Abstract
Oxidative stress is emerging as an important contributor to the pathogenesis of colorectal cancer (CRC), however, the molecular mechanisms by which the disturbed redox balance regulates CRC development remain undefined. Using a liquid chromatography–tandem mass spectrometry-based lipidomics, we found that epoxyketooctadecenoic acid (EKODE), which is a lipid peroxidation product, was among the most dramatically increased lipid molecules in the colon of azoxymethane (AOM)/dextran sodium sulfate (DSS)-induced CRC mice. This is, at least in part, due to increased oxidative stress in colon tumors, as assessed by analyzing gene expression of oxidative markers in AOM/DSS-induced CRC mice and human CRC patients in the Cancer Genome Atlas (TCGA) database. Systemic, short-time treatment with low-dose EKODE increased the severity of DSS-induced colitis, caused intestinal barrier dysfunction and enhanced lipopolysaccharide (LPS)/bacterial translocation, and exacerbates the development of AOM/DSS-induced CRC in mice. Furthermore, treatment with EKODE, at nM doses, induced inflammatory responses via JNK-dependent mechanisms in both colon cancer cells and macrophage cells. Overall, these results demonstrate that the lipid peroxidation product EKODE is an important mediator of colonic inflammation and colon tumorigenesis, providing a novel mechanistic linkage between oxidative stress and CRC development.
Keywords: Oxidative stress, EKODE, Lipidomics, Colonic inflammation, Colorectal cancer
1. Introduction
Colorectal cancer (CRC) is the third most common cancer and the second leading cause of cancer-related death in the United States [1], emphasizing the need for discovery of novel cellular targets which are crucial in the pathogenesis of CRC. Oxidative stress, which is a common feature of many human diseases including CRC, is emerging as an important contributor to the development of CRC [2,3]. However, the molecular mechanisms by which the disturbed redox balance promotes CRC development remain undefined [2]. It is important to better understand the underlying mechanisms, in order to identify novel pathogenic factors of CRC, facilitating development of diagnosis markers and/or intervention targets to reduce the risks of CRC.
Emerging research supports that lipid metabolites, such as enzymatic metabolites of fatty acids (e.g. eicosanoids) and non-enzymatic oxidation products of fatty acids (e.g. lipid peroxidation products), are important signaling molecules that regulate inflammation and tumorigenesis [[4], [5], [6]]. In an effort to systematically analyze how lipid metabolites are deregulated in CRC, we performed a liquid chromatography–tandem mass spectrometry (LC-MS/MS)-based lipidomics, which can measure >100 lipid metabolites derived from enzymatic and non-enzymatic processes, in a well-established azoxymethane (AOM)/dextran sodium sulfate (DSS)-induced CRC model in mice [7]. Here, we found that epoxyketooctadecenoic acid (EKODE), which was a lipid peroxidation-derived compound, was among the most dramatically increased lipid metabolites in the colon tissues of AOM/DSS-induced CRC mice compared to those of the healthy mice (Fig. 1). This leads to our hypothesis that formation of EKODE could provide a novel mechanistic linkage between oxidative stress and CRC development. To date, the effects of EKODE on inflammation and cancer are unknown. To test our hypothesis, here we determined the functional roles of EKODE in regulating colonic inflammation and colon carcinogenesis in both animal and cell culture models.
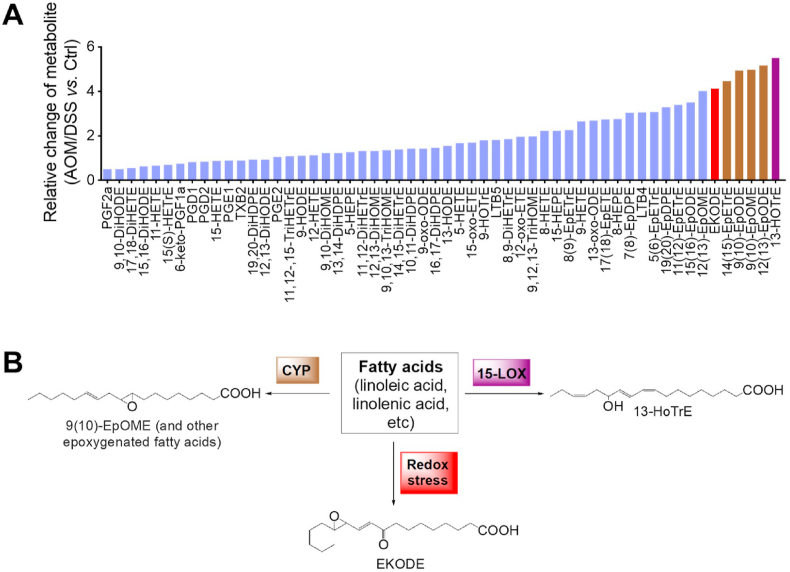
Fig. 1.
Lipidomics analysis showed that EKODE was among the most dramatically increased lipids in the colon of AOM/DSS-induced CRC mice. A, LC-MS/MS profiling of bioactive lipids in the colon, the Y-axis is expressed as relative change of lipid metabolite in the colon of AOM/DSS-induced CRC mice (n = 10 mice per group) vs. control mice (n = 6 mice per group). B, Structures and biosynthetic scheme of 13-HoTrE, epoxygenated fatty acids (using EpOME as an example), and EKODE.
2. Materials and methods
2.1. Animal experiments
All animal experiments were conducted in accordance with the protocols approved by the Institutional Animal Care and Use Committee of the University of Massachusetts Amherst. C57BL/6 male mice (Charles River) were maintained in a standard animal facility at the University of Massachusetts Amherst.
2.2. Animal protocol 1: DSS-induced colitis in mice
C57BL/6 mice (age = 6 weeks) were maintained on a modified AIN-93G diet (see diet composition in Supplementary Table S1) throughout the experiment. After 2 weeks of diet treatment, the mice were treated with drinking water with or without 2% DSS (MP Biomedicals) for 1 week. At end of the experiment, the mice were sacrificed to collect tissues for analysis, as we described [8].
2.3. Animal protocol 2: AOM/DSS-induced CRC in mice
C57BL/6 mice (age = 6 weeks) were maintained on a modified AIN-93G diet throughout the experiment. After 2 weeks of diet treatment, the mice were divided into two groups: (1) the mice in the CRC group were treated with 10 mg/kg AOM (Sigma-Aldrich) via intraperitoneal injection; after 1 week, the mice were stimulated with 2% DSS in drinking water for 1 week; and (2) the mice in the control group were not treated with AOM or DSS. At week 9.5 post the AOM injection, the mice were sacrificed for analysis, as we described [7,8].
2.4. Animal protocol 3: effects of EKODE on DSS-induced colitis in mice
C57BL/6 mice (age = 6 weeks) were maintained on a standard mouse chow, and treated with 2% DSS in drinking water to induce colitis, as well as intraperitoneal injection with EKODE (dose = 1 mg/kg/day, Cayman Chemical) or vehicle DMSO (volume = 20 μL). After 1 week, the mice were sacrificed to collect blood and colon tissues for analysis.
2.5. Animal protocol 4: effects of EKODE on AOM/DSS-induced CRC
C57BL/6 mice (age = 6 weeks) were maintained on a standard mouse chow and treated with 10 mg/kg AOM via intraperitoneal injection. At week 1 post the AOM injection, they were treated with 2% DSS in drinking water for 1 week. At week 3 post the AOM injection, the mice were treated with EKODE (dose = 1 mg/kg/day) or vehicle DMSO (volume = 20 μL) via intraperitoneal injection for 10 days. At week 9.5 post the AOM injection, the mice were sacrificed to collect blood and colon tissues for analysis.
