Abstract
Brain tumors are associated with adverse outcomes despite improvements in radiation therapy, chemotherapy, and photodynamic therapy. However, treatment approaches are evolving, and new biological phenomena are being explored to identify the appropriate treatment of brain tumors. Long non-coding RNAs (lncRNAs), a type of non-coding RNA longer than 200 nucleotides, regulate gene expression at the transcriptional, post-transcriptional, and epigenetic levels and are involved in a variety of biological functions. Recent studies on lncRNAs have revealed their aberrant expression in various cancers, with distinct expression patterns associated with their instrumental roles in cancer. Abnormal expression of lncRNAs has also been identified in brain tumors. Here, we review the potential roles of lncRNAs and their biological functions in the context of brain tumors. We also summarize the current understanding of the molecular mechanisms and signaling pathways related to lncRNAs that may guide clinical trials for brain tumor therapy.
Keywords: Brain disease, Genomics, Glioma, LncRNA, Neurodegenerative disease
Background
The occurrence of brain tumors is increasing worldwide and is more common among men than women [1] (Fig. 1). Glioma, a heterogeneous group of tumors, is one of the most common and major types of malignant brain tumors of the central nervous system. The World Health Organization has stated that according to cellular origin and based on histological subtypes and genetic features, gliomas can be subclassified into astrocytoma, oligoastrocytoma, oligodendroglioma, or glioblastoma. Moreover, each class of gliomas is categorized into grades I, II, III, and IV to assess the degree of malignancy, where grade I is the least malignant and grade IV is the most malignant [2, 3]. In addition, brain tumors afflict a large number of individuals worldwide and are difficult to detect and treat. One problem associated with the treatment of such tumors is the blood–brain barrier, a specific system of endothelial cells that prevents molecules in the circulating blood from non-selectively crossing into the extracellular fluid of the central nervous system to protect the neural cells and preserve neural homeostasis, which makes drug diffusion inefficient or impossible. Although there have been improvements in surgical and other therapeutic approaches, non-specificity of the potentially toxic drugs used to treat the condition remains an issue [4, 5]. Therefore, alternative approaches to treatment of brain tumors targeting different kinds of molecules are actively being pursued.
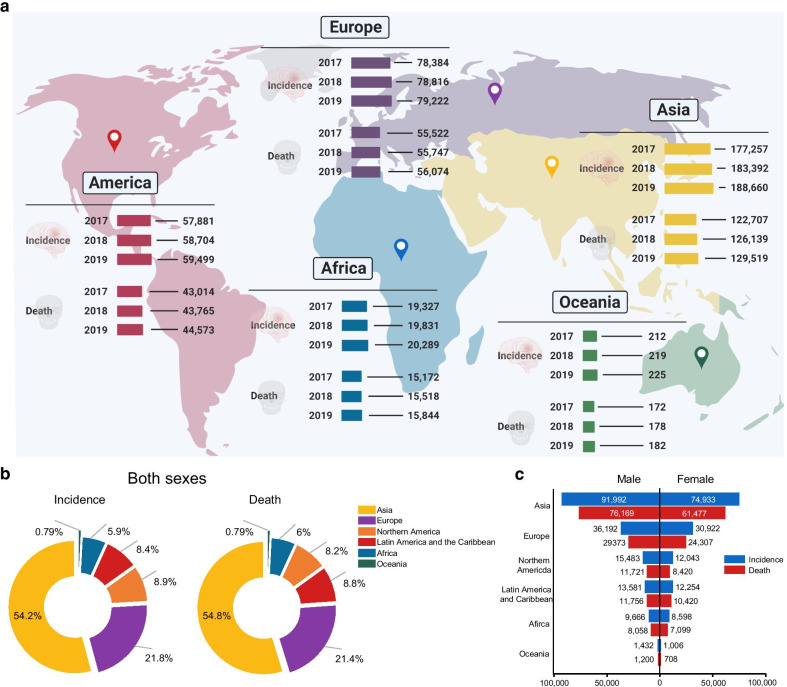
Fig. 1.
Global trends of brain tumor incidence and death. a Global map showing the incidence and deaths related to brain tumor across the world from 2017 to 2019. b Pie charts present the distribution of incidence and deaths related to brain tumor in major world regions in 2020 for both sexes and all ages. c Bar graph reveals the incidence of brain tumor and corresponding deaths in 2020 in males and females. The data sources and methods used for each world region and detailed condition of the estimates are available online from global health data exchange and GLOBALCAN 2020 at the Global Cancer Observatory (GCO)
Previously, non-coding regions of the genome, which do not translate into proteins, were regarded as junk DNA. However, high-throughput technologies have accelerated and improved our understanding of the non-coding genome. Studies have revealed that long non-coding RNAs (lncRNAs), whose transcripts are longer than 200 nucleotides, are involved in a wide range of biological processes [6] (Fig. 2). lncRNAs regulate not only gene expression but also epigenetic, transcriptional, and post-transcriptional modification, and their expression may be closely related to diverse biological functions including cell survival [7–9]. In the past two decades, various roles of lncRNAs have been implicated in tumor biology [10]. The dysregulation of lncRNAs is increasingly being associated with many human diseases, especially cancers. lncRNAs exhibit unique expression patterns in various types of cancers, and play instrumental roles as cancer suppressors or promoters. Additionally, aberrant lncRNA expression in cancers has been linked to modulated expression of genes involved in tumorigenesis, metastasis, and tumor stage progression [11–16]. Moreover, several studies have revealed that lncRNAs control post-translational modification by functioning as molecular sponges or competing endogenous RNAs (ceRNAs) that modulate microRNAs (miRNAs), which in turn regulate the expression of target genes [17–19]. Furthermore, lncRNAs are involved in signal transduction pathways, miRNA silencing, DNA damage, cell cycle control, epigenetic regulation, and hormone-driven disease states, all of which play key roles in cancer progression and metastasis [20].
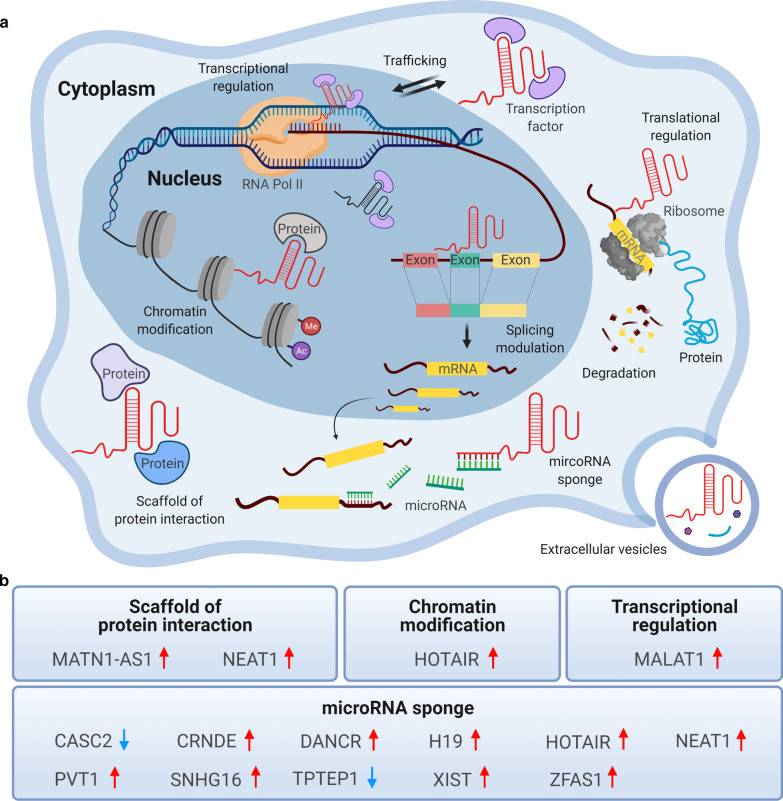
Fig. 2.
Classification of lncRNAs based on their functional mechanisms. a The multiple functional roles of lncRNAs. In the nucleus, lncRNAs are involved in transcription regulation (activation or repression) via acting as transcription factors, splicing of mRNA, and chromatin modification. In the cytoplasm, they regulate translation by interacting with ribosomes, sponging miRNAs, degrading mRNAs, scaffolding protein interactions, and trafficking transcription factors. Moreover, lncRNAs can be enclosed and released by extracellular vesicles, such as exosomes. b The functional contributions and expression levels of reported lncRNAs in brain tumor
Recent studies have shown that aberrantly expressed lncRNAs are closely involved with the pathology and prognosis of gliomas. These studies also identified the molecular mechanisms underlying lncRNA activity, which eventually modulate biological processes in brain tumors [21]. In addition to these molecular insights, advances in genome-wide association technologies have led to the discovery of several types of lncRNAs in brain cancer. Here, we review the current understanding of the functions of lncRNAs in brain tumors by discussing latest findings from diverse mammalian systems.
LncRNAs in signaling pathways and brain tumors
The roles of lncRNAs and their associated pathways in gliomas have been identified, and their interactions with miRNAs have been studied [21, 22]. This section describes the mechanisms of action of lncRNAs in cellular processes and signaling pathways involved in the development of gliomas (Fig. 3).
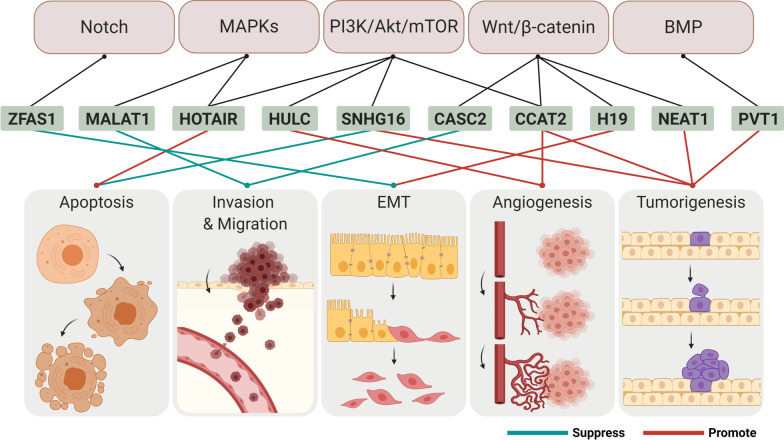
Fig. 3.
LncRNAs regulate cellular processes in the tumor cell through diverse signaling pathways. Based on the genetic characteristics and biochemical studies of lncRNAs, it has been proposed that lncRNAs are required for the coordination of several intracellular pathways, such as PI3K-Akt and Wnt pathways. lncRNAs are generated by transcriptional processes when particular signaling pathways are activated. Moreover, expression of lncRNAs is linked to physiological processes of tumor cells
PI3K/Akt/mTOR pathway
The PI3K/Akt/mTOR pathway is often deregulated in cancer cells, leading to enhanced cell growth, proliferation, and survival of these cells. This pathway also plays a crucial role in apoptosis and autophagy in glioma [23, 24].
The cancer susceptibility candidate 2 (CASC2) lncRNA, which is downregulated gene in various cancers including gliomas, is an important modulator of this pathway. CASC2 has been identified as a tumor suppressor that is located on chromosome 10q26 [25, 26]. CASC2 modulates glioma growth and resistance to temozolomide (TMZ), one of the most commonly used chemotherapeutic agents for glioma, by upregulating the expression of phosphatase and tensin homolog (PTEN), a known tumor suppressor involving in several cancers including gliomas and inhibits Akt signaling pathway and p-Akt pathway via direct inhibition of miR‐181a [27]. Moreover, it negatively regulates miR-193a-5p expression, which in turn reduces TMZ-induced autophagy via upregulation of the mTOR pathway [28].
The differentiation antagonizing non-protein coding RNA (DANCR) lncRNA, a tumor-associated lncRNA located on chromosome 14, is upregulated in several types of human cancers [29]. Aberrant expression of DANCR in various cancers is associated with oncogenic or tumor suppressive mechanisms that are hallmarks of cancer, such as cell proliferation, invasion, and metastasis [29–31]. Additionally, DANCR upregulates AXL, a receptor tyrosine kinase that activates the PI3K/Akt/NF-κB signaling pathway [32, 33] by competitively binding with miR-33a-5p, miRNA-33b-5p, miR-1-3p, miR-206 and miR-613 to mediate the cisplatin resistance in gliomas [34].
The highly upregulated in liver cancer (HULC) lncRNA is located on chromosome 6p24.3 and highly expressed in glioma as an oncogene that contributes to tumor progression [35]. HULC silencing suppresses proliferation, adhesion, migration, invasion, and angiogenesis through the regulation of the cell cycle and anoikis, a Greek word for "homelessness" and a critical mechanism in preventing tumor cell apoptosis via a decrease in cell–matrix interaction, in the glioma by modulating endothelial cell-specific molecule 1 (ESM-1) expression via the PI3K/Akt/mTOR signaling pathway [36].
