Abstract
Background:
Although chronic rhinosinusitis (CRS) is considered the most treatable form of olfactory dysfunction (OD), there has been relatively little clinical attention focused on assessing endotypes as they pertain to olfactory loss.
Objective:
The goal of this study was to explore inflammatory endotypes in CRS using an unsupervised cluster analysis of olfactory cleft (OC) biomarkers in a phenotype-free approach.
Methods:
Patients with CRS were prospectively recruited and psychophysical olfactory testing, Questionnaire of Olfactory Dysfunction (QOD-NS), and bilateral OC endoscopy were obtained. Mucus was collected from the OC and evaluated for 26 biomarkers using principal component analysis (PCA). Cluster analysis was performed using only OC biomarkers and differences in olfactory measures were compared across clusters.
Results:
198 subjects (128 with CRS and 70 controls) were evaluated. Evaluation of OC biomarkers indicated 6 principal components, explaining 69.50% of the variance, with Type 2, mixed Type1/Th17, growth factor, and neutrophil chemo-attractant inflammatory signatures. A total of 10 clusters were identified which differed significantly in frequency of controls, CRSsNP, and CRSwNP across the clusters (LRT χ2(18)=178.64, p<0.001). Olfactory measures differed significantly across clusters, including olfactory testing, QOD-NS, and OC endoscopy (p<0.001 for all).
Conclusion:
Clustering based solely on OC biomarkers can organize patients into clinically meaningful endotypes that discriminate between CRS and controls. Validation studies are necessary to confirm these findings and further refine olfactory endotypes.
MeSH Key words: Cluster analysis, translational medical research, chronic disease, patient reported outcome measures, sinusitis, smell
CAPSULE SUMMARY:
This preliminary study identifies olfactory-specific endotypes in patients with chronic rhinosinusitis based on the underlying inflammatory profile that may guide future personalized treatments.
INTRODUCTION
Chronic rhinosinusitis (CRS) is a highly prevalent condition and impacts up to 12% of the United States (U.S.) population.1 Over the last several years, there has been an increased realization that inflammation characteristic of CRS is not homogeneous. Instead, several studies have shown that patients with CRS likely belong to one of several underlying inflammatory endotypes.2–6 Although patients may appear similar clinically, these underlying inflammatory endotypes differ across patients and could have important treatment implications, particularly in an era of personalized medicine.
Most clinical treatments for CRS are focused on the sinuses in general, and success or failure is usually considered in terms of sinus-specific outcomes such as nasal drainage or congestion. However, a recent study assessed symptom importance in patients presenting for sinus surgery and found that loss of smell rated #2 out of 22 possible symptoms, with 72% considering it “important” or “very important”.7 Unfortunately, quantifiable olfactory dysfunction (OD) is present in up to 60–80% of patients with CRS, which equates to a population prevalence of roughly 30 million Americans including all racial and ethnic groups.8 Because the average age of a patient is roughly 45 years, CRS is the primary cause of OD in young and middle age adults.9
Current treatment of CRS-associated OD most commonly includes anti-inflammatory medications, usually in the form of oral or topical corticosteroids, sometimes combined with endoscopic sinus surgery when initial medications fail.10 For those with nasal polyps, newer biologic medications also appear to improve olfaction, including those targeting both IL4/13 and IgE.11 Despite these treatment options, meta-analyses have found that mean improvement rate ranges from 10.6% to 43.5% and only half of patients will regain normal olfaction.12–14
Although CRS is considered the most treatable form of OD, there has been relatively little clinical attention focused on assessing endotypes as they pertain to olfactory loss. The goal of the current study was to identify inflammatory endotypes in patients with CRS using an unsupervised cluster analysis of olfactory cleft (OC) biomarkers in a phenotype-free approach. Secondary goals were to determine whether clinically-relevant phenotypic features differed across endotypes, particularly as it relates to olfactory measures. Findings would give insights into underlying pathophysiology, as well as the potential for therapeutics to specifically target olfactory loss in patients with CRS.
METHODS
Study enrollment
Patients with CRS were recruited from five centers across the U.S. in a prospective fashion, including the Medical University of South Carolina (MUSC, Charleston, SC), Oregon Health and Science University (OHSU, Portland, OR), the University of Utah (Salt Lake City, UT), the University of Colorado (Aurora, CO), and the University of Virginia (UVA, Charlottesville, VA). All patients met diagnostic criteria for CRS according to the American Academy of Otolaryngology-Head and Neck Surgery, including characteristic symptoms and the presence of visible inflammation on nasal endoscopy and computed tomography (CT).15 Patients with CRS were excluded if they had cystic fibrosis, primary ciliary dyskinesia, systemic inflammatory disease (granulomatosis with polyangiitis, sarcoidosis, eosinophilic granulomatosis with polyangiitis), or had been on systemic corticosteroids within the preceding month. Control subjects without CRS were similarly recruited from the surrounding community. Control subjects were excluded if they were on immunosuppressive medications, had current symptoms fitting diagnostic criteria for CRS, or a history of CRS, Parkinson’s disease, dementia, or systemic inflammatory disease. The Institutional Review Board at each enrollment site provided ethical oversight and study subjects provided informed consent prior to study participation in accordance with good clinical practice guidelines for research on human subjects.
Demographics, comorbidities, and CRS disease severity
Information related to demographics and medical comorbidities was collected directly from subjects via survey and supplemented by the medical record. The presence of allergic rhinitis was determined based on physician’s diagnosis and confirmation with previous positive objective testing. The presence of aspirin exacerbated respiratory disease (AERD) was based on asthma, nasal polyps, >1 respiratory reaction to NSAIDs. Allergic fungal rhinosinusitis was determined by the enrolling rhinologist based on characteristic radiographic findings and presence of allergic mucin on endoscopy.16 Patients with CRS were categorized as chronic rhinosinusitis with nasal polyps (CRSwNP) or without nasal polyps (CRSsNP) based upon the presence of nasal polyps identified during nasal endoscopy, as determined by the enrolling physician. Patients with CRS underwent CT scanning as part of clinical care as indicated. Each CT scan was graded using the standard Lund-Mackay scoring method, with reviewers blinded to all olfaction data.17 Patients with CRS also underwent sinonasal endoscopy and Lund-Kennedy endoscopy scores (LKES) were recorded for each patient.18 Those with CRS also completed the 22-item Sinonasal Outcome Test (SNOT-22; ©2006, Washington University, St. Louis) to capture sinus-specific quality of life (QOL).19
Olfactory-specific assessments
All subjects underwent psychophysical olfactory testing using “Sniffin’ Sticks” (Burghart Messtechnik, Wedel, Germany).20 This examination evaluated three separate domain items of olfactory function including: odorant threshold (T, score range: 1–16), odorant discrimination (D, score range: 0–16), and odorant identification (I, score range: 0–16). Correct responses are summarized into a composite TDI total score (score range: 1–48) with higher scores reflecting superior olfaction.
All study participants were also asked to complete 17 negatively termed questions of the Questionnaire of Olfactory Dysfunction (QOD-NS).21,22 The QOD-NS is a validated, olfactory-specific survey which summarizes Likert scale responses from 0 (“Disagree”) to 3 (“Agree”) whereas higher total scores (score range: 0–51) represent higher global impacts of olfactory impairment.
All subjects also underwent bilateral nasal endoscopy in order to evaluate the OC specifically. Physicians quantified the severity of discharge, edema, polyps, crusting and scarring of the OC using a Likert score from 0–2 for each attribute. Results for each side were recorded separately and combined for a final Olfactory Cleft Endoscopy Scale (OCES) that ranged from 0–20, with higher scores representing increased disease severity.23
For subjects with CRS, a cross-sectional analysis of CT scans was performed in order to evaluate opacification of the OC using OsiriX MD imaging software (Pixmeo, Bernex, Switzerland) as previously reported.24 This analysis required specific formatting with thin (0.6mm.) cuts axial, coronal and sagittal planes for consistency, thus some CT scans were excluded. The region of the OC was demarcated on each section and all pixels in Hounsfield unit range of bone were excluded. Pixels representing soft tissue were then divided by all remaining pixels (soft tissue+air) to determine the percentage opacification for each section.
Olfactory cleft biomarkers
At the time of nasal endoscopy, subjects had mucus collected from the OC. Utilizing rigid nasal endoscopy, a Leukosorb filter paper (Pall Scientific, Port Washington, NY) strip was placed directly into the OC of each side by a treating rhinologist and allowed to dwell for three minutes, as described and validated previously.25–27 A broad array of 26 OC biomarkers was assessed in order to capture the heterogeneity of CRS, including cytokines, chemokines, and growth factors. These biomarkers were chosen for analysis based on previous evidence suggesting a role in CRS endotypes, olfactory dysfunction, or inflammation/remodeling.2,4,6,11,25,26,28–30 All proteins, except those noted below, were quantified by LegendPlex Mix & Match Cytometric Bead Array (BioLegend, San Diego, CA) following the manufacturer’s recommended protocol and read on a Guava easy Cyte 8HT flow cytometer (EMD Millipore, Burlington, MA). Data analysis was performed with LegendPlex software provided by the manufacturer. Total IgE was quantified via ELISA following the kit instructions (GenWay Biotech. Inc, San Diego, CA).