2.6. Flow cytometry analysis of immune cell infiltration in colon tissues
Distal colon tissues were dissected and digested with Hank's-balanced salt solution (Lonza) supplemented with 1 mM dithiothreitol (DTT) and 5 mM EDTA overnight at 4 °C. After filtering through 70 μm cell strainer (BD Biosciences), the single-cell suspensions were stained with FITC-conjugated anti-mouse CD45 antibody, PerCP/Cy5.5-conjugated anti-mouse F4/80 antibody, PE/Cy7-conjugated anti-mouse Ly-6G/Ly-6C (GR-1) antibody, isotype control antibody and Zombie Violet™ dye (BioLegend). Data were acquired using BD LSRFortessa™ cell analyzer (BD Biosciences) and analyzed using FlowJo software (FlowJo LLC). In our analysis, leukocytes were identified as CD45+ cells, macrophages were identifed as CD45+ F4/80+ cells, and neutrophils were identified as CD45+ GR-1+ cells.
2.7. H&E staining and immunohistochemistry
Dissected colon tissues were fixed in formalin (Thermo Fisher Scientific) for 48 h. Then the tissues were embedded in paraffin, sliced (5-μm), dewaxed in serial xylene (Thermo Fisher Scientific) and rehydrated through graded ethanol solutions (Pharmco-Aaper). For H&E staining, the slides are stained with hematoxylin and eosin (SigmaAldrich), and examined with a light microscopy (Nikon Instruments). The histological scores were evaluated by blind observers according to the following measures: crypt architecture, degree of inflammatory cell infiltration, muscle thickening, goblet cell depletion and crypt abscess. The pathological score is the sum of each individual score. For immunohistochemistry, antigen retrieval was performed by heating the sections in 0.01 M citrate buffer (pH 6.0) to 95 °C for 10 min. The slides were incubated with primary antibodies against occludin, proliferating cell nuclear antigen (PCNA) and β-catenin (Cell Signaling Technology) overnight at 4 °C. Horseradish peroxidase (HRP)-conjugated secondary antibodies were then applied to the sections, followed by the chromogen 4-diaminobenzidine staining according to the instructions of HRP/DAB (ABC) Detection IHC Kit (Abcam). Sections were then counterstained with hematoxylin for 1 min and observed under a light microscope (Nikon Instruments).
2.8. qRT-PCR analysis of gene expression in tissues and cells
For tissues analysis, colon tissues from the same locations or spleen tissues were grounded after freezing in liquid nitrogen and then added TRIzol reagent (Ambion). For cell assay, the HCT-116 or RAW 264.7 cells were treated with 300 nM EKODE or DMSO vehicle in complete medium for 24 h, then the cells were washed with cold PBS and added with TRIzol reagent. Total RNA was isolated from tissues or cells based on manufacturer's instructions of TRIzol reagent. The concentration and quality of RNA was measured by NanoDrop Spectrophotometer (Thermo Fisher Scientific), then the RNA was reverse transcribed into cDNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems) according to manufacturer's instructions. qRT-PCR was performed using a DNA Engine Opticon system (Bio-Rad Laboratories) with Maxima SYBR-green Master Mix (Thermo Fisher Scientific). The sequences of primers are listed in Supplementary Table S2. The results of target genes were normalized to glyceraldehyde-3-phosphate dehydrogenase (Gapdh) and expressed to control mice using the 2 -ΔΔCt method.
2.9. qRT-PCR analysis of 16S rRNA in blood and spleen tissues
Blood (~100 μL) and spleen tissues (~30 mg) were collected for analyzing the bacterial load. The total DNA was extracted using QIAamp DNAeasy Blood & Tissue Kit (Qiagen) following the manufacturer's instruction with the addition of a bead-beating step. The quality of the extracted DNA was measured using a NanoDrop Spectrophotometer (Thermo Scientific) and qRT-PCR was performed using the same amount of DNA (5 ng/mL) with the Maxima SYBR green Master Mix (Thermo Fisher Scientific). The sequences of mouse-specific primer are listed in Supplementary Table S2.
2.10. Measurement of cytokines and lipopolysaccharide (LPS) in plasma
The concentration of MCP-1 in plasma was determined using a CBA Mouse Inflammation Kit (BD Biosciences). The concentration of LPS in plasma was quantified using an ELISA kit (MBS261904, MyBioSource) following the manufacturer's instructions.
2.11. Protein extraction and immunoblotting
For total protein extraction, RAW 264.7 or HCT -116 cells were treated with 300 nM EKODE or DMSO vehicle in complete medium for 5–60min, then the cells were washed with cold PBS and lysed by RIPA lysis buffer with a protease inhibitor cocktail (Boston BioProducts). For nuclear protein extraction, nuclear and cytoplasmic extraction reagents (#78833, Thermo Scientific) were used following the manufacturer's instructions. Protein concentrations were determined using BCA protein assay kit (Thermo Scientific). The samples with equal amount of protein (20 μg) were resolved using SDS/PAGE and transferred onto a nitrocellulose membrane (LI-COR). The membrane was blocked in 5% bovine serum albumin (BSA, Thermo Fisher Scientific) buffer for 1 h at room temperature, then incubated with primary antibodies against phospho-JNK, JNK, IκBα, p65, Lamin, IL6 (Cell Signaling Technology) and β-actin (Sigma-Aldrich) in 5% BSA solution at 4 °C overnight. The membrane was then probed with LI-COR IRDye 800 C W goat anti-rabbit and IRDye 680RD goat anti-mouse secondary antibodies and then detected using the Odyssey imaging system (LI-COR). Quantification of immunoblotting was performed using ImageJ Software (NIH). Data are normalized against those of the corresponding β-actin.
2.12. LC-MS/MS analysis
To extract lipid metabolites from colon tissues, ~100 mg tissues were mixed with an antioxidant solution (0.2 mg/mL butylated hydroxytoluene and 0.2 mg/mL triphenylphosphine in methanol), deuterated internal standards, and 400 μL extract solution (0.1% acetic acid with 0.2 mg/mL butylated hydroxytoluene in a methanol solution), and then homogenized; the resulting homogenates were kept in −80 °C overnight. After centrifugation of the homogenates, the pellets were washed with methanol (containing 0.1% butylated hydroxytoluene and 0.1% acetic acid) and then combined with the supernatant. The combined solutions were loaded onto pre-washed Waters® Oasis solid phase extraction (SPE) cartridges, washed with a solution of 95:5 water/methanol with 0.1% acetic acid, the analytes were eluted with methanol and ethyl acetate, dried using a centrifugal vacuum evaporator, then reconstituted in methanol for LC-MS/MS analysis. The LC-MS/MS analysis was carried out using an Agilent 1200SL HPLC system (Agilent, Santa Clara, CA) coupled to a 4000 QTRAP MS/MS (AB Sciex, Foster City, CA) as described in our previous report [7].
2.13. Data analysis
Data are expressed as mean ± SEM. For the comparison between two groups, Shapiro-Wilk test was used to verify the normality of data; when data were normally distributed, statistical significance was determined using two-side t-test; otherwise, significance was determined by Wilcoxon–Mann–Whitney test. Analysis of four groups (e.g. roles of JNK signaling in the proinflammatory effect of EKODE) was performed using two-way ANOVA. The statistical analyses were performed using GraphPad Prism 6 software, and P values less than 0.05 were considered statistically significant. Gene expression data of ROS markers in CRC and nontumor were derived from The Cancer Genome Atlas (TCGA) database through the UCSC Xena dataset (https://xena.ucsc.edu/).
