Summary
Parvalbumin (PV) is a cytosolic Ca2+-binding protein highly expressed in fast skeletal muscle, contributing to an increased relaxation rate. Moreover, PV is an “atrogene” downregulated in most muscle atrophy conditions. Here, we exploit mice lacking PV to explore the link between the two PV functions. Surprisingly, PV ablation partially counteracts muscle loss after denervation. Furthermore, acute PV downregulation is accompanied by hypertrophy and upregulation by atrophy. PV ablation has a minor impact on sarcoplasmic reticulum but is associated with increased mitochondrial Ca2+ uptake, mitochondrial size and number, and contacts with Ca2+ release sites. Mitochondrial calcium uniporter (MCU) silencing abolishes the hypertrophic effect of PV ablation, suggesting that mitochondrial Ca2+ uptake is required for hypertrophy. In turn, an increase of mitochondrial Ca2+ is required to enhance expression of the pro-hypertrophy gene PGC-1α4, whose silencing blocks hypertrophy due to PV ablation. These results reveal how PV links cytosolic Ca2+ control to mitochondrial adaptations, leading to muscle mass regulation.
Keywords: calcium buffer, mitochondria, skeletal muscle
Graphical abstract
Highlights
-
•
PV is downregulated during skeletal muscle atrophy, and its levels affect trophism
-
•
Skeletal muscle mitochondria undergo remodeling in PV knockout mice
-
•
Mitochondria increase cytosolic Ca2+ buffer capacity in PV knockout skeletal muscles
-
•
Increased mitochondrial Ca2+ triggers the PGC-1α4 pathway, inducing muscle growth
Parvalbumin is one of the most important cytosolic Ca2+ buffers in skeletal muscle. Butera et al. demonstrate that the profound mitochondria rearrangement in muscle fibers, caused by parvalbumin ablation, increases mitochondria Ca2+ buffering capacity. This impinges on the PGC-1α4 pathway, counteracting atrophy and inducing muscle fiber hypertrophy.
Introduction
Intracellular Ca2+ modulates many cellular physiological responses such as proliferation, muscle contraction, immune response, gene transcription, and apoptosis. The cytosolic Ca2+ concentration ([Ca2+]cyt), in response to various stimuli, can transiently increase from 10−7 M in the resting state to 10−5 M or more. The ability of Ca2+ to play the role of intracellular signal results from a strict control of its concentration changes in space, time, and amplitude (Berridge et al., 2000, 2003). To perform this precise regulation, cells rely on proteins that are able to generate and modulate [Ca2+]cyt fluctuations, such as pumps, channels, and binding and buffering proteins (Berridge et al., 2003; Rizzuto et al., 2012)
The first Ca2+ buffer of the EF-hand family was discovered more than 50 years ago and named parvalbumin (PV) (Henrotte, 1952). Although principally expressed in GABAergic interneurons, PV is also present in other neuron populations, fast-twitch muscle fibers, parathyroid glands, and distal convolute tubules in nephrons (Henzi and Schwaller, 2015; Schwaller, 2010).
The identification of PV’s role as a cytosolic Ca2+ buffer in skeletal muscle fibers dates back to the 1990s. In relaxed muscle fibers, Ca2+ is stored in the sarcoplasmic reticulum (SR) and PV is in the Ca2+-free conformation, binding Mg2+. When Ca2+ is released from the SR, PV binds Ca2+ and dissociates Mg2+. In this way, PV contributes to lower [Ca2+ ]cyt and facilitates fiber relaxation acting, as a Ca2+ shuttle (Gillis et al., 1982). Müntener et al. reported the first in vivo experiment that addressed the function of PV in a rodent model and showed that PV expression in the rat slow-twitch muscle soleus significantly increases the speed of relaxation, without affecting amplitude and time to peak of contraction (Müntener et al., 1995). When the first knockout (KO) mouse for PV (PV−/−) was generated (Schwaller et al., 1999), the comparison between wild-type (WT) and PV-deficient muscles confirmed the contribution of PV in accelerating the relaxation rate of fast mouse muscles.
More recently, PV has been also described as an “atrogene,” as it belongs to a set of ∼120 genes that are commonly up- or downregulated during disuse and systemic conditions, leading to skeletal muscle atrophy (Lecker et al., 2004; Sacheck et al., 2007). Among these genes, PV is one of the most downregulated (Lecker et al., 2004; Sacheck et al., 2007), suggesting that this protein could also play an important role in skeletal muscle plasticity. Yet, the direct effect and mechanism of PV modulation of muscle trophism have not been clarified.
A clue to understanding the mechanisms has been given by the discovery that PV ablation is accompanied by an increase in mitochondria number and size (Chen et al., 2001, 2006). Mitochondria not only are the powerhouse of the cells but also participate in many regulatory functions, among them Ca2+ homeostasis. Mitochondria can take up Ca2+ through a highly sophisticated channel, the mitochondrial calcium uniporter (MCU) complex (Baughman et al., 2011; Feno et al., 2021; Patron et al., 2014; De Stefani et al., 2011). Furthermore, in the last decade, evidence has emerged showing that mitochondria contribute to the regulation of fiber size (Mammucari et al., 2015).
Here, we tested the hypothesis that mitochondria are the link between the two functions of PV, cytosolic Ca2+ buffering and regulation of muscle trophism. Indeed, we found that PV ablation not only allows mitochondria to accumulate more Ca2+ but also increases both mitochondrial number and volume, as well as the contacts between mitochondria and Ca2+ release units (CRUs). Altogether, these adaptations make the mitochondria able to significantly contribute to cytosolic Ca2+ homeostasis and trigger signaling pathways, ultimately inducing muscle fiber growth.
Results
PV ablation partially protects muscles from denervation atrophy
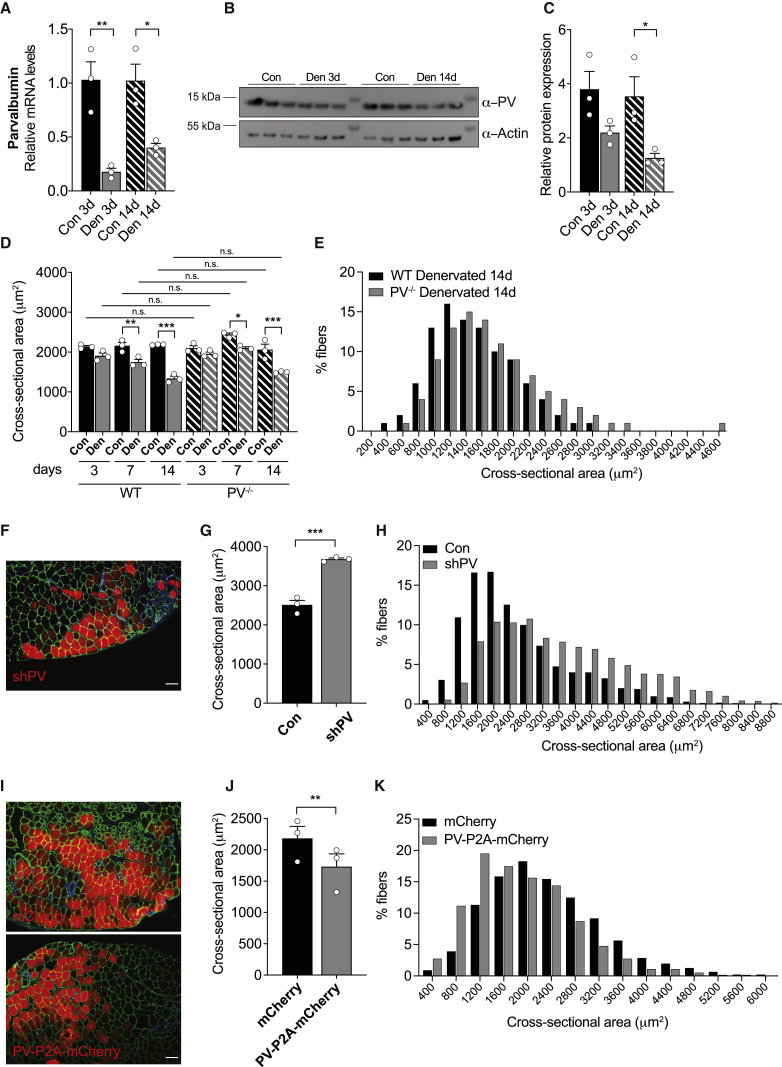
To confirm PV downregulation during atrophy (Lecker et al., 2004; Sacheck et al., 2007), we analyzed PV expression during the progression of skeletal muscle denervation in mice. We thus unilaterally cut the sciatic nerve of WT mice and analyzed denervated and contralateral tibialis anterior (TA) muscles 3 and 14 days after nerve cut. Our data confirmed (see Leberer et al., 1987) that both PV mRNA (Figure 1A) and protein (Figures 1B and 1C) are downregulated during the progression of denervation atrophy.
Figure 1.
PV expression level decreases after denervation and the modulation of its expression affects muscle trophism
(A) mRNA expression levels of PV normalized to contralateral muscles in TA muscles at 3 and 14 days after denervation. Data are normalized to POL2. n = 3.
(B) Representative WB of denervated and contralateral muscles. α-Actin was used as loading control. n = 3.
(C) Quantification of the levels of PV protein obtained as in (B) and normalized to actin. n = 3.
(D) Mean fiber CSA at 3, 7, and 14 days after denervation compared to contralateral muscles of TA muscles from WT and PV−/− mice. At least 1,500 fibers per muscle; n = 3.
(E) Size-frequency distribution of CSA of the experiment as in (D) at 14 days after denervation.
(F) Representative images of muscle cross-sections of TA muscles electroporated with shPV for 7 days. shPV-positive fibers are displayed in red and α-laminin in green. Scale bar, 100 μm.
(G) CSA of shPV-positive fibers compared to non-transfected fibers in the same muscle. More than 200 fibers per muscle; n = 3.
(H) Size-frequency distribution of CSA of the experiment as in (F) and (G).
(I) Representative images of muscle cross-sections of mCherry-transfected muscles (upper panel) and PV-overexpressing muscles (lower panel) for 7 days. α-Laminin is visualized in green. Scale bar, 100 μm.
(J) CSA measurements of control and PV-overexpressing fibers. More than 200 fibers per muscle; n = 3.
(K) Size-frequency distribution of CSA of the experiment as in (J) and (K).
Data are expressed as mean ± SEM. For data analysis, a two-tailed, unpaired Student’s t test (A and C); two-way ANOVA with Bonferroni post hoc (D); or two-tailed, paired Student’s t test (G and J) were performed. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; n.s., not statistically significant. Con, contralateral muscle; Den, denervated muscle.
To investigate whether PV plays a direct role in atrophy, we performed denervation experiments using the PV−/− mouse model (Schwaller et al., 1999; see Figure S1A). Of note, fiber size analysis performed on TA muscles of 3-month-old WT and PV−/− mice reported no difference in muscle fiber size (Figure S1B).
Denervation atrophy was induced by sciatic nerve section in WT and PV−/− mice, and muscle fiber size was evaluated at 3, 7, or 14 days after the denervation (Figure 1D). As expected, denervation induced a progressive atrophy in both WT and PV−/− TA muscles. However, in WT mice, the percentage of muscle mass loss at 3, 7, and 14 days after denervation was 10.9% ± 3.2%, 19.1% ± 2.2%, and 38.9% ± 3.6%, respectively, while in PV−/− mice, it was only the 7.0% ± 0.81%, 13.9% ± 2.3%, and 26.8% ± 5.3% (p = 0.07 compared to WT), respectively (data are expressed as mean ± SEM).
Therefore, PV ablation is associated with a trend of partial protection from denervation-induced atrophy, as also shown by the shift to the right of the size-frequency distribution of cross-sectional area (CSA) of PV−/− fibers compared to WT fibers at 14 days after denervation (Figure 1E).
PV acute silencing and overexpression in vivo affects skeletal muscle trophism
Since constitutive PV ablation does not cause any difference in fiber size and causes a modest but unexpected effect on denervation, as atrophy was limited and not enhanced, we analyzed the impact of an acute downregulation of PV expression on muscle size. To this end, we silenced PV in TA of adult WT mice by electroporating a plasmid expressing a specific short hairpin RNA (shRNA) targeted to PV together with mCherry red fluorescent protein (shPV-mCherry, hereafter indicated as shPV) and evaluating fiber size. Three different shRNAs were designed to silence PV, and the efficiency of silencing was tested on HeLa cells (Figures S2A and S2B). Then, flexor digitorum brevis (FDB) muscles of WT mice were transfected with either non-targeting shRNA (shLUC) or shPV and collected at either 7 or 14 days, and silencing efficiency was tested by RT-PCR (Figure S2C). We finally electroporated TA muscles of adult WT mice with either control shLUC or shPV plasmids, and after 7 days, we collected muscles and measured fiber CSA. Since fibers transfected with shLUC did not show any difference compared with surrounding untransfected fibers (Figure S2D), we decided to use as control for CSA analysis the untransfected fibers of the same muscles electroporated with shPV. Notably, PV silencing induced an increase in fiber size of ∼35% (Figures 1F and 1G) and a clear shift to the right of the size-frequency histogram (Figure 1H). Overexpression of PV gave coherent results. Indeed, we electroporated TA muscles of WT mice with either control or PV-P2A-mCherry plasmids for 7 days and, interestingly, PV-overexpressing fibers showed a significant reduction of fiber size compared to control (Figures 1I and 1J) and a shift to the left of the size-frequency histogram (Figure 1K), suggesting that PV overexpression is sufficient to induce muscle atrophy. Overexpression efficiency was confirmed by RT-PCR and western blotting (WB) on FDB muscles electroporated with either mCherry (control) or PV-P2A-mCherry plasmids for 7 days (Figures S2E and S2F).
