Summary
We have developed a rapid, accurate, and cost-effective serologic test for SARS-CoV-2 virus, which caused the COVID-19 pandemic, on the basis of antibody-dependent agglutination of antigen-coated latex particles. When validated using plasma samples that are positive or negative for SARS-CoV-2, the agglutination assay detected antibodies against the receptor-binding domain of the spike (S-RBD) or the nucleocapsid protein of SARS-CoV-2 with 100% specificity and ∼98% sensitivity. Furthermore, we found that the strength of the S-RBD antibody response measured by the agglutination assay correlated with the efficiency of the plasma in blocking RBD binding to the angiotensin-converting enzyme 2 in a surrogate neutralization assay, suggesting that the agglutination assay might be used to identify individuals with virus-neutralizing antibodies. Intriguingly, we found that >92% of patients had detectable antibodies on the day of a positive viral RNA test, suggesting that the agglutination antibody test might complement RNA testing for the diagnosis of SARS-CoV-2 infection.
Keywords: SARS-CoV-2, COVID-19, antibody test, serology, agglutination assay, rapid testing, neutralizing antibody, spike, nucleocapsid
Graphical abstract
Highlights
-
•
Agglutination assay detects SARS-CoV-2 antibodies with high accuracy
-
•
High sensitivity of the agglutination assay allows for early antibody detection
-
•
RBD-dependent agglutination correlates with neutralizing antibody response
Motivation
As COVID-19 continues to spread around the world, there is urgent need for a rapid yet accurate antibody test to detect individuals' humoral immune responses to the SARS-CoV-2 virus and emerging variants of concern. A simple and reliable antibody test would also allow longitudinal analysis of antibody responses to inform vaccination strategies. We addressed this pressing need by developing a simple, quick, and sensitive antibody test based on latex particle agglutination.
Esmail et al. develop and validate a rapid and cost-effective antibody-dependent agglutination (aggregation) test by using SARS-CoV-2 antigen-coated latex particles. The agglutination assay detects SARS-CoV-2 antibodies with 100% specificity and ∼98% sensitivity. They further show that the agglutination assay might be used to identify neutralizing antibodies and monitor dynamic changes in antibody responses over time.
Introduction
Development of rapid point-of-care coronavirus disease 2019 (COVID-19) diagnostics for use at the community level remains a top priority in the global response to the COVID-19 pandemic (Peeling et al., 2020). Although capacity for detecting SARS-CoV-2 based on the nucleic acid amplification test (NAAT) has grown immensely and enabled effective public health responses, serologic testing for virus-specific antibodies has not gained the same widespread application because of concerns over sensitivity, specificity, cost, and turnaround time (Kruttgen et al., 2020; Peeling et al., 2020). Although NAAT is the current gold standard for diagnosing acute infection, it is not effective in identifying individuals who have recovered from previous infection (Ravi et al., 2020). Given that approximately 40% of infected individuals remain asymptomatic (Amanat et al., 2020; He et al., 2020; Peeling et al., 2020), large-scale antibody testing could help better establish the true extent of the COVID-19 pandemic, identifying disease hotspots and high-risk populations to enable more effective isolation and contact tracing (Peeling et al., 2020; Ravi et al., 2020; Weisberg et al., 2020). Moreover, antibody testing might identify individuals with a strong neutralizing antibody response who might be suitable donors for convalescent plasma or serum therapy for the treatment of those with severe symptoms (Shen et al., 2020).
To date, a number of antibody tests have been approved for emergency use in the United States and Europe. These tests detect the immunoglobulin G (IgG), IgM, or IgA antibody against the spike (including the receptor-binding domain [RBD]) or nucleocapsid (N) protein of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus by enzyme-linked immunosorbent assay (ELISA) (Ravi et al., 2020). ELISA-based antibody tests, which can be qualitative or quantitative, require specialized instruments and are usually performed in a laboratory by a trained technician. The sensitivity and specificity of different ELISA kits vary widely (Jaaskelainen et al., 2020; Knauer et al., 2020; Lisboa Bastos et al., 2020). To enable point-of-care (POC) testing, several rapid diagnostic tests (RDTs) based on lateral flow have been developed. Although the RDTs have reduced the time of the antibody test to 10–30 min from 2–5 h (for ELISA), they generally suffer from decreased sensitivity and specificity compared with ELISA-based assays (Li et al., 2020; Pavlova et al., 2020; Peeling et al., 2020; Whitman et al., 2020).
To address the pressing need for a simple, rapid, yet accurate antibody test (Peeling et al., 2020), we resorted to the tested-and-proven serology method of agglutination that has been used in blood typing and antibody testing (Alves et al., 2020; Gupta and Chaudhary, 2003; Hursh et al., 1989). We show here that the agglutination of red blood cells (RBCs) or latex particles induced by specific antigen-antibody interaction affords a highly sensitive and accurate assay for SARS-CoV-2 antibodies. We validated the antibody assay on the basis of latex particle agglutination by using 169 plasma samples that were tested positive for SARS-CoV-2 by NAAT, 121 samples that were NAAT negative, and 100 SARS-CoV-2-naive plasma samples. The agglutination-based antibody assay produced 100% specificity and 97%–98.2% sensitivity. Because this simple assay requires no instrument and generates results in 2 min, it has the potential to be used as a POC test.
