Abstract
Single-cell omics technologies are currently solving biological and medical problems that earlier have remained elusive, such as discovery of new cell types, cellular differentiation trajectories and communication networks across cells and tissues. Current advances especially in single-cell multi-omics hold high potential for breakthroughs by integration of multiple different omics layers. To pair with the recent biotechnological developments, many computational approaches to process and analyze single-cell multi-omics data have been proposed. In this review, we first introduce recent developments in single-cell multi-omics in general and then focus on the available data integration strategies. The integration approaches are divided into three categories: early, intermediate, and late data integration. For each category, we describe the underlying conceptual principles and main characteristics, as well as provide examples of currently available tools and how they have been applied to analyze single-cell multi-omics data. Finally, we explore the challenges and prospective future directions of single-cell multi-omics data integration, including examples of adopting multi-view analysis approaches used in other disciplines to single-cell multi-omics.
Keywords: Single-cell, Multi-omics, Integration, Clustering
1. Introduction
Recent developments in single-cell omics technologies to measure different modalities such as genome, transcriptome, epigenome, and proteome have enabled unprecedented insight and resolution to cellular phenotypes, biological processes and developmental stages [1], [2], [11], [3], [4], [5], [6], [7], [8], [9], [10]. Single-cell studies can resolve the confounding effects of distinct cell types in heterogeneous samples, that can not be separated with traditional bulk approaches. Recent technological advancements have demonstrated simultaneous assaying of two or more of different omics layers [12], [13], [22], [23], [24], [25], [26], [27], [28], [29], [30], [14], [15], [16], [17], [18], [19], [20], [21]. The multimodal approaches at single-cell resolution are pushing forward a new era of scientific exploration in the field of molecular biology and medicine. Combination of several single-cell omics layers have enabled higher resolution for differentiation processes involved, for instance, in embryonic development [20], [31], development of immune system [32], [33], [34], [35], [36], cancer biology [37], and neuronal development [38], [39]. Additionally, the potential for new translational aspects is high [40]; within known cell-types distinct subpopulations of cells have been discovered to associate with disease versus healthy states, for instance, in the context of somatic cancer evolution [41], heart [42], [43] and neuronal diseases [44], and recurrent miscarriage [45]. Generally, in cancer cells, tumor heterogeneity plays a crucial role in drug resistance, relapse and metastasis. Therefore, accurately identifying tumor subpopulations using multi-omics approaches holds potential in the field of precision medicine. Further, multimodal omics data enables joint analysis of the different players, such as transcripts and proteins, in complex regulatory processes [46].
Computationally, the multimodal single-cell omics profiling has opened up the way for developing models that can relate the interactions and associations among multiple omics layers at single-cell resolution and allows utilization of complementary evidence from the multimodal data [47], [48]. At the core of the single-cell analysis are clustering algorithms that are used to separate cell types or functional cell states, either static or continuous. Strategically, multimodal single-cell data analysis can be roughly divided into three main approaches based on the stage where the integration of the data layers is conducted: early, intermediate, and late integration (Fig. 1). Similar categories have been described earlier in the context of bulk multi-omics data analysis [49], [50]. Early integration concatenates multiple omics data types into one integrated dataset and performs analysis on this data using the same algorithms typically used for the single omics layers. In late integration, analysis is first performed separately on each omics layer and these results are then integrated to determine the final consensus results. In intermediate integration, the multiple omics layers are analyzed together, including integration of sample similarities, joint dimension reduction techniques, and statistical modeling approaches [49].
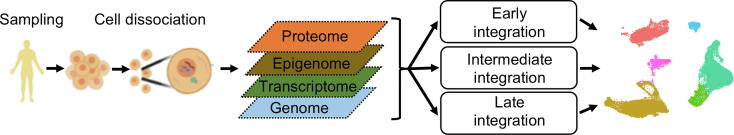
Fig. 1.
Single-cell multi-omics workflow. The first step in the workflow is sample extraction where cells are harvested, for example, from blood or tissues. Next, the extracted cells are dissociated and used to profile multiple layers of omics data from individual cells. In the computational analysis three data integration strategies can be used: early, intermediate and late data integration. In the end, for instance, distinct cell types and cell states can be recognized by clustering.
In this review, we provide a coarse overview of the recent development in different approaches for integrative single-cell multi-omics analysis and clustering. We focus on the basic principles and strategies and provide examples of the available tools and software utilizing the different strategies. We also briefly discuss the challenges and future directions for the method development and application.
2. Single-cell multi-omics data
The single-cell omics datasets can either be matched, i.e. different omics layers have been measured simultaneously from the same individual cell with recent techniques such as Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITE-Seq), RNA expression and protein sequencing assay (REAP-seq), gDNA-mRNA sequencing (DR-seq) or single-cell methylome and transcriptome sequencing (scM&T-seq) [12], [16], [24], [27] with a comprehensive listing in [51], or unmatched, i.e. different omics layers have been measured from different single-cell experimental samples [52]. Compared to matched multimodal data, the unmatched multi-omics datasets have a relatively higher source of variation as the different omics layers originate from different cells and experimental setups [48]. Despite the challenge in addressing different sources of variations and batch effects, the unmatched single-cell multi-omics data integration has large potential to reveal novel biological insights because of the high quantity of single-modality single-cell data generated in recent years. Until recently, measurement of one layer of single-cell data has been economically a far more reachable and easier option than matched multi-omics. Hence, in several cases where related data are available, integrating these is still a viable option for wider research community. Also, several of the current data analysis methods have been developed using unmatched data. The first comparisons to provide details on the increased accuracy of the matched data are only currently emerging. As a very preliminary example, a recent study compared computationally inferred cluster assignments from matched single-cell RNA sequencing (scRNA-seq) and single-cell Assay for Transposase-Accessible Chromatin using sequencing (scATAC-seq) datasets to (de facto) measured couplings and reported highly variable and dataset dependent accuracy (37–75%) for the computational inference; however, the clustering miss-assignments represented related cell types [1].
Single-cell transcriptomic (scRNA-seq) data is by far the most commonly assayed single-cell data type. Epigenomic (scATAC-seq and methylome) data are typically sparser than scRNA-seq data, leading to a situation where the integration strategy should be weighted to take into account the unbalanced information content. One simplistic solution here is to transfer clusters or cell type labels from information-rich scRNA-seq data to another more sparse data layer [53], [54]. On the other hand, when cell surface receptor data is available from CITE-seq, then it may be biologically relevant to use the well-known protein markers to guide the clustering of scRNA-seq results [55].
Considering experimental design, scRNA-seq from whole single cells commonly requires fresh tissues, whereas nuclear samples (nuclear snRNA-seq, ATAC-seq and methylome) can be frozen specimens, which greatly facilitates projects with extensive sampling. Promisingly, although nuclear transcriptome sequencing have less coverage and depth compared to full cell scRNA-seq of mRNA, recent comparisons have suggested that majority of the expression changes can be retrieved from single-nuclei RNA-seq [56], [57], further motivating the use of matched nuclei samples (RNA + ATAC, RNA + methylome etc.) for gene regulation studies. Notably, some assays retrieve matched genomic/chromatin and RNA from nuclei such as the commercially available (10X Genomics) snRNA/ATAC-seq, whereas others such as scM&T-seq or single-cell Chromatin Accessibility and Transcriptome sequencing (scCAT-seq) [16], [58] combine nuclear genomic/chromatin collection with transcriptome assay from cytoplasm. Further, methods to simultaneously assay three omics modalities simultaneously from single cells are currently developed, such as scNOMeRe [59] that collects transcriptome data from cytoplasmic extract and DNA methylation and chromatin accessibility data from nuclei or scNMT-seq [29] that instead of chromatin accessibility collects nucleosome data.
