Abstract
Pharmacologic inhibition of PARP is the primary therapeutic strategy for BRCA mutant ovarian cancer. However, most of patients carry wild-type BRCA1/2 with no significant clinical benefits from PARP inhibitors, calling for the needs to further understanding and developing new strategy when employing PARP inhibitors to treat ovarian cancer. Here, we show that ferroptosis, a form of regulated cell death driven by iron-dependent phospholipid peroxidation, is partly responsible for the efficacy of PARP inhibitor olaparib. Mechanistically, pharmacological inhibition or genetic deletion of PARP downregulates the expression of cystine transporter SLC7A11 in a p53-dependent manner. Consequently, decreased glutathione biosynthesis caused by SLC7A11 repression promotes lipid peroxidation and ferroptosis. Furthermore, ferroptosis perturbation results in significant resistance to olaparib without affecting DNA damage response, while boosting ferroptosis by ferroptosis inducers (FINs) synergistically sensitizes BRCA-proficient ovarian cancer cells and xenografts to PARP inhibitor. Together, our results reveal a previously unappreciated mechanism coupling ferroptosis to PARP inhibition and suggest the combination of PARP inhibitor and FINs in the treatment of BRCA-proficient ovarian cancer.
Keywords: PARP, Ferroptosis, Lipid peroxidation, SLC7A11, Ovarian cancer
Graphical abstract
Highlights
-
•
PARP inhibition promotes ferroptosis by repressing SLC7A11 in ovarian cancer.
-
•
Ferroptosis represents a critical mechanism mediating the efficacy of PARP inhibitor.
-
•
Olaparib-induced ferroptosis is independent of olaparib-induced DNA damage.
-
•
Olaparib combined with FINS is a promising strategy for BRCA wild-type ovarian cancer.
1. Introduction
Poly (ADP-ribose) polymerases (PARP) constitute a family of nuclear and cytoplasmic proteins and are involved in a variety of cellular processes, including DNA repair and the maintenance of genomic integrity, DNA methylation, programmed cell death, transcriptional regulation, and metabolic regulation [[1], [2], [3], [4], [5]]. Among them, PARP1 is a prototype member with most abundance, serving as a significant target for cancer therapy [1,5,6]. Traditionally, PARP inhibition has been considered to be predominantly related to DNA damage and its downstream effect-apoptosis, which is synthetically lethal to cancer cells harboring a deficient homologous recombination (HR) pathway, such as BRCA1/2 mutations [[7], [8], [9], [10], [11]]. However, mechanisms of action of PARP inhibition in cancer cells has not been fully understood, and whether it can suppress tumors through mechanisms that are not directly linked to DNA damage remains unknown.
Ovarian cancer mortality ranks first among malignant tumors of the female reproductive system [12]. Approximately 70% of patients are at an advanced stage when they are first diagnosed [13,14]. Pharmacologic inhibition of PARP is the promising therapeutic strategy for advanced ovarian cancer with HR deficiency [15]. Olaparib, the most classical and effective PARP inhibitor, has been approved by the US Food and Drug Administration (FDA) for treatment and maintenance in advanced ovarian cancer with germline BRCA1/2 mutations [[16], [17], [18]]. However, the clinical benefit of olaparib appears to be restricted because only a small percentage of patients carry mutant BRCA genes [7,15,17]. Thus, it is critical to further explore the unrecognized sensitization mechanism of PARP inhibitor in ovarian cancer with intrinsic resistance to the DNA damage and apoptosis, and to develop novel combination strategies on this basis to expand the utility of PARP inhibitor into BRCA-proficient ovarian cancer patients.
Ferroptosis is a recently identified form of regulated cell death driven by iron-dependent phospholipid peroxidation, and differs from apoptosis in morphology, biochemistry and genetics [19,20]. It is considered a natural tumor suppression mechanism and plays an significant role in the anti-cancer effects of many cancer therapies including radiotherapy, immunotherapy, and certain chemotherapy [[21], [22], [23], [24], [25]]. Previous studies have suggested that ferroptosis-mediated anti-cancer effect does not seems to be involved in DNA damage and repair [26]. Although chemo-resistant cancer cells are commonly caused by enhanced DNA repair capabilities or defects in the apoptosis pathway, they are vulnerable to ferroptosis induction [27,28]. Ferroptosis onset is caused by the accumulation of peroxides of polyunsaturated fatty acid-containing phospholipids (PUFA-PLs), which overwhelm cellular defense systems [29,30]. Solute carrier family 7 member 11 (SLC7A11) is the catalytic subunit of amino acid transporter system xc− that takes up extracellular cystine, and then cystine is reduced to cysteine in the cytosol, serving as the rate-limiting precursor for glutathione (GSH) biosynthesis [20,31]. GSH is a potent lipid peroxide scavenger and acts as the essential cofactor for the selenoenzyme glutathione peroxidase 4 (GPX4) to detoxify phospholipid peroxidation and protect cells from ferroptosis [[32], [33], [34]]. Therefore, SLC7A11-GSH system confers the key cellular mechanism to defend against ferroptosis, and dysfunction of this system by ferroptosis inducers (FINs) to trigger ferroptosis or potentiate chemoradiotherapy-induced ferroptosis is a potential therapeutic strategy for cancers [20,29,35].
In addition to inhibiting DNA single-strand break repair and leading to DNA double-strand breaks, it remains largely unexplored whether ferroptosis is an important contributor to PARP inhibition-mediated cell death and tumor suppression. In this study, we characterized ferroptosis as a previously unappreciated mechanism of cell death and tumor suppression triggered by PARP inhibitor olaparib in ovarian cancer, and implicated that PARP inhibition promotes ferroptosis partially through suppressing SLC7A11-mediated GSH biosynthesis. Furthermore, we established that the combination of olaparib and FINs might be a promising therapeutic strategy for BRCA wild-type ovarian cancer.
2. Results
2.1. PARP inhibition promotes ferroptosis in ovarian cancer cells
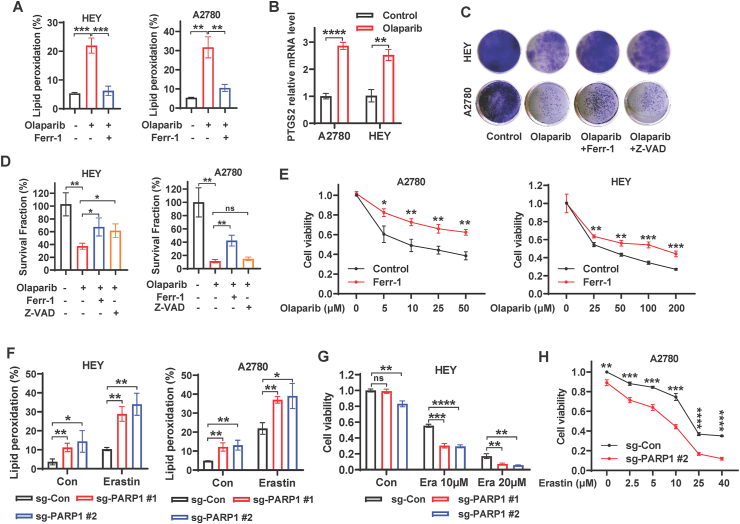
PARP inhibition is generally considered to induce cancer cell death via inhibiting DNA repair and promoting apoptosis [[7], [8], [9], [10], [11]], while it remains unclear whether ferroptosis are underlying toxic mechanism of PARP inhibition in ovarian cancer cells. Given that ferroptosis is driven by accumulation of lipid peroxidation products [20,33], we used BODIPY™ 581/591C11 staining to quantify the percentages of lipid peroxidation in tested cells. We found that pharmacologic inhibition of PARP with olaparib markedly induced lipid peroxidation in the ovarian cancer cell lines HEY and A2780, and that increased accumulation of lipid peroxides following olaparib treatment was abolished by concurrent treatment with ferrostatin-1, the potent ferroptosis inhibitor (Fig. 1A and S1A). We want to point out that lipid peroxidation is measured by BODIPY™ 581/591C11 only in living cells. Therefore, we treated cells with relatively low doses of olaparib to prevent excessive cell death. In this context, the decrease in viability of olaparib-treated HEY cells or A2780 cells was less than 20% or around 30%, respectively (Fig. S1B). We then measured the mRNA levels of PTGS2, the genetic hallmark of ferroptosis [33], in HEY and A2780 cells treated with or without olaparib. Consistently, olaparib significantly induced the expression of PTGS2 (Fig. 1B), suggesting that the PARP inhibitor plays an important role in ferroptosis onset in ovarian cancer cells.
Fig. 1.
PARP inhibition induces ferroptosis in ovarian cancer cells. A Lipid peroxidation levels in HEY and A2780 cells treated with DMSO or olaparib (25 μM for HEY, 5 μM for A2780) or olaparib in combination with ferrostatin-1 (5 μM) for 48 h. B mRNA levels of PTGS2 in A2780 and HEY cells treated with DMSO or olaparib (25 μM for HEY, 5 μM for A2780) for 48 h. C Representative images of clonogenic assay in HEY and A2780 cells treated with DMSO, or olaparib (20 μM for HEY, 10 μM for A2780), or olaparib in combination with ferrostatin-1 (5 μM) or Z-VAD (10 μM). D Quantification of clonogenic survival fractions in HEY and A2780 cells subjected to the indicated treatments. E Cell viability in A2780 and HEY cells treated with indicated concentrations of olaparib with or without ferrostatin-1 (5 μM). F Lipid peroxidation levels in sg-control and sg-PARP1 HEY/A2780 cells treated with DMSO or erastin (HEY, 2.5 μM, 24 h; A2780, 10 μM, 48 h). G-H Cell viability in sg-control and sg-PARP1 HEY/A2780 cells treated with DMSO or indicated concentrations of erastin for 24 (HEY) or 48 (A2780) hours. Data are presented as representative images or as mean ± SD from three independent repeats. Statistical analysis was conducted using 2-tailed unpaired Student t-test.
