Abstract
In the present minireview, we intend to provide a brief history of the field of CD9 involvement in oncogenesis and in the metastatic process of cancer, considering its potential value as a tumor-associated antigenic target. Over the years, CD9 has been identified as a favorable prognostic marker or predictor of metastatic potential depending on the cancer type. To understand its implications in cancer beside its use as an antigenic biomarker, it is essential to know its physiological functions, including its molecular partners in a given cell system. Moreover, the discovery that CD9 is one of the most specific and broadly expressed markers of extracellular membrane vesicles, nanometer-sized entities that are released into extracellular space and various physiological body fluids and play a role in intercellular communication under physiological and pathological conditions, notably the establishment of cancer metastases, has added a new dimension to our knowledge of CD9 function in cancer. Here, we will discuss these issues as well as the possible cancer therapeutic implications of CD9, their limitations, and pitfalls.
Keywords: Antibody, CD9, tetraspanin, cancer, immunotherapy, exosome, extracellular vesicle
Impact statement
Over the last three decades, the relevance of CD9 as a tumor-associated antigen has been evidenced by numerous studies describing its potential role as a prognostic marker and its involvement in cancer progression. However, CD9 has not been targeted in cancer due to its major side effects, particularly platelet aggregation and thrombosis. The discovery of additional roles of CD9 as part of plasma membrane protrusions and extracellular vesicles and the development of new antibodies, including monovalent Fabs and possibly nanobodies, devoid of platelet-related adverse effects, open a new opportunity to reconsider CD9 as a potential therapeutic target.
Introduction
Originally discovered in lymphohematopoietic progenitor and acute lymphoblastic leukemia cells,1,2 the cluster of differentiation 9 (CD9, tetraspanin-29 (TSPAN29), Motility-Related Protein-1, Leukemia-Associated Cell Surface Antigen p24) is expressed in multiple hematopoietic and non-hematopoietic tissues and cell types, including epithelial, endothelial, and stromal cells as well as most types of malignant cells.3–5 Depending on the cell type and its interacting partners, CD9 is involved in numerous cellular processes such as cell–cell contact, cell-extracellular matrix interaction, integrin-dependent cell migration, signaling, membrane fusion, apoptosis, inflammation, proliferation, and differentiation.6–8 In cancer, CD9 has various impacts, either as a tumor suppressor or a promoter of tumorigenic/metastatic activities.9 It has been shown that many steps of the metastatic cascade, such as primary tumor evasion, intravasation, extravasation, colonization and growth of secondary tumors, are influenced by CD9 expression.10 CD9 may also facilitate tumor angiogenesis and lymphangiogenesis.
CD9
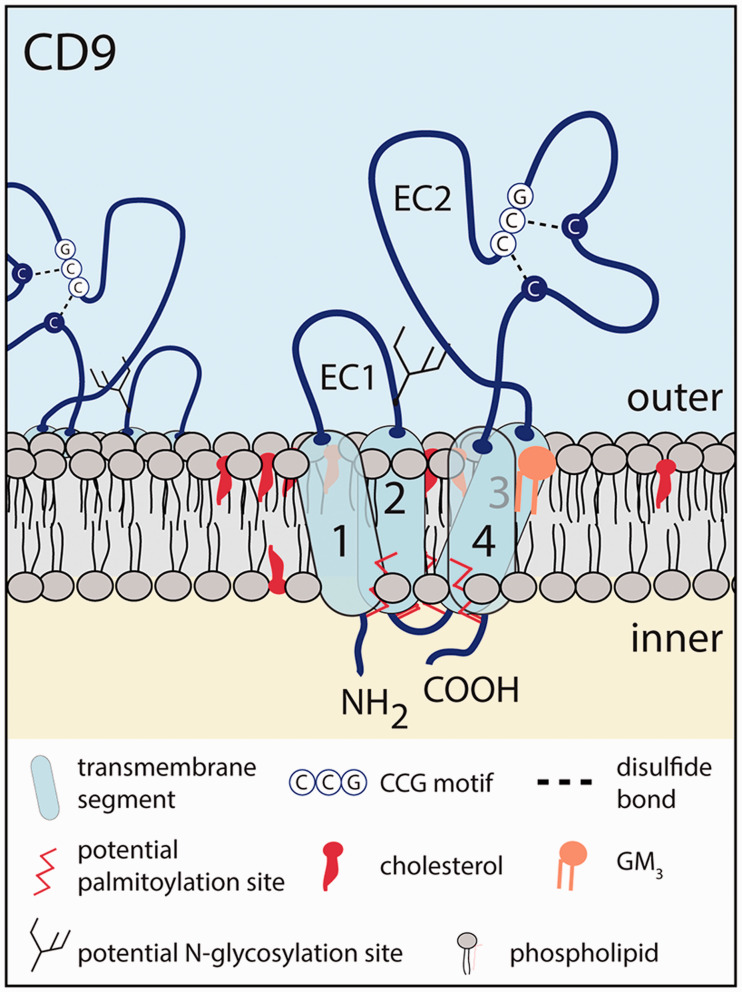
CD9 is an integral 24–27 kDa membrane protein that belongs to the tetraspanin family, which contains 33 distinct proteins in humans. Structurally, tetraspanins have four transmembrane segments, one small and one large extracellular loop (domain EC1 and EC2, respectively) and short cytoplasmic N- and C-terminal domains.11,12 The human 228-amino acid protein CD9 holds a potential N-glycosylation site located in the EC1 domain.13 The EC2 domain (also known as the large extracellular loop (LEL)) contains four cysteine residues with two in a highly conserved Cys-Cys-Gly motif, resulting in the formation of disulfide bridges13,14 (Figure 1). The latter are crucial for the proper folding of the EC2 domain. As for the other tetraspanins, three α-helices within the CD9 LEL define a constant, well-conserved region involved in tetraspanin dimerization and oligomerization, whereas two other α-helices define a variable region, involved in most lateral interactions with other membrane proteins.3 Additional CD9 intra- and inter-molecular interactions are mediated by its conserved residues in transmembrane domains.15,16 The proper interactions and packing of transmembrane domains are essential for the folding and transport of tetraspanin proteins.17 Of note, the amino acid sequence of CD9 is well conserved between species.18–23 CD9 is found at the plasma membrane, endocytic compartment, nucleus, and small extracellular vesicles (EVs), which are released in various body fluids.
Figure 1.
Membrane topology of CD9. The human CD9 protein contains four transmembrane segments (1–4), short cytoplasmic N- and C-protein termini, and two extracellular (EC) domains forming a short and a larger loop. The CD9 EC2 domain is properly folded with two disulfide bridges in which two cysteine residues are found in a conserved CCG motif among all tetraspanins. A potential N-glycosylation site is located in the EC1 domain, close to the second transmembrane domain. Several cysteine residues at the transition of the cytoplasmic domains and transmembrane segments are subject to palmitoylation. The outer and inner leaflets of the plasma membrane are shown. Therein, membrane cholesterol and GM3 ganglioside could alter CD9 function. Note that the structure of CD9 is not represented with the appropriate scale.
CD9 in tetraspanin web and plasma membrane
As an organizer of biological membranes, the multiple interactions of CD9 with other proteins such as tetraspanins (e.g. CD81/Tspan28 and CD151/Tspan24), various members of the immunoglobulin superfamily, in particular EWI-2 (also named IgSF8, CD316) and EWI-F (FPRP, CD9P-1, CD315), a subset of integrins (e.g. α1β1, α2β1, α3β1 (VLA-3), α4β1 (VLA-4), α5β1, α6β1 (VLA-6), α7β1, αIIbβ3) result in the formation of structural and functional units called tetraspanin-enriched microdomains (TEMs) or, more commonly, tetraspanin web.3,24–32 In contrast to lipid rafts,33 TEMs are mainly generated by protein–protein interactions,11,12 which are modulated by differential protein expressions as well as post-translational modifications. Membrane cholesterol could nevertheless play a role in the formation and stabilization of these membrane platforms.34 Cross-talks between the TEMs and lipid rafts are not excluded.35,36 CD9 partners EWI-2 and EWI-F may link TEMs to the underlying actin cytoskeleton through their interactions with proteins of the ezrin-radixin-moesin (ERM) family.37 Interaction, or a close contact, of CD9 with membrane lipids (e.g. cholesterol and gangliosides) could also occur,34,38,39 modulating its binding to protein interactors. It has been shown that GM3 ganglioside promotes the interaction between CD9 and α3 integrins, which has an impact on laminin-5-dependent cell motility.40 The high motility of cancer cells can be controlled by CD9-ganglioside-epidermal growth factor (EGF) receptor (EGFR) complex.41 The palmitoylation of CD9 could also influence its homo- or hetero-clustering with other partners.42,43 For example, non-palmitoylated forms of CD9, EWI-2 and EWI-F are frequently observed in cancer.44 Due to their protein composition, TEMs regulate cell membrane plasticity and influence intercellular and extracellular matrix interactions. Therefore, a downregulation or an upregulation of CD9 can modify the TEM organization, thereby altering cellular properties, including those involved in cancer progression and metastasis such as adhesion and motility as well as fusogenicity, among others.
Cell surface CD9 plays a significant role in various processes regulating cellular trafficking such as the immune response. CD9 is expressed in all major types of leukocytes including B and T cells, natural killer (NK) cells, macrophages, and dendritic cells (reviewed in Reyes et al.3). CD9 supports integrin-mediated signaling at the T cell immunological synapses, as demonstrated by its silencing, which affects the subcellular localization of integrins and alters their downstream targets; the focal adhesion kinase and extracellular signal-related kinase 1/2.45 Furthermore, its association with receptors for the Fc-gamma region of IgG (Fcγ) modifies the signals for phagocytosis and inflammatory responses on macrophages.46 It is also highly expressed by endothelial cells in line with its crucial role in trans-endothelial migration. The inclusion of CD9 and its partners such as the vascular cell adhesion molecule-1 (VCAM-1, CD106) and intercellular adhesion molecule-1 (ICAM-1, CD54) or other immunoglobulin-superfamily members in TEMs contribute to these roles.47 The adhesive function of transmembrane glycoprotein activated leukocyte cell adhesion molecule (ALCAM, CD166) is mediated by CD9 through its cis interaction in a protein complex that includes the metalloproteinase ADAM17 (also named tumor necrosis factor-α converting enzyme).48 CD9 favored the homotypic ALCAM interactions as well as the upregulation of ALCAM at the cell surface by the inhibition of the sheddase activity of ADAM17.49 Thus, CD9-mediated ALCAM-ALCAM interactions modulate interactions between leukocytes (or cancer cells) with endothelial cells, and hence participate in physiological and pathological cell migration, the latter being an important step in cancer and in the development of metastases.50,51 CD9 associates also with transmembrane transforming growth factor (TGF)-α and regulates TGF-α-induced EGFR activation and cell proliferation.52 Notably, the association of CD9 with transmembrane protein TGF-α decreases the ectodomain shedding and the release of soluble TGF-α, and their co-expression confers changes (i) in cytoskeletal organization such as a decrease in actin stress fibers and focal adhesions and (ii) in RhoA and Rac1 GTPase activities. These alterations are reversed by inhibiting the EGFR signaling.53 Moreover, CD9 forms complexes with EGFR and β1 integrin that lead to their colocalization on the cell surface, especially at cell–cell contact sites (see below).54
Within the plasma membrane, CD9 as other tetraspanins (e.g. CD81, CD82) is often concentrated in particular subdomains that have in common to protrude, such as microvillar-like structures and other types of highly curved plasma membrane protrusions (PMPs), including filopodia46,55–58 (Figure 2). The crystal structure of CD9 and the cryo-electron microscopic structure of CD9 in complex with EWI-2 have revealed that the reversed cone-like molecular shape of CD9 could generate membrane curvature, which explains its specific subcellular localization in tubular structures such as PMPs.59,60 Interactions of CD9 with EWI-2/EWI-F-ERM proteins can regulate the formation of microvilli, among others. Since some PMPs are involved in cellular functions such as adhesion, migration, fusion and intercellular communication, the association of CD9 (and other tetraspanins) with these protruding structures somehow explains their influence on various biological processes. For example, we and others have shown that CD9 silencing modifies the microvillus architecture and the leading edge of lamellipodium (see below), which in turn can influence cellular interactions and migration.61,62 The absence of CD9 in leukocytes resulted in the inhibition of microvillus formation, which reduced adhesion and trans-endothelial migration.61 In such cellular system, microvilli host integrins and integrin-associated proteins that play a role in mediating leukocyte adhesion under flow.63
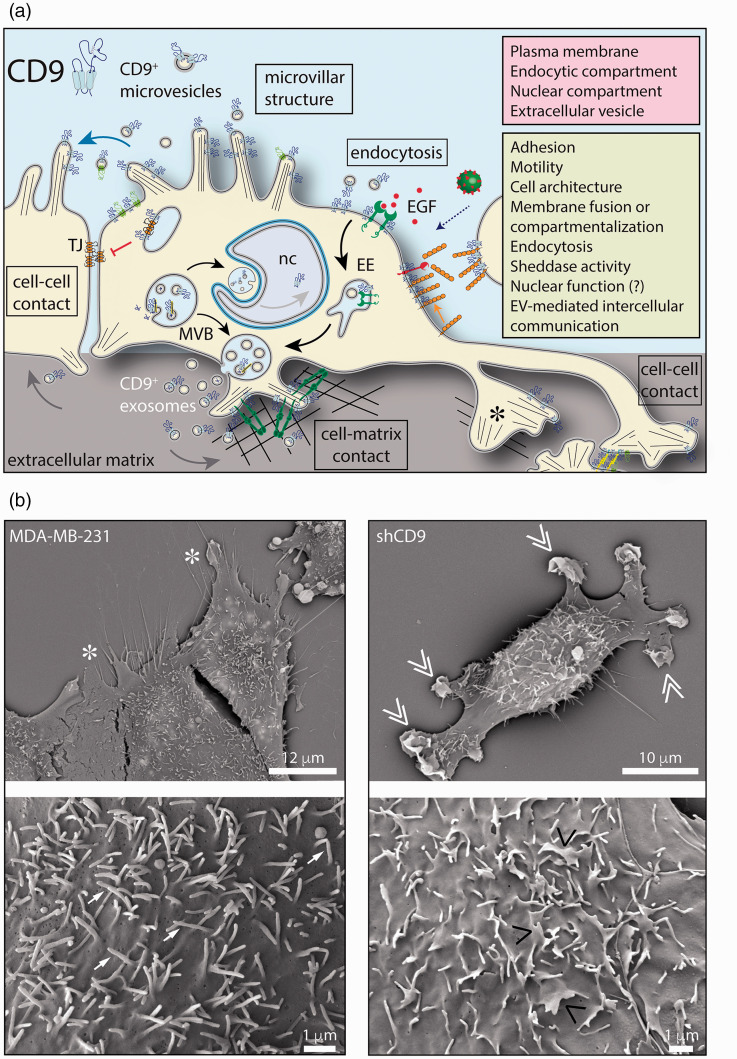
Figure 2.
