Abstract
Objectives
Surveillance of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genomic epidemiology led us to detect several variants since summer 2020. We report the recent spread of a new SARS-CoV-2 spike 501Y variant.
Methods
SARS-CoV-2 sequences obtained from human nasopharyngeal samples by Illumina next-generation sequencing were analysed using Nextclade and an in-house Python script and were compared using BLASTn to the GISAID database. Phylogeny was investigated using the IQ-TREE software.
Results
We identified that SARS-CoV-2 genomes from four patients diagnosed in our institute harboured a new set of amino acid substitutions including L18F, L452R, N501Y, A653V, H655Y, D796Y, G1219V ± Q677H. These spike N501Y genomes are the first of Nextstrain clade 19B. We obtained partial spike gene sequences of this genotype for an additional 43 patients. All patients infected with this genotype were diagnosed since mid-January 2021. We detected 42 other genomes of this genotype in GISAID, which were obtained from samples collected in December 2020 in four individuals and in 2021 in 38 individuals. The 89 sequences obtained in our institute or other laboratories originated from the Comoros archipelago, western European countries (mostly metropolitan France), Turkey and Nigeria.
Conclusion
These findings warrant further studies to investigate the spread, epidemiological and clinical features, and sensitivity to immune responses of this variant.
Keywords: Coronavirus disease 2019, Emergence, Epidemic, N501Y, Severe acute respiratory syndrome coronavirus 2, Spike, Variant
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has now spread worldwide for >1 year (https://www.ecdc.europa.eu/en/geographical-distribution-2019-ncov-cases). The emergence of major viral variants has expanded dramatically since summer 2020, although this only recently came into the spotlight. We have implemented, since the first occurrence of SARS-CoV-2 in France in late February 2020, a surveillance based on their genomes obtained by next-generation sequencing [1]. During summer 2020 we detected the emergence of ten viral variants, concomitantly with the re-increase of SARS-CoV-2 diagnoses [2]. These variants have since been responsible for juxtaposed or successive epidemics and accounted in various proportions over time for the total burden of cases. Since 1 January 2021, we have also performed a systematic screening of variants that predominated or arose in our geographical area with variant-specific in-house quantitative PCR assays (https://www.mediterranee-infection.com/procedure-pour-la-detection-par-qpcr-des-variants-sars-cov-2-marseille-4/; https://www.mediterranee-infection.com/procedure-pour-la-detection-par-qpcr-des-variants-sars-cov-2-n501y/). These variants include the Marseille-4 variant (Nexstrain clade 20A.EU1) [3], which has been the most prevalent strain between August 2020 and January 2021, and the rapidly spreading variants 20I/501Y.V1, 20H/501Y.V2 and 20J/501Y.V3, which harbour in their spike protein the amino acid (aa) substitution N501Y [4]. We describe herein the first SARS-CoV-2 spike 501Y variant belonging to Nextstrain clade 19B.
Materials and methods
SARS-CoV-2 genomes were obtained directly from nasopharyngeal swab fluid by next-generation sequencing with the Illumina Nextera XT paired-end strategy on a MiSeq instrument (Illumina Inc., San Diego, CA, USA), as previously described [5]. Genome consensus sequences were generated through mapping on the SARS-CoV-2 genome GenBank accession no. NC_045512.2 (Wuhan-Hu-1 isolate) with the CLC Genomics workbench v.7. In addition, sequences of the spike gene 5ʹ-region (nucleotides 1–1854 in reference to NC_045512.2) were obtained by next-generation sequencing after PCR amplification with an in-house protocol (see Supplementary material, Data S1). Sequences were analysed using Nextclade tool (https://clades.nextstrain.org/) [6] and an in-house Python script. Other SARS-CoV-2 sequences of this spike 501Y clade 19B genotype were searched using BLASTn [7] among all 674 762 sequences available in the GISAID database (https://www.gisaid.org/) [8] as of 3 March 2021, with genomes obtained in our institute as queries. Phylogeny reconstruction was performed using the IQ-TREE software with the GTR Model and 1000 ultrafast bootstrap repetitions after alignment of genomes using MAFFT v.7; the tree was visualized with the iTOL (Interactive Tree Of Life) software [[9], [10], [11]]. SARS-CoV-2 culture was performed by inoculating nasopharyngeal samples on Vero E6 cells, as previously described [12].
