Abstract
Background:
Antimicrobial usage and stewardship programmes during COVID-19 have been poorly studied. Prescribing practice varies despite national guidelines, and there is concern that stewardship principles have suffered.
Aim:
To analyse antibiotic prescriptions during the COVID-19 pandemic at a teaching hospital and to propose improved approaches to stewardship.
Methods:
We reviewed COVID-19 admissions to medical wards and intensive care units (ICUs) in a London teaching hospital to assess initial antibiotic usage and evidence of bacterial co-infection, and to determine if our current antibiotic guidelines were adhered to.
Findings:
Data from 130 inpatients (76% medical and 24% ICU) were obtained. On admission, 90% were treated with antibiotics. No microbiological samples taken on admission provided definitive evidence of respiratory co-infection. In 13% of cases, antibiotics were escalated, usually without supporting clinical, radiological or laboratory evidence. In 16% of cases, antibiotics were stopped or de-escalated within 72 h. Blood results and chest radiographs were characteristic of COVID-19 in 20% of ward patients and 42% of ICU patients. Overall mortality was 25% at 14 days – similar to rates described for the UK as a whole.
Conclusion:
The majority of patients received antibiotics despite limited evidence of co-infection. Most patients received narrower spectrum antibiotics than recommended by NICE. As understanding of the natural history of COVID-19 infections progresses, stewardship programmes will need to evolve; however, at this point, we feel that a more restrictive antibiotic prescribing approach is warranted. We propose strategies for effective stewardship and estimate the effect this may have on antibiotic consumption.
Keywords: Antimicrobial stewardship, COVID-19, SARS-CoV-2, coronavirus
Background
SARS-CoV-2, the virus responsible for COVID-19, has spread rapidly from first recognition on 31 December 2019. The first cases were detected in the UK on 31 January 2020, in a student and their relative who had recently visited China. At the time of writing, the subsequent outbreak in the UK had caused 40,465 deaths, which is greater than in all other European countries and most other countries in the world (Department of Health and Social Care and Public Health England, 2020)
The pathogenesis of multisystem disease is thought to be related to direct viral invasion via the ACE2 receptor, which is expressed in cells in the lung, renal tract, myocardium and gut. In some individuals, a hyper-inflammatory cascade occurs around 10 days after the onset of symptoms (Colafrancesco et al., 2020). Current experimental treatments target both virus replication (Beigel et al., 2020; Wang et al., 2020) and the ensuing inflammatory cascade (Dimopoulos et al., 2020; Xu et al., 2020).
The role of bacterial co-infection in COVID-19 is not fully elucidated at present, although two recent systematic reviews suggested that bacterial infection was not commonly found (Lansbury et al., 2020; Rawson et al., 2020). There is ongoing work to determine whether atypical infections such as Mycoplasma pneumoniae are more common in the COVID-19 cohort – but this might be due to the COVID-19 pandemic overlapping with a peak in Mycoplasma infections, false-positive antibody results and similar appearances on chest imaging (Lansbury et al., 2020). These findings are in contrast to influenza, where bacterial co-infection is a large contributor to mortality (Joseph et al., 2013).
A review by Clancy and Nguyen (Clancy and Nguyen, 2020) reports higher rates of bacterial infections in those patients with COVID-19 who had worse clinical outcomes, but their study focuses on nosocomial infections. There is little information relating to bacterial infections present on admission, and therefore there are limited data to guide empiric antibiotic use in these patients. Despite this, the majority of guidelines for treating COVID-19 suggest antibiotics for severe infections.
The global health emergency of increasing antimicrobial resistance will likely outlive COVID-19, and reducing unnecessary use of antibiotics in the treatment of this pandemic virus is important. A growing number of reports have appeared suggesting that antimicrobial stewardship has suffered, and that even fundamental principles have been overlooked during the pandemic (Huttner et al., 2020).
