Abstract
Objective
To evaluate the expression of retinoid-related orphan receptor gamma (RORγ) and its potential role in the prognosis of colon cancer.
Methods
The Cancer Genome Atlas and GSE117606 were used to evaluate to RORγ levels in colon cancer, and real-time quantitative polymerase chain reaction was applied for validation. UALCAN and MEXPRESS were used to analyze the associations of RORγ expression with clinical parameters. The survival analysis was conducted in GEPIA.
Results
RORγ expression was significantly lower in colon tumors than in adjacent normal mucosa tissues. RORγ expression was significantly associated with tumor stage, lymph node metastasis, and liver metastasis. The area under the curve for diagnosis was 0.71. Decreased RORγ expression was positively correlated with the incidence of lymphatic invasion, microsatellite instability, the presence of residual tumor, venous invasion, and copy number variation. Overall survival was longer in patients with higher RORγ expression, especially those with microsatellite instability-high features. Methylation analysis revealed that hypermethylation of the RORγ promoter was associated with the colon cancer stage.
Conclusions
RORγ downregulation could be a potential biomarker for colon cancer, especially for predicting prognosis. Decreased RORγ expression in colon tumor may be associated with promoter hypermethylation.
Keywords: Retinoid-related orphan receptor gamma, hypermethylation, prognosis, biomarker, colon cancer, copy number variation, microsatellite instability
Introduction
Colon cancer is a dangerous malignant tumor with high mortality rates.1 In recent years, changes in living habits, aging of the population, and other factors have contributed to an increased incidence of colon cancer.2 According to the treatment guidelines for colon cancer, surgery, chemotherapy, radiotherapy, targeted therapy, and combined therapy are the common treatment strategies for prolonging survival in patients with colon cancer.3,4 However, colon cancer is the second-leading cause of cancer-related death according to the cancer statistics of 2020.1 Clinical data indicate that metastasis and drug resistance contribute to the high mortality of colon cancer.5 Furthermore, because of the absence of early symptoms, most patients with colon cancer are diagnosed in the middle or advanced stages, permitting little opportunity for radical surgery, and palliative treatments are the most common approaches in the clinic.6,7 Hence, novel therapeutic targets and drug discovery have become hotspots in the study of colon cancer.
Retinoid-related orphan receptor gamma (RORγ) is an orphan receptor that is widely expressed in organs, including the pancreas, liver, adipose tissues, and skeletal muscles.8,9 RORγt is a member of retinoid-related orphan receptor (ROR) family, which is involved in immune-related regulation.10 The function of RORγt includes the regulation of thymocytes and development of lymphoid organs.11 When RORγt is deficient, the peripheral mesenteric lymph nodes and Peyer’s patches are disrupted, suggesting that RORγt is indispensable for lymph node organogenesis.12 The transcriptional axis involving Rel–RORγ–RORγt signaling has been reported in the control of Th17 cell function, and some potential ligands have been revealed to regulate biological activities.13,14 In recent years, the role of RORγ in tumorigenesis has attracted substantial attention. In some literature, RORγ acts as a tumor suppressor. Muscat and co-authors reported RORγ that expression is decreased in advanced breast cancers, and RORγ could inhibit the TGFβ and MaSC pathways, which are activated in cancers.15 Other researchers identified that RORγ ligands also function as anti-tumor modulators in the inhibition of migration and cell viability in breast cancer and melanoma.16,17 However, RORγ was identified as an oncogene in liver cancer, lung cancer, prostate cancer, gastric cancer, cervical cancer, and multiple myeloma.18–22 Some researchers also indicated that RORγ could exert pro-tumor effects in breast cancer through regulating LC3,15,23 suggesting that the function of RORγ varies among different cancers.
Relatively few studies have assessed RORγ expression and its underlying function in colon cancer. In this study, RORγ expression was fully studied to reveal its possible role in the progression of colon cancer. Moreover, the association between methylation regulation and abnormal RORγ expression was also examined, and the potential application of RORγ in the prediction of prognosis in patients with colon cancer was discussed. The present study aimed to clarify the expression of RORγ and its potential importance in the development of colon cancer.
