Abstract
Background
Bleeding is associated with a significantly increased morbidity and mortality. Bleeding events are often described in the unstructured text of electronic health records, which makes them difficult to identify by manual inspection.
Objectives
To develop a deep learning model that detects and visualizes bleeding events in electronic health records.
Patients/Methods
Three hundred electronic health records with International Classification of Diseases, Tenth Revision diagnosis codes for bleeding or leukemia were extracted. Each sentence in the electronic health record was annotated as positive or negative for bleeding. The annotated sentences were used to develop a deep learning model that detects bleeding at sentence and note level.
Results
On a balanced test set of 1178 sentences, the best‐performing deep learning model achieved a sensitivity of 0.90, specificity of 0.90, and negative predictive value of 0.90. On a test set consisting of 700 notes, of which 49 were positive for bleeding, the model achieved a note‐level sensitivity of 1.00, specificity of 0.52, and negative predictive value of 1.00. By using a sentence‐level model on a note level, the model can explain its predictions by visualizing the exact sentence in a note that contains information regarding bleeding. Moreover, we found that the model performed consistently well across different types of bleedings.
Conclusions
A deep learning model can be used to detect and visualize bleeding events in the free text of electronic health records. The deep learning model can thus facilitate systematic assessment of bleeding risk, and thereby optimize patient care and safety.
Keywords: decision support systems (clinical), deep learning, electronic health record, hemorrhage, international classification of diseases, machine learning
Essentials.
Bleeding events are difficult to locate in electronic health records.
A deep learning model detects bleeding events and visualizes them to the clinicians.
The model identified 90.0% of bleeding‐positive sentences and 89.6% of negative sentences
The model identified 100% of bleeding‐positive notes and 52.4% of negative notes.
1. INTRODUCTION
Bleeding occurs in 3.2% of medical patients within 14 days of admission, and approximately one‐third of the bleeding events are considered major events. 1 Bleeding is associated with a significantly increased morbidity and mortality. 2 , 3 Furthermore, previous clinically relevant bleeding events are a strong independent risk factor for future bleeding. 1 Hence, knowledge about bleeding history is essential for providing optimal care to patients.
In clinical practice, bleeding risk can be assessed using bleeding risk scores that include information about the patient's bleeding history, for example the HAS‐BLED (hypertension, abnormal renal and liver function, stroke, bleeding, labile international normalized ratio, elderly, drugs or alcohol) score, which is recommended for determining bleeding risk during anticoagulation treatment, 4 , 5 or the IMPROVE (International Medical Prevention Registry on Venous Thromboembolism) score, which is recommended to guide prophylactic anticoagulant treatment for adult medical patients at admission. 1
Although crucial for patient care, bleeding risk is not always systematically evaluated. Studies have shown that a large proportion of hospitalized medical patients do not get appropriate prophylactic anticoagulant treatment during admission. 6 , 7 , 8 One reason is that the recommended scoring systems for assessment of thrombosis and bleeding risk are not always used in clinical practice. 6 , 7 , 8 This could be caused by the fact that risk scores are laborious to obtain because it requires manual work to go through the electronic health record (EHR) for relevant information 7 and that it must be done at the time of admission when health care professionals are busy handling the acute situation.
In recent years, deep learning techniques have achieved state‐of‐the‐art performance on text classification benchmarks. 9 In medicine, various deep learning techniques have been used for text classification including, but not limited to, recurrent neural networks (RNNs), 10 , 11 , 12 convolutional neural networks (CNNs), 13 , 14 and hybrid models combining more than one technique. 15 These techniques have the potential for automatic detection of relevant clinical information in EHR text. This could facilitate the systematic assessment of bleeding risk and thereby optimize patient care and safety as well as freeing up time for health care professionals. To date, only a few studies have used deep learning for finding bleeding events in EHRs. 15 , 16 , 17
A general concern about deep learning is how the models reach their conclusions. It often remains a black box, making the users struggle to assess the basis for results or whether the model answers the questions for which clinicians want assistance. 18 , 19 Therefore, there is a growing awareness that deep learning models need to be self‐explanatory. 20 For text classification models, it means that it is relevant to show the prediction‐supporting part of the text upon request. However, such approaches are lacking in bleeding detection models.
Therefore, the purpose of this study was to establish a deep learning model that automatically detects bleeding events on a sentence level and to visualize the bleeding events to the clinician in the unstructured EHR text.