To understand the potential role of ferroptosis in the olaparib-mediated anticancer effect, we studied the impact of the ferroptosis inhibitor ferrostatin-1 and the apoptosis inhibitor Z-VAD-fmk on the clonogenic survival of cells treated with olaparib. We observed that olaparib treatment expectedly inhibited clonogenic survival, while the combination treatment of olaparib and ferrostatin-1 partially restored survival in A2780 and HEY cells (Fig. 1C and D). Notably, the restoration of cell survival by Z-VAD-fmk was weaker than that by ferrostatin-1 in both cell lines, and it did not even significantly rescue clonogenic survival in A2780 cells (Fig. 1C and D). Similarly, ferrostatin-1 treatment increased cell viability in A2780 and HEY cells upon treatment with various concentrations of olaparib (Fig. 1E), suggesting that ferroptosis is partially responsible for the efficacy of the PARP inhibitor olaparib.
To confirm whether genetic deletion of PARP1 promotes ferroptosis in ovarian cancer cells, we utilized CRISPR/Cas9 system to generate a pool of PARP1-deficient HEY and A2780 cells with 2 independent single guide RNAs (sgRNAs) (Figs. S1C–D). BODIPY™ 581/591C11 staining showed that PARP1 deletion (sgPARP1) promoted lipid peroxidation under basal condition and dramatically augmented erastin (a canonical ferroptosis inducer)-induced lipid peroxidation in both cell lines (Fig. 1F and S1E). Consistently, PARP1 deletion markedly potentiated erastin-induced ferroptosis in HEY and A2780 cells (Fig. 1G–H, S1F), suggesting that PARP1 might serve as a critical mediator in ferroptosis pathway. Collectively, these data strongly suggest that PARP inhibition promotes ferroptosis and that ferroptosis is essential for the efficacy of PARP inhibitor olaparib in ovarian cancer cells.
2.2. PARP inhibition represses SLC7A11 expression in a p53-dependent manner
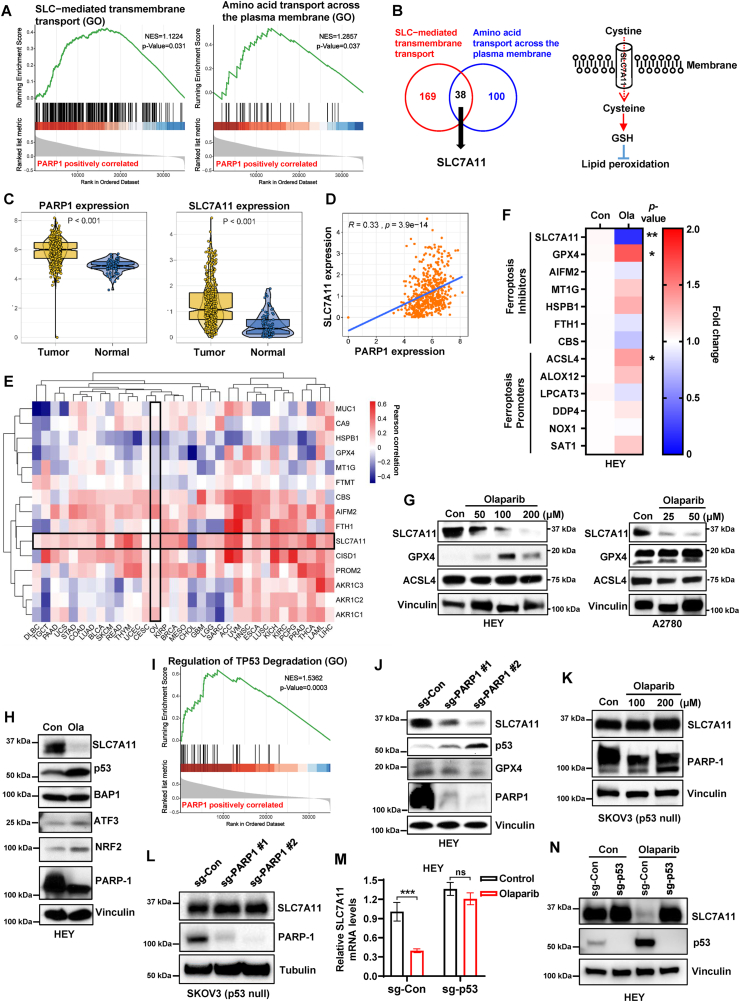
To explore the mechanisms by which PARP inhibition promotes ferroptosis, we evaluated the potential ferroptosis-related biological process enriched in PARP1-regulated genes by gene set enrichment analysis (GSEA) in TCGA ovarian cancer database. Intriguingly, we found that a significant enrichment of gene signatures including “Amino acid transport across the plasma membrane” and “Solute carrier (SLC)-mediated transmembrane transport” in PARP1-regulated genes (Fig. 2A). Integration of both gene sets identified 38 genes that potentially link PARP1 to the regulation of SLC-mediated amino acid transport, wherein SLC7A11 is responsible for the uptake of extracellular cystine to supplement cellular cysteine (the reduced form of cystine), thus promoting glutathione (GSH) biosynthesis to inhibit lipid peroxidation and ferroptosis [20] (Fig. 2B), which prompted us to investigate the regulation of SLC7A11 by PARP1 in ferroptosis pathway of ovarian cancer. To define the correlation between PARP1 and SLC7A11, we examined the expression of both genes in ovarian cancer and normal tissue using TCGA and GTEx databases. We found that the expression of both PARP1 and SLC7A11 were significantly higher in ovarian cancer tissue than those in normal ovarian tissues (Fig. 2C). Further analysis of Pearson correlation coefficient revealed that the expression levels of PARP1 were positively correlated with those of SLC7A11 in ovarian cancer and normal tissues (Fig. 2D). The aforementioned data prompted us to further investigate the connection between PARP1 and ferroptosis defense system in TCGA database. Consistent with in ovarian cancer, the positive correlation of PARP1 with SLC7A11 was observed in most other cancer types; of note, PARP1 was also correlated with some other ferroptosis protectors, such as AIFM2 (also known as FSP1), CBS and HSPB1 in ovarian cancer (Fig. 2E). To define the importance of SLC7A11 in PARP-related ferroptosis regulators, we examined the transcriptional levels of several key components involved in ferroptosis pathway in HEY cells following PARP inhibition. Consistent with the correlations in TCGA database, olaparib obviously repressed the mRNA expression of SLC7A11, whereas there seems to be no significant change in the expression of AIFM2, CBS, HSPB1, and several ferroptosis-executing genes, such as LPCAT3, NOX1 and ALOX12, upon PARP inhibition (Fig. 2F). Consistent with mRNA expression, SLC7A11 protein levels were markedly reduced by olaparib treatment in both HEY and A2780 cells (Fig. 2G). Although ACSL4 and GPX4 mRNA levels were mildly or moderately increased in HEY cells treated with olaparib, the protein expression of ACSL4 was not significantly induced in HEY and A2780 cells, and the alteration of GPX4 protein expression was not consistent between HEY and A2780 cells (Fig. 2G), indicating the prominence of SLC7A11 in the ferroptosis pathway involved in PARP inhibition.
Fig. 2.
PARP inhibition represses SLC7A11 expression in a p53-dependent manner.
A Gene set enrichment pathway analysis (GSEA) showing the significant enrichment of the Gene Ontology (GO) gene sets involving “SLC−mediated transmembrane transport” and “Amino acid transport across the plasma membrane” in PARP1-regulated genes in TCGA ovarian cancer database. B Left panel, the overlap between 169 PARP1-related genes with gene set “SLC−mediated transmembrane transport” and 100 PARP1-related genes with gene set “Amino acid transport across the plasma membrane” in TCGA ovarian cancer database. Right panel, simplified schematic representation of SLC7A11 functioning as a cystine transporter. C Expression levels of PARP1 (left panel) or SLC7A11 (right panel) comparison between ovarian tumor samples and normal tissues from the TCGA and GTEx databases. D Correlation between PARP1 and SLC7A11 expression in ovarian tumor samples and normal tissues in TCGA and GTEx databases. R, Pearson correlation coefficient. E Heatmap showing the Pearson's correlation between expression of PARP1 and ferroptosis-related genes in 33 cancer types from TCGA. The cancer types (in columns) and indicated genes (in rows) are arranged by hierarchical clustering. F Heatmap showing relative mRNA expression of indicated ferroptosis regulators in HEY cells treated with DMSO or olaparib (100 μM) for 48 h. G Protein levels of SLC7A11, GPX4 and ACSL4 were analyzed by western blotting in A2780 and HEY cells treated with DMSO or the indicated concentrations of olaparib for 48 h. H Protein levels of indicated genes were analyzed by western blotting in HEY cells treated with DMSO or olaparib (100 μM) for 48 h. I Gene set enrichment pathway analysis (GSEA) showing the significant enrichment of the Gene Ontology (GO) gene set “Regulation of TP53 Degradation” in PARP1-regulated genes in TCGA ovarian cancer database. J Protein levels of SLC7A11, p53, GPX4 and PARP1 were analyzed by western blotting in sg-control and sg-PARP1 HEY cells. K Protein levels of SLC7A11 and PARP1 were analyzed by western blotting in SKOV3 cells treated with DMSO or indicated concentrations of olaparib for 48 h. L Protein levels of SLC7A11 and PARP1 were analyzed by western blotting in sg-control and sg-PARP1 SKOV3 cells. M mRNA levels of SLC7A11 in sg-control and sg-p53 HEY cells treated with DMSO or olaparib (100 μM) for 48 h. N Protein levels of SLC7A11 and p53 were analyzed by western blotting in sg-control and sg-p53 HEY cells treated with DMSO or olaparib (100 μM) for 48 h. Data are presented as representative images or as mean ± SD from three independent repeats. Statistical analysis was conducted using 2-tailed unpaired Student t-test in c, f, and m. Pearson's correlation (two-sided) analysis was used in d and e.