During the outbreak, we maintained the existing antimicrobial stewardship programme in our 1100-bedded London teaching hospital, where intensive care provision rose from 60 to 144 beds. This includes daily microbiology liaison with all intensive care units (ICUs) and twice-weekly stewardship rounds on the medical wards focussed on reviewing prescriptions for restricted antimicrobials (piperacillin-tazobactam, meropenem and quinolones). Antibiotics prescribed in accordance with hospital guidelines are not routinely reviewed. These guidelines are regularly updated and disseminated through a website and smartphone-based app (MicroGuide). All antibiotic prescriptions were reviewed for ICU patients on a daily basis.
The National Institute for Health and Care Excellence (NICE) in the UK published rapid guidelines for antibiotic prescribing for patients managed in the community (National Institute for Health and Care Excellence, 2020a) and subsequently those admitted to hospital on 1 May 2020 (National Institute for Health and Care Excellence, 2020b). The recommendations for hospitalised patients include co-amoxiclav and cephalosporins as empirical agents. Rapid guidelines are produced in response to emergencies and, as such, the processes for developing these guidelines may differ from standard guidelines.
We undertook a retrospective review to look at compliance with our antimicrobial guidelines and impact of our stewardship activities.
Methods
A total of 130 adult cases (aged over 18 years) were selected from those admitted on or after 1 April 2020 (the date when our COVID-19 antibiotic guidelines were published) from a list of positive SARS-CoV-2 swabs taken from inpatients at St George’s Hospital, London. The list was produced on 25 April 2020, and patients were chosen to represent admissions evenly across the month. Only community-onset, symptomatic patients were included.
Computerised hospital records were examined for clinical, laboratory and radiology data, as well as antibiotic prescriptions. Antibiotics prescribed in the Accident and Emergency Department were not included, because these are prescribed separately on paper charts.
All chest radiographs were reported by consultant or specialty registrar radiologists as part of routine patient care. The majority of radiographs were scored as classical, indeterminate, non-COVID-19 or normal, as per the British Society of Thoracic Imaging (https://www.bsti.org.uk/covid-19-resources/covid-19-bsti-reporting-templates/). A minority of reports described abnormal findings, but without classifying them as classic or indeterminate.
Blood results were deemed to be characteristic of COVID-19 if they demonstrated a lymphopenia (lower limit of normal = 1.1 × 109/L) and an absence of neutrophilia (upper limit of normal = 8.0 × 109/L) (Sharma et al., 2020; Tang et al., 2020).
Microbiological specimens were processed in our UKAS-accredited laboratory, South West London Pathology. SARS-CoV-2 testing was performed on combined nose and throat swabs in-house using the Altona RealStar® assay (detecting the E and S genes) or the Roche assay (which detects the E gene and ORF-1).
A new antibiotic guideline was specifically developed for patients with COVID-19 at the start of the pandemic. It was available on the hospital’s intranet and smartphone app. Infectious diseases doctors were embedded within clinical teams, further supporting the use of the guideline. It recommended antibiotic choices according to a clinical severity grading that was aligned with the grading used for the general management of patients with COVID-19, rather than CURB-65 score. According to this guideline, antibiotics were not indicated for patients without an oxygen requirement; doxycycline alone was recommended for those with low oxygen requirements or tachypnoea, with amoxicillin and doxycycline being recommended for patients requiring higher levels of support. The guideline reinforced the importance of stopping antibiotics if a diagnosis of COVID-19 was made, and that antibiotic prescriptions should be limited to a five-day course. This guideline was released on 1 April 2020; the full guideline is available in Supplementary Information.
Legionella antigen testing and mycoplasma serology was recommended for all intensive care cases but not routinely for ward patients. Blood cultures were advised if pyrexial, as well as sputum culture in patients with a productive cough. Respiratory PCR (including Mycoplasma PCR) was not available due to lack of reagents.
In line with the national ‘Start Smart Then Focus’ campaign, antibiotic prescriptions were classified at 72 h as ‘stopped,’ ‘de-escalated’ if they had been reduced from dual therapy to monotherapy or from intravenous to oral therapy, ‘escalated’ if they had been changed to a broader spectrum antibiotic, or ‘changed’ if a switch was needed due to allergy, intolerance or evidence of another infection. If antibiotics were continued for more than 72 h, then the course was deemed completed. Where patients died within 72 h of admission, this is recorded as such, since there was not sufficient time to assess stewardship decision-making.