2. METHODS
2.1. Population and data set
Data were acquired from the EHR system of the Region of Southern Denmark. To ensure inclusion of EHR notes with a high likelihood of bleeding events in the text, we extracted EHRs from 300 patients with International Classification of Diseases, Tenth Revision (ICD‐10) diagnosis codes for bleeding or leukemia. ICD‐10 codes for bleeding from the following sites were included: eyes, ear‐nose‐throat and respiratory tract, gastrointestinal, urogenital, internal organs, hematoma, and others. EHRs from patients with leukemia were included, as this patient group has a high incidence of bleeding (see Appendix S1 for ICD‐10 codes). 21 Before annotation, we discarded administrative notes, as they would not contain any bleeding events.
Twelve physicians annotated the 300 EHRs. Each EHR was annotated by one physician. To determine the agreement between physicians’ annotation, we calculated the kappa score on a sample of 1328 sentences from randomly chosen EHRs.
The EHRs were annotated 22 on sentence level with two different labels:
Positive: Sentences that indicate any kind of bleeding.
Misinterpretable negative: Sentences that were deemed by the annotator to have a high risk of being misinterpreted by the deep learning model, for example, “The patient is not bleeding.”
All sentences left after annotation of positive and misinterpretable negative sentences were then considered negative sentences. We chose to annotate the misinterpretable negative sentences as a subcategory to the negative category to be able to feed many negative samples that resemble positive samples to the model. This should help the model distinguish for example “the patient has a bleeding” from “the patient might have a bleeding.”
Data were split into a balanced training (80%), validation (10%), and test set (10%) using subsampling of the overrepresented class. 23 The negative sentences consisted of 50% random negatives and 50% misinterpretable negatives. The training set was used to train the models, the validation set was used to tune parameters of the models during training, and the test set was used to evaluate final performance.
Sentences were tokenized using the Stanza sentence tokenizer. 24 Samples were preprocessed by elimination of superfluous spaces, special characters, and duplicate sentences.
2.2. Models for detection of bleeding events on sentence level
2.2.1. Rule‐based classifier
A rule‐based classifier was developed to compare the deep learning models with a traditional approach to text classification.
The rule‐based model was constructed by defining a set of bleeding‐indicating words and modifiers using corpus statistics and manual inspection of the data. Corpus statistics were used to calculate the most frequent words in bleeding‐indicating sentences. A bleeding‐indicating word could for example be bleeding, and a modifier could, for example, be no (bleeding). Next, by evaluating performance on the training data, a window size was defined where a modifier could modify a bleeding‐indicating word. For example, no would modify bleeding in “no sign of bleeding” for a window size of 3. The model uses the indicating and modifying words and the window size to create rules for classifying individual sentences. The rules were iteratively updated during training to improve performance.
2.2.2. Deep learning models
Three different deep learning models were developed: a CNN model, an RNN model, and a hybrid model combining an RNN and a CNN. In deep learning, a model transforms the input to a classification via many layers of processing steps that are learned from labeled data during training. The input to the models is the individual words from each sentence represented as word embeddings. Word embeddings are numerical vector representations of words that encode their meaning with similar words having similar vectors. For this study, 100‐dimensional GloVe word embeddings pretrained on 323 122 Danish EHRs were used. 25 , 26
2.3. Evaluation of internal validity
We performed an internal sensitivity analysis on the best‐performing model to evaluate if it performs equally well on the seven patient groups included in the study.
2.4. Bleeding detection on note level
Because each note may contain multiple positive sentences that often describe the same bleeding event, we calculated the performance of the best model on a note level by classifying all sentences of each note. A positive note is defined as a note that includes at least one bleeding‐positive sentence. The test was performed on seven randomly selected EHRs from patients in the leukemia group not included in the original data set. A total of 100 notes per EHR were collected.
2.5. Visualization of bleeding events in EHR text
Finally, we present how the bleeding‐positive output of the model can be presented to the physician as a visualization of complete notes with the bleeding events highlighted, helping the physician understand the prediction and decreasing the time needed to find a bleeding event in an EHR.
2.6. Statistical analysis
We calculated accuracy, sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and a harmonic mean of sensitivity and positive predictive value (F1) score. For each model, we plotted receiver operating characteristic curves and calculated area under the receiver operating characteristic curve (AUC).
