Abstract
The LOX genes have been identified and characterized in many plant species, but studies on the banana LOX genes are very limited. In this study, we respectively identified 18 MaLOX, 11 MbLOX, and 12 MiLOX genes from the Musa acuminata, M. balbisiana and M. itinerans genome data, investigated their gene structures and characterized the physicochemical properties of their encoded proteins. Banana LOXs showed a preference for using and ending with G/C and their encoded proteins can be classified into 9-LOX, Type I 13-LOX and Type II 13-LOX subfamilies. The expansion of the MaLOXs might result from the combined actions of genome-wide, tandem, and segmental duplications. However, tandem and segmental duplications contribute to the expansion of MbLOXs. Transcriptome data based gene expression analysis showed that MaLOX1, 4, and 7 were highly expressed in fruit and their expression levels were significantly regulated by ethylene. And 11, 12 and 7 MaLOXs were found to be low temperature-, high temperature-, and Fusarium oxysporum f. sp. Cubense tropical race 4 (FocTR4)-responsive, respectively. MaLOX8, 9 and 13 are responsive to all the three stresses, MaLOX4 and MaLOX12 are high temperature- and FocTR4-responsive; MaLOX6 and MaLOX17 are significantly induced by low temperature and FocTR4; and the expression of MaLOX7 and MaLOX16 are only affected by high temperature. Quantitative real-time PCR (qRT-PCR) analysis revealed that the expression levels of several MaLOXs are regulated by MeJA and FocTR4, indicating that they can increase the resistance of banana by regulating the JA pathway. Additionally, the weighted gene co-expression network analysis (WGCNA) of MaLOXs revealed 3 models respectively for 5 (MaLOX7-11), 3 (MaLOX6, 13, and 17), and 1 (MaLOX12) MaLOX genes. Our findings can provide valuable information for the characterization, evolution, diversity and functionality of MaLOX, MbLOX and MiLOX genes and are helpful for understanding the roles of LOXs in banana growth and development and adaptations to different stresses.
Subject terms: Biotechnology, Molecular biology
Introduction
Lipoxygenases (LOXs, EC:1.13.11.12), non-heme iron-containing oxygenases catalyzing the oxygenation of polyunsaturated fatty acids to produce fatty acid hydroperoxides, play important roles in various physiological progresses such as growth and development, signal transduction, abiotic and biotic stress responses of plants1. The N-terminal and C-terminal of LOX respectively contains a conserved PLAT/LH2 (polycystin-1, lipoxygenase, alpha-toxin/lipoxygenase homology) domain and a typical LOX domain2. The PLAT/LH2 domain functions in mediating the interaction between enzyme and biological membranes3. While the LOX domain, existing a histidine (His)-rich region consisted of [His-(X)4-His-(X)4-His-(X)17-His-(X)8-His], is critical for the iron coordination, substrate binding and enzyme activity4. According to their oxygenation sites on the fatty acid carbon chain, LOXs can be further divided into 9-LOX and 13-LOX5. Moreover, 13-LOXs can be further classified into type I 13-LOX and type II 13-LOX subgroups according to the absence (Type I) or presence (Type II) of chloroplast transit peptides in their N-terminals6.
LOXs are ubiquitously distributed in plants and have been isolated from a variety of plant species, such as Arabidopsis7, rice7, tomato8, poplar9, tea10, cotton11, peach12, and radish13. The expression of plant LOXs have been proved to be regulated by some phytohormones and pathogens. For instance, the expression of Arabidopsis AtLOX1 was abscisic acid and JA inducible14, and the rice OsLOX3 was MeJA and Magnaporthe Grisea inducible15. Their diverse functions during plant growth and developmental and stress response processes have also been experimentally confirmed in various plant species. Arabidopsis AtLOX3 and AtLOX4 double mutant plants showed developmental dysfunctions of higher plant height and increased inflorescence shoots and flowers16. AtLOX2 and AtLOX6 are found to be involved in wound induced JA synthesis in leaves17,18. Transgenic plants overexpressing rice OsLOX2 showed shortened seed germination time19. Kiwifruit AdLOXs were involved in the formation of fruit aroma20. Silencing of CaLOX2 in pepper plants resulted in decreased JA accumulation and reduced thrips resistance21. Transgenic tomato plants overexpressing the tomato lipoxygenase D (TomLoxD) gene resulted in enhanced wound-induced JA biosynthesis and increased Helicoverpa armigera and Botrytis cinerea resistance22. Transgenic Arabidopsis plant overexpressing persimmon DkLOX3 showed increased salt tolerance and disease resistance23.
Banana, as one of the most important and popular fruit, is an herbaceous perennial plant belonging to Musa family. Cultivated banana is generally low in stress resistance and is susceptible to external environmental stresses such as low temperature and Fusarium wilt24. There are also several reports on the expression patterns of some banana LOXs using omic techniques, and their roles in banana responses to high temperature, low temperature and Fusarium wilt have been described25–28. Given that LOXs are vital for plant growth and stress resistance and different LOX members’ functions varied, it is of great importance to analyze the LOX gene family from whole genome level for the clarification of their diverse potentials in banana. In the present study, whole genome wide LOX gene family identification was performed based on the M. acuminata, M. balbisiana and M. itinerans genome data. Totally, we identified 18 MaLOX, 11 MbLOX, and 12 MiLOX family members, which were then subjected to series of bioinformatics analysis to show the chromosome location, gene structure and gene duplication events of LOX genes and to reveal the physiological and biochemical characteristics, subcellular localization, and phylogenetic relationship of their encoded proteins. Moreover, the expression patterns of MaLOXs were investigated using quantitative real time PCR (qRT-PCR) and transcriptome data. Our preliminary results can extend the knowledge of banana LOX gene family and can provide insights into their roles in banana growth and development and stress responses.
Materials and methods
Plant materials
In our previous study, ‘Tianbaojiao’ banana (Musa spp., Cavendish, AAA group) plantlets were used for transcriptome profiling to show the transcriptome changes caused by 4 ℃ low temperature in leaves of four-leaf stage plantlets, by 45 ℃ high temperature in leaves of five-stage plantlets, and by FocTR4 inoculation in banana roots. Moreover, transcriptome changes of natural ripening and ethylene treated ‘Tianbaojiao’ banana fruits at 0, 1, 3, and 5 days were also compared. Moreover, to show the influence of MeJA treatment on the expression of banana LOXs, ‘Brizil’ banana (Musa acuminata cv. Brazil) plantlets at six-leaf stage were exposed to 100 mM MeJA solution (containing 0.02% (v/v) Tween 20) treatment9, treated leaves were sampled at 0, 6, 12, 24 h after MeJA treatment. In addition, in order to further explore the expression of MaLOXs in response to FocTR4 treatment, ‘Zhongjiao No.3’ banana (Musa acuminata cv. Brazil) plantlets at six-leaf stage were inoculated with 1 × 107/mL FocTR4 spore suspension according to the inoculation method described by Wang et al.29. Roots were collected 0 day, 4 days, 2 weeks, and 4 weeks after treatment. Banana plantlets showed no visible symptom in corm until 4 weeks after FocTR4 inoculation. All samples were immediately frozen in liquid nitrogen and stored at − 80 °C for further use. For qRT-PCR analysis, three independent replicates were used for each time point of MeJA and FocTR4 treatments. All the banana materials used in this research were harvested from cultivated varieties (‘Tianbaojiao’ banana is a famous traditional cultivar in Tianbao county, Fujian province, China. ‘Brazil’ is one of the most popular banana variety in the world and ‘Zhongjiao No.3’ is a new banana variety selected from ‘Brazil’ by Institute of fruit science, Guangdong Agricultural Academy), and their collections complied with relevant institutional, national, and international guidelines and legislation.
Identification of banana LOX genes
The genomic DNA, CDS, and protein sequence files of M. acuminata var. DH-Pahang, M. balbisiana var. DH PKW and M. itinerans var. Yunnan were downloaded from the banana genome databases (https://banana-genome-hub.southgreen.fr/). HMMER3.0 software was used to search against the banana protein sequences using The Hidden Markov Model file of Lipoxygenase (PF00305) downloaded from the Pfam database (http://pfam.xfam.org/) with E-value ≤ 1 × 10–5 to obtain candidate LOX proteins, which were further submitted to conserved domain database (CDD, https://www.ncbi.nlm.nih.gov/cdd) for the confirmation of the existence of the lipoxygenase and PLAT/LH2 domains10. Sequences without Lipoxygenase domain and/or PLAT/LH2 domain were removed. The remaining banana LOXs are named sequentially according to the chromosomal location of their corresponding genes. ExPASy (https://web.expasy.org/protparam/) was used to analyze the basic physicochemical properties of LOX proteins. Chloroplast transit peptide and subcellular localization were predicted by ChloroP 1.1 Server (http://www.cbs.dtu.dk/services/ChloroP/) and WoLF PSORT (https://wolfpsort.hgc.jp/). The global sequence alignment program Needle (https://www.ebi.ac.uk/Tools/psa/emboss_needle/) in the EMBOSS tool was used to perform pairwise alignment of protein sequences to determine the similarity and identity between LOX members. Gene structure of banana LOXs was drawn by GSDS (http://gsds.cbi.pku.edu.cn/). The conserved motifs of LOXs (20 maximum number of motifs) were analyzed using MEME suite (http://meme-suite.org/tools/meme) and visualized using TBtools software30. The CodonW software (version 1.4.2, http://codonw.sourceforge.net/) was used to calculate the effective number of codons (ENC), codon adaptation index (CAI), relative synonymous codon usage (RSCU), and other codon preference parameters6.
Phylogenetic analysis
The LOX protein sequences of Arabidopsis thaliana, rice, tomato, poplar and some other plants were downloaded from TAIR (https://www.arabidopsis.org/)7, RGAP (http://rice.plantbiology.msu.edu/)7, SGN tomato (https://solgenomics.net/)8, Phytozome (http://www.phytozome.net/), and NCBI (http://www.ncbi.nlm.nih.gov/)9, respectively. After domain verification using CDD, OsLOX2, OsLOX9, OsLOX14, GmLOX2, and PvLOX2c without incomplete Lipoxygenase and PLAT/LH2 domains were removed. Multiple sequence alignment was performed using Muscle software, and phylogenetic tree was constructed by Neighbor-joining method using MEGA 6.06 (Possion mode, complete deletion, and 1000 bootstrap values) and was visualized using EvolView (https://www.evolgenius.info/evolview/).
Chromosome location and gene duplication analysis
Blast software (version 2.10.0, https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to perform self-alignment and pairwise alignment analysis of LOX proteins (E-value ≤ 1 × 10–10). The intra/inter-species gene collinear relationship of the LOX family was analyzed by using MCScanX (version 0.8, http://chibba.pgml.uga.edu/mcscan2/)31. According to the chromosomal location information, the Circos software (version 0.69-9, http://circos.ca/) was used to visualize the syntenic relationships between banana LOXs and LOXs from other plant species32. KaKs_Calclator 2.0 software (https://sourceforge.net/) was used to estimate synonymous (Ks) and nonsynonymous (Ka) substitution rates33. For the timing of duplication events, the formula: T = Ks/2λ × 10–6 Mya was used to calculate divergence time (T) in millions of years (Mya), where λ = 4.5 × 10− 9 represented the evolution rate of Musa34.
