Abstract
Background
The effects of BRAFnon-V600E and BRAFV600E on the outcomes and the molecular characteristics of adult glioma patients are unknown and need to be explored, although BRAFV600E has been extensively studied in pediatric glioma.
Methods
Co-occurring mutations and copy number alterations of associated genes in the MAPK and p53 pathways were investigated using data from The Cancer Genome Atlas (TCGA) public database retrieved by cBioPortal. The prognosis of available adult glioma cohorts with BRAFV600E and BRAFnon-V600E mutations were also investigated.
Results
Ninety patients with BRAFV600E or BRAFnon-V600E were enrolled in this study, and data from 52 nonredundant patients were investigated. Glioblastoma multiform was the most common cancer type, with BRAF non-V600E and BRAFV600E. TP53 (56.00% vs. 7.41%), IDH1/2 (36.00% vs. 3.70%), and ATRX (32.00% vs. 7.41%) exhibited more mutations in BRAFnon-V600E than in BRAFV600E, and TP53 was an independent risk factor (56.00% vs. 7.41%). Both BRAFnon-V600E and BRAFV600E frequently overlapped with CDKN2A/2B homozygous deletions (HDs), but there was no significant difference. Survival analysis showed no difference between the BRAF non-V600E and BRAFV600E cohorts, even after excluding the survival benefit of IDH1/2 mutations and considering the BRAFnon-V600E mutations in the glycine-rich loop (G-loop) and in the activation segment. The estimated mean survival of patients with BRAFnon-V600E & IDH1/2WT with mutations in the G-loop groups was the shortest.
Conclusions
BRAFnon-V600E exhibited a stronger association with IDH1/2 mutations than BRAFV600E, but no survival advantage was found.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12883-021-02224-6.
Keywords: Adult patient with glioma, BRAFnon-V600E, BRAFV600E, IDH1/2
Background
BRAF (v-raf murine sarcoma viral oncogene homolog B1) is a serine-threonine kinase in the Ras/Raf/mitogen-activated protein kinase (MAPK) pathway [1, 2] that transduces mitogenic stimuli after the activation by growth factor receptors that are involved in cell survival, proliferation, and differentiation [3]. MAPK pathway activation is common in various neoplasms. Active RAS mutations have been detected in approximately 15% of malignant human tumors.
Compared with ARAF and RAF1, BRAF plays a critical role in kinase activity [4]. A previous study showed that RAF1 is activated by BRAF through direct interactions between proteins and phosphorylation [5]. BRAF participates in the pathological mechanism of 7% of human neoplasms, especially in patients with melanoma and colorectal, thyroid, and lung cancer [6, 7]. The expression of BRAF is highly restrained [1, 8]. The high expression of BRAF in neural cells indicates that it is a vital MEK kinase in neuronal tissues [9, 10]. BRAF mutations are found in some central nervous system neoplasms. In pediatric low-grade gliomas (LGGs), these alterations correlate with oncogenic senescence, which may contribute to an improved prognosis [11]. The BRAFV600E mutation is rare in adult LGGs and glioblastomas and can only be found in 1 to 5% of samples [12, 13]. While BRAF activation contributes to tumor development and progression in the neural stem cells and progenitor cells of Homo sapiens, BRAF mutations are detected in adult diffuse gliomas and are associated with poor outcomes [14].
Most studies have focused on the BRAFV600E mutation, although more than 70 BRAF mutations have been reported to date. Mutations in BRAF at V600 can activate ERK, which plays a critical role in the G1/S transition by adjusting the expression of cyclin D, cyclin E, and p21Cip1 [15]. The BRAFV600E mutation is the most potent MAPK pathway activator, whereas BRAFnon-V600E mutations are low-activity kinases that slightly stimulate the MAPK pathway [16]. However, these low-activity BRAF mutants could activate MAPK signaling in COS-1 cells to a high level by activating RAF1 [16].
Isocitrate dehydrogenase (IDH) is a frequent mutation associated with a survival benefit in glioma patients and it has been defined as a molecular parameter to define the categories of brain tumors in the updated 2016 edition of the World Health Organization (WHO) Classification of Tumors of the Central Nervous System (CNS) [17]. IDH1 and BRAFV600E mutations are associated with infiltrative gliomas or circumscribed gliomas and glioneuronal tumors, respectively [18, 19], and they are exclusive in most cases [20]. The exact effect of BRAF non-V600E and BRAFV600E on the prognosis of glioma patients and whether there are unique molecular characteristics in their MAPK and p53 pathways remain largely unknown.
In this study, co-occurring mutations and copy number alterations of 35 associated genes in the MAPK and p53 pathways were retrieved and investigated, and the prognosis of the available adult glioma cohorts with BRAFV600E and BRAFnon-V600E were evaluated by using The Cancer Genome Atlas (TCGA) public database. We determined that BRAFnon-V600E exhibited a stronger association with the IDH1/2 mutation than BRAFV600E, but no survival advantage was found.
Methods
Data collection and enrollment
All data were collected and generated from the TCGA public database using the TCGA data mining tool cBioPortal (https://www.cbioportal.org/) [21, 22]. We strictly followed the TCGA publication guidelines (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga/using-tcga/citing-tcga). In multiple patient cohorts of all twenty available CNS/brain studies (6164 samples), the available data were queried, including the gene mutations, copy number alterations, mRNA expression, and protein expression data of patients with BRAF gene mutations. In each study, the mutations were selected for genomic profiles. Samples with mutation data were selected for the patient/case set and entered into three groups: (1) General: Ras-Raf-MEK-ErK/JNK signaling (26 genes), including KRAS, HRAS, BRAF, RAF1, MAP 3 K1, MAP 3 K2, MAP 3 K3, MAP 3 K4, MAP 3 K5, MAP 2 K1, MAP 2 K2, MAP 2 K3, MAP 2 K4, MAP 2 K5, MAPK1, MAPK3, MAPK4, MAPK6, MAPK7, MAPK8, MAPK9, MAPK12, MAPK14, DAB2, RASSF1, and RAB25; (2) General: p53 signaling (6 genes), including TP53, MDM2, MDM4, CDKN2A, CDKN2B, and TP53BP1; (3) Other frequently mutated genes, including IDH1, IDH2, and ATRX, were then submitted for query. Among the downloadable data files, the available data regarding the mutations, copy number alterations, mRNA expression, and protein expression were downloaded. In the type of genetic alterations across all samples, samples harboring the BRAF mutation were chosen. Data regarding mutations and copy number alterations on the summary page and the patient and sample data on the clinical data page were downloaded. All of the data were recorded in a chart for further analysis (Supplementary Dataset S1).
Major characteristics of the BRAFV600E and BRAFnon-V600E cohorts using univariate logistic regression analysis
The enrolled populations were divided into BRAFV600E and BRAFnon-V600E groups. The numbers and percentages of categorical variables were calculated. Their demographic characteristics, including sex, diagnosis age, cancer type, and overall survival status, were analyzed using univariate logistic regression analysis. The odds ratios (ORs) and 95% confidence intervals (CIs) were estimated.
Co-occurring mutations of the BRAFV600E and BRAFnon-V600E cohorts using univariate and multivariate logistic regression analysis
The numbers and percentages of categorical variables were calculated in the BRAFV600E and BRAFnon-V600E groups. The available data for co-occurring mutated genes in these two groups were analyzed using univariate logistic regression analysis. Thereafter, significant variables (P < 0.10) were analyzed using multivariate logistic regression analysis. The ORs and 95% CIs were estimated.
Co-occurring copy number alterations in the BRAFV600E and BRAFnon-V600E cohorts using heatmap and univariate logistic regression analysis
The available copy number alterations of the BRAFV600E and BRAFnon-V600E cohorts were retrieved and displayed using a heatmap by Morpheus (https://software.broadinstitute.org/morpheus). The putative copy-number alterations are as follows: − 2 = homozygous deletion; − 1 = hemizygous deletion; 0 = neutral/no change; 1 = gain; 2 = high-level amplification. Univariate logistic regression analysis was used to calculate the numbers and percentages of CDKN2A homozygous deletion (HD) and CDKN2B HD. The ORs and 95% CIs were estimated.
Crossover analysis with Kaplan–Meier survival curves and the log rank (mantel-Cox) test
The overall survival rates of the BRAFV600E and BRAFnon-V600E cohorts were compared using Kaplan-Meier curves and the log rank (Mantel-Cox) test [23]. To exclude the benefit of IDH1/2 on survival, we referred to the BRAFV600E & IDH1/2WT group as the BRAFV600E group minus those with IDH1/2 mutations, as well the BRAF non-V600E & IDH1/2WT group as the BRAF non-V600E group minus those with IDH1/2 mutations. The survival of the BRAFV600E & IDH1/2WT group was compared with that of the BRAFnon-V600E & IDH1/2WT groups. There were two clusters of mutations, one in the glycine-rich loop (referred to as the G-loop) and the other in the activation segment. To evaluate the effect of the mutation site on survival, we defined two subgroups in the BRAFnon-V600E & IDH1/2WT group. One subgroup was the BRAFnon-V600E & IDH1/2WT group with the mutation site in the G-loop, and the other subgroup was the BRAFnon-V600E & IDH1/2WT group with the mutation site in the activation segment. The BRAFV600E & IDH1/2WT group was compared with those two subgroups. Furthermore, the G-loop BRAFnon-V600E & IDH1/2WT subgroup was compared with the remaining patients in the BRAFnon-V600E & IDH1/2WT group.
Statistical analysis
Major characteristics, co-occurring mutations and copy number alterations of the BRAFV600E and BRAFnon-V600E cohorts were analyzed using univariate logistic regression analysis. Significant variables (P < 0.10) of co-occurring mutations of the BRAFV600E and BRAFnon-V600E cohorts were analyzed using multivariate logistic regression analysis. Kaplan-Meier curves were generated for glioma patients with BRAF mutations and were compared using the log-rank (Mantel-Cox) test. A P value < 0.05 was considered statistically significant.
Results
Data enrollment in the study
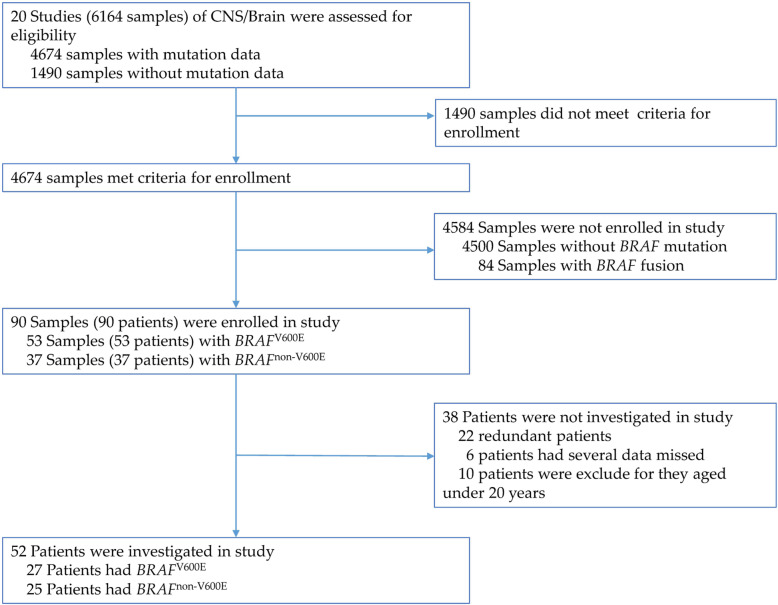
In all 20 CNS/brain studies (6164 samples), 4674 samples with mutation data were queried; 90 samples (90 patients) with BRAF mutations, including 53 samples (53 patients) with BRAFV600E and 37 samples (37 patients) with BRAFnon-V600E, are shown in Table 1. The cancer types of 20 CNS/brain studies included diffuse glioma, glioblastoma, oligodendroglioma, embryonal tumor, encapsulated glioma, and miscellaneous neuroepithelial tumor. The scheme for the final enrolled and investigated data is shown in Fig. 1. Ninety patients with BRAFV600E or BRAFnon-V600E were enrolled in this study, and data from 52 nonredundant patients were investigated. The integrated data of their major patient characteristics, including sex, age, diagnosis age, cancer type, data of co-occurring mutations, copy number alterations, and overall survival time and status, were collected for further analysis.
Table 1.
