Abstract
Coronavirus disease 2019 (COVID-19) is an infectious disease caused by SARS-CoV-2, a newly discovered coronavirus that exhibits many similarities with the severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) coronaviruses (SARS-CoV and MERS-CoV, respectively). The definite pathogenesis and immunological influences of SARS-CoV-2 have not been fully elucidated. Therefore, we constructed a brief summary comparison of SARS-CoV-2, SARS-CoV, and MERS-CoV infections regarding their immunological changes. In addition, we further investigated the immunological differences between severe and nonsevere COVID-19 cases, and we searched for possible immunological predictors of the patient outcome by reviewing case series studies to date. Possible immunological predictors of a poor outcome are leukocytosis, neutrophilia, lymphopenia (both CD4 and CD8 T cells), an increased neutrophil-to-lymphocyte ratio (NLR), and increased levels of pro-inflammatory cytokines (IL-6 and TNF-α), Th1 cytokines (IL-2 and IFN-γ), regulatory T cell cytokines (IL-10) and Th17 cytokines (IL-17). A more precise immunological map needs to be established, which may assist in diagnosing this disease and facilitate immunological precision medicine treatment.
Keywords: COVID-19, Immune, Pandemic, Pneumonia, SARS-CoV-2
Coronavirus disease 2019 (COVID-19) is an infectious disease caused by SARS-CoV-2, a virus closely related to severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV). The world experienced outbreaks of coronavirus infections that threatened to become global pandemics in 2002–2003 for SARS and in 2011 for Middle East respiratory syndrome (MERS). As the world is witnessing the COVID-19 epidemic, the disease caused by the novel coronavirus SARS-CoV-2, emerging genetic evidence suggests it has many similarities to SARS and MERS. To date, there is no available medication for the treatment of SARS-CoV-2 infection. A precise immunological map of SARS-CoV-2 infection is critical to recognize the host defense in patients with different prognoses or outcomes and becomes basis for immunological precision medicine in the treatment of SARS-CoV-2 infection.
Comparison of possible involved systems in SARS-CoV-2, SARS-CoV, and MERS-CoV infections
Coronaviruses can infect humans and many species of animals. Common cold human coronaviruses consist of four viruses that cause worldwide mild upper airway symptoms and are responsible for up to 15% of common cold infections.1 Other coronaviruses such as SARS-CoV, MERS-CoV, and SARS-CoV-2, caused epidemic outbreaks in the 21st century with high infection to fatality ratios. Each of these coronaviruses can cause respiratory, enteric, hepatic, and neurological diseases.2
Due to limited clinical evidence from cell line studies, we reviewed current autopsies and laboratory cell line cultures to identify possible affected systems and cells in SARS-CoV-2 infection (Table 1 ).2, 3, 4, 5, 6, 7, 8, 9, 10 SARS-CoV and SARS-CoV-2 share the same cell surface receptor, angiotensin-converting enzyme 2 (ACE2), which is predominantly expressed on lung type II alveolar cells and minimally expressed in alveolar epithelial cells, type 2 pneumocytes, lung macrophages, and monocytes.2 , 3 The results showed that the respiratory tract with alveolar epithelium cell involvement is the most common and that the immune and digestive systems are also involved. In addition, a positive SARS-CoV-2 antigen with a real-time PCR nucleic acid signal was noted in both the alveolar epithelium and macrophages in one autopsy study.4 Central venous symptoms, including headache, dizziness, change in mental status, and meningeal signs, are also common.6 In addition, gastrointestinal symptoms, including anorexia and abdominal pain, are more common in severe cases.9 In the genital-urinary system, acute kidney injury with renal tubule involvement is the most common symptom.6 Cardiovascular complications most often present with acute myocardial injury.4 , 6
Table 1.
Possible systems involved in SARS-CoV-2, SARS-CoV and MERS-CoV infections.
| Involved system | SARS-CoV-2 | SARS-CoV | MERS-CoV |
|---|---|---|---|
| Cell surface receptor2,3 | Human ACE2 | Human ACE2 | Human DPP4 |
| Mortality rate2 | Lowest | Middle | Highest |
| Immune system | +4‡ | +2†,5† | +2†,5† |
| Alveolar macrophage cells4‡ | Tissue-resident macrophages2†,5† | Tissue-resident macrophages (lung, skeletal muscle)2† | |
| Monocytes5† | |||
| T lymphocytes5† | |||
| Histiocytic cell lines5† | |||
| Respiratory system | +4‡,6‡,7‡ | +2†,5† | +2†,3†,5† |
| Alveolar epithelial cells5‡ | Respiratory alveolar epithelial cells2†,5† | Pneumocytes3† Multinucleated epithelial cells | |
| Bronchial submucosal gland cells2†,3†,5† | |||
| Neurological system | +6‡ | +3† | +5† |
| Neurons in the brain3† | Neurons in the brain5† | ||
| Digestive system | –4‡; +6‡,7‡,8† | +3†,5† | +3†,5† |
| Liver7‡ | Intestinal mucosa3† | Intestinal mucosa3† | |
| Liver epithelium6† | Liver epithelium5† | ||
| Genitourinary system | –4‡; +6‡ | +2†,5† | +2†,5† |
| Renal distal tubule epithelium2† | Renal proximal tubular epithelial cells2† | ||
| Kidney5† | Kidney and prostate5† | ||
| Cardiovascular system | –4‡,6‡,7‡ | +10a | +10a |
Note: Evidence of COVID-19 is presented with autopsy data due to the lack of a recent cell line study.
+: affected according to cell line susceptibility data (in vitro) † or pathological findings on autopsy ‡
–: not affected according to pathological findings on autopsy.