Modulation of PV expression affects cytosolic Ca2+ transients
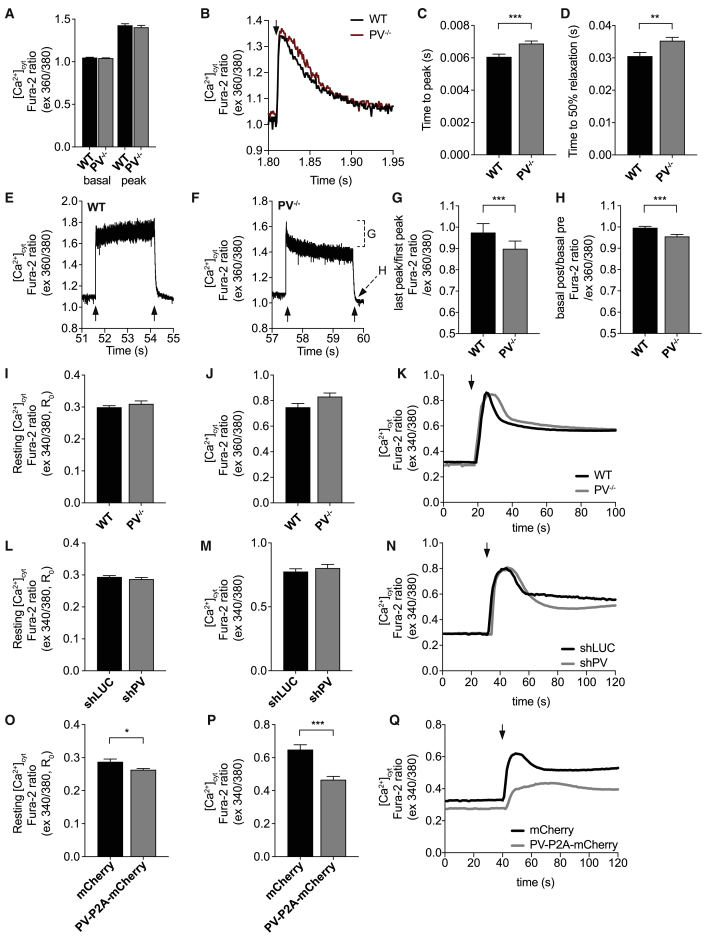
We then studied the effect of PV ablation on Ca2+ dynamics, including all three main subcellular compartments challenged by different kinds of Ca2+ release. First, we measured [Ca2+]cyt in FDB muscle fibers of WT and PV−/− animals with Fura-2 AM. [Ca2+]cyt in the resting condition and upon a single electrical pulse did not show any differences between WT and PV−/− muscle fibers (Figures 2A), whereas the kinetics of Ca2+ transients and Ca2+ clearance were altered in PV−/− muscle fibers; indeed, consistent with the physiological role of PV in buffering cytosolic Ca2+, the time to peak and half-relaxation time were prolonged in PV−/− fibers compared to WT (Figures 2B–2D).
Figure 2.
Cytosolic Ca2+ transients in single isolated FDB muscle fibers of WT and PV−/− mice
(A) [Ca2+]cyt determined with Fura-2 AM (ratio 360/380 nm) in the basal resting condition and at the peak of the transient in WT and PV−/− single FDB fibers with electrical stimulation at 0.5 Hz. n ≥ 38.
(B) Representative traces of the experiment. The arrow indicates stimulation.
(C) Time to the Ca2+ transient peak.
(D) Half-time of the decay of the Ca2+ transient.
(E and F) Representative traces of [Ca2+]cyt transients in WT and PV−/− FDB fibers induced by tetanic stimulation at 60 Hz (2 s) with Fura-2 AM (ratio 360/380 nm). Note that the stimulation frequency is not sufficient for complete fusion. The arrows indicate the beginning and the end of the tetanic stimulation. Segment G, decay from first to last peak; arrow H, dip at the end of relaxation.
(G) Decay of [Ca2+]cyt expressed as the ratio of the last peak to the first. n ≥ 15.
(H) Decay of the basal [Ca2+]cyt 200 ms after the last electrical pulse expressed as the ratio to basal [Ca2+] before the start of the stimulation. n ≥ 15.
(I and J) Resting [Ca2+]cyt (I) and peak cytosolic Ca2+ transients (J) in response to 10 mM caffeine in WT and PV−/− FDB muscle fibers determined with Fura-2 AM (ratio 340/380 nm). n ≥ 43.
(K) Representative traces of the experiment as in (I) and (J).
(L and M) Resting [Ca2+]cyt (L) and peak cytosolic Ca2+ transients (M) in FDB fibers electroporated with either shLUC or shPV determined with Fura-2 AM (ratio 340/380 nm). n ≥ 43.
(N) Representative traces of the experiment as in (L) and (M).
(O and P) Resting [Ca2+]cyt (O) and peak cytosolic Ca2+ transients (P) in FDB fibers electroporated with either mCherry or PV-P2A-mCherry plasmids for 7 days determined as in (L) and (M). n ≥ 28.
(Q) Representative traces of the experiment as in (O) and (P).
Data are expressed as mean ± SEM. For data analysis, a two-tailed, unpaired Student’s t test was performed. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Note that the Fura-2 data were collected with two different set-ups. (A–H) Ionoptix setup (ratio 360/380) with 1-kHz sampling rate; (I–Q) Crisel setup (ratio 340/380) with 20 Hz sampling.
Next, we investigated cytosolic Ca2+ transients during a train of high-frequency electrical stimulation (2 s at 60 Hz), suitable to induce a partially fused tetanic contraction (Paolini et al., 2011). Unexpectedly, PV−/− fibers were unable to sustain high levels of [Ca2+]cyt during a tetanus as WT fibers (Figures 2E and 2F). The ratio between the cytosolic Ca2+ peaks measured at the end and, respectively, the beginning of the tetanic stimulation was lower in PV−/− than in WT muscle fibers, pointing to a significant decrease of [Ca2+]cyt during high-frequency stimulation (Figure 2G). Moreover, in PV−/− muscle fibers, the level of resting basal [Ca2+]cyt after the end of the stimulation train was transiently lower than the level before the beginning of the stimulation (Figure 2H), and the typical slow phase of decline or “tail” of the transients, well detectable in WT fibers (Figure 2E), was lacking in PV−/− muscle fibers (Figure 2F). Finally, the ability of the fibers to withstand a sudden increase in [Ca2+]cyt was tested with a caffeine-induced release of Ca2+. As can be seen (Figures 2I–2K), the peak of [Ca2+]cyt induced by 10 mM caffeine was not different in PV−/− and WT fibers.
To exclude any possible in vivo compensatory effects induced by PV ablation during development in our KO model, we performed PV acute silencing and overexpression in adult WT FDB muscle fibers, and we monitored the ability of the fibers to control the cytosolic Ca2+ wave induced by caffeine, as in the previous experiments. Similarly to what observed in PV−/− muscles, no significant differences were detected in [Ca2+]cyt between WT fibers transfected with shLUC and PV-silenced fibers, both in the basal resting condition (Figure 2L) and upon caffeine-induced release of Ca2+ (Figures 2M and 2N).
Next, we analyzed the effect of PV overexpression on cytosolic Ca2+ transients. WT FDB fibers overexpressing PV exhibited a significant decrease of [Ca2+]cyt both in the basal condition (Figure 2O) and upon caffeine-induced Ca2+ release (Figures 2P and 2Q), suggesting that overexpressed PV enhanced the cytosolic buffering power at rest and in the presence of a large Ca2+ release.
PV ablation does not alter SR Ca2+ concentration but affects mitochondrial Ca2+ handling
The changes in cytosolic Ca2+ transients prompted us to investigate possible changes of Ca2+ concentration in the SR ([Ca2+]SR) using the probe D1ER cameleon (Galla et al., 2019; Palmer et al., 2004) in single FDB fibers. Free [Ca2+]SR measured in resting basal condition was indistinguishable in fibers lacking PV from the WT value (Figure S3A), while during electrical stimulation (2-s train at 60 Hz), the decline in concentration was slightly, but not significantly, greater in PV−/− fibers than in WT fibers (−9.31% and −7.64%, respectively) (Figures S3B–S3D). We determined with RT-PCR the expression of the genes most relevant for SR Ca2+ handling, including Ryanodine Receptor 1 (RyR1), SERCA1, SERCA2, CASQ1, and calreticulin. We also checked the expression levels of STIM1 and ORAI1, which are responsible of store-operated Ca2+ entry. For all the genes analyzed, no significant difference was detected between WT and PV−/− fibers (Figure S3E). We also performed WB analysis of CASQ1, RyR1, IP3R, SERCA1, and SERCA2, and again, no significant differences in protein levels were detected between WT and PV−/− muscles (Figures S3F–S3I), further confirming that the expression of SR proteins involved in Ca2+ homeostasis is not affected by PV deficiency.
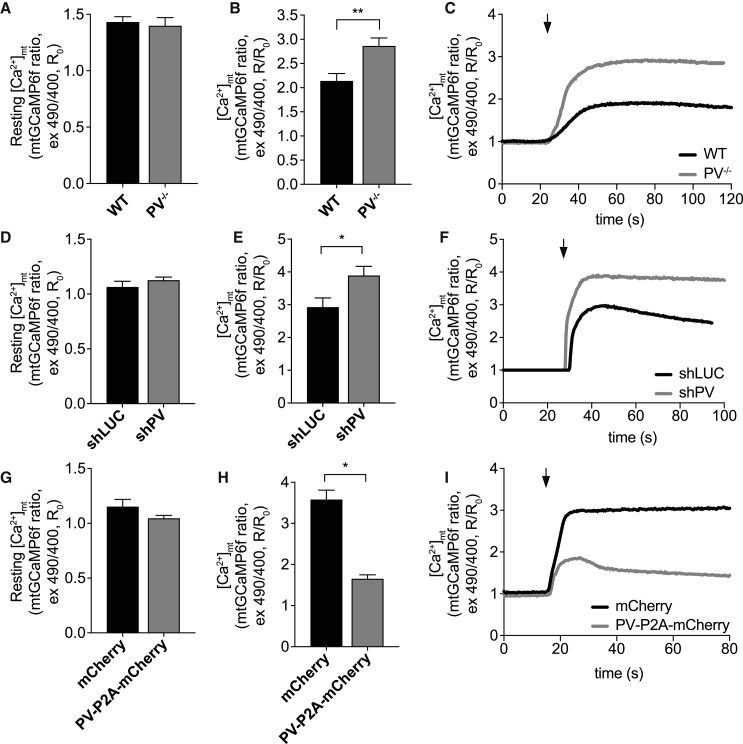
Next, we verified the effect of PV ablation on mitochondrial Ca2+ transients by transfecting adult FDB mouse muscle of WT and PV−/− with plasmids encoding 4mtGCaMP6f, as previously described (Chen et al., 2013; Mammucari et al., 2015; Vecellio Reane et al., 2016). No difference was detected in mitochondrial Ca2+ concentration [Ca2+]mt in the resting condition (Figures 3A and S4A), while mitochondrial Ca2+ uptake during caffeine-induced Ca2+ release and during tetanic stimulation was significantly higher in PV−/− muscle fibers compared to control (Figures 3B, 3C, S4B, and S4C). This effect is specific, since mitochondrial membrane potential was not perturbed by PV ablation (Figure S4D).
Figure 3.
Modulation of PV expression affects mitochondrial Ca2+ uptake in single isolated FDB fibers
(A and B) Resting [Ca2+]mt (A) and peak [Ca2+]mt (B) upon caffeine stimulation in WT and PV−/− FDB muscle fibers. n ≥ 23.
(C) Representative traces of the experiment as in (A) and (B). The arrow indicates the addition of caffeine.
(D and E) Resting [Ca2+]mt (D) and peak [Ca2+]mt (E) upon caffeine stimulation in FDB muscle single fibers electroporated with either shLUC or shPV for 7 days. n ≥ 18.
(F) Representative traces of the experiment as in (D) and (E). The arrow indicates the addition of caffeine.
(G and H) Resting [Ca2+]mt (G) and [Ca2+]mt peak (H) upon caffeine stimulation in FDB muscle single fibers electroporated with either mCherry or PV-P2A-mCherry for 7 days. n ≥ 21.