Results
Agglutination-based serologic testing for SARS-CoV-2 antibodies
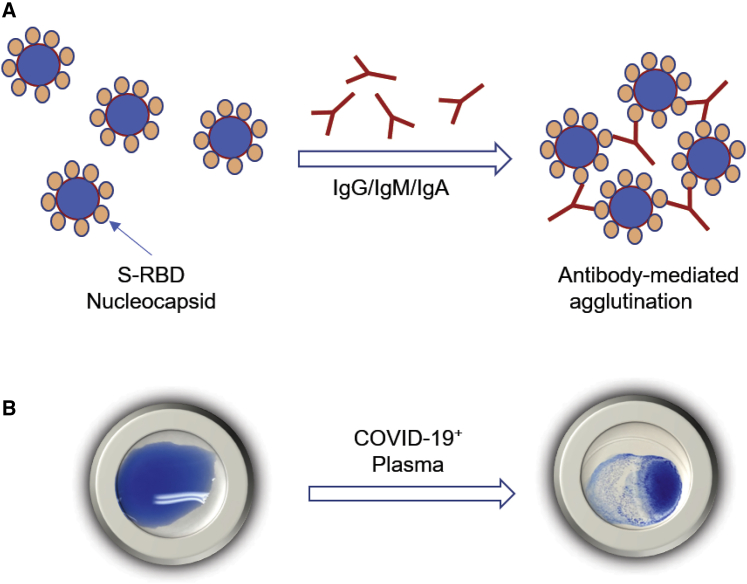
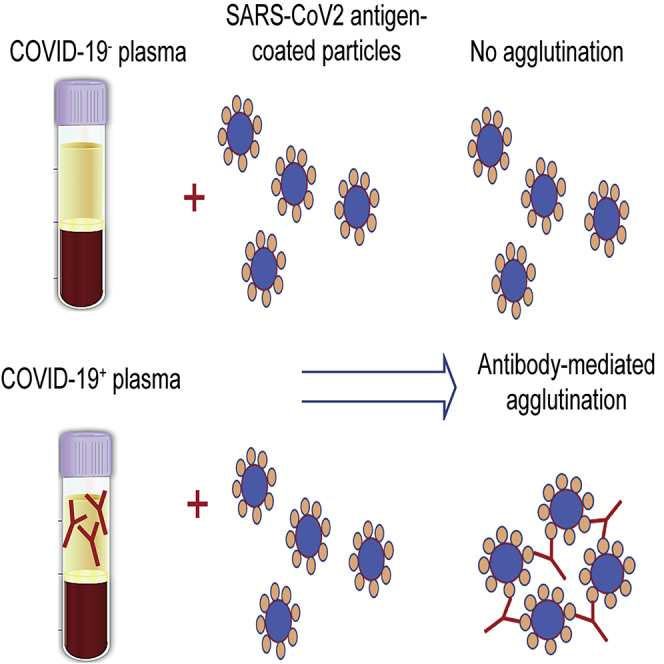
Agglutination of RBCs is widely used in blood typing, whereas latex-particle agglutination assays have been used to detect antibodies against a variety of different viruses. We sought to establish whether either or both approaches could be adapted for SARS-CoV-2 antibody testing. In principle, coating the RBCs or the latex particles with a SARS-CoV-2-specific antigen would enable their respective agglutination by the corresponding antibody (Figure 1). To explore this notion, we labeled Group O (R2R2) RBCs carrying the D antigen with the spike RBD (S-RBD) or the RNA-binding domain of the nucleocapsid (N-RBD) protein through streptavidin-biotin-mediated coupling (Figures S1 and S2; see STAR Methods for details). Incubating the antigen-coated RBCs with COVID-19+ plasma led to robust agglutination, whereas the COVID-19− plasma failed to induce RBC agglutination, suggesting that the aggregation of the S-RBD/N-RBD-coated RBCs might be used to detect antibody response to SARS-CoV-2 (Figure S2).
Figure 1.
Illustration of the principle of agglutination assay for SARS-CoV-2 antibody testing
(A) Latex particles or red blood cells are surface-coated with a SARS-CoV-2 antigen, S-RBD, or nucleocapsid. Incubation with plasma containing antibodies against the coated antigen would induce agglutination of the latex particles or RBCs.
(B) Representative image of the agglutination assay using latex beads coated with S-RBD.
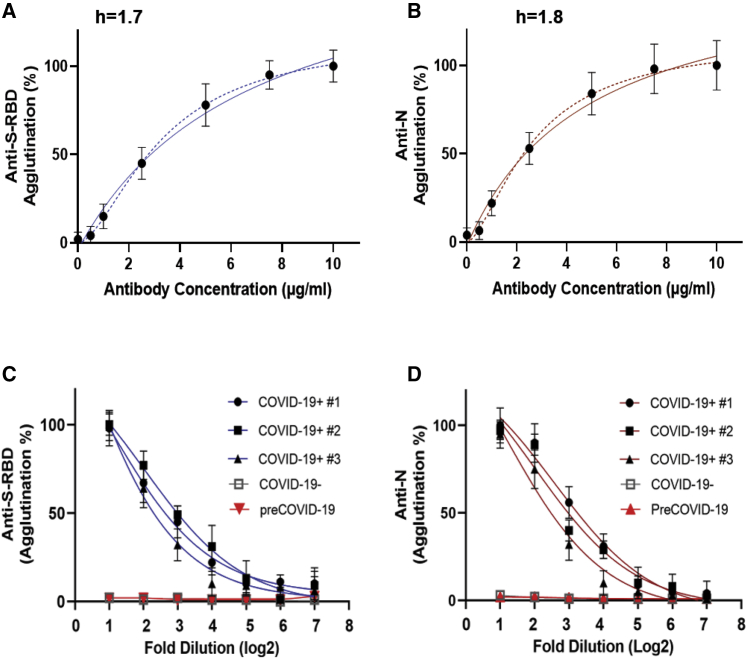
To develop a cost-effective agglutination assay, we next investigated whether latex particles were a suitable substitute for the RBCs. To this end, we coated latex particles with recombinant S-RBD or the full-length nucleocapsid (N) protein (Figure S1). The antigen-coated latex beads were first tested with a monoclonal anti-S-RBD and a polyclonal anti-nucleocapsid antibody. Upon incubating with the corresponding antibody, the antigen-coated latex particles formed clumps within 2 min. Importantly, the area of clump formation grew larger with increasing antibody concentrations (Figure S3). Although latex agglutination is commonly used as a qualitative assay, it is possible to determine the degree of agglutination on the basis of the area of clump formation via image analysis. As shown in Figure 2, the percentage of agglutination for both the S-RBD- and N-coated latex particles increased when an incremental amount of anti-S-RBD or anti-N antibody was added. Fitting the data to Hill's equation yielded Hill's coefficient of 1.7 for the former and 1.8 for the latter. This suggests that the antibody-induced agglutination of latex particles is a cooperative event (Figures 2A and 2B).