The small nucleolar RNA host gene 16 (SNHG16) lncRNA is highly expressed in gliomas and is correlated with short survival and poor clinicopathologic features. Knockdown of SNHG16 suppresses the viability of glioma cells and promotes apoptosis by regulating the expression of B-cell lymphoma 2 family (Bcl-2) proteins and activating the PI3K/Akt signaling pathway. Moreover, SNHG16 promotes glioma tumorigenesis by acting as the ceRNA that upregulates the expression of the miR-4518 targeted gene, protein arginine methyltransferase 5 (PRMT5) by directly sponging miR-4518 [37]. Additionally, high expression of SNHG16 increases cell migration, invasion, and proliferation via the PI3K/Akt pathway and sponging of miR-373, leading to release of epidermal growth factor receptor (EGFR), which is an important receptor tyrosine kinase in tumorigenesis and a target of miR-373. Thus, SNHG16 enhances the tumorigenicity of gliomas through the miR-373/EGFR axis via the PI3K/Akt pathway [38, 39].
The colorectal neoplasia differentially expressed (CRNDE) lncRNA, which is specifically expressed in certain regions of the human brain, is also upregulated in gliomas [40]. This lncRNA negatively regulates miR-136-5p by binding as a ceRNA, thereby inhibiting Bcl-2 and Wnt2, which act downstream of the PI3K/Akt/mTOR signaling pathway, thus enhancing the glioma malignancy [41]. Additionally, CRNDE modulates the mTOR signaling pathway; knockdown of the CRNDE decreases the phosphorylation of P70S6K, a downstream signaling molecule in the mTOR signaling pathway [42].
The HOX transcript antisense RNA (HOTAIR) lncRNA is a ~ 2.2 kb lncRNA belonging to the homeobox superfamily that is transcribed from the HOXC locus at chromosome 12q13. HOTAIR acts as a negative prognostic factor for various cancers, including glioma, and its abundant expression is closely related to tumor progression [43, 44]. Knockdown of HOTAIR inhibits fibroblast growth factor 1 (FGF1) expression and blocks the activity of the PI3K/Akt and MEK1/2 pathways, leading to the suppression of malignant behavior, by upregulating miR-326, which targets FGF1. This intervention induces cell cycle arrest and apoptosis, and inhibits cell migration, invasion, and proliferation in human glioma cells [45].
The X-inactive-specific transcript (XIST) lncRNA, produced by the XIST gene, is a critical regulator of cancer, levels of which correlate with cell differentiation, proliferation, and genome maintenance. Abnormal expression of XIST has been seen in various human cancers, where it plays a pathological function [46, 47]. Moreover, XIST expression levels are increased in glioma and human glioma stem cells. Attenuation of cell growth and glucose metabolism was seen in glioblastoma cells following knockdown of XIST. This effect occurs via regulation of the insulin receptor substrate 1 (IRS1)/PI3K/Akt pathway as XIST functions as the ceRNA of miR-126; in vivo, this effect reduced glioblastoma tumor growth [48].
The cancer susceptibility candidate 2 (CCAT2) lncRNA is an oncogenic 1,752 bp lncRNA located on chromosome 8q24.21 that is upregulated in various human cancers, including glioma [49]. CCAT2 enhances cell proliferation and endothelial angiogenesis in glioma by increasing vascular endothelial growth factor A (VEGFA) through miR-424 and activating the PI3K/Akt signaling pathway[50].
MAPK pathway
Mitogen-activated protein kinases (MAPKs) are a family of signaling molecules, which are activated by growth factors, cytokines, and environmental stress. They control crucial cellular processes, including cell growth, proliferation, migration, and apoptosis [51].
The metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) lncRNA, also called nuclear-enriched transcript 2, is an 8.7 kb lncRNA located on chromosome 11q13. MALAT1 is known as a prognostic indicator in non-small cell lung cancer, but is abnormally expressed in various human cancers; it is upregulated in gliomas, where its expression is associated with the progression of glioma and poor prognosis [52–54]. Additionally, the tumor-suppressive function of MALAT1 is related to inhibition of the extracellular signal-regulated kinase/mitogen-activated protein kinase (ERK/MAPK) signaling pathway, which modulates cell proliferation and invasion, as well as inhibition of matrix metalloproteinase 2 (MMP2), a key enzyme in invasion. Therefore, knockdown of MALAT1 promotes cell proliferation and invasion in glioma [55]. Similarly, downregulation of MALAT1 enhances cell proliferation via activation of the MAPK pathway in glioma stem cells [56].
The downregulation of MATN1 antisense RNA 1 (MATN-AS1) lncRNA is associated with short survival duration and poor prognosis of glioblastoma patients. Upregulation of MATN1-AS2 suppressed the expression of RELA, a group of the Rel proteins family and MAPK pathway, thereby inhibiting cell proliferation and invasion through apoptosis of glioblastoma cells [57].
The transmembrane phosphatase with tensin homology pseudogene 1 (TPTEP1) lncRNA is one of the most downregulated lncRNAs in non-small cell lung cancer, where it inhibits the cell proliferation by competitively sponging miR-328-5p [58]. In glioma, TPTEP1 promotes the P38 MAPK pathway through competitive interactions with miR-106a-5p, thus inhibiting stemness and radioresistance [59].
Wnt/β-catenin pathway
Wnt/β-catenin signaling is essential in embryonic development and tissue homeostasis, and thus controls cell proliferation and cell fate. Hence, activation and mutation of the Wnt/ β-catenin pathway is associated with various human cancers and other diseases [60, 61].
Overexpression of CASC2 significantly suppresses migration, invasion, and proliferation in glioma through the inhibition of Wnt/β-catenin signaling pathway [62].
DANCR, which is upregulated in glioma, that activates the Wnt/β-catenin signaling pathway, leading to increased migration and proliferation of glioma cells. However, this tendency is reduced by si-DANCR [63].
The H19 lncRNA is located downstream of insulin growth factor 2 (IGF2) on chromosome 11p15 in humans and chromosome 7 in mice. Its expression is increased in various cancers including gliomas, where it correlates with cell growth and invasion and it plays an oncogenic role [64–67]. Furthermore, H19 expression is closely related with glioma grade [68]. Conversely, downregulation of H19 suppresses glioma cell progression, including, migration, invasion, and proliferation via suppressing the Wnt/β-catenin signaling pathway [69]. Silencing of H19 suppresses epithelial–mesenchymal transition (EMT), which induces cell migration and decrease intercellular adhesion, and is associated with chemoresistance in various cancers, thus, decreasing chemoresistance to TMZ via suppressing the Wnt/β-catenin signaling pathway [70].
The nuclear-enriched abundant transcript 1 (NEAT1) lncRNA, a 3.7 kb lncRNA found on chromosome 11q13.1, localizes to the nucleus and is an essential core component of the paraspeckle substructure [71, 72]. This lncRNA is upregulated in some cancers, including glioma, where it plays as an oncogene, and is involved in proliferation, migration, invasion, and impaired apoptosis [73–75]. Moreover, an increase in this lncRNA enhances progression of glioblastoma and tumorigenesis via the effects of the EGFR pathway on cell cycle regulation. This involves binding of enhancer of zeste 2 (EZH2) and the Wnt/β-catenin signaling pathway [76].
Wnt inhibitory factor 1 (WIF1) attenuates non-canonical Wnt signaling through downregulation of MALAT1, thereby suppressing migration, but not proliferation, of glioblastoma cells [77].
Similarly, knockdown of CCAT2 suppresses cell migration, proliferation, cell cycle progression, and tumorigenesis in glioma by downregulating the Wnt/β-catenin signaling pathway [78].
Other pathways
In addition to these pathways, several other pathways, including the miRNA axis, bone morphogenetic protein (BMP), Notch, nuclear factor kappa-light-chain-enhancer of activation B cells (NF-κB) pathways are involved in cancer. miRNAs are small non-coding RNAs that are 21–25 nucleotides in length, which mainly regulate expression of gene at the post-transcriptional level, and the expression of protein via binding to their target mRNAs [79]. BMP protein is a part of the transforming growth factor-β (TGF-β) family and plays pivotal roles in the morphogenesis of different organs, including the neural system. BMP signaling has also been reported as a tumor promoter as well as a tumor suppressor in various types of cancer [80]. Notch signaling is a conserved ligand-receptor signaling pathway that is involved in fundamental mechanistic functions, such as cell survival, differentiation, development, and apoptosis. Dysregulation of Notch signaling has been reported in various diseases including cancer [81]. Finally, NF-κB is one of the long-suspected factors that mediates interactions between cancer and inflammation. NF-κB signaling pathway regulates cell migration, proliferation, inflammation, and apoptosis by modulating the expression of anti-apoptotic molecules and pro-inflammatory factors [82].
The overexpression of plasmacytoma variant translocation 1 (PVT1) lncRNA located on chromosome 8q24, has been reported to play an important role in tumor progression and biological processes, such as apoptosis, invasion, mobility, and proliferation in various types of cancer [83–85]. It affects the progression of glioma by targeting miRNAs, such as miR-186 [86], miR-200a [87], miR-424 [88], and miR-140-5p [89]. Knockdown of this lncRNA suppresses malignant behaviors in glioma through the binding and upregulation of miR-190a-5p and miR-488-3p, which target and inhibit myocyte enhancer factor 2C (MEF2C), which is also transcriptionally regulated by the oncogene jagged 1 (JAG1) [90]. Furthermore, this lncRNA modulates Gremlin 1 (GREM1) and BMP downstream signaling pathway through sponging of miR-128-3p, thereby promoting tumorigenesis and progression in gliomas [91].
As mentioned previously, CASC2, which is downregulated in glioma, modulates miRNAs. Overexpression of CASC2 inhibits glioma malignancy, migration, invasion, and proliferation and induces cell apoptosis via negative modulation of miR-21 [92].
The ZNFX1 antisense RNA 1 (ZFAS1) lncRNA located on chromosome 20q13 is another lncRNA involved in progression and metastasis of various cancers [93]. ZFAS1 induces glioma progression by sponging miR‐150‐5p to regulate proteolipid protein 2 (PLP2), which is known as the functional target of miR‐150‐5p [94]. Inhibition of ZFAS1 significantly suppresses proliferation, migration, and invasion of glioma cells. Furthermore, ZFAS1 knockdown reduces the viability of glioma cells and increases their cisplatin chemosensitivity by targeting miR-432-5p [95]. Moreover, the EMT and the Notch signaling pathway are inactivated in the glioma cells after ZFAS1 knockdown. Thus, ZFAS1 exhibits an oncogenic role in glioma progression by regulating the EMT and the Notch signaling pathway [96].
Similarly, DANCR promotes glioma progression by acting as a sponge of miR-634, thus modulating RAB1A, which has an important role in tumor progression [97]. Furthermore, knockdown of DANCR revealed its role as the ceRNA that regulates expression of B-lymphoma Moloney murine leukemia virus insertion region-1 (BMI1), a self-renewal gene that is associated with tumorigenesis and progression of cancer in glioma [98]. Thus, knockdown of DANCR inhibits migration, invasion, and proliferation of glioma cells via sponging of miR-135a-5p [99].
The ceRNA role of SNHG16, which is overexpressed in glioma, also promotes the malignancy through the enhancement of endogenous target gene, E2F1 via sponging of miR-20a-5p [100].
Knockdown of CRNDE promotes the miR-348 expression, leading to inhibition of piwi-like RNA-mediated gene silencing 4 (PIWIL4), which has an oncogenic role in glioma, thus inhibiting the malignant progression of glioma [101]. CRNDE also downregulates miR-186, which is known to have a malignant biological role in glioma stem cells [102].
Glioma malignancy is also promoted by HOTAIR via its interactions with the polycomb repressive complex 2 (PRC2) and certain miRNAs [103]. Also, HOTAIR expression correlates with angiogenesis via modulation of VEGFA expression, one of the most important factors in tumor angiogenesis, and transmission of this factor to endothelial cells through glioma cell-derived extracellular vesicles [104]. HOTAIR also regulates glutaminase levels to enhance glioma progression via binding of miR-126-5p as a ceRNA [105]. High expression of HOTAIR was found to interact with miR-148b-3p and inhibit malignancy in glioma cell [106]. HOTAIR regulates the 3’ untranslated region of spindle and kinetochore associated complex subunit 2 (SKA2), and binds the tumor suppressor miR-141 as an endogenous sponge, thus participating in the pathogenesis and progression of human gliomas [107]. Knockdown of HOTAIR increased the permeability of the blood–tumor barrier (BTB) by connecting with miR-148b-3p as a target RNA. This knockdown also decreased the levels of tight junction-correlated proteins, such as Zonula occludens 1 (ZO-1), occludin, and claudin-5, in glioma microvascular endothelial cells by targeting upstream stimulatory factor 1 (USF1), thus enabling drug delivery [108]. Finally, HOTAIR is regulated by the glioblastoma proliferative pathway, specifically by bromodomain and extra-terminal domain proteins, such as bromodomain-containing 4 (BRD4), which binds to and increases HOTAIR levels in this pathway [109].