Cellular expression of CD9 and its functional roles. (a) The tetraspanin CD9 is associated with plasma membrane notably protruding structures such as microvillar-like projections, filopodia, and lamellipodia. An intracellular fraction of CD9 is also present in the endosomal system, notably the late endosome/multivesicular body (MVB), which correlates to its release in association with exosomes into the extracellular milieu. CD9 is also released from microvilli in association with budding microvesicles. A nuclear pool of CD9 has been reported, but no related function (?) has been described. CD9 can associate with various protein partners and regulate their activities in various cellular processes as indicated. For example, the binding of CD9 to adhesion and/or integrin molecules can suppress or promote cell–cell and cell-matrix interactions as well as cell migration. Similarly, the interaction of CD9 with claudin-1 could affect the formation of the tight junction (TJ), and favor epithelial-mesenchymal transition, or EGFR, and attenuate the EGF-EGFR signaling pathway. CD9 located on the cell surface could promote the endocytosis of CD9-positive EVs or regulate the entry of the virus or cell-cell fusion. EVs can play a role in intercellular communication. CD9 may regulate the sheddase activity of certain cell surface enzymes. (b) Silencing CD9 in the MDA-MB-231 (MDA) breast cancer cell line alters the plasma membrane. Scanning electron microscopy revealed that the cell border and microvillar-like structures at the dorsal membrane of MDA cells (left panels, asterisk and arrow, respectively) are altered in CD9-deficient MDA cells (shCD9, right panels, double and single pointing angle quotation mark, respectively). Cell culture conditions and other methods are described in Rappa et al.62 EE: early endosome; EGF: epidermal growth factor; nc: nucleus. Scale bars are indicated.
In addition to cell adhesion and trafficking, CD9 is also involved in vesicular and cellular fusion (reviewed in Hemler12). Functionally, myotubes lacking both CD9 and CD81 or the CD9 partner EWI-F fuse with a higher frequency than normal myotubes, suggesting that CD9 expression prevents inappropriate fusion of myotubes.64 CD9 has also a regulatory role in canine distemper virus and human immunodeficient virus (HIV)-1-induced cell-cell fusion.65–67 The CD9 downregulation increased the Env-mediated syncytia formation and HIV-1 entry, while overexpression of CD9 rendered cells less susceptible to syncytia formation and viral entry.65 Its expression in oocytes (and spermatozoa) is essential for the occurrence of sperm-egg fusion, a process involving the CD9-associated integrin α6β1 and the sperm ADAM2 (β-fertilin) among other players.68–74 The role of integrin α6β1 was nonetheless challenged.75 Thus, CD9-deficient oocytes do not fuse properly with sperm during fertilization and, as a consequence, CD9 knockout mice have reduced fertility.69,70,76 Altogether, the presence of CD9 in the microvillar membrane of oocytes, its apparent role in maintaining the normal shape of microvilli,55 and the observation that CD9 deletion is associated with a decreased density of microvilli on the oocyte surface77 suggest that microvillus-associated CD9 plays a key role in egg-sperm fusion during mammalian fertilization. At the molecular level, Inoue and colleagues suggested that CD9 is crucial for the surface compartmentalization of GPI-anchored proteins, such as the sperm Izumo1 receptor JUNO, favoring gamete membrane fusion.78 Crystal structure studies of CD9 have revealed that the LEL region of CD9 is critically involved in sperm-egg fusion.59
All in all, these few examples illustrate the wide range of physiological functions mediated by the various interactions of CD9 and its membrane partners. For a more exhaustive list of CD9 protein partners and the physiological impact of these interactions, we invite readers to consult the review by Reyes and colleagues and references therein.3 In addition, as a potential membrane receptor, murine CD9 is thought to act as a receptor for pregnancy-specific glycoproteins 17 and 19, via its EC2 domain.79,80 Similarly, it may act as an alternative receptor for interleukin-16 (IL-16), a pro-inflammatory cytokine promoting cell motility, as recently supported by CRISPR/Cas-mediated ablation of the CD9 gene.8,81,82 CD9 may also stimulate the receptor activity of heparin-binding EGF-like growth factor for diphtheria toxin,83 or play a role in MERS-coronavirus cell entry by bringing the cell surface glycoprotein dipeptidyl peptidase 4 (DPP4, CD26), MERS-coronavirus receptor, to a membrane-fusion activating protease (transmembrane protease serine 2, TMPRSS2), forming a cell surface complex that stimulates rapid and effective infection.84
CD9 and extracellular membrane vesicles
CD9 is found not only at the cell surface, but also at the membrane of EVs85–89 (Figure 2(a)). EVs are classified according to their biogenesis as exosomes when derived from multivesicular bodies or as ectosomes/microvesicles when originating directly from the plasma membrane.90 The heterogeneity of EVs is reflected by their size that varies considerably (from ∼40 nm to few microns), exosomes being the smaller entities (∼40–100 nm).91,92 For more details on the mechanisms underlying the formation of exosomes or microvesicles, readers are invited to consult several excellent reviews.93,94 EVs are implicated in intercellular communication in both healthy and disease states.95–98 They act as vehicles for the intercellular transfer of bioactive membranous and cytosolic molecules such as proteins, lipids, and various types of RNAs.99–101 EVs can be isolated from various body fluids such as blood, urine, saliva, tears, seminal fluid, cerebrospinal fluid, and malignant ascites. In cancer, the amount of EVs associated with a given fluid is often increased.102–104 Cancer-associated EVs are associated with the development of the tumor premetastatic niche, a distant site that has become transformed into a more favorable environment for metastasizing tumor cells to settle and grow in the presence of neo-angiogenesis.105–107 The high abundance of CD9 and other tetraspanin proteins (e.g. CD63, CD81) on the EV membrane has made them classical markers for the characterization of the EVs (notably exosomes) found in various physiological fluids.108 Moreover, they might participate in the formation and the general composition of EVs or their cellular uptake.85 In this context, we have shown that CD9 knockdown in EVs released by breast cancer cells and/or recipient cells strongly reduces EV endocytosis.109 CD9-positive EVs were also reported to play a major role in the transfer of molecules between epididymal cells and spermatozoa, leading to the maturation of the latter.110 In mice, sperm fusion properties are conferred by the CD9-positive EVs released from eggs.111
In agreement with the involvement of TEMs in modulating internalization and recycling of plasma membrane proteins, it has been shown that different members of the tetraspanin family regulate protein sorting in EVs, especially exosomes.85 Through its association with E-cadherin and β-catenin, CD9 is instrumental in the cellular export of β-catenin by exosomes, which could modulate the wnt signaling pathway112 (Figure 2(a)). Similarly, the effective incorporation of CD10 metallopeptidase into EVs can be stimulated by its interaction with CD9.113 The peptidase activity of this enzyme could regulate the extracellular matrix especially when EVs are released into a particular microenvironment such as bone marrow, which is sensitive to cell transformation. Buschow et al. have found that CD9 on EVs, especially exosomes, can influence antigen presentation, possibly through the transfer of MHC-peptide complexes.114 In fact, antigen-loaded dendritic cells generate multivesicular bodies with light intraluminal vesicles carrying MHC II and CD9 that are afterward secreted as exosomes and transferred to interacting T cells.
Cytoplasmic and nuclear CD9
In addition to its location at the cell membrane and EVs, CD9 has been reported in the endocytic compartment and in the nucleus (Figure 2(a)).86,109 The presence of CD9 in early endosomes and late endosomes/multivesicular bodies is consistent with its release in association with exosomes. Increased cytoplasmic CD9 levels in some cancers may reflect an increase in the release of CD9-positive EVs and may be associated with enhanced malignancy. In fact, Houle et al. showed in epithelial ovarian carcinoma a shift in the subcellular localization of CD9 from the plasma membrane to the cytoplasm in grade 3 tumors compared to grade 1 tumors.115 An inverse relationship between tumor grade and CD9 expression was also noted with a low expression in high-grade tumors and metastases. In patients with squamous cell carcinoma of the head and neck, Mhawech et al. reported that the impact of CD9 expression on disease-free survival was greater in the subgroup with both membranous and cytoplasmic patterns compared to the subgroup with only a membranous pattern.116 Along with its cytoplasmic localization, we found that CD9 is also present in cell nuclei as observed in primary ductal breast carcinoma patient specimens.86 Approximately 40% of total CD9 cell fluorescence was associated with the nuclear compartment. Although the function of CD9 inside the nucleus remains unclear, the nuclear pool of CD9 may contribute to mitotic processes since CD9 depletion or exposure of breast cancer cells to an anti-CD9 monoclonal antibody (Ab) has resulted in polynucleation and multipolar mitosis.86 Further research is needed with a larger cohort of samples to determine the potential of the nuclear pool of CD9 as a biological prognostic tool for ductal carcinoma of the breast. The mechanism underlying the delivery of CD9 to the nuclear compartment remains to be characterized; however, our observation showing the transfer of CD9 associated with EVs to the nucleus of recipient cells provided an indication of this translocation. To reach the nuclear compartment, EV-associated CD9 might use a novel intracellular pathway where the endocytosed EVs are transported in the nucleoplasmic reticulum via late endosomes/multivesicular bodies.109,117,118 Divalent anti-CD9 Ab can stimulate the uptake of CD9-positive EVs and CD9 accumulation in the nuclear compartment.109 Although to the best of our knowledge no other study has reported the nuclear localization of CD9, the CD9-binding partner EWI-2 was also found in the nuclear compartment.86,119 As there is an interdependence between these proteins, their co-location in the nucleus is conceivable, and further studies should dissect this issue.
CD9 in neoplastic diseases
CD9 is implicated in various diseases and pathological conditions, such as inflammation, viral and bacterial infections, cancer, and chronic lung allograft dysfunction (reviewed in Brosseau et al.120). In the next sections, we will focus on cancer, where CD9 has been extensively studied.10,121 Our database search was performed using as queries either CD9, motility-related protein-1, anti-CD9, or CD9 antibody alone or in combination with motility, migration, invasion, adhesion, morphology, cell proliferation, differentiation, cancer, tumor, metastasis, angiogenesis, immune, patient, extracellular vesicle, and exosome. Although CD9 was initially considered as a tumor suppressor,9 the pro-tumorigenic and pro-metastatic function of CD9 has recently been established in several cancer models (Table 1).8,122 The association of CD9 with different protein partners and its expression levels within TEMs could explain these diverse and sometimes opposite functions observed in distinct cell types, notably in transformed cells.9
Table 1.
A non-exhaustive list of studies showing tumor suppressor or pro-tumorigenic activities of CD9 in cancer.
| Impact on cancer | Cancer type | Potential mechanism | References |
|---|---|---|---|
| Tumor suppressor activity | Prostate carcinoma | • CD9 downregulation during prostate cancer progression often due to deletions or mutations in its transcript• CD9 expression increased upon androgen therapy• Ablation of CD9 had no detectable effect on de novo primary tumor onset, but increased liver metastases• Inverse relationship of CD9 with CD151 in EVs; CD9low and CD151high EV populations have increased TGF-β-induced protein and several subunits of the proteasome complex | 127–129,145 |
| Lung carcinoma (NSCLC) | • Inhibition of cell motility and invasiveness | 131,132 | |
| Lung carcinoma (SCLC) | • Absence of CD9 contributed to differentiation and MMP-2 production via PI3K/Akt pathway, while ectopic expression of CD9 suppressed neurite-like process outgrowth and promoted apoptotic death• Reduction in cell motility through association with β1 integrin | 130,133 | |
| Colon carcinoma | • Ectopic expression of CD9 enhanced β1 integrin-dependent adhesion and inhibition of cell growth | 134,135 | |
| Breast carcinoma | • CD9 suppression resulted in increased motility • CD9 suppression resulted in decreased spread and increased motility, attributed to specific CD9-mediated control of the localization of talin, a critical regulator of integrin activation, to focal adhesion | 136–141 | |
| Mesothelioma | • CD9: favorable prognostic marker for inhibition of cell migration• CD9 and CD26 co-modulated with each other. Depletion of CD26 led to an increase in CD9 and vice versa. CD9 depletion led to enhanced invasiveness. CD9 negatively regulated tumor cell invasion by reducing the level of CD26-α5β1 integrin complex | 143,144 | |
| Gastric carcinoma | • In metastatic gastric cancer tissues, LSD1 deletion suppressed gastric cancer migration by decreasing intracellular miR-142-5p, which led to the upregulation of CD9 | 205 | |
| Hepatocellular carcinoma | • The upregulation of CD9 suppressed carcinoma development via c-Jun N-terminal kinase (JNK) signaling pathway | 206 | |
| Fibrosarcoma | • Through its association with EWI-2/EWI-F/β1 complex and EGFR pathway, and the activation of Akt and p38 signaling, CD9 promoted cell apoptosis and cell spreading, and inhibited cell adhesion, migration, and cell colony formation | 207 | |
| Cervical carcinoma | • Strong local expression of CD9 at sites of trans-endothelial invasion is associated with progression of cervical carcinomas | 208 | |
| Pro-tumorigenic and metastatic activities | Lung carcinoma (NSCLC) | • CD9 increased cell migration to chemoattractants including IL-16• CD9 is a factor of poor prognosis | 8,209 |
| Ovarian carcinoma | • CD9 overexpression induced TNF-α, IL-6, and IL-8 and activation of the NF-κB signaling pathway• CD9 overexpressed in primary ovarian tumors versus normal human ovarian surface epithelium | 150–152 | |
| Breast carcinoma | • CD9 expression associated with worse overall and disease-free survival in patients with invasive lobular carcinoma• CD9 overexpression promoted the development of bone metastases• CD9 on stromal immune cells was associated with a longer disease-free survival, while CD9 on tumor cells correlated with both lymph node and distant metastases• In breast cancer cell lines, CD9 sequestered and destabilized claudin-1, preventing its association with the tight junctions and resulting in increased epithelial-mesenchymal transition and migration and thus promoting tumor progression• Silencing CD9 in MDA-MB-231 cells affected the proper formation of plasma membrane protrusions, and reduced cell migration• CD9/CD81-silenced cells showed delayed α3β1-dependent cell spreading and impaired directed motility and altered front-rear cell morphology, linked to breast carcinoma progression and metastasis | 62,122,148,149,155,210 | |
| Gastric carcinoma | • CD9 expression was greater in tissues from primary and metastatic gastric carcinoma than in surrounding stroma and higher expression of CD9 correlated with vessel invasion, lymph node metastasis, and advanced stage• CD9 positivity correlated with the highly malignant scirrhous-type, with lymph node metastasis and venous invasion. • CD9-positive EVs from cancer-associated fibroblasts stimulated the migration and invasion of cancer cells | 146,147 | |
| Pancreatic carcinoma | • CD9 identified a subpopulation of pancreatic cancer stem cells capable of initiating the carcinoma and give rise to its heterogeneity. CD9 modulated glutamine metabolism to fuel tumor growth | 211 | |
| Multiple myeloma | • CD9 expression was upregulated in vivo by the close interaction of myeloma cells with bone marrow endothelial cells, resulting in trans-endothelial invasion | 212 |
NSCLC: non-small cell lung cancer; SCLC: small-cell lung cancer.