Ethics
Data have been generated as part of the routine work at Assistance Publique-Hôpitaux de Marseille (Marseille University Hospitals). This study has been approved by the ethics committee of our institution (No. 2020-016-03). According to European General Data Protection Regulation No. 2016/679, patients were informed of the potential use of their medical data and that they could refuse the use of their data. The analysis of collected data followed the MR-004 reference methodology registered under No. 2020-151 and No. 2020-152 in the AP-HM register.
Results
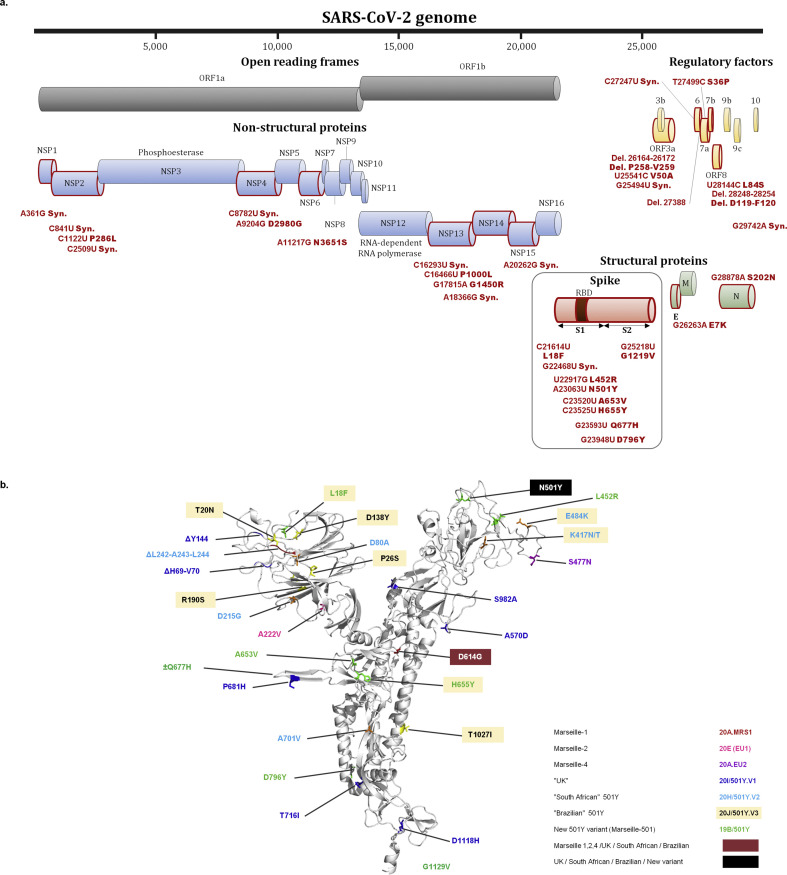
Our SARS-CoV-2 genomic surveillance allowed us to detect a variant harbouring a new combination of eight mutations in the spike protein in four individuals diagnosed with SARS-CoV-2 in our institute. These mutations include aa substitution N501Y, associated with aa substitutions L18F, L452R, A653V, H655Y, D796Y and G1219V, and a synonymous mutation at nucleotide position 22 468 (in reference to NC_045512.2); Fig. 1 a,b); an eighth spike aa susbtitution, Q677H, was identified in one of these genomes. Other nucleotide or aa substitutions or deletions were present in 14 other proteins; these changes notably include deletions in ORF3a and ORF8 genes (Fig. 1a). Overall, 24–31 nucleotide substitutions, 14–19 aa substitutions and three deletions were present in these four SARS-CoV-2 genomes (Fig. 1a; GISAID accession numbers: EPI_ISL_1097023; EPI_ISL_1097024; EPI_ISL_1201045; EPI_ISL_1201046). Viral isolates were obtained by culturing the nasopharyngeal samples of the four individuals, which will allow further analyses of the phenotypic features of this variant. In addition, sequences that span codons 1–618 of the spike gene and allow the identification of this variant by covering four of its hallmark mutations (C21614T (substitution L18F), G22468T, T22917G (L452R), A23063T (N501Y)) and by being devoid of the Nextstrain clade 20 A23403G mutation, were obtained from 43 other patients (deposited at: https://doi.org/10.35081/43r6-sz33).