The H19 lncRNA increases proliferation and invasion via downregulating miR-152 [110], and miR-140, which targets iASPP [111] and driving of the miR-675 [66, 68, 112, 113]. It also regulates glioma angiogenesis by inhibiting miR-29a, which targets and represses the angiogenic factor vasohibin 2 (VASH2) [114]. H19 acts as a ceRNA on miR-138, which targets HIF-1α and modulates the expression of VEGF, thus promoting proliferation, migration, invasion, and angiogenesis in gliomas [115]. In addition, H19 regulates levels of the sex determining region Y-box protein 4 (SOX4) by sponging miR-130a-3p as a ceRNA. These findings indicate that this lncRNA regulates the EMT process, in which epithelial cells lose their original roles and gain migratory and invasion-related roles, becoming mesenchymal cells, as seen in various tumors [116]. H19 stimulation via oxidative stress promotes TMZ resistance through activation of the NF-κB signaling pathway [117].
Similarly, XIST regulates miR-429, thus promoting tumorigenicity and angiogenesis in gliomas [118]. XIST is correlated with miR-29c and modulates TMZ chemoresistance via its effects on DNA mismatch repair-related molecules, such as O6-methylguanine-DNA methyltransferase (MGMT) and specificity protein 1 (SP1) [119]. XIST also has oncogenic functions in glioma; it inhibits miR-137 by acting as a sponge, thereby increasing Ras-related C3 botulinum toxin substrate 1(Rac1) expression [120]. Knockdown of XIST upregulates miR-137, enhancing BTB permeability and decreasing angiogenesis in glioma via inhibition of the transcription factors forkhead box C1 (FOXC1) and ZO-2 [121]. It also upregulates miR-152, suppressing cell cycle progression and increasing apoptosis in glioma stem cells, thus playing a tumor suppressive role [122].
NEAT1 directly targets miR-449b-5p as a molecular sponge and promotes upregulation of c-Met, a direct target of miR-449b-5p, to increase pathogenesis in glioma [123]. Meanwhile, knockdown of NEAT1 activates miR-let-7e, which acts as a cancer inhibitor by decreasing migration, invasion, and proliferation while increasing apoptosis in glioma stem cells and this leads to downregulation of NRAS protein, an isoform of RAS reported to be mutated in several cancers and known to play an oncogenic role in glioma stem cells, thus inhibiting disease progression [124]. Similarly, NEAT1 knockdown targets miR-132 [125], which regulates the oncogene sex determining region Y-box protein 2 (SOX2) and miR-107 [126, 127], a modulator of cyclin-dependent kinase 6 (CDK6) and CDK14; these findings were closely connected with glioma malignancy. NEAT1 knockdown also promotes BTB permeability as it binds to miR-181-5p and targets sex determining region Y-box protein 5 (SOX5). Thus, knockdown of this lncRNA reduces expression of tight junction proteins, such as ZO-1, occludin, and claudin-5 in glioma microvascular endothelial cells [128].
MALAT1 increases the level of zinc fingers and homeoboxes 1 (ZHX1), which plays a role in the carcinogenesis of several cancers including glioblastoma by acting as a transcription repressor via molecular sponging of miR-199a and as a ceRNA, thereby promoting proliferation and progression of glioblastoma [129]. MALAT1 also acts as a tumor promoter, enhancing the tumorigenesis of glioma stem cells via modulation of miR-129 and subsequent suppression of SOX2 [130]. In addition, MALAT1 enhances progression of glioma cells via upregulation of Rap1B, STMN1, RAB5A, and ATG4D by sponging miR-101 [131, 132]. Moreover, MALAT1 promotes chemoresistance to TMZ in glioblastoma cells by repressing miR-101[133] and miR-203 [134]. However, MALAT1 also plays tumor-suppressive roles that involve repression of miR-155 and enhancement of F-box and WD repeat domain-containing 7 (FBXW7) function, which downregulates the expression of numerous oncoproteins in glioma cells [135].
In glioma, MATN1-AS1 enhances the carcinogenesis by sponging the miR-200b/c/429 as the ceRNA to regulate chromodomain helicase DNA-binding protein 1 (CHD1), which has an important role in the determination of cell fate [136].
CCAT2 also acts as a molecular sponge to downregulate miR-424 and upregulates Chk1, thereby enhancing chemotolerance in glioma cells [137].
LncRNAs and brain tumor immunity
The immune system shows responses to pathogens that may be categorized as part of innate or adaptive immunity. Pathogen-infected cells synthesize inflammatory mediators and cytokines via transcriptional and post-transcriptional regulation of genes as part of the immune response. Such immune responses start with discriminative recognition of pathogens by certain receptors, such as toll-like receptors [138]. Toll-like receptors recognize ligands and stimulate synthesis of inflammatory factors and cytokines through transcriptional regulation of genes via transcription factors, such as NF-κB and post-transcriptional regulation of gene expression through miRNAs [138, 139]. These miRNAs regulate the activation of T cells and the release of inflammatory cytokines, such as tumor necrosis factor α, interleukin, and interferon β [140–142]. Additionally, lncRNAs modulate the expression of immunogenes through transcriptional regulation [143]. LncRNAs also play various roles during an immune reaction including immune activation, immune cell migration, infiltration, antigen presentation, and antigen release as part of the immune response against cancer [144, 145]. Therefore, miRNAs and lncRNAs are important regulators involved in the activation of immunocytes during the pathology of tumor [146].
In anaplastic glioma patients, nine immune-related lncRNAs (SNHG8, PGM5-AS1, ST20-AS1, LINC00937, AGAP2-AS1, MIR155HG, TUG1, MAPKAPK5-AS1, and HCG18) have been identified. Among these, PGM5-AS1, ST20-AS1, AGAP2-AS1 and MIR155HG have been identified as risk-related genes, whereas SNHG8, LINC00937, TUGI, MAPKAPK5-AS1 and HCG18 are considered protective genes. The expression of these genes reveals differences in immune status among low- and high-risk groups. These immune-related lncRNAs directly or indirectly regulate several prognostic immune-related mRNAs [147]. Similarly, 11 immune-related lncRNA signatures have been identified in glioma patients: H19, DLGAP1.AS1, AC025171.1, PAXIP1.AS2, FEZF1.AS1, HOTAIRM1, HOXD.AS2, and ARHGEF26.AS1 are upregulated and LINC00205, WDR11.AS1, and THAP7.AS1 are downregulated in a high-risk glioma group compared to a low-risk group [148, 149]. A recent study reported four prognostic lncRNA (AK098425, AL833059, AK056155 and CR613436) that are coexpressed with protein-coding genes and are significantly associated with immune responses, such as the inflammatory response, innate immune response, and B cell-mediated immunity in glioblastomas [150]. Similarly, six immune-related lncRNA (AC005013.5, UBE2R2-AS1, ENTPD1-AS1, RP11-89C21.2, AC073115.6, and XLOC-004803) were found to be closely associated with immune-related biological processes and pathways in glioblastomas [151]. In addition, the lncRNA HOTAIR myeloid-specific 1 (HOTAIRM1), whose expression is upregulated in glioma, enhances malignancy via sponging of miR-129-5p as ceRNA and is also related to immune activation that promotes T cell-mediated immune responses [152]. Thus, these lncRNAs are useful for the development of specific immunotherapies for glioma patients. Targeting the immune response with cancer therapeutics is a promising new strategy for the treatment of gliomas.
Exocytosis and endocytosis of lncRNA in brain tumors
Extracellular vesicles (EVs) are nano-sized membranous and lipid bilayer vesicles. They can be divided into exosomes, ectosomes, microvesicles, and apoptotic bodies based on their biogenesis and size [153]. These vesicles are crucial factors in various disease processes, including in neurodegenerative diseases [154]. Exosomes ranges from 30 to 100 nm in diameter and are generated through the endosomal pathway by the exocytosis of multivesicular bodies from the plasma membrane. Exosomes are released by various types of cells, such as normal, immune, and tumor cells [155]. Released exosomes can be enclosed by neighboring cells or by recipient cells [156, 157]. Moreover, exosomes can transport various types of molecules, such as proteins, mRNAs and lncRNA that play important roles in cell to cell communications. Such exchange of genetic and epigenetic information through intercellular communication promotes epigenetic changes in cells of different tissues [158]. Interestingly, lncRNAs are selectively sorted into exosomes and the released exosomal lncRNAs can reveal differences in expression levels between normal cells and cancer cells. Hence, cancer cells show discrepancies in expression of specific exosomal lncRNAs, which may be involved in various functions [159, 160]. Several recent studies have reported the multiple functions of exosomal lncRNAs in cancer, including in tumorigenesis, angiogenesis, drug resistance, and regulation of tumor microenvironment [154, 161–163].
The POU class 3 homeobox 3 (POU3F3) lncRNA is an oncogene seen in various human cancers, such as cervical cancer, nasopharyngeal carcinoma, and breast cancer [164]. In glioma, expression of this lncRNA is upregulated and enriched exosomes released by glioma cells enhance angiogenesis [165].
Similarly, the SBF2 antisense RNA 1 (SBF2-AS1) lncRNA, located on the 11p15.1 locus, has been identified as a critical regulation factor in cancer progression [166, 167]. In glioblastoma, SBF2-AS1 can be transferred into exosomes and secretion of such oncogenic SBF2-AS1-enriched exosomes promotes chemoresistance to TMZ. SBF2-AS1 acts as a ceRNA molecular sponge of miR-151a-3p, thus promoting oncogenesis [168].
CCAT2-enriched exosomes are also released by glioma cells and may also be involved in human umbilical vein endothelial cells. In these endothelial cells, upregulation of CCAT2 enhances angiogenesis via increasing vascular endothelial growth factor A and transforming growth factor β expression and inhibits apoptosis via increasing Bcl-2 and decreasing Bcl-2-associated X (Bax) and caspase-3 expression. Therefore, the release of exosomes containing the lncRNA CCAT2 by glioma cells promotes angiogenesis and inhibits endothelial cell apoptosis [169].
LncRNA activated by transforming growth factor (TGF)-β (lncRNA-ATB), is abnormally expressed and functions as a progression factor in various cancers, such as hepatocellular carcinoma, gastric cancer, and breast cancer [170]. This exosomal lncRNA is upregulated in glioma and activates astrocytes, but connects and suppresses miR-204-3p in astrocytes. Thus, this lncRNA enhances the invasion of glioma cells [171].
Conclusions and perspectives
In 1955, Georges Palade, the cell biologist, identified the first ncRNA, which was linked to the ribonucleotide protein complex. This was followed by discoveries of regulatory ncRNAs, such as miRNAs, ribosomal RNAs, transfer RNAs, small nuclear RNAs, short interfering RNAs, ceRNAs, enhancer RNAs, piwi-interacting RNAs, and lncRNAs in the both prokaryotic and eukaryotic organisms [172]. However, ncRNAs were considered as ‘junk’ products of gene transcription processes and study of the ncRNAs was hampered by the lack of genetic information in the past decades. However, the roles of ncRNAs have been gradually elucidated to understand the genetic heterogeneity and their diverse roles in biological steps, and emerging findings have revealed crucial functions of ncRNAs in transcriptional regulation. In addition, advances in genome-wide association studies (GWAS) have led to the discovery of new ncRNAs. Among the ncRNAs, diverse biological functions of lncRNAs have been identified, including in tumor biology, such as modulation of transcription and translation by acting as transcription factors, splicing of mRNA, modification of chromatin, interaction of ribosomes, sponging with miRNA, degradation of mRNAs, scaffolding of protein interactions, and trafficking of transcription factors [172]. Recently, several remarkable studies reported the relation of lncRNAs and EVs, which translocate the DNA, RNA, proteins, and lipids to other cells, resulting in diverse phenotypic responses that promote functional modulation of cellular activity in recipient cells [173, 174]. Furthermore, lncRNAs and EVs are closely related with various diseases. In addition, they are associated with tumorigenesis and tumor microenvironment [175–177]. Consequently, these unique functions and biology of the EVs in cell-to-cell communications through the delivery of cargo, including mRNAs, miRNAs, and lncRNAs, make them attractive as potential therapeutics and biomarkers in various diseases [178, 179].
Molecular insights into lncRNAs-mediated intracellular pathways and transcriptional networks have provided answers for several unanswered questions in transcriptional and translational regulation. Recent observations have also revealed the roles of lncRNAs in diverse diseases and responses to various stresses. Besides, abnormal transcription and mutation of lncRNAs have been observed in many sporadic tumors [180]. Indeed, while activation or inactivation of lncRNAs might affect tumor development, cancer cells may also rely on the molecular environment. Given the potential importance of lncRNAs in brain tumors, they are an ideal target for oligonucleotide-based therapeutics. However, several biological issues must be resolved before their successful application as the therapeutic agents. For example, a more rigorous approach for clinical evaluation of lncRNAs for brain tumor therapeutics requires the assessment of blood–brain barrier (BBB) permeability or stability in cerebrospinal fluid exposure [181]. Although circulating lncRNAs are derived from several sources including blood and serum, identification of the lncRNAs in mammalian fluids has been hampered by a lack of genetic information. Moreover, most studies have been considered and preferred to identify miRNAs present in the fluids in diseased state as therapeutic targets compared to lncRNAs in whole blood [182]. Previously, cell-free nucleic acids, such as DNA, mRNA, and miRNA were considered as biomarkers in cancer patients [183]. Recent advances in lncRNA research, which were based on GWAS and biochemical systems, have improved the knowledge of cell-free nucleic acids including circulating lncRNAs [184, 185]. This impressively wide range of findings can facilitate the establishment of preclinical models for identification of potent lncRNAs for application in clinical treatment in cancer [186]. Thus, it is worth studying specific interactions between lncRNAs and brain tumors, which may enable determination of lncRNA-targeted therapeutic strategies that might be used increasingly in the future.