The CNS/brain projects of TCGA database enrolled in the study retrieved by cBioPortal
| Project | All Samples | Samples with mutation data | Samples with BRAFV600E | Samples with BRAFnon-V600E | References |
|---|---|---|---|---|---|
| Diffuse Glioma | |||||
| Brain Lower Grade Glioma (TCGA, Firehose Legacy) | 530 | 286 | 1 | 1 | https://www.cancer.gov |
| Brain Lower Grade Glioma (TCGA, PanCancer Atlas) | 514 | 512 | 1 | 2 | [24–29] |
| Glioma (MSK, Nature 2019) | 91 | 91 | 2 | 1 | https://www.cancer.gov |
| Glioma (MSKCC, Clin Cancer Res 2019) | 1004 | 1004 | 22 | 19 | [30] |
| Low-Grade Gliomas (UCSF, Science 2014) | 61 | 61 | 2 | 0 | [31] |
| Merged Cohort of LGG and GBM (TCGA, Cell 2016) | 1102 | 812 | 5 | 2 | [32] |
| GLIOBLASTOMA | |||||
| Brain Tumor PDXs (Mayo Clinic, 2019) | 95 | 83 | 2 | 1 | https://www.cbioportal.org |
| Glioblastoma (Columbia, Nat Med. 2019) | 42 | 32 | 1 | 1 | [33] |
| Glioblastoma (TCGA, Cell 2013) | 543 | 257 | 3 | 0 | [34] |
| Glioblastoma (TCGA, Nature 2008) | 206 | 91 | 0 | 0 | [35] |
| Glioblastoma Multiforme (TCGA, Firehose Legacy) | 604 | 290 | 5 | 1 | https://www.cancer.gov |
| Glioblastoma Multiforme (TCGA, PanCancer Atlas) | 592 | 397 | 5 | 3 | [24–29, 36] |
| OLIGODENDROGLIOMA | |||||
| Anaplastic Oligodendroglioma and Anaplastic Oligogastrocytoma (MSKCC, Neuro Oncol 2017) | 22 | 22 | 0 | 0 | [37] |
| Embryonal Tumor | |||||
| MEDULLOBLASTOMA | |||||
| Medulloblastoma (Broad, Nature 2012) | 92 | 92 | 0 | 0 | [38] |
| Medulloblastoma (ICGC, Nature 2012) | 125 | 125 | 0 | 0 | [39] |
| Medulloblastoma (PCGP, Nature 2012) | 37 | 37 | 0 | 0 | [40] |
| Medulloblastoma (Sickkids, Nature 2016) | 46 | 46 | 0 | 1 | [41] |
| Encaspulated Glioma | |||||
| PILOCYTIC ASTROCYTOMA | |||||
| Pilocytic Astrocytoma (ICGC, Nature Genetics 2013) | 96 | 96 | 4 | 3 | [42] |
| Miscellaneous Neuroepithelial Tumor | |||||
| Pheochromocytoma and Paraganglioma (TCGA, Cell 2017) | 178 | 178 | 0 | 1 | [43] |
| Pheochromocytoma and Paraganglioma (TCGA, Firehose Legacy) | 184 | 162 | 0 | 1 | https://www.cancer.gov |
Fig. 1.
The scheme of the enrollment and investigation of data. In all 20 CNS/Brain studies, including 6164 samples, 90 patients with BRAFV600E or BRAFnon-V600E were enrolled; 52 nonredundant patients displayed major patient characteristics, including sex, age, cancer type detailed, co-occurring mutations, and copy number alteration genes, and were enrolled for further analysis
Major characteristics of the cohorts with BRAFV600E and BRAFnon-V600E
The study populations were divided into two groups, BRAFV600E and BRAFnon-V600E. The major demographic characteristics and clinical data of the two groups are summarized in Table 2. The patients’ ages ranged from 20 to 85 years and were divided into early adulthood, midlife, mature adulthood, and late adulthood (aged 20–35, 35–50, 50–80, and > 80 years, respectively). The two groups had comparable proportions of male patients, diagnosis age, cancer type, and overall survival status. Glioblastoma multiform was the most common cancer type in both cohorts (74.07% vs. 56.00%; P = 0.175; Table 2).
Table 2.
The major characteristics of cohorts including BRAFV600E and BRAFnon-V600E
| Variables |
BRAFV600E (n = 27) |
BRAFnon-V600E (n = 25) |
Univariate analysis | ||||
|---|---|---|---|---|---|---|---|
| Number | % | Number | % | Odds Ratio | 95% Confidence Interval | P Value | |
| Male | 16 | 59.26 | 18 | 72.00 | 0.566 | 0.177–1.809 | 0.337 |
| Diagnosis Age | |||||||
| Ages 20–35 | 9 | 33.33 | 6 | 24.00 | 1.583 | 0.469–5.350 | 0.459 |
| Ages 36–50 | 9 | 33.33 | 8 | 32.00 | 1.062 | 0.333–3.390 | 0.918 |
| Ages 51–80 | 7 | 25.93 | 11 | 44.00 | 0.445 | 0.139–1.433 | 0.175 |
| Age 80+ | 2 | 7.41 | 0 | 0.00 | 1,615,474,843 | 0.000- | 0.999 |
| Cancer type detailed | |||||||
| Glioblastoma Multiform | 20 | 74.07 | 14 | 56.00 | 2.245 | 0.698–7.219 | 0.175 |
| Astrocytoma | 3 | 11.11 | 6 | 24.00 | 0.396 | 0.087–1.794 | 0.229 |
| Oligoastrocytoma | 1 | 3.70 | 0 | 0.00 | 1,553,341,195 | 0.000- | 1.000 |
| Oligodendroglioma | 0 | 0.00 | 3 | 12.00 | 0.000 | 0.000- | 0.999 |
| Gliosarcoma | 0 | 0.00 | 2 | 8.00 | 0.000 | 0.000- | 0.999 |
| Other glioma | 3 | 11.11 | 0 | 0.00 | 1,682,786,295 | 0.000- | 0.999 |
| Overall survival status | |||||||
| Deceased | 14 | 51.85 | 11 | 44.00 | 1.371 | 0.460–4.087 | 0.572 |
Co-occurring mutations of the BRAFV600E and BRAFnon-V600E cohorts using univariate and multivariate logistic regression analysis
Available co-occurring gene mutations of the BRAFV600E and BRAFnon-V600E cohorts were retrieved, and differences between the two groups were compared; the results are summarized in Table 3. The mutation frequencies of KRAS, HRAS, RAF1, MAP 3 K1, MAP 2 K1, MAP 2 K2, MAP 2 K4, MDM2, MDM4, CDKN2A, and CDKN2B were comparable between the two groups. In contrast, the BRAFnon-V600E group exhibited a significantly higher mutation frequency of TP53 (56.00% vs. 7.41%; P = 0.001), IDH1/2 (36.00% vs. 3.70%; P = 0.015), and ATRX (32.00% vs. 7.41%; P = 0.037) than the BRAFV600E group. The variables with P < 0.10 were analyzed using multivariate logistic regression analysis, and the BRAFnon-V600E group exhibited a significantly higher TP53 mutation frequency (56.00% vs. 7.41%; P = 0.031) than the BRAFV600E group (Table 3).
Table 3.
The co-occurred mutations of BRAFV600E and BRAFnon-V600E cohort using univariate and multivariate logistics regression analysis
| Gene |
BRAFV600E (n = 27) |
BRAFnon-V600E (n = 25) |
Univariate analysis | Multivariate analysis | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Number | % | Number | % | Odds Ratio | 95% Confidence Interval | P Value | Odds Ratio | 95% Confidence Interval | P Value | |
| KRAS | 0 | 0.00 | 1 | 4.00 | 1,817,409,198 | 0.000- | 1.000 | |||
| HRAS | 0 | 0.00 | 1 | 4.00 | 1,817,409,198 | 0.000- | 1.000 | |||
| RAF1 | 0 | 0.00 | 2 | 8.00 | 1,896,426,989 | 0.000- | 0.999 | |||
| MAP 3 K1 | 1 | 3.70 | 6 | 24.00 | 8.211 | 0.911–73.959 | 0.060 | |||
| MAP 2 K1 | 0 | 0.00 | 2 | 8.00 | 1,896,426,989 | 0.000- | 0.999 | |||
| MAP 2 K2 | 0 | 0.00 | 4 | 16.00 | 2,077,039,084 | 0.000- | 0.999 | |||
| MAP 2 K4 | 0 | 0.00 | 2 | 8.00 | 1,896,426,989 | 0.000- | 0.999 | |||
| TP53 | 2 | 7.41 | 14 | 56.00 | 15.909 | 3.078–82.224 | 0.001 | 12.186 | 1.251–118.721 | 0.031 |
| MDM2 | 1 | 3.70 | 3 | 12.00 | 3.545 | 0.344–36.561 | 0.298 | |||
| MDM4 | 0 | 0.00 | 4 | 16.00 | 2,077,039,084 | 0.000- | 0.999 | |||
| CDKN2A | 0 | 0.00 | 3 | 12.00 | 1,982,628,216 | 0.000- | 0.999 | |||
| CDKN2B | 0 | 0.00 | 1 | 4.00 | 1,817,409,198 | 0.000- | 1.000 | |||
| IDH1/2 | 1 | 3.70 | 9 | 36.00 | 14.625 | 1.690–126.537 | 0.015 | 5.498 | 0.512–59.020 | 0.159 |
| ATRX | 2 | 7.41 | 8 | 32.00 | 5.882 | 1.110–31.170 | 0.037 | 0.665 | 0.048–9.188 | 0.761 |
Co-occurring copy number alteration in the BRAFV600E and BRAFnon-V600E cohorts using heatmap and univariate logistic regression analysis
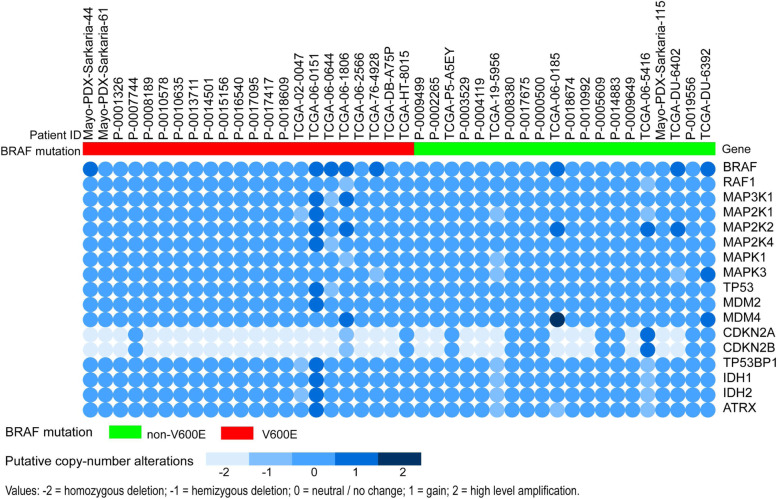
There were no available copy number data for five patients with BRAFV600E and five patients with BRAFnon-V600E. The copy number alterations of the available co-occurring genes included BRAF, RAF1, MAP 3 K1, MAP 2 K1, MAP 2 K2, MAP 2 K4, MAPK1, MAPK3, TP53, MDM2, MDM4, TP53BP1, IDH1, IDH2, ATRX, CDKN2A, and CDKN2B. The HD copy number was frequently retrieved for these two genes, including CDKN2A and CDKN2B (Fig. 2), and the HD of both CDKN2A (77.27.00% vs. 60.00%; P = 0.032) and CDKN2B (77.27.00% vs. 60.00%; P = 0.032) was more frequent in the BRAFV600E cohort than in the BRAFnon-V600E cohort (Table 4).
Fig. 2.
The co-occurring copy number alterations of the BRAFV600E cohort and BRAFnon-V600E cohort using a heatmap. The cohorts of BRAFV600E (red) or BRAFnon-V600E (green) are shown, and putative copy-number alterations change from light to dark with value enhancement
Table 4.
CDKN2A/2B HD of BRAFV600E and BRAFnon-V600E cohort using univariate logistics regression analysis
| Variables |
BRAFV600E (n = 22) |
BRAFnon-V600E (n = 20) |
Univariate analysis | ||||
|---|---|---|---|---|---|---|---|
| Number | % | Number | % | Odds Ratio | 95% Confidence Interval | P Value | |
| CDKN2A | 17 | 77.27 | 12 | 60.00 | 0.193 | 0.043–0.867 | 0.032 |
| CDKN2B | 17 | 77.27 | 12 | 60.00 | 0.193 | 0.043–0.867 | 0.032 |
Crossover analysis using Kaplan–Meier survival curves and the log-rank (mantel-Cox) test
Crossover Kaplan–Meier survival curves and the log-rank (Mantel-Cox) test were performed to explore the difference between the overall survival of glioma patients with BRAFV600E and BRAFnon-V600E. The estimated mean survival time was 51.394 months for patients with BRAFV600E, 89.958 months for patients with BRAFnon-V600E, 44.500 months for patients with BRAFV600E & IDH1/2WT, and 93.821 months for patients with BRAFnon-V600E & IDH1/2WT. There was no difference between the survival of BRAFV600E and BRAFnon-V600E (51.394 vs. 89.958, chi-square 1.130, P = 0.288). In addition, there was no difference between the survival of BRAFV600E & IDH1/2WT and BRAFnon-V600E & IDH1/2WT (44.500 vs. 93.821, chi-square 0.007, P = 0.935), which excluded the survival benefit of IDH1/2. We also evaluated the survival of BRAFnon-V600E & IDH1/2WT with mutations in the G-loop and activation segment. The estimated survival time of these two subgroups was 12.250 months for patients with BRAFnon-V600E & IDH1/2WT with mutations in the G-loop and 34.800 months for patients with BRAFnon-V600E & IDH1/2WT with mutations in the activation segment. In addition, there was no difference between the BRAFV600E & IDH1/2WT cohorts and those of the BRAFnon-V600E & IDH1/2WT cohorts. As shown below, BRAFV600E & IDH1/2WT vs. BRAFnon-V600E & IDH1/2WT had mutations in the G-loop (44.500 vs. 12.250, chi-squared 0.122, P = 0.727), and BRAFV600E & IDH1/2WT vs. BRAFnon-V600E & IDH1/2WT had mutations in the activation segment (44.500 vs. 34.800, chi-square 0.145, P = 0.703). Since the estimated mean survival of BRAFnon-V600E & IDH1/2WT with mutations in the G-loop was the shortest, we compared the BRAFnon-V600E & IDH1/2WT with mutations in the G-loop with the remaining BRAFnon-V600E & IDH1/2WT patients. There was no difference between them (12.250 vs. 95.100, chi-square 0.008, P = 0.927) (Fig. 3). The numbers at risk of Kaplan–Meier survival curves were shown in Supplementary Dataset S2.
