Graphical abstract
Keywords: G-quadruplex, RHAU helicase, DHX36, DEAH-box family, Molecular dynamics, Free energy simulation
Abstract
Because of high stability and slow unfolding rates of G-quadruplexes (G4), cells have evolved specialized helicases that disrupt these non-canonical DNA and RNA structures in an ATP-dependent manner. One example is DHX36, a DEAH-box helicase, which participates in gene expression and replication by recognizing and unwinding parallel G4s. Here, we studied the molecular basis for the high affinity and specificity of DHX36 for parallel-type G4s using all-atom molecular dynamics simulations. By computing binding free energies, we found that the two main G4-interacting subdomains of DHX36, DSM and OB, separately exhibit high G4 affinity but they act cooperatively to recognize two distinctive features of parallel G4s: the exposed planar face of a guanine tetrad and the unique backbone conformation of a continuous guanine tract, respectively. Our results also show that DSM-mediated interactions are the main contributor to the binding free energy and rely on making extensive van der Waals contacts between the GXXXG motifs and hydrophobic residues of DSM and a flat guanine plane. Accordingly, the sterically more accessible 5′-G-tetrad allows for more favorable van der Waals and hydrophobic interactions which leads to the preferential binding of DSM to the 5′-side. In contrast to DSM, OB binds to G4 mostly through polar interactions by flexibly adapting to the 5′-terminal guanine tract to form a number of strong hydrogen bonds with the backbone phosphate groups. We also identified a third DHX36/G4 interaction site formed by the flexible loop missing in the crystal structure.
1. Introduction
G-quadruplexes (G4s) are four-stranded secondary structures of nucleic acids in which four guanines associate by Hoogsteen hydrogen bonds into planar G-tetrads stabilized by monovalent cations [1], [2]. DNA G-quadruplexes are polymorphic and can adopt diverse topologies [3], [4] depending on some intrinsic properties, such as the base composition of the intervening sequences (termed loops) [5], [6] and flanking nucleotides [7], as well as environmental conditions, such as type and concentration of ions [8], [9] and molecular crowding [10], [11]. In contrast, structural diversity of RNA G-quadruplexes is limited to the one dominant parallel topology, in which all four guanine tracts (G-tracts) are oriented in the same direction [12], [13].
Although G4s can be multi-molecular, in cells, they are usually formed by single guanine-rich strands [14], [15] and are enriched in telomeric and regulatory regions of the genome [16], [17], [18]. In fact, a recent genome-wide detection in living cells [19] has revealed that G4s are present in >60% of gene promoters (especially at the transcription start site) and in ~70% of genes. In particular, they are found at promoters of some protooncogenes, ribosomal DNA and immunoglobulin switch regions [15], [20], [21], [22], as well as in replication origins [23]. Even though their biological relevance is not fully understood, some guanine-rich RNA sequences, such as telomeric RNA repeats (TERRA) and 3′- and 5′-untranslated regions of mRNA (UTRs), are also known to form stable G4s [24], [25], [26].
There is mounting evidence that maintaining a delicate balance between folded and unfolded G4 motifs within regulatory genomic regions plays an important role in controlling gene expression and DNA replication [27], [28], [29]. Indeed, it has been found that, on the one hand, G4s present intrinsic obstacles to DNA and RNA synthesis by polymerases [30], but, on the other, their formation might be involved in activation of some DNA replication origins as well as in recruiting transcription factors to promoters [31], [23]. Furthermore, it has also been shown that G4s forming within UTRs increase the stability of mature mRNA and affect translation efficiency [24], [28], [32], while those present in pre-mRNAs’ intronic regions can act as both, splicing enhancers and silencers [33], [34].
The role of G4s as regulatory genomic elements requires that their formation and unwinding is temporally controlled during cell cycle progression [26], [35], [36], [37]. As a result of high thermodynamic stability and slow unfolding rates of G4s, cells have evolved specialized helicases that disrupt DNA and RNA G4s in an ATP-dependent manner [27], [38], [39], [40]. A prominent example is DHX36, a member of the DEAH/RHAU family of helicases [41], [42], which – among other functions – participates in transcriptional regulation [43], [44], [45] and prevents the accumulation of translationally inactive mRNAs with G4-structures in untranslated regions [46]. Even though the coupling between ATP hydrolysis and G4 unfolding mediated by DHX36 are poorly understood at the molecular level, Chen et al. [47] proposed, consistently with previous biochemical studies [48], [49], that a passive destabilization of G4 occurs upon its binding to DHX36 and futher (active) unfolding happens during the major conformational change of the helicase driven by ATP hydrolysis. Alternatively, it has been suggested that DHX36 undergoes ATP-independent conformational transition that exerts force on the 3′-tail of the bound parallel-stranded G4 leading to its unwinding by one nucleotide at a time [50]. Upon ATP hydrolysis, the partially destabilized G-quadruplex is then released from the helicase [51], [50].
Another intriguing question is what underlies the DHX36 high specificity for binding and unfolding DNA and RNA G-quadruplexes with parallel topology. The recent crystal structure of the DHX36/G4 complex does not fully answers this question, as it captures partially-destabilized, “register-shifted” G4 with 5′-tetrad composed of two guanines, adenine and thymine (G · G · A · T) (Fig. 1AB) [50]. Additionally, the reported structure does not contain two fragments, potentially important for interactions with G4: a 20-residue linker between the DHX36-specific motif (DSM) and RecA1 and a 13-residue loop in RecA2, located in the proximity of the G4 binding interface. Nevertheless, the structure provides a compelling molecular explanation why the DSM α1 helix may act as a key recognition element with a high affinity for parallel G4s, in agreement with biochemical findings [48], [52]. Specifically, DSM binds to the solvent-exposed flat surface formed by the G-tetrad at the 5′-end of the G-quadruplex, consistently with the earlier NMR structure of a truncated DSM in complex with G4 [53]. The other essential G4 binding interface identified in the crystal structure is formed by the subdomain of the C-terminal domain showing the oligonucleotide/oligosaccharide-binding like fold (OB), particularly, its OI loop interacting with the sugar phosphate backbone of the bound G-quadruplex [50].
Fig. 1.