No current cell line susceptibility or autopsy data available (only anecdotal evidence).
In contrast to SARS-CoV and SARS-CoV-2, MERS-CoV uses dipeptidyl peptidase 4 (DPP4) as a specific entry receptor, which is widely expressed on epithelial cells in the kidney, alveoli, small intestine, liver, prostate, and leukocytes3; therefore, MERS-CoV has a broader infection range and greater disease severity than other coronaviruses. The mortality rate of SARS-CoV-2 infection (2.15%, data obtained at the World Health Organization website on April 14, 2021) is far lower than that of SARS (9.19%) and MERS (34.4%).2 , 11
Host-pathogen interactions and initial immunological responses in COVID-19
The clinical presentations of SARS-CoV-2, SARS-CoV, and MERS-CoV show similarities and they vary from asymptomatic infection to severe disease. The transmission and penetration of asymptomatic COVID-19 patients may be highest among the three diseases; thus, SARS-CoV-2 can disseminate markedly and cause a pandemic infection if public health isolation policies are not sufficient. Due to the normally benign nature of human coronavirus infection, Lavine et al. proposed that if the endemic phase of COVID-19 with primary childhood exposure is reached, the virulence of SARS-CoV-2 infection would be no more than the common cold.12
In SARS-CoV-2 infection, the virus binds to the host cell via the ACE2 receptor with the help of transmembrane protease serine 2 (TMPRSS2), a serine protease for viral S protein priming. Then, the virus will replicate in the host cell and cause massive destruction of the affected tissues by cell pyroptosis, especially in organs with high ACE2 expression, such as respiratory tract cells.13 , 14 In infected host cells, the virus may be recognized by pattern recognition receptors (PRRs) including Toll-like receptor 3 and 7 (TLRs). The TLR3 response also induces NLR family pyrin domain containing 3 (NLRP3) inflammasome activation, which leads to the production of proinflammatory cytokines including IL-1βand IL-18, and promotes host cell pyroptosis.13
Comparison of the effects of SARS-CoV-2, SARS-CoV, and MERS-CoV infection on the immune cell response and cytokine expression in humans
We summarized several review articles and clinical studies that investigated the differences in the immune responses among SARS-CoV-2, SARS-CoV, and MERS-CoV infections (Table 2 ).1 , 2 , 15, 16, 17, 18, 19 All three coronavirus infection patients had decreasing trends in neutrophils and lymphocytes (including CD4+ T cells and CD8+ T cells). In SARS-CoV-2 infection, 35–63% of patients had lymphopenia (Table 2). The levels of proinflammatory cytokines, especially IL-6 and TNF-α, were significantly increased in all three viral infections. The levels of T helper 1 cytokines, including IL-2 and interferon-γ (IFN-γ), and the Th17 cytokine IL-17, were significantly elevated in all three viral infections. Notably, monocyte chemoattractant protein-1 (MCP-1), macrophage inflammatory proteins alpha (MIP-1α), IL-8, and IL-12 expression were higher in MERS-CoV patients than in SARS-CoV patients.16 The immune system may be important for the elimination of SARS-CoV-2; however, inappropriate immune responses (e.g., a cytokine storm) may result in fatal disease.
Table 2.
Effects of SARS-CoV-2, SARS-CoV and MERS-CoV infections on immune cell responses and cytokine expression in humans.
| SARS-CoV-2 | SARS-CoV | MERS-CoV | |
|---|---|---|---|
| Affected immune cell parameter | |||
| Neutrophil count | ↑ or ↓a | ↓2 | ↓2 |
| Lymphocyte count | – or ↓a | ↓2 | ↓2 |
| CD4+ T cell count | – or ↓a | ↓2 | ↓2 |
| CD8+ T cell count | – or ↓a | ↓2 | ↓1 |
| Natural killer cell count | – or ↓a | ↓16 | NA |
| B cell count | – or ↓a | NA | NA |
| T cell apoptosis | –1 | +1 | +1 |
| Cytokines | |||
| Pro-inflammatory cytokines | |||
| IL-1 | ↑ or – | ↑1, 2, 3 | ↑18 |
| IL-6 | ↑ or –a | ↑1, 2, 3 | ↑1,3 |
| IL-8 | ↑ or – | ↑2,3 | ↑1,3 |
| TNF-α | ↑ or –a | ↑1,3 | ↑1,3 |
| Th1 cytokines | |||
| IL-2 | ↑ or –a | ↑3 | –19 |
| IL-12 | – | ↑1,3 | ↑3 |
| IFN-γ | ↑ or – | ↑1,3 | ↑3 |
| Th2 cytokines | |||
| IL-4 | ↑ or –a | ↑1 | –19 |
| IL-5 | – | ↑1 | –19 |
| Th17 cytokines | |||
| IL-17 | ↑ | ↑1 | ↑1,19 |
| Treg cytokines | |||
| IL-10 | ↑ or –a | ↑1,2 | ↑1 |
| Cytokine storm | Yes2,3 | Yes2,3 | Yes2,3 |
Note: The data on COVID-19 were collected from original case series reports.
↑: increase due to infection.
↓: decrease due to infection.
–: no significant effect due to infection.
NA: data not currently available.
Effect may differ according to disease severity (Table 3).
The possible mechanism of lymphopenia may come from defective T cell activation/proliferation and T cell apoptosis. Diao B et al. reported that serum concentrations of TNF-α, IL-6, and IL-10 were negatively correlated with the numbers of T cells, CD4+, and CD8+ cells in COVID-19 patients, suggesting that high TNF-α, IL-6, and IL-10 expression may negatively regulate T cell survival and proliferation.20 Whether SARS-CoV-2 infections induce T cell apoptosis and contribute to lymphopenia is still unknown.