Materials and methods
Reagents
TRIeasy™ Total RNA extraction reagent (Cat. 10606ES60), DEPC-treated water (DNase- and RNase-free, Cat. 10601ES76), a Hifair™ first-strand cDNA synthesis kit (Cat. 11137ES60), and a qPCR SYBR Green kit (Cat. 11203ES03) were purchased from Yeasen Biotechnology (Shanghai, China). The primers for qPCR were synthesized by Sangon Biotechnology (Shanghai, China). The primers targeted RORγ (forward: 5′-CGTTTTGAGGAACACAGGCA-3′, reverse: 5′-GAGAAGATGTTGGAGCGCTG-3′) and β-actin (forward: 5′-CATCCGCAAAGACCTGTACG-3′, reverse: 5′-CCTGCTTGCTGATCCACATC-3′). Other chemical reagents were of analytical purity and acquired from Sigma-Aldrich (St. Louis, MO, USA).
The Cancer Genome Atlas (TCGA) analysis
The TCGA colon adenocarcinoma dataset analysis was conducted in the UALCAN portal,24 which is a comprehensive and interactive web source for analyzing expression data. The adjacent normal mucosa tissues were used as normal controls, and the correlations with certain clinicopathological features were examined according to RORγ expression, including tumor stage and lymph node metastasis. Furthermore, the epigenetic regulation of gene expression via promoter methylation was also analyzed in this study.
Gene Expression Omnibus (GEO) analysis
Samples from the GSE117606 dataset were analyzed in the GEO database using the GEO2R portal. The gene expression platform was GPL25373, the adjacent normal mucosa tissues comprised the normal control group, and the resected tumor tissues comprised the tumor disease group. The distribution of data for these samples was checked. The data were subjected to log transformation, and statistical analysis was performed with adjustment for the P-value.
Human tissue collection
Clinical tissues including colon tumor tissues and the corresponding adjacent normal mucosa tissues were collected in the Department of Colorectal and Anal Surgery, the Affiliated Hospital of West Anhui Health Vocational College (Lu’an, China). This study was approved by the Ethics Committee of the Affiliated Hospital of West Anhui Health Vocational College (Approval number: LEAY-2019-001). All patients provided written informed consent.
Reverse transcription-quantitative PCR (RT-qPCR)
The resected tissues were stored at −80°C, and total RNA was extracted using RNA extraction reagent (Yeasen Biotechnology, Shanghai, China) according to the manufacturer’s instructions. Total RNA was used as the template, and first-strand cDNA was synthesized using the kit. The collected cDNA was treated with gDNA eraser to remove genomic DNA. The cDNA was used as a template to analyze the mRNA levels of the indicated genes using a qPCR SYBR kit according to the instruction of the manual. ROX was included in the reaction as an internal control. The melting curve was conducted to confirm the specific primer as indicated previously.25 β-actin was used as a housekeeping gene to quantify the relative RORγ mRNA expression.
Receiver operating characteristic (ROC) analysis
ROC analysis is a common method for evaluating potential diagnostic biomarkers.26 Thirty patients were included in the analysis. Colon tumor tissues comprised the patient group, and the corresponding adjacent normal tissues comprised the control group. The Clopper–Pearson method was used for analysis with 95% confidence intervals (CIs). The area under the curve (AUC) was used to identify the efficiency of the indicated gene as a diagnostic biomarker. The Youden index was used as cutoff for predicting the positive (PPV) and negative predictive value (NPV).27
Overall survival (OS) analysis
The survival analysis based on the expression levels of RORγ was conducted in the GEPIA2 portal,28 with RORγ expression divided into quartiles. The highest cutoff was set at 75%, and the lowest cutoff was set at 25%. The results were reported as hazard ratios (HRs) and 95% CIs. The log-rank P-value was used to assess significant differences. Data were analyzed using the Kaplan–Meier method.
Statistical analysis
Data analysis was conducted using GraphPad 8.0 version software (GraphPad, San Diego, CA, USA). Differences between the two groups were analyzed using Student’s t-test. Pearson’s correlation analysis was used to assess correlations between variables. The log-rank test was used to analyze survival data. Significance was indicated by P < 0.05.
Results
Decreased expression of RORγ in colon cancer
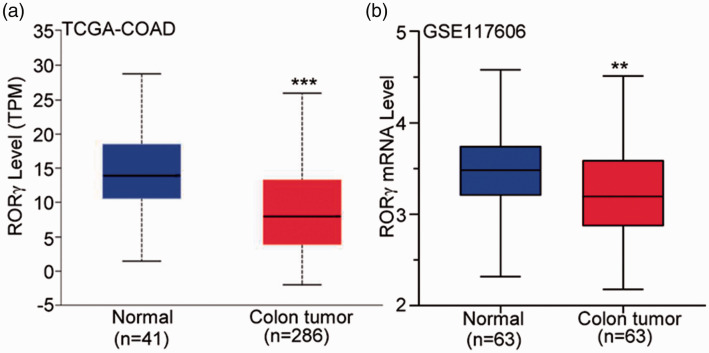
As presented in Figure 1a, RORγ mRNA expression was significantly lower in colon tumor tissues than in adjacent normal mucosa tissues (P < 0.001). Compared with the findings in the corresponding adjacent normal mucosa tissues collected from the same patients, RORγ expression was also downregulated in colon tumor tissues (P < 0.01, Figure 1b). These data suggest that RORγ expression was altered in colon cancer, and RORγ might influence the occurrence and development of colon cancer.