Another developing front is the integration of the above mentioned liquid omics with spatial transcriptomics and other data that preserves the information of tissue structures [60] Also, for translational aspects, single-cell diagnostic such as the detection of circulating tumor cells (CTCs) [61], [62], [63] are emerging, and in the future these may also include integration of several omics layers.
3. Single-cell multi-omics data integration strategies
A primary goal of single-cell analysis is to discover known and novel cell populations. Hence, the data analysis methods to achieve this goal most often use an unsupervised approach. Additionally, some semi-supervised approaches have been suggested [64]. Here, we describe the general single-cell multi-omics integration strategies, divided into early, intermediate and late integration (Fig. 2). While some single-cell applications have used early or late data integration, intermediate integration approach has been most widely used in integrating single-cell multi-omics data.
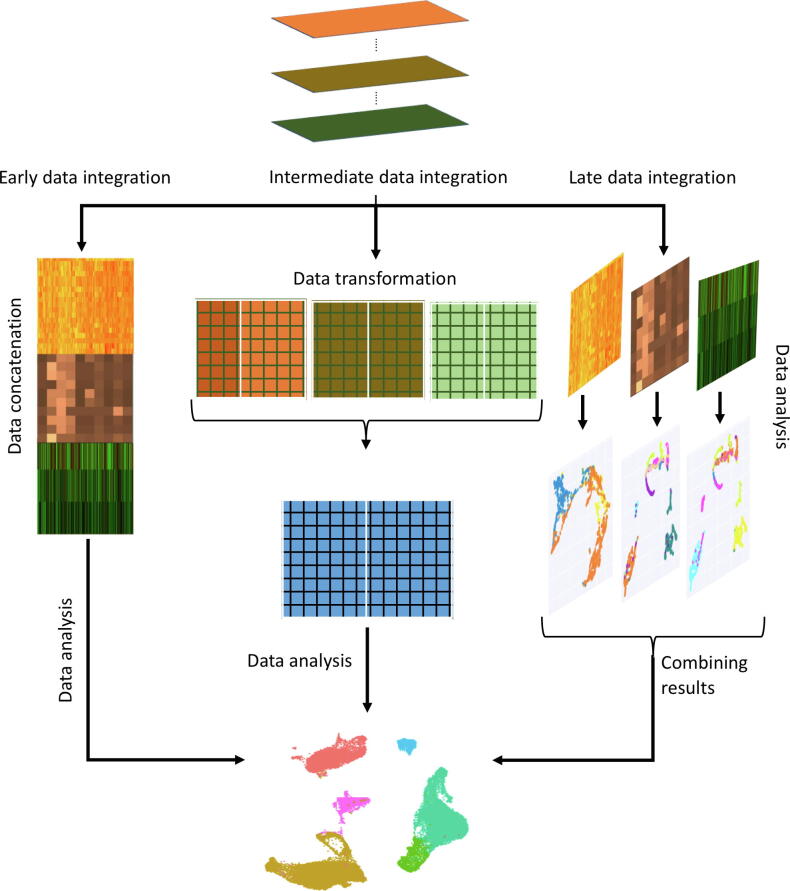
Fig. 2.
Schematic illustration of the early, intermediate and late data integration strategies in single-cell multi-omics analysis. In early data integration, multiple omics datasets are concatenated together for downstream analysis. By default, early integration increases the dimensionality of the data and does not account for the different distribution of the values in each separate omics layer. The intermediate data integration strategy covers a range of techniques to jointly analyze multiple omics datasets. Typically, this is done by transforming the datasets to a single integrated data matrix using, for instance, similarity-based integration, joint dimensionality reduction, or statistical modeling-based approaches. The late data integration strategy first employs the data analysis separately for each omics layer and then integrates these results to create a consensus result.
In early data integration, multiple omics data layers are first concatenated as a single merged data matrix before proceeding into the analysis. A merged data matrix can then be used as an input for machine learning methods that are able to consider any type of dependences between the features [65]. The advantages of this approach include relatively easy application of any method that can utilize a data matrix. However, the merged data matrix increases complexity beyond single omics data and hence early data integration approaches often utilize automatic feature learning, such as dimensionality reduction and representation learning [66]. Feature learning methods, such as autoencoders combine multiple omics layers with variable numbers of features into a compressed data matrix at the hidden layer to create an integrated representation from the multi-omics data. However, as such autoencoders are conceptually closer to intermediate integration approaches. In bulk omics setting, the early integration has been applied, for instance, for tumor subtyping involving jointly all the considered omics [67]. In single-cell omics setting, early data integration has been most commonly applied to combine multiple datasets of the same omics type from different studies, such as scRNAseq data from multiple sources coupled with different normalization and scaling steps [68] A major challenge of early data integration is that the features from multiple omics datasets are often different both in terms of dimension and scale that may lead to more weight on an omics layer with more dimensions unless properly normalized [69]. Furthermore, the sparsity and the high-dimensional nature of omics datasets make it challenging to construct a common representation across multiple datasets. This could be addressed by lower dimensional embedding of an individual datasets retaining the overall structure of the original data followed by the subsequent data integration technique, including several linear [70] and nonlinear [71] methods.
The late integration strategy first employs the analysis separately for each omics data layer and then integrates these results to create a consensus result. These methods have previously been used to integrate separate scRNA-seq experiment or in bulk multi-omics but have not yet been widely applied to single-cell multi-omics. For instance, mixture model ensemble clustering has been applied to combine multiple scRNA-seq clustering results and could readily be applied to create a consensus clustering of multi-omics data [72]. It models the interdependence of local clustering results with the aim to find a robust and improved global clustering solution across multiple data sources through optimization. The SAME-clustering [72] tool implements a mixture model ensemble method for aggregating clustering solutions generated from different clustering algorithms in scRNA-seq data aiming to land in a robust consensus clustering. The Graph Partitioning-Based Cluster Ensemble Method Sc-GPE [73] and the Ensemble Clustering Based on Probability Graphical Model With Graph Regularization EC-PGMGR [74] use graph-based ensemble clustering. These approaches can also be applied for single-cell multi-omics clustering, as they give flexibility to use different omics specific clustering algorithms to generate the best local clustering solutions. On the other hand, potential late integration approaches have been employed in bulk multi-omics, including Cluster-of-clusters analysis (COCA) [75], a two-step integrative clustering algorithm that performs integrative cluster analysis summarizing the clustering results found from multiple omics datasets. In Kernel Learning Integrative Clustering (KLIC) [76] multiple clustering structures are integrated as a multiple kernel learning problem where each of the datasets provide a weighted contribution to the final clustering. Obviously, as late integration algorithms often take a clustering result as an input, they directly fit to a workflow where unmatched single-cell multi-omics datasets are first analyzed separately.”
The intermediate integration covers a range of techniques that aim to jointly analyze the different omics layers together using, for instance, similarity-based integration, joint dimension reduction, or statistical modeling. The similarity-based integration approaches include, for instance, spectral clustering approaches and graph fusion algorithms. The joint dimension reduction techniques aim to find a lower dimensional representation for the single-cell multimodal data layers by projecting them into a common latent space. These include various matrix factorization techniques as well as covariance-based techniques, such as canonical correlation analysis. The statistical modelling techniques for integration utilize, for instance, Bayesian approaches to determine cluster probabilities of cells from multiple omics layers. Representative examples of different tools that apply these approaches are provided in Table 1.
Table 1.