Previous studies revealed that transcriptional, post-transcriptional, or post-translational regulation of SLC7A11 is associated with several genes, such as tumor suppressor p53 or BAP1, and the transcription factor Nrf2 or ATF3 [21,[36], [37], [38]]. To establish how PARP-inhibition represses SLC7A11 expression, we first examined protein levels of its transcriptional regulators under basal and olaparib-treated conditions. As expected, olaparib treatment in HEY cells resulted in the downregulation of PARP1 and SLC7A11; notably, olaparib dramatically upregulated p53 expression without significantly affecting the levels of other SLC7A11 regulators, such as BAP1 and ATF3 (Fig. 2H). NRF2, which is a transcriptional factor promoting SLC7A11 transcription [37], was even upregulated possibly owning to the stress caused by olaparib treatment (Fig. 2H). Further enrichment pathway analysis revealed that PARP1 were significantly positively correlated with p53 degradation (Fig. 2I). We next determined the impact of genetic ablation of PARP1 on the expression of SLC7A11 and p53, in line with the pharmacologic inhibition of PARP with olaparib, PARP1 KO obviously reduced SLC7A11 protein levels and induced p53 expression in HEY cells (Fig. 2J). Interestingly, GPX4 upregulation was not found in HEY cells with genetic ablation of PARP1 (Fig. 2J), suggesting GPX4 induction in HEY cells upon olaparib treatment is probably an adaptive response to oxidative stress, since basal GPX4 level is low in this cell line (Fig. 2G). p53 was found to transcriptionally repress SLC7A11 expression and promote ferroptosis in response to ROS-mediated stress [21]. We then wondered whether p53 activation is involved in PARP inhibition-mediated SLC7A11 suppression. To test this, we treated p53-deficient ovarian cancer cell line SKOV3 with olaparib. We observed that olaparib reduced PARP1 protein levels without affecting SLC7A11 protein levels in SKOV3 cells (Fig. 2K). Similarly, PARP1 deletion failed to suppress SLC7A11 expression in SKOV3 cells (Fig. 2L). To further define whether PARP inhibition suppresses the expression of SLC7A11 in a p53-dependent manner, we deleted p53 in wild-type p53-expressing HEY cells by the CRISPR/Cas9 system. We found that, consistent with p53-deficient SKOV3 cells, olaparib downregulated the mRNA and protein expression of SLC7A11 in control cells, but not in p53 KO cells (Fig. 2M, N). Taken together, our results suggest that PARP inhibition represses SLC7A11 transcription primarily through upregulation of p53 in ovarian cancer cells.
2.3. PARP inhibitor olaparib promotes ferroptosis partially through suppressing SLC7A11-mediated GSH synthesis
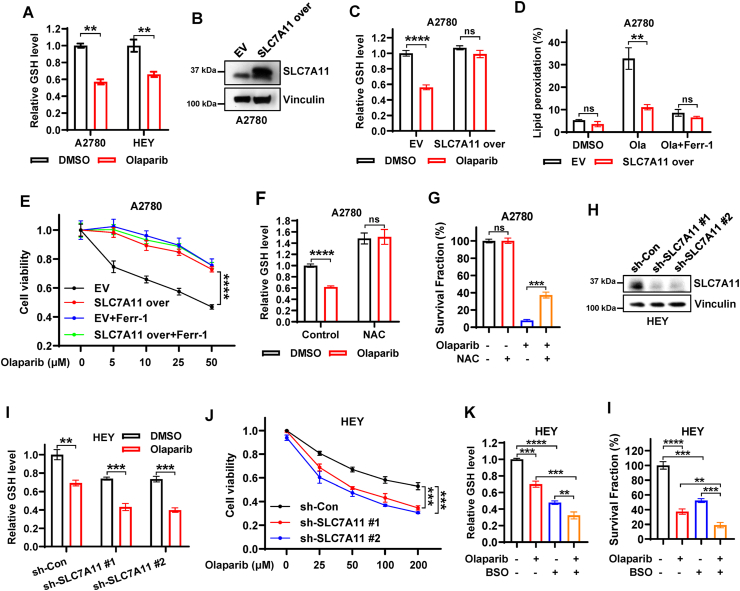
We next sought to functionally link SLC7A11 to olaparib-mediated ferroptosis and efficacy in ovarian cancer. Since SLC7A11 mediates cystine (oxidized dimeric form of cysteine) uptake, and promotes GSH biosynthesis through providing its precursor cysteine [31], we first examined whether the olaparib-repressed expression of SLC7A11 is correlated with the decreased cystine uptake and GSH synthesis in ovarian cancer cells. Consistent with our hypothesis, olaparib treatment indeed inhibit cystine uptake and significantly reduced GSH levels (Figs. S2A and 3A), suggesting olaparib might cause the functional inhibition of SLC7A11. To further confirm this, we generated SLC7A11-overexpressing A2780 cells (Fig. 3B). We observed that, while olaparib decreased GSH levels and promoted lipid peroxidation in control cells, overexpression of SLC7A11 obviously restored GSH levels and mitigated olaparib-induced lipid peroxidation (Fig. 3C–D, S2B). Consistently, SLC7A11 overexpression significantly restored cell viability in olaparib-treated A2780 cells to a level similar to that caused by ferrostatin-1 treatment (Fig. 3E). It should be noted that SLC7A11 overexpression failed to further promote the resistance to olaparib under the condition of ferroptosis perturbation by ferrostatin-1 treatment (Fig. 3E), suggesting that overexpression of SLC7A11 attenuates the efficacy of olaparib mainly through blocking ferroptosis. We then performed propidium iodide (PI) staining to confirm cell death, and we found inhibition of lipid peroxidation and ferroptosis by SLC7A11 overexpression did markedly abrogate olaparib-induced cell death (Fig. S2C). To further link SLC7A11-mediated GSH synthesis to olaparib-induced ferroptosis, we supplemented A2780 cells with N-acetyl cysteine (NAC) to promote GSH synthesis upon olaparib treatment. Similar to SLC7A11 overexpression, NAC treatment increased GSH levels in A2780 cells under both basal and olaparib-treated conditions, and partially restored clonogenic survival in A2780 cells upon olaparib treatment (Fig. 3F and G), suggesting that SLC7A11-promoted GSH biosynthesis represents an important mechanism in olaparib-induced ferroptosis.
Fig. 3.
Olaparib promotes ferroptosis partially through suppressing SLC7A11-mediated GSH synthesis.
A Relative glutathione levels in A2780 and HEY cells treated with DMSO or olaparib (50 μM for HEY, 25 μM for A2780). B The expression of SLC7A11 was examined by western blotting in A2780 cells stably expressing empty vector (EV) or SLC7A11. C Relative glutathione levels in EV- and SLC7A11-expressing A2780 cells treated with DMSO or olaparib (25 μM). D Lipid peroxidation levels in EV- and SLC7A11-expressing A2780 cells treated with DMSO or olaparib (5 μM) or olaparib in combination with ferrostatin-1 (5 μM) for 48 h. E Cell viability in EV- and SLC7A11-expressing A2780 cells treated with indicated concentrations of olaparib with or without ferrostatin-1 (5 μM). F Relative glutathione levels in A2780 cells treated with DMSO or NAC (5 mM) or olaparib (25 μM) or olaparib in combination with NAC. G Quantification of clonogenic survival fractions in A2780 cells treated with DMSO or NAC (5 mM) or olaparib (10 μM) or olaparib in combination with NAC. H The knockdown efficiency of sh-SLC7A11 in HEY cells was examined by western blotting. i Relative glutathione levels in sh-control and sh-SLC7A11 HEY cells treated with DMSO or olaparib (50 μM). J Cell viability in sh-control and sh-SLC7A11 HEY cells treated with indicated concentrations of olaparib for 48 h. K Relative glutathione levels in HEY cells treated with DMSO or olaparib or BSO or olaparib in combination with BSO. I Quantification of clonogenic survival fractions in HEY cells treated with DMSO or olaparib or BSO or olaparib in combination with BSO. Data are presented as representative images or as mean ± SD from three independent repeats. Statistical analysis was conducted using 2-tailed unpaired Student t-test.
We next sought to define whether p53 deletion has similar effect on olaparib-induced ferroptosis as SLC7A11 overexpression. As shown in Figs. S2D–E, p53 KO significantly attenuated olaparib-induced lipid peroxidation, and significantly promoted resistance of HEY cells to olaparib; importantly, ferrostatin-1 treatment partially restored the survival upon olaparib treatment in control cells, but not in p53 KO cells, likely because olaparib-induced ferroptosis is already largely abrogated in p53 KO cells. These data further support our major conclusion that olaparib downregulates the SLC7A11 expression and thus promotes ferroptosis in a p53-dependent manner. We then used shRNA to knock down SLC7A11 in HEY cells (Fig. 3H). Conversely, we found that SLC7A11 knockdown (sh-SLC7A11) further decreased olaparib-depleted GSH levels and significantly reduced cell viability in olaparib-treated HEY cells (Fig. 3I and J), suggesting that SLC7A11 knockdown rendered cells more sensitive to olaparib. In contrast to the NAC treatment, treatment of HEY cells with l-buthionine sulfoximine (BSO) further augmented the inhibition of GSH biosynthesis by olaparib and dramatically sensitized the cells to olaparib (Fig. 3K and L), in line with SLC7A11 knockdown. Together, these results reveal that the PARP inhibitor olaparib promotes ferroptosis at least partly through inhibiting SLC7A11-mediated GSH synthesis, and that the efficacy of olaparib can be modulated by genetically altering SLC7A11 expression or pharmacologically modulating GSH levels in ovarian cancer cells.
