Abstract
Oxidatively generated damage to DNA has been implicated in the pathogenesis of a wide variety of diseases. Increasingly, interest is also focusing upon the effects of damage to the other nucleic acids, RNA and the (2′-deoxy-)ribonucleotide pools, and evidence is growing that these too may have an important role in disease. LC-MS/MS has the ability to provide absolute quantification of specific biomarkers, such as 8-oxo-7,8-dihydro-2′-deoxyGuo (8-oxodG), in both nuclear and mitochondrial DNA, and 8-oxoGuo in RNA. However, significant quantities of tissue are needed, limiting its use in human biomonitoring studies. In contrast, the comet assay requires much less material, and as little as 5 μL of blood may be used, offering a minimally invasive means of assessing oxidative stress in vivo, but this is restricted to nuclear DNA damage only. Urine is an ideal matrix in which to non-invasively study nucleic acid-derived biomarkers of oxidative stress, and considerable progress has been made towards robustly validating these measurements, not least through the efforts of the European Standards Committee on Urinary (DNA) Lesion Analysis. For urine, LC-MS/MS is considered the gold standard approach, and although there have been improvements to the ELISA methodology, this is largely limited to 8-oxodG. Emerging DNA adductomics approaches, which either comprehensively assess the totality of adducts in DNA, or map DNA damage across the nuclear and mitochondrial genomes, offer the potential to considerably advance our understanding of the mechanistic role of oxidatively damaged nucleic acids in disease.
Keywords: Oxidative stress, DNA, RNA, Nucleotide pool, Biomarkers, DNA repair
Graphical abstract
Highlights
-
•
Oxidatively damaged nucleic acids are implicated in the pathogenesis of disease.
-
•
LC-MS/MS, comet assay and ELISA are often used to study oxidatively damaged DNA.
-
•
Urinary oxidatively damaged nucleic acids non-invasively reflect oxidative stress.
-
•
DNA adductomics will aid understanding the role of ROS damaged DNA in disease.
1. Introduction
Normal cellular metabolism, and exposure to the ambient, external environmental, leads to the production of reactive oxygen species (ROS [1,2]). This results in a physiological oxidative stress, recently termed oxidative eustress [3]. Over-production of ROS, depletion of antioxidant defences, or a combination of both, may occur following exposure to xenobiotics, radiation etc, resulting in oxidative stress, or oxidative distress [4]. Interestingly, the concept of physiological and pathological oxidative stress has been used previously in the context of pregnancy, in which pregnancy itself results in a physiological oxidative stress, but levels elevated beyond this may have pathological consequences, such as that associated with fetal growth restriction [5]. At the other end of the lifespan, ageing is linked to oxidative stress in the form of a vicious cycle where oxidative stress may accelerate the ageing process, and older individuals may be more susceptible to oxidative stress due to decreased activity of protective factors such as the antioxidant defence system and DNA repair activity [6]. A consequence of physiological production of ROS is damage to cellular macromolecules, as some ROS will always evade the antioxidant defenses. Levels of damage can be elevated further if exposure to the stressors is increased, or sustained, and/or the antioxidant defenses depleted. Whilst these macromolecules can include lipids and proteins, nucleic acids [7] are of particular interest because, in the case of DNA at least, they must be repaired rather than replaced. For the purpose of this review, “nucleic acids” is defined as DNA, RNA and their precursor 2′-deoxyribonucleotide and ribonucleotide pools.
1.1. Nomenclature
In this review we adhere to previous recommendations for terminology describing oxidative stress-induced damage to nucleic acids [8,9]. The term oxidative [insert name of biomolecule] damage is widespread within the literature e.g., oxidative DNA damage, but the increasingly preferred term is oxidatively damaged [insert name of biomolecule], or oxidatively generated [insert name of biomolecule] damage e.g., oxidatively damaged DNA, or oxidatively generated DNA damage. The reason for this is that oxidative damage implies that the damage itself has the ability to oxidise other substrates, and most forms of damage do not. Also, the widely studied oxidatively modified nucleobase, 8-oxo-7,8-dihydroguanine is abbreviated to 8-oxoGua, and its corresponding ribonucleoside (8-oxo-7,8-dihydroguanosine; 8-oxoGuo) and 2′-deoxyribonucleoside (8-oxo-7,8-dihydro-2′-deoxyguanosine; abbreviated to 8-oxodG, or 8-oxodGuo). Further explanation of this rationale, and other points of accuracy, are outlined by Cooke et al. [8], and in further detail by Cadet et al. [9]. The aim is to achieve greater standardization within the literature, to improve accuracy, clarity, and aid in literature searches.
2. Formation, repair and consequences of damage to nucleic acids
2.1. Formation and consequences of oxidatively generated damage to DNA
The interaction of ROS with DNA leads to the formation of at least 30 nucleobase modifications [10], and over 70 DNA lesions have been identified [11]. This includes single pyrimidine and purine nucleobase lesions, intra- and interstrand cross-links, and DNA-protein adducts, formed by the reactions of either the nucleobases or the 2-deoxyribose moiety with the hydroxyl radical (although a recent report has questioned the importance of •OH, in favour of the carbonate radical cation [12]), one-electron oxidants, singlet oxygen, and hypochlorous acid [11]. Of the nucleobase-derived lesions, 8-oxoGua, and 8-oxodG (Fig. 1) are most frequently measured.
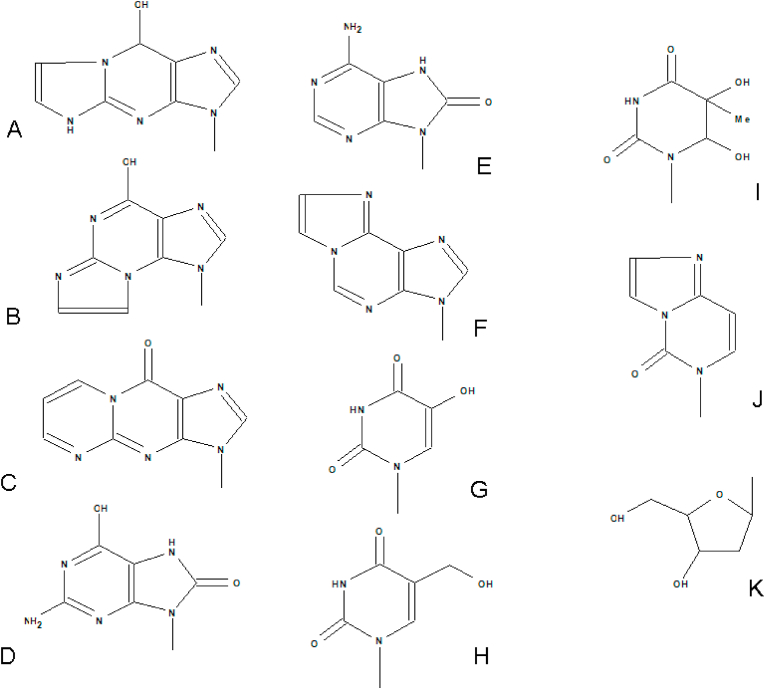
Fig. 1.
Structures of representative DNA nucleobase adducts analyzed in urine. A. 1,N2-εGua, B.N2,3-εGua, C. M1Gua, D. 8-oxoGua, E. 8-oxoAde, F. 1,N6-εAde, G. 5-OHUra, H. 5-HMeUra, I. Tg, J. 3,N4-εCyt, K. 2-deoxyribose which forms a N-glycosidic bond with the N9 in purines and N1 in pyrimidines to form the 2′-deoxyribonucleoside analogues of these nucleobase modifications.
The number of DNA adducts increases further when consideration is given to those adducts formed via the interaction between DNA and reactive intermediates, arising from the ROS modification of lipids and proteins, so-called secondary DNA products of oxidation reactions. These processes give rise to such adducts as 1,N6-etheno-2′-deoxyadenosine (εdA), 3-(2-deoxy-β-D-erythro-pentofuranosyl)pyrimido[1,2-α]purin-10(3H)-one (M1dG; Fig. 1), together with DNA-DNA, and DNA-protein cross-links [13].
Classically, oxidatively generated damage to DNA is thought to be of particular importance simply because it can lead to mutations [14], and hence cancer. However, the effects of non-mutational events, such as the promotion of microsatellite instability, and loss of heterozygosity, influence on gene expression through the interaction with transcription factors, and the acceleration of telomere shortening have also been highlighted [15]. Consequently ROS-induced DNA damage, and its repair, has been implicated in a wide variety of pathological conditions, such as cancer (reviewed in Kasai [16], neurodegenerative and cardiovascular disease, together with aging, reviewed in Ref. [17]), or see the collection of articles referred to by Nelson and Dizdaroglu (2002) [18]. The reported human levels of oxidatively generated damage to nucleic acids in disease and health is explored further under the ‘nucleic acid oxidation in human health and disease’ section.
Most recently attention has focused upon possible epigenetic effects arising from the oxidatively generated modification of nucleobases [[19], [20], [21]] perhaps, in part, mediated by their repair [22,23], and which has been considered in detail elsewhere [[24], [25], [26]].
By giving consideration to the above, various mechanisms together, we are building a better picture of how ‘damage’, or perhaps more accurately ‘modifications’, are involved in the disease process. This is leading to a better understanding of how oxidative stress, DNA damage/genomic instability, gene regulatory mechanisms and DNA repair, can influence the cellular redox status, maintain homeostasis, or contribute to, or prevent, the development or progression of various diseases (discussed in Mikhed et al. [27], and Gorinin et al. [28]), which include cancer [29,30], Alzheimer's disease [31], and other neurodegenerative diseases [32], and also ageing [33,34].
2.2. Nuclear vs. mitochondrial DNA
To date, the majority of reports have focused upon the measurement of oxidatively damaged nuclear DNA, particularly in human studies. However, the proximity of mitochondrial DNA (mtDNA) to sources of ROS production, together with the protection afforded by higher order levels of structure associated with nuclear DNA, makes mtDNA particularly vulnerable to the formation of damage from endogenous and exogenous sources [35,36], despite protective pathways, such as DNA repair [37]. Furthermore, it is well established that such damage to mtDNA, leads to mitochondrial dysfunction, which is a hallmark of a variety of diseases [38], such as Parkinson's Disease [[37], [38], [39]], and other neurological diseases (reviewed in Ref. [40]), together with ageing [41], possibly via mechanisms involving epigenetic regulation [42], for example, via mitochondrial dysfunction influencing the epigenetic profile of the nuclear genome [43]. Due to the small size of the mitochondrial genome, PCR has generally been the approach to study damage in individual and multiple genes in mtDNA [44]. The concept is that during PCR amplification, the presence of a damaged nucleobase, or abasic site, or strand break, will halt the polymerase, resulting in a prematurely truncated PCR product. The result is then corrected for mtDNA contents or mtDNA copy number. The concept of mtDNA content analysis is to determine the relative copy number using the single-copy gene of damaged nDNA and mtDNA versus undamaged [45]. Greater sensitivity can be achieved by using a longer amplicon, thereby increasing the likelihood of the polymerase identifying a lesion, using so-called long-range PCR [44,46], and this is capable of evaluating both deletion mutations, and damage in human mtDNA [47]. However, some forms of DNA damage are not detectable because the polymerase bypasses the damage, rather than being blocked by it [48].
Most recently, there have been two reports of genome-wide mapping approaches which have been applied to study the distribution of damage in both the nuclear and mitochondrial genomes, although one did not study oxidatively generated damage to mtDNA [49]. In the other, Wauchope et al. reported that M1dG is equally distributed across the mitochondrial genome, with no specific sites of enrichment, compared to untreated cells [50]. In contrast, differential levels of ROS-induced damage were noted across four mitochondrial DNA sites (D-Loop, COII/ATPase6/8, ND4/5, and ND1) [51], although levels of oxidised purines appear to be the same across three mitochondrial DNA regions (D-loop, Ori-L, and ND1) [52]. Levels of M1dG were higher in mtDNA, than nuclear DNA, likely reflecting the greater sensitivity of the mitochondrial genome, and/or less effective repair [50]. Global levels of M1dG persisted in mtDNA for at least 24 h; however, a mitochondrial genome-wide analysis was not performed for this part of the study, so it cannot be evaluated whether or not a sequence-specific loss of M1dG occurs. Despite the likely importance of damage to mtDNA, and indeed the crosstalk between the nuclear and mitochondrial genomes [53,54], damage to mtDNA is still not assessed as routinely as nuclear DNA damage.
2.3. Formation and consequences of oxidatively generated damage to the (2’-deoxy)ribonucleotide pools
The nucleic acid precursor (d)NTP pools do not possess the protective features of DNA, such as location in the nucleus, away from primary sources of ROS, nucleohistones, the higher orders of structure, and multiple, overlapping repair pathways – plus free (2′-deoxy)nucleotides are more prone to oxidation than paired polynucleotides [55]. Consequently, the (d)NTP pools, located in both the nucleus/cytosol and mitochondria, and with greater concentrations in the mitochondria [56] represent significant targets for oxidants. Furthermore, the ribonucleotide (NTP) pool is considerably larger than the 2′-deoxyribonucleotide (dNTP) pool (800.8 pmol/cell vs. 10.8 pmol/cell [56]), and hence would be expected to represent a greater target (see below).
Oxidation of the dGTP, for example, can lead to mis-incorporation of 8-oxodGTP into nuclear [57] and mitochondrial DNA [58], and it has been recently shown that 8-oxoGTP can also be mis-incorporated [59], both of which cause replication errors, such as mutations. As noted below, there are mechanisms to prevent this erroneous incorporation (e.g., 8-oxodGTPases), but if this fails, misincorporation into genomic DNA will lead to an increase in levels of the oxidised nucleobase [60], and can result in strand breaks, and cell death [61]. Indeed, this process has focused interest on using inhibition of 8-oxodGTPases as a therapeutic target in cancer [62] as, due to their faster rate of proliferation, and increased intracellular ROS, cancer cells are thought to have greater reliance on enzymes, such as the 8-oxodGTPases [[63], [64], [65]]. However, the results from the various studies exploring this area have been mixed between positive [[66], [67], [68]] and negative [69,70], although this may arise from differences in the inhibitors, the experimental systems being used [71], and redundancy in the pathways being inhibited [72].
Although the role of the (deoxyribo)nucleotide pool has also been studied in cancer [63], and neurodegenerative disease [73] for some time, increasingly it has been implicated in numerous other diseases, such as pulmonary arterial hypertension [74] – and with the current interest in compounds which inhibit the enzymes which sanitise the (d)NTP pools, and their potential application to diseases other than cancer, this area appears to be highly active.
With the focus on the oxidation of dGTP, there has been relatively limited investigation of oxidation of other DNA and RNA synthesis precursors, and their biological impact, and hence the literature is limited, but worth attention. A prime example is the reported 2-OHdATPase activity of hMTH1 [75] which might suggest an anti-mutagenic role in this context, in light of the mis-paring/mis-incorporation potential of 2-OHAde (isoguanine) [76]. Similarly, overexpression of MTH1 also prevents 2-OH-Ado-induced cytotoxicity, and limits the intracellular accumulation of both 2-OH-ATP and 2-OH-Ado in RNA [77]. Additionally, the activity of MTH1 towards modified, and potentially mutagenic, substrates has recently expanded beyond the oxidatively generated lesions, to include alkylation products (e.g., O6-methyl-dGTP and N6-methyl-dATP) [78,79], suggesting a potential role for this enzyme in the production of extracellular alkylated 2′-deoxyribonucleosides. In addition to 8-oxodGTP, the study of the activity of the Nudix hydrolases towards other modifications [80] is clearly warranted, and sanitisation of the (2′-deoxy)ribonucleotide pool is an important, but still largely overlooked, process to prevent the introduction of damage into the nucleic acids.
2.4. Formation and consequences of oxidatively generated damage to RNA and the NTP pools
Until relatively recently, oxidatively generated damage to RNA has received little attention [81], although we highlighted it as an emerging area of importance in 2004 [15]. Due to its abundance, single-stranded structure, relative lack of physically associated proteins, and location, RNA is more easily oxidised than DNA [82,83], and the detection of urinary 8-oxoGuo and 8-oxoGua, suggests that this oxidation does occur in some form in vivo [84].
Previously it was speculated that the persistence of damaged RNA could lead to the formation of error-containing proteins, with the authors reporting that 8-oxoguanine-containing RNA is sequestered to prevent entry into the process of translation [85]. More recently, it has been shown that the accumulation of 8-oxoGua in RNA can indeed alter protein synthesis, and lead to increased cellular production of β amyloid [86], which illustrates just how important RNA oxidation might be in pathogenesis. Supporting the notion that damage to RNA has important consequences for cell function, is evidence for the repair of RNA, for example the repair of alkylated RNA by the AlkB homologues [87,88]. However, the repair of oxidatively generated damage to RNA, in a manner analogous to the hOGG1 repair of DNA, does not yet seem to be have been reported, given their absence from a recent review [89], and our search of the literature. In contrast, an alternative mechanism exists which acts via limiting the cellular availability of oxidised transcripts to the translation machinery. This has been reported to occur via the human Y-box-binding protein 1 (YB-1), which serves a variety of functions associated with transcriptional and translational control and responses to stress [90]. Specifically, the YB-1 protein can bind 8-oxoGua-containing RNA, extracting it from the pool and preventing the production of aberrant proteins [91]. AUF1, and PCBP1 are human proteins which bind to RNA which contains a single 8-oxoGuo, or more than two 8-oxoGuo, respectively, for the purpose of triggering degradation of the RNA or apoptosis, respectively (reviewed in Ref. [89]; Fig. 2). PCBP2, also binds to heavily oxidised RNA but, unlike PCBP1, suppresses apoptosis during oxidative stress [92]. In addition to the direct formation of 8-oxoGuo by oxidation in situ in RNA, 8-oxoGTP can be mis-incorporated into RNA, at least in Escherichia coli, but this can be prevented by MTH1 hydrolysis of 8-oxoGTP [93].
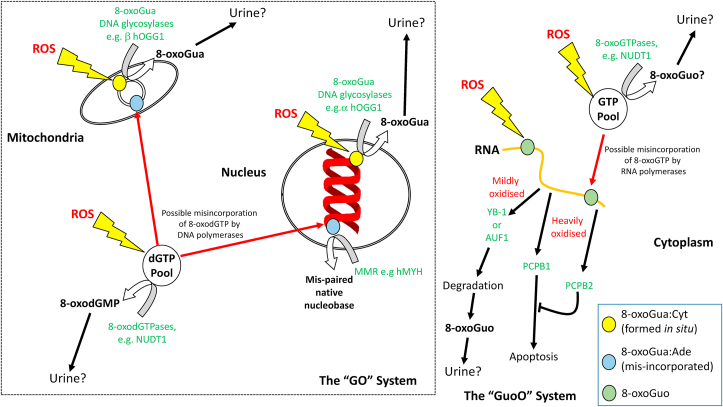
Fig. 2.
The “Gua Oxidation” (GO), and the “Guo Oxidation” (GuoO) Systems for preventing the presence and persistence of 8-oxoGua in DNA and RNA, respectively, together with potential sources of urinary 8-oxoGua, 8-oxodG, and 8-oxoGuo. 8-oxoGua may occur in DNA and RNA either by direct oxidation of Gua in situ, or mis-incorporation of 8-oxodGTP, or 8-oxoGTP, respectively, from the 2′-deoxyribonucleotide, and ribonucleotide pools, respectively.
A key role for RNA oxidation in disease is becoming clear. For example, there is significant RNA oxidation in affected neurones in Alzheimer's disease [94,95], and other neurodegenerative diseases, such as amyotrophic lateral sclerosis [96]. There is evidence that some mRNA species are more susceptible to oxidation than others, in particular [poly(A)+] mRNA, and rRNA [[97], [98], [99]]. In vitro studies with primary cultures, further demonstrated that the presence of oxidised nucleobases in mRNA cause ribosome stalling on the transcripts, resulting in a decrease in protein expression, and neuronal deterioration, providing a mechanistic link [100]. These earlier findings are confirmed by recent data using an exciting new methodology, 8-oxoGua-RNA-immunoprecipitation and RNA sequencing which, given the functional relevance of the oxidised transcripts, led the authors to propose that RNA oxidation is an additional driver of cell physiology, health, and disease [101].
Supportive this proposal there is an increasing number of medical conditions in which 8-oxoGuo in extracellular matrices (mainly urine) has been measured in humans, as a biomarker of RNA oxidation. These include: aging, and related disorders (summarised in Ref. [102]), hemochromatosis [103], diabetes [[104], [105], [106], [107], [108], [109], [110]], and a number of psychiatric disorders, such as schizophrenia [111], depression [112], bipolar disorder [113], psychosis [114], liver injury associated with Hepatitis B virus infection [115], sepsis [116], cerebral infarction [117], traumatic brain injury [118], and spontaneous intra-cerebral haemorrhage [119]. Unfortunately, to date, the mechanistic studies to explain the potential role of RNA oxidation in the above conditions, is less well advanced compared to these observational studies.
3. Methods for measuring nucleic acid biomarkers of oxidative stress
3.1. Artefactual formation of damage
To fully understand the extent to which such DNA lesions are involved in disease, methods for their analysis are essential. Numerous approaches have been applied to the study of oxidatively damaged DNA, including gas chromatography with mass spectrometry (GC/MS [120]), LC with electrochemical detection (LC-EC [121]), LC with single- [122], or tandem [123] mass spectrometry, 32P-post-labelling [124], immunoassay [125,126], alkaline elution [127] and the Comet assay [128], plus other methods based upon the nicking of DNA at oxidised nucleobases [129], using repair enzymes [130]. However, following the publication of a series of findings from the European Standards Committee on Oxidative DNA Damage (ESCODD [[130], [131], [132], [133], [134]]) and others [135,136], DNA extraction and sample workup (e.g., DNA hydrolysis and/or derivatisation) were identified as possible sources for the artefactual formation of damage, and a number of these techniques fell out of favour (reviewed by Guetens et al. [137]), and while the possibility of adventitious oxidation during sample storage and DNA extraction may not have been ruled out entirely, a number of procedures have been optimised to minimise the risk [138]. For example, drying under vacuum or pre-purification of the analyte using manual SPE (e.g., C18 cartridges) could lead to a significant, up to three-fold, increase in the levels of 8-oxodG (from 13 to 42 8-oxodG/106 dG in mouse liver DNA). To effectively prevent the artifacts formed during sample workup, the simplest approach is to use a direct measurement method involving an online enrichment/purification technique (e.g., online solid phase extraction; SPE) [139]. At present, the most widely used assays for the measurement of oxidatively damaged DNA are LC-MS/MS, the comet assay, and various immunochemical formats, e.g., ELISA.