Fig. 3.

Crossover analysis with Kaplan–Meier survival curves and the log-rank (Mantel-Cox) test. a BRAFV600E vs. BRAFnon-V600E (51.394 vs. 89.958, Chi-Square 1.130, P = 0.288); b BRAFV600E & IDH1/2WT vs. BRAFnon-V600E & IDH1/2WT (44.500 vs. 93.821, Chi-Square 0.007, P = 0.935); c BRAFV600E & IDH1/2WT vs. BRAFnon-V600E & IDH1/2WT with mutations in G-loop (44.500 vs. 12.250, Chi-Square 0.122, P = 0.727); d BRAFV600E & IDH1/2WT vs. BRAFnon-V600E & IDH1/2WT with mutations in activation segment (44.500 vs. 34.800, Chi-Square 0.145, P = 0.703); e BRAFnon-V600E & IDH1/2WT with mutations in G-loop vs. the rest BRAFnon-V600E & IDH1/2WT patients (12.250 vs. 95.100, Chi-Square 0.008, P = 0.927)
Discussion
BRAF mutations critically affect cancer growth and progression and are supposed to be a founder event for mutations occurring early in the initiation process of cancer. However, BRAF mutations must cooperate with other mechanisms for a fully cancerous state, as they are insufficient to induce cancer alone [5]. BRAFV600E has been the mutation of interest in previous studies on glioma, especially in pediatric glioma patients, for the available molecule-targeted drugs. However, various BRAFnon-V600E cells exert different activation effects on the MAPK pathway. The exact impact on the clinical prognosis and possible molecular mechanism of associated co-occurring genes with mutations or copy number alterations co-occurring with BRAF mutations remains unclear in adult glioma patients. In this study, the available data of patients with BRAFnon-V600E and BRAFV600E in the TCGA CNS/brain database were investigated to determine the possible mechanisms of BRAF gene mutations in adult glioma patients.
Our data indicated that in adult glioma patients with BRAF mutations, including both BRAFnon-V600E and BRAFV600E cohorts, glioblastoma multiform was the most common cancer type. A previous study showed that all BRAFV600E glioblastomas were primary tumors in both pediatric and adult patients [44]. Tabouret et al. [20] reported a case the co-occurrence of both IDH1 mutation and BRAFV600E although those two mutations are mutually exclusive in glial tumor. The available co-occurring mutated genes in the MAPK and p53 pathways showed that mutated genes frequently co-occurred in the BRAFnon-V600E cohort, and there were more TP53, IDH1/2, and ATRX mutations in BRAFnon-V600E than in BRAFV600E. Lai et al. [45] found that a TP53 point mutation at position 273 (Arg to Cys) was more common than IDH1 mutations at position 132 (Arg to His). They hypothesized that the TP53 mutation (C → T) occurred in the nontranscribed strand, while the IDH1 mutation existed in the transcribed strand, which is a strand asymmetry pattern [46]. Another study indicated that IDH1/2 mutations represent early events in brain tumor formation [47]. Liu et al. [48] found that ATRX alterations correlated with mutations in IDH1/2 and TP53 in glioma of all grades. It has been reported that ATRX deletions/mutations are correlated with TP53 and IDH1 mutations [49, 50]. Somatic TP53, ATRX, and IDH1/2 mutations have been found in adult LGGs [51]. ATRX mutations are detected in adult diffuse gliomas and astrocytomas harboring both TP53 and IDH1/2. The co-occurrence of these three mutated genes, including TP53, IDH1/2, and ATRX, facilitates the growth of an adult diffuse astrocytoma subgroup [48]. All of the studies above indicate that ATRX mutations frequently overlap with IDH1/2 and TP53 mutations. In the present study, we also found the co-occurrence of these three mutations, which were frequently detected in the BRAFnon-V600E cohort but not in the BRAFV600E cohort. Our findings indicated that in adult glioma patients, a possible correlation between BRAFnon-V600E and these three common mutations simultaneously occurred in glioma. Multivariate logistic regression revealed that TP53 was an independent risk factor in the BRAFnon-V600E cohort vs. the BRAFV600E group. Our data demonstrated a correlation between BRAFnon-V600E and TP53 mutations in adult glioma patients.
Previous findings have shown that active Ras can induce heterodimerization of BRAF and RAF1 [52] and that this event may be critical for RAF1 activation [53]. RAF1 directly regulates cell apoptosis, which does not depend on MAPK signaling [54, 55], but occurs through direct interaction with Bcl-2 [54]. TP53 can regulate Bcl-2 by suppressing Bcl-2 transcription [56]. We proposed that the BRAFnon-V600E mutation might activate the BRAF-RAF1 heterodimer, which shows antiapoptotic properties via the activation of Bcl-2 through RAF1 phosphorylation. Mutant TP53, which is frequently accompanied by IDH1/2 mutation by a strand asymmetry mechanism, fails to regulate Bcl-2. Therefore, with both activated RAF1 and mutated TP53, an enhanced antiapoptotic effect, which promotes cancer growth, might be predicted.
Compared to BRAF fusions, BRAFV600E tends to be more aggressive, more likely to be associated with CDKN2A/B deletions, and can transform cancers into higher-grade tumors [57, 58]. Our data showed that CDKN2A and CDKN2B HDs were more frequent in the BRAFV600E cohort than in the BRAFnon-V600E cohort. Concomitant CDKN2A and CDKN2B HDs could be detected in patients with glioblastoma multiform cancer, astrocytoma, and gliosarcoma. A previous report indicated that five of seven pediatric grade II–IV astrocytomas with BRAFV600E had concomitant CDKN2A HD [59] and CDKN2A deletions combined with BRAFV600E alterations, constituting a subgroup of secondary high-grade gliomas [60]. We found that in adult glioma patients, BRAFV600E and BRAFnon-V600E frequently co-occurred with CDKN2A HDs combined with CDKN2B HDs, especially in patients with BRAFV600E. Except for astrocytoma, glioblastoma multiform cancer was the most common cancer type with these combined alterations. Robinson et al. [61] indicated that activated Akt or Ink4a/ARF deletions are necessary for high-grade brain neoplasms with BRAF mutations in a Cre/lox animal model. Our results showed the possible synergy of CDKN2A and CDKN2B HDs with BRAF mutations, especially in adult glioma patients with BRAFV600E and BRAFnon-V600E.
BRAFV600E reportedly enhances BRAF kinase activity 500-fold [62]. According to its kinase viability, BRAFnon-V600E mutations can be classified into three groups: high activity (130–700 times), intermediate activity (1.3–64 times), and impaired activity (30–80%) [16]. Theoretically, the higher the BRAF kinase activity, the worse the prognosis. To clarify whether there is a difference between BRAFV600E and BRAFnon-V600E, we compared the overall survival of these two cohorts, and no statistical significance was found.
In addition, the status of IDH mutations in glioblastomas definitely influences the prognosis of patients with glioblastomas; therefore, IDH-wildtype glioblastomas are defined as primary tumors, while IDH-mutant glioblastomas are classified as secondary tumors [63]. To exclude the benefit of IDH mutations on survival, we compared the BRAFV600E & IDH1/2WT and BRAFnon-V600E & IDH1/2WT cohorts, and no difference was detected. The positions of the G-loop and the activation segment are 458–470 aa and 577–622 aa in BRAF, respectively [64]. Most BRAFnon-V600E mutations exist in the G-loop and the activation segment [16, 64]; therefore, we selected the two cohorts as BRAFnon-V600E & IDH1/2WT with mutations in the G-loop and activation segment. We compared them with BRAFV600E & IDH1/2WT, and no difference was found between the BRAFV600E & IDH1/2WT cohorts and those of the BRAFnon-V600E & IDH1/2WT cohorts. Furthermore, we compared BRAFnon-V600E & IDH1/2WT with mutations in the G-loop with the remaining BRAFnon-V600E & IDH1/2WT patients and found no difference between them. Although there was no statistical significance, the estimated mean survival of BRAFnon-V600E & IDH1/2WT with mutations in the G-loop was the shortest in all cohorts. We propose that a larger sample is necessary for confirmation of this finding. Our data indicated that the BRAFnon-V600E cohort had no survival advantage from co-occurrence with IDH mutations compared with the BRAFnon-V600E cohort of adult patients with glioma.
Limitations
Because the BRAFV600E mutation is rare in adult glioma, there were few patients in both cohorts retrieved from the publicly available data (cBioPortal). In this study, while their apparent survival times were substantially different, they were not significantly different. To prove the mechanism by which BRAF mutations promote cancer growth via an enhanced antiapoptotic effect of Bcl-2, further study using appropriate clinical tissue samples or animal models are necessary.
Conclusions
In conclusion, we found that in adult patients with gliomas, BRAFnon-V600E, rather than BRAFV600E, frequently co-occurs with TP53, IDH1/2, and ATRX mutations. Both BRAFnon-V600E and BRAFV600E frequently overlapped with CDKN2A/2B HDs, whereas there were no significant differences between the two cohorts. Although there were significant differences in co-occurring gene mutations and copy number alterations, no difference was found in survival between cohorts of BRAFnon-V600E and BRAFV600E with and without IDH1/2 favorable effects on survival. We also found that the estimated mean survival of BRAFnon-V600E & IDH1/2WT with mutations in the G-loop was the shortest; however, no difference was observed between that cohort and other cohorts. Due to the poor available mRNA and protein data in the TCGA database we retrieved in this study, no expression data were evaluated. More clinical data or models are necessary to elucidate the mechanism involved in BRAFnon-V600E-associated glioma in the future.
Supplementary Information
Acknowledgements
The results published or shown here are in whole or part based upon public data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Abbreviations
- HD
Homozygous deletion
- BRAF
v-raf murine sarcoma viral oncogene homolog B1
- LGGs
Low-grade gliomas
- TCGA
The Cancer Genome Atlas
- ORs
Odds ratios
- Cis
Confidence intervals
- G-loop
Glycine-rich loop
- MAPK
Mitogen-activated protein kinase
Authors’ contributions
WW: Formal analysis, Writing- Original draft preparation. MW: Writing- Reviewing & Editing. HJ: Investigation. TW: Data curation. RD: Conceptualization, Methodology. All authors read and approved the final manuscript.
Funding
This study was financially supported by the Natural Science Basic Research Program of Shaanxi (Program No. 2018JM7062), and The Project of The First Affiliated Hospital of Xi’an Jiaotong University (XJYFY-2019w33).
