Abstract
Generation of reactive oxygen species and related oxidants is an inevitable consequence of life. Proteins are major targets for oxidation reactions, because of their rapid reaction rates with oxidants and their high abundance in cells, extracellular tissues, and body fluids. Additionally, oxidative stress is able to degrade lipids and carbohydrates to highly reactive intermediates, which eventually attack proteins at various functional sites. Consequently, a wide variety of distinct posttranslational protein modifications is formed by protein oxidation, glycoxidation, and lipoxidation. Reversible modifications are relevant in physiological processes and constitute signaling mechanisms (“redox signaling”), while non-reversible modifications may contribute to pathological situations and several diseases. A rising number of publications provide evidence for their involvement in the onset and progression of diseases as well as aging processes. Certain protein oxidation products are chemically stable and formed in large quantity, which makes them promising candidates to become biomarkers of oxidative damage. Moreover, progress in the development of detection and quantification methods facilitates analysis time and effort and contributes to their future applicability in clinical routine. The present review outlines the most important classes and selected examples of oxidative protein modifications, elucidates the chemistry beyond their formation and discusses available methods for detection and analysis. Furthermore, the relevance and potential of protein modifications as biomarkers in the context of disease and aging is summarized.
Keywords: Protein oxidation, Protein modification, Biomarker, Aging, Oxidative stress
Graphical abstract
Highlights
-
•
Formation and removal of reactive oxygen species is balanced under normal conditions.
-
•
Protein modifications have been linked to a wide range of pathologies.
-
•
Mass spectrometry is the most accurate method to study protein modifications.
-
•
The relevance as biomarkers for disease and aging depends on chemical properties.
Abbreviations
- 3-NT
3-Nitrotyrosine
- 4-HNE
4-Hydroxy-2-nonenal
- AD
Alzheimer's disease
- AGE
Advanced glycation endproduct
- ALC
Alcoholic liver cirrhosis
- ALD
Alcoholic liver disease
- ALE
Advanced lipoxidation endproduct
- ALS
Amyotrophic lateral sclerosis
- AOPP
Advanced oxidation protein product
- CAD
Coronary artery disease
- CHD
Chronic heart disease
- CKD
Chronic kidney disease
- CML
Carboxymethyl lysine
- COPD
Chronic obstructive pulmonary disease
- CSF
Cerebrospinal fluid
- CVD
Cardiovascular disease
- DHP
Dihydropyridine
- DNPH
2,4-Dinitrophenylhydrazine
- DOPA
Dihydroxyphenylalanine
- DTT
Dithiothreitol
- ELISA
Enzyme linked immunosorbent assay
- EPR
Electron paramagnetic resonance
- ESI
Electrospray ionization
- FLD
Fluorescence detection
- HBV
Hepatitis B virus
- HIV
Human immunodeficiency virus
- IHC
Immunohistochemistry
- HPLC
High pressure liquid chromatography
- MALDI
Matrix assisted laser desorption ionization
- MCI
Mild cognitive impairment
- MDA
Malondialdehyde
- MetS
Metabolic syndrome
- MRM
Multiple reaction monitoring
- MS
Mass spectrometry
- NAFLD
Non-alcoholic fatty liver disease
- NOS
Nitric oxide synthase
- PBMC
Peripheral blood mononuclear cells
- PD
Parkinson's disease
- RA
Rheumatoid arthritis
- RCS
Reactive carbonyl species
- RNS
Reactive nitrogen species
- ROS
Reactive oxygen species
- Skin AF
Skin autofluorescence
- T2D
Type 2 diabetes
- TBARS
Thiobarbituric acid reactive substances
- WB
Western Blot
1. Introduction
Biological macromolecules are constantly exposed to oxidants and oxidative damage to cellular components has been increasingly recognized as a significant pathophysiological event leading to disease and aging processes. Free radicals and other oxidizing species are derived from both endogenous sources such as mitochondria, peroxisomes, and phagocytic cells, but also exogenous sources such as tobacco smoke, pollution, alcohol, and certain drugs.
The most important oxidants originate from oxygen (reactive oxygen species, ROS) and nitrogen (reactive nitrogen species, RNS). The oxygen molecule itself is a biradical with two unpaired electrons. Additionally, important primary oxygen-containing compounds with reactive properties are superoxide (O2•-) and the most reactive hydroxyl radical (•OH), formed from O2•- and hydrogen peroxide (H2O2) in the presence of metal ions (Fenton reaction) with a very short half-life of approximately 10−9 s [1]. Less powerful ROS are the alkoxyl radical (RO•) as well as peroxyl radical (ROO•), both key intermediates in lipid peroxidation chain reactions. Nitric oxide (•NO) is a slowly reacting molecule, with a high relevance as signaling molecule. However, nitric oxide reacts with superoxide at a high rate constant to give peroxynitrite (ONOO−), able to decompose spontaneously to yield nitrogen dioxide (•NO2) and hydroxyl radicals [2].
Under normal circumstances, the formation and subsequent degradation of reactive species is regulated by cellular defense systems, including scavenging enzymes able to remove oxidants or their precursors such as superoxide dismutases, catalase, thioredoxin/thioredoxin reductase, and glutathione peroxidase. Non-enzymatic antioxidants such as tocopherols and ascorbic acid, but also metal binding proteins delay oxidation reactions or prevent the development of reactive species. Repair and removal systems such as methionine sulfoxide reductases, disulfide reductases/isomerases, and the ubiquitin-proteasome-system complete the damage defense. However, even if these preventive and repair systems are interconnected and work efficiently, they cannot fully prevent oxidative damage to cellular components. Furthermore, a higher level of damage may arise from increased oxidant generation and/or a decrease or failure of defense systems under several conditions such as diseases and aging [3,4]. This imbalance between the excessive production of reactive species and the ability to detoxify them or repair the resulting damage is termed “oxidative stress” (the full concept on oxidative stress is reviewed in Ref. [5]).
Generally, reactive species can generate damage to all cellular components, including proteins, carbohydrates, lipids, and DNA. It has been estimated that proteins can scavenge a majority (50%–75%) of generated reactive species [6]. Highly reactive species damage numerous sites at side-chains and backbones of proteins, while less reactive species have higher selectivity regarding targeted residues [7]. The variety of reaction sites generates a wide range of posttranslational protein modifications consequently changing composition and folding, the net charge, as well as the hydrophobicity/hydrophilicity of proteins. This affects their functions as receptors, enzymes, carrier or structural proteins [8]. It has been established that reversible modifications are thought to be relevant in physiological processes and constitute signaling mechanisms under appropriate conditions (“redox signaling”), while non-reversible modifications may contribute to pathological situations and several diseases [9].
Since free radicals are very reactive and short-lived, their detection is challenging and techniques with fast response times such as electron paramagnetic resonance (EPR) are needed. However, detection of more stable products, resulting from reactions with free radicals, such as oxidative protein modifications yield more convincing data and often result in higher-quality quantitative data. Over the last years considerable advances have been made in the development of techniques to detect, identify, and quantify protein modifications.
Studies reveal an age-related increase in the level of oxidatively modified proteins [10,11] and many diseases have an oxidative etiology [9,[12], [13], [14]]. It can be therefore assumed that proteins also accumulate evidence of oxidative damage in relation to disease, although some of these are only associations and oxidation is not always causal, but a contributing factor. To function as suitable biomarkers, it is critical that protein oxidation products are stable, accumulate in detectable concentrations, and correlate with disease severity. Sample availability is another important factor limiting the reliability of a biomarker. Protein oxidation products can be determined in blood and urine samples, but also in specific tissue or cell samples.
In this review, we focus on the chemistry of reversible and irreversible protein modifications by oxidants and available methods for detection and analysis. Discussion whether protein oxidation provides suitable biomarkers in the context of disease and aging is another major aim of this review.
2. Mechanisms of oxidative protein modifications
2.1. Oxidation of sulfur-containing amino acids
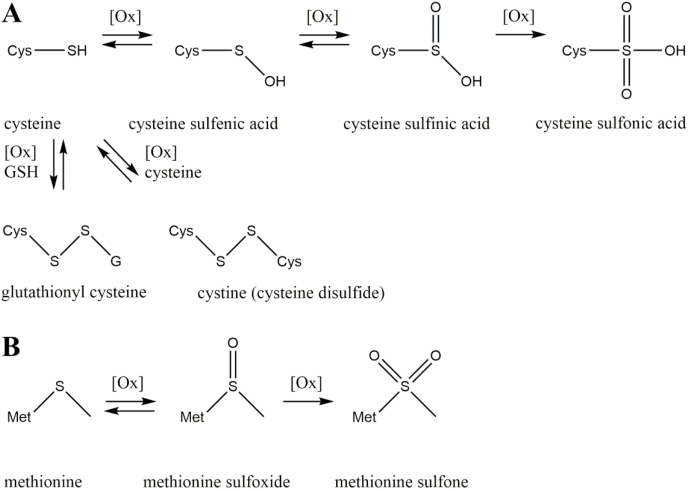
Sulfur-containing moieties such as the sulfhydryl group of cysteine and the thioether group of methionine are targeted by oxidative stress and a plethora of posttranslational protein modifications is formed (Fig. 1) [15]. Formation of cysteine thiyl radical and sulfenic acid is a reversible process and both intermediates are highly unstable. Both serve as precursors for several oxidized cysteine modifications (Fig. 1A) [16].
Fig. 1.
Oxidation of sulfur-containing amino acids. Cysteine is oxidized in a multi-step reaction to the respective sulfenic, sulfinic, and sulfonic acid modifications or oxidatively conjugated with glutathione (GSH) or another cysteine residue (A). Methionine is reversibly oxidized to methionine sulfoxide and irreversibly oxidized to methionine sulfone (B).
One example with huge biological relevance is the formation of disulfide bonds between cysteine and thiols under oxidative conditions. Reaction of cysteine with another cysteine under non-enzymatic as well as enzyme mediated conditions leads to cystine. Cystine is a vitally important modification, because it stabilizes protein structures via intra- and intermolecular disulfide bridges. Formation and cleavage of disulfide bonds is a reversible process, which is controlled by several enzymes in vivo [17]. Cysteine residues can be protected against “overoxidation” by S-glutathionylation, which is the reversible addition of glutathione via disulfide linkage. Overoxidation describes the further oxidation of cysteine sulfenic acid to cysteine sulfinic and finally sulfonic acid [18]. Cysteine sulfenic and sulfinic acid modifications can be reversed by glutaredoxin mediated reduction of sulfinic acid, conjugation of sulfenic acid via S-glutathionylation, and deglutathionylation by glutaredoxin or sulfiredoxin. The overoxidation product cysteine sulfonic acid is irreparably damaged and the affected protein has to be degraded by the proteasome [19].
Another sulfur containing amino acid targeted by oxidative modification is methionine (Fig. 1B). In contrast to the non-enzymatic oxidation of methionine to methionine sulfoxide, the reduction is catalyzed by methionine sulfoxide reductases [20]. Further oxidation results in methionine sulfone formation, which is not targeted by methionine sulfoxide reductases and has to be considered as a stable modification [21].
2.2. Oxidation of aromatic moieties
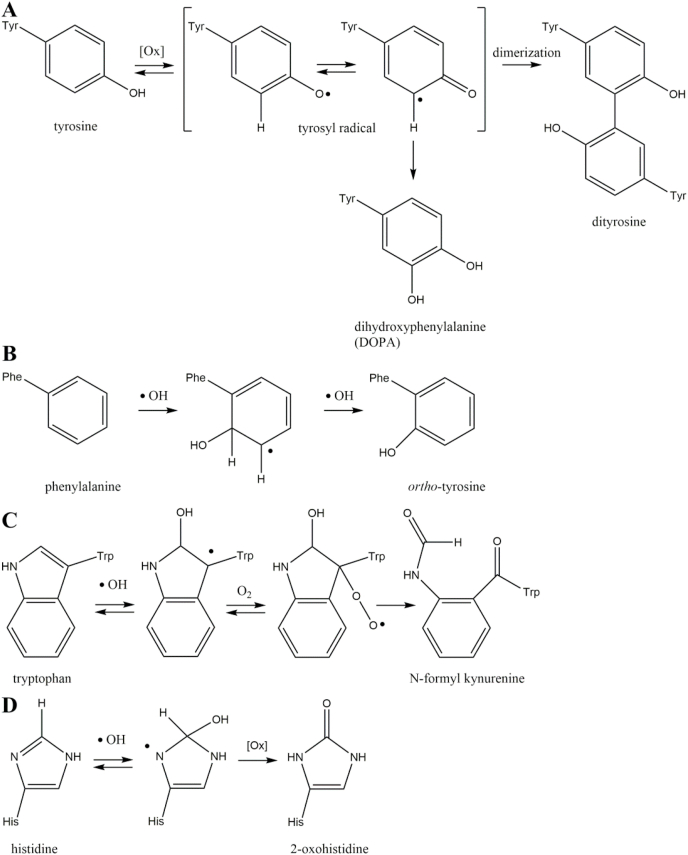
Aromatic functionalities of amino acids are excellent targets of protein oxidation (Fig. 2). In particular, tyrosine is a redox active structure. The phenolic side-chain is easily oxidized, because the intermediary tyrosyl radical is stabilized by mesomeric delocalization of the unpaired electron (Fig. 2A) [22]. The tyrosyl radical can react with another tyrosyl radical. After enolization, the main product of this reaction is the fluorescent protein crosslink ortho,ortho-dityrosine [23].
Fig. 2.
Oxidation of aromatic amino acids. Oxidation of tyrosine is favored by the intermediary tyrosyl radical and leads to the stable modifications dihydroxyphenylalanine and dityrosine (A). Unusual tyrosine isomers like ortho-tyrosine are generated by oxidation of phenylalanine residues (B). Oxidation of tryptophan is the initial step of the N-formyl kynurenine reaction cascade (C). The stable modification 2-oxohistidine is formed by histidine oxidation (D).
Reaction of protein bound tyrosyl radicals with hydroxyl radicals leads to 3-hydroxytyrosine, which is an analogue of the neurotransmitter 3,4-dihydroxyphenylalanine (DOPA) [24]. Beside proteinogenic para-tyrosine, the abnormal isomers ortho- and meta-tyrosine can be formed by oxidation of phenylalanine residues via hydroxyl radicals (Fig. 2B) [25]. Tryptophan is oxidized by hydroxyl radicals to hydroxytryptophan, which is cleaved by oxygen to yield N-formyl kynurenine (Fig. 2C) [26]. Oxidation of histidine by metal catalyzed reaction with hydroxyl radicals leads to 2-oxohistidine (Fig. 2D) [27,28].
2.3. Glycoxidation
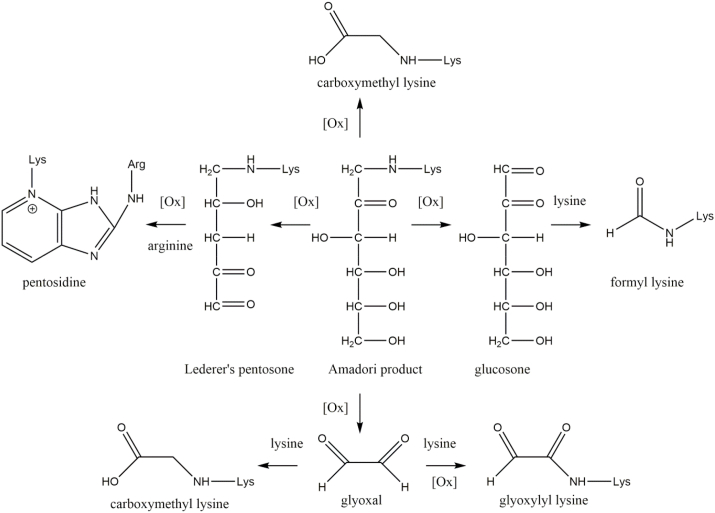
The basic amino acids lysine and arginine are readily modified by glycation. The reaction results in the formation of intermediate Amadori products and subsequently advanced glycation endproducts (AGEs). The protein modifications are formed by the nucleophilic reaction of amino acid residues with reductive sugars or their reactive degradation products (α-dicarbonyl compounds). It is important to note that the formation of AGEs does not generally require oxidative conditions and only selected AGEs are generated by oxidation. This special subset of AGEs is called glycoxidation products, because they are formed by a combination of glycation and oxidation (Fig. 3) [29]. This review focuses on glycoxidation products as feasible markers of combined oxidative and carbonyl stress. A more general overview of AGE structures and underlying mechanisms of the Maillard reaction in vivo can be found elsewhere [30,31].
Fig. 3.
Formation of glycoxidation products. A special subset of advanced glycation endproducts is exclusively formed under oxidative conditions including the prominent modifications carboxymethyl lysine and pentosidine. The novel modifications glyoxylyl and formyl lysine are potential biomarkers of oxidative stress based on their formation pathways.
The most prominent and one of the most abundant AGEs in vivo is carboxymethyl lysine (CML), which was extensively reviewed before [[32], [33], [34]]. CML was the first discovered glycoxidation product and is formed by oxidative degradation of fructoselysine (Amadori product) [35]. An alternative pathway is the reaction between the α-dicarbonyl compound glyoxal and lysine leading to CML formation via an isomerization mechanism [36]. Although the latter mechanism is non-oxidative, the reactive precursor glyoxal is mainly formed by oxidative degradation of carbohydrates, lipids, nucleotides, and serine [37]. The oxidation of the central enaminol intermediate of this isomerization cascade results in the formation of the α-oxoamide AGE glyoxylyl lysine. Compared to CML this structure is an even more sensitive marker of oxidative stress, because the precursor glyoxal and in addition the formation of glyoxylyl lysine itself rely on oxidative processes [38].
Another well-known glycoxidation product is pentosidine, a crosslink between lysine and arginine residues. Despite the rather low abundance of pentosidine, it was one of the first discovered AGEs due to its specific fluorescence [39]. Pentoses were at first postulated as the main source of the C5 backbone in pentosidine [40]. Later studies proved the oxidative cleavage of C6 structures such as glucose to yield pentosidine as well [41]. Beside oxidatively formed glycoxidation products some AGEs are mainly formed by precursors which are generated by oxidation. An excellent example is the oxidation of glucose or Amadori product thereof to glucosone [42]. A major product of lysine mediated cleavage of glucosone is formyl lysine [43]. Recently, formyl lysine was discovered in very high quantities in various murine tissues and histones [44,45]. Alternative pathways of formyl lysine formation such as formyl phosphate [46] and formaldehyde metabolism [47] were reported in the literature. All postulated mechanisms require an oxidation step and first experiments in cell culture give rising evidence of correlation between formyl lysine concentration and oxidative stress [46].
2.4. Lipoxidation
As mentioned above the CML precursor glyoxal can be formed by carbohydrate degradation and lipid oxidation. Hence, CML is not only considered as an AGE, but also as an advanced lipoxidation endproduct (ALE) [48]. ALEs are protein modifications formed by the nucleophilic reaction of proteins with reactive carbonyl species (RCS) originating from oxidative lipid degradation [49]. A complete overview of ALE structures with their respective precursors and mechanisms of formation was previously published [50].
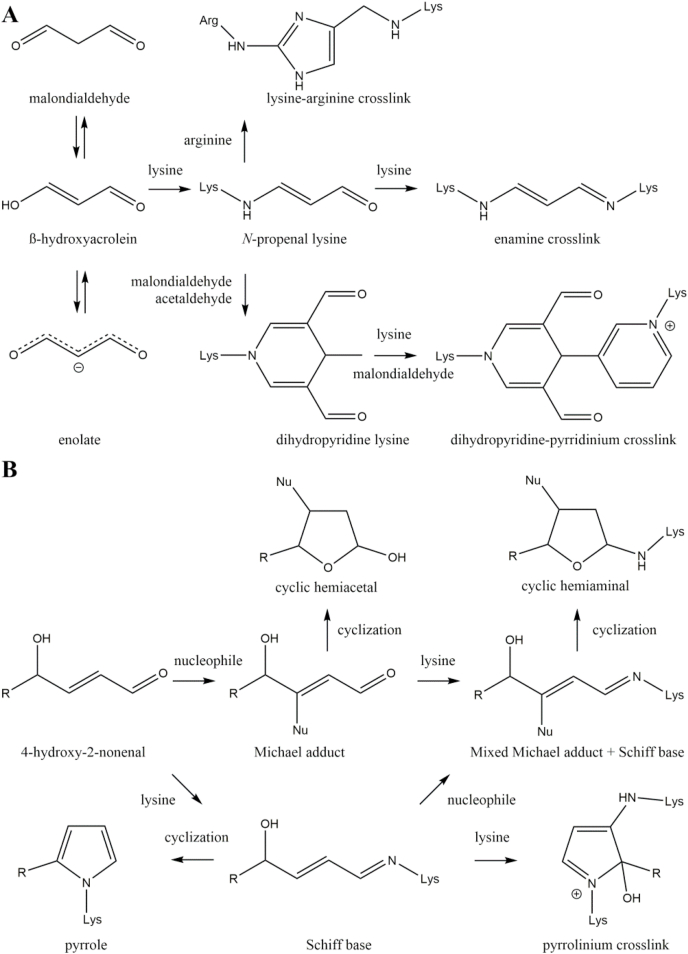
During lipid oxidation, a plethora of distinct RCS is formed, which can be categorized as α/β unsaturated, keto aldehyde, and dialdehyde RCS. One of the most abundant dialdehyde RCS resulting from lipid peroxidation is malondialdehyde (MDA). MDA is less reactive compared to other RCS, because at physiological pH it enolizes to β-hydroxyacrolein and the enolate is formed by loss of a proton [51]. Only the protonated β-hydroxyacrolein is attacked by lysine yielding the key intermediate N-propenal lysine (Fig. 4A) [52]. An enamine crosslink is formed by reaction of N-propenal lysine with a second lysine residue [53]. Alternatively, reaction with a second MDA molecule and acetaldehyde, which is a MDA degradation product, results in formation of the fluorescent dihydropyridine (DHP) product lysine 4-methyl-1,4-dihydro-3,5-dicarbaldehyde [54]. This DHP derivative can react with another lysine residue and MDA to form the fluorescent 3,5-diformyl-1,4-dihydropyridin-4-yl-pyridinium crosslink [55]. Reaction between N-propenal lysine and arginine is the main source of the lysine-arginine crosslink 2-ornithinyl-4-methyl(1ε-lysyl)-1,3-imidazole [56].