4. Measurement of oxidation damage present in cellular nucleic acids
4.1. LC-MS (/MS)
Although already growing in popularity before ESCODD [[140], [141], [142]], the ESCODD studies demonstrated that liquid chromatography, coupled with mass spectrometry is a promising platform with which to study oxidatively damaged DNA [130]. Strengths of this technique include the use of stable, isotopically-labelled internal standards, which could account for matrix effects, and provide internal standardization, for more accurate quantification; the use of quantifier and qualifier ions to provide more reliable identification of the target molecule; a wide range of potential analytes [143]. Weaknesses of the approach include the requirement for significant amounts of biological material e.g., tissue, to obtain sufficient DNA for analysis; the need for extraction and hydrolysis of DNA, with attendant risks of artefact formation, although these have been largely addressed [138].
4.2. The comet assay
The comet assay is a single cell gel electrophoresis technique. The migration of DNA in agarose gels is considered to occur by relaxation of supercoiled DNA due to strand breaks. Importantly, the comet assay is adapted to measure oxidatively damaged DNA in mammalian cells by treatment of the gel-embedded nucleoids with base excision repair enzymes from either bacteria or human cells. As the repair enzymes excise the nucleobase lesions, strand breaks are created and thus add to the DNA migration in the comet assay. These additional lesions are called enzyme-sensitive sites, usually with more specific reference to the type of enzyme that has been used. The most widely used enzymes are formamidopyrimidine DNA glycosylase (Fpg) from bacteria and hOGG1. Both enzymes excise 8-oxoGua from DNA.
The enzyme-modified comet assay has been widely employed to measure levels of oxidatively damaged DNA in cells from animals and humans. An important advantage of the comet assay is that it can be performed on very few cells. For instance, one drop of blood from a finger prick contains enough cells to measure levels of oxidatively damaged DNA by the comet assay. In general, the comet assay is widely used in research fields where oxidative stress is considered to be implicated in health effects, including aging, dietary antioxidants/phytochemicals, air pollution and nanomaterials [[144], [145], [146]].
The standard comet assay was first described in the 1980s [147]. However, the Fpg-modified comet assay came into use in the middle of the 1990s [148], and the hOGG1-modified version a decade later [149]. Importantly, the Fpg-modified comet assay was used in ESCODD in an effort to determine the background level of 8-oxodG in human cells [130]. These studies demonstrated that the enzymic detection of 8-oxoGua by the comet assay, alkaline unwinding and alkaline elution gave similar background levels of 8-oxoGua, whereas this level of 8-oxoGua was approximately one-order of magnitude lower than the levels measured by chromatographic assays [131,132]. The discrepancy was attributed to spurious oxidation of DNA during the processing or analysis of samples in the chromatographic assays, which is not a methodological issue in the comet assay because DNA is not isolated as such.
The ESCODD project demonstrated that the Fpg-modified comet assay worked reasonably well in most laboratories i.e., they were able to detect a difference between cells with high levels of DNA damage (i.e., 8-oxoGua generated by the photosensitizer Ro19-8022 plus visible light) and unexposed cells with low level of 8-oxoGua [132]. However, only three out of ten laboratories could detect a monotonic relationship between Ro19-8022 plus light exposure and Fpg-sensitive sites in cells [132]. Another disturbing observation was that the levels of Fpg-sensitive sites in peripheral blood mononuclear cells (PBMCs) from humans in different laboratories seemed to depend on the comet assay protocol used. This issue was dealt with by the subsequent European Comet Assay Validation Group (ECVAG) study that focused on assessment and reduction in variation related to DNA damage and repair activity in PBMCs [[150], [151], [152]]. Building on the experience gained from the ESCODD ring-trials, the ECVAG studies confirmed that the main source of variation in levels of Fpg-sensitive sites in different laboratories was assay procedures, and implementation of a standard assay procedure in all participating laboratories actually decreased the variation in levels of Fpg-sensitive sites in test samples that had been treated with Ro19-8022 and white light [153,154]. However, the inter-laboratory variation in Fpg-sensitive sites in human PBMCs from different laboratories was not substantially different in the ECVAG study as compared to a similar type of assessment in the ESCODD trial ten years earlier [155]. Currently, the assessment of oxidatively damaged DNA by the enzyme-modified comet assay is quite robust in individual laboratories, but it is still a challenge (and limitation) that a consensus on background levels of DNA damage has not been reached. In fact, the discussion about “realistic levels” of DNA damage by the comet assay has mainly pertained to various primary descriptors such as whether or not the % tail DNA descriptor is more informative than tail length or tail moment, two alternative endpoints. However, one needs to be an expert in the comet assay to interpret this kind of difference, whereas reporting the levels of 8-oxoGua relative to unaltered guanines or base pairs is much easier to understand [156]. Actually, the conversion of primary comet assay descriptors to lesions per million unaltered 2′-dG (or base pairs) were used in both the ESCODD and ECVAG projects by use of calibration using ionizing radiation [157]. It could be argued that one of the failures of the ESCODD and ECVAG projects is they have not been able to convince fellow researchers that the true numerical value of DNA damage is more informative than a comet assay-specific descriptor. However, the contributions and achievements that both ESCODD and ECVAG have made to the field are considered later on in this article.
4.3. Methods for the immunochemical detection of oxidatively damaged DNA
The application of immunochemical methods for the detection of oxidatively damaged DNA depends upon the availability of antibodies against the oxidised nucleobases (and/or their corresponding 2′-dN moieties) and their optimization for the various types of immunochemical approaches. Although numerous types of oxidatively damaged nucleobases have been described, most of the commercially available antibodies recognize 8-oxodG and/or 8-oxoGua, and in some cases also the corresponding ribonucleoside (8-oxoGuo). Therefore, we shall focus on application of these antibodies, and use the term anti-8-oxodG antibodies.
Two forms of immunochemical detection methods are most commonly used: immunohistochemistry/cytochemistry and microplate-based approaches (enzyme-linked immunosorbent assay, ELISA). Immunohistochemistry/cytochemistry requires fixation of the tissue or a cell suspension using formaldehyde or a similar fixative. The samples are then mounted on a microscope slide and permeabilized using detergents (e.g., Triton X-100, Tween 20). Proteins and RNA are digested with proteinases and RNases, and non-specific, antibody binding blocked using a suitable agent [e.g., fetal calf serum (FCS), or bovine serum albumin]. The recognition of oxidised nucleobases in DNA is performed using an appropriate, specific primary antibody. In the case of 8-oxodG, at least, it is important to denature both the chromatin, and the nuclear DNA, to allow the primary antibody access to the lesion [158]. This step is particularly important for cultured cells, and frozen tissue sections, although not formalin-fixed, paraffin-embedded sections [158]. An enzyme-conjugated secondary antibody which leads to the generation of colour/fluorescence. The samples are subsequently analyzed under a microscope [159]. Although this method allows for the localization of oxidised DNA in the samples, it is not suitable for precise quantification of the target/signal and is rather regarded as qualitative/semi-quantitative.
For quantification of oxidised nucleobases in DNA, ELISA is the immunochemical method of choice. Commercially available ELISA kits mostly use the competitive format, in which the competitor, i.e., the compound recognized by the primary antibody (e.g., 8-oxodG), is bound to the bottom of the ELISA plate (plate coating). The plate is blocked with FCS and equal volumes of the samples and the primary anti-8-oxodG antibody are added to the wells. Competition for antibody binding occurs between the 8-oxodG in the sample, and that coating the plate. After incubation and washing off the unbound samples/primary antibody, the secondary, enzyme-conjugated antibody is added to the plate. The detection of the antigen is performed by incubation with a suitable chromogenic substrate and measurement of the signal using a microplate reader. The intensity of the signal is inversely proportional to the concentration of antigen in the sample. The ELISA kits include a calibration curve, comprised of a range of concentrations of 8-oxodG, which are added as competitors for the anti-8-oxodG antibody, and provide greater quantification. In some ELISA experimental protocols, the DNA to be analyzed is hydrolysed to free 2′-dN, e.g., 8-oxodG, prior to incubation with the primary antibody [160], rather than simply using extracted DNA. Given that the calibration curve is in the format of free 2′-dN, inclusion of this step is highly recommended. Using unhydrolyzed, double-, or even single-stranded DNA in the ELISA is unlikely to provide accurate results, as it is not certain that primary antibody will have access to every lesion. Additional accuracy can be provided by separately quantifying the free dG (e.g., by LC-UV), and expressing the 8-oxodG results relative to dG, taking into account the efficiency of hydrolysis, instead of simply ng/mL of 8-oxodG [160]. Finally, as discussed elsewhere in this review, caution must be taken to avoid artefactual oxidation of the samples, particularly when extracting DNA from cells/tissues.
A more recent antibody application is the mapping 8-oxodG locations in genomic DNA. When this approach was first described, antibodies were used for immunodetection of 8-oxodG in metaphase chromosomes, spread on microscope slides and evaluated using fluorescence microscopy [161]. Although this method allowed mapping of the distribution of 8-oxodG in the genome, the results were limited by the low resolution of optical microscopy. However, technological advances have led to an improved approach for the detection of modified nucleobases in genomic DNA, utilizing next generation sequencing after immunoprecipitation of the damaged regions using anti-8-oxodG antibody (reviewed in Ref. [162], and considered more in “The Future” section of this review).
In summary, while immunochemical methods represent a relatively cheap, fast and potentially high-throughput means of detecting oxidatively damaged DNA, they are limited by availability of suitable antibodies, coupled with their specificity and detection limits, which may substantially affect quality of the experimental data.
4.4. Limitations of, and alternatives to, the measurement of solid tissue-derived, nucleic acid biomarkers of oxidative stress
Assessment of damage to DNA by methods requiring invasive procedures, e.g., venepuncture to obtain blood samples or tissue biopsy, imposes severe limitations in (large-scale) human studies, requiring staff with specialist training, decreasing the likelihood of consent, and decreasing access to vulnerable populations, such as the very young, or very old. Furthermore, in the case of whole blood, the blood needs to be diluted, prior to isolation of the PBMCs by density gradient separation (preparation of the buffy coat) – which is a time-consuming process. In an effort to address this, and understanding that the comet assay requires a relatively low number of cells per gel (~30,000 cells are added to the agarose, but only 50–100 comets per gel are scored), we demonstrated that it is possible to add a pin-prick of blood (5 μL) directly to the agarose, and score the leukocytes present in the gel [163]. Furthermore, we discovered that, unlike isolated PBMCs, a cryopreservative did not need to be added to these small volumes of blood (<250 μL) to prevent the formation of artefactual DNA oxidation products upon freezing for up to one month [163]. Longer cryopreservation periods have not been tested for specific DNA oxidation products, although the results show that background levels of DNA strand break in the cells of whole blood samples are not affected after up to five years of storage in the freezer [164,165]. This work led to further investigations on applicability of stored, frozen blood samples to the comet assay. Ladeira et al. (2019) showed that whilst long-term storage of frozen buffy coat samples in cryostraws, with no cryopreservative was not suitable for subsequent comet assay analysis, frozen whole blood samples stored in cryostraws, without cryoprotection (the sample storage method of choice for many biobanks), resists efficiently freezing/thawing artefact, with minimal increases in strand breaks (but no other lesion), for at least three months [166]. Taken together, this indicates that the comet assay and small volumes of whole blood are well suited to human biomonitoring.
5. Measurement of oxidised nucleic acid products in extracellular biological matrices
Examining the products of oxidatively generated damage to nucleic acids in extracellular matrices offers a means by which oxidative stress may be assessed non-, or at least minimally-invasively, and circumvents extraction and the associated risk of artefact formation, offering a number of advantages over examining levels of damage in cellular nucleic acids. To date, the extracellular matrices in which these biomarkers have been studied include: serum/plasma [167,168], saliva [167], amniotic fluid [168], seminal plasma [168], follicular fluid [169,170], breast milk [168], cerebrospinal fluid [171], and, most widely used of all, urine (reviewed in Refs. [172,173]). LC-MS/MS is the main methodology for the majority of these matrices, emphasizing the strength of this approach, although ELISA has been used for serum [125,174,175], saliva [[176], [177], [178], [179]], sputum [180], follicular fluid [169,170], and urine [[181], [182], [183], [184]] (for others, see the ELISA section below).
5.1. Measurement of oxidised nucleic acid products in urine
The methods that have been applied primarily to the analysis of oxidatively damaged DNA lesions in urine are either chromatographic [e.g., LC-GC/MS (liquid chromatography pre-purification prior to GC-MS), LC-MC/MS LC-EC, GC-MS], or immunochemical. The majority of assays focus upon 8-oxodG as the analyte of choice, although the others lesions have been reported to be present in urine, either as nucleobase adducts, or as the corresponding modified 2′-deoxyribonucleoside.
5.1.1. Chromatographic techniques
Urine is a complex matrix, and therefore the common challenge for all chromatographic techniques has been to clean-up the urine sufficiently to simplify analysis, plus this also extends instrument life. At its simplest, column switching has meant that, following chromatographic separation of the urine's constituents, only the fraction containing the compound of interest (e.g., 8-oxodG) is applied to the final separation column and detector, either EC [185,186] or MS [84], and the remaining material is diverted to waste.
Early studies focused upon urinary thymine glycol (ThyGly), using boronate, SPE columns to isolate ThyGly and thymidine glycol (dThyGly) from urine, after which the lesions were chemically reduced back to thymine and thymidine prior to analysis by HPLC with UV detection [187]. However, 8-oxodG offered a one thousand-fold greater detection sensitivity over ThyGly, using LC-EC, plus higher levels in urine [188], which meant that urinary measurements of ThyGly were quickly superseded by 8-oxodG, using a variety of LC-EC formats [185,[189], [190], [191], [192]].
The popularity of LC-EC largely remained, until the development of mass spectrometry, with its benefits of the use of isotopically-labelled internal standards, simplifying quantification and accounting for loss during sample workup (and storage), differences in ionization efficiencies due to matrix effects, and confirmation of analyte identity. Also, more than one analyte could be studied in the same run by LC-MS/MS e.g., 8-oxoGua, 8-oxoGuo and 8-oxodG, together with the native moieties (Gua, Guo and dG) [84]. Alternatively, LC-GC/MS could also be used, with similar benefits to LC-MS/MS e.g., the simultaneous analysis of five urinary, oxidatively modified DNA products including 8-oxoGua, 5-(hydroxymethy)luracil (5-HMUra), 5-hydroxyuracil (5-OHUra), 8-oxodG, and 8-oxo-7,8-dihydroadenine (8-oxoAde) [193], but also Gua, and dG [194,195]. Analysis of 8-oxodG using manual SPE prior to LC-MS/MS [[196], [197], [198]] proved effective, but is time-consuming, and has been largely replaced by online SPE, which offers automation, and higher throughput [199], and is suitable for application to other extracellular matrices, such as plasma and saliva [200]. It is worth noting that, while offline SPE for the analysis of 8-oxodG in DNA has a risk of artefactual formation of 8-oxodG, this is not the case for offline SPE of urine. More recently, (5′R)- and (5′S)-8,5′-cyclo-2′-deoxyadenosines have been identified in urine [201], extending further the repertoire of oxidatively generated lesions present in urine.
5.1.2. Immunoassay
For the detection of oxidatively generated lesions, specifically 8-oxodG, in extracellular matrices, competitive ELISA represents the optimal approach. A diverse range of types of samples can be analyzed by 8-oxodG ELISA. Depending on the ELISA manufacturer, these include e.g., blood plasma, serum, cell lysates, tissue homogenates, urine, and DNA extracted from biological material and other matrices. In the literature, the origin of the ELISAs used has been described either as “in-house”, or as a ready-to use commercial kit. For the in-house assay, a microplate is coated with a suitable antigen-protein conjugate usually prepared by the investigator, and the primary anti-8-oxodG antibody is obtained from a commercial supplier, or other source [202,203]. Commercial kits come with the plates already pre-coated with an antigen-protein conjugate and thus the experimental protocol is usually relatively short, consisting of adding the analyzed samples and the primary antibody to the wells, their incubation, washing, adding the secondary antibody conjugated with an enzyme, then developing, and quantifying the colour, after pipetting substrate into the plate.
Although the in-house ELISA format represents a cost-effective variant of the assay, comparison of the data generated by this approach with chromatographic methods shows rather poor agreement [204,205]. It is therefore advisable to use commercial ELISA kits that provide better standardization and consistency in the results, not least derived from the rigours of the manufacturing process. Currently, 8-oxodG ELISA kits are available from over 25 manufacturers/distributors, including the Japanese Institute for the Control of Ageing (JaICA), Trevigen, R&D Systems, Abcam, Cell Biolabs, Cayman Chemical, Enzo, Oxford Biomedical Research, MyBioSource, Creative Diagnostics, and StressMarq Biosciences. The kits appear to differ in the types of samples recommended as being suitable for analysis (serum, plasma, urine, culture medium, cell lysates, saliva, DNA) and detection limits (ranging from 0.1 ng mL to 3.12 ng/mL) (summarised in Table 1). Cross-reactivity of the primary antibody with other compounds is reported by some, but not all manufacturers. These interfering compounds include e.g., 8-oxoGuo, 8-oxoGua and other 8-oxodG analogues, uric acid, urea, creatinine or creatine. Interestingly, some companies report no significant cross-reactivity with 8-oxodG analogues, whereas others note that the primary antibody recognizes not only free 8-oxodG, but also the nucleoside incorporated in DNA fragments present in complex matrices (e.g., cell lysates, plasma, tissue homogenates). Some kit instructions stress that ELISA data do not agree with that reported by LC-MS/MS. Unfortunately, the name of the clone from which the primary antibody was derived is often not specified, making it difficult to obtain critical information on the specificity and cross-reactivity of the assay. However, given differences in the matrices, structures recognized by the kits’ antibodies and the detection limits, it is reasonable to conclude that several different primary antibodies are used in the kits.
Table 1.