Figure 1.
RORγ expression was decreased in colon tumor tissue in datasets from TCGA and GEO. (a) The TCGA-COAD was used to compare RORγ expression between adjacent normal mucosa and tumor tissues. (b) GSE117606 from the GEO database was used to examine the mRNA expression of RORγ in primary colon tumors and corresponding normal mucosa tissues. **P < 0.01, ***P < 0.001 vs. adjacent normal mucosa tissues (Student’s t-test).
RORγ, retinoid-related orphan receptor gamma; TCGA, The Cancer Genome Atlas: GEO, Gene Expression Omnibus; COAD, colon adenocarcinoma dataset.
RORγ expression is correlated with the development of colon cancer
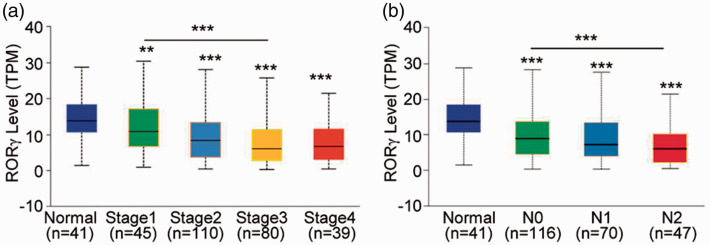
To further understand the potential role of RORγ in the development of colon cancer, RORγ expression was examined in patients with different stages of colon cancer and different lymph node metastasis statuses. As presented in Figure 2a, RORγ expression was remarkably decreased in patients with colon cancer, including stage 1 cancer (P < 0.01). With increasing colon cancer progression, RORγ expression continuously decreased, suggesting that RORγ is significantly downregulated by certain signaling pathways in colon tumors. Furthermore, we evaluated RORγ expression in patients with colon cancer with or without lymph node metastasis. The data illustrated that RORγ expression was lower in patients with lymph node metastasis (P < 0.01). In addition, RORγ expression was lower in tumor tissues in patients without lymph node metastasis than in the adjacent normal mucosa tissues (P < 0.001), which was consistent with the analysis of RORγ expression according to tumor stage (Figure 2b). From these data, we hypothesized that RORγ is correlated with the development of colon cancer, and RORγ could potentially reflect the clinicopathological features of colon cancer.
Figure 2.
RORγ expression was further decreased in patients with advanced colon cancer. (a) RORγ expression was examined in different stages of colon cancer in patients included in TCGA-COAD. (b) The patients from TCGA-COAD were divided into three groups (N0, N1, and N2) according to the lymph node metastasis, and RORγ expression was evaluated. The differences between groups were analyzed using Student’s t-test, **P < 0.01, ***P < 0.001.
RORγ, retinoid-related orphan receptor gamma; TCGA-COAD, The Cancer Genome Atlas colon adenocarcinoma dataset; N0, no regional lymph node metastasis; N1, metastasis in 1–3 axillary lymph nodes; N2, metastasis in 4–9 axillary lymph nodes.
Validation of decreased RORγ in colon cancer
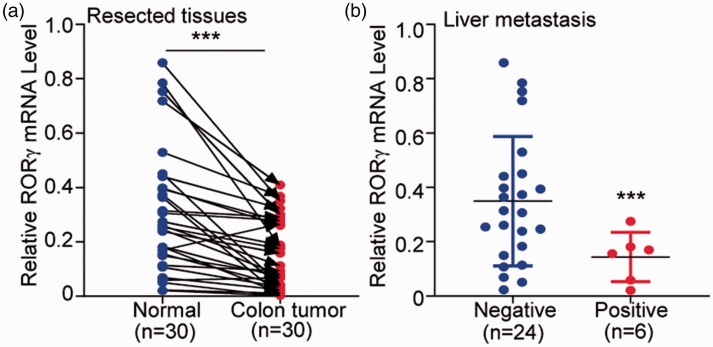
To validate our aforementioned hypothesis, 30 patients with colon cancer were included in our analysis. RORγ expression was significantly lower in colon tumors than in the corresponding adjacent normal mucosa (P < 0.001, Figure 3a). Because liver metastasis is a common poor prognostic factor for colon cancer,29,30 we also evaluated RORγ expression in patients with colon cancer with or without liver metastasis. The result illustrated RORγ expression was lower in the six patients with liver metastasis than in the 24 patients without liver metastasis (P < 0.001, Figure 3b). This experiment further validated the low expression of RORγ in colon cancer and its potential association with the progression of colon cancer.