Fig. 4.
Formation of lipoxidation products. Complex reaction cascades of malondialdehyde (A) and 4-hydroxynonenal (B) are briefly summarized and important protein modifications presented.
Another important example for α/β unsaturated RCS is 4-hydroxy-2-nonenal (4-HNE). The carbonyl function pulls away electrons from the alkene group resulting in electron deficiency at the β-carbon of 4-HNE. This positive partial charge is attacked by nucleophiles such as cysteine and histidine and stable Michael adducts are formed (Fig. 4B) [50]. Nucleophilic attack of lysine forms unstable Michael adducts, which require reduction, e.g., by sodium borohydride prior to analysis [57]. The open forms of Michael adducts of 4-HNE and nucleophiles are in an equilibrium with their favored cyclic hemiacetal form [58]. Beside nucleophilic attack at the alkene group, reversible formation of Schiff bases between the carbonyl function and lysine residues is a common reaction. In analogy to the Michael adduct the Schiff Base is stabilized by cyclization as a pyrrole [59]. Alternatively, attack of nucleophiles results in a mixed Michael adduct and Schiff base intermediate [60], which is stabilized as a cyclic hemiaminal [61]. Oxidation of the Schiff base and attack of a second lysine residue yields a fluorescent pyrrolinium crosslink [62].
2.5. Carbonylation
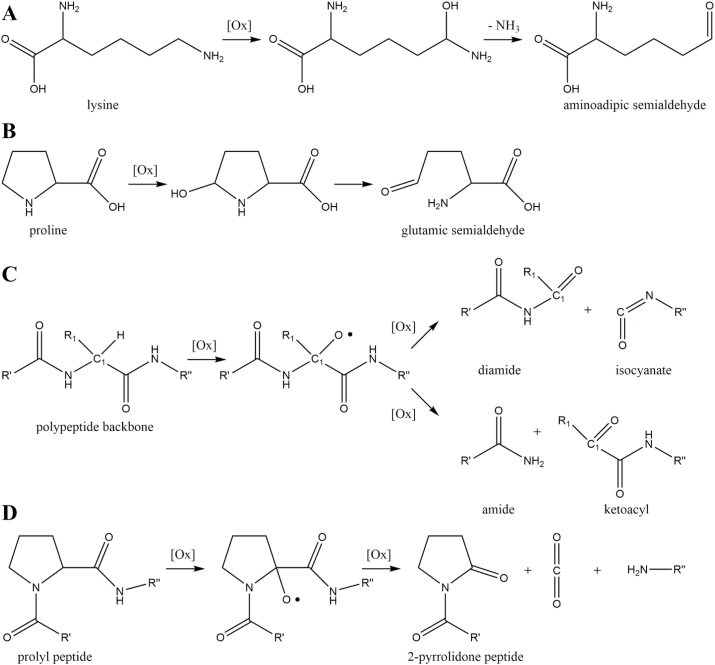
Protein carbonylation is a stable modification induced by ROS via three pathways: direct oxidation of protein-bound amino acids, oxidative cleavage of the protein backbone, and incorporation of carbonyls from glycoxidation or lipoxidation [63]. The latter option has been described in the above chapters. Aminoadipic and glutamic semialdehyde are examples for direct oxidation of amino acids and responsible for about 60% of total protein carbonylation in liver [64]. Formation of aminoadipic semialdehyde requires hydroxyl radical mediated abstraction of the hydrogen located next to the N6-amino function of lysine, metal-catalyzed oxidation of the carbon-centered radical, and hydrolysis of the resulting imine (Fig. 5A). A similar reaction scheme leads to glutamic semialdehyde, in which the proton next to the guanidino function of arginine or the proton next to the nitrogen of the proline pyrrole ring is abstracted, the carbon radical is oxidized, and the intermediate is hydrolyzed (Fig. 5B) [65].
Fig. 5.
Pathways of protein carbonylation. Lysine and proline residues are vitally important precursors of protein carbonylation by oxidative formation of aminoadipic semialdehyde (A) and glutamic semialdehyde (B), respectively. Oxidative cleavage of peptide backbone results in protein carbonylation via diamide and α-amidation pathways (C). Formation of 2-pyrrolidone by oxidation of prolyl containing peptide (D).
Generally, oxidative cleavage of the peptide backbone is initiated by superoxide mediated alkoxyl radical formation at the α-carbon next to a peptide bond. The alkoxyl radical fragments either by the homolytic cleavage of the carbon-carbon bond (diamide pathway) or the carbon-nitrogen bond (α-amidation pathway) (Fig. 5C). The diamide pathway results in a diamide and isocyanate, while a Nα-ketoacyl derivative and an amide are the endproducts of the α-amidation pathway [66]. Additionally, oxidative cleavage of the peptide backbone by specific mechanism can be facilitated by prolyl, glutamyl, and aspartyl residues [67]. As an example, protein bound proline is oxidized to the 2-pyrrolidone peptide under carbon-carbon cleavage (Fig. 5D) [68].
2.6. Nitration/nitrosylation
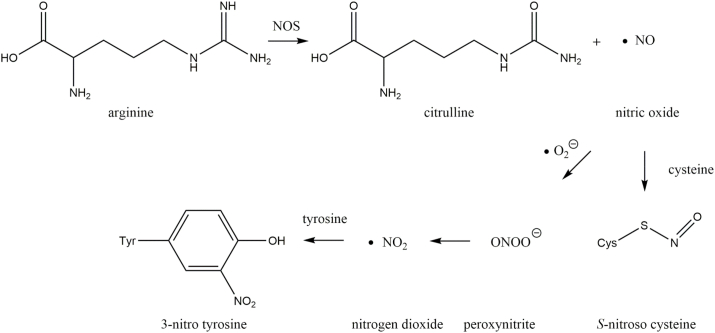
Nitric oxide (•NO) is an important signaling molecule involved in vasodilation and neurotransmission via activation of soluble guanylate cyclase and generation of the second messenger cGMP [69]. Formation of •NO is catalyzed by three isoforms of nitric oxide synthase (NOS) (Fig. 6): endothelial NOS, neuronal NOS or inducible NOS. All NOS isoforms use arginine and oxygen to produce citrulline and •NO [70]. Apart from its role in cell signaling •NO readily reacts with superoxide anions forming RNS such as peroxynitrite and nitrogen dioxide [71]. These RNS are potent oxidizing agents in vivo [72]. Additionally, •NO or higher nitrogen oxides including dinitrogen trioxide (N2O3) can directly and reversibly modify cysteine residues by S-nitrosation [73]. The irreversible modification 3-nitrotyrosine (3-NT) is formed by nitration of tyrosine via attack of the nitrogen dioxide radical at the ortho-position of the aromatic ring [74].
Fig. 6.
Generation of reactive nitrogen species (RNS). Nitric oxide synthase (NOS) catalyzes the generation of nitric oxide, which is the precursor of S-nitrosylation and nitration.
3. Analysis of oxidative protein modifications
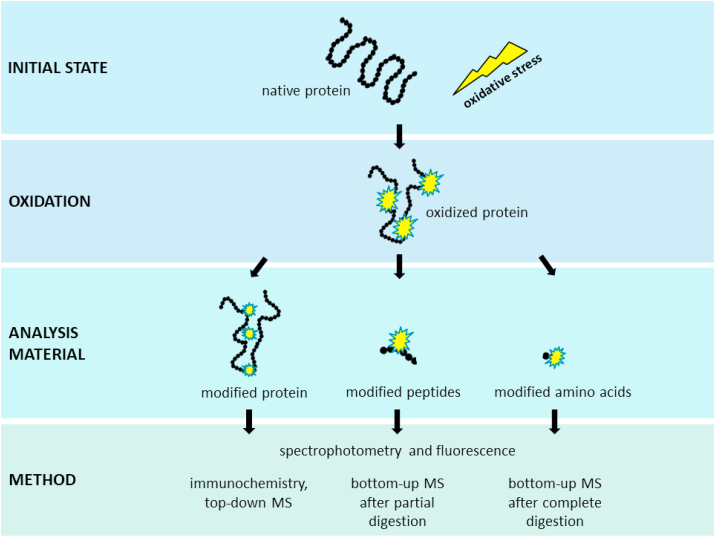
The high chemical diversity and plethora of oxidative protein modifications is reflected in various techniques which are currently used to detect modified proteins. This can be accomplished by analyzing the whole proteome, specific proteins, and even solely certain modifications. The following section focuses on general techniques to analyze protein oxidation which are summarized in Fig. 7.
Fig. 7.
Analysis of oxidative protein modifications. Proteins are oxidized by ROS. Subsequent molecular changes can be analyzed for individual proteins, peptides or modified amino acids. Depending on the choice of analytes, different analysis techniques are available. Among the presented methods, mass spectrometry is the most accurate technique and its application shall be further increased for biological samples to advance the knowledge regarding protein oxidation.
3.1. Sample preparation
Great attention should be paid on sample preparation to ensure reliable and reproducible analysis results. Preservation of biologically oxidized modifications and minimization of artifactual events at the same time are the biggest challenges and of utmost importance. Rogowska-Wrzesinska et al. [75] point out that proteins are not the only target of oxidation processes in biological samples. Oxidized molecules, such as nucleic acids and lipids, can cause high background signals, interfere with analysis procedures or can further promote oxidation reactions. An example for the latter is the formation of AGEs and ALEs from reaction of oxidized carbohydrates or lipids with proteins. Extraction steps might be applied to reduce the number of interfering compounds. Simultaneously, this leads to enrichment of proteins-of-interest which might be of importance for low abundant proteins [76]. Chemicals used for protein extraction should be chosen with caution since some of them can interfere with the protein oxidation analysis. For example, mild reducing agents such as dithiothreitol (DTT) and mercaptoethanol may reduce some protein oxidation products such as disulfides and carbonyl groups, respectively, making them unavailable for detection [77,78]. In addition, atmospheric oxygen and free metal ions can accelerate oxidation. To avoid overestimation of modified proteins due to sample preparation, it might be helpful to use derivatization reagents such as 2,4-dinitrophenylhydrazine (DNPH) or p-aminobenzaldehyde [75]. With derivatization, biological modifications are captured and stabilized and new modifications, resulting from further sample handling, are not contributing to the measured values. Chemical labeling of modifications offers the possibility of enrichment by isolating a specific modification from complex samples by affinity chromatography. Enrichment can be performed based on chemical reactivity, resin capture agents or antibodies for the modification or chemical tag [79]. Depending on the subsequent analysis technique, chemical tags might be removed or can even facilitate downstream analysis (e.g. DNPH as a matrix for matrix assisted laser desorption ionization mass spectrometry (MALDI-MS) of carbonylated proteins) or chemical tags function as reporters during MS analysis [80]. It can be crucial to perform sample preparation as quickly as possible after sampling to prevent changes from the biological oxidation pattern or formation of artifacts. Moreover, standardization of methods and reference material can help to obtain consistent results. There are commercially available oxidized proteins which can be used to regularly check analytical results. However, data on exact degree of modification provided by the supplier is limited. In most cases, the specific types and sites of modifications are not identified.
3.2. Analysis of specific oxidation products
The analysis of specific oxidation products is crucial to reveal the chemistry behind oxidation and give information about the protein involved in oxidation. Furthermore, depending on the method, targeted analysis provides quantitative or at least semi-quantitative data.
3.2.1. Spectrophotometric methods
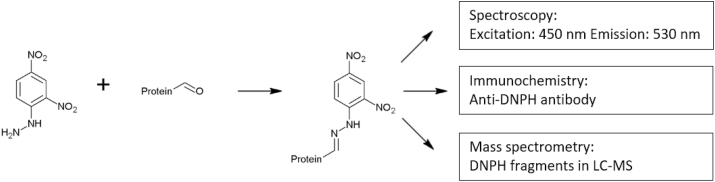
Protein oxidation can lead to the introduction of carbonyl groups into amino acid residues of the protein (see Fig. 5). The derivatization of this functional group with carbonyl-specific reagents provides a method to detect and quantify protein carbonylation [81]. The most common method to analyze protein carbonylation is the reaction with 2,4- dinitrophenylhydrazine (DNPH) (Fig. 8).
Fig. 8.
Quantitation of protein carbonylation. Carbonyl groups are stabilized by derivatization with 2,4-dinitrophenylhydrazine (DNPH). The resulting hydrazones are quantified using a spectrophotometer, specific antibodies, or are analyzed using LC-MS.
The resulting hydrazones are quantified spectrophotometrically or by immunological methods. Sample preparation and derivatization conditions for the determination of protein carbonyls were extensively reviewed elsewhere [82]. In brief, oxidized proteins are incubated with an excess of DNPH for complete derivatization of carbonyl groups. Unbound DNPH needs to be removed from the sample, because it absorbs at the same wavelength as hydrazones (λ = 370 nm). Several washing steps of the protein are required to remove unbound DNPH leading to an inevitable loss of protein of up to 10–15% [75]. An alternative assay to overcome the limitations of the classical DNPH assay was developed by Mesquita et al. [83]. DNPH-derivatized protein solution is neutralized with sodium hydroxide prior to spectrophotometric measurement. Neutralization shifts the absorbance of protein-conjugated hydrazones from 370 nm to 450 nm, thereby eliminating the interference with unbound DNPH. According to the authors, no further washing steps are required. It is important to notice that carbonyl compounds can also be formed during lipid peroxidation and the Maillard reaction, leading to an overestimation of protein oxidation. The accessibility and reactivity of carbonyl groups depends on the tertiary structure of proteins. Protein crosslinking or pronounced tertiary structures can hide DNPH-active carbonyl structures inside of proteins making them unavailable for detection. An important drawback of the DNPH method is its unspecific nature towards other oxidation products besides carbonyl groups. It was mentioned by Hellwig [84] that sulfenic acids, which are not of carbonyl form, still react in the assay.
In 2019, Ma and colleagues described a novel method for the detection of carbonylated proteins by frequency-shift based surface-enhanced Raman spectroscopy immunoassay, eliminating potential interferences by introducing a capture antibody [85]. Besides this, LC-MS/MS has been also used for proteomic analysis to identify carbonylated proteins, providing a more sensitive approach, but reveal limitations for quantitative assessments [86].
3.2.2. Fluorescence
Fluorescence spectroscopy can serve as a sum parameter for overall protein oxidation. It needs to be considered that several oxidative and also glycoxidative modifications of amino acid residues can contribute to fluorescence signals. The limited selectivity is a major drawback of fluorescence spectroscopy for studying protein oxidation products. However, because of its sensitivity, robustness and the minimal instrumental requirements, fluorescence spectroscopy became a popular approach in protein modification analysis. A selection of fluorescent oxidation products is shown in Table 1. Dityrosine and N-formyl kynurenine are formed by oxidation, whereas other compounds, such as pentosidine and the “aging pigment” lipofuscin, are formed by glycation and/or oxidation reactions. Since several fluorophores, each with a different specific fluorescence, contribute to the total fluorescence of biological samples, no quantitative calibration can be achieved. Since fluorescence is not limited to oxidation products, the method without prior chromatographic separation is only of minor importance for biomarker analysis of protein oxidation.
Table 1.
3.2.3. Immunochemical methods
Immunological techniques are widely used to identify modified protein residues. Reduced antibody binding to native epitopes indicates modification of binding sites and can be a marker of oxidative changes. Depending on the degree of modification, oxidative damage can also lead to increased antibody binding. This was shown in several studies examining the effect of hypochlorous acid on extracellular matrix proteins [87,88]. The rather high sensitivity of immunological methods and easy-to-carry-out protocols boosted the number of biological studies working with immunoblotting or enzyme linked immunosorbent assay (ELISA). Various commercially available monoclonal and polyclonal antibodies promise to detect specific protein oxidation products. Due to minimal instrumental requirements and straightforward experimental implementation, immunoassays became very popular for the analysis of protein modifications, especially in clinical applications. Immunological detection can be performed in complex mixtures or after separation, e.g., gel electrophoresis. Full chemical and structural characterization of antibody binding is crucial for the evaluation of its selectivity and sensitivity [89,90]. However, most commercial antibodies lack this characterization and their target epitopes remain unknown. Most antibodies detect different oxidation products which can be explained by poor antigen preparation and characterization. An alternative to the rather unspecific binding of antibodies to oxidation products might be the use of more selective antibodies raised against derivatization reagents. A prominent example is the anti-DNP antibody, which detects hydrazones resulting from the reaction of DNPH and carbonyl groups [91]. Besides structural uncertainty, absolute quantification is problematic with immunochemical methods. Most commercial suppliers of immunoassays provide standards in their kits. Since there is no market wide standardization of reference material, concentration values may vary depending on the used kit. Protein carbonyls, 3-NT and methionine sulfoxide are biomarkers of protein oxidation which are commonly analyzed via immunochemical methods [75,82,92].
3.2.4. Mass spectrometric methods
Mass spectrometry (MS) has been used for many years to identify and characterize proteins. Consequently, MS-based methods were developed to analyze protein oxidation products. Compared to other methods available, MS is currently the gold standard and arguably the most informative technique for protein analysis. It can be divided in “top-down” or “bottom-up” approaches which basically describes the protein treatment prior to analysis. “Top-down” means that intact proteins are introduced in the MS and their fragmentation patterns are assessed, whereas “bottom-up” requires enzymatic digestion of proteins to peptide fragments prior to MS analysis [79]. The latter is more common in proteome research and helped to sequence and identify proteins in biological samples. A special case of the “bottom-up” approach is complete digestion of the protein down to individual (modified) amino acids followed by MS analysis. It was used to investigate a wide range of oxidized amino acids, thus, identifying new oxidation products and allowing total quantification of a particular product. A drawback of complete digestion is the lack of information on the specific protein that has been modified, or the exact site of modification.
3.2.4.1. Top-down
The mass of intact proteins can be analyzed via MALDI (matrix-assisted laser desorption/ionization) or ESI (electrospray ionization) ionization techniques connected to high resolution mass analyzers, such as quadrupole-time-of-flight or orbitrap instruments. ESI results in various m/z ratios of the protein of interest, since more than one charge during ionization is usually acquired. Regarding ionization, ESI is the more common and often sensitive technique since the compilation of peaks can be used to calculate protein mass more accurately. Analysis of intact proteins requires no extensive handling of protein samples prior to MS analysis. However, since experimental spectra are compared to theoretical spectra to determine the modification status of proteins, mostly individual proteins are processed. Purification of complex protein mixture is especially challenging in biological samples. Top-down approaches can be helpful to evaluate the total modification status on an individual protein [[93], [94], [95], [96]]. Small modifications with low abundancies require more sophisticated, high-resolution instruments such as Q-TOFs, Orbitraps or FT-ICR MS. Another option to explore minor modifications occurring on a small percentage of proteins is fragmentation of intact proteins. A selected protein ion (“precursor ion”) can be dissociated resulting in smaller charged fragments (“product ions”). Dissociation methods such as electron capture dissociation (ECD) and electron transfer dissociation (ETD) are particularly good at preserving labile posttranslational protein modifications. Besides recent advances in top-down MS approaches, a number of disadvantages require the additional performance of bottom-up measurements to receive valid data. For example, sequence coverage in biological samples remains quite low using top-down MS due to overlapping and deconvolution effects [79]. Moreover, complex proteins often need to be dissolved in buffer systems or detergents to ensure complete solubility, but ESI mass spectrometers are not compatible with many buffers and especially not with detergents. Thus, buffer salts need to be removed before analysis and if a protein is insoluble in detergent-free solutions, it cannot be analyzed by top-down MS. With regard to biological samples the main drawback of top-down MS is its limitation to analyze mostly individual proteins and its focus on targeted analysis of known proteins. Complex mixtures need to be purified extensively by liquid chromatography before top-down analysis. Finally, top-down analysis requires high amounts of purified proteins to acquire highly resolved peaks of precursor and product ions with sufficient intensity [97]. Taken together, top-down MS remains an attractive technique to evaluate the overall modification status of proteins, but due to its various drawbacks, complementary techniques, such as bottom-up proteomics, need to be employed to acquire a comprehensive understanding of the protein structure.