Biostatistical methods
Data were analyzed using the IBM SPSS 25.0 software package (SPSS Inc., Armonk, NY). For continuous variables, results are expressed as means and standard deviations and modified heat maps. One-way analyses of variance (ANOVAs) were used for between group comparisons; when heterogeneous within-group variances were indicated, Games-Howell and Welch tests were conducted to assess the sensitivity of the ANOVA-based conclusions to this violation.31,32 For categorical variables, likelihood ratio chi-square tests were used to assess differences across groups. Statistical significance was defined as p≤0.050.
Consistent with literature recommendations regarding cluster analysis, the 26 biomarker variables were explored for outlying data values and their pairwise correlations evaluated; outlying data values tend to create numerous small clusters and correlated variables tend to be given greater weight in cluster creation.33 As most of the biomarker distributions had long right tails, a log (base 10) transformation was applied to the 26 biomarker variables prior to analysis; for biomarkers where 0 is a legitimate value, a constant of 10 was added to the variable scores prior to the log transformation. To account for correlations among the 26 log-transformed biomarkers we conducted a principal component analysis (PCA) with orthogonal rotation and Kaiser normalization; component scores which have means of zero, variances of unity, and are pairwise uncorrelated were used an input into the cluster analysis. An additional benefit of this approach is that cluster analysis can give greater weight to input variables with larger variances which are equated using the PCA approach.
We used an agglomerative, hierarchical clustering approach using Ward’s minimum variance method and the squared Euclidian distance metric.34 To determine the number of clusters initially, we assessed the changes in the agglomeration coefficients throughout the clustering process; a large increase in this coefficient from one step to the next indicates that cluster solution prior to the large increase is favorable.33 We evaluated the validity of the cluster solution through comparisons of cluster differences both numerically and graphically using the component scores underlying the cluster analysis and the 26 log-transformed (and standardized) biomarkers, applying Bonferroni adjustments to minimize inflation of Type 1 error.
RESULTS
A total of 198 subjects were enrolled and had complete biomarker data available for cluster analysis, including 128 patients with CRS and 70 controls. There were no differences between cases and controls with regard to age, gender, or race/ethnicity. The population with CRS was comprised of 75 patients with CRSwNP and 53 patients with CRSsNP. Most patients with CRS were on maintenance medical therapy (saline irrigations=70%, topical corticosteroid=62%, antihistamine=47%, leukotriene modifier=24%). An overview of the study population as well as the frequency of notable comorbidities is detailed in Table 1.
Table 1.
Final study cohort
| Characteristics: | Case subjects (n=128) | Control subjects (n=70) | Test statistic | p-value | |
|---|---|---|---|---|---|
| Mean[±SD] | 49.0 [±15.9] | 50.6 [±18.3] | t=0.63 | 0.532 | |
| N (%) | 59 (46%) | 27 (39%) | X2=1.04 | 0.307 | |
| Females | 69 (54%) | 43 (61%) | |||
| White/Caucasian | 110 (86%) | 52 (74%) | X2=4.13 | 0.248 | |
| African American | 16 (13%) | 16 (23%) | |||
| Asian | 1 (1%) | 1 (1%) | |||
| Hispanic/Latino | 7 (6%) | 3 (4%) | X2=0.13 | 0.716 | |
| Allergic rhinitis / response | 70 (55%) | 18 (26%) | X2=15.39 | <0.001 | |
| Asthma | 57 (45%) | 1 (1%) | X2=40.59 | <0.001 | |
| Allergic fungal sinusitis | 7 (6%) | 0 (0%) | X2=3.97 | 0.046 | |
| AERD / ASA sensitivity | 19 (15%) | 0 (0%) | X2=11.49 | 0.001 | |
| Nasal polyposis | 75 (59%) | 0 (0%) | X2=66.03 | <0.001 | |
| Current smoker | 5 (4%) | 4 (6%) | X2=0.34 | 0.559 | |
| Diabetes mellitus (Type I/II) | 14 (11%) | 5 (7%) | X2=0.75 | 0.386 | |
| GERD | 35 (27%) | 6 (9%) | X2=9.71 | 0.002 | |
| CRS metrics | |||||
| Mean[±SD] | 13.6 [±5.6] | ---- | ---- | ---- | |
| LK Endoscopy total score | 7.1 [±3.5] | ---- | ---- | ---- | |
| SNOT-22 total score | 49.8 [±22.2] | ---- | ---- | ---- | |
| Olfactory metrics | |||||
| Sniffin’ Sticks total score | 21.7 [±9.5] | 30.4 [±5.8] | t=6.91 | <0.001 | |
| Threshold (T) score | 3.7 [±3.0] | 6.7 [±2.5] | t=7.36 | <0.001 | |
| Discrimination (D) score | 9.1 [±3.5] | 11.5 [±2.4] | t=5.22 | <0.001 | |
| Identification (I) score | 9.0 [±4.3] | 12.1 [±2.3] | t=5.60 | <0.001 | |
| QOD-NS total score | 12.9 [±11.2] | 4.3 [±5.8] | t= −5.97 | <0.001 | |
| OCES | 4.7 [±3.9] | 0.6 [±1.0] | t= −8.19 | <0.001 | |
| OC Anterior opacification (%) | 57% [±32%] | ---- | ---- | ---- | |
| OC Middle opacification (%) | 63% [±29%] | ---- | ---- | ---- | |
| OC Posterior opacification (%) | 68% [±29%] | ---- | ---- | ---- | |
SD, standard deviation; AERD, aspirin exacerbated respiratory disease; ASA, acetylsalicyclic acid; GERD, gastroesophageal reflux disease; LM CT, Lund-Mackay computed tomography; LK, Lund-Kennedy; SNOT-22, 22-item SinoNasal Outcome Test; QOD-NS, Questionnaire of Olfactory Dysfunction-Negative Statements; OCES, olfactory cleft endoscopy score; OC, olfactory cleft.
OC Biomarker Analysis
Descriptive statistics for all 26 biomarkers are presented in Table E1. The PCA of the 26 log-transformed OC biomarkers indicated 6 principal components (PCs) with eigenvalues greater than unity, explaining 69.50% of the variances in these biomarkers. These PCs are described in Table 2, which demonstrates how each biomarker loads onto a PC. Each of the 26 biomarkers a represented at least once in each PC. To aid interpretation of the PCs, the biomarker variables are sorted within components in descending order based on their component loading; non-salient loadings (less than .30) are suppressed to reduce visual noise. The communalities represent how well the 6 rotated components as a whole relate to each underlying biomarker (interpreted as the percentage of variance in each biomarker explained by the retained and rotated components). Biomarkers with lower communalities will be less well represented in the cluster analysis and will be less likely to differentiate the clusters. Although the statistical relationship among analyzed proteins initially appears complex, these groupings reflect known relationships. For example, PC- 1 includes a number of cytokines typical of a mixed Type 1/Th17 pathway, including IL6, IL10, TNFα, IL17, and IL23. PC-2 is dominated by Type 2 inflammatory cytokines, including IL4, IL5, and IL13. Similarly, PC-3 also includes IL5 and IL13, but includes IgE as well. PC-4 includes growth factors such as VEGF and EGF. PC-5 includes IL 33 and CCL5, but also bFGF. Lastly, PC-6 is dominated by the neutrophil chemo-attractant CXCL1.
Table 2.
Component Loading Matrix and Communalities for the 26 Log-Transformed Cytokine Variables on the First Six Principal Components after Varimax Rotation
| Cytokine Variable | PC-1 | PC-2 | PC-3 | PC-4 | PC-5 | PC-6 | Com. |
|---|---|---|---|---|---|---|---|
| IL6 | .755 | -- | .318 | -- | -- | -- | .757 |
| IL10 | .747 | .362 | -- | -- | -- | -- | .788 |
| CCL3 (MIP1α) | .706 | -- | .394 | -- | -- | -- | .737 |
| TNFα | .699 | .552 | -- | -- | -- | -- | .829 |
| CCL2 (MCP1) | .641 | -- | .450 | -- | -- | -- | .683 |
| CCL20 (MIP3α) | .581 | -- | -- | .388 | -- | −.368 | .736 |
| CXCL5 (ENA-78) | .549 | -- | -- | -- | -- | -- | .473 |
| CXCL9 (MIG) | .538 | -- | -- | .523 | -- | -- | .602 |
| IL17F | -- | .808 | -- | -- | -- | -- | .806 |
| IL4 | -- | .792 | -- | -- | -- | -- | .717 |
| IL2 | -- | .789 | -- | -- | -- | -- | .748 |
| IL23 | .319 | .731 | -- | -- | -- | .304 | .752 |
| IL17A | .541 | .574 | -- | -- | -- | -- | .721 |
| IL5 | -- | .381 | .718 | -- | -- | -- | .797 |
| IL13 | -- | .436 | .672 | -- | -- | -- | .748 |
| IgE | -- | -- | .645 | -- | -- | -- | .513 |
| IL9 | -- | .486 | .627 | -- | -- | -- | .683 |
| VEGF-A | -- | -- | -- | .692 | -- | -- | .576 |
| CCL11 (Eotaxin) | -- | -- | .516 | .679 | -- | -- | .773 |
| EGF | -- | -- | -- | .672 | −.483 | -- | .713 |
| IL8 | -- | -- | -- | .553 | -- | -- | .355 |
| CXCL11 (I-TAC) | .468 | -- | -- | .516 | -- | -- | .593 |
| bFGF | -- | -- | -- | -- | .837 | -- | .812 |
| IL33 | -- | -- | -- | -- | .785 | -- | .684 |
| CCL5 (RANTES) | .562 | -- | -- | -- | .569 | -- | .674 |
| CXCL1 (GROα) | -- | -- | -- | -- | -- | .822 | .798 |
N=198. All cytokine variables are log transformed. PC-1 through PC-6 are the six rotated principal components. Component loadings less than |.30| are suppressed and available on request. Com., Cytokine communalities based on the rotated 6 component solution.