3. Results
3.1. EKODE is increased in the colon tissue of CRC mice
In our previous study, we used an LC-MS/MS-based lipidomics to systematically analyze how lipid metabolites are deregulated in an AOM/DSS-induced CRC model in mice [7]. We re-analyzed the lipidomics data (see heat-map analysis in Supplementary Fig. S1). Among the lipid molecules detected in the colon (56 compounds in total), the most dramatically increased compounds include three classes of lipids: (i) 15-lipoxygegnase (LOX)-derived 13-hydroxyoctadecatrienoic acid (13-HoTrE), (ii) CYP-derived epoxygenated fatty acids including 9 (10)-epoxyoctadecenoic acid (EpOME), 9(10)-, 12(13)-epoxyoctadecadienoic acid (EpODE), and 14 (15)- epoxyeicosatrienoic acid (EET), and (iii) oxidative stress-derived EKODE (Fig. 1A). Previous research by us and others have shown that the 15-LOX- and CYP-derived lipid metabolites are important mediators of CRC [7,9], while the roles of EKODE in CRC are unknown. Therefore, here we focused on EKODE.
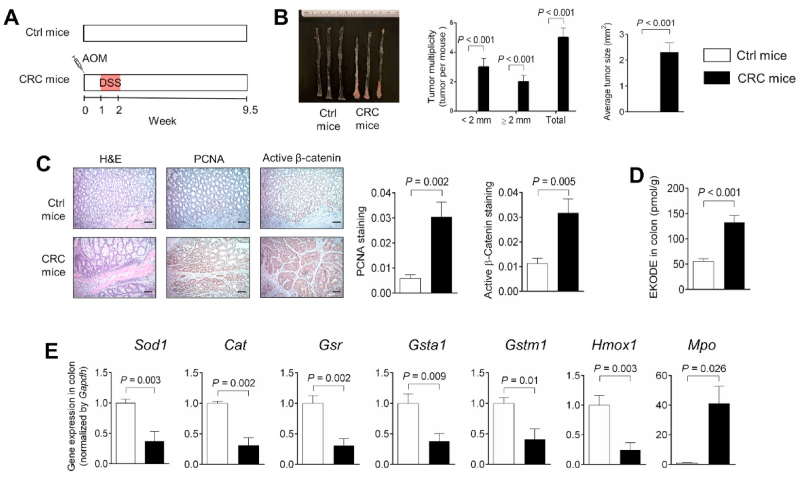
EKODE is produced when reactive oxygen species attack membrane phospholipids [10] (Fig. 1B). We hypothesize that the colon tissues of CRC mice have more severe oxidative stress, leading to higher concentrations of EKODE. To test this hypothesis, we analyzed expression of oxidative markers in the colon of control healthy mice vs. AOM/DSS-induced CRC mice (see scheme of experiment in Fig. 2A). First, we analyzed colon tumorigenesis in the mice. The control healthy mice (not treated with AOM/DSS) had no tumors in the colon, while the AOM/DSS-treated mice had an average of ~5 tumors per mouse (Fig. 2B), with high expression of PCNA and active β-catenin in the colon (Fig. 2C). In agreement with our results above (Fig. 1A), the AOM/DSS-induced CRC mice had higher concentration of EKODE in the colon (Fig. 2D), further supporting that EKODE is increased in CRC. Next, we analyzed expression of oxidative markers in the colon of the mice. Compared with control mice, the CRC mice had lower expression of anti-oxidative genes, including Sod1 (encoding superoxide dismutase 1), Cat (encoding catalase), Gsr (encoding glutathione-disulfide reductase), Gsta1 (encoding glutathione S-transferase A1), Gstm1 (encoding glutathione S-transferase M1), and Hmox1 (encoding heme oxygenase-1) in the colon. In addition, the CRC mice had higher expression of a pro-oxidative gene Mpo (encoding myeloperoxidase) in the colon (Fig. 2E). Overall, these results suggest that the CRC mice have more severe oxidative stress in the colon.
Fig. 2.
Oxidative stress and EKODE are increased in the colon of AOM/DSS-induced CRC mice. A, Scheme of animal experiment. B, Quantification of colon tumor in mice (n = 7–8 mice per group). C, H&E histology and IHC staining of PCNA and β-catenin in colon (n = 7 mice per group, scale bars: 50 μm). D, Concentration of EKODE in colon (n = 6–7 mice per group). E, Gene expression of Sod1, Cat, Gsr, Gsta1, Gstm1, Hmox1 and Mpo in colon (n = 6–7 mice per group). The results are mean ± SEM. The statistical significance of two groups was determined using Student's t-test or Wilcoxon-Mann-Whitney test.
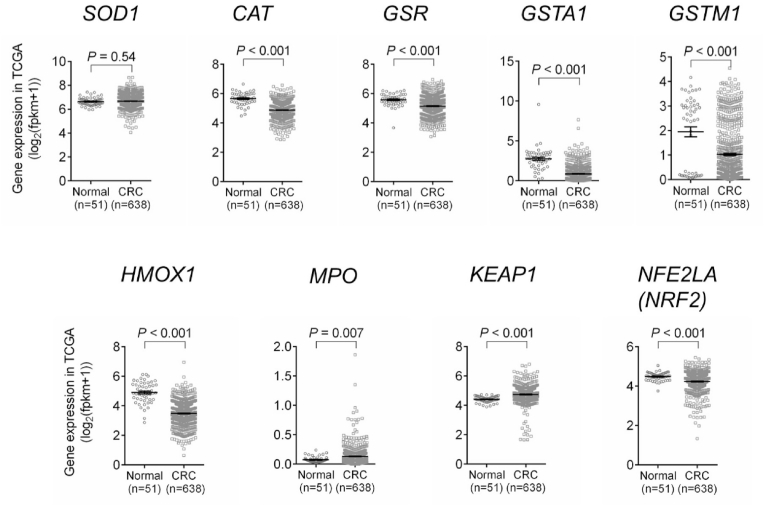
After demonstrating that oxidative markers are altered in the mouse model of CRC, we analyzed their expressions in human CRC patients using the TCGA database. Compared with normal controls, the expression of anti-oxidative genes (CAT, GSR, GSTA1, GSTM1, and HMOX1) were significantly decreased, while the expression of the pro-oxidative gene MPO was increased, in tumor samples of human CRC patients (Fig. 3). Sod1 was reduced in mouse colon tumors (Fig. 2E), but it was not changed in human CRC patients (Fig. 3). We also analyzed other oxidative markers in the TCGA database. Glutathione peroxidase (GPX) is an important redox protein [3]. We found that compared with normal controls, the expressions of GPX1, GPX2, GPX4, GPX7, and GPX8 were increased, while the expression of GPX3 was decreased, GPX5 and GPX6 were not changed, in CRC patients (Fig. S2). Since many of these oxidative markers are regulated by the Nrf2 pathway [3], we also analyzed the expressions of KEAP1 (a negative regulator of Nrf2 pathway) and NRF2. The expression of KEAP1 is increased, while the expression of NRF2 is decreased, in CRC patients compared with controls (Fig. 3). Overall, these results are largely consistent with our mouse data (Fig. 2E), supporting that there is a more severe oxidative microenvironment in colon tumors compared with normal colon tissues.
Fig. 3.
TCGA databased showed that the expressions of antioxidant genes (CAT, GSR, GSTA1, GSTM1, HMOX1, and NRF2) were reduced, while the expressions of pro-oxidant genes (MPO and KEAP1) were increased in CRC patients. The results are mean ± SEM. The statistical significance of two groups was determined using Student's t-test or Wilcoxon-Mann-Whitney test.
3.2. EKODE exacerbates colonic inflammation in mice
To understand the effects of EKODE on colonic inflammation, we determined the effect of EKODE on development of DSS-induced colitis in C57BL/6 mice. We treated mice with DSS, as well as EKODE (dose = 1 mg/kg/day, via intraperitoneal injection) or vehicle (see scheme of animal experiment in Fig. 4A, and validating experiment of the DSS model in Supplementary Fig. S3). We used the dose of 1 mg/kg/day, since a previous study showed that intraperitoneal injection of a similar lipid oxidation compound 4-hydroxynonenal (4-HNE), at a higher dose (5 mg/kg/day), caused no toxic effect in mice [11]. Therefore, this experimental design allows us to study the impact of low-dose EKODE on colitis.