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mercer KE, Pritchard CA. Raf proteins and cancer: B-Raf is identified as a mutational target. Biochim Biophys Acta. 2003;1653(1):25–40. doi: 10.1016/s0304-419x(03)00016-7. [DOI] [PubMed] [Google Scholar]
- 2.Roskoski R., Jr RAF protein-serine/threonine kinases: structure and regulation. Biochem Biophys Res Commun. 2010;399(3):313–317. doi: 10.1016/j.bbrc.2010.07.092. [DOI] [PubMed] [Google Scholar]
- 3.Basto D, Trovisco V, Lopes JM, Martins A, Pardal F, Soares P, et al. Mutation analysis of B-RAF gene in human gliomas. Acta Neuropathol. 2005;109(2):207–210. doi: 10.1007/s00401-004-0936-x. [DOI] [PubMed] [Google Scholar]
- 4.Kaltsas P, Want S, Cohen J. Development of a time-to-positivity assay as a tool in the antibiotic management of septic patients. Clin Microbiol Infect. 2005;11(2):109–114. doi: 10.1111/j.1469-0691.2004.01054.x. [DOI] [PubMed] [Google Scholar]
- 5.Dhomen N, Marais R. New insight into BRAF mutations in cancer. Curr Opin Genet Dev. 2007;17(1):31–39. doi: 10.1016/j.gde.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 6.Network TCGAR. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JWC, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 8.Barnier JV, Papin C, Eychene A, Lecoq O, Calothy G. The mouse B-raf gene encodes multiple protein isoforms with tissue-specific expression. J Biol Chem. 1995;270(40):23381–23389. doi: 10.1074/jbc.270.40.23381. [DOI] [PubMed] [Google Scholar]
- 9.Catling AD, Reuter CW, Cox ME, Parsons SJ, Weber MJ. Partial purification of a mitogen-activated protein kinase kinase activator from bovine brain. Identification as B-Raf or a B-Raf-associated activity. J Biol Chem. 1994;269(47):30014–30021. doi: 10.1016/S0021-9258(18)43982-8. [DOI] [PubMed] [Google Scholar]
- 10.Xie P, Streu C, Qin J, Bregman H, Pagano N, Meggers E, Marmorstein R. The crystal structure of BRAF in complex with an organoruthenium inhibitor reveals a mechanism for inhibition of an active form of BRAF kinase. Biochemistry. 2009;48(23):5187–5198. doi: 10.1021/bi802067u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raabe EH, Lim KS, Kim JM, Meeker A, Mao XG, Nikkhah G, Maciaczyk J, Kahlert U, Jain D, Bar E, Cohen KJ, Eberhart CG. BRAF activation induces transformation and then senescence in human neural stem cells: a pilocytic astrocytoma model. Clin Cancer Res. 2011;17(11):3590–3599. doi: 10.1158/1078-0432.CCR-10-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schindler G, Capper D, Meyer J, Janzarik W, Omran H, Herold-Mende C, Schmieder K, Wesseling P, Mawrin C, Hasselblatt M, Louis DN, Korshunov A, Pfister S, Hartmann C, Paulus W, Reifenberger G, von Deimling A. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol. 2011;121(3):397–405. doi: 10.1007/s00401-011-0802-6. [DOI] [PubMed] [Google Scholar]
- 13.Behling F, Barrantes-Freer A, Skardelly M, Nieser M, Christians A, Stockhammer F, Rohde V, Tatagiba M, Hartmann C, Stadelmann C, Schittenhelm J. Frequency of BRAF V600E mutations in 969 central nervous system neoplasms. Diagn Pathol. 2016;11(1):55. doi: 10.1186/s13000-016-0506-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Behling F, Schittenhelm J. Oncogenic BRAF alterations and their role in brain tumors. Cancers (Basel) 2019;11(6):794. doi: 10.3390/cancers11060794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woods D, Parry D, Cherwinski H, Bosch E, Lees E, McMahon M. Raf-induced proliferation or cell cycle arrest is determined by the level of Raf activity with arrest mediated by p21Cip1. Mol Cell Biol. 1997;17(9):5598–5611. doi: 10.1128/MCB.17.9.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116(6):855–867. doi: 10.1016/S0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 17.Komori T. The 2016 WHO classification of Tumours of the central nervous system: the major points of revision. Neurol Med Chir (Tokyo) 2017;57(7):301–311. doi: 10.2176/nmc.ra.2017-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang C, Moore LM, Li X, Yung WK, Zhang W. IDH1/2 mutations target a key hallmark of cancer by deregulating cellular metabolism in glioma. Neuro-Oncology. 2013;15(9):1114–1126. doi: 10.1093/neuonc/not087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tabouret E, Fina F, Vincentelli F, Nanni I, Figarella-Branger D. New IDH1 I113T mutation associated with BRAF V600E mutation: new driver of gliomagenesis? J Neurol Sci. 2014;342(1–2):204–206. doi: 10.1016/j.jns.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 21.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleinbaum DG, Klein M. Kaplan–Meier Survival Curves and the Log–Rank Test. 1996. [Google Scholar]
- 24.Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V, Akbani R, Bowlby R, Wong CK, Wiznerowicz M, Sanchez-Vega F, Robertson AG, Schneider BG, Lawrence MS, Noushmehr H, Malta TM, Stuart JM, Benz CC, Laird PW, Caesar-Johnson SJ, Demchok JA, Felau I, Kasapi M, Ferguson ML, Hutter CM, Sofia HJ, Tarnuzzer R, Wang Z, Yang L, Zenklusen JC, Zhang J(J), Chudamani S, Liu J, Lolla L, Naresh R, Pihl T, Sun Q, Wan Y, Wu Y, Cho J, DeFreitas T, Frazer S, Gehlenborg N, Getz G, Heiman DI, Kim J, Lawrence MS, Lin P, Meier S, Noble MS, Saksena G, Voet D, Zhang H, Bernard B, Chambwe N, Dhankani V, Knijnenburg T, Kramer R, Leinonen K, Liu Y, Miller M, Reynolds S, Shmulevich I, Thorsson V, Zhang W, Akbani R, Broom BM, Hegde AM, Ju Z, Kanchi RS, Korkut A, Li J, Liang H, Ling S, Liu W, Lu Y, Mills GB, Ng KS, Rao A, Ryan M, Wang J, Weinstein JN, Zhang J, Abeshouse A, Armenia J, Chakravarty D, Chatila WK, de Bruijn I, Gao J, Gross BE, Heins ZJ, Kundra R, la K, Ladanyi M, Luna A, Nissan MG, Ochoa A, Phillips SM, Reznik E, Sanchez-Vega F, Sander C, Schultz N, Sheridan R, Sumer SO, Sun Y, Taylor BS, Wang J, Zhang H, Anur P, Peto M, Spellman P, Benz C, Stuart JM, Wong CK, Yau C, Hayes DN, Parker JS, Wilkerson MD, Ally A, Balasundaram M, Bowlby R, Brooks D, Carlsen R, Chuah E, Dhalla N, Holt R, Jones SJM, Kasaian K, Lee D, Ma Y, Marra MA, Mayo M, Moore RA, Mungall AJ, Mungall K, Robertson AG, Sadeghi S, Schein JE, Sipahimalani P, Tam A, Thiessen N, Tse K, Wong T, Berger AC, Beroukhim R, Cherniack AD, Cibulskis C, Gabriel SB, Gao GF, Ha G, Meyerson M, Schumacher SE, Shih J, Kucherlapati MH, Kucherlapati RS, Baylin S, Cope L, Danilova L, Bootwalla MS, Lai PH, Maglinte DT, van den Berg DJ, Weisenberger DJ, Auman JT, Balu S, Bodenheimer T, Fan C, Hoadley KA, Hoyle AP, Jefferys SR, Jones CD, Meng S, Mieczkowski PA, Mose LE, Perou AH, Perou CM, Roach J, Shi Y, Simons JV, Skelly T, Soloway MG, Tan D, Veluvolu U, Fan H, Hinoue T, Laird PW, Shen H, Zhou W, Bellair M, Chang K, Covington K, Creighton CJ, Dinh H, Doddapaneni HV, Donehower LA, Drummond J, Gibbs RA, Glenn R, Hale W, Han Y, Hu J, Korchina V, Lee S, Lewis L, Li W, Liu X, Morgan M, Morton D, Muzny D, Santibanez J, Sheth M, Shinbrot E, Wang L, Wang M, Wheeler DA, Xi L, Zhao F, Hess J, Appelbaum EL, Bailey M, Cordes MG, Ding L, Fronick CC, Fulton LA, Fulton RS, Kandoth C, Mardis ER, McLellan MD, Miller CA, Schmidt HK, Wilson RK, Crain D, Curley E, Gardner J, Lau K, Mallery D, Morris S, Paulauskis J, Penny R, Shelton C, Shelton T, Sherman M, Thompson E, Yena P, Bowen J, Gastier-Foster JM, Gerken M, Leraas KM, Lichtenberg TM, Ramirez NC, Wise L, Zmuda E, Corcoran N, Costello T, Hovens C, Carvalho AL, de Carvalho AC, Fregnani JH, Longatto-Filho A, Reis RM, Scapulatempo-Neto C, Silveira HCS, Vidal DO, Burnette A, Eschbacher J, Hermes B, Noss A, Singh R, Anderson ML, Castro PD, Ittmann M, Huntsman D, Kohl B, le X, Thorp R, Andry C, Duffy ER, Lyadov V, Paklina O, Setdikova G, Shabunin A, Tavobilov M, McPherson C, Warnick R, Berkowitz R, Cramer D, Feltmate C, Horowitz N, Kibel A, Muto M, Raut CP, Malykh A, Barnholtz-Sloan JS, Barrett W, Devine K, Fulop J, Ostrom QT, Shimmel K, Wolinsky Y, Sloan AE, de Rose A, Giuliante F, Goodman M, Karlan BY, Hagedorn CH, Eckman J, Harr J, Myers J, Tucker K, Zach LA, Deyarmin B, Hu H, Kvecher L, Larson C, Mural RJ, Somiari S, Vicha A, Zelinka T, Bennett J, Iacocca M, Rabeno B, Swanson P, Latour M, Lacombe L, Têtu B, Bergeron A, McGraw M, Staugaitis SM, Chabot J, Hibshoosh H, Sepulveda A, Su T, Wang T, Potapova O, Voronina O, Desjardins L, Mariani O, Roman-Roman S, Sastre X, Stern MH, Cheng F, Signoretti S, Berchuck A, Bigner D, Lipp E, Marks J, McCall S, McLendon R, Secord A, Sharp A, Behera M, Brat DJ, Chen A, Delman K, Force S, Khuri F, Magliocca K, Maithel S, Olson JJ, Owonikoko T, Pickens A, Ramalingam S, Shin DM, Sica G, van Meir EG, Zhang H, Eijckenboom W, Gillis A, Korpershoek E, Looijenga L, Oosterhuis W, Stoop H, van Kessel KE, Zwarthoff EC, Calatozzolo C, Cuppini L, Cuzzubbo S, DiMeco F, Finocchiaro G, Mattei L, Perin A, Pollo B, Chen C, Houck J, Lohavanichbutr P, Hartmann A, Stoehr C, Stoehr R, Taubert H, Wach S, Wullich B, Kycler W, Murawa D, Wiznerowicz M, Chung K, Edenfield WJ, Martin J, Baudin E, Bubley G, Bueno R, de Rienzo A, Richards WG, Kalkanis S, Mikkelsen T, Noushmehr H, Scarpace L, Girard N, Aymerich M, Campo E, Giné E, Guillermo AL, van Bang N, Hanh PT, Phu BD, Tang Y, Colman H, Evason K, Dottino PR, Martignetti JA, Gabra H, Juhl H, Akeredolu T, Stepa S, Hoon D, Ahn K, Kang KJ, Beuschlein F, Breggia A, Birrer M, Bell D, Borad M, Bryce AH, Castle E, Chandan V, Cheville J, Copland JA, Farnell M, Flotte T, Giama N, Ho T, Kendrick M, Kocher JP, Kopp K, Moser C, Nagorney D, O’Brien D, O’Neill BP, Patel T, Petersen G, Que F, Rivera M, Roberts L, Smallridge R, Smyrk T, Stanton M, Thompson RH, Torbenson M, Yang JD, Zhang L, Brimo F, Ajani JA, Gonzalez AMA, Behrens C, Bondaruk, Broaddus R, Czerniak B, Esmaeli B, Fujimoto J, Gershenwald J, Guo C, Lazar AJ, Logothetis C, Meric-Bernstam F, Moran C, Ramondetta L, Rice D, Sood A, Tamboli P, Thompson T, Troncoso P, Tsao A, Wistuba I, Carter C, Haydu L, Hersey P, Jakrot V, Kakavand H, Kefford R, Lee K, Long G, Mann G, Quinn M, Saw R, Scolyer R, Shannon K, Spillane A, Stretch J, Synott M, Thompson J, Wilmott J, al-Ahmadie H, Chan TA, Ghossein R, Gopalan A, Levine DA, Reuter V, Singer S, Singh B, Tien NV, Broudy T, Mirsaidi C, Nair P, Drwiega P, Miller J, Smith J, Zaren H, Park JW, Hung NP, Kebebew E, Linehan WM, Metwalli AR, Pacak K, Pinto PA, Schiffman M, Schmidt LS, Vocke CD, Wentzensen N, Worrell R, Yang H, Moncrieff M, Goparaju C, Melamed