The models were developed in Python 3.6 (Python Software Foundation, Wilmington, DE, USA) using the Tensorflow 2.0 framework.
3. RESULTS
The 300 extracted EHRs contained 88 477 notes. Of those, we filtered out 43 602 as administrative notes. The remaining 44 875 EHR notes were annotated on a sentence level. In total, 6111 sentences were annotated as positive and 5630 as misinterpretable negative. Overall, 3973 notes contained bleeding events and there were 1 to 19 positive sentences per note. The EHRs contained 0 to 108 notes with bleeding per patient.
Among the different patient groups, “gastrointestinal bleeding” had the highest average number of positive sentences per EHR (n = 25) while “Hematomas and other bleedings” had the lowest (n = 8; see Table 1). Although the EHRs were extracted on the basis of ICD‐10 codes for bleeding, 13 EHRs did not contain any information about bleeding (“internal bleedings,” n = 2; “eyes,” n = 5; and “hematomas and others,” n = 6). Another 5 EHRs from leukemia patients did not contain bleeding events.
TABLE 1.
Patient group distribution of extracted EHRs
| Patient group | Number of EHRs | Number of EHR notes | Number of positive EHR notes | Number of positive sentences | Average number of positive sentences per EHR |
|---|---|---|---|---|---|
| Eye bleeding | 65 | 7781 | 771 | 1546 | 24 |
| Ear‐nose‐throat and respiratory tract bleeding | 23 | 3702 | 372 | 532 | 23 |
| Gastrointestinal bleeding | 51 | 6968 | 1,055 | 1,250 | 25 |
| Urogenital bleeding | 45 | 4409 | 499 | 855 | 19 |
| Internal organ bleeding | 45 | 6078 | 753 | 1,082 | 24 |
| Hematoma and other bleeding | 38 | 5597 | 229 | 319 | 8 |
| Leukemia bleeding | 33 | 10 340 | 294 | 527 | 16 |
| Total | 300 | 44 875 | 3973 | 6111 | … |
Abbreviation: EHR, electronic health record.
When assessing agreement among the 12 physicians, they achieved a kappa score of 0.75 on a sample of 1328 sentences from randomly chosen EHRs. This is considered a substantial agreement. 27
3.1. Establishing models
For development of models, we removed duplicate sentences (n = 218), resulting in 5893 positive samples. To create a balanced data set, we randomly subsampled 2947 misinterpretable negative sentences. These were added to 2946 randomly extracted negative samples to give 5893 total negative samples. Together with the 5893 positive samples, they constitute the balanced data set of 11 786 samples.
The balanced data set was divided into training (n = 9430), validation (n = 1178), and test sets (n = 1178). The distribution is seen in Table S1.
3.1.1. Rule‐based results
The bleeding‐indicating and modifying words were aggregated into a stem to capture different conjugations; for example, the Danish word for hemorrhage (hæmoragi) was aggregated to hæm and defined as a bleeding‐indicating word.
The developed rule‐based classification model searched each sentence for a positive word. If no positive words were found, the sentence was classified as negative. If a positive word was found, the model searched its context words in a window of size 4 to look for negative modifiers. If a word from the positive list was not accompanied by a negative modifier, the sample was classified as positive. If all positive words were accompanied by negative modifiers, the sample was classified as negative.
3.1.2. Deep learning
The developed CNN consisted of convolutional layers that extract information from neighboring words. The extracted information was used by a linear classification layer that classifies the sentence as either bleeding present or bleeding absent.
Our RNN model was based on the Bidirectional Gated Recurrent Unit (BiGRU). 28 The model consisted of a single BiGRU layer that extracts information from the input words by processing them sequentially. The extracted information was used by a linear classification layer that classifies the sentence.
The hybrid model used the output from both a CNN and an RNN to classify the sentences. This model was developed to exploit the information extracted from both a CNN and RNN in a final linear classification layer.
A more thorough description of the models can be seen in Appendix S2.
For each deep learning model, the seven versions of the model that performed best on the validation set were selected for an ensemble classifier. The ensemble classifier averages the predictions of each model to a final prediction.
3.2. Performance of models for bleeding detection in EHRs on sentence level
Table 2 shows the performance of the rule‐based and deep learning classifiers on the test set.
TABLE 2.