(I) Representative traces of the experiment as in (G) and (H).
Data are presented as mean ± SEM. For data analysis, two-tailed, unpaired Student’s t tests was performed. ∗p < 0.05; ∗∗p < 0.01. The arrow indicates the addition of caffeine.
See also Figures S3 and S4.
To exclude that the changes in mitochondrial Ca2+ uptake were due to possible in vivo compensatory effects during development following constitutive PV ablation, we performed the same experiment on PV acute silencing and overexpression in adult WT FDB muscle fibers. We first co-transfected WT mouse FDB muscles with shPV together with 4mtGCaMP6f probe. Ex vivo imaging experiments showed that while basal [Ca2+]mt is not affected by the removal of PV (Figure 3D), a marked increase of Ca2+ uptake following caffeine-induced Ca2+ release was detected in shPV-transfected fibers (Figures 3E and 3F). Furthermore, PV acute overexpression had a profound effect on mitochondrial Ca2+ handling, causing a significant decrease in mitochondrial Ca2+ uptake after caffeine-induced Ca2+ release (Figures 3H and 3I) without affecting resting [Ca2+]mt (Figure 3G).
The mitochondrial Ca2+ measurements made us hypothesize that PV ablation could induce an adaptive response of mitochondria. To test this hypothesis, we first focused our attention on the expression levels of the different components of the MCU complex. The results showed that the expression of the pore-forming MCU and the regulatory subunit MICU1 was not changed at the protein level, in contrast with an increased expression at the transcriptional level of not only MCU and MICU1 but also the muscle-specific alternative splicing isoforms MICU1.1 and MICU2 (Figures S4E and S4F).
PV ablation alters mitochondrial morphology and the number and expression of genes involved in mitochondrial biogenesis and dynamics
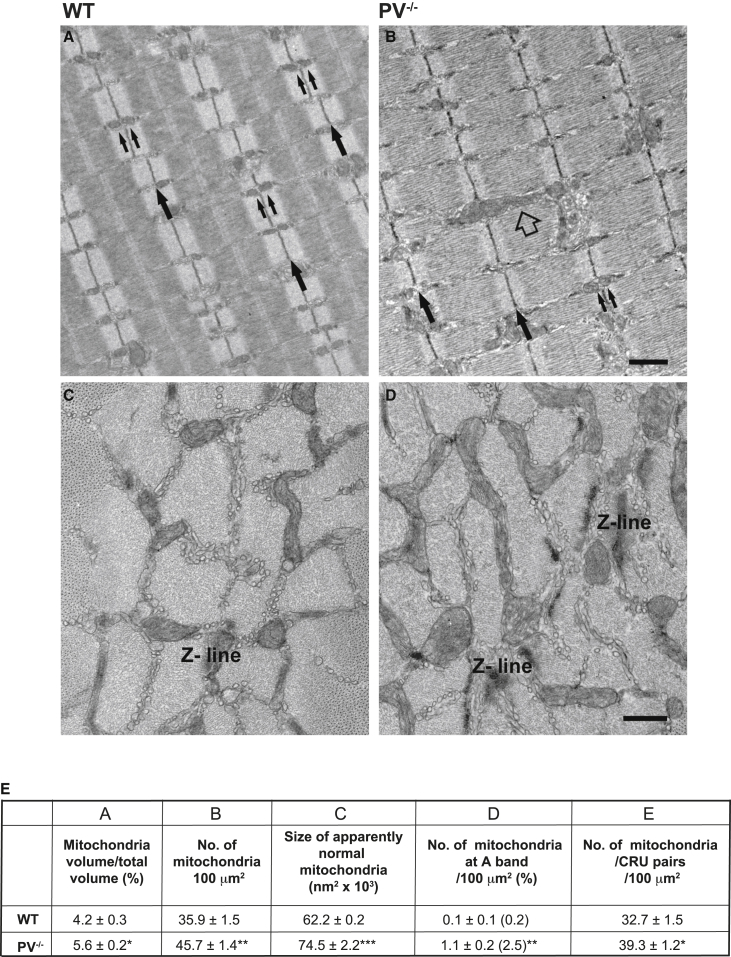
To further investigate mitochondrial adaptation to PV removal, we performed transmission electron microscopy (EM) on transversal and longitudinal sections of extensor digitorum longus (EDL) muscles of WT and PV−/− mice. EM images revealed that while mitochondria in WT EDL fibers (indicated by two small black arrows) are mostly round and almost exclusively positioned at the I band on both sides of the Z lines (indicated by large black arrows) (Figure 4A), mitochondria in PV−/− fibers appear more variable in shape and more often longitudinally oriented between myofibrils (Figure 4B). Images of cross sections in proximity of the Z line show that mitochondria (darker organelles) may occupy a higher percentage of the fiber volume (Figures 4C and 4D). This observation is in full agreement with previous findings by Schwaller and coworkers (Chen et al., 2001).
Figure 4.
PV ablation alters mitochondrial density, volume, and distribution and increases the number of mitochondria-CRU pairs
(A and B) Representative longitudinal sections of EDL fibers from adult WT and PV−/− mice. Small black arrows indicate pairs of mitochondria at the I band, large black arrows point to Z lines, and the empty arrow indicates a longitudinally oriented mitochondrion.
(C and D) Representative images of the distribution of mitochondria in cross-sections of EDL fibers from WT and PV−/− mice. Scale bars represent 1 μm (A and C) and 0.5 μm (B and D). n = 2 mice (WT); n = 3 mice (PV−/−).
(E) Quantitative analyses of mitochondria population. Column A, mitochondria volume; columns B and C, number and size; column D, percentage of mitochondria misplaced at the A band; column E, number of mitochondria/CRU pairs. Data are presented as mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Sample sizes are as follows: column A, 13 fibers from two WT mice and 25 fibers from three PV−/− mice, with two micrographs per fiber; columns B–D, 20 fibers from two WT mice and 30 fibers from three PV−/− mice, with five micrographs per fiber; and column E, 20 fibers from two WT mice and 30 fibers from three PV−/− mice, with five micrographs per fiber.
A further detailed quantitative analysis revealed that in PV−/− EDL fibers, all of the following parameters were increased: (1) the volume occupied by mitochondria relative to total fiber volume (+33%; table in Figure 4E, column A), (2) the number of mitochondria per 100 μm2 of image area (+27%; table in Figure 4E, column B), (3) the size of apparently normal mitochondria (+20%; table in Figure 4E, column C), and (4) the percentage of mitochondria next to the A band (table in Figure 4E, column D). Importantly, the number of mitochondria coupled with CRUs, specialized intracellular junctions (also named triads), which are involved in excitation-contraction (EC) coupling, was significantly increased (+20%; table in Figure 4E, column E).
Regarding mitochondria morphology, we evaluated the form factor (FF), which reflects the degree of their branching, and the aspect ratio (AR), the ratio between the major and minor axis of the mitochondrion (i.e., a measure of its shape). This quantitative assessment revealed that (1) in PV−/− fibers, FF is slightly decreased, even if not significantly, compared to WT, which indicates a lower degree of mitochondrial branching (2.1 ± 0.4 versus 1.9 ± 0.3 in WT and PV−/−, respectively); and (2) in PV−/− fibers, mitochondria shape is more elliptical, the AR value being 3.7 ± 0.3 versus 4.2 ± 0.3 in WT and PV−/−, respectively.
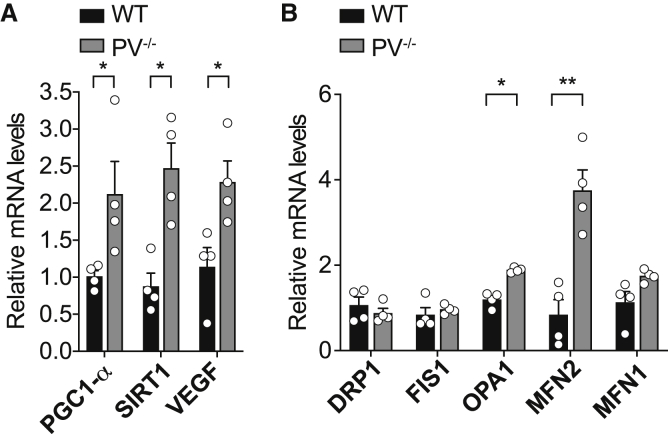
The changes observed in PV−/− fibers in mitochondrial volume density, morphology, and function prompted us to study whether the signaling pathways involved in mitochondrial biogenesis and dynamics were also affected. We thus investigated the expression levels of the master regulator of mitochondrial biogenesis, PGC-1α, and its positive regulator, the deacetylase SIRT1 (Jornayvaz and Shulman, 2010), in TA muscles of WT and PV−/− mice through RT-PCR.
Interestingly, both PGC-1α and SIRT1 mRNA expression was induced in TA of PV−/− mice, and the transcriptional activity was confirmed by the increased expression of vascular endothelial growth factor (VEGF), a known target of PGC-1α (Martínez-Redondo et al., 2016; Figure 5A).
Figure 5.
PV ablation affects the expression of genes involved in mitochondrial biogenesis and fusion
(A) Expression levels of PGC-1α, SIRT1, and VEGF in WT and PV−/− TA muscles.
(B) Expression levels of DRP1, FIS1, OPA1, MFN2, and MFN1 in WT and PV−/− TA muscles.
Data were normalized to WT and are presented as mean ± SEM. Expression levels were normalized to GAPDH. For statistical analysis, a parametric Student’s t test (two tailed, unpaired) was performed. ∗p < 0.05; ∗∗p < 0.01. See also Figure S5.
Furthermore, we asked whether the increase in mitochondrial volume could also be due to an alteration in the fusion/fission processes (Herzig and Martinou, 2008) by performing RT-PCR of the genes involved in mitochondrial dynamics on TA muscle of WT and PV−/− mice. In PV−/− muscles, OPA1 and MFN2, involved in mitochondrial fusion (Twig and Shirihai, 2011; Twig et al., 2008), were upregulated (Figure 5B), while no changes were observed in MFN1 expression (Figure 5B) and the expression levels of the genes involved in mitochondrial fission, DRP1 and FIS1 (Romanello et al., 2010; Westermann, 2012; Figure 5B).
The increase of mitochondrial volume (Figure 4) induced by PV ablation in fast-twitch muscles is reminiscent of the features of slow-type fibers, characterized by high oxidative and low glycolytic capacity (Racay et al., 2006; Schiaffino and Reggiani, 2011). We therefore asked whether PV removal could also induce a fiber-type switch, but no evident alterations of skeletal muscle fiber-type composition were detected in PV−/− muscles (Figure S5A). Indeed, both WT and PV−/− TA muscles had comparable numbers of type IIA, IIB, and IIX fibers, while type I fibers were virtually undetectable (Figures S5B).
Mitochondria act as dynamic Ca2+ buffers in muscle fibers lacking PV
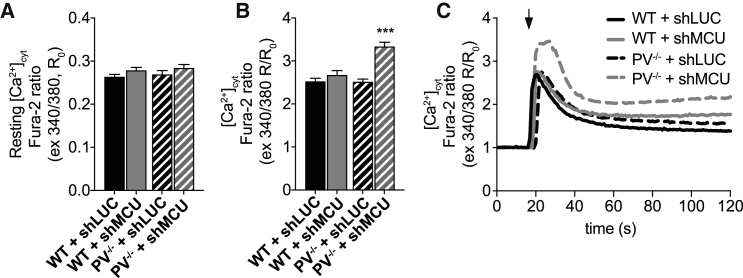
So far, our results suggest a potential role of mitochondria in buffering Ca2+ released from SR in the absence of PV. To verify this hypothesis, we silenced MCU in WT and PV−/− FDB muscle fibers, and we challenged their buffer capacity in response to a large Ca2+ release induced by caffeine. FDB muscles of WT and PV−/− mice were transfected in vivo with plasmids encoding either shLUC or shRNA designed to specifically silence MCU (shMCU), as previously described (Mammucari et al., 2015). One week later, [Ca2+]cyt was measured in single FDB fibers with Fura-2 AM in the basal resting condition and upon caffeine-induced Ca2+ release. In the resting condition, PV−/− and WT MCU-silenced fibers did not show any significant difference in basal [Ca2+]cyt compared to control fibers (PV−/− and WT transfected with shLUC) (Figure 6A). Upon caffeine-induced Ca2+ release, peak [Ca2+]cyt was not affected by the absence of MCU in WT fibers, while in PV−/− fibers, a significant increase in peak [Ca2+]cyt occurred (Figures 6B and 6C), indicating that the profound mitochondrial adaptation induced by PV ablation contributes to prevent excessive [Ca2+]cyt increases. Accordingly, when Ca2+ release was induced by a train of electrical pulses (2 s, 60 Hz), the amplitude of the Ca2+ transient was also increased by MCU silencing in PV−/−, but not WT, fibers (Figure S6A). As expected, MCU silencing blunts the increase of mitochondrial Ca2+ to similar extent in both WT and PV−/− muscle fibers, confirming its essential role in mitochondrial Ca2+ uptake (Figures S6B and S6C).