Figure 2.
Antibody-induced latex particle agglutination correlates with the antibody titer
(A and B) Changes in agglutination in response to increased concentrations of the anti-S-RBD (n = 3) (A) or anti-N antibody (n = 3) (B). Dashed lines represent fitted curves to the Hill equation (h, Hill coefficient).
(C and D) S-RBD (C) or N (D) antibody-induced agglutination decreased with increased dilution of plasma. Shown are agglutination data (in log2 scale) from COVID-19+, COVID-19−, and pre-COVID-19 plasma samples with 1:2 to 1:128 dilution. Data shown are from three replicates per concentration for three samples per group. Error bars represent standard deviation.
We next examined whether the latex agglutination assay could be used to gauge COVID-19 antibody response. Using plasma samples from patients who tested positive for SARS-CoV-2 by NAAT and confirmed for strong antibody response by ELISA, we found that the patient plasma samples were not only capable of inducing agglutination of the S-RBD- or N-coated latex particles, they did so in a concentration-dependent manner. As shown in Figures 2C and 2D, the extent of agglutination decreased as the plasma was diluted, indicating that the agglutination assay might be used to estimate antibody titer as in an ELISA-based antibody test.
The latex agglutination-based antibody assay showed high sensitivity and specificity
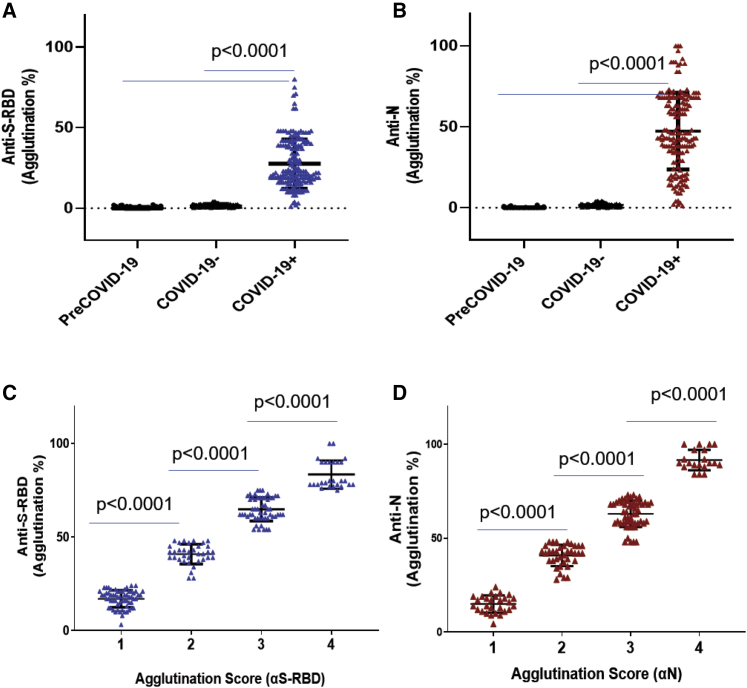
To validate the antibody test based on latex-particle agglutination, we carried out agglutination assays for 290 residual plasma samples from individuals who tested positive (169) or negative (121) for virus RNA by the Roche (Basel, Switzerland) cobas SARS-CoV-2 test (Knauer et al., 2020). To assess specificity, we also included 100 virus-naive samples banked in 2018 in our agglutination assay. None of the 121 SARS-CoV-2− or the 100 pre-COVID-19 plasma samples was capable of promoting the agglutination of either the S-RBD- or N-coated latex particles, indicating 100% specificity for the agglutination assay (Table 1). In contrast, of the 169 SARS-CoV-2+ plasma samples tested, 166 (98.2%) promoted agglutination in response to the S-RBD antigen and 164 (97%) to the N antigen, with overall sensitivity of 98.2%. We compared the latex agglutination assay with the EUROIMMUN (Lübeck, Germany) IgG test for the S antibody and the Roche Elecsys Total assay for the N antibody by using the same set of SARS-CoV-2+ plasma samples and found that the latex agglutination assay outperformed both ELISA-based antibody tests (Table 2). The agglutination assay also exhibited better specificity than either ELISA kit (Table 2). Quantification of the agglutination data showed that the COVID-19+ group is significantly different from the COVID-19− or pre-COVID-19 group, indicating that the latex agglutination assay effectively distinguished SARS-CoV2+ from SARS-CoV2− individuals (Figures 3A and 3B).
Table 1.
Clinical performance of the agglutination-based antibody assay
| Samples | Anti-S-RBD (n) |
Anti-N (n) |
Overall sensitivity |
Overall specificity |
||
|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |||
| SARS-CoV-2 NAAT positive (n = 169) | 166 | 3 | 164 | 5 | 98.2% (166/169) | n/a |
| Days of SARS-CoV-2 NAAT positive | ||||||
| 1 (n = 41) | 38 | 3 | 36 | 5 | 92.7% (38/41) | n/a |
| 2 (n = 31) | 30 | 1 | 31 | 0 | 100% | n/a |
| ≥3 (n = 97) | 97 | 0 | 96 | 1 | 100% | n/a |
| SARS-CoV-2 NAAT negative (n = 121) | 0 | 121 | 0 | 121 | n/a | 100% |
| Pre-COVID-19 (n = 100) | 0 | 100 | 0 | 100 | n/a | 100% |
Table 2.