Computational single-cell multi-omics tools applying intermediate integration approaches and their applicable omic data types.
| Tool | Methodology | Single-cell omics types (designed for matched/unmatched) | Refs. |
|---|---|---|---|
| Similarity-based approaches | |||
| SCHEMA | Metric-learning based method | Multi-omics data (matched) | [77] |
| Spectrum | Weighted-nearest neighbor analysis | Multi-omics data (unmatched) | [78] |
| Seurat4 | Weighted-nearest neighbor analysis | Transcriptome and chromatin accessibility or proteome data (matched) | [79] |
| Dimension reduction-based approaches | |||
| BindSC | Canonical correlation analysis | Transcriptome and chromatin accessibility data (matched) | [80] |
| CoupledNMF | Non-negative matrix factorization | Transcriptome and chromatin accessibility data (unmatched) | [53] |
| LIGER | Non-negative matrix factorization | Transcriptome and spatial gene expression data or DNA methylation (unmatched) | [81] |
| MAGAN | Manifold alignment | Multi-omics data (unmatched) | [82] |
| MATCHER | Manifold alignment | Transcriptome and DNA methylation data (matched) | [83] |
| MMD-MA | Manifold alignment | Multi-omics data (matched) | [84] |
| MOFA+ | Factor analysis | Multi-omics data (matched) | [85] |
| scMVAE | Variational autoencoder | Multi-omics data (matched) | [86] |
| Seurat3 | Canonical correlation analysis | Transcriptome and chromatin accessibility data (unmatched) | [87] |
| totalVI | Deep generative model | Transcriptome and proteome data (matched) | [88] |
| Unicom | Manifold alignment | Multi-omics data (unmatched) | [89] |
| Statistical modeling-based approaches | |||
| BREM-SC | Bayesian mixture model | Transcriptome and proteome data (matched) | [90] |
| Clonealign | Statistical model | Transcriptome and genome data (unmatched) | [91] |
4. Computational tools for intermediate integration of single-cell multi-omics data
4.1. Similarity-based approaches
Spectral clustering utilizes similarity matrices as a basis for clustering. The adoption of the multi-view version of spectral clustering can be used to deal with the multi-omics data. Currently, several methods that are applied in bulk multi-omics data integration are being proposed for single-cell multi-omics integration [92], [93], [94], [95], [96]. For example, Spectrum [78] uses a self-tuning density-aware kernel that enhances the similarity between points that share common nearest neighbours. In addition to bulk data, it has been applied on simulated single-cell data [78]. The Pair-wised Co-regularized Multimodal Spectral Clustering (PC-MSC) [97] implements a co-regularization approach to combine multiple kernels representing the different omics layers. The method has been applied to single-cell transcriptome and protein marker data [94]. SCHEMA [77] implements a metric-learning based method [98], which first determines similarities between cells under each modality and then transforms the primary modality so that it has maximum level of agreement with the other modalities.
Graph fusion algorithms construct graphs from each omics layer and map them to a single fused graph. Recently, several graph fusion algorithms [92], [93], [94], [95], [96] have been proposed for integrating graphs in multi-view clustering domains. Generally, once graphs are integrated from multiple omics layers, any conventional clustering methods can be implemented to partition the joint graph into clusters. Crucial for the accuracy of this approach is that geometric properties of the single data layers are sufficiently maintained in the global presentation. Most notably for single-cell omics, the latest version of the widely used Seurat, Seurat4 [79], implements a weighted-nearest neighbor graph-based integration for cluster analysis. It has been applied, for instance, on CITE-seq data of blood cells to improve the discovery of cell states and cell types.
4.2. Dimension reduction-based approaches
Canonical correlation analysis (CCA) is a correlation-based multivariate analysis method to examine the linear relationship between two datasets [99], [100]. A set of linear combinations of all variables in each of the two datasets is determined so that it maximizes the correlation between them and best explains both within and between dataset variability. The high dimensionality, sparsity and variable feature spaces across the different omics layers pose constraints for the linear combinations limiting the biological applicability of CCA. Generally, to solve these issues variants of CCA including sparse CCA [100] and penalized matrix decomposition (PMD) method [101] have been proposed. For instance Seurat3 [87] implements CCA in order to integrate two single-cell omics datasets. It first jointly reduces the dimensionality of two datasets using the diagonalized CCA followed by a search for a mutual nearest neighbor in lower dimensional space, and then establishes the cellular relationship across the datasets as an anchor. This has been used, for example, to integrate scRNA-seq and scATAC-seq data from the mouse visual cortex and scRNA-seq and surface protein expression from bone marrow [87]. Another recent adaptation of CCA for single-cell multi-omics clustering is bindSC [80] which utilizes bi-order canonical correlation analysis (bi-CCA) that captures the correlated variables from both cells and features between two modalities to formulate the canonical correlation vectors in a latent space. While Seurat3 or bindSC can only be applied to two datasets at a time, multiset CCA [102] aims to simultaneously find multivariate associations between more than two modalities. In multiset CCA, the canonical coefficients of all variables are optimized to maximize the pairwise canonical correlations [103]. Currently, we are not aware of multiset CAA being applied to single-cell multi-omics.
Non-negative matrix factorization (NMF) extracts a low-dimensional non-negative representation of the high-dimensional data that is typically sparse. LIGER (linked inference of genomic experimental relationships) [81] is a recently introduced tool for single-cell multi-omics analysis that utilizes integrative non-negative matrix factorization (iNMF) [104] in order to identify the shared and dataset specific factors across the datasets. It was applied to spatial and scRNA-seq data from mouse brain frontal cortex in order to cluster cell subtypes, and to scRNA-seq and DNA methylation data from mouse cortical to perform integrative cluster analysis [81]. Further, recently [105] extended the iNMF implementation of LIGER to make it an online learning algorithm [106] where multiple datasets are used as mini-batches in a continual cycle allowing fast and memory efficient integration of large multimodal datasets. Another NMF-based implementation for scRNA-seq and scATAC-seq data coupledNMF [53] formulates an optimization problem to couple the information from each dataset during the cluster optimization. The factor analysis-based tool MOFA [107] and its improved version MOFA+ [85], on the other hand, use a variational Bayesian inference framework and have been applied to both bulk and single-cell multi-omics analysis.
Manifold alignment is a class of machine learning algorithms that produce projections between sets of data that lie on a common manifold [108]. The idea is to create a low-dimensional representation (or manifold) for each dataset and then align these representations (manifolds) in a common space where the different datasets are directly comparable. Manifold alignment algorithms can be supervised, semi-supervised, or unsupervised based on the level of available correspondence information among disparate datasets. The currently available manifold alignment tools are unsupervised, such as MATCHER [83] which has been applied on matched and unmatched single-cell transcriptome and DNA methylation data. The method assumes that the variation among cells can be explained mainly by a single latent variable. Another tool, Manifold-Aligning GAN (MAGAN) [82], is a generative adversarial network (GAN) based manifold alignment tool for single-cell multi-omics analysis. It has demonstrated its efficiency in integrating scRNA-seq and proteomic (mass cytometry) datasets. Other manifold tools that have been introduced for single-cell multi-omics include, for example, Unicom [89] and MMD-MA [84].
Autoencoders [109] are neural networks that unfold the underlying nonlinear patterns from multiple high-dimensional datasets by compressing them into a unified lower-dimensional subspace. Architecturally, autoencoders have an input, hidden and output layers with the bottleneck in the middle showing the most compressed form of the input data at subspace. The encoder part of the neural network compresses the input data so as to store the compressed data at the bottleneck layer, whereas the decoder part decompresses the data to regenerate the original input data as an output. The compressed data can then be used for further analysis. Two variations of autoencoders have been recently applied in single-cell multi-omics, variational autoencoders (VAE) [86], [88], and adversarial autoencoders (AAE) [110]. The advantage of variational autoencoders is that they encode the latent attributes of the input in a probabilistic distribution instead of a deterministic single value. This approach has been used in totalVI [88] for jointly transforming the RNA and protein data into joint lower-dimensional cell states. The single-cell multimodal variational autoencoder (scMVAE) was recently used in integrative analysis of scRNA-seq and scATAC-seq data [86]. Additionally, an adversarial autoencoder method [110] was recently developed and applied to integrate scRNA-seq and imaging data. Adversarial autoencoders take advantage of GANs to more accurately integrate the data layers [111].