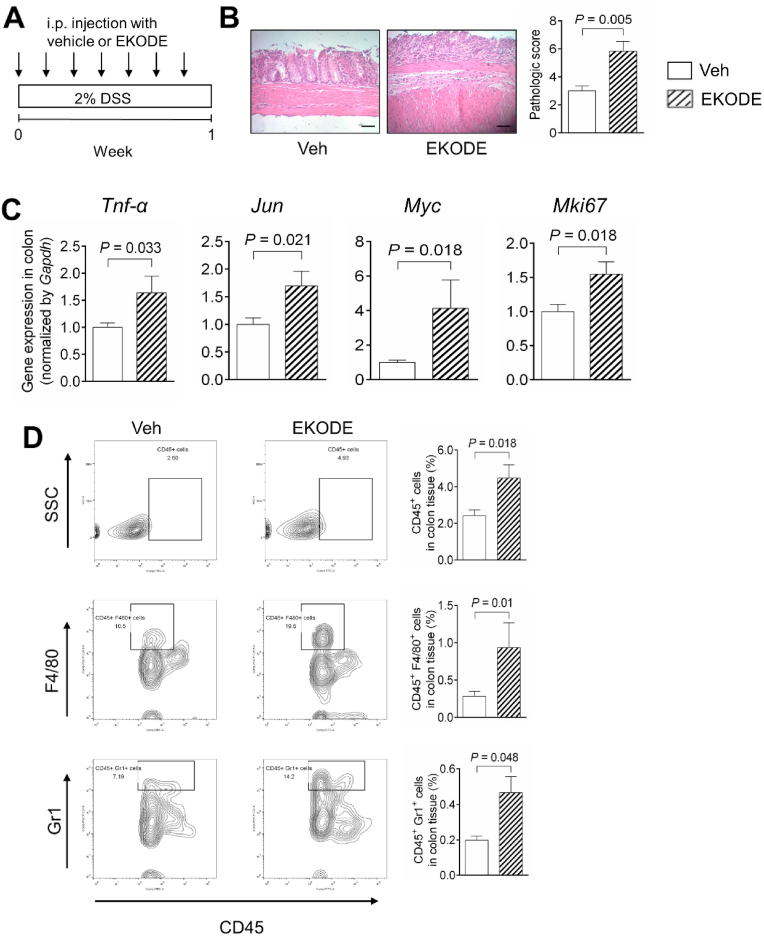
Fig. 4.
EKODE increases DSS-induced colitis in mice. A, Scheme of animal experiment. The dose of EKODE is 1 mg/kg/day, administered via intraperitoneal injection. B, H&E staining of colon (n = 6 mice per group, scale bars: 50 μm). C, Gene expression of Tnf-α, Jun, Myc and Mki67 in colon (n = 4–7 mice per group). D, FACS quantification of immune cells in colon (n = 5–7 mice per group). The results are mean ± SEM. The statistical significance of two groups was determined using Student's t-test or Wilcoxon-Mann-Whitney test.
We found that EKODE treatment exaggerated DSS-induced colitis in mice. Compared with vehicle-treated DSS mice, the EKODE-treated DSS mice had more severe crypt damage in the colon (Fig. 4B), enhanced colonic expression of genes that are associated with inflammation and cell proliferation (Tnf-α, Jun, Myc, and Mki67) (Fig. 4C), higher protein expression levels of phosphorylated JNK in the colon (Fig. S4A), and higher infiltration of immune cells (CD45+ leukocytes, CD45+ F4/80+ macrophages, and CD45+ Gr1+ neutrophils) in the colon (Fig. 4D). Overall, these results demonstrate that treatment with low-dose EKODE increases the severity of DSS-induced colitis in mice, demonstrating its potent colitis-enhancing effect. We also analyzed the effect of EKODE on expression of Hmox1, which is a down-stream target of the Nrf2 pathway [3], and found that EKODE treatment did not change colonic expression of Hmox1 in mice (Fig. S4B).
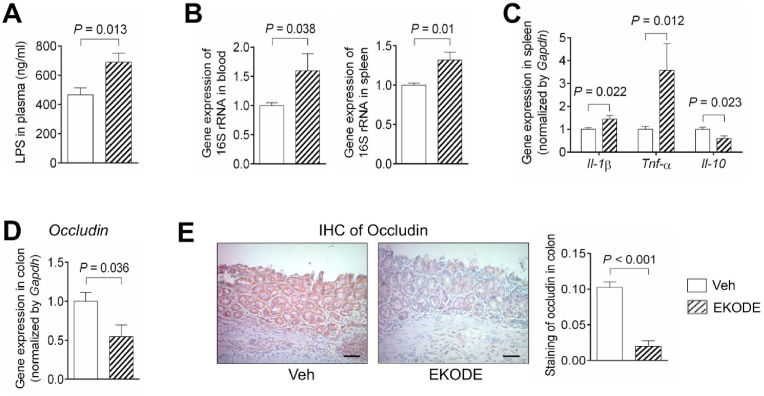
Colitis is associated with intestinal barrier dysfunction, leading to translocation of LPS and bacteria from the gut into bloodstream and other organs [12]. We analyzed whether EKODE treatment exaggerated bacteria/LPS translocation in the DSS-induced colitis model. Compared with vehicle-treated DSS mice, the EKODE-treated DSS mice had a higher concentration of LPS in the plasma (Fig. 5A), and higher levels of bacteria (as assessed by gene expression of 16S rRNA) in the blood and spleen (Fig. 5B), demonstrating that EKODE treatment exaggerated bacteria/LPS translocation. Bacterial invasion into tissues could lead to tissue inflammation [12]. We analyzed whether EKODE treatment exaggerated spleen inflammation. Compared with vehicle-treated DSS mice, the spleen tissues of EKODE-treated DSS mice had increased expression of pro-inflammatory cytokines Tnf-α and Il-1β and reduced expression of an anti-inflammatory cytokine Il-10, demonstrating that EKODE treatment exaggerated spleen inflammation (Fig. 5C). Overall, these results demonstrate that EKODE treatment disrupted intestinal barrier function, leading to enhanced LPS/bacterial translocation and resulting in bacteria invasion-induced tissue inflammation.
Fig. 5.
EKODE induces intestinal barrier dysfunction and increases LPS/bacterial translocation. A, LPS concentration in plasma (n = 6–8 mice per group). B, Gene expression of 16S rRNA gene in blood and spleen (n = 4–6 mice per group). C, Gene expression of Il-1β, Tnf-α and Il-10 in spleen (n = 4–7 mice per group). D, Gene expression of Occludin in colon (n = 5–7 mice per group). E, IHC staining of Occludin in colon (n = 6–7 mice per group, scale bars: 50 μm). The results are mean ± SEM. The statistical significance of two groups was determined using Student's t-test or Wilcoxon-Mann-Whitney test.
To understand the mechanisms by which EKODE induced intestinal barrier dysfunction, we analyzed colonic expression of Occludin, which is a tight-junction protein involved in regulation of intestinal barrier function [13]. We found that EKODE treatment reduced gene expression of Occludin in the colon (Fig. 5D). This finding is further validated by immunohistochemical staining, which showed that EKODE reduced protein expression of Occludin in the colon (Fig. 5E). Overall, these results suggest that EKODE treatment disrupted intestinal barrier function, at least in part, through reducing colonic expression of Occludin.
