Abstract
Rationale and Objectives:
We propose a novel convolutional neural network derived pixel-wise breast cancer risk model using mammographic dataset.
Materials and Methods:
An institutional review board approved retrospective case-control study of 1474 mammographic images was performed in average risk women. First, 210 patients with new incidence of breast cancer were identified. Mammograms from these patients prior to developing breast cancer were identified and made up the case group [420 bilateral craniocaudal mammograms]. The control group consisted of 527 patients without breast cancer from the same time period. Prior mammograms from these patients made up the control group [1054 bilateral craniocaudal mammograms]. A convolutional neural network (CNN) architecture was designed for pixel-wise breast cancer risk prediction. Briefly, each mammogram was normalized as a map of z-scores and resized to an input image size of 256 × 256. Then a contracting and expanding fully convolutional CNN architecture was composed entirely of 3 × 3 convolutions, a total of four strided convolutions instead of pooling layers, and symmetric residual connections. L2 regularization and augmentation methods were implemented to prevent overfitting. Cases were separated into training (80%) and test sets (20%). A 5-fold cross validation was performed. Software code was written in Python using the TensorFlow module on a Linux workstation with NVIDIA GTX 1070 Pascal GPU.
Results:
The average age of patients between the case and the control groups was not statistically different [case: 57.4 years (SD, 10.4) and control: 58.2 years (SD, 10.9), p = 0.33]. Breast Density (BD) was significantly higher in the case group [2.39 (SD, 0.7)] than the control group [1.98 (SD, 0.75), p < 0.0001]. On multivariate logistic regression analysis, both CNN pixel-wise mammographic risk model and BD were significant independent predictors of breast cancer risk (p < 0.0001). The CNN risk model showed greater predictive potential [OR = 4.42 (95% CI, 3.4–5.7] compared to BD [OR = 1.67 (95% CI, 1.4–1.9). The CNN risk model achieved an overall accuracy of 72% (95%CI, 69.8–74.4) in predicting patients in the case group.
Conclusion:
Novel pixel-wise mammographic breast evaluation using a CNN architecture can stratify breast cancer risk, independent of the BD. Larger dataset will likely improve our model.
Keywords: CNN, breast cancer risk, breast density
INTRODUCTION
Breast cancer is a leading cause of death worldwide and is the second most common cause of cancer deaths among women in the United States (1). One in eight women will develop breast cancer, however the risk is not homogeneously distributed throughout the population. While some risk factors have been established, the majority of women diagnosed with breast cancer have no identifiable risk (2). This limits the ability of the medical community to determine high versus low risk women.
The greatest evidence for stratifying the risk of developing breast cancer lies in mammographic breast density, defined as the proportion of radiopaque epithelial and stromal tissue compared to radiolucent fat (3). In 1976, Wolfe was the first to hypothesize breast density as a cancer risk factor, with four distinct classifications based on parenchymal patterns: primarily fat (N1), ductal prominence involving up to one-fourth of the breast (P1), ductal prominence involving more than one-fourth of the breast (P2), and severe ductal prominence (DY) (4).
Later studies described more quantitative categorization of breast density as it relates to cancer predisposition, such as the Tabar classification (5,6). Analogous to Wolfe’s, the American College of Radiology Breast Imaging Reporting and Data System (BI-RADS) defines four categories: entirely fatty, scattered fibroglandular densities, heterogeneously dense, and extremely dense. Several studies have examined the correlation of breast cancer risk and BI-RADS breast density criteria.
A large prospective study by Vacek et al. (7) showed risk to increase with a higher BI-RADS category, with heterogeneously dense breasts (BI-RADS 3) 2.8 and extremely dense breasts (BI-RADS 4) 4.0 times more likely to develop cancer compared to entirely fatty breasts (BI-RADS 1) (7). Similarly, Kerlikowske et al. (8) demonstrated an increase in BI-RADS breast density to correlate with an increased risk of breast cancer over a 3 year follow up. Beyond the correlation of breast density and cancer risk, evidence has shown increased density to be an independent risk factor beyond a masking effect, as it represents the amount of stromal and epithelial tissue from which breast cancer derives (3).