3.2.4.2. Bottom-up
Digestion of proteins to the peptide level prior to analysis is characteristic for bottom-up MS approaches. After digestion, specific labels (also called “tags”) can be introduced into the molecule, whereby a specific modification reacts with a chemical label which can then be analyzed with reporter ions. This technique facilitates the analysis of specific modifications in a peptide and is also able to determine the modification status of peptides. A prominent example for a labeling approach is the tagging of cysteine to monitor the reversible oxidation status [98,99]. However, more common in standard proteomics are unlabeled methods which have the advantage of less sample manipulation.
Bottom-up approaches can be divided in untargeted (“discovery” or “shot-gun”) and targeted approaches. For the detection of oxidative protein modifications, untargeted methods are only of limited help. The selection of precursor ions for subsequent fragmentation is usually performed automatically and depends on the abundancy of the corresponding ion in the MS scan. However, modified peptides often occur only at low abundancies making it unlikely that they are covered by untargeted approaches. New techniques use upstream scanning methods, which identify unique ions of oxidized residues that reveal the presence of oxidative modifications and subsequent fragmentation of the according peptides. In general, untargeted approaches are mostly used to scan for certain proteins in biological samples and check for proteome changes in samples of healthy versus diseased individuals and not to analyze specific protein modifications. The specificity for the identification of oxidative modifications is improved with semi-targeted and targeted methods. In the latter, precursor and product ion are known, whereas in semi-targeted approaches only the product ion can be used as a diagnostic tool. The presence of a specific fragment ion serves as a reporter for a certain modification. Especially on low resolution instruments, data evaluation should be treated with caution since isobaric ions from non-modified peptides can yield false-positive results [100]. At best, it is recommended to confirm modification sites with synthetic modified peptides. Fully targeted analysis of oxidative protein modifications is the strategy with the highest sensitivity and most accurate quantification. This includes single reaction monitoring (SRM) and multiple reaction monitoring (MRM), where both the precursor ion and product ion masses are known for the corresponding analyte. For quantification, the abundance of the analyte ion intensities after appropriate normalization can directly be compared. The modification of a peptide may alter its ionization properties greatly, especially when ionizable groups are introduced or removed. This needs to be considered when comparing ion intensities between a certain peptide and its modified form. The most accurate method is certainly quantification with standard material of the oxidative modification of interest. However, since standard material is often not available, especially not in peptide form, this approach requires synthesis efforts or cannot always be implemented. To gain as much information as possible about the modification status of a protein of interest, it is useful to combine top down and bottom up mass spectrometric approaches and check whether a specific modification analyzed by targeted approaches can be confirmed by the loss of parent structures.
3.3. Analysis of total protein modification
A major challenge in analyzing protein modifications in biological samples is that unmodified reference material is commonly not available. However, the comparison of different sample groups (such as healthy and diseased) can provide important information without examining the exact modification site. One plausible approach is the analysis of protein structure that can provide information concerning the degree of protein modification without analyzing specific oxidation products. For example, aggregate formation can be monitored by gel electrophoresis or biophysical methods, such as light scattering or X-ray crystallography. However, these methods do not give any information about the type or site of modification and are therefore only of limited use for biomarker studies. A method which reveals modification site, but not the type of modification is amino acid analysis. By analyzing hydrolysates of native and modified protein samples, the amino acid composition can provide information about the loss of parent proteins, but also the formation of products [101]. Various protocols for the analysis of amino acids exist using derivatization with dansyl chloride or ortho-phthalaldehyde and subsequent HPLC-FLD [102,103] or HPLC-UV [104,105] or preparation of butyl esters followed by ion-pair LC-MS/MS [106]. For analysis of protein-bound amino acids, hydrolytic cleavage needs to be performed. Acid hydrolysis conditions lead to a loss of cysteine, cystine, and some tryptophan species, whereas alkaline conditions preserve tryptophan species, but breaks down arginine, serine, threonine and cysteine [107]. A mild method for proteolysis is enzymatic cleavage with nonspecific proteases. Depending on the parent protein, it might be useful to apply multistep enzymatic approaches [108]. Quantitative data can easily be achieved by using external calibration and, in case of MS analysis, internal isotope-labeled standards. Thus, amino acid analysis provides valuable information about the modification state of proteins. For detailed information on modification products and the types of proteins, MS analysis of peptides, as described above, is inevitable.
4. The relevance of biomarkers of protein oxidation in the context of human disease and aging
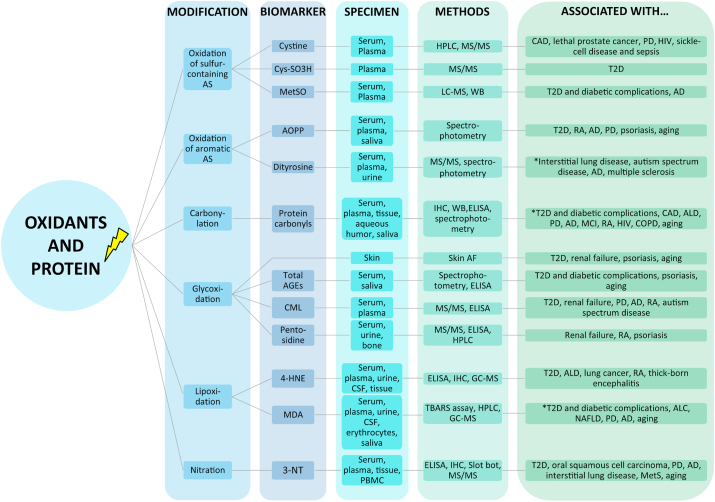
Research over the past decades displayed a substantial impact of protein oxidation in various human diseases, among others diabetes, cardiovascular disease (CVD), cancer, atherosclerosis, arthritis and neurodegenerative disease. It has been shown that most of these pathologic conditions increase as a function of age, indicating that oxidation of proteins occurs simultaneously to aging and age-related diseases [[109], [110], [111]]. Therefore, the identification of valid biomarkers of protein oxidation has an indispensable relevance to improve the understanding of human disease pathogenesis and provide an important tool to capture disease state as well as develop potential treatment strategies. In general, biomarkers for clinical application, typically measured in samples from body fluids and diverse tissues, have to fulfill certain requirements ranging from high sensitivity, specificity, reproducibility to reliability. Moreover, due to high sample throughput in clinical studies, methods should be affordable, convenient and robust as well as rapid and easy to perform [[112], [113], [114], [115]]. Here, we aim to review the most chemically stable and frequently used biomarkers of protein oxidation in routine clinical diagnostics (summarized in Fig. 9), and discuss their strengths and limitations, based on the current state of research. Since several outstanding reviews of single or multiple biomarkers and their clinical and biological relevance have been published previously [110,116,117], we selected and focused on recent clinical studies between 2015 and 2020. The search strategy on PubMed database in November 2020 was composed of search terms of the individual biomarkers plus “human disease”, “human age-related disease”, or “human aging”.
Fig. 9.
Schematic overview of frequently used biomarkers of protein oxidation. Their strengths, limitations, and relevance as biomarkers in human diseases and aging are discussed in sections 4.1–4.6, based on the current state of research. For further details see also Table 2, Table 3, Table 4, Table 5, Table 6, Table 7 3-NT – 3-Nitrotyrosine, 4-HNE – 4-Hydroxy-2-nonenal, AD – Alzheimer's disease, AGE – Advanced glycation endproduct, ALD – Alcoholic liver disease, AOPP – Advanced oxidation protein product, AS – Amino acids, CAD – Coronary artery disease, CML – Carboxymethyl lysine, COPD – Chronic obstructive pulmonary disease, CSF – Cerebrospinal fluid, Cys-SO3H – Cysteine sulfonic acid, ELISA – Enzyme linked immunosorbent assay, HIV – Human immunodeficiency virus, HPLC – High pressure liquid chromatography, IHC – Immunohistochemistry, NAFLD – Non-alcoholic fatty liver disease, MCI – Mild cognitive impairment, MDA – Malondialdehyde, MetS – Metabolic syndrome, MetSO – Methionine sulfoxide, MS – Mass spectrometry, PBMC – Peripheral blood mononuclear cells, PD – Parkinson's disease, RA – Rheumatoid arthritis, Skin AF – Skin autofluorescence, T2D – Type 2 diabetes, TBARS – Thiobarbituric acid reactive substances, WB – Western Blot. *among others.
4.1. Sulfur-containing amino acids in human diseases and aging
Sulfur-containing amino acids, such as cysteine and methionine are highly susceptible to diverse forms of oxidation processes [118,119]. In particular, cysteine residues are major targets of ROS undergoing a large number of posttranslational modifications, leading to functional and structural alterations of proteins. Thus, cysteine and its oxidized forms, such as cystine and further cysteine disulfide species, are considered as potential biomarkers of protein oxidation in human disease [120,121]. However, a strong limitation for the analysis of cysteine species is their instability due to the dynamic nature, since oxidation products of protein cysteine residues can be easily reduced by other thiols [122,123]. Cysteine residues have been detected in serum and plasma samples via HPLC or different combinations of chromatographic techniques with MS or MS/MS. In particular, the latter approaches have been used in recent human studies, providing high sensitivity, specificity and reproducibility, but are limited for routine clinical applications, because of high costs for laboratory equipment and low sample throughput. By using these methods, researchers found higher cystine levels in patients with Parkinson's disease (PD) and human immunodeficiency virus (HIV) compared to healthy individuals as well as elevated serum cystine levels associated with reduced risk for lethal prostate cancer (Table 2). Another promising method has been described in 2019 by Fu and colleagues. They used a UPLC-MS/MS approach for simultaneous quantification of thiols as well as disulfides and found higher levels of cysteine disulfides in plasma of patients with sickle-cell disease and sepsis compared to healthy donors [124]. All of these studies are related to the oxidation of free amino acids or glutathione and can therefore not be considered as studies investigating direct protein oxidation. Quantitative detection of cysteine trioxidation, a stepwise and irreversible cysteine modification generating sulfonic acid motifs, has been recently developed by Paramasivan and colleagues. By using a LC-MS/MS approach, they found elevated cysteine sulfonic acid in human serum albumin of type 2 diabetes (T2D) patients compared to control subjects, indicating the potential of trioxidized peptides as prognostic biomarker of protein oxidation in human disease [125]. The use of cysteine modifications as biomarkers of aging has not been investigated in recent years, but a review by Oliveira and Laurindo in 2018 highlights the association of changes in the thiolred/thiolox redox couple in aging and age-related disease, including cancer, CVD, and neurodegenerative disease [121].
Table 2.
Selected studies using modifications of sulfur-containing amino acids as biomarkers of protein oxidation in clinical settings.
| Disease/aging | Specimen | Study population | Method | Main outcome | Reference |
|---|---|---|---|---|---|
| Cystine and cysteine sulfonic acid | |||||
| Coronary artery disease (CAD) | Plasma | N = 247 patients (of n = 1411) experienced primary outcome of death | HPLC | Higher cystine levels associated with increased risk for all-cause mortality in CAD patients | [208] |
| Lethal prostate cancer | Serum | Control subjects vs. lethal prostate cancer patients (n = 523 per group) | LC-MS/MS | Higher serum cystine levels are associated with reduced risk for lethal prostate cancer | [209] |
| PD | Serum | Control subjects (n = 30) vs. PD patients (n = 20) |
UPLC-MS | Higher cystine levels in PD patients | [210] |
| HIV | Serum | HIV-negative (n = 21) vs. HIV-positive (n = 113) patients | GC-MS | Increased levels of cystine in HIV-positive patients | [211] |
| Sickle-cell disease and sepsis | Plasma | Healthy donors (n = 18) vs. sickle-cell disease (n = 9) and sepsis patients (n = 6) |
UPLC-MS/MS | Elevated levels of cysteine disulfides (protein-bound cysteine) in disease patients | [124] |
| T2D | Plasma | Healthy subjects vs. T2D patients (n = 8 per group) | LC-MS/MS | Higher cysteine sulfonic acid levels in T2D patients | [125] |
| Methionine sulfoxide | |||||
| T2D and diabetic complications | Serum (serum albumin) | Healthy subjects (n = 18) vs. T2D patients with (n = 23) and w/o (n = 12) renal failure | LC-MS | Higher levels of methionine sulfoxide in T2D patients with and w/o renal failure compared to controls | [212] |
| AD | Plasma | Healthy subjects (n = 23) vs. patients with mild-cognitive impairment (MCI) (n = 13) or AD (n = 25) | WB | Elevated levels of methionine sulfoxide in plasma of AD patients compared to other groups | [130] |
Besides cysteine, methionine is another sulfur-containing amino acid that reacts to methionine sulfoxide under oxidative stress conditions. In contrast to other thiol oxidation products, methionine sulfoxide shows higher stability and is easily detectable via a conventional amino acid analyzer [116,126,127]. Only a few publications are available, determining levels of methionine sulfoxide in clinical settings. However, higher methionine sulfoxide levels have been found in plasma and serum samples of T2D patients with and without renal failure and Alzheimer's disease (AD) in comparison to control subjects, measured via immunoblotting or LC-MS (Table 2). To our knowledge, studies investigating methionine sulfoxide levels as a function of age have not been published, recently. The fact that methionine sulfoxide levels increase age-dependently in human skin collagen, as shown earlier [128,129], and are elevated in AD patients, suggests an impact in the aging process [130].
4.2. Aromatic amino acids in human diseases and aging
Aromatic amino acid residues are major targets for several forms of ROS and RNS, leading to the formation of dityrosine-containing crosslinks, classified as AOPPs [131,132]. In clinical settings, AOPPs have been primarily detected via spectrophotometric assays, based on the oxidation reaction of iodide to iodate, as firstly described by Witko-Sarsat and colleagues in 1996 [133]. This technique has been modulated and optimized by different groups in recent years [134,135]. For instance, Taylor and colleagues described a modification by determining total iodide ion oxidizing capacity of plasma and eliminating the effect of sample precipitation that improves accuracy and reproducibility of the assay [135]. Due to the assay's simplicity, allowing for high sample throughput, most researchers used this technique in recent years. However, spectrophotometric assays for the determination of AOPPs are highly unspecific, detecting a wide range of optically absorbing species or total oxidative capacity of plasma and the exact contribution of AOPPs to this reaction remains unknown. Therefore, studies performed with this assay and subsequent conclusions regarding AOPPs should be treated with caution. Nevertheless, groups using this method indicated higher AOPP levels in patients with several diseases, including T2D, rheumatoid arthritis (RA), neurodegenerative disease as well as aging, but conflicting results have been found for psoriasis, a common immune-mediated skin disorder (Table 3). While Haberka et al. and Yazici et al. showed higher concentrations of AOPPs in psoriasis patients compared to healthy subjects, no differences have been found in the study by Skoie and colleagues [[136], [137], [138]]. This might be explained by the inclusion of patients with only mild severity of the disease, generally associated with lower levels of oxidative stress [138]. In line with previous findings [139,140], Maciejczyk et al. and Taylor et al. found elevated AOPP levels in plasma and saliva of humans at advanced age, suggesting their relevance in the aging process [135,141].
Table 3.
Selected clinical studies using AOPP and dityrosine-containing crosslinks as biomarkers of protein oxidation.
| Disease/aging | Specimen | Study population | Method | Main outcome | Reference |
|---|---|---|---|---|---|
| AOPP | |||||
| T2D or rheumatoid arthritis (RA) | Serum | Control subjects (n = 17) vs. T2D or RA patients (n = 20 per group) | Spectrophotometry | Higher AOPP levels in RA and T2D patients | [213] |
| T2D and aging | Plasma | Non-diabetic subjects (n = 38) vs. T2D patients (n = 50) | Spectrophotometry | Age-dependent increase of AOPP levels No differences between non-diabetic subjects and T2D patients |
[135] |
| AD | Plasma | Control subjects (n = 34) vs. AD patients (n = 38) | Spectrophotometry | Higher levels of AOPPs in AD patients | [214] |
| PD | Plasma | Control subjects (n = 46) vs. PD patients (n = 40) | Spectrophotometry | Elevated AOPP levels in PD patients | [185] |
| Mild to moderate psoriasis | Serum | Healthy individuals (n = 40) vs. patients with mild to moderate psoriasis (n = 80) | Spectrophotometry | Increased levels of AOPP in psoriasis patients | [136] |
| Mild psoriasis | Plasma | Healthy subjects vs. patients with mild psoriasis (n = 84 per group) | Spectrophotometry | No difference in AOPP levels | [138] |
| Mild and moderate to severe psoriasis |
Plasma |
Healthy subjects (n = 14) vs. psoriasis patients (n = 29; mild n = 14, moderate to severe n = 15) |
Spectrophotometry |
Higher levels of AOPP in mild and moderate to severe psoriasis patients compared to controls |
[137] |
| Aging | Saliva and plasma | Children (2–14 years), adults (25–45 years), elderly (65–85 years, n = 30 per group) | Spectrophotometry | Higher AOPP levels in saliva and plasma of elderly people compared to other groups | [141] |
| Dityrosine | |||||
| Interstitial lung disease | Plasma | Healthy individuals vs. patients with interstitial lung disease (n = 42 per group) | LC-MS/MS | Elevated levels of dityrosine in patients with interstitial lung disease | [142] |
| Autism spectrum disorders | Plasma, urine | Healthy children (n = 31) vs. children with autism spectrum disorders (n = 38) | LC-MS/MS | Elevated plasma and urine dityrosine levels in children with autism spectrum disorders | [143] |
| AD | Serum | Control subjects (n = 32) vs. AD patients (n = 52) | Spectrofluorometry | Higher dityrosine levels in AD patients | [215] |
| Chronic liver diseases | Plasma | Healthy subjects vs. patients with chronic liver disease (n = 30 per group) | Spectrophotometry | Elevated dityrosine levels in patients with chronic liver diseases | [216] |
| Multiple sclerosis | Plasma | Healthy subjects (n = 11) vs. patients with multiple sclerosis (n = 14) | Spectrophotometry | Estimated dityrosine levels were higher in patients with multiple sclerosis | [217] |
Besides this, crosslinks such as dityrosine are implicated in a number of pathologies and have not been only detected via spectrophotometric or spectrofluorimetric assays, but also by chromatographic separation followed by MS. Although MS approaches are expensive, require special laboratory equipment and are therefore often not applicable in routine clinical diagnostics, MS is the most specific and sensitive technique for the detection of dityrosine. With this, elevated concentrations of dityrosine have been found in plasma and urine samples of patients with interstitial lung disease and autism spectrum disorders [142,143]. Dityrosine levels have not been determined as a function of age in recent years, but in 2011 Campos et al. found a positive correlation between age and levels of dityrosine in urine samples of healthy smoker and non-smoker controls using unspecific spectrofluorometric detection [144]. Moreover, dityrosine crosslinks have been previously observed in amyloid plaques and cerebrospinal fluid (CSF) of AD patients via LC-ESI-MS/MS, indicating a potential role in aging and age-related diseases [145].
4.3. Carbonylation in human diseases and aging
Protein carbonylation is an irreversible protein modification, associated with crucial alterations in their functional and structural integrity, contributing to cellular dysfunction and tissue damage [146,147]. Due to relatively early formation during oxidative stress, higher stability in comparison to other oxidation products and simple analysis methods, protein carbonyls are one of the most commonly analyzed biomarkers in human metabolic and age-related diseases [63,111]. Traditionally, protein carbonyls are determined in plasma, serum, and tissue samples, but also in aqueous humor and saliva (Table 4). The latter provides advantages for patients due to convenient, non-invasive, pain- and infection-free sample collection, but might be affected by oral hygiene [148]. Protein carbonyls can be determined by ELISA, immunoblot techniques, immunohisto- and cytochemistry via specific anti-DNP antibodies, and direct spectrophotometric or HPLC measurements. These methods are based on the derivatization of carbonyl groups with DNPH, forming a stable hydrazone adduct, as described in section 3.2. In 2015, two independent articles compared the available standard methods for the detection of protein carbonylation and highlighted advantages and disadvantages of the individual techniques in detail. Since immunoblotting and LC-MS/MS approaches rather provides insights into molecular mass and the nature of carbonyl modifications, the use of ELISAs has been declared as the method of choice for quantification of protein carbonyls [82,86]. As shown in Table 4, this technique has been primarily used to determine protein carbonyls in clinical studies, because of its suitability for high-throughput analysis, small sample volumes and minimal laboratory requirements. Although Goycheva et al. and Almogbel and colleagues included T2D patients with different diabetic complications (vascular complications vs. diabetic nephropathy), protein carbonyls levels measured by ELISA were comparable in control as well as disease groups [149,150]. Independent of the used methods and specimens, almost all selected studies consistently revealed elevated levels of protein carbonyls in multiple diseases, including T2D and diabetic complications, lung, liver, infectious, or rheumatological disease (Table 4). The study by Sharma and colleagues, however, found lower levels of protein carbonyls in female AD patients compared to healthy control subjects, whereas male AD patients revealed higher concentrations. These sex-specific differences were also observed for other markers of protein oxidation, including AGEs, MDA, or 3-NT, and should be considered in future clinical assessments [151]. Although Weber et al. found no correlation between protein carbonyls and aging in two of three cohorts from the European MARK-AGE study, presumably related to genetic and lifestyle factors, all other recent studies revealed higher concentrations of protein carbonyls in adults and elderly compared to younger individuals (Table 4). In addition, elevated protein carbonyl levels have been found in neurodegenerative disease, such as AD, together indicating the relevance of protein carbonyls in aging and age-related diseases [63,151].