Figure 3.
Real-time quantitative PCR analysis of resected colon tumor tissues validated the decreased expression of RORγ in colon cancer. (a) Thirty patients with colon cancer were included to examine RORγ mRNA in colon tumors and their corresponding adjacent normal mucosa tissues. (b) The patients were divided into two groups according to the presence of liver metastasis, and RORγ mRNA expression was compared between 24 patients without liver metastasis and six patients with liver metastasis. The differences between the groups were analyzed using a paired t-test. ***P < 0.001.
RORγ, retinoid-related orphan receptor gamma.
RORγ is a potential diagnostic marker for colon cancer
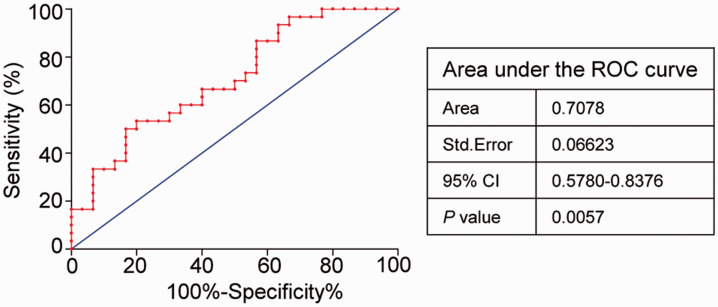
Subsequently, ROC analysis of the 30 aforementioned patients was performed to evaluate the efficiency of RORγ as a diagnostic factor. As highlighted in Figure 4, the AUC was 0.7078 (P = 0.0057), and using the Youden index as the cutoff, the PPV and NPV were 53.3% and 80%, respectively. These data suggest the RORγ has encouraging utility as a diagnostic marker in colon cancer.
Figure 4.
ROC analysis of RORγ expression in patients with colon cancer. In total, 30 patients with colon cancer were subjected to ROC analysis. The corresponding adjacent normal mucosa tissues were used as controls, and the colon tumor tissues comprised the patient group. The Clopper–Pearson method was used for the ROC analysis with 95% CIs. P < 0.05 indicated statistical significance.
ROC, receiver operating characteristic; RORγ, retinoid-related orphan receptor gamma; CI, confidence interval.
RORγ expression is closely correlated with the clinicopathological features of colon cancer
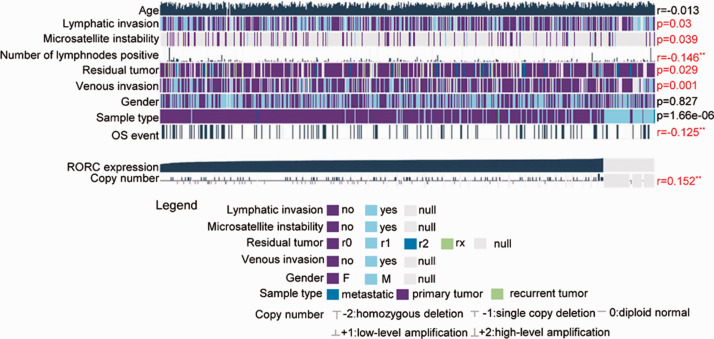
We then further analyzed the correlation between RORγ expression and the clinicopathological features of colon cancer. As illustrated in Figure 5, RORγ expression was closely correlated with the number of metastasis-positive lymph nodes (R = −0.146, P < 0.01) and OS (R = −0.125, P < 0.01). Furthermore, RORγ expression was closely associated with lymphatic invasion (P = 0.03), microsatellite instability (MSI, P = 0.039), residual tumor (P = 0.029), and venous invasion (P = 0.001). Similar to previous findings, RORγ expression had no significant association with patient age (R = −0.013, P > 0.05) and gender (P = 0.827). Interestingly, copy number variation (CNV) of RORγ was positively correlated with RORγ expression (R = 0.152, P < 0.01), suggesting that RORγ was expressed in a copy number-dependent manner. In addition, CNV of RORγ in the genome might contribute to its decreased expression.
