then oxidised by ROS into the fluorescein 2′,7’ - dichlorofluorescein (DCF), which cannot diffuse out of cells. As DCF is fluorescent, it can be detected using fluorescence spectroscopy, with excitation and emission wavelengths of 495 nm and 529 nm, respectively. The level of lipid ROS in cells or tissues was evaluated using flow cytometry after 20 min of staining with 1.5 mM C11-BODIPY (Invitrogen, Carlsbad, CA, USA) [2]. In cell lysates subjected to different treatments, cellular glutathione (GSH) levels were determined using a GSH colorimetric assay kit (BioVision Inc., Milpitas, CA, USA) following the manufacturer's instructions.
2.8. Therapeutic gene loading by nanocarriers and gene delivery assays
The in vivo transfection reagent (Engreen Biosystem, Auckland, New Zealand) was used for the nucleic acid transfection, intravenously administered via tail vein according to the manufacturer's instructions [14]. We prepared a nucleic acid dilution by dissolving 50 μg of miR-101–3p mimic, miR-101–3p inhibitor, or NC miR in 25 μL PBS. Then, 25 μL of PBS was added, and the solution was mixed well. Next, a transfection reagent dilution was prepared by dissolving 25 μL of diluted transfection reagent in 25 μL PBS, with 25 μL PBS, and the solution was mixed well. Finally, the microRNA dilution and transfection reagent dilution were mixed (1:1) to form a solution for in vitro cell transfection or in vivo intravenous injection [28].
To enable confocal laser scanning microscopic (CLSM) observations of the in vitro gene transmission efficiency, miR-101–3p mimic, miR-101–3p inhibitor, and NC miR were labelled with POPO-3 according to the manufacturer's protocols, as previously described [32]. Then, fluorescently labelled nucleic acids were loaded with nanocarriers for 2 h for in vitro transfection. After transfection, cells were washed three times with fresh PBS, fixed, and further incubated with DAPI for nucleic acid staining. CLSM observations were performed using a microscope (Carl Zeiss Co., Ltd., Gottingen, Germany). To observe the efficacy of in vivo nanomedicine delivery to tumours, we used the near-infrared fluorescent (NIRF) dye, CY7, to label the in vivo transfection reagent nanocarrier for intravenous injection. In vivo NIRF fluorescence scanning was performed 2 h after injection as previously reported [32].
2.9. Xenografted tumour model and in vivo treatment assay
BALB/c mice (male, 6 weeks old, 20–22 g) were obtained from Vital River Laboratories (Beijing, China). All animal experiments were executed according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The experimental procedures were approved by the Institutional Animal Care and Use Committees (IACUCs) of First Affiliated Hospital of Sun Yat-sen University. The animals were treated humanely for all experimental procedures. All animals were lawfully acquired, and their care and use were in compliance with federal, state, and local laws and regulations and in accordance with the guidelines of the IACUC guide for the care and use of laboratory animals. Immunodeficient mice were randomly divided into groups (n = 6 per group). A549 cells (1 × 106) were injected subcutaneously into the dorsal left flank of nude mice.
In each group, mice (20–25 g) were administered 100 μL of the nanomedicine working solution through the tail vein, once every 3 days. Tumours were measured every week, with the length (L) and width (W) measured, and the tumour volumes calculated using equation (L × W2)/2. On day 28, the animals were sacrificed, and the tumours were harvested and weighed. For the in vivo BrdU proliferation assay, each animal received an injection of BrdU (80 mg/kg, BD Biosciences) 1 h before sacrifice. Fresh tumour tissue was used for RT-PCR and Western blot assays [30]. To perform the TdT-mediated dUTP nick end labelling (TUNEL) assay, BrdU immunofluorescence assay, and ROS fluorescence assay, tissue samples were harvested immediately after euthanasia and used for pathology sectioning. For lipid ROS detection, tumour tissues were processed as single-cell suspensions for flow cytometry after C11-BODIPY staining. The pathological specimens were assessed using a TUNEL apoptosis detection kit (Merck, USA), BrdU immunofluorescence cell proliferation assay kit (BioVision, USA), or a DCFDA cellular ROS kit. All experiments were performed in triplicate.
2.10. Statistical analysis
Statistical analyses were performed using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). The data are presented as the mean ± SD (standard deviation). Two-tailed Student's t-test and ANOVA were implemented to determine differences. P < 0.01 was considered to indicate a statistically significant difference.
3. Results
3.1. Expression of miR-101–3p was diminished in tumour cell lines and tissue samples
Expression levels of miR-101–3p were determined by RT-PCR in tumour cell lines A549, L78, NCI–H460, GLC-82, SPC-A1, and PC9. In the cell lines, miR-101–3p expression levels were lower than those observed in the human epithelial cell line, BEAS-2B (P < 0.01, Fig. 1A), encouraging us to further explore this phenomenon in clinical tissues besides immortalised cell lines.
Fig. 1.
Exploring the possible clinical role of miR-101–3p in tumour cells and tissues. (A) The expression levels of miR-101–3p in tumour cell lines A549, L78, NCI–H460, GLC-82, SPC-A1, and PC9 were compared with those in the human normal cell line, BEAS-2B. (B) The expression levels of miR-101–3p in tumour tissues and adjacent normal tissues (ANT) were compared by RT-PCR assay. (C) The expression levels of miR-101–3p in tumour tissues of diameter (ϕ) > 3 cm and ≤3 cm were compared. (D) The expression levels of miR-101–3p in tumour tissues of TNM I + II and III + IV stage patients were compared by RT-PCR assay. (*P < 0.01 v.s. BEAS-2B cells, ANT tissues, ϕ ≤ 3 cm group tissues or TNM I + II group tissue). miR, microRNA.
Therefore, the discrepancy between the expression of miR-101–3p in 32 lung cancer (LC) clinical tissues and homologous adjacent normal tissues (ANT) was explored. As shown in Fig. 1B, expression levels of miR-101–3p in LC tissues were lower than those in ANT tissues (P < 0.01). The relationship between the expression levels of miR-101–3p in 32 LC patients and corresponding patient clinicopathological factors was further investigated to determine its possible biological function in LC. The decreased miR-101–3p level in tumour tissues was found to be significantly associated with larger tumour size (>3 cm) and more severe TNM stage (stage III or IV) (P < 0.01; Fig. 1C and D). These findings suggested that decreased miR-101–3p expression may be associated with tumorigenesis in LC, but this relationship needs to be comprehensively explored.
3.2. Promoting miR-101–3p expression may inhibit tumour cell proliferation and enhance apoptosis
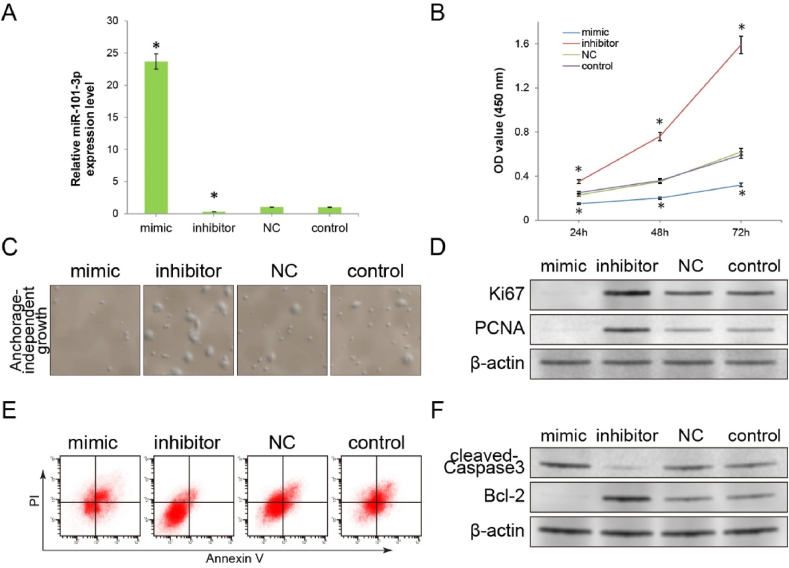
To further investigate the role of miR-101–3p in tumour cell proliferation and apoptosis, miR-101–3p mimics or inhibitors were transfected into A549 cells. A549 cells transfected with NC miR and untransfected control A549 cells were used as negative and blank controls, respectively. RT-qPCR results showed that miR-101–3p mimics significantly elevated miR-101–3p levels when compared with control groups (NC miR group or control group). Simultaneously, miR-101–3p inhibitors significantly reduced miR-101–3p levels (all P < 0.01). As clinical studies have indicated that miR-101–3p is associated with tumour size (Fig. 1C), we initially assessed the proliferative capacity of A549 cells transfected with miR-101–3p mimics or inhibitors to investigate its role. The proliferative capacity of each group of cells was determined using CCK-8 assays. The results of the CCK8 assay showed that the OD values for the miR-101–3p mimic group were significantly reduced at 24 h, 48 h, and 72 h when compared with those of the control groups, indicating that the proliferation capacity was inhibited, whereas OD values were elevated in the miR-101–3p inhibitor group (P < 0.01; Fig. 2B).