A Structure of the DHX36 helicase in complex with the register-shifted partially unfolded G-quadruplex (DNAmyc-G4) with parallel topology, taken from the X-ray structure [50] with the missing parts modeled in as described in Methods. Individual domains of DHX36 are marked with different colors (NTD and CTD are the N- and C-terminal domains, respectively). Darker shades show the binding interfaces with the G-quadruplex with LK−Q labeling a new interface identified in our work. B, C Structures of DNAmyc-G4 in the register-shifted partially unfolded (puG4) and fully folded states (ffG4), respectively. Potassium ions (K+) are shown in semi-transparent sphere representation. D Free energy profiles for binding of the DSM α1 helix to puG4 (yellow) and ffG4 (cyan). For convergence of the free energy profiles see Fig. S2, and for the biased distributions of the reaction coordinate Fig. S3.
Despite these important structural insights, the mechanism by which DHX36 recognizes parallel-stranded G4s is not fully understood. Since the structure of DHX36 in complex with a fully folded G4 has not yet been obtained, it is not clear whether the DSM binding mode and binding energy depend on the G4 state and, consequently, whether DSM act merely as an anchor for G4s or rather it actively participates in the unfolding process [54]. A strong preference of DSM to bind to the 5′-terminal side of parallel G4s also requires clarification at the molecular level [53]. Furthermore, it is not known, given the flexible attachment of DSM to the helicase core, what is the relative importance of DSM and OB in recognizing G4s and whether G4 binding to both these subdomains is independent. Finally, the role of individual protein residues in the recognition process and their relative contribution to the binding energy is yet to be established.
Here, to address the above questions, we studied the recognition of parallel-type G-quadruplexes by DHX36 with all-atom molecular dynamics simulations. Using a set of model systems based on the crystal structure of the DHX36/G4 complex [50], we computed the relative contributions to the binding free energy due to the two main recognition subdomains, DSM and OB, and found that G4 binding to these sites is cooperative, i.e., they mutually enhance each other’s affinity for G4. Detailed description of the binding interfaces in terms of polar and non-polar interactions provided molecular-level explanation of the observed affinities and cooperativity, as well as preferential binding of DSM to the 5′-terminal G-tetrad. Our simulations also revealed that the 13-residue flexible loop in the RecA2 domain might form a third binding interface that can provide additional stabilization of the complex with G4.
2. Materials and methods
2.1. Molecular systems
The simulated systems were constructed on the basis of the co-crystal structure of the bovine DHX36 bound to a parallel G-quadruplex with 3′-end single-stranded (ss) DNA segment (PDB id: 5VHE) [50]. As opposed to 5VHE, the other solved structures of DHX36 [47], [52] do not contain bound G4 and therefore cannot be directly used as a starting point for the analysis of the recognition process. Two missing fragments in the crystal structure, a 20-residue linker between the DHX36-specific motif (DSM) and RecA1 and a 13-residue loop in RecA2, were rebuilt using Modeller [55] (highlighted in Fig. S1), obtaining the full-length helicase structure. Further, we have discarded the 3′-ssDNA tail because of the following reasons: (1) our primary interest was to explore the recognition process of only all-parallel-G4, (2) it is known from the experiment that removal of 3′-ssDNA tail does not substantially affect the binding of G4 to the helicase [47], [48], (3) the flexible ssDNA tail can present an obstacle to the enforced G4 dissociation, and thus slow down the convergence of free energy computations, preventing us from drawing firm conclusions. Therefore, we kept only the first 17 nucleotides corresponding to the G4 sequence from the human c-myc promoter (DNAmyc-G4: [A(G)3T(G)3TA(G)3T(G)3]). For the purpose of dissecting the role of individual DHX36 subdomains in the G4 recognition, we simulated two variants of the helicase: i) the complete structure involving all interfaces with G4 and ii) the structure with the DSM motif removed along with the entire N-terminal linker region and a several initial residues of RecA1 forming an overhanging loop in equilibrium MD (residues Pro57–Ile200).
To examine the dependence of DSM binding on a specific folded state adopted by the parallel G4 and to understand the preference of DSM for binding to the 5′-G-tetrad, we prepared three additional systems containing only the truncated DSM α1 helix (22 residues, Pro57–Lys78) and DNAmyc-G4 in solution (which also allowed for better free energy convergence). Specifically, in these DSM/G4 systems the DSM α1 helix is bound to (1) the register-shifted partially unfolded G4 (puG4) captured in the co-crystal structure (PDB id: 5VHE), (2) the 5′-G-tetrad of the native fully folded DNAmyc-G4 (ffG4) taken from its solution NMR structure (PDB id:1XAV) [56] and (3) the 3′-G-tetrad of ffG4. The initial structure of the DSM/G4 complex for system (1) was taken directly from the crystallographic data, whereas for systems (2) and (3) was obtained by fitting the NMR structure of ffG4 to puG4 in the system (1) using non–hydrogen backbone atoms of four nucleotides forming the G-tetrad bound to DSM. As a result, in all three systems, the initial orientation and positioning of the DSM α1 helix in relation to the respective G-quadruplexes was the same.
All prepared protein-DNAmyc-G4 complexes were solvated with TIP3P water molecules [57] in a dodecahedron box with dimensions ensuring at least 1.2 nm distance from the solute to the box edges. The number of K+ and Cl- ions were adjusted to maintain a physiological salt concentration of 0.15 M and neutralize the net charge of the system. Two of K+ ions were stably bound in the central channel of DNAmyc-G4.
2.2. Simulation details
All MD simulations were performed using Gromacs 5 [58] in conjunction with the PLUMED 2.2.3 plugin [59] in the isothermal-isobaric (NPT) ensembles with the temperature kept at 300 K using the v-rescale thermostat [60] and pressure kept at 1 bar using Parrinello-Rahman barostat [61]. The AMBER parmbsc1 force field [62] was used for the DNA, proteins and ions. Periodic boundary conditions were applied in 3D, and electrostatic interactions were calculated using the particle mesh Ewald (PME) method [63] with a real-space cutoff of 1.2 nm and a Fourier grid spacing of 0.12 nm. A cutoff of 1.2 nm was used for Lennard-Jones interactions. Bond lengths were constrained by P-LINCS [64] for the DNA and protein molecules and SETTLE [65] for water. The equations of motion were integrated using the leap-frog algorithm with a 2 fs time step. Initially, each system was subjected to at least 500 ns of relaxation via unbiased MD simulation.
2.3. Free energy simulations
To investigate the interplay between the individual DHX36 subdomains in G4 recognition, we calculated the set of relevant free energy profiles using umbrella sampling (US) approach [66], as described below.
To dissociate the complexes of puG4 with DHX36, either including or not DSM (systems (i) and (ii), respectively, as described above), we performed steered MD (SMD) simulations, in which, starting from the bound state, puG4 was pulled away from the helicase during 500-ns-long runs. This was done using a moving harmonic potential with a force constant of 179.42 and 119.61 kcal/(mol·nm2) (for system (i) and (ii), respectively) applied to the coordinate defined as the separation distance (r) between the centers of mass (COM) of the heavy atoms of the guanine core and the Cα atoms of the OB subdomain (see Fig. 4A).