Comparison of sensitivity and specificity between the agglutination-based and ELISA-based antibody assays
| Anti-S (RBD) | Agglutination assay (n) |
ELISA (Knauer et al., 2020) EUROIMMUN (n) |
Sensitivity (agglutination) | Sensitivity (EUROIMMUN) | Specificity (agglutination) | Specificity (EUROIMMUN) | ||
|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |||||
| SARS-CoV-2 NAAT positive (n = 121) | 118 | 3 | 90 | 31 | 97.5% (118/121) | 74.4% (90/121) | N/A | |
| SARS-CoV-2 NAAT negative and pre-COVID-19 | 0 | 221 | 11 | 194 | N/A | 100% (221/221) | 94.6% (194/205) | |
| Anti-N | Agglutination assay (n) |
ELISA (Knauer et al., 2020) Roche Elecsys (n) |
Sensitivity (agglutination) | Sensitivity (Roche) | Specificity (agglutination) | Specificity (Roche) | ||
|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |||||
| SARS-CoV-2 NAAT positive (n = 69) | 68 | 1 | 66 | 2 | 98.6% (68/69) | 95.7% (66/69) | NA | |
| SARS-CoV-2 NAAT negative and pre-COVID-19 | 0 | 221 | 1 | 135 | N/A | 100% (221/221) | 99% (135/136) | |
N/A, no data available.
Figure 3.
Agglutination assay distinguished COVID-19+ from COVID-19− samples
(A and B) Comparison of S-RBD (A) and N antibody (B) responses between COVID-19+ (n = 169), COVID-19− (n = 121), and pre-COVID-19 (n = 100) samples determined by the aggregation assay.
(C and D) The strength of the S-RBD (C) and N antibody (D) response in the COVID-19+ (n = 169) plasma samples could be determined semi-quantitatively by the aggregation score (1–4 denotes weak to strong antibody response). Statistical analyses were performed using unpaired Student's t test with Welch's correction (p values shown on graph). Error bars represent standard deviation.
On the basis of the background signals of the COVID-19− samples (0%–4% agglutination for both S-RBD- and N-coated latex particles), we set 5% agglutination as the cutoff for antibody positivity. To facilitate the use of the latex agglutination assay as a simple semi-quantitative antibody test, we developed a numerical scoring system for antibody response. We assigned the scores 1, 2, 3, and 4 to samples that produced 5%–25%, 25%–50%, 50%–75%, and >75% agglutination, respectively (Figure S4). We found that this scoring scheme effectively distinguished samples with strong antibody response from those with medium or weak ones (Figures 3C and 3D). The agglutination score might be readily assigned by visual inspection and comparing with reference wells containing a predetermined amount of pure anti-S-RBD or anti-N antibody (Figure S3). The specificity of the agglutination assay was further validated by using latex particles without immobilized antigens, which showed minimal background aggregation (<5%) in the presence of the COVID-19+, COVID-19−, or pre-COVID-19 plasma (Figure S5).
The S-RBD antibody response correlated with neutralizing antibody titer
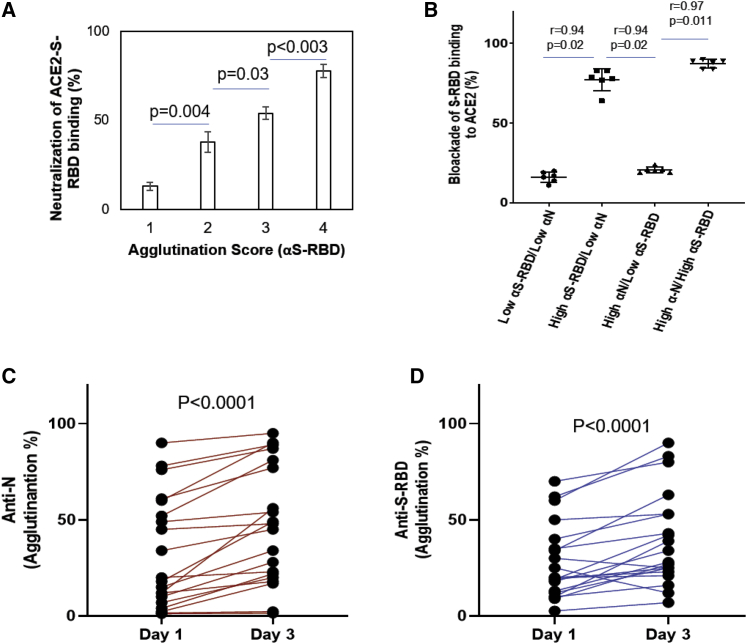
Because neutralizing antibodies play a pivotal role in the humoral immune response to the virus (Jiang et al., 2020), we next examined whether the S-RBD antibody response determined by the agglutination assay correlated with neutralization efficiency. We developed a surrogate neutralization assay by measuring the efficacy of patient plasma in blocking S-RBD binding to its host receptor, angiotensin-converting enzyme 2 (ACE2), in vitro. Similar approaches have been used by others to evaluate neutralization efficiency of patient plasma or therapeutic antibodies (Abe et al., 2020; Tortorici MA et al., 2020). In brief, binding of biotinylated ACE2 to immobilized S-RBD is detected by ELISA through horseradish peroxidase-conjugated streptavidin. The presence of neutralizing antibody would block this interaction, resulting in reduction of the ELISA signal. Using this surrogate neutralization assay, we found that the neutralization efficiency increased with the agglutination score for the S-RBD antibody (Figure 4A). Intriguingly, comparison of samples with distinct S-RBD and N antibody responses indicated that the neutralization efficiency was significantly correlated with the S-RBD, but not the N antibody strength. This is not surprising given that the nucleocapsid is not involved in mediating virus entry into the host cells via ACE2. Nevertheless, it remains to be determined whether the plasma with strong N antibody response would confer immunity by inhibiting virus replication in vivo.
Figure 4.
Using the latex agglutination assay to determine neutralizing antibody titer and dynamic changes in antibody response
(A) Neutralization antibody response correlated significantly with the agglutination score. p values were calculated by unpaired Student’s t test (n = 10 for each group).
(B) Spearman (r) correlation of efficiency of neutralization and S-RBD or N antibody response (n = 25).
(C and D) Dynamic changes in antibody responses in COVID-19 patients. n = 20 for day 1 and n = 20 for day 3; p values were calculated by unpaired Student’s t test with Welch's correction. Error bars represent standard deviation.