Fig. 2.
Preliminary exploration of the inhibitory effect of miR-101–3p on tumour cells in vitro. (A) A549 cells were transfected with the miR-101–3p mimic, miR-101–3p inhibitor or negative control (NC) miR. The expression levels of miR-101–3p in the different A549 cell groups were compared by RT-PCR assay. Untransfected blank control cells were used as a control group. (B) The CCK8 assay was performed to compare the influence on the proliferation of different A549 cell groups. (C) The results of anchorage-independent growth assay revealed the extent of influence on the clonal formation capacity of A549 cells in each group. (50 × magnification). (D) The expression levels of proliferation markers (Ki67 and PCNA) were detected using the Western blot assay. (E) Apoptosis rates of different cell groups were detected by the Annexin V/PI flow cytometry assay. (F) The expression levels of apoptosis markers (cleaved-Caspase3 and Bcl-2) were detected by Western blot assay. (*P < 0.01 v.s. control group). miR, microRNA; CCK8, Cell Counting Kit-8; OD, optical density.
Anchorage-independent growth assays corroborated the proliferative function results obtained using the CCK-8 assay. Fig. 2C shows that the number and size of colonies formed by the miR-101–3p mimic cell group on soft agar were significantly decreased, while miR-101–3p inhibitor transfection resulted in a significant increase in the colony-forming ability of cells. The detection of proliferation markers, Ki67 and PCNA, provided further evidence, as these were downregulated in the miR-101–3p mimic cell group, with significant upregulation observed in the miR-101–3p inhibitor group (Fig. 2D). These results suggested that the expression of miR-101–3p is related to the proliferative potential of tumour cells and that miR-101–3p is an important regulator of cell proliferation.
Apoptosis is closely related to the proliferative potential of tumour cells. Therefore, we investigated whether the regulation of proliferation by miR-101–3p is associated with apoptosis. To assess the relationship between the quantity of miR-101–3p expression and the level of apoptosis in A549 cells, an Annexin V/PI flow cytometric assay was performed. The flow cytometric results showed an increased apoptosis rate in the mimic group and a decreased apoptosis rate in the inhibitor group when compared with the control group (Fig. 2E).
To confirm the mechanism underlying changes in the proportion of apoptosis, western blotting was used to determine the expression levels of apoptotic markers, cleaved-Caspase3 and Bcl-2. As shown in Fig. 2F, cleaved-Caspase3 expression was upregulated in the miR-101–3p mimic group, whereas Bcl-2 expression was downregulated, when compared with NC or control cells. In contrast, the expression of cleaved-Caspase3 in the miR-101–3p inhibitor group was significantly downregulated and the expression of Bcl-2 was significantly upregulated. Thus, our findings demonstrated that miR-101–3p could inhibit proliferation by promoting apoptosis. With its critical function in tumour cell inhibition, miR-101–3p could be used as a target to inhibit apoptosis-associated proliferation. However, before utilisation in antineoplastic therapy, the molecular mechanisms underlying the therapeutic effects must be carefully explored.
3.3. miR-101–3p inhibits tumour cells by directly targeting TBLR1
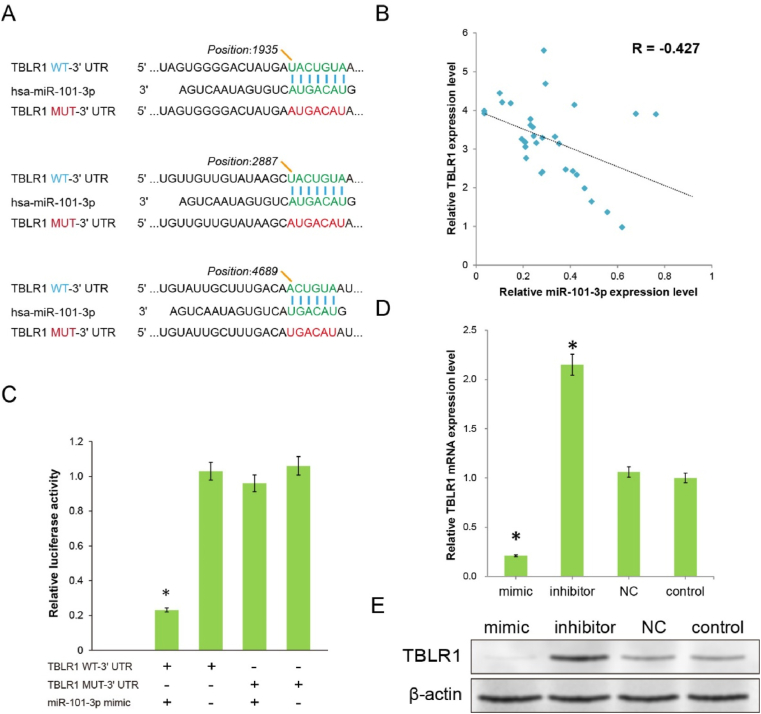
Although the role of miR-101–3p in the development and progression of various types of cancers, including leukaemia, colon, and cervical cancer, has been previously reported, the mechanism through which miR-101–3p inhibits pulmonary cancer has not been comprehensively investigated. Using MIRBASE and TargetScan programs, miR-101–3p was predicted to possess multiple hypothetical binding sites, simultaneously presenting within the 3′UTR of TBLR1 (Fig. 3A).
Fig. 3.
Initial exploration of the target through which miR-101–3p produces therapeutic effects. (A) Multiple binding sites predicted the binding of miR-101–3p to the wild-type (WT) TBLR1 3′UTR, and the corresponding mutant (MUT) 3′UTR was designed for validation. (B) Correlation between the expression levels of miR-101–3p and relative TBLR1 mRNA in clinical tumour specimens by RT-PCR assay. (C) The reporter activities of A549 cells cotransfected by miR-101–3p mimics with pmirGLO-TBLR1-WT-3′UTR, or pmirGLO-TBLR1-MUT-3′UTR plasmids, were determined by luciferase activity assay. (D) The relative mRNA expression levels of TBLR1 in the different A549 cell groups were determined by RT-PCR assay. (E) The protein expression levels of TBLR1 in the different cell groups were determined by western blotting. β-actin was used as an internal control. (*P < 0.01 v.s. the pmirGLO-TBLR1-WT transfected alone cells or control cells).