Fig. 4.
A Free energy profiles for binding of the parallel G-quadruplex (puG4) to the OB subdomain of DHX36 in the absence of DSM. For convergence of the free energy profile see Fig. S7. The inset shows the comparison of this profile (−DSM), in the bound-state minimum region, to the one obtained in the presence of DSM bound to the 5′-G-tetrad (+DSM), indicating ~4 kcal/mol deepening of the global minimum in presence of DSM. B Representative structure of puG4 the tightly bound to the OB subdomain, corresponding to the 1.8–2.1 nm range of r in panel A. The equilibrium probabilities of a given OB residue making a contact with the G4 are color-coded using a red scale. C Representative structure of the tightly-bound native complex of puG4 with the OB subdomain showing the most probable complex-stabilizing hydrogen bonds (as dotted lines), along with their formation probabilities (two views of the same snapshot). D Equilibrium probabilities of forming a contact (minimum distance < 0.4 nm; left) or a hydrogen bonds (right) between pairs of residues across the puG4/OB binding interface in the absence (top) and presence (bottom) of the DSM subdomain. For nucleotide residues, the nucleobase (NB) and sugar-phosphate moieties (BB) are shown separately.
From thus obtained enforced-dissociation trajectories, we extracted initial configurations for the US simulations. In particular, we used 23 uniformly distributed, 0.15-nm separated US windows in the range of r from 1.7 to 5.0 nm. In each of these windows, the system was simulated for 500 ns, using the harmonic potential with a similar force constant (used during SMD) to restrain the system along the reaction coordinate r.
To dissociate the three complexes of the DSM α1 helix, formed either with the 5′-tetrad of puG4, the 5′-G-tetrad of ffG4 or the 3′-G-tetrad of ffG4 (systems (1), (2) and (3), respectively, as described above), we performed steered MD (SMD) simulations, in which DSM was pulled away from G4 during 500 ns run. This was done using a moving harmonic potential with a force constant of 119.61 kcal/(mol·nm2) applied to the coordinate defined as the separation distance (r) between the COMs of the guanine-core heavy atoms and the Cα atoms of DSM. From these enforced-dissociation trajectories, we extracted initial configurations for the US simulations. In particular, we used 22 uniformly distributed 0.15-nm separated US windows in the range of r from 0.85 to 4.0 nm. In each of these windows the system was simulated for 500 ns, using the harmonic potential with a force constant of 56.80 kcal/(mol·nm2) to restrain the system along the reaction coordinate r. Due to the tendency of the DSM α1 helix to spontaneously unfold in the absence of G4, in all simulated systems, we preserved the DSM helicity using the ALPHARMSD coordinate [67] and a force constant of 59.8 kcal/mol·nm2 applied.
The free energy profiles were determined after discarding the first 10% of thus obtained trajectories using standard weighted histogram analysis (WHAM) method [68]. Uncertainties were estimated using bootstrap error analysis taking into account the correlation in the analyzed time series. For the purpose of interaction analysis, the original (biased) umbrella sampling data were reweighted to recover the unbiased probabilities, using weights of the form: , where is the applied biasing potential and the free energy constant associated with the bias as calculated by the WHAM algorithm, in the i-th US window.
3. Results and discussion
DSM plays a major role in recognition of parallel G4s.
Since the available structural data provide only an indirect insight into the recognition of native G-quadruplexes by DEAH/RHAU helicases [50], [53], we first examined how the G4-binding affinity of DSM, the main recognition motif of DHX36, and the bound-state structure are affected by a helicase-mediated one-nucleotide register shift in the G-quadruplex (Fig. 1B). To this end, we computed the free energy profiles for binding of the isolated DSM α1 helix (residues Pro57–Lys78) to the G-quadruplex from the c-myc promoter (DNAmyc-G4) in the native fully folded state (ffG4; Fig. 1C), as well as in the register-shifted partially unfolded state extracted from the co-crystallized complex with DHX36 [50] (puG4; Fig. 1B). As a reaction coordinate, we used the center-of-mass distance between the DSM α1 helix and the guanine cores of the respective G-quadruplexes (Fig. 1D). To retain the extended α-helical conformation of DSM found in the DHX36/G4 complex, the helicity of DSM was restrained for the purpose of our free energy calculations (see Methods).
The well-pronounced ~20 kcal/mol deep minimum in the resulting free energy curves in Fig. 1D at 0.8–1.1 nm correspond to DSM tightly bound at the surface of the G-quadruplexes (Fig. 2A,B). This deep bound state minimum is consistent with high affinities of DSM for parallel G4s measured in quantitative gel electrophoresis [50], [53] and thus support a primary role of DSM in the recognition process [69]. It should be noted, however, that due to force field inaccuracies and undersampling of the partially-bound and unbound states in our umbrella sampling approach, the obtained profiles should mostly be used for comparative analysis, as the absolute binding free energies point to a considerable overstabilization of the DHX36/G4 complexes.
Fig. 2.
A, B Representative structures of the DSM α1 helix tightly bound to either puG4 or ffG4, respectively, corresponding to the 0.8–1.1 nm range in Fig. 1D. The equilibrium probabilities of a given DSM residue making a contact with the G4 are color-coded using a blue scale. Most probable hydrogen bonds are shown with black dotted lines along with their formation probabilities. Glycine residues in the two adjacent GXXXG motifs in DSM are marked in orange. C Equilibrium probabilities of forming a contact (minimum distance < 0.4 nm; left) or a hydrogen bonds (right) between pairs of residues across the DSM/G4 binding interface for the complexes involving either puG4 (top) or ffG4 (bottom). For nucleotide residues, the nucleobase (NB) and sugar-phosphate moieties (BB) are shown separately.
Importantly, similar binding free energies predicted for both G-quadruplex folds (<1.5 kcal/mol difference between ffG4 and puG4 in Fig. 1D) indicate that the strength of DSM binding to G4 virtually does not depend on the base composition of the outer tetrad. Rather, the DSM motif seems to be especially suited for recognition of all-parallel G4 structures characterized by easily accessible flat surfaces. Therefore, it acts as a specialized anchor for G4s as previously found by Srinivasan et al. [52] who observed that DSM shows preference for G4 DNA structures over the canonical double helix. Furthermore, very high affinities for the parallel G4s are consistent with the experimentally established capability of DSM to promote G-quadruplex remodeling towards the all-parallel fold, even though definitive conclusions would require computing the binding free energy for other G4 forms as well [48], [52], [54].