2.4. DNA damage response is not involved in ferroptosis-mediated efficacy of olaparib
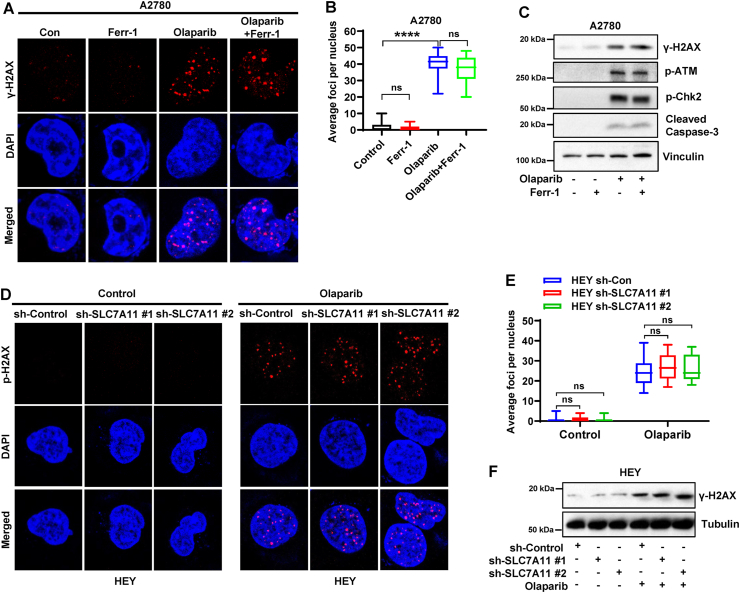
The mechanism of the olaparib-mediated cellular lethality was previously considered to be mainly derived from the inhibition of PARP catalytic activity to prevent DNA single-strand break repair and promote DNA double-strand breaks [7,11]. Unexpectedly, our above-mentioned data suggested that olaparib induces ferroptosis in ovarian cancer cells and ferroptosis represents a part of the olaparib-mediated anticancer effect. We next sought to study whether there is a crosstalk between ferroptosis and DNA damage responses following olaparib treatment. Immunofluorescence analysis showed that phosphorylated (γ) H2AX foci (a marker for DNA damage) were markedly increased by olaparib treatment in A2780 cells, while ferrostatin-1 treatment, although it rescued cell survival following olaparib (Fig. 1D and E), did not affect the number of observed γH2AX foci in the cells with or without olaparib treatment (Fig. 4A and B). Consistently, western blotting showed that olaparib induced phosphorylation of H2AX and other regulators in DNA-repair pathway including ATM and Chk2, and increased the expression of cleaved caspase-3 (a marker for apoptosis), whereas ferrostatin-1 treatment did not obviously affect these markers in A2780 cells with or without olaparib treatment (Fig. 4C), suggesting that ferroptosis inhibition has no significant effect on DNA damage response and its downstream effect-apoptosis following olaparib treatment.
Fig. 4.
DNA damage response is not involved in ferroptosis-mediated efficacy of olaparib.
A Representative immunofluorescence images of γ-H2AX foci in A2780 cells treated with DMSO or ferrostatin-1 (5 μM) or olaparib (5 μM) or olaparib in combination with ferrostatin-1 for 48 h. B Quantification of γ-H2AX foci in each nucleus based on immunofluorescence in A2780 cells subjected to the indicated treatments for 48 h. C Protein levels of indicated regulators of DNA damage response were analyzed by western blotting in A2780 cells treated with DMSO or ferrostatin-1 (5 μM) or olaparib (5 μM) or olaparib in combination with ferrostatin-1 for 48 h. D Representative immunofluorescence images of γ-H2AX foci in sh-control and sh-SLC7A11 HEY cells treated with DMSO or olaparib (25 μM) for 48 h. E Quantification of γ-H2AX foci in each nucleus based on immunofluorescence in sh-control and sh-SLC7A11 HEY cells subjected to the indicated treatments for 48 h. F Protein levels of γ-H2AX were analyzed by western blotting in sh-control and sh-SLC7A11 HEY cells treated with DMSO or olaparib (25 μM) for 48 h. Data are presented as representative images or as mean ± SD from twenty independent repeats. Statistical analysis was conducted using 2-tailed unpaired Student t-test.
Since we found that olaparib induced ferroptosis partially through down-regulating SLC7A11, to further genetically verify that olaparib-induced ferroptosis does not affect DNA damage responses, we examined the γH2AX foci or protein levels in control and SLC7A11 knockdown HEY cells with or without olaparib treatment. As expected, olaparib increased the number of γH2AX foci and induced the phosphorylation of H2AX in control cells, while SLC7A11 knockdown did not further alter either of them (Fig. 4D–F), although it potentiated olaparib-induced ferroptosis (Fig. 3J). Together, these results suggest that DNA damage or repair is neither involved in ferroptosis perturbation-mediated olaparib-resistance nor in ferroptosis augmentation-mediated olaparib-sensitization.
2.5. FINs synergistically sensitize BRCA proficient ovarian cancer cells to olaparib
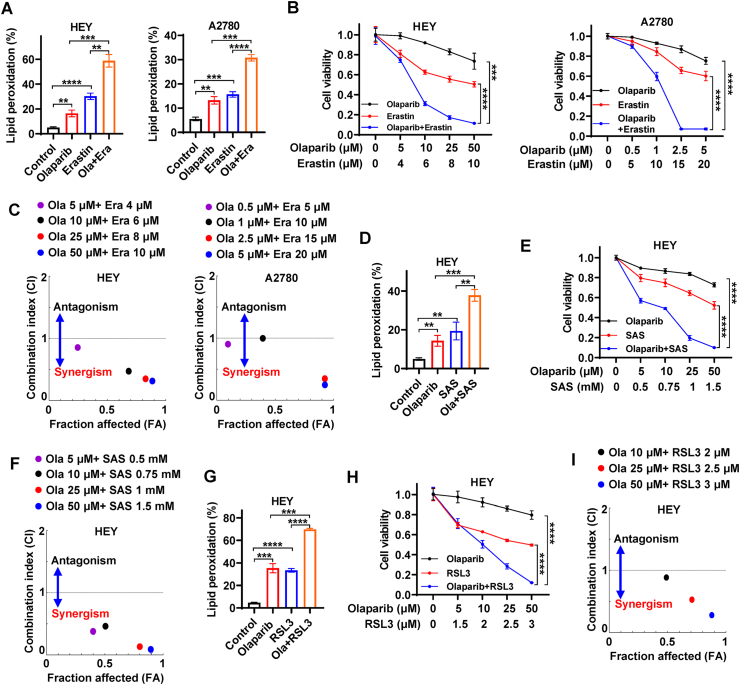
Olaparib failed to provide substantial clinical benefit in BRCA proficient ovarian cancer, since wild-type BRCA can repair olaparib-induced DNA damage and cause olaparib-resistance [7,10]. Given that BRCA mutation frequency only varies from 3% to 27% in ovarian cancer [17], it is clinically urgent to extend the application of PARP inhibitors to BRCA proficient ovarian cancer. Our aforementioned data revealed that ferroptosis significantly contributed to the efficacy of olaparib (Fig. 1d, e), facilitating us to explore whether olaparib in combination with ferroptosis inducers (FINs), such as erastin, sulfasalazine, or RSL3, could synergistically potentiate ferroptosis and thus would sensitize BRCA proficient ovarian cancer cells to olaparib. Since knockdown of SLC7A11 enhanced the efficacy of olaparib (Fig. 3J), we first checked whether erastin targeting SLC7A11 would result in a similar effect in BRCA wild-type ovarian cancer cells. BODIPY™ 581/591C11 staining showed that erastin not only markedly triggered lipid peroxidation, but also synergistically potentiated olaparib-induced lipid peroxidation in BRCA wild-type HEY and A2780 cells (Fig. 5A, S3A). To test whether the synergistic increase in lipid peroxidation enhances the efficacy of olaparib, we treated HEY and A2780 cells with various concentrations of olaparib, erasrtin, or olaparib in combination with erasrtin. Consistent with our hypothesis, HEY and A2780 cells was relatively resistant to olaparib, while erastin treatment reduced the cell viability and dramatically sensitized HEY and A2780 cells to olaparib (Fig. 5B), suggesting that BRCA wild-type ovarian cancer cells are sensitive to ferroptosis onset and that the erastin overcomes olaparib resistance in BRCA wild-type ovarian cancer cells. To confirm that the combination effectiveness of olaparib and erastin was synergistic rather than additive or antagonistic, we calculated combination index using CompuSyn software based on Chou-Talalay methodology. It should be noted that combination indexes at indicated concentrations of FINs and olaparib were less than 1 in both HEY and A2780 cells (Fig. 5C, S3B), suggesting that erastin synergize with olaparib. We obtained similar observations in sulfasalazine treatment, an SLC7A11 inhibitor commonly used in clinic [35,39], namely that sulfasalazine promoted olaparib-induced lipid peroxidation and sensitized HEY cells to olaparib in a synergistic manner (Fig. 5D–F, S3C-D).
Fig. 5.
FINs synergistically sensitize BRCA proficient ovarian cancer cells to olaparib.
A Lipid peroxidation levels in HEY and A2780 cells treated with DMSO or olaparib (10 μM) or erastin (6 μM) or olaparib in combination with erastin. B Cell viability in HEY and A2780 cells treated with olaparib and/or erastin at indicated concentrations. C The Chou-Talalay plot showing the combination effect of indicated treatments. The purple or black or red or blue dots in the plot represent the combination of olaparib and erastin at indicated concentrations. CI values less than, equal to, or greater than 1 indicate synergistic, additive, or antagonistic effects, respectively. D Lipid peroxidation levels in HEY cells treated with DMSO or olaparib (10 μM) or sulfasalazine (0.75 mM) or olaparib in combination with sulfasalazine. E Cell viability in HEY cells treated with olaparib and/or sulfasalazine at indicated concentrations. F The Chou-Talalay plot showing the combination effect of indicated treatments. The purple or black or red or blue dots in the plot represent the combination of olaparib and sulfasalazine at indicated concentrations. CI values less than, equal to, or greater than 1 indicate synergistic, additive, or antagonistic effects, respectively. G Lipid peroxidation levels in HEY cells treated with DMSO or olaparib (50 μM) or RSL3 (3 μM) or olaparib in combination with RSL3. H Cell viability in HEY cells treated with olaparib and/or RSL3 at indicated concentrations. I The Chou-Talalay plot showing the combination effect of indicated treatments. The black or red or blue dots in the plot represent the combination of olaparib and RSL3 at indicated concentrations. CI values less than, equal to, or greater than 1 indicate synergistic, additive, or antagonistic effects, respectively. Data are presented as representative images or as mean ± SD from three independent repeats. Statistical analysis was conducted using 2-tailed unpaired Student t-test. Data are presented as representative images or as mean ± SD from twenty independent repeats. Statistical analysis was conducted using 2-tailed unpaired Student t-test. . (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Our above-mentioned western blotting showed that GPX4 expression was mildly or moderately induced in HEY cells upon olaparib treatment (Fig. 3C). GPX4 is on the downstream of SLC7A11, utilizing GSH to detoxify lipid peroxides and prevent ferroptosis. Therefore, its induction may represent an adaptive response to tackle the olaparib-mediated ferroptosis stress in some ovarian cancer cells, which may weaken anticancer effect of olaparib to a certain extent. We then wondered whether the combination of FINs targeting GPX4 and olaparib would overcome this adaptive response. We found that RSL3 (inactivating GPX4) treatment augmented olaparib-induced lipid peroxidation in HEY cells and significantly sensitized HEY cells to olaparib in a synergistic manner (Fig. 5G–I, S3E-F). Consistently, PI staining showed that olaparib or FINs (erastin or RSL3) mildly or moderately induced cell death, whereas olaparib significantly potentiated FINs-induced cell death in HEY or A2780 cells (Figs. S3G–H).