On the other hand, T cells undergo functional exhaustion in COVID-19 patients, which diminishes host antiviral activity. The inhibitory factor PD-1 was highly expressed in T cells of COVID-19 patients compared to T cells of healthy controls. PD1+CD8+ T cell levels were significantly increased in intensive care unit (ICU) patients compared with those in non-ICU patients and healthy controls.20 Several T cell functional molecules, such as IFNγ and TNFα in CD4+ T cells were lower in the severe group than in the mild group.21 The levels of granzyme B and perforin in CD8+ T cells were higher in the severe group than in the mild group.21 This finding indicates that SARS-CoV-2 damages the function of CD4+ T cells and promotes excessive activation and possibly subsequent exhaustion of CD8+ T cells. Moreover, the levels of multifunctional CD4+ T cells significantly decreased in the severe group, whereas the proportion of nonfunctional subsets increased. The elevated exhaustion level and reduced functional diversity of T cells may serve as a predictive marker for severe progression in COVID-19 patients.21 In both SARS-CoV and SARS-CoV-2 infection, the levels of multifunctional CD4+ T cells were increased in severe cases compared with moderate cases.21 , 22 The reduced functional diversity of T cells may be a unique immune response to SARS-CoV-2 infection compared to that of other coronaviruses.
Cytokine profiles and cytokine storms in SARS-CoV-2 infection
In SARS-CoV and MERS-CoV infection, delayed or suppressed type I IFN induction in host cells has been found to be an early important immunopathological feature, and this phenomenon is also observed in SARS-CoV 2 infection. These coronaviruses employ multiple strategies to interfere with the signaling leading to type I IFN production and/or the signaling downstream of the interferon-α/β receptor (IFNAR).1 , 13 This dampening strategy is closely associated with disease severity.13 As a consequence, strategies to boost immune responses (antisera or pegylated IFNα) at an early stage may be important.
For the development of an endogenous protective immune response in the incubation and nonsevere stages, the host should be in good general health and have an appropriate genetic background (e.g., HLA) that elicits specific antiviral immunity. However, when the protective immune response is impaired, the virus will propagate, and massive destruction of the affected tissues will occur, especially in organs that have high ACE2 expression, such as the intestine and kidney.
A cytokine storm is a potentially fatal hyperrelease of inflammatory mediators and cytokines in response to stimulation of T cells and macrophages by pathogens. An early rapid increase in the serum levels of proinflammatory cytokines was also observed in SARS-CoV and MERS-CoV infection, suggesting a potential similar cytokine storm-mediated disease severity.23 The T cell response in SARS-CoV infection was extensively investigated, and the data show that strong T cell responses were correlated significantly with increased neutralizing antibody levels, while higher serum Th2 cytokine levels (IL-4, IL-5, and IL-10) were detected in the fatal group.22 In MERS-CoV infection, an early rise in CD8+ T cells correlated with disease severity, and in the convalescent phase, Th1 cells were dominant.24
Cytokine storms also play an important role in fatal COVID-19. Forty-one hospitalized patients with high levels of proinflammatory cytokines, including IL-2, IL-7, IL-10, interferon gamma-induced protein 10 (IP-10), MCP-1, MIP-1α, and TNF-α, were observed in severe COVID-19 cases; these findings are in line with SARS and MERS in the presence of lymphopenia.23 Cytokine storms can initiate viral sepsis and inflammatory-induced lung injury, which leads to other complications, including pneumonitis, acute respiratory distress syndrome, respiratory failure, shock, organ failure, and potentially death.15 , 23 Further autopsy or biopsy studies are necessary to understand additional details.
Relationship between the immune reaction and disease severity in COVID-19 patients
We performed a literature review in PubMed of confirmed COVID-19 case series to investigate the relationship between immune reactions and disease severity in COVID-19 patients (Table 3 ).10 , 23 , 25, 26, 27, 28, 29, 30 Compared to nonsevere cases, severe COVID-19 cases had a significant increase in leukocyte and neutrophil counts; on the other hand, decreased total lymphocyte (both CD4+ and CD8+ T cell) and NK cell and normal or decreased B cell and eosinophil counts were also found. Elevated levels of proinflammatory cytokines (IL-6 and TNF-α), Th1 cytokines (IL-2 and IFN-γ), regulatory T cell cytokines (IL-10), and Th17 cytokines (IL-17) were also observed. Th2 cytokines (IL-4) elevation may be noted occasionally. In conclusion, these findings suggest that Th1 and Th17 responses have an important role in COVID-19 severity.
Table 3.
Relation between in immune reaction and disease severity in COVID 19 patients.