The range 1.3–2.5 nm of r in Fig. 1D corresponds to non-native DSM/G4 complexes. We found that, in all these partially formed intermediates, the DSM helix interacts with the 5′-tetrad, yet in an energetically sub-optimal manner, again pointing to specialization of DSM in sensing flat molecular surfaces. For example, a local minimum at ~1.6 nm in the case of puG4 corresponds to the state in which the DSM helix is shifted along the tetrad while its Tyr69 and Ile65 residues interact favorably with the groove region of the G-quadruplex (Fig. S4).
DSM recognizes parallel G4s through extensive van der Waals and hydrophobic interactions with a planar G-tetrad surface.
To investigate in more detail the molecular determinants of the observed high affinities of DSM for G4 and characterize the role of the individual interfacial residues in the recognition process, we further calculated the probabilities of formation of van der Waals (vdW) contacts (with minimal distance cutoff of 0.4 nm) and hydrogen bonds (h-bonds) between the DSM α1 helix and the two G-quadruplex folds in the bound state minima (see Fig. 2A,B). To this end, the original (biased) US data were reweighted to recover the unbiased equilibrium distribution (see Methods). The results are presented as h-bond and contact matrices in Fig. 2C.
Contact matrices presented in Fig. 2C show that numerous (>10) DSM residues make extensive and stable (prob. ≈1) contacts with the nucleobase and/or sugar-phosphate moieties, especially of those nucleotides that form the 5′-tetrad (G2 · G6 · G11(A10) · G15(T14)). A large contact area between DSM and G4 is possible due the presence of two adjacent GXXXG motifs (see Fig. 2AB) that form a smooth surface on one side of the DSM helix and allow it to associate closely with a flat surface of the tetrad. Indeed, a detailed inspection of the contact matrices reveals that four DSM glycine residues (58, 62, 66 and 74) make a direct stable vdW contact with G4. As a result of this intimate interaction, a number of other, mostly hydrophobic residues, also establish contacts with the 5′-tetrad, e.g., Tyr69 that is involved in π-stacking interactions with G2 and G6 (>90% prob.), Ala70 interacts with A1 and G2 (>70% prob.), Leu67 with A1 (>70% prob.) and Ile65 with G6 (>90% prob.).
Notably, the contact patterns with ffG4 and puG4 are very similar, reflecting the same alignment of DSM at the G4 surface (see also Fig. S5), with differences resulting mostly from the DHX36-induced sequence shift in the 5′-tetrad (G11 -> A10 and G15 -> T14; G2 and G6 stay intact). Also in both cases, the A1 base from the 5′-overhang wraps around the DSM helix and makes with it multiple stable contacts. Given similar binding affinities (Fig. 1D), the matching contact patterns found here for puG4 and ffG4 strongly suggest that the identified flat-surface-to-flat-surface binding mode underlies the essential role of DSM in recognizing and binding parallel G4s with high specificity [53], [69].
We further checked whether polar interactions in the form of hydrogen bonds also contribute to the stability of the complexes formed by DSM with G-quadruplexes. H-bond matrices in Fig. 2C reveal the formation of only a few hydrogen bonds across the DSM-G4 interface. In particular, in the puG4 case, we only found one significant hydrogen bond (>40 % prob.), between Asn77 and the G3 phosphate group (Fig. 2A). Lack of hydrogen bonds suggests that the high binding free energy observed for the DSM/G4 complexes results from the van der Waals and hydrophobic attractive forces between the two extended apolar surfaces. Because the second loop in ffG4 is 1 nt longer, a few additional h-bonds are formed in this case between the phosphate groups of G11 and G6 and basic groups of DSM (Lys61, Arg63, Lys76 and N-terminal Pro57) with probabilities well exceeding 50 %. They might account for a slightly higher affinity for ffG4 predicted by our simulations.
Preferential binding of DSM to the 5′-G-tetrad results from a tighter contact between the surfaces.
Next, we investigated the molecular basis of the known preference of DSM for binding to the 5′-G-tetrad of parallel G4s [50], [53]. To this end, we used the same approach as above to obtain the free energy profile for binding of the DSM α1 helix to DNAmyc-G4, in this case, however, to the most plausible alternative binding site, i.e., the 3′-G-tetrad. The resulting profile (3′ffG4) is compared to the original one describing the 5′-G-tetrad binding (5′ffG4) in Fig. 3A.
Fig. 3.
A Comparison of the free energy profiles for binding of the DSM α1 helix to the 3′- (3′ffG4, red)) and 5′-G-tetrad (5′ffG4, cyan) of ffG4. For convergence of the free energy simulations see Figs. S2 and S6B, C Representative structures of the tightly bound complex of the DSM α1 helix and 3′ffG4, corresponding to the 0.8–1.1 nm range in Fig. 3A. The equilibrium probabilities of a given DSM residue making a contact with the G4 are color-coded using a blue scale. Most probable hydrogen bonds are shown with black dotted lines along with their formation probabilities. D Equilibrium probabilities of forming a contact (minimum distance < 0.4 nm; left) or a hydrogen bonds (right) between pairs of residues across the DSM/G4 binding interface for the complexes involving 3′ffG4. For nucleotide residues, the nucleobase (NB) and sugar-phosphate moieties (BB) are shown separately.
As can be seen, the binding of DSM to the 3′-G-tetrad of G4 (3′ffG4) is also characterized by a well-defined global minimum indicative of a stable complex. However, the DSM affinity for the 3′-G-tetrad is predicted to be ~5 kcal/mol lower than for the 5′-G-tetrad, consistently with earlier NMR results, which demonstrated that binding of DSM to the 3′-G-tetrad only occurs at significantly higher peptide concentrations [53] or when it is enforced by G4 dimerization through the 5′-G-tetrads [70]. Given the key role of DSM in G4 recognition, its strong preference for the 5′-G-tetrad might be critical for dictating the overall binding mode, and hence for properly orienting G4 for interactions with other DHX36 subdomains involved in the unfolding.
To gain a better understanding of the observed preference for the 5′-G-tetrad, we characterized the 3′-G-tetrad binding mode (3′ffG4) by analyzing complex-stabilizing vdW contacts and hydrogen bonds, using the same approach as above for the 5′ffG4 mode. The resulting contact matrix shown in Fig. 3D reveals that, also in the 3′ffG4 case, DSM makes extensive contacts with the G-tetrad accounting for a relatively high affinity. Identical DSM residues involved in the interaction with G4 imply that, similarly as at the 5′-G-tetrad, the DSM helix is oriented with its flat side towards the 3′-G-tetrad (Fig. 3BC). Therefore, the contact pattern is essentially the same as before with the difference that the nucleotides are replaced by the ones forming the 3′-G-tetrad (G4 · G8 · G13 · G17).