In this review, we have summarized the current understanding of the roles of lncRNAs in brain tumors (Table 1). Understanding the various functions of gene transcripts has led to advancements in identification of biomarkers and drug development. We have reviewed recent insights into biological functions of lncRNAs that were gleaned through genetic and biochemical studies, though many unanswered questions remain. For example, the genetic or epigenetic mechanisms that regulate the expression, localization, and activity of lncRNAs are still unclear. In addition, a lack of certain transcription factors in lncRNA pathways raises the question of whether bidirectional mechanisms other than genetic functions of sense or antisense transcripts exist. Besides, it is not clear whether lncRNAs are fundamental molecules in brain tumor progression. The development of brain tumor involves several complex mechanisms regulated by multiple molecules and pathways. Cells are constantly challenged by diverse abnormalities caused by exogenous and endogenous factors. In addition, genomic instability caused by various factors is related to tumor evolution and patient outcomes [187]. In general, tumors are ascribed to genetic heterogeneity resulting from changes in processes, such as transcriptional or epigenetic regulation [188]. The biological features of lncRNAs are heterogenous; they are associated with cis- or trans-acting transcriptional regulation and they are involved in modulation of targeted RNAs or proteins [6]. Since the discovery of the biological roles of lncRNAs, there have been significant advances in the field of tumorigenesis [189]. Therefore, novel therapeutic approaches might also include targeting active lncRNAs that are essential for cell death or survival in brain tumors. High-throughput screening studies across several decades have focused on identifying non-coding RNAs that could serve as genomic signatures in biological systems [190]. Using this approach, the importance of lncRNAs as therapeutic targets has emerged in various cancers [189]. The diverse roles of lncRNAs in tumorigenesis could shed greater light on other undiscovered functions of lncRNAs and potentially provide new therapeutic approaches for treating brain tumors.
Table 1.
Signaling pathway and role of lncRNA-target genes in brain tumors
| lncRNA | Target | Signaling pathway | Activity | Reference |
|---|---|---|---|---|
| PVT1 | miR-128-3p | BMP pathway | Promote tumorigenesis and progression | [91] |
| miR-186 | Promote cell proliferation, migration, and angiogenesis | [86] | ||
| miR-200a | Promote cell proliferation and invasion | [87] | ||
| miR-424 | Promote cell viability, migration invasion and tumor growth | [88] | ||
| miR-140-5p | Promote cell proliferation, invasion, and aerobic glycolysis | [89] | ||
| miR-190a-5p/MEF2C | Promote malignant behaviors | [90] | ||
| miR-488-3p/MEF2C | ||||
| CASC2 | miR-181a | PTEN and Akt pathway | Inhibit cell growth and resistance to TMZ | [27] |
| miR-193a-5p | mTOR pathway | Promote TMZ cytotoxicity through autophagy inhibition | [28] | |
| Wnt/β-catenin pathway | Inhibit cell proliferation, migration, and invasion | [62] | ||
| miR-21 | Inhibit cell proliferation, migration and invasion and promote apoptosis | [92] | ||
| ZFAS1 | Notch pathway | Promote progression via regulating EMT | [96] | |
| miR-150-5p | Promote progression | [94] | ||
| miR-432-5p | Promote viability and promote cisplatin cytotoxicity | [95] | ||
| DANCR | Wnt/β-catenin pathway | Promote progression through activation EMT | [63] | |
|
miR-33a-5p miR-33b-5p miR-1-3p miR-206 miR-613a |
PI3K/Akt/NF-κB pathway | Promote cisplatin resistance in vitro and in vivo | [34] | |
| miR-634/RAB1A | Promote progression | [97] | ||
| miR-135a-5p/BMI1 | Promote proliferation, migration, and invasion | [99] | ||
| HULC | ESM-1 | PI3K/Akt/mTOR pathway | Promote angiogenesis by regulated ESM-1 | [36] |
| SNHG16 | miR-4518/PRMT5 | PI3K/Akt pathway | Inhibit apoptosis and promote tumorigenesis via upregulating PRMT5 | [37] |
| miR-373/EGFR | PI3K/Akt pathway | Promote tumorigenesis via releasing EGFR | [38] | |
| miR-20a-5p/E2F1 | Promote malignancy through increase E2F1 | [100] | ||
| CRNDE | miR-136-5p/Bcl-2, Wnt2 | PI3K/Akt/mTOR pathway | Promote malignancy via inhibiting Bcl-2 and Wnt2 | [41] |
| P70S6K | mTOR pathway | Promote cell growth and invasion via increasing phosphorylation of P70S6K | [42] | |
| miR-348/PIWIL4 | Promote malignant progression via enhancing PIWIL4 | [101] | ||
| miR-186 | Promote cell proliferation, migration, invasion, and tumor growth by inhibiting apoptosis | [102] | ||
| HOTAIR | miR326/FGF1 | PI3K/Akt and MEK1/2 pathway | Promote apoptosis and cell cycle via inhibiting FGF1 | [45] |
| PRC2 | Promote cell cycle progression via interacting PRC2 | [103] | ||
| VEGFA | Promote angiogenesis via increasing VEGFA | [104] | ||
| miR-126-5p/GLS | Promote progression via regulating GLS | [105] | ||
| miR-148-3p | Inhibit malignant biological behaviors | [106] | ||
| miR-141/SKA2 | Promote progression and tumor growth | [107] | ||
| miR-148-3p/USF1 | Inhibit BTB permeability via promoting USF1 | [108] | ||
| BRD4 | Promote proliferation by regulation of BRD4 | [109] | ||
| H19 | NF-κB pathway | Promote TMZ resistance | [117] | |
| Wnt/β-catenin pathway | Promote proliferation, migration, and invasion | [69] | ||
| Wnt/β-catenin pathway | Promote TMZ resistance through suppressed EMT | [70] | ||
| miR-152 | Promote proliferation and invasion | [110] | ||
| miR-140/iASPP | Promote proliferation and invasion by targeting iASPP | [111] | ||
| miR-675 | Promote proliferation, migration invasion and tumor growth | [66, 68, 112, 113] | ||
| miR-29a/VASH2 | Promote angiogenesis | [114] | ||
| miR-138/HIF-1α | Promote proliferation, migration, invasion, and angiogenesis by targeting HIF-1α | [115] | ||
| miR-130a-3p/SOX4 | Promote migration, invasion and EMT via regulating SOX4 | [116] | ||
| XIST | miR-126/IRS1 | PI3K/Akt pathway | Promote cell growth, glucose metabolism and tumor growth via regulating IRS1 | [48] |
| miR-429 | Promote angiogenesis and tumorigenicity | [118] | ||
| miR-29c/SP1/MGMT | Promote proliferation and TMZ resistance through the DNA mismatch repair pathway | [119] | ||
| miR-137/Rac1 | Promote progression by upregulated Rac1 | [120] | ||
| miR-137/FOXC1/ZO-2 | Inhibit BTB permeability and promote angiogenesis by increasing FOXC1 and ZO-2 | [121] | ||
| miR-152 | Promote progression and decrease apoptosis | [122] | ||
| NEAT1 | EGFR | Wnt/β-catenin pathway | Promote progression and tumorigenesis through EGFR | [76] |
| miR-449b-5p/c-Met | Promote pathogenesis by upregulated c-Met | [123] | ||
| miR-let-7e/NRAS | Promote proliferation migration and invasion while inhibit apoptosis by upregulated NRAS | [124] | ||
| miR-132/SOX2 | Promote migration and invasion through upregulation of SOX2 | [125] | ||
| miR-107/CDK6, 14 | Promote malignant progression by promoted CDK6, 14 | [126, 127] | ||
| miR-181-5p/SOX5 | Inhibit BTB permeability via upregulating SOX5 | [128] | ||
| MALAT1 | WIF1 | Non-canonical Wnt pathway | Promote migration via regulating of WIF1 | [77] |
| ERK/MAPK pathway | Inhibit proliferation and invasion | [55, 56] | ||
| miR-199a/ZHX1 | Promote proliferation and progression by increase ZHX1 | [129] | ||
| miR-129/SOX2 | Promote tumorigenesis via facilitating of SOX2 | [130] | ||
| miR-101 | Promote progression and TMZ resistance | [131–133] | ||
| miR-203 | Promote TMZ resistance | [134] | ||
| miR-155/FBXW7 | Inhibit cell viability by upregulated FBXW7 | [135] | ||
| MATN1-AS1 | RELA | MAPK pathway | Inhibit cell proliferation and invasion through the apoptosis | [57] |
| miR-200b/c/429/CHD1 | Promote glioma progression | [136] | ||
| TPTEP1 | miR-106a-5p | P38 MAPK pathway | Inhibit stemness and radio resistance | [59] |
| CCAT2 | miR-424/VEGFA | PI3K/Akt pathway | Promote proliferation and angiogenesis via increasing VEGFA | [50] |
| Wnt/β-catenin pathway | Promote proliferation, migration, cell cycle and tumorigenesis | [78] | ||
| miR-424/Chk1 | Promote chemodrug resistance by upregulated Chk1 | [137] | ||
| Promote angiogenesis and inhibit endothelial cell apoptosis through release CCAT2-enriched exosomes | [169] | |||
| PGM5-AS1 | risk-related genes | Regulate several prognostic immune-related mRNA | [147] | |
| ST20-AS1 | ||||
| AGAP2-AS1 | ||||
| MIR155HG | ||||
| SNHG8 | protective genes | Regulate several prognostic immune-related mRNA | ||
| LINC00937 | ||||
| TUGI | ||||
| MAPKAPK5-AS1 | ||||
| HCG18 | ||||
| AK098425 | protein coding gene | Associate immune-related biological immunity | [150] | |
| AL833059 | ||||
| AK056155 | ||||
| CR613436 | ||||
| AC005013.5 | protein coding gene | Associate immune-related biological process and pathway | [151] | |
| UBE2R2-AS1 | ||||
| RP11-89C21.2 | ||||
| AC073115.6 | ||||
| XLOC-004803 | ||||
| HOTAIRM1 | miR-129-5p | Promote malignancy and relate immune activation | [152] | |
| POU3F3 | Promote angiogenesis and release POU3F3-enriched exosomes | [165] | ||
| SBF2-AS1 | miR-151a-3p | Promote TMZ resistance and secret SBF2-AS1-enriched exosomes | [168] | |
| ATB | miR-204-3p | Promote cell activation and invasion through release ATB-enriched exosomes | [171] |
Acknowledgements
All graphic figures were made with biorender.com. We would like to thank Editage for English language editing.
Abbreviations
- ATB
Activated by transforming growth factor-β
- Bcl-2
B-cell lymphoma 2 family
- Bax
Bcl-2-associated X
- BTB
Blood–tumor barrier
- BBB
Blood–brain barrier
- BMI1
B-lymphoma Moloney murine leukemia virus insertion region-1
- BMP
Bone morphogenetic protein
- BRD4
Bromodomain-containing 4
- CASC2
Cancer susceptibility candidate 2
- CCAT2
Cancer susceptibility candidate 2
- CHD1
Chromodomain helicase DNA-binding protein 1
- CRNDE
Colorectal neoplasia differentially expressed
- ceRNAs
Competing endogenous RNAs
- CDK6
Cyclin-dependent kinase 6
- DANCR
Differentiation antagonizing non-protein coding RNA
- ESM-1
Endothelial cell-specific molecule 1
- EZH2
Enhancer of zeste 2
- EGFR
Epidermal growth factor receptor
- EMT
Epithelial–mesenchymal transition
- ERK
Extracellular signal-regulated kinase
- EVs
Extracellular vesicles
- FBXW7
F-box and WD repeat domain-containing 7
- FGF1
Fibroblast growth factor 1
- FOXC1
Forkhead box C1
- GWAS
Genome-wide association studies
- GREM1
Gremlin 1
- HULC
Highly upregulated in liver cancer
- HOTAIR
HOX transcript antisense RNA
- IGF2
Insulin growth factor 2
- IRS1
Insulin receptor substrate 1
- JAG1
Jagged 1
- LncRNAs
Long non-coding RNAs
- MATN-AS1
MATN1 antisense RNA 1
- MMP2
Matrix metalloproteinase 2
- MALAT1
Metastasis-associated lung adenocarcinoma transcript 1
- miRNAs
MicroRNAs
- MAPKs
Mitogen-activated protein kinases
- MEF2C
Myocyte enhancer factor 2C
- NF-κB
Nuclear factor kappa-light-chain-enhancer of activation B cells
- NEAT1
Nuclear-enriched abundant transcript 1
- MGMT
O6-methylguanine-DNA methyltransferase
- PTEN
Phosphatase and tensin homolog
- PIWIL4
Piwi-like RNA-mediated gene silencing 4
- PVT1
Plasmacytoma variant translocation 1
- PRC2
Polycomb repressive complex 2
- POU3F3
POU class 3 homeobox 3
- PRMT5
Protein arginine methyltransferase 5
- PLP2
Proteolipid protein 2
- Rac1
Ras-related C3 botulinum toxin substrate 1
- SBF2-AS1
SBF2 antisense RNA 1
- SOX2
Sex determining region Y-box protein 2
- SOX4
Sex determining region Y-box protein 4
- SOX5
Sex determining region Y-box protein 5
- SNHG16
Small nucleolar RNA host gene 16
- SP1
Specificity protein 1
- SKA2
Spindle and kinetochore associated complex subunit 2
- TMZ
Temozolomide
- TGF-β
Transforming growth factor-β
- TPTEP1
Transmembrane phosphatase with tensin homology pseudogene 1
- USF1
Upstream stimulatory factor 1
- VEGFA
Vascular endothelial growth factor A
- VASH2
Vasohibin 2
- WIF1
Wnt inhibitory factor 1
- XIST
X-inactive-specific transcript;
- ZHX1
Zinc fingers and homeoboxes 1
- ZFAS1
ZNFX1 antisense RNA 1
- ZO-1
Zonula occludens 1
Authors’ contributions
S.-H.K., K.-H.L., S.Y., and J.-Y.J. wrote the paper. S.-H.K. and S.Y. created the figures and tables. S.-H.K. and K.-H.L. have contributed equally to this work. J.-Y.J. provided guidance and direction for manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by Korea Brain Research Institute (KBRI) basic research program through KBRI funded by the Ministry of Science and ICT (21-BR-02–09), and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2019R1F1A1059595).