Figure 5.
RORγ expression and its relationship with clinical TCGA-COAD data were determined using MEXPRESS. TCGA-COAD samples were grouped according to RORγ expression, and clinical information, including age, lymphatic invasion, microsatellite instability, the number of positive lymph nodes, the presence of residual tumor, venous invasion, gender, OS events, and RORγ copy number, were examined using Wilcoxon’s rank-sum test and Pearson’s correlation analysis. *P < 0.05, **P < 0.01.
RORγ, retinoid-related orphan receptor gamma; OS, overall survival; RORC, another name for RORγ.
Decreased RORγ expression portended a worse prognosis in colon cancer
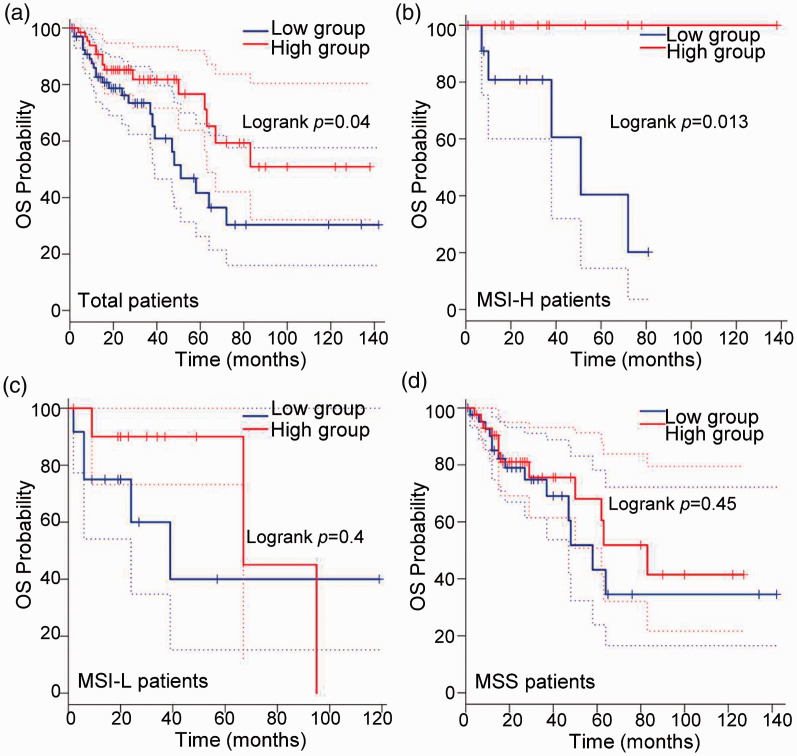
To further evaluate the effect of RORγ on the prognosis of colon cancer, Kaplan–Meier analysis was performed to evaluate the prognostic value of RORγ for patients with colon cancer. As presented in Figure 6a, patients with higher RORγ expression had longer OS, consistent with the result in Figure 5. From these data, we could conclude that decreased RORγ expression might be a poor prognostic marker in colon cancer. As indicated in Figure 5, RORγ was closely correlated with MSI. Because MSI is an important feature of colon cancer that contributes to genomic alteration,31,32 we analyzed the effect of the association of RORγ and MSI on OS. As indicated in Figure 6b–d, RORγ could represent a prognostic marker for patients with MSI-high (MSI-H) colon cancer (P = 0.013), but not those with MSI-low (MSI-L, P = 0.04) or microsatellite-stable (MSS) colon cancer (P = 0.45). These results suggest that RORγ is a probable prognostic marker for patients with colon cancer, especially those with MSI-H features.
Figure 6.
The overall survival of patients in TCGA-COAD was analyzed using GEPIA according to RORγ expression. (a) Patients in TCGA-COAD were analyzed according to RORγ expression, and patients in the highest and lowest quartiles for RORγ comprised the high and low RORγ expression groups, respectively. OS was determined via survival curve analysis. Patients with MSI-H (b), MSI-L (c), and MSS features (d) were included in the OS analysis, and patients in the highest and lowest quartiles for RORγ comprised the high and low RORγ expression groups, respectively. Statistical analysis was performed using the log-rank method, and P < 0.05 indicated statistical significance.
TCGA-COAD, The Cancer Genome Atlas colon adenocarcinoma dataset; RORγ, retinoid-related orphan receptor gamma; MSI-H, microsatellite instability-high; MSI-L, microsatellite instability-low; MSS, microsatellite-stable.