However, in the 3′ffG4 case, the calculated equilibrium contact probabilities are almost uniformly lower than the corresponding ones for 5′ffG4 (Fig. 2C). This systematic shift in contact stabilities suggests that the observed preference of DSM for binding to the 5′-end of the G-quadruplex is a result of more favorable vdW and hydrophobic interactions with the G-tetrad. A detailed analysis of the binding mode indicates that a weaker vdW attraction to the 3′-G-tetrad is caused by a slightly more protruding sugar-phosphate moieties that sterically prevent the bulky DSM helix from optimally adsorbing at the flat surface of the guanine plane. Differences in the arrangement of the sugar phosphate backbone between the 3′- and 5′-ends of parallel G-quadruplexes arise from DNA strand polarity and were discussed before in the context of G4 binding preferences [71], [72].
H-bond matrix in Fig. 3D demonstrates the existence of only one strong hydrogen bond between Arg63 and T9 phosphate group again pointing to a marginal role of polar interactions in driving the binding of DSM to G4.
OB cooperatively enhances binding affinity for G4 via specific polar interactions with the 5′-end segment of the DNA backbone.
Previous in vitro binding studies as well as in vivo assays have shown that the DSM motif is essential but not sufficient for high-affinity G-quadruplex binding by DHX36 [52], [69], [73]. It has also been reported that DHX36 is able to bind G4 even after deletion of DSM, though with lower affinity [48]. Therefore, prompted by the recent crystal structure demonstrating that also the OB subdomain is implicated in the interaction with G4 [50], we set out to clarify the relative contribution of OB to G4 recognition. With this purpose, we first obtained the model consisting of the puG4 G-quadruplex bound to DHX36 with the DSM motif removed (see Methods) and determined the free energy profile for the G4 binding to the OB subdomain only (Fig. 4A). As a reaction coordinate, we used the center-of-mass distance between the OB Cα atoms and the guanine core of G4.
A deep free energy minimum of ~16 kcal/mol, found at 1.8–2.1 nm, corresponds to the OB/G4 bound mode identical to that in the full complex (see Fig. 4B,C), while the other noticeable local minima (at 2.5 and 3.1 nm) characterize loosely-bound intermediates of the mostly intact puG4 (for a detailed description see Figs. S8 and S9). Thus, the obtained profile clearly confirms the important role of the OB subdomain in G4 recognition; however, the OB contribution to the binding free energy is predicted to be markedly lower than that of DSM (~20 kcal/mol as indicated by the deep global minimum in Fig. 1D).
Further, to determine whether the binding free energy of G4 to OB depends on DSM being already bound to 5′-G-tetrad, i.e., whether the G4 binding by DHX36 is cooperative in nature, we recomputed the OB/G4 free energy in the context of the full-length DHX36, that is in the presence of DSM helix bound to the 5′-G-tetrad (+DSM in the inset in Fig. 4A). To make sure DSM interacts optimally with G4 regardless of the OB-G4 distance, we focused on the local response of the free energy profile by limiting the range of the reaction coordinate to 1.8–3.1 nm. Indeed, comparison of the DSM-G4 contacts and h-bonds observed in this simulation (Fig. S10) with those determined above for the DSM/puG4 complex (Fig. 2C) confirms that DSM remains optimally bound to the 5′-G-tetrad over the entire considered range of the reaction coordinate.
By comparing the two curves in the inset of Fig. 4A, we see that the presence of DSM does not change the general character of the binding free energy profile, however, the minimum corresponding to the native complex is over 4 kcal/mol more stable. This significant enhancement of OB affinity for G4 in the presence of the 5′-G-tetrad-bound DSM allows us to conclude that both DHX36 subdomains bind to G4 cooperatively. Structurally, this finding can be understood by considering that the DSM α1 helix and OB together form continuous binding interfaces that embrace both sides of the G-quadruplex and thereby can mutually stabilize each other (see Fig. S11). This conclusion is additionally supported by the free energy profile describing the simultaneous binding of G4 to both G4-interacting sites, in the context of the full-length DXH36 (Fig. S12). Even though this profile suffers from similar bound state overstabilization as those discussed above, it again points to a synergistic effect, as the free energy gain due to G4 binding to both sites at the same time (−48 kcal/mol) significantly exceeds the sum of individual gains (−20 and −16 kcal/mol for isolated DSM and OB, respectively).
To provide a molecular-level description of OB/G4 binding energetics and its dependence on the presence of DSM, we subsequently analyzed complex-stabilizing vdW contacts and hydrogen bonds, using the same approach as described above. The contact matrices in Fig. 4D show that the OB/G4 interaction is mediated by several protein residues distributed throughout the OB domain, most of which, however, belong to the Asn851–Lys860 reverse turn motif, referred to as the OI loop; (see bottom part of the matrix in the Fig. 4D and Fig. S13). As can be also seen in Fig. 4B, the OI residues make stable contacts with the first three 5′-terminal guanine nucleotides (G2, G3 and G4) as well as with the G4’s 3′-end, mainly G16 and G17. The observed contact pattern arises because the OI loop binds to the 3′-end and, at the same time, aligns itself parallel to the 5′-terminal part of the sugar-phosphate backbone (the first G-tract) to form with it several specific polar interactions.
Indeed, h-bond matrices in the Fig. 4D demonstrate that, in contrast to DSM, the OB subdomain is involved in strong h-bonding interactions with G4. In particular, Leu852, Gly853 and Asn851 were found to form high-probability h-bonds with the phosphate group of G4 (see Fig. 4C, right), while Lys860 and Tyr862 interact stably with the G3 phosphate (Fig. 4C, left). At the same time, Ser898, Gly853 and Ile871 make weaker h-bonds with the 3′-terminal guanine nucleotides, G16 and G17.
Comparison of the interaction matrices between the +DSM and −DSM systems (Fig. 4D) reveals that the binding of DSM domain to 5′-G-tetrad does not affect the general OB/G4 contact and h-bond pattern, however, it leads to a visible increase in the stability of the hydrogen bonds, mainly those with the 5′-end G-tract. This enhancement of hydrogen bonds is associated with pronounced conformational changes of the side chains involved (see Fig. S13).