Figure 6.
Mitochondria act as dynamic Ca2+ buffers in PV−/− muscle fibers
(A and B) Resting [Ca2+]cyt (A) and [Ca2+]cyt peak (B) upon caffeine stimulation in FDB muscle fibers from WT and PV−/− mice and electroporated with either control shLUC or shMCU.
(C) Representative traces of the experiment as in (A) and (B).
Data are presented as mean ± SEM n ≥ 20. For data analysis, two-way ANOVA with post hoc Bonferroni was performed. ∗∗∗p < 0.001. See also Figure S6.
PV regulates muscle mass by affecting mitochondrial Ca2+ and the expression of PGC-1α4
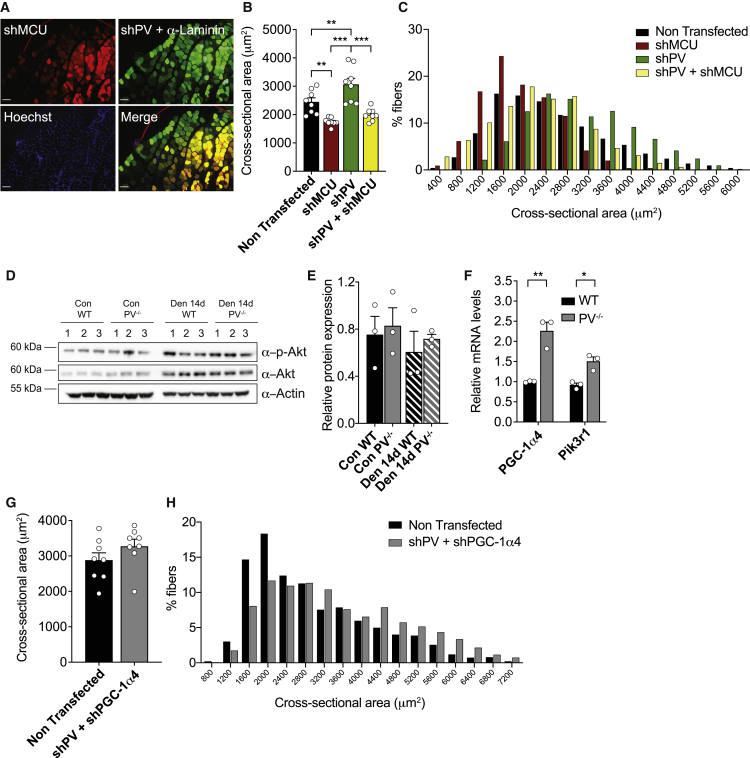
Mitochondrial Ca2+ uptake has been demonstrated to positively regulate skeletal muscle trophism (Mammucari et al., 2015). Moreover, our data suggest that PV ablation leads to an increased mitochondrial Ca2+ uptake and, at same time, to a partial protection against denervation-induced atrophy (Figures 1D and 1E). We thus decided to check whether the changes in mitochondrial Ca2+ handling in PV−/− muscles were causally related to the effect of PV on fiber size. We thus silenced both PV and MCU in adult WT mice and then analyzed fiber CSA variations. For this purpose, we co-transfected adult WT TA muscles with shRNA against MCU fused with mCherry (shMCU-mCherry, hereafter indicated as shMCU) and shRNA against PV fused with zsGreen (shPV-zsGreen, hereafter indicated as shPV) for 7 days (Figure 7A). Consistent with the data reported by Mammucari et al. (2015), shMCU-positive fibers show a 25% decrease in fiber size (Figure 7B; Mammucari et al., 2015). In accordance with data shown in Figures 1F–1H, shPV-positive fibers were characterized by a significant increase in fiber size (Figure 7B). Fibers co-expressing shPV and shMCU were smaller compared to shPV-expressing fibers and showed a shift to the left of the size-frequency distribution (Figure 7C), thus suggesting a causative role of the changes in mitochondrial Ca2+ homeostasis on the effects of PV downregulation on fiber size.
Figure 7.
PV controls muscle fiber size by modulating mitochondrial Ca2+ and PGC-1α4 expression
(A) Representative images of muscle cross-sections of TA muscles electroporated with shMCU and shPV for 7 days. shMCU-positive fibers are in red, shPV-positive fibers in green, and α-laminin in green; Hoechst staining marks the nuclei. Scale bars, 100 μm.
(B) CSA measurements of non-transfected fibers, shMCU-transfected fibers, shPV-transfected fibers, and shMCU- and shPV-co-transfected fibers. More than 100 fibers per muscle; n = 8.
(C) Size-frequency distribution of CSA of the experiment as in (A) and (B).
(D) Representative WB of indicated antibodies on denervated (Den) and contralateral (Con) TA muscle from WT and PV−/− mice. n = 3.
(E) Quantification of the levels of p-Akt protein levels obtained as in (D) and normalized to total Akt.
(F) Expression levels of PGC-1α4 and Pik3r1 in WT and PV−/− TA muscles. Data were normalized to WT. Expression levels were normalized to POL2. n = 3.
(G) CSA measurements of non-transfected and shPV- and shPGC-1α4-double-positive fibers from the same TA muscle transfected for 7 days. More than 100 fibers per muscle; n = 8.
(H) Size-frequency distribution of CSA of the experiment as in (G).
Data are shown as mean ± SEM. For data analysis, one-way ANOVA with Tukey’s post hoc (B and E) or two-tailed, unpaired Student’s t tests (F and G) were used. ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p < 0.001. See also Figure S7.
The Insulin Like Growth Factor 1-AKT Serine/Threonine Kinase/Protein Kinase B (IGF1-AKT/PKB) pathway, a major signaling pathway that positively controls protein synthesis (Bodine et al., 2001; Schiaffino and Mammucari, 2011), is not involved as suggested by the lack of difference in in the phosphorylation levels of AKT in WT and PV−/− control and 14-day-denervated muscles for 14 days (Figures 7D and 7E).
Recently, PGC-1α4, a splice variant of PGC-1α, was discovered as a key player in triggering muscle hypertrophy as an adaptive response to exercise (Ruas et al., 2012). Importantly, its expression is positively regulated by mitochondrial Ca2+ (Mammucari et al., 2015). Interestingly, we observed a significant induction of PGC-1α4 expression and its target gene, Pik3r1 (Martínez-Redondo et al., 2016), in PV−/− muscles compared to WT (Figure 7F), strongly suggesting that PV ablation, via an increase of mitochondrial Ca2+ uptake, activates PGC-1α4-dependent pathways, thus controlling muscle trophism. Coherently, we observed the same induction of PGC-1α4 gene upon acute PV silencing on TA muscles (Figure S7A), while PGC-1α4 gene expression was strongly suppressed upon PV overexpression (Figure S7B). Furthermore, to assess that mitochondrial Ca2+ uptake is required to activate the PGC-1α4-dependent pathways in our PV−/− model, we demonstrated that MCU silencing represses PGC-1α4 expression (Figure S7C). Finally, to confirm the causative role of PGC-1α4 in the phenotype of muscles lacking PV, we silenced PGC-1α4 in vivo (Figure S7D), together with shPV. As shown in Figures 7G and 7H, PGC-1α4 downregulation completely blunts the fiber hypertrophic response induced by the removal of PV.
Discussion
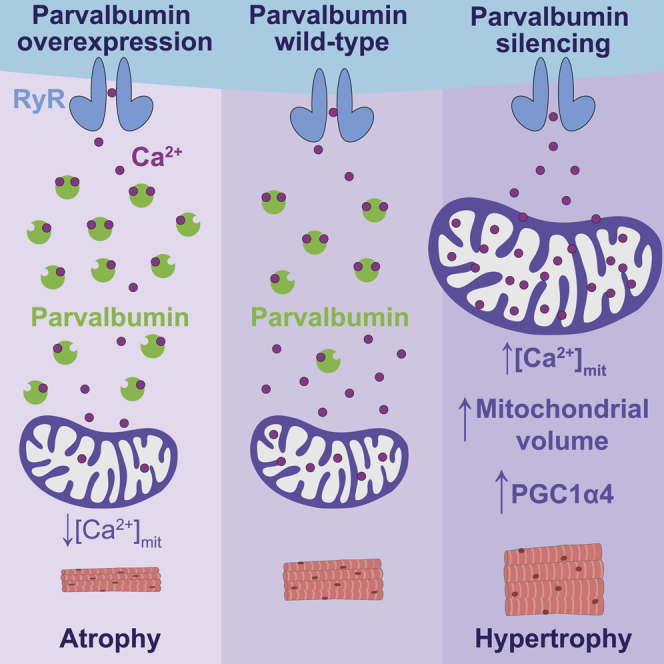
PV is one of the most downregulated “atrogenes,” the family of genes commonly up- and downregulated in both systemic and disuse-induced muscle atrophy (Lecker et al., 2004; Sacheck et al., 2007). Here, we confirmed the downregulation of both PV mRNA and protein in a model of denervation atrophy in mice. However, we found not only that PV ablation has a partial protective anti-atrophy effect in denervated muscles, but also that PV acute downregulation is followed by fiber hypertrophy. Coherently, PV overexpression has the opposite effect, inducing atrophy. The observation of the impact of PV removal on intramitochondrial Ca2+ opened the way to uncover the link between the two functions of PV, cytosolic Ca2+ binding protein and atrogene.
The analysis of Ca2+ dynamics of the three intracellular compartments (Canato et al., 2019; Marcucci et al., 2018) confirmed the prolongation of the cytosolic transients in response to a single stimulus in PV−/− muscle fibers (Schwaller et al., 1999). This is consistent with the view that PV facilitates the removal of Ca2+ from the cytosol playing not only the role of buffer, which requires time for Mg2+ replacement (Hou et al., 1991), but also the role of shuttle between myofibrils and the SR (Raymackers et al., 2000). The cytosolic Ca2+ transients during unfused tetani (60 Hz) in PV−/− fibers showed the disappearance of the prolonged slow decay of [Ca2+]cyt after the end of the stimulation train, indicated as the “tail.” This is consistent with the view of a slow release of Ca2+ from PV after the end of the electrical stimulation (Westerblad and Allen, 1994). Instead of the tail, at the end of the tetanus transient, a dip in [Ca2+]cyt below the pre-stimulation level was detectable. Moreover, the Ca2+ transients induced by 60-Hz stimulation showed a further unexpected feature, i.e., a slow decay of the peak amplitudes during the stimulation train. However, only the kinetics and not the amplitude of the [Ca2+]cyt transient was affected by PV ablation. This was true also when the cytosol buffering power was challenged with a large release of Ca2+ induced by caffeine.
While no significant changes were detected in free [Ca2+]SR at rest and during electrical stimulation and in SR protein expression, mitochondria were responding to PV ablation with profound functional and morphological adaptations. At rest, the levels of free [Ca2+]mt were similar in PV−/− and in WT, but following a Ca2+ release from the SR, induced by either electrical or caffeine stimulation, the increase was much greater in PV−/− than in WT. This suggests that mitochondria might remove from cytosol a substantially greater amount of Ca2+ in the absence of PV. The increased uptake was confirmed also when PV levels were acutely reduced by silencing with shRNA, and the opposite effect was observed upon acute PV overexpression. The overall increased capacity of mitochondria to remove Ca2+ from the cytosol was promoted by the increased number of contacts between mitochondria and CRUs and further magnified by the increased mitochondrial volume density, the latter in accordance with the “mitochondria-parvalbumin antagonism” model (Chen et al., 2001; Ducreux et al., 2012; Racay et al., 2006). The increased mitochondrial number and size was accompanied by increased expression of the master mitochondrial biogenesis regulator PGC-1α and genes involved in mitochondrial fusion OPA1 and MFN2. The similarity of this response to that observed in kidney tubular cells and neurons (Chen et al., 2001, 2006; Ducreux et al., 2012; Henzi and Schwaller, 2015; Racay et al., 2006) suggests the induction of a precise transcriptional program, triggered either by the lack of PV or by the altered Ca2+ dynamics in the absence of PV, that orchestrates both molecular and morphological adaptations of mitochondria. In skeletal muscle fibers, even minor increases in cytosolic Ca2+, such as those caused by RyR leakage in muscle training (Ivarsson et al., 2019) or cold adaptation (Bruton et al., 2010), have been proven sufficient to trigger mitochondrial biogenesis via PGC1-α expression. Published evidence points to a link between cytosolic Ca2+ and PGC1-α expression based on calmodulin and Calcium/Calmodulin Dependent Protein Kinase (CAMK) or calcineurin activation (Maurya et al., 2018; Wright et al., 2007). Moreover, a persistent increase of mitochondrial Ca2+ transient amplitude could lead to an altered NAD(+)/NADH ratio, thus stimulating the expression and activity of SIRT1 (Marcu et al., 2014), which is relevant for PGC-1α transcriptional activity.