| Disease severity | Non-severe | Severe | ||||
|---|---|---|---|---|---|---|
| Immune cell | ||||||
| WBC count | ↑NA | –10,25, 26, 27, 28, 29 | ↓23,30 | ↑23,25, 26, 27, 28 | –29,30 | ↓NA |
| Neutrophil count | ↑NA | –25, 26, 27, 28,30 | ↓23,28 | ↑23,25,26,28,30 | –29 | ↓NA |
| Lymphocyte count | ↑NA | –29 | ↓23,25, 26, 27, 28,30 | ↑NA | –29 | ↓23,25, 26, 27, 28,30 |
| CD 4+ T cell count | ↑NA | –28 | ↓25,26,30 | ↑NA | –NA | ↓25,26,30a |
| CD 8+ T cell count | ↑NA | –28 | ↓25,26,30 | ↑NA | –NA | ↓25,26a,30a |
| NK cell count | ↑NA | –25 | ↓26,30,a | ↑NA | –25 | ↓26a,30 |
| NLR | ↑26,30 | –NA | ↓NA | ↑26,30 | –NA | ↓NA |
| Regulatory T cells | ↑30 | –NA | ↓25,26 | ↑30 | –NA | ↓25,26 |
| B cell count | ↑NA | –25,26 | ↓30,a | ↑NA | –25,26 | ↓30 |
| Monocyte count | ↑NA | –26,28 | ↓NA | ↑NA | –26,28 | ↓NA |
| Basophil | ↑NA | –26 | ↓NA | ↑NA | –26 | ↓NA |
| Eosinophil | ↑NA | –27 | ↓26,28 | ↑NA | –27 | ↓26,28 |
| Cytokine | ||||||
| Pro-inflammatory Cytokine | ||||||
| IL-1β | ↑23 | –25,26,29 | ↓NA | ↑23a | –25,26,29 | ↓NA |
| IL-6 | ↑23,25,26,28,a | –30 | ↓NA | ↑23,25,26,28,29,30,a | –NA | ↓NA |
| IL-8 | ↑23 | –26,29 | ↓NA | ↑23a | –26,29 | ↓NA |
| TNF-α | ↑23,26 | –25,28, 29, 30 | ↓NA | ↑23,25,26,30 | –,28,29 | ↓NA |
| Th 1 Cytokine | ||||||
| IL-2R | ↑23,26 | –25,28, 29, 30 | ↓NA | ↑23,25,26,30 | –28,29 | ↓NA |
| IL-12 | ↑NA | –23 | ↓NA | ↑NA | –23 | ↓NA |
| IFN-γ | ↑23 | –30 | ↓NA | ↑23 | –30 | ↓NA |
| Th 2 Cytokine | ||||||
| IL-4 | ↑26,30 | –23,28 | ↓NA | ↑23a | –28,30 | ↓NA |
| IL-5 | ↑NA | –NA | ↓NA | ↑NA | –23 | ↓NA |
| IL-9 | ↑23 | –NA | ↓NA | ↑23a | –NA | ↓NA |
| Th 17 Cytokine | ||||||
| IL-17 | ↑23a | –NA | ↓NA | ↑23a | –NA | ↓NA |
| T reg Cytokine | ||||||
| IL-10 | ↑23,28,29a | –25,26,29 | ↓NA | ↑23,25,28,30 | –26,29 | ↓NA |
| Immunoglobulins | ||||||
| IgG | ↑NA | –26 | ↓NA | ↑NA | –26 | ↓NA |
| IgA | ↑NA | –26 | ↓NA | ↑NA | –26 | ↓NA |
| IgM | ↑NA | –26 | ↓NA | ↑NA | –26 | ↓NA |
| Complement | ||||||
| C3 | ↑NA | –26 | ↓NA | ↑NA | –26 | ↓NA |
| C4 | ↑NA | –26 | ↓NA | ↑NA | –26 | ↓NA |
Note: Data about COVID-19 infection are collected from original case series with reference number properly cited.
WBC: White blood cell.
NLR: Neutrophil to lymphocyte ratio.
Regulatory T cells (CD3+CD4+CD25 + CD127low+) T cell.
NA: no study available.
Definition of COVID-19 severe case.
1. Respiration rate ≥30 times/min; at rest, oxygen saturation ≤93%; arterial partial pressure of oxygen (PaO2)/fraction of inspired oxygen (FiO2) ≤ 300 mmHg).
2. Patient admitted to ICU.
↑: significant increased compared with healthy people and non-severe patients.
↓: significant decreased compared with both healthy people and non-severe patients.
–: no significant difference compared with healthy people and non-severe patients.
Increased compared with healthy people, but no significant difference compared with non-severe patients.
In Th17 responses, increased IL-17 secretion can further induce the production of proinflammatory cytokines IL-1β, IL-6, and TNF-α, with chemokines. Furthermore, proinflammatory cytokines such as IL-1β and TNF-α could promote the Th17 response in turn. Th17 cells can promote eosinophil production and recruitment into the lungs to induce lung allergic disease. Furthermore, studies have demonstrated that IL-6 is essential in SARS lung pathology; therefore, this finding could explain the important role of the Th17 response in COVID-19 lung disease.31
Zhou P reported a clinical analysis of 99 cases in Wuhan that showed increased total neutrophils, reduced total lymphocytes, increased serum IL-6, and increased C-reactive protein (CRP).32 In severe or lethal cases of SARS-CoV or MERS-CoV infection, increased neutrophil and monocyte-macrophage influx have been consistently observed. Another report also revealed significantly increased total neutrophils and decreased total lymphocytes in patients receiving ICU care compared to those in patients receiving non-ICU care; furthermore, increased neutrophil and decreased lymphocyte counts also correlate with disease severity and death.33 Since lymphocytopenia is often seen in severe COVID-19 patients, the severe and fatal cases caused by SARS-CoV-2 infection may be mediated by leukocytes other than T cells. Furthermore, the neutrophil-to-lymphocyte ratio (NLR) and lymphocyte-to-C-reactive protein ratio (LCR) are established inflammation markers that reflect the systemic inflammatory response, and both tests are available in almost all hospital laboratories. NLR values increased significantly in severe COVID-19 patients, while LCR values decreased significantly. Increased NLR values and decreased LCR levels may indicate a poor prognosis.23 , 27
A recent review of antibody mediated immunity to coronaviruses revealed that a slower antibody response may be associated with more severe disease in MERS-CoV and SARS-CoV-2 infections.34 In addition, the median time to detect different antibodies was shortest for SARS-CoV-2, followed by SARS-CoV, and the longest time was seen for MERS-CoV infection.34 These findings could partially explain the differences in the disease severity among these three coronaviruses. Therefore, a delayed antibody response may be considered a risk factor for a poor prognosis.