To further support the proposed mechanism that PARP inhibition promotes ferroptosis and synergizes with FINs via repressing SLC7A11 in a p53-dependent manner, we treated control and p53 KO HEY cells, EV- and SLC7A11-overexpressed A2780 cells with olaparib or/and FINs. As shown in Figs. S3I–J, p53 KO resulted in pronounced resistance of HEY cells to olaparib, erastin and SAS; notably, p53 KO significantly counteracted the synergy of olaparib combined with erastin or SAS. Consistently, SLC7A11 overexpression not only contributed to the significant resistance to olaparib and erastin, but also dramatically neutralized the synergy of olaparib combined with erastin in A2780 cells (Fig. S3K). Together, our data robustly support that FINs synergistically sensitize BRCA wild-type ovarian cancer cells without p53 deficiency to olaparib, suggesting that the utility of olaparib might be extended to BRCA wild-type ovarian cancer in combination with FINs.
2.6. Ferroptosis correlates with olaparib-mediated tumor suppression in vivo
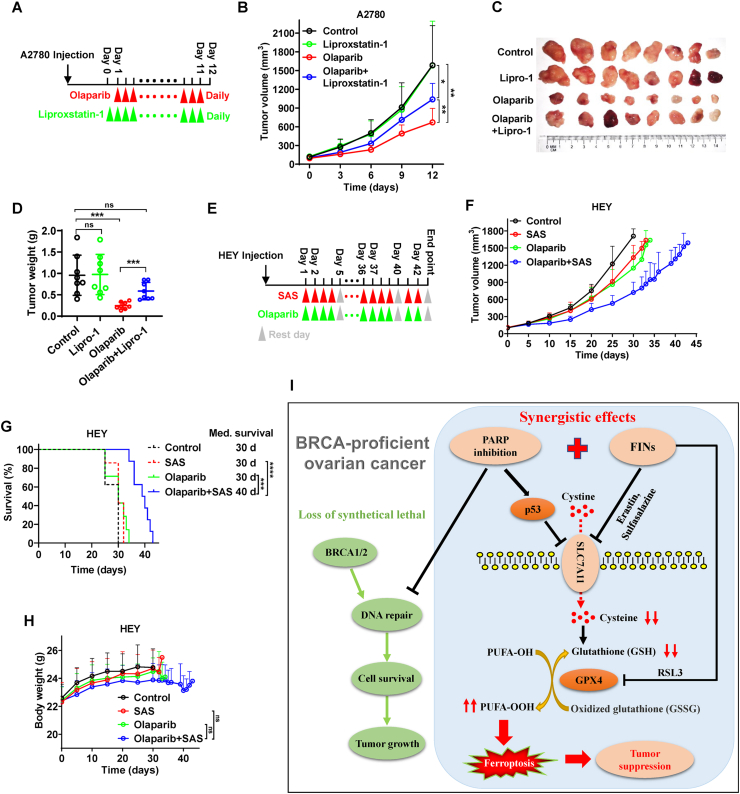
To confirm the potential relevance of ferroptosis in the efficacy of olaparib in vivo, we inoculated A2780 cells into nude mice and treated the mice with olaparib, liproxstatin-1 (a stable ferroptosis antagonist exhibited excellent in vivo efficacy), or both agents concurrently (Fig. 6A). Consistent with our in vitro results, olaparib significantly reduced tumor growth. Treatment with liproxstatin-1 alone did not affect tumor growth, but partially abrogated the efficacy of olaparib (Fig. 6B–D), suggesting that olaparib induces ferroptosis in tumors and that ferroptosis inhibition results in olaparib-resistance in vivo.
Fig. 6.
Ferroptosis correlates with olaparib-mediated tumor suppression in vivo. A Treatment schema for nude mice bearing A2780 xenograft. Liproxstatin-1 treatment initiated on day 0 and was administered daily until experimental endpoints. Olaparib treatment initiated on day 1 and was administered daily until experimental endpoints. B Volume of A2780 xenografts treated with olaparib and/or liproxstatin-1 at different time points. Error bars are presented as mean ± SD from 8 independent repeats. P values determined using 2-way ANOVA. C Representative images of A2780 xenograft tumors treated with olaparib and/or liproxstatin-1 at experimental endpoints. D Weights of A2780 xenograft tumors treated with olaparib and/or liproxstatin-1 at different time points. Error bars are presented as mean ± SD from 8 independent repeats. P values calculated using 2-tailed unpaired Student t-test. E Treatment schema for nude mice bearing HEY xenograft. Olaparib and/or sulfasalazine treatment initiated on day 1, and every 5 days was a treatment cycle until experimental endpoints, in which drug treatment was administered daily for the first 4 days, and the last day is a rest day. F Volume of HEY xenografts treated with olaparib and/or sulfasalazine at different time points. 8 mice per group at the beginning and error bars are presented as mean ± SD. P values determined using 2-way ANOVA. G Kaplan–Meier survival curves of nude mice bearing HEY xenograft treated with olaparib and/or sulfasalazine. P values calculated by log-rank test. H Body weights of nude mice bearing HEY xenograft treated with olaparib and/or sulfasalazine. P values calculated by 2-tailed unpaired Student's t-test. I The working model depicting the role of ferroptosis in PARP inhibition-mediated tumor suppression in BRCA-proficient ovarian cancer. BRCA proficient ovarian cancers are resistant to PARP inhibition due to the inability to efficiently induce DNA damage, while PARP inhibition can activate p53 to repress SLC7A11 expression, leading to a decrease in GSH levels, thereby promoting ferroptosis. On this basis, PARP inhibitors in combination with FINS targeting SLC7A11 or GPX4 synergistically augment ferroptosis, resulting in potent tumor suppression in BRCA-proficient ovarian cancer.
To verify whether augmenting ferroptosis with FINs can sensitize BRCA proficient ovarian cancer to olaparib in vivo, we inoculated HEY cells into nude mice and treated the mice with olaparib, sulfasalazine, or both agents concurrently (Fig. 6E). Sulfasalazine, an FDA-approved clinical drug to treat chronic inflammatory disease, is a commonly used FINs that targeting SLC7A11 in vivo [35,39], which might allow for faster translation of our regimen into clinical ovarian cancer therapies. As expected, tumors derived from HEY cells were resistant to olaparib treatment. Although treatment with sulfasalazine alone did not dramatically reduce tumor growth or prolong mice survivals, it did remarkably sensitize these HEY-derived tumors to olaparib, leading to potent tumor suppression and survival benefits (Fig. 6F, G, S4). Notably, body weights of mice were not significantly decreased by the combination treatment compared to that by other treatments (Fig. 6H), suggesting that toxicities of olaparib in combination with sulfasalazine is tolerated in vivo. Collectively, our data suggest that the combination of olaparib and sulfasalazine is a promising therapeutic strategy for patients with BRCA wild-type ovarian cancer.
3. Discussion
PARP inhibition has long been thought to be primarily associated with inhibition of DNA single-strand repair and DNA double-strand breaks, and the plight of PARP inhibitor in BRCA wild-type ovarian cancer has been attributed to insufficient DNA damage [10,11], promoting us to seek alternative mechanisms that are not directly related to DNA damage upon PARP inhibition (Fig. 6I). Ferroptosis is driven by accumulation of lipid peroxides generated in phospholipid membranes and damages cell membranes, and multiple lines of evidence seem to distinguish ferroptosis from DNA damage and apoptosis. First, ferroptotic cells exhibited none of the morphological features associated with nuclear alterations. Karyorhexis, margination of chromatin, and DNA fragmentation were not visible in erastin-treated cells [19,40]. Second, apoptosis and necroptosis, generally linked to DNA damage, inhibitors failed to protect cells from ferroptosis [19,40]. Third, a study by Brent Stockwell's group showed that imidazole ketone erastin (IKE) or RSL3 neither induced DNA damage nor increased radiation-induced DNA damage by comet assay and γ-H2AX foci detection. Further, cytoplasmic irradiation but not nuclear irradiation generated excessive lipid peroxidation without inducing DNA damage [26]. Our study reveals an important tumor suppression mechanism linking ferroptosis to PARP inhibition. To be specific, we suggest that pharmacological inhibition or genetic deletion of PARP promotes lipid peroxidation and ferroptosis in ovarian cancer cells, and that ferroptosis represents an important mechanism mediating the efficacy of olaparib (classical and effective PARP inhibitor) in vitro and in vivo. We further demonstrate that ferroptosis inhibition or enhancement, while modulating the efficacy of olaparib, does not affect the olaparib-induced DNA damage response, suggesting that DNA damage or repair is neither involved in ferroptosis perturbation-mediated olaparib-resistance nor in ferroptosis augmentation-mediated olaparib-sensitization. Collectively, our results, together with above-mentioned studies, suggest a lack of association between lipid peroxidation and DNA damage. Indeed, we acknowledge that some studies seem to indicate that lipid peroxidation can induce DNA damage, and we speculate that this may be involved in different contexts, the exact reasons for which require our further investigation.