The agglutination assay allowed for early antibody detection and tracking of dynamic antibody response
We noted that the agglutination assay detected antibody response in >92% of plasma samples collected on the day of SARS-CoV2+ diagnosis by NAAT and in 100% of samples on day 2 and afterward. This is in stark contrast to the 47%–83% sensitivity for ELISA-based antibody tests on samples collected within 7 days of positive NAAT (Knauer et al., 2020). The superb sensitivity of the latex agglutination assay suggests that it might be used to detect antibody response in the early stage of virus infection and monitor its dynamic changes over time (Qu et al., 2020; Robbiani et al., 2020; Whitman et al., 2020). Using serial blood samples from SARS-CoV-2+ patients, we compared the changes in the S-RBD and N antibody responses between days 1 and 3 (Figures 4C and 4D). We found that the majority of patients showed detectable S-RBD and/or N antibodies on NAAT+ day 1. Moreover, the antibody titer increased significantly on day 3 compared with on day 1. Because it is not possible to determine how long these patients had contracted the virus prior to the NAAT test, seroconversion might have occurred for some on the day of diagnosis. Nevertheless, we were able to detect anti-S-RBD or anti-N antibodies on day 3 for several patients who showed no detectable antibody on day 1, suggesting that seroconversion occurred rapidly in these patients (Amanat et al., 2020). Altogether, these data suggest that the latex agglutination assay might be used to diagnose active infection in conjunction with NAAT. Combined antibody and RNA testing might increase the sensitivity of the latter. Moreover, the agglutination-based antibody test might be used to monitor the evolution of humoral immune reaction in infected individuals over time.
Discussion
Antibody testing offers an additional and much-needed tool for managing the COVID-19 pandemic, which might allow for rapid and cost-effective POC diagnosis to facilitate treatment and public health responses. Furthermore, antibody testing might play an important role in identifying individuals who have gained protective immunity from previous exposure or immunization programs. Although the clinical trial results for several vaccine candidates are encouraging, seroconversion is unlikely to occur for all vaccinated individuals (Thanh Le et al., 2020). A highly sensitive and specific antibody test would allow for accurate assessment of humoral response to a vaccine, and post-vaccination serial testing could indicate the duration of humoral immunity against SARS-CoV-2. Such information will be invaluable to inform public health decisions for both directing resource allocation and determining response to vaccination.
Although most serology tests used in the clinic are specific for a specific antibody isoform, IgG, IgM, or IgA, the latex agglutination antibody assay is, in principle, isotope independent (Pavlova et al., 2020). This might contribute to its high sensitivity, especially on samples collected in the early phase of infection when IgG isotope switching has not yet occurred. It has been shown that the IgM antibody response precedes that of IgG (Pavlova et al., 2020; Ravi et al., 2020), and the pentameric architecture of IgM might make it a stronger promoter of agglutination than IgG. Moreover, the agglutination assay detects antibodies against the spike and the nucleocapsid in parallel, whereas most existing assays are specific for either protein. These factors explain why the agglutination assay could detect antibody in >92% of plasma samples collected on the day of NAAT+ diagnosis and 100% on day 2 and onward, whereas other antibody tests were less sensitive. The ability of our assay to detect antibodies at the early stages of infection suggests that it might be used to complement or confirm the diagnosis based on NAAT, which is prone to false positives or false negatives especially when done only once (Jarrom et al., 2020).
Several features of the agglutination assay make the latex agglutination assay a potential candidate for POC antibody testing. First, the assay is highly sensitive and accurate, with 100% specificity and ∼98% sensitivity on the samples tested in this study. Importantly, it detected antibodies in >92% of COVID-19 patients on the day of diagnosis. This sensitivity rivals that of NAAT tests (Jarrom et al., 2020). Second, the agglutination test is fast. It takes 2 min from mixing the plasma with the latex particles to obtain the result. Third, the agglutination assay is simple to run and instrument free. As the formation of latex bead clumps is easy to identify by the naked eye, the test can be performed without extensive specialized training. Fourth, because the agglutination assay takes only ∼5 μL of plasma, it can be developed into a finger-prick blood test suitable for use at home. Finally, the low cost of the latex agglutination assay makes it universally affordable and ideal for antibody testing at the community or population level in high- and low-resource settings alike. By combining with gel cards, the latex agglutination assay might be readily adopted in clinical labs (Alves et al., 2020). It might also be possible to develop the latex agglutination assay as an at-home antibody test by a combination of a finger-prick blood sample and a smartphone app (Mertz, 2020). Such a test would make it possible for individuals to monitor their antibody responses to vaccines over time.
Limitations of the study
The assignment of agglutination score based on imaging analysis makes it a challenge to apply in practice without training. Although it is possible to develop computer software to facilitate and simplify this process, a large number of samples (e.g., >1,000), including convalescent plasma with different levels of SARS-CoV-2 antibodies determined by an orthogonal method, would have to be tested by the agglutination assay to obtain sufficient data for training. The latex agglutination assay in the current setup showed significant background signals when unprocessed blood was used. This issue has to be resolved before the agglutination assay can be developed into a home test.
Data availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Shawn S.-C. Li (sli@uwo.ca)
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Spike-RBD antibody | Novus Biologicals | NBP2-90980 |
| Anti-nucleocapsid antibody | ThermoFisher Scientific | Cat# PA5-81794; RRID: AB_2788968 |
| Goat anti-human IgG HRP antibody | Millipore Sigma | Cat# AP113P; RRID: AB_92443 |
| Goat anti-rabbit IgG HRP | Bio-Rad | Cat# 170-6515; RRID: AB_11125142 |
| Chemicals, reagents and recombinant protein | ||
| Blue dyed polystyrene latex beads, 0.8 μm | Sigma Aldrich | L1398 |
| biotin using EZ-Link Sulfo-NHS-LC-LC-Biotin | ThermoFisher Scientific | A35358 |
| ChonBlock ELISA blocking and antibody dilution buffer | Chondrex Inc | 9068 |
| TMB substrate (3,3’,5,5’-Tetramethylbenzidine) | ThermoFisher Scientific | N301 |
| Spike Receptor Binding Domain (RBD) (14μM) | ThermoFisher Scientific | RP-87678 |
| Nucleocapsid (full length) (5μM) | RayBiotech | 230-01104 |
| ACE2 protein | This Study | |
| Software | ||
| Qupath | https://qupath.github.io/ | v0.1.2 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by Shawn Li (sli@uwo.ca).