The current climate of changing breast cancer screening recommendations by the United States Preventive Services Task Force and American Cancer Society has demonstrated a consistent trend toward later, less frequent screening, unless a woman is considered to be high risk. This makes the challenge of defining the high risk group within the general population even more important (9,10). According to the Breast Cancer Surveillance Consortium database, almost half (47%) of the population falls into the category of dense breasts (BiRADS 3 and 4) and therefore can be classified as high risk (11). Clearly a more individualized stratification is needed to appropriately predict breast cancer risk and therefore designate the most appropriate screening regimen.
While advances in imaging technology have provided high quality mammograms with increased clarity, the question remains: is there something beyond the amount of breast density that is not appreciated by the human eye? Recent advances in technology have allowed machine learning to address this complex clinical question. Specifically, a subset of machine learning through artificial neural network such as convolutional neural network (CNN) has shown significant promise in advancing visual tasks. CNN synthetically learns from the input image itself through multiple increasingly complex layers. This new technology has surpassed traditional machine learning, which relies on human extracted pattern-recognition and input (12).
We propose a novel convolutional neural network derived pixel-wise breast cancer risk model using mammographic dataset to stratify patients into personalized breast cancer risk categories beyond just breast density.
METHODS
An institutional review board approved case-control study was performed retrospectively utilizing our institution’s screening mammogram database from 1/2011 to 1/2017. Average risk screening women were evaluated by excluding women who have personal history of breast cancer, family history of breast cancer, and any known genetic mutation that increases the risk for breast cancer. After applying the exclusion criteria, 210 patients were identified consecutively with a new first time diagnosis of breast cancer. Mammograms from these patients, at least 2 years (median 3.3 years, range 2.0–5.3 years) prior to developing breast cancer, were identified and made up the “high risk” case group composed of the bilateral craniocaudal mammographic dataset (420 total). The control group consisted of 527 patients without breast cancer from the same time period. Prior mammograms from these patients made up the “low risk” control group composed of the bilateral craniocaudal mammographic dataset (1054 total). These 527 patients in the control group had documented negative follow-up mammogram for at least 2 years (median 3.1 years, range 2.0–4.8 years).
From each patient, the age and the BI-RADS mammographic density assessment was recorded on a 4-point scale (1-fatty, 2-scattered, 3-heterogeneously dense, and 4-extremely dense) by one of five breast fellowship trained radiologists. Mammograms at our institution were performed on dedicated mammography units (Senographe Essential, GE Healthcare). Of patients who developed breast cancer histologic subtype was recorded based on the World Health Organization classification (13). Statistical analysis was performed using the IBM SPSS software (version 24).
Originally introduced by Long et al. (14) fully convolutional neural networks are implemented by a series of upsampling convolutional transpose operators performed on the deepest network layers, resulting in a dense classification matrix equal in dimension to the original image size for each forward pass. Ronneberger et al. (15) elaborated on this technique by proposing a symmetric contracting and expanding topology that efficiently combines low- and high-level features. This study further adapts these previous approaches by replacing concatenation operations with residual connections (and associated projection matrices as needed to match feature layer dimensions) (Fig 1). Originally described by He et al. (16), residual neural networks are able to stabilize gradients during backpropagation, leading to improved optimization and facilitating greater network depth. Furthermore in a symmetric contracting and expanding topology, residual connections allow the network to learn the appropriate feature depth, as contributions from the deepest, large field-of-view feature maps can be selectively eliminated through identity mappings.
Figure 1.