Overview and basic characteristics of 8-oxodG ELISA kits currently available on the market. Information obtained from webpages of individual manufacturers. N.B., We have largely retained the nomenclature used by the manufacturer to describe the kit, which may not be accurate, or indeed correct. Furthermore, the same kit may be distributed by multiple companies, but it is often impossible to establish whether this is the case, if the kit has been re-branded. Where possible, we have aimed to identify this.
| Kit name | Sample type | Sensitivity | Specificity/cross-reactivity/other note | Manufacturer | Kit price | Comment |
|---|---|---|---|---|---|---|
| 8-oxo-dG ELISA Kit | DNA, plasma, urine, saliva samples | 0.57 ng/mL | Cross-reactivity: 8-hydroxydeoxyguanosine 100%, 8-hydroxyguanosine ~30%, 8-hydroxyguanine ~20%, 8-mercatoguanosine ~4%, 8-bromoguanosine CMP, guanosine (Guo), guanine (Gua), 2′-deoxyinosine, N2-methylguanosine | Creative Diagnostics, New York, NY, USA | Upon inquiry | No information on experimental protocol was available at the manufacturer's website |
| New 8-OHdG Check ELISA | Urine from human and animals | 0.5 ng/mL | Antibody N45.1. Specific for 8-OHdG, tested against 8-OHdG analogues: Guo, 7-methyl-G, 6-SH-G, 8-bromo-G, dA, dC, dT, dI, dU, dG, O6-methyl-dG, 8-OHdA, Gua, O6-methyl-Gua, 8-OH-Gua, uric acid, urea, creatine, creatinine, 8-sulfhydryl-G, 8-OH-G. | Japan Institute for the Control of Aging (JaICA), Fukuroi, Shizuoka, Japan | US$ 450 | A commonly used kit that includes the well characterized antibody N45.1. Manufacturer recommends for analysis of urine samples, and was used with our improved methodology for urine. |
| Highly sensitive 8-OHdG Check ELISA | Urine, serum, tissue and cultured cells | 0.125 ng/mL | Antibody N45.1. Specific for 8-OHdG, tested against 8-OHdG analogues: Guo, 7-methyl-G, 6-SH-G, 8-bromo-G, dA, dC, dT, dI, dU, dG, O6-methyl-dG, 8-OHdA, Gua, O6-methyl-Gua, 8-OH-Gua, uric acid, urea, creatine, creatinine, 8-sulfhydryl-G, 8-OH-G. | Japan Institute for the Control of Aging (JaICA), Fukuroi, Shizuoka, Japan | US$ 450 | A kit similar to the “New 8-OHdG Check ELISA”, but with higher sensitivity, manufacturer specifically recommends for analysis of serum, tissue and cultured cells. |
| High Sensitivity 8-Hydroxydeoxyguanosine (8-OHdG) ELISA Assay Kit | Plasma and DNA extracts from cells or tissue | 0.125 ng/mL | Antibody N45.1 (see above). | Northwest Life Science Specialities, Vancouver, WA, USA | US$ 945 | This kit appears to be similar/identical to “Highly sensitive 8-OHdG Check ELISA” by JaICA |
| DNA Damage (8-OHdG) ELISA kit | Cell lysates, plasma, urine | 0.59 ng/mL | Cross-reactivity: 8-hydroxy-2-deoxyguanosine (8-OHdG): 100%. 8-hydroxyguanosine (8-OHG): 23%. 8-hydroxyguanine (8-oxoG): 23%. guanosine: <0.01%. | StressMarq Biosciences Inc., Victoria, BC, Canada |
US$ 399 | No information on the primary antibody. No detailed protocol available. Incubation time of the samples 1 h, but temperature is not specified (probably room temperature). |
| DNA/RNA Oxidative Damage (High Sensitivity) ELISA Kit | Urine, plasma, serum, culture media, cell lysates, tissue samples, saliva | 0.30 ng/mL | Cross Reactivity: 8-hydroxy-2′-deoxyguanosine: 100%, 8-hydroxyguanosine: 38%, 8-hydroxyguanine: 23%, guanosine: <0.01% | Cayman Chemical Ann Arbor, MI, USA |
Upon inquiry | This kit is also recommended, by the manufacturer, for detection of oxidatively damaged RNA. A competitive ELISA that uses 8-OH-dG-acetylcholinesterase conjugate. No information on primary antibody provided. |
| 8-hydroxy 2 deoxyguanosine ELISA Kit | Cell culture supernatant, cell lysate, plasma, saliva, serum, tissue extracts, urine | 0.59 ng/mL | 8-OHdG antibody used in this assay recognizes both free 8-OHdG and DNA-incorporated 8-OHdG, concentrations of 8-OHdG reported by ELISA methodology will not coincide with those reported by LC-MS | Abcam, Cambridge, UK | € 795 | No information on primary antibody. The manufacturer explicitly mentions discrepancy between ELISA and chromatography. Incubation of the samples with primary antibody 1 h at room temperature. (N.B. Evidence suggests that this will decrease the kit's potential specificity, as incubation at 4 °C overnight improves Ab binding specificity.) |
| DNA Damage (8-OHdG) ELISA kit | Urine, cell culture, plasma and other sample matrices. | 0.59 ng/mL | 8-OHdG antibody used in this assay recognizes both free 8-OHdG and 8-OHdG incorporated into DNA. Since complex samples such as plasma, cell lysates, and tissues are comprised of mixtures of DNA fragments and free 8-OHdG, concentrations of 8-OHdG reported by ELISA methodology will not coincide with those reported by LC-MS where the single nucleoside is typically measured. This should be kept in mind when analyzing and interpreting experimental results. | Agrisera, Vännäs, Sweden | € 669 | The kit's description suggests similarity with “8-hydroxy 2 deoxyguanosine ELISA Kit” by Abcam. Incubation of the samples with primary antibody 1 h at room temperature (see above comment). |
| 8 Hydroxyguanosine (8-OHdG) ELISA Kit | Urine, cell culture supernatant, serum, plasma, saliva and cell/tissue lysate | 0.59 ng/mL | 8-OHdG antibody used in this assay recognizes both free 8-OHdG and DNA-incorporated 8-OHdG. Since complex samples such as plasma, cell lysates, and tissues are comprised of mixtures of DNA fragments and free 8-OHdG, concentrations of 8-OHdG reported by ELISA methodology will not coincide with those reported by LC-MS where the single nucleoside is typically measured. This should be kept in mind when analyzing and interpreting experimental results. | Arigobio, Hsinchu City, ROC, Taiwan | Upon inquiry | The kit description suggests similarity with “8-hydroxy 2 deoxyguanosine ELISA Kit” by Abcam. Incubation of the samples with primary antibody 1 h at room temperature (see above comment). |
| DNA damage ELISA kit | Plasma, seminal fluids, cell lysates, culture supernatants and DNA extracts | 0.59 ng/mL | The DNA Damage ELISA also detects 8-hydroxyguanosine (product of oxidative RNA damage) and 8-hydroxyguanine (product of oxidative DNA damage by hydroxyl radicals). | Enzo Life Sciences, Inc., New York, NY, USA | Upon inquiry | No information on primary antibody. Incubation of the samples with primary antibody for 1 h at room temperature (see above comment). |
| ELISA Kit for 8-Hydroxydeoxyguanosine (8-OHdG) | Serum, plasma and other biological fluids | 0.74 ng/mL | This assay has high sensitivity and excellent specificity for detection of 8-Hydroxydeoxyguanosine (8-OHdG). No significant cross-reactivity or interference between 8-Hydroxydeoxyguanosine (8-OHdG) and analogues was observed. | Cloud-Clone Corp., Katy, TX, USA | US$ 796 | Primary antibody not specified. Requires incubation of the samples with primary antibody 1 h at 37 °C (see above comment). |
| 8-hydroxy-2′-deoxyguanosine (8-OHdG) ELISA Kit | Serum, plasma, tissue homogenates, culture supernatants and other biological fluids | 0.469 ng/mL | Cross Reactivity: No significant cross-reactivity or interference between this analyte and its analogues was observed. | BioTrend, BioVision Inc., Köln, Germany | Upon inquiry | No information on primary antibody. The samples are incubated with the primary antibody for 45 min at 37 °C (see above comment). |
| ELISA Kit for 8-Hydroxydeoxyguanosine (8-OHdG) | Serum, plasma and other biological fluids | 0.74 ng/mL | This assay has high sensitivity and excellent specificity for detection of 8-Hydroxydeoxyguanosine (8-OHdG). No significant cross-reactivity or interference between 8-Hydroxydeoxyguanosine (8-OHdG) and analogues was observed. | USCN, Wuhan USCN Business Co., Ltd., Hubei, China | US$ 796 | The kit appears to be identical to “ELISA Kit for 8-Hydroxydeoxyguanosine (8-OHdG)” by Cloud-Clone Corp. |
| HT 8-oxo-dG ELISA Kit II | Plasma, urine, saliva samples and cultured cells | 0.57 ng/mL | N/A | R&D Systems, Inc., Minneapolis, MN, USA | US$ 962 | Primary antibody not specified. Information on possible interference with “factors” in the samples compromising the assay. Samples incubated with primary antibody 1 h at room temperature (see above comment). |
| The HT 8-oxo-dG ELISA Kit II quantifies 8-hydroxy-2′-deoxyguanosine (8-OHdG) | DNA, plasma, urine and saliva samples | 0.57 ng/mL | N/A | Trevigen, Gaithersburg, MD, USA | US$ 962 | The kit is identical to “HT 8-oxo-dG ELISA Kit II” by R&D Systems, Inc. |
| HT 8-oxo-dG ELISA Kit II | DNA, plasma, urine and saliva samples | 0.57 ng/mL | N/A | Bio-Techne, Minneapolis, MN, USA | US$ 962 | The kit is identical to “HT 8-oxo-dG ELISA Kit II” by R&D Systems, Inc. |
| Human 8-Hydroxyguanine (8-oxo-dG) ELISA Kit | Serum, plasma, cell culture supernatants, cell lysates, tissue homogenates | 2.89 ng/mL | N/A | MyBioSource, San Diego, CA, USA | US$ 405 | A non-standard sandwich ELISA. No information on anti-8-oxodG antibodies provided. Samples incubation 1 h at 37 °C (see above comment). |
| 8-OHdG DNA Damage ELISA | Urine, serum, cells or tissues | 0.10 ng/mL | N/A | Cell Biolabs Inc., San Diego, CA, USA | US$ 820 | No information on primary antibody, no specific samples pre-treatment recommended. Samples should be incubated with primary antibody for 1 h at room temperature (see above comment). |
| 8OHDG ELISA Kit | Serum, plasma, tissue homogenates, cell culture supernatants and other biological fluids | 0.27 ng/mL | N/A | CliniSciences, Nanterre, France | Upon inquiry | The kit is manufactured by Aviva Systems Biology – see below for more information. |
| 8-OHdG (8-Hydroxydeoxyguanosine) ELISA Kit | Serum, plasma and other biological fluids | 0.94 ng/mL | N/A | Elabscience, Houston, TX, USA | US$ 495 | A competitive format that uses biotin-avidin-horseradish peroxidase for detection. Samples incubation: 45 min at 37 °C (see above comment). |
| 8-Hydroxy-2' -Deoxyguanosine (8-OHdG) ELISA Kit | Serum, plasma and other biological fluids. | 0.94 ng/mL | N/A | Abbexa Ltd, Cambridge, UK | € 669 | The kit appears to be identical with “8-OHdG(8-Hydroxydeoxyguanosine) ELISA Kit” by Elabscience |
| Human 8-Hydroxydeoxyguanosine (8-OHdG) ELISA kit | Serum, urine | 3.12 ng/mL | N/A | CUSABIO, Houston, TX, USA | Upon inquiry | No information on primary antibody. Np specific instructions for urine samples processing, apart from centrifugation. The samples are incubated with primary antibody for 30 min at 37 °C (see above comment). |
| DNA Damage (8-OHdG) ELISA Kit | Cell lysates, plasma, urine | 0.59 ng/mL | 8-OHdG antibody used in this assay recognizes both free 8-OHdG and DNA-incorporated 8-OHdG. Since complex samples such as plasma, cell lysates, and tissues are comprised of mixtures of DNA fragments and free 8-OHdG, concentrations of 8-OHdG reported by ELISA methodology will not coincide with those reported by LC-MS where the single nucleoside is typically measured. This should be kept in mind when analyzing and interpreting experimental results. | BOSTERBIO, Pleasanton, CA, USA | US$ 390 | The kit description suggests similarity with “8-hydroxy 2 deoxyguanosine ELISA Kit” by Abcam. |
| 8OHDG ELISA Kit | Serum, plasma, tissue homogenates, cell culture supernatants and other biological fluids | 0.27 ng/mL | N/A | Aviva Systems Biology, San Diego, CA, USA | US$ 559 | The clone of primary antibody not specified. No specific information on urine samples processing. Kit uses 8-OHdG-biotin complex that should be incubated with samples for 1 h at room temperature (see above comment). |
| 8-OHdG Assay Kit | Tissue, serum, plasma or urine samples | NA | N/A | Oxford Biomedical Research, Oxford, MI, UK | US$ 1130 |
No information on experimental protocol available at the manufacturer's website |
N/A, information not available.
The kits manufactured by JaICA (i.e., the “New 8-oxodG Check ELISA”, and the “Highly sensitive 8-oxodG Check ELISA”) use an antibody named N45.1, which was initially characterized by Toyokuni et al. [203]. The specificity and usefulness of this antibody has been studied in a number of individual, and interlaboratory comparisons between ELISA and chromatographic methods [197,198,[204], [205], [206], [207], [208], [209]]. Thus, kits based on the N45.1 antibody have known, but well characterized, limitations which make them the best candidates for studies in which a reasonable agreement with chromatographic techniques is required.
When compared with chromatographic methods, in particular those utilizing mass spectrometry, the ELISA has several advantages. It is easy to perform, does not require expensive equipment, and allows the detection of DNA lesions in various matrices, usually without the need for any specific pre-treatment, other than centrifugation to remove larger contaminants (although, as we note elsewhere in this review, under ‘Thawing’, the use of centrifugation should be adopted with some caution). Finally, the method is high throughput allowing analysis of a significant number of samples per day.
The greatest limitation of ELISA stems from antibody cross-reactivity with components structurally similar to the target molecule. This applies not only to urine (see ‘Inter-laboratory comparisons and best practice’, below), but also to other matrices. Thus, although ELISA appears to detect 8-oxodG in many matrices, without proper identification of cross-reacting compounds, the data should be interpreted with some caution.
5.2. Analysis of secondary DNA products of oxidation reactions in urine
Methodology for analysing secondary products of DNA oxidation reaction have developed alongside those for primary products, and have shared similar challenges. For example, the origins of these modified 2′-dN in urine has not been fully demonstrated (discussed in the section entitled: ‘Improve our understanding of the source(s) of adducts in the urine’). A number of methods have been developed for the measurement of lipid peroxidation-induced etheno-DNA adducts in human urine. These include LC-electrospray ionization (ESI)-MS/MS, and isotope dilution-gas chromatography-negative ion chemical ionization/mass spectrometry (GC-NICI/MS) for urinary 1,N6-ethenoadenine (ϵAde) [210], and the corresponding 2′dN (εdA), using LC-fluorescence detection [211,212], and LC-MS/MS [213].
3,N4-ethenocytosine (ϵCyt), 3,N4-ethenodeoxycytidine (ϵdC) have been detected using GC-NICI/MS [214,215], with LC-MS/MS also detecting ϵdC [216]. Low levels of 1,N2-ethenoguanine have been detected in the urine of healthy individuals, using LC-ESI-MS/MS, with higher levels reported in cigarette smokers [217]. There is some evidence to suggest that the principal metabolite of M1dG, 6-oxo-M1dG, might be a suitable urinary biomarker of in vivo oxidative stress [218], although a recent search of the literature failed to identify a report of its measurement in urine. However, there is one report of urinary M1dG, using immune-affinity purification, prior to chemical reduction with NaBH4 and LC combined with atmospheric pressure chemical ionization MS/MS [219]. In a limited number of individuals (n = 5), the authors reported an average 24 h excretion rate of M1dG is about 12±3.8 fmol/kg.
To date, urinary 8-oxodG remains the most widely studied, non-invasive biomarker of oxidative stress-induced damage to nucleic acids, and in light of this, it will be the basis for much of the discussion to come, as it is the matrix and biomarker about which most is known. It may be considered prototypical for other nucleic acid-derived biomarkers in extracellular matrices.
5.3. Risk of artefact formation and stability of nucleic acid biomarkers in urine
The analysis of biomarkers in extracellular matrices circumvents the risk of artefactual production of damage, largely through avoiding DNA extraction, minimising sample workup. Furthermore, other sources of artefact, such as exposure of nucleic acids, or their constituents, to metabolic enzymes, or other oxidants, after release into the systemic circulation, or in the urine (as significant concentrations of hydrogen peroxide have been reported to be present in urine [220]), appear not to occur (at least for the DNA moieties studied: dG, 8-oxodG, ThyGly, and dThyGly) [187,196,221,222]. In contrast, M1G and M1dG both appear to undergo further oxidative metabolism in rat liver cytosol, with the nucleobase adduct being a better substrate for such enzymic oxidation than the 2′-deoxyribonucleoside adduct [[223], [224], [225]]. There is also some evidence to suggest that M1G is further oxidised when administered intravenously, although M1dG was not examined [223].
Urine samples are widely stored, as part of biobanks, and other applications, and as such represent a significant resource for studying oxidative stress – if 8-oxodG (and other biomarkers) are stable. This stability was confirmed in a study where urine samples, stored at −20 °C, underwent repeat measurements over a 15 y period, with no observable increase in 8-oxodG [226]. Also reported is that 8-oxodG is stable in urine at 25 °C for up to 24 h [227], a finding confirmed by ourselves (Karbaschi et al. unpublished results), which is useful if study logistics ‘in the field’, do not allow immediate storage at low temperature. Stability of 8-oxodG and 5-HMUra has also been confirmed for over two years [227], and up to four months [228], respectively, when stored at −80 °C. As for 8-oxoGua, although it is not stable and may undergo decomposition in a basic solution (<2 d), at −20 °C, it is relatively stable in urine for at least 60 days [229].
In summary, it would appear that 8-oxodG, ThyGly and dThyGly, and probably 8-oxoGua, are not formed artefactually in urine samples of mammals, including humans, and there is no published evidence for their degradation upon release. In contrast, there is strong evidence that M1G and M1dG may undergo oxidation in vivo, either enzymically, or by some other process in the systemic circulation. Given the availability of multiple methods of analysis, the benefits of using extracellular matrices, and the stability of the analyte(s), the measurement of urinary 8-oxodG is widely reported within the literature. However, some discrepancies between levels of urinary 8-oxodG reported by some techniques were evident from the literature, and a number of academic researchers, and small-to-medium enterprises, across the globe, formed a consortium called the European Standards Committee on Urinary DNA Lesion Analysis (ESCULA), to address this issue.
5.4. European Standards Committee on Urinary DNA Lesion Analysis
Analogous to ESCODD, the task of ESCULA was to identify, and where necessary/possible, address major factors associated with the measurement of urinary DNA lesions, using 8-oxodG as the model lesion. The main goals of ESCULA were to:
-
(1)
Examine inter-laboratory, and intra-technique variation in urinary 8-oxodG measurement, and provide recommendations for preferred techniques and best practice.
-
(2)
Improve our understanding of the source(s) of adducts in the urine.
The following sections relate to efforts from ESCULA, and others, to address the above critical issues:
5.4.1. Inter-laboratory comparisons and best practice
A principal source of criticism of ELISA derives from the discrepancy between chromatographic methods and immunoassay in the determination of ‘background’ or baseline levels of urinary 8-oxodG, in healthy individuals [230]. This difference can be anywhere between four and ten-fold, although recent improvements in the JaICA kit (narrowing the range of the calibration curve, recommendations for strict temperature control, sample pre-treatment using SPE), have reduced this margin [204,205]. For the most part, however, 8-oxodG measurements by the two approaches have shown significant correlation reaching up to r = 0.98, p < 0.001, although this is not always the case (summarised in Ref. [205]). These studies suggest that both techniques share a common analyte, with the ELISA recognising additional compounds. As this discrepancy primarily arises from specificity, or lack thereof, of the antibody used in the ELISA, it is critical to identify, and therefore report, the name (i.e., clone) of the antibody used in all studies, to allow for full data interpretation – some antibodies have greater specificity than others. For example, the widely used antibody N45.1 is highly specific for 8-oxodG, rather than 8-oxoGua or 8-oxoGuo [203]. This specificity derives from discrimination between the C6 carbonyl group of 8-oxoGua (and the C2 NH2), and the C6 amino group of 8-oxoAde. Furthermore, unlike many antibodies, N45.1 displays preference for the 2′-deoxyribose moiety of 8-oxodG. The closest identified competitor for this antibody is 8-oxoGuo, which needs to be present in concentrations two orders of magnitude higher than 8-oxodG, in order to compete to the same extent [231]. As the concentration of 8-oxoGuo in urine is similar to that of 8-oxodG [84], this compound is unlikely to be a significant competitor [231]. Similarly, whilst 8-oxoGua is present in urine at significant concentrations (136±12 nmol/24 h), the risk of reactivity of N45.1 towards the modified nucleobase seems to be negligible, due to its specificity for the 2-deoxyribose moiety.
Identifying the basis of the discrepancy between ELISA and chromatographic techniques has proven to be challenging. We have speculated that, as chromatographic assays can only detect ‘free’ 8-oxodG, other compounds structurally related to 8-oxodG, and recognized by N45.1, may exist and be present in urine [231]. For example, creatinine, a substance present in urine at various concentrations, has been found to be recognized by N45.1 [204], as has urea [209], which led to further discussion of what anti-8-oxodG ELISAs detect [232]. By profiling the organic compounds present in the urine samples for which there is greatest discrepancy between ELISA and chromatography, the particular presence of compounds with aromatic and heterocyclic rings was noted, as well as a high concentration of saccharides, such as beta-d-galactose and alfa-d-glucose [205]. It is currently unknown whether these compounds originate from a specific (environmental) exposure of the subjects who provided the urine samples, or if they are related to some individual variability in metabolism. As these, albeit not conclusively identified, components are present in the urine samples even after SPE purification and discrepancy between the techniques remained despite application of the steps recommended to improve ELISA (see below), obtaining a reasonable inter-technique correlation for such urine samples will be impossible.
Several attempts have been made to improve the agreement between ELISA and chromatographic methods [198,204,[206], [207], [208], [209]]. The most promising approach included a combination of sample pre-purification by SPE, strict temperature control during incubation of the samples with the anti-8-oxodG antibody and normalization of 8-oxodG levels based on creatinine content [204], for spot urines at least, 24 h collections have not been tested. To achieve optimal inter-laboratory agreement, commercial kits from a single manufacturer that use antibody N45.1 were recommended as, to date, other kits have not been tested [205]. We published a protocol summarizing the optimization steps and recommendations for the improved 8-oxodG ELISA in 2013 [204]. Three years later we reported the results of an inter-laboratory comparison study in which the improved protocol was tested [205]. Between 2013 and 2020, these articles were cited thirty times, however, of these citations only four studies [[233], [234], [235], [236]] actually reported the adoption of the recommended optimization steps. In contrast, a PubMed search for a string “(8-oxodG or 8-oxodG) AND ELISA AND urine” returned 56 articles published between 2013 and 2020, that used the non-improved ELISA for detection of 8-oxodG in urine – clearly the steps recommended for improving the reliability of the ELISA are being overlooked. This is a concern as the scientific literature will continue to accumulate potentially unreliable data on the detection of oxidatively damaged DNA. The authors, reviewers and readers of such studies should be cautious when interpreting the data and making conclusions on 8-oxodG ELISA results, as the result of using a non-specific biomarker of oxidative stress in molecular epidemiology is attenuation bias due to non-differential misclassification, as sources other than oxidative stress occur randomly in the study population.
5.4.1.1. Normalization of biomarker levels to account for variation in urine concentration
It is widely agreed that 24 h collections are the gold standard for adjusting urinary biomarker levels [237], although this is practically and logistically challenging. As a result, 8-oxodG excretion is usually assessed using spot urine samples, which are then adjusted for urinary flow using creatinine concentration or specific gravity (SG) [238,239]. ESCULA made a concerted effort to examine normalization, and inter-individual variation in urinary 8-oxodG levels, and reported a number of recommendations [207]: spot urine samples are acceptable for analysis, with timed samples used, where possible, and expressing the results as excretion rates (nmol/h). If this is not possible, correction for creatinine is the next preferred option, which also decreases intra-individual variability. If first void samples are used, adjustment for specific gravity is also a good alternative. When comparing populations, or examining predictors or treatment effects, creatinine concentrations should be included in the model for creatinine-adjusted levels. These recommendations further substantiated earlier findings [240].