Materials availability
Most of the materials used in this study are available from commercial sources. Materials generated specifically for this study are available upon request.
Data and code availability
This study did not generate sequence data or code.
Experimental model and subject details
Information about sex and the age/developmental stage of the patients donating samples are not disclosed, as the samples were de-identified.
Blood sample collection
Blood samples were collected following a protocol (study number: 116284) approved by the Research Ethics Board (REB) of Western University. The plasma samples were de-identified prior to transfer from the Laboratory of Clinical Medicine (London Health Sciences Center, London, Canada) to a biosafety Level 3 (CL3) lab (ImPaKT, Western University) following Transportation of Dangerous Goods (TDG) guidelines. All plasma samples were heat-inactivated at 56 oC for 30 minutes at the ImPaKT CL3 facility as per Western university biosafety regulation. Heat inactivated plasma samples were then transferred to the testing laboratory. We tested the effect of heat inactivation on SARS-CoV-2 antibody titer and found no significant impact of heat-inactivation (see also Figure S6).
Recombinant protein production and purification
The expression plasmid for human ACE2 cloned into the mammalian expression vector pαH (residues 1−615 with a C-terminal HRV3C protease cleavage site, a TwinStrepTag and an 8XHisTag) was a generous gift by Dr. McLellan. SARS-CoV-2 S-RBD cloned into pCAGGs the expression vector was received from Dr. Harding's lab. SARS-CoV-2 N-RBD was cloned into the pMCSG53 prokaryotic expression vector (residues 47-173-terminal 6x-His tag + TEV protease cleavage site).
Recombinant ACE2 and S-RBD proteins were produced by transient transfection of Expi293F cells (ThermoFisher Scientific, A14527) with a corresponding expression vectors and FectoPRO® DNA transfection reagent (Polyplus-transfection® SA, Cat. #116-010). Supernatants from transfected cells were harvested after 96 hours of the post-transfection time by centrifugation of the culture at 65,000 RPM for 30 min at 4oC. Cleared supernatant was then incubated with 4 ml TALON® Metal Affinity Resin (Takara Bio USA, Inc. Cat#635652) for 2h at 40C. Ni-NTA chromatography was used to purify His-tagged recombinant proteins. Each protein was dialyzed and concentrated in Amicon centrifugal units (EMD Millipore) in a final buffer of 20mM HEPES (pH 7.5, 200mM NaCl, pH 7.5, 5% glycerol).
Recombinant N-RBD was expressed in E. coli BL21(DE3)-Gold. Ni-NTA chromatography and size exclusion chromatography-Superdex S200 was used to purify N-RBD. The tag was cleaved using TEV followed by dialysis. The N-RBD protein was resuspended in 0.1 M NaCl, 20 mM HEPES pH 7.5 and stored at -80 oC until use. Recombinant nucleocapsid (residues 1-419) was obtained from RayBiotech (Cat #230-01104). Protein purity was confirmed by SDS-PAGE (Figure S1).
Method details
Preparation of SARS-CoV-2 antigen coated latex particles
Blue dyed polystyrene latex beads, 0.8 μm in diameter, were purchased from Sigma Aldrich (L1398). Prior to use, the latex beads were washed according to the manufacturer’s instructions with some modifications. Briefly, 2.5mL of 5% (w/v) latex suspension was washed twice in 10 mL PBS buffer (135 mM NaCl, 2.6 mM KCl, 8 mM Na2HPO4, and 1.5 mM KH2PO4, pH 7.4) by mixing and centrifuging the latex suspension at 3,000g for 10 minutes at room temperature. The beads were then resuspended with 2.5 ml 0.025M MES buffer (2-(N-Morpholino) ethanesulfonic acid, pH 6.0) to obtain 5% (w/v) suspension.
SARS-CoV-2 antigen-latex particle conjugates were prepared by passive adsorption following the procedures described by Mahat et al.(Mahat et al., 2014), with some modifications. Briefly, 0.4 mL of 5% (w/v) latex suspension was centrifuged at 3,000 g for 5 minutes at room temperature, and the supernatant was discarded. The beads were incubated with 200 μg recombinant Receptor Binding Domain of the SARS-CoV-2 spike protein (S-RBD) (Structural Genomics Consortium, University of Toronto) or the Nucleocapsid protein (N protein) (RayBiotech, 230-01104) in 4 mL MES buffer. The mixture was allowed to incubate for 24 hours at 4°C with periodic mixing. After conjugation, the antigen-latex bead conjugate was centrifuged, and the supernatant was kept for determination of unabsorbed protein concentration (Bio-Rad protein assay kit). The antigen-bead conjugate was washed twice with PBS and blocked for 30 min at room temperature in PBS containing 3% bovine serum albumin (BSA). The conjugate was then resuspended at 2.5% (w/v) in PBS containing 1% BSA and stored at 4°C until use.
Agglutination assay for SARS-CoV-2 antibody testing and data interpretation
For the agglutination assay, 5 μl plasma was mixed with 25 μl antigen-coated beads (2.5%, w/v) per assay. The agglutination was allowed to proceed for 2 min at room temperature before imaging with a camera. The relative degree of agglutination induced by the SARS-CoV-2 antibody was measured by the area of clump formation based on the corresponding image. Agglutination data analyses were performed using qualitative and semi-quantitative assessments. For semi-quantification of agglutination, the image analysis software Qupath (v0.1.2) was used (https://qupath.github.io/) and quantification was done by calculating the percentage of agglutination based on estimated agglutination/clumps area (mm2) relative to the total latex reaction area. In qualitative assessments, agglutination intensity was inspected visually, and agglutination score was assigned (i.e. 1, 2, 3 and 4). Specifically, 1 corresponds to small clumps with ∼25% agglutination, 2 (∼50% agglutination), 3 (∼75% agglutination), and 4 (large clumps that forms in less than 1 min with ∼100% agglutination).
