Abstract
The nucleotide analogue prodrug remdesivir remains the only FDA-approved antiviral small molecule for the treatment of infection with SARS-CoV-2. Biochemical studies revealed that the active form of the drug targets the viral RNA-dependent RNA polymerase and causes delayed chain-termination. Delayed chain-termination is incomplete, but the continuation of RNA synthesis enables a partial escape from viral proofreading. Remdesivir becomes embedded in the copy of the RNA genome that later serves as a template. Incorporation of an incoming nucleotide triphosphate is now inhibited by the modified template. Knowledge on the mechanism of action matters. Enzymatic inhibition links to antiviral effects in cell cultures, animal models and viral load reduction in patients, which provides the logical chain that is expected for a direct acting antiviral. Hence, remdesivir also serves as a benchmark in current drug development efforts that will hopefully lead to orally available treatments to the benefit of a broader population.
Current Opinion in Virology 2021, 49:81–85
This review comes from a themed issue on Engineering for viral resistance
Edited by Richard Plemper
For complete overview about the section, refer Engineering for viral resistance
Available online 12th May 2021
https://doi.org/10.1016/j.coviro.2021.04.014
1879-6257/Crown Copyright © 2021 Published by Elsevier B.V. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Introduction
Hundreds if not thousands of investigational antiviral agents have been tested for the treatment of infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes coronavirus disease 2019 (Covid-19). One year into the pandemic, only a single small molecule compound has been given green light by the US Food and Drug Administration (FDA). On October 22, 2020, the FDA approved remdesivir (Veklury) for the treatment of Covid-19 in hospitalized patients who are 12 years of age or older [1]. An emergency use authorization (EUA) was already granted on May 1, 2020. The approval is essentially based on the Adaptive Covid-19 Treatment Trial (ACTT-1) by the National Institute of Allergy and Infectious Diseases (NIAID). ACCT-1 is a Randomized Clinical Trial (RCT) that showed a reduced median time to recovery from 15 days in the placebo group to 10 days in the remdesivir group [2••]. In contrast, the Solidarity trial sponsored by the World Health Organization (WHO) did not reveal such a benefit [3••]. It has been argued that differences in the design of the two trials may account for this discrepancy [1]. However, the consequences are profound. Largely based on Solidarity, the WHO recommended against the use of remdesivir. But can we really blame the drug? As an antiviral agent, remdesivir should decrease the viral load in SARS-CoV-2 infected patients. Neither ACTT-1 nor Solidarity have addressed this question. In this opinion article it is intended to discuss the value of biochemical studies. It has been stated before that the mechanism matters in drug development and approval processes [4].
Mechanism of action: why bother?
The approval of a new drug by the FDA does not require detailed knowledge on the mechanism of action. RCTs are the gold standard for the assessment of potential therapeutics. So why bother with biochemical studies that are designed to provide knowledge on how a given compound is working? This question might be even more relevant during a pandemic when time is a critical factor. A focus on RCTs that determine patient outcomes for promising drug candidates seems to be a very logical approach under these circumstances. However, variations in study design, different patient populations, timing of treatment and the specific endpoints may give rise to dissenting results as described above. The first year in the Covid-19 pandemic has been a roller coaster for several investigational therapeutics that were prematurely touted as ‘game changers’ or dismissed as ‘flops’. The reality is often more nuanced and some of the relevant compounds deserve a spot between the two bookends. In the face of clinical data that are difficult to reconcile it might be helpful to go back to the basics and ask what can we actually expect from a drug like remdesivir? The value of biochemical studies is here discussed in the context of a large body of pre-clinical data that justified clinical trials in humans, which ultimately led to its approval for the treatment of infection with SARS-CoV-2.
What is the target for remdesivir?