5.4.1.2. Effect of urine thawing on biomarker levels
The formation of uncharacterized precipitates frequently occurs in urine samples upon storage at low temperature (e.g., 4, −20 or −80 °C). It is a long neglected issue whether the uncharacterized precipitates present in urine affect the analysis of biomarkers of oxidative stress. In the literature, only two studies have investigated this issue. Bogdanov et al. (1999) showed that the 8-oxodG can be significantly underestimated, by up to 90%, if the precipitate was not fully re-dissolved [192]. Ten years later, this work was followed up by Shih et al. who further demonstrated that the precipitates in frozen urine samples could lead to a significant underestimation up to ~100% for 8-oxoGua, and ~86% for both 8-oxodGuo and 8-oxoGuo, and so recommendations for careful thawing of the samples to avoid this were made [241].
5.4.2. Improve our understanding of the source(s) of adducts in the urine
The presence of both modified nucleobases and modified (2′-deoxy)ribonucleosides in urine, and other extracellular matrices, raises the question of their origins, and the processes that give rise to the presence in urine. We put forward the following possible sources, using 8-oxodG and 8-oxoGua as examples, with the possibility that they may be regarded as prototypical for other adducts, such as the etheno-adducts, malondialdehyde-derived adducts, ThyGly and dThyGly etc, and their ribonucleoside equivalents, should they exist.
5.4.2.1. DNA repair
The DNA repair sources of oxidatively modified DNA nucleobases in extracellular matrices seems clear. The DNA N-glycosylases responsible for removing oxidatively modified nucleobases are increasingly well defined [17], and have been discussed thoroughly elsewhere [10]. 8-oxoguanine DNA glycosylase 1 (OGG1) is the major enzyme involved in the removal of 8-oxoGua from cellular DNA (Fig. 2), and its significant contribution to urinary 8-oxoGua levels has been demonstrated [242], although it is clearly not the only contributor. However, the presence of 2′-deoxyribonucleoside lesions in extracellular matrices is very much less well defined, as there are no reports of a single DNA repair enzyme whose activity yields 8-oxodG. Based upon existing evidence, we have identified a number of possible DNA repair activities (Fig. 2) which represent potential contributors to the presence of oxidatively-modified 2′-deoxyribonucleosides in urine:
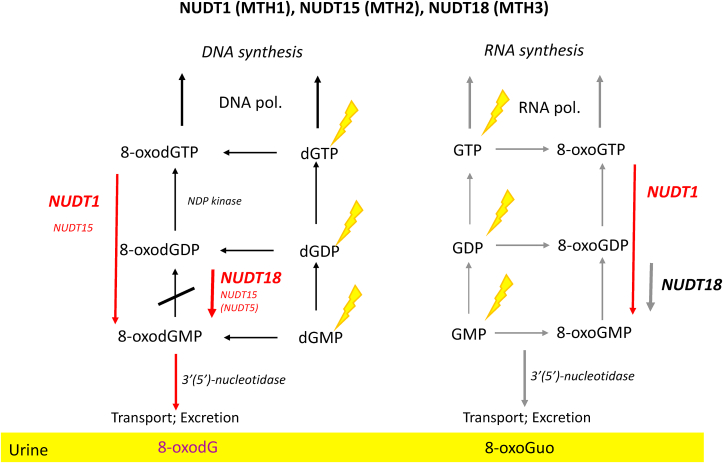
(A) Nudix hydrolases. Given the low discrimination of DNA polymerases for modified dNTPs there is an imperative for preventing modified DNA precursors from being incorporated into the genome (Fig. 2). The best characterised enzyme which performs such a role is the 8-oxo-2′-deoxyguanosine triphosphatase (8-oxodGTPase [243]) activity of NUDT1 (aka. MTH1), which hydrolyses 8-oxodGTP to 8-oxodGMP, and 8-oxoGTP to 8-oxoGMP [244] (Fig. 3). It has been suggested that further processing, perhaps by 5′(3′)-nucleotidases [243], may give rise to 8-oxodG which, given that it is not charged, we propose can be removed from the cell, ultimately appearing in the urine. Indeed, there is evidence that NUDT1 activity is a major source of extracellular 8-oxodG [245], although studies with various MTH1 inhibitors strongly suggest backup pathways exist [72], which is consistent with other repair pathways, such as BER. Indeed, the precise roles of other Nudix hydrolases such as NUDT15 (MTH2) and NUDT5, which include 8-oxodGTP and 8-oxodGDP (and 8-oxodADP [246]) amongst their substrates respectively (Fig. 3), remain to be defined fully [[247], [248], [249]], although the activity of MTH2 towards 8-oxodGTP seems to have been largely ruled out by some authors [80,250]. NUDT18 protein (aka MTH3) further extends the activity of the Nudix hydrolases to include ribonucleoside moieties, and it degrades 8-oxo-Gua-containing nucleoside diphosphates to the corresponding monophosphates [251] (Fig. 3). Considering other modified (d)NTPs, recent evidence shows that the activity of NUDT1/MTH1 is broader than simply 8-oxodGTP, and other substrates include O6-methyl-dGTP [78], N6-methyl-dATP [79], Fapy-dGTP [252], supporting our proposal that the activity of the Nudix hydrolases, and other Nudix hydrolase-like activities, could be responsible for the presence of other modified (2′-deoxy)ribonucloside products in extracellular matrices.
Fig. 3.
The potential roles of the Nudix hydrolases, identified to date, that may contribute to the sanitisation of the 2′-deoxyribonucleotide, and ribonucleotide pools. Non-oxidised nucleotide precursors may be oxidised, and may then phosphorylated, prior to incorporation into DNA or RNA, by DNA and RNA polymerases, respectively. 8-oxodGMP cannot be phosphorylated, and therefore when generated as the result of NUDT1 activity, acts as a block, preventing misincorporation of 8-oxodGTP into DNA. The corresponding pathways for GTP, are less well established. Note: recent evidence suggests that the contribution of NUDT5 and NUDT15, as backups for the primary 8-oxodGTPase NUDT1, is negligible.
(B) Nucleotide excision repair. Despite appearing to be directed principally towards bulky lesions, such as cyclobutane thymine dimers (T<>T), there is some evidence that NER may act upon non-bulky lesions such as 8-oxoGua [253]. Indeed, the rate of 8-oxoGua removal has been reported to be comparable to that for T<>T [253], seemingly generating a lesion-containing oligomer, approximately 24–32 nucleotides long [254], appear to be processed further to a smaller, adduct-containing moiety prior to excretion [255], as T<>T-containing moieties have been detected in urine [181,[256], [257], [258]]. Similarly, (5′R)- and (5′S)-8,5′-cyclo-2′-deoxyadenosines are present in urine as modified 2′-dN, and are reported to be removed from DNA by NER [259,260], albeit with the potential involvement of the BER enzyme Nei-like protein 1 (NEIL1) [261]. Seemingly confirming NER as a potential source of certain adducts in urine. However, 8-oxoGua-containing oligomers have yet to be shown to be present in urine [262] implying that further processing occurs, perhaps ultimately yielding 8-oxodG, or that they do not exist. However, under normal circumstances, the role of NER in the removal of 8-oxoGua, and perhaps other small oxidatively generated DNA lesions, would appear to be negligible [[263], [264], [265], [266], [267]], although results from NER-deficient xeroderma pimentosum cell lines have not entirely excluded the possibility [253,264,268,269], and a recent review suggests that some proteins involved in NER may facilitate BER of oxidatively damaged DNA through a form of chromatin remodelling involving nucleosomes [270]. In a study to more specifically address the contribution of NER to urinary 8-oxodG, we noted that levels of urinary 8-oxodG were the same in wild type and mice deficient in GG-NER and TC-NER, leading us to conclude that these pathways do not contribute to the presence of 8-oxodG in urine [271]. Unfortunately, the results with MTH1−/− were inconclusive, preventing us from answering this key question fully [271]. Nevertheless, NER might be a credible source of urinary 2′dN adducts, if those adducts are significant substrates for this repair pathway.
(C) Endonuclease(s). A poorly characterised endonuclease has been reported which, because it lacks a glycosylase activity, is predicted to release 3′,5′-8-oxodGDP [272]. We have previously proposed that this may be subsequently hydrolysed to 8-oxodG by nucleotidase(s) [273], although since there appears to have been no further reports related to this activity, since our last review [172], the importance of this pathway cannot be determined.
5.4.2.2. Cell death/turnover
It has been stated previously that urinary 8-oxodG does not reflect DNA repair, as it is not a product of BER [274], which is correct, but it was proposed to be a product of non-specific nucleases, acting upon DNA released from cell death, liberating dG which is subsequently oxidised [274]. This overlooks the potential existence of repair pathways, other than BER, which may yield 8-oxodG. The evidence against a contribution from cell turnover has been largely anecdotal, such as the reports in which urinary 8-oxodG has been measured in patients undergoing chemotherapy, with an absence in a concomitant increase in urinary 8-oxodG, which would be expected if urinary 8-oxodG arose from cell death/turnover (reviewed in Cooke et al. [231]). To date, the most decisive argument against the contribution of cell death to urinary levels of 8-oxodG and 8-oxoGua comes from the Olinski group [275], the findings of which are supported by the conclusions of Weimann et al. which state that the limited excretion of oligonucleotides into urine argues against oligonucleotides, or indeed nucleosides, originating from cell death [262].
5.4.2.3. Diet as a source of 8-oxodG in urine
Urine is the extracellular matrix upon which dietary influence has been most studied because diet has the potential to make a major impact on the interpretation of results if the urinary excretion of oxidised nucleic acid products merely reflects the intake of the same compounds from dietary products. The initial popularity of measuring 8-oxodG in urine, arose from early work that shows that diet can affect levels of urinary 8-oxoGua, but not 8-oxodG (discussed in detail by Cooke et al. [231]). More recent evidence shows that, in humans, neither 8-oxoGua, nor 8-oxodG, are affected by diet [276], which was confirmed by the results of a study in which healthy, male volunteers were fed heavily oxidised [15N]-DNA, and first void, mid-stream urine samples were collected for up to 14 days later [195]. Neither [15N5]-8-oxodG, nor [15N5]-8-oxoGua were detected in urine, by LC-GC/MS, and nor was [15N5]-dG and [15N5]-Gua, or indeed [15N5]-uric acid, the final product of purine metabolism. Combined, these results provide a compelling argument that diet is not a significant contributor to 8-oxoGua and 8-oxodG levels in human urine, consistent with the animal data relating to other 2′-deoxyribonucleoside lesions [221], including dThyGly [187], but in disagreement with early data for 8-oxoGua and ThyGly [187,277]. Species differences may account for this discrepancy, although the focus of our attention must be upon the more relevant, human data, and earlier animal studies should be repeated using the highly specific and sensitive mass spectrometric techniques.
6. Nucleic acid oxidation in human health and disease
There are a large number of pathological conditions in which oxidative stress has been proposed to have a role. Urinary biomarkers of oxidative stress offer the means by which this stress could be monitored via a non-invasive route and they may also have the potential to act as biomarkers of disease development risk, or assess the efficacy of therapy. Therefore, accurate and straightforward measurement of these biomarkers would be of great benefit to achieve a better understanding of the role of this damage in disease, as well as to establish reference ranges for clinical application. Table 2 summarises the urinary levels of oxidative stress biomarkers (8-oxodG, 8-oxoGua, and 8-oxoGuo) in healthy subjects and patients with various diseases, as reported in the recent literature. The urinary levels of 8-oxodG, 8-oxoGua and 8-oxoGuo were chosen mainly because oxidative stress induces oxidation of DNA and RNA predominantly at the guanine moiety. These data were included from the works published in recent years and measured with LC-MS/MS. From the studies noted in Table 2, the reference ranges in urine of healthy subjects were 2.03–7.83 ng/mg creatinine for 8-oxodG, 13.6–36.3 ng/mg creatinine for 8-oxoGua and 3.60–12.2 ng/mg creatinine for 8-oxoGuo. Despite that the same analytical technique was used, some variations were still noted, which may be attributed to differences in race, age, gender and smoking status. When compared to the healthy subjects, the patients with various diseases had about 1.1–3.7 times the levels of 8-oxodGuo, 1.1–3.0 times of 8-oxoGua and 1.5–5.6 times of 8-oxoGuo higher levels in urine. Notably, of these three biomarkers, the biggest difference between healthy subjects and patients was seen for urinary 8-oxoGuo level, implying that 8-oxoGuo exhibited a superior ability to discriminate between the patients and the healthy subjects, compared to 8-oxodG or 8-oxoGua. Nevertheless, to date, there are no clear cut-off values for the above biomarkers for active diseases, partially owing to the considerable inter-individual variation observed in the urinary levels of adducts.
Table 2.
Urinary levels of oxidatively generated damage (8-oxodG, 8-oxoGua, 8-oxoGuo) in healthy subjects and patients with various diseases. As the precise sources of 8-oxodG, and 8-oxoGua (which could be from DNA-, or RNA-related sources, or both) in urine have yet to be defined, we have broadly labelled their origin as ‘DNA’. 8-oxoGuo is from RNA-related source(s). UPLC = ultra performance liquid chromatography.
| Healthy controls | Patients | Methods | Ref. | |
|---|---|---|---|---|
| ‘DNA’ oxidation | 3.68 (n = 61) | 4.58 (n = 57; unipolar and bipolar disorders) | LC-MS/MS | [308] |
| 8-oxodG (ng/mg creatinine) | ||||
| 2.53 (n = 70) | 3.46 (n = 148; bipolar I disorder) | UPLC-MS/MS | [309] | |
| 3.90 (n = 128) | 4.35 (n = 87; COPD); 14.6 (n = 100; patients with mechanical ventilation) |
LC-MS/MS | [241] | |
| 4.13 (n = 16) | 5.23 (n = 46; renal disease) | LC-MS/MS | [310] | |
| 2.80 (n = 73) | 4.71 (n = 60; breast cancer) | UPLC-MS/MS | [311] | |
| -a | 5.41 (n = 12; colorectal cancer) | UPLC-MS/MS | [312] | |
| – | 6.76 (n = 508; cardiovascular disease) | UPLC-MS/MS | [313] | |
| – | 4.62 (n = 2721; type 2 diabetes) | UPLC-MS/MS | [314] | |
| – | 5.27 (n = 609; type 2 diabetes) | UPLC-MS/MS | [109] | |
| 2.03 (n = 60) | – | UPLC-MS/MS | [315] | |
| 7.83 (n = 132) | – | LC-MS/MS | [316] | |
| 8-oxoGua (ng/mg creatinine) | 13.6 (n = 128) | 14.7 (n = 87; COPD); 40.2 (n = 100; patients with mechanical ventilation) |
LC-MS/MS | [241] |
| 36.3 (n = 132) | – | LC-MS/MS | [316] | |
| RNA oxidation | ||||
| 8-oxoGuo (ng/mg creatinine) | 3.60 (n = 70) | 5.44 (n = 148; bipolar I disorder) | UPLC-MS/MS | [309] |
| 4.90 (n = 128) | 8.0 (n = 87; COPD); 27.6 (n = 100; patients with mechanical ventilation) |
LC-MS/MS | [241] | |
| 6.80 (n = 16) | 15.2 (n = 46; renal disease) | LC-MS/MS | [310] | |
| – | 9.87 (n = 508; cardiovascular disease) | UPLC-MS/MS | [313] | |
| – | 7.82 (n = 2721; type 2 diabetes) | UPLC-MS/MS | [314] | |
| – | 9.58 (n = 608; type 2 diabetes) | UPLC-MS/MS | [109] | |
| 5.89 (n = 60) | – | UPLC-MS/MS | [315] | |
| 12.2 (n = 132) | – | LC-MS/MS | [316] | |
Not available.
In addition to the urinary ranges for healthy adults, the urinary ranges of oxidative stress biomarkers in children have also been reported, although there are fewer of these studies in the literature. The reference urinary ranges are 2.63–4.61 ng/mg creatinine for 8-oxodGuo [[278], [279], [280]], 5.58–12.5 ng/mg creatinine for 8-oxoGuo [[280], [281], [282]], and 11.5–17.8 ng/mg creatinine for 8-oxoGua [281,283].
7. Perspectives on the interpretation of biomarkers of oxidatively generated damage to nucleic acids in surrogate tissues or biological matrices
It is often difficult, or impossible, to measure a particular biomarker(s) in a target tissue. In this instance, their measurement in surrogate tissues, or biological matrices is often used, although the validity of this approach does not appear to have been widely studied. This is perhaps due to the potential risks of invasively collecting biopsies from healthy humans. Additionally, tissues obtained from patients may not be representative of “normal” tissue, even if it is obtained from a site adjacent to the pathologic tissue.
7.1. Surrogate vs. target tissue
The use of surrogate tissues is justifiable if there is proportionality (i.e., a correlation) between the biomarker levels in the target tissue and surrogate. A correlation between levels of biomarkers in the surrogate and target tissues would be expected in situations where inter-individual variation is mainly driven by external exposures to damaging agents (e.g., chemicals or smoking), in which the surrogate tissue is as likely to be damaged as the target. In this case, absolute values in biomarkers may differ between the surrogate and target tissue(s), but there would be proportionality between the levels in surrogate and target tissue.
7.2. Use of a surrogate tissue/matrix to assess generalised, whole body stress
Under baseline, healthy conditions, i.e., in the absence of a single organ, or organ system being under “pathological” oxidative stress, as a result of pathogenesis, the measurement of biomarkers in certain surrogate tissues, or matrices, seems to be justifiable as proxies for whole body, or systemic effects. For example, PBMC spend a significant proportion of their lifetime outside of the systemic circulation, and in the peripheral tissues. In this regard, they are likely to be exposed to the oxidative stress within those tissues, and hence likely to represent systemic oxidative stress, rather than that derived from a particular organ.
In the case of nucleic acids, or DNA at least, and as discussed herein, it is understood that the products of the repair of oxidative stress-induced damage (from all tissues) ultimately appear in urine. In this regard, urinary biomarkers will also represent systemic oxidative stress (or eustress, under baseline, healthy conditions), rather than that derived from a particular organ. What seems more difficult to determine is whether, under these conditions, one organ contributes more to the systemic measure of oxidative stress, than another. However, there is some evidence that the skin (under ambient UV exposure) may be a significant contributor [284,285], but this subject has not been studied extensively.
7.3. Is it possible for “local”, pathological oxidative stress in a single organ to influence systemic levels?
Under disease conditions, it seems possible that increased oxidative stress in a particular organ could increase the systemic levels of oxidative stress e.g., in PBMC or urine. Size of the organ, and extent of the oxidative stress in that organ, will be among the factors influencing whether localized oxidative stress impacts systemic measures. This is demonstrated in many studies in the literature, for example the presence of chronic lung inflammation (e.g., chronic obstructive pulmonary disease, COPD) has been found to significantly increase the urinary levels of oxidative stress biomarkers up to two times [241,286], and critically ill patients on mechanical ventilation in intensive care units have approximately three times higher levels of oxidised nucleic acids products in urine, compared to COPD patients [241].
When it comes to interpretation of elevated PBMC or urinary levels of biomarkers of oxidative stress, knowledge of a pre-existing disease, can lead to the assumption that the disease is responsible for the elevation. Indeed, this has led to various reports in the literature where successful correlations between surrogate or systemic levels of biomarkers and target tissue levels have been made (e.g. Refs. [[287], [288], [289]]), apparently validating this approach. However, in the absence of that knowledge of pre-existing disease, at the present it seems impossible to ascribe elevated levels to any particular organ.
8. The future
8.1. The legacy of ECVAG
The ECVAG trials culminated in the general notion that the Fpg-modified comet assay is a robust way of measuring oxidatively damaged DNA, but there is inter-laboratory variation in the reported levels of Fpg-sensitive sites in human PBMCs and other cells and tissues. Especially, it has been clear that advances in comet assay research arise as a consequence multi-laboratory projects. One such project was the ComNet, which was launched in 2012 as a network of researchers using the comet assay in biomonitoring [290,291]. This was superseded by the “comet assay in human biomonitoring” (hCOMET) consortium that among a number of objectives aimed to:
-
(1)
collect results of individual human population studies and create a unified database of comet assay data relating to human health and disease,
-
(2)
determine the experimental factors affecting the performance of the assay and therefore its reliability and reproducibility,
-
(3)
provide guidelines for best practice in design of human population studies, and in performance of the comet assay, and
-
(4)
encourage the use of standard protocols to facilitate the comparison of results from different studies.
The work of hCOMET has led to technical recommendations of the enzyme-modified comet assay [292], and detailed reviews on the use of the comet assay for assessment of oxidatively damaged DNA and BER activity in human biomonitoring studies [293,294]. A rather striking outcome is also the Minimum Information for Reporting Comet Assay (MIRCA) recommendations, which has been fostered to increase the quality and impact of comet assay results, including oxidatively damaged DNA [295]. Alongside this effort, there has been a gentle push for comet assay researchers to use and report assay controls in their experimental procedure as only about 20% of published articles mention the use, or show results from assay controls [296,297]. One of the problems has been that positive controls for the assessment of oxidatively damaged DNA by the comet assay have not been thoroughly tested. The photosensitizer Ro19-8022, which was used by the ESCODD and ECVAG ring trials, is not widely commercially available, and can be relatively difficult to find. Work performed after the ECVAG studies has identified potassium bromate as a good positive assay control for the hOGG1-and Fpg-modified comet assay [298]. The stability of potassium bromate-induced DNA damage in cryopreserved samples is currently being investigated, which would be a crucial aspect of any human biomonitoring or clinical study [298].
8.2. The legacy of ESCULA
Further to the publication by ESCULA in 2008 [172], it is useful to review what progress has been made. There remains a pressing need for non- or minimally-invasive biomarkers of oxidative stress that are sufficiently validated to be applied to large scale, molecular epidemiology studies. For 8-oxodG, as a prototypical nucleic acid-derived biomarker of oxidative stress, ESCULA delivered on recommendations for the most robust and reproducible methods for analysis (LC-MS/MS), but acknowledged that there had been much improvement in some ELISA measures [207]. Using these techniques, ESCULA demonstrated that good inter-laboratory agreement was achievable, and also made recommendation for the optimal means of correcting biomarker values for variation in urine concentration [207].