As the immune system contributes to the prevention of tumor formation, growth, and metastasis, the broad expression of CD9 in different immune cell types and its participation in TEM organization result in CD9 playing an important role in shaping anti-tumor immunity. Its potential immuno-suppressive or immuno-stimulatory role depends on the different types of immune cells present in the tumor niche.3,123 Thus, in patients with metastatic melanoma, CD9 expression on NK cells was observed to correlate with serum levels of TGF-β, while CD9 was absent on NK cells in healthy controls.124 In addition, CD9 upregulation on NK cells after TGF-β incubation led to immunosuppressive NK cells, suggesting that CD9 has an immunosuppressive role and its targeting may be beneficial to cancer patients.124 CD9 is also a phenotypic marker for B-regulatory cells (Bregs) that are known to produce large amounts of IL-10 and TGF-β, which inhibit effector immune cells, thereby suppressing antitumor immunity.125,126 Although the relationship between CD9, IL-10, and TGF-β is unclear, targeting CD9 on specific immune subsets in the tumor microenvironment may have therapeutic potential.
CD9 as tumor growth and progression suppressor
Earlier clinical studies based on immunohistochemistry and patient prognosis as well as experimental studies where CD9 levels were manipulated in tumor cell lines and/or animal models have correlated CD9 expression levels with cancer aggressiveness.9 Here we will summarize studies conducted in different types of prostate, lung, colon, breast, pancreatic, and mesothelioma cancers that have suggested a role for CD9 as a suppressor of tumor growth and progression, primarily by inhibiting cancer cell motility and promoting adhesion to the surrounding cells and extracellular matrix (Table 1).
In patients with prostate carcinoma, cancer progression has been accompanied by a decrease in CD9 levels due to deletions or mutations in its transcript with a significant difference between clinically localized and advanced disease, confirming in a clinical setting that CD9 inactivation may play an important role in prostate cancer progression.127 Increase in prostate cancer CD9 expression was observed in patients undergoing androgen therapy.128 To decipher the effects of endogenous CD9 on both prostate cancer initiation and progression, Copeland et al. crossed CD9 knockout mice with transgenic adenocarcinoma of mouse prostate (TRAMP) mice—a model of de novo developing and spontaneously metastasizing prostate cancer. The ablation of CD9 significantly increased liver metastasis, demonstrating the role of CD9 as a progression suppressor.129
Several studies in lung carcinoma patients have found evidence of CD9-induced suppression of tumor growth and/or progression, although many of them did not investigate the molecular mechanism(s) responsible for those effects. Lung cancer is generally classified into non-small cell lung cancer (NSCLC) and highly malignant small cell lung cancer (SCLC). Compared with NSCLC patients, the prognosis of SCLC patients is poor because their tumors metastasize extremely early.130 Utilizing an orthotopic NSCLC model, Takeda et al. found that overexpression of CD9 in epidermoid Lewis lung carcinoma cells inhibited lymph node metastasis without suppressing growth at the implantation site, suggesting that inhibition of cell motility was responsible for the observed effects.131 The CD9 ectopic expression in the human lung adenocarcinoma cell line MAC10 suppressed cell motility and inhibited tumor growth, with the effect on cell motility being dependent on the CD9 expression level.132 Contrary to the less aggressive NSCLC, in most SCLC lines analyzed by Funakoshi et al., CD9 was absent or expressed at low levels and its ectopic expression suppressed the integrin β1-dependent motility.133 Similarly, it was reported that the CD9 ectopic overexpression suppressed neurite-like process outgrowth and promoted apoptotic death of SCLC cells, while its absence contributed to post-adhesive morphologic differentiation, survival, and matrix metalloprotease-2 (MMP-2) production via phosphoinositide 3-kinase (PI3K)/Akt pathway.130
Other studies in colon carcinoma found a potential tumor suppressor role for CD9. In human colon carcinoma, CD9 suppresses the primary growth by increasing the integrin-mediated cell adhesion through a mechanism involving β1 integrin clustering,134 while the examination of tumor surgical samples revealed that CD9 was strongly expressed at the primary site, compared to the low levels in metastases.135 In the latter study, CD9 expression levels were correlated with cell motility, with cancer cells derived from the primary site showing a higher migration potential than cells derived from metastasis. Likewise, the decrease in CD9 expression induced by the transfection of miR-518f-5p in the triple-negative breast cancer cell line MDA-MB-231 (hereafter MDA) increased cell migration in vitro.136 Powner et al. also reported that CD9 deficiency in MDA cells was correlated with decreased cell spread and increased motility. They attributed these observations to a specific CD9-mediated control of the subcellular localization of talin, a critical regulator of integrin activation, to focal adhesions.137 In patients with ductal carcinoma of the breast, CD9 expression in tumor tissues was inversely associated with clinical stage, and was lower in metastatic lymph nodes than in primary breast cancers.138 The overall and disease-free survival rates of patients with CD9high breast cancers were significantly higher than those of patients with CD9low.139 In addition, CD9 positivity correlated better with disease-free survival than estrogen receptor, tumor, and lymph node status.139 The authors suggested that CD9 screening could identify patients with node-negative breast cancer who are at high risk for early disease recurrence. The inverse correlation between CD9 level and stage of ductal breast carcinoma was confirmed,140 showing a significantly higher relapse‐free survival in CD9‐positive cases than in CD9-negative cases. The same study revealed a repression of CD9 expression in metastatic cells compared to that of the corresponding primary tumor cells. Finally, low CD9 expression was linked to poor prognosis with recurrence in breast cancer patients.141
In adenocarcinomas of the pancreas, CD9 gene expression has been associated with lymph node and pathological status and inversely associated with histopathological grading.142 The survival rate of patients with tumors with reduced CD9 mRNA level was lower than that of patients with CD9‐positive tumors and, by multivariate analysis, CD9 status was found to be the most significant together with CD82 transcripts.142 CD9 expression was also found to be a favorable prognostic marker for patients with malignant mesothelioma.143 This is probably related to the observation that CD26, whose expression correlates with disease aggressiveness and invasive potential of selected malignancies, and CD9 co-modulated each other in malignant mesothelioma cell lines.144 The inter-dependence of CD9 and CD26 was shown by the depletion of CD26 that led to an increase in CD9 and vice versa.144 The overexpression of CD26 or CD9 gene depletion led to enhanced invasiveness, while CD26 gene depletion resulted in reduced invasive potential.144 The authors suggested that CD26 potentiates tumor cell invasion through its interaction with α5β1 integrin, and CD9 negatively regulates this process by reducing the level of CD26-α5β1 integrin complex through an inverse correlation between CD9 and CD26 expression.144
In addition to a role for plasma membrane-associated CD9 in cancer inhibition, EV-associated CD9 should also be considered. It was found that EVs from prostate cells with reduced CD9 or increased CD151 enhanced migratory and invasive capabilities of non-tumorigenic prostate cells.145 The authors found that changes in CD9 and CD151 abundance caused a significant alteration in the EV proteome. In particular, in CD9low and CD151high EV populations, an increased expression of TGF-β-induced protein ig-h3 (TGFBI), an integrin-binding partner able to regulate cell attachment to the extracellular matrix, and the β subunits (4, 5, 6, and 7) of the proteasome complex was observed.145 Since the degradation of extracellular matrix components is important for the formation of the premetastatic niche, the selective enrichment of proteins of the proteasome degradation pathways in tumorigenic EVs points towards a suppressive role of CD9. Since tetraspanins influence the sorting of cargo into EVs (see above), the authors suggested that these functional differences between CD9low and CD151high EVs may be attributed to a selective recruitment of cargo molecules that may act on EV target cells to affect migration and invasion, two functions important for the metastatic process.145
CD9 as a cancer promoter
Clinical and experimental studies have demonstrated that CD9 leads to increased proliferation, migration, and survival of several histotypes of cancer cells, presumably through CD9-induced changes in the organization of TEMs and/or affecting specific signaling pathways10,121 (Table 1). In contrast to its role as a tumor suppressor, as discussed in the previous section, CD9 expression has been described as a poor prognostic factor in NSCLC patients. Blake et al. found that CD9 increased cell migration towards chemoattractants, including IL-16, in a model of NSCLC, the A549 cell line.8 Prostate cancer progression to castration refractory disease is associated with anomalous transcriptional activity of the androgen receptor. CD9 knockdown inhibited the expression of androgen receptor-responsive genes, leading the authors to hypothesize that CD9 is involved in the activation of ligand-independent activity of the androgen-receptor.119 An inter-dependence between CD9 and EWI-2 has been also observed, suggesting that activation of CD9 by the EWI-2 down-regulation may be involved in the development of castration-resistant prostate cancer.119
Two independent reports have revealed a cancer promoter role of CD9 in gastric carcinoma. In the first study, it was found that CD9 expression was greater in tissues from primary and metastatic gastric carcinoma than in surrounding stromal non-cancerous areas from the same patient, and that higher expression of CD9 correlated with vessel invasion, lymph node metastasis, and advanced stage.146 In the second study with a large number of cases of patients with gastric cancer, CD9 positivity was significantly correlated with the highly malignant scirrhous-type, with lymph node metastasis and venous invasion.147 Interestingly, CD9-positive EVs from cancer-associated fibroblasts stimulated the migration and invasion of scirrhous-type gastric cancer cells, the latter effects being prevented by CD9 small interfering RNA or anti-CD9 Abs.147 Neither of the two studies investigated the molecular interactions of CD9 resulting in the described pro-metastatic effects.
The CD9 overexpression in human breast cancer was found to promote the development of bone metastases.148 Since the expression of CD9 on tumor cells and on those in the local tumor microenvironment may have different effects on tumor growth and progression, Kwon et al. analyzed independently CD9 levels in tumor and stromal immune cells of patients with invasive breast carcinoma.149 They found that CD9 on stromal immune cells was associated with a longer disease-free survival, while CD9 on tumor cells correlated with both lymph node and distant metastases. In particular, subgroup analysis revealed that CD9 expression on tumor cells was a biomarker for poor prognosis in luminal A invasive breast carcinoma, whereas CD9 expression on stromal immune cells was a marker of good prognosis in luminal B (HER2-negative) tumors. In patients with invasive lobular carcinoma, CD9 expression was associated with worse overall and disease-free survival compared to patients without CD9 expression, indicating that in patients with invasive lobular carcinoma, CD9 may be a significant and independent prognostic factor.122
Specific signaling pathways also contributed to the pro-tumorigenic function of CD9. The overexpression of CD9 in ovarian carcinoma tissues and cell lines was shown to induce the expression of the pro-inflammatory cytokine tumor necrosis factor (TNF)-α, IL-6 and IL-8, and activation of the nuclear factor kappa B (NF-κB)-signaling pathway.150 A comprehensive analysis of gene expression in primary human ovarian tumors versus normal ovarian surface epithelium has revealed a series of overexpressed potential markers, including CD9. These data were confirmed by other studies, where using immunohistochemistry, CD9 was found to be overexpressed in ovarian carcinoma tissues compared with benign ovarian surface epithelium and benign cysts.151,152 Interestingly, Murayama et al. demonstrated the physical and functional association of CD9 with EGFR in the MKN-28 gastric cancer cell line and in numerous others, including CD9/EGFR-transfected ones.54 They provided evidences that CD9 increased the internalization of cell surface EGFR and reduced the EGF-EGFR-induced signals, suggesting that CD9 not only associates with EGFR but also affects EGF-induced signaling in EGF-responsive cancer cells54 (Figure 2(a)).
Other molecular and cellular mechanisms for the pro-malignant function of CD9 have been proposed. One of them relies on the interaction of CD9 with claudin-1, an important component of tight junctions153 (reviewed in Zhou et al.154). In breast cancer cell lines such as MDA and MCF-7, CD9-claudin-1 interaction seems to occur in the cytoplasmic compartment (may be in the endosomes; Figure 2(a)), where claudin-1 is sequestered and destabilized, preventing its association with the tight junctions.153 The latter could lead to an increase of epithelial-mesenchymal transition and migration, thereby promoting tumor progression.155 The CD9-mediated malignant transformation can also be based on its impact on the general morphology of cells, including PMPs such as filopodia, magnupodia, and lamellipodia that modulate the communication between cancer cells and local stroma as well as the cellular processes of intra- and extra-vasation, regulating the formation of cancer metastases.156–159 For forward movement to occur, a complex, dynamic, and coordinated phenomenon must happen under the action of the actin network located beneath the plasma membrane. The formation of cellular projections (e.g. filopodia, lamellipodia) on the front edge, adhesion to the underlying extracellular matrix and detachment of the rear pole would contribute, together with the contraction of the cytoskeleton, to pull the cell forward. In that context, our group reported that CD9-positive PMPs play a role in the invasiveness of breast carcinoma cells when co-cultured with bone marrow-derived multipotent mesenchymal stromal cells. CD9-positive PMPs and CD9 itself could also promote the formation of cancer/mesenchymal stromal cell hybrid cells,62 which may confer chemoresistance in breast cancer via a CD9-dependent mechanism.160,161 The small interfering RNA-mediated silencing of CD9 or Ab-mediated inhibition of its activities reduced the invasive capacity in vitro and suppressed the metastatic capacity of MDA cells in mouse xenografts.62 While MDA cells were spread and created lamellipodia and filopodia in close contact with the extracellular matrix, we observed that CD9-deficient cells showed an adhesion impairment, especially at their cell border where numerous membrane ruffle-like structures were observed62 (Figure 2(b)). In addition, the individual microvillus-like structures that cover the surface of MDA cells were altered, as they appeared as small ruffles at the dorsal membrane, consistent with the involvement of CD9 in the morphogenesis of microvilli as observed in leukocytes and oocytes (Figure 2(b)). The molecular mechanisms underlying these alterations remain to be identified. Interestingly, a morphological comparison of CD9-positive PMPs in three breast cancer cell lines MDA, MA-11 and MCF-7 revealed that the less metastatic MCF-7 cells had shorter, and less frequent CD9-positive PMPs, than MDA cells, while the MA-11 cells, which have an intermediate metastatic potential, had more CD9-positive PMPs than MCF-7 cells.62 Collectively, these observations suggest that CD9 is a key protein for PMP-mediated invasion of cancer cells and, with its association with EVs derived from cancer cells or cancer-associated fibroblasts, there may be a link between these membrane structures, leading to the acquisition of invasive properties of breast cancer cells.62,86,162 The latter issue would require further investigation.
Although many reports have shown that CD9 has an impact on cancer, it is worth mentioning that few other studies have found no relationship between CD9 and clinical outcomes (reviewed in Romanska and Berditchevski163). For instance, a study using a larger cohort of patients with metastatic ductal breast cancer found no correlation between loss of CD9 expression alone in the invasive component and lymph node status.164 Likewise, an immunohistochemical examination of breast carcinoma concluded that CD9 is unlikely to provide useful prognostic information for routine use,165 questioning the clinical relevance of CD9 as a useful indicator of high risk for early disease recurrence in node‐negative breast cancer patients.