J, Pass H, Botnariuc N, Caraman I, Cernat M, Chemencedji I, Clipca A, Doruc S, Gorincioi G, Mura S, Pirtac M, Stancul I, Tcaciuc D, Albert M, Alexopoulou I, Arnaout A, Bartlett J, Engel J, Gilbert S, Parfitt J, Sekhon H, Thomas G, Rassl DM, Rintoul RC, Bifulco C, Tamakawa R, Urba W, Hayward N, Timmers H, Antenucci A, Facciolo F, Grazi G, Marino M, Merola R, de Krijger R, Gimenez-Roqueplo AP, Piché A, Chevalier S, McKercher G, Birsoy K, Barnett G, Brewer C, Farver C, Naska T, Pennell NA, Raymond D, Schilero C, Smolenski K, Williams F, Morrison C, Borgia JA, Liptay MJ, Pool M, Seder CW, Junker K, Omberg L, Dinkin M, Manikhas G, Alvaro D, Bragazzi MC, Cardinale V, Carpino G, Gaudio E, Chesla D, Cottingham S, Dubina M, Moiseenko F, Dhanasekaran R, Becker KF, Janssen KP, Slotta-Huspenina J, Abdel-Rahman MH, Aziz D, Bell S, Cebulla CM, Davis A, Duell R, Elder JB, Hilty J, Kumar B, Lang J, Lehman NL, Mandt R, Nguyen P, Pilarski R, Rai K, Schoenfield L, Senecal K, Wakely P, Hansen P, Lechan R, Powers J, Tischler A, Grizzle WE, Sexton KC, Kastl A, Henderson J, Porten S, Waldmann J, Fassnacht M, Asa SL, Schadendorf D, Couce M, Graefen M, Huland H, Sauter G, Schlomm T, Simon R, Tennstedt P, Olabode O, Nelson M, Bathe O, Carroll PR, Chan JM, Disaia P, Glenn P, Kelley RK, Landen CN, Phillips J, Prados M, Simko J, Smith-McCune K, VandenBerg S, Roggin K, Fehrenbach A, Kendler A, Sifri S, Steele R, Jimeno A, Carey F, Forgie I, Mannelli M, Carney M, Hernandez B, Campos B, Herold-Mende C, Jungk C, Unterberg A, von Deimling A, Bossler A, Galbraith J, Jacobus L, Knudson M, Knutson T, Ma D, Milhem M, Sigmund R, Godwin AK, Madan R, Rosenthal HG, Adebamowo C, Adebamowo SN, Boussioutas A, Beer D, Giordano T, Mes-Masson AM, Saad F, Bocklage T, Landrum L, Mannel R, Moore K, Moxley K, Postier R, Walker J, Zuna R, Feldman M, Valdivieso F, Dhir R, Luketich J, Pinero EMM, Quintero-Aguilo M, Carlotti CG, Jr, Dos Santos JS, Kemp R, Sankarankuty A, Tirapelli D, Catto J, Agnew K, Swisher E, Creaney J, Robinson B, Shelley CS, Godwin EM, Kendall S, Shipman C, Bradford C, Carey T, Haddad A, Moyer J, Peterson L, Prince M, Rozek L, Wolf G, Bowman R, Fong KM, Yang I, Korst R, Rathmell WK, Fantacone-Campbell JL, Hooke JA, Kovatich AJ, Shriver CD, DiPersio J, Drake B, Govindan R, Heath S, Ley T, van Tine B, Westervelt P, Rubin MA, Lee JI, Aredes ND, Mariamidze A. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of Cancer. Cell. 2018;173(2):291–304. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellrott K, Bailey MH, Saksena G, Covington KR, Kandoth C, Stewart C, Hess J, Ma S, Chiotti KE, McLellan M, Sofia HJ, Hutter C, Getz G, Wheeler D, Ding L, Caesar-Johnson SJ, Demchok JA, Felau I, Kasapi M, Ferguson ML, Hutter CM, Sofia HJ, Tarnuzzer R, Wang Z, Yang L, Zenklusen JC, Zhang J(J), Chudamani S, Liu J, Lolla L, Naresh R, Pihl T, Sun Q, Wan Y, Wu Y, Cho J, DeFreitas T, Frazer S, Gehlenborg N, Getz G, Heiman DI, Kim J, Lawrence MS, Lin P, Meier S, Noble MS, Saksena G, Voet D, Zhang H, Bernard B, Chambwe N, Dhankani V, Knijnenburg T, Kramer R, Leinonen K, Liu Y, Miller M, Reynolds S, Shmulevich I, Thorsson V, Zhang W, Akbani R, Broom BM, Hegde AM, Ju Z, Kanchi RS, Korkut A, Li J, Liang H, Ling S, Liu W, Lu Y, Mills GB, Ng KS, Rao A, Ryan M, Wang J, Weinstein JN, Zhang J, Abeshouse A, Armenia J, Chakravarty D, Chatila WK, de Bruijn I, Gao J, Gross BE, Heins ZJ, Kundra R, la K, Ladanyi M, Luna A, Nissan MG, Ochoa A, Phillips SM, Reznik E, Sanchez-Vega F, Sander C, Schultz N, Sheridan R, Sumer SO, Sun Y, Taylor BS, Wang J, Zhang H, Anur P, Peto M, Spellman P, Benz C, Stuart JM, Wong CK, Yau C, Hayes DN, Parker, Wilkerson MD, Ally A, Balasundaram M, Bowlby R, Brooks D, Carlsen R, Chuah E, Dhalla N, Holt R, Jones SJM, Kasaian K, Lee D, Ma Y, Marra MA, Mayo M, Moore RA, Mungall AJ, Mungall K, Robertson AG, Sadeghi S, Schein JE, Sipahimalani P, Tam A, Thiessen N, Tse K, Wong T, Berger AC, Beroukhim R, Cherniack AD, Cibulskis C, Gabriel SB, Gao GF, Ha G, Meyerson M, Schumacher SE, Shih J, Kucherlapati MH, Kucherlapati RS, Baylin S, Cope L, Danilova L, Bootwalla MS, Lai PH, Maglinte DT, van den Berg DJ, Weisenberger DJ, Auman JT, Balu S, Bodenheimer T, Fan C, Hoadley KA, Hoyle AP, Jefferys SR, Jones CD, Meng S, Mieczkowski PA, Mose LE, Perou AH, Perou CM, Roach J, Shi Y, Simons JV, Skelly T, Soloway MG, Tan D, Veluvolu U, Fan H, Hinoue T, Laird PW, Shen H, Zhou W, Bellair M, Chang K, Covington K, Creighton CJ, Dinh H, Doddapaneni HV, Donehower LA, Drummond J, Gibbs RA, Glenn R, Hale W, Han Y, Hu J, Korchina V, Lee S, Lewis L, Li W, Liu X, Morgan M, Morton D, Muzny D, Santibanez J, Sheth M, Shinbrot E, Wang L, Wang M, Wheeler DA, Xi L, Zhao F, Hess J, Appelbaum EL, Bailey M, Cordes MG, Ding L, Fronick CC, Fulton LA, Fulton RS, Kandoth C, Mardis ER, McLellan MD, Miller CA, Schmidt HK, Wilson RK, Crain D, Curley E, Gardner J, Lau K, Mallery D, Morris S, Paulauskis J, Penny R, Shelton C, Shelton T, Sherman M, Thompson E, Yena P, Bowen J, Gastier-Foster JM, Gerken M, Leraas KM, Lichtenberg TM, Ramirez NC, Wise L, Zmuda E, Corcoran N, Costello T, Hovens C, Carvalho AL, de Carvalho AC, Fregnani JH, Longatto-Filho A, Reis RM, Scapulatempo-Neto C, Silveira HCS, Vidal DO, Burnette A, Eschbacher J, Hermes B, Noss A, Singh R, Anderson ML, Castro PD, Ittmann M, Huntsman D, Kohl B, le X, Thorp R, Andry C, Duffy ER, Lyadov V, Paklina O, Setdikova G, Shabunin A, Tavobilov M, McPherson C, Warnick R, Berkowitz R, Cramer D, Feltmate C, Horowitz N, Kibel A, Muto M, Raut CP, Malykh A, Barnholtz-Sloan JS, Barrett W, Devine K, Fulop J, Ostrom QT, Shimmel K, Wolinsky Y, Sloan AE, de Rose A, Giuliante F, Goodman M, Karlan BY, Hagedorn CH, Eckman J, Harr J, Myers J, Tucker K, Zach LA, Deyarmin B, Hu H, Kvecher L, Larson C, Mural RJ, Somiari S, Vicha A, Zelinka T, Bennett J, Iacocca M, Rabeno B, Swanson P, Latour M, Lacombe L, Têtu B, Bergeron A, McGraw M, Staugaitis SM, Chabot J, Hibshoosh H, Sepulveda A, Su T, Wang T, Potapova O, Voronina O, Desjardins L, Mariani O, Roman-Roman S, Sastre X, Stern MH, Cheng F, Signoretti S, Berchuck A, Bigner D, Lipp E, Marks J, McCall S, McLendon R, Secord A, Sharp A, Behera M, Brat DJ, Chen A, Delman K, Force S, Khuri F, Magliocca K, Maithel S, Olson JJ, Owonikoko T, Pickens A, Ramalingam S, Shin DM, Sica G, van Meir EG, Zhang H, Eijckenboom W, Gillis A, Korpershoek E, Looijenga L, Oosterhuis W, Stoop H, van Kessel KE, Zwarthoff EC, Calatozzolo C, Cuppini L, Cuzzubbo S, DiMeco F, Finocchiaro G, Mattei L, Perin A, Pollo B, Chen C, Houck J, Lohavanichbutr P, Hartmann A, Stoehr C, Stoehr R, Taubert H, Wach S, Wullich B, Kycler W, Murawa D, Wiznerowicz M, Chung K, Edenfield WJ, Martin J, Baudin E, Bubley G, Bueno R, de Rienzo A, Richards WG, Kalkanis S, Mikkelsen T, Noushmehr H, Scarpace L, Girard N, Aymerich M, Campo E, Giné E, Guillermo AL, van Bang N, Hanh PT, Phu BD, Tang Y, Colman H, Evason K, Dottino PR, Martignetti JA, Gabra H, Juhl H, Akeredolu T, Stepa S, Hoon D, Ahn K, Kang KJ, Beuschlein F, Breggia A, Birrer M, Bell D, Borad M, Bryce AH, Castle E, Chandan V, Cheville J, Copland JA, Farnell M, Flotte T, Giama N, Ho T, Kendrick M, Kocher JP, Kopp K, Moser C, Nagorney D, O’Brien D, O’Neill BP, Patel T, Petersen G, Que F, Rivera M, Roberts L, Smallridge R, Smyrk T, Stanton M, Thompson RH, Torbenson M, Yang JD, Zhang L, Brimo F, Ajani JA, Angulo Gonzalez AM, Behrens C, Bondaruk J, Broaddus R, Czerniak B, Esmaeli B, Fujimoto J, Gershenwald J, Guo C, Lazar AJ, Logothetis C, Meric-Bernstam F, Moran C, Ramondetta L, Rice D, Sood A, Tamboli P, Thompson T, Troncoso P, Tsao A, Wistuba I, Carter C, Haydu L, Hersey P, Jakrot V, Kakavand H, Kefford R, Lee K, Long G, Mann G, Quinn M, Saw R, Scolyer R, Shannon K, Spillane A, Stretch J, Synott M, Thompson J, Wilmott J, al-Ahmadie H, Chan TA, Ghossein R, Gopalan A, Levine DA, Reuter V, Singer S, Singh B, Tien NV, Broudy T, Mirsaidi C, Nair P, Drwiega P, Miller J, Smith J, Zaren H, Park JW, Hung NP, Kebebew E, Linehan WM, Metwalli AR, Pacak K, Pinto PA, Schiffman M, Schmidt LS, Vocke CD, Wentzensen N, Worrell R, Yang H, Moncrieff M, Goparaju C, Melamed J, Pass H, Botnariuc N, Caraman I, Cernat M, Chemencedji I, Clipca A, Doruc S, Gorincioi G, Mura S, Pirtac M, Stancul I, Tcaciuc D, Albert M, Alexopoulou I, Arnaout A, Bartlett J, Engel J, Gilbert S, Parfitt J, Sekhon H, Thomas G, Rassl DM, Rintoul RC, Bifulco C, Tamakawa R, Urba W, Hayward N, Timmers H, Antenucci A, Facciolo F, Grazi G, Marino M, Merola R, de Krijger R, Gimenez-Roqueplo AP, Piché A, Chevalier S, McKercher G, Birsoy K, Barnett G, Brewer C, Farver C, Naska T, Pennell NA, Raymond D, Schilero C, Smolenski K, Williams F, Morrison C, Borgia JA, Liptay MJ, Pool M, Seder CW, Junker K, Omberg L, Dinkin M, Manikhas G, Alvaro D, Bragazzi MC, Cardinale V, Carpino G, Gaudio E, Chesla D, Cottingham S, Dubina M, Moiseenko F, Dhanasekaran R, Becker KF, Janssen KP, Slotta-Huspenina J, Abdel-Rahman MH, Aziz D, Bell S, Cebulla CM, Davis A, Duell R, Elder JB, Hilty J, Kumar B, Lang J, Lehman NL, Mandt R, Nguyen P, Pilarski R, Rai K, Schoenfield L, Senecal K, Wakely P, Hansen P, Lechan R, Powers J, Tischler A, Grizzle WE, Sexton KC, Kastl A, Henderson J, Porten S, Waldmann J, Fassnacht M, Asa SL, Schadendorf D, Couce M, Graefen M, Huland H, Sauter G, Schlomm T, Simon R, Tennstedt P, Olabode O, Nelson M, Bathe O, Carroll PR, Chan JM, Disaia P, Glenn P, Kelley RK, Landen CN, Phillips J, Prados M, Simko J, Smith-McCune K, VandenBerg S, Roggin K, Fehrenbach A, Kendler A, Sifri S, Steele R, Jimeno A, Carey F, Forgie I, Mannelli M, Carney M, Hernandez B, Campos B, Herold-Mende C, Jungk C, Unterberg A, von Deimling A, Bossler A, Galbraith J, Jacobus L, Knudson M, Knutson T, Ma D, Milhem M, Sigmund R, Godwin AK, Madan R, Rosenthal HG, Adebamowo C, Adebamowo SN, Boussioutas A, Beer D, Giordano T, Mes-Masson AM, Saad F, Bocklage T, Landrum L, Mannel R, Moore K, Moxley K, Postier R, Walker J, Zuna R, Feldman M, Valdivieso F, Dhir R, Luketich J, Mora Pinero EM, Quintero-Aguilo M, Carlotti CG, Jr, Dos Santos JS, Kemp R, Sankarankuty A, Tirapelli D, Catto J, Agnew K, Swisher E, Creaney J, Robinson B, Shelley CS, Godwin EM, Kendall S, Shipman C, Bradford C, Carey T, Haddad A, Moyer J, Peterson L, Prince M, Rozek L, Wolf G, Bowman R, Fong KM, Yang I, Korst R, Rathmell WK, Fantacone-Campbell JL, Hooke JA, Kovatich AJ, Shriver CD, DiPersio J, Drake B, Govindan R, Heath S, Ley T, van Tine B, Westervelt P, Rubin MA, Lee JI, Aredes ND, Mariamidze A. Scalable Open Science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst. 2018;6(3):271–281. doi: 10.1016/j.cels.