Proving or excluding bacterial co-infection is difficult. Clinical features such as fever are non-specific and changing oxygen requirements are multifactorial. Chest radiographs are frequently abnormal, particularly for patients in the ICU. We proposed a definition of bacterial co-infection when at least two of the following were present: imaging suggestive of lobar pneumonia; productive cough or purulent respiratory secretions on suction; and raised peripheral blood neutrophil count. This is based on our expert opinion.
Results
Demographic characteristics, diagnostic findings and outcomes of the 130 patients included in this study are shown in Table 1.
Table 1.
Demographic characteristics, diagnostic findings and outcomes for patients.
| Wards (n = 99) | ICU (n = 31) | Total (n = 130) | |
|---|---|---|---|
| Gender | |||
| Male | 60 (61) | 19 (61) | 79 (61) |
| Female | 39 (39) | 12 (39) | 51 (39) |
| Age (years) | 73 (59.5–82) | 58 (45–66.5) | 67 (54.5–77.75) |
| CURB-65 score | |||
| 0 | 17 (17) | 2 (7) | 19 (15) |
| 1 | 21 (21) | 14 (45) | 35 (27) |
| 2 | 28 (28) | 7 (23) | 35 (27) |
| 3 | 21 (21) | 6 (19) | 27 (21) |
| 4 | 9 (9) | 2 (7) | 11 (8) |
| 5 | 3 (3) | 0 (0) | 3 (2) |
| Productive cough | 20 (20) | 6 (20) | 26 (20) |
| CXR findings | |||
| Classic COVID-19 | 41 (41) | 22 (71) | 63 (48) |
| Indeterminate | 23 (23) | 3 (10) | 26 (20) |
| Focal consolidation | 7 (7) | 1 (3) | 8 (6) |
| Clear lung fields | 8 (8) | 2 (6) | 10 (8) |
| Other | 20 (20) | 3 (10) | 23 (18) |
| Biochemical results | |||
| Lymphopenia | 60 (60) | 25 (81) | 85 (65) |
| Neutrophilia | 30 (30) | 11 (35) | 41 (32) |
| Composite results | |||
| Classic CXR, no neutrophilia | 30 (30) | 15 (48) | 45 (35) |
| Classic CXR, lymphopenia, no neutrophilia | 20 (20) | 13 (42) | 33 (25) |
| Outcome at 14 days | |||
| Died | 21 (21) | 12 (39) | 33 (25) |
| Ventilated | 0 (0) | 11 (35) | 11 (8) |
| On oxygen | 6 (6) | 1 (3) | 7 (5) |
| Still admitted, off oxygen | 13 (13) | 2 (6) | 15 (12) |
| Discharged home | 58 (59) | 5 (16) | 63 (48) |
| Discharged to hospice | 1 (1) | 0 (0) | 1 (1) |
Values are given as n (%) or median (IQR).
CXR, chest X-ray; IQR, interquartile range.
Only 20% of patients had coughs described as productive, which was consistent across the medical wards and ICU. It was often unclear if this represented a new and purulent cough – that is, whether this finding would support a diagnosis of bacterial infection – because this was poorly recorded in the patient record.
Legionella and pneumococcal antigen testing was performed in 11% and 12% of ward patients, respectively, and 71% and 45% of ICU patients, respectively. Mycoplasma IgM was measured in 2% of ward patients and 3% of ICU patients. All these pathogen-specific tests were negative, except for one low-positive Mycoplasma IgM – and we interpreted this as a false-positive.
Sputum was sent for culture from only 1 of the 99 (1%) ward patients within 48 h of admission, and this grew normal upper respiratory tract flora only. Out of 31 ICU patients, 3 (10%) had a sputum sample sent within 48 h of admission, with Proteus hauseri (which we interpreted as colonising rather than infecting) and upper respiratory tract flora being isolated.