4.3. Statistical modeling-based approaches
Bayesian framework allows probabilistic modeling of multi-omics data. For instance, Dirichlet mixture model can be used to construct a context-dependent Bayesian clustering framework that can be used for clustering multiple omics datasets on the level of individual omics, while also simultaneously extracting global multi-omics structure [112]. The probabilistic model-based algorithm BREM-SC [90] utilizes Dirichlet multinomial distribution and introduces specific random effects in order to correlate between different omics layers. It was recently applied on gene expression and surface protein expression data. Clonealign [91] also implements a statistical framework for integrating gene expression and copy number profiles from unmatched single-cell RNA-seq and scDNA-seq data to assign gene expression states to cancer clones. The inference is done using a mean field variational Bayes approach. Other Bayesian frameworks for integrative model-based clustering have been proposed for clustering multi-omics data in bulk studies [113], [114]. Such methods can be a useful asset to be tested in the context of single-cell multimodal cluster analysis.
5. Summary and outlook
Single-cell technology is having enormous impact on the discovery of novel cell-types and defining more accurate cell differentiation trajectories, as well as translational effects on precision medicine. Clustering is a widely used unsupervised machine learning method used for analyzing cellular heterogeneity in both single-cell mono- and multi-omics analysis. In the multi-omics analysis, we discussed early, intermediate and late data integration strategies together with recently introduced single-cell multi-omics analytical tools. These tools apply algorithms and analysis methods that have previously been developed in a wider framework of multi-view analysis [115], [116] in different fields, such as text mining [54], image/video analysis [116], [117] and bulk multi-omics analysis [49], [50], [118]. Many of these methods still remain unexplored in single-cell multi-omics analysis and we expect them to be intensively examined in that context in the near future. Here we expand our previous description of the specific tools already used in the field of single-cell multi-omics by discussing multi-view approaches that have been utilized in other fields not yet applied to single-cell multi-omics.
Currently the most widely used multi-omics integration approaches reduce the datasets to a single data matrix from multiple omics datasets using CCAs, manifold alignments, graph-based integration techniques, or autoencoders before performing cluster analysis. The CCAs, that have most often been used via for instance Seurat3, could in the future be further developed to take into account the potential advances of sparse CCA [100], [101], [119]. The non-linearity aspect of the high-dimensional single-cell multi-omics data could be also dealt with other CCA variants, such as kernel CCA [103], [120] or deep CCA [121]. Further, importantly, new unified distributional embedding methods, such as Multi-view Neighborhood Embedding (MvNE) [50] are potentially relevant additions in single-cell omics.
In general, data integration approaches where each of the omics datasets are jointly used for optimization can be considered to have advantage. For this there still remains a variety of clustering implementations for multi-view data in a co-training fashion that have not been properly tested for single-cell multi-omics clustering, while their utility in other disciplines such text mining is more established. For example, multi-view k-means clustering has proved its effectiveness in the fields of image analysis [122], [123], [124], [125], [126], [127], whereas Cluster-of-clusters analysis (COCA) [75], Kernel Learning Integrative Clustering (KLIC) [76] and perturbation-based clustering [128] have been used in bulk multi-omics cluster analysis but have not yet been widely applied for single-cell multi-omics. The benefit of late integration approaches, on the other hand, is the flexibility for the different algorithms that are used at each of the individual omics layers before integration into an ensemble solution.
Currently, several single-cell multi-omics tools have been developed to address the integration and clustering of multi-omics datasets (Table 1), but comprehensive and objective comparison and benchmarking of these recent methods is yet to be conducted and in high demand. Additionally, the current multi-modal analysis tools mostly focus on integrative clustering of multi-omics data with the aim to identify the shared cell type heterogeneity. More tools are needed that are capable of addressing various biological questions from matched single-cell multi-omics data, such as integrative motif discovery and inference of gene regulatory networks or combining spatial expression patterns with liquid based sequencing results.
The future is likely to bring more robust and improved technological advancement in the area of single-cell multi-modal profiling, enabling multitudes of omics and other data such as imaging from a single cell. This will open up new opportunities in finding novel insights in relation to the biological mechanisms answering key questions related to diseases and advances in personalized medicine. Future developments include advanced simultaneous assays for three [59] or more omics modalities, and more solutions for preserved samples in order to enhance practicality of wet-lab and the possibility to study large clinical cohorts. Also, there remain challenges in relation to data storage, management and analytical aspects. In terms of data storage and management, there are few efforts to aggregate the multi-omics data in bulk setups [34], [129], [130], [131], [132]. So far, however, there is no unified single-cell multi-omics platform that encompasses the multi-modal single-cell omics data in a repository, except some efforts taken by the recent activities under the human cell atlas project [133]. Therefore, gathering the growing multi-modal single-cell multi-omics data in a unified repository would facilitate a collaborative work towards computational multi-omics analysis. In terms of cluster analytics, the multi-modal single-cell analysis has already benefited from the recently advanced multi-view machine learning methodologies [54], [134], [135] and these will continue to advance the computational analysis of single-cell multi-omics data.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
NA has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No.: 675395. LLE reports grants from the European Research Council ERC (677943), European Union's Horizon 2020 research and innovation programme (675395), Academy of Finland (296801, 304995, 310561, 314443, and 329278), and Sigrid Juselius Foundation during the conduct of the study. Our research is also supported by University of Turku Graduate School (UTUGS), Biocenter Finland, and ELIXIR Finland. Work of KTR was also supported by Eemil Aaltonen Foundation, Juhani Aho Foundation and Waldemar von Frenckell Foundation.
Contributor Information
Kalle T. Rytkönen, Email: katury@utu.fi.
Laura L. Elo, Email: laura.elo@utu.fi.