Multisystem inflammatory syndrome in children (MIS-C) with SARS-CoV-2 infection
During the worldwide SARS-CoV-2 pandemic, multisystem inflammatory syndrome was first diagnosed as hyperinflammatory shock. MIS-C shares partially similar symptoms but is distinct from typical Kawasaki disease. A systemic review of 953 MIS-C cases worldwide showed that MIS-C has a higher median age of onset than typical KD. MIS-C is distinct from KD, with a higher rate of shock, intensive care treatment, coronary dilatation, aneurysm formation, and mortality rates.35
MIS-C presents with more systemic inflammation (higher leukocyte counts and CRP), more frequent lymphocytopenia with thrombocytopenia, and higher myocardial injury markers including troponin I, BNP (B-type natriuretic peptide), and coagulopathy (D-dimers), than typical KD.35 The immunological characteristics of MIS-C are similar to those of KD, including elevated IL-6 and CXCL-10 levels, which contribute to most cytokine storms.36 However, significantly elevated IL-17 A was only observed in KD but not MIS-C.36
SARS-CoV-2 infection may be associated with the development of systemic autoimmune disease
Recent studies have revealed that autoantibodies may contribute to the immunopathology of SARS-CoV-2 infection. In a Greek study, a high percentage (68.7%) of COVID-19 patients admitted to the intensive care unit without previous systemic autoimmune disease had systemic autoantibodies, including antinuclear antibodies (34.5%), anti-neutrophil cytoplasmic antibodies (13%), anti-cardiolipin (aCL, 24%), and antibodies against b2-glycoprotein I (anti-b2-GPI, 34.5%).37 This observation suggests a link between SARS-CoV-2 infection and autoimmune activation. Several possible mechanisms have been proposed, including molecular mimicry, epitope spreading, and bystander activation of autoreactive T or B cells, which may contribute to this autoimmune reactivity.38
Current medications for COVID-19 treatment
Hydroxychloroquine and chloroquine
Hydroxychloroquine (HCQ) and chloroquine (CQ) are antimalarial drugs successfully applied to treat parasite and viral infections; HCQ has also been used as an antirheumatic agent for systemic lupus erythematosus and rheumatoid arthritis.39 HCQ and CQ inhibit viral infection by altering the glycosylation of ACE2 receptors, which are used as SARS-CoV inhibitors. However, there is no consistent clinical evidence to support HCQ and CQ as standard treatments for COVID-19. The use of HCQ and CQ for SARS-CoV-2 therapy should be considered carefully due to lethal adverse effects such as cardiotoxicity and QTc interval prolongation.39
Dexamethasone
Dexamethasone is a corticosteroid with anti-inflammatory and immunosuppressant activity; its anti-inflammatory effect may reduce the collection of exudates in lung alveoli to prevent alveolar damage and inhibit acute respiratory distress. Currently, there are no available clinical trials of dexamethasone for the treatment of SARS-CoV-2 infection. However, there is evidence that the use of dexamethasone may significantly decrease the mortality rate in critical COVID-19 patients on mechanical ventilation.40
Remdesivir
Remdesivir is a nucleoside analog used as the substrate of viral RNA-dependent enzyme RNA polymerase to inhibit viral RNA production. In the past, remdesivir was successfully used to treat coronaviral infections, including endemic human-CoVs, SARS-CoV, and MERS-CoV. Some clinical trials have shown that remdesivir reduces symptoms and mortality rates in severe COVID-19 patients.39 , 41 However, a recent randomized, placebo-controlled trial revealed that remdesivir had no clinical benefits in severe COVID-19 patients.42 Although the efficacy and safety of remdesivir still need to be validated, remdesivir is a promising drug for treating severe COVID-19 patients requiring mechanical ventilation and extracorporeal membrane oxygenation (ECMO).43 Therefore, remdesivir is currently the only drug approved by the United States Food and Drug Administration (US-FDA) to treat hospitalized COVID-19 patients.
Lopinavir/ritonavir
Lopinavir and ritonavir are antiretroviral protease inhibitors used in the treatment of HIV and are considered candidates for the treatment of COVID-19. In one clinical trial, the combination of lopinavir and ritonavir had no effects on clinical improvement or mortality rates. Furthermore, serious adverse effects, including respiratory failure, acute kidney injury, and secondary infection, were observed in the group treated with lopinavir and ritonavir.44 So far, there have been only limited clinical trials to support the efficacy of lopinavir and ritonavir against COVID-19 infections.