3.3. EKODE exacerbates colon tumorigenesis in mice
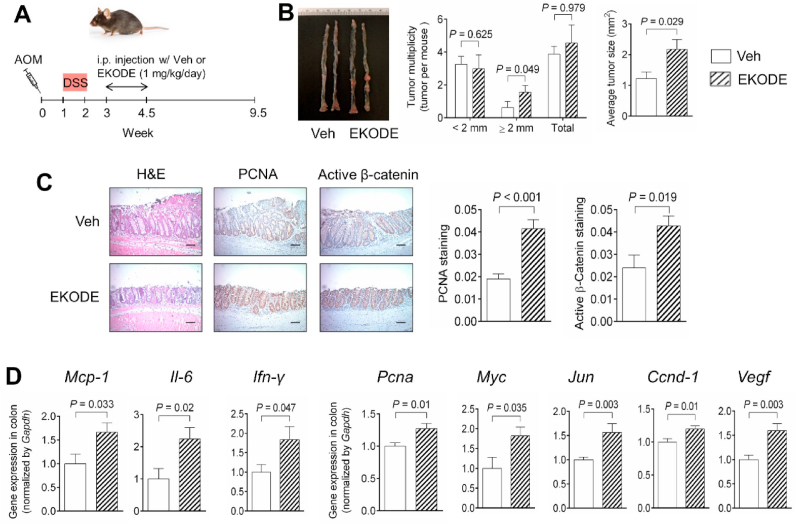
We determined the effect of EKODE on development of AOM/DSS-induced colon tumorigenesis in C57BL/6 mice. To do so, we stimulated the mice with AOM and DSS to initiate colon tumorigenesis, then treated the mice with EKODE (dose = 1 mg/kg/day, via intraperitoneal injection, the dose is the same as our colitis experiment as above in Fig. 4) or vehicle during week 3 to week 4.5 post the AOM injection (see scheme of animal experiment in Fig. 6A). This experimental design allows us to determine the extent to which systemic, short-time, treatment with low-dose EKODE modulates the development of CRC.
Fig. 6.
Treatment with EKODE exaggerates AOM/DSS-induced colon tumorigenesis in mice. A, Scheme of animal experiment (dose of EKODE = 1 mg/kg/day). B, Quantification of colon tumor in mice (n = 8–9 mice per group). C, H&E histology and IHC staining of PCNA and β-catenin in colon (n = 8–9 mice per group, scale bars: 50 μm). D, Gene expression of Mcp-1, Il-6, Ifn-γ, Pcna, Myc, Jun, Ccnd-1 and Vegf in colon (n = 8–9 mice per group). The results are expressed as means ± SEM. The statistical significance of two groups was determined using Student's t-test or Wilcoxon-Mann-Whitney test.
We found that treatment with EKODE exaggerated AOM/DSS-induced colon tumorigenesis in mice. EKODE increased the number of large-size (diameter > 2 mm) tumors, though it did not significantly increase the number of small-size (diameter < 2 mm) tumors or the number of total tumors (Fig. 6B). Furthermore, EKODE treatment significantly increased average tumor size in mice (Fig. 6B). Immunohistochemical staining showed that EKODE treatment increased expression of CRC markers, such as PCNA and active β-catenin, in the colon (Fig. 6C). In addition, we found that EKODE treatment increased expression of pro-inflammatory genes (Mcp-1, Il-6, and Ifn-γ) and pro-tumorigenic genes (Pcna, Myc, Jun, Ccnd-1, and Vegf) in the colon (Fig. 6D), enhanced protein expression levels of IL-6 and phosphorylated JNK in the colon (Figs. S5A–B), and higher concentration of MCP-1 in plasma (Fig. S5C), demonstrating that EKODE exacerbated tumor inflammation and colon tumorigenesis. Consistent with our result in Fig. S4C, EKODE treatment did not change colonic expression of Hmox1 (Fig. S5D). Overall, these results demonstrate that EKODE has potent CRC-enhancing effects.
3.4. EKODE induces inflammatory responses and activates NF-κB signaling in both colon cancer cells and macrophage cells
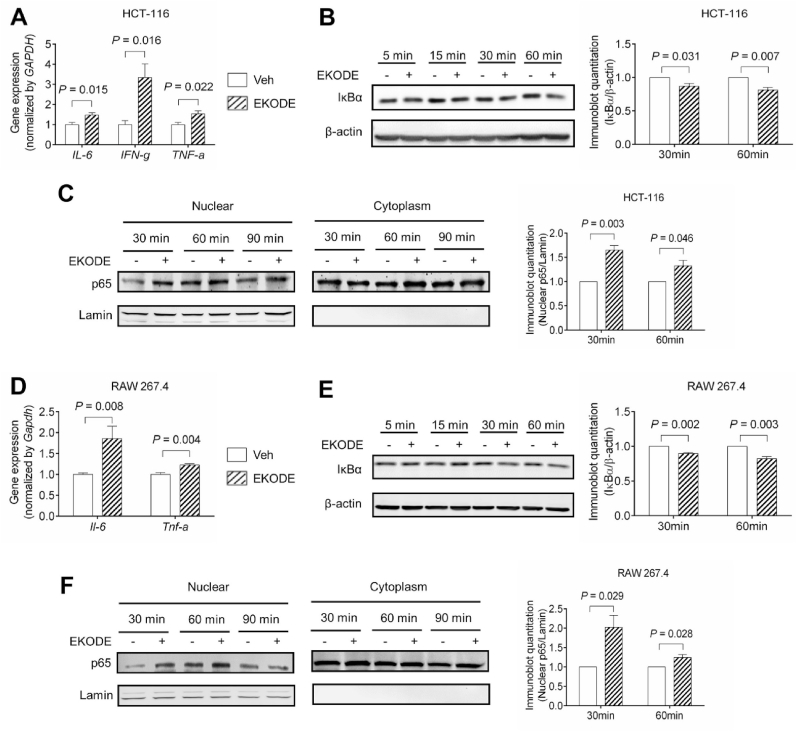
After demonstrating that systemic treatment with EKODE increased colitis and tumor inflammation in vivo, we tested whether EKODE directly acted on colon cancer cells or immune cells to induce inflammation. To do so, we treated colon cancer (HCT-116) cells or macrophage (RAW 264.7) cells with 300 nM EKODE, then examined inflammatory responses. We have determined the concentration of 300 nM, since this is similar to the concentrations of endogenous EKODE in the colon of AOM/DSS-induced CRC mice (Fig. 2D).
In HCT-116 cells, treatment with EKODE induced gene expression of pro-inflammatory cytokines (IL-6, IFN-γ, TNF-α) after 24-h treatment, demonstrating its potent pro-inflammatory effect (Fig. 7A). Next, we tested the effect of EKODE on NF-κB, which is an important signaling pathway involved in inflammation [14]. After 30–60 min treatment, EKODE induced degradation of IκB-α and enhanced nuclear translocation of p65, demonstrating that it activated the NF-κB signaling pathway (Fig. 7B–C). A similar result was also observed in RAW 264.7 cells (Fig. 7D–F). Overall, these results demonstrate that treatment with EKODE, at nM doses, induced inflammatory responses and activated NF-kB pathway in both colon cancer cells and macrophage cells, illustrating its potent pro-inflammatory effect in vitro. We also tested the effects of EKODE on expression of Hmox1, which is a down-stream target of the Nrf2 pathway [3], and found that 24-h treatment with 300 nM EKODE had little effect on Hmox1 expression in vitro (Fig. S6).
Fig. 7.
EKODE induces inflammation in human colon cancer HCT-116 cells and mouse macrophage RAW 264.7 cells. The cells were treated with 300 nM EKODE or vehicle (DMSO). A, EKODE increased gene expression of pro-inflammatory cytokines in HCT-116 cells after 24-h treatment (n = 5 per group). B, EKODE enhanced IκBα degradation in HCT-116 cells (n = 3 per group). C, EKODE increased nuclear translocation of p65 in HCT-116 cells (n = 3 per group). D, EKODE increased gene expression of pro-inflammatory cytokines in RAW 264.7 cells after 24-h treatment (n = 5 per group). B, EKODE enhanced IκBα degradation in RAW 264.7 cells (n = 3 per group). C, EKODE increased nuclear translocation of p65 in RAW 264.7 cells (n = 3 per group). The results are mean ± SEM. The statistical significance of two groups was determined using Student's t-test or Wilcoxon-Mann-Whitney test. The cell culture experiments were performed with at least 3 independent repeats.