Remdesivir (RDV, formerly GS-5734) is a 1′-cyano-substituted nucleotide analogue prodrug [5]. The base moiety is adenosine-like. A C—C bond links sugar and base, while uracil base pairing properties are retained. The phosphoramidate substitution at the 5′-hydroxyl group facilitates delivery into the cell, where hydrolysis yields the monophosphate version of the drug. As a nucleotide analogue, RDV is designed to inhibit viral RNA synthesis. Intracellular phosphorylation to the triphosphate generates the active form that is eventually accommodated by viral RNA-dependent RNA polymerases (RdRp). Antiviral activity of the corresponding nucleoside analogue was originally demonstrated against several positive-strand RNA viruses, including HCV and SARS-CoV [6]. The triphosphate showed inhibition of HCV RdRp with IC50 values (5.6 μM) comparable with the cell-based EC50 measurements (4.1 μM). RDV was later shown to exhibit a broader spectrum of antiviral activities that included also negative-strand RNA viruses like respiratory syncytial virus (RSV), Nipah virus (NiV) and Ebola virus (EBOV) [7]. Biochemical assays with purified polymerase complexes indicated that RDV-TP can cause inhibition of RNA synthesis a few nucleotides downstream from its initial site of incorporation. Such a pattern is commonly referred to as ‘delayed chain-termination’ [8•,9,10]. In 2016 it has been demonstrated that RDV-TP is indeed formed in various human cell types and antiviral activity was also confirmed in a rhesus monkey model of Ebola virus disease (EVD) [8•]. The therapeutic efficacy of RDV was then evaluated in a trial during the 2018/19 Ebola outbreak in the Democratic Republic of the Congo (DRC) [11]. Although two different antibody therapies were more efficacious than RDV, human safety data were now available for this drug. At around the same time it has also been shown that RDV is active against diverse human coronaviruses including SARS-CoV and MERS-CoV [12]. In vitro selection experiments with the mouse hepatitis virus (MHV) revealed two resistance-conferring mutations in the non-structural protein 12 (nsp12), which represents the RdRp enzyme [13•]. Moreover, the virus-associated proofreading exonuclease activity (ExoN) was also shown to decrease susceptibility to RDV. This data suggested that the nucleotide analogue inhibitor is indeed incorporated into the growing RNA chain, but the ExoN activity can partially decrease its potency.
Efficiency as substrate
The pioneering work by Bruno Canard and colleagues after the SARS outbreak in 2002 provided a strong basis for biochemical and structural studies on related coronaviruses [14•]. Major findings indicated that RdRp activity requires a complex of nsp7, nsp8 and nsp12. Using cryo-electron microscopy, Kirchdoerfer and Ward reported the first structure of the SARS-CoV polymerase nsp12 with its cofactors ns7 and nsp8 [15]. This knowledge enabled several successful structural studies on the SARS-CoV-2 complex within the first couple of months in the pandemic [16•,17•,18,19]. The nsp7/8/12 complex can be reconstituted from the three independently expressed proteins or independently expressed nsp12 and linked nsp7-nsp8. In early 2020, we devised a MERS-CoV construct that contained the nsp5 protease with nsp7, nsp8, and nsp12 [20••]. Expression in insect cells yielded an active polymerase complex. Steady-state kinetic measurements of single nucleotide incorporations revealed that RDV-TP is an excellent substrate for the RdRp. The ratio of the efficiency of incorporation (V max /K m) of ATP over the efficiency of incorporation RDV-TP defines the ‘selectivity’. A value below 1 suggests that the analogue is more efficiently used than its natural counterpart ATP. We measured a selectivity value of 0.35 for the MERS-CoV RdRp, 0.32 for SARS-CoV RdRp and 0.28 for SARS-CoV-2 [20••,21••]. A pre-state kinetic approach later revealed almost exactly the same results for the SARS-CoV-2 RdRp complex (0.57) [22]. Together this data suggests that RDV-TP competes efficiently with ATP. For sofosbuvir-TP (SOF-TP), a nucleotide analogue for the treatment of infection with the HCV, we measured a selectivity value of ∼1000 suggesting that the inhibitor is heavily outcompeted by its natural counterpart UTP [21••]. Moreover, human mitochondrial RNA polymerase shows a desirable preference for ATP over RDV-TP. We measured a selectivity value of ∼500, which demonstrates effective discrimination against RDV-TP [9].