Table 4.
Selected studies using protein carbonyls as a biomarker of protein oxidation in clinical settings.
| Disease/aging | Specimen | Study population | Method | Main outcome | Reference |
|---|---|---|---|---|---|
| T2D with vascular complications | Serum | Healthy individuals (n = 94) vs. T2D patients with vascular complications (n = 94) |
ELISA | Higher protein carbonyl levels in T2D patients with vascular complications | [149] |
| T2D with NASH | Serum | Control subjects (n = 50) vs. T2D patients with NASH (n = 60) or T2D w/o NAFLD (n = 50) | ELISA | Elevated protein carbonyl levels in T2D patients with NASH compared to other groups | [218] |
| T2D with and w/o CKD | Serum | Healthy subjects vs. T2D patients with and w/o CKD (n = 50 per group) | Spectrophotometry/HPLC | Higher protein carbonyl levels in T2D patients with and w/o CKD compared to control subjects | [219] |
| Diabetic nephropathy | Serum | Control subjects (n = 142) vs. T2D patients with diabetic nephropathy (n = 153) |
ELISA | Elevated levels of protein carbonylation in patients with diabetic nephropathy | [150] |
| CAD | Plasma | Control subjects vs. premature CAD or normal CAD (n = 30 per group) | ELISA | Higher plasma protein carbonyl levels in premature and normal CAD patients compared to control groups | [220] |
| Acute coronary syndrome or chronic periodontitis | Saliva | Control subjects vs. patients with acute coronary syndrome, chronic periodontitis or both (n = 24 per group) | Spectrophotometry | Higher protein carbonyl levels in all disease groups compared to controls | [221] |
| Alcoholic liver cirrhosis (ALC) | Serum | Healthy control subjects (n = 130) vs. ALC patients (n = 57) | Spectrophotometry | No difference between control and ALC groups | [222] |
| Normal and end stage ALD | Hepatic tissue | Normal ALD (n = 8) vs. end stage ALD (n = 9) |
IHC, WB | Increased protein carbonylation in liver sample of end stage ALD patients | [223] |
| PD and AD | Plasma | Control subjects (n = 34) vs. AD (n = 40) and PD patients (n = 70) | ELISA | Higher protein carbonyl levels in male AD patients compared to PD patients and controls | [151] |
| MCI, AD | Plasma | Healthy control (n = 24) vs. MCI (n = 24) and AD patients (n = 18 mild AD, n = 15 moderate AD, n = 14 severe AD) | WB | Higher levels of protein carbonyls in all AD groups compared to control and MCI groups | [224] |
| RA | Serum | Healthy control subjects (n = 41) vs. RA patients (n = 29) | Spectrophotometry | Higher levels of protein carbonyls in RA patients | [225] |
| RA | Plasma | Control subjects (n = 53) vs. RA patients (n = 120) | Spectrophotometry | Higher protein carbonyl levels in RA patients | [226] |
| Acute rheumatic fever in children | Serum | Healthy control (n = 31) vs. patients with acute rheumatic fever (n = 31) | Spectrophotometry/HPLC | Higher serum protein carbonyl levels in patients with acute rheumatic fever | [227] |
| HIV | Plasma | Control group (n = 300) vs. HIV infected Antiviral Therapy (ART) naive (n = 100), on first line ART (n = 100), on second line ART (n = 100) | ELISA | Higher protein carbonyl content in HIV-infected patients on first- and second-line ART | [228] |
| Chronic obstructive pulmonary disease (COPD) | Plasma | Control patients (n = 15) vs. COPD patients (n = 42) | ELISA | Elevated levels of protein carbonyls in COPD patients | [229] |
| MetS | Serum | Healthy women (n = 77) vs. women with MetS (n = 106) | Spectrophotometry | Higher levels or protein carbonyls in women with MetS | [230] |
| Leptospirosis | Serum | Control subjects (n = 30) vs. mild (n = 50), severe (n = 60) Leptospirosis or Dengue Leptospirosis (n = 30) |
ELISA, Spectrophotometry | Higher serum protein carbonyl levels in severe Leptospirosis patients compared to all other groups | [231] |
| Glaucoma | Serum, aqueous humor | Control subjects (n = 64) vs. glaucoma patients (n = 96) | ELISA | Higher levels of protein carbonyls in plasma and aqueous humor of glaucoma patients | [232] |
| Aging | Saliva, plasma | Young (n = 104) vs. middle-aged (n = 90) and elderly subjects (n = 79) | ELISA | Age-dependent increase in saliva and plasma protein carbonyl levels | [148] |
| Serum | Adolescents (n = 30, mean age 17.1 years) vs. adults (n = 34, mean age 33.3 years) with bipolar disorder | ELISA | Higher protein carbonyl content in adults | [233] | |
| Plasma | Healthy individuals divided into 4 groups: group 30 (28–34 years), group 40 (35–45 years), group 50 (47–54 years) and group above 60 (57–79 years) | ELISA | Higher protein carbonyl levels in groups 40, 50 and 60 compared to group 30 | [234] | |
| Plasma | 3 study groups from MARK-AGE project: RASIG (n = 794), GO (n = 493), SGO (n = 272) Participants divided in 4 groups: 55-59, 60–64, 65–69 and 70–75 years |
ELISA | Positive correlation between protein carbonyl levels and age in RASIG group No correlation between protein carbonyl levels and age in GO and SGO groups |
[194] | |
4.4. Glycoxidation in human diseases and aging
The formation and accumulation of glycoxidation products occurs during normal, but especially under certain physiological conditions, such as hyperglycemia, hyperlipidemia, and oxidative stress. This indicates their relevance in the progression of metabolic diseases, including neurodegenerative disease, chronic kidney disease (CDK), CVD, and cancer, but in particular, T2D and related complications as reviewed elsewhere [152]. Moreover, a growing body of evidence reveals that levels of glycoxidation products increase as a function of age in several tissues, dependent on the dietary intake and the presence of hyperglycemic conditions [152,153]. One major difficulty in detection of glycoxidation products for standardized clinical research is their structural complexity and heterogeneity, deriving from various formation mechanisms [154,155]. Among glycoxidation structures, CML and pentosidine are most frequently measured in clinical studies [156,157]. Several methods for their detection in blood, urine, and tissue samples have been established in recent years, such as spectrophotometry, immunohistochemistry (IHC), immunoblot, and ELISA techniques, whereas the latter has been most commonly used. Although ELISAs are easy to perform and ensure high sample throughput, their specificity and reproducibility have been criticized, especially by the limited description of applied antibodies, associated with different outcomes [158]. This becomes obvious by the comparison of two studies. Guerin-Duborg and colleagues used an AGE-ELISA Kit preferentially quantifying CML and revealed no differences between non-diabetic subjects and T2D patients as well as T2D patients with and without vascular diseases. In contrast, Farhan et al. found elevated AGE levels comparing the latter groups by using a competitive ELISA Kit without specification of detected glycoxidation products [159,160]. AGEs can be also detected via non-invasive skin autofluorescence (skin AF) readers [161]. The measurement correlates well with the total amount of AGEs in skin biopsies and has been applied in clinical settings, indicating elevated AGE levels in several diseases and aging, as shown in Table 5. However, this technique is highly unspecific to capture glycoxidation products since several other fluorescent structures, such as non-oxidatively formed AGEs or nicotinamide adenine dinucleotide are detected simultaneously. The method has to be standardized in terms of using the same (tattoo-free) body region as well as multiple determination and might be limited by the use of body creams, bronzer as well as solarium [162]. The most sensitive approaches for detection of glycoxidation products are methods based on chromatography, including HPLC und UPLC that are mainly combined with MS/MS. These methods represent the gold standard for the identification of glycoxidation structures and detection in free and protein-bound forms, but their use in routine clinical studies is limited, in particular, due to high costs [158,163,164]. For a detailed review of advantages and disadvantages of various immunochemical, bioanalytical, and biochemical methods for the measurement of AGEs, including glycoxidation products, see Ashraf et al. [158]. Studies from recent years, using UPLC- or LC-MS/MS approaches show very heterogenous outcomes in different diseases, depending on the detected structure. This emphasizes the importance of discrimination and selective analysis of the various structures formed by glycoxidation reactions and the limited usefulness of some methods in this field. For instance, O'Grady et al. found higher CML levels in plasma of T2D patients compared to healthy subjects but similar pentosidine concentrations [165]. The latter finding is in contrast to previous investigations, using ELISA or HPLC techniques, revealing higher serum pentosidine levels in diabetes patients compared to healthy controls [166,167]. Moreover, Wetzels and colleagues as well as Teichert et al. show similar plasma levels of various AGE structures, including free and protein-bound CML between non-demented subjects and patients with multiple sclerosis or non-diabetic and pre-diabetic women, respectively [168,169]. In line with earlier findings on glycoxidation levels in aging [153], studies from 2019 suggest an age-related increase in skin AF, pentosidine levels and total amount of AGEs [141,170,171]. In particular, Kida and colleagues performed their analysis in different specimens of spinal surgery patients and found strong correlations between serum, urine, bone, and skin pentosidine levels with age [170]. The fundamental role of AGEs, including glycoxidation products, in human aging, but also in aging of vertebrate and invertebrate model organism has been reviewed in 2018 by Chaudhuri and colleagues [153], together indicating the relevance of AGEs as biomarkers of aging and age-related diseases.
Table 5.
Selected studies using glycoxidation products as biomarkers of protein oxidation in clinical settings.
| Disease | Specimen | Study population | Method | Main outcome | Reference |
|---|---|---|---|---|---|
| T2D with or w/o vascular disease | Plasma | Non-diabetic controls (n = 31) vs. T2D patients (n = 75) with (n = 44) or w/o (n = 31) vascular disease |
Spectrophotometry, ELISA | No difference in fluorescent-AGEs or AGE levels between non-diabetic subjects and T2D patients No differences in AGE levels between T2D patients with or w/o vascular disease Higher fluorescent-AGE levels in T2D patients with vascular disease compared to T2D patients w/o vascular disease |
[160] |
| T2D with or w/o vascular complications | Serum | Healthy subjects (n = 20) vs. T2D patients with or w/o vascular complications (n = 25 per group) | ELISA | Higher AGE levels in T2D patients with and w/o vascular complications Highest AGE levels in T2D patients with vascular complications |
[159] |
| Renal failure or T2D | Serum | Control individuals vs. patients with renal failure (n = 30 per group) Control subjects (n = 49) vs. T2D patients (n = 95) |
Spectrophotometry, skin AF, LC-MS/MS |
Higher total AGE, pentosidine and free CML levels in patients with renal failure Elevated free CML but not pentosidine and total AGE levels in T2D patients Higher skin AF in patients with renal failure and T2D compared to controls |
[165] |
| Prediabetes | Plasma | Women with normal fasting glucose vs. women with impaired fasting glucose (n = 30 per group) | LC-MS/MS | No difference between groups in free CML levels | [169] |
| CKD | Plasma | Patients with CKD (n = 150) | LC-MS/MS, ELISA |
No association between pentosidine levels and CKD | [235] |
| PD and AD | Plasma | Control subjects (n = 34) vs. AD (n = 40) and PD patients (n = 70) | UPLC-MS/MS | Higher protein-bound CML levels in AD and PD patients compared to controls | [151] |
| Schizophrenia | Red blood cell lysates | Control subjects vs. schizophrenia patients (n = 23 per group) | WB | No differences in pentosidine levels | [236] |
| Autism spectrum disorders | Plasma | Healthy children (n = 31) vs. children with autism spectrum disorders (n = 38) | LC-MS/MS | Increased plasma CML levels in children with autism spectrum disorders | [143] |
| RA | Serum | Control subjects (n = 30) vs. RA patients (n = 80) | ELISA | Higher CML and pentosidine levels in RA patients | [237,238] |
| Psoriasis | Skin, serum | Healthy individuals and psoriasis patients (n = 40 per group) | Skin AF, ELISA |
Higher skin AF, total AGEs and pentosidine levels in patients with psoriasis | [239] |
| Multiple sclerosis |
CSF, plasma, brain tissue |
Non-demented control subjects (n = 10) vs. multiple sclerosis patients (n = 15) |
UPLC-MS/MS, IHC |
No difference in plasma free and protein-bound CML levels |
[168] |
| >Aging (and Diabetes) | Skin | Age groups: group 20–30 years, n = 18 group 30–40 years, n = 10 group 40–50 years, n = 14 group 50–60 years, n = 12 group >60 years, n = 11 diabetes group, n = 13 |
Skin AF | Skin AF increases with age Higher skin AF in diabetes patients compared to groups aged <50 years |
[171] |
| Skin, urine, serum, bone | Patients with spine disease | Skin AF, ELISA, HPLC |
Age-related increase in serum, urine, bone and skin pentosidine levels | [170] | |
| Saliva, plasma | Children (2–14 years), adults (25–45 years), elderly (65–85 years, n = 30 per group) | Spectrophotometry | Higher AGE levels in saliva and plasma of elderly people compared to children and adults | [141] |
4.5. Lipoxidation in human diseases and aging
The biological process of lipid peroxidation has been implicated in a high number of human pathologic conditions [172,173]. Recent clinical studies focused mainly on reactive intermediates 4-HNE and MDA, which are prominent precursors of ALE formation [174]. Qualitative and semiquantitative immunological methods, such as ELISA and IHC, using commercially available antibodies, have been primarily performed to detect 4-HNE in biological samples of various human diseases, as shown in Table 6 and reviewed elsewhere [175]. These antibodies are raised against protein-bound 4-HNE, ensuring relatively high specificity. In general, detection by IHC is valuable for the evaluation of morphological alterations in tissue samples, but not advisable for parallel quantitative assessments due to several limitations, such as the use of different fixatives, potential background staining, quality of dyes or humidity [176]. A more sensitive approach has been used by Luczaj and colleagues via GC-MS, but also ELISA, measuring free 4-HNE and protein-bound 4-HNE, respectively, in two independent studies [177,178]. For instance, RA patients consistently revealed higher levels of free and protein-bound 4-HNE in urine and plasma compared to control subjects [177]. To our knowledge, levels of 4-HNE have not been determined as a function of age in recent years. However, 4-HNE is activating or inhibiting certain age-related signaling pathways, including NF-ĸB, Nrf2 or mTOR pathways, and might be therefore contributing to the aging process [179,180].
Table 6.
Selected clinical studies using 4-HNE and MDA as biomarkers in different diseases.
| Disease | Specimen | Study population | Method | Main outcome | Reference |
|---|---|---|---|---|---|
| 4-HNE | |||||
| T2D | Serum | Control subjects (n = 9) vs. T2D patients (n = 11) | ELISA | Higher levels of 4-HNE in patients with T2D | [240] |
| Alcoholic liver disease (ALD) | Human hepatic tissue | Normal ALD (n = 8) vs. end stage ALD (n = 9) |
IHC | Elevated 4-HNE in liver sample of ALD patients | [223] |
| Lung cancer | Serum | Control subjects (n = 40) vs. lung cancer patients (n = 92) | ELISA | Higher levels of 4-HNE in male and female lung cancer patients | [183] |
| RA | Plasma, urine | Healthy subjects vs. RA patients (n = 73 per group) | GC-MS, ELISA | Elevated levels of free and protein-bound 4-HNE in RA patients | [177] |
| Thick-born encephalitis | CSF, plasma, urine | Healthy subjects (n = 56) vs. patients with thick-born encephalitis (n = 60) | GC-MS, ELISA | Elevated free and protein-bound 4-HNE levels in plasma of thick-born encephalitis patients | [178] |
| MDA | |||||
| T2D and NAFLD | Serum | T2D patients with (n = 73) and w/o NAFLD (n = 51) | TBARS assay | Higher MDA levels in T2D patients with NAFLD | [241] |
| T2D and CKD | Plasma | Healthy subjects vs. T2D patients with or w/o CKD (n = 50 per group) | TBARS assay | Higher MDA levels in T2D patients with and w/o CKD compared to control subjects | [219] |
| T2D with vascular complications | Plasma | Healthy individuals (n = 94) vs. T2D patients with vascular complications (n = 93) |
TBARS assay | Higher levels of MDA in T2D patients with vascular complications | [149] |
| ALC | Serum | Healthy subjects (n = 130) vs. ALC patients (n = 57) | TBARS assay | Higher MDA levels in patients with ALC | [222] |
| NAFLD | Plasma | Healthy subjects (n = 40) vs. NAFLD patients (n = 67) | TBARS assay | Higher MDA levels in NAFLD patients | [85] |
| Lung cancer | Serum | Control subjects (n = 40) vs. lung cancer patients (n = 92) | TBARS assay | No differences in MDA levels between the groups | [183] |
| PD | Serum | Control subjects (n = 46) vs. PD patients (n = 40) | TBARS assay | Elevated MDA levels in PD patients | [185] |
| PD and AD | Plasma | Control subjects (n = 34) vs. AD (n = 40) and PD patients (n = 70) | RP-HPLC | Lower MDA levels in AD and PD patients compared to control subjects | [151] |
| RA | Plasma, urine | Healthy subjects vs. RA patients (n = 73 per group) | GC-MS | Elevated levels of MDA in plasma and urine of RA patients | [177] |
| Thick-born encephalitis |
CSF, plasma, urine |
Healthy subjects (n = 56) vs. patients with thick-born encephalitis (n = 60) |
GC-MS |
Elevated MDA levels in all specimens of thick-born encephalitis patients |
[178] |
| Aging | Plasma | Healthy individuals divided into 4 groups: group 30 (28–34 years), group 40 (35–45 years), group 50 (47–54 years) and group above 60 (57–79 years) | HPLC | No differences between the groups | [234] |
| Serum | Adults (Age 20–50 years) vs. elderly (Age ≤ 60 years, n = 30 per group) | TBARS assay | Higher MDA levels in elderly | [242] | |
| Erythrocytes | Healthy subjects aged ≤65 years (n = 27) and ≥65 years (n = 30) | TBARS assay | No differences between the groups | [243] | |
| Saliva, plasma | Children (2–14 years), adults (25–45 years), elderly (65–85 years, n = 30 per group) | TBARS assay | Higher MDA levels in saliva and plasma of elderly people compared to children and adults | [141] | |
| Plasma | 3 study groups from MARK-AGE project: RASIG (n = 794), GO (n = 493), SGO (n = 272) Participants divided in 4 groups: 55-59, 60–64, 65–69 and 70–75 years |
RP-HPLC | No correlation in all groups between MDA levels and age | [194] | |
To our knowledge, MDA modified proteins have not been used as biomarkers in recent clinical investigations although antibodies are commercially available and highly selective MS methods have been established, previously [57]. Nevertheless, free MDA potentially leading to stable protein modifications, has been quantified in several studies via batch thiobarbituric acid reactive substances (TBARS) assay. This approach is the method of choice for the quantification of free MDA due to simplicity and low costs, but is often criticized as being unspecific and rather measuring total oxidative stress instead of MDA alone [181,182]. Except of Zablocka-Slowinska and colleagues, showing no differences between healthy subjects and patients with lung cancer [183], all clinical investigations using the TBARS assay consistently found elevated MDA levels in diverse human metabolic disease, as shown in Table 6. Beside this assay, further methods have been developed in order to improve the specificity of MDA measurement, such as HPLC combined with UV or fluorescence detection [182,184]. Reversed-phase HPLC coupled with fluorescence detection has been further used by Sharma and colleagues, revealing lower levels of MDA in AD and PD patients compared to controls [151]. This is in contrast to the findings by Medeiros and colleagues, indicating higher MDA concentrations in PD patients that might be explained by the use of different methods, but also potential differences in the study populations [185]. The GC-MS approach in the study by Luczaj and colleagues as mentioned above, has been also used for the measurement of MDA, revealing elevated levels in different specimens of RA and thick-born encephalitis patients compared to controls [177,178]. Independent of the applied method, three of five studies found no correlation between MDA and advanced age although they partially included large cohorts, e.g., from the European MARK-AGE study (Table 6). In combination with lower MDA levels in elderly male and female AD and PD patients compared to healthy subjects as well as unchanged concentrations between lung cancer patients and control individuals at advanced age [151,183], this suggests that MDA is no suitable marker for human aging and age-related diseases.
