Abstract
Renal cell carcinoma (RCC) is a most common type of urologic neoplasms; it accounts for 3% of malignant tumors, with high rates of relapse and mortality. The most common types of renal cancer are clear cell carcinoma (ccRCC), papillary renal cell carcinoma (pRCC), and chromophobe renal carcinoma (chRCC), which account for 90%, 6–15%, and 2–5%, respectively, of all renal malignancies. Although surgical resection, chemotherapy, and radiotherapy are the most common treatment method for those diseases, their effects remain dissatisfactory. Furthermore, recent research shows that the treatment efficacy of checkpoint inhibitors in advanced RCC patients is widely variable. Hence, patients urgently need a new molecular biomarker for early diagnosis and evaluating the prognosis of RCC. MicroRNAs (miRNAs) belong to a family of short, non-coding RNAs that are highly conserved, have long half-life evolution, and post-transcriptionally regulate gene expression; they have been predicted to play crucial roles in tumor metastasis, invasion, angiogenesis, proliferation, apoptosis, epithelial-mesenchymal transition, differentiation, metabolism, cancer occurrence, and treatment resistance. Although some previous papers demonstrated that miRNAs play vital roles in renal cancer, such as pathogenesis, diagnosis, and prognosis, the roles of miRNAs in kidney cancer are still unclear. Therefore, we reviewed studies indexed in PubMed from 2017 to 2020, and found several studies suggesting that there are more than 82 miRNAs involved in renal cancers. The present review describes the current status of miRNAs in RCC and their roles in progression, diagnosis, therapy targeting, and prognosis of RCC.
Keywords: Review, Diagnosis, Kidney Neoplasms, MicroRNAs, Prognosis
Background
Renal cell carcinoma (RCC) is a typical malignant kidney lesion which represents 3% of all malignant tumors and has a high rate of relapse and a mortality rate of over 40%. In the last 20 years, there has been an annual increase of 2% in RCC incidence both worldwide and in Europe, leading to approximately 99 200 new RCC cases and 39 100 kidney cancer-related deaths within the European Union (EU) in 2018 [1]. Clear cell renal cell carcinoma (ccRCC) is the most common renal malignancy and accounts for approximately 90% of all kidney malignancy. Other subtypes, including papillary RCC (pRCC) and chromophobe RCC (chRCC), account for 6–15% and 2–5% of renal cancer cases, respectively [2]. Although modern ultrasound and CT technologies enable the diagnosis of kidney tumors in the early stages, imaging diagnosis does not allow precise differentiation between benign and malignant tumors [3]. Furthermore, laparoscopy and robotic surgery procedures have been developed, and biologic response modifiers have been applied in patients with metastatic RCC. However, the prognosis of terminal cancer cases remains poor, with a 5-year survival rate of 5–10% [4]. Tumor recurrence is mainly due to RCC resistance to chemotherapy and radiotherapy, and 5–10% of ccRCC cases extend into the renal vein or the inferior vena cava (IVC) [5]. Although anti-PD-L1 therapy can improve the overall survival (OS) of RCC patients, a recent study demonstrate that the treatment efficacy of checkpoint inhibitors is widely variable in advanced RCC patients [6]. Therefore, a novel molecular biomarker that can be used for early diagnosis and evaluating the prognosis of RCC, and even serve as a novel therapeutic target, is urgently needed.
MicroRNAs(miRNAs) are a family of short, non-coding RNAs that are highly conserved and have a long half-life, and they have been predicted to regulate numerous protein-coding genes. Mature miRNAs exist as 20–24 nucleotide miRNA duplexes comprising an miRNA guide strand and its complementary passenger strand [7]. The miRNA guide strand is subsequently integrated into the RNA-induced silencing complex (RISC), through which it carries out its regulatory function on target mRNAs. The miRNA-loaded RISC suppresses gene expression by interacting with complementary sequences in the 3′ untranslated regions (3′-UTRs) and coding sequences of mRNAs, inducing translational repression [8]. A recent study newly identified ubiquitin ligases as a new type of regulator of target-directed microRNA degradation that can function independently of the trimming and tailing processes, implying that controlling the microRNA decay pathway will become a strategy for treating diseases and cancers [9].
Accumulating evidence has revealed that miRNAs derived from guide strands are pivotal regulators of all hallmarks of cancer, including cell growth and cell cycle control, apoptosis, invasion, and metastasis [10]. Although some previous studies have illustrated the role of miRNAs in renal cancer, their functions remain unclear [11]. Indeed, a recent study posits that some miRNAs generated from passenger strands, such as miR-144-5p, miR-145-3p, and miR-199a-3p, also induce antitumor effects via their targeting of oncogenes in several cancers [12]. The aim of the present review was to describe miRNAs profile in RCC and their roles in the progression, diagnosis, therapeutic targeting, and prognostication of RCC.
miRNAs Act as Tumor Suppressors in Renal Cancer
RAS/MAPK Signaling Pathway
Accumulated evidence has confirmed that the Ras-Raf-MEK-ERK signaling pathway plays a vital role in the development of cancer [13]. For instance, astrocyte-elevated gene-1 (AEG-1), a downstream gene of Ha-ras, is highly expressed in RCC cells and increases cell growth and invasion, while its effect can be reversed by miR-384 [14]. Another study demonstrated that p21-activated kinase 5 (PAK5), a downstream target of Rho GTPases, is upregulated in renal cancer, and impairs the repression of RCC metastasis induced by miR-106a-5p [15]. Kirsten rat sarcoma viral oncogene (KRAS) and Rho-associated protein kinase 1 (Rock1), which are RAS GTPases, are highly expressed in RCC cells and facilitate tumor progression, whereas these effects can be suppressed by miR-199a and miR-532-5p, respectively. Furthermore, miR-532-5p represses tumor growth and inhibits the expression of P-ERK and ETS1 in vivo, and ETS1 act as an oncogene gene in multiple cancers [16,17] (Table 1).