The demonstration that the increase of mitochondrial Ca2+ uptake contributes to control cytosolic Ca2+ increases came from experiments of MCU silencing. We observed a higher peak of cytosolic Ca2+ in MCU-silenced PV−/− fibers, but not in MCU-silenced WT fibers challenged with Ca2+ release induced by caffeine or electrical stimulation. Apparently, when PV is removed or even downregulated, mitochondria achieve a more relevant role in controlling [Ca2+]cyt.
The most intriguing effect of the silencing of MCU was the suppression of the hypertrophic response induced by PV acute downregulation. If the enhanced mitochondrial Ca2+ uptake was inhibited by MCU silencing, the signal to increase fiber size disappeared. This finding is reminiscent of the study by Mammucari and coworkers showing that MCU overexpression induces dramatic skeletal muscle hypertrophy and, coherently, that MCU silencing is sufficient to induce atrophy (Mammucari et al., 2015). The modulation of MCU expression influences the expression of PGC-1α4, whose overexpression has been shown to induce muscle hypertrophy (Ruas et al., 2012). Our present results provide a direct demonstration that the control exerted by PV on fiber size depends on intramitochondrial [Ca2+]. Indeed, simultaneously silencing MCU and PV is sufficient to completely switch off the impact of PV downregulation on fiber size, thus preventing hypertrophy. The IGF1-AKT/PKB pathway seems not to be directly involved. In contrast, PV ablation promoted the expression of PGC-1α4 through the modulation of mitochondrial [Ca2+]. Of note, this was confirmed in acutely PV-silenced muscles, and the opposite (i.e., PGC-1α4 downregulation) was observed in PV-overexpressing fibers. Thus, PGC-1α4 emerges as the mechanism by which PV controls fiber size, and this is further confirmed by the finding that PGC-1α4 silencing prevents the hypertrophic effects induced by the lack of PV.
The pro-hypertrophy effect of PV ablation was, to some extent, unexpected considering its downregulation in atrophy (Lecker et al., 2004; Sacheck et al., 2007). Many members of the atrogene family are typically upregulated during the development of atrophy, the ubiquitin ligases being the best-known examples, as they actively contribute to determine loss of muscle mass (Sandri, 2013). Some other genes of the same family are downregulated, a well-known example being JunB, which encodes for a protein inhibiting the transcription of ubiquitin ligases and therefore promoting atrophy when downregulated (Raffaello et al., 2010). Here, our results show that PV is in itself promoting atrophy, and its downregulation, which occurs in several atrophy models, plays in favor of hypertrophy. This apparently paradoxical behavior is reminiscent of that of bone morphogenetic protein 14 (BMP14 or Gdf5), whose expression is induced during denervation atrophy (Sartori et al., 2013). Indeed, activation of BMP signaling promotes hypertrophy and counteracts atrophy, and, conversely, when this pathway is blocked, muscles undergo atrophy (Sartori et al., 2013). Thus, our results give support the view that during muscle atrophy, some pathways are activated or inhibited to counteract muscle wasting, among them PV, in parallel with the other pathways that directly promote muscle wasting.
In the portrait of PV that emerges from this study, another important point needs to be underlined. PV buffers free [Ca2+]cyt and, by doing this, it subtracts Ca2+ to myofibrils and mitochondria. The impact on myofibrils is the accelerated relaxation, making contractile response faster, which is convenient for fast fibers. The impact on mitochondria is to avoid Ca2+ accumulation in the matrix and, thus, keep under control the potential of a mitochondria-dependent stimulation of muscle trophism. Therefore, our results provide the demonstration that PV, acting as a cytosolic Ca2+ buffer, can play two distinct but connected roles. On the one hand, PV regulates contractile parameters, and on the other hand, PV contributes to muscle fiber size regulation. As to the latter role, our results support the view that the fading of the buffering action of PV on cytosolic Ca2+ activates a pathway with two branches. Along the first branch, the transitory increases in cytosolic Ca2+ are sufficient to trigger mitochondrial biogenesis and mitochondrial fusion, thus leading to a greater overall mitochondrial volume. Along the second branch, all mitochondria are left free to take up more Ca2+, helped in this by the closer contacts with CRUs. The enhanced mitochondrial Ca2+ uptake not only partially compensates for the lack of PV but also activates a pathway involving PGC-1α4 overexpression, leading to muscle fiber size increase. We believe that our results further underline that the mitochondrial pathway and its regulation are of utmost importance to uncover the molecular mechanisms of muscle atrophy.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Actin (C-2) | Santa Cruz Biotechnology | Cat#sc-8432; RRID: AB 626630 |
| Akt (pan) (C67E7) | Cell Signaling | Cat#4691S; RRID: AB_915783 |
| Phospho-Akt (Ser473) (D9E) | Cell Signaling | Cat#4060S; RRID: AB_2315049 |
| Calsequestrin | Thermo Fisher Scientific | Cat# PA1-913; RRID: AB_2071461 |
| F(ab’)2-Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | Thermo Fisher Scientific | Cat#A-21430; RRID: AB_2535851 |
| F(ab’)2-Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11070; RRID: AB_142134 |
| Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-21235; RRID: AB_2535804 |
| GAPDH (G-9) | Santa Cruz Biotechnology | sc-365062; RRID: AB_10847862 |
| GRP75 (D-9) | Santa Cruz Biotechnology | sc-133137; RRID: AB_2120468 |
| IP3R-I/II/III (B-2) | Santa Cruz Biotechnology | sc-377518; RRID: AB_2637028 |
| Laminin | Merk | Cat#L9393; RRID: AB_477163 |
| Monoclonal ANTI-FLAG® M2 antibody produced in mouse | Merk | Cat# F3165; RRID: AB_259529 |
| Myosin Heavy Chain Type IIA antibody (α-SC-71) | DSHB | Cat#SC-71; RRID: AB_2147165 |
| Myosin Heavy Chain Type IIB antibody (α-BF-F3) | DSHB | Cat#BF-F3; RRID: AB_2266724 |
| Myosin Heavy Chain Type I antibody (α-BAD5) | DSHB | Cat#BA-D5; RRID: AB_2235587 |
| Parvalbumin | Abcam | Cat#ab11427; RRID: AB_298032 |
| SERCA1 ATPase Monoclonal Antibody VE121G9 | Thermo Fisher Scientific | Cat# MA3-912; RRID: AB_2061281 |
| SERCA2 ATPase Polyclonal Antibody | Thermo Fisher Scientific | Cat#PA5-78837; RRID: AB_2745953 |
| MCU | Merk | Cat#HPA016480; RRID: AB_2071893 |
| MICU1 | Merk | Cat#HPA037479; RRID: AB_2675495 |
| Goat Anti-Mouse IgG (H L)-HRP Conjugate antibody | Bio-Rad | Cat# 170-6516, RRID:AB_11125547 |
| Goat Anti-Rabbit IgG (H L)-HRP Conjugate antibody | Bio-Rad | Cat# 170-6515, RRID:AB_11125142 |
| Bacterial and virus strains | ||
| E.Coli DH10 MultiBac | Geneva Biotech | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Caffeine | Merk | Cat#58-08-2 |
| DMEM, high glucose, HEPES | Thermo Fisher Scientific | Cat#42430-082 |
| DMEM, high glucose, pyruvate | Thermo Fisher Scientific | Cat#41966052 |
| Penicillin/streptomycin | Euroclone | Cat#ECB3001 |
| Fetal Bovine Serum, qualified, heat inactivated, E.U.-approved, South America Origin | Thermo Fisher Scientific | Cat#10500064 |
| PBS, pH 7.2 | Thermo Fisher Scientific | Cat#20012068 |
| Trypsin 1x EDTA | Euroclone | Cat#ECB3053D |
| TMRM | Thermo Fisher Scientific | Cat#T668 |
| Carbonyl cyanide 3-chlorophenylhydrazone (CCCP) | Merk | Cat#C2759 |
| Bisbenzimide H 33258 Fluorochrome, Trihydrochloride | Merk | Cat#23491-45-4 |
| Fura-2, AM, cell permeant | Thermo Fisher Scientific | Cat#F1221 |
| BTS-Calbiochem, N-Benzyl- p-toluenesulphonamide | Merk | Cat#1576-37-0 |
| Hyaluronidase from bovine testes | Merk | Cat#H4272 |
| Laminin | Merk | Cat#11243217001 |
| Collagenase A | Merk | Cat#10103586001 |
| Tyrode’s Solution, Acidic | Merk | Cat#T1788 |
| ProLong Glass Antifade Mountant | Thermo Fisher Scientific | Cat#P36982 |
| TriZOL | Thermo Fisher Scientific | Cat#15596026 |
| SuperScript® II Reverse Transcriptase | Thermo Fisher Scientific | Cat#18064014 |
| Oligo(dT)12-18 Primer | Thermo Fisher Scientific | Cat#18418012 |
| cOmplete Protease Inhibitor Cocktail | Merk | Cat#4693159001 |
| PhosSTOP | Merk | Cat#4906845001 |
| Mowiol® 4-88 | Merk | Cat#9002-89-5 |
| Critical commercial assays | ||
| Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#23225 |
| Applied Biosystems Power SYBR Green PCR Master Mix | Thermo Fisher Scientific | Cat#4368708 |
| NuPAGE 10%, Bis-Tris, 1.0 mm, Mini Protein Gel, 10-well | Thermo Fisher Scientific | Cat#NP0301BOX |
| NuPAGE 3 to 8%, Tris-Acetate, 1.0 mm, Mini Protein Gel, 10-well | Thermo Fisher Scientific | Cat#EA0375BOX |
| Nitrocellulose Membranes, 0.45 μm, 8 × 12 cm | Thermo Fisher Scientific | Cat# 77010 |
| M.O.M.® (Mouse on Mouse) Immunodetection Kit | Vector Laboratories | Cat#BMK-2202 |
| Experimental models: cell lines | ||
| HeLa | ATCC | Cat#CCL-2; RRID: CVCL_0030 |
| Experimental models: organisms/strains | ||
| CD-1 IGS mouse | Charles River Laboratories | Cat#Crl:CD1(ICR) |
| C57BL/6J mouse | Charles River Laboratories | Cat#C57BL/6J (JAX Mice Strain) |
| C57BL/6J PV −/− mouse | N/A | Schwaller et al., 1999) |
| Oligonucleotides | ||
| Primers for RT-PCR, see Table S1 | This paper | N/A |
| Recombinant DNA | ||
| pcDNA3.1-mtGCaMP6f | Mammucari et al., 2015 | N/A |
| pcDNA3.1-D1ER | Palmer et al., 2004 | N/A |
| pmCherry-N1 | Clontech | Cat#632523 |
| PV-P2A-mCherry | This paper | N/A |
| PV-flag-P2A-mCherry | This paper | N/A |
| pZac-U6-shLuc-ZsGreen | University of Pennsylvania Vector Core (Philadelphia, PA, United States) | N/A |
| pZac-U6-shLuc-mCherry | Mammucari et al., 2015 | N/A |
| pZac-U6-shPV-mCherry | This paper | N/A |
| pZac-U6-shPV-ZsGreen | This paper | N/A |
| pZac-U6-shMCU-mCherry | Mammucari et al., 2015 | N/A |
| Software and algorithms | ||
| Fiji | ImageJ | Schindelin et al., 2012 |
| Prism® | GraphPad | RRID:SCR_002798 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by Anna Raffaello (lead contact, anna.raffaello@unipd.it).
Materials availability
The plasmids generated for this manuscript are available upon requests directed to Anna Raffaello (lead contact, anna.raffaello@unipd.it).
Data and code availability
This study did not generate or analyze datasets or code.
Experimental model and subject details
HeLa cells
HeLa cells were cultured in DMEM (Thermo Fisher Scientific) supplemented with 10% FBS (Thermo Fisher Scientific) containing penicillin (100 U/mL) and streptomycin (100 μg/mL).
Mice
Adult male CD1 or C57/BL6J (2-3 months old) were purchased from Charles River. PV−/− mice were generated in the laboratory of Professor Beat Schwaller (University of Friburg, Switzerland). Disruption of the parvalbumin (PV) gene was obtained by homologous recombination (Schwaller et al., 1999).
PV−/− mice were crossed with C57/BL6 WT mice. Crossing of the heterozygous animals yielded homozygous mice and WT littermates. The frequency of homozygous animals was not statistically different from the expected Mendelian transmission of autosomal gene. Therefore, the deletion of the functional PV gene is neither lethal to embryos nor significantly affects embryonic development. The three different genotypes (WT, PV−/− and heterozygous) were characterized by genotyping procedure.
All mice were housed in a temperature-controlled facility under a 12-hour light/dark cycle, with standard environmental enrichment and ad libitum access to standard rodent chow and water. All experiments were conducted with male mice 8-12 weeks old. All the experiments involving animals were approved by the Animal Care Office at the University of Padova and performed in accordance with the Italian law D. L. n.26/2014.