4.6. Nitration in human diseases and aging
3-NT, the stable endproduct of peroxynitrite-mediated oxidation formed via nitration of free or protein-bound tyrosine, has been primarily used for the determination of protein nitration and nitrosative stress in clinical settings [186,187]. Already in 2008, Pacher et al. published an outstanding and detailed review about the role of peroxynitrite and 3-NT formation in human health and disease, indicating their relevance as diagnostic biomarker for several pathologic conditions [188]. Elevated levels of 3-NT have been also found in a variety of human diseases in recent research, including T2D, cancer, and neurodegenerative diseases, but the available data are partially conflicting (Table 7). For instance, while Ravassa et al. found higher 3-NT levels in T2D patients with additional cardiovascular events, no differences were observed by Segre and colleagues although baseline parameters of the study populations and the used methods were similar [189,190]. Moreover, recent research in AD patients revealed higher 3-NT levels compared to healthy volunteers, whereas no differences were found in a previous investigation by Ryberg and colleagues, but these studies are only partially comparable since different specimens and methodological approaches have been used [151,[191], [192], [193]]. To our knowledge, only Weber et al. determined 3-NT as a function of age in three different cohorts of the MARK-AGE study [194]. Two of three cohorts showed no differences between the age groups, but elevated 3-NT levels in age-related diseases, such as AD and PD, indicate the impact of 3-NT accumulation in human aging [191,192,195]. The measurement of 3-NT can be performed in plasma, serum as well as tissue samples by different methods, including IHC, ELISA, HPLC or the combination of LC with MS/MS, whereas MS approaches have been considered as gold standard. Nevertheless, commercially available ELISAs are preferentially used for 3-NT quantification in clinical studies due to standardization, easy sample preparation, the need of conventional equipment, and high sample throughput, as described for other biomarkers. In turn, several limitations have been highlighted in the literature, such as low sensitivity, minor specificity of applied antibodies, the susceptibility to errors, or missing detection of free 3-NT [[196], [197], [198], [199]]. However, Weber et al. provided a simple and fast indirect ELISA method for the measurement of protein-bound 3-NT in human serum and plasma samples. High detection limits, small sample volumes, and the opportunity for high throughput-screening makes this method useful for clinical applications [196]. Moreover, Tramutola and colleagues used a slot blot approach to detect total 3-NT levels in PBMC (peripheral blood mononuclear cells) of AD patients, followed by a MS/MS-based proteomic analysis of 3-NT-modified proteins [191]. In 2016, Ng et al. demonstrated a novel detection scheme of 3-NT by using a nickel-doped graphene synthetic receptor combined with a localized surface plasmon resonance biosensor that might be of potential relevance for the diagnosis of 3-NT in clinical settings. As declared by the authors, this approach provides advantages compared to other techniques, such as immunoassays, due to label-free detection, higher specificity and capture of 3-NT by direct chemisorption [200]. The most accurate detection of 3-NT, however, requires the combination of LC with MS/MS providing the opportunity to distinguish between free, protein-bound, or total 3-NT, but the use of this method is not feasible for clinical research, because of high costs and low throughput [198].
Table 7.
Selected studies using 3-NT as biomarker of protein oxidation in clinical studies.
| Disease | Specimen | Study population | Method | Main outcome | Reference |
|---|---|---|---|---|---|
| T2D | Serum | Non-diabetic subjects (n = 14) vs. T2D patients (n = 72) T2D patients with (n = 14) or w/o (n = 46) cardiovascular events |
ELISA | Elevated levels of 3-NT in T2D patients Higher 3-NT levels in T2D patients with cardiovascular events |
[189] |
| T2D | Serum | Non-diabetic subjects (n = 35) vs. T2D patients (n = 60) | ELISA | Higher levels of protein-bound 3-NT in T2D patients | [244] |
| T2D and CAD | Plasma | T2D patients with (n = 36) and w/o (n = 32) CAD | ELISA | No differences between the groups | [190] |
| Mortality in CHD patients | Serum | Patients with acute coronary syndrome (n = 342) | ELISA | No relationship between 3-NT and mortality during 4-years of follow-up | [245] |
| Oral squamous cell carcinoma | Tissue samples (oral mucosa) | Normal oral mucosa (n = 16) vs. oral leukoplakia (n = 42) or oral squamous cell carcinoma with (n = 46) and w/o (n = 39) metastases | IHC | Higher 3-NT levels in oral squamous cell carcinoma samples with metastases compared to samples w/o metastases | [246] |
| PD and AD | Plasma | Control subjects (n = 34) vs. AD (n = 40) and PD patients (n = 70) | ELISA | Higher 3-NT levels in male AD patients compared to PD patients and controls | [151] |
| AD | PBMC | Non-demented subjects vs. AD patients | Slot blot | Increased total 3-NT levels in PBMC of AD patients | [191] |
| AD | Plasma | Healthy subjects (n = 37) vs. AD patients (n = 48) | ELISA | Higher plasma 3-NT concentrations in AD patients | [192] |
| Interstitial lung disease | Plasma | Healthy individuals vs. patients with interstitial lung disease (n = 42 per group) | MS/MS | Elevated levels of 3-NT in patients with interstitial lung disease | [142] |
| MetS | Plasma | Healthy vs. MetS subjects (n = 25 per group) | LC-ESI-MS/MS | Higher protein-bound 3-NT levels in MetS subjects | [247] |
| Aging | Plasma | 3 study groups from MARK-AGE project: RASIG (n = 794), GO (n = 493), SGO (n = 272) Participants divided in 4 groups: 55-59, 60–64, 65–69 and 70–75 years |
ELISA | Positive correlation between 3-NT levels and age in RASIG group No correlation in GO and SGO groups between 3-NT levels and age |
[194] |
5. Conclusion
Reactive oxygen and nitrogen species are ubiquitous side-products of metabolism. Impaired balance between their generation and degradation in aging and disease causes oxidative and nitrosative stress. As a result, an incredible number of distinct protein modifications is formed. These modifications are generally more stable compared to their reactive precursors and, thus, important biomarkers for monitoring medium-to long-term oxidative and nitrosative stress. Selection of the appropriate technique to study protein oxidation in biological samples is the responsibility of the user and depends on the experimental question and the available equipment. Many biological disciplines use immunoassays as routine methods. Immunological methods are often based on monoclonal antibodies or polyclonal antisera obtained after immunization with immunogens, lacking full chemical or structural characterization. In many cases, the epitopes recognized by the antibodies remain unknown. To advance the knowledge in protein oxidation and its structural mysteries, a further increase of MS analysis for biological samples would be desirable. Among the presented methods, MS is certainly the most accurate and informative technique, capable of resolving remaining questions regarding protein modifications.
Since many frequently used routine methods are less than optimal, the clinical relevance of most protein modifications as biomarkers is questionable, although protein oxidation seems to play a critical role in several pathologies and in the aging process. Reversible modifications, including cysteine disulfide and methionine sulfoxide have been associated with various human and age-related diseases, but in most studies only oxidized free amino acids have been determined, not reflecting the extent of protein oxidation. Also, the widely used spectrophotometric detection of AOPPs is highly unspecific and detects a wide range of oxidative species, therefore, these studies should be treated with caution. The determination of specific crosslinks such as dityrosine by chromatographic separation followed by MS analysis is more reliable. Moreover, drawing a final conclusion on glycoxidation products as relevant biomarkers of protein oxidation is hardly possible based on the included studies published in the last years. The selection of specific and reliable analytes and detection methods is of utmost importance and can influence the study outcome significantly. Thus, further longitudinal studies with standardized methods and conditions are needed to determine the relationship between these oxidation products and human diseases in more detail. Regarding advanced lipoxidation endproducts, recent studies mainly focused on the reactive intermediates 4-HNE and MDA, which presumably correlate with the amount of protein modifications formed by reaction with this carbonyl species. However, future studies should focus on measuring the corresponding stable protein modifications. Despite partially inconsistent results in studies of recent years, elevated 3-NT levels are associated with multiple diseases and aging, indicating its potential as biomarker for protein oxidation in clinical research. Also, elevated levels of protein carbonyls have been observed in several human diseases and are associated with the aging process. Therefore, protein carbonyl content can be seen as a biomarker of global oxidative protein damage, with the advantage of early formation and long circulation periods compared to other parameters of oxidative stress.
Acknowledgement
This work was supported by the German Ministry of Education and Research (BMBF) and the State of Brandenburg (DZD grant 82DZD00302).
Contributor Information
Richard Kehm, Email: Richard.Kehm@dife.de.
Tim Baldensperger, Email: Tim.Baldensperger@dife.de.
Jana Raupbach, Email: Jana.Raupbach@dife.de.
Annika Höhn, Email: Annika.Hoehn@dife.de.
References
- 1.Sies H. Strategies of antioxidant defense. Eur. J. Biochem. 1993;215:213–219. doi: 10.1111/j.1432-1033.1993.tb18025.x. [DOI] [PubMed] [Google Scholar]
- 2.Hughes M.N., Nicklin H.G. A possible role for the species peroxonitrite in nitrification. Biochim. Biophys. Acta. 1970;222:660–661. doi: 10.1016/0304-4165(70)90192-3. [DOI] [PubMed] [Google Scholar]
- 3.Kregel K.C., Zhang H.J. An integrated view of oxidative stress in aging: basic mechanisms, functional effects, and pathological considerations. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007;292:R18–R36. doi: 10.1152/ajpregu.00327.2006. [DOI] [PubMed] [Google Scholar]
- 4.Liguori I., Russo G., Curcio F., Bulli G., Aran L., Della-Morte D., Gargiulo G., Testa G., Cacciatore F., Bonaduce D., Abete P. Oxidative stress, aging, and diseases. Clin. Interv. Aging. 2018;13:757–772. doi: 10.2147/CIA.S158513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sies H., Berndt C., Jones D.P. Oxidative stress. Annu. Rev. Biochem. 2017;86:715–748. doi: 10.1146/annurev-biochem-061516-045037. [DOI] [PubMed] [Google Scholar]
- 6.Davies M.J., Fu S., Wang H., Dean R.T. Stable markers of oxidant damage to proteins and their application in the study of human disease. Free Radic. Biol. Med. 1999;27:1151–1163. doi: 10.1016/s0891-5849(99)00206-3. [DOI] [PubMed] [Google Scholar]
- 7.Davies M.J. Protein oxidation and peroxidation. Biochem. J. 2016;473:805–825. doi: 10.1042/BJ20151227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Droge W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 9.Spickett C.M., Pitt A.R. Protein oxidation: role in signalling and detection by mass spectrometry. Amino Acids. 2012;42:5–21. doi: 10.1007/s00726-010-0585-4. [DOI] [PubMed] [Google Scholar]
- 10.Gil L., Siems W., Mazurek B., Gross J., Schroeder P., Voss P., Grune T. Age-associated analysis of oxidative stress parameters in human plasma and erythrocytes. Free Radic. Res. 2006;40:495–505. doi: 10.1080/10715760600592962. [DOI] [PubMed] [Google Scholar]
- 11.Mutlu-Turkoglu U., Ilhan E., Oztezcan S., Kuru A., Aykac-Toker G., Uysal M. Age-related increases in plasma malondialdehyde and protein carbonyl levels and lymphocyte DNA damage in elderly subjects. Clin. Biochem. 2003;36:397–400. doi: 10.1016/s0009-9120(03)00035-3. [DOI] [PubMed] [Google Scholar]
- 12.Smallwood M.J., Nissim A., Knight A.R., Whiteman M., Haigh R., Winyard P.G. Oxidative stress in autoimmune rheumatic diseases. Free Radic. Biol. Med. 2018;125:3–14. doi: 10.1016/j.freeradbiomed.2018.05.086. [DOI] [PubMed] [Google Scholar]
- 13.Uttara B., Singh A.V., Zamboni P., Mahajan R.T. Oxidative stress and neurodegenerative diseases: a review of upstream and downstream antioxidant therapeutic options. Curr. Neuropharmacol. 2009;7:65–74. doi: 10.2174/157015909787602823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bickers D.R., Athar M. Oxidative stress in the pathogenesis of skin disease. J. Invest. Dermatol. 2006;126:2565–2575. doi: 10.1038/sj.jid.5700340. [DOI] [PubMed] [Google Scholar]
- 15.Ryan B.J., Nissim A., Winyard P.G. Oxidative post-translational modifications and their involvement in the pathogenesis of autoimmune diseases. Redox Biol. 2014;2:715–724. doi: 10.1016/j.redox.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trujillo M., Alvarez B., Radi R. One-and two-electron oxidation of thiols: mechanisms, kinetics and biological fates. Free Radic. Res. 2016;50:150–171. doi: 10.3109/10715762.2015.1089988. [DOI] [PubMed] [Google Scholar]
- 17.Tu B.P., Weissman J.S. Oxidative protein folding in eukaryotes mechanisms and consequences. JCB (J. Cell Biol.) 2004;164:341–346. doi: 10.1083/jcb.200311055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Curis E., Nicolis I., Moinard C., Osowska S., Zerrouk N., Bénazeth S., Cynober L. Almost all about citrulline in mammals. Amino Acids. 2005;29:177. doi: 10.1007/s00726-005-0235-4. [DOI] [PubMed] [Google Scholar]
- 19.Xiong Y., Uys J.D., Tew K.D., Townsend D.M. S-glutathionylation: from molecular mechanisms to health outcomes. Antioxidants Redox Signal. 2011;15:233–270. doi: 10.1089/ars.2010.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vogt W. Oxidation of methionyl residues in proteins: tools, targets, and reversal. Free Radic. Biol. Med. 1995;18:93–105. doi: 10.1016/0891-5849(94)00158-g. [DOI] [PubMed] [Google Scholar]
- 21.Ahmed N., Thornalley P. Portland Press Ltd; 2003. Quantitative Screening of Protein Biomarkers of Early Glycation, Advanced Glycation, Oxidation and Nitrosation in Cellular and Extracellular Proteins by Tandem Mass Spectrometry Multiple Reaction Monitoring. [DOI] [PubMed] [Google Scholar]
- 22.Correia M., Neves-Petersen M.T., Jeppesen P.B., Gregersen S., Petersen S.B. UV-light exposure of insulin: pharmaceutical implications upon covalent insulin dityrosine dimerization and disulphide bond photolysis. PLoS One. 2012;7 doi: 10.1371/journal.pone.0050733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinecke J.W., Li W., Francis G.A., Goldstein J.A. Tyrosyl radical generated by myeloperoxidase catalyzes the oxidative cross-linking of proteins. J. Clin. Invest. 1993;91:2866–2872. doi: 10.1172/JCI116531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hensley K., Maidt M., Pye Q., Stewart C., Wack M., Tabatabaie T., Floyd R. Quantitation of protein-bound 3-nitrotyrosine and 3, 4-dihydroxyphenylalanine by high-performance liquid chromatography with electrochemical array detection. Anal. Biochem. 1997;251:187–195. doi: 10.1006/abio.1997.2281. [DOI] [PubMed] [Google Scholar]
- 25.Mujika J.I., Uranga J., Matxain J.M. Computational study on the attack of. OH radicals on aromatic amino acids. Chem. A Eur. J. 2013;19:6862–6873. doi: 10.1002/chem.201203862. [DOI] [PubMed] [Google Scholar]
- 26.Josimović L., Janković I., Jovanović S. Radiation induced decomposition of tryptophan in the presence of oxygen. Radiat. Phys. Chem. 1993;41:835–841. [Google Scholar]
- 27.Schöneich C. Mechanisms of metal-catalyzed oxidation of histidine to 2-oxo-histidine in peptides and proteins. J. Pharmaceut. Biomed. Anal. 2000;21:1093–1097. doi: 10.1016/s0731-7085(99)00182-x. [DOI] [PubMed] [Google Scholar]
- 28.Uchida K., Kawakishi S. 2‐Oxo‐histidine as a novel biological marker for oxidatively modified proteins. FEBS Lett. 1993;332:208–210. doi: 10.1016/0014-5793(93)80632-5. [DOI] [PubMed] [Google Scholar]
- 29.Lima M., Baynes J. 2013. Glycation. [Google Scholar]
- 30.Ledl F., Schleicher E. New aspects of the Maillard reaction in foods and in the human body. Angew Chem. Int. Ed. Engl. 1990;29:565–594. [Google Scholar]
- 31.Henning C., Glomb M.A. Pathways of the Maillard reaction under physiological conditions. Glycoconj. J. 2016;33:499–512. doi: 10.1007/s10719-016-9694-y. [DOI] [PubMed] [Google Scholar]
- 32.Thorpe S.R., Baynes J.W. vol. 1245. Elsevier; 2002. CML: a brief history; pp. 91–99. (International Congress Series). [Google Scholar]
- 33.Nguyen H.T., Van der Fels-Klerx H., Van Boekel M. N ε-(carboxymethyl) lysine: a review on analytical methods, formation, and occurrence in processed food, and health impact. Food Rev. Int. 2014;30:36–52. [Google Scholar]
- 34.Delgado-Andrade C. Carboxymethyl-lysine: thirty years of investigation in the field of AGE formation. Food Funct. 2016;7:46–57. doi: 10.1039/c5fo00918a. [DOI] [PubMed] [Google Scholar]
- 35.Ahmed M.U., Thorpe S.R., Baynes J.W. Identification of N epsilon-carboxymethyllysine as a degradation product of fructoselysine in glycated protein. J. Biol. Chem. 1986;261:4889–4894. [PubMed] [Google Scholar]
- 36.Glomb M.A., Monnier V.M. Mechanism of protein modification by glyoxal and glycolaldehyde, reactive intermediates of the Maillard reaction. J. Biol. Chem. 1995;270:10017–10026. doi: 10.1074/jbc.270.17.10017. [DOI] [PubMed] [Google Scholar]
- 37.Rabbani N., Thornalley P.J. Dicarbonyl stress in cell and tissue dysfunction contributing to ageing and disease. Biochem. Biophys. Res. Commun. 2015;458:221–226. doi: 10.1016/j.bbrc.2015.01.140. [DOI] [PubMed] [Google Scholar]
- 38.Baldensperger T., Jost T., Zipprich A., Glomb M.A. Novel α-oxoamide advanced-glycation endproducts within the N 6-carboxymethyl lysine and N 6-carboxyethyl lysine reaction cascades. J. Agric. Food Chem. 2018;66:1898–1906. doi: 10.1021/acs.jafc.7b05813. [DOI] [PubMed] [Google Scholar]
- 39.Sell D.R., Nagaraj R.H., Grandhee S.K., Odetti P., Lapolla A., Fogarty J., Monnier V.M. Pentosidine: a molecular marker for the cumulative damage to proteins in diabetes, aging, and uremia. Diabetes Metab. Rev. 1991;7:239–251. doi: 10.1002/dmr.5610070404. [DOI] [PubMed] [Google Scholar]
- 40.Sell D.R., Monnier V.M. Structure elucidation of a senescence cross-link from human extracellular matrix. Implication of pentoses in the aging process. J. Biol. Chem. 1989;264:21597–21602. [PubMed] [Google Scholar]
- 41.Dyer D.G., Blackledge J.A., Thorpe S.R., Baynes J.W. Formation of pentosidine during nonenzymatic browning of proteins by glucose. Identification of glucose and other carbohydrates as possible precursors of pentosidine in vivo. J. Biol. Chem. 1991;266:11654–11660. [PubMed] [Google Scholar]
- 42.Baker J.R., Zyzak D.V., Thorpe S.R., Baynes J.W. Chemistry of the fructosamine assay: D-glucosone is the product of oxidation of Amadori compounds. Clin. Chem. 1994;40:1950–1955. [PubMed] [Google Scholar]