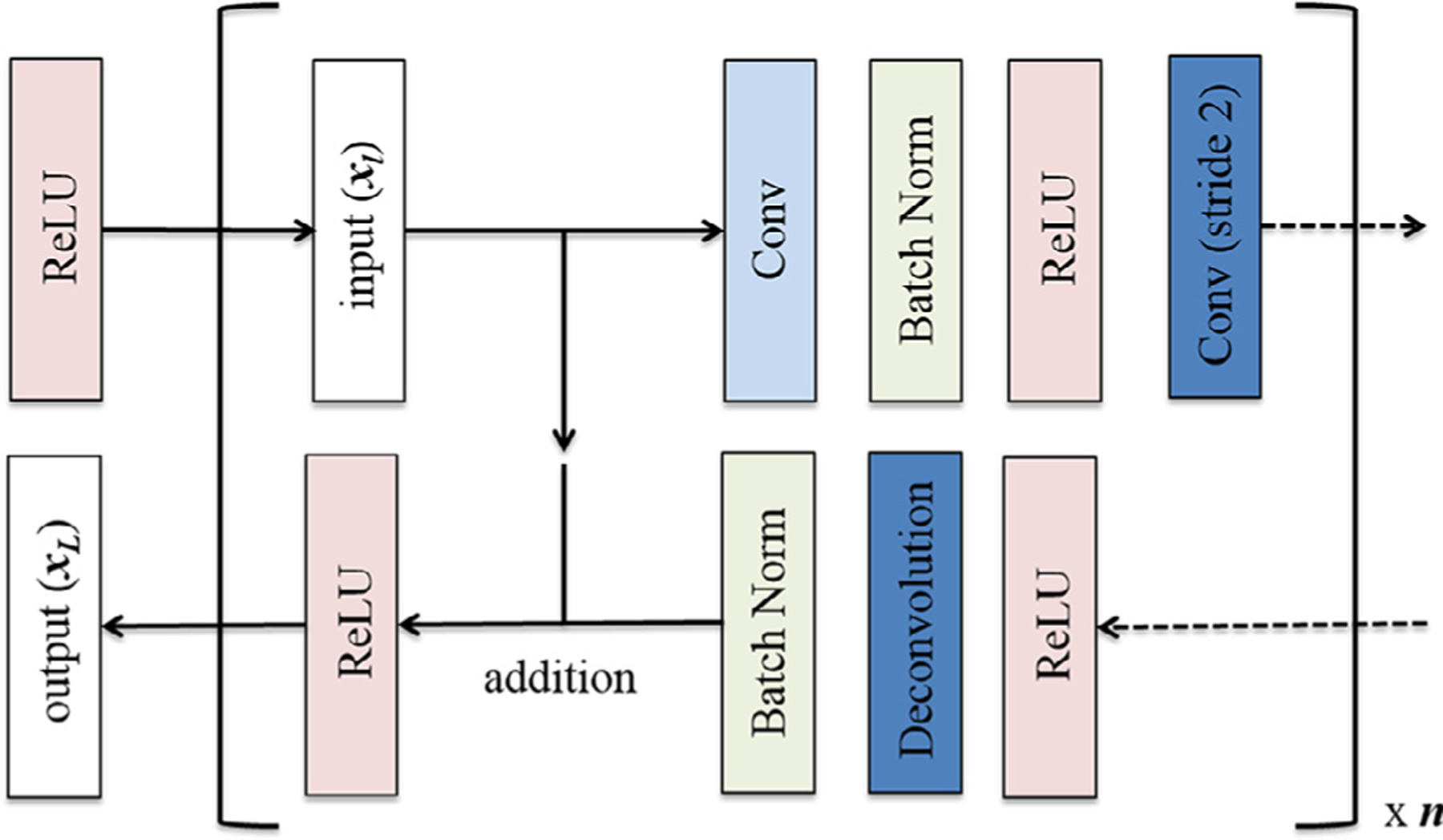
Residual connections implemented by means of a simple addition operation inserted after batch normalization and before nonlinearity (ReLU) of the corresponding layer within the expanding arm of the symmetric, fully convolutional architecture.
The overall network architecture is shown in Figure 2. The CNN is implemented completely by series of 3 × 3 convolutional kernels to prevent overfitting (17–20). No pooling layers are used; instead downsampling is implemented simply by means of a 3 × 3 convolutional kernel with stride length of 2 to decrease the feature maps by 75% in size. All nonlinear functions are modeled by the rectified linear unit (17–20). Batch normalization is used between the convolutional and rectified linear unit layers to limit drift of layer activations during training (21). In successively deeper layers the number of feature channels gradually increases from 16, 32, 64, 128, and 256, reflecting increasing representational complexity.
Figure 2.