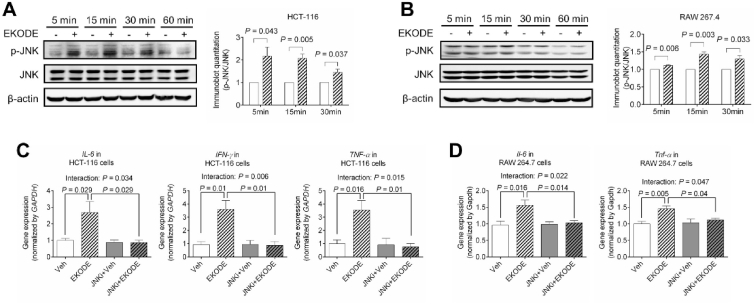
3.5. EDKOE induces inflammation via JNK-dependent mechanisms in vitro
To understand the mechanisms underlying the pro-inflammatory effect of EKODE, we focused on JNK pathway, which plays critical roles in regulating inflammation [15]. In both HCT-116 and RAW 264.7 cells, a 5–15 min treatment with 300 nM EKODE caused phosphorylation of JNK (Fig. 8A–B), demonstrating that EKODE induced a rapid activation of JNK signaling pathway. To determine the roles of JNK signaling in the effect of EKODE in vitro, we tested the extent to which co-administration of a JNK inhibitor, SP600125, attenuates the pro-inflammatory effects of EKODE. We found that co-administration of the JNK inhibitor abolished the pro-inflammatory effect of EKODE in both HCT-116 and RAW 264.7 cells (Fig. 8C–D). Two-way ANOVA analysis showed that there is a statistically significant interaction between EKODE treatment (vehicle vs. EKODE) and JNK inhibitor treatment (with or without the JNK inhibitor) on inflammatory responses in both HCT-116 and RAW 264.7 cells (Fig. 8C–D). Overall, these results support a potential role of the JNK pathway in the pro-inflammatory actions of EKODE in vitro.
Fig. 8.
EKODE increases inflammation via JNK-dependent mechanisms in vitro. A-B, EKODE (concentration = 300 nM) increased JNK phosphorylation in HCT-116 and RAW 264.7 cells (n = 3 per group). C-D, Co-administration of a JNK inhibitor (SP600125, concentration = 100 nM) attenuated the pro-inflammatory effects of EKODE (300 nM) in HCT-116 and RAW 264.7 cells (n = 4–5 per group). The results are mean ± SEM. The statistical significance of two groups was determined using Student's t-test or Wilcoxon-Mann-Whitney test. Analysis of four groups was performed by two-way ANOVA. The cell culture experiments were performed with at least 3 independent repeats.
4. Discussion
Previous studies showed that many oxidative markers, such as nitric oxide, 8-Oxo-2′-deoxyguanosine (8-oxodG), lipid oxidation compounds (e.g. 4-hydroxynonenal and malondialdehyde), and antioxidant or pro-oxidative proteins (e.g. catalase and myeloperoxidase), are altered in CRC, demonstrating that a more severe oxidative stress in CRC [2,3]. Oxidative stress has been suggested to contribute to the pathogenesis of CRC, however, the molecular mechanisms by which the disturbed redox homeostasis regulates the development of CRC are undefined [2]. Here our central finding is that compared with control healthy mice, EKODE, which is a redox stress-derived lipid signaling molecule [10], was significantly increased in the colon of AOM/DSS-induced CRC mice. Systemic, short-time treatment with low-dose EKODE increased the severity of DSS-induced colitis and exaggerated the development of AOM/DSS-induced CRC in mice, supporting that EKODE contributes to the pathogenesis of colonic inflammation and colon tumorigenesis. Regarding the mechanisms for the actions of EKODE, we found that EKODE can directly target colon cancer cells and macrophage cells to induce inflammation via JNK-dependent mechanisms. Overall, these results support that EKODE is an important endogenous mediator of colonic inflammation and colon tumorigenesis, and could contribute to the mechanisms by which oxidative stress promotes CRC development.
The disturbed redox homeostasis in CRC could contribute to the high concentrations of EKODE in colon tumors. We found that, in both mice and humans, the expression of anti-oxidative markers, including catalase (encoding by Cat), glutathione-disulfide reductase (encoding by Gsr), glutathione S-transferase A1 (encoding by Gsta1), glutathione S-transferase M1 (encoding by Gstm1), and heme oxygenase-1 (encoding by Hmox1), are decreased, while the expression of a pro-oxidative marker, myeloperoxidase (encoding by Mpo), is increased, in colon tumors. These results demonstrate that compared with normal colon tissue, the colon tumors have more severe redox stress. The disturbed redox homeostasis in the colon tumor could contribute to the high colonic concentration of EKODE through several possible mechanisms. First, the increased redox stress, notably the reduced expression of catalase and increased expression of myeloperoxidase, is linked with increased colonic production of reactive oxygen species [16,17], which can attack membrane phospholipids and lead to increased production of lipid peroxidation-derived compounds including EKODE [10]. Second, the decreased expression of glutathione S-transferases in colon tumors could also lead to decreased metabolism of EKODE and thus contribute to its high abundance in colon tumors. Previous research showed that glutathione S-transferases are major enzymes involved in metabolism of lipid peroxidation-derived α,β-unsaturated carbonyl compounds, such as 4-hydroxynonenal (4-HNE) and acrolein, converting these compounds to their glutathione conjugates that are thought to be rapidly secreted [18]. It is feasible that EKODE, which shares a similar α,β-unsaturated carbonyl moiety, could also be metabolized by glutathione S-transferases, though biochemical studies are needed to validate this. Our previous study showed that compared with control healthy mice, the plasma concentration of EKODE was not significantly increased in AOM/DSS-induced CRC mice [7]. This could be due to the low chemical and metabolic stability of EKODE in the circulation. Overall, our results demonstrate that there is a more severe oxidative microenvironment in colon tumor compared with normal colon tissue, leading to increased production and/or decreased metabolism of EKODE and resulting in increased concentration of EKODE in colon tumors. Our finding is in agreement with previous studies which showed that oxidative stress and associated lipid peroxidation compounds are increased in CRC [19].
Here, we showed that systemic, short-time, treatment with low-dose EDKODE exaggerated DSS-induced colitis and AOM/DSS-induced CRC in mice, supporting that EKODE is an important mediator of colonic inflammation and CRC. In our animal experiments, we treated mice with EKODE via intraperitoneal injection not oral administration, since EKODE is chemically reactive and could be degraded in the upper gastrointestinal tract and fail to reach the colon after oral administration. We found that EKODE has potent and direct pro-inflammatory effects, since treatment with nM EKODE induced expression of pro-inflammatory cytokines and activated NF-κB signaling in cultured colon cancer cells and macrophage cells. Overall, these results support a model that during development of CRC, the oxidative stress in colon tumors leads to increased colonic concentration of EKODE, which can target intestinal epithelial cells and immune cells to cause exaggerated colonic inflammation, resulting in exacerbated development of CRC. Thus, EKODE is an important endogenous mediator of colonic inflammation and CRC and could contribute to the mechanisms by which oxidative stress regulates CRC development. Besides intestinal epithelial cells and immune cells, the CRC-generated EKODE, as well as other lipid oxidation-derived compounds, could directly interact with bacterial cells that reside in the colon, leading to alteration of gut microbiota and contributing to the development of CRC [20]. Further studies are needed to better understand how CRC-associated redox environment interacts with gut microbiota to affect the development of CRC.