Flexible loop in the RecA2 domain might help to anchor a G-quadruplex.
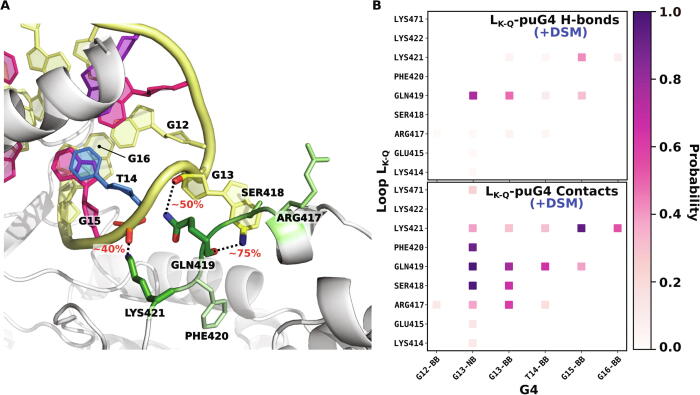
During our MD-based refinement of the completed, full-length DHX36/G4 complex, the flexible 13-residue loop of the Rec2 domain, missing in the crystal structure (Lys414–Gln426; LK−Q in Fig. 1A, see also Fig. S1), showed a clear tendency to interact with the G-quadruplex loop, and hence may provide a third interface involved in G4 recognition. This finding agrees with the results of shorter MD simulations of a modeled complex of parallel G4 with DHX36 from D. melanogaster [47], in which the same missing loop was observed to interact with G4. Here, we examined this interaction in more detail applying the same analysis of vdW contacts and h-bonds as above to the 1 μs-long unbiased trajectory of the DHX36/G4 complex.
Both contact and h-bond probabilities in Fig. 5 consistently show that the interaction of LK−Q with G4 is mostly polar in nature, with the most stable hydrogen bonds forming between Gln419 and the nucleobase and backbone moieties of G13 in the G4’s third loop (Fig. 5A). Since this guanine nucleotide is pulled out of the 5′-G-tetrad during the DHX36-mediated unfolding, it could be speculated that the observed interaction with Gln419 is important for stabilizing the partially unfolded state of the G-quadruplex. The remaining residues of LK−Q contributing to the interaction with G4 include Lys421 and Arg417 that form moderately stable h-bond-enhanced ion pairs with the phosphate groups of G15 and G13, respectively. Note that the h-bond stability is low enough for the loop to remain flexible (as indicated by the root mean square fluctuations in Fig. S14), such that it apparently is not resolvable by X-ray crystallography.
Fig. 5.
A Representative structures of a the binding interface between puG4 and the flexible LK−Q loop (Lys414–Gln426) identified in our simulations of the full DHX36/puG4 complex. The equilibrium probabilities of a given LK−Q residue making a contact with the G4 are color-coded using a green scale. Most probable hydrogen bonds are shown with black dotted lines along with their formation probabilities. B Equilibrium probabilities of forming a contact (minimum distance < 0.4 nm; top) or a hydrogen bonds (bottom) between pairs of residues across the LK−Q/G4 binding interface in the complex formed by the full-length DHX36. For nucleotide residues, the nucleobase (NB) and sugar-phosphate moieties (BB) are shown separately.
To determine whether the LK−Q-mediated interaction with G4 also shows cooperativity with other DHX36 subdomains, we repeated the same interaction analysis taking as an input a 1 μs unbiased trajectory of the DHX36/G4 complex with the entire DSM region removed (Fig. S15). By comparing Fig. 5B and Fig. S15, we found that virtually all LK−Q-G4 hydrogen bonds are destabilized in response to the removal of DSM. In particular, h-bonds formed by Gln419 have their probabilities decreased ~7-fold, while those formed by Lys421- and Arg417 are affected less heavily. This finding is again strongly indicative of a cooperative nature of G4 binding by DHX36.
4. Conclusions
DHX36, an ATP-dependent DEAH-box helicase, is known to bind and resolve RNA and DNA G-quadruplex (G4) substrates, providing polymerases and ribosomes with unfolded single-stranded templates [41], [42]. In this work we investigated the molecular basis of DHX36 specificity for parallel-type G4s, using molecular dynamics-based free energy calculations along with the recently solved crystal structure of DHX36 in complex with the G-quadruplex from the c-myc promoter [50].
By computing binding free energy profiles, we found that the two main DHX36 subdomains involved in the specific recognition of G4s, DSM and OB, separately exhibit a high affinity for parallel-stranded G4s, in agreement with the reported high stability of the DHX36/G4 complex [50], [53]. We also concluded that, in the full-length helicase, G4 binding to DSM and OB sites is cooperative and, consistently with previous structural data [50], that the two subdomains are optimized for sensing two distinctive features of parallel G-quadruplexes, i.e., the exposed planar face of a G-tetrad and the specific backbone conformation of a G-tract, respectively. Since these features are common to all parallel G4s, this binding mode might explain why the unfolding activity of DHX36 does not strongly depend on the loop length [74]. In fact, when the telomeric, parallel-stranded G4 is superimposed on the studied DHX36/G4 complex, only minimal steric clashes are found between the longer G4 loops and DNA-binding interfaces of the helicase (Fig. S16).
In particular, our simulations indicated that the interaction of the flexible DSM α1 helix with the 5′-G-tetrad is the main contributor to the binding free energy, explaining the essential role of DSM in G4 binding and subsequent processing. Importantly, the DSM motif does not discriminate between the fully folded and DHX36-induced partially unfolded conformations of G4, indicating that the binding affinity does not depend on a particular base composition. Rather it relies solely on extensive contacts between the two adjacent GXXXG motifs as well as hydrophobic residues of DSM with the guanine plane. Furthermore, we found that the previously observed preferential binding of DSM to the 5′-G-tetrad [53] can be attributed to the fact that this face of parallel G4s is sterically more accessible and thus allows for more favorable van der Waals and hydrophobic interactions between the two flat surfaces.
In contrast to DSM, OB binds to G4 mostly through polar interactions. More specifically, its OI reverse turn motif tends to align with the 5′-terminal G-tract to form a number of strong hydrogen bonds with the backbone phosphate groups. The OB contribution to the binding free energy is overall lower than that of DSM and was predicted to depend on whether or not DSM is already bound to the 5′-G-tetrad, reflecting the cooperative nature of the G4 recognition process.