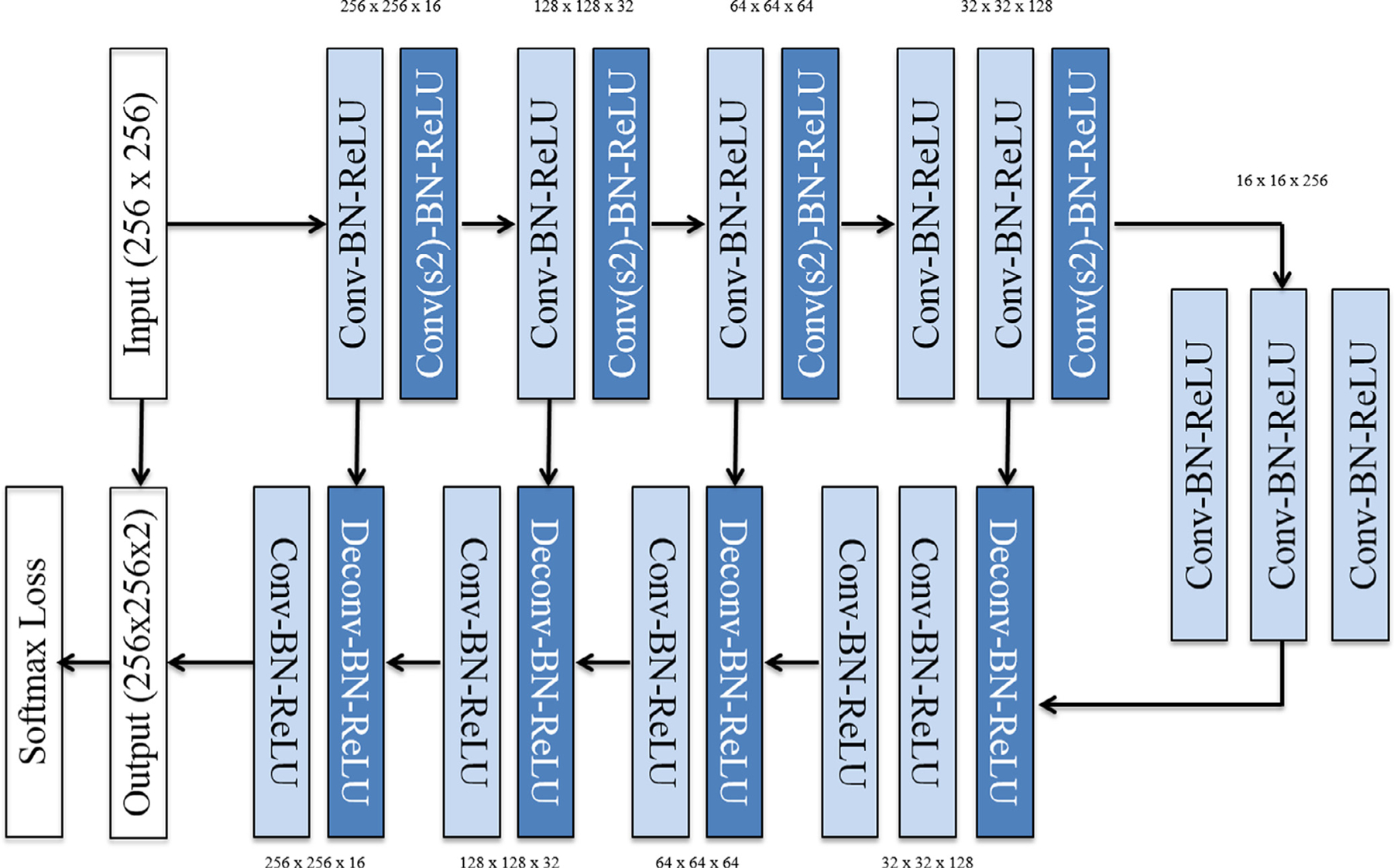
The contracting and expanding fully convolutional CNN architecture is composed entirely of 3 × 3 convolutions, a total of four strided convolutions (and convolutional transpose operations) instead of pooling layers and symmetric residual connections.
Each mammogram was normalized as a map of z-scores and resized to an input image size of 256 × 256. Data augmentation employed by this study involves a number of real-time modifications to the source images at the time of training. Specifically, 50% of all images in a mini-batch were modified randomly by means of: (1) addition across all pixels of a scalar between [¡0.1, 0.1]; (2) random affine transformation of the original mammogram. Given a two-dimensional affine matrix,
the random affine transformation was initialized with random uniform distributions of interval s1, s2 ∈ [0.8, 1.2], t1, t2 ∈ [−0.3, 0.3] and r1, r2 ∈ [−128, 128].
Training was implemented using the Adam optimizer, an algorithm for first-order gradient-based optimization of stochastic objective functions, based on adaptive estimates of lower-order moments (17–20). Parameters are initialized using the heuristic described by He et al. (16). L2 regularization is implemented to prevent over-fitting of data by limiting the squared magnitude of the kernel weights. To account for training dynamics, the learning rate is annealed and the mini-batch size is increased whenever training lost plateaus. Furthermore, a normalized gradient algorithm is employed to allow for locally adaptive learning rates that adjust according to changes in the input signal (17–20). The overall training time was 6 hours.