Although ESCULA has achieved the goals that required action by the entire cohort of laboratories, efforts in smaller groups of laboratories continue. Improvement to the ELISA approach have been made, that achieve greater agreement with values obtained by the gold standard, LC-MS/MS [204,205], and it is recommended that these method improvements are incorporated into ELISA for 8-oxodG generally. Work on-going from ESCULA's aim to define the provenance of urinary 8-oxodG, is narrowing down potential sources, seemingly, to the dNTP pool, by ruling out contributions from TC-NER and GG-NER [271]. Work related to the aims of ESCULA has effectively ruled out other potential sources, diet [195,276], and cell turnover [262,275].
In hindsight, it seems odd that so many reports in the literature continue to measure a biomarker for which we are, only now, getting some idea of from where it comes. This issue makes it challenging to define urinary 8-oxodG. Many in the literature describe it as a biomarker of DNA damage, but in the absence of any reported process that generates 8-oxodG from DNA, this does not seem accurate. In contrast, the sanitisation of the dGTP pool seems more likely, albeit not yet a proven source.
The work of ESCULA collectively, individual members, and related studies, has done much to advance the analysis and understanding of urinary 8-oxodG, and other adducts in urine. Indeed, the lessons learnt have informed on an emerging, new generation of analysis, that of urinary DNA adductomics.
8.3. Urinary DNA adductomics and the targeted analyses of biomarkers of oxidative stress
As is evident from this review, currently the study of oxidative stress is comprised of targeted analyses – one, or several, biomarkers are analyzed simultaneously. However, given that, in the case of DNA, over 70 forms of oxidatively generated damage exist, clearly a lot of information is being lost. To see the bigger picture, a more comprehensive analysis must be performed. The untargeted analysis of DNA damage (DNA adductomics [299]) means that there is no preconception as to what adduct(s) is important, and therefore all of the adducts present must be measured. The principle of cellular DNA adductomics works to use mass spectrometry to filter and detect all damaged 2′-deoxyribonucleosides (2′-dN), from the undamaged, following enzymatic hydrolysis of the DNA [299]. But, for any adducts that can be formed artefactually during DNA extraction and workup, such as oxidatively generated damage, this approach has all the potential limitations of, and need for precautions as, targeted analysis.
Given our experience with urinary 8-oxodG, 8-oxoGuo, and 8-oxoGua, we applied our methodology for cellular DNA adductomics [300] to urine. As noted above, the benefits of using urine are clear: non-invasive, easily collected, transported and stored with low biological hazard, no pre-processing is needed prior to storage, small volumes are required, and adduct stability is high, allowing the use of biobanked samples. Additionally, sample workup is simpler than for DNA, no extraction, hydrolysis or risk of artefactual formation of damage. Urine therefore represents a matrix which contains the totality of adducts, arising from repair in the cells of the entire body, producing levels significantly higher than in individual tissues, plus reflecting a totality of exposures. Our initial method comprised the analysis of urinary damaged 2′-dN only. Noting that we were missing damaged nucleobases (the products of BER) from our analyses, we extended the method for their inclusion [301].
Urinary DNA adductomics provides a more comprehensive picture of whole-body levels of DNA damage, and its repair, than targeted approaches, and clearly has numerous strengths. However, we feel that, for now at least, it will not replace targeted analyses of biomarkers of nucleic acid oxidation.
8.4. Mapping oxidatively damaged DNA across the genome (DNA adductomics)
There is good evidence to support the proposal that critical DNA modifications (e.g., oxidatively damaged DNA/RNA) may occur at specific DNA sequences or chromatin structures. Present MS-based methods all require the hydrolysis of DNA to allow for the analysis of the modified (2′-deoxy)nucleosides, and therefore any information regarding the sites of the modifications is lost. It has been suggested that targeted DNA adduct detection or non-targeted detection (i.e., DNA adductomics) should be combined with genomic-wide sequencing to correlate DNA adduct formation with biologically important mutations [302], and other downstream effects of the presence of adducts. As noted above, under “Immunochemical Techniques”, antibodies have been used to detect site-specific damage, at varying nucleotide resolution, and more recently a number of next generation sequencing (NGS)-based methods have emerged for detecting various types of DNA damage and repair across the entire genome at nucleotide resolution for example [303], and reviewed in Ref. [304]. Nevertheless, NGS-based methods are based on short-read sequencing approaches (~50–300 bp reads) and generally need enrichment of the damage DNA regions, and PCR amplification. In contrast, the development of the third-generation sequencing technologies (also called long-read sequencing), such as Oxford nanopore sequencing, can give extremely long reads (~500 bp to the current record of 2.3 Mb [305]) and do not require PCR amplification. Lately, such an approach has undergone preliminary testing to detect the oxidation of cytosine, adenine and guanine [306,307]. These new sequencing techniques still need further development and await experimental verification. For example, nucleobases are identified by measuring changes in the electric current across the surface of the nanopores. In order to distinguish not only the four canonical nucleobases (Ade, Thy, Gua, Cyt), but potentially other types of nucleobases (including modified ones), it is necessary to synthesize various oxidatively modified DNA nucleobases in a DNA strand to train the device to recognize/characterize the features (ionic current signature) of the modified DNA nucleobases. Such machine learning processes will also involve the use high-resolution mass spectrometry for the identification of the oxidatively generated DNA adducts, and base-calling algorithm (Fig. 4). Overall, these sequencing advances have led us to believe that in the future we will be able to detect the DNA lesions together with their precise locations within the entire genome, and thereby look into the black box which hides the mechanisms linking damage with disease.
Fig. 4.
Our proposed concept of a nucleotide-resolution, sequencing platform for mapping and identification of oxidatively generated DNA lesions by integrated, nanopore technology, cellular DNA adductomic analysis, and machine learning algorithms.
9. Conclusions
The measurement of biomarkers of oxidatively generated damage to nucleic acids, in DNA/RNA, and blood/urine (and other extracellular matrices) is well established, and a number of techniques have emerged as gold standards, principally LC-MS/MS and the comet assay, depending upon the investigator's needs, and matrix of choice. The study of these biomarkers has established a potential role for oxidatively generated damage to nucleic acids in a wide variety of diseases, and the aging process. However, there is a “black box” that prevents the demonstration of how the formation of damage leads to disease. Emerging adductomics approaches, i.e., mapping damage across the genome, and studying the totality of adducts in nucleic acids, offers the potential to better understand how damage leads to downstream events that lead to pathogenesis.
Declaration of competing interest
There is no declaration of interest.
Acknowledgements
MSC acknowledges support from the National Institute of Environmental Health Sciences of the National Institutes of Health under award numbers R15ES027196, and R01ES030557. M-RC acknowledges support from the Ministry of Science and Technology, Taiwan under award number MOST 109-2314-B-040-018-MY3. C-WH acknowledges support from the National Institute of Environmental Health Sciences of the National Institutes of Health under award number R01ES030557. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
PR acknowledges support from the Ministry of Education, Youth and Sports of the Czech Republic project Healthy Aging in Industrial Environment HAIE (CZ.02.1.01/0.0/0.0/16_019/0000798), which is co-financed by the European Union (European Structural and Investment funds; Operation Programme Research, Development and Education).
MSC declares the following patents:
High throughput approach for processing slides for all variations of the single cell gel electrophoresis assay. GB2531983, and US10,401,323.
Devices and methods for high-throughput assay. US 9,897,572.
Novel chilling apparatus. US 10,690,574 B2.
References
- 1.Egea J. European contribution to the study of ROS: a summary of the findings and prospects for the future from the COST action BM1203 (EU-ROS) Redox Biol. 2017;13:94–162. doi: 10.1016/j.redox.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Meo S. Role of ROS and RNS Sources in Physiological and pathological conditions. Oxid. Med. Cell longev. 2016;2016:1245049. doi: 10.1155/2016/1245049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sies H., Berndt C., Jones D.P. Oxidative stress. Annu. Rev. Biochem. 2017;86:715–748. doi: 10.1146/annurev-biochem-061516-045037. [DOI] [PubMed] [Google Scholar]
- 4.Sies H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: oxidative eustress. Redox Biol. 2017;11:613–619. doi: 10.1016/j.redox.2016.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Potdar N. First-trimester increase in oxidative stress and risk of small-for-gestational-age fetus. BJOG. 2009;116(5):637–642. doi: 10.1111/j.1471-0528.2008.02096.x. [DOI] [PubMed] [Google Scholar]
- 6.Birch-Machin M.A., Bowman A. Oxidative stress and ageing. Br. J. Dermatol. 2016;175(Suppl 2):26–29. doi: 10.1111/bjd.14906. [DOI] [PubMed] [Google Scholar]
- 7.Di Mascio P. Singlet molecular oxygen reactions with nucleic acids, lipids, and proteins. Chem. Rev. 2019;119(3):2043–2086. doi: 10.1021/acs.chemrev.8b00554. [DOI] [PubMed] [Google Scholar]
- 8.Cooke M.S. Recommendations for standardized description of and nomenclature concerning oxidatively damaged nucleobases in DNA. Chem. Res. Toxicol. 2010;23(4):705–707. doi: 10.1021/tx1000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cadet J. Biologically relevant oxidants and terminology, classification and nomenclature of oxidatively generated damage to nucleobases and 2-deoxyribose in nucleic acids. Free Radic. Res. 2012;46(4):367–381. doi: 10.3109/10715762.2012.659248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans M.D., Dizdaroglu M., Cooke M.S. Oxidative DNA damage and disease: induction, repair and significance. Mutat. Res. 2004;567(1):1–61. doi: 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 11.Cadet J. Formation and repair of oxidatively generated damage in cellular DNA. Free Radic. Biol. Med. 2017;107:13–34. doi: 10.1016/j.freeradbiomed.2016.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleming A.M., Burrows C.J. On the irrelevancy of hydroxyl radical to DNA damage from oxidative stress and implications for epigenetics. Chem. Soc. Rev. 2020;49(18):6524–6528. doi: 10.1039/d0cs00579g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans M.D., Cooke M.S. Lipid- and Protein-Mediated Oxidative Damage to DNA. In: Singh K.K., editor. Oxidative Stress, Disease and Cancer. Imperial College Press; London: 2006. pp. 201–252. [Google Scholar]
- 14.Moriya M. Single-stranded shuttle phagemid for mutagenesis studies in mammalian cells: 8-oxoguanine in DNA induces targeted G.C-->T.A transversions in simian kidney cells. Proc. Natl. Acad. Sci. U. S. A. 1993;90(3):1122–1126. doi: 10.1073/pnas.90.3.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans M.D., Cooke M.S. Factors contributing to the outcome of oxidative damage to nucleic acids. Bioessays. 2004;26(5):533–542. doi: 10.1002/bies.20027. [DOI] [PubMed] [Google Scholar]
- 16.Kasai H. What causes human cancer? Approaches from the chemistry of DNA damage. Gene Environ. 2016;38:19. doi: 10.1186/s41021-016-0046-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooke M.S. Oxidative DNA damage: mechanisms, mutation, and disease. Faseb. J. 2003;17(10):1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 18.Nelson B.C., Dizdaroglu M. Implications of DNA damage and DNA repair on human diseases. Mutagenesis. 2020;35(1):1–3. doi: 10.1093/mutage/gez048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore S.P., Toomire K.J., Strauss P.R. DNA modifications repaired by base excision repair are epigenetic. DNA Repair. 2013;12(12):1152–1158. doi: 10.1016/j.dnarep.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Zarakowska E. Are 8-oxoguanine (8-oxoGua) and 5-hydroxymethyluracil (5-hmUra) oxidatively damaged DNA bases or transcription (Epigenetic) marks? Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2014:764–765. doi: 10.1016/j.mrgentox.2013.09.002. 58–63. [DOI] [PubMed] [Google Scholar]
- 21.Olinski R., Starczak M., Gackowski D. Enigmatic 5-hydroxymethyluracil: oxidatively modified base, epigenetic mark or both? Mutat. Res. Rev. Mutat. Res. 2016;767:59–66. doi: 10.1016/j.mrrev.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Allgayer J. Widespread transcriptional gene inactivation initiated by a repair intermediate of 8-oxoguanine. Nucleic Acids Res. 2016;44(15):7267–7280. doi: 10.1093/nar/gkw473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ba X., Boldogh I. 8-Oxoguanine DNA glycosylase 1: Beyond repair of the oxidatively modified base lesions. Redox Biol. 2018;14:669–678. doi: 10.1016/j.redox.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleming A.M., Burrows C.J. 8-Oxo-7,8-dihydroguanine, friend and foe: epigenetic-like regulator versus initiator of mutagenesis. DNA Repair. 2017;56:75–83. doi: 10.1016/j.dnarep.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleming A.M., Ding Y., Burrows C.J. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. U. S. A. 2017;114(10):2604–2609. doi: 10.1073/pnas.1619809114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seifermann M., Epe B. Oxidatively generated base modifications in DNA: not only carcinogenic risk factor but also regulatory mark? Free Radic. Biol. Med. 2017;107:258–265. doi: 10.1016/j.freeradbiomed.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 27.Mikhed Y. Redox regulation of genome stability by effects on gene expression, epigenetic pathways and DNA damage/repair. Redox Biol. 2015;5:275–289. doi: 10.1016/j.redox.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorini F. Towards a comprehensive view of 8-oxo-7,8-dihydro-2’-deoxyguanosine: highlighting the intertwined roles of DNA damage and epigenetics in genomic instability. DNA Repair. 2021:97. doi: 10.1016/j.dnarep.2020.103027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dziaman T. Characteristic profiles of DNA epigenetic modifications in colon cancer and its predisposing conditions-benign adenomas and inflammatory bowel disease. Clin. Epigenet. 2018;10:72. doi: 10.1186/s13148-018-0505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olinski R., Gackowski D., Cooke M.S. Endogenously generated DNA nucleobase modifications source, and significance as possible biomarkers of malignant transformation risk, and role in anticancer therapy. Biochim. Biophys. Acta Rev. Canc. 2018;1869(1):29–41. doi: 10.1016/j.bbcan.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 31.Zawia N.H., Lahiri D.K., Cardozo-Pelaez F. Epigenetics, oxidative stress, and Alzheimer disease. Free Radic. Biol. Med. 2009;46(9):1241–1249. doi: 10.1016/j.freeradbiomed.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coppede F., Migliore L. DNA damage in neurodegenerative diseases. Mutat. Res. 2015;776:84–97. doi: 10.1016/j.mrfmmm.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 33.Zarakowska E. Oxidation products of 5-methylcytosine are decreased in senescent cells and tissues of progeroid mice. J. Gerontol. A Biol. Sci. Med. Sci. 2018;73(8):1003–1009. doi: 10.1093/gerona/gly012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashapkin V.V., Kutueva L.I., Vanyushin B.F. Aging as an epigenetic phenomenon. Curr. Genom. 2017;18(5):385–407. doi: 10.2174/1389202918666170412112130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akhmedov A.T., Marin-Garcia J. Mitochondrial DNA maintenance: an appraisal. Mol. Cell. Biochem. 2015;409(1–2):283–305. doi: 10.1007/s11010-015-2532-x. [DOI] [PubMed] [Google Scholar]
- 36.Roubicek D.A., Souza-Pinto N.C. Mitochondria and mitochondrial DNA as relevant targets for environmental contaminants. Toxicology. 2017;391:100–108. doi: 10.1016/j.tox.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 37.Zhao L., Sumberaz P. Mitochondrial DNA damage: prevalence, biological consequence, and emerging pathways. Chem. Res. Toxicol. 2020;33(10):2491–2502. doi: 10.1021/acs.chemrestox.0c00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gustafson M.A., Sullivan E.D., Copeland W.C. Consequences of compromised mitochondrial genome integrity. DNA Repair. 2020;93:102916. doi: 10.1016/j.dnarep.2020.102916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bender A. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat. Genet. 2006;38(5):515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 40.Arun S., Liu L., Donmez G. Mitochondrial biology and neurological diseases. Curr. Neuropharmacol. 2016;14(2):143–154. doi: 10.2174/1570159X13666150703154541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kowalska M. Mitochondrial and nuclear DNA oxidative damage in physiological and pathological aging. DNA Cell Biol. 2020;39(8):1410–1420. doi: 10.1089/dna.2019.5347. [DOI] [PubMed] [Google Scholar]
- 42.Cencioni C. Oxidative stress and epigenetic regulation in ageing and age-related diseases. Int. J. Mol. Sci. 2013;14(9):17643–17663. doi: 10.3390/ijms140917643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minocherhomji S., Tollefsbol T.O., Singh K.K. Mitochondrial regulation of epigenetics and its role in human diseases. Epigenetics. 2012;7(4):326–334. doi: 10.4161/epi.19547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lehle S. LORD-Q: a long-run real-time PCR-based DNA-damage quantification method for nuclear and mitochondrial genome analysis. Nucleic Acids Res. 2014;42(6):p. e41. doi: 10.1093/nar/gkt1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rooney J.P. PCR based determination of mitochondrial DNA copy number in multiple species. Methods Mol. Biol. 2015;1241:23–38. doi: 10.1007/978-1-4939-1875-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhigalina D.I. FISH Diagnostics of chromosomal Translocation with the Technology of Synthesis of locus-specific DNA probes Based on long-range PCR. Russ. J. Genet. 2020;56(6):739–746. [Google Scholar]
- 47.Hanna R. Optimised detection of mitochondrial DNA strand breaks. Mitochondrion. 2019;46:172–178. doi: 10.1016/j.mito.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 48.Mansouri A. An alcoholic binge causes massive degradation of hepatic mitochondrial DNA in mice. Gastroenterology. 1999;117(1):181–190. doi: 10.1016/s0016-5085(99)70566-4. [DOI] [PubMed] [Google Scholar]
- 49.Alhegaili A.S. Genome-wide adductomics analysis reveals Heterogeneity in the Induction and Loss of cyclobutane thymine Dimers across Both the Nuclear and mitochondrial genomes. Int. J. Mol. Sci. 2019;20(20) doi: 10.3390/ijms20205112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wauchope O.R. Oxidative stress increases M1dG, a major peroxidation-derived DNA adduct, in mitochondrial DNA. Nucleic Acids Res. 2018;46(7):3458–3467. doi: 10.1093/nar/gky089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rothfuss O., Gasser T., Patenge N. Analysis of differential DNA damage in the mitochondrial genome employing a semi-long run real-time PCR approach. Nucleic Acids Res. 2010;38(4):p. e24. doi: 10.1093/nar/gkp1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chimienti G. Increased TFAM binding to mtDNA damage hot spots is associated with mtDNA loss in aged rat heart. Free Radic. Biol. Med. 2018;124:447–453. doi: 10.1016/j.freeradbiomed.2018.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaniak-Golik A., Skoneczna A. Mitochondria-nucleus network for genome stability. Free Radic. Biol. Med. 2015;82:73–104. doi: 10.1016/j.freeradbiomed.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 54.Saki M., Prakash A. DNA damage related crosstalk between the nucleus and mitochondria. Free Radic. Biol. Med. 2017;107:216–227. doi: 10.1016/j.freeradbiomed.2016.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kamiya H., Kasai H. Formation of 2-hydroxydeoxyadenosine triphosphate, an oxidatively damaged nucleotide, and its incorporation by DNA polymerases. Steady-state kinetics of the incorporation. J. Biol. Chem. 1995;270(33):19446–19450. doi: 10.1074/jbc.270.33.19446. [DOI] [PubMed] [Google Scholar]