Finally, our simulations also revealed the existence of a third DHX36/G4 interaction site formed by the 13-residue flexible region in RecA2 domain (LK−Q), missing in the crystal structure [50].Detailed interaction analysis suggested that this region may participate in both an initial cooperative anchoring of G4 as well as the stabilization of its partially unfolded conformation, consistently with the previous MD simulations of a model of the dmDHX36/G4 complex [47].
CRediT authorship contribution statement
Kazi Amirul Hossain: Methodology, Formal analysis, Investigation, Writing - original draft, Visualization. Michal Jurkowski: Writing - original draft, Visualization. Jacek Czub: Conceptualization, Formal analysis, Writing. Mateusz Kogut: Conceptualization, Investigation, Supervision, Writing - original draft, Visualization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Science Centre, Poland (Grant No. 2019/35/B/ST4/03559). This research was supported in part by PL-Grid Infrastructure. We thank the Academic Computer Centre TASK (Gdansk, Poland), ICM (Warsaw, Poland) and WCSS (Wroclaw, Poland) for granting CPU time.
Footnotes
Supplementary data associated with this article can be found, in the online version, athttps://doi.org/10.1016/j.csbj.2021.04.039.
Supplementary data
The following are the Supplementary data to this article:
References
- 1.Sen D., Gilbert W. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 2.Davis J.T. Angew Chem Int Ed. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 3.Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S. Nucl Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lane A.N., Chaires J.B., Gray R.D., Trent J.O. Nucl Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hazel P., Huppert J., Balasubramanian S., Neidle S. J Am Chem Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 6.Rachwal P.A., Findlow I.S., Werner J.M., Brown T., Fox K.R. Nucl Acids Res. 2007;35:4214–4222. doi: 10.1093/nar/gkm316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatzakis E., Okamoto K., Yang D. Biochemistry. 2010;49:9152–9160. doi: 10.1021/bi100946g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhattacharyya D., Mirihana Arachchilage G., Basu S. Front Chem. 2016;4:38. doi: 10.3389/fchem.2016.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujii T., Podbevšek P., Plavec J., Sugimoto N. J Inorg Biochem. 2017;166:190–198. doi: 10.1016/j.jinorgbio.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Miyoshi D., Karimata H., Sugimoto N. J Am Chem Soc. 2006;128:7957–7963. doi: 10.1021/ja061267m. [DOI] [PubMed] [Google Scholar]
- 11.Miyoshi D., Fujimoto T., Sugimoto N. Springer; 2012. Quadruplex Nucleic Acids; pp. 87–110. [Google Scholar]
- 12.Phan A.T. FEBS J. 2010;277:1107–1117. doi: 10.1111/j.1742-4658.2009.07464.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhang D.-H., Fujimoto T., Saxena S., Yu H.-Q., Miyoshi D., Sugimoto N. Biochemistry. 2010;49:4554–4563. doi: 10.1021/bi1002822. [DOI] [PubMed] [Google Scholar]
- 14.Rezler E.M., Bearss D.J., Hurley L.H. Curr Opin Pharmacol. 2002;2:415–423. doi: 10.1016/s1471-4892(02)00182-0. [DOI] [PubMed] [Google Scholar]
- 15.Lipps H.J., Rhodes D. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Biffi G., Tannahill D., McCafferty J., Balasubramanian S. Nat Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hänsel-Hertsch R., Di Antonio M., Balasubramanian S. Nat Rev Mol Cell Biol. 2017;18:279–284. doi: 10.1038/nrm.2017.3. [DOI] [PubMed] [Google Scholar]
- 18.Spiegel J., Adhikari S., Balasubramanian S. Trends Chem. 2020;2:123–136. doi: 10.1016/j.trechm.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng K.-W., Zhang J.-Y., He Y.-D., Gong J.-Y., Wen C.-J., Chen J.-N., Hao Y.-H., Zhao Y., Tan Z. Nucl Acids Res. 2020;48:11706–11720. doi: 10.1093/nar/gkaa841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simonsson T., Kubista M., Pecinka P. Nucl Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai J., Dexheimer T.S., Chen D., Carver M., Ambrus A., Jones R.A., Yang D. J Am Chem Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu C.-H., Teulade-Fichou M.-P., Olsthoorn R.C. Nucl Acids Res. 2014;42:1887–1892. doi: 10.1093/nar/gkt1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prorok P., Artufel M., Aze A., Coulombe P., Peiffer I., Lacroix L., Guédin A., Mergny J.-L., Damaschke J., Schepers A. Nat Commun. 2019;10:1–16. doi: 10.1038/s41467-019-11104-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumari S., Bugaut A., Huppert J.L., Balasubramanian S. Nat Chem Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collie G.W., Haider S.M., Neidle S., Parkinson G.N. Nucl Acids Res. 2010;38:5569–5580. doi: 10.1093/nar/gkq259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kharel P., Becker G., Tsvetkov V., Ivanov P. Nucl Acids Res. 2020;48:12534–12555. doi: 10.1093/nar/gkaa1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Y., Shin-ya K., Brosh R.M. Mol Cell Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arora A., Dutkiewicz M., Scaria V., Hariharan M., Maiti S., Kurreck J. Rna. 2008;14:1290–1296. doi: 10.1261/rna.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai J.C., Ponti S., Pan D., Kohler H., Skoda R.C., Matthias P., Nagamine Y. Blood. 2012;119:4291–4300. doi: 10.1182/blood-2011-08-362954. [DOI] [PubMed] [Google Scholar]
- 30.Han H., Hurley L.H., Salazar M. Nucl Acids Res. 1999;27:537–542. doi: 10.1093/nar/27.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Besnard E., Babled A., Lapasset L., Milhavet O., Parrinello H., Dantec C., Marin J.-M., Lemaitre J.-M. Nat Struct Mol Biol. 2012;19:837. doi: 10.1038/nsmb.2339. [DOI] [PubMed] [Google Scholar]
- 32.Morris M.J., Negishi Y., Pazsint C., Schonhoft J.D., Basu S. J Am Chem Soc. 2010;132:17831–17839. doi: 10.1021/ja106287x. [DOI] [PubMed] [Google Scholar]