For statistical analysis, cases were separated into 80% training (590/737) and 20% test sets (147/737). A 5-fold cross validation was performed. A final softmax score threshold of 0.5 from the average of raw logits from each pixel was used for two class classification. Software code for this study was written in Python using the TensorFlow module (1.0.0). Experiments and CNN training was done on a Linux workstation with NVIDIA GTX 1070 Pascal GPU with 8 GB on chip memory, i7 CPU, and 32 GB RAM.
This was an institutional review board approved, Health Insurance Portability and Accountability Act-compliant study. IRB-AAAR5142 Protocol. Approved on 8/3/2017 by our institution.
RESULTS
The average age of patients between the case and the control groups was not statistically different [case: 57.4 years (SD, 10.4) and control: 58.2 years (SD, 10.9), p = 0.33]. All 210 patients had unilateral breast cancers; 69.5% (146/210) had invasive ductal carcinoma; 19% (40/210) had ductal carcinoma in situ; 7.1% (15/210) had invasive lobular carcinoma; 4.3% (9/210) had mixed lobular and ductal invasive carcinoma, and 17.6% (37/210) of the patients had multifocal disease.
Breast Density (BD) was significantly higher in the case group [2.39 (SD, 0.7)] than the control group [1.98 (SD, 0.75), p < 0.0001]. On multivariate logistic regression analysis, both CNN pixel-wise mammographic risk model and BD were significant independent predictors of breast cancer risk (p < 0.0001). The CNN risk model showed greater predictive potential [OR = 4.42 (95% CI, 3.4–5.7] compared to BD [OR = 1.67 (95% CI, 1.4–1.9).
Overall there was a strong signification correlation of CNN pixel-wise mammographic risk results between the left and right breast (Pearson correlation, r = 0.90, n = 737). In the case group, there was a signification correlation between the left and right breast (Pearson correlation, r = 0.86, n = 210). In the control group, there was a signification correlation between the left and right breast (Pearson correlation, r = 0.86, n = 527).
The CNN risk model achieved an overall accuracy of 72% (95% CI, 69.8–74.4%) in predicting patients in the case group. Heat maps were generated by color-coding the final softmax scores on a pixel-wise basis (Fig 3). Intuitively these maps can be interpreted as subregions within the mammogram that are most commonly encountered in normal (blue) and high cancer risk (red) patients.
Figure 3.
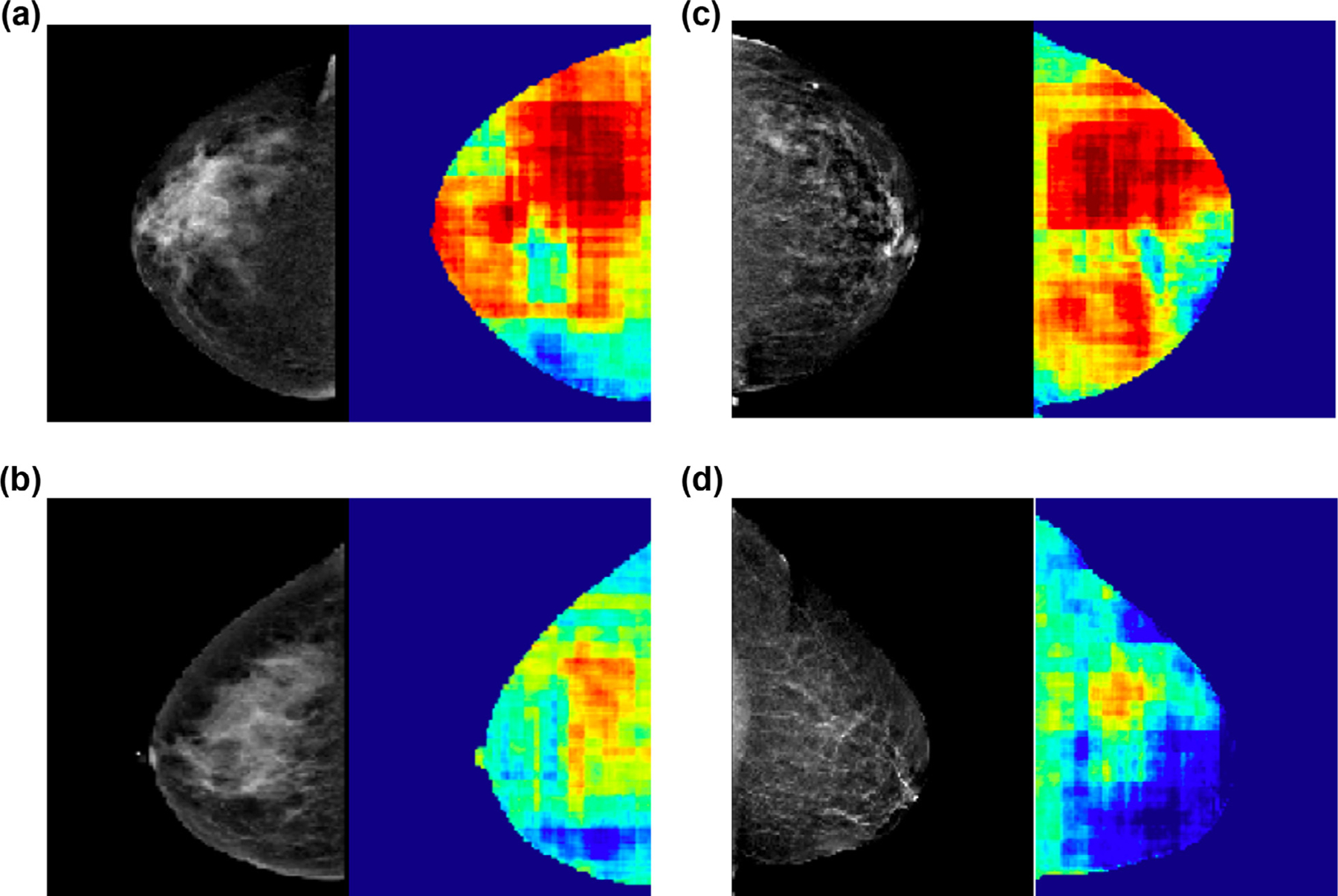