The cut-off value (5%) for positivity in the agglutination assay using either S-RBD- or N-coated latex particles was based on testing results using pre-COVID-19 (n=100, collected in 2018) and COVID-19- (n=121, NAAT negative) plasma samples and background binding signals obtained using latex particles without immobilized antigens. The degree of agglutination for both the pre-COVID-19 and COVID-19- samples was in the range of 0-4% (Figures 3A and 3B). The non-specific micro-agglutination for the latex particles without antigens was determined to be <5% based on image analysis (n=390; Figure S5).
Preparation of red blood cells conjugated with SARS-CoV-2 antigen
The recombinant spike receptor-binding domain (S-RBD) or the nucleocapsid RNA-binding domain (N-RBD) was conjugated in 30-fold molar excess biotin using EZ-Link Sulfo-NHS-LC-LC-Biotin (Thermo Scientific, A35358). Excess unbound biotin was removed using ZebaTM Spin Desalting Columns, 7KMWCO (Thermo Scientific, 89890). Anti-D-IgG was purified from Immucor Anti-D Series 4 (IgG & IgM monoclonal blend) by using protein A magnetic affinity purification (G8782, Promega). The purified anti-D-IgG was then concentrated (3mg/ml) and stored at 4°C until use. Anti-D was then conjugated with streptavidin according to manufacturer instruction (ab102921, abcam).
Bioconjugation of Anti-D-IgG-streptavidin with Reagent Red Blood Cells (RRBC) [0.8% R2R2; blood group O; Rh/D-antigen+] (Ortho-Clinical Diagnostics SELECTOGEN, 6902315) was done by incubating the anti-D-IgG-streptavidin with RRBC for 30 min at room temperature. The RRBC-anti-D-streptavidin complex was then washed twice with low ionic strength RBC diluent (MTS™ Diluent 2 PLUS; Micro Typing Inc., MTS9330S). The complex was centrifuged at 1000g for 2 min to remove unbound anti-D-IgG streptavidin and was then resuspended in the same RBC diluent. RBC-anti-D-IgG-streptavidin was then conjugated with either biotin-S-RBD or biotin-N-RBD for 15 min at room temperature. The RRBC-anti-D-sterptaviding-biotin-S-RBD/N-RBD was stored at 4°C until use. The RRBC agglutination assay was carried out in the same way as for latex agglutination described above.
S-RBD-ACE2 binding ELISA and surrogate neutralization assay
ELISA plate Coating and blocking
S-RBD was dissolved (5 μg/ml) in Tris buffer saline (TBS) (20 mM Tris, 150 mM NaCl, pH7.4) and 100 μl of the S-RBD solution was added to each well of an ELISA plate and incubate at 4°C overnight with slow shaking. The antigen-coated wells were washed 3 times with TBS-tween (TBST) (20 mM Tris, 150 mM NaCl, 0.1% Tween 20).
The S-RBD coated wells were blocked by 100 μl of the ChonBlock™ blocking/sample dilution ELISA buffer (Chondrex, Inc., 9068) for 1 hour at room temperature with slow shaking followed washing 3 times with TBST.
ACE2:S-RBD binding assay
ACE2 was biotinylated as described above. Biotin-ACE2 (1μg/m) was added to S-RBD-coated plate after blocking and incubated for 1hour at room temperature. The wells were washed 3 times with TBST to remove unbound biotin-ACE2. Streptavidin-HRP (1000-fold dilution with Chonblock blocking buffer) was then added to each well and incubated for 1hour at room temperature. The wells were washed 3 times with TBST and TMB substrate (3,3’,5,5’-Tetramethylbenzidine, Thermo Scientific, N301) was added for reaction development and 0.18 M H2SO4 was used to stop reaction. Absorbance at 450nm was measured to detect the S-RBD bound ACE2.
SARS-CoV-2 antibody neutralization assay
Plasma was diluted 1:100 and incubated with S-RBD-coated wells (blocked) for 1hour at room temperature. The wells were washed three times with TBST. Biotin-ACE2 was then added to the wells and incubated for 1 hour at room temperature followed by washing, reaction development and detection as described above.
Quantification and statistical analysis
All statistical analyses were done using the GraphPad Prism9 software. Specifically, the Hill coefficient (h) was calculated from fitting agglutination data obtained using anti-S-RBD and anti-N antibodies to the Hill equation. COVID-19+ samples with distinct agglutination scores and COVID-19− samples were analyzed using unpaired t-test with Welch's correction (no assumption of equal SD between two groups). Changes in agglutination for samples before and after heat-inactivation were analyzed by paired t-test. Spearman’s correlation rank was done to study correlation between antibody titter and ACE2:S-RBD neutralization efficiency. The number of biological and technical replicates are indicated in the figure legends.
Acknowledgments
We thank the Center for Structural Genomics of Infectious Diseases (CDGID) for the N-RBD expression plasmid, Dr. Jason McLellan for expression plasmid for human ACE2, Dr. Shane Harding for the S-RBD expression construct, and Yanjun Li for technical assistance. This work was supported by a grant (to S.S.-C.L., I.C.-Y., M.J.K., B.C.-Y., and V.B.) from Ontario Ministry of Colleges and Universities COVID-19 Rapid Research Fund and by the Toronto COVID-19 Action Fund. S.E. was supported by a Post-Doctoral Fellowship from the National Science and Engineering Council of Canada. S.S.-C.L. held a Canada Research Chair in Molecular and Epigenetic Basis of Cancer.