- 56.Bestwick R.K., Moffett G.L., Mathews C.K. Selective expansion of mitochondrial nucleoside triphosphate pools in antimetabolite-treated HeLa cells. J. Biol. Chem. 1982;257(16):9300–9304. [PubMed] [Google Scholar]
- 57.Mo J.Y., Maki H., Sekiguchi M. Hydrolytic elimination of a mutagenic nucleotide, 8-oxodGTP, by human 18-kilodalton protein: sanitization of nucleotide pool. Proc. Natl. Acad. Sci. U. S. A. 1992;89(22):11021–11025. doi: 10.1073/pnas.89.22.11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pursell Z.F. Trace amounts of 8-oxo-dGTP in mitochondrial dNTP pools reduce DNA polymerase gamma replication fidelity. Nucleic Acids Res. 2008;36(7):2174–2181. doi: 10.1093/nar/gkn062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith M.R. Molecular and structural characterization of oxidized ribonucleotide insertion into DNA by human DNA polymerase beta. J. Biol. Chem. 2020;295(6):1613–1622. doi: 10.1074/jbc.RA119.011569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abbas H.H.K. MTH1 deficiency selectively increases non-cytotoxic oxidative DNA damage in lung cancer cells, more bad news than good? BMC Canc. 2018;18(1):423. doi: 10.1186/s12885-018-4332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoshimura D. An oxidized purine nucleoside triphosphatase, MTH1, suppresses cell death caused by oxidative stress. J. Biol. Chem. 2003;278(39):37965–37973. doi: 10.1074/jbc.M306201200. [DOI] [PubMed] [Google Scholar]
- 62.Warpman Berglund U. Validation and development of MTH1 inhibitors for treatment of cancer. Ann. Oncol. 2016;27(12):2275–2283. doi: 10.1093/annonc/mdw429. [DOI] [PubMed] [Google Scholar]
- 63.Nakabeppu Y. Cellular levels of 8-oxoguanine in either DNA or the nucleotide pool play pivotal roles in carcinogenesis and survival of cancer cells. Int. J. Mol. Sci. 2014;15(7):12543–12557. doi: 10.3390/ijms150712543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rai P. Enhanced elimination of oxidized guanine nucleotides inhibits oncogenic RAS-induced DNA damage and premature senescence. Oncogene. 2011;30(12):1489–1496. doi: 10.1038/onc.2010.520. [DOI] [PubMed] [Google Scholar]
- 65.McPherson L.A. Increased MTH1-specific 8-oxodGTPase activity is a hallmark of cancer in colon, lung and pancreatic tissue. DNA Repair. 2019;83:102644. doi: 10.1016/j.dnarep.2019.102644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gad H. MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature. 2014;508(7495):215–221. doi: 10.1038/nature13181. [DOI] [PubMed] [Google Scholar]
- 67.Rudd S.G. MTH1 inhibitor TH588 disturbs mitotic Progression and induces mitosis-dependent Accumulation of genomic 8-oxodG. Canc. Res. 2020;80(17):3530–3541. doi: 10.1158/0008-5472.CAN-19-0883. [DOI] [PubMed] [Google Scholar]
- 68.Huber K.V. Stereospecific targeting of MTH1 by (S)-crizotinib as an anticancer strategy. Nature. 2014;508(7495):222–227. doi: 10.1038/nature13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ellermann M. Novel class of potent and cellularly active inhibitors devalidates MTH1 as broad-spectrum cancer target. ACS Chem. Biol. 2017;12(8):1986–1992. doi: 10.1021/acschembio.7b00370. [DOI] [PubMed] [Google Scholar]
- 70.Kettle J.G. Potent and selective inhibitors of MTH1 probe its role in cancer cell survival. J. Med. Chem. 2016;59(6):2346–2361. doi: 10.1021/acs.jmedchem.5b01760. [DOI] [PubMed] [Google Scholar]
- 71.Samaranayake G.J., Huynh M., Rai P. MTH1 as a chemotherapeutic target: the Elephant in the room. Cancers. 2017;9(5) doi: 10.3390/cancers9050047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Samaranayake G.J. The existence of MTH1-independent 8-oxodGTPase activity in cancer cells as a compensatory mechanism against on-target effects of MTH1 inhibitors. Mol. Canc. Therapeut. 2020;19(2):432–446. doi: 10.1158/1535-7163.MCT-19-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nakabeppu Y., Ohta E., Abolhassani N. MTH1 as a nucleotide pool sanitizing enzyme, Friend or foe? Free Radic. Biol. Med. 2017;107:151–158. doi: 10.1016/j.freeradbiomed.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 74.Vitry G. Oxidized DNA precursors Cleanup by NUDT1 Contributes to vascular Remodeling in PAH. Am. J. Respir. Crit. Care Med. 2020 doi: 10.1164/rccm.202003-0627OC. [DOI] [PubMed] [Google Scholar]
- 75.Fujikawa K. The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein. J. Biol. Chem. 1999;274(26):18201–18205. doi: 10.1074/jbc.274.26.18201. [DOI] [PubMed] [Google Scholar]
- 76.Kamiya H., Kasai H. 2-Hydroxyadenine (isoguanine) as oxidative DNA damage: its formation and mutation inducibility. Nucleic Acids Symp. Ser. 1995;(34):233–234. [PubMed] [Google Scholar]
- 77.Asada S. 2-Oxoadenosine induces cytotoxicity through intracellular accumulation of 2-oxo-ATP and depletion of ATP but not via the p38 MAPK pathway. Sci. Rep. 2017;7(1):6528. doi: 10.1038/s41598-017-06636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jemth A.S. MutT homologue 1 (MTH1) catalyzes the hydrolysis of mutagenic O6-methyl-dGTP. Nucleic Acids Res. 2018;46(20):10888–10904. doi: 10.1093/nar/gky896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Scaletti E.R. MutT homologue 1 (MTH1) removes N6-methyl-dATP from the dNTP pool. J. Biol. Chem. 2020;295(15):4761–4772. doi: 10.1074/jbc.RA120.012636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carreras-Puigvert J. A comprehensive structural, biochemical and biological profiling of the human NUDIX hydrolase family. Nat. Commun. 2017;8(1):1541. doi: 10.1038/s41467-017-01642-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kong Q., Lin C.L. Oxidative damage to RNA: mechanisms, consequences, and diseases. Cell. Mol. Life Sci. 2010;67(11):1817–1829. doi: 10.1007/s00018-010-0277-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hofer T. Hydrogen peroxide causes greater oxidation in cellular RNA than in DNA. Biol. Chem. 2005;386(4):333–337. doi: 10.1515/BC.2005.040. [DOI] [PubMed] [Google Scholar]
- 83.Yan L.L., Zaher H.S. How do cells cope with RNA damage and its consequences? J. Biol. Chem. 2019;294(41):15158–15171. doi: 10.1074/jbc.REV119.006513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weimann A., Belling D., Poulsen H.E. Quantification of 8-oxo-guanine and guanine as the nucleobase, nucleoside and deoxynucleoside forms in human urine by high-performance liquid chromatography-electrospray tandem mass spectrometry. Nucleic Acids Res. 2002;30(2):E7. doi: 10.1093/nar/30.2.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hayakawa H., Kuwano M., Sekiguchi M. Specific binding of 8-oxoguanine-containing RNA to polynucleotide phosphorylase protein. Biochemistry. 2001;40(33):9977–9982. doi: 10.1021/bi010595q. [DOI] [PubMed] [Google Scholar]
- 86.Dai D.P. Transcriptional mutagenesis mediated by 8-oxoG induces translational errors in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 2018;115(16):4218–4222. doi: 10.1073/pnas.1718363115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Aas P.A. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421(6925):859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 88.Bellacosa A., Moss E.G. RNA repair: damage control. Curr. Biol. 2003;13(12):R482–R484. doi: 10.1016/s0960-9822(03)00408-1. [DOI] [PubMed] [Google Scholar]
- 89.Ishii T., Sekiguchi M. Two ways of escaping from oxidative RNA damage: selective degradation and cell death. DNA Repair. 2019;81:102666. doi: 10.1016/j.dnarep.2019.102666. [DOI] [PubMed] [Google Scholar]
- 90.Kohno K. The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays. 2003;25(7):691–698. doi: 10.1002/bies.10300. [DOI] [PubMed] [Google Scholar]
- 91.Hayakawa H. Binding capacity of human YB-1 protein for RNA containing 8-oxoguanine. Biochemistry. 2002;41(42):12739–12744. doi: 10.1021/bi0201872. [DOI] [PubMed] [Google Scholar]
- 92.Ishii T. PCBP1 and PCBP2 both bind heavily oxidized RNA but cause opposing outcomes, suppressing or increasing apoptosis under oxidative conditions. J. Biol. Chem. 2020;295(34):12247–12261. doi: 10.1074/jbc.RA119.011870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Taddei F. Counteraction by MutT protein of transcriptional errors caused by oxidative damage. Science. 1997;278(5335):128–130. doi: 10.1126/science.278.5335.128. [DOI] [PubMed] [Google Scholar]
- 94.Nunomura A. Neuronal RNA oxidation in Alzheimer's disease and Down's syndrome. Ann. N. Y. Acad. Sci. 1999;893:362–364. doi: 10.1111/j.1749-6632.1999.tb07855.x. [DOI] [PubMed] [Google Scholar]
- 95.Nunomura A. RNA oxidation is a prominent feature of vulnerable neurons in Alzheimer's disease. J. Neurosci. 1999;19(6):1959–1964. doi: 10.1523/JNEUROSCI.19-06-01959.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kong Q. RNA oxidation: a contributing factor or an epiphenomenon in the process of neurodegeneration. Free Radic. Res. 2008;42(9):773–777. doi: 10.1080/10715760802311187. [DOI] [PubMed] [Google Scholar]
- 97.Shan X., Tashiro H., Lin C.L. The identification and characterization of oxidized RNAs in Alzheimer's disease. J. Neurosci. 2003;23(12):4913–4921. doi: 10.1523/JNEUROSCI.23-12-04913.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shan X., Lin C.L. Quantification of oxidized RNAs in Alzheimer's disease. Neurobiol. Aging. 2006;27(5):657–662. doi: 10.1016/j.neurobiolaging.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 99.Chang Y. Messenger RNA oxidation occurs early in disease pathogenesis and promotes motor neuron degeneration in ALS. PloS One. 2008;3(8):e2849. doi: 10.1371/journal.pone.0002849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shan X., Chang Y., Lin C.L. Messenger RNA oxidation is an early event preceding cell death and causes reduced protein expression. Faseb. J. 2007;21(11):2753–2764. doi: 10.1096/fj.07-8200com. [DOI] [PubMed] [Google Scholar]
- 101.Gonzalez-Rivera J.C. RNA oxidation in chromatin modification and DNA-damage response following exposure to formaldehyde. Sci. Rep. 2020;10(1):16545. doi: 10.1038/s41598-020-73376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xu Z. Current perspectives on the clinical implications of oxidative RNA damage in aging research: challenges and opportunities. Geroscience. 2020 doi: 10.1007/s11357-020-00209-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Broedbaek K. Urinary excretion of biomarkers of oxidatively damaged DNA and RNA in hereditary hemochromatosis. Free Radic. Biol. Med. 2009;47(8):1230–1233. doi: 10.1016/j.freeradbiomed.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 104.Broedbaek K. Long-term effects of Irbesartan treatment and smoking on nucleic acid oxidation in patients with type 2 diabetes and microalbuminuria: an Irbesartan in patients with type 2 diabetes and Microalbuminuria (IRMA 2) substudy. Diabetes Care. 2011;34(5):1192–1198. doi: 10.2337/dc10-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Broedbaek K. Urinary markers of nucleic acid oxidation and cancer in type 2 diabetes. Redox Biol. 2015;4:34–39. doi: 10.1016/j.redox.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Broedbaek K. Association between urinary markers of nucleic acid oxidation and mortality in type 2 diabetes: a population-based cohort study. Diabetes Care. 2013;36(3):669–676. doi: 10.2337/dc12-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Broedbaek K. Urinary markers of nucleic acid oxidation and long-term mortality of newly diagnosed type 2 diabetic patients. Diabetes Care. 2011;34(12):2594–2596. doi: 10.2337/dc11-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Broedbaek K. Urinary 8-oxo-7,8-dihydro-2’-deoxyguanosine as a biomarker in type 2 diabetes. Free Radic. Biol. Med. 2011;51(8):1473–1479. doi: 10.1016/j.freeradbiomed.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 109.Kjaer L.K. The effect of structured personal care on RNA oxidation: a 19-year follow-up of the randomized trial Diabetes Care in General Practice (DCGP) J. Diabet. Complicat. 2019;33(3):202–207. doi: 10.1016/j.jdiacomp.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 110.Kjaer L.K. Indicator of RNA oxidation in urine for the prediction of mortality in patients with type 2 diabetes and microalbuminuria: a post-hoc analysis of the Steno-2 trial. Free Radic. Biol. Med. 2018;129:247–255. doi: 10.1016/j.freeradbiomed.2018.09.030. [DOI] [PubMed] [Google Scholar]
- 111.Jorgensen A. Increased systemic oxidatively generated DNA and RNA damage in schizophrenia. Psychiatr. Res. 2013;209(3):417–423. doi: 10.1016/j.psychres.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 112.Jorgensen A. Systemic oxidatively generated DNA/RNA damage in clinical depression: associations to symptom severity and response to electroconvulsive therapy. J. Affect. Disord. 2013;149(1–3):355–362. doi: 10.1016/j.jad.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 113.Munkholm K. A multisystem composite biomarker as a preliminary diagnostic test in bipolar disorder. Acta Psychiatr. Scand. 2019;139(3):227–236. doi: 10.1111/acps.12983. [DOI] [PubMed] [Google Scholar]
- 114.Nordholm D. Systemic oxidative DNA and RNA damage are not increased during early phases of psychosis: a case control study. Psychiatr. Res. 2016;241:201–206. doi: 10.1016/j.psychres.2016.04.062. [DOI] [PubMed] [Google Scholar]
- 115.Xu X.M. Increased oxidative damage of RNA in liver injury caused by hepatitis B virus (HBV) infection. Free Radic. Res. 2018;52(4):426–433. doi: 10.1080/10715762.2018.1439165. [DOI] [PubMed] [Google Scholar]
- 116.Lorente L. Association between DNA and RNA oxidative damage and mortality in septic patients. J. Crit. Care. 2019;54:94–98. doi: 10.1016/j.medin.2019.07.00. [DOI] [PubMed] [Google Scholar]
- 117.Lorente L. DNA and RNA oxidative damage are associated to mortality in patients with cerebral infarction. Med. Intensiva. 2019 doi: 10.1016/j.medin.2019.07.008. [DOI] [PubMed] [Google Scholar]
- 118.Lorente L. Association between DNA and RNA oxidative damage and mortality of patients with traumatic brain injury. Neurocrit. Care. 2020;32(3):790–795. doi: 10.1007/s12028-019-00800-w. [DOI] [PubMed] [Google Scholar]
- 119.Lorente L. High serum DNA and RNA oxidative damage in non-surviving patients with spontaneous intracerebral hemorrhage. Neurocrit. care. 2020;33(1):90–96. doi: 10.1007/s12028-019-00864-8. [DOI] [PubMed] [Google Scholar]
- 120.Halliwell B., Dizdaroglu M. The measurement of oxidative damage to DNA by HPLC and GC/MS techniques. Free Radic. Res. Commun. 1992;16(2):75–87. doi: 10.3109/10715769209049161. [DOI] [PubMed] [Google Scholar]
- 121.Floyd R.A. Hydroxyl free radical adduct of deoxyguanosine: sensitive detection and mechanisms of formation. Free Radic. Res. Commun. 1986;1(3):163–172. doi: 10.3109/10715768609083148. [DOI] [PubMed] [Google Scholar]
- 122.Dizdaroglu M., Jaruga P., Rodriguez H. Measurement of 8-hydroxy-2’-deoxyguanosine in DNA by high-performance liquid chromatography-mass spectrometry: comparison with measurement by gas chromatography-mass spectrometry. Nucleic Acids Res. 2001;29(3):E12. doi: 10.1093/nar/29.3.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ravanat J.L. Isotope dilution high-performance liquid chromatography-electrospray tandem mass spectrometry assay for the measurement of 8-oxo-7,8-dihydro-2'-deoxyguanosine in biological samples. J. Chromatogr. B Biomed. Sci. Appl. 1998;715(2):349–356. doi: 10.1016/s0378-4347(98)00259-x. [DOI] [PubMed] [Google Scholar]
- 124.Bartsch H., Nair J. Ultrasensitive and specific detection methods for exocylic DNA adducts: markers for lipid peroxidation and oxidative stress. Toxicology. 2000;153(1–3):105–114. doi: 10.1016/s0300-483x(00)00307-3. [DOI] [PubMed] [Google Scholar]
- 125.Cooke M.S. Novel repair action of vitamin C upon in vivo oxidative DNA damage. FEBS Lett. 1998;439(3):363–367. doi: 10.1016/s0014-5793(98)01403-3. [DOI] [PubMed] [Google Scholar]
- 126.Shimoi K. Comparison between high-performance liquid chromatography and enzyme-linked immunosorbent assay for the determination of 8-hydroxy-2’-deoxyguanosine in human urine. Canc. Epidemiol. Biomarkers Prev. 2002;11(8):767–770. [PubMed] [Google Scholar]
- 127.Pflaum M. DNA oxidation products determined with repair endonucleases in mammalian cells: types, basal levels and influence of cell proliferation. Free Radic. Res. 1998;29(6):585–594. doi: 10.1080/10715769800300631. [DOI] [PubMed] [Google Scholar]
- 128.Collins A.R. The comet assay for DNA damage and repair: principles, applications, and limitations. Mol. Biotechnol. 2004;26(3):249–261. doi: 10.1385/MB:26:3:249. [DOI] [PubMed] [Google Scholar]
- 129.Muller N. An automated Fpg-based FADU method for the detection of oxidative DNA lesions and screening of antioxidants. Toxicology. 2013;310:15–21. doi: 10.1016/j.tox.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gedik C.M., Collins A. Escodd, Establishing the background level of base oxidation in human lymphocyte DNA: results of an interlaboratory validation study. Faseb. J. 2005;19(1):82–84. doi: 10.1096/fj.04-1767fje. [DOI] [PubMed] [Google Scholar]
- 131.Escodd Comparative analysis of baseline 8-oxo-7,8-dihydroguanine in mammalian cell DNA, by different methods in different laboratories: an approach to consensus. Carcinogenesis. 2002;23(12):2129–2133. doi: 10.1093/carcin/23.12.2129. [DOI] [PubMed] [Google Scholar]
- 132.European Standards Committee on Oxidative Measurement of DNA oxidation in human cells by chromatographic and enzymic methods. Free Radic. Biol. Med. 2003;34(8):1089–1099. doi: 10.1016/s0891-5849(03)00041-8. [DOI] [PubMed] [Google Scholar]
- 133.Lunec J. ESCODD: European standards Committee on oxidative DNA damage. Free Radic. Res. 1998;29(6):601–608. doi: 10.1080/10715769800300651. [DOI] [PubMed] [Google Scholar]
- 134.Riis B. Comparison of results from different laboratories in measuring 8-oxo-2'-deoxyguanosine in synthetic oligonucleotides. Free Radic. Res. 2002;36(6):649–659. doi: 10.1080/10715760290029047. [DOI] [PubMed] [Google Scholar]
- 135.Claycamp H.G. Phenol sensitization of DNA to subsequent oxidative damage in 8-hydroxyguanine assays. Carcinogenesis. 1992;13(7):1289–1292. doi: 10.1093/carcin/13.7.1289. [DOI] [PubMed] [Google Scholar]
- 136.Finnegan M.T. Evidence for sensitisation of DNA to oxidative damage during isolation. Free Radic. Biol. Med. 1996;20(1):93–98. doi: 10.1016/0891-5849(95)02003-9. [DOI] [PubMed] [Google Scholar]
- 137.Guetens G. Oxidative DNA damage: biological significance and methods of analysis. Crit. Rev. Clin. Lab Sci. 2002;39(4–5):331–457. doi: 10.1080/10408360290795547. [DOI] [PubMed] [Google Scholar]
- 138.Ravanat J.L. Cellular background level of 8-oxo-7,8-dihydro-2'-deoxyguanosine: an isotope based method to evaluate artefactual oxidation of DNA during its extraction and subsequent work-up. Carcinogenesis. 2002;23(11):1911–1918. doi: 10.1093/carcin/23.11.1911. [DOI] [PubMed] [Google Scholar]
- 139.Chao M.R., Yen C.C., Hu C.W. Prevention of artifactual oxidation in determination of cellular 8-oxo-7,8-dihydro-2'-deoxyguanosine by isotope-dilution LC-MS/MS with automated solid-phase extraction. Free Radic. Biol. Med. 2008;44(3):464–473. doi: 10.1016/j.freeradbiomed.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 140.Frelon S. High-performance liquid chromatography–tandem mass spectrometry measurement of radiation-induced base damage to isolated and cellular DNA. Chem. Res. Toxicol. 2000;13(10):1002–1010. doi: 10.1021/tx000085h. [DOI] [PubMed] [Google Scholar]
- 141.Cadet J. Assessment of oxidative base damage to isolated and cellular DNA by HPLC-MS/MS measurement. Free Radic. Biol. Med. 2002;33(4):441–449. doi: 10.1016/s0891-5849(02)00820-1. [DOI] [PubMed] [Google Scholar]
- 142.Dizdaroglu M. Free radical-induced damage to DNA: mechanisms and measurement. Free Radic. Biol. Med. 2002;32(11):1102–1115. doi: 10.1016/s0891-5849(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 143.Dizdaroglu M., Coskun E., Jaruga P. Measurement of oxidatively induced DNA damage and its repair, by mass spectrometric techniques. Free Radic. Res. 2015;49(5):525–548. doi: 10.3109/10715762.2015.1014814. [DOI] [PubMed] [Google Scholar]