SLC7A11 functions as an important amino acid transporter that imports extracellular cystine in exchange for intracellular glutamate, thereby promoting cysteine supply and GSH synthesis [31,41]. As such, it is a core regulator of the cellular ferroptosis defense system, which serves as a promising therapeutic target in cancer therapy [20,35,42]. Our study uncovers the important relevance of PARP inhibition to SLC7A11 regulation and ferroptosis. At the mechanistic level, PARP inhibition activates p53 to repress SLC7A11 expression, whereas loss or genetic deletion of p53 prevents the downregulation of SLC7A11 expression upon PARP inhibition (Fig. 6I). We further functionally connect SLC7A11 to PARP inhibition-mediated ferroptosis (Fig. 6I). Olaparib treatment partially depletes GSH, while SLC7A11 overexpression or cysteine supplementation abrogates olaparib-mediated GSH depletion and lipid peroxidation, and thus protects cancer cells from olaparib-induced ferroptosis. Conversely, SLC7A11 knockdown or restriction of GSH synthesis (by BSO) significantly potentiates the above cellular processes. Notably, in this study, we focused on the role of SLC7A11 and p53 in PARP inhibition-promoted ferroptosis, and we do not exclude the possibility that PARP inhibition may affect other ferroptosis regulators. It was observed that low dose olaparib induced lipid peroxidation (Fig. 1A) without potent SLC7A11 inhibition (Fig. 2G). We speculate that the possible reason that higher doses are required to achieve SLC7A11 inhibition and GSH depletion is the presence of robust adaptive responses in tumor cells attempting to protect themselves from olaparib-induced stress, especially at low doses in a long-time treatment assay. With increasing dose, these adaptive responses failed to counteract olaparib-mediated cellular effects, thus exhibiting significant SLC7A11 repression and GSH depletion. The adaptive responses for SLC7A11 expression have been observed under several stress-induced contexts, and in terms of our study, such a response in combating olaparib-mediated SLC7A11 inhibition and GSH depletion may be linked to NRF2 activation, which was observed upon olaparib treatment (Fig. 2H).
The possible sources of ROS responsible for lipid peroxidation at low olaparib doses remain interesting. The induction of ROS by PARP inhibitors has been identified by several studies, even at low doses (10 μM or less), with possible sources being attributed to mitochondrial ROS or NAD(P)H oxidases (NOXs). However, the conclusions regarding sources of ROS in these studies remain somewhat controversial. One proposes that PARP inhibition generates mitochondrial ROS through regulation of mitochondrial function and oxidative metabolism, whereas a NOX-specific inhibitor failed to suppress olaparib-induced ROS [43,44]. Conversely, others argue that PARP inhibition increases ROS by upregulating NOXs [45,46] and that olaparib-induced ROS cannot be reduced with mitochondrial ROS scavengers alone [45]. Collectively, these studies together with our results confirm PARP inhibition induces ROS, and our study further links PARP inhibition to lipid peroxidation and ferroptosis through an unrecognized mechanism (repression of SLC7A11 and depletion of GSH); notably, multiple mechanisms likely contribute to PARP inhibition-induced ROS and lipid peroxidation, and dissecting such mechanisms will remain an important area for future PARP inhibition-ferroptosis developments.
Intriguingly, we note that HEY cells were more resistant to olaparib than A2780 cells, and the difference in GPX4 protein levels between these two cells did not seem to be consistent with the difference in sensitivity to olaparib (low GPX4 in HEY cells and high GPX4 in A2780 cells). To better understand the differences between HEY and A2780 cells in response to olaparib-induced ferroptosis, we first measured the absolute GSH concentrations in both cell lines under basal condition. We found that the GSH levels were significantly higher in HEY cells than that in A2780 cells (Fig. S5A), which could be an important reason for the higher resistance to olaparib in HEY cells than in A2780 cells. we then examined the expression of another core ferroptosis regulator FSP1, a recently identified ferroptosis suppressor parallel to GPX4 against ferroptosis [47,48]. Interestingly, FSP1 expression was significantly higher in HEY cells than in A2780 cells under basal conditions (Fig. S5B). Notably, olaparib significantly increased FSP1 expression in HEY cells, but not in A2780 cells (Figs. S5C–D), which may represent another critical reason for the resistance of HEY cells to olaparib. Therefore, low GPX4-expressing HEY cells were characterized by more GSH levels and FSP1 expression/induction compared to A2780 cells, rendered HEY cells more resistant to olaparib than A2780 cells, further suggesting that ferroptosis is the important mechanism responsible for olaparib induced cell death. Certainly, we do not exclude that DNA damage still plays an essential role in this context. As shown in Fig. 4B and E, the γ-H2AX foci induced by 10 μM olaparib in A2780 cells even exceeded those induced by 25 μM olaparib in HEY cells, suggesting that A2780 cells are more susceptible to DNA damage than HEY cells.
Previous olaparib sensitization in BRCA wild-type ovarian cancer was based on the inhibition of homologous recombination or enhancement of olaparib-mediated DNA damage [[49], [50], [51], [52]], but cancer cells are potentially susceptible to ferroptosis when exhibiting specific mechanisms of resistance to DNA damage and downstream cell death forms (e.g., apoptosis) [25]. For instance, cancer cells undergoing epithelial mesenchymal transformation or chemoresistance are vulnerable to multiple FINs [28,53]. We provide the first evidence that BRCA wild-type ovarian cancer is sensitive to FINs targeting SLC7A11 or GPX4 and that augmentation of ferroptosis by knocking down SLC7A11 or FINs targeting SLC7A11 can significantly potentiate the efficacy of olaparib (Fig. 6I). Interestingly, several studies have shown that erastin or sulfasalazine, while inhibiting the function of SLC7A11, upregulates the expression of SLC7A11 in cells antagonizing erastin or sulfasalazine as an adaptive response [38,54]. It is likely that the synergistic inhibition of SLC7A11 by olaparib in combination with erastin or sulfasalazine could underlie the potently enhanced lipid peroxidation and ferroptosis. In addition, in some contexts, olaparib also induces GPX4 expression as an adaptive response to attenuate olaparib-mediated ferroptosis, in which case FINs targeting GPX4 can synergistically sensitize olaparib through abrogation of this adaptive response (Fig. 6I). Thus, we suggest an important role for ferroptosis in the efficacy of olaparib and inventively provide a promising combination strategy for sensitizing BRCA wild-type ovarian cancer to olaparib treatment. Notably, we show that sensitization of olaparib by boosting ferroptosis does not affect olaparib-mediated DNA damage, suggesting that ferroptosis-mediated olaparib sensitization is likely parallel to DNA damage-mediated olaparib sensitization, which provides novel insights into the synergistic sensitization of olaparib in BRCA wild-type ovarian cancer: co-regulation of ferroptosis and DNA damage responses. In the future, the efficacy and safety of triple therapy combining olaparib, FINs and DNA damage-promoting drugs in the treatment of BRCA wild-type ovarian cancer deserve further exploration.
We acknowledge that a limitation in our animal studies is that we did not use the same cell line-derived xenografts to demonstrate the effects of liproxtatin-1 cotreatment with olaparib and olaparib cotreatment with SAS. We initially sought to demonstrate that olaparib significantly induces ferroptosis and that ferroptosis represents an important mechanism for olaparib-mediated tumor suppression in ovarian cancer, so we chose the A2780 cell-derived xenografts to investigate whether liproxstatin-1 could attenuate the efficacy of olaparib. Subsequently, we used HEY cells that were relatively resistant to olaparib to investigate whether SAS could sensitize olaparib and overcome its resistance in ovarian cancer. However, the optimal option to demonstrate that olaparib induces ferroptosis and synergizes with FINs in vivo is using the same cell line derived xenografts, which will be conducted in our future studies.
In summary, our study reveals that PARP inhibition dramatically promotes ferroptosis in ovarian cancer, suggests that ferroptosis is responsible to PARP inhibition-mediated antitumor effects, and establishes that FINs synergistically sensitize BRCA proficient ovarian cancer cells to the PARP inhibitor olaparib, providing a promising therapeutic strategy for the application of PARP inhibitor in BRCA-proficient ovarian cancer.
4. Materials and methods
4.1. Cancer cell lines
The human ovarian cancer cell lines HEY, A2780, SKOV3 and human embryonic kidney cell line HEK293T obtained from the Cancer Institute, Central South University. HEY, SKOV3 and HEK293T cells were cultured in Dulbecco's modified Eagle medium (Gibco) in a 37 °C incubator with an atmosphere of 5% CO2. A2780 cells were cultured in RPMI-1640 medium (Gibco) in the same incubator with same condition. All the medium supplemented with 10% fetal bovine serum and 10,000 U/mL of penicillin-streptomycin. Cells within the eight passages between thawing and use were used for all experiments.
4.2. Establishment of indicated genes overexpression, knockdown, or CRISPR/Cas9 knockout cell lines
To generated CRISPR/Cas9-mediated PARP1 or p53 knockout cell lines, single guide RNAs (sgRNAs) were cloned into the lentiCRISPR v2 vector (Addgene) and then mixed with the packaging plasmids psPAX2 (Addgene) and PMD2.G (Addgene). These plasmids were subsequently cotransfected into HEK293T cells using Lipofectamine 3000 (Invitrogen) to generate the lentivirus particles. Lentivirus was harvested and used to infect A2780 or HEY cells in the presence of polybrene (GenePharma) at 48 h after transfection. Positive cells were then selected with puromycin (Beyotime) at 24 h after infection to obtain pooled CRISPR/Cas9 KO cells, followed by western blotting examination. The primer sequences of designed sgRNAs are listed in Supplemental Table. To establish SLC7A11 overexpression or knockdown cells, vector containing SLC7A11 cDNA or short hairpin RNA (shRNA) vectors targeting human SLC7A11 were used to generate SLC7A11 stable overexpression or SLC7A11 knockdown cells, respectively. Lentiviral transduction, puromycin selection and western blotting examination were performed as described above.
4.3. Clonogenic survival assay
To assess the effect of BSO (Selleckchem), NAC (Selleckchem), or cell death inhibitors including ferrostatin-1 (Selleckchem) and Z-VAD-FMK (Selleckchem) on the efficacy of olaparib (Selleckchem), cells with appropriate density per well were incubated in triplicate in 6-well or 12-well plates for 24 h. Cells were then treated with DMSO, BSO, NAC, olaparib, or olaparib in combination with BSO or NAC or indicated cell death inhibitors at appropriate concentrations for 48 h. Olaparib was then removed, and the medium was replaced every 48 h with fresh medium containing DMSO, BSO, NAC, or indicated cell death inhibitors. Cells were stained with 0.5% crystal violet (Sigma) dissolved in 20% methanol following 1–2 weeks of incubation, after which the number of colonies were counted visually (or crystal violet was redissolved in methanol and then absorbance was measured at 570 nm). The survival fraction was calculated using GraphPad Prism 6 and normalized to that of control (DMSO) cells.