Analysis of cis-acting elements and transcription factor binding sites in the promoters of banana LOX genes
The 1500 bp upstream of the start codon of each banana LOX gene was extracted from the banana genome database. Due to the presence of large numbers of CTT repeat sequences on MaLOX5 promoter region from the genome data, PCR was used to verify its true sequence. It was found that CTT repeat sequences were absent, thus the corrected sequence was used for subsequent analysis. The cis-acting elements of the promoter were predicted using the PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). PlantTFDB (http:// planttfdb.cbi.pku.edu.cn/) was used to predict the transcription factor binding sites (TFBSs) on promoters with the parameter set of p-value ≤ 1e−6. The promoter regions were partitioned to proximal promoter region (500 bp upstream), median promoter region (501–1000 bp upstream) and distal promoter region (1001–1500 bp upstream).
Gene expression analysis using transcriptome data and qRT-PCR
The expression patterns of banana LOX genes under low temperature, high temperature and FocTR4 treatments were analyzed using our previous transcriptome data. The expression values of banana LOX family genes were extracted from the transcriptome data, and heatmap was drawn using HemI1.0 software (http://hemi.biocuckoo.org/). qRT-PCR was used to show the expression patterns of all the banana LOX genes under JA treatment. Total RNA was extracted using RNAprep Pure Plant Kit (TIANGEN, China) according to the manufacturer’s instructions. A total of 1 μg RNA was used for cDNA synthesis using PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) (Takara, China). CDNA was diluted tenfold for subsequent experiments. The PCR reaction conditions used were 95 °C for 30 s, 95 °C for 5 s, and 60 °C for 34 s (40 cycles). Relative gene expression levels were determined using the 2-∆∆Ct method by using MaCAC as an internal reference35. Primers were designed using Oligo 7.0, and their specificity was checked using information obtained from the NCBI website. All primers used in this study are listed in Supplemen Table S1. Statistical analysis and figure drawing were conducted using SPSS 25.0 and GraphPad Prism 6.0 software, respectively.
Weighted gene co-expression network analysis (WGCNA)
Genes with FPKM value greater than 10 in at least one RNA-Seq sample were subjected to WGCNA (version 1.68) analysis to construct and identify co-expressed gene clusters with MaLOXs36. The parameters were set as follows: The optimal β (soft thresholding power) value was 12; the minModuleSize was 30 and the mergeCutHeight was 0.25. Finally, we extracted the co-expression network of all MaLOXs and filtered out the edges with weights below 0.4. We visualized the network connections using the Cytoscape (version 3.8.0, https://cytoscape.org/) program37. The functional enrichment analysis of MaLOXs and co-expressed genes was performed using Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases.
Results
Identification and characterization of banana LOX gene family members
Totally, 18, 11, and 12 LOX genes were identified from M. acuminata, M. balbisiana, and M. itinerans genome, respectively (Table 1, Supplementary Table S2). According to their chromosomal location information, the 18 MaLOXs were defined as MaLOX1-MaLOX18, respectively. Among these MaLOXs, MaLOX5 had two transcripts, which was named as MaLOX5a and MaLOX5b, respectively. MbLOXs and MiLOXs were named in concordance with their MaLOXs homologous (Supplementary Figure S1).
Table 1.
The information of LOX gene family in banana.
| Species | Gene ID | Transcript ID | Gene name | Chromosome location | CDS/bp | Size/aa | Molecular weight/kD | PI | Chloroplast transit peptides | Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|---|
| Musa acuminata | Ma01_g16400 | Ma01_t16400.1 | MaLOX1 | chr01:11878658..11882747 (+) | 2619 | 872 | 98.62 | 6.03 | − | CP |
| Ma01_g18020 | Ma01_t18020.1 | MaLOX2 | chr01:13322958..13326238 (−) | 2544 | 847 | 95.49 | 6.33 | − | CP | |
| Ma01_g18040 | Ma01_t18040.1 | MaLOX3 | chr01:13344016..13347394 (−) | 2544 | 847 | 95.19 | 6.31 | − | CP | |
| Ma01_g18060 | Ma01_t18060.1 | MaLOX4 | chr01:13358464..13361737 (−) | 2586 | 861 | 96.38 | 6.05 | − | CP | |
| Ma02_g07800 | Ma02_t07800.1 | MaLOX5a | chr02:18320819..18324221 (−) | 2082 | 693 | 77.57 | 9.53 | − | CP | |
| Ma02_t07800.2 | MaLOX5b | 2562 | 853 | 97.04 | 7.00 | − | CP | |||
| Ma03_g07770 | Ma03_t07770.1 | MaLOX6 | chr03:5495340..5499190 (−) | 2745 | 914 | 10.30 | 7.71 | Y | Chl | |
| Ma03_g11520 | Ma03_t11520.1 | MaLOX7 | chr03:8935641..8940695 (−) | 2724 | 907 | 10.20 | 6.37 | Y | Chl | |
| Ma06_g26840 | Ma06_t26840.1 | MaLOX8 | chr06:28772817..28775490 (+) | 2061 | 686 | 76.33 | 5.90 | − | ER | |
| Ma06_g26850 | Ma06_t26850.1 | MaLOX9 | chr06:28828834..28832329 (+) | 2622 | 873 | 97.33 | 5.78 | − | CP | |
| Ma06_g26870 | Ma06_t26870.1 | MaLOX10 | chr06:28849493..28860078 (+) | 3396 | 1131 | 126.29 | 6.26 | − | CP | |
| Ma06_g26890 | Ma06_t26890.1 | MaLOX11 | chr06:28883457..28886953 (+) | 2622 | 873 | 97.34 | 6.00 | − | ER | |
| Ma06_g30170 | Ma06_t30170.1 | MaLOX12 | chr06:31521537..31528304 (−) | 2739 | 912 | 102.66 | 7.37 | Y | Chl | |
| Ma08_g23400 | Ma08_t23400.1 | MaLOX13 | chr08:36811891..36815961 (+) | 2736 | 911 | 102.55 | 6.41 | Y | Chl | |
| Ma09_g12090 | Ma09_t12090.1 | MaLOX14 | chr09:8177556..8181866 (−) | 2712 | 903 | 101.93 | 6.31 | Y | Chl | |
| Ma09_g15420 | Ma09_t15420.1 | MaLOX15 | chr09:10750253..10754168 (−) | 2733 | 910 | 102.29 | 6.61 | Y | Chl | |
| Ma09_g19130 | Ma09_t19130.1 | MaLOX16 | chr09:19917117..19920868 (−) | 2856 | 951 | 107.51 | 6.31 | − | CP | |
| Ma09_g19140 | Ma09_t19140.1 | MaLOX17 | chr09:19917453..19921016 (−) | 2568 | 855 | 96.01 | 5.72 | − | CP | |
| Ma10_g17560 | Ma10_t17560.1 | MaLOX18 | chr10:28903758..28908078 (−) | 2760 | 919 | 102.84 | 8.15 | Y | CP | |
| Musa balbisiana | Mba01_g25910 | Mba01_g25910.1 | MbLOX1 | Bchr01:20001697…20005726 (+) | 2610 | 869 | 98.35 | 6.05 | − | CP |
| Mba03_g07670 | Mba03_g07670.1 | MbLOX6 | Bchr03:5604842…5608862 (−) | 1998 | 665 | 75.15 | 9.40 | Y | Chl | |
| Mba06_g26190 | Mba06_g26190.1 | MbLOX10 | Bchr06:31900379…31903722 (+) | 2580 | 859 | 96.02 | 5.68 | − | CP | |
| Mba06_g26200 | Mba06_g26200.1 | MbLOX9 | Bchr06:31926218…31929597 (+) | 2727 | 908 | 101.59 | 6.20 | − | CP | |
| Mba06_g26210 | Mba06_g26210.1 | MbLOX8 | Bchr06:31964229…31967505 (+) | 2670 | 889 | 99.00 | 5.97 | − | Chl | |
| Mba06_g26220 | Mba06_g26220.1 | MbLOX11 | Bchr06:31978370…31981642 (+) | 2682 | 893 | 99.41 | 6.41 | − | Chl | |
| Mba08_g23020 | Mba08_g23020.1 | MbLOX13 | Bchr08:36835632…36839686 (+) | 2742 | 913 | 102.96 | 6.56 | Y | Chl | |
| Mba09_g11450 | Mba09_g11450.1 | MbLOX14 | Bchr09:8300639…8304936 (−) | 2172 | 723 | 82.14 | 6.28 | Y | Chl | |
| Mba09_g14640 | Mba09_g14640.1 | MbLOX15 | Bchr09:10993723…10997549 (−) | 2286 | 761 | 85.39 | 7.64 | Y | Chl | |
| Mba09_g18010 | Mba09_g18010.1 | MbLOX16 | Bchr09:16550622…16556265 (+) | 2568 | 855 | 95.98 | 5.68 | − | CP | |
| Mba10_g15430 | Mba10_g15430.1 | MbLOX18 | Bchr10:32779595…32784030 (−) | 2658 | 885 | 98.85 | 8.38 | Y | CP | |
| Musa itinerans | Mi_g004153 | Mi_g004153 | MiLOX1 | scaffold1338:259261…263079 (+) | 2532 | 843 | 94.99 | 6.12 | − | CP |
| Mi_g014015 | Mi_g014015 | MiLOX5 | scaffold2542:171670…174847 (+) | 2472 | 823 | 93.78 | 6.62 | − | CP | |
| Mi_g017218 | Mi_g017218 | MiLOX18 | scaffold3004:214416…218434 (−) | 2712 | 903 | 101.07 | 8.45 | − | CP | |
| Mi_g017373 | Mi_g017373 | MiLOX13 | scaffold3031:97765…101524 (−) | 2631 | 877 | 98.92 | 6.35 | Y | Chl | |
| Mi_g018964 | Mi_g018964 | MiLOX10 | scaffold34:234414…237513 (−) | 2535 | 845 | 94.59 | 5.89 | − | CP | |
| Mi_g021392 | Mi_g021392 | MiLOX15 | scaffold406:321838…325445 (+) | 2619 | 873 | 98.22 | 6.79 | Y | CP | |
| Mi_g027370 | Mi_g027370 | MiLOX16 | scaffold647:55264…58996 (−) | 2847 | 949 | 107.52 | 6.57 | − | CP | |
| Mi_g027442 | Mi_g027442 | MiLOX7 | scaffold649:208052…213703 (−) | 2541 | 847 | 95.23 | 6.92 | Y | Chl | |
| Mi_g027551 | Mi_g027551 | MiLOX14 | scaffold655:11648…15808 (+) | 2532 | 843 | 95.23 | 6.53 | Y | Chl | |
| Mi_g028690 | Mi_g028690 | MiLOX12 | scaffold7089:49982…56244 (−) | 2613 | 870 | 97.84 | 7.36 | Y | Chl | |
| Mi_g029482 | Mi_g029482 | MiLOX6 | scaffold768:345315…348813 (−) | 2487 | 829 | 93.83 | 6.66 | − | Chl | |
| Mi_g030630 | Mi_g030630 | MiLOX4 | scaffold829:57403…60534 (−) | 2568 | 855 | 96.06 | 5.95 | − | CP |
Chloroplast transit peptides: Y: yes. Subcellular localization: CP: Cytoplasm; Chl: Chloroplast; ER: Endoplasmic reticulum.
The CDS length of MaLOXs ranged from 2061 to 3396 bp. Their deduced proteins contained 686–1131 amino acids (aa) with theoretical isoelectric points ranged from 5.72 to 9.53. The molecular weight of MaLOXs ranged from 76.33 to 126.29 kD. MbLOX proteins contained 665 to 913 aa with molecular weight ranged from 75.15 to 102.96 kD. MiLOX proteins contained 823 to 949 aa, with molecular weight ranged from 93.78 to 107.52 kD. Their theoretical isoelectric point ranged from 5.68 to 9.40 and from 5.89 to 8.45 for MbLOXs and MiLOXs, respectively. Chloroplast transit peptides were identified in 7 MaLOX (MaLOX6-7, 12–15, and 18), 5 MbLOX (MbLOX6, 13, 14, 15, and 18), and 5 MiLOX (MiLOX7, 12, 13, 14, and 15) members, respectively. The MaLOXs were predicted to located in different cell parts, most of which were cytoplasm located, while MaLOX6-7 and 12–15 were chloroplast located. Six of 11 MbLOX proteins were located in the cytoplasm, and 5 proteins were located in the chloroplast. In addition, 7 MiLOXs were located in the cytoplasm and 5 in the chloroplast.