- 144.Loft S. Antioxidant vitamins and cancer risk: is oxidative damage to DNA a relevant biomarker? Eur. J. Nutr. 2008;47(Suppl 2):19–28. doi: 10.1007/s00394-008-2004-0. [DOI] [PubMed] [Google Scholar]
- 145.Moller P. Applications of the comet assay in particle toxicology: air pollution and engineered nanomaterials exposure. Mutagenesis. 2015;30(1):67–83. doi: 10.1093/mutage/geu035. [DOI] [PubMed] [Google Scholar]
- 146.Moller P. Effect of age and sex on the level of DNA strand breaks and oxidatively damaged DNA in human blood cells. Mutat Res Genet Toxicol Environ Mutagen. 2019;838:16–21. doi: 10.1016/j.mrgentox.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 147.Moller P. The comet assay: ready for 30 more years. Mutagenesis. 2018;33(1):1–7. doi: 10.1093/mutage/gex046. [DOI] [PubMed] [Google Scholar]
- 148.Collins A.R. Oxidative damage to DNA: do we have a reliable biomarker? Environ. Health Perspect. 1996;104(Suppl 3):465–469. doi: 10.1289/ehp.96104s3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Smith C.C., O'Donovan M.R., Martin E.A. hOGG1 recognizes oxidative damage using the comet assay with greater specificity than FPG or ENDOIII. Mutagenesis. 2006;21(3):185–190. doi: 10.1093/mutage/gel019. [DOI] [PubMed] [Google Scholar]
- 150.Moller P. Assessment and reduction of comet assay variation in relation to DNA damage: studies from the European Comet Assay Validation Group. Mutagenesis. 2010;25(2):109–111. doi: 10.1093/mutage/gep067. [DOI] [PubMed] [Google Scholar]
- 151.Forchhammer L. Variation in the measurement of DNA damage by comet assay measured by the ECVAG inter-laboratory validation trial. Mutagenesis. 2010;25(2):113–123. doi: 10.1093/mutage/gep048. [DOI] [PubMed] [Google Scholar]
- 152.Godschalk R.W. DNA-repair measurements by use of the modified comet assay: an inter-laboratory comparison within the European Comet Assay Validation Group (ECVAG) Mutat. Res. 2013;757(1):60–67. doi: 10.1016/j.mrgentox.2013.06.020. [DOI] [PubMed] [Google Scholar]
- 153.Ersson C. An ECVAG inter-laboratory validation study of the comet assay: inter-laboratory and intra-laboratory variations of DNA strand breaks and FPG-sensitive sites in human mononuclear cells. Mutagenesis. 2013;28(3):279–286. doi: 10.1093/mutage/get001. [DOI] [PubMed] [Google Scholar]
- 154.Forchhammer L. Inter-laboratory variation in DNA damage using a standard comet assay protocol. Mutagenesis. 2012;27(6):665–672. doi: 10.1093/mutage/ges032. [DOI] [PubMed] [Google Scholar]
- 155.Godschalk R.W. Variation of DNA damage levels in peripheral blood mononuclear cells isolated in different laboratories. Mutagenesis. 2014;29(4):241–249. doi: 10.1093/mutage/geu012. [DOI] [PubMed] [Google Scholar]
- 156.Moller P., Loft S. Statistical analysis of comet assay results. Front. Genet. 2014;5:292. doi: 10.3389/fgene.2014.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Moller P. Harmonising measurements of 8-oxo-7,8-dihydro-2'-deoxyguanosine in cellular DNA and urine. Free Radic. Res. 2012;46(4):541–553. doi: 10.3109/10715762.2011.644241. [DOI] [PubMed] [Google Scholar]
- 158.Ohno M., Oka S., Nakabeppu Y. Quantitative analysis of oxidized guanine, 8-oxoguanine, in mitochondrial DNA by immunofluorescence method. Methods Mol. Biol. 2009;554:199–212. doi: 10.1007/978-1-59745-521-3_13. [DOI] [PubMed] [Google Scholar]
- 159.Rossner P., Jr., Sram R.J. Immunochemical detection of oxidatively damaged DNA. Free Radic. Res. 2012;46(4):492–522. doi: 10.3109/10715762.2011.632415. [DOI] [PubMed] [Google Scholar]
- 160.Cooke M.S., Herbert K. Immunochemical detection of 8-oxodeoxyguanosine in DNA. In: Lunec J., Griffiths H.R., editors. Measuring in Vivo Oxidative Damage : A Practical Approach. 2000. pp. 63–68. John Wiley & Sons Ltd: Chichester. [Google Scholar]
- 161.Ohno M. A genome-wide distribution of 8-oxoguanine correlates with the preferred regions for recombination and single nucleotide polymorphism in the human genome. Genome Res. 2006;16(5):567–575. doi: 10.1101/gr.4769606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mingard C. Next-generation DNA damage sequencing. Chem. Soc. Rev. 2020;49(20):7354–7377. doi: 10.1039/d0cs00647e. [DOI] [PubMed] [Google Scholar]
- 163.Al-Salmani K. Simplified method for the collection, storage, and comet assay analysis of DNA damage in whole blood. Free Radic. Biol. Med. 2011;51(3):719–725. doi: 10.1016/j.freeradbiomed.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 164.Milic M. Alkaline comet assay results on fresh and one-year frozen whole blood in small volume without cryo-protection in a group of people with different health status. Mutat. Res. 2019;843:3–10. doi: 10.1016/j.mrgentox.2019.03.009. [DOI] [PubMed] [Google Scholar]
- 165.Gajski G. Application of the comet assay for the evaluation of DNA damage from frozen human whole blood samples: implications for human biomonitoring. Toxicol. Lett. 2020;319:58–65. doi: 10.1016/j.toxlet.2019.11.010. [DOI] [PubMed] [Google Scholar]
- 166.Ladeira C. The comet assay for human biomonitoring: effect of cryopreservation on DNA damage in different blood cell preparations. Mutat. Res. 2019;843:11–17. doi: 10.1016/j.mrgentox.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 167.Hu C.W. 8-Oxo-7,8-dihydroguanine and 8-oxo-7,8-dihydro-2’-deoxyguanosine concentrations in various human body fluids: implications for their measurement and interpretation. Arch. Toxicol. 2015;89(2):201–210. doi: 10.1007/s00204-014-1255-1. [DOI] [PubMed] [Google Scholar]
- 168.Lam P.M. Rapid measurement of 8-oxo-7,8-dihydro-2'-deoxyguanosine in human biological matrices using ultra-high-performance liquid chromatography-tandem mass spectrometry. Free Radic. Biol. Med. 2012;52(10):2057–2063. doi: 10.1016/j.freeradbiomed.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Da Broi M.G. Increased concentration of 8-hydroxy-2'-deoxyguanosine in follicular fluid of infertile women with endometriosis. Cell Tissue Res. 2016;366(1):231–242. doi: 10.1007/s00441-016-2428-4. [DOI] [PubMed] [Google Scholar]
- 170.Varnagy A. Levels of total antioxidant capacity and 8-hydroxy-2'-deoxyguanosine of serum and follicular fluid in women undergoing in vitro fertilization: focusing on endometriosis. Hum. Fertil. 2020;23(3):200–208. doi: 10.1080/14647273.2018.1535719. [DOI] [PubMed] [Google Scholar]
- 171.Weimann A., Simonsen A.H., Poulsen H.E. Measurement of 8-oxo-7,8-dihydro-2'-deoxyguanosine and 8-oxo-7,8-dihydro-guanosine in cerebrospinal fluid by ultra performance liquid chromatography-tandem mass spectrometry. J. Chromatogr. B Analyt Technol. Biomed. Life Sci. 2018;1073:110–117. doi: 10.1016/j.jchromb.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 172.Cooke M.S. Measurement and meaning of oxidatively modified DNA lesions in urine. Canc. Epidemiol. Biomark. Prev. 2008;17(1):3–14. doi: 10.1158/1055-9965.EPI-07-0751. [DOI] [PubMed] [Google Scholar]
- 173.Cadet J., Poulsen H. Measurement of oxidatively generated base damage in cellular DNA and urine. Free Radic. Biol. Med. 2010;48(11):1457–1459. doi: 10.1016/j.freeradbiomed.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 174.Pour Khavari A. Serum 8-Oxo-dG as a Predictor of Sensitivity and Outcome of Radiotherapy and Chemotherapy of upper gastrointestinal tumours. Oxid. Med. Cell. Longev. 2018;2018:4153574. doi: 10.1155/2018/4153574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ma J. A pilot study of biomarkers of oxidative stress in serum and schizophrenia. Psychiatr. Res. 2020;284:112757. doi: 10.1016/j.psychres.2020.112757. [DOI] [PubMed] [Google Scholar]
- 176.Onder C. Impact of non-surgical periodontal therapy on saliva and serum levels of markers of oxidative stress. Clin. Oral Invest. 2017;21(6):1961–1969. doi: 10.1007/s00784-016-1984-z. [DOI] [PubMed] [Google Scholar]
- 177.Ongoz Dede F. The effect of initial periodontal treatment on plasma, gingival crevicular fluid and salivary levels of 8-hydroxy-deoxyguanosine in obesity. Arch. Oral Biol. 2016;62:80–85. doi: 10.1016/j.archoralbio.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 178.Dede F.O., Ozden F.O., Avci B. 8-hydroxy-deoxyguanosine levels in gingival crevicular fluid and saliva in patients with chronic periodontitis after initial periodontal treatment. J. Periodontol. 2013;84(6):821–828. doi: 10.1902/jop.2012.120195. [DOI] [PubMed] [Google Scholar]
- 179.Takane M. New biomarker evidence of oxidative DNA damage in whole saliva from clinically healthy and periodontally diseased individuals. J. Periodontol. 2002;73(5):551–554. doi: 10.1902/jop.2002.73.5.551. [DOI] [PubMed] [Google Scholar]
- 180.Kulkarni N., Cooke M.S., Grigg J. Neutrophils in induced sputum from healthy children: role of interleukin-8 and oxidative stress. Respir. Med. 2007;101(10):2108–2112. doi: 10.1016/j.rmed.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 181.Cooke M.S. Induction and excretion of ultraviolet-induced 8-oxo-2'-deoxyguanosine and thymine dimers in vivo: implications for PUVA. J. Invest. Dermatol. 2001;116(2):281–285. doi: 10.1046/j.1523-1747.2001.01251.x. [DOI] [PubMed] [Google Scholar]
- 182.Svecova V. Urinary 8-oxodeoxyguanosine levels in children exposed to air pollutants. Mutat. Res. 2009;662(1–2):37–43. doi: 10.1016/j.mrfmmm.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 183.Serdar B. Short-term markers of DNA damage among roofers who work with hot asphalt. Environ. Health. 2016;15(1):99. doi: 10.1186/s12940-016-0182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Nagao M. Urinary 8-hydroxy-2’-deoxyguanosine Levels and cardiovascular disease Incidence in Japan. J. Atheroscler. Thromb. 2020;27(10):1086–1096. doi: 10.5551/jat.51664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kasai H. A new automated method to analyze urinary 8-hydroxydeoxyguanosine by a high-performance liquid chromatography-electrochemical detector system. J. Radiat. Res. 2003;44(2):185–189. doi: 10.1269/jrr.44.185. [DOI] [PubMed] [Google Scholar]
- 186.Loft S., Poulsen H.E. Markers of oxidative damage to DNA: antioxidants and molecular damage. Methods Enzymol. 1999;300:166–184. doi: 10.1016/s0076-6879(99)00124-x. [DOI] [PubMed] [Google Scholar]
- 187.Cathcart R. Thymine glycol and thymidine glycol in human and rat urine: a possible assay for oxidative DNA damage. Proc. Natl. Acad. Sci. U. S. A. 1984;81(18):5633–5637. doi: 10.1073/pnas.81.18.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bergtold D.S. Urine biomarkers for oxidative DNA damage. Basic Life Sci. 1988;49:483–489. doi: 10.1007/978-1-4684-5568-7_75. [DOI] [PubMed] [Google Scholar]
- 189.Tagesson C. Increased urinary excretion of the oxidative DNA adduct, 8-hydroxydeoxyguanosine, as a possible early indicator of occupational cancer hazards in the asbestos, rubber, and azo-dye industries. Pol. J. Occup. Med. Environ. Health. 1993;6(4):357–368. [PubMed] [Google Scholar]
- 190.Loft S. Oxidative DNA damage estimated by 8-hydroxydeoxyguanosine excretion in humans: influence of smoking, gender and body mass index. Carcinogenesis. 1992;13(12):2241–2247. doi: 10.1093/carcin/13.12.2241. [DOI] [PubMed] [Google Scholar]
- 191.Pilger A. 8-Hydroxydeoxyguanosine in leukocyte DNA and urine of quartz-exposed workers and patients with silicosis. Int. Arch. Occup. Environ. Health. 2000;73(5):305–310. doi: 10.1007/s004200000117. [DOI] [PubMed] [Google Scholar]
- 192.Bogdanov M.B. A carbon column-based liquid chromatography electrochemical approach to routine 8-hydroxy-2'-deoxyguanosine measurements in urine and other biologic matrices: a one-year evaluation of methods. Free Radic. Biol. Med. 1999;27(5–6):647–666. doi: 10.1016/s0891-5849(99)00113-6. [DOI] [PubMed] [Google Scholar]
- 193.Ravanat J.L. Simultaneous determination of five oxidative DNA lesions in human urine. Chem. Res. Toxicol. 1999;12(9):802–808. doi: 10.1021/tx980194k. [DOI] [PubMed] [Google Scholar]
- 194.Rozalski R. Diet is not responsible for the presence of several oxidatively damaged DNA lesions in mouse urine. Free Radic. Res. 2004;38(11):1201–1205. doi: 10.1080/10715760400017350. [DOI] [PubMed] [Google Scholar]
- 195.Cooke M.S. DNA repair is responsible for the presence of oxidatively damaged DNA lesions in urine. Mutat. Res. 2005;574(1–2):58–66. doi: 10.1016/j.mrfmmm.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 196.Lin H.S. A high-throughput and sensitive methodology for the quantification of urinary 8-hydroxy-2'-deoxyguanosine: measurement with gas chromatography-mass spectrometry after single solid-phase extraction. Biochem. J. 2004;380(Pt 2):541–548. doi: 10.1042/BJ20040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Cooke M.S. Evaluation of enzyme-linked immunosorbent assay and liquid chromatography-tandem mass spectrometry methodology for the analysis of 8-oxo-7,8-dihydro-2'-deoxyguanosine in saliva and urine. Free Radic. Biol. Med. 2006;41(12):1829–1836. doi: 10.1016/j.freeradbiomed.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 198.Evans M.D. Analysis of urinary 8-oxo-7,8-dihydro-purine-2'-deoxyribonucleosides by LC-MS/MS and improved ELISA. Free Radic. Res. 2008;42(10):831–840. doi: 10.1080/10715760802506323. [DOI] [PubMed] [Google Scholar]
- 199.Hu C.W. Clinical-scale high-throughput analysis of urinary 8-oxo-7,8-dihydro-2'-deoxyguanosine by isotope-dilution liquid chromatography-tandem mass spectrometry with on-line solid-phase extraction. Clin. Chem. 2006;52(7):1381–1388. doi: 10.1373/clinchem.2005.063735. [DOI] [PubMed] [Google Scholar]
- 200.Hu C.W. Correlation between concentrations of 8-oxo-7,8-dihydro-2'-deoxyguanosine in urine, plasma and saliva measured by on-line solid-phase extraction LC-MS/MS. Clin. Chim. Acta. 2010;411(17–18):1218–1222. doi: 10.1016/j.cca.2010.04.029. [DOI] [PubMed] [Google Scholar]
- 201.Jaruga P., Dizdaroglu M. Identification and quantification of (5'R)- and (5'S)-8,5'-cyclo-2'-deoxyadenosines in human urine as putative biomarkers of oxidatively induced damage to DNA. Biochem. Biophys. Res. Commun. 2010;397(1):48–52. doi: 10.1016/j.bbrc.2010.05.050. [DOI] [PubMed] [Google Scholar]
- 202.Yin B. Determination of 8-hydroxydeoxyguanosine by an immunoaffinity chromatography-monoclonal antibody-based ELISA. Free Radic. Biol. Med. 1995;18(6):1023–1032. doi: 10.1016/0891-5849(95)00003-g. [DOI] [PubMed] [Google Scholar]
- 203.Toyokuni S. Quantitative immunohistochemical determination of 8-hydroxy-2'-deoxyguanosine by a monoclonal antibody N45.1: its application to ferric nitrilotriacetate-induced renal carcinogenesis model. Lab. Invest. 1997;76(3):365–374. [PubMed] [Google Scholar]
- 204.Rossner P., Jr. Urinary 8-oxo-7,8-dihydro-2'-deoxyguanosine values determined by a modified ELISA improves agreement with HPLC-MS/MS. Biochem. Biophys. Res. Commun. 2013;440(4):725–730. doi: 10.1016/j.bbrc.2013.09.133. [DOI] [PubMed] [Google Scholar]
- 205.Rossner P., Jr. Urinary 8-oxo-7,8-dihydro-2'-deoxyguanosine analysis by an improved ELISA: An inter-laboratory comparison study. Free Radic. Biol. Med. 2016;95:169–179. doi: 10.1016/j.freeradbiomed.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 206.European Standards Committee on Urinary Lesion Toward consensus in the analysis of urinary 8-oxo-7,8-dihydro-2'-deoxyguanosine as a noninvasive biomarker of oxidative stress. Faseb. J. 2010;24(4):1249–1260. doi: 10.1096/fj.09-147124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Barregard L. Human and methodological sources of variability in the measurement of urinary 8-oxo-7,8-dihydro-2'-deoxyguanosine. Antioxidants Redox Signal. 2013;18(18):2377–2391. doi: 10.1089/ars.2012.4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Haghdoost S. Extracellular 8-oxo-dG as a sensitive parameter for oxidative stress in vivo and in vitro. Free Radic. Res. 2005;39(2):153–162. doi: 10.1080/10715760500043132. [DOI] [PubMed] [Google Scholar]
- 209.Song M.F. Urea, the most abundant component in urine, cross-reacts with a commercial 8-OH-dG ELISA kit and contributes to overestimation of urinary 8-OH-dG. Free Radic. Biol. Med. 2009;47(1):41–46. doi: 10.1016/j.freeradbiomed.2009.02.017. [DOI] [PubMed] [Google Scholar]
- 210.Chen H.J., Chang C.M. Quantification of urinary excretion of 1,N6-ethenoadenine, a potential biomarker of lipid peroxidation, in humans by stable isotope dilution liquid chromatography-electrospray ionization-tandem mass spectrometry: comparison with gas chromatography-mass spectrometry. Chem. Res. Toxicol. 2004;17(7):963–971. doi: 10.1021/tx0341963. [DOI] [PubMed] [Google Scholar]
- 211.Nair J. Etheno DNA-base adducts from endogenous reactive species. Mutat. Res. 1999;424(1–2):59–69. doi: 10.1016/s0027-5107(99)00008-1. [DOI] [PubMed] [Google Scholar]
- 212.Lee K.H. Effect of short-term fasting on urinary excretion of primary lipid peroxidation products and on markers of oxidative DNA damage in healthy women. Carcinogenesis. 2006;27(7):1398–1403. doi: 10.1093/carcin/bgi337. [DOI] [PubMed] [Google Scholar]
- 213.Hillestrom P.R. Quantification of 1,N6-etheno-2'-deoxyadenosine in human urine by column-switching LC/APCI-MS/MS. Free Radic. Biol. Med. 2004;36(11):1383–1392. doi: 10.1016/j.freeradbiomed.2004.02.068. [DOI] [PubMed] [Google Scholar]
- 214.Chen H.J. Effect of cigarette smoking on urinary 3,N4-ethenocytosine levels measured by gas chromatography/mass spectrometry. Toxicol. Sci. 2003;76(2):321–327. doi: 10.1093/toxsci/kfg219. [DOI] [PubMed] [Google Scholar]
- 215.Chen H.J. Urinary excretion of 3,N4-etheno-2'-deoxycytidine in humans as a biomarker of oxidative stress: association with cigarette smoking. Chem. Res. Toxicol. 2004;17(7):896–903. doi: 10.1021/tx0342013. [DOI] [PubMed] [Google Scholar]
- 216.Hillestrom P.R., Weimann A., Poulsen H.E. Quantification of urinary etheno-DNA adducts by column-switching LC/APCI-MS/MS. J. Am. Soc. Mass Spectrom. 2006;17(4):605–610. doi: 10.1016/j.jasms.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 217.Chen H.J., Chiu W.L. Association between cigarette smoking and urinary excretion of 1,N2-ethenoguanine measured by isotope dilution liquid chromatography-electrospray ionization/tandem mass spectrometry. Chem. Res. Toxicol. 2005;18(10):1593–1599. doi: 10.1021/tx050145p. [DOI] [PubMed] [Google Scholar]
- 218.Knutson C.G. Monitoring in vivo metabolism and elimination of the endogenous DNA adduct, M1dG {3-(2-deoxy-beta-D-erythro-pentofuranosyl)pyrimido[1,2-alpha]purin-10(3H)-one}, by accelerator mass spectrometry. Chem. Res. Toxicol. 2008;21(6):1290–1294. doi: 10.1021/tx800049v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Hoberg A.M. Measurement of the malondialdehyde-2'-deoxyguanosine adduct in human urine by immuno-extraction and liquid chromatography/atmospheric pressure chemical ionization tandem mass spectrometry. J. Mass Spectrom. 2004;39(1):38–42. doi: 10.1002/jms.547. [DOI] [PubMed] [Google Scholar]
- 220.Long L.H., Evans P.J., Halliwell B. Hydrogen peroxide in human urine: implications for antioxidant defense and redox regulation. Biochem. Biophys. Res. Commun. 1999;262(3):605–609. doi: 10.1006/bbrc.1999.1263. [DOI] [PubMed] [Google Scholar]
- 221.Shigenaga M.K., Gimeno C.J., Ames B.N. Urinary 8-hydroxy-2'-deoxyguanosine as a biological marker of in vivo oxidative DNA damage. Proc. Natl. Acad. Sci. U. S. A. 1989;86(24):9697–9701. doi: 10.1073/pnas.86.24.9697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Loft S. Oxidative DNA damage after transplantation of the liver and small intestine in pigs. Transplantation. 1995;59(1):16–20. doi: 10.1097/00007890-199501150-00004. [DOI] [PubMed] [Google Scholar]
- 223.Knutson C.G. Metabolism in vitro and in vivo of the DNA base adduct, M1G. Chem. Res. Toxicol. 2007;20(3):550–557. doi: 10.1021/tx600334x. [DOI] [PubMed] [Google Scholar]
- 224.Knutson C.G. Metabolism and elimination of the endogenous DNA adduct, 3-(2-deoxy-beta-D-erythropentofuranosyl)-pyrimido[1,2-alpha]purine-10(3H)-one, in the rat. J. Biol. Chem. 2007;282(50):36257–36264. doi: 10.1074/jbc.M706814200. [DOI] [PubMed] [Google Scholar]
- 225.Otteneder M.B. In vivo oxidative metabolism of a major peroxidation-derived DNA adduct, M1dG. Proc. Natl. Acad. Sci. U. S. A. 2006;103(17):6665–6669. doi: 10.1073/pnas.0602017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Loft S. Prospective study of 8-oxo-7,8-dihydro-2'-deoxyguanosine excretion and the risk of lung cancer. Carcinogenesis. 2006;27(6):1245–1250. doi: 10.1093/carcin/bgi313. [DOI] [PubMed] [Google Scholar]