Agglomerative cluster analysis was performed with only the 6 principal components as input. Figure 1 provides a graph of the changes in the agglomeration coefficients. Very little change occurs in the early steps of the cluster analysis and large changes occur in the last several steps. In particular, a relatively large step occurs between the 10 and 9 cluster solution (i.e., a change in the agglomeration coefficient of 36.08), indicating that changes in the agglomeration coefficient are minimal beyond the 10-cluster solution.
Figure 1.
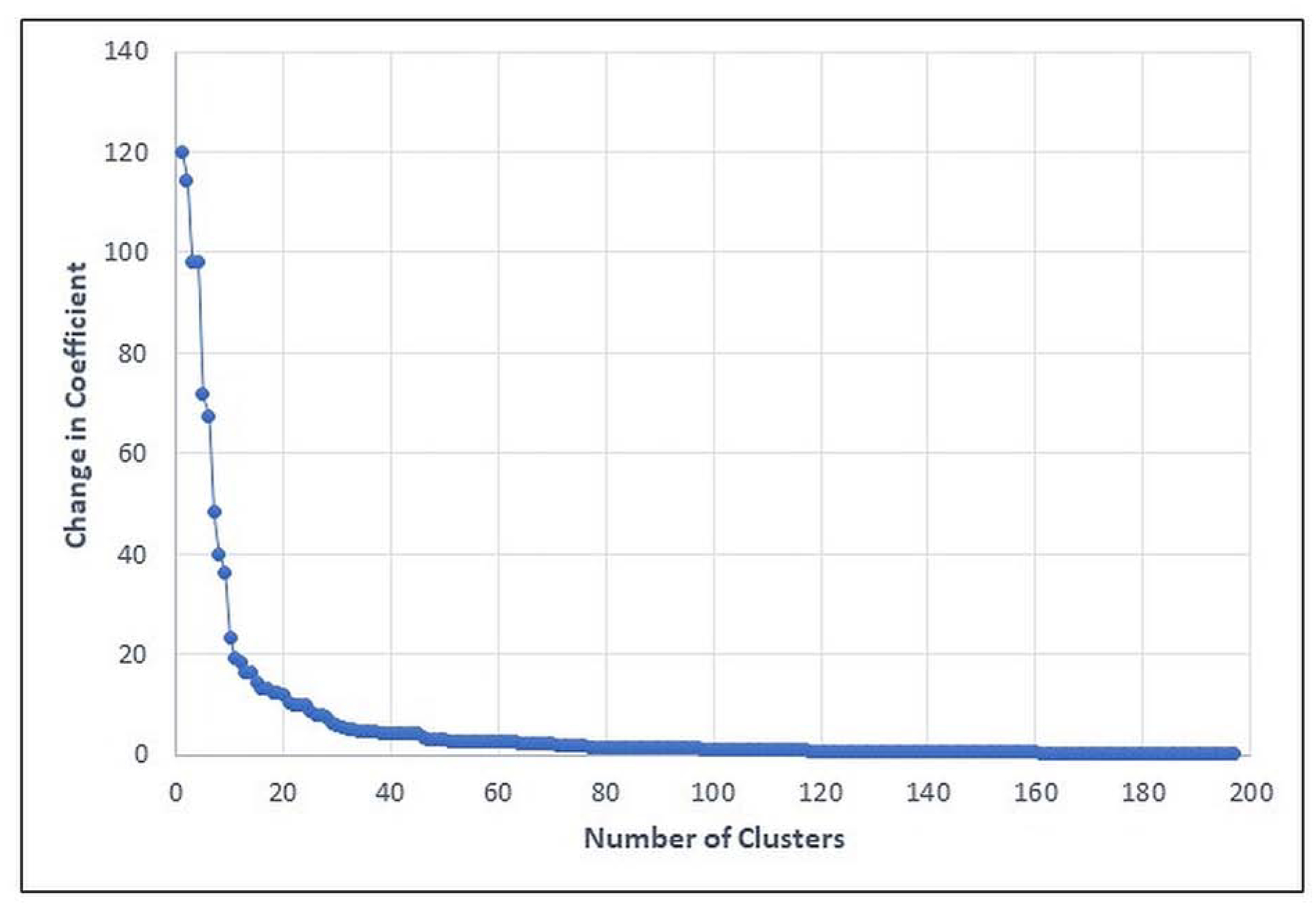
Change in Agglomeration Coefficients in Adjacent Agglomerative Steps.
The ten clusters can be represented visually in the 3-dimensional cluster plot (Figure 2), although it should be kept in mind that the PC analysis actually utilizes 6 dimensions. Figure 3 provides the mean standardized scores of the 6 PCs across the ten clusters. Heat map shading is provided to differentiate various degrees of extremity. The mean levels of all 6 PCs was significantly different across the ten clusters (p<0.001 for all). To aid cluster interpretation further, the 26 log-transformed OC biomarkers were standardized to have a mean of zero and standard deviation of unity. Figure 4 provides the mean standardized scores of the OC biomarkers across the ten clusters, with heat map shading provided to differentiate various degrees of extremity. The mean levels of all OC biomarkers was significantly different across the ten clusters (p<0.001). From these heat maps it can be seen that the relationships are complex, but do reflect known patterns. For example, Clusters 3 and 4 have very low levels of inflammatory cytokines. Clusters 1, 2 and 10 have the strongest classic Type 2 signature with elevations in IL5, IL13, and IgE. In addition to type 2 mediators, Cluster 1 also has broad elevations in other pro-inflammatory cytokines, including those typical of Type 1 and Th17. In contrast, clusters 5, 6, and 7 do not have elevations in these classic type 2 cytokines. Interestingly, cluster 9 also does not have elevation of classic Type 2 cytokines, but rather has elevated IL33 and RANTES demonstrating some Type 2 skewing via unique mediators. Post-hoc power analysis based on the smallest effect reported in Table 4 demonstrates a power of >.80.
Figure 2.
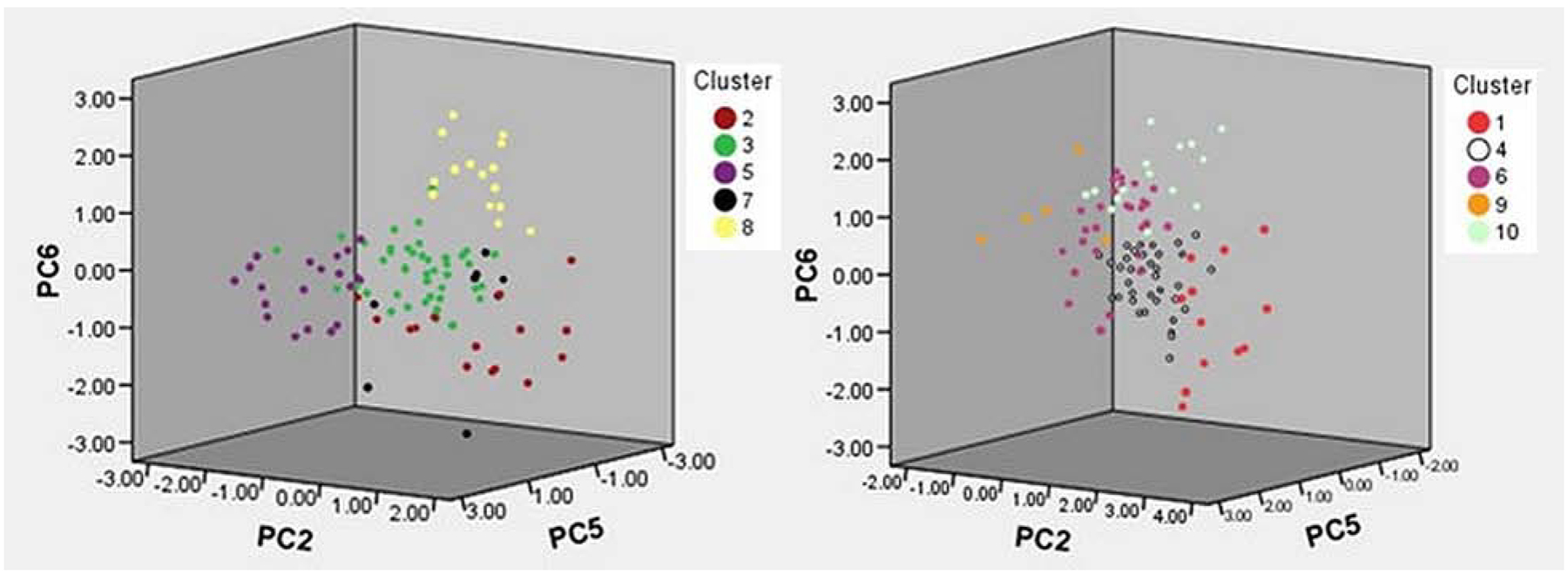
3-Dimensional Cluster Plots Showing biomarkers separated by PCs 2, 5 and 6. LEFT) shows Clusters 2, 3, 5, 7, and 8. RIGHT) shows Clusters 1,4,6,9,10.