Abnormal methylation level of the RORγ promoter in colon cancer
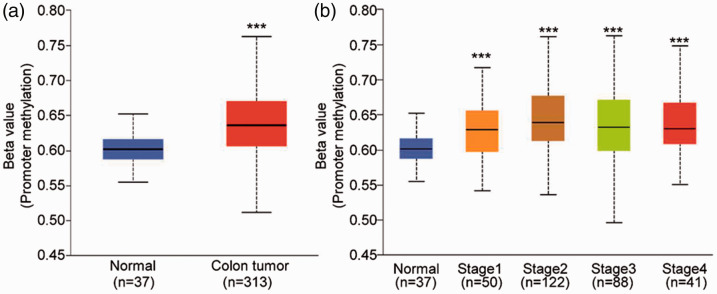
Considering the importance of CNV in the genome on the occurrence and development of cancer, the regulation of epigenetic features has a close association with CNV, and DNA methylation is a significant marker in colon cancer.33,34 Therefore, we examined the methylation level of RORγ in the colon. As presented in Figure 7a, the promoter methylation level of RORγ was remarkably increased in colon tumor tissues (P < 0.001). In addition, the extent of RORγ promoter methylation increased with increasing colon cancer progression (Figure 7b). The RORγ gene expression results mentioned in Figure 2a indicated that RORγ expression decreased with increasing cancer progression. Because of the common negative regulation between gene expression and methylation levels, the gene levels of RORγ were consistent with the results of promoter methylation levels in colon cancer. Thus, abnormal promoter methylation alters RORγ expression.
Figure 7.
The promoter methylation level of RORγ was analyzed in TCGA-COAD. (a) In total, 313 primary colon tumor tissues and 37 adjacent normal mucosa tissues were included to compare the promoter methylation level of RORγ. (b) Patients in TCGA-COAD with promoter methylation data were included and divided into four groups according to the tumor stage, and the promoter methylation level of RORγ was analyzed. ***P < 0.001 vs. adjacent normal mucosa tissues (Student’s t-test).
RORγ, retinoid-related orphan receptor gamma; TCGA-COAD, The Cancer Genome Atlas colon adenocarcinoma dataset.
Discussion
Colon cancer represents a key risk to human health, and it is the second leading cause of cancer-related death.1 Despite the use of surgery, chemotherapy, radiotherapy, targeted therapy, and combination therapy, the prognosis of colon cancer remains poor because of poor diagnostic accuracy and drug resistance.35 Thus, screening novel molecules is important work for improving colon cancer therapy.
As an important orphan receptor, RORγ has received increasing attention, but it has rarely been analyzed in colon cancer. RORγ is overexpressed in some cancer types. Huang reported that RORγ is highly expressed in liver tumor tissues, and its expression is closely associated with HBV infection, suggesting that RORγ is an oncogene involved in the proliferation and migration of hepatocellular carcinoma.18 Some reports also described RORγ overexpression in lung cancer, breast cancer, and melanoma. As a subtype of RORγ, RORγt was reported to inhibit the development of colon cancer,36 and RORγt expression in colon cancer remains controversial. Thus, we first evaluated RORγ expression in this study, and our data indicated that RORγ expression was significantly decreased in colon cancer (Figures 1 and 3a). In addition, the extent of RORγ downregulation was correlated with the progression of colon cancer, including the tumor stage (Figure 2a), lymph node metastasis (Figure 2b), and liver metastasis (Figure 3b), suggesting that RORγ is involved in the occurrence and development of colon cancer. RORγ possibly acts as an important mediator in the regulation of colon cancer. To further understand the importance of RORγ in colon cancer, the correlation between RORγ expression and certain clinicopathological features was evaluated, and the evidence indicated that RORγ expression is significantly correlated with lymphatic invasion, MSI, the presence of residual tumor, venous invasion, and OS (Figure 5).