Method details
In vivo DNA transfection of mouse skeletal muscles
FDB muscle electroporation
2-3 months old male CD1, C57/BL6J or PV−/− male mice were used in all experiments. The animals were anesthetized with intraperitoneal injection of Zoletil - Sedaxilan mixture 15 mg/kg. Hyaluronidase solution (2 mg/mL) was injected under the hind limb footpads. After 30 minutes, either 10 or 20 μg of plasmid DNA was injected with the same procedure of the hyaluronidase. After 10 minutes, one gold-plated acupuncture needle was placed under the skin at the level of the heel, and a second one at the base of the toes. The electrodes are oriented parallel to each other and perpendicular to the long axis of the foot. The electrodes were connected to the BTX porator (Harvard apparatus). Muscles were electroporated by applying 20 pulses, 20 ms each, 1 s of interval to yield an electric field of 100 V. Isolation of single fibers was carried out 7 days later.
TA muscle electroporation
2-3 months old male CD1 mice were used in all experiments. First, animals were anesthetized with intraperitoneal injection of Zoletil - Sedaxilan mixture (15 mg/kg). 40 μg of plasmid DNA was injected along the TA muscle length. Gel G008 ECO & IPL was then spread on the hind limb. Electric pulses were then applied by two stainless steel electrodes that were placed on each side of the hind limb (5 pulses, 200 ms each of 110 Volts) using a BTX porator (Harvard apparatus). Animals were allowed to recover for 7 days. Next, TA muscles were harvested and frozen in liquid nitrogen-cooled isopentane.
Denervation
Right hind limbs of 2-3 months old male C57/BL6J or PV−/− mice were denervated by unilaterally cutting the sciatic nerve. Animals were anesthetized by an intraperitoneal injection of Zoletil – Sedaxilan mixture (15 mg/kg intraperitoneal) and a small portion of sciatic nerve was excised. The contralateral non-denervated hind limb served as an internal control in subsequent analyses, which were carried out 3, 7, or 14 days after the cut of the nerve. This procedure does not affect the animal ability to ambulate. At the time point indicated, TA muscles were dissected from the denervated and non-denervated contralateral hind limbs, frozen in liquid nitrogen and stored at –80°C. The muscles were utilized for CSA measurements in cryosections, gene expression studies and for protein analysis.
Electron microscopy (EM)
Preparation and analysis of samples for electron microscopy (EM)
EDL muscles of 2-3 months old male C57/BL6J or PV−/− mice were dissected, fixed at room temperature with 3.5% glutaraldehyde in 0.1 M NaCaco buffer (pH 7.4), and kept at 4°C in fixative until further use. Small bundles of fixed muscles were then post-fixed, embedded, stained en-block, and sectioned as previously described (Pietrangelo et al., 2015, 2019). For EM, ultrathin sections (∼50 nm) were examined with a Morgagni Series 268D electron microscope (FEI Company, Brno, Czech Republic), equipped with Megaview III digital camera (Olympus equipped with Soft Imaging System) at 60 kV.
EM quantitative analyses
-
1)
Total mitochondrial volume was determined using the well-established stereology point-counting technique (Loud, 1968; Mobley and Eisenberg, 1975) in micrographs taken from transversal sections at 8.900 × of magnification. In each sample, 13-25 fibers were analyzed, and in each fiber 2 micrographs were randomly collected from non-overlapping regions.
-
2)
Number of mitochondria, CRUs, mitochondria-CRU pairs / area and mitochondrial positioning were evaluated in micrographs taken from longitudinal sections at 14.000 × of magnification and reported as average number/100 μm2 (see Boncompagni et al., 2009 for additional details). In each sample, 10 fibers were analyzed, and in each fiber 5 micrographs were randomly collected from non-overlapping regions.
-
3)
The average area of apparently normal mitochondria was also measured in the same set of micrographs as in 2) using the Soft Imaging System. Only mitochondria which were entirely visualized in the micrograph were measured. A total of 112 WT and 192 PV−/− mitochondria areas were measured in each specimen and reported as average area in nm2 × 103 ± SEM
-
4)
Surface area, perimeter, major axis and minor axis of mitochondria from 5 micrographs/sample were also measured using the Soft Imaging System. Only mitochondria which were entirely visualized in the micrograph were measured. All these values were used to evaluate mitochondrial shape descriptors and size, e.g., aspect ratio (AR) and form factor (FF) of mitochondria, as described in Picard et al. (2013). Aspect ratio (AR) is computed as the ratio between major axis and minor axis of the ellipse equivalent to the mitochondrion, reflects the “length-to-width ratio” [(major axis)/(minor axis)] and is a measure of mitochondrial shape; form factor (FF) [(perimeter2)/(4π·surface area)] reflects the complexity and branching aspect of mitochondria. For AR and FF evaluation micrographs were taken from transversal sections at 8.900 × of magnification. In each sample, 5 fibers were analyzed, and in each fiber 2 micrographs were randomly collected from non-overlapping regions.
RNA extraction, reverse transcription, and quantitative real-time PCR (RT-PCR)
Total RNA was extracted from TA and FDB muscles using tissue homogenization in TRIZOL reagent, following manufacturer instructions. The RNA was quantified with Nanodrop (Thermo Fisher Scientific). Complementary DNA was generated from 500 nmol of total RNA with a cDNA Synthesis Kit SuperScript II. Oligo(dT)12–18 primers were used as primer for first-strand cDNA synthesis with reverse transcriptase. The cDNA was analyzed by RT-PCR using the IQ5 thermocycler and the SYBR green chemistry (Bio-Rad). The primers were designed and analyzed with Primer3 (Rozen and Skaletsky, 2000) and their efficiency was between 95 and 100%. The housekeeping genes as GAPDH, GRP75 or POL2 were used for cDNA normalization. For quantification, expression levels were calculated by using the 2-ΔΔCT Method (Rao et al., 2013). Primers for RT-PCR were used at the final concentration of 20 μM and their sequences are reported in Table S1.
Western blotting and antibodies
To determine protein levels, frozen muscles were pulverized using the QIAGEN Tissue Lyser and protein extracts were prepared in a lysis buffer. This buffer contains 50 mM Tris pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 10% glycerol, 2% SDS, 1% Triton X-100, Roche Complete Protease Inhibitor Cocktail, 1 mM PMSF, 1 mM NaVO3, 5 mM NaF and 3 mM β-glycerophosphate. 40 μg of total proteins were loaded, according to BCA quantification. Proteins were separated by SDS-PAGE electrophoresis in 4%–12% acrylamide gels and transferred onto nitrocellulose membranes by semi-dry or wet electrophoretic transfer. Blots were blocked for 1 hour at room temperature with 5% non-fat dry milk (Bio-Rad) in TBS-tween buffer (50 mM Tris, 150 mM NaCl and 0.01% Tween, pH 7.4) and incubated at 4°C with the primary antibodies. Secondary antibodies were incubated for 1 hour at room temperature. Please refer to the Key Resource Table section for the antibodies used in the study.
CSA measurements
20 μm-thick muscle cryo-sections of TA muscles from CD1 mice or from denervated and contralateral hind limbs of WT and PV−/− animals were fixed in 4% PFA for 20 minutes. Next, the slides were quenched with 50 mM NH4Cl in PBS and blocked in PBS containing 0.5% BSA for 20 minutes, as previously described (Mammucari et al., 2015). Sections were then incubated with α-laminin primary antibody at the dilution of 1:100 in PBS with 0.5% BSA over night at 4°C to label the sarcolemma of muscle fibers. Muscle cryosections were washed 3 times with PBS. The sections were then incubated for 1 hour at room temperature with an α-rabbit Alexa Fluor 488-conjugated secondary antibody at the dilution of 1:500 in PBS with 0.5% BSA. Nuclei were identified by Hoechst 33342. Tissue sections were then mounted with aqueous mounting media Mowiol. CSA was manually measured by using ImageJ software.
FDB fiber dissociation and culture
FDB fibers were isolated 7 days after in vivo transfection. Muscles were digested in collagenase A (4 mg/mL) (Roche) dissolved in Tyrode’s salt solution (pH 7.4) containing 10% FBS. Single fibers were mechanically dissociated, plated on laminin-coated glass coverslips and cultured in DMEM with HEPES, supplemented with 10% FBS, containing penicillin (100 U/mL), streptomycin (100 μg/mL) at 37°C with 5% CO2.
Mitochondrial [Ca2+] measurement in FDB fibers
FDB muscles were electroporated with a plasmid encoding 4mtGCaMP6f (Chen et al., 2013; Raffaello et al., 2013; De Stefani et al., 2011; Vecellio Reane et al., 2016). 7 days after electroporation, single fibers were isolated as described above. During the experiments, myofibers were maintained in Krebs-Ringer modified buffer (135 mM NaCl, 5 mM KCl, 1 mM MgCl2, 20 mM HEPES, 1 mM MgSO4,0.4 mM KH2PO4, 1 mM CaCl2, 5.5 mM glucose, pH 7.4) at room temperature, in the presence of 75 μM N-benzyl-P-toluenesulfonamide (BTS) to avoid fiber contraction. Experiments were performed on a Zeiss Axiovert 200 microscope equipped with a 40 × /1.3 N.A. PlanFluor objective. Excitation was performed with a DeltaRAM V high-speed monochromator (Photon Technology International) equipped with a xenon arc lamp. Images were captured with a high sensitivity Evolve 512 Delta EMCCD (Photometrics). The system was controlled by MetaMorph 7.5 (Molecular Devices) and was assembled by Crisel Instruments.
Mitochondrial Ca2+ measurements were performed as previously reported (Logan et al., 2014; Mammucari et al., 2015; Vecellio Reane et al., 2016). Briefly, 4mtGCaMP6f was alternatively excited through a 475/20 and a 410/20 nm band-pass excitation filters and images were acquired through an emission filter (535/30 nm) (Chroma). Exposure time was set to 20 ms for fibers stimulated with an electrical pulse and 50 ms for fibers stimulated with 10 mM caffeine. Acquisition was performed at binning 1 with 200 of EM gain. Image analysis was performed with Fiji distribution of the ImageJ software (Schindelin et al., 2012). Images were background-corrected frame by frame by subtracting the mean pixel value of a cell-free region of interest. Changes in Ca2+ levels (475/410 nm fluorescence ratio) were expressed as R/R0, where R is the ratio at time t and R0 is the ratio at the beginning of the experiment. Mitochondrial Ca2+ peak was expressed as R/R0.
To study mitochondrial Ca2+ uptake, Ca2+ release was induced either with caffeine 10 mM administration or with electrical stimulation with trains at high frequency (60 Hz, 5 seconds duration). Master-8 pulsing stimulator (A.M.P.I, Jerusalem, Israel) was used to induce electrical stimulations and the Master-8 software was used to control stimulation parameters (number of pulses per train, pulse duration and interval time) in accordance with the experiments.
Mitochondrial membrane potential determination with TMRM
Isolated FDB muscle fibers were incubated with 20 nM tetramethyl rhodamine methyl ester dye (TMRM) together with 75 μM BTS in Krebs-Ringer buffer for 20 min at 37°C. Data acquisition was done with Leica TCS-SP5-II equipped with a PlanApo 100 × /1.4 N.A. objective. The TMRM fluorescence was excited by the 543 nm HeNe laser and its emission was collected in the 555-700 nm range. 10μM carbonyl cyanide 3-chlorophenylhydrazone (CCCP) was added at the end of the experiment to completely collapse the Δψ. Data are expressed as difference of background corrected TMRM fluorescence before and after CCCP depolarization.
SR [Ca2+ ] measurement in FDB fibers
To measure [Ca2+]SR, we took advantage of the SR-targeted Cameleon D1ER (Palmer et al., 2004). The probe was expressed in FDB muscles by electroporation of plasmid vectors as described above. SR free [Ca2+] levels were determined from the YFP/CFP ratio of the cameleon probe using an inverted fluorescence microscope (Eclipse-Ti; Nikon Instruments, Firenze, Italy), equipped with the perfect focus system (PFS). Fibers were placed in a chamber containing imaging buffer (125 mM NaCl, 5 mM KCl, 1 mM MgSO4, 1 mM KH2PO4, 5.5 mM glucose, 1 mM CaCl2, 20 mM HEPES with 50 μM BTS, mounted on the movable stage of the microscope and temperature was set at 25°C. Excitation of the fluorophore was performed by means of a Hg arc lamp using a 435-nm filter (10 nm bandwidth). YFP and CFP intensities were recorded by means of a cooled CCD camera (C9100-13; Hamamatsu-Photonics Italia, Roma, Italy) equipped with a 515-nm dichroic mirror at 535 (40 nm bandwidth) and 480 nm (30-nm bandwidth), respectively. Two images of 256 × 128 pixels each, corresponding, respectively, to YFP (acceptor) and CFP (donor) light emissions, were collected with a time resolution of 9 ms. Quantification of FRET measurements was performed using ImageJ software (Schindelin et al., 2012). YFP and CFP intensities were corrected for background and the ratio (R) was defined as follows: R = (YFP fiber – YFP background)/(CFP fiber – CFP background). Electrically induced release of Ca2+ from the SR, was achieved by electrical stimulation trains at high frequency (60 Hz, 2 s) via Grass S88 Dual Output Square Pulse Stimulator. The basal ratio level was measured just at the beginning of the recordings and the amplitude of the transients elicited by electrical stimulation was measured at their peaks.