Table 1.
miRNAs act as tumor suppressors in renal cancer. A summary of miRNAs name, specimen types, targeted messenger RNAs, functions, and clinical application is provided.
| MicroRNA | Specimen | Biological function | Clinical application | Target | Pathways | Ref. |
|---|---|---|---|---|---|---|
| miR-122-5p and miR-206 | In serum | liquid biopsy | [3] | |||
| miR-144-5p | In vitro | Suppress cell proliferation, migration and invasion | DFS | SDC3 | [12] | |
| miR-384 | In vitro | Inhibit cell proliferation, colony formation and invasion | AEG-1 | RAS signaling pathway | [14] | |
| miR-106a-5p | In vitro and in vivo | Inhibit tumor metastasis | Diagnosis, potential therapeutic target | PAK5 | RAS signaling pathway | [15] |
| miR-532-5p | In vitro and in vivo | Inhibit tumor growth and decrease expression of KRAS, NAP1L1, P-ERK and ETS1 | KRAS, NAP1L1 and ETS1 | MAPK signaling pathway | [16] | |
| miR-199a | In vitro | Suppress cell proliferation, migration and invasion | ROCK1 | RAS signaling pathway | [17] | |
| miR-622 | In vitro | Suppress cell migration and invasion and decrease levels of P-ERK | CCL18 | MAPK signaling pathway | [18] | |
| miR-200b | In vitro and in vivo | Inhibit tumor metastasis and decrease levels of P-ERK | LAMA4 | MAPK signaling pathway | [19] | |
| miR-363 | In vitro and in vivo | Suppress cell proliferation, migration and invasion, decrease level of STAT3, JAK2, VEGF, p-STAT3/JAK2/ERK, PDGF-A/B, N-cadherin, vimentin and ZEB1 | GHR, S1PR1 | MAPK/VEGF signaling pathway | [20, 38] | |
| miR-10a-5p | In vitro | Suppresses cell proliferation, migration and invasion, reduce p-ERK1/2, AKT, FAK and SRC | Potential therapeutic target | SKA1 | MAPK and AKT signaling pathway | [21] |
| miR-149 | In vitro | Inhibit cell migration, invasion and proliferation | FOXM1 | PI3K/AKT signaling pathway | [23] | |
| miR-320a | In vitro and in vivo | Reduce tumor growth | OS, diagnosis | FoxM1 | PI3K/AKT signaling pathway | [24] |
| miR-338-3p | In vitro | Increase cell proliferation and invasion | p-AKT and PI3K | PI3K/AKT signaling pathway | [25] | |
| miR-15a | In vitro | Inhibit cell proliferation, invasion and induce apoptosis, decrease expression of P13K, p-AKT, mTOR, cyclin D1, cyclin E, Bax, c-Myc and MMP3 | eIF4E | P13K/AKT/mTOR signaling pathway | [26] | |
| miR-488 | In vitro and in vivo | Reduce tumor growth and decrease expression of N-cadherin, vimentin, p-AKT, p-mTOR, and P13K | Potential therapeutic target | HMGN5 | P13K/AKT/mTOR signaling pathway | [27] |
| miR-520c-3p/ 372-3p/373-3p | In vitro and in vivo | Decrease tumor growth, metastasis and increase the expression of E-cadherin and PTEN | SPOP | PI3K/AKT signaling pathway | [28] | |
| miR-203 | In vitro and in vivo | Decrease tumor growth, metastasis and increase the expression of E-cadherin, PTEN, p21 and p27 | PI3K/AKT signaling pathway | [29] | ||
| miR-148a | In vitro and in vivo | Reduce tumor growth and decrease p-Akt/mTOR, improve sensitivity to TRAIL and cisplatin | Potential therapeutic target | AKT2 and Rab14 | AKT signaling pathway | [30, 31] |
| miR-766-3p | In vitro and in vivo | Reduce tumor growth and decrease P-AKT and P-ERK | OS | SF2 | AKT and MAPK signaling pathway | [32] |
| miR-375 | In vitro | Inhibits cell proliferation, migration and invasion, while induce cell apoptosis | PDK1 | [33] | ||
| miR-100 | In vitro | Inhibit cell invasion, migration and increase autophagy, reduce expression of mTOR, MMP-2 and MMP-9, whereas improve level of LC3 and LC3-II/LC3-I | NOX4 | mTOR signaling pathway | [34] | |
| miR-205-5p | In vitro and in vivo | Repress tumor growth, inhibit expression of p-PI3K/Akt/-mTOR, increase sensitivity of cell to sunitinib, paclitaxel, 5-FU and oxaliplatin | OS, potential therapeutic target | VEGFA | VEGFA and Pl3k/AKT signaling pathway | [36] |
| miR-299 | In vitro and in vivo | Suppress tumor growth and inhibit expression of vimentin and N-cadherin | VEGF | VEGF signaling pathway | [37] | |
| miR-218 | In vitro and in vivo | Decreases the expression of VEGFA, p-PI3K/p-Akt/p-mTOR diminish tumor angiogenesis | OS | GAB2 | VEGFA and Pl3k/AKT/mTOR signaling pathway | [39] |
| miR-125a-3p | In vitro | Inhibit the expression of VEGF and tube numbers formed by HUVECs | OS, DFS | VEGF | VEGF signaling pathway | [122] |
| miR-148b-3p | in vitro and in vivo | Suppress tumor growth, tube formation of HUVECs and inhibit expression of HIF-1a, VEGF-A, PDGF-BB, and PDGF-D | FGF2 | VEGF signaling pathway | [41] | |
| miR-486-5p | in vitro | Inhibit cell proliferation and induce apoptosis, decrease apoptosis resistance induced by CCL2 | TAK1 | TGF-β signaling pathway | [43] | |
| miR-328 | In vitro | Inhibit cell proliferation | ITGA5 | TGF-β signaling pathway | [44] | |
| miR-186 | In vitro | Inhibit cell proliferation, invasion and induce apoptosis, decrease level of p-IkBa and p-p65 | SENP1 | NF-κB signaling pathway | [46] | |
| miR-765 | In vitro | Suppress tumor growth and inhibit expression of VEGFA and Ki67 and eliminate lipids accumulation | PLP2 | Metabolic related mechanism | [48] | |
| miR-409-3p | In vitro | Decrease cell extracellular acidification rate, ATP production and increased oxygen consumption rate | PDK1 | Metabolic related mechanism | [50] | |
| miR-497-5p | In vitro | Inhibit cell proliferation, migration and increase apoptosis | OS | PD-L1 | Immunity related mechanism | [51] |