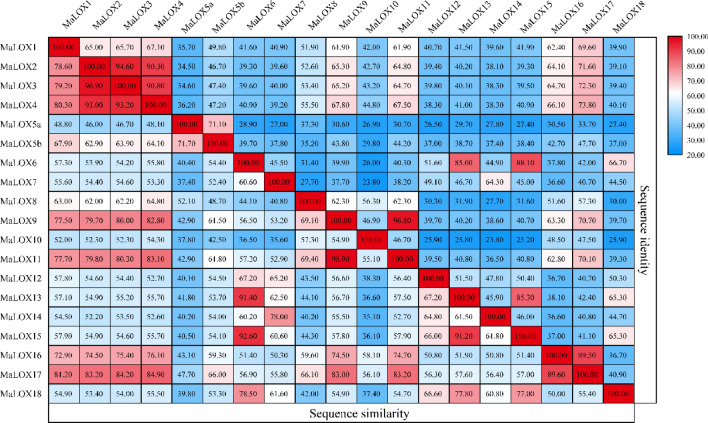
Protein sequence alignment result revealed that the sequence similarity among MaLOXs ranged from 35.10 to 98.90%, and the sequence identity ranged from 23.80 to 96.80% (Fig. 1). The similarity and identity between MaLOX5a and other members are relatively low, and MaLOX9 and MaLOX11 showed the highest similarity and identity, while the similarity and identity between MaLOX10 and MaLOX14 was the lowest (Fig. 1). Besides, the sequence similarity among MbLOXs and MiLOXs was 37.60–95.2% and 47.30–91.00%, and sequence identity was 27.60–93.70% and 33.90–84.70%, respectively (Supplementary Figures S2, S3).
Figure 1.
Sequence identities and similarities (%) among the MaLOXs.
Phylogenetic relationship of banana LOX protein family
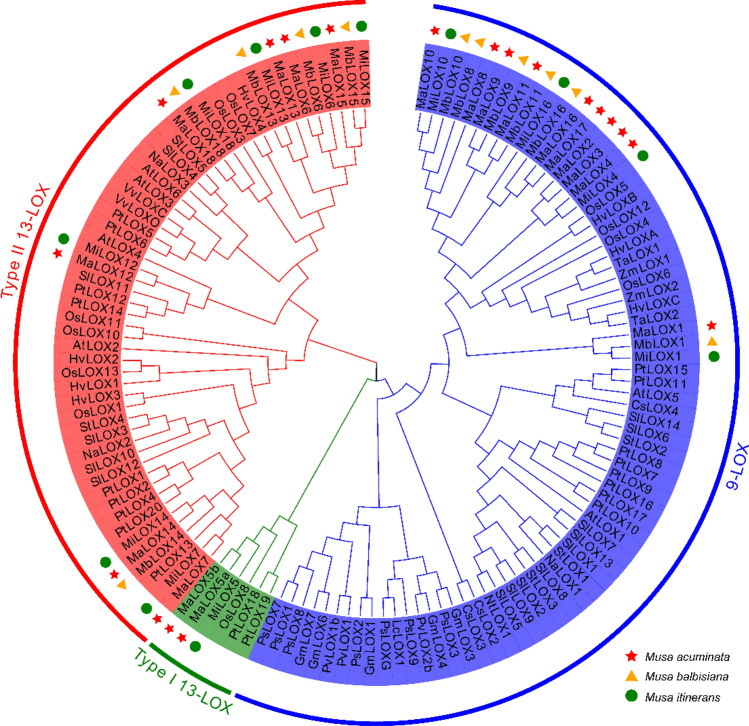
To determine the phylogenetic relationship among MaLOXs, MbLOXs, and MiLOXs, the LOX protein sequences from Arabidopsis (6), rice (11), tomato (14), poplar (19), banana (42) and other plants (41) were used for phylogenetic analysis. All LOX proteins could be classified into two subfamilies, 9-LOX and 13-LOX. And 13-LOX can be further divided into Type I and Type II (Fig. 2). The 9-LOX subfamily includes 10 MaLOXs (MaLOX1-4, 8–11, 16, and 17), 6 MbLOXs (MbLOX1, 8, 9, 11, and 16), and 4 MiLOXs (MiLOX1, 4, 10, and 16), respectively. Seven MaLOXs, 5 MbLOXs and 7 MiLOXs belong to Type II 13-LOX subfamily. MaLOX5a, MaLOX5b, and MiLOX5 belong to the Type I 13-LOX subfamily, and this subfamily only contains banana, rice and poplar LOXs.
Figure 2.
Phylogenetic tree of LOX proteins from M. acuminata, M. balbisiana, M. itinerans and some other plant species.
Analysis of gene structure and conserved domain
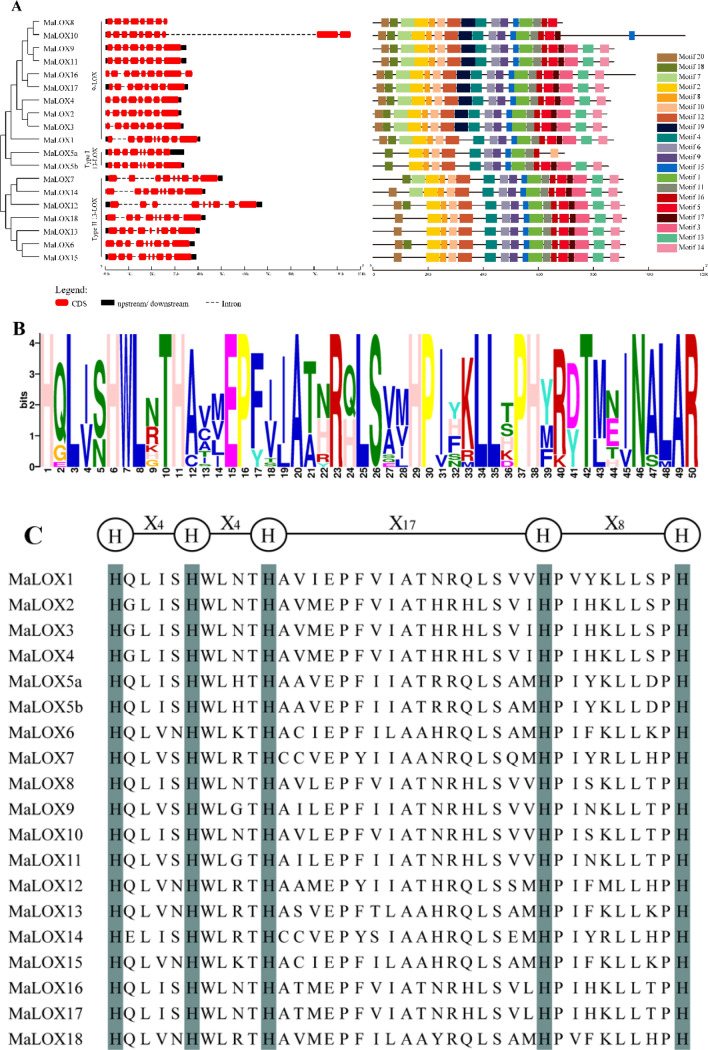
GSDS was used to show the gene structure diagram of MaLOXs, MbLOXs, and MiLOXs. As shown in Fig. 3A, MaLOXs have 8–10 exons, of which MaLOX10 has the largest numbers of exons. The exons of 9-LOX subfamily genes are very similar in length and distribution, suggesting that this subfamily may originate from the same ancestor gene. Most MaLOX members have gDNA lengths between 3 and 5 kb, except MaLOX10 and MaLOX12, whose gDNA length is about 11 kb and 7 kb, respectively. MbLOXs and MiLOXs have similar gene structures with MaLOXs. MbLOXs contain 6–10 exons, and MiLOXs have 8–10 exons (Supplementary Figures S4A, S5A). In addition, most banana LOX genes within the same subfamily presented similar exon–intron distribution patterns.
Figure 3.
Gene structures and conserved motifs (A), Motif 1 sequence (B) and the 38 conserved residues of MaLOX proteins of MaLOXs or their encoded proteins. The dark color in C shows highly conserved histidine (His).
Conserved motif analysis showed that most banana LOXs contained similar types and arrangements of conserved motifs (Fig. 3A, Supplementary Figures S4A, S5A). Eleven conserved motifs (motif 1, 2, 4, 6, 8–12, 15 and 16) were found in all MaLOXs. In addition to the common motifs, the 9-LOX subfamily proteins also contain motifs 5, 7, 18, and 20, and the Type II 13-LOX subfamily also contains motifs 3, 5, 13–14, and 17. MbLOXs and MiLOXs have similar conserved motifs with MaLOXs. The histidine (His)-rich Motif 1, plays an important role in the biological activity of lipoxygenase, is highly conserved among banana LOX family members (Fig. 3B, Supplementary Figures S4B, S5B). A typical domain of the banana LOXs is consisted of 38 amino acids of [His-(X)4-His-(X)4-His-(X)17-His-(X)8-His] (Fig. 3C, Supplementary Figures S4C, S5C).
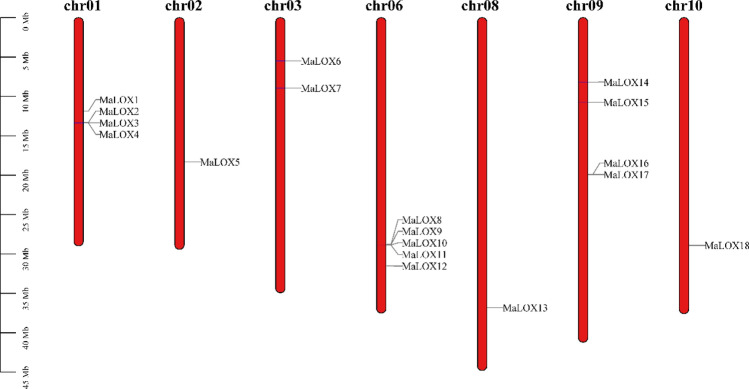
Chromosome location and gene duplication
As shown in Fig. 4, MaLOX genes are randomly and unevenly distributed on 7 chromosomes (chr). The highest number of MaLOXs was observed in chr06, with 5 members, follow by chr01, 03 and 09 with 4, 2, and 4 members, respectively. Eleven MbLOX genes were located on 6 of the 11 chromosomes (Bchr) and exhibited uneven distributions (Supplementary Figure S6). Bchr06 contained the highest number of MbLOX genes (4, 36.36%), followed by Bchr09 (3, 27.27%), while minimum genes were distributed on Bchr01, 03, 08, and 10 (1, 9.09%). M. itinerans genome was only assembled to the scaffold level (S). The 12 MiLOX genes are identified from 12 different scaffolds (S1338, S2542, S3004, S3031, S34, S406, S647, S649, S655, S7089, S768, S829) (Supplementary Figure S7).
Figure 4.
Chromosome localization of MaLOX genes.