Pixel-wise heat maps. Heat maps generated by color-coding the final softmax scores on a pixel-wise basis, demonstrating subregions within the mammogram that are most commonly encountered in normal (blue) and high cancer risk (red) patients. Mammograms in (A) and (B) illustrate similar breast densities (heterogeneously dense) and mammograms in (C) and (D) illustrate similar breast densities (scattered) but the corresponding heat maps are different with patient A with significantly higher mammographic regions containing red and correctly identifying high risk. Similarly, patient C with significantly higher mammographic regions containing red and correctly identifying high risk.
The CNN was trained for a total of 144,000 iterations (approximately 1170 epochs with a batch size of 12) before convergence. A single forward pass through during test time for classification of new cases can be achieved in 0.063 seconds.
DISCUSSION
The CNN algorithm in this study applied a novel approach of pixel-wise cancer risk assessment using mammogram to define risk on an individual basis. In this preliminary study, we achieved an overall accuracy of 72% in predicting high versus low cancer risk mammograms. CNN, a subset of machine learning used in our study, has recently gained popularity throughout medicine. The transition from human-extracted pattern recognition to synthetic learning from raw input data facilitates analysis of complex visual tasks, such as mammographic individualized cancer risk stratification (12).
With ever changing screening guidelines, it is paramount to better define an individual’s risk for breast cancer. While mammographic breast density categorization schemes exist, accurate identification of who is at high risk remains a challenge. Using heat maps, our study illustrates breast cancer risk heterogeneity among mammographic breast density categories. For example, not all heterogeneously dense breasts are high risk, with a subset demonstrating a stronger resemblance to a low risk pattern. Similarly, not all breasts with the scattered fibroglandular density demonstrate a low risk pattern. While approximately half the population is categorized as having dense breasts (BI-RADS 3 and 4) (11), our study challenges the uniform presumption of associated high cancer risk.
Our CNN algorithm did not show any significant bias toward the cancer side. In addition, we observed significant correlation between the two breasts (the side that developed cancer and the contralateral noncancer side), indicating that the CNN algorithm in this study predicts risk for breast cancer based on features that are largely conserved on an individual basis. The red areas on the pixel map (Fig 3) indicate regions within the breast that have the most overlapping mammographic features with patients who subsequently developed cancer. The overlapping features come from both breasts (the side that developed cancer and the contralateral side that never developed cancer). While it is possible that the cancer may arise from the red areas, our pixel map was not designed to predict specific areas of breast that will develop breast cancer.