- 43.Henning C., Smuda M., Girndt M., Ulrich C., Glomb M.A. Molecular basis of maillard amide-advanced glycation end product (AGE) formation in vivo. J. Biol. Chem. 2011;286:44350–44356. doi: 10.1074/jbc.M111.282442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baldensperger T., Sanzo S.D., Ori A., Glomb M.A. Quantitation of reactive acyl-CoA species mediated protein acylation by HPLC–MS/MS. Anal. Chem. 2019;91:12336–12343. doi: 10.1021/acs.analchem.9b02656. [DOI] [PubMed] [Google Scholar]
- 45.Baldensperger T., Eggen M., Kappen J., Winterhalter P.R., Pfirrmann T., Glomb M.A. Comprehensive analysis of posttranslational protein modifications in aging of subcellular compartments. Sci. Rep. 2020;10:1–11. doi: 10.1038/s41598-020-64265-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang T., Zhou X., Taghizadeh K., Dong M., Dedon P.C. N-formylation of lysine in histone proteins as a secondary modification arising from oxidative DNA damage. Proc. Natl. Acad. Sci. Unit. States Am. 2007;104:60–65. doi: 10.1073/pnas.0606775103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edrissi B., Taghizadeh K., Dedon P.C. Quantitative analysis of histone modifications: formaldehyde is a source of pathological N 6-formyllysine that is refractory to histone deacetylases. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu M.-X., Requena J.R., Jenkins A.J., Lyons T.J., Baynes J.W., Thorpe S.R. The advanced glycation end product, N-(carboxymethyl) lysine, is a product of both lipid peroxidation and glycoxidation reactions. J. Biol. Chem. 1996;271:9982–9986. doi: 10.1074/jbc.271.17.9982. [DOI] [PubMed] [Google Scholar]
- 49.Gianazza E., Brioschi M., Fernandez A.M., Banfi C. Lipoxidation in cardiovascular diseases. Redox Biol. 2019;23:101119. doi: 10.1016/j.redox.2019.101119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vistoli G., De Maddis D., Cipak A., Zarkovic N., Carini M., Aldini G. Advanced glycoxidation and lipoxidation end products (AGEs and ALEs): an overview of their mechanisms of formation. Free Radic. Res. 2013;47:3–27. doi: 10.3109/10715762.2013.815348. [DOI] [PubMed] [Google Scholar]
- 51.Esterbauer H., Schaur R.J., Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic. Biol. Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 52.Uchida K., Sakai K., Itakura K., Osawa T., Toyokuni S. Protein modification by lipid peroxidation products: formation of malondialdehyde-derivednε-(2-propenal) lysine in proteins. Arch. Biochem. Biophys. 1997;346:45–52. doi: 10.1006/abbi.1997.0266. [DOI] [PubMed] [Google Scholar]
- 53.Nair V., Vietti D.E., Cooper C.S. Degenerative chemistry of malondialdehyde. Structure, stereochemistry, and kinetics of formation of enaminals from reaction with amino acids. J. Am. Chem. Soc. 1981;103:3030–3036. [Google Scholar]
- 54.Kikugawa K., Ido Y. Studies on peroxidized lipids. V. Formation and characterization of 1, 4-dihydropyridine-3, 5-dicarbaldehydes as model of fluorescent components in lipofuscin. Lipids. 1984;19:600–608. doi: 10.1007/BF02534718. [DOI] [PubMed] [Google Scholar]
- 55.Itakura K., Uchida K., Osawa T. A novel fluorescent malondialdehyde-lysine adduct. Chem. Phys. Lipids. 1996;84:75–79. [Google Scholar]
- 56.Slatter D.A., Avery N.C., Bailey A.J. Identification of a new cross-link and unique histidine adduct from bovine serum albumin incubated with malondialdehyde. J. Biol. Chem. 2004;279:61–69. doi: 10.1074/jbc.M310608200. [DOI] [PubMed] [Google Scholar]
- 57.Requena J.R., Fu M.X., Ahmed M.U., Jenkins A.J., Lyons T.J., Baynes J.W., Thorpe S.R. Quantification of malondialdehyde and 4-hydroxynonenal adducts to lysine residues in native and oxidized human low-density lipoprotein. Biochem. J. 1997;322:317–325. doi: 10.1042/bj3220317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nadkarni D.V., Sayre L.M. Structural definition of early lysine and histidine adduction chemistry of 4-hydroxynonenal. Chem. Res. Toxicol. 1995;8:284–291. doi: 10.1021/tx00044a014. [DOI] [PubMed] [Google Scholar]
- 59.Sayre L.M., Arora P.K., Iyer R.S., Salomon R.G. Pyrrole formation from 4-hydroxynonenal and primary amines. Chem. Res. Toxicol. 1993;6:19–22. doi: 10.1021/tx00031a002. [DOI] [PubMed] [Google Scholar]
- 60.Uchida K., Stadtman E. Covalent attachment of 4-hydroxynonenal to glyceraldehyde-3-phosphate dehydrogenase. A possible involvement of intra-and intermolecular cross-linking reaction. J. Biol. Chem. 1993;268:6388–6393. [PubMed] [Google Scholar]
- 61.Cohn J.A., Tsai L., Friguet B., Szweda L.I. Chemical characterization of a protein-4-hydroxy-2-nonenal cross-link: immunochemical detection in mitochondria exposed to oxidative stress. Arch. Biochem. Biophys. 1996;328:158–164. doi: 10.1006/abbi.1996.0156. [DOI] [PubMed] [Google Scholar]
- 62.Xu G., Sayre L.M. Structural characterization of a 4-hydroxy-2-alkenal-derived fluorophore that contributes to lipoperoxidation-dependent protein cross-linking in aging and degenerative disease. Chem. Res. Toxicol. 1998;11:247–251. doi: 10.1021/tx980003d. [DOI] [PubMed] [Google Scholar]
- 63.Dalle-Donne I., Giustarini D., Colombo R., Rossi R., Milzani A. Protein carbonylation in human diseases. Trends Mol. Med. 2003;9:169–176. doi: 10.1016/s1471-4914(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 64.Requena J.R., Chao C.-C., Levine R.L., Stadtman E.R. Glutamic and aminoadipic semialdehydes are the main carbonyl products of metal-catalyzed oxidation of proteins. Proc. Natl. Acad. Sci. Unit. States Am. 2001;98:69–74. doi: 10.1073/pnas.011526698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Akagawa M., Sasaki D., Ishii Y., Kurota Y., Yotsu-Yamashita M., Uchida K., Suyama K. New method for the quantitative determination of major protein carbonyls, α-aminoadipic and γ-glutamic semialdehydes: investigation of the formation mechanism and chemical nature in vitro and in vivo. Chem. Res. Toxicol. 2006;19:1059–1065. doi: 10.1021/tx060026p. [DOI] [PubMed] [Google Scholar]
- 66.Stringfellow H.M., Jones M.R., Green M.C., Wilson A.K., Francisco J.S. Selectivity in ROS-induced peptide backbone bond cleavage. J. Phys. Chem. 2014;118:11399–11404. doi: 10.1021/jp508877m. [DOI] [PubMed] [Google Scholar]
- 67.Stadtman E., Levine R. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids. 2003;25:207–218. doi: 10.1007/s00726-003-0011-2. [DOI] [PubMed] [Google Scholar]
- 68.Uchida K., Kato Y., Kawakishi S. A novel mechanism for oxidative cleavage of prolyl peptides induced by the hydroxyl radical. Biochem. Biophys. Res. Commun. 1990;169:265–271. doi: 10.1016/0006-291x(90)91463-3. [DOI] [PubMed] [Google Scholar]
- 69.Murad F. Cyclic guanosine monophosphate as a mediator of vasodilation. J. Clin. Invest. 1986;78:1–5. doi: 10.1172/JCI112536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hess D.T., Matsumoto A., Kim S.-O., Marshall H.E., Stamler J.S. Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 71.White P.J., Charbonneau A., Cooney G.J., Marette A. Nitrosative modifications of protein and lipid signaling molecules by reactive nitrogen species. Am. J. Physiol. Endocrinol. Metab. 2010;299:E868–E878. doi: 10.1152/ajpendo.00510.2010. [DOI] [PubMed] [Google Scholar]
- 72.Radi R. Oxygen radicals, nitric oxide, and peroxynitrite: redox pathways in molecular medicine. Proc. Natl. Acad. Sci. Unit. States Am. 2018;115:5839–5848. doi: 10.1073/pnas.1804932115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hogg N. The biochemistry and physiology of S-nitrosothiols. Annu. Rev. Pharmacol. Toxicol. 2002;42:585–600. doi: 10.1146/annurev.pharmtox.42.092501.104328. [DOI] [PubMed] [Google Scholar]
- 74.Bartesaghi S., Radi R. Fundamentals on the biochemistry of peroxynitrite and protein tyrosine nitration. Redox Biol. 2018;14:618–625. doi: 10.1016/j.redox.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rogowska-Wrzesinska A., Wojdyla K., Nedic O., Baron C.P., Griffiths H.R. Analysis of protein carbonylation--pitfalls and promise in commonly used methods. Free Radic. Res. 2014;48:1145–1162. doi: 10.3109/10715762.2014.944868. [DOI] [PubMed] [Google Scholar]
- 76.Wang X., Liang S. Sample preparation for the analysis of membrane proteomes by mass spectrometry. Protein Cell. 2012;3:661–668. doi: 10.1007/s13238-012-2062-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Held J.M., Gibson B.W. Regulatory control or oxidative damage? Proteomic approaches to interrogate the role of cysteine oxidation status in biological processes. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.R111.013037. R111 013037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wong C.M., Cheema A.K., Zhang L., Suzuki Y.J. Protein carbonylation as a novel mechanism in redox signaling. Circ. Res. 2008;102:310–318. doi: 10.1161/CIRCRESAHA.107.159814. [DOI] [PubMed] [Google Scholar]
- 79.Verrastro I., Pasha S., Jensen K.T., Pitt A.R., Spickett C.M. Mass spectrometry-based methods for identifying oxidized proteins in disease: advances and challenges. Biomolecules. 2015;5:378–411. doi: 10.3390/biom5020378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Amoresano A., Chiappetta G., Pucci P., D'Ischia M., Marino G. Bidimensional tandem mass spectrometry for selective identification of nitration sites in proteins. Anal. Chem. 2007;79:2109–2117. doi: 10.1021/ac0620361. [DOI] [PubMed] [Google Scholar]
- 81.Levine R.L., Garland D., Oliver C.N., Amici A., Climent I., Lenz A.G., Ahn B.W., Shaltiel S., Stadtman E.R. Determination of carbonyl content in oxidatively modified proteins. Methods Enzymol. 1990;186:464–478. doi: 10.1016/0076-6879(90)86141-h. [DOI] [PubMed] [Google Scholar]
- 82.Weber D., Davies M.J., Grune T. Determination of protein carbonyls in plasma, cell extracts, tissue homogenates, isolated proteins: focus on sample preparation and derivatization conditions. Redox Biol. 2015;5:367–380. doi: 10.1016/j.redox.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mesquita C.S., Oliveira R., Bento F., Geraldo D., Rodrigues J.V., Marcos J.C. Simplified 2,4-dinitrophenylhydrazine spectrophotometric assay for quantification of carbonyls in oxidized proteins. Anal. Biochem. 2014;458:69–71. doi: 10.1016/j.ab.2014.04.034. [DOI] [PubMed] [Google Scholar]
- 84.Hellwig M. The chemistry of protein oxidation in food. Angew Chem. Int. Ed. Engl. 2019;58:16742–16763. doi: 10.1002/anie.201814144. [DOI] [PubMed] [Google Scholar]
- 85.Swiderska M., Maciejczyk M., Zalewska A., Pogorzelska J., Flisiak R., Chabowski A. Oxidative stress biomarkers in the serum and plasma of patients with non-alcoholic fatty liver disease (NAFLD). Can plasma AGE be a marker of NAFLD? Oxidative stress biomarkers in NAFLD patients. Free Radic. Res. 2019;53:841–850. doi: 10.1080/10715762.2019.1635691. [DOI] [PubMed] [Google Scholar]
- 86.Augustyniak E., Adam A., Wojdyla K., Rogowska-Wrzesinska A., Willetts R., Korkmaz A., Atalay M., Weber D., Grune T., Borsa C., Gradinaru D., Chand Bollineni R., Fedorova M., Griffiths H.R. Validation of protein carbonyl measurement: a multi-centre study. Redox Biol. 2015;4:149–157. doi: 10.1016/j.redox.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ahmad P., Tantry I.Q., Ali A., Siddiqui S.A., Rehman S.U., Waris S., Jairajpuri M.A. Structural alteration in hypochlorous acid modified antithrombin indicates generation of neo-epitopes. Arch. Biochem. Biophys. 2020;685:108332. doi: 10.1016/j.abb.2020.108332. [DOI] [PubMed] [Google Scholar]
- 88.Olszowski S., Olszowska E., Kusior D., Piwowarczyk M., Stelmaszynska T. Hypochlorite action on plasma fibronectin promotes its extended conformation in complexes with antibodies. J. Protein Chem. 2003;22:449–456. doi: 10.1023/b:jopc.0000005460.94172.1d. [DOI] [PubMed] [Google Scholar]
- 89.Mitsuhashi T., Li Y.M., Fishbane S., Vlassara H. Depletion of reactive advanced glycation endproducts from diabetic uremic sera using a lysozyme-linked matrix. J. Clin. Invest. 1997;100:847–854. doi: 10.1172/JCI119600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Seo Y.H., Carroll K.S. Profiling protein thiol oxidation in tumor cells using sulfenic acid-specific antibodies. Proc. Natl. Acad. Sci. U. S. A. 2009;106:16163–16168. doi: 10.1073/pnas.0903015106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Buss H., Chan T.P., Sluis K.B., Domigan N.M., Winterbourn C.C. Protein carbonyl measurement by a sensitive ELISA method. Free Radic. Biol. Med. 1997;23:361–366. doi: 10.1016/s0891-5849(97)00104-4. [DOI] [PubMed] [Google Scholar]
- 92.Teixeira D., Fernandes R., Prudencio C., Vieira M. 3-Nitrotyrosine quantification methods: current concepts and future challenges. Biochimie. 2016;125:1–11. doi: 10.1016/j.biochi.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 93.Anderson D.M., Nye-Wood M.G., Rose K.L., Donaldson P.J., Grey A.C., Schey K.L. MALDI imaging mass spectrometry of beta- and gamma-crystallins in the ocular lens. J. Mass Spectrom. 2020;55:e4473. doi: 10.1002/jms.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jin Y., Diffee G.M., Colman R.J., Anderson R.M., Ge Y. Top-down mass spectrometry of Sarcomeric protein post-translational modifications from non-human primate skeletal muscle. J. Am. Soc. Mass Spectrom. 2019;30:2460–2469. doi: 10.1007/s13361-019-02139-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Melby J.A., Jin Y., Lin Z., Tucholski T., Wu Z., Gregorich Z.R., Diffee G.M., Ge Y. Top-down proteomics reveals myofilament proteoform heterogeneity among various rat skeletal muscle tissues. J. Proteome Res. 2020;19:446–454. doi: 10.1021/acs.jproteome.9b00623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Srzentic K., Fornelli L., Tsybin Y.O., Loo J.A., Seckler H., Agar J.N., Anderson L.C., Bai D.L., Beck A., Brodbelt J.S., van der Burgt Y.E.M., Chamot-Rooke J., Chatterjee S., Chen Y., Clarke D.J., Danis P.O., Diedrich J.K., D'Ippolito R.A., Dupre M., Gasilova N., Ge Y., Goo Y.A., Goodlett D.R., Greer S., Haselmann K.F., He L., Hendrickson C.L., Hinkle J.D., Holt M.V., Hughes S., Hunt D.F., Kelleher N.L., Kozhinov A.N., Lin Z., Malosse C., Marshall A.G., Menin L., Millikin R.J., Nagornov K.O., Nicolardi S., Pasa-Tolic L., Pengelley S., Quebbemann N.R., Resemann A., Sandoval W., Sarin R., Schmitt N.D., Shabanowitz J., Shaw J.B., Shortreed M.R., Smith L.M., Sobott F., Suckau D., Toby T., Weisbrod C.R., Wildburger N.C., Yates J.R., 3rd, Yoon S.H., Young N.L., Zhou M. Interlaboratory study for characterizing monoclonal antibodies by top-down and middle-down mass spectrometry. J. Am. Soc. Mass Spectrom. 2020;31:1783–1802. doi: 10.1021/jasms.0c00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Siuti N., Kelleher N.L. Decoding protein modifications using top-down mass spectrometry. Nat. Methods. 2007;4:817–821. doi: 10.1038/nmeth1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mannaa A., Hanisch F.G. Redox proteomes in human physiology and disease mechanisms. J. Proteome Res. 2020;19:1–17. doi: 10.1021/acs.jproteome.9b00586. [DOI] [PubMed] [Google Scholar]
- 99.Garcia-Santamarina S., Boronat S., Domenech A., Ayte J., Molina H., Hidalgo E. Monitoring in vivo reversible cysteine oxidation in proteins using ICAT and mass spectrometry. Nat. Protoc. 2014;9:1131–1145. doi: 10.1038/nprot.2014.065. [DOI] [PubMed] [Google Scholar]
- 100.Abello N., Kerstjens H.A., Postma D.S., Bischoff R. Protein tyrosine nitration: selectivity, physicochemical and biological consequences, denitration, and proteomics methods for the identification of tyrosine-nitrated proteins. J. Proteome Res. 2009;8:3222–3238. doi: 10.1021/pr900039c. [DOI] [PubMed] [Google Scholar]
- 101.Nambo-Venegas R., Valdez-Vargas C., Cisneros B., Palacios-Gonzalez B., Vela-Amieva M., Ibarra-Gonzalez I., Cerecedo-Zapata C.M., Martinez-Cruz E., Cortes H., Reyes-Grajeda J.P., Magana J.J. Altered plasma acylcarnitines and amino acids profile in Spinocerebellar ataxia type 7. Biomolecules. 2020;10 doi: 10.3390/biom10030390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jambor A., Molnar-Perl I. Quantitation of amino acids in plasma by high performance liquid chromatography: simultaneous deproteinization and derivatization with 9-fluorenylmethyloxycarbonyl chloride. J. Chromatogr. A. 2009;1216:6218–6223. doi: 10.1016/j.chroma.2009.06.083. [DOI] [PubMed] [Google Scholar]
- 103.Li Y., Tang A.G., Mu S. HPLC-FLD determination of serum aromatic amino acids: application in chronic kidney disease patients. Clin. Chim. Acta. 2011;412:1032–1035. doi: 10.1016/j.cca.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 104.Bartolomeo M.P., Maisano F. Validation of a reversed-phase HPLC method for quantitative amino acid analysis. J. Biomol. Tech. 2006;17:131–137. [PMC free article] [PubMed] [Google Scholar]
- 105.Tapuhi Y., Schmidt D.E., Lindner W., Karger B.L. Dansylation of amino acids for high-performance liquid chromatography analysis. Anal. Biochem. 1981;115:123–129. doi: 10.1016/0003-2697(81)90534-0. [DOI] [PubMed] [Google Scholar]
- 106.Harder U., Koletzko B., Peissner W. Quantification of 22 plasma amino acids combining derivatization and ion-pair LC-MS/MS. J Chromatogr B Analyt Technol Biomed Life Sci. 2011;879:495–504. doi: 10.1016/j.jchromb.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 107.Fountoulakis M., Lahm H.W. Hydrolysis and amino acid composition of proteins. J. Chromatogr. A. 1998;826:109–134. doi: 10.1016/s0021-9673(98)00721-3. [DOI] [PubMed] [Google Scholar]
- 108.Chiu C.J., Rabbani N., Rowan S., Chang M.L., Sawyer S., Hu F.B., Willett W., Thornalley P.J., Anwar A., Bar L., Kang J.H., Taylor A. Studies of advanced glycation end products and oxidation biomarkers for type 2 diabetes. Biofactors. 2018;44:281–288. doi: 10.1002/biof.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stadtman E.R. Protein oxidation in aging and age-related diseases. Ann. N. Y. Acad. Sci. 2001;928:22–38. doi: 10.1111/j.1749-6632.2001.tb05632.x. [DOI] [PubMed] [Google Scholar]
- 110.Garcia-Garcia A., Rodriguez-Rocha H., Madayiputhiya N., Pappa A., Panayiotidis M.I., Franco R. Biomarkers of protein oxidation in human disease. Curr. Mol. Med. 2012;12:681–697. doi: 10.2174/156652412800792543. [DOI] [PubMed] [Google Scholar]
- 111.Dalle-Donne I., Rossi R., Colombo R., Giustarini D., Milzani A. Biomarkers of oxidative damage in human disease. Clin. Chem. 2006;52:601–623. doi: 10.1373/clinchem.2005.061408. [DOI] [PubMed] [Google Scholar]
- 112.Mayeux R. Biomarkers: potential uses and limitations. NeuroRx. 2004;1:182–188. doi: 10.1602/neurorx.1.2.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dudley J.T., Butte A.J. Identification of discriminating biomarkers for human disease using integrative network biology. Pac Symp Biocomput. 2009:27–38. [PMC free article] [PubMed] [Google Scholar]