Availability of data and materials
Not applicable.
Declarations
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing financial interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sung-Hyun Kim and Key-Hwan Lim have contributed equally to this work.
References
- 1.Miranda-Filho A, Pineros M, Soerjomataram I, Deltour I, Bray F. Cancers of the brain and CNS: global patterns and trends in incidence. Neuro Oncol. 2017;19(2):270–280. doi: 10.1093/neuonc/now166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wesseling P, Capper D. WHO 2016 classification of gliomas. Neuropathol Appl Neurobiol. 2018;44(2):139–150. doi: 10.1111/nan.12432. [DOI] [PubMed] [Google Scholar]
- 3.Behnan J, Finocchiaro G, Hanna G. The landscape of the mesenchymal signature in brain tumours. Brain. 2019;142(4):847–866. doi: 10.1093/brain/awz044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jovcevska I, Kocevar N, Komel R. Glioma and glioblastoma—how much do we (not) know? Mol Clin Oncol. 2013;1(6):935–941. doi: 10.3892/mco.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poon CC, Sarkar S, Yong VW, Kelly JJP. Glioblastoma-associated microglia and macrophages: targets for therapies to improve prognosis. Brain. 2017;140(6):1548–1560. doi: 10.1093/brain/aww355. [DOI] [PubMed] [Google Scholar]
- 6.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei CW, Luo T, Zou SS, Wu AS. The role of long noncoding rnas in central nervous system and neurodegenerative diseases. Front Behav Neurosci. 2018;12:175. doi: 10.3389/fnbeh.2018.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bali KK, Kuner R. Noncoding RNAs: key molecules in understanding and treating pain. Trends Mol Med. 2014;20(8):437–448. doi: 10.1016/j.molmed.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pickard MR, Mourtada-Maarabouni M, Williams GT. Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim Biophys Acta. 2013;1832(10):1613–1623. doi: 10.1016/j.bbadis.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333(2):213–221. doi: 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 13.Sun W, Wu Y, Yu X, Liu Y, Song H, Xia T, Xiao B, Guo J. Decreased expression of long noncoding RNA AC096655.1-002 in gastric cancer and its clinical significance. Tumour Biol. 2013;34(5):2697–2701. doi: 10.1007/s13277-013-0821-0. [DOI] [PubMed] [Google Scholar]
- 14.Thai P, Statt S, Chen CH, Liang E, Campbell C, Wu R. Characterization of a novel long noncoding RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer cell lines. Am J Respir Cell Mol Biol. 2013;49(2):204–211. doi: 10.1165/rcmb.2013-0159RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ge X, Chen Y, Liao X, Liu D, Li F, Ruan H, Jia W. Overexpression of long noncoding RNA PCAT-1 is a novel biomarker of poor prognosis in patients with colorectal cancer. Med Oncol. 2013;30(2):588. doi: 10.1007/s12032-013-0588-6. [DOI] [PubMed] [Google Scholar]
- 16.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3(10):1113–1121. doi: 10.1158/2159-8290.CD-13-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang C, Wu D, Gao L, Liu X, Jin Y, Wang D, Wang T, Li X. Competing endogenous RNA networks in human cancer: hypothesis, validation, and perspectives. Oncotarget. 2016;7(12):13479–13490. doi: 10.18632/oncotarget.7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li F, Li Q, Wu X. Construction and analysis for differentially expressed long non-coding RNAs and MicroRNAs mediated competing endogenous RNA network in colon cancer. PLoS ONE. 2018;13(2):e0192494. doi: 10.1371/journal.pone.0192494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahu A, Singhal U, Chinnaiyan AM. Long noncoding RNAs in cancer: from function to translation. Trends Cancer. 2015;1(2):93–109. doi: 10.1016/j.trecan.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Zhu Y, Wang H, Ji X. Targeting long noncoding RNA in glioma: a pathway perspective. Mol Ther Nucleic Acids. 2018;13:431–441. doi: 10.1016/j.omtn.2018.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi J, Dong B, Cao J, Mao Y, Guan W, Peng Y, Wang S. Long non-coding RNA in glioma: signaling pathways. Oncotarget. 2017;8(16):27582–27592. doi: 10.18632/oncotarget.15175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi EJ, Cho BJ, Lee DJ, Hwang YH, Chun SH, Kim HH, Kim IA. Enhanced cytotoxic effect of radiation and temozolomide in malignant glioma cells: targeting PI3K-AKT-mTOR signaling, HSP90 and histone deacetylases. BMC Cancer. 2014;14:17. doi: 10.1186/1471-2407-14-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willems L, Tamburini J, Chapuis N, Lacombe C, Mayeux P, Bouscary D. PI3K and mTOR signaling pathways in cancer: new data on targeted therapies. Curr Oncol Rep. 2012;14(2):129–138. doi: 10.1007/s11912-012-0227-y. [DOI] [PubMed] [Google Scholar]
- 25.Baldinu P, Cossu A, Manca A, Satta MP, Sini MC, Rozzo C, Dessole S, Cherchi P, Gianfrancesco F, Pintus A, et al. Identification of a novel candidate gene, CASC2, in a region of common allelic loss at chromosome 10q26 in human endometrial cancer. Hum Mutat. 2004;23(4):318–326. doi: 10.1002/humu.20015. [DOI] [PubMed] [Google Scholar]
- 26.Palmieri G, Paliogiannis P, Sini MC, Manca A, Palomba G, Doneddu V, Tanda F, Pascale MR, Cossu A. Long non-coding RNA CASC2 in human cancer. Crit Rev Oncol Hematol. 2017;111:31–38. doi: 10.1016/j.critrevonc.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Liao Y, Shen L, Zhao H, Liu Q, Fu J, Guo Y, Peng R, Cheng L. LncRNA CASC2 interacts with miR-181a to modulate glioma growth and resistance to TMZ through PTEN pathway. J Cell Biochem. 2017;118(7):1889–1899. doi: 10.1002/jcb.25910. [DOI] [PubMed] [Google Scholar]
- 28.Jiang C, Shen F, Du J, Fang X, Li X, Su J, Wang X, Huang X, Liu Z. Upregulation of CASC2 sensitized glioma to temozolomide cytotoxicity through autophagy inhibition by sponging miR-193a-5p and regulating mTOR expression. Biomed Pharmacother. 2018;97:844–850. doi: 10.1016/j.biopha.2017.10.146. [DOI] [PubMed] [Google Scholar]
- 29.Thin KZ, Liu X, Feng X, Raveendran S, Tu JC. LncRNA-DANCR: A valuable cancer related long non-coding RNA for human cancers. Pathol Res Pract. 2018;214(6):801–805. doi: 10.1016/j.prp.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 30.Jin L, Fu H, Quan J, Pan X, He T, Hu J, Li Y, Li H, Yang Y, Ye J, et al. Overexpression of long non-coding RNA differentiation antagonizing non-protein coding RNA inhibits the proliferation, migration and invasion and promotes apoptosis of renal cell carcinoma. Mol Med Rep. 2017;16(4):4463–4468. doi: 10.3892/mmr.2017.7135. [DOI] [PubMed] [Google Scholar]
- 31.Sha S, Yuan D, Liu Y, Han B, Zhong N. Targeting long non-coding RNA DANCR inhibits triple negative breast cancer progression. Biol Open. 2017;6(9):1310–1316. doi: 10.1242/bio.023135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elkabets M, Pazarentzos E, Juric D, Sheng Q, Pelossof RA, Brook S, Benzaken AO, Rodon J, Morse N, Yan JJ, et al. AXL mediates resistance to PI3Kalpha inhibition by activating the EGFR/PKC/mTOR axis in head and neck and esophageal squamous cell carcinomas. Cancer Cell. 2015;27(4):533–546. doi: 10.1016/j.ccell.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stitt TN, Conn G, Gore M, Lai C, Bruno J, Radziejewski C, Mattsson K, Fisher J, Gies DR, Jones PF, et al. The anticoagulation factor protein S and its relative, Gas6, are ligands for the Tyro 3/Axl family of receptor tyrosine kinases. Cell. 1995;80(4):661–670. doi: 10.1016/0092-8674(95)90520-0. [DOI] [PubMed] [Google Scholar]
- 34.Ma Y, Zhou G, Li M, Hu D, Zhang L, Liu P, Lin K. Long noncoding RNA DANCR mediates cisplatin resistance in glioma cells via activating AXL/PI3K/Akt/NF-kappaB signaling pathway. Neurochem Int. 2018;118:233–241. doi: 10.1016/j.neuint.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 35.Yang YR, Jung JH, Kim SJ, Hamada K, Suzuki A, Kim HJ, Lee JH, Kwon OB, Lee YK, Kim J, et al. Forebrain-specific ablation of phospholipase Cgamma1 causes manic-like behavior. Mol Psychiatry. 2017;22(10):1473–1482. doi: 10.1038/mp.2016.261. [DOI] [PubMed] [Google Scholar]
- 36.Zhu Y, Zhang X, Qi L, Cai Y, Yang P, Xuan G, Jiang Y. HULC long noncoding RNA silencing suppresses angiogenesis by regulating ESM-1 via the PI3K/Akt/mTOR signaling pathway in human gliomas. Oncotarget. 2016;7(12):14429–14440. doi: 10.18632/oncotarget.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu YF, Cai XL, Li ZZ, Lv J, Xiang YA, Chen JJ, Chen WJ, Sun WY, Liu XM, Chen JB. LncRNA SNHG16 functions as an oncogene by sponging MiR-4518 and up-regulating PRMT5 expression in glioma. Cell Physiol Biochem. 2018;45(5):1975–1985. doi: 10.1159/000487974. [DOI] [PubMed] [Google Scholar]
- 38.Zhou XY, Liu H, Ding ZB, Xi HP, Wang GW. lncRNA SNHG16 promotes glioma tumorigenicity through miR-373/EGFR axis by activating PI3K/AKT pathway. Genomics. 2020;112(1):1021–1029. doi: 10.1016/j.ygeno.2019.06.017. [DOI] [PubMed] [Google Scholar]
- 39.Lee JH, Liu R, Li J, Zhang C, Wang Y, Cai Q, Qian X, Xia Y, Zheng Y, Piao Y, et al. Stabilization of phosphofructokinase 1 platelet isoform by AKT promotes tumorigenesis. Nat Commun. 2017;8(1):949. doi: 10.1038/s41467-017-00906-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ellis BC, Molloy PL, Graham LD. CRNDE: a long non-coding RNA involved in cancer, neurobiology, and development. Front Genet. 2012;3:270. doi: 10.3389/fgene.2012.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li DX, Fei XR, Dong YF, Cheng CD, Yang Y, Deng XF, Huang HL, Niu WX, Zhou CX, Xia CY, et al. The long non-coding RNA CRNDE acts as a ceRNA and promotes glioma malignancy by preventing miR-136-5p-mediated downregulation of Bcl-2 and Wnt2. Oncotarget. 2017;8(50):88163–88178. doi: 10.18632/oncotarget.21513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Wang Y, Li J, Zhang Y, Yin H, Han B. CRNDE, a long-noncoding RNA, promotes glioma cell growth and invasion through mTOR signaling. Cancer Lett. 2015;367(2):122–128. doi: 10.1016/j.canlet.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y, Zhang K, Luo Z, Liu L, Wu L, Liu J. Circulating long non-coding HOX transcript antisense intergenic ribonucleic acid in plasma as a potential biomarker for diagnosis of breast cancer. Thorac Cancer. 2016;7(6):627–632. doi: 10.1111/1759-7714.12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang JX, Han L, Bao ZS, Wang YY, Chen LY, Yan W, Yu SZ, Pu PY, Liu N, You YP, et al. HOTAIR, a cell cycle-associated long noncoding RNA and a strong predictor of survival, is preferentially expressed in classical and mesenchymal glioma. Neuro Oncol. 2013;15(12):1595–1603. doi: 10.1093/neuonc/not131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ke J, Yao YL, Zheng J, Wang P, Liu YH, Ma J, Li Z, Liu XB, Li ZQ, Wang ZH, et al. Knockdown of long non-coding RNA HOTAIR inhibits malignant biological behaviors of human glioma cells via modulation of miR-326. Oncotarget. 2015;6(26):21934–21949. doi: 10.18632/oncotarget.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weakley SM, Wang H, Yao Q, Chen C. Expression and function of a large non-coding RNA gene XIST in human cancer. World J Surg. 2011;35(8):1751–1756. doi: 10.1007/s00268-010-0951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349(6304):38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 48.Cheng Z, Luo C, Guo Z. LncRNA-XIST/microRNA-126 sponge mediates cell proliferation and glucose metabolism through the IRS1/PI3K/Akt pathway in glioma. J Cell Biochem. 2020;121(3):2170–2183. doi: 10.1002/jcb.29440. [DOI] [PubMed] [Google Scholar]