Figure 3.
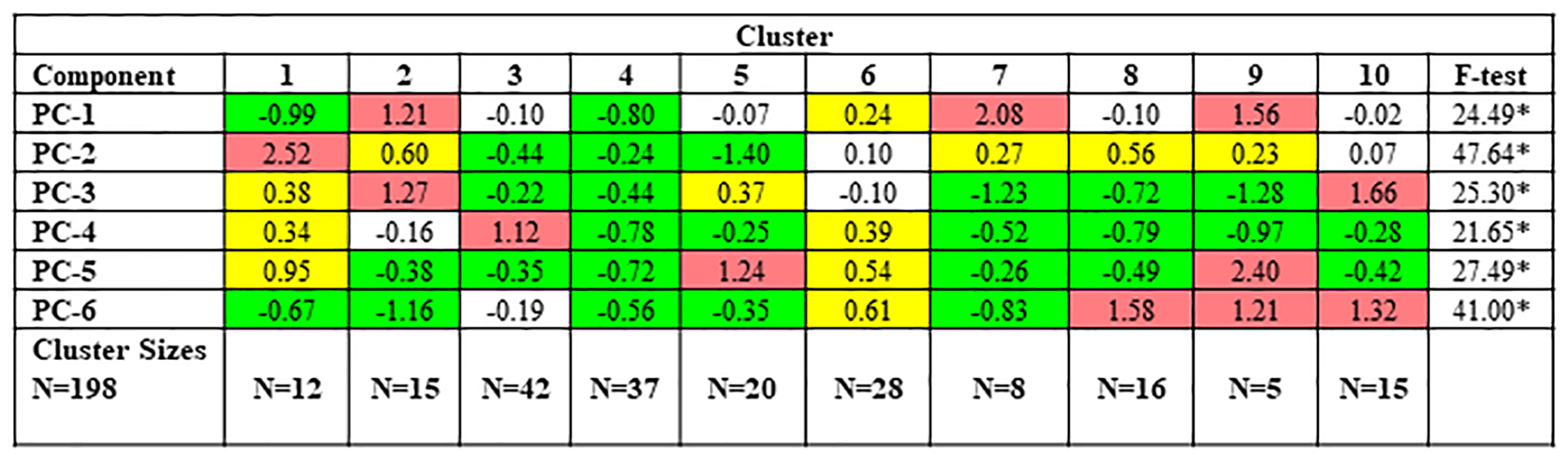
Mean Standardized Scores for each Principal Component Across the Ten Clusters. Red shading denotes a mean standardized score >1.00 and high relative levels of component/cytokine (more than 1 SD above overall sample average); yellow shading indicates elevated levels but not high relative of that component/cytokine (between 0.20 and 1.0 SD above the overall sample average); green shading indicates low relative levels of that component/cytokine (more than −.20 standard deviations below the overall sample average); no shading indicates a mean standardized score between −.20 and .20; SD, standard deviation. *p<0.050.
Figure 4.
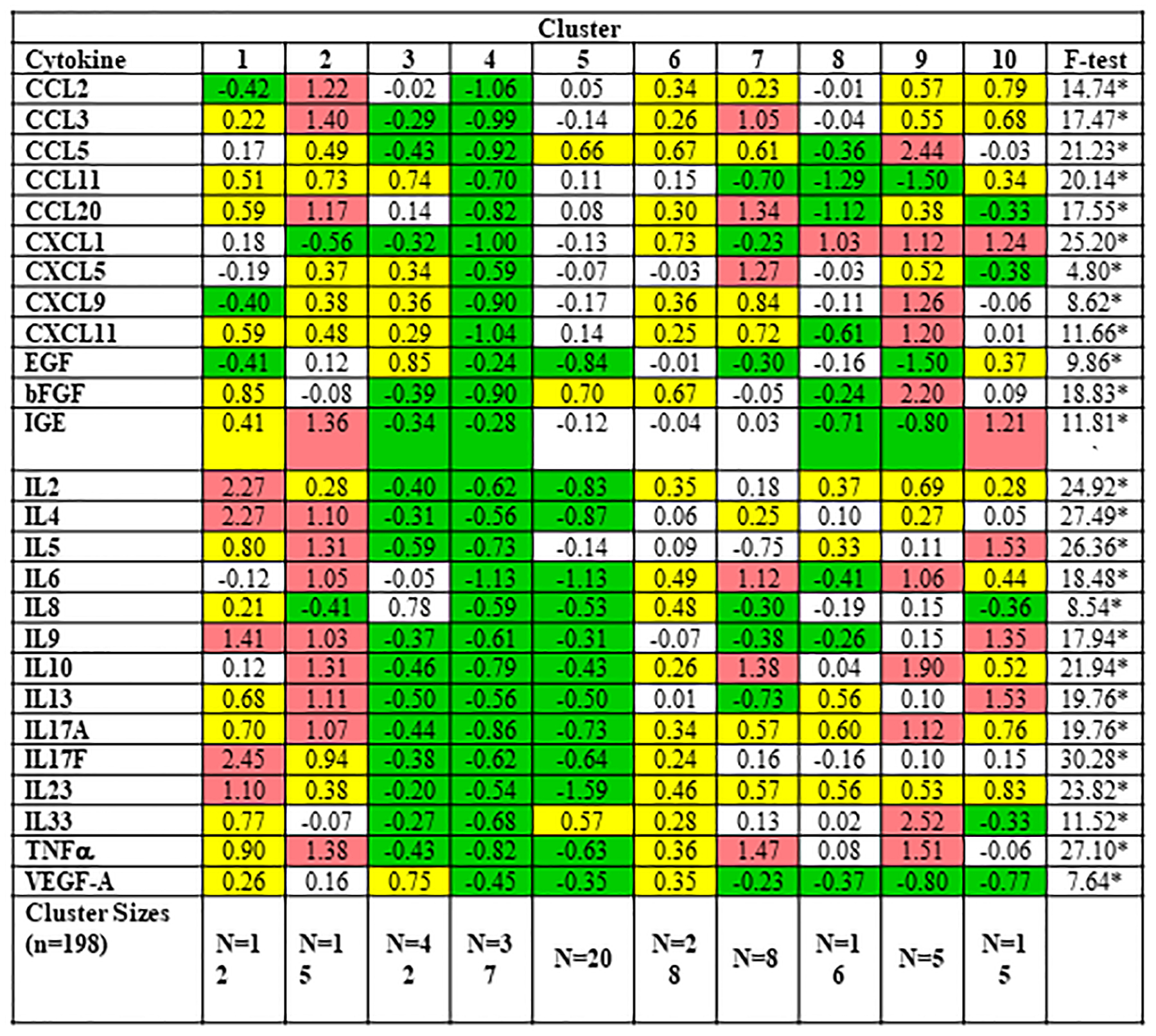
Mean Standardized Scores for 26 Log-Transformed Cytokine Scores Across the Ten Clusters. Red shading denotes a mean standardized score >1.00 and high relative levels of component/cytokine (more than 1 SD above overall sample average); yellow shading indicates elevated levels but not high relative of that component/cytokine (between 0.20 and 1.0 SD above the overall sample average); green shading indicates low relative levels of that component/cytokine (more than −.20 standard deviations below the overall sample average); no shading indicates a mean standardized score between −.20 and .20; SD, standard deviation. Note that the F-test values vary across the cytokines. In this case, the higher the F-statistic, the more that biomarker differentiates cluster membership. *p <0.05.
Table 4.