- 114.Strimbu K., Tavel J.A. What are biomarkers? Curr. Opin. HIV AIDS. 2010;5:463–466. doi: 10.1097/COH.0b013e32833ed177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Davis K.D., Aghaeepour N., Ahn A.H., Angst M.S., Borsook D., Brenton A., Burczynski M.E., Crean C., Edwards R., Gaudilliere B., Hergenroeder G.W., Iadarola M.J., Iyengar S., Jiang Y., Kong J.T., Mackey S., Saab C.Y., Sang C.N., Scholz J., Segerdahl M., Tracey I., Veasley C., Wang J., Wager T.D., Wasan A.D., Pelleymounter M.A. Discovery and validation of biomarkers to aid the development of safe and effective pain therapeutics: challenges and opportunities. Nat. Rev. Neurol. 2020;16:381–400. doi: 10.1038/s41582-020-0362-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Frijhoff J., Winyard P.G., Zarkovic N., Davies S.S., Stocker R., Cheng D., Knight A.R., Taylor E.L., Oettrich J., Ruskovska T., Gasparovic A.C., Cuadrado A., Weber D., Poulsen H.E., Grune T., Schmidt H.H., Ghezzi P. Clinical relevance of biomarkers of oxidative stress. Antioxidants Redox Signal. 2015;23:1144–1170. doi: 10.1089/ars.2015.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Marrocco I., Altieri F., Peluso I. Measurement and clinical significance of biomarkers of oxidative stress in humans. Oxid Med Cell Longev. 2017 doi: 10.1155/2017/6501046. 2017, 6501046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bin P., Huang R., Zhou X. Oxidation resistance of the sulfur amino acids: methionine and cysteine. BioMed Res. Int. 2017;2017:9584932. doi: 10.1155/2017/9584932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brosnan J.T., Brosnan M.E. The sulfur-containing amino acids: an overview. J. Nutr. 2006;136:1636S–1640S. doi: 10.1093/jn/136.6.1636S. [DOI] [PubMed] [Google Scholar]
- 120.Paul B.D., Sbodio J.I., Snyder S.H. Cysteine metabolism in neuronal redox homeostasis. Trends Pharmacol. Sci. 2018;39:513–524. doi: 10.1016/j.tips.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Oliveira P.V.S., Laurindo F.R.M. Implications of plasma thiol redox in disease. Clin. Sci. (Lond.) 2018;132:1257–1280. doi: 10.1042/CS20180157. [DOI] [PubMed] [Google Scholar]
- 122.Poole L.B. The basics of thiols and cysteines in redox biology and chemistry. Free Radic. Biol. Med. 2015;80:148–157. doi: 10.1016/j.freeradbiomed.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Turell L., Zeida A., Trujillo M. Mechanisms and consequences of protein cysteine oxidation: the role of the initial short-lived intermediates. Essays Biochem. 2020;64:55–66. doi: 10.1042/EBC20190053. [DOI] [PubMed] [Google Scholar]
- 124.Fu X., Cate S.A., Dominguez M., Osborn W., Ozpolat T., Konkle B.A., Chen J., Lopez J.A. Cysteine disulfides (Cys-ss-X) as sensitive plasma biomarkers of oxidative stress. Sci. Rep. 2019;9:115. doi: 10.1038/s41598-018-35566-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Paramasivan S., Adav S.S., Ngan S.C., Dalan R., Leow M.K., Ho H.H., Sze S.K. Serum albumin cysteine trioxidation is a potential oxidative stress biomarker of type 2 diabetes mellitus. Sci. Rep. 2020;10:6475. doi: 10.1038/s41598-020-62341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gu S.X., Stevens J.W., Lentz S.R. Regulation of thrombosis and vascular function by protein methionine oxidation. Blood. 2015;125:3851–3859. doi: 10.1182/blood-2015-01-544676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lim J.M., Kim G., Levine R.L. Methionine in proteins: it's not just for protein initiation anymore. Neurochem. Res. 2019;44:247–257. doi: 10.1007/s11064-017-2460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Beisswenger P.J., Howell S., Mackenzie T., Corstjens H., Muizzuddin N., Matsui M.S. Two fluorescent wavelengths, 440(ex)/520(em) nm and 370(ex)/440(em) nm, reflect advanced glycation and oxidation end products in human skin without diabetes. Diabetes Technol. Therapeut. 2012;14:285–292. doi: 10.1089/dia.2011.0108. [DOI] [PubMed] [Google Scholar]
- 129.Wells-Knecht M.C., Lyons T.J., McCance D.R., Thorpe S.R., Baynes J.W. Age-dependent increase in ortho-tyrosine and methionine sulfoxide in human skin collagen is not accelerated in diabetes. Evidence against a generalized increase in oxidative stress in diabetes. J. Clin. Invest. 1997;100:839–846. doi: 10.1172/JCI119599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Deng Y., Marsh B.M., Moskovitz J. Increased levels of protein-methionine sulfoxide in plasma correlate with a shift from a mild cognitive impairment to an alzheimer's disease stage. Innov Clin Neurosci. 2019;16:29–31. [PMC free article] [PubMed] [Google Scholar]
- 131.Zhao Y., Zhang L., Ouyang X., Jiang Z., Xie Z., Fan L., Zhu D., Li L. Advanced oxidation protein products play critical roles in liver diseases. Eur. J. Clin. Invest. 2019 doi: 10.1111/eci.13098. [DOI] [PubMed] [Google Scholar]
- 132.Martinez-Sanchez G., Giuliani A., Perez-Davison G., Leon-Fernandez O.S. Oxidized proteins and their contribution to redox homeostasis. Redox Rep. 2005;10:175–185. doi: 10.1179/135100005X57382. [DOI] [PubMed] [Google Scholar]
- 133.Witko-Sarsat V., Friedlander M., Capeillere-Blandin C., Nguyen-Khoa T., Nguyen A.T., Zingraff J., Jungers P., Descamps-Latscha B. Advanced oxidation protein products as a novel marker of oxidative stress in uremia. Kidney Int. 1996;49:1304–1313. doi: 10.1038/ki.1996.186. [DOI] [PubMed] [Google Scholar]
- 134.Gelisgen R., Genc H., Kayali R., Oncul M., Benian A., Guralp O., Uludag S., Cakatay U., Albayrak M., Uzun H. Protein oxidation markers in women with and without gestational diabetes mellitus: a possible relation with paraoxonase activity. Diabetes Res. Clin. Pract. 2011;94:404–409. doi: 10.1016/j.diabres.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 135.Taylor E.L., Armstrong K.R., Perrett D., Hattersley A.T., Winyard P.G. Optimisation of an advanced oxidation protein products assay: its application to studies of oxidative stress in diabetes mellitus. Oxid Med Cell Longev. 2015:496271. doi: 10.1155/2015/496271. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Haberka M., Banska-Kisiel K., Bergler-Czop B., Biedron M., Brzezinska-Wcislo L., Okopien B., Gasior Z. Mild to moderate psoriasis is associated with oxidative stress, subclinical atherosclerosis, and endothelial dysfunction. Pol. Arch. Intern. Med. 2018;128:434–439. doi: 10.20452/pamw.4301. [DOI] [PubMed] [Google Scholar]
- 137.Yazici C., Kose K., Utas S., Tanrikulu E., Taslidere N. A novel approach in psoriasis: first usage of known protein oxidation markers to prove oxidative stress. Arch. Dermatol. Res. 2016;308:207–212. doi: 10.1007/s00403-016-1624-0. [DOI] [PubMed] [Google Scholar]
- 138.Skoie I.M., Dalen I., Omdal R., Jonsson G. Malondialdehyde and advanced oxidation protein products are not increased in psoriasis: a controlled study. Arch. Dermatol. Res. 2019;311:299–308. doi: 10.1007/s00403-019-01903-2. [DOI] [PubMed] [Google Scholar]
- 139.Komosinska-Vassev K., Olczyk P., Winsz-Szczotka K., Kuznik-Trocha K., Klimek K., Olczyk K. Age- and gender-related alteration in plasma advanced oxidation protein products (AOPP) and glycosaminoglycan (GAG) concentrations in physiological ageing. Clin. Chem. Lab. Med. 2012;50:557–563. doi: 10.1515/cclm.2011.789. [DOI] [PubMed] [Google Scholar]
- 140.Qing Z., Ling-Ling E., Dong-Sheng W., Hong-Chen L. Relationship of advanced oxidative protein products in human saliva and plasma: age- and gender-related changes and stability during storage. Free Radic. Res. 2012;46:1201–1206. doi: 10.3109/10715762.2012.700113. [DOI] [PubMed] [Google Scholar]
- 141.Maciejczyk M., Zalewska A., Ladny J.R. Salivary antioxidant barrier, redox status, and oxidative damage to proteins and lipids in healthy children, adults, and the elderly. Oxid Med Cell Longev. 2019 doi: 10.1155/2019/4393460. 2019, 4393460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pennathur S., Vivekanandan-Giri A., Locy M.L., Kulkarni T., Zhi D., Zeng L., Byun J., de Andrade J.A., Thannickal V.J. Oxidative modifications of protein tyrosyl residues are increased in plasma of human subjects with interstitial lung disease. Am. J. Respir. Crit. Care Med. 2016;193:861–868. doi: 10.1164/rccm.201505-0992OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Anwar A., Abruzzo P.M., Pasha S., Rajpoot K., Bolotta A., Ghezzo A., Marini M., Posar A., Visconti P., Thornalley P.J., Rabbani N. Advanced glycation endproducts, dityrosine and arginine transporter dysfunction in autism - a source of biomarkers for clinical diagnosis. Mol. Autism. 2018;9:3. doi: 10.1186/s13229-017-0183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Campos C., Guzman R., Lopez-Fernandez E., Casado A. Urinary biomarkers of oxidative/nitrosative stress in healthy smokers. Inhal. Toxicol. 2011;23:148–156. doi: 10.3109/08958378.2011.554460. [DOI] [PubMed] [Google Scholar]
- 145.Al-Hilaly Y.K., Williams T.L., Stewart-Parker M., Ford L., Skaria E., Cole M., Bucher W.G., Morris K.L., Sada A.A., Thorpe J.R., Serpell L.C. A central role for dityrosine crosslinking of Amyloid-beta in Alzheimer's disease. Acta Neuropathol Commun. 2013;1:83. doi: 10.1186/2051-5960-1-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cecarini V., Gee J., Fioretti E., Amici M., Angeletti M., Eleuteri A.M., Keller J.N. Protein oxidation and cellular homeostasis: emphasis on metabolism. Biochim. Biophys. Acta. 2007;1773:93–104. doi: 10.1016/j.bbamcr.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 147.Nguyen A.T., Donaldson R.P. Metal-catalyzed oxidation induces carbonylation of peroxisomal proteins and loss of enzymatic activities. Arch. Biochem. Biophys. 2005;439:25–31. doi: 10.1016/j.abb.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 148.Wang Z., Wang Y., Liu H., Che Y., Xu Y., E L. Age-related variations of protein carbonyls in human saliva and plasma: is saliva protein carbonyls an alternative biomarker of aging? Age (Dordr) 2015;37:9781. doi: 10.1007/s11357-015-9781-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Goycheva P., Nikolova G., Ivanova M., Kundurdzhiev T., Gadjeva V. Predictive value of some pro-oxidants in type 2 diabetes mellitus with vascular complications. Biosci Trends. 2019;13:168–175. doi: 10.5582/bst.2019.01020. [DOI] [PubMed] [Google Scholar]
- 150.Almogbel E., Rasheed N. Elevated levels of protein carbonylation in patients with diabetic nephropathy: therapeutic and diagnostic prospects. Am. J. Med. Sci. 2019;358:26–32. doi: 10.1016/j.amjms.2019.03.011. [DOI] [PubMed] [Google Scholar]
- 151.Sharma A., Weber D., Raupbach J., Dakal T.C., Fliessbach K., Ramirez A., Grune T., Wullner U. Advanced glycation end products and protein carbonyl levels in plasma reveal sex-specific differences in Parkinson's and Alzheimer's disease. Redox Biol. 2020;34:101546. doi: 10.1016/j.redox.2020.101546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nowotny K., Jung T., Hohn A., Weber D., Grune T. Advanced glycation end products and oxidative stress in type 2 diabetes mellitus. Biomolecules. 2015;5:194–222. doi: 10.3390/biom5010194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chaudhuri J., Bains Y., Guha S., Kahn A., Hall D., Bose N., Gugliucci A., Kapahi P. The role of advanced glycation end products in aging and metabolic diseases: bridging association and causality. Cell Metabol. 2018;28:337–352. doi: 10.1016/j.cmet.2018.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Perrone A., Giovino A., Benny J., Martinelli F. Advanced glycation end products (AGEs): biochemistry, signaling, analytical methods, and epigenetic effects. Oxid Med Cell Longev. 2020;2020:3818196. doi: 10.1155/2020/3818196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nedic O., Rattan S.I., Grune T., Trougakos I.P. Molecular effects of advanced glycation end products on cell signalling pathways, ageing and pathophysiology. Free Radic. Res. 2013;47(Suppl 1):28–38. doi: 10.3109/10715762.2013.806798. [DOI] [PubMed] [Google Scholar]
- 156.Goldberg T., Cai W., Peppa M., Dardaine V., Baliga B.S., Uribarri J., Vlassara H. Advanced glycoxidation end products in commonly consumed foods. J. Am. Diet Assoc. 2004;104:1287–1291. doi: 10.1016/j.jada.2004.05.214. [DOI] [PubMed] [Google Scholar]
- 157.Poulsen M.W., Hedegaard R.V., Andersen J.M., de Courten B., Bugel S., Nielsen J., Skibsted L.H., Dragsted L.O. Advanced glycation endproducts in food and their effects on health. Food Chem. Toxicol. 2013;60:10–37. doi: 10.1016/j.fct.2013.06.052. [DOI] [PubMed] [Google Scholar]
- 158.Ashraf J.M., Ahmad S., Choi I., Ahmad N., Farhan M., Tatyana G., Shahab U. Recent advances in detection of AGEs: immunochemical, bioanalytical and biochemical approaches. IUBMB Life. 2015;67:897–913. doi: 10.1002/iub.1450. [DOI] [PubMed] [Google Scholar]
- 159.Farhan S.S., Hussain S.A. Advanced glycation end products (AGEs) and their soluble receptors (sRAGE) as early predictors of reno-vascular complications in patients with uncontrolled type 2 diabetes mellitus. Diabetes Metab Syndr. 2019;13:2457–2461. doi: 10.1016/j.dsx.2019.06.019. [DOI] [PubMed] [Google Scholar]
- 160.Guerin-Dubourg A., Cournot M., Planesse C., Debussche X., Meilhac O., Rondeau P., Bourdon E. Association between fluorescent advanced glycation end-products and vascular complications in type 2 diabetic patients. BioMed Res. Int. 2017 doi: 10.1155/2017/7989180. 2017, 7989180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Stirban A., Heinemann L. Skin autofluorescence - a non-invasive measurement for assessing cardiovascular risk and risk of diabetes. Eur. Endocrinol. 2014;10:106–110. doi: 10.17925/EE.2014.10.02.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Da Moura Semedo C., Webb M., Waller H., Khunti K., Davies M. Skin autofluorescence, a non-invasive marker of advanced glycation end products: clinical relevance and limitations. Postgrad Med J. 2017;93:289–294. doi: 10.1136/postgradmedj-2016-134579. [DOI] [PubMed] [Google Scholar]
- 163.Thornalley P.J., Rabbani N. Detection of oxidized and glycated proteins in clinical samples using mass spectrometry--a user's perspective. Biochim. Biophys. Acta. 2014;1840:818–829. doi: 10.1016/j.bbagen.2013.03.025. [DOI] [PubMed] [Google Scholar]
- 164.de Vos L.C., Lefrandt J.D., Dullaart R.P., Zeebregts C.J., Smit A.J. Advanced glycation end products: an emerging biomarker for adverse outcome in patients with peripheral artery disease. Atherosclerosis. 2016;254:291–299. doi: 10.1016/j.atherosclerosis.2016.10.012. [DOI] [PubMed] [Google Scholar]
- 165.O'Grady K.L., Khosla S., Farr J.N., Bondar O.P., Atkinson E.J., Achenbach S.J., Eckhardt B.A., Thicke B.S., Tweed A.J., Volkman T.L., Drake M.T., Hines J.M., Singh R.J. Development and application of mass spectroscopy assays for nepsilon-(1-carboxymethyl)-L-lysine and pentosidine in renal failure and diabetes. J Appl Lab Med. 2020;5:558–568. doi: 10.1093/jalm/jfaa023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yoshida N., Okumura K., Aso Y. High serum pentosidine concentrations are associated with increased arterial stiffness and thickness in patients with type 2 diabetes. Metabolism. 2005;54:345–350. doi: 10.1016/j.metabol.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 167.Ghanem A.A., Elewa A., Arafa L.F. Pentosidine and N-carboxymethyl-lysine: biomarkers for type 2 diabetic retinopathy. Eur. J. Ophthalmol. 2011;21:48–54. doi: 10.5301/ejo.2010.4447. [DOI] [PubMed] [Google Scholar]
- 168.Wetzels S., Vanmierlo T., Scheijen J., van Horssen J., Amor S., Somers V., Schalkwijk C.G., Hendriks J.J.A., Wouters K. Methylglyoxal-derived advanced glycation endproducts accumulate in multiple sclerosis lesions. Front. Immunol. 2019;10:855. doi: 10.3389/fimmu.2019.00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Teichert T., Hellwig A., Pessler A., Hellwig M., Vossoughi M., Sugiri D., Vierkotter A., Schulte T., Freund J., Roden M., Hoffmann B., Schikowski T., Luckhaus C., Kramer U., Henle T., Herder C. Association between advanced glycation end products and impaired fasting glucose: results from the SALIA study. PLoS One. 2015;10 doi: 10.1371/journal.pone.0128293. e0128293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kida Y., Saito M., Shinohara A., Soshi S., Marumo K. Non-invasive skin autofluorescence, blood and urine assays of the advanced glycation end product (AGE) pentosidine as an indirect indicator of AGE content in human bone. BMC Muscoskel. Disord. 2019;20:627. doi: 10.1186/s12891-019-3011-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Paolillo F.R., Mattos V.S., Borghi-Silva A., Bagnato V.S., de Castro Neto J.C. Advanced glycation endproducts as biomarkers for risk of diabetes and cardiovascular diseases by skin autofluorescence: a noninvasive optical screening. Photobiomodul Photomed Laser Surg. 2019;37:168–174. doi: 10.1089/photob.2018.4563. [DOI] [PubMed] [Google Scholar]
- 172.Tsikas D. Assessment of lipid peroxidation by measuring malondialdehyde (MDA) and relatives in biological samples: analytical and biological challenges. Anal. Biochem. 2017;524:13–30. doi: 10.1016/j.ab.2016.10.021. [DOI] [PubMed] [Google Scholar]
- 173.Ramana K.V., Srivastava S., Singhal S.S. Lipid peroxidation products in human health and disease 2019. Oxid Med Cell Longev. 2019;2019:7147235. doi: 10.1155/2019/7147235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Negre-Salvayre A., Coatrieux C., Ingueneau C., Salvayre R. Advanced lipid peroxidation end products in oxidative damage to proteins. Potential role in diseases and therapeutic prospects for the inhibitors. Br. J. Pharmacol. 2008;153:6–20. doi: 10.1038/sj.bjp.0707395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Castro J.P., Jung T., Grune T., Siems W. 4-Hydroxynonenal (HNE) modified proteins in metabolic diseases. Free Radic. Biol. Med. 2017;111:309–315. doi: 10.1016/j.freeradbiomed.2016.10.497. [DOI] [PubMed] [Google Scholar]
- 176.Zarkovic K., Jakovcevic A., Zarkovic N. Contribution of the HNE-immunohistochemistry to modern pathological concepts of major human diseases. Free Radic. Biol. Med. 2017;111:110–126. doi: 10.1016/j.freeradbiomed.2016.12.009. [DOI] [PubMed] [Google Scholar]
- 177.Luczaj W., Gindzienska-Sieskiewicz E., Jarocka-Karpowicz I., Andrisic L., Sierakowski S., Zarkovic N., Waeg G., Skrzydlewska E. The onset of lipid peroxidation in rheumatoid arthritis: consequences and monitoring. Free Radic. Res. 2016;50:304–313. doi: 10.3109/10715762.2015.1112901. [DOI] [PubMed] [Google Scholar]
- 178.Luczaj W., Moniuszko A., Jarocka-Karpowicz I., Pancewicz S., Andrisic L., Zarkovic N., Skrzydlewska E. Tick-borne encephalitis--lipid peroxidation and its consequences. Scand. J. Clin. Lab. Invest. 2016;76:1–9. doi: 10.3109/00365513.2015.1084040. [DOI] [PubMed] [Google Scholar]
- 179.Zhang H., Forman H.J. 4-hydroxynonenal-mediated signaling and aging. Free Radic. Biol. Med. 2017;111:219–225. doi: 10.1016/j.freeradbiomed.2016.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shoeb M., Ansari N.H., Srivastava S.K., Ramana K.V. 4-Hydroxynonenal in the pathogenesis and progression of human diseases. Curr. Med. Chem. 2014;21:230–237. doi: 10.2174/09298673113209990181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Niki E. Biomarkers of lipid peroxidation in clinical material. Biochim. Biophys. Acta. 2014;1840:809–817. doi: 10.1016/j.bbagen.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 182.Grotto D., Santa Maria L.D., Boeira S., Valentini J., Charao M.F., Moro A.M., Nascimento P.C., Pomblum V.J., Garcia S.C. Rapid quantification of malondialdehyde in plasma by high performance liquid chromatography-visible detection. J. Pharmaceut. Biomed. Anal. 2007;43:619–624. doi: 10.1016/j.jpba.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 183.Zablocka-Slowinska K., Placzkowska S., Skorska K., Prescha A., Pawelczyk K., Porebska I., Kosacka M., Grajeta H. Oxidative stress in lung cancer patients is associated with altered serum markers of lipid metabolism. PLoS One. 2019;14 doi: 10.1371/journal.pone.0215246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Domijan A.M., Ralic J., Radic Brkanac S., Rumora L., Zanic-Grubisic T. Quantification of malondialdehyde by HPLC-FL - application to various biological samples. Biomed. Chromatogr. 2015;29:41–46. doi: 10.1002/bmc.3361. [DOI] [PubMed] [Google Scholar]