CD9 as a target of tumor angiogenesis and cancer
Although only a relatively small proportion of tetraspanin proteins is exposed on the cell surface, these molecules have been investigated as therapeutic immunotargets in cancer (see Hemler166), as shown by the targeting of CD37 in clinical trials for B-cell malignancies.167,168 While experimental data suggest that several tetraspanins (CD37, CD151, and tetraspanin 8) are promising candidate therapeutic targets,10,167 the relationship between CD9 and the cancer processes is more complex, as highlighted in the previous sections. Moreover, severe side effects, including platelet activation and aggregation, may impede the use of particular divalent Abs against CD9 (see below). Consequently, the CD9 targeting for the treatment of cancer is still in the pre-clinical phase. We will refer here to CD9 studies using cancer cell lines and/or mouse tumor xenograft models, which have demonstrated the possibility of CD9 targeting (Table 2).
Table 2.
Anti-angiogenic and anti-cancer effects of CD9-targeting antibodies.
| Impact on cancer | Pre-clinical model | Effectsa | References |
|---|---|---|---|
| Anti-angiogenic therapy | In vitro wound repair | • Ab anti-CD9 Ab (ALMA.1) inhibited endothelial cell migration | 176 |
| Gastric cancer xenograft | • Anti-CD9 Ab (ALB6) decreased growth and angiogenesis | 178 | |
| Laser-induced choroidal neovascularization in mice | • Intravitreal injection of a small interfering RNA or anti-CD9 Ab (ALB6) inhibited laser-induced neovascularization | 179 | |
| Anti-cancer cell therapy | Gastrointestinal cell lines | • Ab anti-CD9 Ab (ALB6) stimulated the endocytosis of EGFR and attenuated EGFR signaling• Anti-CD9 Ab (ALB6 or TP82) stimulated apoptosis through the MAP kinase pathway and caspase cascade | 54 |
| Breast cancer cell lines | • Anti-CD9 Ab (P1/33/2) inhibited the invasion of stromal cells by the breast cancer cells | 62 | |
| Colon carcinoma xenograft and cell lines | • Anti-CD9 Ab (PAINS-13) that disrupts the association of CD9 with β1 integrin inhibited the growth of a human colon carcinoma xenograft more effectively than another anti-CD9 Ab (VJI/20) or anti-integrin Ab• Anti-CD9 Ab (ALB6) inhibited cell motility without affecting cellular adhesion | 134,135 | |
| Ovarian carcinoma | • Anti-CD9 Ab (ALB6) reduced the growth of cancer xenografts and impact on the activation of the NF-κB signaling pathway | 150 | |
| Melanoma cell line | • Anti-CD9 Ab (VJI/10, VJI/20, and GR2110) inhibited the trans-endothelial migration of melanoma cells | 180 | |
| Patients with B-acute lymphoblastic leukemia | • Anti-CD9 Ab (AT1412) induced Ab-dependent cellular cytotoxicity in all B-ALL samples to which it bound and in none of the T-ALL samples• Anti-CD9 Ab (AT1412) did not induce thrombosis | 198,200 | |
| Leukemic cell line | • Anti-CD9 Ab (PAINS-13) recognized a conformation-dependent epitope and disrupted the association of CD9 with β1 integrin | 173 | |
| Acute lymphoblastic leukemia xenografts | • Anti-CD9 Ab (ALB6) suppressed disease progression in NOD/SCID mice xenografted with CD9-positive cell lines and primary leukemic blasts from patients with high-risk and refractory ALL through inhibition of leukemic cell proliferation and activation of p38. In combination with chemotherapeutic agents, anti-CD9 Ab increased apoptotic death | 213 |
The name/clone of anti-CD9 Ab is indicated in brackets.
The neutralization of CD9 can be achieved by silencing its expression by different means such as RNA interference or intercepting its protein function with specific nucleic acid-based aptamers or Ab. Of note, the recombinant soluble EC2/LEL domain of CD9 has been shown to inhibit gamete fusion upon its incubation with oocytes or HIV infection of macrophages.169–171 The soluble CD9 LEL may interfere with its homo- and/or hetero-oligomerization with other tetraspanins, thereby disrupting TEM organization and membrane fusion. Similarly, the application of anti-CD9 Ab can interfere with CD9 functions. In some cases, however, Ab mimics the natural ligand-partner; therefore, it may enhance, rather than inhibit, CD9 function.172 Over the last decades, many anti-CD9 Abs have been developed. It should be noted that the use of different anti-CD9 Abs that recognize distinctive epitopes, either linear or conformation-dependent, could lead to different outcomes, including side effects.173 They can disrupt (or stimulate) the dimerization of CD9, its interactions with other membrane proteins notably integrin and adhesion molecules, affect its internalization, and stimulate the apoptosis in target cancer cells. Expression levels of CD9 partners and the TEM organization could also impact the anti-CD9 Ab targeting strategies. In the following section, the name/clone of the anti-CD9 Ab is indicated in brackets. The dual impact of CD9 on cancer progression and tumor angiogenesis has opened the door to the possibility that CD9 is a novel therapeutic target for both events.174 The implication of CD9 in angiogenesis was suggested by studies in murine models. Thus, mouse CD9 associated with lymphatic endothelial cells facilitated tumor growth in a lung carcinoma xenograft model, as demonstrated after intrathoracic implantation of Lewis lung carcinoma cells in CD9 knockout animals.175 Therein, lymph node metastases were decreased and accompanied by decreased lymphangiogenesis compared to wild-type mice. Moreover, the human CD9 stimulated the endothelial cell migration in vitro, a process inhibited by anti-CD9 Ab (clone ALMA.1).176 The intravenous administration of Ab (ALB6, see Boucheix et al.177) directed against CD9 successfully inhibited tumor progression via antiproliferative, proapoptotic, and antiangiogenetic effects in human gastric cancer cell xenograft.178 Similarly, the intravitreal injection of a small interfering RNA or anti-CD9 Ab (ALB6) was effective to inhibit the laser-induced choroidal neovascularization in mice, suggesting that targeting CD9 may lead to antiangiogenic therapies.179 Anti-CD9 Abs (VJ1/10, VJ1/20, and GR2110) were found to specifically inhibit the trans-endothelial migration of human melanoma cells; the inhibitory effect was likely caused by a strengthening of CD9-mediated interactions between melanoma and endothelial cells.180 Indirectly, targeting the CD9-interacting partner EWI-F may also find some application for inhibition of tumor-associated angiogenesis and tumor growth.181
The tumor suppressor activity of CD9 can be stimulated by addition of anti-CD9 Ab. As described above in cases of colon carcinoma, anti-CD9 Ab (VJ1/20) can stimulate the β1 integrin clustering, alter morphological characteristics of cells, and thus increase cell adhesion, resulting in inhibition of cell proliferation and tumorigenic capacity, as observed in nude mice.134 The anti-CD9 Ab (ALB6) can also prevent cell migration without affecting cell adhesion.135 Thus, anti-CD9 Ab can act at different levels during the metastasis process, including the motility of cancer cells involved in local dissemination from the primary cancer site and adhesive properties for the extravasation to distant sites. The anti-CD9 Ab (ALB6) can also induce apoptosis in human gastrointestinal cancer cells among others.182 As a possible mechanism of ALB6-induced apoptosis, the authors proposed that anti-CD9 Ab activates c-Jun NH2-terminal kinase/stress-activated protein kinase (JNK/SAPK), p38 mitogen-activated-protein kinase (MAPK), and caspase-3. However, the same anti-CD9 Ab (or antisense oligonucleotides directed against CD9) can abrogate human myeloma cell susceptibility to IL-2-activated T cells and NK cells-mediated cytolysis.183 In ovarian tumors, Hwang et al. found that ALB6 Ab injected into the peritoneum reduced tumor growth in human cancer xenografts.150 They attributed the effects to the negative impact of Ab on the oncogenic function of CD9, which was overexpressed, and linked to the induction of expression of pro-inflammatory cytokines and NF-κB-signaling pathway (see above). Altogether, these few examples have highlighted the potential use of anti-CD9 Ab as a biological tool in cancer treatment. More studies are described in Table 2.
In addition to being a direct cellular target, CD9 can be indirectly used to interfere with EV-mediated intercellular communication between cancer cells and their surrounding microenvironment. Given that anti-CD9 Ab can favor the CD9 dimerization and its internalization, oligomerization with its interacting partners or promote the ligation between cells (see below),54,109,184,185 we designed a new strategy to block the endocytosis of cancer-derived CD9-positive EVs by recipient/receptor cells notably stromal cells, the target cells in the tumor microenvironment. To that aim, we generated an antigen-binding fragment (Fab fragment) from an anti-CD9 Ab (5H9).184 The latter, at doses achievable in vivo,186 can saturate CD9 molecules present on the surface of EVs and host cells, including primary mesenchymal stromal cells, and thus interfere with EV endocytosis.184 The effective response was corroborated by the levels of CD9 expression, particularly on the target/receptor cells. The use of distinct anti-CD9 Fabs produced from different anti-CD9 Ab recognizing diverse CD9 epitopes, could synergistically stimulate the inhibitory effect. Thus, the interception of intercellular communication in the tumor niche using an anti-CD9 Fab, combined with the direct targeting of cancer cells, could lead to the development of novel anti-cancer therapeutic strategies.
Clinical use of anti-CD9 antibodies: Potential toxicity and perspectives
Caution should be exercised when divalent anti-CD9 Ab are proposed to disrupt CD9 cancer promoter function or as delivery vehicles, targeted therapeutic agents, for the potential treatment of cancer, especially with the Ab ligation phenomenon.177,187 As the major platelet cell surface protein,188,189 CD9, in combination with fibrinogen receptor α2 bβ3 integrin (GPIIb-IIIa)) and other tetraspanins, can trigger platelet activation, aggregation, or lysis dependent on the subclass of Ab employed.29,30,190–195 Fc receptor found at the surface of platelets can explain this specificity.196 Kawakatsu et al. demonstrated in a primate model that anti-CD9 Ab promotes platelet aggregation leading to fatal pulmonary thrombosis.197 Therefore, the clinical development of such biological tools should exclude the occurrence of potential toxic events including severe thrombocytopenia and/or thrombocyte aggregation. The broad expression of CD9 in various tissues and organs must also be considered when CD9 is employed as a potential tumor-associated antigenic molecular target. Although its level of expression may be upregulated in some cancers relative to its physiological expression in the host, the use of anti-CD9 Ab, especially when conjugated with toxins or cytotoxic agents, could be harmful.
The development of divalent anti-CD9 Ab that do not induce platelet aggregation or the use of bispecific Abs, monovalent Fab Ab or nanobodies could be beneficial for such therapeutic application and limit the pitfalls of most current anti-CD9 Ab.177,198,199 A promising divalent anti-CD9 Ab is the AT1412 clone isolated from survivors of stage IV metastatic melanoma that did not induce platelet aggregation or thrombosis.198,200,201 Although not experimentally evaluated, it is conceivable (and perhaps advantageous) that the development of bispecific Abs targeting CD9 and another tumor-associated antigen or one of its interacting partners (e.g. EWI-2,24 EWI-F,202 integrins), notably those involved in cell adhesion or migration, could improve the specific targeting and potentially interfere with cancer progression. It should be noted that such an approach could limit off-target effects. Obviously, the dual targeting of CD9 and its interacting partner should not stimulate but inhibit their joint action. Due to the tumor suppressor role that CD9 frequently plays in certain tumor histotypes, it is also essential to consider that targeting CD9 may actually favor rather than inhibit tumor growth and progression.
It is important to mention that the presence of CD9 on EVs could have a negative impact on the use of anti-CD9 Ab at different levels. First, these nanometer-sized CD9-positive particles can neutralize the injected anti-CD9 Abs (i.e. divalent and monovalent Ab), thus limiting their effect on the target cells. Second, divalent anti-CD9 Ab, in contrast to monovalent anti-CD9-Fab, may enhance the endocytosis of CD9-positive cancer EVs as well as the nuclear translocation of their cargo in host cells,109,184 thus promoting the pro-metastatic effect of EV-mediated tumor-stroma communication.203 Conversely, CD9-positive EVs may facilitate (and possibly stimulate) internalization of divalent anti-CD9 Ab in non-cancerous host cells and, when conjugated to cytotoxic agents, these tools will be harmful as mentioned above. Overall, EV-associated phenomena would be counterproductive, even fatal, in some circumstances.
In sum, it becomes essential to increase our basic knowledge of CD9, and therefore to continue to dissect its molecular and cellular characteristics in view of developing new clinical and safety tools against this tetraspanin protein. Among these, it will be interesting to fully determine the impact of post-translational modifications (e.g. glycosylation and lipidation) on CD9 and its binding to proteins and lipid interactors.199,204 Similarly, the exact level of CD9 expression in a given cell type and under specific conditions would require further study, as a defined threshold could determine its relationship with TEMs and its involvement in certain molecular and cellular pathways, including the formation and possibly the composition of PMPs and EVs.
Conclusions
Although the degree of involvement of CD9 in carcinogenesis is not yet clear or even contradictory, a growing body of evidence suggests that CD9 may have clinical significance in at least some malignancies where its role as a promoter is highlighted. In this context, the multiple lateral interactions of CD9 within TEMs that regulate cell and extracellular matrix adhesion and migration need to be further investigated. It is conceivable that CD9-based clinical targeting of cancer using tools against CD9 and perhaps specific interacting partners will need to be personalized. The association of CD9 with EVs should be considered when applying a monovalent or divalent anti-CD9 Ab as a therapeutic agent. This latter concern is also valid for other tetraspanins associated with EVs.
ACKNOWLEDGMENTS
We apologize to the individuals whose studies were not specifically cited in this minireview due to reference limitations. The authors acknowledge the constant support of E. Findlay, C. Vanier, and W. Gilliar at Touro University Nevada College of Medicine.