References
- 1.Ma S., Zhang B., LaFave L.M., Earl A.S., Chiang Z., Hu Y. Chromatin Potential Identified by Shared Single-Cell Profiling of RNA and Chromatin. Cell. 2020;183(4):1103–1116.e20. doi: 10.1016/j.cell.2020.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hwang B., Lee J.H., Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med. 2018;50(8):1–14. doi: 10.1038/s12276-018-0071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanter I., Kalisky T. Single cell transcriptomics: Methods and applications. Front Oncol. 2015:5. doi: 10.3389/fonc.2015.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kester L., van Oudenaarden A. Single-Cell Transcriptomics Meets Lineage Tracing. Cell Stem Cell. 2018;23(2):166–179. doi: 10.1016/j.stem.2018.04.014. [DOI] [PubMed] [Google Scholar]
- 5.Schwartzman O., Tanay A. Single-cell epigenomics: Techniques and emerging applications. Nat Rev Genet. 2015;16(12):716–726. doi: 10.1038/nrg3980. [DOI] [PubMed] [Google Scholar]
- 6.Ai S., Xiong H., Li C.C., Luo Y., Shi Q., Liu Y. Profiling chromatin states using single-cell itChIP-seq. Nat Cell Biol. 2019;21(9):1164–1172. doi: 10.1038/s41556-019-0383-5. [DOI] [PubMed] [Google Scholar]
- 7.Pott S., Lieb J.D. Single-cell ATAC-seq: Strength in numbers. Genome Biol. 2015;16(1) doi: 10.1186/s13059-015-0737-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smallwood S.A., Lee H.J., Angermueller C., Krueger F., Saadeh H., Peat J. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods. 2014;11(8):817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marx V. A dream of single-cell proteomics. Nat Methods. 2019;16(9):809–812. doi: 10.1038/s41592-019-0540-6. [DOI] [PubMed] [Google Scholar]
- 10.Su Y., Shi Q., Wei W. Single cell proteomics in biomedicine: High-dimensional data acquisition, visualization, and analysis. Proteomics. 2017;17(3-4):1600267. doi: 10.1002/pmic.v17.3-410.1002/pmic.201600267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nam AS, Dusaj N, Izzo F, Murali R, Mouhieddine TH, Myers RM, et al. Single-Cell Multi-Omics in Human Clonal Hematopoiesis Reveals That DNMT3A R882 Mutations Perturb Early Progenitor States through Selective Hypomethylation. Blood 2020. Doi: 10.1182/blood-2020-142574. [DOI] [PMC free article] [PubMed]
- 12.Dey S.S., Kester L., Spanjaard B., Bienko M., van Oudenaarden A. Integrated genome and transcriptome sequencing of the same cell. Nat Biotechnol. 2015;33(3):285–289. doi: 10.1038/nbt.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han K.Y., Kim K.-T., Joung J.-G., Son D.-S., Kim Y.J., Jo A. SIDR: simultaneous isolation and parallel sequencing of genomic DNA and total RNA from single cells. Genome Res. 2018;28(1):75–87. doi: 10.1101/gr.223263.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macaulay I.C., Haerty W., Kumar P., Li Y.I., Hu T.X., Teng M.J. G&T-seq: Parallel sequencing of single-cell genomes and transcriptomes. Nat Methods. 2015;12:519–522. doi: 10.1038/nmeth.3370. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Meira A., Buck G., Clark S.-A., Povinelli B.J., Alcolea V., Louka E. Unravelling Intratumoral Heterogeneity through High-Sensitivity Single-Cell Mutational Analysis and Parallel RNA Sequencing. Mol Cell. 2019;73(6):1292–1305.e8. doi: 10.1016/j.molcel.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Angermueller C., Clark S.J., Lee H.J., Macaulay I.C., Teng M.J., Hu T.X. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat Methods. 2016;13(3):229–232. doi: 10.1038/nmeth.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Y., Huang K., An Q., Du G., Hu G., Xue J. Simultaneous profiling of transcriptome and DNA methylome from a single cell. Genome Biol. 2016;17(1) doi: 10.1186/s13059-016-0950-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao J., Cusanovich D.A., Ramani V., Aghamirzaie D., Pliner H.A., Hill A.J. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science. 2018;361(6409):1380–1385. doi: 10.1126/science:aau0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen S., Lake B.B., Zhang K. High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nat Biotechnol. 2019;37(12):1452–1457. doi: 10.1038/s41587-019-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo F., Li L., Li J., Wu X., Hu B., Zhu P. Single-cell multi-omics sequencing of mouse early embryos and embryonic stem cells. Cell Res. 2017;27(8):967–988. doi: 10.1038/cr.2017.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pott S. Simultaneous measurement of chromatin accessibility, DNA methylation, and nucleosome phasing in single cells. Elife 2017;6. Doi: 10.7554/eLife.23203. [DOI] [PMC free article] [PubMed]
- 22.Kochan J., Wawro M., Kasza A. Simultaneous detection of mRNA and protein in single cells using immunofluorescence-combined single-molecule RNA FISH. Biotechniques. 2015;59:209–221. doi: 10.2144/000114340. [DOI] [PubMed] [Google Scholar]
- 23.Nestorowa S, Hamey FK, Pijuan Sala B, Diamanti E, Shepherd M, Laurenti E, et al. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood 2016;128:e20–31. Doi: 10.1182/blood-2016-05-716480. [DOI] [PMC free article] [PubMed]
- 24.Peterson V.M., Zhang K.X., Kumar N., Wong J., Li L., Wilson D.C. Multiplexed quantification of proteins and transcripts in single cells. Nat Biotechnol. 2017;35(10):936–939. doi: 10.1038/nbt.3973. [DOI] [PubMed] [Google Scholar]
- 25.Soh K.T., Tario J.D., Colligan S., Maguire O., Pan D., Minderman H. Simultaneous, single-cell measurement of messenger RNA, cell surface proteins, and intracellular proteins. Curr Protoc Cytom. 2016;75(1) doi: 10.1002/0471142956.2016.75.issue-110.1002/0471142956.cy0745s75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frei A.P., Bava F.-A., Zunder E.R., Hsieh E.W.Y., Chen S.-Y., Nolan G.P. Highly multiplexed simultaneous detection of RNAs and proteins in single cells. Nat Methods. 2016;13(3):269–275. doi: 10.1038/nmeth.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoeckius M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P.K., Swerdlow H. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14(9):865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou Y.u., Guo H., Cao C., Li X., Hu B., Zhu P. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 2016;26(3):304–319. doi: 10.1038/cr.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark S.J., Argelaguet R., Kapourani C.-A., Stubbs T.M., Lee H.J., Alda-Catalinas C. ScNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells e. Nat Commun. 2018;9(1) doi: 10.1038/s41467-018-03149-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu L., Liu C., Quintero A., Wu L., Yuan Y., Wang M. Deconvolution of single-cell multi-omics layers reveals regulatory heterogeneity. Nat Commun. 2019;10(1) doi: 10.1038/s41467-018-08205-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L., Guo F., Gao Y., Ren Y., Yuan P., Yan L. Single-cell multi-omics sequencing of human early embryos. Nat Cell Biol. 2018;20(7):847–858. doi: 10.1038/s41556-018-0123-2. [DOI] [PubMed] [Google Scholar]
- 32.Gaiti F., Chaligne R., Gu H., Brand R.M., Kothen-Hill S., Schulman R.C. Epigenetic evolution and lineage histories of chronic lymphocytic leukaemia. Nature. 2019;569(7757):576–580. doi: 10.1038/s41586-019-1198-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gomes T., Teichmann S.A., Talavera-López C. Immunology Driven by Large-Scale Single-Cell Sequencing Unraveling the Immune System One Cell at a Time Trends in Immunology. Trends Immunol. 2019;40(11):1011–1021. doi: 10.1016/j.it.2019.09.004. [DOI] [PubMed] [Google Scholar]