| miR-216a | In vitro and in vivo | Reduce tumor growth | TLR4 | Immunity related mechanism | [52] | |
| miR-211-5p | In vitro and in vivo | Decrease tumor growth and metastasis | DFS, potential therapeutic target | SNAI1 | EMT program | [58] |
| miR-124/203 | In vitro | Inhibit cell proliferation and migration | ZEB2 | EMT program | [59] | |
| miR-101-5p | In vitro | Inhibit cell proliferation, invasion and induce apoptosis | slug | EMT program | [60] | |
| miR-490-3p | In vitro and in vivo | Inhibit tumor growth and metastasis, decrease VM formation | TR4 | [61] | ||
| miR-32-5p | In vitro and in vivo | Inhibit tumor metastasis and repress expression of TR4, HGF and p-Met | TR4 | [62] | ||
| miR-451a | In vitro | Suppresses cell migration and invasion | PMM2 | [121] | ||
| miR-200a-3p | In vitro and in vivo | Suppress tumor growth | CBL | [54] | ||
| miR-182-5p | In vitro and in vivo | Inhibit tumor growth and metastasis, increase expression of P53 | [56] | |||
| miR-376b-3p | In rcc tissues | PFS, diagnosis | [101] | |||
| miR-9-5p | In rcc tissues | Diagnosis | [102] | |||
| miR-10a-5p/ 10b-5p/106a-5p/142-5p | In rcc tissues | Diagnosis | [107] | |||
| miR-1208 | In vitro | Inhibits cell proliferation and promote apoptosis, sensitizes cisplatin-induced apoptosis and TRAIL-induced apoptosis | Potential therapeutic target | TBCK | [111] | |
| miR-99a-3p | In vitro | Inhibit cell proliferation and facilitate apoptosis, induce S phase arrest and increase sunitinib sensitivity | Potential therapeutic target | RRM2 | [112] | |
| miR-126 | In vitro | Decrease cell migration and lactate production, inhibit expression of p-mTOR, and sensitize the cancer cells tocisplatin or X-ray treatment | Potential therapeutic target | SERPINE1 | mTOR signaling pathway | [113] |
| miR-378a-5p | In vitro | Inhibit cell proliferation, migration, invasion and promote apoptosis | OS | [119] | ||
| miR-31-5p | In vitro | Suppress cell proliferation, migration and invasion | OS | CDK1 | [120] | |
| miR-22/24/99a/ 194/214/ 335/339/708 | Biomarker | [6] |
miRNAs – microRNAs; DFS – disease-free survival; SDC3 – syndecan-3; AEG – 1-astrocyte-elevated gene-1; RAS – rat sarcoma; PAK5 – p21-activated kinase 5; KRAS – Kirsten rat sarcoma viral oncogene; p-ERK – phosphorylate extracellular signal regulated kinase; ETS1 – E26 transformation-specific-1; MAPK – mitogen-activated protein kinases; ROCK1 – Rho-associated coiled-coil-forming protein kinase 1; CCL18/2 – C-C motif chemokine 18/2; LAMA4 – laminin subunit alpha-4; STAT3 – signal transducer and activator of transcription 3; JAK2 – Janus kinases 2; VEGF – vascular endothelial growth factor; PDGF – platelet-derived growth factor; ZEB1 – zinc finger E-box binding homeobox 1; GHR – growth hormone receptor; S1PR1 – sphingosine-1-phosphate receptor 1; AKT – protein kinase B; FAK – focal adhesion kinase; SKA1 – spindle and kinetochore-associated protein 1; FOXM1 – forkhead box M1; OS – overall survival; PI3K – phosphatidylinositol 3-kinase; mTOR – mammalian target of rapamycin; MMP3 – matrix metalloproteinase-3; eIF4E – eukaryotic initiation factor 4E; HMGN5 – high-mobility group nucleosome binding domain 5; PTEN – phosphatase and tensin homolog deleted on chromosome 10; SPOP – speckle-type POZ protein; TRAIL – tumor necrosis factor-related apoptosis inducing ligand; Rab14 – ras-related protein 14; SF2 – splicing factor 2; PDK1 – phosphoinositide-dependent kinase 1; NOX4 – NADPH oxidase 4; LC3 – microtubule-associated protein 1 light chain 3; GAB2 – GRB2-associated binding protein 2; HUVECs – human umbilical vein endothelial cells; FGF2 – fibroblast growth factor 2; TAK1 – TGF-beta-activated kinase 1; ITGA5 – integrin alpha5; SENP1 – sentrin specific peptidase1; PLP2 – proteolipid protein 2; PD-L1 – programmed death ligand 1; TLR4 – toll-like receptor 4; SNAI1 – snail family transcriptional repressor 1; TR4 – testicular nuclear receptor 4; VM – vasculogenic mimicry; HGF – hepatocyte growth factor; PMM2 – phosphomannomutase 2; CBL – casitas B-lineage lymphoma; PFS – progression-free survival; TBCK – TBC1-domain-containing kinase; RRM2 – ribonucleotide reductase regulatory subunit m2; SLC7A5 – solute carrier family 7 member 5; HIF1a/2a – hypoxia inducible factor 1a/2a; SERPINE1 – serine protease inhibitor clade E member 1.
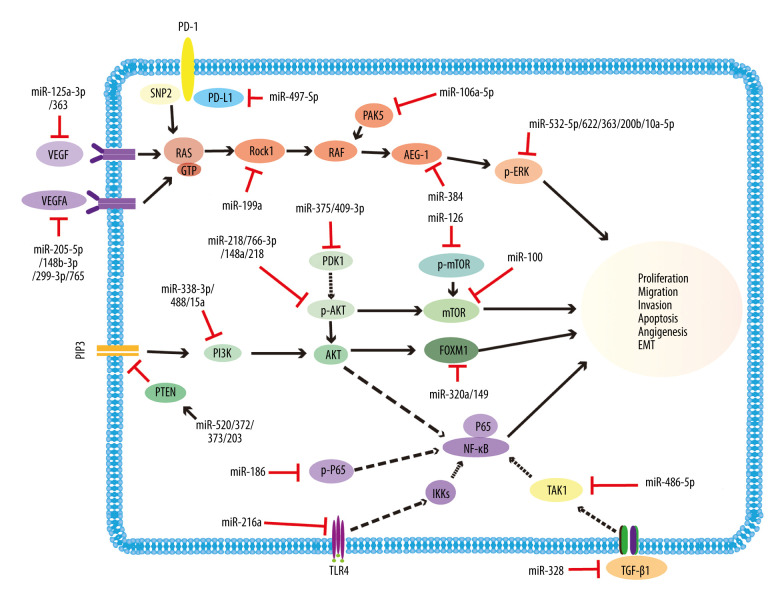
The protein level of phosphorylation ERK(p-ERK), which is associated with the MAPK signaling pathway, can be increased by CCL18/LAMA4 and reduced by miR-622/200b in RCC. In addition, high CCL18 and LAMA4 expression in kidney cancer facilitates tumor progression, while its effect can be reversed by miR-622, and C-C motif chemokine 18(CCL18) and laminin subunit alpha-4 (LAMA4) play key roles in tumor progression [18,19]. Furthermore, overexpression of miR-363 impairs tumor growth and decreases the expression of p-ERK, N-cadherin, vimentin, and ZEB1 in ccRCC [20]. Likewise, SKA1 increases the levels of p-ERK1/2 and p-AKT, enhances tumor development, and also decreases the effect of tyrosine kinase inhibitor (TKI) treatment in renal cancer, whereas these effects can be reversed by miR-10a-5p, and spindle and kinetochore-associated protein 1 (SKA1) has been reported to be an oncogene in multiple cancers [21] (Figure 1).