Author contributions
S.S.-C.L., I.C.-Y., and S.E. conceived and designed the study. S.E. performed the agglutination and neutralization assay and data analysis. M.J.K. and H.A. conducted serological antibody testing by ELISA. C.V. helped with protein and plasma sample quality control. P.S., A.S., A.H., F.Y., T.S., E.E., and S.A. contributed to subcloning, expression, and purification of SARS-CoV-2 proteins and human ACE2. V.B., M.J.K., L.L., H.A., and B.D.H. contributed to patient recruitment, blood sample collection, and NAAT and ELISA testing. S.E., S.S.-C.L., I.C.-Y., and B.C.-Y. interpreted the data. S.S.-C.L. and S.E. wrote the manuscript with input from B.C.-Y., I.C.-Y., and M.J.K.
Declaration of interests
S.E., C.V., and S.S.-C.L. are co-inventors in a US Provisional patent application on the agglutination assay submitted by Western University.
Published: June 21, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2021.100011.
Supplemental information
References
- Abe K.T., Li Z., Samson R., Samavarchi-Tehrani P., Valcourt E.J., Wood H., Budylowski P., Dupuis A.P., 2nd, Girardin R.C., Rathod B., et al. A simple protein-based surrogate neutralization assay for SARS-CoV-2. JCI Insight. 2020;5 doi: 10.1172/jci.insight.142362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves D., Curvello R., Henderson E., Kesarwani V., Walker J.A., Leguizamon S.C., McLiesh H., Raghuwanshi V.S., Samadian H., Wood E.M., et al. Rapid gel card agglutination assays for serological analysis following SARS-CoV-2 infection in humans. ACS Sens. 2020;5:2596–2603. doi: 10.1021/acssensors.0c01050. [DOI] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Chaudhary V.K. Whole-blood agglutination assay for on-site detection of human immunodeficiency virus infection. J. Clin. Microbiol. 2003;41:2814–2821. doi: 10.1128/JCM.41.7.2814-2821.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X., et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Hursh D.A., Sun R., Iltis J.P., Rice D.H., Gleaves C.A. Evaluation of a latex particle agglutination assay for the detection of cytomegalovirus antibody in patient serum. J. Clin. Microbiol. 1989:2878–2879. doi: 10.1128/jcm.27.12.2878-2879.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaaskelainen A.J., Kuivanen S., Kekalainen E., Ahava M.J., Loginov R., Kallio-Kokko H., Vapalahti O., Jarva H., Kurkela S., Lappalainen M. Performance of six SARS-CoV-2 immunoassays in comparison with microneutralisation. J. Clin. Virol. 2020;129:104512. doi: 10.1016/j.jcv.2020.104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrom D., Elston L., Washington J., Prettyjohns M., Cann K., Myles S., Groves P. Effectiveness of tests to detect the presence of SARS-CoV-2 virus, and antibodies to SARS-CoV-2, to inform COVID-19 diagnosis: a rapid systematic review. BMJ Evid. Based Med. 2020 doi: 10.1136/bmjebm-2020-111511. [DOI] [PubMed] [Google Scholar]
- Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer M.J., Hedley B.D., Bhayana V., Payne M., Chin-Yee I., Delport J. Interim analysis of the clinical performance of five SARS-Cov-2 serology assays. Clin. Biochem. 2020 doi: 10.1016/j.clinbiochem.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruttgen A., Cornelissen C.G., Dreher M., Hornef M., Imohl M., Kleines M. Comparison of four new commercial serologic assays for determination of SARS-CoV-2 IgG. J. Clin. Virol. 2020;128:104394. doi: 10.1016/j.jcv.2020.104394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W., et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020 doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisboa Bastos M., Tavaziva G., Abidi S.K., Campbell J.R., Haraoui L.P., Johnston J.C., Lan Z., Law S., MacLean E., Trajman A., et al. Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ. 2020;370:m2516. doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahat M., Abdullah W.Z., Che Hussin C.M. Conventional rapid latex agglutination in estimation of von Willebrand factor: method revisited and potential clinical applications. J. Immunol. Res. 2014;2014 doi: 10.1155/2014/850810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz L. New, at-home antibody test for detecting, tracking COVID-19. IEEE Pulse. 2020;11:28–31. doi: 10.1109/MPULS.2020.3022203. [DOI] [PubMed] [Google Scholar]
- Pavlova I.P., Nair S.S., Kyprianou N., Tewari A.K. The rapid coronavirus antibody test: can we improve accuracy? Front Med. (Lausanne) 2020;7:569. doi: 10.3389/fmed.2020.00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeling R.W., Wedderburn C.J., Garcia P.J., Boeras D., Fongwen N., Nkengasong J., Sall A., Tanuri A., Heymann D.L. Serology testing in the COVID-19 pandemic response. Lancet Infect. Dis. 2020;20:e245–e249. doi: 10.1016/S1473-3099(20)30517-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J., Wu C., Li X., Zhang G., Jiang Z., Li X., Zhu Q., Liu L. Profile of IgG and IgM antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect Dis. 2020 doi: 10.1093/cid/ciaa489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi N., Cortade D.L., Ng E., Wang S.X. Diagnostics for SARS-CoV-2 detection: a comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens. Bioelectron. 2020;165:112454. doi: 10.1016/j.bios.2020.112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 infection in convalescent individuals. bioRxiv. 2020 doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., Wang F., Li D., Yang M., Xing L., et al. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanh Le T., Andreadakis Z., Kumar A., Gomez Roman R., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- Tortorici MA B.M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., McCallum M., Bowen J., Minola A., Jaconi S., Zatta F., et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020:eabe3354. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg S.P., Connors T.J., Zhu Y., Baldwin M.R., Lin W.H., Wontakal S., Szabo P.A., Wells S.B., Dogra P., Gray J., et al. Distinct antibody responses to SARS-CoV-2 in children and adults across the COVID-19 clinical spectrum. Nat. Immunol. 2020 doi: 10.1038/s41590-020-00826-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman J.D., Hiatt J., Mowery C.T., Shy B.R., Yu R., Yamamoto T.N., Rathore U., Goldgof G.M., Whitty C., Woo J.M., et al. Test performance evaluation of SARS-CoV-2 serological assays. medRxiv. 2020 doi: 10.1101/2020.04.25.20074856. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Shawn S.-C. Li (sli@uwo.ca)
This study did not generate sequence data or code.