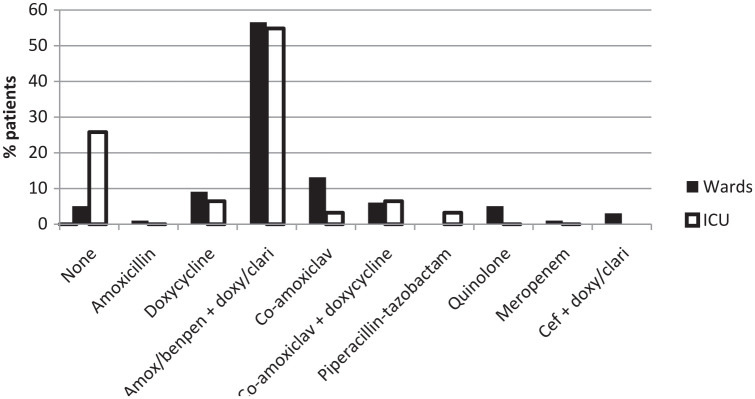
Initial antibiotic regimens used are shown in Figure 1. Of the patients, 56% received dual therapy with either benzylpenicillin or amoxicillin (i.e. a narrow-spectrum beta-lactam antibiotic) together with doxycycline or clarithromycin (to target atypical pathogens). The next most common regimen was doxycycline monotherapy, accounting for 8% of admissions. All quinolone and cefuroxime prescriptions, accounting for 7% of ward patients, were for patients with a documented beta-lactam allergy and were appropriate within our guidelines. More patients admitted to the ICUs received no antibiotic on admission and for the first 48 h (26%) compared to patients admitted to the general medical wards (5%).
Figure 1.
Initial antibiotic regimens. Amox, amoxicillin; Benpen, benzylpenicillin; Cef, cefuroxime or ceftriaxone; Clari, Clarithromycin; Doxy, doxycycline.
The antibiotic outcomes at 72 h are shown in Figure 2. The initial antibiotic was stopped or reduced to monotherapy within 72 h in 16% of cases (26% on ICU and 13% on medical wards). The median duration of the initial antibiotic course was seven days on the general medical wards and three days on the ICUs. Where antibiotics were escalated, this occurred at a median of 5.5 days (interquartile range [IQR] = 2.5–6.0) on the general medical wards and three days (IQR = 2.5–3.5) on the ICUs. Despite careful review of case notes, it generally proved difficult to identify clear evidence of bacterial super-infection as a reason for escalation but was generally due to a worsening of the clinical condition. There were a small number of hospital-acquired bloodstream infections and ventilator-associated pneumonias. The reasons for changes to antibiotic prescriptions were inconsistently documented, but specialist microbiology input was more reliably documented for patients on the ICUs.
Figure 2.
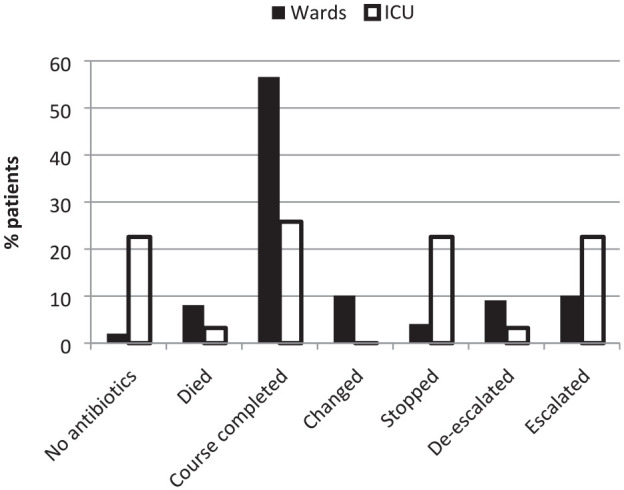
Changes to initial antibiotic prescriptions within 72 h.
Of the patients, 48% had classic COVID-19 chest radiograph changes; 19% had indeterminate imaging findings and 8% had clear lung fields; and 6% had focal changes that could be consistent with bacterial pneumonia. Patients admitted to the ICUs more frequently had classic radiograph appearances and these patients were also more likely to be lymphopenic.