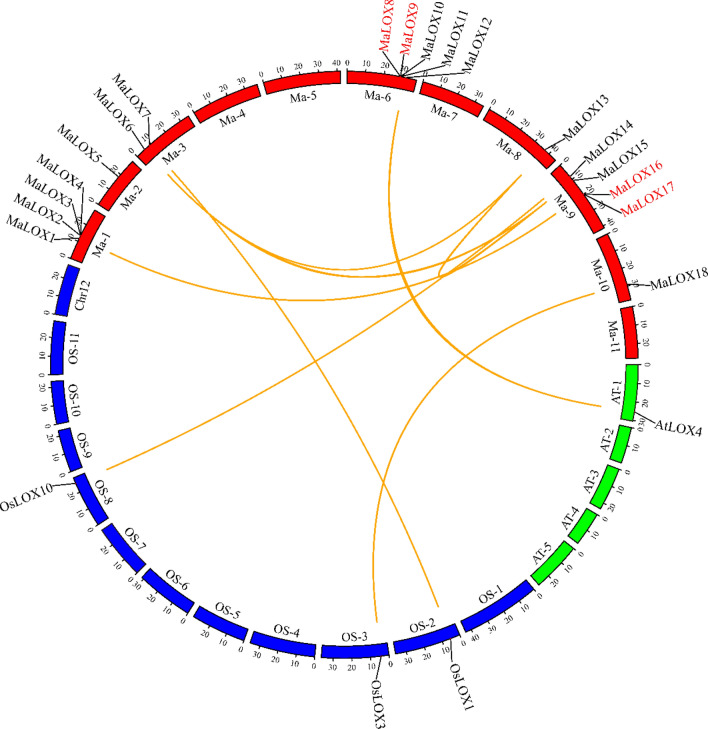
In order to explore the gene duplication events of the LOX family, we investigated the collinearity relationships between banana LOXs as well as pairwise relationships analysis of LOXs from M. acuminata, M. balbisiana, M. itinerans, Arabidopsis, and rice (Fig. 5; Supplementary Figures S8, S9, S10; Table 2; Supplementary Tables S3, S4). There are 2 tandem duplicate pairs (MaLOX8/MaLOX9 and MaLOX16/MaLOX17) and 4 segmental duplicate pairs (MaLOX1/MaLOX16, MaLOX6/MaLOX13, MaLOX6/MaLOX15, and MaLOX13/MaLOX15) in the MaLOX gene family. MbLOX gene family has 3 tandem duplicate pairs (MbLOX10/MbLOX9, MbLOX9/MbLOX8, and MbLOX8/MbLOX11) and 3 segmental duplicate pairs (MbLOX6/MbLOX13, MbLOX6/MbLOX15, and MbLOX13/MbLOX15) (Supplementary Figure S8; Supplementary Table S3). However, MiLOX gene family does not contain any duplicated pairs (Supplementary Figure S9). In addition, three OsLOX genes had a syntenic relationship with three MaLOXs (OsLOX1/MaLOX7, OsLOX3/MaLOX18, and OsLOX10/MaLOX14) and 1 collinear pair (AtLOX4/MaLOX12) was identified between M. acuminata and Arabidopsis.
Figure 5.
Collinear distribution of MaLOX genes. The orange line indicates the collinearity between the MaLOXs, and the gene names in red are tandem replication genes.
Table 2.
LOX gene family intraspecific and interspecific gene replication events.
| Gene name | Gene ID | Gene name | Gene ID | Ka | Ks | Ka/Ks | Duplication date /Mya | Duplication type |
|---|---|---|---|---|---|---|---|---|
| MaLOX1 | Ma01_g16400 | MaLOX16 | Ma09_g19130 | 0.2021 | 0.9918 | 0.2038 | 110.20 | Segmental duplication |
| MaLOX6 | Ma03_g07770 | MaLOX13 | Ma08_g23400 | 0.0760 | 0.4850 | 0.1566 | 53.89 | Segmental duplication |
| MaLOX6 | Ma03_g07770 | MaLOX15 | Ma09_g15420 | 0.0596 | 0.3439 | 0.1733 | 38.21 | Segmental duplication |
| MaLOX13 | Ma08_g23400 | MaLOX15 | Ma09_g15420 | 0.0751 | 0.5039 | 0.1490 | 55.99 | Segmental duplication |
| MaLOX8 | Ma06_g26840 | MaLOX9 | Ma06_g26850 | 0.1298 | 0.5639 | 0.2302 | 62.65 | Tandem duplication |
| MaLOX16 | Ma09_g19130 | MaLOX17 | Ma09_g19140 | 0.0028 | 0.0008 | 3.5461 | 0.09 | Tandem duplication |
| AtLOX4 | AT1G67560 | MaLOX12 | Ma06_g30170 | 0.3223 | 2.9750 | 0.1083 | Segmental duplication | |
| OsLOX1 | LOC_Os02g10120 | MaLOX7 | Ma03_g11520 | 0.3320 | 0.9242 | 0.3593 | Segmental duplication | |
| OsLOX3 | LOC_Os03g08220 | MaLOX18 | Ma10_g17560 | 0.1624 | 1.0741 | 0.1512 | Segmental duplication | |
| OsLOX10 | LOC_Os08g39840 | MaLOX14 | Ma09_g12090 | 0.3114 | 1.4462 | 0.2153 | Segmental duplication |
The collinearity relationships between three banana species are shown in Supplementary Figure S10. Fourteen orthologous gene pairs between M. acuminata and M. balbisiana were identified, 12 orthologous gene pairs were found between M. acuminata and M. itinerans, and 9 orthologous gene pairs existed between M. balbisiana and M. itinerans. Moreover, some LOX genes are relatively conserved between banana species. For example, MaLOX1, 6, 9, 13–15, and 18 have collinearity with their orthologs in M. balbisiana and M. itinerans.
To further understand whether the genes of the LOX family have been subjected to natural selection pressures during the evolution process and to trace the duplication time of banana LOXs, we calculated the ratios of nonsynonymous (Ka) versus synonymous (Ks) mutation of orthologous gene pairs. As shown in Table 2, Supplementary Table S3, and Supplementary Table S4, the Ka/Ks ratios of MaLOX16/MaLOX17 is more than 1, which may have experienced strong positive selection. In addition, the Ka/Ks ratios of other duplicate pairs less than 1, suggesting that these pairs have undergone purifying selection pressure during evolution.
Based on the divergence rate of 4.5 × 10–9 synonymous mutations per synonymous site year proposed for banana, we estimated the time of occurrence of duplicating events of the paralogous LOX gene pairs. The results showed that MaLOX1/MaLOX16 and MaLOX16/MaLOX17 occurred at about 110.20 and 0.09 million years ago (Mya), while other MaLOX gene pairs occurred between 38.21 to 62.65 Mya (Table 2). The estimated divergence time of the duplicated gene pairs of MbLOX family varies from 47.54 to 74.29 Mya (Supplementary Table S3). Furthermore, the replication times for syntenic genes between MaLOX and MbLOX, between MaLOX and MiLOX, and between MbLOX and MiLOX was 3.10–107.17 Mya, 2.38–70.73 Mya, and 3.39–50.98 Mya, respectively (Supplementary Table S4).
Cis-acting elements prediction results of banana LOX gene promoters
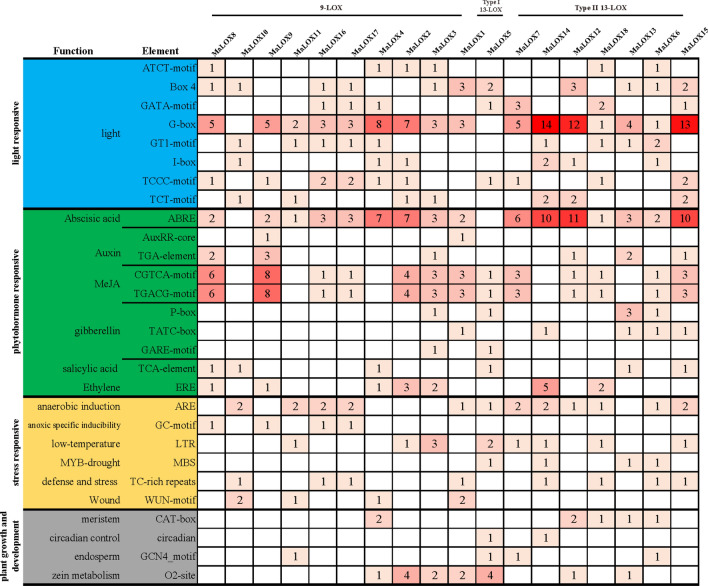
In order to further explore the possible expression regulation patterns in the members of the banana LOX gene family, we extracted the promoter sequences of their family members for cis-acting element prediction analysis. In total, four categories of cis-acting elements were identified, including light responsiveness, phytohormone responsiveness, stress responsiveness, and plant growth and development-related elements (Fig. 6, Supplementary Figures S11, S12). Therefore, it is speculated that the expression of banana LOXs may be regulated by multiple factors.
Figure 6.
The identified cis-acting elements in MaLOX gene family promoters.
Many light responsive elements are present in the promoters of LOX genes from three banana species, of which the number of G-box elements is the largest. Banana LOXs contain at least a cis-acting element involved in phytohormone responsiveness classification. Further analysis of the phytohormone responsiveness elements revealed that the number of elements related to abscisic acid was the largest, followed by MeJA. All promoters contain abscisic acid responsiveness elements (ABRE) except MaLOX5, MaLOX10, MbLOX9, MbLOX10, MiLOX1, and MiLOX5. MeJA (TGACG-motif, CGTCA-motif) responsive elements were found in the 30 banana LOX gene promoters (MaLOX1-3, 5–9, 12, and 15–18; MbLOX8-10, 13–15, 16, and 18; MiLOX4, 5, 7, 10, 12, 14–16, and 18). Furthermore, auxin (TGA-element, AuxRR-core), gibberellin (P-box, TATC-box, and GARE-motif), salicylic acid (TCA-element), and ethylene (ERE) responsive elements are also present on banana LOX promoters.
Besides, the promoters also contain several types of stress responsiveness elements, including anaerobic induction (ARE), anoxic specific inducibility (GC-motif), low temperature (LTR), MYB drought-inducibility binding site (MBS), defense and stress (TC-rich repeats), and wound (WUN-motif) responsive elements. Additionally, plant growth and development related cis-elements in charge of meristem expression (CAT-box), circadian (circadian), endosperm expression (GCN4_motif), and Zein metabolism (O2-site) regulation were found in the promoter regions of MaLOXs, MbLOXs, and MiLOXs.