Figure 1.
Representative diagram of miRNAs acting as tumor suppressors, and their associated signaling pathways in renal cancer. The drawing mainly illustrates that miR-125a-3p/363 and miR-205-5p/148b-3p/299-3p/765 inhibit the VEGF and VEGFA signaling pathways through decreasing the expression of VEGF and VEGFA, respectively. miR-199a, miR-106a-5p, miR-384, and miR-532-5p/622/363/200b/10a-5p restrain RAS/MAPK signaling pathways by decreasing the levels of Rock1, PAK5, AEG-1, and p-ERK, respectively. Furthermore, miR-338-3p/488/15a, miR-218/766-3p/148a/218, miR-375/409-3p, miR-126, miR-100, and miR-320a/149 influence the PI3k/AKT/mTOR signaling pathways by decreasing the levels of PI3k, p-AKT, PDK1, p-mTOR, mTOR, and FOXM1, respectively. In contrast, miR-520/372/373/203 increases the expression of PTEN and exerts the same effect. miR-328 and miR-486-5p inhibit the TGF-β signaling pathways through decreasing the expression of TGF-β1 and TAK1, respectively. miR-186 and miR-216a influence NF-κB signaling pathways by decreasing the levels of p-p65 and TLR4, respectively. In addition, miR-497-5p targets PD-L1, thus influencing immunity-related mechanisms. VEGF – vascular endothelial growth factor; Rock1 – Rho-associated coiled-coil-forming protein kinase; PAK5 – p21-activated kinase 5; AEG-1 – AEG-1-astrocyte-elevated gene-1; p-ERK – phosphorylate extracellular signal regulated kinase; PI3k – phosphatidylinositol 3-kinase; p-AKT – p-protein kinase B; PDK1 – phosphoinositide-dependent kinase 1; mTOR – mammalian target of rapamycin; FOXM1 – forkhead box M1; PTEN – phosphatase and tensin homolog deleted on chromosome 10; TGF-β1 – transforming growth factor-β 1; TAK1 – TGF-beta-activated kinase 1; PLP2 – proteolipid protein 2; INF; TLR4 – toll-like receptor 4; PD-L1 – programmed death ligand 1.
PI3K/AKT/mTOR Signaling Pathway
The PI3K/Akt/mTOR signaling pathway is frequently dysregulated in renal cancer [22]. Forkhead box protein M1(FOXM1) belongs to the Forkhead box family, which is a downstream target of the PI3K/Akt pathway. FOXM1 is highly expressed in RCC and enhances tumor development, and these effects can be reversed by miR-149 and miR-320a [23,24]. The expression of PI3K and Akt can be downregulated by miR-338-3p, miR-15a, and miR-488 or upregulated by KIFC (Kinesin family member C1), eIF4E (eukaryotic initiation factor 4E), and HMGN5 (High-mobility group nucleosome binding domain 5) in RCC. In addition, overexpression of miR-338-3p, miR-15a, and miR-488 leads to repression of renal cancer progression induced by KIFC1, eIF4E, and HMGN5, respectively. KIFC1, eIF4E, and HMGN5 have been reported to act as oncogenes in various cancers [25–27] Moreover, the expression of PTEN, which is a master regulator of the PI3K/Akt pathway, could be augmented by miR-520/372/373 and miR-203 or diminished by SPOP in RCC. Overexpression of miR-520/372/373 results in attenuated renal tumor development by impairing SPOP, and speckle-type POZ protein (SPOP) has been reported to act as an oncogene in renal cancer [28,29] (Table 1).
AKT2 belongs to the Akt family, is highly expressed in RCC, and abolishes the inhibition of cell growth and mobility induced by miR-148a. In addition, miR-148a decreases the p-Akt and mTOR levels in renal cancer and boosts the expression of TRAIL (tumor necrosis factor-related apoptosis inducing ligand) and increases the drug sensitivity of RCC cells to cisplatin by regulating Rab14, Rab14 (Rab14 GTPase) as a member of the RAS oncogene family [30,31]. Additionally, the expression of P-Akt and P-ERK can be improved by splicing factor 2 (SF2) and reduced by miR-766-3p in RCC. Overexpression of miR-766-3p leads to suppression of tumor growth by regulating SF2, and SF2 belongs to the splicing factor family and promotes carcinoma formation [32]. Furthermore, PDK1 (phosphoinositide-dependent kinase 1), an important molecule in the Akt pathway, is also highly expressed in kidney cancer cells and increases cell proliferation, while its effect can be reversed by miR-375 [33]. Interestingly, miR-100 attenuates the expression of mTOR and NOX4, and augments the levels of LC3 and LC3-II/LC3-I in RCC, consequently impairing the aggressiveness of RCC cells and improving autophagy. NOX4 (NADPH Oxidase 4) and LC3 (microtubule-associated protein 1 light chain 3) act as vital regulators of autophagy [34] (Figure 1).
VEGF Signaling Pathway
Vascular endothelial growth factor (VEGF), a cytokine secreted by tumor cells, plays a pivotal role in tumor development [35]. For example, VEGFA is highly expressed in RCC and enhances tumor progression, while this effect can be reversed by miR-205-5p and miR-299-3p. miR-205-5p increases the sensitivity of RCC cells to sunitinib, paclitaxel, 5-FU, and oxaliplatin by downregulating the levels of VEGFA and p-PI3K/p-Akt/p-mTOR [36,37]. Another study showed that the expression of VEGF is upregulated by GHR, which boosts tumor mobility of RCC cells, whereas these effects can be reversed by miR-363, and GHRH was verified to be positively correlated with the proliferation of renal cell carcinoma [38]. Similarly, miR-218 not only decreases the expression of VEGFA and p-PI3K/p-Akt/p-mTOR and restrains the migration ability of HUVECs (human umbilical vein endothelial cells), but also diminishes tumor angiogenesis in RCC by downregulating GRB2-associated binding protein 2 (GAB2). GAB2 is an important member of the Gabs family and acts as an oncogene in multiple cancers [39]. Previous evidence has shown that FGF (fibroblast growth factor) and VEGF play equal roles in angioblast induction and migration during vascular development [40]. Indeed, FGF2 augments the tube formation and invasion of HUVECs in RCC, but its effect can be reversed by miR-148b-3p. In addition miR-148b-3p impairs the expression of VEGF-A and platelet-derived growth factor-BB/D(PDGF-BB/D) in RCC, and PDGF-BB/D act as pro-angiogenic actors in multiple cancers [41] (Table 1).