Favipiravir
Favipiravir, a purine nucleic acid analog that inhibits viral RNA replication by binding to RNA-dependent RNA polymerase (RdRp), is used to treat influenza and several other RNA viruses.41 Currently, there are limited clinical trials of favipiravir in the treatment of SARS-CoV-2 infection, but some encouraging results with higher survival rates and shortened durations of symptoms were observed in limited clinical trials.45
Tocilizumab
As IL-6 induced cytokine storms play important roles in both innate and adaptive immune responses and inflammatory diseases, tocilizumab (a human recombinant IL-6 receptor antibody) has been used for rheumatoid arthritis, multiple myeloma, and life-threatening cytokine storms.41 In two retrospective clinical trials, the use of tocilizumab in severe COVID-19 patients reduced the risk of invasive mechanical ventilation and the mortality rate.46 , 47
Barcitinib
JAK pathway plays an important role in IFN-stimulated gene expression and promotes IFN-I production in acute viral infection.13 Barcitinib is a selective Janus kinase (JAK) inhibitor currently used to treat rheumatoid arthritis.41 Barcitinib may reduce the severity of SARS-CoV-2 infections via inhibiting viral endocytosis and proinflammatory cytokine release.41 In a retrospective study, barcitinib showed significance in reducing intensive care unit admissions and mortality rates. Therefore, Barcitinib is considered to be one of the possible candidates in the treatment of SARS-CoV-2 infection.48
Risk factors related to severe SARS-CoV-2 infection or a poor prognosis
Recent evidence has revealed that old age, sex, and comorbid conditions are important risk factors for severe infection and a poor prognosis in COVID-19.49 Possible hypotheses for age-related vulnerability have been proposed: 1) overexpression of ACE2 receptors and TMPRSS2, 2) pre-existing immunity due to previous common coronavirus infection, 3) immunosenescence and inflammaging, and 4) comorbidities including obesity and diabetes.49 In age-related immunosenescence, immune function declines, such as a weakened type 1 IFN response and impaired CD4+ T and CD8+ T cell development, which may lead to decreased SARS-CoV-2 clearance and result in high mortality in the elderly group.13 , 49 In comparison, in children, a stronger innate immunity with lower T cell activation, which means better virus clearance and more frequent viral infections, including commonly circulating human CoV, may help in fighting SARS-CoV-2 infections; these findings could explain why children generally have milder symptoms than elderly adults.49 Male sex is another risk factor for SARS-CoV-2 infections; due to the stronger type I IFN response in virus clearance, women have higher survival rates than men.13
A recent systemic review revealed that a delayed antibody response and seroconversion are correlated with disease severity in SARS-CoV-2 and MERS-CoV; slower antibody responses and lower seroconversion rates are found in severe cases.34 The antibody-dependent enhancement effect is a phenomenon in which the binding of nonneutralizing antibodies to viruses can enhance viral invasion into specific cells and promote further viral infection. The combination of virus surface proteins and antibodies can promote FcγR-related endocytosis in phagocytic cells, leading to enhanced viral infection. In previous research on SARS-CoV and MERS-CoV, ADEs promoted infections and affected the treatment results of vaccination, especially in MERS-CoV patients.34 Potential ADEs may also exist in SARS-CoV-2 due to the similarities between these coronaviruses. Thus, ADE has been considered as the potential mechanisms resulting in severe SARS-CoV-2 infection.13 However, there is limited clinical evidence to clarify the role of ADEs in SARS-CoV-2 infection.
Vaccine-associated hypersensitivity reaction (VAH) is another important vaccine-related issue due to several reports of hypersensitivity reactions after COVID-19 vaccine injection. Due to the rarity of allergic reactions, including anaphylaxis, the European Academy of Allergy and Clinical Immunology (EAACI) statement suggested that vaccinations are contraindicated only when there is an allergy to one of the vaccine components or if there was a severe allergic reaction to the first dose.50
Precision medicine and artificial intelligence by immunological mapping in SARS-CoV-2 infection
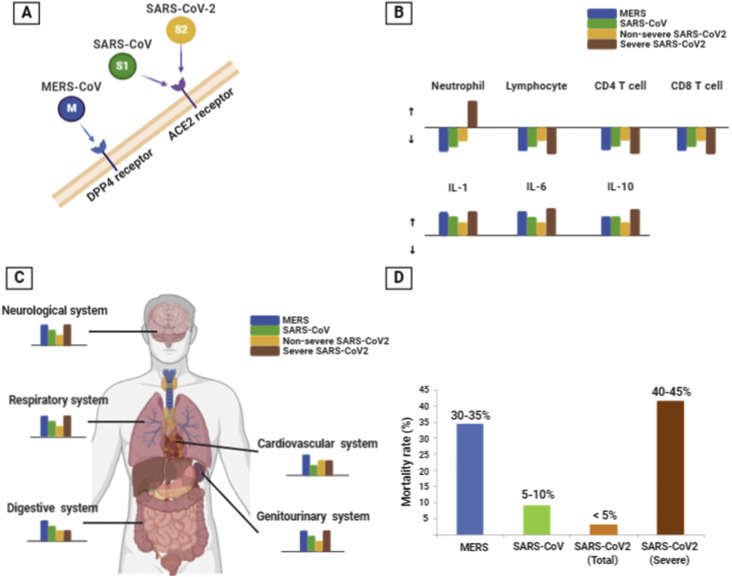
Based on our data regarding severe COVID-19 cases, poor prognostic factors of immune cell changes may be related to leukocytosis, neutrophilia, lymphopenia (both CD4 and CD8 T cells), and an increased NLR. In addition, elevated proinflammatory cytokines (IL-6 and TNF-α), Th1 cytokines (IL-2 and IFN-γ), regulatory T cell cytokines (IL-10), and Th17 cytokines (IL-17) may also be considered as predictors of a poor outcome. Patients needing ICU care had elevated plasma levels of many innate cytokines, such as IP-10, MCP-1, MIP-1A, and TNF-α. These clinical features suggested the likelihood of the involvement of highly proinflammatory conditions in both disease progression and severity. Due to the hyperinflammatory status in SARS-CoV-2 infection, immunosuppressing agents such as anakinra (anti- IL-1) or tocilizumab (anti- IL-6) may be taken into consideration. A summary of the immunological maps of MERS-CoV, SARS-CoV and SARS-CoV-2 infections is shown in Fig. 1 .
Figure 1.