Delayed chain-termination
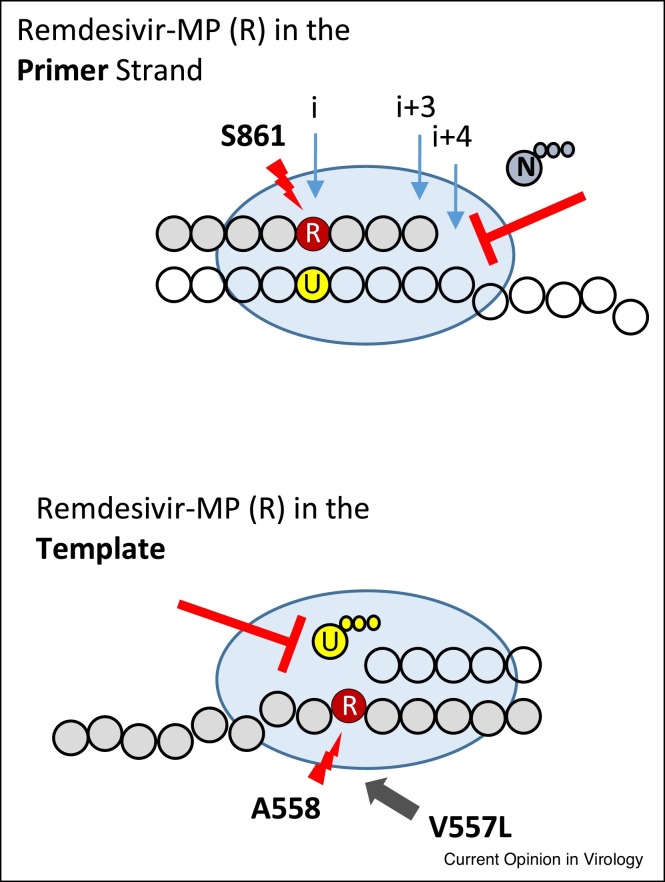
Efficient incorporation of a nucleotide analogue is usually a prerequisite for inhibition, but this is not a sufficient requirement. The presence of the 3′-hydroxyl group of the incorporated RDV-MP at position i allows the incorporation of the next NTP substrate by the coronavirus RdRp. RNA synthesis continues and is inhibited at position i + 3 (Figure 1 , top) [21••]. This type of inhibition has previously been observed with other compounds and other viral polymerases. In general, nucleotide analogues that do not block DNA or RNA synthesis at the point of incorporation, but only after additional natural nucleotides are added to the growing chain, are referred to as ‘delayed chain-terminators’ [23]. Inhibition is not necessarily absolute and longer reaction times may overcome the blockage. The hepatitis B virus (HBV) and human immunodeficiency virus type 1 (HIV-1) reverse transcriptase inhibitor entecavir [24], and the human cytomegalovirus (HCMV) DNA polymerase inhibitor ganciclovir are prominent example in this regard [25]. For SARS-CoV-2, modeling studies predicted a steric clash between the 1′-cyano group of RDV and the side chain of S861 [21••]. The RNA primer with the incorporated RDV-MP can translocate without obstruction from position i through i + 3; however, the clash is evident at position i + 4. It was therefore predicted that mutant enzymes with smaller side chains should diminish the clash and support translocation. While the S861A mutant shows a subtle reduction in delayed chain-termination [16•,27••], RNA synthesis is not inhibited with the S861G mutant [27••]. These results guided structural studies that confirmed the original model [28,29].
Figure 1.
How does RDV work against SARS-CoV-2?