In our previous studies, TBLR1 was revealed as an oncogene in liver cancer and cervical cancer. Furthermore, evidence from a few studies has suggested its oncogenic role in pulmonary cancer. Therefore, to initially validate the bioinformatics prediction, we investigated the correlation between miR-101–3p and TBLR1 expression levels in each tumour specimen from clinical samples using RT-PCR. The results showed that the expression level of miR-101–3p significantly and negatively correlated with the mRNA expression levels of TBLR1 (R = −0.427, Fig. 3B). These interesting findings prompted us to confirm this direct regulatory relationship through further investigation. To assess whether TBLR1 is a direct target of miR-101–3p, we designed a TBLR1-mutation (MUT)-3′UTR plasmid containing mutations at three predicted sites (sites 1935, 2887, and 4689). The miR-101–3p mimics or NC miR were further compared with a pmirGLO-TBLR1-wild type (WT)-3′UTR plasmid or pmirGLO-TBLR1-MUT-3′UTR plasmid cotransfected into A549 cells. (Fig. 3A). As shown in Fig. 3C, the miR-101–3p mimic and pmirGLO-TBLR1-WT-3′UTR cotransfected cells demonstrated significantly reduced reporter levels when compared with cells transfected with only pmirGLO-TBLR1-WT (P < 0.01), indicating the presence of a targeting relationship between miR and 3′UTR. Moreover, cells cotransfected with the miR-101–3p mimic and pmirGLO-TBLR1-MUT-3′UTR, or pmirGLO-TBLR1-MUT-3′UTR only, demonstrated no obvious change in reporter levels (P > 0.01). The luciferase activity of wild-type TBLR1-3′UTR reporters was reduced by miR-101–3p, but not the designed mutant TBLR1-3′UTR reporters, indicating that target recognition and regulation occurred at the position predicted. To further confirm the regulatory effect of miR-101–3p on TBLR1 at the protein blotting level, Western blot analysis was performed. The results showed that with the promotion of miR-101–3p expression by the mimics, the mRNA and protein quantity of TBLR1 were significantly decreased, while miR-101–3p inhibitor transfection significantly increased TBLR1 expression when compared with control treatments (Fig. 3D, E, P < 0.01). These results indicated that miR-101–3p may directly control the expression level of TBLR1 by binding to the 3′UTR region of TBLR1.
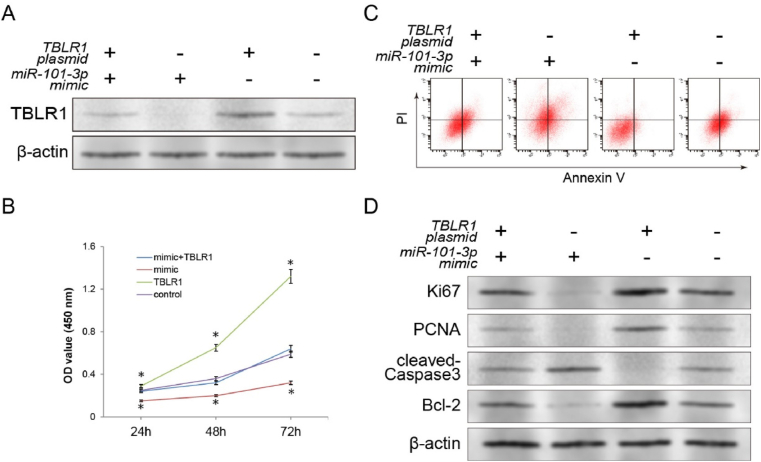
To confirm whether the regulatory effect of miR-101–3p on proliferation and apoptosis depends on binding to TBLR1 3′UTR, a pCMV-TBLR1 eukaryotic overexpression plasmid was synthesised and cotransfected with the miR-101–3p mimic. As control groups, cells transfected with the miR-101–3p mimic, TBLR1 overexpression plasmids, or untransfected cells were used. The protein expression level of TBLR1 was significantly increased in the TBLR1 overexpression plasmid-transfected group when compared with the blank control group. More importantly, compared with the miR-101–3p mimic-transfected group alone, the TBLR1 expression level in the pCMV-TBLR1 plasmid/miR-101–3p mimic cotransfected group was significantly restored (Fig. 4A). The CCK-8 assay was performed again to investigate the effect of TBLR1 overexpression on the inhibitory ability of miR-101–3p on proliferation. Compared with the miR-101–3p mimic transfection group, the pCMV-TBLR1 overexpression plasmid cotransfection group revealed a significant rescue effect on the proliferation capacity (P < 0.01; Fig. 4B). Flow cytometry was used to evaluate the role of TBLR1 on the apoptotic promotion activities of miR-101–3p. The apoptotic regulatory effect of the miR-101–3p mimic was significantly suppressed after cotransfection with the pCMV-TBLR1 plasmid (P < 0.01; Fig. 4C). The findings regarding the proliferation and apoptosis markers in the cotransfection group of A549 cells further supported the conclusion that TBLR1, as a direct target, is involved in the miR-101–3p killing of tumour cells. Western blot results showed that the ectopic expression of TBLR1 induced by co-transfection significantly rescued the inhibition of Ki67/PCNA/Bcl-2 and promotion of cleaved-Caspase3 during tumour inhibition by the miR-101–3p mimic. The aforementioned experiments demonstrated that miR-101–3p is a highly efficient regulator of tumour cell proliferation and apoptosis, and these therapeutic effects were largely produced through the direct inhibition of TBLR1 protein expression.
Fig. 4.
In vitro functional validation of target of miR-101–3p generating therapeutic action. (A) Western blotting examined the rescue effect of TBLR1 eukaryotic expression plasmids on miR-101–3p inhibition of TBLR1 expression. (B) The antagonistic effect of ectopic TBLR1 expression on the inhibition of miR-101–3p induced proliferation was examined by CCK-8 assay. (C) The antagonistic effect of ectopic TBLR1 expression on the ability of miR-101–3p to promote apoptosis was examined by Annexin V/PI flow cytometry assay. (D) The antagonistic effects on expression levels of proliferation markers (Ki67 and PCNA) and apoptosis markers (cleaved-Caspase3 and Bcl-2) were detected by Western blot assay. (n = 3, *P < 0.01 v.s. blank control cells).
3.4. The miR-101–3p/TBLR1 axis regulates ferroptosis and serves as a promising target for cancer therapy
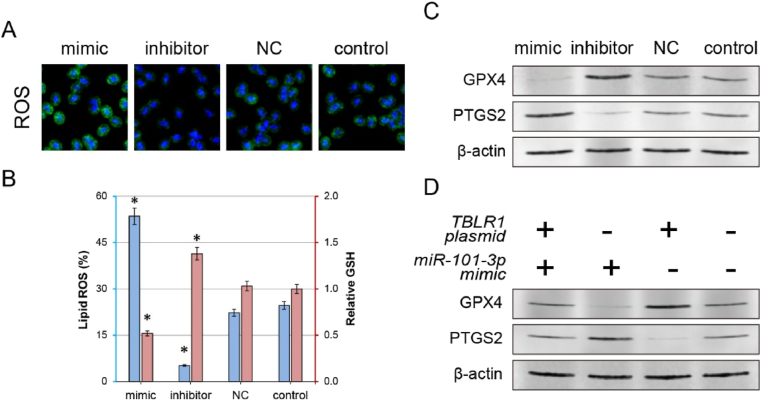
Typically, mutations in cancer cells significantly impede the activation of apoptosis. Therefore, we aimed to explore the in-depth molecular mechanisms through which the miR-101–3p/TBLR1 axis activates apoptosis, and thus inhibits proliferation [15]. Our previous studies have demonstrated that inhibition of TBLR1 expression significantly reduces the activity of the nuclear factor kappa B (NF-κB) pathway. Reportedly, the NF-κB pathway regulates ferroptosis in tumour cells through key proteins such as GSH peroxidase 4 (GPX4) [7] or prostaglandin-endoperoxide synthase 2 (PTGS2) [4]. Therefore, we explored the ferroptosis activity of cells presenting an altered miR-101–3p/TBLR1 axis activity. Laser confocal microscopy revealed that the total ROS level of cells was significantly increased after miR-101–3p mimic treatment. The total ROS level is one of the most important and recognisable markers of intracellular ferroptosis. However, compared with the control groups, the miR-101–3p inhibitor further decreased the total cellular ROS level (Fig. 5A). The C11-BODIPY assay revealed similar changes in lipid ROS, with a significant increase in the biomembrane oxidation level of cells in the mimic group and a significant decrease in the inhibitor group. In the regulation of ferroptosis, GSH is a substrate for GPX4, which protects cells from lipid peroxidation. The intracellular levels of GSH were significantly decreased or increased after treatment with miR-101–3p mimic or inhibitor, respectively (Fig. 5B).