- 227.Matsumoto Y. The stability of the oxidative stress marker, urinary 8-hydroxy-2'- deoxyguanosine (8-OHdG), when stored at room temperature. J. Occup. Health. 2008;50(4):366–372. doi: 10.1539/joh.l7144. [DOI] [PubMed] [Google Scholar]
- 228.Bianchini F. Monitoring urinary excretion of 5-hydroxymethyluracil for assessment of oxidative DNA damage and repair. Biomarkers. 1996;1(3):178–184. doi: 10.3109/13547509609079354. [DOI] [PubMed] [Google Scholar]
- 229.Hu C.W., Chao M.R., Sie C.H. Urinary analysis of 8-oxo-7,8-dihydroguanine and 8-oxo-7,8-dihydro-2'-deoxyguanosine by isotope-dilution LC-MS/MS with automated solid-phase extraction: study of 8-oxo-7,8-dihydroguanine stability. Free Radic. Biol. Med. 2010;48(1):89–97. doi: 10.1016/j.freeradbiomed.2009.10.029. [DOI] [PubMed] [Google Scholar]
- 230.Ellegaard P.K., Poulsen H.E. Tobacco smoking and oxidative stress to DNA: a meta-analysis of studies using chromatographic and immunological methods. Scand. J. Clin. Lab. Invest. 2016;76(2):151–158. doi: 10.3109/00365513.2015.1127407. [DOI] [PubMed] [Google Scholar]
- 231.Cooke M.S., Lunec J., Evans M.D. Progress in the analysis of urinary oxidative DNA damage. Free Radic. Biol. Med. 2002;33(12):1601–1614. doi: 10.1016/s0891-5849(02)01146-2. [DOI] [PubMed] [Google Scholar]
- 232.Cooke M.S. A commentary on "Urea, the most abundant component in urine, cross-reacts with a commercial 8-OH-dG ELISA kit and contributes to overestimation of urinary 8-OH-dG". What is ELISA detecting? Free Radic. Biol. Med. 2009;47(1):30–31. doi: 10.1016/j.freeradbiomed.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 233.Guo X. Protective effect of folic acid on oxidative DNA damage: a randomized, double-blind, and placebo controlled clinical trial. Medicine. 2015;94(45):e1872. doi: 10.1097/MD.0000000000001872. Baltimore. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Ambroz A. Impact of air pollution on oxidative DNA damage and lipid peroxidation in mothers and their newborns. Int. J. Hyg Environ. Health. 2016;219(6):545–556. doi: 10.1016/j.ijheh.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 235.Franken C. Phthalate-induced oxidative stress and association with asthma-related airway inflammation in adolescents. Int. J. Hyg Environ. Health. 2017;220(2 Pt B):468–477. doi: 10.1016/j.ijheh.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 236.Wei Y.P. Serum cholesterol positively associated with oxidative DNA damage: a propensity score-matched analysis. Free Radic. Res. 2019;53(4):411–417. doi: 10.1080/10715762.2019.1595613. [DOI] [PubMed] [Google Scholar]
- 237.Poulsen H.E. Detection and interpretation of 8-oxodG and 8-oxoGua in urine, plasma and cerebrospinal fluid. Biochim. Biophys. Acta. 2014;1840(2):801–808. doi: 10.1016/j.bbagen.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 238.Pilger A. Longitudinal study of urinary 8-hydroxy-2'-deoxyguanosine excretion in healthy adults. Free Radic. Res. 2001;35(3):273–280. doi: 10.1080/10715760100300811. [DOI] [PubMed] [Google Scholar]
- 239.Poulsen H.E. Oxidative DNA damage in vivo: relationship to age, plasma antioxidants, drug metabolism, glutathione-S-transferase activity and urinary creatinine excretion. Free Radic. Res. 1998;29(6):565–571. doi: 10.1080/10715769800300601. [DOI] [PubMed] [Google Scholar]
- 240.Andreoli R. Quantitative determination of urinary 8-oxo-7,8-dihydro-2'-deoxyguanosine, 8-oxo-7,8-dihydroguanine, 8-oxo-7,8-dihydroguanosine, and their non-oxidized forms: daily concentration profile in healthy volunteers. Biomarkers. 2010;15(3):221–231. doi: 10.3109/13547500903434501. [DOI] [PubMed] [Google Scholar]
- 241.Shih Y.M. Clinical relevance of guanine-derived urinary biomarkers of oxidative stress, determined by LC-MS/MS. Redox Biol. 2019;20:556–565. doi: 10.1016/j.redox.2018.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Rozalski R. Substantial decrease of urinary 8-oxo-7,8-dihydroguanine, a product of the base excision repair pathway, in DNA glycosylase defective mice. Int. J. Biochem. Cell Biol. 2005;37(6):1331–1336. doi: 10.1016/j.biocel.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 243.Hayakawa H. Generation and elimination of 8-oxo-7,8-dihydro-2'-deoxyguanosine 5'-triphosphate, a mutagenic substrate for DNA synthesis, in human cells. Biochemistry. 1995;34(1):89–95. doi: 10.1021/bi00001a011. [DOI] [PubMed] [Google Scholar]
- 244.Hayakawa H. Metabolic fate of oxidized guanine ribonucleotides in mammalian cells. Biochemistry. 1999;38(12):3610–3614. doi: 10.1021/bi982361l. [DOI] [PubMed] [Google Scholar]
- 245.Haghdoost S. The nucleotide pool is a significant target for oxidative stress. Free Radic. Biol. Med. 2006;41(4):620–626. doi: 10.1016/j.freeradbiomed.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 246.Arimori T. Diverse substrate recognition and hydrolysis mechanisms of human NUDT5. Nucleic Acids Res. 2011;39(20):8972–8983. doi: 10.1093/nar/gkr575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Cai J.P. Mouse MTH2 protein which prevents mutations caused by 8-oxoguanine nucleotides. Biochem. Biophys. Res. Commun. 2003;305(4):1073–1077. doi: 10.1016/s0006-291x(03)00864-7. [DOI] [PubMed] [Google Scholar]
- 248.Ishibashi T., Hayakawa H., Sekiguchi M. A novel mechanism for preventing mutations caused by oxidation of guanine nucleotides. EMBO Rep. 2003;4(5):479–483. doi: 10.1038/sj.embor.embor838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Ito R. Cleavage of oxidized guanine nucleotide and ADP sugar by human NUDT5 protein. J. Biochem. 2011;149(6):731–738. doi: 10.1093/jb/mvr028. [DOI] [PubMed] [Google Scholar]
- 250.Carter M. Crystal structure, biochemical and cellular activities demonstrate separate functions of MTH1 and MTH2. Nat. Commun. 2015;6:7871. doi: 10.1038/ncomms8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Takagi Y. Human MTH3 (NUDT18) protein hydrolyzes oxidized forms of guanosine and deoxyguanosine diphosphates: comparison with MTH1 and MTH2. J. Biol. Chem. 2012;287(25):21541–21549. doi: 10.1074/jbc.M112.363010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Smith M.R. A guardian residue hinders insertion of a Fapy*dGTP analog by modulating the open-closed DNA polymerase transition. Nucleic Acids Res. 2019;47(6):3197–3207. doi: 10.1093/nar/gkz002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Reardon J.T. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc. Natl. Acad. Sci. U. S. A. 1997;94(17):9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Sancar A. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 255.Cooke M.S. DNA nucleotide excision repair, where do all the cyclobutane pyrimidine dimers go? Cell Cycle. 2013 doi: 10.4161/cc.24701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Ahmad J. Urinary thymine dimers and 8-oxo-2'-deoxyguanosine in psoriasis. FEBS Lett. 1999;460(3):549–553. doi: 10.1016/s0014-5793(99)01402-7. [DOI] [PubMed] [Google Scholar]
- 257.Narbutt J. Children sustain high levels of skin DNA photodamage, with a modest increase of serum 25-hydroxyvitamin D3, after a summer holiday in Northern Europe. Br. J. Dermatol. 2018;179(4):940–950. doi: 10.1111/bjd.16668. [DOI] [PubMed] [Google Scholar]
- 258.Petersen B. Sun and ski holidays improve vitamin D status, but are associated with high levels of DNA damage. J. Invest. Dermatol. 2014;134(11):2806–2813. doi: 10.1038/jid.2014.223. [DOI] [PubMed] [Google Scholar]
- 259.Brooks P.J. The oxidative DNA lesion 8,5'-(S)-cyclo-2'-deoxyadenosine is repaired by the nucleotide excision repair pathway and blocks gene expression in mammalian cells. J. Biol. Chem. 2000;275(29):22355–22362. doi: 10.1074/jbc.M002259200. [DOI] [PubMed] [Google Scholar]
- 260.Kuraoka I. Removal of oxygen free-radical-induced 5',8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc. Natl. Acad. Sci. U. S. A. 2000;97(8):3832–3837. doi: 10.1073/pnas.070471597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Jaruga P. Evidence for the involvement of DNA repair enzyme NEIL1 in nucleotide excision repair of (5'R)- and (5'S)-8,5'-cyclo-2'-deoxyadenosines. Biochemistry. 2010;49(6):1053–1055. doi: 10.1021/bi902161f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Weimann A., Riis B., Poulsen H.E. Oligonucleotides in human urine do not contain 8-oxo-7,8-dihydrodeoxyguanosine. Free Radic. Biol. Med. 2004;36(11):1378–1382. doi: 10.1016/j.freeradbiomed.2004.02.070. [DOI] [PubMed] [Google Scholar]
- 263.Dianov G. Repair pathways for processing of 8-oxoguanine in DNA by mammalian cell extracts. J. Biol. Chem. 1998;273(50):33811–33816. doi: 10.1074/jbc.273.50.33811. [DOI] [PubMed] [Google Scholar]
- 264.Jaiswal M. Efficient in vitro repair of 7-hydro-8-oxodeoxyguanosine by human cell extracts: involvement of multiple pathways. Nucleic Acids Res. 1998;26(9):2184–2191. doi: 10.1093/nar/26.9.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Cappelli E., Degan P., Frosina G. Comparative repair of the endogenous lesions 8-oxo-7, 8-dihydroguanine (8-oxoG), uracil and abasic site by mammalian cell extracts: 8-oxoG is poorly repaired by human cell extracts. Carcinogenesis. 2000;21(6):1135–1141. [PubMed] [Google Scholar]
- 266.Pascucci B. Reconstitution of the base excision repair pathway for 7,8-dihydro-8-oxoguanine with purified human proteins. Nucleic Acids Res. 2002;30(10):2124–2130. doi: 10.1093/nar/30.10.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Sokhansanj B.A. A quantitative model of human DNA base excision repair. I. Mechanistic insights. Nucleic Acids Res. 2002;30(8):1817–1825. doi: 10.1093/nar/30.8.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Lipinski L.J. Repair of oxidative DNA base lesions induced by fluorescent light is defective in xeroderma pigmentosum group A cells. Nucleic Acids Res. 1999;27(15):3153–3158. doi: 10.1093/nar/27.15.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Klein J.C. Repair and replication of plasmids with site-specific 8-oxodG and 8-AAFdG residues in normal and repair-deficient human cells. Nucleic Acids Res. 1992;20(17):4437–4443. doi: 10.1093/nar/20.17.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Kumar N., Raja S., Van Houten B. The involvement of nucleotide excision repair proteins in the removal of oxidative DNA damage. Nucleic Acids Res. 2020;48(20):11227–11243. doi: 10.1093/nar/gkaa777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Evans M.D. Nucleotide excision repair of oxidised genomic DNA is not a source of urinary 8-oxo-7,8-dihydro-2'-deoxyguanosine. Free Radic. Biol. Med. 2016;99:385–391. doi: 10.1016/j.freeradbiomed.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 272.Bessho T. Evidence for two DNA repair enzymes for 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) in human cells. J. Biol. Chem. 1993;268(26):19416–19421. [PubMed] [Google Scholar]
- 273.Cooke M.S. Urinary 8-oxo-2'-deoxyguanosine–source, significance and supplements. Free Radic. Res. 2000;32(5):381–397. doi: 10.1080/10715760000300391. [DOI] [PubMed] [Google Scholar]
- 274.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362(6422):709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 275.Siomek A. Severe oxidatively damaged DNA after cisplatin treatment of cancer patients. Int. J. Canc. 2006;119(9):2228–2230. doi: 10.1002/ijc.22088. [DOI] [PubMed] [Google Scholar]
- 276.Gackowski D. 8-Oxo-7,8-dihydroguanine and 8-oxo-7,8-dihydro-2’-deoxyguanosine levels in human urine do not depend on diet. Free Radic. Res. 2001;35(6):825–832. doi: 10.1080/10715760100301321. [DOI] [PubMed] [Google Scholar]
- 277.Fraga C.G. Oxidative damage to DNA during aging: 8-hydroxy-2'-deoxyguanosine in rat organ DNA and urine. Proc. Natl. Acad. Sci. U. S. A. 1990;87(12):4533–4537. doi: 10.1073/pnas.87.12.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Chao M.R. Children are particularly vulnerable to environmental tobacco smoke exposure: evidence from biomarkers of tobacco-specific nitrosamines, and oxidative stress. Environ. Int. 2018;120:238–245. doi: 10.1016/j.envint.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 279.Barreto M. Urinary and exhaled biomarkers of exercise-induced bronchoconstriction in atopic asthmatic children. Pediatr. Pulmonol. 2019;54(9):1447–1456. doi: 10.1002/ppul.24419. [DOI] [PubMed] [Google Scholar]
- 280.Li Z. Classification and temporal variability in urinary 8-oxodG and 8-oxoGuo: analysis by UHPLC-MS/MS. Sci. Rep. 2019;9(1):8187. doi: 10.1038/s41598-019-44240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Andreoli R. Urinary biomarkers of exposure and of oxidative damage in children exposed to low airborne concentrations of benzene. Environ. Res. 2015;142:264–272. doi: 10.1016/j.envres.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 282.Kloppenborg J.T. Urinary markers of nucleic acid oxidation in Danish overweight/obese children and youths. Free Radic. Res. 2016;50(7):691–697. doi: 10.3109/10715762.2016.1164310. [DOI] [PubMed] [Google Scholar]
- 283.Szaflarska-Poplawska A. Oxidatively damaged DNA/oxidative stress in children with celiac disease. Canc. Epidemiol. Biomark. Prev. 2010;19(8):1960–1965. doi: 10.1158/1055-9965.EPI-10-0295. [DOI] [PubMed] [Google Scholar]
- 284.Shih B.B. Fractional sunburn threshold UVR doses generate equivalent vitamin D and DNA damage in skin types I-vi but with epidermal DNA damage gradient correlated to skin darkness. J. Invest. Dermatol. 2018;138(10):2244–2252. doi: 10.1016/j.jid.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Shih B.B. Influence of skin melanisation and ultraviolet radiation on biomarkers of systemic oxidative stress. Free Radic. Biol. Med. 2020;160:40–46. doi: 10.1016/j.freeradbiomed.2020.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Malic Z. Oxidative stress and genetic variants of xenobiotic-metabolising enzymes associated with COPD development and severity in Serbian adults. COPD. 2017;14(1):95–104. doi: 10.1080/15412555.2016.1199667. [DOI] [PubMed] [Google Scholar]
- 287.Dziaman T. 8-Oxo-7,8-dihydroguanine and uric acid as efficient predictors of survival in colon cancer patients. Int. J. Canc. 2014;134(2):376–383. doi: 10.1002/ijc.28374. [DOI] [PubMed] [Google Scholar]
- 288.Collins A.R. Oxidative DNA damage measured in human lymphocytes: large differences between sexes and between countries, and correlations with heart disease mortality rates. Faseb. J. 1998;12(13):1397–1400. [PubMed] [Google Scholar]
- 289.Lenton K.J. Glutathione and ascorbate are negatively correlated with oxidative DNA damage in human lymphocytes. Carcinogenesis. 1999;20(4):607–613. doi: 10.1093/carcin/20.4.607. [DOI] [PubMed] [Google Scholar]
- 290.Collins A. Launch of the ComNet (comet network) project on the comet assay in human population studies during the International Comet Assay Workshop meeting in Kusadasi, Turkey. Mutagenesis. 2012;27(4):385–386. doi: 10.1093/mutage/ges014. 13-16. [DOI] [PubMed] [Google Scholar]
- 291.Collins A. The comet assay as a tool for human biomonitoring studies: the ComNet project. Mutat. Res. Rev. Mutat. Res. 2014;759:27–39. doi: 10.1016/j.mrrev.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 292.Azqueta A. Technical recommendations to perform the alkaline standard and enzyme-modified comet assay in human biomonitoring studies. Mutat. Res. 2019;843:24–32. doi: 10.1016/j.mrgentox.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 293.Azqueta A. DNA repair as a human biomonitoring tool: Comet assay approaches. Mutat. Res. 2019;781:71–87. doi: 10.1016/j.mrrev.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 294.Azqueta A. Application of the comet assay in human biomonitoring: an hCOMET perspective. Mutat. Res. 2020;783:108288. doi: 10.1016/j.mrrev.2019.108288. [DOI] [PubMed] [Google Scholar]
- 295.Moller P. Minimum Information for Reporting on the Comet Assay (MIRCA): recommendations for describing comet assay procedures and results. Nat. Protoc. 2020;15:3817–3826. doi: 10.1038/s41596-020-0398-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Moller P. Searching for assay controls for the Fpg- and hOGG1-modified comet assay. Mutagenesis. 2018;33(1):9–19. doi: 10.1093/mutage/gex015. [DOI] [PubMed] [Google Scholar]
- 297.Moller P., Stopper H., Collins A.R. Measurement of DNA damage with the comet assay in high-prevalence diseases: current status and future directions. Mutagenesis. 2020;35(1):5–18. doi: 10.1093/mutage/gez018. [DOI] [PubMed] [Google Scholar]
- 298.Moller P. Potassium bromate as positive assay control for the Fpg-modified comet assay. Mutagenesis. 2020;35(4):341–348. doi: 10.1093/mutage/geaa011. [DOI] [PubMed] [Google Scholar]
- 299.Kanaly R.A. Development of the adductome approach to detect DNA damage in humans. Antioxidants Redox Signal. 2006;8(5–6):993–1001. doi: 10.1089/ars.2006.8.993. [DOI] [PubMed] [Google Scholar]
- 300.Chang Y.J. Novel approach to integrated DNA adductomics for the assessment of in vitro and in vivo environmental exposures. Arch. Toxicol. 2018;92(8):2665–2680. doi: 10.1007/s00204-018-2252-6. [DOI] [PubMed] [Google Scholar]
- 301.Cooke M.S. Urinary DNA adductomics - a novel approach for exposomics. Environ. Int. 2018;121(Pt 2):1033–1038. doi: 10.1016/j.envint.2018.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Villalta P.W., Balbo S. The future of DNA adductomic analysis. Int. J. Mol. Sci. 2017;18(9) doi: 10.3390/ijms18091870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Cao B. Nick-seq for single-nucleotide resolution genomic maps of DNA modifications and damage. Nucleic Acids Res. 2020;48(12):6715–6725. doi: 10.1093/nar/gkaa473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Li W., Sancar A. Methodologies for detecting environmentally induced DNA damage and repair. Environ. Mol. Mutagen. 2020;61(7):664–679. doi: 10.1002/em.22365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Payne A. BulkVis: a graphical viewer for Oxford nanopore bulk FAST5 files. Bioinformatics. 2019;35(13):2193–2198. doi: 10.1093/bioinformatics/bty841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.An N. Nanopore detection of 8-oxoguanine in the human telomere repeat sequence. ACS Nano. 2015;9(4):4296–4307. doi: 10.1021/acsnano.5b00722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Alvarez J.R. Mapping base modifications in DNA by transverse-current sequencing. Phys. Rev. Applied. 2018;9 024024. [Google Scholar]
- 308.Ceylan D. Alterations in levels of 8-Oxo-2'-deoxyguanosine and 8-Oxoguanine DNA glycosylase 1 during a current episode and after remission in unipolar and bipolar depression. Psychoneuroendocrinology. 2020;114:104600. doi: 10.1016/j.psyneuen.2020.104600. [DOI] [PubMed] [Google Scholar]
- 309.Jacoby A.S. Increased DNA and RNA damage by oxidation in patients with bipolar I disorder. Transl. Psychiatry. 2016;6(8):e867. doi: 10.1038/tp.2016.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Mao Y.H. Levels of 8-oxo-dGsn and 8-oxo-Gsn in random urine are consistent with 24 h urine in healthy subjects and patients with renal disease. Free Radic. Res. 2017;51(6):616–621. doi: 10.1080/10715762.2017.1346249. [DOI] [PubMed] [Google Scholar]
- 311.Guo C. Discriminating patients with early-stage breast cancer from benign lesions by detection of oxidative DNA damage biomarker in urine. Oncotarget. 2017;8(32):53100–53109. doi: 10.18632/oncotarget.17831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Mao L., Guo C., Zheng S. Elevated urinary 8-oxo-7,8-dihydro-2'-deoxyguanosine and serum uric acid are associated with progression and are prognostic factors of colorectal cancer. OncoTargets Ther. 2018;11:5895–5902. doi: 10.2147/OTT.S175112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Liang Y.D. Urinary 8-oxo-7,8-dihydroguanosine as a potential biomarker of frailty for elderly patients with cardiovascular disease. Free Radic. Biol. Med. 2020;152:248–254. doi: 10.1016/j.freeradbiomed.2020.03.011. [DOI] [PubMed] [Google Scholar]
- 314.Sorensen A.S. The effect of smoking on the urinary excretion of 8-oxodG and 8-oxoGuo in patients with type 2 diabetes. Scand. J. Clin. Lab. Invest. 2017;77(4):253–258. doi: 10.1080/00365513.2017.1299208. [DOI] [PubMed] [Google Scholar]
- 315.Zhao G. A simple method for the determination of 8-oxoguanosine, 8-oxo-2'-deoxyguanosine and 8-iso-prostaglandin F2 alpha. Chromatographia. 2017;80:401–408. [Google Scholar]
- 316.Tranfo G. Levels of urinary biomarkers of oxidatively generated damage to DNA and RNA in different groups of workers compared to general population. Int. J. Environ. Res. Publ. Health. 2019;16(16) doi: 10.3390/ijerph16162995. [DOI] [PMC free article] [PubMed] [Google Scholar]