(Top) The viral polymerase (blue oval) uses RDV-TP as a substrate and incorporates RDV-MP (red) into the RNA primer strand (green bubbles) opposite UMP in the template (yellow/clear bubbles). A clash between the incorporated RDV-MP and S861 blocks translocation of the polymerase, which inhibits its ability to bind the next nucleoside triphosphate (N) at position i + 4. (Bottom) Inhibition at position i + 3 is not complete, the primer is eventually extended and is later used as a template (now green). Here, RDV-MP clashes with A558, which reduces binding of UTP(U). V557L is a mutation in the polymerase that can in part neutralize this effect. This mutation is associated with low-level resistance to RDV in MHV. Adapted from Ref. [27••].
Template-dependent inhibition
Delayed chain-termination is also reduced by increasing the concentration of the NTP that would be incorporated at position i + 4 [20••,21••]. These are conditions that drive translocation as demonstrated for HIV-1 reverse transcriptase using site-specific footprinting techniques [30,31]. NTP concentrations in the low micromolar range are sufficient to obtain the full-length product. This can compromise inhibition in a cellular environment with NTP concentrations in the low milimolar range. Overcoming delayed chain-termination will yield RNA strands with embedded RDV-MP residues, and RDV-MP in the template inhibits incorporation of the complementary UTP (Figure 1, bottom) [27••]. Such template-dependent blockage provides a second opportunity for inhibition and in this case inhibition is less sensitive to the concentration of the next nucleotide. Of note, one of the RDV resistance-associated mutations previously selected with MHV diminishes specifically the template-dependent inhibition mechanism [27••]. In SARS-CoV-2, the equivalent mutation (V557L) would be in contact with the base moiety of RDV-MP in the template. However, there is currently no evidence that RDV selects for this mutation in SARS-CoV-2. Resistance in the context of SARS-CoV-2 remains elusive at this point [32,33].
Antiviral effects
In February 2020, Wang et al. published the first study on the antiviral effects of RDV against a clinical isolate of SARS-CoV-2 in Vero cells [26]. In July 2020, Pruijssers et al. demonstrated that RDV inhibits SARS-CoV-2 in human lung cell cultures and the potency dependent on cell-type-specific concentrations of RDV-TP [34•]. EC50 values were as low as 10 nM, which indicated high potency. Reductions in viral load have also been shown in various animal models [34•,35]. Most importantly, a case report has shown that the use of RDV in an immunocompromised patient correlated with clinical resolution and viral clearance [36•]. This patient never developed severe disease, which likely expanded the window for the effective use of an antiviral treatment. The observation of antiviral effects in these various settings can be explained by the aforementioned biochemical inhibition studies. The active form of RDV inhibits viral RNA synthesis when incorporated in the primer strand and/or embedded in the template strand. The collective body of evidence supports the notion that RDV is a potent direct acting antiviral (DAA) [21••]. Thus, the drug does what it is expected to do. That said, a recently published retrospective clinical study confirmed that treatment with remdesivir is associated with a significant reduction in the time to recovery [37••].
Future directions
The SARS-CoV-2 replication transcription complex (RTC) is composed of nsp7/8/12 and several other non-structural proteins involved in activities that include RNA capping, RNA unwinding and proofreading [38]. Nsp14 contains the ExoN activity that can reduce susceptibility to nucleotide analogue inhibitors [39]. Nsp10 acts as a cofactor and increases the ExoN activity of nsp14 [40]. Parameters that trigger the transfer of the 3′-end of the primer from the polymerase to the ExoN active site remain to be defined. In this context it is important to note that MERS-CoV nsp14 is in vivo not active against the mutagenic nucleotide analogue β-d-N 4-hydroxycytidine (NHC) [41]. Its prodrug molnupiravir (MK-4482/EIDD-2801) is currently evaluated in a clinical phase 3 trial for the treatment of SARS-Cov-2 infection in outpatients. The guanosine nucleotide analogue AT-527 has also advanced into clinical trials; however, a potential mechanism of action has yet to be elucidated for this compound [42]. Future biochemical studies aimed at investigational nucleotide analogues would ideally include the nsp14/nsp10 complex and perhaps also other components of the RTC, including the helicase nsp13. A recent structural study suggested that nsp13 can move along the template to cause backtracking of the RdRp complex, which in turn liberates the 3′-end of the primer strand and makes it potentially accessible for proofreading [43].