4.4. Cell viability assay and treatment combination analysis
Cells at appropriate density per well were plated in triplicate in 96-well plates and allowed to adhere for 24 h. To assess the rescue effect of ferrostatin-1 on cells treated with olaparib, cells were treated with DMSO, ferrostatin-1, olaparib, or olaparib in combination with ferrostatin-1 at indicated concentrations for 48 (A2780) or 72 (HEY) hours, followed by incubation in fresh medium containing DMSO or ferrostatin-1 (DMSO or ferrostatin-1 was refreshed daily). To assess the sensitization of erastin (Selleckchem), sulfasalazine (Selleckchem) or RSL3 (Selleckchem) to olaparib, cells were first treated with DMSO or olaparib for 24 h and then co-treated with erastin/sulfasalazine/RSL3 and DMSO/olaparib for 24 h. Then the medium was replaced with 100 μL fresh medium containing 10 μL Cell Counting Kit-8 (CCK8) reagent (Beyotime Biotechnology, Nantong, China), and cells were incubated in a humidified incubator at 37 °C, 5% CO2 for 1.5 h. Cell viability was measured at 450 nm absorbance using a Spectra Max 250 spectrophotometer (Molecular Devices, Sunnyvale, CA, USA) and calculated according to the manufacturer's instructions.
To evaluate the combination effect of olaparib and FINs (erastin, sulfasalazine, or RSL3) at indicated concentrations, combination indexes (CI) at indicated fraction affected (FA) levels were calculated by CompuSyn software using Chou-Talalay method with nonconstant-ratio combinations. CI values less than, equal to, or greater than 1 indicate synergistic, additive, or antagonistic effects, respectively.
4.5. Cell death measurement
Cells were seeded in triplicate in 12-well plates for 24 h, followed by treatment with test compounds for the indicated times. After trypsinization into a cell suspension, cells were washed with PBS by centrifugation, followed by staining with 2 μM propidium iodide (PI) dye. The fluorescence intensity of cells with PI positive staining was measured by flow cytometry (Beckman Coulter) on the FL3 detector.
4.6. Lipid peroxidation assay
Published protocols were used for flow cytometry analysis [55,56]. Briefly, cells were seeded in triplicate in 12-well plates for 24 h, followed by treatment with test compounds for the indicated times. Cells in each well were then incubated with fresh medium containing 2 μM BODIPY 581/591C11 dye (Invitrogen) at 37 °C for 20 min. After trypsinization into a cell suspension, cells were washed with PBS by centrifugation. The fluorescence intensity of cells with BODIPY 581/591C11 staining was measured by flow cytometry (Beckman Coulter) on the FL1 detector that records only live cells using a gating technique. Oxidation of BODIPY™ 581/591C11 results in a shift of the fluorescence emission peak from −590nm (FL2 or FL3) to −510nm (FL1). The relative lipid peroxidation level is indicated by the percentage of cells gated by the black solid line based on the fluorescence intensity in FL1 channel, and the values obtained from three independent replicates for each condition per cell line were used to generate bar graph as quantitative data.
4.7. Quantitative real-time PCR
qRT-PCR was performed as previously described [57,58]. Briefly, total RNA was isolated from cells using TRIzol reagent (Invitrogen) and reversely transcribed into cDNA using Revert Aid First Strand cDNA Synthesis Kit (K1622, Thermoscientific, USA). Quantitative real-time PCR was performed using SYBR Premix Ex TaqII (TaKaRa), and triplicate samples were run on the RocheLightCycler® 96 instrument with the software (05815916001, Lifescience). The primers used are listed in Supplemental Table. The threshold cycle (Ct) value of target gene was normalized to those of β-actin, and relative expression of the target genes was calculated using 2−ΔΔCt.
4.8. Western blotting analysis
Western blotting was performed as previously described according to the standard protocol [57,58]. The primary antibodies and concentrations used for western blotting were: PARP1 (Cell Signaling Technology, 1:1000 dilution), SLC7A11/xCT (D2M7A) (Cell Signaling Technology, 1:1000 dilution), p53 (Cell Signaling Technology, 1:1000), GPX4 (Abcam-ab125066, 1:1000 dilution), BAP1 (Cell Signaling Technology, 1:1000), ATF3 (Cell Signaling Technology, 1:1000), NRF2 (Cell Signaling Technology, 1:1000), Vinculin (Cell Signaling Technology, 1:3000), Tubulin (Cell Signaling Technology, 1:3000), phospho-histone H2A.X (Ser139) (Cell Signaling Technology, 1:1000), phospho-Chk2 (Thr68) (Cell Signaling Technology, 1:1000), phospho-ATM (Ser1981) (Cell Signaling Technology, 1:1000), cleaved Caspase-3(Cell Signaling Technology, 1:1000). The secondary antibodies used were: horseradish peroxidase-conjugated anti-rabbit IgG (Cell Signaling Technology, 1:5000 dilution), horseradish peroxidase-conjugated anti-mouse IgG (Cell Signaling Technology, 1:5000 dilution). Proteins were visualized with the ECL Western blotting substrate (32,109, ThermoScientific, USA).
4.9. Immunofluorescence
Immunofluorescence were performed as previously described [57]. Briefly, after fixation in 3.7% formaldehyde in PBS and permeabilization in 0.1% TritonX-100/PBS, cells were blocked in 5% bovine serum albumin in 0.1% Triton/PBS for 1 h. Cells were then stained with phospho-histone H2A.X (Ser139) mouse antibody (Life Technology, USA, 1:500 dilution) at 4 °C for 12 h, followed by incubation with Alexa 594 anti-mouse fluorescent secondary antibodies (
Cell Signaling Technology, USA) in the dark. After counterstained with DAPI (C1005, Beyotime), cells were examined under a fluorescent microscope. Foci of phospho-H2AX were captured and quantified by a confocal laser scanning fluorescence microscope (LSM 700; Zeiss, Oberkochen, Germany).
4.10. Glutathione (GSH) assay
5000 cells per well were plated in triplicate in 96-well plates and allowed to adhere for 24 h, followed by treatment with the test compound (NAC, or BSO, or olaparib, or olaparib + NAC, or olaparib + BSO) for indicated time. Next, the medium was carefully replaced with fresh medium containing 100 μl of prepared 1 × GSH-Glo Reagent (Promega) and then incubated at room temperature for 30 min. Then 100 μl of reconstituted Luciferin Detection Reagent (Promega) was added to each well, shaking plate briefly, followed by incubation at room temperature for 15 min. Luminescence was detected and then normalized to cell viability. Relative GSH levels in cells treated with the test compound were normalized to those in control (DMSO) cells.
4.11. Animal experiments
Xenograft mouse model experiments were conducted in accordance with a protocol reviewed and approved by the Animal Care Committee of Hunan Cancer Hospital and the Affiliated Cancer Hospital of Xiangya School of Medicine (Changsha, China). Female 4- to 6-week-old BALB/c nude mice were purchased from SLA Laboratory Animal (Changsha, China) and housed in a specific pathogen-free facility. 2 × 106 A2780 or 1 × 106 HEY cells were injected subcutaneously into mice to grow tumors up to approximately 100 mm3. Mice were then intraperitoneally injected olaparib (100 mg/kg) or/and liproxstatin-1 (10 mg/kg, A2780) or/and sulfasalazine (250 mg/kg, HEY) until the endpoint indicated in the corresponding figures. Mice were monitored daily and the tumor volume calculated according to the equation volume = length × width [2] × 1/2.
4.12. Gene expression comparison and correlation in TCGA and GTEx databases
The expression data of PARP1, SLC7A11 and other ferroptosis-related genes in 33 cancer types were obtained from TCGA. The expression data of PARP1 and SLC7A11 in ovarian normal tissues were obtained from the GTEx database. Pearson's correlation coefficient was calculated to determine the expression correlation between PARP1 and SLC7A11 in ovarian tumor samples and normal tissues, or between PARP1 and ferroptosis-related genes in 33 cancer types.
4.13. Gene set enrichment analysis
All Gene Ontology (GO) gene sets were downloaded from the MSigDB database. Gene set enrichment analysis (GSEA) were performed using the clusterProfiler R package. All transcripts were ranked by correlation coefficient of PARP1 in TCGA-OV. The overrepresentation of indicated GO gene sets in the ranked gene lists is presented by the normalized enrichment score (NES).
4.14. Statistical analysis
Data are presented as means ± standard deviation (SD). Statistical significance (P values) was calculated by two-tailed unpaired Student t-test, or log-rank test using R software or GraphPad Prism 7.0. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; n.s., non-significant. All results of in vitro experiments were collected from at least 3 independent biological replicates.
Author contribution
T.H. performed most of the experiments with assistance from N.W. and X.Z. T.H. and G.L. conducted the data analysis. J.W., Y.Z. and G.L. designed the experiments and supervised the study. T.H. and G.L. wrote most of the manuscript. J.W. and Y.Z. revised the manuscript.
Declaration of competing interest
No competing interests.
Acknowledgements
This work was supported by the research grants from the National Natural Science Foundation of China (81972836 and 82003190), National Key R&D Program of China (2016YFC1303703), and Hunan Province Key R&D Program Projects (2020SK2120).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.101928.
Contributor Information
Yilei Zhang, Email: zhangyilei@xjtu.edu.cn.