Performance of models for detecting bleeding in electronic health records on sentence level
| Rule‐based | CNN | RNN | Hybrid | |
|---|---|---|---|---|
| Accuracy | 0.80 | 0.89 | 0.89 | 0.90 |
| Sensitivity | 0.86 | 0.90 | 0.89 | 0.90 |
| Specificity | 0.72 | 0.89 | 0.88 | 0.90 |
| Positive predictive value | 0.76 | 0.89 | 0.88 | 0.90 |
| Negative predictive value | 0.84 | 0.90 | 0.90 | 0.90 |
| F1 score | 0.81 | 0.89 | 0.89 | 0.90 |
| AUC | 0.79 | 0.89 | 0.89 | 0.90 |
Abbreviations: AUC, area under the receiver operating characteristic curve; CNN, convolutional neural network; F1, harmonic mean of sensitivity and positive predictive value; RNN, recurrent neural network.
Figure 1 shows the ROC curves of the hybrid, CNN, RNN, and rule‐based models with their corresponding AUC.
FIGURE 1.
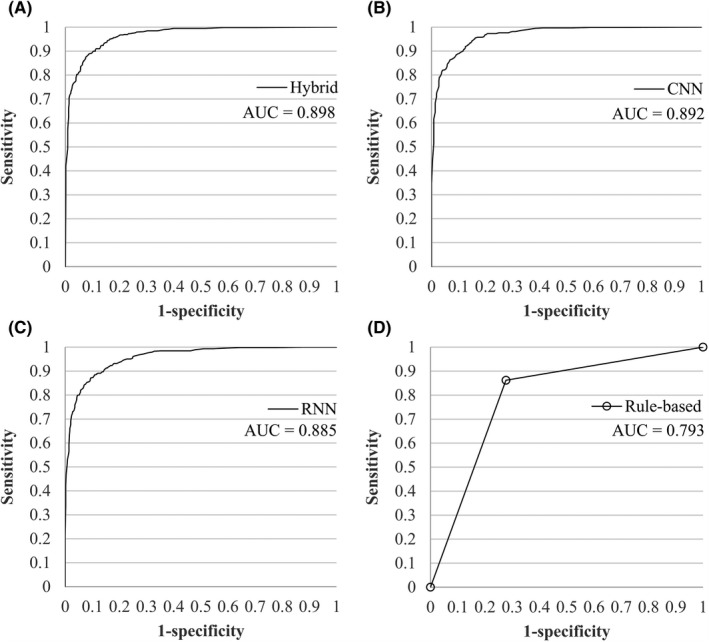
ROC curves and AUC for all models on sentence level. (A) Hybrid model. (B) CNN model. (C) RNN model. (D) Rule‐based model. AUC, area under the curve; CNN, convolutional neural network; RNN, recurrent neural network; ROC, receiver operating characteristic
Overall, the performance of the hybrid model was the best. It achieved an F1 score of 0.90, a sensitivity of 0.90, a specificity of 0.90, a PPV of 0.90, and an NPV of 0.90. The CNN model achieved equally high sensitivity of 0.90 but performed slightly worse on the additional metrics, while the RNN performed consistently worse on all metrics against both the hybrid and CNN model. The rule‐based model performed worse than all deep learning models.
3.3. Test of internal validity
We evaluated the hybrid model’s performance within each of the different patient groups on sentence level (Figure 2). It was calculated on the full data set including training, validation, and test sets. The model shows an almost equal performance for all patient groups, highest for “eyes” at 0.98, and lowest for “leukemia” at 0.95.
FIGURE 2.
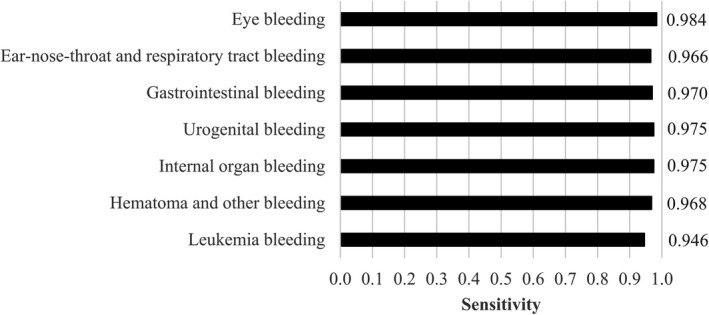
Internal validity for detection of bleeding on note level for the hybrid model
3.4. Performance of the hybrid model on note level
We further tested the performance of the hybrid model on a note level by classifying all sentences and aggregating the result to the full note. The seven EHRs contained 700 notes, of which 49 were positive. The hybrid model achieved a sensitivity of 1.00, a specificity of 0.52, a PPV of 0.14, an NPV of 1.00, an F1 score of 0.24, and an AUC of 0.76.