Cytosolic [Ca2+] measurement in FDB fibers
FDB fibers were enzymatically dissociated and plated as described above. For the experiment, fibers were loaded with 2 μM Fura-2/AM diluted in Krebs-Ringer modified buffer (described above) containing 0.02% pluronic acid for 20 min at 37°C and then washed with Krebs-Ringer modified buffer in presence of 75 μM BTS to avoid the fiber contraction. Ca2+ release was induced either with electrical field stimulation or with 10 mM caffeine treatment. Supramaximal electrical pulses (1 ms duration) were delivered via platinum electrodes at 0.5 Hz for 5 minutes, then trains of 2 s duration and 60 Hz frequency were delivered. A Grass S88 Dual Output Square Pulse Stimulator was used to generate electrical pulses. Caffeine was added in a single shot to reach the final concentration (10 mM). The experiments were performed on two different set-ups depending on the sampling rate required for data analysis. Experiments on transients induced by electrical stimulation were performed on the Ionoptix system (sampling rate 1 kHz) to be able to measure kinetics parameters. Ca2+ signals were recorded using a dual-beam excitation fluorescence photometry setup (IonOptix Corp.) at 25°C, mounted on a Nikon Eclipse TS100, equipped with a Xenon Short Arc Lamp. Measurements were expressed as the ratio of the emission at 510 nm with excitation at 360 nm and 380 nm. Recordings were analyzed with the Ionwizard software (IonOptix Corp.).
Experiments of caffeine stimulation were performed on a Zeiss Axiovert 200 microscope equipped with a 40 × /1.3 N.A. PlanFluor objective. Excitation was performed with a DeltaRAM V high-speed monochromator (Photon Technology International) equipped with a 75 W xenon arc lamp. Images were captured with a high-sensitivity Evolve 512 Delta EMCCD (Photometrics). The system was controlled by MetaMorph 7.5 (Molecular Devices) and was assembled by Crisel Instruments. Images were collected by alternatively exciting the fluorophore at 340 and 380 nm and fluorescence emission recorded through a 515/30 nm band-pass filter (Semrock). Exposure time was set to 50 ms (Sampling rate 20 Hz). Acquisition was performed at binning 1 with 200 of EM gain. Image analysis was performed with Fiji distribution of the ImageJ software after background-subtraction.
Determination of fiber type distribution
For fiber type measurements, 20 μm thick cryosections were blocked in M.O.M. working solution, following manufacturer instructions. Sections were then incubated with primary antibody α-SC-71 1:100 in 0.5% BSA in PBS to label the type IIA myosin for 1 hour at 37°C and washed 3 times in PBS. Alexa Fluor 488 conjugated secondary antibody was used. Then, sections were incubated with primary antibody α-BF-F3 1:100 in 0.5% BSA in PBS to label the type IIB myosin for 1 hour at 37°C and washed 3 times in PBS. Alexa Fluor 555 conjugated secondary antibody was used. Finally, sections were then incubated with primary antibody α-BAD5 1:100 in 0.5% BSA in PBS to label the type I myosin for 1 hour at 37°C and washed 3 times in PBS. Alexa Fluor 647 conjugated secondary antibody was used. Sections then were mounted with the glycerol-based (antifade) mounting media ProLongs that prevents photo-bleaching and preserves the signals of fluorescently labeled target molecules for long-term storage and analysis. Fiber type analyses were performed with the Fiji distribution of ImageJ.
Confocal microscopy
Samples were imaged using a confocal microscope (Leica TCS SP5, Leica microsystems, Germany), equipped with 40 × 1.4NA and 63 × , 1.4NA oil immersion objectives. The microscope is equipped with four different laser lines for excitation (405 nm, 488 nm, 561 nm and 633 nm) and three tenable emission filters. Images were digitally acquired and stored in a PC station, and analysis was performed using the image analysis software ImageJ.
Cell culture and transfection
HeLa cells were cultured in DMEM supplemented with 10% FBS containing penicillin (100 U/mL) and streptomycin (100 μg/mL). Cells were transfected with plasmids expressing 1 μg of either pcDNA or pcDNA3.1-PV-Flag together with 7 μg of either shLUC, shPV1, shPV2 or shPV3. Cells were transfected with a standard Ca2+-phosphate procedure as previously described (Raffaello et al., 2013; De Stefani et al., 2011). All experiments were carried out 48 hours after transfection.
shRNA and constructs
Three potential target sites on murine PV cDNA sequence were selected by the absence of BLAST matches, absence of overlap with a region of SNP and absence of multiple binding targets within the gene. We used as control a shRNA targeting the luciferase gene (shLUC). The plasmid containing the shLUC and shPV expresses also mCherry or zsGreen fluorescent protein under the control of CMV promoter. The red or, where indicated, the green fluorescent proteins were used to visualize the shRNA efficacy of transfection in vitro in HeLa cells and to visualize the transfected fibers in vivo.
The shRNA sequences used in the experiments are reported in Table S1.
For PV overexpression in muscle fibers, pmCherry-P2A-N1 was obtained from pmCherry-N1 (Clonotech) by cloning in frame the P2A sequence (GSGATNFSLLKQAGDVEENPGP) to the N-end of mCherry.
Fw: 5′GATCCAGGTTCCGGCGCAACAAACTTCTCTCTGCTGAAACAAGCCGGAGATGTCGAAGAGAATCCTGGACCG 3′
Rv: 5′GGCCCGGTCCAGGATTCTCTTCGACATCTCCGGCTTGTTTCAGCAGAGAGAAGTTTGTTGCGCCGGAACCTG 3′
The aligned oligos were cloned into BamHI and AgeI sites of pmCherry in frame with the mCherry coding sequence.
Mouse PV was amplified from mouse skeletal muscle cDNA.
For cloning PV-Flag in pmCherry-P2A the following primers were used:
Fw: 5′ AATTTCTCGAGGCCACCATGTCGATGACAGACGTGCT 3′
Rev: 5′ AAATTGGATCCCGCTTATCGTCGTCATCCTTGT 3′
The PCR fragment were cloned into XhoI and BamHI sites in pmCherry-P2A-N1.
The expression of multiple polypeptides from a single mRNA is made possible by inclusion of a short viral 2A self-cleaving peptide coding sequence, allowing to co-express the gene of interest along with a reporter, such as a fluorescence gene or a resistance gene (Daniels et al., 2014; Doronina et al., 2008; Kim et al., 2011). In mitochondrial Ca2+ handling experiments, FDB muscle fibers were co-transfected with the plasmid expressing PV and the mitochondrial Ca2+ probe. Only fibers positive for both 4mtGCaMP6f and mCherry were analyzed.
Quantification and statistical analysis
For EM quantitative analyses, 5-25 fibers were analyzed, and, in each fiber, 2-5 different images were used to determine morphometric parameters. For additional details, please refer to the Electron Microscopy (EM) section.
For all the other data analysis, Excel® and GraphPad Prism® were used. Statistical data are presented as mean ± SEM, unless otherwise specified. All Ca2+ measurements were performed at least 3 times and averaged values were shown in the bar diagrams and used for statistical analysis. For the other experiments, the experiments were performed at least 2 times and bar diagrams show the results of one representative experiment. Depending on the experiments, a parametric Student’s t test (two-tailed, unpaired), one-way ANOVA with Dunnett’s or Turkey’s multiple comparison test and two-way ANOVA with Bonferroni post hoc test were applied. p ≤ 0.05 was considered statistically significant.
Acknowledgments
We thank Professor Beat Schwaller for providing the PV−/− animals and Dr. Gaia Gherardi for helping with the silencing experiments in vivo. This research was supported with funding from the Italian Association for Cancer Research (AIRC) (IG18633 to R.R.), the University of Padova (STARS@UNIPD WiC grant 2017 to R.R.), the Italian Telethon Association (GGP16029 to R.R., GGP16026 to A.R., and GGP19231 to F.P.), the Italian Ministry of Health (RF-2016-02363566 to R.R. and GR-2016-02362779 to A.R.), the Cariparo Foundation (R.R.), and the Italian MIUR (PRIN 2015ZZR4W3 to F.P.).
Author contributions
Conceptualization, A.R. and C.R.; methodology, G.B., D.V.R., M.C., and L.P.; investigation, G.B., D.V.R., M.C., and L.P.; writing – original draft, A.R. and C.R.; writing – review & editing, A.R. and C.R.; funding acquisition, A.R., R.R., and C.R.; resources, A.R., C.R., R.R., S.B., and F.P.; supervision, S.B., F.P., and R.R.
Declaration of interests
The authors declare no competing interests.
Published: May 4, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109087.
Contributor Information
Carlo Reggiani, Email: carlo.reggiani@unipd.it.
Anna Raffaello, Email: anna.raffaello@unipd.it.
Supplemental information
References
- Baughman J.M., Perocchi F., Girgis H.S., Plovanich M., Belcher-Timme C.A., Sancak Y., Bao X.R., Strittmatter L., Goldberger O., Bogorad R.L. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476:341–345. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge M.J., Lipp P., Bootman M.D. The Versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- Berridge M.J., Bootman M.D., Roderick H.L. Calcium signalling: dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- Bodine S.C., Stitt T.N., Gonzalez M., Kline W.O., Stover G.L., Bauerlein R., Zlotchenko E., Scrimgeour A., Lawrence J.C., Glass D.J., Yancopoulos G.D. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 2001;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- Boncompagni S., Rossi A.E., Micaroni M., Beznoussenko G.V., Polishchuk R.S., Dirksen R.T., Protasi F. Mitochondria are linked to calcium stores in striated muscle by developmentally regulated tethering structures. Mol. Biol. Cell. 2009;20:1058–1067. doi: 10.1091/mbc.E08-07-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruton J.D., Aydin J., Yamada T., Shabalina I.G., Ivarsson N., Zhang S.-J., Wada M., Tavi P., Nedergaard J., Katz A., Westerblad H. Increased fatigue resistance linked to Ca2+-stimulated mitochondrial biogenesis in muscle fibres of cold-acclimated mice. J. Physiol. 2010;588:4275–4288. doi: 10.1113/jphysiol.2010.198598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canato M., Capitanio P., Cancellara L., Leanza L., Raffaello A., Reane D.V., Marcucci L., Michelucci A., Protasi F., Reggiani C. Excessive Accumulation of Ca2 + in Mitochondria of Y522S-RYR1 Knock-in Mice: A Link Between Leak From the Sarcoplasmic Reticulum and Altered Redox State. Front. Physiol. 2019;10:1142. doi: 10.3389/fphys.2019.01142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Carroll S., Racay P., Dick J., Pette D., Traub I., Vrbova G., Eggli P., Celio M., Schwaller B. Deficiency in parvalbumin increases fatigue resistance in fast-twitch muscle and upregulates mitochondria. Am. J. Physiol. Cell Physiol. 2001;281:C114–C122. doi: 10.1152/ajpcell.2001.281.1.C114. [DOI] [PubMed] [Google Scholar]
- Chen G., Racay P., Bichet S., Celio M.R., Eggli P., Schwaller B. Deficiency in parvalbumin, but not in calbindin D-28k upregulates mitochondrial volume and decreases smooth endoplasmic reticulum surface selectively in a peripheral, subplasmalemmal region in the soma of Purkinje cells. Neuroscience. 2006;142:97–105. doi: 10.1016/j.neuroscience.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Chen T.-W., Wardill T.J., Sun Y., Pulver S.R., Renninger S.L., Baohan A., Schreiter E.R., Kerr R.A., Orger M.B., Jayaraman V. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels R.W., Rossano A.J., Macleod G.T., Ganetzky B. Expression of multiple transgenes from a single construct using viral 2A peptides in Drosophila. PLoS ONE. 2014;9:e100637. doi: 10.1371/journal.pone.0100637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Stefani D., Raffaello A., Teardo E., Szabò I., Rizzuto R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 2011;476:336–340. doi: 10.1038/nature10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doronina V.A., Wu C., de Felipe P., Sachs M.S., Ryan M.D., Brown J.D. Site-specific release of nascent chains from ribosomes at a sense codon. Mol. Cell. Biol. 2008;28:4227–4239. doi: 10.1128/MCB.00421-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducreux S., Gregory P., Schwaller B. Inverse regulation of the cytosolic Ca2+ buffer parvalbumin and mitochondrial volume in muscle cells via SIRT1/PGC-1α axis. PLoS ONE. 2012;7:e44837. doi: 10.1371/journal.pone.0044837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feno S., Rizzuto R., Raffaello A., Vecellio Reane D. The molecular complexity of the Mitochondrial Calcium Uniporter. Cell Calcium. 2021;93:102322. doi: 10.1016/j.ceca.2020.102322. [DOI] [PubMed] [Google Scholar]