Summary of the immunologic map in MERS-CoV, SARS-CoV, and SARS-CoV-2 infections. A: MERS-CoV, SARS-CoV, and SARS-CoV-2 invade human cells via different cell surface receptors. SARS-CoV and SARS-CoV-2 invade via angiotensin-converting enzyme 2 (ACE2), and MERS-CoV invades via dipeptidyl peptidase 4 (DPP4). B: The viral influences of MERS-CoV, SARS-CoV, and SARS-CoV-2 on immune cells and cytokines. The upward arrow means increased level and the downward arrow means decreased level. The length of each color bar is correlated with the influence degree of each infection. C: The viral influences of MERS-CoV, SARS-CoV, and SARS-CoV-2 on organ system. The length of each color bar is correlated with the influence degree of each infection. D: The mortality rate of MERS-CoV, SARS-CoV, and SARS-CoV-2 infection (SARS-CoV-2 mortality rate data were obtained at the world health organization website on March 15, 2021).
Novel biologic agents that target specific and critical pathophysiological pathways have been developed to better manage these severe patients by targeting the key cytokines of cytokine storms in severe COVID-19 cases. With biologics, defining biomarkers would facilitate the selection of patients with severe COVID-19 cases that would likely respond therapeutically. Artificial intelligence (AI) is necessary to help select and identify important cytokines in severe or fatal COVID-19 cases. Once AI establishes the immunological map of severe COVID-19, the use of AIs to select effective biologics for immunological precision medicine could rapidly enter clinical use.
Impact statements
This study summarized a brief comparison of SARS-CoV-2, SARS-CoV, and MERS-CoV infections regarding immunological changes. The immunological map offers the basis for immunological precision medicine in SARS-CoV-2 infection.
Acknowledgments
This work was supported partially by the Research Center for Environmental Medicine, Kaohsiung Medical University, Kaohsiung, Taiwan from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education in Taiwan and by Kaohsiung Medical University Research Center Grant (KMU-TC109A01).
References
- 1.Sariol A., Perlman S. Lessons for COVID-19 immunity from other coronavirus infections. Immunity. 2020;53:248–263. doi: 10.1016/j.immuni.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020;38:1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 3.Liu J., Zheng X., Tong Q., Li W., Wang B., Sutter K., et al. Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV. J Med Virol. 2020;92:491–494. doi: 10.1002/jmv.25709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yao X.H., Li T.Y., He Z.C., Ping Y.F., Liu H.W., Yu S.C., et al. [A pathological report of three COVID-19 cases by minimal invasive autopsies] Zhonghua Bing Li Xue Za Zhi. 2020;49:411–417. doi: 10.3760/cma.j.cn112151-20200312-00193. [DOI] [PubMed] [Google Scholar]
- 5.Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., et al. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019:11. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al Nemer A. Histopathologic and autopsy findings in patients diagnosed with coronavirus disease 2019 (COVID-19): what we know So far based on correlation with clinical, morphologic and pathobiological aspects. Adv Anat Pathol. 2020;27:363–370. doi: 10.1097/PAP.0000000000000276. [DOI] [PubMed] [Google Scholar]
- 7.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wurtz N., Penant G., Jardot P., Duclos N., La Scola B. Culture of SARS-CoV-2 in a panel of laboratory cell lines, permissivity, and differences in growth profile. Eur J Clin Microbiol Infect Dis. 2021;40:477–484. doi: 10.1007/s10096-020-04106-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian Y., Rong L., Nian W., He Y. Review article: gastrointestinal features in COVID-19 and the possibility of faecal transmission. Aliment Pharmacol Ther. 2020;51:843–851. doi: 10.1111/apt.15731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang G., Zhang J., Wang B., Zhu X., Wang Q., Qiu S. Analysis of clinical characteristics and laboratory findings of 95 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a retrospective analysis. Respir Res. 2020;21:74. doi: 10.1186/s12931-020-01338-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sohrabi C., Alsafi Z., O'Neill N., Khan M., Kerwan A., Al-Jabir A., et al. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19) Int J Surg. 2020;76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lavine J.S., Bjornstad O.N., Antia R. Immunological characteristics govern the transition of COVID-19 to endemicity. Science. 2021;371:741–745. doi: 10.1126/science.abe6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brodin P. Immune determinants of COVID-19 disease presentation and severity. Nat Med. 2021;27:28–33. doi: 10.1038/s41591-020-01202-8. [DOI] [PubMed] [Google Scholar]
- 14.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y.Y., Li B.R., Ning B.T. The comparative immunological characteristics of SARS-CoV, MERS-CoV, and SARS-CoV-2 coronavirus infections. Front Immunol. 2020;11:2033. doi: 10.3389/fimmu.2020.02033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.İnandıklıoğlu N., Akkoc T. Immune responses to SARS-CoV, MERS-CoV and SARS-CoV-2. Adv Exp Med Biol. 2020;1288:5–12. doi: 10.1007/5584_2020_549. [DOI] [PubMed] [Google Scholar]