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor AM, Shih J, Ha G, Gao GF, Zhang X, Berger AC, Schumacher SE, Wang C, Hu H, Liu J, Lazar AJ, Cherniack AD, Beroukhim R, Meyerson M, Caesar-Johnson SJ, Demchok JA, Felau I, Kasapi M, Ferguson ML, Hutter CM, Sofia HJ, Tarnuzzer R, Wang Z, Yang L, Zenklusen JC, Zhang J(J), Chudamani S, Liu J, Lolla L, Naresh R, Pihl T, Sun Q, Wan Y, Wu Y, Cho J, DeFreitas T, Frazer S, Gehlenborg N, Getz G, Heiman DI, Kim J, Lawrence MS, Lin P, Meier S, Noble MS, Saksena G, Voet D, Zhang H, Bernard B, Chambwe N, Dhankani V, Knijnenburg T, Kramer R, Leinonen K, Liu Y, Miller M, Reynolds S, Shmulevich I, Thorsson V, Zhang W, Akbani R, Broom BM, Hegde AM, Ju Z, Kanchi RS, Korkut A, Li J, Liang H, Ling S, Liu W, Lu Y, Mills GB, Ng KS, Rao A, Ryan M, Wang J, Weinstein JN, Zhang J, Abeshouse A, Armenia J, Chakravarty D, Chatila WK, de Bruijn I, Gao J, Gross BE, Heins ZJ, Kundra R, la K, Ladanyi M, Luna A, Nissan MG, Ochoa A, Phillips SM, Reznik E, Sanchez-Vega F, Sander C, Schultz N, Sheridan R, Sumer SO, Sun Y, Taylor BS, Wang J, Zhang H, Anur P, Peto M, Spellman P, Benz C, Stuart JM, Wong CK, Yau C, Hayes DN, Parker JS, Wilkerson MD, Ally A, Balasundaram M, Bowlby R, Brooks D, Carlsen R, Chuah E, Dhalla N, Holt R, Jones SJM, Kasaian K, Lee D, Ma Y, Marra MA, Mayo M, Moore RA, Mungall AJ, Mungall K, Robertson AG, Sadeghi S, Schein JE, Sipahimalani P, Tam A, Thiessen N, Tse K, Wong T, Berger AC, Beroukhim R, Cherniack AD, Cibulskis C, Gabriel SB, Gao GF, Ha G, Meyerson M, Schumacher SE, Shih J, Kucherlapati MH, Kucherlapati RS, Baylin S, Cope L, Danilova L, Bootwalla MS, Lai PH, Maglinte DT, van den Berg DJ, Weisenberger DJ, Auman JT, Balu S, Bodenheimer T, Fan C, Hoadley KA, Hoyle AP, Jefferys SR, Jones CD, Meng S, Mieczkowski PA, Mose LE, Perou AH, Perou CM, Roach J, Shi Y, Simons JV, Skelly T, Soloway MG, Tan D, Veluvolu U, Fan H, Hinoue T, Laird PW, Shen H, Zhou W, Bellair M, Chang K, Covington K, Creighton CJ, Dinh H, Doddapaneni HV, Donehower LA, Drummond J, Gibbs RA, Glenn R, Hale W, Han Y, Hu J, Korchina V, Lee S, Lewis L, Li W, Liu X, Morgan M, Morton D, Muzny D, Santibanez J, Sheth M, Shinbrot E, Wang L, Wang M, Wheeler DA, Xi L, Zhao F, Hess J, Appelbaum EL, Bailey M, Cordes MG, Ding L, Fronick CC, Fulton LA, Fulton RS, Kandoth C, Mardis ER, McLellan MD, Miller CA, Schmidt HK, Wilson RK, Crain D, Curley E, Gardner J, Lau K, Mallery D, Morris S, Paulauskis J, Penny R, Shelton C, Shelton T, Sherman M, Thompson E, Yena P, Bowen J, Gastier-Foster JM, Gerken M, Leraas KM, Lichtenberg TM, Ramirez NC, Wise L, Zmuda E, Corcoran N, Costello T, Hovens C, Carvalho AL, de Carvalho AC, Fregnani JH, Longatto-Filho A, Reis RM, Scapulatempo-Neto C, Silveira HCS, Vidal DO, Burnette A, Eschbacher J, Hermes B, Noss A, Singh R, Anderson ML, Castro PD, Ittmann M, Huntsman D, Kohl B, le X, Thorp R, Andry C, Duffy ER, Lyadov V, Paklina O, Setdikova G, Shabunin A, Tavobilov M, McPherson C, Warnick R, Berkowitz R, Cramer D, Feltmate C, Horowitz N, Kibel A, Muto M, Raut CP, Malykh A, Barnholtz-Sloan JS, Barrett W, Devine K, Fulop J, Ostrom QT, Shimmel K, Wolinsky Y, Sloan AE, de Rose A, Giuliante F, Goodman M, Karlan BY, Hagedorn CH, Eckman J, Harr J, Myers J, Tucker K, Zach LA, Deyarmin B, Hu H, Kvecher L, Larson C, Mural RJ, Somiari S, Vicha A, Zelinka T, Bennett J, Iacocca M, Rabeno B, Swanson P, Latour M, Lacombe L, Têtu B, Bergeron A, McGraw M, Staugaitis SM, Chabot J, Hibshoosh H, Sepulveda A, Su T, Wang T, Potapova O, Voronina O, Desjardins L, Mariani O, Roman-Roman S, Sastre X, Stern MH, Cheng F, Signoretti S, Berchuck A, Bigner D, Lipp E, Marks J, McCall S, McLendon R, Secord A, Sharp A, Behera M, Brat DJ, Chen A, Delman K, Force S, Khuri F, Magliocca K, Maithel S, Olson JJ, Owonikoko T, Pickens A, Ramalingam S, Shin DM, Sica G, van Meir EG, Zhang H, Eijckenboom W, Gillis A, Korpershoek E, Looijenga L, Oosterhuis W, Stoop H, van Kessel KE, Zwarthoff EC, Calatozzolo C, Cuppini L, Cuzzubbo S, DiMeco F, Finocchiaro G, Mattei L, Perin A, Pollo B, Chen C, Houck J, Lohavanichbutr P, Hartmann A, Stoehr C, Stoehr R, Taubert H, Wach S, Wullich B, Kycler W, Murawa D, Wiznerowicz M, Chung K, Edenfield WJ, Martin J, Baudin E, Bubley G, Bueno R, de Rienzo A, Richards WG, Kalkanis S, Mikkelsen T, Noushmehr H, Scarpace L, Girard N, Aymerich M, Campo E, Giné E, Guillermo AL, van Bang N, Hanh PT, Phu BD, Tang Y, Colman H, Evason K, Dottino PR, Martignetti JA, Gabra H, Juhl H, Akeredolu T, Stepa S, Hoon D, Ahn K, Kang KJ, Beuschlein F, Breggia A, Birrer M, Bell D, Borad M, Bryce AH, Castle E, Chandan V, Cheville J, Copland JA, Farnell M, Flotte T, Giama N, Ho T, Kendrick M, Kocher JP, Kopp K, Moser C, Nagorney D, O’Brien D, O’Neill BP, Patel T, Petersen G, Que F, Rivera M, Roberts L, Smallridge R, Smyrk T, Stanton M, Thompson RH, Torbenson M, Yang JD, Zhang L, Brimo F, Ajani JA, Angulo Gonzalez AM, Behrens C, Bondaruk J, Broaddus R, Czerniak B, Esmaeli B, Fujimoto J, Gershenwald J, Guo C, Lazar AJ, Logothetis C, Meric-Bernstam F, Moran C, Ramondetta L, Rice D, Sood A, Tamboli P, Thompson T, Troncoso P, Tsao A, Wistuba I, Carter C, Haydu L, Hersey P, Jakrot V, Kakavand H, Kefford R, Lee K, Long G, Mann G, Quinn M, Saw R, Scolyer R, Shannon K, Spillane A, Stretch J, Synott M, Thompson J, Wilmott J, al-Ahmadie H, Chan TA, Ghossein R, Gopalan A, Levine DA, Reuter V, Singer S, Singh B, Tien NV, Broudy T, Mirsaidi C, Nair P, Drwiega P, Miller J, Smith J, Zaren H, Park JW, Hung NP, Kebebew E, Linehan WM, Metwalli AR, Pacak K, Pinto PA, Schiffman M, Schmidt LS, Vocke CD, Wentzensen N, Worrell R, Yang H, Moncrieff M, Goparaju C, Melamed J, Pass H, Botnariuc N, Caraman I, Cernat M, Chemencedji I, Clipca A, Doruc S, Gorincioi G, Mura S, Pirtac M, Stancul I, Tcaciuc D, Albert M, Alexopoulou I, Arnaout A, Bartlett J, Engel J, Gilbert S, Parfitt J, Sekhon H, Thomas G, Rassl DM, Rintoul RC, Bifulco C, Tamakawa R, Urba W, Hayward N, Timmers H, Antenucci A, Facciolo F, Grazi G, Marino M, Merola R, de Krijger R, Gimenez-Roqueplo AP, Piché A, Chevalier S, McKercher G, Birsoy K, Barnett G, Brewer C, Farver C, Naska T, Pennell NA, Raymond D, Schilero C, Smolenski K, Williams F, Morrison C, Borgia JA, Liptay MJ, Pool M, Seder CW, Junker K, Omberg L, Dinkin M, Manikhas G, Alvaro D, Bragazzi MC, Cardinale V, Carpino G, Gaudio E, Chesla D, Cottingham S, Dubina M, Moiseenko F, Dhanasekaran R, Becker KF, Janssen KP, Slotta-Huspenina J, Abdel-Rahman MH, Aziz D, Bell S, Cebulla CM, Davis A, Duell R, Elder JB, Hilty J, Kumar B, Lang J, Lehman NL, Mandt R, Nguyen P, Pilarski R, Rai K, Schoenfield L, Senecal K, Wakely P, Hansen P, Lechan R, Powers J, Tischler A, Grizzle WE, Sexton KC, Kastl A, Henderson J, Porten S, Waldmann J, Fassnacht M, Asa SL, Schadendorf D, Couce M, Graefen M, Huland H, Sauter G, Schlomm T, Simon R, Tennstedt P, Olabode O, Nelson M, Bathe O, Carroll PR, Chan JM, Disaia P, Glenn P, Kelley RK, Landen CN, Phillips J, Prados M, Simko J, Smith-McCune K, VandenBerg S, Roggin K, Fehrenbach A, Kendler A, Sifri S, Steele R, Jimeno A, Carey F, Forgie I, Mannelli M, Carney M, Hernandez B, Campos B, Herold-Mende C, Jungk C, Unterberg A, von Deimling A, Bossler A, Galbraith J, Jacobus L, Knudson M, Knutson T, Ma D, Milhem M, Sigmund R, Godwin AK, Madan R, Rosenthal HG, Adebamowo C, Adebamowo SN, Boussioutas A, Beer D, Giordano T, Mes-Masson AM, Saad F, Bocklage T, Landrum L, Mannel R, Moore K, Moxley K, Postier R, Walker J, Zuna R, Feldman M, Valdivieso F, Dhir R, Luketich J, Mora Pinero EM, Quintero-Aguilo M, Carlotti CG, Jr, Dos Santos JS, Kemp R, Sankarankuty A, Tirapelli D, Catto J, Agnew K, Swisher E, Creaney J, Robinson B, Shelley CS, Godwin EM, Kendall S, Shipman C, Bradford C, Carey T, Haddad A, Moyer J, Peterson L, Prince M, Rozek L, Wolf G, Bowman R, Fong KM, Yang I, Korst R, Rathmell WK, Fantacone-Campbell JL, Hooke JA, Kovatich AJ, Shriver CD, DiPersio J, Drake B, Govindan R, Heath S, Ley T, van Tine B, Westervelt P, Rubin MA, Lee JI, Aredes ND, Mariamidze A. Genomic and functional approaches to understanding Cancer aneuploidy. Cancer Cell. 2018;33(4):676–689. doi: 10.1016/j.ccell.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Q, Liang WW, Foltz SM, Mutharasu G, Jayasinghe RG, Cao S, Liao WW, Reynolds SM, Wyczalkowski MA, Yao L, Yu L, Sun SQ, Chen K, Lazar AJ, Fields RC, Wendl MC, van Tine BA, Vij R, Chen F, Nykter M, Shmulevich I, Ding L, Caesar-Johnson SJ, Demchok JA, Felau I, Kasapi M, Ferguson ML, Hutter CM, Sofia HJ, Tarnuzzer R, Wang Z, Yang L, Zenklusen JC, Zhang J(J), Chudamani S, Liu J, Lolla L, Naresh R, Pihl T, Sun Q, Wan Y, Wu Y, Cho J, DeFreitas T, Frazer S, Gehlenborg N, Getz G, Heiman DI, Kim J, Lawrence MS, Lin P, Meier S, Noble MS, Saksena G, Voet D, Zhang H, Bernard B, Chambwe N, Dhankani V, Knijnenburg T, Kramer R, Leinonen K, Liu Y, Miller M, Reynolds S, Shmulevich I, Thorsson V, Zhang W, Akbani R, Broom BM, Hegde AM, Ju Z, Kanchi RS, Korkut A, Li J, Liang H, Ling S, Liu W, Lu Y, Mills GB, Ng KS, Rao A, Ryan M, Wang J, Weinstein JN, Zhang J, Abeshouse A, Armenia J, Chakravarty D, Chatila WK, de Bruijn I, Gao J, Gross BE, Heins ZJ, Kundra R, la K, Ladanyi M, Luna A, Nissan MG, Ochoa A, Phillips SM, Reznik E, Sanchez-Vega F, Sander C, Schultz N, Sheridan R, Sumer SO, Sun Y, Taylor BS, Wang J, Zhang H, Anur P, Peto M, Spellman P, Benz C, Stuart JM, Wong CK, Yau C, Hayes DN, Parker JS, Wilkerson MD, Ally A, Balasundaram M, Bowlby R, Brooks D, Carlsen R, Chuah E, Dhalla N, Holt R, Jones SJM, Kasaian K, Lee D, Ma Y, Marra MA, Mayo M, Moore RA, Mungall AJ, Mungall K, Robertson AG, Sadeghi S, Schein JE, Sipahimalani P, Tam A, Thiessen N, Tse K, Wong T, Berger AC, Beroukhim R, Cherniack AD, Cibulskis C, Gabriel SB, Gao GF, Ha G, Meyerson M, Schumacher SE, Shih J, Kucherlapati MH, Kucherlapati RS, Baylin S, Cope L, Danilova L, Bootwalla MS, Lai PH, Maglinte DT, van den Berg DJ, Weisenberger DJ, Auman JT, Balu S, Bodenheimer T, Fan C, Hoadley KA, Hoyle AP, Jefferys SR, Jones CD, Meng S, Mieczkowski PA, Mose LE, Perou AH, Perou CM, Roach