TGF-β/NF-κB signaling pathway
The Transforming growth factor-β (TGFβ) and nuclear factor kappa B(NF-κB) pathways play pivotal roles in renal disease [42]. Indeed, TAK1 is highly expressed in RCC and increases tumor progression, while its effect can be reversed by miR-486-5p, and TGF-beta-activated protein kinase 1 (TAK1) is a critical regulator of the TGF-beta pathway [43]. Another study showed that TGF-β1 promotes tumor development by inhibiting miR-328, thus enhancing the expression of integrin α5 (ITGA5). miR-328 reduces cell proliferation of RCC cells by downregulating ITGA5, and ITGA5 belongs to the integrin family [44]. Interestingly, SNIP1 (Smad nuclear interacting protein 1) can negatively regulate the transcription of the NF-κB signaling pathways [45]. Indeed, the expression of p-IkBa and p-p65, which are components of the NF-κB signaling pathways, can be boosted by SENP1 and impaired by miR-186 in renal cancer. In addition, miR-186 suppresses RCC cell progression by targeting SENP1 in vitro [46] (Figure 1).
Metabolism/Immunity-Related Mechanism
Metabolic changes in the tumor micro-environment, inhibit the antitumor immunity by producing immunosuppressive metabolites [47]. For example, PLP2 high expression in ccRCC promotes tumor growth and mobility, and increases the expression of VEGFA and enhances lipid accumulation, while this effect can be reversed by miR-765. PLP2 is a novel member of the Cd-up-regulated genes and has been reported to act as an oncogene in breast cancer [48]. Previous research has shown that RCC cells, like multiple other types of cancers cells, have aberrant HIF stabilization and are dependent on aerobic glycolysis for ATP production [49]. Indeed, PDK1 high expression in ccRCC cells boosts ECAR (extracellular acidification rate) under hypoxic conditions, improves ATP production, and diminishes the oxygen consumption rate (OCR) of tumor cells, whereas these effects can be reversed by miR-409-3p [50]. Tumor immune escape is a common topic of research and is a hallmark of cancer. For instance, PD-L1, a ligand of PD-1 (Programmed death 1), is overexpressed in renal cancer and facilitates tumor progression, but its effect can be reversed by miR-497-5p [51]. Furthermore, Toll-like receptors (TLRs) mediate the innate immune response, which has been shown to participate in tumor development; for example, miR-216a suppresses RCC growth in vitro and in vivo by targeting TLR4 [52] (Table 1).
Other Mechanisms
Depending on its specific substrate, an E3 ligase can either promote or inhibit cancer development. For example, CbL, a RING finger E3 ubiquitin ligase, has been identified as a critical regulator of cancer metastasis [53], and is overexpressed in RCC, which boosts tumor progression by downregulating miR-200a-3p [54]. E3 ubiquitin ligases usually retain an p53 inactive state in multiple cancers [55], whereas miR-182-5p causes cell cycle arrest at the G2/M phase, thus repressing ccRCC progression by upregulating p53 [56]. The abnormal activation of transcription factors promotes the proliferation and differentiation of tumor cells [57]. For instance, snail family transcriptional repressor 1 (SNAI1), zinc finger E-box binding homeobox 2 (ZEB2), and slug, as transcription factors related to EMT, are highly expressed in RCC and increase tumor development, while their effects can be reversed by miR-211-5p, miR-124/miR-203, and miR-101-5p, respectively [58–60]. Another study showed that testicular nuclear receptor 4 (TR4) promotes vasculogenic mimicry (VM) formation and metastasis of ccRCC and augments the expression of vimentin, whereas these effects can be reversed by miR-490-3p and miR-32-5p, respectively [61,62]. TR4 is a transcriptional factor and is positively associated with the progression of prostate cancer [63] (Table 1).
miRNAs Act as Oncogenes in Renal Cancer
mTOR/Metabolic Pathway
Mammalian target of rapamycin (mTOR) is a protein kinase regulating cell growth and metabolism in various cancers [64]. miR-92b-3p decreases the protein expression of TSC complex subunit 1 (TSC1) and increases the phosphorylation of p70S6 kinase, which is downstream of TSC1, consequently activating the mTOR pathway and promoting ccRCC progression. TSC1 is an inhibitor of mTORC1 [65]. Another study demonstrated that overexpression of miR-501-5p increases cell autophagy through activating p-mTOR, leading to p53 degradation in renal cancer, thus facilitating tumor progression of RCC [66]. Recent research suggests that IMPA2 (inositol monophosphotase 2) leads to decreasing p-mTORC1 levels in ccRCC cells, and thus could be a biomarker for guiding the use of mTOR inhibitors to combat metastatic ccRCC in clinical practice [67]. Indeed, IMPA2 underexpression in ccRCC diminishes the expression of N-cadherin and Slug, and sabotages tumor metastasis by downregulating miR-25-3p [68]. Dysregulated cellular energetics is one of the hallmarks of RCC and of multiple cancers [47]. For example, upregulation of the pentose phosphate pathway (PPP) is a key feature of the dysregulated metabolism of RCC cells, but G6PD is a rate-limiting enzyme of the PPP, and its inhibition attenuates the survival of RCC cells. Furthermore, upregulation of miR-146a-5p increases the expression of G6PD and transketolase (TKT), facilitating proliferation of RCC cells [69] (Table 2).
Table 2.
miRNAs act as oncogenes in renal cancer. A summary of miRNAs name, specimen types, targeted messenger RNAs, functions and clinical applicant is provided.