We believe these data further demonstrate that RORγ downregulation is closely associated with the development of colon cancer. In addition, we found that RORγ downregulation might represent a good diagnostic marker with high sensitivity and specificity (Figure 4). Interestingly, we also found that CNV of RORγ was positively correlated with RORγ gene expression (Figure 5), suggesting that decreased RORγ expression in the colon reflects copy number-dependent gene expression, and CNV of RORγ is a potential explanation for RORγ downregulation. Based on the positive correlations of RORγ expression with clinicopathological features, we hypothesized that RORγ might be a good prognostic marker for the evaluation of patients with colon cancer. To confirm the hypothesis, OS was compared between patients with high and low RORγ expression, and the results demonstrated that lower RORγ expression portended worse survival (Figure 6a). Inspired by the significant correlation between RORγ expression and MSI, we further analyzed the prognostic value of RORγ in MSI-H, MSI-L, and MSS colon cancer, concluding that decreased RORγ expression might be a potential poor prognostic factor only in patients with MSI-H cancer (Figure 6b–d). Another issue deserving our attention was the potential mechanism of RORγ downregulation. As previously mentioned, RORγ expression might be copy number-dependent, and because of the importance of CNV in genomic alternation and the role of epigenetic regulation in the abnormal gene expression, we evaluated the correlation between RORγ methylation and RORγ expression. The level of RORγ promoter methylation was significantly higher in colon cancer, and RORγ hypermethylation is closely correlated with the progression of colon cancer (Figure 7), consistent with the increasing gene expression changes with cancer progression. In summary, we evaluated the expression and potential role of RORγ in this study, and RORγ downregulation is potentially involved in the progression of colon cancer, suggesting a possible role of RORγ in the diagnosis and prognosis of this malignancy.
Footnotes
Declaration of conflicting interest: The authors declare that there is no conflict of interest.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The authors acknowledge support from the Affiliated Hospital of West Anhui Health Vocational College.
ORCID iD: Xiaofei Pan https://orcid.org/0000-0002-4475-2795
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020; 70: 7–30. [DOI] [PubMed] [Google Scholar]
- 2.Wu H, Liu J, Yin Y, et al. Therapeutic Opportunities in Colorectal Cancer: Focus on Melatonin Antioncogenic Action. Biomed Res Int 2019; 2019: 9740568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang W, Yan Y, Chen M, et al. Aspirin enhances the sensitivity of colon cancer cells to cisplatin by abrogating the binding of NF-κB to the COX-2 promoter. Aging (Albany NY) 2020; 12: 611–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jalilzadeh N, Samadi N, Salehi R, et al. Novel nano-vehicle for delivery and efficiency of anticancer auraptene against colon cancer cells. Sci Rep 2020; 10: 1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pothuraju R, Rachagani S, Krishn SR, et al. Molecular implications of MUC5AC-CD44 axis in colorectal cancer progression and chemoresistance. Mol Cancer 2020; 19: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okugawa Y, Toiyama Y, Toden S, et al. Clinical significance of SNORA42 as an oncogene and a prognostic biomarker in colorectal cancer. Gut 2017; 66: 107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hector S, Rehm M, Schmid J, et al. Clinical application of a systems model of apoptosis execution for the prediction of colorectal cancer therapy responses and personalisation of therapy. Gut 2012; 61: 725–733. [DOI] [PubMed] [Google Scholar]
- 8.Billon C, Murray MH, Avdagic A, et al. RORγ regulates the NLRP3 inflammasome. J Biol Chem 2019; 294: 10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mühlbauer E, Bazwinsky-Wutschke I, Wolgast S, et al. Differential and day-time dependent expression of nuclear receptors RORα, RORβ, RORγ and RXRα in the rodent pancreas and islet. Mol Cell Endocrinol 2013; 365: 129–138. [DOI] [PubMed] [Google Scholar]
- 10.Yordanova IA, Cortés A, Klotz C, et al. RORγt(+) Treg to Th17 ratios correlate with susceptibility to Giardia infection. Sci Rep 2019; 9: 20328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Z, Unutmaz D, Zou YR, et al. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science 2000; 288: 2369–2373. [DOI] [PubMed] [Google Scholar]