J, Shi Y, Simons JV, Skelly T, Soloway MG, Tan D, Veluvolu U, Fan H, Hinoue T, Laird PW, Shen H, Zhou W, Bellair M, Chang K, Covington K, Creighton CJ, Dinh H, Doddapaneni HV, Donehower LA, Drummond J, Gibbs RA, Glenn R, Hale W, Han Y, Hu J, Korchina V, Lee S, Lewis L, Li W, Liu X, Morgan M, Morton D, Muzny D, Santibanez J, Sheth M, Shinbrot E, Wang L, Wang M, Wheeler DA, Xi L, Zhao F, Hess J, Appelbaum EL, Bailey M, Cordes MG, Ding L, Fronick CC, Fulton LA, Fulton RS, Kandoth C, Mardis ER, McLellan MD, Miller CA, Schmidt HK, Wilson RK, Crain D, Curley E, Gardner J, Lau K, Mallery D, Morris S, Paulauskis J, Penny R, Shelton C, Shelton T, Sherman M, Thompson E, Yena P, Bowen J, Gastier-Foster JM, Gerken M, Leraas KM, Lichtenberg TM, Ramirez NC, Wise L, Zmuda E, Corcoran N, Costello T, Hovens C, Carvalho AL, de Carvalho AC, Fregnani JH, Longatto-Filho A, Reis RM, Scapulatempo-Neto C, Silveira HCS, Vidal DO, Burnette A, Eschbacher J, Hermes B, Noss A, Singh R, Anderson ML, Castro PD, Ittmann M, Huntsman D, Kohl B, le X, Thorp R, Andry C, Duffy ER, Lyadov V, Paklina O, Setdikova G, Shabunin A, Tavobilov M, McPherson C, Warnick R, Berkowitz R, Cramer D, Feltmate C, Horowitz N, Kibel A, Muto M, Raut CP, Malykh A, Barnholtz-Sloan JS, Barrett W, Devine K, Fulop J, Ostrom QT, Shimmel K, Wolinsky Y, Sloan AE, de Rose A, Giuliante F, Goodman M, Karlan BY, Hagedorn CH, Eckman J, Harr J, Myers J, Tucker K, Zach LA, Deyarmin B, Hu H, Kvecher L, Larson C, Mural RJ, Somiari S, Vicha A, Zelinka T, Bennett J, Iacocca M, Rabeno B, Swanson P, Latour M, Lacombe L, Têtu B, Bergeron A, McGraw M, Staugaitis SM, Chabot J, Hibshoosh H, Sepulveda A, Su T, Wang T, Potapova O, Voronina O, Desjardins L, Mariani O, Roman-Roman S, Sastre X, Stern MH, Cheng F, Signoretti S, Berchuck A, Bigner D, Lipp E, Marks J, McCall S, McLendon R, Secord A, Sharp A, Behera M, Brat DJ, Chen A, Delman K, Force S, Khuri F, Magliocca K, Maithel S, Olson JJ, Owonikoko T, Pickens A, Ramalingam S, Shin DM, Sica G, van Meir EG, Zhang H, Eijckenboom W, Gillis A, Korpershoek E, Looijenga L, Oosterhuis W, Stoop H, van Kessel KE, Zwarthoff EC, Calatozzolo C, Cuppini L, Cuzzubbo S, DiMeco F, Finocchiaro G, Mattei L, Perin A, Pollo B, Chen C, Houck J, Lohavanichbutr P, Hartmann A, Stoehr C, Stoehr R, Taubert H, Wach S, Wullich B, Kycler W, Murawa D, Wiznerowicz M, Chung K, Edenfield WJ, Martin J, Baudin E, Bubley G, Bueno R, de Rienzo A, Richards WG, Kalkanis S, Mikkelsen T, Noushmehr H, Scarpace L, Girard N, Aymerich M, Campo E, Giné E, Guillermo AL, van Bang N, Hanh PT, Phu BD, Tang Y, Colman H, Evason K, Dottino PR, Martignetti JA, Gabra H, Juhl H, Akeredolu T, Stepa S, Hoon D, Ahn K, Kang KJ, Beuschlein F, Breggia A, Birrer M, Bell D, Borad M, Bryce AH, Castle E, Chandan V, Cheville J, Copland JA, Farnell M, Flotte T, Giama N, Ho T, Kendrick M, Kocher JP, Kopp K, Moser C, Nagorney D, O’Brien D, O’Neill BP, Patel T, Petersen G, Que F, Rivera M, Roberts L, Smallridge R, Smyrk T, Stanton M, Thompson RH, Torbenson M, Yang JD, Zhang L, Brimo F, Ajani JA, Gonzalez AMA, Behrens C, Bondaruk J, Broaddus R, Czerniak B, Esmaeli B, Fujimoto J, Gershenwald J, Guo C, Lazar AJ, Logothetis C, Meric-Bernstam F, Moran C, Ramondetta L, Rice D, Sood A, Tamboli P, Thompson T, Troncoso P, Tsao A, Wistuba I, Carter C, Haydu L, Hersey P, Jakrot V, Kakavand H, Kefford R, Lee K, Long G, Mann G, Quinn M, Saw R, Scolyer R, Shannon K, Spillane A, Stretch J, Synott M, Thompson J, Wilmott J, al-Ahmadie H, Chan TA, Ghossein R, Gopalan A, Levine DA, Reuter V, Singer S, Singh B, Tien NV, Broudy T, Mirsaidi C, Nair P, Drwiega P, Miller J, Smith J, Zaren H, Park JW, Hung NP, Kebebew E, Linehan WM, Metwalli AR, Pacak K, Pinto PA, Schiffman M, Schmidt LS, Vocke CD, Wentzensen N, Worrell R, Yang H, Moncrieff M, Goparaju C, Melamed J, Pass H, Botnariuc N, Caraman I, Cernat M, Chemencedji I, Clipca A, Doruc S, Gorincioi G, Mura S, Pirtac M, Stancul I, Tcaciuc D, Albert M, Alexopoulou I, Arnaout A, Bartlett J, Engel J, Gilbert S, Parfitt J, Sekhon H, Thomas G, Rassl DM, Rintoul RC, Bifulco C, Tamakawa R, Urba W, Hayward N, Timmers H, Antenucci A, Facciolo F, Grazi G, Marino M, Merola R, de Krijger R, Gimenez-Roqueplo AP, Piché A, Chevalier S, McKercher G, Birsoy K, Barnett G, Brewer C, Farver C, Naska T, Pennell NA, Raymond D, Schilero C, Smolenski K, Williams F, Morrison C, Borgia JA, Liptay MJ, Pool M, Seder CW, Junker K, Omberg L, Dinkin M, Manikhas G, Alvaro D, Bragazzi MC, Cardinale V, Carpino G, Gaudio E, Chesla D, Cottingham S, Dubina M, Moiseenko F, Dhanasekaran R, Becker KF, Janssen KP, Slotta-Huspenina J, Abdel-Rahman MH, Aziz D, Bell S, Cebulla CM, Davis A, Duell R, Elder JB, Hilty J, Kumar B, Lang J, Lehman NL, Mandt R, Nguyen P, Pilarski R, Rai K, Schoenfield L, Senecal K, Wakely P, Hansen P, Lechan R, Powers J, Tischler A, Grizzle WE, Sexton KC, Kastl A, Henderson J, Porten S, Waldmann J, Fassnacht M, Asa SL, Schadendorf D, Couce M, Graefen M, Huland H, Sauter G, Schlomm T, Simon R, Tennstedt P, Olabode O, Nelson M, Bathe O, Carroll PR, Chan JM, Disaia P, Glenn P, Kelley RK, Landen CN, Phillips J, Prados M, Simko J, Smith-McCune K, VandenBerg S, Roggin K, Fehrenbach A, Kendler A, Sifri S, Steele R, Jimeno A, Carey F, Forgie I, Mannelli M, Carney M, Hernandez B, Campos B, Herold-Mende C, Jungk C, Unterberg A, von Deimling A, Bossler A, Galbraith J, Jacobus L, Knudson M, Knutson T, Ma D, Milhem M, Sigmund R, Godwin AK, Madan R, Rosenthal HG, Adebamowo C, Adebamowo SN, Boussioutas A, Beer D, Giordano T, Mes-Masson AM, Saad F, Bocklage T, Landrum L, Mannel R, Moore K, Moxley K, Postier R, Walker J, Zuna R, Feldman M, Valdivieso F, Dhir R, Luketich J, Pinero EMM, Quintero-Aguilo M, Carlotti CG, Jr, Dos Santos JS, Kemp R, Sankarankuty A, Tirapelli D, Catto J, Agnew K, Swisher E, Creaney J, Robinson B, Shelley CS, Godwin EM, Kendall S, Shipman C, Bradford C, Carey T, Haddad A, Moyer J, Peterson L, Prince M, Rozek L, Wolf G, Bowman R, Fong KM, Yang I, Korst R, Rathmell WK, Fantacone-Campbell JL, Hooke JA, Kovatich AJ, Shriver CD, DiPersio J, Drake B, Govindan R, Heath S, Ley T, van Tine B, Westervelt P, Rubin MA, Lee JI, Aredes ND, Mariamidze A. Driver fusions and their implications in the development and treatment of human cancers. Cell Rep. 2018;23(1):227–238. doi: 10.1016/j.celrep.2018.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An integrated TCGA pan-Cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):400–416. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic signaling pathways in the Cancer genome atlas. Cell. 2018;173(2):321–337. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jonsson P, Lin AL, Young RJ, DiStefano NM, Hyman DM, Li BT, et al. Genomic correlates of disease progression and treatment response in prospectively characterized gliomas. Clin Cancer Res. 2019;25(18):5537–5547. doi: 10.1158/1078-0432.CCR-19-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, Morozova O, Newton Y, Radenbaugh A, Pagnotta SM, Anjum S, Wang J, Manyam G, Zoppoli P, Ling S, Rao AA, Grifford M, Cherniack AD, Zhang H, Poisson L, Carlotti CG, Jr, Tirapelli DPC, Rao A, Mikkelsen T, Lau CC, Yung WKA, Rabadan R, Huse J, Brat DJ, Lehman NL, Barnholtz-Sloan JS, Zheng S, Hess K, Rao G, Meyerson M, Beroukhim R, Cooper L, Akbani R, Wrensch M, Haussler D, Aldape KD, Laird PW, Gutmann DH, Noushmehr H, Iavarone A, Verhaak RGW, Anjum S, Arachchi H, Auman JT, Balasundaram M, Balu S, Barnett G, Baylin S, Bell S, Benz C, Bir N, Black KL, Bodenheimer T, Boice L, Bootwalla MS, Bowen J, Bristow CA, Butterfield YSN, Chen QR, Chin L, Cho J, Chuah E, Chudamani S, Coetzee SG, Cohen ML, Colman H, Couce M, D’Angelo F, Davidsen T, Davis A, Demchok JA, Devine K, Ding L, Duell R, Elder JB, Eschbacher JM, Fehrenbach A, Ferguson M, Frazer S, Fuller G, Fulop J, Gabriel SB, Garofano L, Gastier-Foster JM, Gehlenborg N, Gerken M, Getz G, Giannini C, Gibson WJ, Hadjipanayis A, Hayes DN, Heiman DI, Hermes B, Hilty J, Hoadley KA, Hoyle AP, Huang M, Jefferys SR, Jones CD, Jones SJM, Ju Z, Kastl A, Kendler A, Kim J, Kucherlapati R, Lai PH, Lawrence MS, Lee S, Leraas KM, Lichtenberg TM, Lin P, Liu Y, Liu J, Ljubimova JY, Lu Y, Ma Y, Maglinte DT, Mahadeshwar HS, Marra MA, McGraw M, McPherson C, Meng S, Mieczkowski PA, Miller CR, Mills GB, Moore RA, Mose LE, Mungall AJ, Naresh R, Naska T, Neder L, Noble MS, Noss A, O’Neill BP, Ostrom QT, Palmer C, Pantazi A, Parfenov M, Park PJ, Parker JS, Perou CM, Pierson CR, Pihl T, Protopopov A, Radenbaugh A, Ramirez NC, Rathmell WK, Ren X, Roach J, Robertson AG, Saksena G, Schein JE, Schumacher SE, Seidman J, Senecal K, Seth S, Shen H, Shi Y, Shih J, Shimmel K, Sicotte H, Sifri S, Silva T, Simons JV, Singh R, Skelly T, Sloan AE, Sofia HJ, Soloway MG, Song X, Sougnez C, Souza C, Staugaitis SM, Sun H, Sun C, Tan D, Tang J, Tang Y, Thorne L, Trevisan FA, Triche T, van den Berg DJ, Veluvolu U, Voet D, Wan Y, Wang Z, Warnick R, Weinstein JN, Weisenberger DJ, Wilkerson MD, Williams F, Wise L, Wolinsky Y, Wu J, Xu AW, Yang L, Yang L, Zack TI, Zenklusen JC, Zhang J, Zhang W, Zhang J, Zmuda E. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao J, Chen AX, Gartrell RD, Silverman AM, Aparicio L, Chu T, Bordbar D, Shan D, Samanamud J, Mahajan A, Filip I, Orenbuch R, Goetz M, Yamaguchi JT, Cloney M, Horbinski C, Lukas RV, Raizer J, Rae AI, Yuan J, Canoll P, Bruce JN, Saenger YM, Sims P, Iwamoto FM, Sonabend AM, Rabadan R. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat Med. 2019;25(3):462–469. doi: 10.1038/s41591-019-0349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cancer Genome Atlas Research N Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhandari V, Hoey C, Liu LY, Lalonde E, Ray J, Livingstone J, Lesurf R, Shiah YJ, Vujcic T, Huang X, Espiritu SMG, Heisler LE, Yousif F, Huang V, Yamaguchi TN, Yao CQ, Sabelnykova VY, Fraser M, Chua MLK, van der Kwast T, Liu SK, Boutros PC, Bristow RG. Molecular landmarks of tumor hypoxia across cancer types. Nat Genet. 2019;51(2):308–318. doi: 10.1038/s41588-018-0318-2. [DOI] [PubMed] [Google Scholar]