Previous studies showed that EKODE can stimulate production of dehydroepiandrosterone and corticosterone and activate Nrf2 signaling in cultured cells [[21], [22], [23], [24], [25], [26]]. The concentrations required by EKODE to induce these effects are in high-μM range. For example, Wang et al. showed that EDKOE at 10 μM activated Nrf2 signaling, while it did not have such effects at lower concentrations [24]. This is higher than the concentration of EKODE observed in our studies: for example, the concentration of EKODE in the colon of AOM/DSS-induced CRC mice is ~150 pmol/g tissue (~0.15 μM). Therefore, the Nrf2-inducing activity of EKODE may have a limited contribution to its impacts on development of inflammation and CRC as observed in this study. In support of this notion, we found that treatment with 300 nM EKODE induced gene expression of pro-inflammatory cytokines and activated NF-κB signaling in vitro, while it had little effect on expression of Hmox1 (encoding heme oxygenase-1), which is a down-stream target of the Nrf2 pathway [3]. In addition, we found that in both DSS-induced colitis model and AOM/DSS-induced CRC model, EKODE treatment did not alter colonic expression of Hmox1 in mice. This could be, at least in part, due to the low dose of EKODE (1 mg/kg/day) used in our animal experiments. Our results are largely consistent with previous studies, which showed that EKODE did not activate Nrf2 pathway at low doses [24].
Our results support that EKODE induces inflammation through JNK-dependent mechanisms in vitro. We found that EDKOE induces a rapid activation of JNK in both colon cancer (HCT-116) and macrophage (RAW 264.7) cells; and co-administration of 100 nM of SP600125, a JNK inhibitor, abolishes the pro-inflammatory effects of EKODE in these two cell lines. Previous study has shown that SP600125 is a selective JNK inhibitor: it inhibits JNK1, JNK2, and JNK3 with IC50 = 40–90 nM, and inhibits other proteins at much higher concentrations [27]. Overall, these results support a potential role of JNK signaling in the pro-inflammatory effect of EKODE in vitro. We showed that EKODE increased DSS-induced colitis and AOM/DSS-induced CRC in mouse models, and further studies are needed to characterize the roles of JNK signaling in the effects of EKODE in vivo. Previous studies showed that treatment with JNK inhibitors (e.g. SP600125) attenuated DSS-induced colitis in rodent models [[28], [29], [30]] and play critical roles in regulating colon homeostasis [31], however, genetic ablation of JNK1 or JNK2 increased DSS-induced colitis in mice [[32], [33], [34]]. Regarding its roles in CRC, JNK overexpression exacerbated AOM/DSS-induced CRC, but had little impact on tumorigenesis triggered by Apc mutation [35]. Treatment with JNK inhibitors attenuated early stage of CRC under high-fat dietary conditions [36], as well as Apc mutation-induced CRC [37]. These results suggest a complicated mechanism of JNK in mediating the pathogenesis of colitis and CRC.
In this study, a major limitation is that due to the limited amount of colon tissues from mice, we did not measure the protein expression levels of some markers. For example, we showed that compared with control mice, the expressions of anti-oxidant genes, such as Sod1, Cat, and Gsr, were reduced in the colon of the AOM/DSS-induced CRC mice, but we did not measure the protein expression levels of these markers in the mouse colon tissues. Further studies are needed to further validate the gene expression results.
In summary, here we showed that EKODE, which is an endogenous lipid peroxidation-derived compound, is increased in a mouse model of CRC and exaggerates the development of colitis and CRC in mice. These results support that EKODE could be a novel mechanistic linkage between oxidative stress and CRC development.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgement
This research is supported by USDA NIFA 2016-67017-24423, 2019-67017-29248, 2020-67017-30844, and USDA/Hatch MAS00556 (to G. Zhang).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.101880.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics. Ca - Cancer J. Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. 2018. [DOI] [PubMed] [Google Scholar]
- 2.Perse M. Oxidative stress in the pathogenesis of colorectal cancer: cause or consequence? BioMed Res. Int. 2013;2013:725710. doi: 10.1155/2013/725710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sies H., Berndt C., Jones D.P. Oxidative stress. Annu. Rev. Biochem. 2017;86(1):715–748. doi: 10.1146/annurev-biochem-061516-045037. [DOI] [PubMed] [Google Scholar]
- 4.Wang D., Dubois R.N. Eicosanoids and cancer. Nat. Rev. Canc. 2010;10(3):181–193. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayala A., Muñoz M.F., Argüelles S. Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid Med Cell Longev. 2014;2014 doi: 10.1155/2014/360438. 360438-360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murase R., Taketomi Y., Miki Y., Nishito Y., Saito M., Fukami K., Yamamoto K., Murakami M. Group III phospholipase A2 promotes colitis and colorectal cancer. Sci. Rep. 2017;7(1):12261. doi: 10.1038/s41598-017-12434-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang W., Yang J., Edin M.L., Wang Y., Luo Y., Wan D., Yang H., Song C.Q., Xue W., Sanidad K.Z., Song M., Bisbee H.A., Bradbury J.A., Nan G., Zhang J., Shih P.B., Lee K.S.S., Minter L.M., Kim D., Xiao H., Liu J.Y., Hammock B.D., Zeldin D.C., Zhang G. Targeted metabolomics identifies the cytochrome P450 monooxygenase eicosanoid pathway as a novel therapeutic target of colon tumorigenesis. Canc. Res. 2019;79(8):1822–1830. doi: 10.1158/0008-5472.CAN-18-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang H., Wang W., Romano K.A., Gu M., Sanidad K.Z., Kim D., Yang J., Schmidt B., Panigrahy D., Pei R., Martin D.A., Ozay E.I., Wang Y., Song M., Bolling B.W., Xiao H., Minter L.M., Yang G.Y., Liu Z., Rey F.E., Zhang G. A common antimicrobial additive increases colonic inflammation and colitis-associated colon tumorigenesis in mice. Sci. Transl. Med. 2018;10(443) doi: 10.1126/scitranslmed.aan4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Il Lee S., Zuo X., Shureiqi I. 15-Lipoxygenase-1 as a tumor suppressor gene in colon cancer: is the verdict in? Canc. Metastasis Rev. 2011;30(3–4):481–491. doi: 10.1007/s10555-011-9321-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin D., Zhang J., Sayre L.M. Synthesis of six epoxyketooctadecenoic acid (EKODE) isomers, their generation from nonenzymatic oxidation of linoleic acid, and their reactivity with imidazole nucleophiles. J. Org. Chem. 2007;72(25):9471–9480. doi: 10.1021/jo701373f. [DOI] [PubMed] [Google Scholar]
- 11.Nishikawa A., Furukawa F., Kasahara K., Ikezaki S., Itoh T., Suzuki T., Uchida K., Kurihara M., Hayashi M., Miyata N., Hirose M. Trans-4-hydroxy-2-nonenal, an aldehydic lipid peroxidation product, lacks genotoxicity in lacI transgenic mice. Canc. Lett. 2000;148(1):81–86. doi: 10.1016/s0304-3835(99)00318-3. [DOI] [PubMed] [Google Scholar]