Individualized breast cancer risk stratification has the potential to significantly impact clinical management. If validated, this risk assessment could be implemented into screening guidelines. In the setting of later and less frequent evolving screening guidelines for average risk women, accurately categorized high risk women may benefit from earlier and more frequent screening.
Previous studies support the results of our investigation of utilizing digital features in mammogram to predict breast cancer risk (22–25). However, all of these studies involved hand crafted features based on extracted patterns. In contrast, our study utilizes neural networks, allowing the computer to automatically construct predictive statistical models, tailored to solve a specific problem subset. Instead of laborious task of human engineers inputting specific patterns to be recognized, we used CNN to self-optimize and discriminate through increasingly complex layers by inputting curated data.
Beyond screening, individualized risk assessment has potential utilization for chemoprevention strategies. The American Society of Clinical Oncology, National Comprehensive Cancer Network, and United States Preventive Services Task Force recommend counseling high risk women above the age of 35 on pharmacologic interventions for breast cancer risk reduction (26–28). Two selective estrogen receptor modulators, tamoxifen and raloxifene, approved for chemoprevention in the US, show up to a 50% cancer risk reduction. Additionally, two aromatase inhibitors, exemestane and anastrozole, not yet approved for use in the US, have shown significant chemopreventive potential in preliminary studies (29). Individualized breast cancer risk assessment has potential to aid in selection of high risk patients and counseling on chemoprevention.
Our study has a few limitations. It is a retrospective study in a single institution with relatively small dataset. Therefore, improved risk stratification is likely to be generated by an even larger mammogram dataset. Additionally, long training time is an intrinsic limitation of CNN. In comparison to traditional machine learning, increased algorithm complexity of CNN requires a long training time, with the benefit of a much shorter testing time. Finally, the breast density classification can be subjective and prone to intra and interobserver variability, especially if taken from radiology reports (30–32). However, given that the case and control groups are randomly distributed with equal likelihood of each radiologist giving the breast density assessment, we feel that the potential impact is limited.
In conclusion, our novel pixel-wise mammographic breast evaluation using a CNN architecture can stratify breast cancer risk, independent of the mammographic BD. The CNN risk model showed greater predictive potential compared to mammographic BD in our study. Validation by a prospective randomized study is needed to potentially implement our individualized risk stratification scheme into screening and chemoprevention guidelines.
ACKNOWLEDGMENTS
Peter Chang and Richard Ha received grants from National Institutes of Health (NIBIB) T32 Training Grant, T32EB001631 and Nvidia corporation, GPU donated by Nvidia GPU Grant, respectively.
Footnotes
Work originated from Columbia University Medical Center.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin 2017; 67:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Madigan MP, Ziegler RG, Benichou J, et al. Proportion of breast cancer cases in the United States explained by well-established risk factors. J Natl Cancer Inst 1995; 87:1681–1685. [DOI] [PubMed] [Google Scholar]
- 3.Freer PE. Mammographic breast density: impact on breast cancer risk and implications for screening. Radiographics 2015; 35:302–315. [DOI] [PubMed] [Google Scholar]
- 4.Wolfe JN. Breast patterns as an index of risk for developing breast cancer. Am J Roentgenol 1976; 126:1130–1137. [DOI] [PubMed] [Google Scholar]
- 5.Boyd NF, Byng JW, Jong RA, et al. Quantitative classification of mammographic densities and breast cancer risk: results from the Canadian National Breast Screening Study. J Natl Cancer Inst 1995; 87:670–675. [DOI] [PubMed] [Google Scholar]
- 6.Gram IT, Funkhouser E, Tabar L. The Tabar classification of mammographic parenchymal patterns. Eur J Radiol 1997; 24:131–136. [DOI] [PubMed] [Google Scholar]
- 7.Vacek PM, Geller BM. A prospective study of breast cancer risk using routine mammographic breast density measurements. Cancer Epidemiol Biomark Prev 2004; 13:715–722. [PubMed] [Google Scholar]