- 12.Kurebayashi S, Ueda E, Sakaue M, et al. Retinoid-related orphan receptor gamma (RORgamma) is essential for lymphoid organogenesis and controls apoptosis during thymopoiesis. Proc Natl Acad Sci USA 2000; 97: 10132–10137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin L, Martynowski D, Zheng S, et al. Structural basis for hydroxycholesterols as natural ligands of orphan nuclear receptor RORgamma. Mol Endocrinol 2010; 24: 923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slominski AT, Kim TK, Hobrath JV, et al. Endogenously produced nonclassical vitamin D hydroxy-metabolites act as “biased” agonists on VDR and inverse agonists on RORα and RORγ. J Steroid Biochem Mol Biol 2017; 173: 42–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh TG, Wang SCM, Acharya BR, et al. The Nuclear Receptor, RORγ, Regulates Pathways Necessary for Breast Cancer Metastasis. EBioMedicine 2016; 6: 59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slominski AT, Kim TK, Hobrath JV, et al. Characterization of a new pathway that activates lumisterol in vivo to biologically active hydroxylumisterols. Sci Rep 2017; 7: 11434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffiths WJ, Abdel-Khalik J, Hearn T, et al. Current trends in oxysterol research. Biochem Soc Trans 2016; 44: 652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y, Liang H, He C, et al. Hepatitis B Virus X Protein-Induced RORγ Expression to Promote the Migration and Proliferation of Hepatocellular Carcinoma. Biomed Res Int 2019; 2019: 5407126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan J, Lv Z, Yang G, et al . Retinoic Acid Receptor-Related Orphan Receptors: Critical Roles in Tumorigenesis. Front Immunol. 2018; 9: 1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Zou JX, Xue X, et al. ROR-γ drives androgen receptor expression and represents a therapeutic target in castration-resistant prostate cancer. Nat Med 2016; 22: 488–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahalingam D, Wang JS, Hamilton EP, et al. Phase 1 Open-Label, Multicenter Study of First-in-Class RORγ Agonist LYC-55716 (Cintirorgon): Safety, Tolerability, and Preliminary Evidence of Antitumor Activity. Clin Cancer Res 2019; 25: 3508–3516. [DOI] [PubMed] [Google Scholar]
- 22.Wang RX, Liu H, Xu L, et al. Melatonin downregulates nuclear receptor RZR/RORγ expression causing growth-inhibitory and anti-angiogenesis activity in human gastric cancer cells in vitro and in vivo. Oncol Lett 2016; 12: 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oh TG, Bailey P, Dray E, et al. PRMT2 and RORγ expression are associated with breast cancer survival outcomes. Mol Endocrinol 2014; 28: 1166–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017; 19: 649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng H, Fan X, Wu C, et al. Cyclodextrin-Based Star-Like Amphiphilic Cationic Polymer as a Potential Pharmaceutical Carrier in Macrophages. Macromol Rapid Commun 2019; 40: e1800207. [DOI] [PubMed] [Google Scholar]
- 26.Cheng Y, Xu T, Li S, et al. GPX1, a biomarker for the diagnosis and prognosis of kidney cancer, promotes the progression of kidney cancer. Aging (Albany NY) 2019; 11: 12165–12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao L, Cheng H, Jiang Q, et al. APEX1 is a novel diagnostic and prognostic biomarker for hepatocellular carcinoma. Aging (Albany NY) 2020; 12: 4573–4591. 10.18632/aging.102913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019; 47: W556–W560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lafitte M, Sirvent A, Roche S. Collagen Kinase Receptors as Potential Therapeutic Targets in Metastatic Colon Cancer. Front Oncol 2020; 10: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snyder RA, Hao S, Irish W, et al. Thirty-Day Morbidity after Simultaneous Resection of Colorectal Cancer and Colorectal Liver Metastasis: American College of Surgeons NSQIP Analysis. J Am Coll Surg 2020; 230: 617–627.e9. [DOI] [PubMed] [Google Scholar]
- 31.Song Y, Wang L, Ran W, et al. Effect of Tumor Location on Clinicopathological and Molecular Markers in Colorectal Cancer in Eastern China Patients: An Analysis of 2,356 Cases. Front Genet 2020; 11: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allan RE, Luis RP, Juan P. Microsatellite instability in Costa Rican patients with colorectal adenocarcinoma and its association with overall survival and response to fluoropyrimidine-based chemotherapy. Cancer Epidemiol 2020; 65: 101680. [DOI] [PubMed] [Google Scholar]
- 33.Yang C, Zhang Y, Xu X, et al. Molecular subtypes based on DNA methylation predict prognosis in colon adenocarcinoma patients. Aging (Albany NY) 2019; 11: 11880–11892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kerachian MA, Javadmanesh A, Azghandi M, et al. Crosstalk between DNA methylation and gene expression in colorectal cancer, a potential plasma biomarker for tracing this tumor. Sci Rep 2020; 10: 2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Nie L, Xu K, et al. SIRT6, a novel direct transcriptional target of FoxO3a, mediates colon cancer therapy. Theranostics 2019; 9: 2380–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kathania M, Khare P, Zeng M, et al. Itch inhibits IL-17-mediated colon inflammation and tumorigenesis by ROR-γt ubiquitination. Nat Immunol 2016; 17: 997–1004. [DOI] [PubMed] [Google Scholar]