- 34.Gomez-Cabrero D., Tarazona S., Ferreirós-Vidal I., Ramirez R.N., Company C., Schmidt A. STATegra, a comprehensive multi-omics dataset of B-cell differentiation in mouse. Sci Data. 2019;6(1) doi: 10.1038/s41597-019-0202-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samir J., Rizzetto S., Gupta M., Luciani F. Exploring and analysing single cell multi-omics data with VDJView. BMC Med Genomics. 2020;13(1) doi: 10.1186/s12920-020-0696-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park J.-E., Botting R.A., Domínguez Conde C., Popescu D.-M., Lavaert M., Kunz D.J. A cell atlas of human thymic development defines T cell repertoire formation. Science. 2020;367(6480):eaay3224. doi: 10.1126/science:aay3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng A., Mao X., Zhong J., Fan S., Hu Y. Single-Cell Multi-Omics and Its Prospective Application in Cancer Biology. Proteomics. 2020;20(13):1900271. doi: 10.1002/pmic.v20.1310.1002/pmic.201900271. [DOI] [PubMed] [Google Scholar]
- 38.Golomb S.M., Guldner I.H., Zhao A., Wang Q., Palakurthi B., Aleksandrovic E.A. Multi-modal Single-Cell Analysis Reveals Brain Immune Landscape Plasticity during Aging and Gut Microbiota Dysbiosis. Cell Rep. 2020;33(9):108438. doi: 10.1016/j.celrep.2020.108438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayer S., Chen J., Velmeshev D., Mayer A., Eze U.C., Bhaduri A. Multimodal Single-Cell Analysis Reveals Physiological Maturation in the Developing Human Neocortex. Neuron. 2019;102(1):143–158.e7. doi: 10.1016/j.neuron.2019.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bock C., Farlik M., Sheffield N.C. Multi-Omics of Single Cells: Strategies and Applications. Trends Biotechnol. 2016;34(8):605–608. doi: 10.1016/j.tibtech.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nam A.S., Chaligne R., Landau D.A. Integrating genetic and non-genetic determinants of cancer evolution by single-cell multi-omics. Nat Rev Genet. 2021;22(1):3–18. doi: 10.1038/s41576-020-0265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jia G., Preussner J., Chen X.i., Guenther S., Yuan X., Yekelchyk M. Single cell RNA-seq and ATAC-seq analysis of cardiac progenitor cell transition states and lineage settlement. Nat Commun. 2018;9(1) doi: 10.1038/s41467-018-07307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yifan C., Fan Y., Jun P. Visualization of cardiovascular development, physiology and disease at the single-cell level: Opportunities and future challenges. J Mol Cell Cardiol. 2020;142:80–92. doi: 10.1016/j.yjmcc.2020.03.005. [DOI] [PubMed] [Google Scholar]
- 44.Lake B.B., Chen S., Sos B.C., Fan J., Kaeser G.E., Yung Y.C. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat Biotechnol. 2018;36(1):70–80. doi: 10.1038/nbt.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lucas E.S., Vrljicak P., Muter J., Diniz-da-Costa M.M., Brighton P.J., Kong C.-S. Recurrent pregnancy loss is associated with a pro-senescent decidual response during the peri-implantation window. Commun Biol. 2020;3(1) doi: 10.1038/s42003-020-0763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Behjati Ardakani F, Kattler K, Heinen T, Schmidt F, Feuerborn D, Gasparoni G, et al. Prediction of single-cell gene expression for transcription factor analysis. Gigascience 2020;9. Doi: 10.1093/gigascience/giaa113. [DOI] [PMC free article] [PubMed]
- 47.Efremova M., Teichmann S.A. Computational methods for single-cell omics across modalities. Nat Methods. 2020;17(1):14–17. doi: 10.1038/s41592-019-0692-4. [DOI] [PubMed] [Google Scholar]
- 48.Ma A., McDermaid A., Xu J., Chang Y., Ma Q. Integrative Methods and Practical Challenges for Single-Cell Multi-omics. Trends Biotechnol. 2020;38(9):1007–1022. doi: 10.1016/j.tibtech.2020.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rappoport N., Shamir R. Multi-omic and multi-view clustering algorithms: review and cancer benchmark. Nucleic Acids Res. 2018;46:10546–10562. doi: 10.1093/nar/gky889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mitra S., Saha S., Li Y. A multiobjective multi-view cluster ensemble technique: Application in patient subclassification. PLoS ONE. 2019;14(5):e0216904. doi: 10.1371/journal.pone.0216904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu C., Preissl S., Ren B. Single-cell multimodal omics: the power of many. Nat Methods. 2020;17(1):11–14. doi: 10.1038/s41592-019-0691-5. [DOI] [PubMed] [Google Scholar]
- 52.Macaulay I.C., Ponting C.P., Voet T. Single-Cell Multiomics: Multiple Measurements from Single Cells. Trends Genet. 2017;33(2):155–168. doi: 10.1016/j.tig.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duren Z., Chen X.i., Zamanighomi M., Zeng W., Satpathy A.T., Chang H.Y. Integrative analysis of single-cell genomics data by coupled nonnegative matrix factorizations. Proc Natl Acad Sci U S A. 2018;115(30):7723–7728. doi: 10.1073/pnas.1805681115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu X, Prof JSV, Chairman CV, Moor PB De, Prof P, Prof JSV, et al. Learning from multi-view data: clustering algorithm and text mining application; 2011.
- 55.Wang X., Xu Z., Zhou X., Zhang Y., Huang H., Ding Y. SECANT: A biology-guided semi-supervised method for clustering, classification, and annotation of single-cell multi-omics. BioRxiv. 2020 doi: 10.1101/2020.11.06.371849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ding J., Adiconis X., Simmons S.K., Kowalczyk M.S., Hession C.C., Marjanovic N.D. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat Biotechnol. 2020;38(6):737–746. doi: 10.1038/s41587-020-0465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Slyper M., Porter C.B.M., Ashenberg O., Waldman J., Drokhlyansky E., Wakiro I. A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nat Med. 2020;26(5):792–802. doi: 10.1038/s41591-020-0844-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu Y, Zhong J, Xiao Y, Xing Z, Sheu K, Fan S, et al. ScCAT-seq: Single-cell identification and quantification of mRNA isoforms by cost-effective short-read sequencing of cap and tail. BioRxiv 2019:2019.12.11.873505. Doi: 10.1101/2019.12.11.873505.
- 59.Wang Y., Yuan P., Yan Z., Yang M., Huo Y., Nie Y. Single-cell multiomics sequencing reveals the functional regulatory landscape of early embryos. Nat Commun. 2021;12(1) doi: 10.1038/s41467-021-21409-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waylen L.N., Nim H.T., Martelotto L.G., Ramialison M. From whole-mount to single-cell spatial assessment of gene expression in 3D. Commun Biol. 2020;3:602. doi: 10.1038/s42003-020-01341-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zeune L.L., Boink Y.E., van Dalum G., Nanou A., de Wit S., Andree K.C. Deep learning of circulating tumour cells. Nat Mach Intell. 2020;2(2):124–133. doi: 10.1038/s42256-020-0153-x. [DOI] [Google Scholar]
- 62.Miccio L., Cimmino F., Kurelac I., Villone M.M., Bianco V., Memmolo P. Perspectives on liquid biopsy for label-free detection of “circulating tumor cells” through intelligent lab-on-chips. View. 2020;1(3):20200034. doi: 10.1002/viw2.v1.310.1002/VIW.20200034. [DOI] [Google Scholar]
- 63.Zhang R.u., Le B., Xu W., Guo K., Sun X., Su H. Magnetic “Squashing” of Circulating Tumor Cells on Plasmonic Substrates for Ultrasensitive NIR Fluorescence Detection. Small Methods. 2019;3(2):1800474. doi: 10.1002/smtd.v3.210.1002/smtd.201800474. [DOI] [Google Scholar]
- 64.Chen L., He Q., Zhai Y., Deng M. Single-cell RNA-seq data semi-supervised clustering and annotation via structural regularized domain adaptation. Bioinformatics. 2020 doi: 10.1093/bioinformatics/btaa908. [DOI] [PubMed] [Google Scholar]
- 65.Zitnik M, Nguyen F, Wang B, Leskovec J, Goldenberg A, Hoffman MM. Machine learning for integrating data in biology and medicine: Principles, practice, and opportunities. Inf Fusion 2019;50:71–91. Doi: 10.1016/j.inffus.2018.09.012. [DOI] [PMC free article] [PubMed]
- 66.Vincent P., Larochelle H., Lajoie I., Bengio Y., Manzagol P.A. Stacked denoising autoencoders: Learning Useful Representations in a Deep Network with a Local Denoising Criterion. J Mach Learn Res. 2010;11:3371–3408. [Google Scholar]