| MicroRNA | Specimen | biological Function | clinical application | Target | Pathways | Refs. |
|---|---|---|---|---|---|---|
| miR-154-5p | In vitro | Promote cell proliferation, migration, invasion and inhibit apoptosis | OS | [2] | ||
| miR-92b-3p | In vitro | Promote cell proliferation, migration and invasion, decrease expression of TSC1 and enhance p-p70S6 kinase | OS | TSC1 | mTOR signaling pathway | [65] |
| miR-501-5p | In vitro and in vivo | Increase cell autophagy, growth, migration and activate mTOR kinase | mTOR signaling pathway | [66] | ||
| miR-25-3p | In vitro | Enhance cell migration and increase expression of N-cadherin and Slug | OS | IMPA2 | [68] | |
| miR-146a-5p | In vitro | Increase cell proliferation and improve expression of G6PD and TKT | Metabolic related mechanism | [69] | ||
| miR-193a-3p and -224 | In vitro | Promote cell proliferation, invasion, migration and inhibit apoptosis, improve expression of PI3k and p-Akt | ST3GalIV | PI3K/Akt signaling pathway | [71] | |
| miR-19 | In vitro | Enhance cell proliferation and inhibit expression of FRK and PTEN | FRK and PTEN | PI3K/Akt signaling pathway | [72] | |
| miR-122 | In vitro and in vivo | Promote tumor growth, enhance expression of ZEB1 and ZEB2, p-Erk1/2 and p38 | PFS | FOXO3 and occludin | PI3K/Akt, MAPK signaling pathway | [73, 91] |
| miR-142-5p | In vitro | Increase cell proliferation and migration | BTG3 | [75] | ||
| miR-452-5p | In vitro and in vivo | Enhance tumor metastasis | OS, target | SMAD4 | TGF-β signaling pathway | [77] |
| miR-1274a | In vitro | Promote cell proliferation and inhibit apoptosis | BMPR1B | [79] | ||
| miR-543 | In vitro and in vivo | Facilitate tumor growth, metastasis and increase expression β-catenin and p-GSK-3β, while inhibit expression of p21 | DKK1, KLF6 | Wnt signaling pathway | [82, 86] | |
| miR-125b | In vitro and in vivo | Promote tumor growth and metastasis, inhibit sensitivity of cells to doxorubicin and sunitinib | DKK3 | Wnt signaling pathway | [83] | |
| miR-146b-5p | In vitro and in vivo | Inhibit expression of p65 and TRAF6 | TRAF6 | NF-κB signaling pathway | [84] | |
| miR-381-3p | In vitro | Inhibit TNF-induced cell apoptosis and necroptosis | OS | RIPK3 | [89] | |
| miR-223-3p | In vitro | Promote cell proliferation, migration, invasion and increase expression of KRAS | OS | SLC4A4 | RAS signaling pathway | [90] |
| miR-21 | In vitro and in serum | Increase cell proliferation, invasion, migration and reduce apoptosis, decrease expression of p53 and p21, Bax, cyclin E2, VEGFA and p-c-Jun | Diagnosis, target | PTEN, PDCD4 | PI3K/AKT and NF-κB signaling pathway | [8,93,94] |
| miR-204-5p | In urine/mice/RCC tissues | Liquid biopsy | [96] | |||
| miR-301a-3p and -1293 | In plasma | Liquid biopsy | [97] | |||
| miR-19b-3p | In vitro and in exosomes | Enhance cell migration and invasion, while impair expression of E-cadherin and PTEN | Diagnosis | [99] | ||
| miR-130b/18a/ 223 | In RCC tissues | Diagnosis | [108] | |||
| miR-15a/ 182/138/200c/16/ 210/34a/155 | In RCC tissues | Diagnosis | [104] | |||
| miR-3199-2/ 1293 | In RCC tissues | Diagnosis | [105] | |||
| miR-21/142/150/ 155 | In RCC tissues | Diagnosis | [106] | |||
| miR-489-3p/630 | In vitro | Promote cell proliferation and chemoresistance to oxaliplatin | OCT2 | [110] | ||
| miR-720 | In vitro and in vivo | Promote tumor growth | OS, diagnosis | E-cadherin and αE-catenin | [118] | |
| miR-23a-3p | In vitro | Promote cell proliferation, migration and invasion, while inhibit apoptosis | OS | PNRC2 | [117] | |
| miR-572 | In vitro | Promote cell proliferation, migration, invasion and inhibit cell apoptosis | OS | [4] | ||
| miR-221-5p | In vitro | Promote cell proliferation, migration, invasion and inhibit apoptosis | OS | [114] | ||
| miR-566 | In vitro | Promote cell proliferation, migration, invasion and inhibit apoptosis | OS | [115] | ||
| miR-663a | In vitro | Promote cell proliferation, invasion, migration and inhibit apoptosis | OS | [116] | ||
| miR-155-5p/ 210-3p | In RCC tissues | Biomarker of recurrence | [123] |
OS – overall survival; TSC1 – tuberous sclerosis complex subunit 1; mTOR – mammalian target of rapamycin; IMPA2 – myo-inositol monophosphatase 2; G6PD – glucose-6-phosphate dehydrogenase; TKT – transketolase; ST3GalIV – alpha-2,3-sialyltransferase IV; PI3K – phosphatidylinositol 3-kinase; AKT – protein kinase B; FRK – fyn-related kinase; PTEN – phosphatase and tensin homolog deleted on chromosome 10; ZEB1 – zinc finger E-box binding homeobox 1; Erk – extracellular signal regulated kinase; PFS – progression-free survival; FOXO3 – forkhead box O3; BTG3 – B-cell translocation gene 3; TGF-β – transforming growth factor-β; SMAD4 – SMAD family member 4; BMPR1B – bone morphogenetic protein receptor type 1B; DKK1/3 – Dickkopf1/3; KLF6 – Kruppel-like factor 6; TRAF6 – TNF receptor associated factor 6; RIPK3 – receptor-interacting protein kinase 1; SLC4A4 – solute carrier family 4; PDCD4 – programmed cell death 4; VEGFA – vascular endothelial growth factor A; OCT2 – octamer binding transcription factor 2; pnrc2 – proline-rich nuclear receptor co-activator 2.
PI3K/AKT Signaling Pathway
The PI3K/Akt pathway is commonly mutated and highly activated in RCC, representing a tumorigenic characteristic [70]. The expression of p-PI3K and p-Akt is upregulated by miR-193a-3p and miR-224 and downregulated by ST3GalIV in RCC. miR-193a-3p and miR-224 promote tumor progression by targeting ST3GalIV (alpha-2,3-SialyltransferaseIV). ST3GalIV can catalyze the synthesis of α-2,3-sialic acid on the cell surface, which is closely related to tumor metastasis potential [71]. Another Study demonstrated that the level of p-PTEN was improved by FRK (Fyn-related kinase) because it is a substrate of FRK, but this effect can be impaired by miR-19 in ccRCC. In addition, miR-19 overexpression facilitates cell proliferation of renal cancer by modifying FRK and PTEN [72]. FOXO3 belongs to the Forkhead box family, which is downstream of the PI3K-Akt signaling pathway; it shows high expression in ccRCC and is negatively regulated by miR-122. Overexpression of miR-122 promotes tumor development and increases the expression of E-cadherin in kidney cancer [73]. Intriguingly, BTG3 (B-cell translocation gene 3) diminishes p-Akt levels and acts as a tumor suppressor in prostate cancer cells [74]. Indeed, BTG3 inhibits RCC cell proliferation by negatively mediating miR-142-5p [75] (Figure 2).
Figure 2.