- 17.The involvement of natural killer cells in the pathogenesis of severe acute respiratory syndrome. Am J Clin Pathol. 2004;121:507–511. doi: 10.1309/WPK7Y2XKNF4CBF3R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alosaimi B., Hamed M.E., Naeem A., Alsharef A.A., AlQahtani S.Y., AlDosari K.M., et al. MERS-CoV infection is associated with downregulation of genes encoding Th1 and Th2 cytokines/chemokines and elevated inflammatory innate immune response in the lower respiratory tract. Cytokine. 2020;126:154895. doi: 10.1016/j.cyto.2019.154895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahallawi W.H., Khabour O.F., Zhang Q., Makhdoum H.M., Suliman B.A. MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine. 2018;104:8–13. doi: 10.1016/j.cyto.2018.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng H.Y., Zhang M., Yang C.X., Zhang N., Wang X.C., Yang X.P., et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol. 2020;17:541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C.K., Wu H., Yan H., Ma S., Wang L., Zhang M., et al. T cell responses to whole SARS coronavirus in humans. J Immunol. 2008;181:5490–5500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin H.S., Kim Y., Kim G., Lee J.Y., Jeong I., Joh J.S., et al. Immune responses to Middle East respiratory syndrome coronavirus during the acute and convalescent phases of human infection. Clin Infect Dis. 2019;68:984–992. doi: 10.1093/cid/ciy595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., et al. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in wuhan, China. Clin Infect Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J.J., Dong X., Cao Y.Y., Yuan Y.D., Yang Y.B., Yan Y.Q., et al. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020;75:1730–1741. doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- 28.Wang Z., Yang B., Li Q., Wen L., Zhang R. Clinical features of 69 cases with coronavirus disease 2019 in wuhan, China. Clin Infect Dis. 2020;71:769–777. doi: 10.1093/cid/ciaa272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L., Liu H.G., Liu W., Liu J., Liu K., Shang J., et al. [Analysis of clinical features of 29 patients with 2019 novel coronavirus pneumonia] Zhonghua Jiehe He Huxi Zazhi. 2020;43:203–208. doi: 10.3760/cma.j.issn.1001-0939.2020.03.013. [DOI] [PubMed] [Google Scholar]
- 30.Tan M., Liu Y., Zhou R., Deng X., Li F., Liang K., et al. Immunopathological characteristics of coronavirus disease 2019 cases in Guangzhou, China. Immunology. 2020;160:261–268. doi: 10.1111/imm.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hotez P.J., Bottazzi M.E., Corry D.B. The potential role of Th17 immune responses in coronavirus immunopathology and vaccine-induced immune enhancement. Microb Infect. 2020;22:165–167. doi: 10.1016/j.micinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang A.T., Garcia-Carreras B., Hitchings M.D.T., Yang B., Katzelnick L.C., Rattigan S.M., et al. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat Commun. 2020;11:4704. doi: 10.1038/s41467-020-18450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoste L., Van Paemel R., Haerynck F. Multisystem inflammatory syndrome in children related to COVID-19: a systematic review. Eur J Pediatr. 2021:1–16. doi: 10.1007/s00431-021-03993-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Consiglio C.R., Cotugno N., Sardh F., Pou C., Amodio D., Rodriguez L., et al. The Immunology of multisystem inflammatory syndrome in children with COVID-19. Cell. 2020;183:968–981.e7. doi: 10.1016/j.cell.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vlachoyiannopoulos P.G., Magira E., Alexopoulos H., Jahaj E., Theophilopoulou K., Kotanidou A., et al. Autoantibodies related to systemic autoimmune rheumatic diseases in severely ill patients with COVID-19. Ann Rheum Dis. 2020;79:1661–1663. doi: 10.1136/annrheumdis-2020-218009. [DOI] [PubMed] [Google Scholar]
- 38.Lakota K., Perdan-Pirkmajer K., Hočevar A., Sodin-Semrl S., Rotar Ž., Čučnik S., et al. COVID-19 in association with development, course, and treatment of systemic autoimmune rheumatic diseases. Front Immunol. 2020;11:611318. doi: 10.3389/fimmu.2020.611318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kichloo A., Albosta M., Kumar A., Aljadah M., Mohamed M., El-Amir Z., et al. Emerging therapeutics in the management of COVID-19. World J Virol. 2021;10:1–29. doi: 10.5501/wjv.v10.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., Linsell L., et al. Dexamethasone in hospitalized patients with covid-19. N Engl J Med. 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naik R.R., Shakya A.K. Therapeutic strategies in the management of COVID-19. Front Mol Biosci. 2020;7:636738. doi: 10.3389/fmolb.2020.636738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhatraju P.K., Ghassemieh B.J., Nichols M., Kim R., Jerome K.R., Nalla A.K., et al. Covid-19 in critically ill patients in the seattle region - case series. N Engl J Med. 2020;382:2012–2022. doi: 10.1056/NEJMoa2004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., et al. A trial of lopinavir-ritonavir in adults hospitalized with severe covid-19. N Engl J Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cai Q., Yang M., Liu D., Chen J., Shu D., Xia J., et al. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering. 2020;6:1192–1198. doi: 10.1016/j.eng.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kewan T., Covut F., Al-Jaghbeer M.J., Rose L., Gopalakrishna K.V., Akbik B. Tocilizumab for treatment of patients with severe COVID-19: a retrospective cohort study. EClinicalMedicine. 2020;24:100418. doi: 10.1016/j.eclinm.2020.100418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guaraldi G., Meschiari M., Cozzi-Lepri A., Milic J., Tonelli R., Menozzi M., et al. Tocilizumab in patients with severe COVID-19: a retrospective cohort study. Lancet Rheumatol. 2020;2:e474–e484. doi: 10.1016/S2665-9913(20)30173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cantini F., Niccoli L., Nannini C., Matarrese D., Natale M.E.D., Lotti P., et al. Beneficial impact of Baricitinib in COVID-19 moderate pneumonia; multicentre study. J Infect. 2020;81:647–679. doi: 10.1016/j.jinf.2020.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y., Klein S.L., Garibaldi B.T., Li H., Wu C., Osevala N.M., et al. Aging in COVID-19: vulnerability, immunity and intervention. Ageing Res Rev. 2021;65:101205. doi: 10.1016/j.arr.2020.101205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sokolowska M., Eiwegger T., Ollert M., Torres M.J., Barber D., Del Giacco S., et al. 2021. EAACI statement on the diagnosis, management and prevention of severe allergic reactions to COVID-19 vaccines. Allergy. [DOI] [PMC free article] [PubMed] [Google Scholar]