- 67.Lock E.F., Hoadley K.A., Marron J.S., Nobel A.B. Joint and individual variation explained (JIVE) for integrated analysis of multiple data types. Ann Appl Stat. 2013;7:523–542. doi: 10.1214/12-AOAS597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin Y., Ghazanfar S., Wang K.Y.X., Gagnon-Bartsch J.A., Lo K.K., Su X. ScMerge leverages factor analysis, stable expression, and pseudoreplication to merge multiple single-cell RNA-seq datasets. Proc Natl Acad Sci U S A. 2019;116(20):9775–9784. doi: 10.1073/pnas.1820006116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sharifi-Noghabi H., Zolotareva O., Collins C.C., Ester M. MOLI: Multi-omics late integration with deep neural networks for drug response prediction. Bioinformatics. 2019;35:i501–i509. doi: 10.1093/bioinformatics/btz318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maćkiewicz A., Ratajczak W. Principal components analysis (PCA) Comput Geosci. 1993;19(3):303–342. doi: 10.1016/0098-3004(93)90090-R. [DOI] [Google Scholar]
- 71.Roweis S.T., Saul L.K. Nonlinear dimensionality reduction by locally linear embedding. Science. 2000;290:2323–2326. doi: 10.1126/science.290.5500.2323. [DOI] [PubMed] [Google Scholar]
- 72.Huh R., Yang Y., Jiang Y., Shen Y., Li Y. SAME-clustering: Single-cell Aggregated Clustering via Mixture Model Ensemble. Nucleic Acids Res. 2020;48:86–95. doi: 10.1093/nar/gkz959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu X., Li J., Li H.D., Xie M., Sc-GPE W.J. A Graph Partitioning-Based Cluster Ensemble Method for Single-Cell. Front Genet. 2020;11. doi: 10.3389/fgene.2020.604790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu Y., Zhang D.-X., Zhang X.-F., Yi M., Ou-Yang L.e., Wu M. Ensemble Clustering Based on Probability Graphical Model With Graph Regularization for Single-Cell RNA-seq Data. Front Genet. 2020;11:11. doi: 10.3389/fgene.2020.572242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koboldt D.C., Fulton R.S., McLellan M.D., Schmidt H., Kalicki-Veizer J., McMichael J.F. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cabassi A, Kirk PDW. Multiple kernel learning for integrative consensus clustering of omic datasets. Bioinformatics 2020;36:4789–96. Doi: 10.1093/bioinformatics/btaa593. [DOI] [PMC free article] [PubMed]
- 77.Singh R., Narayan A., Hie B., Berger B. Schema: A general framework for integrating heterogeneous single-cell modalities. BioRxiv. 2019 doi: 10.1101/834549. [DOI] [Google Scholar]
- 78.John C.R., Watson D., Barnes M.R., Pitzalis C., Lewis M.J. Spectrum: fast density-aware spectral clustering for single and multi-omic data. Bioinformatics. 2019;36:1159–1166. doi: 10.1093/bioinformatics/btz704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., Zheng S., Butler A. Integrated analysis of multimodal single-cell data. BioRxiv. 2020 doi: 10.1101/2020.10.12.335331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dou J., Liang S., Mohanty V., Cheng X., Kim S., Choi J. Unbiased integration of single cell multi-omics data. BioRxiv. 2020 doi: 10.1101/2020.12.11.422014. [DOI] [Google Scholar]
- 81.Martin C., Welch J., Kozareva V., Ferreira A., Vanderburg C., Martin C. Integrative inference of brain cell similarities and differences from single-cell genomics. BioRxiv. 2018 doi: 10.1101/459891. [DOI] [Google Scholar]
- 82.Amodio M, Krishnaswamy S. MAGAN: Aligning biological manifolds. 35th Int. Conf. Mach. Learn. ICML 2018, vol. 1, 2018, p. 327–35.
- 83.Welch J.D., Hartemink A.J., Prins J.F. Manifold alignment reveals correspondence between single cell transcriptome and epigenome dynamics. Genome Biol. 2017;18(1) doi: 10.1186/s13059-017-1269-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu J., Huang Y., Singh R., Vert J.P., Noble W.S. Jointly embedding multiple single-cell omics measurements. Leibniz Int. Proc. Informatics, LIPIcs. 2019 doi: 10.4230/LIPIcs.WABI.2019.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Argelaguet R., Arnol D., Bredikhin D., Deloro Y., Velten B., Marioni J.C. MOFA+: A statistical framework for comprehensive integration of multi-modal single-cell data. Genome Biol. 2020;21(1) doi: 10.1186/s13059-020-02015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zuo C., Chen L. Deep-joint-learning analysis model of single cell transcriptome and open chromatin accessibility data. Brief Bioinform. 2020 doi: 10.1093/bib/bbaa287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M. Comprehensive Integration of Single-Cell Data. Cell. 2019;177(7):1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gayoso A., Steier Z., Lopez R., Regier J., Nazor K.L., Streets A. Joint probabilistic modeling of paired transcriptome and proteome measurements in single cells. BioRxiv. 2020 doi: 10.1101/2020.05.08.083337. [DOI] [Google Scholar]
- 89.Cao K., Bai X., Hong Y., Wan L. Unsupervised Topological Alignment for Single-Cell Multi-Omics Integration. Bioinformatics. 2020 doi: 10.1101/2020.02.02.931394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang X, Sun Z, Zhang Y, Xu Z, Xin H, Huang H, et al. BREM-SC: a bayesian random effects mixture model for joint clustering single cell multi-omics data. Nucleic Acids Res 2020;48:5814–24. Doi: 10.1093/nar/gkaa314. [DOI] [PMC free article] [PubMed]
- 91.Campbell K.R., Steif A., Laks E., Zahn H., Lai D., McPherson A. Clonealign: Statistical integration of independent single-cell RNA and DNA sequencing data from human cancers. Genome Biol. 2019;20(1) doi: 10.1186/s13059-019-1645-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhan K., Niu C., Chen C., Nie F., Zhang C., Yang Y.i. Graph Structure Fusion for Multiview Clustering. IEEE Trans Knowl Data Eng. 2019;31(10):1984–1993. doi: 10.1109/TKDE.6910.1109/TKDE.2018.2872061. [DOI] [Google Scholar]
- 93.Kang Z., Shi G., Huang S., Chen W., Pu X., Zhou J.T. Multi-graph fusion for multi-view spectral clustering. Knowledge-Based Syst. 2020;189:105102. doi: 10.1016/j.knosys.2019.105102. [DOI] [Google Scholar]
- 94.Wang H., Yang Y., Liu B. GMC: Graph-Based Multi-View Clustering. IEEE Trans Knowl Data Eng. 2020;32(6):1116–1129. doi: 10.1109/TKDE.6910.1109/TKDE.2019.2903810. [DOI] [Google Scholar]
- 95.Huang Z, Zhou JT, Peng X, Zhang C, Zhu H, Lv J. Multi-view spectral clustering network. IJCAI Int. Jt. Conf. Artif. Intell., vol. 2019- Augus, 2019, p. 2563–9. Doi: 10.24963/ijcai.2019/356.
- 96.Nie F, Li J, Li X. Parameter-free auto-weighted multiple graph learning: A framework for multiview clustering and semi-supervised classification. IJCAI Int. Jt. Conf. Artif. Intell., vol. 2016- Janua, 2016, p. 1881–7.
- 97.Kumar A, Rai P, Daumé H. Co-regularized multi-view spectral clustering. Adv. Neural Inf. Process. Syst. 24 25th Annu. Conf. Neural Inf. Process. Syst. 2011, NIPS 2011, 2011.
- 98.Xing E.P., Ng A.Y., Jordan M.I., Russell S. Distance metric learning, with application to clustering with side-information. Adv. Neural Inf. Process. Syst. 2003 [Google Scholar]
- 99.Ter Braak C.J.F. Canonical correspondence analysis: a new eigenvector technique for multivariate direct gradient analysis. Ecology. 1986;67:1167–1179. doi: 10.2307/1938672. [DOI] [Google Scholar]
- 100.Hardoon D.R., Shawe-Taylor J. Sparse canonical correlation analysis. Mach Learn. 2011;83(3):331–353. doi: 10.1007/s10994-010-5222-7. [DOI] [Google Scholar]
- 101.Witten D.M., Tibshirani R., Hastie T. A penalized matrix decomposition, with applications to sparse principal components and canonical correlation analysis. Biostatistics. 2009;10(3):515–534. doi: 10.1093/biostatistics/kxp008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.KETTENRING J.R. Canonical analysis of several sets of variables. Biometrika. 1971;58(3):433–451. doi: 10.1093/biomet/58.3.433. [DOI] [Google Scholar]
- 103.Zhuang X., Yang Z., Cordes D. A technical review of canonical correlation analysis for neuroscience applications. Hum Brain Mapp. 2020;41(13):3807–3833. doi: 10.1002/hbm.v41.1310.1002/hbm.25090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang Z., Michailidis G. A non-negative matrix factorization method for detecting modules in heterogeneous omics multi-modal data. Bioinformatics. 2016;32:1–8. doi: 10.1093/bioinformatics/btv544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gao C, Welch JD. Iterative refinement of cellular identity from single-cell data using online learning. Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics), vol. 12074 LNBI, 2020, p. 248–50. https://doi.org/10.1007/978-3-030-45257-5_24.