Representative diagram of miRNAs acting as oncogenes and their correlation with signaling pathways in renal cancer. The picture mainly demonstrates that miR-224/193a-3p and miR-501-5p improve PI3K/AKT and mTOR signaling pathway through increasing the level of PI3K, AKT, and p-mTOR, respectively. On the contrary, miR-19, miR-122, and miR-92b-3p inhibit the expression of PTEN, and FOXO3 and TSC1 exert the same effect. miR-21 and miR-223-3p facilitate the VEGF and RAS/MAPK signaling pathway by augmenting the levels of VEGFA/c-jun and KRAS, respectively. Moreover, miR-125b and miR-543 increase the Wnt signaling pathway by decreasing the expression of DKK3 and DDK1, respectively. miR-452-5p activates the TGF-β signaling pathway by decreasing the expression of SMAD4. In addition, miR-146a-5p and miR-146b-5p target G6PD and TARL6, and thus are involved in PPP metabolism and inflammation mechanism, respectively. PI3k – phosphatidylinositol 3-kinase; p-AKT – p-protein kinase B; mTOR – mammalian target of rapamycin; PTEN – phosphatase and tensin homolog deleted on chromosome 10; FOXO3 – forkhead box O3; TSC1 – tuberous sclerosis complex subunit 1; VEGFA – vascular endothelial growth factor A; KRAS – Kirsten rat sarcoma viral oncogene; DKK1/3 – Dickkopf1/3; SMAD4 – SMAD family member 4; G6PD – glucose-6-phosphate dehydrogenase; TRAF6 – TNF receptor associated factor 6.
TGF-β/Wnt Signaling Pathway
TGF-β and Wnt regulate numerous developmental events and participate in the development of numerous cancers [76]. For example, SMAD family member 4 (SMAD4) is a critical component of TGF-β signaling and low expression in RCC, and suppresses tumor metastasis in vitro, while its effect can be reversed by miR-452-5p. In addition, miR-452-5p impairs sensitivity of renal cancer cells in TKI treatment by regulating SMAD4 [77]. BMPR1B (bone morphogenetic protein receptor type 1B) is a member of the TGF-β superfamily, and participates in the progression of numerous cancers [78]. Similarly, miR-1274a increases cell proliferation and decreases apoptosis of ccRCC by downregulating BMPR1B [79]. Previous evidence has verified that Dickkopf1 (DKK1) and DKK3 belong to the extracellular Wnt inhibitor family and act as tumor suppressors in renal cancer [80,81]. Likewise, miR-543 and miR-125b facilitate tumor growth through negatively regulating DKK1 and DKK3, respectively, in RCC. In addition, overexpression of miR-125b leads to decreasing sensitivity to doxorubicin and sunitinib in renal cancer cells [82,83] (Figure 2).
NF-κB signaling pathway
The protein levels of NF-κB (p65) and TRAF6 are decreased by miR-146b-5p in renal cancer. In addition, miR-146b-5p increases tumor growth by regulating TRAF6 (TNF receptor associated factor 6) and impairs the serum level of IFN-γ. TRAF6 is a signal transducer in the NF-κB pathway, and IFN-γ has been applied to treat patients with ovarian cancer [84]. Interestingly, KLF6 reduces the localization of p65 and inhibits cancer progression in glioblastoma [85]. Likewise, overexpression KLF6 increases the level of p21 and represses tumor progression in ccRCC, while its effect can be reversed by miR-543 [86], and p21 has been confirmed inactivate the NF-κB pathway in prostate cancer [87]. Previous evidence suggested that TNF induced cell apoptosis and necroptosis by deactivating the NF-κB pathway [88]. In contrast, miR-381-3p inhibits TNF-induced apoptosis and necroptosis through downregulating caspase-8, caspase-3, and RIPK3 (receptor-interacting protein kinase 1) in renal cancer, whereas it has no effect on TNF-induced NF-κB activation, thus facilitating tumor progression and implying a poor outcome for papillary RCC patients; these findings suggest that the NF κB pathway has different functions in different cells. RIPK3 is a key regulatory protein for programmed cell necroptosis [89] (Figure 2).
RAS/MAPK Signaling Pathway
The expression of KRAS can be attenuated by solute carrier family 4 (SLC4A4) and increased by miR-223-3p in ccRCC. In addition, SLC4A4 restrains ccRCC cell progression by targeting KRAS, whereas its effect can be reversed by miR-223-3p. SLC4A4 has been reported to inhibit the development of ccRCC [90]. Another study showed that the level of p-Erk1/2 and cell migration can be reduced by occludin and enhanced by miR-122 in ccRCC, consequently promoting tumor progression. Occludin has been reported to act as a tumor suppressor in ccRCC [91]. Furthermore, the levels of p-c-Jun, which is part of the MAPK pathway, and VEGFA can be increased by miR-21 or diminished by programmed cell death 4 (PDCD4) in RCC. In addition, PDCD4 reduces the number of tubes and tube junctions of HMEC-1 cells and impairs RCC cell mobility, while its effect can be reversed by miR-21. PDCD4 act as a tumor suppressor in RCC [92, 93]. Additionally, miR-21 boosts cell proliferation and decreases the levels of p53, p21, cyclin E2, and Bax, which inhibits the p53 pathway to facilitate the progression of renal cancer [94] (Figure 2).
miRNAs Act as Biomarkers for the Diagnosis of RCC
Liquid Biopsies
Liquid biopsy is an important research area and has been used in multiple cancers, including renal cancer [95]. For instance, in PRCC-TFE3 Tg mice and translational RCC (tRCC) patients, miR-204-5p levels are significantly increased in urinary exosomes samples taken before and after tumor development; thus, miR-204-5p can be used as a marker to diagnose patients with Xp11 tRCC at an early stage [96]. Another study found ccRCC patients have a high level of plasma miR-1293 and a low level of plasma miR-301a-3p after surgery, and miR-301a-3p and miR-1293 are derived from extracellular vesicles. In other words, these miRNAs can serve as markers of the effect of surgical resection [97]. In addition, miR-92a-1-5p, miR-149-3p, and miR-424-3p in plasma exosomes can be used to distinguish RCC patients from healthy patients, with sensitivities of 87.5%, 75%, and 75%, and specificities of 77.3%,72.7%, and 81.8%, respectively [98]. Furthermore, the levels of serum miR-122-5p and miR-206 are significantly reduced in ccRCC patients. The level of miR-122-5p is correlated with RCC metastasis and grade, and the level of miR-206 is correlated with pT-stage and metastasis [3]. Intriguingly, miRNAs exist in both cancer exosomes and CSC (cancer stem cells). For example, the expression of miR-19b-3p in CSC (cancer stem cells) exosomes is significantly higher than that in renal cancer exosomes. Overexpression of miR-19b-3p impairs the expression of PTEN in ccRCC cells, promoting tumor cell metastasis [99] (Table 2).