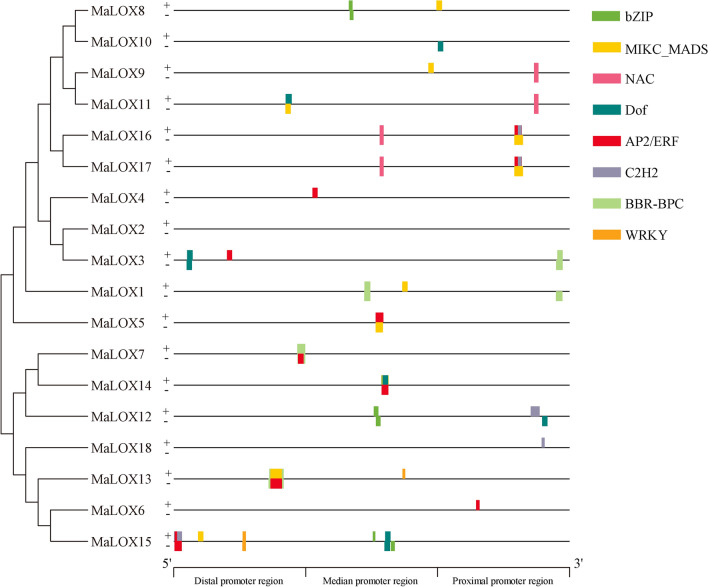
Transcription factor binding site (TFBS) prediction
To investigate the regulation of transcription factors (TFs) on the expression of banana LOXs, transcription factor binding sites (TFBSs) on the promoter were predicted using PlantTFDB online tool. A total of 8 TF families (AP2/ERF, BBR-BPC, bZIP, C2H2, Dof, MIKC_MADS, NAC and WRKY) were identified in the MaLOX promoters, which covers 10, 4, 3, 5, 7, 11, 4, and 2 members, respectively (Fig. 7). BBR-BPC family has the largest number of binding sites (51), while WRKY has the least number of binding sites (6). Besides, MbLOX and MiLOX gene promoters contain six identical TF families, which are AP2/ERF, BBR-BPC, Dof, GATA, MIKC_MADS, and MYB (Supplementary Figure S13A, B). Meanwhile, there are also ARF on the MbLOX promoters, and C2H2 and TALE are present in MiLOX promoter sequences. In addition, there are certain differences in the TFBS types, number and distribution in the banana LOX gene promoters. For instance, 6 types of TF binding sites were found in MaLOX15, while MaLOX2, MbLOX14, and MiLOX5 were devoid of any TF families. MaLOX13 has the largest number of TFBS (62), but only 1 TFBS in the promoters of MaLOX4, 10, and 18, MbLOX6, 11, and 13, MiLOX1 and 6.
Figure 7.
Transcription factor binding sites predicted in the promoters of MaLOXs. Boxes of different colors represent different transcription factor families. " + " and "−" represents positive and negative strand, respectively.
Codon usage bias of MaLOX genes
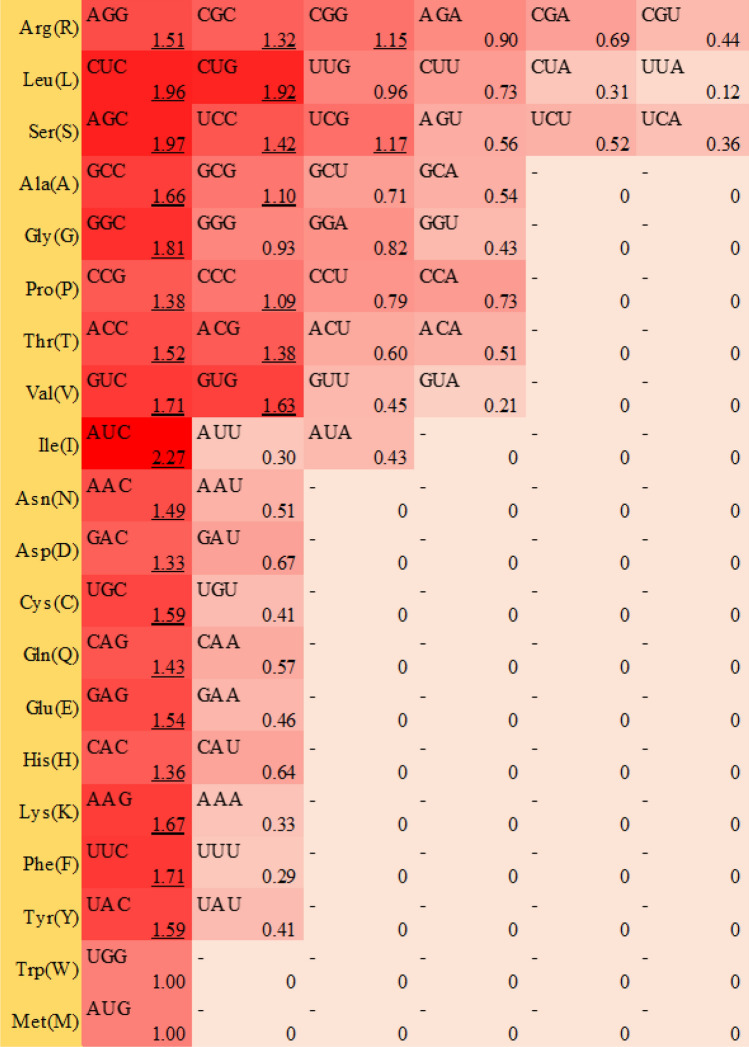
The CodonW software was used to analyze the codon usage bias of the banana LOX gene family. Results showed that the effective number of codons (ENC) values of MaLOXs, MbLOXs and MiLOXs are respectively 42.51–56.39, 42.46–54.57, and 42.99–56.70, with an average value of 46.93, 45.81, and 46.94, indicating that the gene expression levels of banana LOX genes were relatively low (Table 3, Supplementary Table S5). The codon adaptation index (CAI) value of MaLOXs, MbLOXs and MiLOXs ranged respectively from 0.18 to 0.26, from 0.18 to 0.26, and from 0.19 to 0.26, with a mean value of 0.23, 0.23, and 0.22, suggesting that the codon bias of banana LOXs was weak. With the exception of MiLOX12, the average content of C3s and G3s was significantly higher than that of A3s and T3s, and the average content of GC and GC3s was greater than 0.5, which indicated that the banana LOX codons generally prefer to use and end with G/C. Relative synonymous codon usage (RSCU) can intuitively reflect the degree to which specified codons deviate from synonymous codons, and RSCU > 1 indicates that the codons are used more frequently than expected. 27 codons showed strong preference for GC-ending codons based on the above criterion in MaLOXs, MbLOXs, and MiLOXs, respectively (Fig. 8, Supplementary Figures S14, S15). Among these, 11 codons end in G and 16 codons end in C.
Table 3.
Codon preference parameters of MaLOX family genes.
| Gene name | T3s | C3s | A3s | G3s | CAI | ENC | GC3s | GC |
|---|---|---|---|---|---|---|---|---|
| MaLOX1 | 0.14 | 0.58 | 0.09 | 0.45 | 0.26 | 42.51 | 0.81 | 0.59 |
| MaLOX2 | 0.13 | 0.54 | 0.11 | 0.48 | 0.26 | 43.76 | 0.80 | 0.60 |
| MaLOX3 | 0.14 | 0.54 | 0.12 | 0.46 | 0.25 | 44.44 | 0.79 | 0.60 |
| MaLOX4 | 0.14 | 0.53 | 0.13 | 0.45 | 0.24 | 45.86 | 0.78 | 0.59 |
| MaLOX5a | 0.23 | 0.40 | 0.22 | 0.37 | 0.18 | 55.23 | 0.62 | 0.54 |
| MaLOX5b | 0.18 | 0.49 | 0.19 | 0.40 | 0.20 | 50.59 | 0.70 | 0.56 |
| MaLOX6 | 0.17 | 0.46 | 0.13 | 0.48 | 0.20 | 45.90 | 0.75 | 0.60 |
| MaLOX7 | 0.16 | 0.50 | 0.13 | 0.48 | 0.24 | 45.68 | 0.77 | 0.59 |
| MaLOX8 | 0.15 | 0.53 | 0.16 | 0.41 | 0.25 | 46.04 | 0.75 | 0.58 |
| MaLOX9 | 0.14 | 0.54 | 0.11 | 0.46 | 0.25 | 46.03 | 0.79 | 0.59 |
| MaLOX10 | 0.16 | 0.50 | 0.18 | 0.41 | 0.21 | 48.06 | 0.73 | 0.59 |
| MaLOX11 | 0.14 | 0.54 | 0.11 | 0.46 | 0.25 | 45.91 | 0.79 | 0.59 |
| MaLOX12 | 0.33 | 0.29 | 0.31 | 0.32 | 0.19 | 56.39 | 0.47 | 0.48 |
| MaLOX13 | 0.16 | 0.45 | 0.14 | 0.49 | 0.21 | 46.10 | 0.75 | 0.60 |
| MaLOX14 | 0.22 | 0.41 | 0.20 | 0.43 | 0.21 | 53.68 | 0.66 | 0.55 |
| MaLOX15 | 0.13 | 0.51 | 0.11 | 0.51 | 0.22 | 42.70 | 0.81 | 0.61 |
| MaLOX16 | 0.16 | 0.54 | 0.15 | 0.40 | 0.23 | 47.11 | 0.75 | 0.57 |
| MaLOX17 | 0.14 | 0.57 | 0.13 | 0.41 | 0.24 | 42.93 | 0.78 | 0.59 |
| MaLOX18 | 0.13 | 0.52 | 0.11 | 0.49 | 0.24 | 42.81 | 0.80 | 0.61 |
| Average | 0.17 | 0.50 | 0.15 | 0.44 | 0.23 | 46.93 | 0.74 | 0.58 |
T3s, C3s, A3s, G3s, and GC3s indicate that the third base of the codon is the content of T, C, A, G, and G + C. GC: total GC content in of CDS. CAI: codon adaptation index. ENC: effective number of codons.
Figure 8.
Relative usage of synonymous codons in MaLOX gene family members. The underlined data indicate that the MaLOX genes preferentially to use this codon.
Expression pattern of MaLOX genes under different stresses
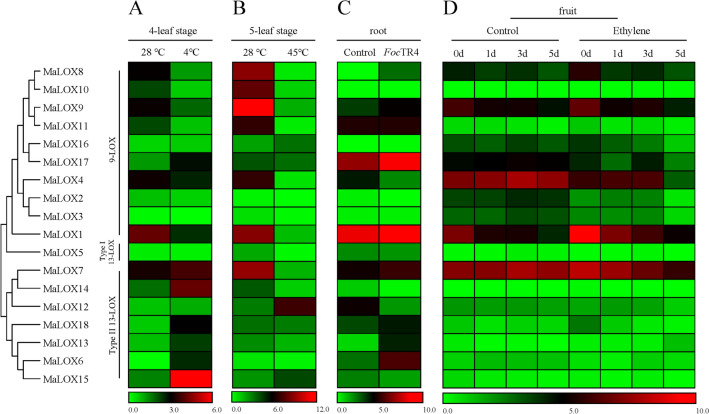
As shown in Fig. 9, MaLOXs showed divergent expression patterns across different tissues (Supplementary Table S6). MaLOX1 was found to be a highly expressed gene in banana leaves, roots, and fruits. MaLOX7 was highly expressed in fruits and leaves. The expression of MaLOX4 in fruits is higher than in leaves and roots. MaLOX17 was predominantly expressed in the root.
Figure 9.
Diagram for the expression of all the MaLOX gene family members. (A) Leaf transcriptome data of 4-leaf stage ‘Tianbaojiao’ banana treated with 24 h 4 °C low temperature and 28 ℃ control; (B) leaf transcriptome data of 5-leaf stage ‘Tianbaojiao’ banana treated with 3 days 45 °C high temperature and 28 °C control; (C) root transcriptome data of FocTR4 treated ‘Tianbaojiao’ banana; (D) fruit transcriptome data of ‘Tianbaojiao’ banana at natural ripening and ethylene induced ripening stages.
Under low temperature treatment, 6 MaLOX members (33.33%) were upregulated and 5 members (27.78%) were downregulated. The expression of most members of the 9-LOX subfamily was inhibited, however, MaLOX17 was significantly induced (Fig. 9A). Most members of TypeII 13-LOX were upregulated by low temperature, with MaLOX15 being particularly significant. Under high temperature stress, 3 members (16.67%), including MaLOX12, 15, and 16, were upregulated, in which MaLOX12 was significantly induced (Fig. 9B), and 9 members were downregulated. The expression of MaLOX6, 8, 9, 13, and 17 increased greatly under FocTR4 treatment, while the expression of MaLOX4 and 12 declined (Fig. 9C).