- 37.Thomas AA, Abrey LE, Terziev R, Raizer J, Martinez NL, Forsyth P, Paleologos N, Matasar M, Sauter CS, Moskowitz C, Nimer SD, DeAngelis LM, Kaley T, Grimm S, Louis DN, Cairncross JG, Panageas KS, Briggs S, Faivre G, Mohile NA, Mehta J, Jonsson P, Chakravarty D, Gao J, Schultz N, Brennan CW, Huse JT, Omuro A. Multicenter phase II study of temozolomide and myeloablative chemotherapy with autologous stem cell transplant for newly diagnosed anaplastic oligodendroglioma. Neuro-Oncology. 2017;19(10):1380–1390. doi: 10.1093/neuonc/nox086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488(7409):106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488(7409):100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, Chalhoub N, Baker SJ, Huether R, Kriwacki R, Curley N, Thiruvenkatam R, Wang J, Wu G, Rusch M, Hong X, Becksfort J, Gupta P, Ma J, Easton J, Vadodaria B, Onar-Thomas A, Lin T, Li S, Pounds S, Paugh S, Zhao D, Kawauchi D, Roussel MF, Finkelstein D, Ellison DW, Lau CC, Bouffet E, Hassall T, Gururangan S, Cohn R, Fulton RS, Fulton LL, Dooling DJ, Ochoa K, Gajjar A, Mardis ER, Wilson RK, Downing JR, Zhang J, Gilbertson RJ. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488(7409):43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morrissy AS, Garzia L, Shih DJ, Zuyderduyn S, Huang X, Skowron P, et al. Divergent clonal selection dominates medulloblastoma at recurrence. Nature. 2016;529(7586):351–357. doi: 10.1038/nature16478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones DT, Hutter B, Jager N, Korshunov A, Kool M, Warnatz HJ, et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45(8):927–932. doi: 10.1038/ng.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fishbein L, Leshchiner I, Walter V, Danilova L, Robertson AG, Johnson AR, Lichtenberg TM, Murray BA, Ghayee HK, Else T, Ling S, Jefferys SR, de Cubas AA, Wenz B, Korpershoek E, Amelio AL, Makowski L, Rathmell WK, Gimenez-Roqueplo AP, Giordano TJ, Asa SL, Tischler AS, Pacak K, Nathanson KL, Wilkerson MD, Akbani R, Ally A, Amar L, Amelio AL, Arachchi H, Asa SL, Auchus RJ, Auman JT, Baertsch R, Balasundaram M, Balu S, Bartsch DK, Baudin E, Bauer T, Beaver A, Benz C, Beroukhim R, Beuschlein F, Bodenheimer T, Boice L, Bowen J, Bowlby R, Brooks D, Carlsen R, Carter S, Cassol CA, Cherniack AD, Chin L, Cho J, Chuah E, Chudamani S, Cope L, Crain D, Curley E, Danilova L, de Cubas AA, de Krijger RR, Demchok JA, Deutschbein T, Dhalla N, Dimmock D, Dinjens WNM, Else T, Eng C, Eschbacher J, Fassnacht M, Felau I, Feldman M, Ferguson ML, Fiddes I, Fishbein L, Frazer S, Gabriel SB, Gardner J, Gastier-Foster JM, Gehlenborg N, Gerken M, Getz G, Geurts J, Ghayee HK, Gimenez-Roqueplo AP, Giordano TJ, Goldman M, Graim K, Gupta M, Haan D, Hahner S, Hantel C, Haussler D, Hayes DN, Heiman DI, Hoadley KA, Holt RA, Hoyle AP, Huang M, Hunt B, Hutter CM, Jefferys SR, Johnson AR, Jones SJM, Jones CD, Kasaian K, Kebebew E, Kim J, Kimes P, Knijnenburg T, Korpershoek E, Lander E, Lawrence MS, Lechan R, Lee D, Leraas KM, Lerario A, Leshchiner I, Lichtenberg TM, Lin P, Ling S, Liu J, LiVolsi VA, Lolla L, Lotan Y, Lu Y, Ma Y, Maison N, Makowski L, Mallery D, Mannelli M, Marquard J, Marra MA, Matthew T, Mayo M, Méatchi T, Meng S, Merino MJ, Mete O, Meyerson M, Mieczkowski PA, Mills GB, Moore RA, Morozova O, Morris S, Mose LE, Mungall AJ, Murray BA, Naresh R, Nathanson KL, Newton Y, Ng S, Ni Y, Noble MS, Nwariaku F, Pacak K, Parker JS, Paul E, Penny R, Perou CM, Perou AH, Pihl T, Powers J, Rabaglia J, Radenbaugh A, Ramirez NC, Rao A, Rathmell WK, Riester A, Roach J, Robertson AG, Sadeghi S, Saksena G, Salama S, Saller C, Sandusky G, Sbiera S, Schein JE, Schumacher SE, Shelton C, Shelton T, Sheth M, Shi Y, Shih J, Shmulevich I, Simons JV, Sipahimalani P, Skelly T, Sofia HJ, Sokolov A, Soloway MG, Sougnez C, Stuart J, Sun C, Swatloski T, Tam A, Tan D, Tarnuzzer R, Tarvin K, Thiessen N, Thorne LB, Timmers HJ, Tischler AS, Tse K, Uzunangelov V, van Berkel A, Veluvolu U, Vicha A, Voet D, Waldmann J, Walter V, Wan Y, Wang Z, Wang TS, Weaver J, Weinstein JN, Weismann D, Wenz B, Wilkerson MD, Wise L, Wong T, Wong C, Wu Y, Yang L, Zelinka T, Zenklusen JC, Zhang J(J), Zhang W, Zhu J, Zinzindohoué F, Zmuda E. Comprehensive molecular characterization of Pheochromocytoma and Paraganglioma. Cancer Cell. 2017;31(2):181–193. doi: 10.1016/j.ccell.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dahiya S, Emnett RJ, Haydon DH, Leonard JR, Phillips JJ, Perry A, Gutmann DH. BRAF-V600E mutation in pediatric and adult glioblastoma. Neuro-Oncology. 2014;16(2):318–319. doi: 10.1093/neuonc/not146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodin SN, Rodin AS. Strand asymmetry of CpG transitions as indicator of G1 phase-dependent origin of multiple tumorigenic p53 mutations in stem cells. Proc Natl Acad Sci U S A. 1998;95(20):11927–11932. doi: 10.1073/pnas.95.20.11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai A, Kharbanda S, Pope WB, Tran A, Solis OE, Peale F, Forrest WF, Pujara K, Carrillo JA, Pandita A, Ellingson BM, Bowers CW, Soriano RH, Schmidt NO, Mohan S, Yong WH, Seshagiri S, Modrusan Z, Jiang Z, Aldape KD, Mischel PS, Liau LM, Escovedo CJ, Chen W, Nghiemphu PL, James CD, Prados MD, Westphal M, Lamszus K, Cloughesy T, Phillips HS. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol. 2011;29(34):4482–4490. doi: 10.1200/JCO.2010.33.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dunn GP, Andronesi OC, Cahill DP. From genomics to the clinic: biological and translational insights of mutant IDH1/2 in glioma. Neurosurg Focus. 2013;34(2):E2. doi: 10.3171/2012.12.FOCUS12355. [DOI] [PubMed] [Google Scholar]
- 48.Liu XY, Gerges N, Korshunov A, Sabha N, Khuong-Quang DA, Fontebasso AM, Fleming A, Hadjadj D, Schwartzentruber J, Majewski J, Dong Z, Siegel P, Albrecht S, Croul S, Jones DTW, Kool M, Tonjes M, Reifenberger G, Faury D, Zadeh G, Pfister S, Jabado N. Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol. 2012;124(5):615–625. doi: 10.1007/s00401-012-1031-3. [DOI] [PubMed] [Google Scholar]
- 49.Cai J, Zhang C, Zhang W, Wang G, Yao K, Wang Z, Li G, Qian Z, Li Y, Jiang T, Jiang C. ATRX, IDH1-R132H and Ki-67 immunohistochemistry as a classification scheme for astrocytic tumors. Oncoscience. 2016;3(7–8):258–265. doi: 10.18632/oncoscience.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Modrek AS, Golub D, Khan T, Bready D, Prado J, Bowman C, Deng J, Zhang G, Rocha PP, Raviram R, Lazaris C, Stafford JM, LeRoy G, Kader M, Dhaliwal J, Bayin NS, Frenster JD, Serrano J, Chiriboga L, Baitalmal R, Nanjangud G, Chi AS, Golfinos JG, Wang J, Karajannis MA, Bonneau RA, Reinberg D, Tsirigos A, Zagzag D, Snuderl M, Skok JA, Neubert TA, Placantonakis DG. Low-grade astrocytoma mutations in IDH1, P53, and ATRX cooperate to block differentiation of human neural stem cells via repression of SOX2. Cell Rep. 2017;21(5):1267–1280. doi: 10.1016/j.celrep.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kannan K, Inagaki A, Silber J, Gorovets D, Zhang J, Kastenhuber ER, et al. Whole-exome sequencing identifies ATRX mutation as a key molecular determinant in lower-grade glioma. Oncotarget. 2012;3(10):1194–1203. doi: 10.18632/oncotarget.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weber CK, Slupsky JR, Kalmes HA, Rapp UR. Active Ras induces heterodimerization of cRaf and BRaf. Cancer Res. 2001;61(9):3595–3598. [PubMed] [Google Scholar]
- 53.Mizutani S, Inouye K, Koide H, Kaziro Y. Involvement of B-Raf in Ras-induced Raf-1 activation. FEBS Lett. 2001;507(3):295–298. doi: 10.1016/S0014-5793(01)02992-1. [DOI] [PubMed] [Google Scholar]
- 54.Wang HG, Rapp UR, Reed JC. Bcl-2 targets the protein kinase Raf-1 to mitochondria. Cell. 1996;87(4):629–638. doi: 10.1016/S0092-8674(00)81383-5. [DOI] [PubMed] [Google Scholar]
- 55.Wang HG, Takayama S, Rapp UR, Reed JC. Bcl-2 interacting protein, BAG-1, binds to and activates the kinase Raf-1. Proc Natl Acad Sci U S A. 1996;93(14):7063–7068. doi: 10.1073/pnas.93.14.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hemann MT, Lowe SW. The p53-Bcl-2 connection. Cell Death Differ. 2006;13(8):1256–1259. doi: 10.1038/sj.cdd.4401962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ryall S, Tabori U, Hawkins C. A comprehensive review of paediatric low-grade diffuse glioma: pathology, molecular genetics and treatment. Brain Tumor Pathol. 2017;34(2):1–11. doi: 10.1007/s10014-017-0282-z. [DOI] [PubMed] [Google Scholar]
- 58.Lassaletta A, Zapotocky M, Mistry M, Ramaswamy V, Tabori U. Therapeutic and prognostic implications of BRAF V600E in pediatric low-grade gliomas. J Clin Oncol. 2017;35(25):JCO2016718726. doi: 10.1200/JCO.2016.71.8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schiffman JD, Hodgson JG, VandenBerg SR, Flaherty P, Polley MY, Yu M, et al. Oncogenic BRAF mutation with CDKN2A inactivation is characteristic of a subset of pediatric malignant astrocytomas. Cancer Res. 2010;70(2):512–519. doi: 10.1158/0008-5472.CAN-09-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mistry M, Zhukova N, Merico D, Rakopoulos P, Krishnatry R, Shago M, Stavropoulos J, Alon N, Pole JD, Ray PN, Navickiene V, Mangerel J, Remke M, Buczkowicz P, Ramaswamy V, Guerreiro Stucklin A, Li M, Young EJ, Zhang C, Castelo-Branco P, Bakry D, Laughlin S, Shlien A, Chan J, Ligon KL, Rutka JT, Dirks PB, Taylor MD, Greenberg M, Malkin D, Huang A, Bouffet E, Hawkins CE, Tabori U. BRAF mutation and CDKN2A deletion define a clinically distinct subgroup of childhood secondary high-grade glioma. J Clin Oncol. 2015;33(9):1015–1022. doi: 10.1200/JCO.2014.58.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson JP, VanBrocklin MW, Guilbeault AR, Signorelli DL, Brandner S, Holmen SL. Activated BRAF induces gliomas in mice when combined with Ink4a/Arf loss or Akt activation. Oncogene. 2010;29(3):335–344. doi: 10.1038/onc.2009.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Putlyaeva LV, Demin DE, Uvarova AN, Zinevich LS, Prokofjeva MM, Gazizova GR, Shagimardanova EI, Schwartz AM. PTPN11 knockdown prevents changes in the expression of genes controlling cell cycle, chemotherapy resistance, and oncogene-induced senescence in human thyroid cells overexpressing BRAF V600E oncogenic protein. Biochemistry (Mosc) 2020;85(1):108–118. doi: 10.1134/S0006297920010101. [DOI] [PubMed] [Google Scholar]
- 63.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 64.Wellbrock C, Karasarides M, Marais R. The RAF proteins take Centre stage. Nat Rev Mol Cell Biol. 2004;5(11):875–885. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.