Fig. 5.
Anti-tumour effects of the miR-101–3p/TBLR1 axis resulting from ferroptosis modulation. (A) Laser confocal microscopy was used to observe cellular ROS levels in each group. (B) Comparison of lipid ROS and GSH levels after treatment with miR-101–3p in each cell group. (*P < 0.01 v.s. control group) (C) Detection of changes in the expression of ferroptosis markers (GPX4 and PTGS2) after miR-101–3p treatment. (D) The antagonistic effects of TBLR1 eukaryotic expression plasmids on expression levels of ferroptosis markers (GPX4 and PTGS2) were detected by western blotting. ROS, reactive oxygen species; GSH, glutathione; GPX4, glutathione peroxidase 4; PTGS2, prostaglandin-endoperoxide synthase 2.
Additionally, we explored expression changes in ferroptosis protein markers (GPX4 and PTGS2) to further clarify the relationship between observed changes in proliferation/apoptosis during treatment and ferroptosis. miR-101–3p mimic treatment resulted in a significant decrease in the expression of GPX4 protein and an increase in the expression of PTGS2 protein; inhibitor treatment showed opposite changes in the two proteins when compared with the control groups (Fig. 5C). Following TBLR1 overexpression, the ferroptosis marker detection assay further validated the regulatory function of the miR-101–3p/TBLR1 axis in ferroptosis. Ectopic overexpression of TBLR1 significantly rescued the regulatory effect of miR-101–3p on the mentioned ferroptosis markers. Compared with cells transfected with miR-101–3p alone, elevated expression levels of GPX4 and reduced expression levels of PTGS2 were observed in TBLR1/miR-101–3p cotransfected cells (Fig. 5D). These findings suggest that activation of the miR-101–3p/TBLR1 axis directly recovers tumour cell ferroptosis, which may be responsible for therapeutic effects such as inhibition of proliferation and enhancement of apoptosis. Thus, it is known that miR-101–3p has a powerful tumour-suppressive effect, and once in vivo distribution to tumour cells is achieved, the development of an efficient therapeutic drug will be highly attainable.
3.5. Nanocarriers enable efficient distribution of miR-101–3p for in vivo tumour treatment
Nano-genecarriers can form nanometer-sized drugs with therapeutic nucleic acids to ensure the stability of therapeutic nucleic acids during in vivo treatment, and then achieve tumour tissue distribution through the EPR effect (enhanced permeability and retention effect). We intend to use nanocarrier for gene delivery and gene therapy with miR-101–3p in tumour cells in vivo [27]. We used a previously reported method to label microRNA using the fluorescent dye POPO-3, then complexed with nanocarriers. Laser confocal experiments showed that after nanocarrier loading, the transfection efficiency of POPO-3 (red fluorescence)-labelled plasmids into cells was significantly improved. However, in the control group, the fluorescently labelled plasmid itself demonstrated difficulty entering cells efficiently (Fig. 6A). After staining nanocarrier using the NIRF dye, CY7, and administration via the tail vein, a significant concentration of the nanocarrier was observed in the tumour area using the in vivo fluorescence assay. This signal concentration in the tumour region was not significantly related to CY7 itself (Fig. 6B). These tumour tissues were further harvested to obtain fast-frozen pathology sections, which were then used for fluorescence microscopy. Significant fluorescence aggregation was observed in tumour cells in the nanocarrier group when compared with the control groups (Fig. 6C).
Fig. 6.
Molecular image tracing technology to investigate the possibility of using nanocarriers to target tumour ferroptosis for in vivo therapy. (A) Molecular imaging examined the ability of plasmids transported by nanocarriers (red, labelled with POPO-3 fluorescent dye) for endografting into cells. The transfection effect of POPO3 fluorescence-labelled plasmids without nanocarriers was used as a control. (B) In vivo imaging was used to investigate the distribution of the nanocarrier labelled with the near-infrared dye, CY7, after intravenous injection to cancerous animals. The in vivo imaging effect of an equal amount of CY7 fluorescent dye was used as a control. (C) Laser confocal experiments assessed pathological sections of the tumour tissue after intravenous injection of CY7-labelled (green) nanocarriers, directly demonstrating the efficiency of in vivo nanocarrier distribution into tumour cells. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
This ex vivo experimental result directly demonstrates the aggregation of nanocarriers in the tumour region, as observed in the in vivo fluorescence staining. The aforementioned study illustrates that the nanocarriers can efficiently carry plasmids into the tumour region, and thus facilitate the in vivo transfection of tumour cells. Therefore, we were keen on utilising this nanocarrier to carry miR-101–3p mimics, which has been shown to possess definitive ferroptosis-promoting and tumour-killing effects in in vitro experiments for in vivo tumour therapy.
3.6. miR-101–3p nanomedicine inhibits in vivo tumour growth
To further investigate the in vivo tumour suppression function of nanocarriers carrying nanomedicines composed of miR-101–3p, we developed a subcutaneous tumour model. Tumour cells were subcutaneously injected into nude mice (4 weeks), and then the mice were treated via a tail vein injection using nanocarrier, containing miR-101–3p mimic, miR-101–3p inhibitor, or NC miR [33]. The same volume of PBS was injected into the blank control group. Tumour growth was evaluated by continuously monitoring the tumour volume. The tumour growth rate was suppressed following treatment with the miR-101–3p mimic-loaded nanomedicine when compared with the control treatments. However, the tumour growth rate was accelerated following treatment with the miR-101–3p inhibitor-loaded nanomedicine (Fig. 7A). Ex vivo measurements of the tumour weights at the end of the experiment further confirmed results of the tumour growth rate assay (Fig. 7B). To determine whether the variations in tumour nodule growth were attributed to changes in the quantity of miR-101–3p under the influence of nanomedicine, we used RT-PCR to detect the relative expression levels of miR-101–3p in tumour tissues from different groups. We observed that compared with the control groups, the miR-101–3p quantity was significantly increased in the miR-101–3p mimic group; however, it was decreased in the miR-101–3p inhibitor group (Fig. 7C). Thus, these results indicated that miR-101–3p mimic-loaded nanomedicine inhibits the growth of tumours in vivo. A BrdU immunofluorescence staining assay was performed to directly detect the proliferation of tumour cells in vivo. In the mimic group tumour nodules, the BrdU incorporation rate of cancer cells was lower than that in the control group. However, the BrdU incorporation rate of tumour cells in the inhibitor group tumour nodules was significantly higher (Fig. 7D).
Fig. 7.
Effective gene delivery for miR-101–3p produces therapeutic effects on tumours in vivo by promoting ferroptosis. (A) The growth rate of tumours in each mouse group was continuously measured after intravenous injection of nanomedicines regulating miR-101–3p expression once every 3 days. (B) Tumour volumes were observed on day 28, after continuous nanomedicine treatment. (C) Expression levels of miR-101–3p in tumour tissues after in vivo treatment with each nanomedicine group were detected by RT-PCR assay. (D) Pathological sections of tumour specimens were observed by BrdU immunofluorescence assay, TUNEL immunohistochemistry assay, and ROS fluorescence assay to detect the proliferation potential, apoptosis level, and ferroptosis level of lung cancer cells in tumour nodes in vivo, respectively. (E) Detection of the protein expression levels of the target protein of miR-101–3p (TBLR1), proliferation markers (Ki67 and PCNA), apoptosis markers (cleaved-Caspase3 and Bcl-2) and ferroptosis markers (GPX4 and PTGS2) in each group of lung cancer tissue by western blotting. (F) Detection of lipid ROS and GSH levels after nanomedicine treatment in each group of tumour tissues. (n = 6, *P < 0.01 v.s. control group). TUNEL, TdT-mediated dUTP nick end labelling assay; ROS, reactive oxygen species; GSH, glutathione; GPX4, Glutathione peroxidase 4; PTGS2, prostaglandin-endoperoxide synthase 2.