Authors’ contributions: AL, MLR, JK, DC, and GP conceived and wrote the manuscript. JK prepared the figures.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iDs: Aurelio Lorico https://orcid.org/0000-0003-0644-7375
Giuseppe Pizzorno https://orcid.org/0000-0001-7087-2806
References
- 1.Kersey JH, LeBien TW, Abramson CS, Newman R, Sutherland R, Greaves M. P-24: a human leukemia-associated and lymphohemopoietic progenitor cell surface structure identified with monoclonal antibody. J Exp Med 1981; 153:726–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones NH, Borowitz MJ, Metzgar RS. Characterization and distribution of a 24,000-molecular weight antigen defined by a monoclonal antibody (DU-ALL-1) elicited to common acute lymphoblastic leukemia (cALL) cells. Leuk Res 1982; 6:449–64 [DOI] [PubMed] [Google Scholar]
- 3.Reyes R, Cardeñes B, Machado-Pineda Y, Cabañas C. Tetraspanin CD9: a key regulator of cell adhesion in the immune system. Front Immunol 2018; 9:863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucheix C, Duc GH, Jasmin C, Rubinstein E. Tetraspanins and malignancy. Expert Rev Mol Med 2001; 2001:1–17 [DOI] [PubMed] [Google Scholar]
- 5.Horiguchi K, Yoshida S, Tsukada T, Nakakura T, Fujiwara K, Hasegawa R, Takigami S, Ohsako S. Expression and functions of cluster of differentiation 9 and 81 in rat mammary epithelial cells. J Reprod Dev 2020. doi: 10.1262/jrd.2020-082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hemler ME. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu Rev Cell Dev Biol 2003; 19:397–422 [DOI] [PubMed] [Google Scholar]
- 7.Machado-Pineda Y, Cardeñes B, Reyes R, López-Martín S, Toribio V, Sánchez-Organero P, Suarez H, Grötzinger J, Lorenzen I, Yáñez-Mó M, Cabañas C. CD9 controls integrin alpha5beta1-mediated cell adhesion by modulating its association with the metalloproteinase ADAM17. Front Immunol 2018; 9:2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blake DJ, Martiszus JD, Lone TH, Fenster SD. Ablation of the CD9 receptor in human lung cancer cells using CRISPR/cas alters migration to chemoattractants including IL-16. Cytokine 2018; 111:567–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zöller M. Tetraspanins: push and pull in suppressing and promoting metastasis. Nat Rev Cancer 2009; 9:40–55 [DOI] [PubMed] [Google Scholar]
- 10.Hemler ME. Tetraspanin proteins promote multiple cancer stages. Nat Rev Cancer 2014; 14:49–60 [DOI] [PubMed] [Google Scholar]
- 11.Seigneuret M, Delaguillaumie A, Lagaudriere-Gesbert C, Conjeaud H. Structure of the tetraspanin main extracellular domain. A partially conserved fold with a structurally variable domain insertion. J Biol Chem 2001; 276:40055–64 [DOI] [PubMed] [Google Scholar]
- 12.Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol 2005; 6:801–11 [DOI] [PubMed] [Google Scholar]
- 13.Boucheix C, Benoit P, Frachet P, Billard M, Worthington RE, Gagnon J, Uzan G. Molecular cloning of the CD9 antigen. A new family of cell surface proteins. J Biol Chem 1991; 266:117–22 [PubMed] [Google Scholar]
- 14.Boucheix C, Rubinstein E. Tetraspanins. Cell Mol Life Sci 2001; 58:1189–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovalenko OV, Metcalf DG, DeGrado WF, Hemler ME. Structural organization and interactions of transmembrane domains in tetraspanin proteins. BMC Struct Biol 2005; 5:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stipp CS, Kolesnikova TV, Hemler ME. Functional domains in tetraspanin proteins. Trends Biochem Sci 2003; 28:106–12 [DOI] [PubMed] [Google Scholar]
- 17.Toyo-Oka K, Yashiro-Ohtani Y, Park CS, Tai XG, Miyake K, Hamaoka T, Fujiwara H. Association of a tetraspanin CD9 with CD5 on the T cell surface: role of particular transmembrane domains in the association. Int Immunol 1999; 11:2043–52 [DOI] [PubMed] [Google Scholar]
- 18.Martín-Alonso JM, Hernando N, Ghosh S, Coca-Prados M. Molecular cloning of the bovine CD9 antigen from ocular ciliary epithelial cells. J Biochem 1992; 112:63–7 [DOI] [PubMed] [Google Scholar]
- 19.Rubinstein E, Billard M, Plaisance S, Prenant M, Boucheix C. Molecular cloning of the mouse equivalent of CD9 antigen. Thromb Res 1993; 71:377–83 [DOI] [PubMed] [Google Scholar]
- 20.Willett BJ, Neil JC. cDNA cloning and eukaryotic expression of feline CD9. Mol Immunol 1995; 32:417–23 [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi T. Molecular cloning and characterization of chick CD9. Kurume Med J 2000; 47:151–7 [DOI] [PubMed] [Google Scholar]
- 22.Yubero N, Jimenez-Marin A, Yerle M, Morera L, Barbancho MJ, Llanes D, Garrido JJ. Molecular cloning, expression pattern and chromosomal mapping of pig CD9 antigen. Cytogenet Genome Res 2003; 101:143–6 [DOI] [PubMed] [Google Scholar]
- 23.Xing WJ, Wang LQ, Wu Q, Ren SC, Bao XH, Bou S. Molecular cloning and characterization of CD9 cDNA from sheep and cashmere goat. Reprod Domest Anim 2010; 45:383–92 [DOI] [PubMed] [Google Scholar]
- 24.Stipp CS, Kolesnikova TV, Hemler ME. EWI-2 is a major CD9 and CD81 partner and member of a novel Ig protein subfamily. J Biol Chem 2001; 276:40545–54 [DOI] [PubMed] [Google Scholar]
- 25.Charrin S, Le Naour F, Oualid M, Billard M, Faure G, Hanash SM, Boucheix C, Rubinstein E. The major CD9 and CD81 molecular partner. Identification and characterization of the complexes. J Biol Chem 2001; 276:14329–37 [DOI] [PubMed] [Google Scholar]
- 26.Berditchevski F. Complexes of tetraspanins with integrins: more than meets the eye. J Cell Sci 2001; 114:4143–51 [DOI] [PubMed] [Google Scholar]
- 27.Levy S, Shoham T. The tetraspanin web modulates immune-signalling complexes. Nat Rev Immunol 2005; 5:136–48 [DOI] [PubMed] [Google Scholar]
- 28.Zuidscherwoude M, Gottfert F, Dunlock VM, Figdor CG, van den Bogaart G, van Spriel AB. The tetraspanin web revisited by super-resolution microscopy. Sci Rep 2015; 5:12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slupsky JR, Seehafer JG, Tang SC, Masellis-Smith A, Shaw AR. Evidence that monoclonal antibodies against CD9 antigen induce specific association between CD9 and the platelet glycoprotein IIb-IIIa complex. J Biol Chem 1989; 264:12289–93 [PubMed] [Google Scholar]
- 30.Indig FE, Diaz-Gonzalez F, Ginsberg MH. Analysis of the tetraspanin CD9-integrin alphaIIbbeta3 (GPIIb-IIIa) complex in platelet membranes and transfected cells. Biochem J 1997; 327:291–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lozahic S, Christiansen D, Manie S, Gerlier D, Billard M, Boucheix C, Rubinstein E. CD46 (membrane cofactor protein) associates with multiple beta1 integrins and tetraspans. Eur J Immunol 2000; 30:900–7 [DOI] [PubMed] [Google Scholar]
- 32.Oosterheert W, Xenaki KT, Neviani V, Pos W, Doulkeridou S, Manshande J, Pearce NM, Kroon-Batenburg LM, Lutz M, van Bergen En Henegouwen PM, Gros P. Implications for tetraspanin-enriched microdomain assembly based on structures of CD9 with EWI-F. Life Sci Alliance 2020; 3:e202000883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simons K, Ikonen E. Functional rafts in cell membranes. Nature 1997; 387:569–72 [DOI] [PubMed] [Google Scholar]
- 34.Charrin S, Manie S, Thiele C, Billard M, Gerlier D, Boucheix C, Rubinstein E. A physical and functional link between cholesterol and tetraspanins. Eur J Immunol 2003; 33:2479–89 [DOI] [PubMed] [Google Scholar]
- 35.Claas C, Stipp CS, Hemler ME. Evaluation of prototype transmembrane 4 superfamily protein complexes and their relation to lipid rafts. J Biol Chem 2001; 276:7974–84 [DOI] [PubMed] [Google Scholar]
- 36.Israels SJ, McMillan-Ward EM. Platelet tetraspanin complexes and their association with lipid rafts. Thromb Haemost 2007; 98:1081–7 [PubMed] [Google Scholar]
- 37.Sala-Valdés M, Ursa A, Charrin S, Rubinstein E, Hemler ME, Sánchez-Madrid F, Yáñez-Mó M. EWI-2 and EWI-F link the tetraspanin web to the actin cytoskeleton through their direct association with ezrin-radixin-moesin proteins. J Biol Chem 2006; 281:19665–75 [DOI] [PubMed] [Google Scholar]
- 38.Zimmerman B, Kelly B, McMillan BJ, Seegar TCM, Dror RO, Kruse AC, Blacklow SC. Crystal structure of a full-length human tetraspanin reveals a cholesterol-binding pocket. Cell 2016; 167:1041–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ono M, Handa K, Sonnino S, Withers DA, Nagai H, Hakomori S. GM3 ganglioside inhibits CD9-facilitated haptotactic cell motility: coexpression of GM3 and CD9 is essential in the downregulation of tumor cell motility and malignancy. Biochemistry 2001; 40:6414–21 [DOI] [PubMed] [Google Scholar]
- 40.Kawakami Y, Kawakami K, Steelant WF, Ono M, Baek RC, Handa K, Withers DA, Hakomori S. Tetraspanin CD9 is a "proteolipid," and its interaction with alpha 3 integrin in microdomain is promoted by GM3 ganglioside, leading to inhibition of laminin-5-dependent cell motility. J Biol Chem 2002; 277:34349–58 [DOI] [PubMed] [Google Scholar]
- 41.Park SY, Yoon SJ, Freire-de-Lima L, Kim JH, Hakomori SI. Control of cell motility by interaction of gangliosides, tetraspanins, and epidermal growth factor receptor in A431 versus KB epidermoid tumor cells. Carbohydr Res 2009; 344:1479–86 [DOI] [PubMed] [Google Scholar]
- 42.Charrin S, Manie S, Oualid M, Billard M, Boucheix C, Rubinstein E. Differential stability of tetraspanin/tetraspanin interactions: role of palmitoylation. FEBS Lett 2002; 516:139–44 [DOI] [PubMed] [Google Scholar]
- 43.Israels SJ, McMillan-Ward EM. Palmitoylation supports the association of tetraspanin CD63 with CD9 and integrin alphaIIbbeta3 in activated platelets. Thromb Res 2010; 125:152–8 [DOI] [PubMed] [Google Scholar]
- 44.Yang XH, Kovalenko OV, Kolesnikova TV, Andzelm MM, Rubinstein E, Strominger JL, Hemler ME. Contrasting effects of EWI proteins, integrins, and protein palmitoylation on cell surface CD9 organization. J Biol Chem 2006; 281:12976–85 [DOI] [PubMed] [Google Scholar]
- 45.Rocha-Perugini V, González-Granado JM, Tejera E, López-Martín S, Yáñez-Mó M, Sánchez-Madrid F. Tetraspanins CD9 and CD151 at the immune synapse support T-cell integrin signaling. Eur J Immunol 2014; 44:1967–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaji K, Takeshita S, Miyake K, Takai T, Kudo A. Functional association of CD9 with the Fc gamma receptors in macrophages. J Immunol 2001; 166:3256–65 [DOI] [PubMed] [Google Scholar]
- 47.Barreiro O, Yáñez-Mó M, Sala-Valdés M, Gutiérrez-Lopez MD, Ovalle S, Higginbottom A, Monk PN, Cabañas C, Sánchez-Madrid F. Endothelial tetraspanin microdomains regulate leukocyte firm adhesion during extravasation. Blood 2005; 105:2852–61 [DOI] [PubMed] [Google Scholar]
- 48.Gilsanz A, Sánchez-Martín L, Gutiérrez-Lopez MD, Ovalle S, Machado-Pineda Y, Reyes R, Swart GW, Figdor CG, Lafuente EM, Cabañas C. ALCAM/CD166 adhesive function is regulated by the tetraspanin CD9. Cell Mol Life Sci 2013; 70:475–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gutiérrez-López MD, Gilsanz A, Yáñez-Mó M, Ovalle S, Lafuente EM, Dominguez C, Monk PN, Gonzalez-Alvaro I, Sánchez-Madrid F, Cabañas C. The sheddase activity of ADAM17/TACE is regulated by the tetraspanin CD9. Cell Mol Life Sci 2011; 68:3275–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Münsterberg J, Loreth D, Brylka L, Werner S, Karbanová J, Gandrass M, Schneegans S, Besler K, Hamester F, Robador JR, Bauer AT, Schneider SW, Wrage M, Lamszus K, Matschke J, Vashist Y, Uzunoglu G, Steurer S, Horst AK, Oliveira-Ferrer L, Glatzel M, Schinke T, Corbeil D, Pantel K, Maire C, Wikman H. ALCAM contributes to brain metastasis formation in non-small-cell lung cancer through interaction with the vascular endothelium. Neuro Oncol 2020; 22:955–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weidle UH, Eggle D, Klostermann S, Swart GW. ALCAM/CD166: cancer-related issues. Cancer Genom Proteom 2010; 7:231–43 [PubMed] [Google Scholar]
- 52.Shi W, Fan H, Shum L, Derynck R. The tetraspanin CD9 associates with transmembrane TGF-alpha and regulates TGF-alpha-induced EGF receptor activation and cell proliferation. J Cell Biol 2000; 148:591–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Imhof I, Gasper WJ, Derynck R. Association of tetraspanin CD9 with transmembrane TGF{alpha} confers alterations in cell-surface presentation of TGF{alpha} and cytoskeletal organization. J Cell Sci 2008; 121:2265–74 [DOI] [PubMed] [Google Scholar]
- 54.Murayama Y, Shinomura Y, Oritani K, Miyagawa J, Yoshida H, Nishida M, Katsube F, Shiraga M, Miyazaki T, Nakamoto T, Tsutsui S, Tamura S, Higashiyama S, Shimomura I, Hayashi N. The tetraspanin CD9 modulates epidermal growth factor receptor signaling in cancer cells. J Cell Physiol 2008; 216:135–43 [DOI] [PubMed] [Google Scholar]
- 55.Runge KE, Evans JE, He ZY, Gupta S, McDonald KL, Stahlberg H, Primakoff P, Myles DG. Oocyte CD9 is enriched on the microvillar membrane and required for normal microvillar shape and distribution. Dev Biol 2007; 304:317–25 [DOI] [PubMed] [Google Scholar]
- 56.Wang HX, Kolesnikova TV, Denison C, Gygi SP, Hemler ME. The C-terminal tail of tetraspanin protein CD9 contributes to its function and molecular organization. J Cell Sci 2011; 124:2702–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peñas PF, García-Díez A, Sánchez-Madrid F, Yáñez-Mó M. Tetraspanins are localized at motility-related structures and involved in normal human keratinocyte wound healing migration. J Invest Dermatol 2000; 114:1126–35 [DOI] [PubMed] [Google Scholar]