Jing Wang, Email: wangjing0081@hnca.org.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Krishnakumar R., Kraus W.L. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol. Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaitanya G.V., Alexander J.S., Babu P.P. PARP-1 cleavage fragments: signatures of cell-death proteases in neurodegeneration. Cell Commun. Signal. 2010;8:31. doi: 10.1186/1478-811X-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiu L.-Y., Ho F.-M., Shiah S.-G., Chang Y., Lin W.-W. Oxidative stress initiates DNA damager MNNG-induced poly (ADP-ribose) polymerase-1-dependent parthanatos cell death. Biochem. Pharmacol. 2011;81:459–470. doi: 10.1016/j.bcp.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 4.Bai P., Cantó C. The role of PARP-1 and PARP-2 enzymes in metabolic regulation and disease. Cell Metabol. 2012;16:290–295. doi: 10.1016/j.cmet.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 5.Gibson B.A., Kraus W.L. New insights into the molecular and cellular functions of poly (ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 6.Feng F.Y., De Bono J.S., Rubin M.A., Knudsen K.E. Chromatin to clinic: the molecular rationale for PARP1 inhibitor function. Mol. Cell. 2015;58:925–934. doi: 10.1016/j.molcel.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sonnenblick A., De Azambuja E., Azim H.A., Jr., Piccart M. An update on PARP inhibitors—moving to the adjuvant setting. Nat. Rev. Clin. Oncol. 2015;12:27. doi: 10.1038/nrclinonc.2014.163. [DOI] [PubMed] [Google Scholar]
- 8.Engert F., Schneider C., Weiβ L.M., Probst M., Fulda S. PARP inhibitors sensitize Ewing sarcoma cells to temozolomide-induced apoptosis via the mitochondrial pathway. Mol. Canc. Therapeut. 2015;14:2818–2830. doi: 10.1158/1535-7163.MCT-15-0587. [DOI] [PubMed] [Google Scholar]
- 9.Huang X. Leveraging an NQO1 bioactivatable drug for tumor-selective use of poly (ADP-ribose) polymerase inhibitors. Canc. Cell. 2016;30:940–952. doi: 10.1016/j.ccell.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pommier Y., O'Connor M.J., de Bono J. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci. Transl. Med. 2016;8 doi: 10.1126/scitranslmed.aaf9246. 362ps317-362ps317. [DOI] [PubMed] [Google Scholar]
- 11.Lord C.J., Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355:1152–1158. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siegel R., PhD M.K.A.J.D. Cancer statistics. CA A Cancer J. Clin. 2018;68:11. 2018. [Google Scholar]
- 13.Vaughan S. Rethinking ovarian cancer: recommendations for improving outcomes. Nat. Rev. Canc. 2011;11:719–725. doi: 10.1038/nrc3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torre L.A. Ovarian cancer statistics. CA A Cancer J. Clin. 2018;68:284–296. doi: 10.3322/caac.21456. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konstantinopoulos P.A., Ceccaldi R., Shapiro G.I., D'Andrea A.D. Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Canc. Discov. 2015;5:1137–1154. doi: 10.1158/2159-8290.CD-15-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Audeh M.W. Oral poly (ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 17.Alsop K. BRCA mutation frequency and patterns of treatment response in BRCA mutation–positive women with ovarian cancer: a report from the Australian Ovarian Cancer Study Group. J. Clin. Oncol. 2012;30:2654. doi: 10.1200/JCO.2011.39.8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore K. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N. Engl. J. Med. 2018;379:2495–2505. doi: 10.1056/NEJMoa1810858. [DOI] [PubMed] [Google Scholar]
- 19.Dixon S.J. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stockwell B.R. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang L. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo J. Ferroptosis: a novel anti-tumor action for cisplatin. Cancer research and treatment. official journal of Korean Cancer Association. 2018;50:445. doi: 10.4143/crt.2016.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W. CD8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569:270–274. doi: 10.1038/s41586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lang X. Radiotherapy and immunotherapy promote tumoral lipid oxidation and ferroptosis via synergistic repression of SLC7A11. Canc. Discov. 2019;9:1673–1685. doi: 10.1158/2159-8290.CD-19-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Angeli J.P.F., Krysko D.V., Conrad M. Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat. Rev. Canc. 2019;19:405–414. doi: 10.1038/s41568-019-0149-1. [DOI] [PubMed] [Google Scholar]
- 26.Ye L.F. Radiation-induced lipid peroxidation triggers ferroptosis and synergizes with ferroptosis inducers. ACS Chem. Biol. 2020;15:469–484. doi: 10.1021/acschembio.9b00939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roh J.-L., Kim E.H., Jang H.J., Park J.Y., Shin D. Induction of ferroptotic cell death for overcoming cisplatin resistance of head and neck cancer. Canc. Lett. 2016;381:96–103. doi: 10.1016/j.canlet.2016.07.035. [DOI] [PubMed] [Google Scholar]
- 28.Hangauer M.J. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature. 2017;551:247–250. doi: 10.1038/nature24297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hassannia B., Vandenabeele P., Berghe T.V. Vol. 35. Cancer Cell; 2019. Targeting Ferroptosis to Iron Out Cancer; pp. 830–849. [DOI] [PubMed] [Google Scholar]
- 30.Stockwell B.R. A powerful cell-protection system prevents cell death by ferroptosis. Nature. 2019;575:597–598. doi: 10.1038/d41586-019-03145-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conrad M., Sato H. The oxidative stress-inducible cystine/glutamate antiporter, system x c−: cystine supplier and beyond. Amino Acids. 2012;42:231–246. doi: 10.1007/s00726-011-0867-5. [DOI] [PubMed] [Google Scholar]
- 32.Friedmann Angeli J.P. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 2014;16:1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang W.S. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao J.Y. A genome-wide haploid genetic screen identifies regulators of glutathione abundance and ferroptosis sensitivity. Cell Rep. 2019;26:1544–1556. doi: 10.1016/j.celrep.2019.01.043. e1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dixon S.J. Pharmacological inhibition of cystine–glutamate exchange induces endoplasmic reticulum stress and ferroptosis. Elife. 2014;3 doi: 10.7554/eLife.02523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Affar E.B., Carbone M. BAP1 regulates different mechanisms of cell death. Cell Death Dis. 2018;9:1151. doi: 10.1038/s41419-018-1206-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de la Vega M.R., Chapman E., Zhang D.D. NRF2 and the hallmarks of cancer. Canc. Cell. 2018;34:21–43. doi: 10.1016/j.ccell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L. ATF3 promotes erastin-induced ferroptosis by suppressing system Xc–. Cell Death Differ. 2020;27:662–675. doi: 10.1038/s41418-019-0380-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gout P., Buckley A., Simms C., Bruchovsky N. Sulfasalazine, a potent suppressor of lymphoma growth by inhibition of the xc− cystine transporter: a new action for an old drug. Leukemia. 2001;15:1633–1640. doi: 10.1038/sj.leu.2402238. [DOI] [PubMed] [Google Scholar]
- 40.Dolma S., Lessnick S.L., Hahn W.C., Stockwell B.R. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Canc. Cell. 2003;3:285–296. doi: 10.1016/s1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- 41.Sato H., Tamba M., Ishii T., Bannai S. Cloning and expression of a plasma membrane cystine/glutamate exchange transporter composed of two distinct proteins. J. Biol. Chem. 1999;274:11455–11458. doi: 10.1074/jbc.274.17.11455. [DOI] [PubMed] [Google Scholar]
- 42.Hu K. Suppression of the SLC7A11/glutathione axis causes synthetic lethality in KRAS-mutant lung adenocarcinoma. J. Clin. Invest. 2020;130:1752–1766. doi: 10.1172/JCI124049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Q. PARP-1 inhibition with or without ionizing radiation confers reactive oxygen species-mediated cytotoxicity preferentially to cancer cells with mutant TP53. Oncogene. 2018;37:2793–2805. doi: 10.1038/s41388-018-0130-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bai P. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metabol. 2011;13:461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marcar L. Acquired resistance of EGFR-mutated lung cancer to tyrosine kinase inhibitor treatment promotes PARP inhibitor sensitivity. Cell Rep. 2019;27:3422–3432. doi: 10.1016/j.celrep.2019.05.058. e3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hou D. Increased oxidative stress mediates the antitumor effect of PARP inhibition in ovarian cancer. Redox biology. 2018;17:99–111. doi: 10.1016/j.redox.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doll S. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575:693–698. doi: 10.1038/s41586-019-1707-0. [DOI] [PubMed] [Google Scholar]
- 48.Bersuker K. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575:688–692. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang L. Repression of BET activity sensitizes homologous recombination–proficient cancers to PARP inhibition. Sci. Transl. Med. 2017;9:eaal1645. doi: 10.1126/scitranslmed.aal1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun C. BRD4 inhibition is synthetic lethal with PARP inhibitors through the induction of homologous recombination deficiency. Canc. Cell. 2018;33:401–416. doi: 10.1016/j.ccell.2018.01.019. e408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ali R. Targeting PARP1 in XRCC1-deficient sporadic invasive breast cancer or preinvasive ductal carcinoma in situ induces synthetic lethality and chemoprevention. Canc. Res. 2018;78:6818–6827. doi: 10.1158/0008-5472.CAN-18-0633. [DOI] [PubMed] [Google Scholar]
- 52.Karakashev S. EZH2 inhibition sensitizes CARM1-high, homologous recombination proficient ovarian cancers to PARP inhibition. Canc. Cell. 2020;37:157–167. doi: 10.1016/j.ccell.2019.12.015. e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Viswanathan V.S. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature. 2017;547:453–457. doi: 10.1038/nature23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang L.-C. Heme oxygenase-1 mediates BAY 11–7085 induced ferroptosis. Canc. Lett. 2018;416:124–137. doi: 10.1016/j.canlet.2017.12.025. [DOI] [PubMed] [Google Scholar]
- 55.Martinez A.M., Kim A., Yang W.S. Immune Mediators in Cancer. 2020. Detection of ferroptosis by BODIPYTM 581/591 C11; pp. 125–130. [Google Scholar]
- 56.Ameziane-El-Hassani R., Dupuy C. Detection of intracellular reactive oxygen species (CM-H2DCFDA) Bio-protocol. 2013;3:e313. [Google Scholar]
- 57.Liu L. TRPM7 promotes the epithelial–mesenchymal transition in ovarian cancer through the calcium-related PI3K/AKT oncogenic signaling. J. Exp. Clin. Canc. Res. 2019;38:1–15. doi: 10.1186/s13046-019-1061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bing X. Circular RNA circTNPO3 regulates paclitaxel resistance of ovarian cancer cells by miR-1299/NEK2 signaling pathway. Mol. Ther. Nucleic Acids. 2020;21:780–791. doi: 10.1016/j.omtn.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.