In the patients who had their antibiotic regime de-escalated or stopped, only one patient from the general medical wards and three patients from the ICUs had classic chest radiograph findings and blood results, suggesting that other parameters were useful in making prescribing decisions.
Discussion
Our hospital maintained an antimicrobial stewardship programme even at the peak of the epidemic. We show that nearly all patients admitted with COVID-19 to our hospital had antibiotics prescribed. This is more than we believe is necessary, based on the limited evidence of bacterial co-infection in this viral infection. Antibiotic courses on the general wards were, on average, longer than most guidelines suggest. Increasing evidence suggests that there is little co-infection contributing to the clinical picture (Rawson et al., 2020) and cytokine-driven storm is likely to be important in deteriorating patients. Outcomes for our patients, at least those on the ICUs, are broadly similar to a large, national cohort (Docherty et al., 2020).
More patients in the ICU were prescribed no antibiotics on admission than in the general wards. We believe this is due to a more classic presentation in cases of severe COVID-19, and this is evidenced by a higher proportion of typical chest radiographs and laboratory findings.
Antibiotic prescriptions were more frequently changed in the ICUs than in the general wards, which likely reflects daily microbiology liaison. Additional resources should be focussed on patients who are not in the intensive care settings.
If antibiotics had been stopped in all patients with a classic chest radiograph, lymphopenia and absent neutrophilia, this would have resulted in stopping antibiotics in 25% of patients. If antibiotics had been stopped in patients with a classic chest radiograph and absent neutrophilia (but allowing a normal lymphocyte count), this figure rises to 35%. When we applied our proposed definition for bacterial super-infection (see ‘Methods’), only eight ward patients and two ICU patients met these criteria. Further, it is notable that on admission radiographs, only 6% were reported as having a focal or lobar consolidation.
Currently, international antibiotic guidelines are in conflict with our guidelines in the UK. Specialty guidance from NHS England states that antibiotics should not be used for patients with uncomplicated COVID-19 in intensive care, although it is unclear what ‘uncomplicated’ means for such a patient. We found no microbiological evidence to support the routine use of co-amoxiclav or antibiotics with broader spectra on admission to hospital. On the contrary, we believe that a significant proportion of patients who were prescribed antibiotics did not have a bacterial infection.
Although UK national guidelines recommend considering Legionella testing in all cases, none of our patients tested positive, and antibiotic cover for atypical organisms may not be necessary. We suggest that routine Legionella testing should be reserved for ICU patients.
A major challenge in developing antimicrobial stewardship programmes is identifying which patients do not require antibiotics. In the first instance, a key factor in guiding the need for an antibiotic is the history, and particularly the nature of any cough including the production of purulent sputum. Rapid virological confirmation of the COVID-19 diagnosis is also key in confirming the primary pathology and guiding the assessment on the value of ongoing empirical antibiotic therapy. The turnaround time for SARS-CoV-2 testing for our hospital reduced from up to seven days to under 24 h during the course of the outbreak, when in-house testing was commenced before 1 April, which we found helpful in stewarding antibiotics. Routine blood test results may help exclude bacterial infection. For example, a normal neutrophil count associated with lymphopenia is seen as being classic of COVID-19 that is not complicated by bacterial infection. Lymphopenia is more common in severe disease, and one-third of our patients were not lymphopenic. Therefore, while it may be useful in diagnosing and prognosticating in COVID-19, lymphocyte count may be less useful as a tool for antimicrobial stewardship.
Biomarkers have been used to distinguish bacterial from viral infection. Procalcitonin (PCT) is particularly topical. However, PCT testing is not universally available, and its role in COVID-19 is still to be fully elucidated. Several studies have correlated COVID-19 severity with PCT levels (Lippi and Plebani, 2020; Liu et al., 2020), with high PCT levels even in the absence of bacterial co-infection. NICE does not endorse the use of PCT but encourages further research due a lack of evidence. Indeed, before the global COVID-19 pandemic, the World Health Organization EML Antibiotic Working Group had rejected PCT as an essential in vitro diagnostic test (Procalcitonin (PCT) test (immunoassay) Submission to the WHO EDL 2019) for similar reasons. A Cochrane systematic review showed no difference in mortality, reinfection or duration of antibiotic therapy in the PCT versus non-PCT groups (Andriolo et al., 2017). The Infectious Diseases Society of America guidelines do, however, advocate the use of PCT in antimicrobial stewardship programs, albeit that this is described as a weak recommendation (Barlam et al., 2016).