- 106.Mairal J., Bach F., Ponce J., Sapiro G. Online learning for matrix factorization and sparse coding. J Mach Learn Res. 2010 doi: 10.1145/1756006.1756008. [DOI] [Google Scholar]
- 107.Argelaguet R., Velten B., Arnol D., Dietrich S., Zenz T., Marioni J.C. Multi-Omics Factor Analysis—a framework for unsupervised integration of multi-omics data sets. Mol Syst Biol. 2018;14(6) doi: 10.15252/msb:20178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jang SJ, Ham MS, Lee JM, Chung SK, Lee HJ, Kim JH, et al. New integration vector using a cellulase gene as a screening marker for Lactobacillus. FEMS Microbiol Lett 2003;224:191–5. Doi: 10.1016/S0378-1097(03)00422-1. [DOI] [PubMed]
- 109.Hinton G.E., Salakhutdinov R.R. Reducing the dimensionality of data with neural networks. Science. 2006;313:504–507. doi: 10.1126/science.1127647. [DOI] [PubMed] [Google Scholar]
- 110.Yang K.D., Belyaeva A., Venkatachalapathy S., Damodaran K., Katcoff A., Radhakrishnan A. Multi-domain translation between single-cell imaging and sequencing data using autoencoders. Nat Commun. 2021;12(1) doi: 10.1038/s41467-020-20249-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Makhzani A, Shlens J, Jaitly N, Goodfellow I, Frey B. Adversarial Autoencoders 2015.
- 112.Gabasova E., Reid J., Wernisch L., Morris Q. Integrative context-dependent clustering for heterogeneous datasets. PLoS Comput Biol. 2017;13.;13(10):e1005781. doi: 10.1371/journal.pcbi.1005781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lock E.F., Dunson D.B. Bayesian consensus clustering. Bioinformatics. 2013;29:2610–2616. doi: 10.1093/bioinformatics/btt425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kirk P., Griffin J.E., Savage R.S., Ghahramani Z., Wild D.L. Bayesian correlated clustering to integrate multiple datasets. Bioinformatics. 2012;28:3290–3297. doi: 10.1093/bioinformatics/bts595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bickel S, Scheffer T. Multi-view clustering. Proc. - Fourth IEEE Int. Conf. Data Mining, ICDM 2004, 2004, p. 19–26. Doi: 10.1109/ICDM.2004.10095.
- 116.Y. Yang H. Wang Multi-view clustering: A survey Big Data Min Anal 1 2018 83 107 Doi: 10.26599/BDMA.2018.9020003.
- 117.Cao X, Zhang C, Fu H, Liu S, Zhang H. Diversity-induced Multi-view Subspace Clustering. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit., vol. 07-12- June, 2015, p. 586–94. Doi: 10.1109/CVPR.2015.7298657.
- 118.Mitra Sayantan, Saha Sriparna, Hasanuzzaman Mohammed. Multi-view clustering for multi-omics data using unified embedding. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-70229-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Parkhomenko E, Tritchler D, Beyene J. Sparse canonical correlation analysis with application to genomic data integration. Stat Appl Genet Mol Biol 2009;8. Doi: 10.2202/1544-6115.1406. [DOI] [PubMed]
- 120.Yoshida Kosuke, Yoshimoto Junichiro, Doya Kenji. Sparse kernel canonical correlation analysis for discovery of nonlinear interactions in high-dimensional data. BMC Bioinf. 2017;18(1) doi: 10.1186/s12859-017-1543-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Andrew G, Arora R, Bilmes J, Livescu K. Deep canonical correlation analysis. 30th Int. Conf. Mach. Learn. ICML 2013, 2013, p. 2284–92.
- 122.Hu J, Pan Y, Li T, Yang Y. TW-Co-MFC: Two-level weighted collaborative multi-view fuzzy clustering based on maximum entropy. Proc. - 2019 7th Int. Conf. Adv. Cloud Big Data, CBD 2019, 2019, p. 303–8. Doi: 10.1109/CBD.2019.00061.
- 123.Jiang B., Qiu F., Wang L. Multi-view clustering via simultaneous weighting on views and features. Appl Soft Comput J. 2016;47:304–315. doi: 10.1016/j.asoc.2016.06.010. [DOI] [Google Scholar]
- 124.Shen R., Olshen A.B., Ladanyi M. Integrative clustering of multiple genomic data types using a joint latent variable model with application to breast and lung cancer subtype analysis. Bioinformatics. 2009;25:2906–2912. doi: 10.1093/bioinformatics/btp543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xu Jinglin, Han Junwei, Nie Feiping, Li Xuelong. Re-weighted discriminatively embedded K-means for multi-view clustering. IEEE Trans Image Process. 2017;26(6):3016–3027. doi: 10.1109/TIP.2017.2665976. [DOI] [PubMed] [Google Scholar]
- 126.Xu Y.M., Wang C.D., Lai J.H. Weighted Multi-view Clustering with Feature Selection. Pattern Recognit. 2016;53:25–35. doi: 10.1016/j.patcog.2015.12.007. [DOI] [Google Scholar]
- 127.Zhao X., Evans N., Dugelay J.L. A subspace co-training framework for multi-view clustering. Pattern Recognit Lett. 2014;41:73–82. doi: 10.1016/j.patrec.2013.12.003. [DOI] [Google Scholar]
- 128.Nguyen Tin, Tagett Rebecca, Diaz Diana, Draghici Sorin. A novel approach for data integration and disease subtyping. Genome Res. 2017;27(12):2025–2039. doi: 10.1101/gr.215129.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Conesa Ana, Beck Stephan. Making multi-omics data accessible to researchers. Sci Data. 2019;6(1) doi: 10.1038/s41597-019-0258-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fernandez-Banet Julio, Esposito Anthony, Coffin Scott, Horvath Istvan Boerner, Estrella Heather, Schefzick Sabine. OASIS: Web-based platform for exploring cancer multi-omics data. Nat Methods. 2016;13(1):9–10. doi: 10.1038/nmeth.3692. [DOI] [PubMed] [Google Scholar]
- 131.Vasaikar S.V., Straub P., Wang J., Zhang B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46:D956–D963. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhu J., Shi Z., Wang J., Zhang B. Empowering biologists with multi-omics data: Colorectal cancer as a paradigm. Bioinformatics. 2015;31:1436–1443. doi: 10.1093/bioinformatics/btu834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, et al. The human cell atlas. Elife 2017;6. Doi: 10.7554/eLife.27041. [DOI] [PMC free article] [PubMed]
- 134.Nguyen ND, Wang D. Multiview learning for understanding functional multiomics. PLoS Comput Biol 2020;16. Doi: 10.1371/journal.pcbi.1007677. [DOI] [PMC free article] [PubMed]
- 135.Serra A, Galdi P, Tagliaferri R. Multiview learning in biomedical applications. Artif Intell Age Neural Networks Brain Comput; 2018. Doi: 10.1016/B978-0-12-815480-9.00013-X.