- Galla L., Pizzo P., Greotti E. Exploiting Cameleon Probes to Investigate Organelles Ca2+ Handling. Methods Mol. Biol. 2019;1925:15–30. doi: 10.1007/978-1-4939-9018-4_2. [DOI] [PubMed] [Google Scholar]
- Gillis J.M., Thomason D., Lefèvre J., Kretsinger R.H., Schwaller B., Dick J., Dhoot G., Carroll S., Vrbova G., Nicotera P. Parvalbumins and muscle relaxation: a computer simulation study. J. Muscle Res. Cell Motil. 1982;3:377–398. doi: 10.1007/BF00712090. [DOI] [PubMed] [Google Scholar]
- Henrotte J.G. A crystalline constituent from myogen of carp muscles. Nature. 1952;169:968–969. doi: 10.1038/169968b0. [DOI] [PubMed] [Google Scholar]
- Henzi T., Schwaller B. Antagonistic regulation of parvalbumin expression and mitochondrial calcium handling capacity in renal epithelial cells. PLoS ONE. 2015;10:e0142005. doi: 10.1371/journal.pone.0142005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig S., Martinou J.-C. Mitochondrial dynamics: to be in good shape to survive. Curr. Mol. Med. 2008;8:131–137. doi: 10.2174/156652408783769625. [DOI] [PubMed] [Google Scholar]
- Hou T.T., Johnson J.D., Rall J.A. Parvalbumin content and Ca2+ and Mg2+ dissociation rates correlated with changes in relaxation rate of frog muscle fibres. J. Physiol. 1991;441:285–304. doi: 10.1113/jphysiol.1991.sp018752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivarsson N., Mattsson C.M., Cheng A.J., Bruton J.D., Ekblom B., Lanner J.T., Westerblad H. SR Ca2+ leak in skeletal muscle fibers acts as an intracellular signal to increase fatigue resistance. J. Gen. Physiol. 2019;151:567–577. doi: 10.1085/jgp.201812152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jornayvaz F.R., Shulman G.I. Regulation of mitochondrial biogenesis. Essays Biochem. 2010;47:69–84. doi: 10.1042/bse0470069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Lee S.R., Li L.H., Park H.J., Park J.H., Lee K.Y., Kim M.K., Shin B.A., Choi S.Y. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS ONE. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E., Klug G.A., Seedorf U., Pette D. Regulation of parvalbumin concentration in mammalian muscle. Methods Enzymol. 1987;139:763–776. doi: 10.1016/0076-6879(87)39125-6. [DOI] [PubMed] [Google Scholar]
- Lecker S.H., Jagoe R.T., Gilbert A., Gomes M., Baracos V., Bailey J., Price S.R., Mitch W.E., Goldberg A.L. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- Logan C.V., Szabadkai G., Sharpe J.A., Parry D.A., Torelli S., Childs A.-M., Kriek M., Phadke R., Johnson C.A., Roberts N.Y., UK10K Consortium Loss-of-function mutations in MICU1 cause a brain and muscle disorder linked to primary alterations in mitochondrial calcium signaling. Nat. Genet. 2014;46:188–193. doi: 10.1038/ng.2851. [DOI] [PubMed] [Google Scholar]
- Loud A.V. A quantitative stereological description of the ultrastructure of normal rat liver parenchymal cells. J. Cell Biol. 1968;37:27–46. doi: 10.1083/jcb.37.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammucari C., Gherardi G., Zamparo I., Raffaello A., Boncompagni S., Chemello F., Cagnin S., Braga A., Zanin S., Pallafacchina G. The mitochondrial calcium uniporter controls skeletal muscle trophism in vivo. Cell Rep. 2015;10:1269–1279. doi: 10.1016/j.celrep.2015.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcu R., Wiczer B.M., Neeley C.K., Hawkins B.J. Mitochondrial matrix Ca2+ accumulation regulates cytosolic NAD+/NADH metabolism, protein acetylation, and sirtuin expression. Mol. Cell. Biol. 2014;34:2890–2902. doi: 10.1128/MCB.00068-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci L., Canato M., Protasi F., Stienen G.J.M., Reggiani C. A 3D diffusional-compartmental model of the calcium dynamics in cytosol, sarcoplasmic reticulum and mitochondria of murine skeletal muscle fibers. PLoS ONE. 2018;13:e0201050. doi: 10.1371/journal.pone.0201050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Redondo V., Jannig P.R., Correia J.C., Ferreira D.M.S., Cervenka I., Lindvall J.M., Sinha I., Izadi M., Pettersson-Klein A.T., Agudelo L.Z. Peroxisome Proliferator-activated Receptor γ Coactivator-1 α Isoforms Selectively Regulate Multiple Splicing Events on Target Genes. J. Biol. Chem. 2016;291:15169–15184. doi: 10.1074/jbc.M115.705822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurya S.K., Herrera J.L., Sahoo S.K., Reis F.C.G., Vega R.B., Kelly D.P., Periasamy M. Sarcolipin Signaling Promotes Mitochondrial Biogenesis and Oxidative Metabolism in Skeletal Muscle. Cell Rep. 2018;24:2919–2931. doi: 10.1016/j.celrep.2018.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley B.A., Eisenberg B.R. Sizes of components in frog skeletal muscle measured by methods of stereology. J. Gen. Physiol. 1975;66:31–45. doi: 10.1085/jgp.66.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müntener M., Käser L., Weber J., Berchtold M.W. Increase of skeletal muscle relaxation speed by direct injection of parvalbumin cDNA. Proc. Natl. Acad. Sci. USA. 1995;92:6504–6508. doi: 10.1073/pnas.92.14.6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer A.E., Jin C., Reed J.C., Tsien R.Y. Bcl-2-mediated alterations in endoplasmic reticulum Ca2+ analyzed with an improved genetically encoded fluorescent sensor. Proc. Natl. Acad. Sci. USA. 2004;101:17404–17409. doi: 10.1073/pnas.0408030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolini C., Quarta M., D’Onofrio L., Reggiani C., Protasi F. Differential effect of calsequestrin ablation on structure and function of fast and slow skeletal muscle fibers. J. Biomed. Biotechnol. 2011;2011:634075. doi: 10.1155/2011/634075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patron M., Checchetto V., Raffaello A., Teardo E., Vecellio Reane D., Mantoan M., Granatiero V., Szabò I., De Stefani D., Rizzuto R. MICU1 and MICU2 finely tune the mitochondrial Ca2+ uniporter by exerting opposite effects on MCU activity. Mol. Cell. 2014;53:726–737. doi: 10.1016/j.molcel.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard M., White K., Turnbull D.M. Mitochondrial morphology, topology, and membrane interactions in skeletal muscle: a quantitative three-dimensional electron microscopy study. J Appl Physiol (1985) 2013;114:161–171. doi: 10.1152/japplphysiol.01096.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrangelo L., D’Incecco A., Ainbinder A., Michelucci A., Kern H., Dirksen R.T., Boncompagni S., Protasi F. Age-dependent uncoupling of mitochondria from Ca2+ release units in skeletal muscle. Oncotarget. 2015;6:35358–35371. doi: 10.18632/oncotarget.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrangelo L., Michelucci A., Ambrogini P., Sartini S., Guarnier F.A., Fusella A., Zamparo I., Mammucari C., Protasi F., Boncompagni S. Muscle activity prevents the uncoupling of mitochondria from Ca2+ Release Units induced by ageing and disuse. Arch. Biochem. Biophys. 2019;663:22–33. doi: 10.1016/j.abb.2018.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racay P., Gregory P., Schwaller B. Parvalbumin deficiency in fast-twitch muscles leads to increased ‘slow-twitch type’ mitochondria, but does not affect the expression of fiber specific proteins. FEBS J. 2006;273:96–108. doi: 10.1111/j.1742-4658.2005.05046.x. [DOI] [PubMed] [Google Scholar]
- Raffaello A., Milan G., Masiero E., Carnio S., Lee D., Lanfranchi G., Goldberg A.L., Sandri M. JunB transcription factor maintains skeletal muscle mass and promotes hypertrophy. J. Cell Biol. 2010;191:101–113. doi: 10.1083/jcb.201001136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaello A., De Stefani D., Sabbadin D., Teardo E., Merli G., Picard A., Checchetto V., Moro S., Szabò I., Rizzuto R. The mitochondrial calcium uniporter is a multimer that can include a dominant-negative pore-forming subunit. EMBO J. 2013;32:2362–2376. doi: 10.1038/emboj.2013.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao X., Huang X., Zhou Z., Lin X. An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinforma. Biomath. 2013;3:71–85. [PMC free article] [PubMed] [Google Scholar]
- Raymackers J.M., Gailly P., Schoor M.C., Pette D., Schwaller B., Hunziker W., Celio M.R., Gillis J.M. Tetanus relaxation of fast skeletal muscles of the mouse made parvalbumin deficient by gene inactivation. J. Physiol. 2000;527:355–364. doi: 10.1111/j.1469-7793.2000.00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R., De Stefani D., Raffaello A., Mammucari C. Mitochondria as sensors and regulators of calcium signalling. Nat. Rev. Mol. Cell Biol. 2012;13:566–578. doi: 10.1038/nrm3412. [DOI] [PubMed] [Google Scholar]
- Romanello V., Guadagnin E., Gomes L., Roder I., Sandri C., Petersen Y., Milan G., Masiero E., Del Piccolo P., Foretz M. Mitochondrial fission and remodelling contributes to muscle atrophy. EMBO J. 2010;29:1774–1785. doi: 10.1038/emboj.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Ruas J.L., White J.P., Rao R.R., Kleiner S., Brannan K.T., Harrison B.C., Greene N.P., Wu J., Estall J.L., Irving B.A. A PGC-1α isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell. 2012;151:1319–1331. doi: 10.1016/j.cell.2012.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacheck J.M., Hyatt J.-P.K., Raffaello A., Jagoe R.T., Roy R.R., Edgerton V.R., Lecker S.H., Goldberg A.L. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J. 2007;21:140–155. doi: 10.1096/fj.06-6604com. [DOI] [PubMed] [Google Scholar]
- Sandri M. Protein breakdown in muscle wasting: role of autophagy-lysosome and ubiquitin-proteasome. Int. J. Biochem. Cell Biol. 2013;45:2121–2129. doi: 10.1016/j.biocel.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori R., Schirwis E., Blaauw B., Bortolanza S., Zhao J., Enzo E., Stantzou A., Mouisel E., Toniolo L., Ferry A. BMP signaling controls muscle mass. Nat. Genet. 2013;45:1309–1318. doi: 10.1038/ng.2772. [DOI] [PubMed] [Google Scholar]
- Schiaffino S., Mammucari C. Regulation of skeletal muscle growth by the IGF1-Akt/PKB pathway: insights from genetic models. Skelet. Muscle. 2011;1:4. doi: 10.1186/2044-5040-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiaffino S., Reggiani C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 2011;91:1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaller B. Cytosolic Ca2+ buffers. Cold Spring Harb. Perspect. Biol. 2010;2:a004051. doi: 10.1101/cshperspect.a004051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaller B., Dick J., Dhoot G., Carroll S., Vrbova G., Nicotera P., Pette D., Wyss A., Bluethmann H., Hunziker W., Celio M.R. Prolonged contraction-relaxation cycle of fast-twitch muscles in parvalbumin knockout mice. Am. J. Physiol. 1999;276:C395–C403. doi: 10.1152/ajpcell.1999.276.2.C395. [DOI] [PubMed] [Google Scholar]
- Twig G., Shirihai O.S. The interplay between mitochondrial dynamics and mitophagy. Antioxid. Redox Signal. 2011;14:1939–1951. doi: 10.1089/ars.2010.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G., Elorza A., Molina A.J.A., Mohamed H., Wikstrom J.D., Walzer G., Stiles L., Haigh S.E., Katz S., Las G. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecellio Reane D., Vallese F., Checchetto V., Acquasaliente L., Butera G., De Filippis V., Szabò I., Zanotti G., Rizzuto R., Raffaello A. A MICU1 Splice Variant Confers High Sensitivity to the Mitochondrial Ca2+ Uptake Machinery of Skeletal Muscle. Mol. Cell. 2016;64:760–773. doi: 10.1016/j.molcel.2016.10.001. [DOI] [PubMed] [Google Scholar]
- Westerblad H., Allen D.G. Relaxation, [Ca2+]i and [Mg2+]i during prolonged tetanic stimulation of intact, single fibres from mouse skeletal muscle. J. Physiol. 1994;480:31–43. doi: 10.1113/jphysiol.1994.sp020338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B. Bioenergetic role of mitochondrial fusion and fission. Biochim. Biophys. Acta. 2012;1817:1833–1838. doi: 10.1016/j.bbabio.2012.02.033. [DOI] [PubMed] [Google Scholar]
- Wright D.C., Han D.-H., Garcia-Roves P.M., Geiger P.C., Jones T.E., Holloszy J.O. Exercise-induced mitochondrial biogenesis begins before the increase in muscle PGC-1alpha expression. J. Biol. Chem. 2007;282:194–199. doi: 10.1074/jbc.M606116200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate or analyze datasets or code.