The role of imaging in confidently identifying classic COVID-19-related changes is being increasingly understood, and the role for artificial intelligence in diagnosing COVID-19 from plain chest radiographs is also being investigated (Murphy et al., 2020; Wong et al., 2019). Our revised antibiotic guidelines will take into account classic findings of COVID-19 on the chest radiograph.
We believe that a simple risk stratification for identifying possible cases of bacterial co-infection could be developed. This could be based on our proposed criteria that is used on tests that are already routinely performed.
CRB-65 has been recommended by NICE to assess the severity of COVID-19 in the community (National Institute for Health and Care Excellence, 2020a), although it has not been validated in this condition. The CURB-65 score, for use in hospitals, has not been recommended by NICE; indeed, strong evidence has emerged that link other risk factors such as male gender, age, ethnicity and defined co-morbid conditions, such as diabetes and hypertension, to worse outcomes. However, severity of infection does not equate to risk of bacterial infection, and we would advise caution in using it as a basis on which to guide antimicrobial therapy in a primary viral infection (Shi et al., 2020).
Our COVID-19 antibiotic guideline recommends amoxicillin in place of benzylpenicillin, which we had previously used for community-acquired pneumonia. This has several advantages. It requires fewer administrations of an intravenous antibiotic and reduces nursing time spent preparing and delivering drugs. It also reduces the number of infusion pumps required to deliver antibiotics, which had been in short supply. We have also witnessed an increased number of bloodstream infections in our cohort (Sturdy et al., 2020), which we postulate is due to difficulties in maintaining good infection control and line care practices when enhanced personal protective equipment is worn. Reducing the need for intravenous access will reduce nosocomial infection – principles that are well established and have been proven to reduce bloodstream infections (Bion et al., 2013; Pronovost et al., 2006).
An additional, well-recognised challenge of antimicrobial stewardship programmes is adherence to guidelines. We found a significant number of prescriptions that did not follow our guidelines, and this could be improved by better dissemination and education which, after the peak of infection, we are now undertaking.
One limitation of our study is the difficulty in definitively excluding bacterial co-infection in the deteriorating patient. The vast majority of patients with COVID-19 do not have bronchoalveolar lavages undertaken, even in the ICU, though these may provide better microbiological samples. However, several studies have reached the same conclusion as us that bacterial co-infection is not commonly seen in COVID-19. Furthermore, assessing antibiotic use in COVID-19 is complicated because patients commonly acquire unrelated nosocomial infections.
In conclusion, despite high levels of antibiotic prescribing, this retrospective study is useful in supporting the use of narrow spectrum antibiotics, which are rapidly discontinued when the clinical picture and diagnostic work-up makes bacterial involvement unlikely. A proactive review of prescriptions is important to ensure courses are restricted to the minimum required.
Supplemental Material
Supplemental material, sj-pptx-1-bji-10.1177_1757177420976813 for Antibiotic usage and stewardship in patients with COVID-19: too much antibiotic in uncharted waters? by Terry John Evans, Harriet Claire Davidson, Jen Mae Low, Marina Basarab and Amber Arnold in Journal of Infection Prevention
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Peer review statement: Not commissioned; blind peer-reviewed.
ORCID iDs: Terry John Evans  https://orcid.org/0000-0001-8976-5197
https://orcid.org/0000-0001-8976-5197
Harriet Claire Davidson  https://orcid.org/0000-0002-4222-4358
https://orcid.org/0000-0002-4222-4358
Supplemental material: Supplemental material for this article is available online.