3.5. Visualization of bleeding events in EHR text
In this study, we chose to use the sentence‐level model on a note level because it makes the model capable of explaining its predictions. The model outputs all notes with predicted bleeding events, highlighting the sentence(s) found to indicate bleeding (representative example translated to English in Figure 3, original in Figure S1).
FIGURE 3.

Example of the visualization of bleeding events in an electronic health record note. To keep the original format, the text is translated directly from Danish to English, which results in incorrect sentence structures
4. DISCUSSION
We present a deep learning model that automatically detects bleeding events in EHRs with a sensitivity of 0.90 on sentence level and 1.00 on note level. This enables clinicians to receive automatic visualization of EHR notes with bleeding events. The hybrid model, combining an RNN and a CNN, performed best for bleeding detection on sentence level (F1 = 0.90).
In congruence with our study, others have found that machine learning can be used for finding bleeding in EHRs. Rumeng et al. 15 used a deep learning model to detect bleeding events in sentences of EHRs (F1 = 0.94). The study comprised a data set of 2902 sentences extracted from 878 notes from patients with cardiovascular events. Taggart et al. 17 detected bleeding events at a note level with a rule‐based approach (F1 = 0.74) and a CNN (F1 = 0.40) on a test set consisting of 660 notes. The rule‐based model was trained on 990 notes and the CNN was trained on 450 notes.
In contrast to our study, Taggart et al. found that their rule‐based approach performed better than their CNN but it may, however, be due to the limited amount of training data for the CNN, which is a well‐known limitation in machine learning. 29 , 30 Rumeng et al. also used a small data set, and moreover, the data used were exclusively from patients with cardiovascular events. Therefore, in the above studies, the data sets might not be representative of bleeding in all sites and the model might not be generalizable to other patient groups. The model presented in our study used a data set of 11 786 sentences extracted from 44 875 notes representative for multiple types of bleeding. In the internal validity test, we found that our model generalizes well to different types of bleeding.
Lee et al. 16 used a rule‐based (sensitivity = 0.83), machine learning (sensitivity = 1.00), and score function (sensitivity = 0.98) approach to find clopidogrel‐induced bleedings in EHRs. They defined bleeding events as the presence of specific ICD, Ninth Revision (ICD‐9) codes, specific keywords, and unique identifiers of the Unified Medical Language System related to bleeding. Thus, the bleeding definition was simplified to specific words, which is a limitation for use in clinical practice, as bleeding can be reported with numerous different phrases in EHRs. In agreement, we found thousands of different sentences that corresponded to bleeding according to the physicians involved. Moreover, the construction of keyword and rule lists requires manual effort that is difficult to scale because of the unstructured and noisy nature of the clinical notes (eg, grammatical ambiguity, synonyms, term abbreviation, misspelling, or negation of concepts). 31
Additionally, validation of ICD‐9 and ICD‐10 diagnosis codes has shown that they are not always accurate. 32 , 33 However, the major concern is that diagnosis coding requires manual collection of the patient history to choose the codes of relevance and that bleeding events that are not a major contributing cause of admittance are not registered with a code for bleeding. The present study provides an attractive alternative by leveraging the information‐rich yet unstructured text data in clinical notes in EHRs, which are currently often omitted when developing models. 34
In the present approach, we established a deep learning model that points out relevant information in the EHRs on sentence level. The advantage of making a sentence‐level classifier is that it enables the model to explain its predictions on a note level by showing the prediction‐supporting part of the text. We were, therefore, able to visualize where in the notes the model has detected a bleeding event, which enables us to point out relevant sentence(s) in the long unstructured EHR text for the physician. A fast overview of patient bleeding history facilitates clinical decision making. Accordingly, studies have shown that clinical practice may improve when decision support systems give automatic recommendations where the decision is interpretable and understandable for the physician. 35 , 36
An automatic summary of bleeding history may be valuable in clinical practice to diagnose, monitor disease, or address treatment options. The presented approach can be extended to include other symptoms and findings. Information regarding specific past events, for example, bleeding events during medical procedures, is important when planning a new medical procedure. Thus, the information may have an impact on patient safety because, for example, procedure and operation bleeding risk and medication side effects can be monitored effectively. It may also prove useful for health care statistics and resource management. Finally, the approach may save time because a focused review of an EHR to find all past bleeding events is very time consuming. Thus, it provides more time for direct patient care.