Global differences in average olfactory measures and CRS-specific measures across the 10 Clusters
| Cluster | Test statistic | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Olfactory measures: | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| Total TDI score | 26.08 (10.43) | 18.17 (10.03) | 28.57 (6.67) | 30.78 (5.45) | 20.48 (9.11) | 21.53 (10.25) | 25.03 (8.22) | 26.14 (9.04) | 18.90 (9.63) | 17.13 (7.64) | F=7.12** |
| Threshold (T) score | 5.67 (3.27) | 2.77 (2.77) | 6.11 (2.87) | 6.59 (2.80) | 3.13 (2.82) | 3.94 (3.12) | 4.66 (2.39) | 4.41 (2.97) | 2.70 (2.41) | 2.53 (2.11) | F=6.05** |
| Discrimination (D) score | 10.50 (4.06) | 7.60 (3.92) | 11.00 (2.56) | 11.51 (2.18) | 8.80 (3.76) | 9.41 (3.65) | 9.88 (2.75) | 10.56 (3.41) | 7.60 (2.79) | 7.27 (2.40) | F=4.63** |
| Identification (I) score | 9.92 (3.94) | 7.80 (4.40) | 11.45 (2.88) | 12.68 (1.96) | 8.55 (3.93) | 8.19 (4.60) | 10.50 (4.24) | 11.19 (3.94) | 8.60 (4.83) | 7.33 (4.17) | F=5.88** |
| QOD-NS | 7.75 (6.65) | 15.40 (13.42) | 6.50 (7.49) | 5.38 (8.02) | 14.85 (13.37) | 8.56 (8.49) | 9.00 (9.49) | 10.88 (11.66) | 21.80 (14.94) | 17.33 (8.67) | F=4.57** |
| OCES Total | 2.00 (1.73) | 5.67 (3.92) | 1.60 (2.86) | 0.45 (0.83) | 4.22 (3.93) | 3.62 (3.99) | 4.00 (2.00) | 6.50 (5.00) | 4.00 (5.29) | 6.20 (2.78) | F=7.07** |
| CRS-specific measures: | |||||||||||
| LM-CT total score | 14.13 (5.59) | 14.85 (4.39) | 10.07 (6.08) | 10.00 (3.47) | 14.47 (5.79) | 12.04 (5.85) | 13.71 (3.82) | 15.45 (5.72) | 14.00 (7.65) | 17.00 (3.98) | F=2.02* |
| OC Anterior (%) | 38.67 (29.74) | 62.69 (31.14) | 47.43 (32.20) | 38.00 (16.52) | 61.55 (30.38) | 54.13 (34.44) | 46.14 (26.28) | 51.36 (35.55) | 58.33 (34.02) | 78.71 (27.69) | F=1.33 |
| OC Middle (%) | 47.33 (31.53) | 63.77 (29.86) | 48.43 (32.68) | 85.67 (5.51) | 65.00 (31.67) | 62.96 (26.99) | 55.57 (23.05) | 53.00 (32.59) | 72.00 (23.43) | 85.64 (20.80) | F=1.98* |
| OC Posterior (%) | 56.00 (32.74) | 74.85 (22.90) | 52.71 (30.90) | 89.67 (9.07) | 68.00 (30.04) | 67.71 (29.33) | 54.43 (24.54) | 61.27 (33.86) | 77.33 (34.96) | 85.93 (22.13) | F=1.72 |
| LK Endoscopy total score | 5.63 (3.34) | 8.00 (2.48) | 5.93 (4.01) | 2.67 (1.16) | 7.76 (3.75) | 7.07 (3.36) | 6.25 (2.92) | 7.94 (4.37) | 7.33 (3.06) | 8.13 (2.92) | F=1.38 |
| SNOT-22 total score | 43.00 (20.71) | 54.79 (25.16) | 45.00 (19.05) | 59.67 (26.65) | 54.35 (28.34) | 44.84 (22.21) | 44.85 (43.50) | 47.88 (17.96) | 63.80 (13.33) | 55.60 (18.20) | F=0.95 |
| CRS subtypes: | |||||||||||
| No AERD (N = 109; 85.2%) | 8 (100%) | 13 (92.9%) | 13 (86.7%) | 3 (100%) | 15 (88.2%) | 23 (85.2%) | 8 (100%) | 14 (87.5%) | 2 (40.0%) | 10 (66.7%) | ꭕ2=15.68 |
| AERD (N = 19; 14.8%) | 0 (0.0%) | 1 (7.1%) | 2 (13.3%) | 0 (0.0%) | 2 (11.8%) | 4 (14.8%) | 0 (0.0%) | 2 (12.5%) | 3 (60.0%) | 5 (33.3%) | |
| No AFRS (N = 121; 94.5%) | 7 (87.5%) | 12 (85.7%) | 15 (100%) | 3 (100%) | 17 (100%) | 27 (100%) | 7 (87.5%) | 15 (93.8%) | 4 (80.0%) | 14 (93.3%) | ꭕ2=10.92 |
| AFRS (N = 7; 5.5%) | 1 (12.5%) | 2 (14.3%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (12.5%) | 1 (6.3%) | 1 (20.0%) | 1 (6.7%) | |
| CRS Patients (N = 128) | N=8 | N=14 | N=15 | N=3 | N=17 | N=27 | N=8 | N=16 | N=5 | N=15 | |
N=198. SD, standard deviation; T, Sniffin’ Sticks threshold score; D, Sniffin’ Sticks discrimination score; I, Sniffin’ Sticks identification score; QOD-NS, Questionnaire of Olfactory Dysfunction-Negative Statements; OCES, olfactory cleft endoscopy score; OCES total scores were only available for N=159 patients; OCES total scores were available for N=159 patients. CRS, chronic rhinosinusitis; LM-CT, Lund-Mackay computed tomography; OC, olfactory cleft; LK, Lund-Kennedy; SNOT-22, 22-item SinoNasal Outcome Test; AERD, aspirin exacerbated respiratory disease; AFRS, allergic fungal rhinosinusitis; Analyses based on the subset of patients with CRS with sample sizes in each cluster in the last row.
indicates significant differences (p≤0.001) between any two cluster groups using F-test or chi-square (ꭕ2) statistics.
Clinical Measures of Disease
Disease status, demographics, and comorbidities were then compared across the 10 clusters defined solely by OC biomarkers (Table 3). There was a significant difference in the frequency of controls, CRSsNP, and CRSwNP across the clusters (LRT χ2(18) = 178.64, p<0.001). Nearly all of the control subjects (n=61/70=87.1%) belonged to either Cluster 3 or Cluster 4, the two clusters defined by the lowest inflammatory cytokine profile. In contrast, only 4% (n=3/75) of the patients with CRSwNP belonged to one of these control clusters. Most of the CRSwNP patients were spread across clusters 2, 5, 6, 8, 10. Of these groups, clusters 2 and 10 are dominated by Type 2 cytokines, whereas clusters 5 and 6 have relatively low levels of IL5, IL13, or IgE. In patients with CRSsNP, 28.3% (n=12/53) belonged to either cluster 3 or 4, sharing a similar biomarker profile with control subjects. The largest percentage of patients with CRSsNP belonged to cluster 6, which is characterized by moderately elevated levels of IL6, IL8, IL17, and TNFα. Of note, the remainder of patients with CRSsNP were quite heterogeneous and spread out across the other clusters. A total of 34.0% (n=18/53) of patients with CRSsNP belonged to clusters that included elevations in Type 2 cytokines.
Table 3.
Global differences in patient disease status and comorbid factors across the 10 Clusters
| Cluster | Test statistic | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||
| Patient Disease Status: | |||||||||||
| Non-CRS controls (N = 70; 35.4%) | 4 (33.3%) | 1 (6.7%) | 27 (64.3%) | 34 (91.9%) | 3 (15.0%) | 1 (3.6%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | ꭕ2=178.64** |
| CRSsNP (N = 53; 26.8%) | 6 (50.0%) | 0 (0.0%) | 12 (28.6%) | 3 (8.1%) | 6 (30.0%) | 14 (50.0%) | 4 (50.0%) | 5 (31.3%) | 2 (40.0%) | 1 (6.7%) | |
| CRSwNP (N = 75; 37.9%) | 2 (16.7%) | 14 (93.3%) | 3 (7.1%) | 0 (0.0%) | 11 (55.0%) | 13 (46.4%) | 4 (50.0%) | 11 (68.8%) | 3 (60.0%) | 14 (93.3%) | |
| Comorbidity: | |||||||||||
| No Asthma (N = 140; 70.7%) | 10 (83.3%) | 6 (40.0%) | 36 (85.7%) | 34 (91.9%) | 15 (75.0%) | 12 (42.9%) | 7 (87.5%) | 9 (56.3%) | 4 (80.0%) | 7 (46.7%) | ꭕ2=38.78** |
| Asthma (N = 58; 29.3%) | 2 (16.7%) | 9 (60.0%) | 6 (14.3%) | 3 (8.1%) | 5 (25.0%) | 16 (57.1%) | 1 (12.5%) | 7 (43.8%) | 1 (20.0%) | 8 (53.5%) | |
| No allergic rhinitis / response (N = 113; 57.1%) | 9 (75.0%) | 6 (40.0%) | 24 (57.1%) | 29 (78.4%) | 10 (50.0%) | 13 (46.4%) | 5 (62.5%) | 8 (50.0%) | 3 (60.0%) | 3 (20.0%) | ꭕ2=21.45* |
| Allergic rhinitis / response (N = 85; 42.9%) | 3 (25.0%) | 9 (60.0%) | 18 (42.9%) | 8 (21.6%) | 10 (50.0%) | 15 (53.6%) | 3 (37.5%) | 8 (50.0%) | 2 (40.0%) | 12 (80.0%) | |
| Not Current Smoker (N = 189; 95.5%) | 12 (100%) | 15 (100%) | 38 (90.5%) | 36 (97.3%) | 17 (85.0%) | 28 (100%) | 7 (87.5%) | 16 (100%) | 5 (100%) | 15 (100%) | ꭕ2= 14.68 |
| Current Smoker (N = 9; 4.5%) | 0 (0.0%) | 0 (0.0%) | 4 (9.5%) | 1 (2.7%) | 3 (15.0%) | 0 (0.0%) | 1 (12.5%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | |
| No Diabetes (N = 179; 90.4%) | 11 (97.7%) | 14 (93.3%) | 39 (92.9%) | 34 (91.9%) | 18 (90.0%) | 25 (89.3%) | 7 (87.5%) | 13 (81.3%) | 5 (100%) | 13 (86.7%) | ꭕ2= 15.25 |
| Insulin Dependent Diabetes (N = 8; 4.0%) | 1 (8.3%) | 0 (0.0%) | 1 (2.4%) | 1 (2.7%) | 2 (10.0%) | 2 (7.1%) | 1 (12.5%) | 0 (0.0%) | 0 (0.0%) | 1 (6.7%) | |
| Non-Insulin Dependent Diabetes (N = 11, 5.6%) | 0 (0.0%) | 1 (6.7%) | 2 (4.8%) | 2 (5.4%) | 0 (0.0%) | 1 (3.6%) | 0 (0.0%) | 3 (18.8%) | 0 (0.0%) | 1 (6.7%) | |
| No GERD (N = 157; 79.3%) | 9 (75.0%) | 10 (66.7%) | 33 (78.6%) | 34 (91.9%) | 18 (90.0%) | 20 (71.4%) | 5 (62.5%) | 13 (81.3%) | 5 (100%) | 10 (66.7%) | ꭕ2=13.29 |
| GERD (N = 41; 20.7%) | 3 (25.0%) | 5 (33.3%) | 9 (21.4%) | 3 (8.1%) | 2 (10.0%) | 8 (28.6%) | 3 (37.5%) | 3 (18.8%) | 0 (0.0%) | 5 (33.3%) | |
| Overall (N = 198) | N=12 | N=15 | N=42 | N=37 | N=20 | N=28 | N=8 | N=16 | N=5 | N=15 | |
Percentages (in parentheses) are relative to the total number of patients in that cluster. CRS, chronic rhinosinusitis; GERD, gastroesophageal reflux disease. CRSsNP, chronic rhinosinusitis without nasal polyposis; CRSwNP, chronic rhinosinusitis with nasal polyposis; ꭕ2, chi-square test statistic.