References
- Andriolo BNG, Andriolo RB, Salomao R, Atallah AN. (2017) Effectiveness and safety of procalcitonin evaluation for reducing mortality in adults with sepsis, severe sepsis or septic shock. Cochrane Database of Systematic Reviews 1: CD010959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlam TF, Cosgrove SE, Abbo LM, MacDougall C, Schuetz AN, Septimus EJ, Srinivasan A, Dellit TH, Falck-Ytter YT, Fishman NO, Hamilton CW, Jenkins TC, Lipsett PA, Malani PN, May LS, Moran GJ, Neuhauser MM, Newland JG, Ohl CA, Samore MH, Seo SK, Trivedi KK. (2016) Implementing an antibiotic stewardship program: Guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clinical Infectious Diseases 62: e51–e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AC, Hohmann E, Chu HY, Luetkemeyer A, Kline S, Lopez de Castilla D, Finberg RW. (2020) Remdesivir for the Treatment of Covid-19 - Preliminary Report. New England Journal of Medicine 383: 1813–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bion J, Richardson A, Hibbert P, Beer J, Abrusci T, McCutcheon M, Cassidy J, Eddleston J, Gunning K, Bellingan G, Patten M, Harrison D. and Matching Michigan Collaboration & Writing Committee. (2013) Matching Michigan: A 2-year stepped interventional programme to minimise central venous catheterblood stream infections in intensive care units in England. BMJ Quality and Safety 22(2): 110–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy CJ, Nguyen MH. (2020) COVID-19, superinfections and antimicrobial development: What can we expect? Clinical Infectious Diseases. DOI: 10.1093/cid/ciaa524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colafrancesco S, Alessandri C, Conti F, Priori R. (2020) COVID-19 gone bad: A new character in the spectrum of the hyperferritinemic syndrome? Autoimmunity Reviews 19: 102573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Department of Health and Social Care and Public Health England. (2020) Number of coronavirus (COVID-19) cases and risk in the UK - GOV.UK. Available at: https://www.gov.uk/guidance/coronavirus-covid-19-information-for-the-public (accessed 4 June 2020).
- Dimopoulos G, de Mast Q, Markou N, Theodorakopoulou M, Komnos A, Mouktaroudi M, Netea MG, Spyridopoulos T, Verheggen RJ, Hoogerwerf J, Lachana A, van de, Veerdonk FL, Giamarellos-Bourboulis EJ. (2020) Favorable Anakinra Responses In Severe Covid-19 Patients With Secondary Hemophagocytic Lymphohistiocytosis. Cell Host & Microbe 28: 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docherty AB, Harrison EM, Green CA, Hardwick HE, Pius R, Holden KA, Read JM, Dondelinger F, Carson G, Merson L, Lee J, Plotkin D, Sigfrid L, Halpin S, Jackson C, Gamble C, Horby PW, Nguyen-Van-Tam JS, Ho A, Russell CD, Dunning J, Openshaw PJ, Baillie JK, Semple MG. and ISARIC4C Investigators. (2020) Features of 20 133 UK patients in hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: prospective observational cohort study. BMJ 369: m1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttner BD, Catho G, Rano-Pardo JR, Pulcini C, Schouten J. (2020) COVID-19: don’t neglect antimicrobial stewardship principles! Clinical Microbiology and Infection 26: 808–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph C, Togawa Y, Shindo N. (2013) Bacterial and viral infections associated with influenza. Influenza and other Respiratory Viruses 7 (Suppl. 