Conclusion
So what is the value of biochemical studies? The mechanism of action of RDV is the basis of a logical chain from demonstrated enzymatic inhibition to antiviral effects in cell culture, antiviral effects in animal models, reduction in viral load in patients and ultimately improved patient outcomes. Insufficient data at each of these steps would raise further questions with respect to both safety and efficacy. RDV is now the benchmark and investigational antivirals are compared against this antiviral drug. Biochemical data provide necessary tools to improve properties of nucleotide analogue inhibitors, which will hopefully contribute to the development of novel treatments that can be used earlier in the disease to the benefit of more patients. Remdesivir is administered intravenously and oral antivirals that could be used in outpatients are not yet approved. Finally, knowledge on the mechanism of action generates trust between doctors and patients and the FDA included the available information on the mechanism of action of RDV in the fact sheet for healthcare providers.
Conflict of interest statement
MG received funding from Gilead Sciences for the mechanistic studies on remdesivir.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
The work on remdesivir in my laboratory has been supported by the Canadian Institutes of Health Research (CIHR), The Alberta Ministry for Jobs, Economy and Innovation, and Gilead Sciences, Inc. (Foster City, US). I would like to thank all members in my group for their dedication and hard work.
References
- 1.Rubin D., Chan-Tack K., Farley J., Sherwat A. FDA approval of remdesivir — a step in the right direction. N Engl J Med. 2020;383:2598–2600. doi: 10.1056/NEJMp2032369. [DOI] [PubMed] [Google Scholar]
- 2••.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., et al. Remdesivir for the treatment of Covid-19-final report. N Engl J Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]; First Randomized Clinical Trial that demonstrates reduced time of recovery with remdesivir.
- 3••.Consortium W.S.T. Repurposed antiviral drugs for Covid-19-interim WHO solidarity trial results. N Engl J Med. 2021;384:497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]; Large Randomized Clinical Trial that fails to demonstrate benefit of repurposed antivirals for the treatment of Covid-19.
- 4.Mechanism matters. Nat Med. 2010;16:347. doi: 10.1038/nm0410-347. [DOI] [PubMed] [Google Scholar]
- 5.Eastman R.T., Roth J.S., Brimacombe K.R., Simeonov A., Shen M., Patnaik S., Hall M.D. Remdesivir: a review of its discovery and development leading to emergency use authorization for treatment of COVID-19. ACS Cent Sci. 2020;6:672–683. doi: 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho A., Saunders O.L., Butler T., Zhang L.J., Xu J., Vela J.E., Feng J.Y., Ray A.S., Kim C.U. Synthesis and antiviral activity of a series of 1’-substituted 4-aza-7,9-dideazaadenosine C-nucleosides. Bioorg Med Chem Lett. 2012;22:2705–2707. doi: 10.1016/j.bmcl.2012.02.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lo M.K., Jordan R., Arvey A., Sudhamsu J., Shrivastava-Ranjan P., Hotard A.L., Flint M., McMullan L.K., Siegel D., Clarke M.O., et al. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci Rep. 2017;7 doi: 10.1038/srep43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8•.Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C., et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]; First comprehensive study on the antiviral effects of remdesivir in Ebola virus disease models.
- 9.Tchesnokov E.P., Feng J.Y., Porter D.P., Gotte M. Mechanism of inhibition of Ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019;11 doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jordan P.C., Liu C., Raynaud P., Lo M.K., Spiropoulou C.F., Symons J.A., Beigelman L., Deval J. Initiation, extension, and termination of RNA synthesis by a paramyxovirus polymerase. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mulangu S., Dodd L.E., Davey R.T., Mbaya O.T., Proschan M., Mukadi D., Manzo M.L., Nzolo D., Oloma A.T., Ibanda A., et al. A randomized, controlled trial of Ebola virus disease therapeutics. N Engl J Med. 2019;381:2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., Leist S.R., Pyrc K., Feng J.Y., Trantcheva I., et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13•.Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R., et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9 doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]; In vitro selection of remdesivir resistance in coronaviruses.