- 58.Bari R, Guo Q, Xia B, Zhang YH, Giesert EE, Levy S, Zheng JJ, Zhang XA. Tetraspanins regulate the protrusive activities of cell membrane. Biochem Biophys Res Commun 2011; 415:619–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Umeda R, Satouh Y, Takemoto M, Nakada-Nakura Y, Liu K, Yokoyama T, Shirouzu M, Iwata S, Nomura N, Sato K, Ikawa M, Nishizawa T, Nureki O. Structural insights into tetraspanin CD9 function. Nat Commun 2020; 11:1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang XA, Huang C. Tetraspanins and cell membrane tubular structures. Cell Mol Life Sci 2012; 69:2843–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Franz J, Brinkmann BF, Konig M, Huve J, Stock C, Ebnet K, Riethmüller C. Nanoscale imaging reveals a tetraspanin-CD9 coordinated elevation of endothelial ICAM-1 clusters. PLoS One 2016; 11:e0146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rappa G, Green TM, Karbanová J, Corbeil D, Lorico A. Tetraspanin CD9 determines invasiveness and tumorigenicity of human breast cancer cells. Oncotarget 2015; 6:7970–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abitorabi MA, Pachynski RK, Ferrando RE, Tidswell M, Erle DJ. Presentation of integrins on leukocyte microvilli: a role for the extracellular domain in determining membrane localization. J Cell Biol 1997; 139:563–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Charrin S, Latil M, Soave S, Polesskaya A, Chrétien F, Boucheix C, Rubinstein E. Normal muscle regeneration requires tight control of muscle cell fusion by tetraspanins CD9 and CD81. Nat Commun 2013; 4:1674. [DOI] [PubMed] [Google Scholar]
- 65.Gordón-Alonso M, Yáñez-Mó M, Barreiro O, Álvarez S, Muñoz-Fernández MA, Valenzuela-Fernandez A, Sánchez-Madrid F. Tetraspanins CD9 and CD81 modulate HIV-1-induced membrane fusion. J Immunol 2006; 177:5129–37 [DOI] [PubMed] [Google Scholar]
- 66.Loffler S, Lottspeich F, Lanza F, Azorsa DO, ter Meulen V, Schneider-Schaulies J. CD9, a tetraspan transmembrane protein, renders cells susceptible to canine distemper virus. J Virol 1997; 71:42–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singethan K, Müller N, Schubert S, Lüttge D, Krementsov DN, Khurana SR, Krohne G, Schneider-Schaulies S, Thali M, Schneider-Schaulies J. CD9 clustering and formation of microvilli zippers between contacting cells regulates virus-induced cell fusion. Traffic 2008; 9:924–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vjugina U, Evans JP. New insights into the molecular basis of mammalian sperm-egg membrane interactions. Front Biosci 2008; 13:462–76 [DOI] [PubMed] [Google Scholar]
- 69.Le Naour F, Rubinstein E, Jasmin C, Prenant M, Boucheix C. Severely reduced female fertility in CD9-deficient mice. Science 2000; 287:319–21 [DOI] [PubMed] [Google Scholar]
- 70.Kaji K, Oda S, Shikano T, Ohnuki T, Uematsu Y, Sakagami J, Tada N, Miyazaki S, Kudo A. The gamete fusion process is defective in eggs of Cd9-deficient mice. Nat Genet 2000; 24:279–82 [DOI] [PubMed] [Google Scholar]
- 71.Chen MS, Tung KS, Coonrod SA, Takahashi Y, Bigler D, Chang A, Yamashita Y, Kincade PW, Herr JC, White JM. Role of the integrin-associated protein CD9 in binding between sperm ADAM 2 and the egg integrin alpha6beta1: implications for murine fertilization. Proc Natl Acad Sci U S A 1999; 96:11830–5 USA [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jankovičová J, Neuerová Z, Sečová P, Bartóková M, Bubeníčková F, Komrsková K, Postlerová P, Antaliková J. Tetraspanins in mammalian reproduction: spermatozoa, oocytes and embryos. Med Microbiol Immunol 2020; 209:407–25 [DOI] [PubMed] [Google Scholar]
- 73.Frolikova M, Manaskova-Postlerova P, Cerny J, Jankovicova J, Simonik O, Pohlova A, Secova P, Antalikova J. Dvorakova-Hortova K. CD9 and CD81 interactions and their structural modelling in sperm prior to fertilization. Int J Mol Sci 2018; 19:1236–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ziyyat A, Rubinstein E, Monier-Gavelle F, Barraud V, Kulski O, Prenant M, Boucheix C, Bomsel M, Wolf JP. CD9 controls the formation of clusters that contain tetraspanins and the integrin alpha 6 beta 1, which are involved in human and mouse gamete fusion. J Cell Sci 2006; 119:416–24 [DOI] [PubMed] [Google Scholar]
- 75.Miller BJ, Georges-Labouesse E, Primakoff P, Myles DG. Normal fertilization occurs with eggs lacking the integrin alpha6beta1 and is CD9-dependent. J Cell Biol 2000; 149:1289–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miyado K, Yamada G, Yamada S, Hasuwa H, Nakamura Y, Ryu F, Suzuki K, Kosai K, Inoue K, Ogura A, Okabe M, Mekada E. Requirement of CD9 on the egg plasma membrane for fertilization. Science 2000; 287:321–4 [DOI] [PubMed] [Google Scholar]
- 77.Benammar A, Ziyyat A, Lefevre B, Wolf JP. Tetraspanins and mouse oocyte microvilli related to fertilizing ability. Reprod Sci 2017; 24:1062–9 [DOI] [PubMed] [Google Scholar]
- 78.Inoue N, Saito T, Wada I. Unveiling a novel function of CD9 in surface compartmentalization of oocytes. Development 2020; 147:dev189985. [DOI] [PubMed] [Google Scholar]
- 79.Waterhouse R, Ha C, Dveksler GS. Murine CD9 is the receptor for pregnancy-specific glycoprotein 17. J Exp Med 2002; 195:277–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ha CT, Waterhouse R, Warren J, Zimmermann W, Dveksler GS. N-glycosylation is required for binding of murine pregnancy-specific glycoproteins 17 and 19 to the receptor CD9. Am J Reprod Immunol 2008; 59:251–8 [DOI] [PubMed] [Google Scholar]
- 81.Qi JC, Wang J, Mandadi S, Tanaka K, Roufogalis BD, Madigan MC, Lai K, Yan F, Chong BH, Stevens RL, Krilis SA. Human and mouse mast cells use the tetraspanin CD9 as an alternate interleukin-16 receptor. Blood 2006; 107:135–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yadav S, Shi Y, Wang H. IL-16 effects on A549 lung epithelial cells: dependence on CD9 as an IL-16 receptor? J Immunotoxicol 2010; 7:183–93 [DOI] [PubMed] [Google Scholar]
- 83.Iwamoto R, Higashiyama S, Mitamura T, Taniguchi N, Klagsbrun M, Mekada E. Heparin-binding EGF-like growth factor, which acts as the diphtheria toxin receptor, forms a complex with membrane protein DRA. P27/CD9, which up-regulates functional receptors and diphtheria toxin sensitivity. EMBO J 1994; 13:2322–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Earnest JT, Hantak MP, Li K, McCray PB, Jr., Perlman S, Gallagher T. The tetraspanin CD9 facilitates MERS-coronavirus entry by scaffolding host cell receptors and proteases. PLoS Pathog 2017; 13:e1006546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Andreu Z, Yáñez-Mó M. Tetraspanins in extracellular vesicle formation and function. Front Immunol 2014; 5:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rappa G, Green TM, Lorico A. The nuclear Pool of tetraspanin CD9 contributes to mitotic processes in human breast carcinoma. Mol Cancer Res 2014; 12:1840–50 [DOI] [PubMed] [Google Scholar]
- 87.Wei P, Wu F, Kang B, Sun X, Heskia F, Pachot A, Liang J, Li D. Plasma extracellular vesicles detected by single molecule array technology as a liquid biopsy for colorectal cancer. J Extracell Vesicles 2020; 9:1809765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hoshino A, Kim HS, Bojmar L, Gyan KE, Cioffi M, Hernandez J, Zambirinis CP, Rodrigues G, Molina H, Heissel S, Mark MT, Steiner L, Benito-Martin A, Lucotti S, Di Giannatale A, Offer K, Nakajima M, Williams C, Nogues L, Pelissier Vatter FA, Hashimoto A, Davies AE, Freitas D, Kenific CM, Ararso Y, Buehring W, Lauritzen P, Ogitani Y, Sugiura K, Takahashi N, Aleckovic M, Bailey KA, Jolissant JS, Wang H, Harris A, Schaeffer LM, Garcia-Santos G, Posner Z, Balachandran VP, Khakoo Y, Raju GP, Scherz A, Sagi I, Scherz-Shouval R, Yarden Y, Oren M, Malladi M, Petriccione M, De Braganca KC, Donzelli M, Fischer C, Vitolano S, Wright GP, Ganshaw L, Marrano M, Ahmed A, DeStefano J, Danzer E, Roehrl MHA, Lacayo NJ, Vincent TC, Weiser MR, Brady MS, Meyers PA, Wexler LH, Ambati SR, Chou AJ, Slotkin EK, Modak S, Roberts SS, Basu EM, Diolaiti D, Krantz BA, Cardoso F, Simpson AL, Berger M, Rudin CM, Simeone DM, Jain M, Ghajar CM, Batra SK, Stanger BZ, Bui J, Brown KA, Rajasekhar VK, Healey JH, de Sousa M, Kramer K, Sheth S, Baisch J, Pascual V, Heaton TE, La Quaglia MP, Pisapia DJ, Schwartz R, Zhang H, Liu Y, Shukla A, Blavier L, DeClerck YA, LaBarge M, Bissell MJ, Caffrey TC, Grandgenett PM, Hollingsworth MA, Bromberg J, Costa-Silva B, Peinado H, Kang Y, Garcia BA, O'Reilly EM, Kelsen D, Trippett TM, Jones DR, Matei IR, Jarnagin WR, Lyden D. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell 2020; 182:1044–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ding XQ, Wang ZY, Xia D, Wang RX, Pan XR, Tong JH. Proteomic profiling of serum exosomes from patients with metastatic gastric cancer. Front Oncol 2020; 10:1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 2013; 200:373–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ciardiello C, Cavallini L, Spinelli C, Yang J, Reis-Sobreiro M, de Candia P, Minciacchi VR, Di Vizio D. Focus on extracellular vesicles: new frontiers of cell-to-cell communication in cancer. Int J Mol Sci 2016; 17:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Corbeil D, Lorico A. Exosomes, microvesicles, and their friends in solid tumors. In: L Edelstein, J Smythies, P Quesenberry, D Noble. (eds) Exosomes: a clinical compendium. Cambridge, MA: Academic Press, 2020, Chapter 3, pp.39–80 [Google Scholar]
- 93.van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 2018; 19:213–28 [DOI] [PubMed] [Google Scholar]
- 94.Tkach M, Théry C. Communication by extracellular vesicles: where we are and where we need to go. Cell 2016; 164:1226–32 [DOI] [PubMed] [Google Scholar]
- 95.Buzas EI, Gyorgy B, Nagy G, Falus A, Gay S. Emerging role of extracellular vesicles in inflammatory diseases. Nat Rev Rheumatol 2014; 10:356–64 [DOI] [PubMed] [Google Scholar]
- 96.Wang Y, Zhao M, Liu S, Guo J, Lu Y, Cheng J, Liu J. Macrophage-derived extracellular vesicles: diverse mediators of pathology and therapeutics in multiple diseases. Cell Death Dis 2020; 11:924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fu S, Zhang Y, Li Y, Luo L, Zhao Y, Yao Y. Extracellular vesicles in cardiovascular diseases. Cell Death Discov 2020; 6:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Feng R, Ullah M, Chen K, Ali Q, Lin Y, Sun Z. Stem cell-derived extracellular vesicles mitigate ageing-associated arterial stiffness and hypertension. J Extracell Vesicles 2020; 9:1783869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ratajczak J, Wysoczynski M, Hayek F, Janowska-Wieczorek A, Ratajczak MZ. Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia 2006; 20:1487–95 [DOI] [PubMed] [Google Scholar]
- 100.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007; 9:654–9 [DOI] [PubMed] [Google Scholar]
- 101.Ullah M, Qiao Y, Concepcion W, Thakor AS. Stem cell-derived extracellular vesicles: role in oncogenic processes, bioengineering potential, and technical challenges. Stem Cell Res Ther 2019; 10:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Soekmadji C, Riches JD, Russell PJ, Ruelcke JE, McPherson S, Wang C, Hovens CM, Corcoran NM. Australian prostate cancer collaboration B, hill MM, Nelson CC. Modulation of paracrine signaling by CD9 positive small extracellular vesicles mediates cellular growth of androgen deprived prostate cancer. Oncotarget 2017; 8:52237–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Soekmadji C, Corcoran NM, Oleinikova I, Jovanovic L, Australian Prostate Cancer Collaboration B, Ramm GA, Nelson CC, Jenster G, Russell PJ. Extracellular vesicles for personalized therapy decision support in advanced metastatic cancers and its potential impact for prostate cancer. Prostate 2017; 77:1416–23 [DOI] [PubMed] [Google Scholar]
- 104.Inubushi S, Kawaguchi H, Mizumoto S, Kunihisa T, Baba M, Kitayama Y, Takeuchi T, Hoffman RM, Tanino H, Sasaki R. Oncogenic miRNAs identified in tear exosomes from metastatic breast cancer patients. Anticancer Res 2020; 40:3091–6 [DOI] [PubMed] [Google Scholar]
- 105.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, Zhang H, Hernandez J, Weiss JM, Dumont-Cole VD, Kramer K, Wexler LH, Narendran A, Schwartz GK, Healey JH, Sandstrom P, Labori KJ, Kure EH, Grandgenett PM, Hollingsworth MA, de Sousa M, Kaur S, Jain M, Mallya K, Batra SK, Jarnagin WR, Brady MS, Fodstad O, Muller V, Pantel K, Minn AJ, Bissell MJ, Garcia BA, Kang Y, Rajasekhar VK, Ghajar CM, Matei I, Peinado H, Bromberg J, Lyden D. Tumour exosome integrins determine organotropic metastasis. Nature 2015; 527:329–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fujita Y, Yoshioka Y, Ochiya T. Extracellular vesicle transfer of cancer pathogenic components. Cancer Sci 2016; 107:385–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xu R, Rai A, Chen M, Suwakulsiri W, Greening DW, Simpson RJ. Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat Rev Clin Oncol 2018; 15:617–38 [DOI] [PubMed] [Google Scholar]