MaLOXs expression pattern analysis during natural ripening and ethylene induced ripening was also performed. The expression of the MaLOX1 was downregulated and MaLOX8 showed fluctuation change as fruit ripens (Fig. 9D). MaLOX1, 7, 8, and 18 were upregulated, while MaLOX2 and MaLOX4 were downregulated by ethylene at 0 day compared with the control group, but they were downregulated at following timepoints in comparison to the postharvest naturally ripening stage.
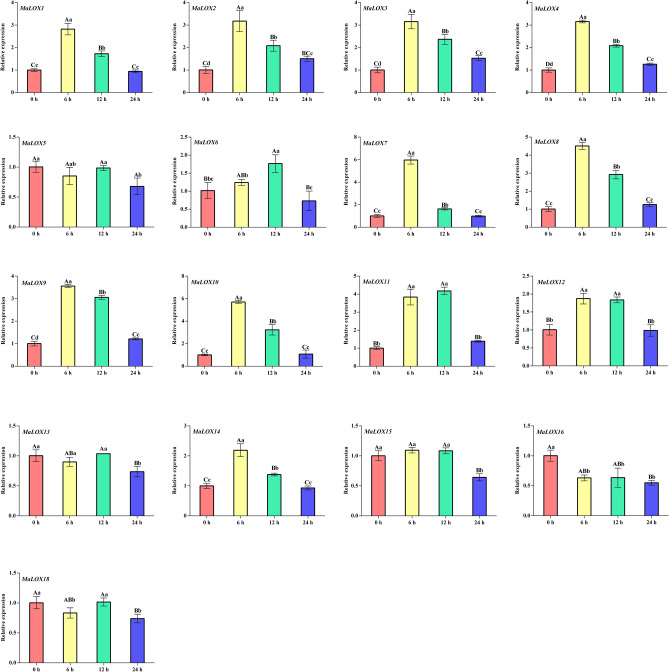
Expression patterns of MaLOX genes under MeJA treatment
qRT-PCR was performed to determine the responses of the MaLOXs to MeJA treatment (Supplementary Table 7). The expression level of MaLOX17 is too low that its relative expression level was not shown in Fig. 10. The expression of MaLOX2-4 and MaLOX9 significantly increased after MeJA treatment, while 5 MaLOX members (MaLOX5, 13, 15, 16, and 18) declined significantly. Eight MaLOX members (MaLOX1-4 and 7–10) were significantly induced by MeJA, and their relative expression peaked at 6 h, then began to decline sharply. MaLOX6 was significantly upregulated at 12 h post MeJA treatment. MaLOX12 was dramatically upregulated at 6 h and 12 h and restored to its basal levels during the later periods. The expression level of MaLOX13, 15, and 18 did not change significantly at 6 h and 12 h, but significantly downregulated at 24 h. However, unlike those genes, the expression of MaLOX14 was significantly induced at 6 h, but afterwards its expression level gradually recovered to the basic level.
Figure 10.
Expression analysis result of MaLOX genes under MeJA treatment. Uppercase and lowercase letters are used to indicate significantly differences at P < 0.01 and 0.05, respectively.
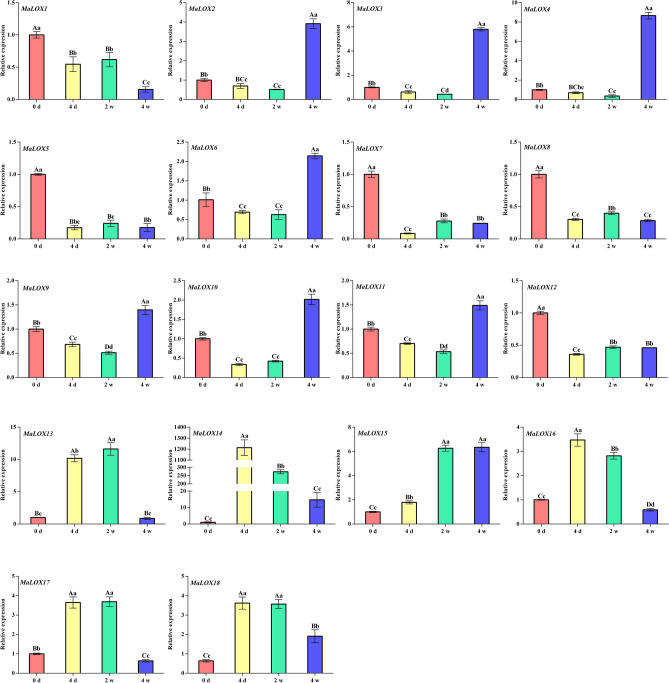
Expression patterns of MaLOX genes under FocTR4 treatment
Gene expression levels of MaLOX genes in response to FocTR4 infection were analyzed using qRT-PCR (Fig. 11, Supplementary Table 8). Within 2 weeks of FocTR4 treatment, the expression level of MaLOX1-12 decreased significantly, among which 4 members (MaLOX7, 8, 10, and 12) showed the lowest expression level at 4 days, and MaLOX3, 9, and 11 reached their lowest level of expression at 2 weeks. FocTR4 significantly induced the expression of MaLOX13-18, where the expression of MaLOX13 and 15 gradually increased, and MaLOX14 and 16 reached their peak at 4 d, and then began to decline. Compared with the 2 weeks, at 4 weeks post FocTR4 treatment, the expression of 7 MaLOX members (MaLOX2-4, 6, and 9–10) was significantly upregulated and reached the maximum, and the expression of 7 members (MaLOX5, 8, 13, 14, and 16–18) was significantly downregulated.
Figure 11.
Expression analysis of MaLOX genes after FocTR4 treatment. Uppercase and lowercase letters are used to indicate significantly differences at P < 0.01 and 0.05, respectively.
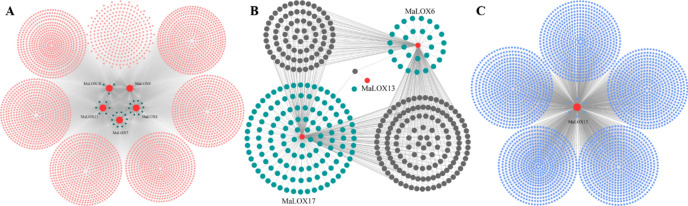
Weighted gene co-expression network analysis (WGCNA) of MaLOXs
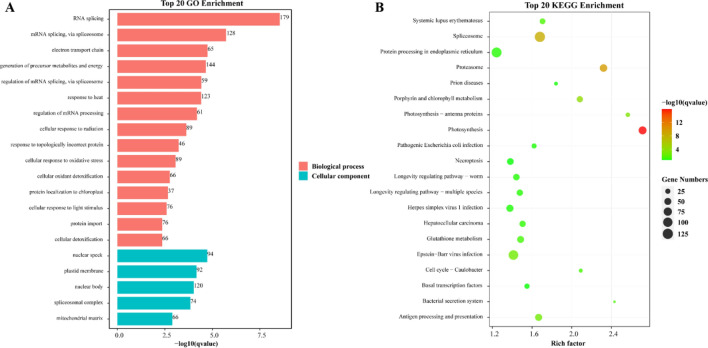
To explore the potential interaction and functions between co-expressed genes, WGCNA was applied to construct the co-expression network based on 4 different transcriptome datas, including banana fruit ripening stages, leaves response to high and low temperature, and roots inoculated with FocTR4. We only keep edges with strong connections with weight values ≥ 0.4. A total of 7629 genes were co-expressed with nine MaLOXs. Visualization using Cytoscape software, three co-expression networks models, respectively containing 5 (MaLOX7-11), 3 (MaLOX6, 13, and 17), and 1 (MaLOX12) MaLOX, were constructed (Fig. 12). GO enrichment analysis result revealed that, from the aspect of biological process, the MaLOXs co-expressed genes were mainly enriched in RNA splicing, mRNA splicing via spliceosome, electron transport chain, generation of precursor metabolites and energy, regulation of mRNA splicing via spliceosome, and response to heat (Fig. 13A); from the aspect of molecular function, nuclear speck, plastid membrane, nuclear body, and spliceosomal complex related co-expressed genes were enriched. According to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment results, these MaLOX co-expressed genes were found to be enriched in photosynthesis, proteasome, spliceosome, porphyrin and chlorophyll metabolism, and photosynthesis—antenna proteins (Fig. 13B).
Figure 12.
Co-expression network for MaLOX genes. The red nodes indicate MaLOX genes and all other color nodes indicate co-expressed genes with MaLOXs. (A) Model 1, (B) Model 2, and (C) Model 3.
Figure 13.
GO and KEGG analysis of the genes in the co-expression network of the MaLOX genes. (A) GO functional annotation; (B) KEGG pathway enrichment.
Discussion
Comprehensive genome-wide identification of LOXs in banana
Lipoxygenase is a crucial restriction enzyme in the LOX pathway, which catalyzes the fatty acid metabolism of plant, actively participates in growth and development, and resists extreme external environmental conditions38. In this study, we identified 18 MaLOX, 11 MbLOX, and 12 MiLOX genes from M. acuminata, M. balbisiana, and M. itinerans genome, respectively. MaLOXs have more members than Arabidopsis (6), rice (14), and tomato (14), but are the same as grapes (18)39 and melon (18)40. Besides, the number of LOX in banana from most to least is MaLOXs > MiLOXs > MbLOXs, which is consistent with their genome size (501.5 MB for M. acuminata, 430 MB for M. balbisiana and 462.1 MB for M. itinerans).
Phylogenetic analysis showed that the banana LOXs family could be further divided into three subfamilies, including 9-LOX, Type I 13-LOX, and Type II 13-LOX, which was consistent with the results of poplar9 and tea plant10. The sequence similarity among Type I LOX members ranged from 26.90 to 98.90%. Type II 13-LOX members contained chloroplast transit peptides except MiLOX6 and MiLOX18, and their sequence similarity ranged from 44.90 to 92.60%. Our results are not completely consistent with classification method of Shibata et al.41, who put forward that Type I LOX genes exhibit high sequence similarity (more than 75%) and lack of chloroplast transit peptide, while Type II LOX genes show moderate overall sequence similarity (up to 35%) and exist chloroplast transit peptide. But our result was consistent with the melon LOXs40, which may be related to the diversity of the evolution process of the LOX genes. The prediction of subcellular localization showed that MaLOX18, MbLOX18 and MiLOX18 were localized in the cytoplasm, while other Type II 13-LOX members were all localized in the chloroplast. This may be due to the poor conservation of the amino acid sequence of the chloroplast transit peptide of banana LOX1842,43. The members of this subfamily have similar gene structure and conserved motifs, indicating that the gene function of banana LOX members from the same subfamily showed certain degree of conservativeness. Our study found that codon bias of banana LOXs was weak, preferring to use and end with G/C, which is consistent with the codon preference characteristics of monocotyledon plants44 and banana genome45. Thus, it was hypnotized that in order to cope with environmental pressures, different banana species have formed unique codon usage bias during evolution.
LOXs may play special roles in banana evolution
Gene duplication is a major factor responsible for the amplification in family gene numbers, in which whole genome duplication (WGD) is considered to be an important driving force for expansion and an important source of gene function diversification4. There are three pairs of segmental duplication genes and three tandem duplication gene clusters in the poplar LOX family genes9. Five tandem repeat pairs were observed in tomato LOX family, and no segmental duplicate pairs8. In this study, the four pairs of segmental duplication genes and two pairs of tandem repeat genes were found in MaLOX gene family, accounting for 27.78% (4/18) and 22.22% (5/18), respectively. Banana is speculated to undergo three whole genome duplication events during the evolution, which were α, β, and γ events, respectively46. The duplication events of the MaLOX genes were supposed to originate from 0.09 to 110.20 Mya, of which MaLOX1/MaLOX16 dated the duplication event at 110.20 Mya, corresponding to the γ event. The tandem repeat event of MaLOX8/MaLOX9 occurred at 62.65 Mya, corresponding to the α or β event. It was suspected that the whole genome, segmental, and tandem duplication together contributed together to the expansion of MaLOX gene family. Moreover, MbLOX gene family also contains three pairs of segmental duplication genes and three pairs of tandem repeat genes, accounting for 27.27% (3/11) and 36.36% (4/11), which indicates that tandem duplications and segmental duplications together play a role in the expansion of MbLOX gene family.