To summarize the main points of the discussion, comparing the related studies, the current study used the largest annotated corpus, providing an advantage to the deep learning model. This study also included many different types of bleeding, and it evaluated model accuracy by type of bleeding. In contrast to Taggart et al., we found that a deep learning approach works better than a rule‐based approach. We additionally show a simple approach to visualizing the sentences indicating bleeding to physicians, allowing for interpretation of the deep learning model.
4.1. Limitations
The rule‐based algorithm may have been further optimized by being more specific on search terms with inclusion of more words and their common misspellings instead of using more global terms to group words; for example, the Danish stem hæm may find words with various meanings that do not imply bleeding. Another limitation is that the study included only EHRs with an ICD‐10 code of bleeding, which does not capture all EHRs with bleeding events. Additionally, we did not validate the algorithm on an independent cohort. Of note, we found a high sensitivity for bleeding in EHRs from patients with leukemia, who comprise a patient group experiencing bleeding from different organ systems. 21 It thus suggests that the model performs well on EHRs without ICD‐10 for bleeding. It is crucial that the text that we used for training the model is representative for any way that bleeding can be reported in the EHR. It is a limitation to the study that we cannot guarantee this, and it would be beneficial to include a larger and more general data set. Nevertheless, this approach clearly showed that it is feasible to automatically extract and visualize bleeding events in EHRs. Future research shall focus on developing a model on data including even more bleeding types and optimizing the strategy, which includes differentiation between clinically relevant versus trivial bleedings and surgical versus medical bleeding.
5. CONCLUSION
We have developed a deep learning model that identifies bleeding events in EHRs with a sensitivity of 0.90 on sentence level and 1.00 on note level. Further, we have shown how bleeding‐positive notes can be visualized to physicians, making the model easily interpretable to the clinician.
AUTHOR CONTRIBUTIONS
MSL and JSP contributed equally in the analysis of data and production of results. PJV, TRS, and SAJ contributed to the design and conception of the study. PJV, RSH, ABA, KVB, IMK, CG, AFT, ESA, CBN, and LCA annotated the electronic health records.
RELATIONSHIP DISCLOSURE
The authors declare no conflicts of interest.
Supporting information
Supplementary Material
Figure S1
Figure S2
Figure S3
Figure S4
ACKNOWLEDGMENTS
The authors thank Helene Kirkegaard, Camilla Brødsgaard Nielsen, and Kristina Bjerg Appel for contributing to the annotation process, as well as Jan Hellden for extracting data.
Jannik S. Pedersen and Martin S. Laursen are co‐first authors.