- 33.Wong M.S., Wright W.E., Shay J.W. Trends Genet. 2014;30:430–438. doi: 10.1016/j.tig.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcel V., Tran P.L., Sagne C., Martel-Planche G., Vaslin L., Teulade-Fichou M.-P., Hall J., Mergny J.-L., Hainaut P., Van Dyck E. Carcinogenesis. 2011;32:271–278. doi: 10.1093/carcin/bgq253. [DOI] [PubMed] [Google Scholar]
- 35.Yu Z., Schonhoft J.D., Dhakal S., Bajracharya R., Hegde R., Basu S., Mao H. J Am Chem Soc. 2009;131:1876–1882. doi: 10.1021/ja806782s. [DOI] [PubMed] [Google Scholar]
- 36.Rhodes D., Lipps H.J. Nucl Acids Res. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsumura K., Kawasaki Y., Miyamoto M., Kamoshida Y., Nakamura J., Negishi L., Suda S., Akiyama T. Oncogene. 2017;36:1191–1199. doi: 10.1038/onc.2016.282. [DOI] [PubMed] [Google Scholar]
- 38.Creacy S.D., Routh E.D., Iwamoto F., Nagamine Y., Akman S.A., Vaughn J.P. J Biol Chem. 2008;283:34626–34634. doi: 10.1074/jbc.M806277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Byrd A.K., Bell M.R., Raney K.D. J Biol Chem. 2018;293:17792–17802. doi: 10.1074/jbc.RA118.004499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paul T., Voter A.F., Cueny R.R., Gavrilov M., Ha T., Keck J.L., Myong S. Nucl Acids Res. 2020;48:6640–6653. doi: 10.1093/nar/gkaa442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He Y., Andersen G.R., Nielsen K.H. EMBO Rep. 2010;11:180–186. doi: 10.1038/embor.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sauer M., Paeschke K. Biochem Soc Trans. 2017;45:1173–1182. doi: 10.1042/BST20170097. [DOI] [PubMed] [Google Scholar]
- 43.Iwamoto F., Stadler M., Chalupníková K., Oakeley E., Nagamine Y. Exp Cell Res. 2008;314:1378–1391. doi: 10.1016/j.yexcr.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 44.Booy E.P., Howard R., Marushchak O., Ariyo E.O., Meier M., Novakowski S.K., Deo S.R., Dzananovic E., Stetefeld J., McKenna S.A. Nucl Acids Res. 2014;42:3346–3361. doi: 10.1093/nar/gkt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao X, Ma W, Nie J, Zhang C, Zhang J, Yao G, Han J, Xu J, Hu B, Du Y, et al. Cell Death Dis 2015;6:e1610–e1610. [DOI] [PMC free article] [PubMed]
- 46.Sauer M., Juranek S.A., Marks J., De Magis A., Kazemier H.G., Hilbig D., Benhalevy D., Wang X., Hafner M., Paeschke K. Nat Commun. 2019;10:1–15. doi: 10.1038/s41467-019-10432-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen W.-F., Rety S., Guo H.-L., Dai Y.-X., Wu W.-Q., Liu N.-N., Auguin D., Liu Q.-W., Hou X.-M., Dou S.-X. Structure. 2018;26:403–415. doi: 10.1016/j.str.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 48.You H., Lattmann S., Rhodes D., Yan J. Nucl Acids Res. 2017;45:206–214. doi: 10.1093/nar/gkw881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prabu J.R., Müller M., Thomae A.W., Schüssler S., Bonneau F., Becker P.B., Conti E. Mol cell. 2015;60:487–499. doi: 10.1016/j.molcel.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 50.Chen M.C., Tippana R., Demeshkina N.A., Murat P., Balasubramanian S., Myong S., Ferré-D’Amaré A.R. Nature. 2018;558:465–469. doi: 10.1038/s41586-018-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen M.C., Murat P., Abecassis K., Ferré-D’Amaré A.R., Balasubramanian S. Nucl Acids Res. 2015;43:2223–2231. doi: 10.1093/nar/gkv051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Srinivasan S., Liu Z., Chuenchor W., Xiao T.S., Jankowsky E. J Mol Biol. 2020;432:2217–2231. doi: 10.1016/j.jmb.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heddi B., Cheong V.V., Martadinata H., Phan A.T. Proc Natl Acad Sci. 2015;112:9608–9613. doi: 10.1073/pnas.1422605112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang-Gu B., Bradburna D., Yangyuorub P.M., Russell R. Biol Chem. 2021 doi: 10.1515/hsz-2020-0302. 000010151520200302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Webb B., Sali A. Curr Protocols Bioinf. 2016;54:5–6. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ambrus A., Chen D., Dai J., Jones R.A., Yang D. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 57.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 58.Hess B., Kutzner C., Van Der Spoel D., Lindahl E. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 59.Tribello G.A., Bonomi M., Branduardi D., Camilloni C., Bussi G. Comput Phys Commun. 2014;185:604–613. [Google Scholar]
- 60.Bussi G., Donadio D., Parrinello M. J Chem Phys. 2007;126 doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 61.Parrinello M., Rahman A. J Appl Phys. 1981;52:7182–7190. [Google Scholar]
- 62.Ivani I, Dans PD, Noy A, Pérez A, Faustino A, Hospital A, Walther J, Andrio P, Goñi R, Balaceanu A, et al. Nat Methods 2016;13:55. [DOI] [PMC free article] [PubMed]
- 63.Darden T., York D., Pedersen L. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 64.Hess B. J Chem Theory Comput. 2008;4:116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 65.Miyamoto S., Kollman P.A. J Comput Chem. 1992;13:952–962. [Google Scholar]
- 66.Torrie G.M., Valleau J.P. J Comput Phys. 1977;23:187–199. [Google Scholar]
- 67.Pietrucci F., Laio A. J Chem Theory Comput. 2009;5:2197–2201. doi: 10.1021/ct900202f. [DOI] [PubMed] [Google Scholar]
- 68.Kumar S., Rosenberg J.M., Bouzida D., Swendsen R.H., Kollman P.A. J Comput Chem. 1995;16:1339–1350. [Google Scholar]
- 69.Lattmann S., Giri B., Vaughn J.P., Akman S.A., Nagamine Y. Nucl Acids Res. 2010;38:6219–6233. doi: 10.1093/nar/gkq372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heddi B., Cheong V.V., Schmitt E., Mechulam Y., Phan A.T. J Struct Biol. 2020;209 doi: 10.1016/j.jsb.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 71.Parkinson G.N., Lee M.P., Neidle S. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 72.Kogut M., Kleist C., Czub J. PLoS Comput Biol. 2019;15 doi: 10.1371/journal.pcbi.1007383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chalupníková K., Lattmann S., Selak N., Iwamoto F., Fujiki Y., Nagamine Y. J Biol Chem. 2008;283:35186–35198. doi: 10.1074/jbc.M804857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yangyuoru P.M., Bradburn D.A., Liu Z., Xiao T.S., Russell R. J Biol Chem. 2018;293:1924–1932. doi: 10.1074/jbc.M117.815076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.