To further investigate whether the proliferation-regulating effects of the miR-101–3p mimics/inhibitor-loaded nanomedicine on tumour nodules correlated with variations in TBLR1 protein expression in vivo, we used western blotting to detect the levels of TBLR1, Ki67, and PCNA proteins in tumour tissues. We observed that the protein expression levels of TBLR1 and proliferation markers Ki67 and PCNA were decreased in the miR-101–3p mimic group, but increased in the miR-101–3p inhibitor group (Fig. 7E). These results directly demonstrate that the miR-101–3p/TBLR1 axis has a definite inhibitory effect on proliferation in tumour cells in vivo.
These interesting in vivo inhibitory proliferation results prompted us to explore the therapeutic effect of the miR-101–3p/TBLR1 axis. As shown in Fig. 7D, the TUNEL assay performed using pathological sections showed that the tumour tissue harvested from the miR-101–3p mimic-loaded nanomedicine group presented an increased number of apoptotic cells, whereas the tumour tissue harvested from the miR-101–3p inhibitor-loaded nanomedicine group presented a decreased number of apoptotic cells when compared with that of the control groups. Then, the expression levels of the apoptotic markers, cleaved-Caspase3 and Bcl-2, in the tumour tissue from the 4 groups were determined by western blotting. Compared with that of the NC or blank control groups, tumour tissues from the miR-101–3p mimic group showed lower Bcl-2 expression levels and higher cleaved-Caspase3 expression levels. Conversely, tumour tissues from the miR-101–3p inhibitor group affected Bcl-2 and cleaved-Caspase3 protein expression levels, which was in contrast to that observed in the miR-101–3p mimic group (Fig. 7E). Accordingly, the miR-101–3p/TBLR1 axis promoted apoptosis in LC cells in vitro and in vivo.
As suggested by the in vitro results, the in vivo therapeutic effect demonstrated by this miR-101–3p mimic-loaded nanomedicine could be attributed to the regulation of tumour cell ferroptosis by the miR-101–3p/TBLR1 axis. Therefore, we detected changes in total ROS, lipid ROS, and GSH levels in tumour tissues in vivo to assess changes in ferroptosis levels after nanomedicine treatment. Fluorescence microscopy revealed that the total ROS levels in the tumour tissues of the miR-101–3p mimic group were significantly increased, and the total ROS levels in the tumour tissues of the inhibitor group were significantly decreased when compared with the control groups (Fig. 7D). Similarly, compared with the control group, lipid ROS levels were significantly elevated in tumour tissues in the mimic group and significantly reduced in the inhibitor group (Fig. 7F). The detection of tumour GSH levels revealed that in the mimic group, the GSH level in tumour tissues was significantly lower, whereas the GSH level in the inhibitor group was significantly higher than that in the control group (Fig. 7F). We further examined the expression levels of ferroptosis markers in tumour tissues and observed the same trends in the in vitro experimental studies. Compared with the control group, the expression levels of GPX4 were significantly lower and PTGS2 was significantly higher in the mimic group, while the expression levels of these two proteins showed opposite changes in the inhibitor group (Fig. 7E). As shown above, the miR-101–3p mimic-loaded nanomedicine we constructed can promote ferroptosis by activating the miR-101–3p/TBLR1 axis activity of tumour cells in vivo.
In summary, the results of in vivo and in vitro tumour therapeutic assays of nanomedicines were consistent with the results of in vitro experiments, both revealing that the miR-101–3p/TBLR1 axis inhibits proliferation by promoting ferroptosis and apoptosis in tumour cells, and revealing the potential of constructing nanomedicines for tumour therapy through this mechanism.
4. Discussion
It has been postulated that efficient gene therapy for tumours could be achieved through a two-step process: first, exploring efficient gene targets for tumour therapy, and second, achieving therapeutic efficacy by enabling the specific delivery of therapeutic genes into tumour cells in vivo. However, currently, it remains challenging to screen the most effective therapeutic targets among numerous mutated genes owing to the accumulation of multiple mutations during tumorigenesis. Conversely, even if the most effective therapeutic targets are screened, traditional gene delivery methods fail to achieve effective in vivo cancer gene therapy. Researchers can validate therapeutic functions in vivo only by establishing cell lines that stably express or knockout specific therapeutic targets, thereby creating animal models. This widely varies from what is expected during in vivo cancer drug therapy. By simultaneously using both therapeutic gene screening and a user-friendly nanogene carrier technology, this study proposes a possible strategy of drug development in tumor pharmacology. More significantly, this miRNA-based therapeutic nanomedicine was shown to possess a definitive ferroptosis regulation function, and this important therapeutic mechanism is achieved in tumours in vivo through gene delivery by nanocarriers, ensuring its tumour-suppressive effect.
In this study, we first identified a therapeutic gene demonstrating a potential tumour ferroptosis recovery function, miR-101–3p, through rigorous clinical specimen analysis and in-depth functional validation. Pulmonary cancer is one of the most malignant cancers among malignancies worldwide. Although surgery, chemotherapy, radiotherapy, and other clinical techniques have been developed in the past decade, LC continues to present the highest morbidity and mortality rates among tumours. The refractoriness of LC is largely attributed to the enormous proliferative capacity of the primary lesions and metastatic foci. However, the mechanism of LC proliferation has been poorly understood. Apoptosis is now considered a critical event that influences LC proliferation. However, the mechanism influencing the apoptosis of LC cells is not comprehensively elucidated. miRNAs are small non-coding RNAs involved in the posttranscriptional regulation of gene expression. miRNAs are reportedly associated with the development of several cancer types, either as oncogenes or tumour suppressor genes [24]. Previous studies have reported atypical expression levels of various miRNAs, including miR-101–3p. Reportedly, miR-101–3p demonstrates reduced expression in several cancer types such as ovarian cancer [20], cervical cancer [36] and hepatocellular carcinoma [29]. Furthermore, its therapeutic effects and molecular biological mechanisms in these tumour types have been well elucidated. The abnormal expression of miR-101–3p has been correlated with the proliferation of several types of tumours. These studies suggest that miR-101–3p may be an important tumour-associated gene, with potential in drug development. In LC, miR-101–3p has generally been studied as a common targeting site for long-noncoding-RNA MALAT1 [8] or SNHG6 [16], implying that it may have an important therapeutic role. However, its expression level and actual therapeutic effect in LC, as well as its oncological therapeutic mechanism, have not been reported. Therefore, systematic research is needed to clarify the status and mechanism of miR-101–3p in LC.
In the present study, before evaluating the function of miR-101–3p in tumour tissues, we first determined its expression level in the LC cell lines and tissues, demonstrating that its levels were lower when compared with those in non-tumour cells and tissues (Fig. 1A and B). Additionally, significant associations were revealed between the tissue expression levels of miR-101–3p and the tumour size and TNM stage in LC patients, indicating that miR-101–3p plays a remarkable role in LC proliferation (Fig. 1C and D).