- 14•.Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci U S A. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]; First demonstration that coronavirus RNA polymerase activity requires nsp7, nsp8 and nsp12.
- 15.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun. 2019;10 doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., Ji W., Yan L., Zhu Y., Zhu C., et al. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182:417–428.e3. doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]; Structural models of SARS-CoV-2 RNA-dependent RNA polymerase with RNA.
- 17•.Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584:154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]; Structural models of SARS-CoV-2 RNA-dependent RNA polymerase with RNA.
- 18.Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., Maruthi K., Eng E.T., Vatandaslar H., Chait B.T., et al. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell. 2020;182:1560–1573.e13. doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin W.C., Mao C.Y., Luan X.D., Shen D.D., Shen Q.Y., Su H.X., Wang X.X., Zhou F.L., Zhao W.F., Gao M.Q., et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20••.Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Gotte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J Biol Chem. 2020;295:4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]; First report on the mechanism of action of remdesivir against cornavirus RNA polymerase.
- 21••.Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Gotte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]; First report on remdesivir-mediated delayed chain-termination against SARS-CoV-2 RNA polymerase.
- 22.Dangerfield T.L., Huang N.Z., Johnson K.A. Remdesivir is effective in combating COVID-19 because it is a better substrate than ATP for the viral RNA-dependent RNA polymerase. iScience. 2020;23 doi: 10.1016/j.isci.2020.101849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyer P.L., Julias J.G., Marquez V.E., Hughes S.H. Fixed conformation nucleoside analogs effectively inhibit excision-proficient HIV-1 reverse transcriptases. J Mol Biol. 2005;345:441–450. doi: 10.1016/j.jmb.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 24.Tchesnokov E.P., Obikhod A., Schinazi R.F., Gotte M. Delayed chain termination protects the anti-hepatitis B virus drug entecavir from excision by HIV-1 reverse transcriptase. J Biol Chem. 2008;283:34218–34228. doi: 10.1074/jbc.M806797200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H., Beardsley G.P., Coen D.M. Mechanism of ganciclovir-induced chain termination revealed by resistant viral polymerase mutants with reduced exonuclease activity. Proc Natl Acad Sci U S A. 2014;111:17462–17467. doi: 10.1073/pnas.1405981111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. Epub 2020 Feb 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27••.Tchesnokov E.P., Gordon C.J., Woolner E., Kocinkova D., Perry J.K., Feng J.Y., Porter D.P., Gotte M. Template-dependent inhibition of coronavirus RNA-dependent RNA polymerase by remdesivir reveals a second mechanism of action. J Biol Chem. 2020;295:16156–16165. doi: 10.1074/jbc.AC120.015720. [DOI] [PMC free article] [PubMed] [Google Scholar]; First report on template-dependent inhibition of SARS-CoV-2 RNA polymerase.