- 108.Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Théry C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A 2016; 113:E968–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rappa G, Santos MF, Green TM, Karbanová J, Hassler J, Bai Y, Barsky SH, Corbeil D, Lorico A. Nuclear transport of cancer extracellular vesicle-derived biomaterials through nuclear envelope invagination-associated late endosomes. Oncotarget 2017; 8:14443–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Caballero JN, Frenette G, Belleannee C, Sullivan R. CD9-positive microvesicles mediate the transfer of molecules to bovine spermatozoa during epididymal maturation. PLoS One 2013; 8:e65364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Miyado K, Yoshida K, Yamagata K, Sakakibara K, Okabe M, Wang X, Miyamoto K, Akutsu H, Kondo T, Takahashi Y, Ban T, Ito C, Toshimori K, Nakamura A, Ito M, Miyado M, Mekada E, Umezawa A. The fusing ability of sperm is bestowed by CD9-containing vesicles released from eggs in mice. Proc Natl Acad Sci U S A 2008; 105:12921–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chairoungdua A, Smith DL, Pochard P, Hull M, Caplan MJ. Exosome release of beta-catenin: a novel mechanism that antagonizes wnt signaling. J Cell Biol 2010; 190:1079–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mazurov D, Barbashova L, Filatov A. Tetraspanin protein CD9 interacts with metalloprotease CD10 and enhances its release via exosomes. FEBS J 2013; 280:1200–13 [DOI] [PubMed] [Google Scholar]
- 114.Buschow SI, Nolte-'t Hoen EN, van Niel G, Pols MS, ten Broeke T, Lauwen M, Ossendorp F, Melief CJ, Raposo G, Wubbolts R, Wauben MH, Stoorvogel W. MHC II in dendritic cells is targeted to lysosomes or T cell-induced exosomes via distinct multivesicular body pathways. Traffic 2009; 10:1528–42 [DOI] [PubMed] [Google Scholar]
- 115.Houle CD, Ding XY, Foley JF, Afshari CA, Barrett JC, Davis BJ. Loss of expression and altered localization of KAI1 and CD9 protein are associated with epithelial ovarian cancer progression. Gynecol Oncol 2002; 86:69–78 [DOI] [PubMed] [Google Scholar]
- 116.Mhawech P, Dulguerov P, Tschanz E, Verdan C, Ares C, Allal AS. Motility-related protein-1 (MRP-1/CD9) expression can predict disease-free survival in patients with squamous cell carcinoma of the head and neck. Br J Cancer 2004; 90:471–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Santos MF, Rappa G, Karbanová J, Kurth T, Corbeil D, Lorico A. VAMP-associated protein-A and oxysterol-binding protein-related protein 3 promote the entry of late endosomes into the nucleoplasmic reticulum. J Biol Chem 2018; 293:13834–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Corbeil D, Santos MF, Karbanová J, Kurth T, Rappa G, Lorico A. Uptake and fate of extracellular vesicles: nucleoplasmic reticulum-associated late endosomes as a new gate to intercellular communication. Cells 2020; 9:1931–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Levina E, Ji H, Chen M, Baig M, Oliver D, Ohouo P, Lim CU, Schools G, Carmack S, Ding Y, Broude EV, Roninson IB, Buttyan R, Shtutman M. Identification of novel genes that regulate androgen receptor signaling and growth of androgen-deprived prostate cancer cells. Oncotarget 2015; 6:13088–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Brosseau C, Colas L, Magnan A, Brouard SC. Tetraspanin A new pathway for the regulation of inflammation? Front Immunol 2018; 9:2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Charrin S, Jouannet S, Boucheix C, Rubinstein E. Tetraspanins at a glance. J Cell Sci 2014; 127:3641–8 [DOI] [PubMed] [Google Scholar]
- 122.Baek J, Jang N, Choi JE, Kim JR, Bae YK. CD9 expression in tumor cells is associated with poor prognosis in patients with invasive lobular carcinoma. J Breast Cancer 2019; 22:77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schaper F, van Spriel AB. Antitumor immunity is controlled by tetraspanin proteins. Front Immunol 2018; 9:1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Holtan SG, Creedon DJ, Thompson MA, Nevala WK, Markovic SN. Expansion of CD16-negative natural killer cells in the peripheral blood of patients with metastatic melanoma. Clin Dev Immunol 2011; 2011:316314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sun J, Wang J, Pefanis E, Chao J, Rothschild G, Tachibana I, Chen JK, Ivanov II, Rabadan R, Takeda Y, Basu U. Transcriptomics identify CD9 as a marker of murine IL-10-competent regulatory B cells. Cell Rep 2015; 13:1110–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Matsushita T, Le Huu D, Kobayashi T, Hamaguchi Y, Hasegawa M, Naka K, Hirao A, Muramatsu M, Takehara K, Fujimoto M. A novel splenic B1 regulatory cell subset suppresses allergic disease through phosphatidylinositol 3-kinase-Akt pathway activation. J Allergy Clin Immunol 2016; 138:1170–82 [DOI] [PubMed] [Google Scholar]
- 127.Wang JC, Begin LR, Berube NG, Chevalier S, Aprikian AG, Gourdeau H, Chevrette M. Down-regulation of CD9 expression during prostate carcinoma progression is associated with CD9 mRNA modifications. Clin Cancer Res 2007; 13:2354–61 [DOI] [PubMed] [Google Scholar]
- 128.Chuan Y, Pang ST, Bergh A, Norstedt G, Pousette A. Androgens induce CD-9 in human prostate tissue. Int J Androl 2005; 28:291–6 [DOI] [PubMed] [Google Scholar]
- 129.Copeland BT, Bowman MJ, Boucheix C, Ashman LK. Knockout of the tetraspanin Cd9 in the TRAMP model of de novo prostate cancer increases spontaneous metastases in an organ-specific manner. Int J Cancer 2013; 133:1803–12 [DOI] [PubMed] [Google Scholar]
- 130.Saito Y, Tachibana I, Takeda Y, Yamane H, He P, Suzuki M, Minami S, Kijima T, Yoshida M, Kumagai T, Osaki T, Kawase I. Absence of CD9 enhances adhesion-dependent morphologic differentiation, survival, and matrix metalloproteinase-2 production in small cell lung cancer cells. Cancer Res 2006; 66:9557–65 [DOI] [PubMed] [Google Scholar]
- 131.Takeda T, Hattori N, Tokuhara T, Nishimura Y, Yokoyama M, Miyake M. Adenoviral transduction of MRP-1/CD9 and KAI1/CD82 inhibits lymph node metastasis in orthotopic lung cancer model. Cancer Res 2007; 67:1744–9 [DOI] [PubMed] [Google Scholar]
- 132.Ikeyama S, Koyama M, Yamaoko M, Sasada R, Miyake M. Suppression of cell motility and metastasis by transfection with human motility-related protein (MRP-1/CD9) DNA. J Exp Med 1993; 177:1231–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Funakoshi T, Tachibana I, Hoshida Y, Kimura H, Takeda Y, Kijima T, Nishino K, Goto H, Yoneda T, Kumagai T, Osaki T, Hayashi S, Aozasa K, Kawase I. Expression of tetraspanins in human lung cancer cells: frequent downregulation of CD9 and its contribution to cell motility in small cell lung cancer. Oncogene 2003; 22:674–87 [DOI] [PubMed] [Google Scholar]
- 134.Ovalle S, Gutiérrez-López MD, Olmo N, Turnay J, Lizarbe MA, Majano P, Molina-Jiménez F, López-Cabrera M, Yáñez-Mó M, Sánchez-Madrid F, Cabañas C. The tetraspanin CD9 inhibits the proliferation and tumorigenicity of human colon carcinoma cells. Int J Cancer 2007; 121:2140–52 [DOI] [PubMed] [Google Scholar]
- 135.Cajot JF, Sordat I, Silvestre T, Sordat B. Differential display cloning identifies motility-related protein (MRP1/CD9) as highly expressed in primary compared to metastatic human colon carcinoma cells. Cancer Res 1997; 57:2593–7 [PubMed] [Google Scholar]
- 136.Bond DR, Kahl R, Brzozowski JS, Jankowski H, Naudin C, Pariyar M, Avery-Kiejda KA, Scarlett CJ, Boucheix C, Muller WJ, Ashman LK, Cairns MJ, Roselli S, Weidenhofer J. Tetraspanin CD9 is regulated by miR-518f-5p and functions in breast cell migration and in vivo tumor growth. Cancers 2020; 12:795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Powner D, Kopp PM, Monkley SJ, Critchley DR, Berditchevski F. Tetraspanin CD9 in cell migration. Biochem Soc Trans 2011; 39:563–7 [DOI] [PubMed] [Google Scholar]
- 138.Miyake M, Nakano K, Ieki Y, Adachi M, Huang CL, Itoi S, Koh T, Taki T. Motility related protein 1 (MRP-1/CD9) expression: inverse correlation with metastases in breast cancer. Cancer Res 1995; 55:4127–31 [PubMed] [Google Scholar]
- 139.Miyake M, Nakano K, Itoi SI, Koh T, Taki T. Motility-related protein-1 (MRP-1/CD9) reduction as a factor of poor prognosis in breast cancer. Cancer Res 1996; 56:1244–9 [PubMed] [Google Scholar]
- 140.Mimori K, Kataoka A, Yoshinaga K, Ohta M, Sagara Y, Yoshikawa Y, Ohno S, Barnard GF, Mori M. Identification of molecular markers for metastasis-related genes in primary breast cancer cells. Clin Exp Metastasis 2005; 22:59–67 [DOI] [PubMed] [Google Scholar]
- 141.Huang CI, Kohno N, Ogawa E, Adachi M, Taki T, Miyake M. Correlation of reduction in MRP-1/CD9 and KAI1/CD82 expression with recurrences in breast cancer patients. Am J Pathol 1998; 153:973–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sho M, Adachi M, Taki T, Hashida H, Konishi T, Huang CL, Ikeda N, Nakajima Y, Kanehiro H, Hisanaga M, Nakano H, Miyake M. Transmembrane 4 superfamily as a prognostic factor in pancreatic cancer. Int J Cancer 1998; 79:509–16 [DOI] [PubMed] [Google Scholar]
- 143.Amatya VJ, Takeshima Y, Aoe K, Fujimoto N, Okamoto T, Yamada T, Kishimoto T, Morimoto C, Inai K. CD9 expression as a favorable prognostic marker for patients with malignant mesothelioma. Oncol Rep 2013;21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Okamoto T, Iwata S, Yamazaki H, Hatano R, Komiya E, Dang NH, Ohnuma K, Morimoto C. CD9 negatively regulates CD26 expression and inhibits CD26-mediated enhancement of invasive potential of malignant mesothelioma cells. PLoS One 2014; 9:e86671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Brzozowski JS, Bond DR, Jankowski H, Goldie BJ, Burchell R, Naudin C, Smith ND, Scarlett CJ, Larsen MR, Dun MD, Skelding KA, Weidenhofer J. Extracellular vesicles with altered tetraspanin CD9 and CD151 levels confer increased prostate cell motility and invasion. Sci Rep 2018; 8:8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hori H, Yano S, Koufuji K, Takeda J, Shirouzu K. CD9 expression in gastric cancer and its significance. J Surg Res 2004; 117:208–15 [DOI] [PubMed] [Google Scholar]
- 147.Miki Y, Yashiro M, Okuno T, Kitayama K, Masuda G, Hirakawa K, Ohira M. CD9-positive exosomes from cancer-associated fibroblasts stimulate the migration ability of scirrhous-type gastric cancer cells. Br J Cancer 2018; 118:867–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kischel P, Bellahcene A, Deux B, Lamour V, Dobson R, Dep E, Clezardin P, Castronovo V. Overexpression of CD9 in human breast cancer cells promotes the development of bone metastases. Anticancer Res 2012; 32:5211–20 [PubMed] [Google Scholar]
- 149.Kwon HJ, Choi JE, Kang SH, Son Y, Bae YK. Prognostic significance of CD9 expression differs between tumour cells and stromal immune cells, and depends on the molecular subtype of the invasive breast carcinoma. Histopathology 2017; 70:1155–65 [DOI] [PubMed] [Google Scholar]
- 150.Hwang JR, Jo K, Lee Y, Sung BJ, Park YW, Lee JH. Upregulation of CD9 in ovarian cancer is related to the induction of TNF-alpha gene expression and constitutive NF-kappaB activation. Carcinogenesis 2012; 33:77–83 [DOI] [PubMed] [Google Scholar]
- 151.Peters DG, Kudla DM, Deloia JA, Chu TJ, Fairfull L, Edwards RP, Ferrell RE. Comparative gene expression analysis of ovarian carcinoma and normal ovarian epithelium by serial analysis of gene expression. Cancer Epidemiol Biomarkers Prev 2005; 14:1717–23 [DOI] [PubMed] [Google Scholar]
- 152.Drapkin R, Crum CP, Hecht JL. Expression of candidate tumor markers in ovarian carcinoma and benign ovary: evidence for a link between epithelial phenotype and neoplasia. Hum Pathol 2004; 35:1014–21 [DOI] [PubMed] [Google Scholar]
- 153.Kovalenko OV, Yang XH, Hemler ME. A novel cysteine cross-linking method reveals a direct association between claudin-1 and tetraspanin CD9. Mol Cell Proteom 2007; 6:1855–67 [DOI] [PubMed] [Google Scholar]
- 154.Zhou B, Moodie A, Blanchard AA, Leygue E, Myal Y. Claudin 1 in breast cancer: new insights. J Clin Med 2015; 4:1960–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Oku N, Sasabe E, Ueta E, Yamamoto T, Osaki T. Tight junction protein claudin-1 enhances the invasive activity of oral squamous cell carcinoma cells by promoting cleavage of laminin-5 gamma2 chain via matrix metalloproteinase (MMP)-2 and membrane-type MMP-1. Cancer Res 2006; 66:5251–7 [DOI] [PubMed] [Google Scholar]
- 156.Takebe N, Warren RQ, Ivy SP. Breast cancer growth and metastasis: interplay between cancer stem cells, embryonic signaling pathways and epithelial-to-mesenchymal transition. Breast Cancer Res 2011; 13:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013; 19:1423–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Machesky LM. Lamellipodia and filopodia in metastasis and invasion. FEBS Lett 2008; 582:2102–11 [DOI] [PubMed] [Google Scholar]
- 159.Khuon S, Liang L, Dettman RW, Sporn PH, Wysolmerski RB, Chew TL. Myosin light chain kinase mediates transcellular intravasation of breast cancer cells through the underlying endothelial cells: a three-dimensional FRET study. J Cell Sci 2010; 123:431–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ullah M, Akbar A, Ng NN, Concepcion W, Thakor AS. Mesenchymal stem cells confer chemoresistance in breast cancer via a CD9 dependent mechanism. Oncotarget 2019; 10:3435–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ullah M, Akbar A, Thakor AS. An emerging role of CD9 in stemness and chemoresistance. Oncotarget 2019; 10:4000–1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Heusermann W, Hean J, Trojer D, Steib E, von Bueren S, Graff-Meyer A, Genoud C, Martin K, Pizzato N, Voshol J, Morrissey DV, Andaloussi SE, Wood MJ, Meisner-Kober NC. Exosomes surf on filopodia to enter cells at endocytic hot spots, traffic within endosomes, and are targeted to the ER. J Cell Biol 2016; 213:173–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Romanska HM, Berditchevski F. Tetraspanins in human epithelial malignancies. J Pathol 2011; 223:4–14 [DOI] [PubMed] [Google Scholar]