Fig. 1.
Map of the nucleotide and amino acid (aa) substitutions and deletions of the new variant along the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genome (a) and three-dimensional structure of the spike protein showing aa substitutions and deletions in various spike N501Y variants (b). In (a), genes that harbour mutations are indicated by a red frame. For (b) see Supplementary material, Methods S1 for structure prediction.
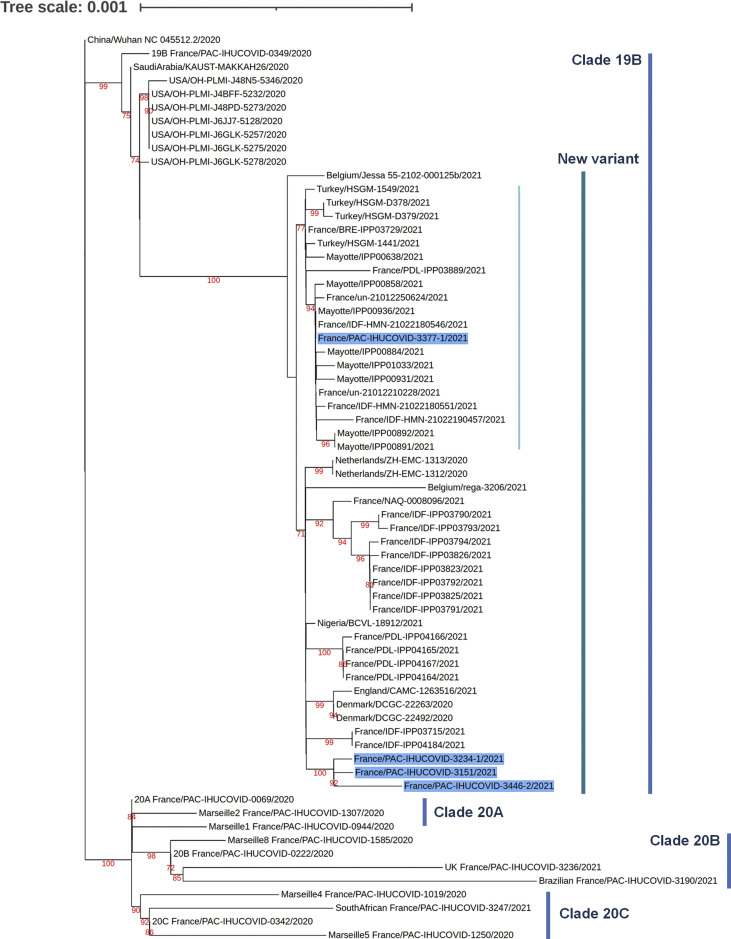
This variant, named Marseille-501, was first detected in our institute in a patient sampled on 18 January 2021, and its number increased from week 5 (see Supplementary material, Fig. S1). Between 18 January and 23 February (37 days), the SARS-CoV-2 genotype was obtained by sequencing for 1016 patients. We identified in the GISAID database 42 other SARS-CoV-2 genomes, all from humans, with the same spike aa substitution set (Fig. 2 ). Eight originated from Mayotte, a French overseas department in the Comoros archipelago. The others originated from France (n = 22), Denmark (n = 2), the Netherlands (n = 2), Belgium (n = 2), England (n = 1), Turkey (n = 4) and Nigeria (n = 1). The oldest samples were collected mid-December in Denmark and late December in the Netherlands. The other 38 SARS-CoV-2 genomes were obtained from samples collected in 2021 (see Supplementary material, Fig. S1).
Fig. 2.