To further explore the role of miR-101–3p in tumour proliferation, a miR-101–3p mimic or inhibitor was individually transfected into A549 cells, and NC or blank control A549 cells were used as control cells. Compared with control cells, the expression levels of miR-101–3p increased significantly in A549 cells transfected with the miR-101–3p mimic, whereas the expression levels of miR-101–3p decreased with the transfection of the miR-101–3p inhibitor (Fig. 2A). With increased levels of miR-101–3p in the mimic group, the proliferative ability of A549 cells was inhibited when compared with that of control groups (Fig. 2B and C). Simultaneously, the expression levels of proliferation markers in LC cells were inhibited by the miR-101–3p mimic (Fig. 2D).
After demonstrating its exact inhibitory effect on proliferation, we further explored the mechanism of these observed effects. Apoptosis serves as a fundamental regulatory function during cancer proliferation. Evidence suggests that miRNAs are involved in proliferation and apoptosis regulation in numerous types of cancer cells [6]. The apoptotic cell ratio in the mimic group was significantly increased and was accompanied by changes in apoptotic markers. In the inhibitor group, cancer cells showed the opposite effects, and apoptosis was inhibited (Fig. 2E and F). Therefore, it was concluded that miR-101–3p potentially suppresses the proliferation process in tumour cells by inducing apoptosis.
miRNAs invariably function by binding to the 3′UTR of their target genes. Reportedly, miR-101–3p produces therapeutic effects through diverse target proteins in different types of tumours. For example, Liang et al. have observed that miR-101–3p inhibits EMT and invasive metastasis in ovarian cancer by regulating the expression of ZEB1 [12]. Fan et al. have identified miR-101–3p as a therapeutic target of ZEB1 in cervical cancer [18]. However, Yan et al. have reported that miR-101–3p has an entirely different target site in liver cancer, possibly SOX9 [23] or Beclin-1 [26]. Although miR-101–3p exhibits universal antitumor effects, it may produce therapeutic effects in different tumours through distinct mechanisms. Therefore, the specific targets of the aforementioned therapeutic effects observed in tumour cells need to be specifically explored.
Bioinformatic analysis has revealed that miR-101–3p may bind to multiple 3′UTR sites of TBLR1 (Position 1935, 2887, and 4689), which may induce the definitive inhibition of protein expression (Fig. 3A). TBLR1 is an oncogene that is reportedly crucial in tumours such as cervical cancer [15] and liver cancer [32]. However, bioinformatic predictions do not necessarily correspond to clinical gene expression. Therefore, we detected the expression level of TBLR1 mRNA in LC clinical specimens (Fig. 1B) and analysed its relationship with the expression level of miR-101–3p. Our findings revealed that the miR-101–3p expression levels significantly and negatively correlated with mRNA expression levels of TBLR1 (Fig. 3B). These results further confirmed the bioinformatic prediction. Therefore, we were interested in further exploring whether TBLR1 is a direct target of miR-101–3p in LC. Using a cell luciferase reporter and eukaryotic plasmid transfection assay, TBLR1 was revealed as a direct target gene of miR-101–3p (Fig. 3C–E). However, functional rescue experiments are needed to confirm whether TBLR1 is associated with changes in proliferation and apoptosis induced by miR-101–3p. The rescue of TBLR1 expression by a eukaryotic expression vector (Fig. 4A) transfected with miR-101–3p may, to some extent, reverse the effects on proliferation caused by increased miR-101–3p expression (Fig. 4B). Furthermore, the increase in TBLR1 expression partially rescued the apoptosis-promoting effect of miR-101–3p (Fig. 4C and D). Additionally, these results confirmed that miR-101–3p inhibited the proliferation of tumour cells by targeting TBLR1.
Recent studies have shown that reduced ferroptotic levels are a critical mechanism for tumorigenesis [5] and may dominate the regulation of apoptosis in tumour cells [22]. Therefore, we further explored whether the LC inhibitory effect of the miR-101–3p/TBLR1 axis arises from the restoration of ferroptosis. Ferroptosis-related functional experiments revealed that miR-101–3p elevated total ROS levels and lipid ROS levels in cells, decreasing GSH levels (Fig. 5A and B). ROS are the predominant class of free radicals, consisting of free oxygen radicals such as superoxide anions (-O2-) and hydroxyl radicals (-OH), and non-free radical ROS such as hydrogen peroxide (H2O2), as well as organic hydroperoxides. Generally, intracellular ROS levels are in a dynamic balance of constant generation and elimination. However, following ferroptosis, the concentration of ROS increases dramatically and can non-specifically attack biomolecules such as DNA, lipids, and proteins, causing apoptosis and inhibiting cell proliferation, ultimately producing therapeutic effects in cancer. GSH regulates redox processes in vivo and binds to ROS to counteract its killing effects. Frequently, elevated GSH levels indicate the inhibition of ferroptosis in tumour cells. Lipid peroxidation end products are reduced to the corresponding alcohols by GPX4 using GSH. The expression levels of ferroptosis markers, GPX4 and PTGS2, were further examined to demonstrate that changes in cellular ROS and GSH were directly related to ferroptosis. Following miR-101–3p treatment, characteristic ferroptosis marker alterations were observed, with decreased GPX4 expression and increased PTGS2 expression (Fig. 5C). Additional rescue experiments revealed that TBLR1 is a direct target of the miR-101–3p regulation of ferroptosis (Fig. 5D). The aforementioned results demonstrate that miR-101–3p produces a restorative effect on ferroptosis through direct inhibition of TBLR1, which, in turn, promotes apoptosis, inhibits proliferation, and ultimately exerts a definitive tumour-killing effect. It must be acknowledged that some scholars believe that there are presently no good definitive markers of ferroptosis. Therefore, changes in GPX4 and PTGS2 may only indicate the degree of ferroptosis.
The definitive tumour-killing effect exhibited by miR-101–3p renders it easier to develop targeted gene therapeutics through effective tumour delivery. It is well known that nucleic acids, including miRNAs, cannot be directly treated in vivo through commonly utilised clinical strategies such as intravenous or oral administration. Gene delivery via viral vectors carries a huge risk of infection or immunological side effects [41]. Therefore, we attempted to use a commercial non-viral nanocarrier to achieve miR-101–3p delivery. Before observing the in vivo therapeutic effect of the nanomedicine, we confirmed that this nanocarrier could promote the in vitro transfection of nucleic acids. Then facilitate their intra-tumoural distribution using fluorescence tracer experiments (Fig. 6A–C). The confirmation that nanocarriers can achieve definitive transfection of tumour cells in vivo holds promise for tumour therapy using screened ferroptosis suppressor genes. In vivo studies were conducted to further clarify the therapeutic effects of miR-101–3p nanomedicines. The in vivo experiments confirmed that the growth rates of subcutaneous tumours were reduced by increasing the expression of miR-101–3p (Fig. 7A–C). As classical pathological methods [35], BrdU immunofluorescent staining and TUNEL immunohistochemical staining showed that the miR-101–3p/TBLR1 axis was closely related to the proliferation and apoptosis of LC cells in vivo. Ferroptosis level assays for total ROS, lipid ROS, and GSH revealed that this in vivo therapeutic effect was attributed to the recovery of ferroptosis in tumour cells in vivo (Fig. 7D, F). Tumour tissues from each group were evaluated for proliferation, apoptosis, and ferroptosis marker expression levels in vivo, and the results further supported the findings of previous in vitro and in vivo studies. Thus, our study provides strong evidence highlighting the effect and mechanism of nanomedicines targeting the miR-101–3p/TBLR1 axis in LC treatment in vivo. More encouragingly, as the reduced expression and well-defined therapeutic targets of miR-101–3p in ovarian cancer [20] and hepatocellular carcinoma [29] have been identified, we attempted in vivo therapeutic experiments with this nanomedicine and observed that it also produced an inhibitory effect on subcutaneous grafts in ovarian and hepatocellular carcinoma (data not shown).
5. Conclusions
In conclusion, we designed a new gene therapy strategy to enable in vivo tumour suppression via the regulation of ferroptosis. Restoration of miR-101–3p expression in tumour cells via reliable nano-transfection agent may be a potential candidate therapeutic technique in various tumours.