The mechanism of gene and genome evolution can be understood through a comparative analysis of relatively close between-species genome. This study has found that there are a high conservation level and have a close homology relationship among MaLOX, MbLOX, and MiLOX genes. The ancestor of M. acuminata and M. itinerans diverged with M. balbisiana ancestor about 8.3 Mya, and M. acuminata and M. itinerans diverged about 5.8 Mya, while the divergence time was about 5.4 Mya for the M. acuminata and M. balbisiana47,48. We found that 5 of 14, 8 of 12, and 6 of 9 orthologous gene pairs appeared after the divergence of M. acuminata/M. balbisiana, M. acuminata/M. itinerans, and M. balbisiana/M. itinerans. M. acuminata and M. balbisiana shared less orthologous gene pairs with M. balbisiana, which may be explained that M. balbisiana genome exhibited less expansion and more contraction of gene families after divergence and M. acuminata and M. itinerans have relatively higher similarity47,48.
Besides, evolutionary selection pressure analysis of banana LOX duplication genes showed that MaLOX16/MaLOX17 experienced strong positive selection pressure, indicating that functional differentiation occurred. And other duplication genes were subject to purification selection pressure and limited functional differentiation5.
Functional prediction of MaLOXs
The cis-acting elements of the promoter combine with specific transcription factors to form transcription initiation complex and initiate gene specific expression49. Four types of cis-regulatory elements were identified at the banana LOX promoters, including light, phytohormone, stress, growth and development-related, which is consistent with the report about the functional diversity of LOX genes50. Besides, a variety of kinds of TFBSs were found in the banana LOX promoters. Recent research demonstrated that TFs play an important role in banana growth and adversity stress51–53, and it is further speculated that banana LOX expression is regulated by many TFs.
Lipoxygenase is a kind of oxygenase widely distributed in various organs of plants, and its expression levels in different parts and developmental stages of plants differed, which are closely related to physiological processes such as plant growth, development, maturity, and senescence10,37. In this study, each member of the MaLOX family was expressed in at least one organ. MaLOX1 was highly expressed in leaf, root, and fruit, which suggests that the function of MaLOX1 may be diverse. The expression of MaLOX4 in fruit is higher than in leaf and root, and MaLOX7 is highly expressed in fruit and leaf, which means that the functions of different MaLOXs members varied in different organs.
Low temperature can inhibit the transcriptional level of LOX in banana fruit, reduce the banana volatiles, and the inhibition effect is more obvious as the temperature decreases25. Under high temperature, there is an overall decrease in the amount of LOX proteins in banana peel26,27. Li et al.28 found that the high expression of LOX was related to higher FocTR4 resistance of resistant mutant. LOX1.1–3 and LOX2.3 were significantly induced in resistant variety (Musa yunnanensis) during early infection with FocTR454. In this study, the analysis of transcriptome data under low temperature, high temperature, and FocTR4 treatment revealed that the expression patterns of MaLOXs under different stresses differed. MaLOX8, 9, and 13 responded significantly to the above three stresses. The expressions of MaLOX1, 8, 10, 11, 14, and 15 were regulated by high and low temperature; MaLOX6 and 17 were induced by low temperature and FocTR4; MaLOX4 and MaLOX12 responded to high temperature and FocTR4. MaLOX7 and 16 were differentially expressed at high temperature and MaLOX18 was only induced by low temperature. In addition, this study also found that in the early stage of FocTR4 infection, each member of MaLOXs responded to varying degrees.
WGCNA is an effective way to identify clusters of highly correlated genes and can better preserve the characteristics of biological networks and reflect the relationship among functions and different biological processes55,56. Most of the adjacent genes of MaLOXs in their co-expression network were related to RNA splicing, generation of precursor metabolites and energy, heat stress, photosynthesis, and proteasome. Besides, the promoter regions of these differentially expressed genes contain a large number of stress-related cis-acting elements and TFBSs. These results indicated that MaLOXs are widely involved in banana growth and development and various stress responses.
LOX regulates the processes of plant ripening and senescence by participating in the synthesis of ethylene or catalyzing polyunsaturated fatty acids to generate superoxide radicals and destroying cell membrane structure40,57. And the roles of LOX in fruit ripening and flavor formation have been confirmed in tomato8, apple58, peach12 and kiwi59. Our study found that MaLOX1 was downregulated during fruit ripening and 6 members (MaLOX1, 2, 4, 7, 8, and 18) were found to be ethylene responsive. It was reported that under ethylene and high-temperature treatment, the content of LOXA, LOX4, and LOX5 (corresponding to MaLOX4, MaLOX8, and MaLOX1 in this study, respectively) decreased in banana fruit peel26. During banana fruit ripening, the expression of MaLOX (or named as BanLOX) decreased60. After ethylene treatment, however, it was upregulated in the pulp while it did not change significantly in the peel60, which is similar to the results of this study. Moreover, MaLOX1, 4, and 7 were predominantly and specifically expressed during fruit ripening and were regulated by ethylene. Therefore, we speculated that these LOX genes may be the candidate genes involved in banana fruit ripening and flavor formation.
MeJA/JA, as a signal molecule that affects biological and abiotic reactions in plants, plays an important role in dealing with various external stresses. It was found that the application of exogenous MeJA can induce endogenous JA biosynthesis in plants61, and JA biosynthesis mainly depends on the substrate and expression of the genes at the critical steps of the synthesis pathway, such as LOX, AOC, AOS, and OPR62. In this study, with the exception of MaLOX11, 16, and 17, most 9-LOX subfamily MaLOX genes were upregulated and reached the maximum expression level at 6 h. And the expression trend of MaLOX14, belonging to Type II 13-LOX subfamily, also showed similar expression pattern. The expression of MaLOX16 is suppressed by MeJA, while MaLOX13, 15, 18 were significantly upregulated at 24 h. We also found that most MeJA responsive MaLOXs contain MeJA-responsive elements in their promoters. The expression of poplar9, Panax ginseng63, pepper64, and tomato8 LOX genes were found to be regulated by MeJA to some extent, which is consistent with our results. In addition, external application of MeJA can induce the expression of MaLOX1 and MaLOX2, enhance the content of endogenous JA, and alleviate banana chilling injury partially62. The above results indicate that MeJA can cause the up-regulation of LOX genes, which can increase the content of endogenous JA, thus improve the stress resistance of plants.
Conclusions
In this study, 18, 11, and 12 family members were respectively identified from M. acuminata, M. balbisiana, and M. itinerans genome, which encoded proteins with conserved domains and mainly located in the cytoplasm or chloroplast. The condon usage in banana LOX family members prefer to use and end with G/C. Four segmental duplications and 2 tandem duplications as well as 3 segmental duplications and 3 tandem duplications occurred respectively during M. acuminata and M. balbisiana evolution. Banana LOXs can be divided into three subfamilies, including 9-LOX, Type I 13-LOX, and Type II 13-LOX, and the sequence characteristics between each subfamily members are conservative. The expression of MaLOXs showed certain tissue specificity, and showed different response patterns to MeJA, high temperature, low temperature, and FocTR4 treatments. Moreover, the potential function analysis of the protomer region and the co-expression network of MaLOXs was constructed using WGCNA indicated that MaLOXs might participate in the growth and development and various stress responses in banana. Our present study can extend the knowledge of banana LOX gene family and provide basis for future exploration of their functions.
Supplementary Information
Acknowledgements
This work was supported by the National Key R&D Program of China (2019YFD1000901), the “outstanding young scientists” project of Fujian Agriculture and Forestry University (xjq201721), the National Natural Science Foundation of China (31701900; 31601713), the Construction of Plateau Discipline of Fujian Province (102/71201801101), and the National Special Fund Project for the Construction of Modern Agricultural Technology System (CARS-31-15).
Author contributions
C.Z.C, Y.J.H. and P.T.L conceived the study and design the experimental study. F.L., H.L., J.W.W, B.W., N.T. and J.P.L. performed the experiments. F.L., H.L., X.L.S. and H.W. analyzed the data. F.L. wrote the original draft of this paper. C.Z.C. revised the paper. All authors have read and approved the final version.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Fan Liu and Hua Li.
Contributor Information
Yuji Huang, Email: yjhuang2004@163.com.
Peitao Lü, Email: ptlv@fafu.edu.cn.
Chunzhen Cheng, Email: ld0532cheng@126.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-89211-6.