- 49.Xin Y, Li Z, Zheng H, Chan MTV, Ka Kei Wu W. CCAT2: a novel oncogenic long non-coding RNA in human cancers. Cell Prolif. 2017;50(3):1–4. doi: 10.1111/cpr.12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun SL, Shu YG, Tao MY. LncRNA CCAT2 promotes angiogenesis in glioma through activation of VEGFA signalling by sponging miR-424. Mol Cell Biochem. 2020;468(1–2):69–82. doi: 10.1007/s11010-020-03712-y. [DOI] [PubMed] [Google Scholar]
- 51.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26(22):3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 52.Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 53.Yoshimoto R, Mayeda A, Yoshida M, Nakagawa S. MALAT1 long non-coding RNA in cancer. Biochim Biophys Acta. 2016;1859(1):192–199. doi: 10.1016/j.bbagrm.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 54.Ma KX, Wang HJ, Li XR, Li T, Su G, Yang P, Wu JW. Long noncoding RNA MALAT1 associates with the malignant status and poor prognosis in glioma. Tumour Biol. 2015;36(5):3355–3359. doi: 10.1007/s13277-014-2969-7. [DOI] [PubMed] [Google Scholar]
- 55.Han Y, Wu Z, Wu T, Huang Y, Cheng Z, Li X, Sun T, Xie X, Zhou Y, Du Z. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death Dis. 2016;7:e2123. doi: 10.1038/cddis.2015.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han Y, Zhou L, Wu T, Huang Y, Cheng Z, Li X, Sun T, Zhou Y, Du Z. Downregulation of lncRNA-MALAT1 affects proliferation and the expression of stemness markers in glioma stem cell line SHG139S. Cell Mol Neurobiol. 2016;36(7):1097–1107. doi: 10.1007/s10571-015-0303-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han N, Yang L, Zhang X, Zhou Y, Chen R, Yu Y, Dong Z, Zhang M. LncRNA MATN1-AS1 prevents glioblastoma cell from proliferation and invasion via RELA regulation and MAPK signaling pathway. Ann Transl Med. 2019;7(23):784. doi: 10.21037/atm.2019.11.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cao F, Wang Z, Feng Y, Zhu H, Yang M, Zhang S, Wang X. lncRNA TPTEP1 competitively sponges miR3285p to inhibit the proliferation of nonsmall cell lung cancer cells. Oncol Rep. 2020;43(5):1606–1618. doi: 10.3892/or.2020.7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang T, Wang LX, Yang ML, Zhang RM. lncRNA TPTEP1 inhibits stemness and radioresistance of glioma through miR106a5pmediated P38 MAPK signaling. Mol Med Rep. 2020;22(6):4857–4867. doi: 10.3892/mmr.2020.11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gong A, Huang S. FoxM1 and Wnt/beta-catenin signaling in glioma stem cells. Cancer Res. 2012;72(22):5658–5662. doi: 10.1158/0008-5472.CAN-12-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17(1):9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang R, Li Y, Zhu G, Tian B, Zeng W, Yang Y, Li Z. Long noncoding RNA CASC2 predicts the prognosis of glioma patients and functions as a suppressor for gliomas by suppressing Wnt/beta-catenin signaling pathway. Neuropsychiatr Dis Treat. 2017;13:1805–1813. doi: 10.2147/NDT.S137171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li J, Zhou L. Overexpression of lncRNA DANCR positively affects progression of glioma via activating Wnt/beta-catenin signaling. Biomed Pharmacother. 2018;102:602–607. doi: 10.1016/j.biopha.2018.03.116. [DOI] [PubMed] [Google Scholar]
- 64.Raveh E, Matouk IJ, Gilon M, Hochberg A. The H19 Long non-coding RNA in cancer initiation, progression and metastasis - a proposed unifying theory. Mol Cancer. 2015;14:184. doi: 10.1186/s12943-015-0458-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gabory A, Ripoche MA, Yoshimizu T, Dandolo L. The H19 gene: regulation and function of a non-coding RNA. Cytogenet Genome Res. 2006;113(1–4):188–193. doi: 10.1159/000090831. [DOI] [PubMed] [Google Scholar]
- 66.Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14(7):659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiang X, Yan Y, Hu M, Chen X, Wang Y, Dai Y, Wu D, Wang Y, Zhuang Z, Xia H. Increased level of H19 long noncoding RNA promotes invasion, angiogenesis, and stemness of glioblastoma cells. J Neurosurg. 2016;2016(1):129–136. doi: 10.3171/2014.12.JNS1426.test. [DOI] [PubMed] [Google Scholar]
- 68.Shi Y, Wang Y, Luan W, Wang P, Tao T, Zhang J, Qian J, Liu N, You Y. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS ONE. 2014;9(1):e86295. doi: 10.1371/journal.pone.0086295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guan N, Wang R, Guo WS, Lai YJ, Zhang YD, Cheng YY. Long non-coding RNA H19 regulates the development of gliomas through the Wnt/beta-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23(10):4243–4253. doi: 10.26355/eurrev_201905_17929. [DOI] [PubMed] [Google Scholar]
- 70.Jia L, Tian Y, Chen Y, Zhang G. The silencing of LncRNA-H19 decreases chemoresistance of human glioma cells to temozolomide by suppressing epithelial-mesenchymal transition via the Wnt/beta-Catenin pathway. Onco Targets Ther. 2018;11:313–321. doi: 10.2147/OTT.S154339. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 71.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33(6):717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Naganuma T, Hirose T. Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol. 2013;10(3):456–461. doi: 10.4161/rna.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu Y, Wang H. LncRNA NEAT1 promotes dexamethasone resistance in multiple myeloma by targeting miR-193a/MCL1 pathway. J Biochem Mol Toxicol. 2018;32(1):1–6. doi: 10.1002/jbt.22008. [DOI] [PubMed] [Google Scholar]
- 74.Fang J, Qiao F, Tu J, Xu J, Ding F, Liu Y, Akuo BA, Hu J, Shao S. High expression of long non-coding RNA NEAT1 indicates poor prognosis of human cancer. Oncotarget. 2017;8(28):45918–45927. doi: 10.18632/oncotarget.17439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K, Kossai M, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen Q, Cai J, Wang Q, Wang Y, Liu M, Yang J, Zhou J, Kang C, Li M, Jiang C. Long noncoding RNA NEAT1, regulated by the EGFR pathway, contributes to glioblastoma progression through the WNT/beta-catenin pathway by scaffolding EZH2. Clin Cancer Res. 2018;24(3):684–695. doi: 10.1158/1078-0432.CCR-17-0605. [DOI] [PubMed] [Google Scholar]
- 77.Vassallo I, Zinn P, Lai M, Rajakannu P, Hamou MF, Hegi ME. WIF1 re-expression in glioblastoma inhibits migration through attenuation of non-canonical WNT signaling by downregulating the lncRNA MALAT1. Oncogene. 2016;35(1):12–21. doi: 10.1038/onc.2015.61. [DOI] [PubMed] [Google Scholar]
- 78.Guo H, Hu G, Yang Q, Zhang P, Kuang W, Zhu X, Wu L. Knockdown of long non-coding RNA CCAT2 suppressed proliferation and migration of glioma cells. Oncotarget. 2016;7(49):81806–81814. doi: 10.18632/oncotarget.13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 80.Fukuda T, Fukuda R, Tanabe R, Koinuma D, Koyama H, Hashizume Y, Moustakas A, Miyazono K, Heldin CH. BMP signaling is a therapeutic target in ovarian cancer. Cell Death Discov. 2020;6(1):139. doi: 10.1038/s41420-020-00377-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li L, Tang P, Li S, Qin X, Yang H, Wu C, Liu Y. Notch signaling pathway networks in cancer metastasis: a new target for cancer therapy. Med Oncol. 2017;34(10):180. doi: 10.1007/s12032-017-1039-6. [DOI] [PubMed] [Google Scholar]
- 82.Taniguchi K, Karin M. NF-kappaB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol. 2018;18(5):309–324. doi: 10.1038/nri.2017.142. [DOI] [PubMed] [Google Scholar]
- 83.Takahashi Y, Sawada G, Kurashige J, Uchi R, Matsumura T, Ueo H, Takano Y, Eguchi H, Sudo T, Sugimachi K, et al. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers. Br J Cancer. 2014;110(1):164–171. doi: 10.1038/bjc.2013.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guan Y, Kuo WL, Stilwell JL, Takano H, Lapuk AV, Fridlyand J, Mao JH, Yu M, Miller MA, Santos JL, et al. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res. 2007;13(19):5745–5755. doi: 10.1158/1078-0432.CCR-06-2882. [DOI] [PubMed] [Google Scholar]
- 85.Ding C, Yang Z, Lv Z, Du C, Xiao H, Peng C, Cheng S, Xie H, Zhou L, Wu J, et al. Long non-coding RNA PVT1 is associated with tumor progression and predicts recurrence in hepatocellular carcinoma patients. Oncol Lett. 2015;9(2):955–963. doi: 10.3892/ol.2014.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ma Y, Wang P, Xue Y, Qu C, Zheng J, Liu X, Ma J, Liu Y. PVT1 affects growth of glioma microvascular endothelial cells by negatively regulating miR-186. Tumour Biol. 2017;39(3):1010428317694326. doi: 10.1177/1010428317694326. [DOI] [PubMed] [Google Scholar]
- 87.Zhang Y, Yang G, Luo Y. Long non-coding RNA PVT1 promotes glioma cell proliferation and invasion by targeting miR-200a. Exp Ther Med. 2019;17(2):1337–1345. doi: 10.3892/etm.2018.7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Han Y, Li X, He F, Yan J, Ma C, Zheng X, Zhang J, Zhang D, Meng C, Zhang Z, et al. Knockdown of lncRNA PVT1 inhibits glioma progression by regulating miR-424 expression. Oncol Res. 2019;27(6):681–690. doi: 10.3727/096504018X15424939990246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shao Y, Chen HT, Ma QR, Zhang YW, He YQ, Liu J. Long non-coding RNA PVT1 regulates glioma proliferation, invasion, and aerobic glycolysis via miR-140-5p. Eur Rev Med Pharmacol Sci. 2020;24(1):274–283. doi: 10.26355/eurrev_202001_19922. [DOI] [PubMed] [Google Scholar]
- 90.Xue W, Chen J, Liu X, Gong W, Zheng J, Guo X, Liu Y, Liu L, Ma J, Wang P, et al. (2018) PVT1 regulates the malignant behaviors of human glioma cells by targeting miR-190a-5p and miR-488–3p. Biochim Biophys Acta Mol Basis Dis. 1864;5 Pt A:1783–1794. doi: 10.1016/j.bbadis.2018.02.022. [DOI] [PubMed] [Google Scholar]
- 91.Fu C, Li D, Zhang X, Liu N, Chi G, Jin X. LncRNA PVT1 facilitates tumorigenesis and progression of glioma via regulation of MiR-128-3p/GREM1 axis and BMP signaling pathway. Neurotherapeutics. 2018;15(4):1139–1157. doi: 10.1007/s13311-018-0649-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 92.Wang P, Liu YH, Yao YL, Li Z, Li ZQ, Ma J, Xue YX. Long non-coding RNA CASC2 suppresses malignancy in human gliomas by miR-21. Cell Signal. 2015;27(2):275–282. doi: 10.1016/j.cellsig.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 93.Feng LL, Shen FR, Zhou JH, Chen YG. Expression of the lncRNA ZFAS1 in cervical cancer and its correlation with prognosis and chemosensitivity. Gene. 2019;696:105–112. doi: 10.1016/j.gene.2019.01.025. [DOI] [PubMed] [Google Scholar]
- 94.Li X, Luo Y, Liu L, Cui S, Chen W, Zeng A, Shi Y, Luo L. The long noncoding RNA ZFAS1 promotes the progression of glioma by regulating the miR-150-5p/PLP2 axis. J Cell Physiol. 2020;235(3):2937–2946. doi: 10.1002/jcp.29199. [DOI] [PubMed] [Google Scholar]
- 95.Yang G, Han B, Feng T. ZFAS1 knockdown inhibits viability and enhances cisplatin cytotoxicity by up-regulating miR-432-5p in glioma cells. Basic Clin Pharmacol Toxicol. 2019;125(6):518–526. doi: 10.1111/bcpt.13286. [DOI] [PubMed] [Google Scholar]