- 28.Bravo J.P.K., Dangerfield T.L., Taylor D.W., Johnson K.A. Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication. Mol Cell. 2021;81:1548–1552.e4. doi: 10.1016/j.molcel.2021.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kokic G., Hillen H.S., Tegunov D., Dienemann C., Seitz F., Schmitzova J., Farnung L., Siewert A., Hobartner C., Cramer P. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat Commun. 2021;12:279. doi: 10.1038/s41467-020-20542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchand B., Gotte M. Site-specific footprinting reveals differences in the translocation status of HIV-1 reverse transcriptase. Implications for polymerase translocation and drug resistance. J Biol Chem. 2003;278:35362–35372. doi: 10.1074/jbc.M304262200. [DOI] [PubMed] [Google Scholar]
- 31.Marchand B., Tchesnokov E.P., Gotte M. The pyrophosphate analogue foscarnet traps the pre-translocational state of HIV-1 reverse transcriptase in a Brownian ratchet model of polymerase translocation. J Biol Chem. 2007;282:3337–3346. doi: 10.1074/jbc.M607710200. [DOI] [PubMed] [Google Scholar]
- 32.Martin R., Li J., Parvangada A., Perry J., Cihlar T., Mo H., Porter D., Svarovskaia E. Genetic conservation of SARS-CoV-2 RNA replication complex in globally circulating isolates and recently emerged variants from humans and minks suggests minimal pre-existing resistance to remdesivir. Antiviral Res. 2021;188 doi: 10.1016/j.antiviral.2021.105033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinot M., Jary A., Fafi-Kremer S., Leducq V., Delagreverie H., Garnier M., Pacanowski J., Mekinian A., Pirenne F., Tiberghien P., et al. Remdesivir failure with SARS-CoV-2 RNA-dependent RNA-polymerase mutation in a B-cell immunodeficient patient with protracted Covid-19. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1474. ciaa1474. Published online 2020 Sep 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34•.Pruijssers A.J., George A.S., Schafer A., Leist S.R., Gralinksi L.E., Dinnon K.H., 3rd, Yount B.L., Agostini M.L., Stevens L.J., Chappell J.D., et al. Remdesivir inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107940. [DOI] [PMC free article] [PubMed] [Google Scholar]; First comprehensive study on the antiviral activity of remdesivir in SARS-CoV-2 disease models.
- 35.Williamson B.N., Feldmann F., Schwarz B., Meade-White K., Porter D.P., Schulz J., van Doremalen N., Leighton I., Yinda C.K., Perez-Perez L., et al. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature. 2020;585:273–276. doi: 10.1038/s41586-020-2423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36•.Buckland M.S., Galloway J.B., Fhogartaigh C.N., Meredith L., Provine N.M., Bloor S., Ogbe A., Zelek W.M., Smielewska A., Yakovleva A., et al. Treatment of COVID-19 with remdesivir in the absence of humoral immunity: a case report. Nat Commun. 2020;11 doi: 10.1038/s41467-020-19761-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; First demonstration of viral load reductions in a patient treated with remdesivir.
- 37••.Garibaldi B.T., Wang K., Robinson M.L., Zeger S.L., Bandeen-Roche K., Wang M.C., Alexander G.C., Gupta A., Bollinger R., Xu Y. Comparison of time to clinical improvement with vs without remdesivir treatment in hospitalized patients with COVID-19. JAMA Netw Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]; Restrospective study demonstrating reduced time to recovery with remdesivir in a cohort of predominantly non-White patients.
- 38.Robson F., Khan K.S., Le T.K., Paris C., Demirbag S., Barfuss P., Rocchi P., Ng W.L. Coronavirus RNA proofreading: molecular basis and therapeutic targeting. Mol Cell. 2020;80:1136–1138. doi: 10.1016/j.molcel.2020.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pruijssers A.J., Denison M.R. Nucleoside analogues for the treatment of coronavirus infections. Curr Opin Virol. 2019;35:57–62. doi: 10.1016/j.coviro.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bouvet M., Imbert I., Subissi L., Gluais L., Canard B., Decroly E. RNA 3 ’-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc Natl Acad Sci U S A. 2012;109:9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheahan T.P., Sims A.C., Zhou S.T., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schafer A., Dinnon K.H., Stevens L.J., et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med. 2020;12 doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Good S.S., Westover J., Jung K.H., Zhou X.J., Moussa A., La Colla P., Collu G., Canard B., Sommadossi J.P. AT-527, a double prodrug of a guanosine nucleotide analog, is a potent inhibitor of SARS-CoV-2 in vitro and a promising oral antiviral for treatment of COVID-19. Antimicrob Agents Chemother. 2021;65 doi: 10.1128/AAC.02479-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malone B., Chen J., Wang Q., Llewellyn E., Choi Y.J., Olinares P.D.B., Cao X., Hernandez C., Eng E.T., Chait B.T., et al. Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2102516118. [DOI] [PMC free article] [PubMed] [Google Scholar]