There were no significant difference in demographics across the clusters, including age, gender, education, or race/ethnicity. Differences were seen across clusters with respect to medical comorbidities known to be associated with CRS, including allergic rhinitis [LRT ꭕ2(9) = 21.541, p=0.010] and asthma [LRT ꭕ2(9) = 38.770, p<0.001]. No significant difference was seen for current smoking status, diabetes mellitus, or gastroesophageal reflux disease.
Olfactory Measures
Olfactory-specific measures were then compared across the 10 clusters. Psychophysical olfactory scores were significantly different across the groups, including composite Sniffin’ Sticks TDI, as well as threshold, discrimination, and identification scores (p<0.001 for all; Table 4). The highest TDI scores, indicating the best olfactory function, were found in Cluster 3 (TDI=28.57±6.67) and Cluster 4 (TDI=30.78±5.45), which were dominated by subjects with the lowest inflammatory profile. Cluster 8 also had relatively better olfaction (TDI=26.14±9.04) despite being comprised mostly of patients with CRSwNP. The lowest olfactory scores were seen in Cluster 2 (TDI=18.17±10.03) and Cluster 10 (TDI=17.13±7.64), both clusters which are dominated by IL5, 13, and IgE. Olfactory-specific QOL scores also significantly differed across the clusters (p<0.001), with QOD-NS scores mirroring closely the overall TDI scores. This suggests that members of different clusters not only had different objective olfaction, but these differences translated into real-world impacts perceived by individual subjects. Lastly, we compared endoscopic appearance of the OC across clusters. There was a significant difference in total OCES scores across clusters (p<0.001). As might be expected, higher OCES scores, indicating greater inflammation, were seen in clusters with the worst objective olfactory scores.
CRS-Specific Measures
CRS-specific measures were then compared across patients with CRS (Table 4). Significant differences in Lund-Mackay CT scores were seen across clusters, ranging from 10.0±3.5 to 17.0±4.0 (p=0.043). The percent opacification of the middle portion of the OC on CT scan was also significantly different across clusters, ranging from 47.3%±31.5% to 85.6%±20.8% (p=0.050). In contrast, no significant difference was seen for overall Lund-Kennedy endoscopy scores, nor were any differences seen across CRS-specific QOL as measured by the SNOT-22 (p=0.487).
DISCUSSION
Findings from this study demonstrate that clustering based solely on OC biomarkers can organize patients into clinically meaningful endotypes. These endotypes not only discriminate between CRS and controls, but also differed on clinically-relevant measures including objective olfactory function, olfactory-specific QOL, and opacification of the OC assessed by both CT scan and endoscopy. These data support the idea that inflammation in CRS is complex and heterogenous, but can be organized into endotypes whose membership is not entirely obvious based on clinical phenotype alone. These findings need to be validated, but have important implications as we continue to develop targeted therapeutics and personalized approaches to clinical decision-making.
This study is the first to endotype patients based on mucus collected from the OC and utilized a unique array of cytokines, chemokines, and growth factors. Therefore, findings cannot be directly compared to prior research. However, over the last 4 years several studies have attempted to endotype patients with CRS, including groups from Europe2, New Zealand4, China3 and the U.S.5,6 Methodologies vary significantly between these studies, with some using phenotype-free approaches and others clustering based on both biomarkers and clinical features. All but one of these studies used sinus mucosal tissue, usually collected at the time of surgery, and most studies similarly included patients on maintenance medical therapy. With regard to cluster numbers, the European study reported by Tomassen et al. also found 10 clusters.2 Similarly sized studies in New Zealand and China described 8 and 7 clusters, respectively.3,4 The inflammatory makeup of clusters across studies likely differs based on the array of biomarkers included for each study, geographic makeup of patients, and possibly maintenance medical therapies. However, all studies support the idea that the inflammatory profile is not homogenous across patients with CRS, nor is it homogenous across patients stratified by polyp status alone. Our data, together with these other studies, suggest that a richness of underlying inflammatory endotype exists in CRS and that the number of subtypes is probably much closer to 10 than it is to the current simple clinical dichotomy based on polyp status.
Understanding a patient’s inflammatory endotype may have important treatment implications with regard to olfaction. At present, most patients with CRS-related olfactory loss are treated initially with corticosteroids, either topical or oral, and there are reasonable data showing modest efficacy with these medications.10 Given their broad anti-inflammatory effect, it makes sense based on our findings that most CRS patients, with the exception of those in Cluster 3 or 4, might have the potential to improve with corticosteroids. However, newer medications such as biologics have a much more precise anti-inflammatory mechanism of action. Biologics are currently available for CRSwNP that target IL4/13 (e.g. dupilumab), and IgE (e.g. omalizumab), with data from randomized clinical trials showing significant improvement in objective olfaction as compared with placebo.11,35 Monoclonal antibodies that target IL5 are also in phase 3 clinical trials in patients with CRS.36,37 At present, these medications have only been studied in CRSwNP and thus have approval by the U.S. Food and Drug Administration only for patients with CRSwNP. Within our cohort, many patients with CRSwNP did belong to clusters with elevated IL4, IL5, IL13, or IgE and might be expected to improve with current biologics. Cluster 2 and Cluster 10 were particularly elevated in these cytokines. One distinguishing feature of Cluster 2 versus 10 was that Cluster 2 was associated with elevations in chemokines that promote monocyte/macrophage recruitment (CCL2, CCL3, CCL20) as well as TNFα, which is commonly produced by macrophages. This supports prior reports demonstrating that elevated TNFα can drive olfactory loss.38 Additionally, IL-6 was also elevated in cluster 2, elevations of which have also been shown to be associated with olfactory loss.39,40 Consistent with the role of inflammation in driving olfactory dysfunction, cluster 8 was composed of largely CRSwNP patients with normal olfactory function, but significantly less inflammation than the other CRSwNP-dominated clusters. However, there was a notable portion of patients with CRSwNP that fell into clusters that had low levels of these cytokines and thus theoretically might not respond as well. This included those in Clusters 3–7 and encompassed 41.3% (n=31/75) of all CRSwNP patients. Worldwide, the percentage of patients with CRSwNP that fall into specific clusters likely varies based on country of origin and thus the racial and ethnic makeup of our cohort should be kept in mind when interpreting these results.
Notably, patients with CRSsNP were spread out across a number of clusters with markedly different inflammatory profiles. In patients with CRSsNP, 28.3% belonged to clusters with very little inflammation, suggesting that anti-inflammatory medications of any type may not be efficacious with regard to olfaction. Importantly, 34% of CRSsNP patients belonged to clusters that included elevations in Type 2 cytokines, suggesting currently available biologics could have efficacy in these patients despite lack of visible polyposis on endoscopy. Although Type 2 cytokines are often highlighted given currently available biologics, there remains clusters characterized by other noteworthy cytokines. Clusters 8 and 9 include elevations of IL6 and TNFα, which are known to decrease in respiratory mucosa after macrolide treatment.30 Clusters 1 and 9 are enriched in IL-33, a stronger promoter of Type 2 inflammation, which is a target of biologics under development.41,42
Biomarkers used to endotype subjects in this study were all collected in a non-invasive fashion from mucus. This is in contrast to most other endotyping studies which utilized tissue samples, often done at the time of surgery.2–4,6 Prior work has demonstrated significant correlations between mucus and tissue levels of various cytokines.43 We have also previously shown that OC mucus proteins correlate with olfactory function in a separate cohort of CRS patients.25,26 One other study has used mucus biomarkers collected from the middle meatus to endotype patients with CRS and was able to show differences in olfaction across clusters.5 Taken together, these studies are proof of concept that mucus biomarkers can be used to identify clinically-relevant endotypes in patients with CRS. Mucus sampling has the notable advantage that it can readily be performed in clinic without the need for an invasive biopsy. This advantage is noteworthy as it would allow a patient’s endotype to be assessed regardless of whether surgery was being contemplated. The patient’s endotype could then factor into clinical decision-making as patients and providers weigh various options, particularly those with significant costs such as surgery or biologic therapy.