- 12.Bischoff S.C., Barbara G., Buurman W., Ockhuizen T., Schulzke J.D., Serino M., Tilg H., Watson A., Wells J.M. Intestinal permeability--a new target for disease prevention and therapy. BMC Gastroenterol. 2014;14:189. doi: 10.1186/s12876-014-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feldman G.J., Mullin J.M., Ryan M.P. Occludin: structure, function and regulation. Adv. Drug Deliv. Rev. 2005;57(6):883–917. doi: 10.1016/j.addr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 14.Baeuerle P.A., Henkel T. Function and activation of NF-kappa B in the immune system. Annu. Rev. Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 15.Ip Y.T., Davis R.J. Signal transduction by the c-Jun N-terminal kinase (JNK)--from inflammation to development. Curr. Opin. Cell Biol. 1998;10(2):205–219. doi: 10.1016/s0955-0674(98)80143-9. [DOI] [PubMed] [Google Scholar]
- 16.Davies M.J. Myeloperoxidase-derived oxidation: mechanisms of biological damage and its prevention. J. Clin. Biochem. Nutr. 2011;48(1):8–19. doi: 10.3164/jcbn.11-006FR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goth L., Rass P., Pay A. Catalase enzyme mutations and their association with diseases. Mol. Diagn. 2004;8(3):141–149. doi: 10.1007/BF03260057. [DOI] [PubMed] [Google Scholar]
- 18.Allocati N., Masulli M., Di Ilio C., Federici L. Glutathione transferases: substrates, inihibitors and pro-drugs in cancer and neurodegenerative diseases. Oncogenesis. 2018;7(1):8. doi: 10.1038/s41389-017-0025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skrzydlewska E., Sulkowski S., Koda M., Zalewski B., Kanczuga-Koda L., Sulkowska M. Lipid peroxidation and antioxidant status in colorectal cancer. World J. Gastroenterol. 2005;11(3):403–406. doi: 10.3748/wjg.v11.i3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sekirov I., Russell S.L., Antunes L.C., Finlay B.B. Gut microbiota in health and disease. Physiol. Rev. 2010;90(3):859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 21.Bruder E.D., Raff H., Goodfriend T.L., Aurora St G. Luke's Medical Center Adrenal Tumor Study, an oxidized derivative of linoleic acid stimulates dehydroepiandrosterone production by human adrenal cells. Horm. Metab. Res. 2006;38(12):803–806. doi: 10.1055/s-2006-956182. [DOI] [PubMed] [Google Scholar]
- 22.Bruder E.D., Ball D.L., Goodfriend T.L., Raff H. An oxidized metabolite of linoleic acid stimulates corticosterone production by rat adrenal cells. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2003;284(6):R1631–R1635. doi: 10.1152/ajpregu.00753.2002. [DOI] [PubMed] [Google Scholar]
- 23.Vangaveti V.N., Jansen H., Kennedy R.L., Malabu U.H. Hydroxyoctadecadienoic acids: oxidised derivatives of linoleic acid and their role in inflammation associated with metabolic syndrome and cancer. Eur. J. Pharmacol. 2016;785:70–76. doi: 10.1016/j.ejphar.2015.03.096. [DOI] [PubMed] [Google Scholar]
- 24.Wang R., Kern J.T., Goodfriend T.L., Ball D.L., Luesch H. Activation of the antioxidant response element by specific oxidized metabolites of linoleic acid. Prostagl. Leukot. Essent. Fat. Acids. 2009;81(1):53–59. doi: 10.1016/j.plefa.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodfriend T.L., Ball D.L., Egan B.M., Campbell W.B., Nithipatikom K. Epoxy-keto derivative of linoleic acid stimulates aldosterone secretion. Hypertension. 2004;43(2):358–363. doi: 10.1161/01.HYP.0000113294.06704.64. [DOI] [PubMed] [Google Scholar]
- 26.Goodfriend T.L., Ball D.L., Gardner H.W. An oxidized derivative of linoleic acid affects aldosterone secretion by adrenal cells in vitro. Prostagl. Leukot. Essent. Fat. Acids. 2002;67(2–3):163–167. doi: 10.1054/plef.2002.0414. [DOI] [PubMed] [Google Scholar]
- 27.Bennett B.L., Sasaki D.T., Murray B.W., O'Leary E.C., Sakata S.T., Xu W., Leisten J.C., Motiwala A., Pierce S., Satoh Y., Bhagwat S.S., Manning A.M., Anderson D.W. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl. Acad. Sci. U. S. A. 2001;98(24):13681–13686. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Assi K., Pillai R., Gómez-Muñoz A., Owen D., Salh B. The specific JNK inhibitor SP600125 targets tumour necrosis factor-alpha production and epithelial cell apoptosis in acute murine colitis. Immunology. 2006;118(1):112–121. doi: 10.1111/j.1365-2567.2006.02349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitsuyama K., Suzuki A., Tomiyasu N., Tsuruta O., Kitazaki S., Takeda T., Satoh Y., Bennett B.L., Toyonaga A., Sata M. Pro-inflammatory signaling by Jun-N-terminal kinase in inflammatory bowel disease. Int. J. Mol. Med. 2006;17(3):449–455. [PubMed] [Google Scholar]
- 30.Kersting S., Behrendt V., Kersting J., Reinecke K., Hilgert C., Stricker I., Herdegen T., Janot M.S., Uhl W., Chromik A.M. The impact of JNK inhibitor D-JNKI-1 in a murine model of chronic colitis induced by dextran sulfate sodium. J. Inflamm. Res. 2013;6:71–81. doi: 10.2147/JIR.S40092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang R., Kwon I.K., Singh N., Islam B., Liu K., Sridhar S., Hofmann F., Browning D.D. Type 2 cGMP-dependent protein kinase regulates homeostasis by blocking c-Jun N-terminal kinase in the colon epithelium. Cell Death Differ. 2014;21(3):427–437. doi: 10.1038/cdd.2013.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chromik A.M., Müller A.M., Körner J., Belyaev O., Holland-Letz T., Schmitz F., Herdegen T., Uhl W., Mittelkötter U. Genetic deletion of JNK1 and JNK2 aggravates the DSS-induced colitis in mice. J. Invest. Surg. 2007;20(1):23–33. doi: 10.1080/08941930601126140. [DOI] [PubMed] [Google Scholar]
- 33.Kersting S., Reinecke K., Hilgert C., Janot M.S., Haarmann E., Albrecht M., Muller A.M., Herdegen T., Mittelkotter U., Uhl W., Chromik A.M. Knockout of the c-Jun N-terminal Kinase 2 aggravates the development of mild chronic dextran sulfate sodium colitis independently of expression of intestinal cytokines TNFalpha, TGFB1, and IL-6. J. Inflamm. Res. 2013;6:13–23. doi: 10.2147/JIR.S36415. (1178-7031 (Print)) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mandic A.D., Bennek E., Verdier J., Zhang K., Roubrocks S., Davis R.J., Denecke B., Gassler N., Streetz K., Kel A., Hornef M., Cubero F.J., Trautwein C., Sellge G. c-Jun N-terminal kinase 2 promotes enterocyte survival and goblet cell differentiation in the inflamed intestine. Mucosal Immunol. 2017;10(5):1211–1223. doi: 10.1038/mi.2016.125. [DOI] [PubMed] [Google Scholar]
- 35.Sancho R., Nateri A.S., de Vinuesa A.G., Aguilera C., Nye E., Spencer-Dene B., Behrens A. JNK signalling modulates intestinal homeostasis and tumourigenesis in mice. EMBO J. 2009;28(13):1843–1854. doi: 10.1038/emboj.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Endo H., Hosono K., Fujisawa T., Takahashi H., Sugiyama M., Yoneda K., Nozaki Y., Fujita K., Yoneda M., Inamori M., Wada K., Nakagama H., Nakajima A. Involvement of JNK pathway in the promotion of the early stage of colorectal carcinogenesis under high-fat dietary conditions. Gut. 2009;58(12):1637. doi: 10.1136/gut.2009.183624. [DOI] [PubMed] [Google Scholar]
- 37.Fujishita T., Aoki M., Taketo M.M. JNK signaling promotes intestinal tumorigenesis through activation of mTOR complex 1 in Apc(Δ716) mice. Gastroenterology. 2011;140(5):1556–1563. doi: 10.1053/j.gastro.2011.02.007. e6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.