Declaration of competing interest
There are no potential conflicts of interest to disclose.
Acknowledgments
We thank the patients, study investigators, and staff who participated in this study. This study was supported by grants from the National Natural Science Foundation of China (No. 82072050, 81902482, 81974364, 1671805, 81602723, 81302550, 81801703, 81770608, 82072029), Basic Research Funds for the Universities of the State Education Ministry, Key Young Teacher Incubation Project (20ykzd04), the National Science Fund for Distinguished Young Scholars (No. 81825013), the National high level talents special support plan-“Ten thousand plan”-Young top-notch talent support program, the Natural Science Foundation of Guangdong Province (No. S2012020011070), the Science and Technology Project of Guangdong Province (No. 2016A020215214, 2017A020215125, 2014A020212152), Special Support Program of Guangdong Province, Science and technology innovation youth talent support program (No. 201627015), the Pearl River Science and Technology New Talent of Guangzhou City (No. 201806010076), the Postdoctoral Science Foundation of China (No. 2013M530382), the Guangdong Medical Research Foundation (No. A2015130), Guangzhou Health Science and Technology Project (No. 20201A011040) and AGILE - KeLin New Talent Program of The First Affiliated Hospital of Sun Yat-sen University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.101908.
Contributor Information
Jing Wang, Email: wangj288@mail.sysu.edu.cn.
Zhenwei Peng, Email: pzhenw@mail.sysu.edu.cn.
Xintao Shuai, Email: shuaixt@mail.sysu.edu.cn.
Yu Guo, Email: guoyu35@mail.sysu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Battaglia A.M., Chirillo R., Aversa I., Sacco A., Costanzo F., Biamonte F. Ferroptosis and cancer: mitochondria meet the "iron maiden" cell death. Cells. 2020;9 doi: 10.3390/cells9061505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez A.M., Kim A., Yang W.S. Detection of ferroptosis by BODIPY 581/591 C11. Methods Mol. Biol. 2020;2108:125–130. doi: 10.1007/978-1-0716-0247-8_11. [DOI] [PubMed] [Google Scholar]
- 3.Jie B., Juan Z., Jing W. MicroRNA-101 inhibits epithelial-mesenchymal transition in human liver carcinoma MHCC97H cells via USP22. Organ Transplantation. 2017;8:209–214. doi: 10.3969/j.issn.1674-7445.2017.03.007. [DOI] [Google Scholar]
- 4.Oh B.M., Lee S.J., Park G.L., Hwang Y.S., Lim J., Park E.S., Lee K.H., Kim B.Y., Kwon Y.T., Cho H.J., Lee H.G. Erastin inhibits septic shock and inflammatory gene expression via suppression of the NF-kappaB pathway. J. Clin. Med. 2019;8 doi: 10.3390/jcm8122210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stockwell B.R., Friedmann A.J., Bayir H., Bush A.I., Conrad M., Dixon S.J., Fulda S., Gascon S., Hatzios S.K., Kagan V.E., Noel K., Jiang X., Linkermann A., Murphy M.E., Overholtzer M., Oyagi A., Pagnussat G.C., Park J., Ran Q., Rosenfeld C.S., Salnikow K., Tang D., Torti F.M., Torti S.V., Toyokuni S., Woerpel K.A., Zhang D.D. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He C., Luo B., Jiang N., Liang Y., He Y., Zeng J., Liu J., Zheng X. Leuk Lymphoma; 2018. OncomiR or antioncomiR: role of miRNAs in acute myeloid leukemia; pp. 1–12. [DOI] [PubMed] [Google Scholar]
- 7.Li C., Deng X., Zhang W., Xie X., Conrad M., Liu Y., Angeli J., Lai L. Novel allosteric activators for ferroptosis regulator glutathione peroxidase 4. J. Med. Chem. 2019;62:266–275. doi: 10.1021/acs.jmedchem.8b00315. [DOI] [PubMed] [Google Scholar]
- 8.Luan C., Li Y., Liu Z., Zhao C. Long noncoding RNA MALAT1 promotes the development of colon cancer by regulating miR-101-3p/STC1 Axis. OncoTargets Ther. 2020;13:3653–3665. doi: 10.2147/OTT.S242300. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Wang C., Guo Y., Wang J., Min Z. The suppressive role of SOX7 in hepatocarcinogenesis. PloS One. 2014;9 doi: 10.1371/journal.pone.0097433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang C., Guo Y., Wang J., Min Z. Annexin A2 knockdown inhibits hepatoma cell growth and sensitizes hepatoma cells to 5fluorouracil by regulating betacatenin and cyclin D1 expression. Mol. Med. Rep. 2015;11:2147–2152. doi: 10.3892/mmr.2014.2906. [DOI] [PubMed] [Google Scholar]
- 11.Chow E.K., Ho D. Cancer nanomedicine: from drug delivery to imaging. Sci. Transl. Med. 2013;5:214r–216r. doi: 10.1126/scitranslmed.3005872. [DOI] [PubMed] [Google Scholar]
- 12.Liang H., Yu T., Han Y., Jiang H., Wang C., You T., Zhao X., Shan H., Yang R., Yang L., Shan H., Gu Y. LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression. Mol. Canc. 2018;17:119. doi: 10.1186/s12943-018-0870-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao H., Guo Y., Li B., Li X., Wang Y., Han S., Cheng D., Shuai X. M2-Like tumor-associated macrophage-targeted codelivery of STAT6 inhibitor and IKKbeta siRNA induces M2-to-M1 repolarization for cancer immunotherapy with low immune side effects. ACS Cent. Sci. 2020;6:1208–1222. doi: 10.1021/acscentsci.9b01235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J., Shen J.X., He, Zhang G.J. Bioluminescence imaging for monitoring miR-200c expression in breast cancer cells and its effects on epithelial-mesenchymal transition progress in living animals. Mol. Imag. Biol. 2018;20:761–770. doi: 10.1007/s11307-018-1180-4. [DOI] [PubMed] [Google Scholar]
- 15.Wang J., Ou J., Guo Y., Dai T., Li X., Liu J., Xia M., Liu L., He M. TBLR1 is a novel prognostic marker and promotes epithelial-mesenchymal transition in cervical cancer. Br. J. Canc. 2014;111:112–124. doi: 10.1038/bjc.2014.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li K., Jiang Y., Xiang X., Gong Q., Zhou C., Zhang L., Ma Q., Zhuang L. Long non-coding RNA SNHG6 promotes the growth and invasion of non-small cell lung cancer by downregulating miR-101-3p. Thorac Cancer. 2020;11:1180–1190. doi: 10.1111/1759-7714.13371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu L., Beckebaum S., Iacob S., Wu G., Kaiser G.M., Radtke A., Liu C., Kabar I., Schmidt H.H., Zhang X., Lu M., Cicinnati V.R. MicroRNA-101 inhibits human hepatocellular carcinoma progression through EZH2 downregulation and increased cytostatic drug sensitivity. J. Hepatol. 2014;60:590–598. doi: 10.1016/j.jhep.2013.10.028. [DOI] [PubMed] [Google Scholar]