2): 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbury L, Lim B, Baskaran V, Lim WS. (2020) Co-infections in People With COVID-19: A Systematic Review and Meta-Analysis. Journal of Infection 81: 266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippi G, Plebani M. (2020) Procalcitonin in patients with severe coronavirus disease 2019 (COVID-19): A meta-analysis. Clinica Chimica Acta 505: 190–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Li L, Xu M, Wu J, Luo D, Zhu Y, Li B, Song B, Zhou X. (2020) Prognostic value of interleukin-6, C-reactive protein, and procalcitonin in patients with COVID-19. Journal of Clinical Virology 127: 104370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Smits H, Knoops AJG, Korst MBJM, Samson T, Scholten ET, Schalekamp S, Schaefer-Prokop CM, Philipsen RHHM, Meijers A, Melendez J, van Ginniken B, Rutten M. (2020) COVID-19 on the Chest Radiograph: A Multi-Reader Evaluation of an AI System. Radiology 296: E166–E171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institute for Health and Care Excellence. (2020. a) COVID-19 rapid guideline : managing suspected or confirmed pneumonia in adults in the community. Available at: https://www.nice.org.uk/guidance/ng165. [PubMed]
- National Institute for Health and Care Excellence. (2020. b) COVID-19 rapid guideline: antibiotics for pneumonia in adults in hospital. Available at: https://www.nice.org.uk/guidance/NG173. [PubMed]
- Procalcitonin (PCT) test (immunoassay) Submission to the WHO EDL 2019 (no date). [Google Scholar]
- Pronovost P, Needham D, Berenholtz S, Sinopoli D, Chu H, Cosgrove S, Sexton B, Hyzy R, Welsh R, Roth G, Bander J, Kepros J, Goeschel C. (2006) An intervention to decrease catheter-related bloodstream infections in the ICU. New England Journal of Medicine 355(26): 2725–2732. [DOI] [PubMed] [Google Scholar]
- Rawson TM, Moore LSP, Zhu N, Ranganathan N, Skolimowska K, Gilchrist M, Satta G, Cooke G, Holmes A. (2020) Bacterial and fungal co-infection in individuals with coronavirus: A rapid review to support COVID-19 antimicrobial prescribing. Clinical Infectious Diseases. DOI: 10.1093/cid/ciaa530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, Agarwal M, Gupta M, Somendra S, Saxena SK. (2020) Clinical Characteristics and Differential Clinical Diagnosis of Novel Coronavirus Disease 2019. (COVID-19). In: Saxena SK. (ed.) Coronavirus Disease 2019 (COVID-19). Singapore: Springer Nature, pp. 55–70. [Google Scholar]
- Shi Y, Yu X, Zhao H, Wang H, Zhao R, Sheng J. (2020) Host susceptibility to severe COVID-19 and establishment of a host risk score: Findings of 487 cases outside Wuhan. Critical Care 24(1): 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturdy A, Basarab M, Cotter M, Hager K, Shakespeare D, Shah N, Randall P, Spray D, Arnold A. (2020) Severe COVID-19 and healthcare-associated infections on the ICU: time to remember the basics? Journal of Hospital Infection 105(4): 593–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Comish P, Kang R. (2020) The hallmarks of COVID-19 disease. PLoS Pathogens 16(5): e1008536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang D, Du G, Du R, Zhao J, Jin Y, Fu S, Gao L, Cheng Z, Lu Q, Hu Y, Luo G, Wang K, Lu Y, Li H, Wang S, Ruan S, Yang C, Mei C, Wang Y, Ding D, Wu F, Tang X, Ye X, Ye Y, Liu B, Yang J, Yin W, Wang A, Fan G, Zhou F, Liu Z, Gu X, Xu J, Shang L, Zhang Y, Cao L, Guo T, Wan Y, Qin H, Jiang Y, Jaki T, Hayden FG, Horby PW, Cao B, Wang C. (2020) Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet 395(10236): 1569–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HYF, Lam HYS, Fong AH, Leung ST, Chin TW, Lo CSY, Lui MM, Lee JCY, Chiu KW, Chung TW, Lee EYP, Wan EYF, Hung IFN, Lam TPW, Kuo MD, Ng MY. (2019) Frequency and Distribution of Chest Radiographic Findings in COVID-19 Positive Patients. Radiology 296: E72–E78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Han M, Li T, Sun W, Wang D, Fu B, Zhou Y, Zheng X, Yang Y, Li X, Zhang X, Pan A, Wei H. (2020) Effective treatment of severe COVID-19 patients with tocilizumab. Proceedings of the National Academy of Sciences 117(20): 202005615. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pptx-1-bji-10.1177_1757177420976813 for Antibiotic usage and stewardship in patients with COVID-19: too much antibiotic in uncharted waters? by Terry John Evans, Harriet Claire Davidson, Jen Mae Low, Marina Basarab and Amber Arnold in Journal of Infection Prevention