- 185.Medeiros M.S., Schumacher-Schuh A., Cardoso A.M., Bochi G.V., Baldissarelli J., Kegler A., Santana D., Chaves C.M., Schetinger M.R., Moresco R.N., Rieder C.R., Fighera M.R. Iron and oxidative stress in Parkinson's disease: an observational study of injury biomarkers. PLoS One. 2016;11 doi: 10.1371/journal.pone.0146129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Corpas F.J., Palma J.M., Del Rio L.A., Barroso J.B. Protein tyrosine nitration in higher plants grown under natural and stress conditions. Front. Plant Sci. 2013;4:29. doi: 10.3389/fpls.2013.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Radi R. Protein tyrosine nitration: biochemical mechanisms and structural basis of functional effects. Acc. Chem. Res. 2013;46:550–559. doi: 10.1021/ar300234c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Pacher P., Beckman J.S., Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol. Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ravassa S., Beaumont J., Huerta A., Barba J., Coma-Canella I., Gonzalez A., Lopez B., Diez J. Association of low GLP-1 with oxidative stress is related to cardiac disease and outcome in patients with type 2 diabetes mellitus: a pilot study. Free Radic. Biol. Med. 2015;81:1–12. doi: 10.1016/j.freeradbiomed.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 190.Segre C.A., Hueb W., Garcia R.M., Rezende P.C., Favarato D., Strunz C.M., Sprandel Mda C., Roggerio A., Carvalho A.L., Maranhao R.C., Ramires J.A., Kalil Filho R. Troponin in diabetic patients with and without chronic coronary artery disease. BMC Cardiovasc. Disord. 2015;15:72. doi: 10.1186/s12872-015-0051-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Tramutola A., Abate G., Lanzillotta C., Triani F., Barone E., Iavarone F., Vincenzoni F., Castagnola M., Marziano M., Memo M., Garrafa E., Butterfield D.A., Perluigi M., Di Domenico F., Uberti D. Protein nitration profile of CD3(+) lymphocytes from Alzheimer disease patients: novel hints on immunosenescence and biomarker detection. Free Radic. Biol. Med. 2018;129:430–439. doi: 10.1016/j.freeradbiomed.2018.10.414. [DOI] [PubMed] [Google Scholar]
- 192.Zhao Z., Zhou H., Peng Y., Qiu C.H., Sun Q.Y., Wang F., Xie H.N. Expression and significance of plasma 3-NT and ox-LDL in patients with Alzheimer's disease. Genet. Mol. Res. 2014;13:8428–8435. doi: 10.4238/2014.October.20.19. [DOI] [PubMed] [Google Scholar]
- 193.Ryberg H., Soderling A.S., Davidsson P., Blennow K., Caidahl K., Persson L.I. Cerebrospinal fluid levels of free 3-nitrotyrosine are not elevated in the majority of patients with amyotrophic lateral sclerosis or Alzheimer's disease. Neurochem. Int. 2004;45:57–62. doi: 10.1016/j.neuint.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 194.Weber D., Stuetz W., Toussaint O., Debacq-Chainiaux F., Dolle M.E.T., Jansen E., Gonos E.S., Franceschi C., Sikora E., Hervonen A., Breusing N., Sindlinger T., Moreno-Villanueva M., Burkle A., Grune T. Associations between specific redox biomarkers and age in a large European cohort: the MARK-AGE project. Oxid Med Cell Longev. 2017:1401452. doi: 10.1155/2017/1401452. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Yeo W.S., Kim Y.J., Kabir M.H., Kang J.W., Ahsan-Ul-Bari M., Kim K.P. Mass spectrometric analysis of protein tyrosine nitration in aging and neurodegenerative diseases. Mass Spectrom. Rev. 2015;34:166–183. doi: 10.1002/mas.21429. [DOI] [PubMed] [Google Scholar]
- 196.Weber D., Kneschke N., Grimm S., Bergheim I., Breusing N., Grune T. Rapid and sensitive determination of protein-nitrotyrosine by ELISA: application to human plasma. Free Radic. Res. 2012;46:276–285. doi: 10.3109/10715762.2011.652627. [DOI] [PubMed] [Google Scholar]
- 197.Rabbani N., Thornalley P.J. Nitric Oxide, Part F. 2008. Assay of 3‐nitrotyrosine in tissues and body fluids by liquid chromatography with tandem mass spectrometric detection; pp. 337–359. [DOI] [PubMed] [Google Scholar]
- 198.Tsikas D., Duncan M.W. Mass spectrometry and 3-nitrotyrosine: strategies, controversies, and our current perspective. Mass Spectrom. Rev. 2014;33:237–276. doi: 10.1002/mas.21396. [DOI] [PubMed] [Google Scholar]
- 199.Khan J., Brennand D.M., Bradley N., Gao B., Bruckdorfer R., Jacobs M. 3-Nitrotyrosine in the proteins of human plasma determined by an ELISA method. Biochem. J. 1998;330(Pt 2):795–801. doi: 10.1042/bj3300795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ng S.P., Qiu G., Ding N., Lu X., Wu C.L. Label-free detection of 3-nitro-l-tyrosine with nickel-doped graphene localized surface plasmon resonance biosensor. Biosens. Bioelectron. 2017;89:468–476. doi: 10.1016/j.bios.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 201.Malencik D.A., Anderson S.R. Dityrosine as a product of oxidative stress and fluorescent probe. Amino Acids. 2003;25:233–247. doi: 10.1007/s00726-003-0014-z. [DOI] [PubMed] [Google Scholar]
- 202.Fukunaga Y., Katsuragi Y., Izumi T., Sakiyama F. Fluorescence characteristics of kynurenine and N'-formylkynurenine. Their use as reporters of the environment of tryptophan 62 in hen egg-white lysozyme. J. Biochem. 1982;92:129–141. doi: 10.1093/oxfordjournals.jbchem.a133909. [DOI] [PubMed] [Google Scholar]
- 203.Jung T., Hohn A., Grune T. Lipofuscin: detection and quantification by microscopic techniques. Methods Mol. Biol. 2010;594:173–193. doi: 10.1007/978-1-60761-411-1_13. [DOI] [PubMed] [Google Scholar]
- 204.Monnier V.M., Kohn R.R., Cerami A. Accelerated age-related browning of human collagen in diabetes mellitus. Proc. Natl. Acad. Sci. U. S. A. 1984;81:583–587. doi: 10.1073/pnas.81.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Schmitt A., Gasic-Milenkovic J., Schmitt J. Characterization of advanced glycation end products: mass changes in correlation to side chain modifications. Anal. Biochem. 2005;346:101–106. doi: 10.1016/j.ab.2005.07.035. [DOI] [PubMed] [Google Scholar]
- 206.Lo T.W., Westwood M.E., McLellan A.C., Selwood T., Thornalley P.J. Binding and modification of proteins by methylglyoxal under physiological conditions. A kinetic and mechanistic study with N alpha-acetylarginine, N alpha-acetylcysteine, and N alpha-acetyllysine, and bovine serum albumin. J. Biol. Chem. 1994;269:32299–32305. [PubMed] [Google Scholar]
- 207.Shipanova I.N., Glomb M.A., Nagaraj R.H. Protein modification by methylglyoxal: chemical nature and synthetic mechanism of a major fluorescent adduct. Arch. Biochem. Biophys. 1997;344:29–36. doi: 10.1006/abbi.1997.0195. [DOI] [PubMed] [Google Scholar]
- 208.Patel R.S., Ghasemzadeh N., Eapen D.J., Sher S., Arshad S., Ko Y.A., Veledar E., Samady H., Zafari A.M., Sperling L., Vaccarino V., Jones D.P., Quyyumi A.A. Novel biomarker of oxidative stress is associated with risk of death in patients with coronary artery disease. Circulation. 2016;133:361–369. doi: 10.1161/CIRCULATIONAHA.115.019790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Huang J., Mondul A.M., Weinstein S.J., Derkach A., Moore S.C., Sampson J.N., Albanes D. Prospective serum metabolomic profiling of lethal prostate cancer. Int. J. Canc. 2019;145:3231–3243. doi: 10.1002/ijc.32218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Picca A., Calvani R., Landi G., Marini F., Biancolillo A., Gervasoni J., Persichilli S., Primiano A., Urbani A., Bossola M., Bentivoglio A.R., Cesari M., Landi F., Bernabei R., Marzetti E., Lo Monaco M.R. Circulating amino acid signature in older people with Parkinson's disease: a metabolic complement to the EXosomes in Parkinson Disease (EXPAND) study. Exp. Gerontol. 2019;128:110766. doi: 10.1016/j.exger.2019.110766. [DOI] [PubMed] [Google Scholar]
- 211.Sitole L.J., Tugizimana F., Meyer D. Multi-platform metabonomics unravel amino acids as markers of HIV/combination antiretroviral therapy-induced oxidative stress. J. Pharmaceut. Biomed. Anal. 2019;176:112796. doi: 10.1016/j.jpba.2019.112796. [DOI] [PubMed] [Google Scholar]
- 212.Suzuki S., Kodera Y., Saito T., Fujimoto K., Momozono A., Hayashi A., Kamata Y., Shichiri M. Methionine sulfoxides in serum proteins as potential clinical biomarkers of oxidative stress. Sci. Rep. 2016;6:38299. doi: 10.1038/srep38299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ungurianu A., Margina D., Gradinaru D., Bacanu C., Ilie M., Tsitsimpikou C., Tsarouhas K., Spandidos D.A., Tsatsakis A.M. Lipoprotein redox status evaluation as a marker of cardiovascular disease risk in patients with inflammatory disease. Mol. Med. Rep. 2017;15:256–262. doi: 10.3892/mmr.2016.5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Altunoglu E., Guntas G., Erdenen F., Akkaya E., Topac I., Irmak H., Derici H., Yavuzer H., Gelisgen R., Uzun H. Ischemia-modified albumin and advanced oxidation protein products as potential biomarkers of protein oxidation in Alzheimer's disease. Geriatr. Gerontol. Int. 2015;15:872–880. doi: 10.1111/ggi.12361. [DOI] [PubMed] [Google Scholar]
- 215.Atukeren P., Cengiz M., Yavuzer H., Gelisgen R., Altunoglu E., Oner S., Erdenen F., Yuceakin D., Derici H., Cakatay U., Uzun H. The efficacy of donepezil administration on acetylcholinesterase activity and altered redox homeostasis in Alzheimer's disease. Biomed. Pharmacother. 2017;90:786–795. doi: 10.1016/j.biopha.2017.03.101. [DOI] [PubMed] [Google Scholar]
- 216.Montes-Cortes D.H., Olivares-Corichi I.M., Rosas-Barrientos J.V., Manuel-Apolinar L., Martinez-Godinez M.L.A., Hernandez-Lopez J.C., Cruz-Dominguez M.D.P. Characterization of oxidative stress and ammonia according to the different grades of hepatic encephalopathy. Dig. Dis. 2020;38:240–250. doi: 10.1159/000503097. [DOI] [PubMed] [Google Scholar]
- 217.Adamczyk-Sowa M., Galiniak S., Zyracka E., Grzesik M., Naparlo K., Sowa P., Bartosz G., Sadowska-Bartosz I. Oxidative modification of blood serum proteins in multiple sclerosis after interferon beta and melatonin treatment. Oxid Med Cell Longev. 2017 doi: 10.1155/2017/7905148. 2017, 7905148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Casoinic F., Sampelean D., Buzoianu A.D., Hancu N., Baston D. Serum levels of oxidative stress markers in patients with type 2 diabetes mellitus and non-alcoholic Steatohepatitis. Rom. J. Intern. Med. 2016;54:228–236. doi: 10.1515/rjim-2016-0035. [DOI] [PubMed] [Google Scholar]
- 219.Neelofar K., Arif Z., Arafat M.Y., Alam K., Ahmad J. A study on correlation between oxidative stress parameters and inflammatory markers in type 2 diabetic patients with kidney dysfunction in north Indian population. J. Cell. Biochem. 2019;120:4892–4902. doi: 10.1002/jcb.27763. [DOI] [PubMed] [Google Scholar]
- 220.Musthafa Q.A., Abdul Shukor M.F., Ismail N.A.S., Mohd Ghazi A., Mohd Ali R., If M.N., Dimon M.Z., Wan Ngah W.Z. Oxidative status and reduced glutathione levels in premature coronary artery disease and coronary artery disease. Free Radic. Res. 2017;51:787–798. doi: 10.1080/10715762.2017.1379602. [DOI] [PubMed] [Google Scholar]
- 221.Nguyen T.T., Ngo L.Q., Promsudthi A., Surarit R. Salivary oxidative stress biomarkers in chronic periodontitis and acute coronary syndrome. Clin. Oral Invest. 2017;21:2345–2353. doi: 10.1007/s00784-016-2029-3. [DOI] [PubMed] [Google Scholar]
- 222.Galicia-Moreno M., Rosique-Oramas D., Medina-Avila Z., Alvarez-Torres T., Falcon D., Higuera-de la Tijera F., Bejar Y.L., Cordero-Perez P., Munoz-Espinosa L., Perez-Hernandez J.L., Kershenobich D., Gutierrez-Reyes G. Behavior of oxidative stress markers in alcoholic liver cirrhosis patients. Oxid Med Cell Longev. 2016 doi: 10.1155/2016/9370565. 2016, 9370565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Shearn C.T., Orlicky D.J., Saba L.M., Shearn A.H., Petersen D.R. Increased hepatocellular protein carbonylation in human end-stage alcoholic cirrhosis. Free Radic. Biol. Med. 2015;89:1144–1153. doi: 10.1016/j.freeradbiomed.2015.10.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Perrotte M., Le Page A., Fournet M., Le Sayec M., Rassart E., Fulop T., Ramassamy C. Blood-based redox-signature and their association to the cognitive scores in MCI and Alzheimer's disease patients. Free Radic. Biol. Med. 2019;130:499–511. doi: 10.1016/j.freeradbiomed.2018.10.452. [DOI] [PubMed] [Google Scholar]
- 225.Garcia-Gonzalez A., Gaxiola-Robles R., Zenteno-Savin T. Oxidative stress in patients with rheumatoid arthritis. Rev. Invest. Clin. 2015;67:46–53. [PubMed] [Google Scholar]
- 226.Mateen S., Moin S., Khan A.Q., Zafar A., Fatima N. Increased reactive oxygen species formation and oxidative stress in rheumatoid arthritis. PLoS One. 2016;11 doi: 10.1371/journal.pone.0152925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Gozu Pirinccioglu A., Alyan O., Akin A., Kizil G., Isik F.B. Oxidative stress parameters in children with acute rheumatic fever. Pediatr. Int. 2019;61:962–966. doi: 10.1111/ped.13983. [DOI] [PubMed] [Google Scholar]
- 228.Kolgiri V., Patil V.W. Protein carbonyl content: a novel biomarker for aging in HIV/AIDS patients. Braz. J. Infect. Dis. 2017;21:35–41. doi: 10.1016/j.bjid.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Lee K.Y., Chen T.T., Chiang L.L., Chuang H.C., Feng P.H., Liu W.T., Chen K.Y., Ho S.C. Proteasome activity related with the daily physical activity of COPD patients. Int. J. Chronic Obstr. Pulm. Dis. 2017;12:1519–1525. doi: 10.2147/COPD.S132276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Khalfa A., Tiali A., Zemour L., Fatah A., Mekki K. Prevalence of metabolic syndrome and its association with lifestyle and cardiovascular biomarkers among postmenopausal women in western Algeria. Int. J. Gynaecol. Obstet. 2017;138:201–206. doi: 10.1002/ijgo.12206. [DOI] [PubMed] [Google Scholar]
- 231.Fernando N., Wickremesinghe S., Niloofa R., Rodrigo C., Karunanayake L., de Silva H.J., Wickremesinghe A.R., Premawansa S., Rajapakse S., Handunnetti S.M. Protein carbonyl as a biomarker of oxidative stress in severe Leptospirosis, and its usefulness in differentiating Leptospirosis from dengue infections. PLoS One. 2016;11 doi: 10.1371/journal.pone.0156085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Hondur G., Goktas E., Yang X., Al-Aswad L., Auran J.D., Blumberg D.M., Cioffi G.A., Liebmann J.M., Suh L.H., Trief D., Tezel G. Oxidative stress-related molecular biomarker candidates for glaucoma. Invest. Ophthalmol. Vis. Sci. 2017;58:4078–4088. doi: 10.1167/iovs.17-22242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hatch J., Andreazza A., Olowoyeye O., Rezin G.T., Moody A., Goldstein B.I. Cardiovascular and psychiatric characteristics associated with oxidative stress markers among adolescents with bipolar disorder. J. Psychosom. Res. 2015;79:222–227. doi: 10.1016/j.jpsychores.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 234.Abdul Sani N.F., Ahmad Damanhuri M.H., Amir Hamzah A.I.Z., Abu Bakar Z.H., Tan J.K., Nor Aripin K.N., Mohd Rani M.D., Noh N.A., Shamaan N.A., Razali R., Mohd Yusof Y.A., Mazlan M., Makpol S., Wan Ngah W.Z. DNA damage and protein oxidation associated with ageing correlate with cognitive dysfunction in a Malaysian population. Free Radic. Res. 2018;52:1000–1009. doi: 10.1080/10715762.2018.1506877. [DOI] [PubMed] [Google Scholar]
- 235.Tezuka Y., Nakaya I., Nakayama K., Nakayama M., Yahata M., Soma J. Methylglyoxal as a prognostic factor in patients with chronic kidney disease. Nephrology (Carlton) 2019;24:943–950. doi: 10.1111/nep.13526. [DOI] [PubMed] [Google Scholar]
- 236.Ishida Y.I., Kayama T., Kibune Y., Nishimoto S., Koike S., Suzuki T., Horiuchi Y., Miyashita M., Itokawa M., Arai M., Ogasawara Y. Identification of an argpyrimidine-modified protein in human red blood cells from schizophrenic patients: a possible biomarker for diseases involving carbonyl stress. Biochem. Biophys. Res. Commun. 2017;493:573–577. doi: 10.1016/j.bbrc.2017.08.150. [DOI] [PubMed] [Google Scholar]
- 237.Knani I., Bouzidi H., Zrour S., Bergaoui N., Hammami M., Kerkeni M. Increased serum concentrations of N(varepsilon)-carboxymethyllysine are related to the presence and the severity of rheumatoid arthritis. Ann. Clin. Biochem. 2018;55:430–436. doi: 10.1177/0004563217733500. [DOI] [PubMed] [Google Scholar]
- 238.Knani I., Bouzidi H., Zrour S., Bergaoui N., Hammami M., Kerkeni M. Methylglyoxal: a relevant marker of disease activity in patients with rheumatoid arthritis. Dis. Markers. 2018 doi: 10.1155/2018/8735926. 2018, 8735926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Papagrigoraki A., Del Giglio M., Cosma C., Maurelli M., Girolomoni G., Lapolla A. Advanced glycation end products are increased in the skin and blood of patients with severe psoriasis. Acta Derm. Venereol. 2017;97:782–787. doi: 10.2340/00015555-2661. [DOI] [PubMed] [Google Scholar]
- 240.Lou B., Boger M., Bennewitz K., Sticht C., Kopf S., Morgenstern J., Fleming T., Hell R., Yuan Z., Nawroth P.P., Kroll J. Elevated 4-hydroxynonenal induces hyperglycaemia via Aldh3a1 loss in zebrafish and associates with diabetes progression in humans. Redox Biol. 2020;37:101723. doi: 10.1016/j.redox.2020.101723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Vanjiappan S., Hamide A., Ananthakrishnan R., Periyasamy S.G., Mehalingam V. Nonalcoholic fatty liver disease in patients with type 2 diabetes mellitus and its association with cardiovascular disease. Diabetes Metab Syndr. 2018;12:479–482. doi: 10.1016/j.dsx.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 242.Yavuzer H., Yavuzer S., Cengiz M., Erman H., Doventas A., Balci H., Erdincler D.S., Uzun H. Biomarkers of lipid peroxidation related to hypertension in aging. Hypertens. Res. 2016;39:342–348. doi: 10.1038/hr.2015.156. [DOI] [PubMed] [Google Scholar]
- 243.Pawluk H., Pawluk R., Robaczewska J., Kedziora-Kornatowska K., Kedziora J. Biomarkers of antioxidant status and lipid peroxidation in elderly patients with hypertension. Redox Rep. 2017;22:542–546. doi: 10.1080/13510002.2017.1372072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Stankova T.R., Delcheva G.T., Maneva A.I., Vladeva S.V. Serum levels of carbamylated LDL, nitrotyrosine and soluble lectin-like oxidized low-density lipoprotein receptor-1 in poorly controlled type 2 diabetes mellitus. Folia Med. (Plovdiv) 2019;61:419–425. doi: 10.3897/folmed.61.e39343. [DOI] [PubMed] [Google Scholar]
- 245.Quidim A.V.L., Bruno T., Leocadio P.C.L., Santos I.S., Alvarez-Leite J.I., Dos Reis Menta P.L., Lotufo P.A., Bensenor I.M., Goulart A.C. The prognostic value of nitrotyrosine levels in coronary heart disease: long-term evaluation in the Acute Coronary Syndrome Registry Strategy (ERICO study) Clin. Biochem. 2019;66:37–43. doi: 10.1016/j.clinbiochem.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 246.Silva Servato J.P., Ueira Vieira C., de Faria P.R., Cardoso S.V., Loyola A.M. The importance of inducible nitric oxide synthase and nitrotyrosine as prognostic markers for oral squamous cell carcinoma. J. Oral Pathol. Med. 2019;48:967–975. doi: 10.1111/jop.12942. [DOI] [PubMed] [Google Scholar]
- 247.Pennathur S., Jaiswal M., Vivekanandan-Giri A., White E.A., Ang L., Raffel D.M., Rubenfire M., Pop-Busui R. Structured lifestyle intervention in patients with the metabolic syndrome mitigates oxidative stress but fails to improve measures of cardiovascular autonomic neuropathy. J. Diabet. Complicat. 2017;31:1437–1443. doi: 10.1016/j.jdiacomp.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]