- 8.Kerlikowske K, Ichikawa L, Miglioretti DL, et al. Longitudinal measurement of clinical mammographic breast density to improve estimation of breast cancer risk. J Natl Cancer Inst 2007; 99:386–395. [DOI] [PubMed] [Google Scholar]
- 9.Siu AL. U.S. Preventive Services Task Force. Screening for breast cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med 2016; 164:279–296. [DOI] [PubMed] [Google Scholar]
- 10.Oeffinger KC, Fontham ETH, Etzioni R, et al. Breast cancer screening for women at average risk: 2015 guideline update from the American Cancer Society. JAMA. 2015; 314:1599–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerlikowske K, Zhu W, Hubbard RA, et al. Outcomes of screening mammography by frequency, breast density, and postmenopausal hormone therapy. JAMA Intern Med 2013; 173:807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LeChun Y, Bengio T, Hinton G. Deep learning. Nature 2015; 521:436–444. [DOI] [PubMed] [Google Scholar]
- 13.American Joint Committee on Cancer. AJCC Cancer Staging Manual. 7th ed. New York, NY: Springer, 2010. [Google Scholar]
- 14.Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. In: In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2015. [DOI] [PubMed] [Google Scholar]
- 15.Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2015. [Google Scholar]
- 16.He K, Zhang X, Ren S, et al. Delving deep into rectifiers: surpassing human-level performance on ImageNet classification. Comput Vision Pattern Recognit arXiv:1502.01852 [Google Scholar]
- 17.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. Int Conf Learning Represent 2015. : 1–14. [Google Scholar]
- 18.Nair V, Hinton GE. Rectified linear units improve restricted Boltzmann machines. In: In: Proceedings of the 27th International Conference on Machine Learning, Haifa, Israel; 2010. [Google Scholar]
- 19.Srivastava N, Hinton GE, Krizhevsky A, et al. Dropout : a simple way to prevent neural networks from overfitting. J Mach Learn Res 2014; 15:1929–1958. [Google Scholar]
- 20.Kingma DP, Ba J. Adam: a method for stochastic optimization. Machine Learning arXiv:1412.6980. [Google Scholar]
- 21.Ioffe S, Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift. In: International Conference on Machine Learning; 2015. [Google Scholar]
- 22.Wei J, Chan HP, Wu YT, et al. Association of computerized mammographic parenchymal pattern measure with breast cancer risk: a pilot case-control study. Radiology 2011; 260:42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heidari M, Khuzani AZ, Hollingsworth AB, et al. Prediction of breast cancer risk using a machine learning approach embedded with a locality preserving projection algorithm. Phys Med Biol 2018; 63:035020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan M, Pu J, Cheng S, et al. Assessment of a four-view mammographic image feature based fusion model to predict near-term breast cancer risk. Ann Biomed Eng 2015; 43:2416–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Fan M, Cheng H, et al. Assessment of global and local region-based bilateral mammographic feature asymmetry to predict short-term breast cancer risk. Phys Med Biol 2018; 63:025004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Visvanathan K, Hurley P, Bantug E, et al. Use of pharmacologic interventions for breast cancer risk reduction: American Society of Clinical Oncology clinical practice guideline. J Clin Oncol 2013; 31:2942–2962. [DOI] [PubMed] [Google Scholar]
- 27.National Comprehensive Cancer Network. The NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines) Breast Cancer Risk Reduction (version 1.2014). www.NCCN.org; 2014.
- 28.Moyer VA. Medications to decrease the risk for breast cancer in women: recommendations from the U.S. Preventive Services Task Force recommendation statement. Ann Intern Med 2013; 159:698–708. [DOI] [PubMed] [Google Scholar]
- 29.Pruthi A, Heisey RE, Bevers TB. Chemoprevention for breast cancer. Ann Surg Oncol 2015; 22:3230–3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ciatto S, Houssami N, Apruzzese A, et al. Categorizing breast mammographic density: intra- and interobserver variability of BI-RADS density categories. Breast 2005; 14:269–275. [DOI] [PubMed] [Google Scholar]
- 31.Ciatto S, Houssami N, Apruzzese A, et al. Reader variability in reporting breast imaging according to BI-RADS assessment categories (the Florence experience). Breast 2006; 15:44–51. [DOI] [PubMed] [Google Scholar]
- 32.Ooms EA, Zonderland HM, Eijkemans MJ, et al. Mammography: interobserver variability in breast density assessment. Breast 2007; 16:568–576. [DOI] [PubMed] [Google Scholar]