- 96.Gao K, Ji Z, She K, Yang Q, Shao L. Long non-coding RNA ZFAS1 is an unfavourable prognostic factor and promotes glioma cell progression by activation of the Notch signaling pathway. Biomed Pharmacother. 2017;87:555–560. doi: 10.1016/j.biopha.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 97.Xu D, Yu J, Gao G, Lu G, Zhang Y, Ma P. LncRNA DANCR functions as a competing endogenous RNA to regulate RAB1A expression by sponging miR-634 in glioma. Biosci Rep. 2018;38(1):1–9. doi: 10.1042/BSR20171664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Godlewski J, Nowicki MO, Bronisz A, Williams S, Otsuki A, Nuovo G, Raychaudhury A, Newton HB, Chiocca EA, Lawler S. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68(22):9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 99.Feng L, Lin T, Che H, Wang X. Long noncoding RNA DANCR knockdown inhibits proliferation, migration and invasion of glioma by regulating miR-135a-5p/BMI1. Cancer Cell Int. 2020;20:53. doi: 10.1186/s12935-020-1123-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yang BY, Meng Q, Sun Y, Gao L, Yang JX. Long non-coding RNA SNHG16 contributes to glioma malignancy by competitively binding miR-20a-5p with E2F1. J Biol Regul Homeost Agents. 2018;32(2):251–261. [PubMed] [Google Scholar]
- 101.Zheng J, Liu X, Wang P, Xue Y, Ma J, Qu C, Liu Y. CRNDE promotes malignant progression of glioma by attenuating miR-384/PIWIL4/STAT3 Axis. Mol Ther. 2016;24(7):1199–1215. doi: 10.1038/mt.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 102.Zheng J, Li XD, Wang P, Liu XB, Xue YX, Hu Y, Li Z, Li ZQ, Wang ZH, Liu YH. CRNDE affects the malignant biological characteristics of human glioma stem cells by negatively regulating miR-186. Oncotarget. 2015;6(28):25339–25355. doi: 10.18632/oncotarget.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang K, Sun X, Zhou X, Han L, Chen L, Shi Z, Zhang A, Ye M, Wang Q, Liu C, et al. Long non-coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget. 2015;6(1):537–546. doi: 10.18632/oncotarget.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ma X, Li Z, Li T, Zhu L, Li Z, Tian N. Long non-coding RNA HOTAIR enhances angiogenesis by induction of VEGFA expression in glioma cells and transmission to endothelial cells via glioma cell derived-extracellular vesicles. Am J Transl Res. 2017;9(11):5012–5021. [PMC free article] [PubMed] [Google Scholar]
- 105.Liu L, Cui S, Wan T, Li X, Tian W, Zhang R, Luo L, Shi Y. Long non-coding RNA HOTAIR acts as a competing endogenous RNA to promote glioma progression by sponging miR-126-5p. J Cell Physiol. 2018;233(9):6822–6831. doi: 10.1002/jcp.26432. [DOI] [PubMed] [Google Scholar]
- 106.Wang G, Li Z, Tian N, Han L, Fu Y, Guo Z, Tian Y. miR-148b-3p inhibits malignant biological behaviors of human glioma cells induced by high HOTAIR expression. Oncol Lett. 2016;12(2):879–886. doi: 10.3892/ol.2016.4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bian EB, Ma CC, He XJ, Wang C, Zong G, Wang HL, Zhao B. Epigenetic modification of miR-141 regulates SKA2 by an endogenous 'sponge' HOTAIR in glioma. Oncotarget. 2016;7(21):30610–30625. doi: 10.18632/oncotarget.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sa L, Li Y, Zhao L, Liu Y, Wang P, Liu L, Li Z, Ma J, Cai H, Xue Y. The Role of HOTAIR/miR-148b-3p/USF1 on Regulating the Permeability of BTB. Front Mol Neurosci. 2017;10:194. doi: 10.3389/fnmol.2017.00194. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 109.Pastori C, Kapranov P, Penas C, Peschansky V, Volmar CH, Sarkaria JN, Bregy A, Komotar R, St Laurent G, Ayad NG, et al. The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc Natl Acad Sci USA. 2015;112(27):8326–8331. doi: 10.1073/pnas.1424220112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen L, Wang Y, He J, Zhang C, Chen J, Shi D. Long non-coding RNA H19 promotes proliferation and invasion in human glioma cells by downregulating miR-152. Oncol Res. 2018;26:1–10. doi: 10.3727/096504016X14772410356982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhao H, Peng R, Liu Q, Liu D, Du P, Yuan J, Peng G, Liao Y. The lncRNA H19 interacts with miR-140 to modulate glioma growth by targeting iASPP. Arch Biochem Biophys. 2016;610:1–7. doi: 10.1016/j.abb.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 112.Zhang T, Wang YR, Zeng F, Cao HY, Zhou HD, Wang YJ. LncRNA H19 is overexpressed in glioma tissue, is negatively associated with patient survival, and promotes tumor growth through its derivative miR-675. Eur Rev Med Pharmacol Sci. 2016;20(23):4891–4897. [PubMed] [Google Scholar]
- 113.Li C, Lei B, Huang S, Zheng M, Liu Z, Li Z, Deng Y. H19 derived microRNA-675 regulates cell proliferation and migration through CDK6 in glioma. Am J Transl Res. 2015;7(10):1747–1764. [PMC free article] [PubMed] [Google Scholar]
- 114.Jia P, Cai H, Liu X, Chen J, Ma J, Wang P, Liu Y, Zheng J, Xue Y. Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer Lett. 2016;381(2):359–369. doi: 10.1016/j.canlet.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 115.Liu ZZ, Tian YF, Wu H, Ouyang SY, Kuang WL. LncRNA H19 promotes glioma angiogenesis through miR-138/HIF-1alpha/VEGF axis. Neoplasma. 2020;67(1):111–118. doi: 10.4149/neo_2019_190121N61. [DOI] [PubMed] [Google Scholar]
- 116.Hu Q, Yin J, Zeng A, Jin X, Zhang Z, Yan W, You Y. H19 functions as a competing endogenous RNA to regulate EMT by sponging miR-130a-3p in Glioma. Cell Physiol Biochem. 2018;50(1):233–245. doi: 10.1159/000494002. [DOI] [PubMed] [Google Scholar]
- 117.Duan S, Li M, Wang Z, Wang L, Liu Y. H19 induced by oxidative stress confers temozolomide resistance in human glioma cells via activating NF-kappaB signaling. Onco Targets Ther. 2018;11:6395–6404. doi: 10.2147/OTT.S173244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cheng Z, Li Z, Ma K, Li X, Tian N, Duan J, Xiao X, Wang Y. Long non-coding RNA XIST smiR-429. J Cancer. 2017;8(19):4106–4116. doi: 10.7150/jca.21024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Du P, Zhao H, Peng R, Liu Q, Yuan J, Peng G, Liao Y. LncRNA-XIST interacts with miR-29c to modulate the chemoresistance of glioma cell to TMZ through DNA mismatch repair pathway. Biosci Rep. 2017;37(5):1–12. doi: 10.1042/BSR20170696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang Z, Yuan J, Li L, Yang Y, Xu X, Wang Y. Long non-coding RNA XIST exerts oncogenic functions in human glioma by targeting miR-137. Am J Transl Res. 2017;9(4):1845–1855. [PMC free article] [PubMed] [Google Scholar]
- 121.Yu H, Xue Y, Wang P, Liu X, Ma J, Zheng J, Li Z, Li Z, Cai H, Liu Y. Knockdown of long non-coding RNA XIST increases blood-tumor barrier permeability and inhibits glioma angiogenesis by targeting miR-137. Oncogenesis. 2017;6(3):e303. doi: 10.1038/oncsis.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yao Y, Ma J, Xue Y, Wang P, Li Z, Liu J, Chen L, Xi Z, Teng H, Wang Z, et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015;359(1):75–86. doi: 10.1016/j.canlet.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 123.Zhen L, Yun-Hui L, Hong-Yu D, Jun M, Yi-Long Y. Long noncoding RNA NEAT1 promotes glioma pathogenesis by regulating miR-449b-5p/c-Met axis. Tumour Biol. 2016;37(1):673–683. doi: 10.1007/s13277-015-3843-y. [DOI] [PubMed] [Google Scholar]
- 124.Gong W, Zheng J, Liu X, Ma J, Liu Y, Xue Y. Knockdown of NEAT1 restrained the malignant progression of glioma stem cells by activating microRNA let-7e. Oncotarget. 2016;7(38):62208–62223. doi: 10.18632/oncotarget.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhou K, Zhang C, Yao H, Zhang X, Zhou Y, Che Y, Huang Y. Knockdown of long non-coding RNA NEAT1 inhibits glioma cell migration and invasion via modulation of SOX2 targeted by miR-132. Mol Cancer. 2018;17(1):105. doi: 10.1186/s12943-018-0849-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhen Y, Nan Y, Guo S, Zhang L, Li G, Yue S, Liu X. Knockdown of NEAT1 repressed the malignant progression of glioma through sponging miR-107 and inhibiting CDK14. J Cell Physiol. 2019;234(7):10671–10679. doi: 10.1002/jcp.27727. [DOI] [PubMed] [Google Scholar]
- 127.Yang X, Xiao Z, Du X, Huang L, Du G. Silencing of the long non-coding RNA NEAT1 suppresses glioma stem-like properties through modulation of the miR-107/CDK6 pathway. Oncol Rep. 2017;37(1):555–562. doi: 10.3892/or.2016.5266. [DOI] [PubMed] [Google Scholar]
- 128.Guo J, Cai H, Zheng J, Liu X, Liu Y, Ma J, Que Z, Gong W, Gao Y, Tao W, et al. Long non-coding RNA NEAT1 regulates permeability of the blood-tumor barrier via miR-181d-5p-mediated expression changes in ZO-1, occludin, and claudin-5. Biochim Biophys Acta Mol Basis Dis. 2017;1863(9):2240–2254. doi: 10.1016/j.bbadis.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 129.Liao K, Lin Y, Gao W, Xiao Z, Medina R, Dmitriev P, Cui J, Zhuang Z, Zhao X, Qiu Y, et al. Blocking lncRNA MALAT1/miR-199a/ZHX1 axis inhibits glioblastoma proliferation and progression. Mol Ther Nucleic Acids. 2019;18:388–399. doi: 10.1016/j.omtn.2019.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Xiong Z, Wang L, Wang Q, Yuan Y. LncRNA MALAT1/miR-129 axis promotes glioma tumorigenesis by targeting SOX2. J Cell Mol Med. 2018;22:1–12. doi: 10.1111/jcmm.13667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li Z, Xu C, Ding B, Gao M, Wei X, Ji N. Long non-coding RNA MALAT1 promotes proliferation and suppresses apoptosis of glioma cells through derepressing Rap1B by sponging miR-101. J Neurooncol. 2017;134(1):19–28. doi: 10.1007/s11060-017-2498-5. [DOI] [PubMed] [Google Scholar]
- 132.Fu Z, Luo W, Wang J, Peng T, Sun G, Shi J, Li Z, Zhang B. Malat1 activates autophagy and promotes cell proliferation by sponging miR-101 and upregulating STMN1, RAB5A and ATG4D expression in glioma. Biochem Biophys Res Commun. 2017;492(3):480–486. doi: 10.1016/j.bbrc.2017.08.070. [DOI] [PubMed] [Google Scholar]
- 133.Cai T, Liu Y, Xiao J. Long noncoding RNA MALAT1 knockdown reverses chemoresistance to temozolomide via promoting microRNA-101 in glioblastoma. Cancer Med. 2018;7(4):1404–1415. doi: 10.1002/cam4.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chen W, Xu XK, Li JL, Kong KK, Li H, Chen C, He J, Wang F, Li P, Ge XS, et al. MALAT1 is a prognostic factor in glioblastoma multiforme and induces chemoresistance to temozolomide through suppressing miR-203 and promoting thymidylate synthase expression. Oncotarget. 2017;8(14):22783–22799. doi: 10.18632/oncotarget.15199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cao S, Wang Y, Li J, Lv M, Niu H, Tian Y. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by suppressing miR-155 expression and activating FBXW7 function. Am J Cancer Res. 2016;6(11):2561–2574. [PMC free article] [PubMed] [Google Scholar]
- 136.Zhu J, Gu W, Yu C. MATN1-AS1 promotes glioma progression by functioning as ceRNA of miR-200b/c/429 to regulate CHD1 expression. Cell Prolif. 2020;53(1):e12700. doi: 10.1111/cpr.12700. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 137.Ding J, Zhang L, Chen S, Cao H, Xu C, Wang X. lncRNA CCAT2 enhanced resistance of glioma cells against chemodrugs by disturbing the normal function of miR-424. Onco Targets Ther. 2020;13:1431–1445. doi: 10.2147/OTT.S227831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11(5):373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 139.Sonkoly E, Stahle M, Pivarcsi A. MicroRNAs: novel regulators in skin inflammation. Clin Exp Dermatol. 2008;33(3):312–315. doi: 10.1111/j.1365-2230.2008.02804.x. [DOI] [PubMed] [Google Scholar]
- 140.Primo MN, Bak RO, Schibler B, Mikkelsen JG. Regulation of pro-inflammatory cytokines TNFalpha and IL24 by microRNA-203 in primary keratinocytes. Cytokine. 2012;60(3):741–748. doi: 10.1016/j.cyto.2012.07.031. [DOI] [PubMed] [Google Scholar]