Handling Editor: Dr Lana Castellucci
REFERENCES
- 1. Decousus H, Tapson VF, Bergmann JF et al. Factors at admission associated with bleeding risk in medical patients: findings from the IMPROVE investigators. Chest. 2011;139(1):69‐79. 10.1378/chest.09-3081 [DOI] [PubMed] [Google Scholar]
- 2. Berger JS, Bhatt DL, Steg PG et al. Bleeding, mortality, and antiplatelet therapy: Results from the Clopidogrel for High Atherothrombotic Risk and Ischemic Stabilization, Management, and Avoidance (CHARISMA) trial. Am Heart J. 2011;162(1):98‐105.e1. 10.1016/j.ahj.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 3. Cook DJ, Griffith LE, Walter SD et al. The attribute mortality and length of intensive care unit stay of clinically important gastrointestinal bleeding in critically ill patients. Crit Care. 2001;5(6):368‐375. 10.1186/cc1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pisters R, Lane DA, Nieuwlaat R et al. A novel user‐friendly score (HAS‐BLED) to assess 1‐year risk of major bleeding in patients with atrial fibrillation: The Euro Heart Survey. Chest. 2010;138(5):1093‐1100. 10.1378/chest.10-0134. [DOI] [PubMed] [Google Scholar]
- 5. Rodeghiero F, Tosetto A, Abshire T et al. ISTH/SSC bleeding assessment tool: a standardized questionnaire and a proposal for a new bleeding score for inherited bleeding disorders. J Thromb Haemost. 2010;8(9):2063‐2065. 10.1111/j.1538-7836.2010.03975.x. [DOI] [PubMed] [Google Scholar]
- 6. Amin A, Stemkowski S, Lin J, Yang G. Thromboprophylaxis rates in US medical centers: success or failure? J Thromb Haemost. 2007;5(8):1610‐1616. 10.1111/j.1538-7836.2007.02650.x. [DOI] [PubMed] [Google Scholar]
- 7. Rwabihama JP, Audureau E, Laurent M et al. Prophylaxis of venous thromboembolism in geriatric settings: a cluster‐randomized multicomponent interventional trial. J Am Med Dir Assoc. 2018;19(6):497‐503. 10.1016/j.jamda.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 8. Amin A, Stemkowski S, Lin J, Yang G. Appropriate thromboprophylaxis in hospitalized cancer patients. Clin Adv Hematol Oncol. 2008;6(12):910‐920. [PubMed] [Google Scholar]
- 9. Minaee S, Kalchbrenner N, Cambria E, Nikzad N, Chenaghlu M, Gao J. Deep learning based text classification: a comprehensive review. arXiv Prepr. Published online 2020. https://arxiv.org/pdf/2004.03705.pdf#:~:text=Deeplearningbasedmodelshave,answering%2Candnaturallanguageinference
- 10. Santiso S, Pérez A, Casillas A. Exploring joint AB‐LSTM with embedded lemmas for adverse drug reaction discovery. IEEE J Biomed Heal Informatics. 2019;23(5):2148‐2155. 10.1109/JBHI.2018.2879744. [DOI] [PubMed] [Google Scholar]
- 11. Chen D, Qian G, Pan Q. Breast cancer classification with electronic medical records using hierarchical attention bidirectional networks. In: Proceedings ‐ 2018 IEEE International Conference on Bioinformatics and Biomedicine, BIBM 2018; 2019. 10.1109/BIBM.2018.8621479 [DOI]
- 12. Deng Y, Dolog P, Gass JM, Denecke K. Obesity entity extraction from real outpatient records: When learning‐based methods meet small imbalanced medical data sets. In: Proceedings ‐ IEEE Symposium on Computer‐Based Medical Systems. 2019. 10.1109/CBMS.2019.00087 [DOI]
- 13. Rajput K, Chetty G, Davey R. Deep neural models for chronic disease status detection in free text clinical records. In: IEEE International Conference on Data Mining Workshops, ICDMW. 2019. 10.1109/ICDMW.2018.00127 [DOI]
- 14. Hughes M, Li I, Kotoulas S, Suzumura T. Medical text classification using convolutional neural networks. Stud Health Technol Inform. 2017;235:246‐250. 10.3233/978-1-61499-753-5-246. [DOI] [PubMed] [Google Scholar]
- 15. Li R, Hu B, Liu F et al. Detection of bleeding events in electronic health record notes using convolutional neural network models enhanced with recurrent neural network autoencoders: deep learning approach. J Med Internet Res. 2019;7(1):e10788. 10.2196/10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee H‐J, Jiang M, Wu Y et al. A comparative study of different methods for automatic identification of clopidogrel‐induced bleedings in electronic health records. AMIA Jt Summits Transl Sci proceedings AMIA Jt Summits Transl Sci. Published online 2017. [PMC free article] [PubMed]