Phylogenetic analysis of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) full-length genomes of the new variant recovered from the IHU Méditerranée Infection sequence database and from the GISAID sequence database. The SARS-CoV-2 phylogenetic tree is based on the full-length SARS-CoV-2 genomes. The SARS-CoV-2 sequences obtained by next-generation sequencing from four patients are indicated by a bold font and a dark blue background. The sequences with the highest BLAST scores recovered from the NBCI GenBank nucleotide sequence databases (http://www.ncbi.nlm.nih.gov/nucleotide/) including the 42 that are clustered with these four query sequences, were incorporated in the phylogeny reconstruction. Additional sequences include the genome of the Wuhan-Hu-1 isolate and genomes obtained in our institute and classified as predominant variants. The 67 genomes of SARS-CoV-2 were aligned using MAFFT version 7 with default parameters [10]. Phylogenetic tree was performed by using iQ-TREE under the GTR model and visualized with iTOL [[9], [11]]. GISAID accession numbers: IHUCOVID-3234: EPI_ISL_1097023; IHUCOVID-3151: EPI_ISL_1097024; IHUCOVID-3377: EPI_ISL_1201045; IHUCOVID-3446: EPI_ISL_1201046.
Phylogenetic analysis conducted with the four genomes from our institute and the 42 other GISAID genomes showed that all 46 sequences were clustered and delineated a strongly supported (bootstrap value: 100%) 19B subclade that stands apart from other clade 19B genomes (Fig. 2). Moreover, 21 genomes, one from our institute, the eight from Mayotte and 12 other genomes from France or Turkey, were clustered and all harboured the spike Q677H substitution.
The 47 patients we diagnosed as infected with this variant had a mean age of 39 ± 19 years (range 16–92 years); 26 (55%) were male. Epidemiological and/or clinical characteristics were available for 12 patients (see Supplementary material, Table S1): three patients had travelled to or originated from the Comoros, another originated from Guinea-Conakry but had not travelled abroad recently (see Supplementary material, Table S1). Clinical symptoms were available for nine patients; four were asymptomatic and 34 (85%) had mild symptoms including fever, rhinorrhoea, cough, headache, asthenia or myalgia. A 94-year-old male patient developed SARS-CoV-2-associated hypoxaemic pneumonia 4 days after the second administration of the Pfizer/BioNTech vaccine and died 7 days later. Another patient with severe obesity was admitted to an intensive care unit.
Discussion
We report herein the spread of the first SARS-CoV-2 501Y variant belonging to clade 19B. Its spike protein harbours several aa substitutions recently reported to emerge in various lineages and/or to be associated with immune escape (L18F, L452R, N501Y, H655Y, Q677H) (see Supplementary material, Data S1). For instance, N501Y is part of several variants that belong to distinct lineages and have been detected worldwide in association with various combinations of mutations [4], whereas L18F has been reported in 20E strains in England, in a 20I/N501Y.V1 substrain with an increased replicative fitness, in 20H/501Y.V2 strains with a faster propagation in the presence of convalescent plasma, and in a majority of 20J/501Y.V3 variants [13].
It is currently unresolved how such strains that harbour new blocks of mutations in the spike emerge. Overlooked genome evolution following transmission to minks and back to humans [3], or promotion and selection of mutations during administration of remdesivir or convalescent plasma [14] have been suspected. Also, recombinations known to be common among coronaviruses may occur between SARS-CoV-2 strains [15]. This new 501Y variant is another example that the same aa substitutions can occur in distinct lineages and follow convergent evolution [4]. Epidemiologically, this new variant infected patients in diverse countries, but with a predominance in the Comoros archipelago (11/47 documented cases) and in metropolitan France. Based on the limited data available, it is unknown if this new variant is more or less transmissible compared with other strains or if it is associated with any particular clinical feature.
Overall, these data highlight the need for genomic epidemiology surveillance of SARS-CoV-2 strains. The incidence of this new variant and its epidemiological and clinical features deserve to be closely monitored. Moreover, considering its particular association of numerous spike aa substitutions, its sensitivity to neutralizing antibodies and to plasma from convalescent or vaccinated persons should be investigated, which will be done for the isolates cultured in our institute.