Although the endotypes identified in this cluster analysis have characteristic inflammatory profiles, it remains unknown whether cluster membership actually predicts treatment response with regard to olfaction. We also don’t know the degree to which endotypes are stable over time. Future studies will need to reproduce these results in a separate validation cohort, determine whether treatment efficacy truly varies across endotypes for any given therapeutic, and establish whether endotypes can change over time. Much of the above discussion is premised on the targeted mechanism of action of newer medications like biologics. If monoclonal antibodies truly work through their putative mechanisms, then one would expect that therapeutic effect would be greatest in patients whose inflammation is characterized by those specific cytokines and lesser in those without elevations. Future studies will need to explore outcomes across endotypes for each specific biologic.36 Because each biologic works through a specific mechanism, theoretically an individual endotype might respond better to one biologic than another. Therefore, comparative effectiveness studies should be informed by underlying endotypes. Although personalized medicine is often focused on medications, the same logic applies to surgical treatment. The efficacy of endoscopic sinus surgery could vary across endotypes; or, certain endotypes could require variations in surgical technique in order to achieve equal efficacy. These concepts will need to be tested in vivo in future clinical studies, and data from this study provide some of the framework needed to design these studies.
If endotypes are ultimately shown to impact outcomes for specific treatments, the question remains how to translate these findings into actionable tools in a real-world clinical setting. In our opinion, mucus biomarkers offer more promise than tissue markers given ease of collection. However, collection techniques and assays will need to be standardized so that results are reproducible and generalizable.44 Although mucus was collected from the OC in a targeted fashion, the exact source of this mucus remains unknown. Yoshikawa et al. simultaneously collected mucus from the OC and anterior nasal cavity in healthy adults using a similar endoscopic-guided technique and were able to show that OC mucus had a unique proteomic profile.45 Whether mucus produced in the sinuses in patients with CRS can traffic into and mix with olfactory mucus is unknown and cannot be determined with this study design. Additionally, the array of biomarkers necessary to make specific decisions will need to be determined and discriminant analyses developed that appropriately assign endotype membership to individual patients. Inherently, these arrays will be influenced not only by underlying endotype, but also on available treatments. As newer targeted treatments are developed, arrays will need to include those biomarkers which characterize responsiveness to that specific treatment.
CONCLUSION
Findings from this initial study suggest that olfactory cleft biomarkers can be obtained in a non-invasive fashion and utilized to cluster patients into endotypes that are clinically meaningful. These endotypes not only discriminate between CRS and controls, but also differ on clinically-relevant measures including objective olfactory function, olfactory-specific QOL, and opacification of the OC assessed by both CT scan and endoscopy. These data add to the evolving conceptual framework that support that while inflammation in CRS is complex and heterogenous, biomarkers can be used to organize CRS into endotypes whose membership is not predicated on clinical phenotype alone. If validated, these concepts have important treatment implications for CRS generally and to CRS-related OD as we continue to develop targeted therapies and refine personalized therapeutic approaches.
Supplementary Material
Clinical implications:
Olfactory biomarkers can be utilized to organize chronic rhinosinusitis into endotypes that reflect underlying inflammation. These endotypes are likely to be the foundation of personalized treatment for CRS-related olfactory dysfunction.
Funding Disclosures:
ZMS, RJS, TEB, JAA, VRR, JKM, JCM, and TLS are supported by grant mechanisms from the National Institute on Deafness and Other Communication Disorders (NIDCD), one of the National Institutes of Health, Bethesda, MD (R03 DC013651-01; PI: ZMS; R01 DC005805; PIs: TLS and ZMS; K23DC014747; PI: VRR). JKM is also supported by grants provided by the National Institute of Allergy and Infectious Disease (R01 AI34698 and R01 AI144364). These funding organizations did, in no way, contribute to the design or conduct of this study; preparation, review, approval or decision to submit this manuscript for publication.
Abbreviations:
- CRS
chronic rhinosinusitis
- OD
olfactory dysfunction
- OC
olfactory cleft
- U.S.
United States
- CT
computed tomography
- AERD
aspirin exacerbated respiratory disease
- NSAIDs
nonsteroidal anti-inflammatory drugs
- CRSwNP
chronic rhinosinusitis with nasal polyps
- CRSsNP
chronic rhinosinusitis without nasal polyps
- LKES
Lund Kennedy Endoscopy Score
- SNOT-22
22-item Sinonasal Outcomes Test
- QOL
quality of life
- TDI
Threshold Discrimination Identification
- QOD-NS
Questionnaire of Olfactory Dysfunction-Negative Statements
- OCES
Olfactory Cleft Endoscopy Scale
- ELISA
enzyme-linked immunosorbent assay
- ANOVA
analyses of variance
- PCA
principal component analysis
- PCs
principal components
- CCL2
C-C Motif Chemokine Ligand 2 (monocyte chemoattractant protein 1)
- CCL3
C-C Motif Chemokine Ligand 3 (macrophage inflammatory protein 1-alpha)
- CCL5
C-C Motif Chemokine Ligand 5 (RANTES)
- CCL11
C-C Motif Chemokine Ligand 11 (eotaxin 1)
- CCL20
C-C Motif Chemokine Ligand 20 (macrophage inflammatory protein 3)
- CXCL
chemokine (C-X-C motif) ligand
- EGF
Epidermal growth factor
- bFGF
Basic fibroblast growth factor
- IGE
Immunoglobulin E
- IL
interleukin
- TNFα
Tumor necrosis factor alpha
- VEGF-A
Vascular endothelial growth factor A
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Potential Conflict of Interest Disclosures: None of the following consultancy positions or grant support are affiliated with this investigation or manuscript: Z.M.S: Consultant for Olympus Medical Systems, OptiNose US Inc., Genentech, Lyra Therapeutics, and Sinusonic. R.J.S.: Consultant for ENT Stryker, Medtronic Systems Inc., Healthy Humming, GlaxoSmithKline, Sanofi, and Optinose US Inc. Supported from grants from: ENT Stryker, Healthy Humming, GlaxoSmithKline, Sanofi, and Optinose US Inc. T.E.B: No potential conflicts of interest to disclose. J.A.A.: Consultant for OptiNose US Inc., Medtronic Inc. and GlycoMira Therapeutics, Inc. V.R.R.: Consultant for OptiNose US Inc. and Medtronic Systems Inc. Advisory board member for Genentech, Novartis, and GlaxoSmithKline. J.L.M: No potential conflicts of interest to disclose. J.K.M: No potential conflicts of interest to disclose. J.C.M: No potential conflicts of interest to disclose. T.L.S: No potential conflicts of interest to disclose.
CLINICAL TRIAL REGISTRATION: Public clinical trial registration [www.clinicaltrials.gov] identification #NCT02720653.