- 17. Taggart M, Chapman WW, Steinberg BA et al. Comparison of 2 natural language processing methods for identification of bleeding among critically ill patients. JAMA Netw Open. 2018;1(6):e183451. 10.1001/jamanetworkopen.2018.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shortliffe EH, Sepúlveda MJ. Clinical decision support in the era of artificial intelligence. JAMA. 2018;320(21):2199. 10.1001/jama.2018.17163. [DOI] [PubMed] [Google Scholar]
- 19. Wang F, Kaushal R, Khullar D. Should health care demand interpretable artificial intelligence or accept “black box” medicine? Ann Intern Med. 2020;172(1):59. 10.7326/M19-2548. [DOI] [PubMed] [Google Scholar]
- 20. Rudin C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat Mach Intell. 2019;1(5):206‐215. 10.1038/s42256-019-0048-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Webert KE, Cook RJ, Sigouin CS, Rebulla P, Heddle NM. The risk of bleeding in thrombocytopenic patients with acute myeloid leukemia. Haematologica. 2006;91(11):1530‐1537. [PubMed] [Google Scholar]
- 22. Pustejovsky J, Stubbs A. Natural Language Annotation for Machine Learning: A Guide to Corpus‐Building for Applications. 1005 Gravenstein Highway North, Sebastopol, CA 95472: O’Reilly Media, Inc.; 2012. [Google Scholar]
- 23. Mazurowski MA, Habas PA, Zurada JM, Lo JY, Baker JA, Tourassi GD. Training neural network classifiers for medical decision making: The effects of imbalanced datasets on classification performance. Neural Networks. 2008;21(2‐3):427‐436. 10.1016/j.neunet.2007.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qi P, Zhang Y, Zhang Y, Bolton J, Manning CD. Stanza: a python natural language processing toolkit for many human languages. In; 2020. 10.18653/v1/2020.acl-demos.14 [DOI]
- 25. Rasmussen M, Berggrein N. Named entity recognition and disambiguation in Danish electronic health records. Published online. 2019. https://www.derczynski.com/itu/docs/Named_Entity_Recognition_and_Disambiguation_MSc_Thesis%202019.pdf, Accesed 4 april 2021
- 26. Pantazos K, Lauesen S, Lippert S. Preserving medical correctness, readability and consistency in de‐identified health records. Health Informatics J. 2017;23(4):291‐303. 10.1177/1460458216647760. [DOI] [PubMed] [Google Scholar]
- 27. Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159. 10.2307/2529310. [DOI] [PubMed] [Google Scholar]
- 28. Cho K, Van Merriënboer B, Gulcehre C et al. Learning phrase representations using RNN encoder‐decoder for statistical machine translation. In: EMNLP 2014 ‐ 2014 Conference on Empirical Methods in Natural Language Processing, Proceedings of the Conference. 2014. 10.3115/v1/d14-1179 [DOI]
- 29. Sun C, Shrivastava A, Singh S, Gupta A. Revisiting unreasonable effectiveness of data in deep learning era. In: Proceedings of the IEEE International Conference on Computer Vision; 2017. 10.1109/ICCV.2017.97 [DOI]
- 30. Banko M, Brill E. Mitigating the paucity‐of‐data problem: exploring the effect of training corpus size on classifier performance for natural language processing. Comput Linguist. 2001. 10.3115/1072133.1072204. [DOI] [Google Scholar]
- 31. Zeng Z, Deng Y, Li X, Naumann T, Luo Y. Natural language processing for EHR‐based computational phenotyping. IEEE/ACM Trans Comput Biol Bioinforma. 2019;16(1):139‐153. 10.1109/TCBB.2018.2849968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Valkhoff VE, Coloma PM, Masclee GMC et al. Validation study in four health‐care databases: upper gastrointestinal bleeding misclassification affects precision but not magnitude of drug‐related upper gastrointestinal bleeding risk. J Clin Epidemiol. 2014;67(8):921‐931. 10.1016/j.jclinepi.2014.02.020 [DOI] [PubMed] [Google Scholar]
- 33. Øie LR, Madsbu MA, Giannadakis C et al. Validation of intracranial hemorrhage in the Norwegian patient registry. Brain Behav. 2018;8(2):e00900. 10.1002/brb3.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Esteva A, Robicquet A, Ramsundar B et al. A guide to deep learning in healthcare. Nat Med. 2019;25(1):24‐29. 10.1038/s41591-018-0316-z. [DOI] [PubMed] [Google Scholar]
- 35. Berner ES, La Lande TJ. (2017). Overview of clinical decision support systems. In Clinical decision support systems. (pp. 3‐22). Springer, New York, NY.. 10.1007/978-3-319-31913-1_1 [DOI] [Google Scholar]
- 36. Kawamoto K, Houlihan CA, Balas EA, Lobach DF. Improving clinical practice using clinical decision support systems: a systematic review of trials to identify features critical to success. Br Med J. 2005;330(7494):765. 10.1136/bmj.38398.500764.8F [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Figure S1
Figure S2
Figure S3
Figure S4