References
- 1.Chauvin A, Lenglet A, Wolfender J, Farmer E. Paired hierarchical organization of 13-lipoxygenases in Arabidopsis. Plants. 2016;5:16. doi: 10.3390/plants5020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng B, et al. Molecular analysis of lipoxygenase (LOX) genes in common wheat and phylogenetic investigation of LOX proteins from model and crop plants. J. Cereal Sci. 2010;52:387–394. doi: 10.1016/j.jcs.2010.06.019. [DOI] [Google Scholar]
- 3.Newcomer ME, Brash AR. The structural basis for specificity in lipoxygenase catalysis. Protein Sci. 2015;24:298–309. doi: 10.1002/pro.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teng LH, et al. Evolution and expansion of the prokaryote-like lipoxygenase family in the brown alga Saccharina japonica. Front. Plant Sci. 2017;8:2018. doi: 10.3389/fpls.2017.02018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Z, et al. Retention and molecular evolution of lipoxygenase genes in modern rosid plants. Front. Genet. 2016;7:176. doi: 10.3389/fgene.2016.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song H, et al. Identification of lipoxygenase (LOX) genes from legumes and their responses in wild type and cultivated peanut upon Aspergillus flavus infection. Sci. Rep. 2016;6:35245. doi: 10.1038/srep35245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Umate P. Genome-wide analysis of lipoxygenase gene family in Arabidopsis and rice. Plant Signal. Behav. 2011;6:335–338. doi: 10.4161/psb.6.3.13546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Upadhyay RK, Mattoo AK. Genome-wide identification of tomato (Solanum lycopersicum L.) lipoxygenases coupled with expression profiles during plant development and in response to methyl-jasmonate and wounding. J. Plant Physiol. 2018;231:318–328. doi: 10.1016/j.jplph.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Chen Z, et al. The lipoxygenase gene family in poplar: Identification, classification, and expression in response to MeJA treatment. PLoS ONE. 2015;10:e125526. doi: 10.1371/journal.pone.0125526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu JY, et al. Characterization and alternative splicing profiles of the lipoxygenase gene family in tea plant (Camellia sinensis) Plant Cell Physiol. 2018;59:1765–1781. doi: 10.1093/pcp/pcy091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaban M, Ahmed MM, Sun H, Ullah A, Zhu LF. Genome-wide identification of lipoxygenase gene family in cotton and functional characterization in response to abiotic stresses. BMC Genomics. 2018;19:599. doi: 10.1186/s12864-018-4985-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo SL, Song ZZ, Ma RJ, Yang Y, Yu ML. Genome-wide identification and expression analysis of the lipoxygenase gene family during peach fruit ripening under different postharvest treatments. Acta Physiol. Plant. 2017;39:111. doi: 10.1007/s11738-017-2409-6. [DOI] [Google Scholar]
- 13.Wang JL, et al. Bioinformatics analysis of the lipoxygenase gene family in radish (Raphanus sativus) and functional characterization in response to abiotic and biotic stresses. Int. J. Mol. Sci. 2019;20:6095. doi: 10.3390/ijms20236095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melan MA, et al. An Arabidopsis thaliana lipoxygenase gene can be induced by pathogens, abscisic acid, and methyl jasmonate. Plant Physiol. 1993;101:441–450. doi: 10.1104/pp.101.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marla SS, Singh VK. LOX genes in blast fungus (Magnaporthe grisea) resistance in rice. Funct. Integr. Genomic. 2012;12:265–275. doi: 10.1007/s10142-012-0268-1. [DOI] [PubMed] [Google Scholar]
- 16.Caldelari D, Wang GG, Farmer EE, Dong XN. Arabidopsis lox3 lox4 double mutants are male sterile and defective in global proliferative arrest. Plant Mol. Biol. 2011;75:25–33. doi: 10.1007/s11103-010-9701-9. [DOI] [PubMed] [Google Scholar]
- 17.Bell E, Creelman RA, Mullet JE. A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA. 1995;92:8675–8679. doi: 10.1073/pnas.92.19.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chauvin A, Caldelari D, Wolfender J, Farmer EE. Four 13-lipoxygenases contribute to rapid jasmonate synthesis in wounded Arabidopsis thaliana leaves: A role for lipoxygenase 6 in responses to long-distance wound signals. New Phytol. 2013;197:566–575. doi: 10.1111/nph.12029. [DOI] [PubMed] [Google Scholar]
- 19.Huang JX, et al. OsLOX2, a rice type I lipoxygenase, confers opposite effects on seed germination and longevity. Transgenic Res. 2014;23:643–655. doi: 10.1007/s11248-014-9803-2. [DOI] [PubMed] [Google Scholar]
- 20.Zhang B, et al. Lipoxygenase gene expression in ripening kiwifruit in relation to ethylene and aroma production. J. Agric. Food Chem. 2009;57:2875–2881. doi: 10.1021/jf9000378. [DOI] [PubMed] [Google Scholar]
- 21.Sarde SJ, et al. Involvement of sweet pepper CaLOX2 in jasmonate-dependent induced defence against Western flower thrips. J. Integr. Plant Biol. 2019;61:1085–1098. doi: 10.1111/jipb.12742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan LH, et al. Role of tomato lipoxygenase D in wound-induced jasmonate biosynthesis and plant immunity to insect herbivores. PLoS Genet. 2013;9:e1003964. doi: 10.1371/journal.pgen.1003964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou YL, et al. Overexpression of persimmon 9-lipoxygenase DkLOX3 confers resistance to Pseudomonas syringae pv. tomato DC3000 and Botrytis cinerea in Arabidopsis. Plant Growth Regul. 2018;84:179–189. doi: 10.1007/s10725-017-0331-y. [DOI] [Google Scholar]
- 24.Cheng CZ, et al. Identification of Fusarium oxysporum f. sp. cubense tropical race 4 (FocTR4) responsive miRNAs in banana root. Sci. Rep. 2019;9:13682. doi: 10.1038/s41598-019-50130-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu XY, et al. Low temperature storage reduces aroma-related volatiles production during shelf-life of banana fruit mainly by regulating key genes involved in volatile biosynthetic pathways. Postharvest Biol. Technol. 2018;146:68–78. doi: 10.1016/j.postharvbio.2018.08.015. [DOI] [Google Scholar]
- 26.Du LN, et al. Proteome changes in banana fruit peel tissue in response to ethylene and high-temperature treatments. Hort. Res. 2016;3:16012. doi: 10.1038/hortres.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamdee C, Ketsa S, Doorn WGV. Effect of heat treatment on ripening and early peel spotting in cv. Sucrier banana. Postharvest Biol. Technol. 2009;52:288–293. doi: 10.1016/j.postharvbio.2008.12.003. [DOI] [Google Scholar]
- 28.Li CY, et al. Transcriptome profiling of resistant and susceptible Cavendish banana roots following inoculation with Fusarium oxysporum f. sp. cubense tropical race 4. BMC Genom. 2012;13:374. doi: 10.1186/1471-2164-13-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang D, et al. Secretome analysis of the banana Fusarium wilt fungi Foc R1 and Foc TR4 reveals a new effector OASTL required for full pathogenicity of Foc TR4 in banana. Biomolecules. 2020;10:1430. doi: 10.3390/biom10101430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen CJ, et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 31.Wang YP, et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krzywinski M, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang DP, Zhang YB, Zhang Z, Zhu J, Yu J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinform. 2010;8:77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lescot M, et al. Insights into the Musa genome: Syntenic relationships to rice and between Musa species. BMC Genom. 2008;9:58. doi: 10.1186/1471-2164-9-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L, et al. Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta. 2011;234:377–390. doi: 10.1007/s00425-011-1410-3. [DOI] [PubMed] [Google Scholar]
- 36.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang XY, Jiang WJ, Yu HJ. The expression profiling of the lipoxygenase (LOX) family genes during fruit development, abiotic stress and hormonal treatments in cucumber (Cucumis sativus L.) Int. J. Mol. Sci. 2012;13:2481–2500. doi: 10.3390/ijms13022481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Podolyan A, White J, Jordan B, Winefield C. Identification of the lipoxygenase gene family from Vitis vinifera and biochemical characterisation of two 13-lipoxygenases expressed in grape berries of Sauvignon Blanc. Funct. Plant Biol. 2010;37:767–784. doi: 10.1071/FP09271. [DOI] [Google Scholar]
- 40.Zhang C, et al. The phylogeny and expression profiles of the lipoxygenase (LOX) family genes in the melon (Cucumis melo L.) genome. Sci. Hortic. 2014;170:94–102. doi: 10.1016/j.scienta.2014.03.005. [DOI] [Google Scholar]
- 41.Shibata D, Slusarenko A, Casey R, Hildebrand D, Bell E. Lipoxygenases. Plant Mol. Biol. Rep. 1994;12:S41–S42. doi: 10.1007/BF02671567. [DOI] [Google Scholar]
- 42.Sakamoto W, Miyagishima S, Jarvis P. Chloroplast biogenesis: Control of plastid development, protein Import, division and inheritance. Arabidopsis Book. 2008;6:e110. doi: 10.1199/tab.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Millar AH, Whelan J, Small I. Recent surprises in protein targeting to mitochondria and plastids. Curr. Opin. Plant Biol. 2006;9:610–615. doi: 10.1016/j.pbi.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 44.Liu HM, et al. Analysis of synonymous codon usage in Zea mays. Mol. Biol. Rep. 2010;37:677–684. doi: 10.1007/s11033-009-9521-7. [DOI] [PubMed] [Google Scholar]
- 45.CléMent Y, et al. Evolutionary forces affecting synonymous variations in plant genomes. PLoS Genet. 2017;13:e1006799. doi: 10.1371/journal.pgen.1006799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.D'Hont A, et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature. 2012;488:213–217. doi: 10.1038/nature11241. [DOI] [PubMed] [Google Scholar]
- 47.Wang Z, et al. Musa balbisiana genome reveals subgenome evolution and functional divergence. Nat Plants. 2019;5:810–821. doi: 10.1038/s41477-019-0452-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu W, et al. Whole genome sequencing of a banana wild relative Musa itinerans provides insights into lineage-specific diversification of the Musa genus. Sci. Rep. 2016;6:31586. doi: 10.1038/srep31586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindemose S, O'Shea C, Jensen MK, Skriver K. Structure, function and networks of transcription factors involved in abiotic stress responses. Int. J. Mol. Sci. 2013;14:5842–5878. doi: 10.3390/ijms14035842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Porta H, Rocha-Sosa M. Plant lipoxygenases. Physiological and molecular features. Plant Physiol. 2002;130:15–21. doi: 10.1104/pp.010787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu W, et al. Genome-wide analyses of the bZIP family reveal their involvement in the development, ripening and abiotic stress response in banana. Sci. Rep. 2016;6:30203. doi: 10.1038/srep30203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu JH, et al. Genome-wide analysis of banana MADS-box family closely related to fruit development and ripening. Sci. Rep. 2017;7:3467. doi: 10.1038/s41598-017-03897-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han YC, Fu CC. Cold-inducible MaC2H2s are associated with cold stress response of banana fruit via regulating MaICE1. Plant Cell Rep. 2019;38:673–680. doi: 10.1007/s00299-019-02399-w. [DOI] [PubMed] [Google Scholar]
- 54.Li WB, Li CQ, Sun JB, Peng M. Metabolomic, biochemical, and gene expression analyses reveal the underlying responses of resistant and susceptible banana species during early infection with Fusarium oxysporum f. sp. cubense. Plant Dis. 2017;101:534–543. doi: 10.1094/PDIS-09-16-1245-RE. [DOI] [PubMed] [Google Scholar]
- 55.He X, et al. Comprehensive analyses of the annexin (ANN) gene family in Brassica rapa, Brassica oleracea and Brassica napus reveals their roles in stress response. Sci. Rep. 2020;10:4295. doi: 10.1038/s41598-020-59953-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Z, et al. Genome-wide analysis of basic helix-loop-helix transcription factors to elucidate Candidate genes related to fruit ripening and stress in banana (Musa acuminata L. AAA Group, cv. Cavendish) Front. Plant Sci. 2020;11:650. doi: 10.3389/fpls.2020.00650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han MY, Zhang T, Zhao CP, Zhi JH. Regulation of the expression of lipoxygenase genes in Prunus persica fruit ripening. Acta Physiol. Plant. 2011;33:1345–1352. doi: 10.1007/s11738-010-0668-6. [DOI] [Google Scholar]
- 58.Vogt J, Schiller D, Ulrich D, Schwab W, Dunemann F. Identification of lipoxygenase (LOX) genes putatively involved in fruit flavour formation in apple (Malus × domestica) Tree Genet. Genomes. 2013;9:1493–1511. doi: 10.1007/s11295-013-0653-5. [DOI] [Google Scholar]
- 59.Zhang-BBB B, Yin XR, Shen JY, Chen KS. Volatiles production and lipoxygenase gene expression in kiwifruit peel and flesh during fruit ripening. J. Agric. Food Chem. 2009;134:472–477. doi: 10.1021/jf9000378. [DOI] [PubMed] [Google Scholar]
- 60.Yang XT, Song J, Fillmore S, Pang XQ, Zhang ZQ. Effect of high temperature on color, chlorophyll fluorescence and volatile biosynthesis in green-ripe banana fruit. Postharvest Biol. Technol. 2011;62:246–257. doi: 10.1016/j.postharvbio.2011.06.011. [DOI] [Google Scholar]
- 61.Zhao J, Davis LC, Verpoorte R. Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol. Adv. 2005;23:283–333. doi: 10.1016/j.biotechadv.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 62.Zhao LM, et al. Induction of jasmonate signalling regulators MaMYC2s and their physical interactions with MaICE1 in methyl jasmonate-induced chilling tolerance in banana fruit. Plant Cell Environ. 2013;36:30–51. doi: 10.1111/j.1365-3040.2012.02551.x. [DOI] [PubMed] [Google Scholar]
- 63.Bae K, et al. Molecular characterization of lipoxygenase genes and their expression analysis against biotic and abiotic stresses in Panax ginseng. Eur. J. Plant Pathol. 2016;145:331–343. doi: 10.1007/s10658-015-0847-9. [DOI] [Google Scholar]
- 64.Sarde SJ, Kumar A, Remme RN, Dicke M. Genome-wide identification, classification and expression of lipoxygenase gene family in pepper. Plant Mol. Biol. 2018;98:375–387. doi: 10.1007/s11103-018-0785-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.