REFERENCES
- 1.Blackwell DL, Lucas JW, Clarke TC. Summary health statistics for U.S. adults: national health interview survey, 2012. Vital Health Stat 2014; 10: 1–161. [PubMed] [Google Scholar]
- 2.Tomassen P, Vandeplas G, Van Zele T, Cardell LO, Arebro J, Olze H, et al. Inflammatory endotypes of chronic rhinosinusitis based on cluster analysis of biomarkers. J Allergy Clin Immunol 2016; 137:1449–56. [DOI] [PubMed] [Google Scholar]
- 3.Liao B, Liu J-X, Li Z-Y, Zhen Z, Cao P-P, Yao Y, et al. Multidimensional endotypes of chronic rhinosinusitis and their association with treatment outcomes. Allergy 2018; 73:1459–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoggard M, Waldvogel-Thurlow S, Zoing M, Chang K, Radcliff FJ, Mackenzie BW, et al. Inflammatory endotypes and microbial associations in chronic rhinosinusitis. Front Immunol 2018; 9:2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner JH, Chandra RK, Li P, Bonnet K, Schlundt DG. Identification of clinically relevant chronic rhinosinusitis endotypes using cluster analysis of mucus cytokines. J Allergy Clin Immunol 2018; 141:1895–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Divekar R, Rank M, Squillace D, Kita H, Lal D. Unsupervised network mapping of commercially available immunoassay yields three distinct chronic rhinosinusitis endotypes. Int Forum Allergy Rhinol 2017; 7:373–79. [DOI] [PubMed] [Google Scholar]
- 7.Mattos JL, Rudmik L, Schlosser RJ, Smith TL, Mace JC, Alt JA, et al. Symptom importance, patient expectations, and satisfaction in chronic rhinosinusitis. Int Forum Allergy Rhinol 2019; 9(6):593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohli P, Naik AN, Harruff EE, Nguyen SA, Schlosser RJ, Soler ZM. The prevalence of olfactory dysfunction in chronic rhinosinusitis. Laryngoscope 2017; 127:309–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hastan D, Fokkens WJ, Bachert C, Newson RB, Bislimovska J, Bockelbrink A, et al. Chronic rhinosinusitis in Europe--an underestimated disease. A GA(2)LEN study. Allergy 2011; 66(9):1216–23. [DOI] [PubMed] [Google Scholar]
- 10.Banglawala SM, Oyer SL, Lohia S, Psaltis AJ, Soler ZM, Schlosser RJ. Olfactory outcomes in chronic rhinosinusitis with nasal polyposis after medical treatments: a systematic review and meta-analysis. Int Forum Allergy Rhinol 2014; 4:986–94. [DOI] [PubMed] [Google Scholar]
- 11.Bachert C, Han JK, Desrosiers M, Hellings PW, Amin N, Lee SE, et al. Efficacy and safety of dupilumab in patients with severe chronic rhinosinusitis with nasal polyps (LIBERTY NP SINUS-24 and LIBERTY NP SINUS-52): results from two multicentre, randomised, double-blind, placebo-controlled, parallel-group phase 3 trials. Lancet 2019; 394(10209):1638–50. [DOI] [PubMed] [Google Scholar]
- 12.Haxel BR. Recovery of olfaction after sinus surgery for chronic rhinosinusitis: A review. Laryngoscope 2019; 129:1053–59. [DOI] [PubMed] [Google Scholar]
- 13.Kohli P, Naik AN, Farhood Z, Ong AA, Nguyen SA, Soler ZM, et al. Olfactory outcomes after endoscopic sinus surgery for chronic rhinosinusitis: a meta-analysis. Otolaryngol Head Neck Surg 2016; 155(6):936–48. [DOI] [PubMed] [Google Scholar]
- 14.Soler ZM, Smith TL, Alt JA, Ramakrishnan VR, Mace JC, Schlosser RJ. Olfactory-specific quality of life outcomes after endoscopic sinus surgery. Int Forum Allergy Rhinol 2016; 6:407–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenfeld RM, Piccirillo JF, Chandrasekhar SS, Brook I, Kumar KA, Kramper M, et al. Clinical practice guideline (update): adult sinusitis. Otolaryngol Head Neck Surg 2015; 152(2 Suppl):S1–39. [DOI] [PubMed] [Google Scholar]
- 16.Saravanan K, Panda NK, Chakrabarti A, Das A, Bapuraj RJ. Allergic fungal rhinosinusitis: an attempt to resolve the diagnostic dilemma. Arch Otolaryngol Head Neck Surg. 2006; 132:173–8. [DOI] [PubMed] [Google Scholar]
- 17.Lund VJ, Mackay IS. Staging in rhinosinusitus. Rhinology 1993; 31:183–4. [PubMed] [Google Scholar]
- 18.Lund VJ, Kennedy DW. Staging for rhinosinusitis. Otolaryngol Head Neck Surg 1997; 117:S35–40. [DOI] [PubMed] [Google Scholar]
- 19.Hopkins C, Gillett S, Slack R, Lund VJ, Browne JP. Psychometric validity of the 22-item Sinonasal Outcome Test. Clin Otolaryngol 2009; 34:447–54. [DOI] [PubMed] [Google Scholar]
- 20.Hummel T, Sekinger B, Wolf SR, Pauli E, Kobal G. ‘Sniffin’ sticks’: olfactory performance assessed by the combined testing of odor identification, odor discrimination and olfactory threshold. Chem Senses 1997; 22:39–52. [DOI] [PubMed] [Google Scholar]
- 21.Frasnelli J, Hummel T. Olfactory dysfunction and daily life. Eur Archives Otorhinolaryngol 2005; 262:231–35. [DOI] [PubMed] [Google Scholar]
- 22.Simopoulos E, Katotomichelakis M, Gouveris H, Tripsianis G, Livaditis M, Danielides V. Olfaction-associated quality of life in chronic rhinosinusitis: adaptation and validation of an olfaction-specific questionnaire. Laryngoscope 2012; 122(7):1450–54. [DOI] [PubMed] [Google Scholar]
- 23.Soler ZM, Hyer JM, Karnezis TT, Schlosser RJ. The olfactory cleft endoscopy scale correlates with olfactory metrics in patients with chronic rhinosinusitis. Int Forum Allergy Rhinol 2016; 6(3):293–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loftus C, Schlosser RJ, Smith TL, Alt JA, Ramakrishnan VR, Mattos JL, et al. Olfactory cleft and sinus opacification differentially impact olfaction in chronic rhinosinusitis. Laryngoscope 2020; 130(10):2311–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soler ZM, Yoo F, Schlosser RJ, Mulligan JK, Ramakrishnan VR, Beswick DM, et al. Correlation of mucus inflammatory proteins and olfaction in chronic rhinosinusitis. Int Forum Allergy Rhinol 2020; 10(3):343–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlosser RJ, Mulligan JK, Hyer JM, Karnezis TT, Gudis DA, Soler ZM. Mucous cytokine levels in chronic rhinosinusitis-associated olfactory loss. JAMA Otolaryngol Head Neck Surg. 2016; 142:731–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoo F, Soler ZM, Mulligan JK, Storck KA, Lamira JM, Pasquini WN, et al. Olfactory cleft mucus proteins associated with olfactory dysfunction in a cohort without chronic rhinosinusitis. Int Forum Allergy Rhinol 2019; 9(10):1151–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bachert C, Zhang N, Hellings PW, Bousquet J. Endotype-driven care pathways in patients with chronic rhinosinusitis. J Allergy Clin Immunol 2018; 141:1543–51. [DOI] [PubMed] [Google Scholar]
- 29.Staudacher AG, Peters AT, Kato A, Stevens WW. Use of endotypes, phenotypes, and inflammatory markers to guide treatment decisions in chronic rhinosinusitis. Ann Allergy Asthma Immunol 2020; 124(4):318–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zimmermann P, Ziesenitz VC, Curtis N, Ritz N. The immunomodulatory effects of macrolides-a systematic review of the underlying mechanisms. Front Immunol 2018; 9:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Games PA, Howell JF. Pair wise multiple comparison procedures with unequal N’s and/or variances. J Ed Stat 1976; 1(2):113–25. [Google Scholar]
- 32.Welch BL. On the comparison of several mean values: an alternative approach. Biometrika 1951; 38(3/4):330–6. [Google Scholar]
- 33.Hair JF, Black WC. Cluster analysis (pp: 147–205). In Grimm LG and Yarnold PR (Eds.), Reading and understanding more multivariate statistics. Washington D.C.: American Psychological Association; 2000. [Google Scholar]
- 34.Ward JH Jr. Hierarchical grouping to optimize an objective function. J Am Stat Assoc 1963; 58:236–44. [Google Scholar]
- 35.Gevaert P, Omachi TA, Corren J, Mullol J, Han J, Lee SE, et al. Efficacy and safety of omalizumab in nasal polyposis: 2 randomized phase 3 trials. J Allergy Clin Immunol 2020. 146(3):595–605. [DOI] [PubMed] [Google Scholar]
- 36.Naclerio R, Baroody F, Bachert C, Bleier B, Borish L, Brittain E, et al. Clinical research needs for the management of chronic rhinosinusitis with nasal polyps in the new era of biologics: A National Institute of Allergy and Infectious Diseases workshop. J Allergy Clin Immunol Pract 2020; 8(5):1532–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roland LT, Smith TL, Schlosser RJ, Soler ZM, Peters AT, Laidlaw TM, et al. Guidance for contemporary use of biologics in management of chronic rhinosinusitis with nasal polys: a discussion from a National Institutes of Health-sponsored workshop. Int Forum Allergy Rhinol 2020. 10(9):1037–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia DS, Chen M, Smith AK, Lazarini PR, Lane AP. Role of the type-I tumor necrosis factor receptor in inflammation-associated olfactory dysfunction. Int Forum Allergy Rhinol 2017; 7(2):160–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu J, Chandra RK, Li P, Hull BP, Turner JH. Olfactory and middle meatal cytokine levels correlate with olfactory function in chronic rhinosinusitis. 2018; 128(9):E304–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumari E, Velloso FJ, Nasuhidehnavi A, Somasundaram A, Savanur VH, Buono KD, et al. Developmental IL-6 exposure favors production of PDGF-responsive multipotential progenitors at the expense of neural stem cells and other progenitors. Stem Cell Reports 2020; 14(5):861–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allinne J, Scott G, Lim WK, Birchard D, Erjefalt JS, Sanden C, et al. IL-33 blockade affects mediators of persistence and exacerbation in a model of chronic airway inflammation. J Allergy Clin Immunol 2019; 144(6):1624–37. [DOI] [PubMed] [Google Scholar]
- 42.Donovan C, Hansbro PM. IL-33 in Chronic Respiratory Disease: From Preclinical to Clinical Studies. ACS Pharmacol Transl Sci 2019; 3(1):56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oyer SL, Mulligan JK, Psaltis AJ, Henriquez OA, Schlosser RJ. Cytokine correlation between sinus tissue and nasal secretions among chronic rhinosinusitis and controls. Laryngoscope 2013; 123(12):E72–8. [DOI] [PubMed] [Google Scholar]
- 44.Massey CJ, Diaz Del Valle F, Abuzeid WM, Levy JM, Mueller S, Levine CG, et al. Sample collection for laboratory-based study of the nasal airway and sinuses: a research compendium. Int Forum Allergy Rhinol 2020; 10(3):303–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshikawa K, Wang H, Jaen C, Haneoka M, Saito N, Nakamura J, et al. The human olfactory cleft mucus proteome and its age-related changes. Sci Rep 2018; 8(1):17170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