- 141.Witwer KW, Sisk JM, Gama L, Clements JE. MicroRNA regulation of IFN-beta protein expression: rapid and sensitive modulation of the innate immune response. J Immunol. 2010;184(5):2369–2376. doi: 10.4049/jimmunol.0902712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kroesen BJ, Teteloshvili N, Smigielska-Czepiel K, Brouwer E, Boots AM, van den Berg A, Kluiver J. Immuno-miRs: critical regulators of T-cell development, function and ageing. Immunology. 2015;144(1):1–10. doi: 10.1111/imm.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Geng H, Tan XD. Functional diversity of long non-coding RNAs in immune regulation. Genes Dis. 2016;3(1):72–81. doi: 10.1016/j.gendis.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Heerboth S, Lapinska K, Snyder N, Leary M, Rollinson S, Sarkar S. Use of epigenetic drugs in disease: an overview. Genet Epigenet. 2014;6:9–19. doi: 10.4137/GEG.S12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yu WD, Wang H, He QF, Xu Y, Wang XC. Long noncoding RNAs in cancer-immunity cycle. J Cell Physiol. 2018;233(9):6518–6523. doi: 10.1002/jcp.26568. [DOI] [PubMed] [Google Scholar]
- 146.Xu Z, Li P, Fan L, Wu M. The potential role of circRNA in tumor immunity regulation and immunotherapy. Front Immunol. 2018;9:9. doi: 10.3389/fimmu.2018.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang W, Zhao Z, Yang F, Wang H, Wu F, Liang T, Yan X, Li J, Lan Q, Wang J, et al. An immune-related lncRNA signature for patients with anaplastic gliomas. J Neurooncol. 2018;136(2):263–271. doi: 10.1007/s11060-017-2667-6. [DOI] [PubMed] [Google Scholar]
- 148.Xia P, Li Q, Wu G, Huang Y. An immune-related lncRNA signature to predict survival in glioma patients. Cell Mol Neurobiol. 2020;41:1–11. doi: 10.1007/s10571-020-00857-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Li X, Meng Y. Survival analysis of immune-related lncRNA in low-grade glioma. BMC Cancer. 2019;19(1):813. doi: 10.1186/s12885-019-6032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Boselli M, Lee BH, Robert J, Prado MA, Min SW, Cheng C, Silva MC, Seong C, Elsasser S, Hatle KM, et al. An inhibitor of the proteasomal deubiquitinating enzyme USP14 induces tau elimination in cultured neurons. J Biol Chem. 2017;292(47):19209–19225. doi: 10.1074/jbc.M117.815126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhou M, Zhang Z, Zhao H, Bao S, Cheng L, Sun J. An immune-related six-lncRNA signature to improve prognosis prediction of glioblastoma multiforme. Mol Neurobiol. 2018;55(5):3684–3697. doi: 10.1007/s12035-017-0572-9. [DOI] [PubMed] [Google Scholar]
- 152.Liang Q, Li X, Guan G, Xu X, Chen C, Cheng P, Cheng W, Wu A. Long non-coding RNA, HOTAIRM1, promotes glioma malignancy by forming a ceRNA network. Aging (Albany NY) 2019;11(17):6805–6838. doi: 10.18632/aging.102205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tkach M, Thery C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164(6):1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 154.Zhao W, Liu Y, Zhang C, Duan C. Multiple roles of exosomal long noncoding RNAs in cancers. Biomed Res Int. 2019;2019:1460572. doi: 10.1155/2019/1460572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dragomir M, Chen B, Calin GA. Exosomal lncRNAs as new players in cell-to-cell communication. Transl Cancer Res. 2018;7(Suppl 2):S243–S252. doi: 10.21037/tcr.2017.10.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, Garcia-Santos G, Ghajar C, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18(6):883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhao C, Gao F, Weng S, Liu Q. Pancreatic cancer and associated exosomes. Cancer Biomark. 2017;20(4):357–367. doi: 10.3233/CBM-170258. [DOI] [PubMed] [Google Scholar]
- 158.Mittelbrunn M, Sanchez-Madrid F. Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol. 2012;13(5):328–335. doi: 10.1038/nrm3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sun Z, Yang S, Zhou Q, Wang G, Song J, Li Z, Zhang Z, Xu J, Xia K, Chang Y, et al. Emerging role of exosome-derived long non-coding RNAs in tumor microenvironment. Mol Cancer. 2018;17(1):82. doi: 10.1186/s12943-018-0831-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, Liang M, Dittmar RL, Liu Y, Liang M, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Xie Y, Dang W, Zhang S, Yue W, Yang L, Zhai X, Yan Q, Lu J. The role of exosomal noncoding RNAs in cancer. Mol Cancer. 2019;18(1):37. doi: 10.1186/s12943-019-0984-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wang M, Zhou L, Yu F, Zhang Y, Li P, Wang K. The functional roles of exosomal long non-coding RNAs in cancer. Cell Mol Life Sci. 2019;76(11):2059–2076. doi: 10.1007/s00018-019-03018-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Naderi-Meshkin H, Lai X, Amirkhah R, Vera J, Rasko JEJ, Schmitz U. Exosomal lncRNAs and cancer: connecting the missing links. Bioinformatics. 2019;35(2):352–360. doi: 10.1093/bioinformatics/bty527. [DOI] [PubMed] [Google Scholar]
- 164.Chang S, Sun L, Feng G. SP1-mediated long noncoding RNA POU3F3 accelerates the cervical cancer through miR-127-5p/FOXD1. Biomed Pharmacother. 2019;117:109133. doi: 10.1016/j.biopha.2019.109133. [DOI] [PubMed] [Google Scholar]
- 165.Lang HL, Hu GW, Chen Y, Liu Y, Tu W, Lu YM, Wu L, Xu GH. Glioma cells promote angiogenesis through the release of exosomes containing long non-coding RNA POU3F3. Eur Rev Med Pharmacol Sci. 2017;21(5):959–972. [PubMed] [Google Scholar]
- 166.Gao F, Feng J, Yao H, Li Y, Xi J, Yang J. LncRNA SBF2-AS1 promotes the progression of cervical cancer by regulating miR-361-5p/FOXM1 axis. Artif Cells Nanomed Biotechnol. 2019;47(1):776–782. doi: 10.1080/21691401.2019.1577883. [DOI] [PubMed] [Google Scholar]
- 167.Zhang YT, Li BP, Zhang B, Ma P, Wu QL, Ming L, Xie LM. LncRNA SBF2-AS1 promotes hepatocellular carcinoma metastasis by regulating EMT and predicts unfavorable prognosis. Eur Rev Med Pharmacol Sci. 2018;22(19):6333–6341. doi: 10.26355/eurrev_201810_16044. [DOI] [PubMed] [Google Scholar]
- 168.Zhang Z, Yin J, Lu C, Wei Y, Zeng A, You Y. Exosomal transfer of long non-coding RNA SBF2-AS1 enhances chemoresistance to temozolomide in glioblastoma. J Exp Clin Cancer Res. 2019;38(1):166. doi: 10.1186/s13046-019-1139-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lang HL, Hu GW, Zhang B, Kuang W, Chen Y, Wu L, Xu GH. Glioma cells enhance angiogenesis and inhibit endothelial cell apoptosis through the release of exosomes that contain long non-coding RNA CCAT2. Oncol Rep. 2017;38(2):785–798. doi: 10.3892/or.2017.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Fu XM, Guo W, Li N, Liu HZ, Liu J, Qiu SQ, Zhang Q, Wang LC, Li F, Li CL. The expression and function of long noncoding RNA lncRNA-ATB in papillary thyroid cancer. Eur Rev Med Pharmacol Sci. 2017;21(14):3239–3246. [PubMed] [Google Scholar]
- 171.Bian EB, Chen EF, Xu YD, Yang ZH, Tang F, Ma CC, Wang HL, Zhao B. Exosomal lncRNAATB activates astrocytes that promote glioma cell invasion. Int J Oncol. 2019;54(2):713–721. doi: 10.3892/ijo.2018.4644. [DOI] [PubMed] [Google Scholar]
- 172.Yang S, Lim KH, Kim SH, Joo JY. Molecular landscape of long noncoding RNAs in brain disorders. Mol Psychiatry. 2021;26(4):1060–1074. doi: 10.1038/s41380-020-00947-5. [DOI] [PubMed] [Google Scholar]
- 173.Liang Y, Huang S, Qiao L, Peng X, Li C, Lin K, Xie G, Li J, Lin L, Yin Y, et al. Characterization of protein, long noncoding RNA and microRNA signatures in extracellular vesicles derived from resting and degranulated mast cells. J Extracell Vesicles. 2020;9(1):1697583. doi: 10.1080/20013078.2019.1697583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.O'Brien K, Breyne K, Ughetto S, Laurent LC, Breakefield XO. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat Rev Mol Cell Biol. 2020;21(10):585–606. doi: 10.1038/s41580-020-0251-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wang W, Han Y, Jo HA, Lee J, Song YS. Non-coding RNAs shuttled via exosomes reshape the hypoxic tumor microenvironment. J Hematol Oncol. 2020;13(1):67. doi: 10.1186/s13045-020-00893-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ma P, Pan Y, Li W, Sun C, Liu J, Xu T, Shu Y. Extracellular vesicles-mediated noncoding RNAs transfer in cancer. J Hematol Oncol. 2017;10(1):57. doi: 10.1186/s13045-017-0426-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang WL, Liu Y, Jiang J, Tang YJ, Tang YL, Liang XH. Extracellular vesicle long non-coding RNA-mediated crosstalk in the tumor microenvironment: Tiny molecules, huge roles. Cancer Sci. 2020;111(8):2726–2735. doi: 10.1111/cas.14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Born LJ, Harmon JW, Jay SM. Therapeutic potential of extracellular vesicle-associated long noncoding RNA. Bioeng Transl Med. 2020;5(3):e10172. doi: 10.1002/btm2.10172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sahoo S, Adamiak M, Mathiyalagan P, Kenneweg F, Kafert-Kasting S, Thum T. Therapeutic and diagnostic translation of extracellular vesicles in cardiovascular diseases: roadmap to the clinic. Circulation. 2021;143(14):1426–1449. doi: 10.1161/CIRCULATIONAHA.120.049254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Carlevaro-Fita J, Lanzos A, Feuerbach L, Hong C, Mas-Ponte D, Pedersen JS, Drivers P, Functional Interpretation G, Johnson R, Consortium P. Cancer LncRNA Census reveals evidence for deep functional conservation of long noncoding RNAs in tumorigenesis. Commun Biol. 2020;3(1):56. doi: 10.1038/s42003-019-0741-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Cecchelli R, Berezowski V, Lundquist S, Culot M, Renftel M, Dehouck MP, Fenart L. Modelling of the blood-brain barrier in drug discovery and development. Nat Rev Drug Discov. 2007;6(8):650–661. doi: 10.1038/nrd2368. [DOI] [PubMed] [Google Scholar]
- 182.Qi P, Zhou XY, Du X. Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol Cancer. 2016;15(1):39. doi: 10.1186/s12943-016-0524-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11(6):426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 184.Anfossi S, Babayan A, Pantel K, Calin GA. Clinical utility of circulating non-coding RNAs - an update. Nat Rev Clin Oncol. 2018;15(9):541–563. doi: 10.1038/s41571-018-0035-x. [DOI] [PubMed] [Google Scholar]
- 185.Wang Z, Gu J, Han T, Li K. High-throughput sequencing profile of laryngeal cancers: analysis of co-expression and competing endogenous RNA networks of circular RNAs, long non-coding RNAs, and messenger RNAs. Ann Transl Med. 2021;9(6):483. doi: 10.21037/atm-21-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Arun G, Diermeier SD, Spector DL. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol Med. 2018;24(3):257–277. doi: 10.1016/j.molmed.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501(7467):338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 188.Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 2018;15(2):81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 189.Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108(12):2419–2425. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Battle A, Khan Z, Wang SH, Mitrano A, Ford MJ, Pritchard JK, Gilad Y. Genomic variation. Impact of regulatory variation from RNA to protein. Science. 2015;347(6222):664–667. doi: 10.1126/science.1260793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.