- 18.Fan M.J., Zou Y.H., He P.J., Zhang S., Sun X.M., Li C.Z. Long non-coding RNA SPRY4-IT1 promotes epithelial-mesenchymal transition of cervical cancer by regulating the miR-101-3p/ZEB1 axis. Biosci. Rep. 2019;39 doi: 10.1042/BSR20181339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gasparri M.L., Besharat Z.M., Farooqi A.A., Khalid S., Taghavi K., Besharat R.A., Sabato C., Papadia A., Panici P.B., Mueller M.D., Ferretti E. MiRNAs and their interplay with PI3K/AKT/mTOR pathway in ovarian cancer cells: a potential role in platinum resistance. J. Canc. Res. Clin. Oncol. 2018;144(12):2313–2318. doi: 10.1007/s00432-018-2737-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiczling P., Daghir-Wojtkowiak E., Kaliszan R., Markuszewski M.J., Limon J., Koczkowska M., Stukan M., Kuzniacka A., Ratajska M. Bayesian multilevel model of micro RNA levels in ovarian-cancer and healthy subjects. PloS One. 2019;14 doi: 10.1371/journal.pone.0221764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo R., Wu Z., Wang J., Li Q., Shen S., Wang W., Zhou L., Wang W., Cao Z., Guo Y. Development of a non-coding-RNA-based EMT/CSC inhibitory nanomedicine for in vivo treatment and monitoring of HCC. Adv. Sci. 2019;6:1801885. doi: 10.1002/advs.201801885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thermozier S., Hou W., Zhang X., Shields D., Fisher R., Bayir H., Kagan V., Yu J., Liu B., Bahar I., Epperly M.W., Wipf P., Wang H., Huq M.S., Greenberger J.S. Anti-ferroptosis drug enhances total-body irradiation mitigation by drugs that block Apoptosis and necroptosis. Radiat. Res. 2020;193:435–450. doi: 10.1667/RR15486.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan S., Shan X., Chen K., Liu Y., Yu G., Chen Q., Zeng T., Zhu L., Dang H., Chen F., Ling J., Huang A., Tang H. LINC00052/miR-101-3p axis inhibits cell proliferation and metastasis by targeting SOX9 in hepatocellular carcinoma. Gene. 2018;679:138–149. doi: 10.1016/j.gene.2018.08.038. [DOI] [PubMed] [Google Scholar]
- 24.Zhang T., Liu W., Zeng X.C., Jiang N., Fu B.S., Guo Y., Yi H.M., Li H., Zhang Q., Chen W.J., Chen G.H. Down-regulation of microRNA-338-3p promoted angiogenesis in hepatocellular carcinoma. Biomed. Pharmacother. 2016;84:583–591. doi: 10.1016/j.biopha.2016.09.056. [DOI] [PubMed] [Google Scholar]
- 25.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun W., Zhang Q., Wu Z., Xue N. miR-101-3p sensitizes hepatocellular carcinoma cells to oxaliplatin by inhibiting Beclin-1-mediated autophagy. Int. J. Clin. Exp. Pathol. 2019;12:2056–2065. [PMC free article] [PubMed] [Google Scholar]
- 27.Wei W., Dong Z., Gao H., Zhang Y.Y., Shao L.H., Jin L.L., Lv Y.H., Zhao G., Shen Y.N., Jin S.Z. MicroRNA-9 enhanced radiosensitivity and its mechanism of DNA methylation in non-small cell lung cancer. Gene. 2019;710:178–185. doi: 10.1016/j.gene.2019.05.050. [DOI] [PubMed] [Google Scholar]
- 28.Xie W., Lu Q., Wang K., Lu J., Gu X., Zhu D., Liu F., Guo Z. miR-34b-5p inhibition attenuates lung inflammation and apoptosis in an LPS-induced acute lung injury mouse model by targeting progranulin. J. Cell. Physiol. 2018;233:6615–6631. doi: 10.1002/jcp.26274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X., Zhang N. Downregulation of lncRNA NEAT1_2 radiosensitizes hepatocellular carcinoma cells through regulation of miR-101-3p/WEE1 axis. Cell Biol. Int. 2019;43:44–55. doi: 10.1002/cbin.11077. [DOI] [PubMed] [Google Scholar]
- 30.Ye X., Guo Y., Zhang Q., Chen W., Hua X., Liu W., Yang Y., Chen G. βKlotho suppresses tumor growth in hepatocellular carcinoma by regulating Akt/GSK-3β/Cyclin D1 signaling pathway. PloS One. 2013;8 doi: 10.1371/journal.pone.0055615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo Y., Wang J., Li H., Liu W., Chen D., Zhao K., Liang X., Zhang Q., Yang Y., Chen G. Mediator subunit 23 overexpression as a novel target for suppressing proliferation and tumorigenesis in hepatocellular carcinoma. J. Gastroenterol. Hepatol. 2015;30:1094–1103. doi: 10.1111/jgh.12923. [DOI] [PubMed] [Google Scholar]
- 32.Guo Y., Wang J., Zhang L., Shen S., Guo R., Yang Y., Chen W., Wang Y., Chen G., Shuai X. Theranostical nanosystem-mediated identification of an oncogene and highly effective therapy in hepatocellular carcinoma. Hepatology. 2016;63:1240–1255. doi: 10.1002/hep.28409. [DOI] [PubMed] [Google Scholar]
- 33.Guo Y., Chen W., Wang W., Shen J., Guo R., Gong F., Lin S., Cheng D., Chen G., Shuai X. Simultaneous diagnosis and gene therapy of immuno-rejection in rat allogeneic heart transplantation model using a T-cell-targeted theranostic nanosystem. ACS Nano. 2012;6:10646–10657. doi: 10.1021/nn3037573. [DOI] [PubMed] [Google Scholar]
- 34.Guo Y., Liang X., Lu M., Weng T., Liu Y., Ye X. Mammalian target of rapamycin as a novel target in the treatment of hepatocellular carcinoma. Hepato-Gastroenterology. 2009;57:913–918. [PubMed] [Google Scholar]
- 35.Guo Y., Wu Z., Shen S., Guo R., Wang J., Wang W., Zhao K., Kuang M., Shuai X. Nanomedicines reveal how PBOV1 promotes hepatocellular carcinoma for effective gene therapy. Nat. Commun. 2018;9:3430. doi: 10.1038/s41467-018-05764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mei Y., Jiang P., Shen N., Fu S., Zhang J. Identification of miRNA-mRNA regulatory network and construction of prognostic signature in cervical cancer. DNA Cell Biol. 2020;39:1023–1040. doi: 10.1089/dna.2020.5452. [DOI] [PubMed] [Google Scholar]
- 37.Xu Y., An Y., Wang Y., Zhang C., Zhang H., Huang C., Jiang H., Wang X., Li X. miR-101 inhibits autophagy and enhances cisplatin-induced apoptosis in hepatocellular carcinoma cells. Oncol. Rep. 2013;29:2019–2024. doi: 10.3892/or.2013.2338. [DOI] [PubMed] [Google Scholar]
- 38.Wu Y.F., Liang X.J., Liu Y.Y., Gong W., Liu J.X., Wang X.P., Zhuang Z.Q., Guo Y., Shen H.Y. +Antisense oligonucleotide targeting survivin inhibits growth by inducing apoptosis in human osteosarcoma cells MG-63. Neoplasma. 2010;57:501–506. doi: 10.4149/neo_2010_06_501. [DOI] [PubMed] [Google Scholar]
- 39.Wu Z., Zhao J., Qiu M., Mi Z., Meng M., Guo Y., Wang H., Yuan Z. CRISPR/Cas9 mediated GFP knock-in at the MAP1LC3B locus in 293FT cells is better for bona fide monitoring cellular autophagy. Biotechnol. J. 2018 doi: 10.1002/biot.201700674. [DOI] [PubMed] [Google Scholar]
- 40.Wu Z., Qiu M., Mi Z., Meng M., Guo Y., Jiang X., Fang J., Wang H., Zhao J., Liu Z., Qian D., Yuan Z. WT1-interacting protein inhibits cell proliferation and tumorigenicity in non-small-cell lung cancer via the AKT/FOXO1 axis. Mol Oncol. 2019;13:1059–1074. doi: 10.1002/1878-0261.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Z., Liu X., Zhu D., Wang Y., Zhang Z., Zhou X., Qiu N., Chen X., Shen Y. Nonviral cancer gene therapy: delivery cascade and vector nanoproperty integration. Adv. Drug Deliv. Rev. 2017;115:115–154. doi: 10.1016/j.addr.2017.07.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.