Author contributions
PC and DR conceived and designed the experiments; PC, AL, JD, LP, PD, CD, PEF, BLS and JCL contributed materials/analysis tools; PC, AL, JD, JCL, BLS and DR analysed the data; and PC and DR wrote the paper.
Transparency declaration
This work was supported by the French Government under the ‘Investments for the Future’ programme managed by the National Agency for Research (ANR), Méditerranée-Infection 10-IAHU-03 and was also supported by Région Provence Alpes Côte d’Azur and European funding FEDER PRIMMI (Fonds Européen de Développement Régional-Plateformes de Recherche et d'Innovation Mutualisées Méditerranée Infection), FEDER PA 0000320 PRIMMI.
The authors have no conflicts of interest to declare. Funding sources had no role in the design and conduct of the study; collection, management, analysis and interpretation of the data; and preparation, review or approval of the manuscript.
Acknowledgements
We are grateful to Marielle Bedotto and Ludivine Brechard for their technical help.
Editor: L. Leibovici
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2021.05.006.
FigureS1. Number of sequences available in the GISAID and the IHU Méditerranée Infection sequence databases.
Table S1. Virological, epidemiological and clinical features of cases of infections with the new SARS-CoV-2 variant.
Data S1. Frequency of amino acid substitutions present in the spike of the new 501Y variant in the GISAID and the IHU Méditerranée Infection sequence databases, and reported data on amino acid substitutions present in the spike of the new 501Y variant.
MethodsS1. Partial sequencing of the SARS-CoV-2 spike gene, and structure prediction for spike protein.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Colson P., Levasseur A., Delerce J., Chaudet H., Bossi V., Ben Khedher M., et al. IHU Preprints; 2020. Dramatic increase in the SARS-CoV-2 mutation rate and low mortality rate during the second epidemic in summer in Marseille. [DOI] [Google Scholar]
- 2.Fournier P.E., Colson P., Levasseur A., Gautret P., Luciani L., Bedotto M., et al. Genome sequence analysis enabled deciphering the atypical evolution of COVID-19 in Marseille, France. medRxiv. 2021 doi: 10.35088/kmct-tj43. [DOI] [Google Scholar]
- 3.Fournier P.E., Colson P., Levasseur A., Devaux C.A., Gautret P., Bedotto M., et al. Emergence and outcomes of the SARS-CoV-2 ‘Marseille-4’ variant. Int J Infect Dis. 2021;106:228–236. doi: 10.1016/j.ijid.2021.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-Beltran W.F., Lam E.C., Denis K.S., Nitido A.D., Garcia Z.H., Hauser B.M., et al. Circulating SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. medRxiv. 2021 doi: 10.1101/2021.02.14.21251704. [DOI] [Google Scholar]
- 5.Colson P., Levasseur A., Gautret P., Fenollar F., Hoang V.T., Delerce J., et al. Introduction into the Marseille geographical area of a mild SARS-CoV-2 variant originating from sub-Saharan Africa. Travel Med Infect Dis. 2021;40:101980. doi: 10.1016/j.tmaid.2021.101980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 8.Shu Y., McCauley J. GISAID: Global initiative on sharing all influenza data – from vision to reality. Euro Surveill. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen L.T., Schmidt H.A., von H.A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Letunic I., Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.La Scola B., Le Bideau M., Andreani J., Hoang V.T., Grimaldier C., Colson P., et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grabowski F., Kochanczyk M., Lipniacki T. L18F substrain of SARS-CoV-2 VOC-202012/01 is rapidly spreading in England. medRxiv. 2021 doi: 10.1101/2021.02.07.21251262. [DOI] [Google Scholar]
- 14.Kemp S.A., Collier D.A., Datir R.P., Ferreira I.A.T.M., Gayed S., Jahun A., et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021 doi: 10.1038/s41586-021-03291-y. Feb 5; Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varabyou A., Pockrandt C., Salzberg S.L., Pertea M. Rapid detection of inter-clade recombination in SARS-CoV-2 with Bolotie. bioRxiv. 2020 doi: 10.1101/2020.09.21.300913. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.