- 164.Arihiro K, Kaneko M, Fujii S, Inai K. Loss of CD9 with expression of CD31 and VEGF in breast carcinoma, as predictive factors of lymph node metastasis. Breast Cancer 1998; 5:131–8 [DOI] [PubMed] [Google Scholar]
- 165.Jamil F, Peston D, Shousha S. CD9 immunohistochemical staining of breast carcinoma: unlikely to provide useful prognostic information for routine use. Histopathology 2001; 39:572–7 [DOI] [PubMed] [Google Scholar]
- 166.Hemler ME. Targeting of tetraspanin proteins–potential benefits and strategies. Nat Rev Drug Discov 2008; 7:747–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Betrian S, Ysebaert L, Heider KH, Delord JP, Fournie JJ, Quillet-Mary A. Idelalisib improves CD37 antibody BI 836826 cytotoxicity against chemo-resistant/relapse-initiating CLL cells: a rationale for combination treatment. Blood Cancer J 2016; 6:e496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hicks SW, Lai KC, Gavrilescu LC, Yi Y, Sikka S, Shah P, Kelly ME, Lee J, Lanieri L, Ponte JF, Sloss CM, Romanelli A. The antitumor activity of IMGN529, a CD37-targeting antibody-drug conjugate, is potentiated by rituximab in non-Hodgkin lymphoma models. Neoplasia 2017; 19:661–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhu GZ, Miller BJ, Boucheix C, Rubinstein E, Liu CC, Hynes RO, Myles DG, Primakoff P. Residues SFQ (173-175) in the large extracellular loop of CD9 are required for gamete fusion. Development 2002; 129:1995–2002 [DOI] [PubMed] [Google Scholar]
- 170.Higginbottom A, Takahashi Y, Bolling L, Coonrod SA, White JM, Partridge LJ, Monk PN. Structural requirements for the inhibitory action of the CD9 large extracellular domain in sperm/oocyte binding and fusion. Biochem Biophys Res Commun 2003; 311:208–14 [DOI] [PubMed] [Google Scholar]
- 171.Ho SH, Martin F, Higginbottom A, Partridge LJ, Parthasarathy V, Moseley GW, Lopez P, Cheng-Mayer C, Monk PN. Recombinant extracellular domains of tetraspanin proteins are potent inhibitors of the infection of macrophages by human immunodeficiency virus type 1. J Virol 2006; 80:6487–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Murayama Y, Oritani K, Tsutsui S. Novel CD9-targeted therapies in gastric cancer. World J Gastroenterol 2015; 21:3206–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gutiérrez-López MD, Ovalle S, Yáñez-Mó M, Sánchez-Sánchez N, Rubinstein E, Olmo N, Lizarbe MA, Sánchez-Madrid F, Cabañas C. A functionally relevant conformational epitope on the CD9 tetraspanin depends on the association with activated beta1 integrin. J Biol Chem 2003; 278:208–18 [DOI] [PubMed] [Google Scholar]
- 174.Vasse M, Colin S, Guilmain W, Creoff E, Muraine M, Vannier JP, Al-Mahmood S. [Tetraspanins: a new target for antiangiogenic therapy?] Ann Pharm Fr 2015; 73:100–7 [DOI] [PubMed] [Google Scholar]
- 175.Iwasaki T, Takeda Y, Maruyama K, Yokosaki Y, Tsujino K, Tetsumoto S, Kuhara H, Nakanishi K, Otani Y, Jin Y, Kohmo S, Hirata H, Takahashi R, Suzuki M, Inoue K, Nagatomo I, Goya S, Kijima T, Kumagai T, Tachibana I, Kawase I, Kumanogoh A. Deletion of tetraspanin CD9 diminishes lymphangiogenesis in vivo and in vitro. J Biol Chem 2013; 288:2118–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Klein-Soyer C, Azorsa DO, Cazenave JP, Lanza F. CD9 participates in endothelial cell migration during in vitro wound repair. Arterioscler Thromb Vasc Biol 2000; 20:360–9 [DOI] [PubMed] [Google Scholar]
- 177.Boucheix C, Soria C, Mirshahi M, Soria J, Perrot JY, Fournier N, Billard M, Rosenfeld C. Characteristics of platelet aggregation induced by the monoclonal antibody ALB6 (acute lymphoblastic leukemia antigen p 24). Inhibition of aggregation by ALB6Fab. FEBS Lett 1983; 161:289–95 [DOI] [PubMed] [Google Scholar]
- 178.Nakamoto T, Murayama Y, Oritani K, Boucheix C, Rubinstein E, Nishida M, Katsube F, Watabe K, Kiso S, Tsutsui S, Tamura S, Shinomura Y, Hayashi N. A novel therapeutic strategy with anti-CD9 antibody in gastric cancers. J Gastroenterol 2009; 44:889–96 [DOI] [PubMed] [Google Scholar]
- 179.Kamisasanuki T, Tokushige S, Terasaki H, Khai NC, Wang Y, Sakamoto T, Kosai K. Targeting CD9 produces stimulus-independent antiangiogenic effects predominantly in activated endothelial cells during angiogenesis: a novel antiangiogenic therapy. Biochem Biophys Res Commun 2011; 413:128–35 [DOI] [PubMed] [Google Scholar]
- 180.Longo N, Yáñez-Mó M, Mittelbrunn M, de la Rosa G, Muñoz ML, Sánchez-Madrid F, Sánchez-Mateos P. Regulatory role of tetraspanin CD9 in tumor-endothelial cell interaction during transendothelial invasion of melanoma cells. Blood 2001; 98:3717–26 [DOI] [PubMed] [Google Scholar]
- 181.Colin S, Guilmain W, Creoff E, Schneider C, Steverlynck C, Bongaerts M, Legrand E, Vannier JP, Muraine M, Vasse M, Al-Mahmood SA. Truncated form of CD9-partner 1 (CD9P-1), GS-168AT2, potently inhibits in vivo tumour-induced angiogenesis and tumour growth. Br J Cancer 2011; 105:1002–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Murayama Y, Miyagawa J, Oritani K, Yoshida H, Yamamoto K, Kishida O, Miyazaki T, Tsutsui S, Kiyohara T, Miyazaki Y, Higashiyama S, Matsuzawa Y, Shinomura Y. CD9-mediated activation of the p46 shc isoform leads to apoptosis in cancer cells. J Cell Sci 2004; 117:3379–88 [DOI] [PubMed] [Google Scholar]
- 183.Shallal S, Kornbluth J. CD9 expression enhances the susceptibility of myeloma cell lines to cell-mediated cytolysis. Blood 2000; 96:224–33 [PubMed] [Google Scholar]
- 184.Santos MF, Rappa G, Karbanová J, Vanier C, Morimoto C, Corbeil D, Lorico A. Anti-human CD9 antibody fab fragment impairs the internalization of extracellular vesicles and the nuclear transfer of their cargo proteins. J Cell Mol Med 2019; 23:4408–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Letarte M, Seehafer JG, Greaves A, Masellis-Smith A, Shaw AR. Homotypic aggregation of pre-B leukemic cell lines by antibodies to VLA integrins correlates with their expression of CD9. Leukemia 1993; 7:93–103 [PubMed] [Google Scholar]
- 186.Oude Munnink TH, Henstra MJ, Segerink LI, Movig KL, Brummelhuis-Visser P. Therapeutic drug monitoring of monoclonal antibodies in inflammatory and malignant disease: translating TNF-alpha experience to oncology. Clin Pharmacol Ther 2016; 99:419–31 [DOI] [PubMed] [Google Scholar]
- 187.Rubinstein E, Boucheix C, Worthington RE, Carroll RC. Anti-platelet antibody interactions with Fc gamma receptor. Semin Thromb Hemost 1995; 21:10–22 [DOI] [PubMed] [Google Scholar]
- 188.Blair TA, Michelson AD, Frelinger AL. 3rd Mass cytometry reveals distinct platelet subtypes in healthy subjects and novel alterations in surface glycoproteins in glanzmann thrombasthenia. Sci Rep 2018; 8:10300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kaprielian Z, Cho KO, Hadjiargyrou M, Patterson PH. CD9, a major platelet cell surface glycoprotein, is a ROCA antigen and is expressed in the nervous system. J Neurosci 1995; 15:562–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Carroll RC, Rubinstein E, Worthington RE, Boucheix C. Extensive C1q-complement initiated lysis of human platelets by IgG subclass murine monoclonal antibodies to the CD9 antigen. Thromb Res 1990; 59:831–9 [DOI] [PubMed] [Google Scholar]
- 191.Higashihara M, Maeda H, Shibata Y, Kume S, Ohashi T. A monoclonal anti-human platelet antibody: a new platelet aggregating substance. Blood 1985; 65:382–91 [PubMed] [Google Scholar]
- 192.Miller JL, Kupinski JM, Hustad KO. Characterization of a platelet membrane protein of low molecular weight associated with platelet activation following binding by monoclonal antibody AG-1. Blood 1986; 68:743–51 [PubMed] [Google Scholar]
- 193.Hato T, Ikeda K, Yasukawa M, Watanabe A, Kobayashi Y. Exposure of platelet fibrinogen receptors by a monoclonal antibody to CD9 antigen. Blood 1988; 72:224–9 [PubMed] [Google Scholar]
- 194.Jennings LK, Fox CF, Kouns WC, McKay CP, Ballou LR, Schultz HE. The activation of human platelets mediated by anti-human platelet p24/CD9 monoclonal antibodies. J Biol Chem 1990; 265:3815–22 [PubMed] [Google Scholar]
- 195.Israels SJ, McMillan-Ward EM, Easton J, Robertson C, McNicol A. CD63 associates with the alphaIIb beta3 integrin-CD9 complex on the surface of activated platelets. Thromb Haemost 2001; 85:134–41 [PubMed] [Google Scholar]
- 196.Worthington RE, Carroll RC, Boucheix C. Platelet activation by CD9 monoclonal antibodies is mediated by the Fc gamma II receptor. Br J Haematol 1990; 74:216–22 [DOI] [PubMed] [Google Scholar]
- 197.Kawakatsu T, Suzuki M, Kido H, Sakane H, Hada S, Yamaguchi K, Fukuroi T, Yanabu M, Nagata H, Nomura S, Kokawa T, Yasunaga K. Antithrombotic effect of an anti-glycoprotein IIB/IIIA antibody in primate lethal thrombosis. Thromb Res 1993; 70:245–54 [DOI] [PubMed] [Google Scholar]
- 198.De Jong G, Levie SE, Schotte R, Pos W, Go D, Yasuda E, Cercel M, van Hal-van Veen SE, Frankin E, Szabó A, Villaudy J, van Helden PM, van Eenennaam H, Spits H, Rijneveld A, Hazenberg MD. AT1412, a patient-derived antibody in development for the treatment of CD9 positive precursor B-acute lymphoblastic leukemia. Blood 2019; 134:4461 [Google Scholar]
- 199.Neviani V, van Deventer S, Worner TP, Xenaki KT, van de Waterbeemd M, Rodenburg RNP, Wortel IMN, Kuiper JK, Huisman S, Granneman J, van Bergen En Henegouwen PMP, Heck AJR, van Spriel AB, Gros P. Site-specific functionality and tryptophan mimicry of lipidation in tetraspanin CD9. FEBS J 2020. doi: 10.1111/febs.15295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Villaudy J, Schotte R, Gd J, Neviani V, Pos W, Levie S, Yasuda E, Cercel M, Szabó A, Fatmawati C, Kedde M, Horbach S, Verdegaal EME, Pv H, SHvd B, Rijneveld A, Gros P, Spits H, Hazenberg M, Hv E. Preclinical development of AT1412, a patient derived CD9 antibody that does not induce thrombosis for treatment of precursor B ALL. Ann Oncol 2020; 31:S493 [Google Scholar]
- 201.Schotte R, Villaudy J, Kedde M, Pos W, Wagner K, Go D, Fatmawati C, Moiset G, Yasuda E, Cercel M, Frankin E, SvH-V V, Pv H, Verdegaal EME, Hv E. Burg SHvd, spits H. 580P AT1412, a patient-derived CD9 antibody promotes tumour immune infiltration and induces tumour rejection. Ann Oncol 2020; 31:S492 [Google Scholar]
- 202.Chambrion C, Le Naour F. The tetraspanins CD9 and CD81 regulate CD9P1-induced effects on cell migration. PLoS One 2010; 5:e11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Peinado H, Alečković M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, García-Santos G, Ghajar C, Nitadori-Hoshino A, Hoffman C, Badal K, Garcia BA, Callahan MK, Yuan J, Martins VR, Skog J, Kaplan RN, Brady MS, Wolchok JD, Chapman PB, Kang Y, Bromberg J, Lyden D. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 2012; 18:883–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ono M, Handa K, Withers DA, Hakomori S. Motility inhibition and apoptosis are induced by metastasis-suppressing gene product CD82 and its analogue CD9, with concurrent glycosylation. Cancer Res 1999; 59:2335–9 [PubMed] [Google Scholar]
- 205.Zhao LJ, Fan QQ, Li YY, Ren HM, Zhang T, Liu S, Maa M, Zheng YC, Liu HM. LSD1 deletion represses gastric cancer migration by upregulating a novel miR-142-5p target protein CD9. Pharmacol Res 2020; 159:104991. [DOI] [PubMed] [Google Scholar]
- 206.Li Y, Yu S, Li L, Chen J, Quan M, Li Q, Gao Y. KLF4-mediated upregulation of CD9 and CD81 suppresses hepatocellular carcinoma development via JNK signaling. Cell Death Dis 2020; 11:299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chen S, Sun Y, Jin Z, Jing X. Functional and biochemical studies of CD9 in fibrosarcoma cell line. Mol Cell Biochem 2011; 350:89–99 [DOI] [PubMed] [Google Scholar]
- 208.Sauer G, Windisch J, Kurzeder C, Heilmann V, Kreienberg R, Deissler H. Progression of cervical carcinomas is associated with down-regulation of CD9 but strong local re-expression at sites of transendothelial invasion. Clin Cancer Res 2003; 9:6426–31 [PubMed] [Google Scholar]
- 209.Higashiyama M, Taki T, Ieki Y, Adachi M, Huang CL, Koh T, Kodama K, Doi O. Miyake M. Reduced motility related protein-1 (MRP-1/CD9) gene expression as a factor of poor prognosis in non-small cell lung cancer. Cancer Res 1995; 55:6040–4 [PubMed] [Google Scholar]
- 210.Gustafson-Wagner E, Stipp CS. The CD9/CD81 tetraspanin complex and tetraspanin CD151 regulate alpha3beta1 integrin-dependent tumor cell behaviors by overlapping but distinct mechanisms. PLoS One 2013; 8:e61834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wang VM, Ferreira RMM, Almagro J, Evan T, Legrave N, Zaw Thin M, Frith D, Carvalho J, Barry DJ, Snijders AP, Herbert E, Nye EL, MacRae JI, Behrens A. CD9 identifies pancreatic cancer stem cells and modulates glutamine metabolism to fuel tumour growth. Nat Cell Biol 2019; 21:1425–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.De Bruyne E, Andersen TL, De Raeve H, Van Valckenborgh E, Caers J, Van Camp B, Delaisse JM, Van Riet I, Vanderkerken K. Endothelial cell-driven regulation of CD9 or motility-related protein-1 expression in multiple myeloma cells within the murine 5T33MM model and myeloma patients. Leukemia 2006; 20:1870–9 [DOI] [PubMed] [Google Scholar]
- 213.Leung KT, Zhang C, Chan KYY, Li K, Cheung JTK, Ng MHL, Zhang XB, Sit T, Lee WYW, Kang W, To KF, Yu JWS, Man TKF, Wang H, Tsang KS, Cheng FWT, Lam GKS, Chow TW, Leung AWK, Leung TF, Yuen PMP, Ng PC, LC. CD9 blockade suppresses disease progression of high-risk pediatric B-cell precursor acute lymphoblastic leukemia and enhances chemosensitivity. Leukemia 2020; 34:709–20 [DOI] [PubMed] [Google Scholar]