Assessment of the treatment effect
Sunitinib is routinely used as first-line therapy for RCC, although 10–20% of advanced RCC patients are inherently refractory to sunitinib therapy [100]. Recent research demonstrated that miR-376b-3p enables the prediction of the response to Sunitinib therapy and the identification of patients who are likely to experience a long-term response (progression-free survival >12 months), with a sensitivity of 83% and specificity of 67%. The expression of miR-9-5p is also a marker of the effect of sunitinib treatment in RCC [101,102]. Nivolumab significantly improved the median OS benefit of patients with RCC, which led to regulatory approval in both the EU and the USA, but biomarkers to identify patient subgroups for immune-checkpoint treatment are not yet available [103]. A recent study demonstrated the expression of miRNAs, including miR-22/24/99a/194/214/335/339/708, in peripheral lymphocytes can be increased by anti-PD-1 treatment, implying that these miRNAs can be used to predict which patients are likely to have a long-lasting response to nivolumab treatment [6] (Table 1).
Differentiating Subtype
Although distinction of renal cancer subtypes depends on biopsy in clinical practice, some research demonstrates that miRNAs can also exert the same effect. For instance, miRNAs can distinguish TC-RCC (tubulocystic renal cell carcinoma) from CCPRCC (clear cell papillary renal cell carcinoma) and PRCC (papillary renal cell carcinoma), including miR-15a/182/138/200c/16, which are overexpressed, while miR-210/34a/155 are underexpressed in TC-RCC. Likewise, miR-3199-2 and miR-1293 can be used to distinguish patients with PRCC or other types of RCC from healthy patients [104,105]. Furthermore, miR-21 and miR-142 are significantly upregulated in ccRCC and sarcomatoid RCC, whereas miR-150 is overexpressed in chromophobe tumors. In contrast, miR-155 is downregulated in oncocytoma compared with all RCC subtypes [106] (Table 2).
Discrimination Between Benign and Malignant in RCC
Similarly, some evidence shows that miRNAs can be used be to discriminate malignant tissues from adjacent non-tumor tissues in kidney cancer. For instance, the expression of miR-10a-5p, miR-10b-5p, miR-106a-5p, and miR-142-5p is decreased in RCC nephrectomy specimens and has a sensitivity of 91.7% and specificity of 94% for distinguishing cancer from benign tissues [107]. Another report suggested that miR-130b, miR-18a, and miR-223 can distinguish patients with ccRCC from healthy controls, with a sensitivity of 83.1% and a specificity of 82.5% [108].
Target of Therapy
Accumulating evidence suggests that miRNAs levels correlate well with the effects of chemotherapy and radiotherapy in renal cancer changes [5]. Topotecan inhibits the function of mature miR-21, improves chemosensitivity and therapeutic response in renal cancer, and increases the expression of PDCD4 and PTEN by negatively mediating miR-21 [8]. Repression of octamer binding transcription factor 2 (OCT2) has been verified to drive oxaliplatin resistance in RCC [109]. Indeed, overexpression of miR-489-3p and miR-630 in cells and exosomes of ccRCC promotes tumor growth, and boosts chemoresistance to oxaliplatin by targeting OCT2 [110]. In contrast, the apoptosis of kidney cancer cells induced by cisplatin and TRAIL can be enhanced by miR-1208 via activation of the caspase pathway, thus impairing RCC cell growth, while these effects can be reversed by TBCK (TBC1 domain containing kinase). TBCK has been verified to affect mTOR signaling pathway transduction [111]. Furthermore, miR-99a-3p suppresses RCC development and facilitates TKI treatment by modifying RRM2 (ribonucleotide reductase regulatory subunit M2). RRM2 acts as an oncogene in gastric adenocarcinoma and breast cancer [112]. In addition, migration and lactate production of cells, and the expression of p-mTOR, can be interfered with by miR-126 in RCC. In addition, miR-126 augments the sensitivity of renal cancer cells to cisplatin and X-ray treatment [113] (Figure 1).
Prognosis
Although the outcome of patients with renal cancer depends on TNM stage, accumulating evidence shows that miRNAs can also predict outcome in RCC patients. For instance, miR-221-5p enables the evaluation of the OS of RCC patients, with a specificity and sensitivity of 44% and 63%, respectively [114]. Another study showed that patients with high expression of miR-154-5p/566/663a/572/23a-3p/720 had poor OS because these miRNAs act as oncogenes and promote RCC progression [2,4,115–118]. On the contrary, miR-378a-5p/31-5p/451a/125a-3p act as tumor suppressors and inhibit RCC development; thus, RCC patients with overexpression of miR-378a-5/31-5p/451a/125a-3p have better OS than those with low expression of these miRNAs [119–122]. Also, miR-144-5p, which is derived from the passenger strands, suppresses RCC development by modifying syndecan-3(SDC3). SDC3 has been reported to act as an oncogene in prostate cancer. In addition, RCC patients with high expression of miR-144-5p have better disease-free survival (DFS) than those with low expression of miR-144-5p [12]. miRNAs also can predict the relapse of renal cancer patients; for example, patients with high levels of miR-155-5p have a 2.64-fold increased risk of ccRCC recurrence (95% CI, 1.49 to 4.70; P=0.0009), and a similar result was found for miR-210-3p (HR, 1.80; 95% CI, 1.04 to 3.12; P=0.036) [123] (Table 1).
Conclusions
MicroRNAs are a class of short non-coding RNAs with highly conserved evolution that regulate genes expression through directly degrading or inhibiting the translation of mRNA [7]. Accumulating evidence confirms that miRNAs, which are derived from guide strands, passenger strands, or both strands, play a vital role in cancer progression [10,12]. We also found that miRNAs have considerable potential effects in cancers, including acting as oncomiRs, functioning as biomarkers for diagnosis, serving as potential therapeutic targets, and serving as markers for predicting prognosis. Their potential effects as biomarkers in liquid biopsies and as targets of therapy for RCC are especially intriguing. Although recent studies in the non-coding RNA field have focused on lncRNAs and circular RNAs, those studies also used targeting of miRNAs or sponging miRNAs to assess function [124]. Indeed, miRNA research is still an important topic in research on cancer and other diseases. For instance, 2 new tools use exosomal miRNAs levels to diagnose multiple cancers [125,126]. In addition, researchers designed a novel material and a small molecule compound that could mediate the level of miRNAs and have anticancer effects in vivo [127,128]. Hence, clinical trials using RNA therapies and liquid biopsy-based are currently beginning, and it is likely that within the next few years, the results of these trials will influence treatment of renal cancer.
Footnotes
Conflict of interests
None.
Source of support: The study was supported by the National Natural Science Foundation of China (No. 81760462 and 81860456) and the Science and Technology Project Fund of the Education Department of Jiangxi Province (No. GJJ180789 and GJJ180788)
References
- 1.Ljungberg B, Albiges L, Abu-Ghanem Y, et al. The 2019 update. Eur Urol. 2019;75(5):799–810. doi: 10.1016/j.eururo.2019.02.011. [DOI] [PubMed] [Google Scholar]
- 2.Lin C, Li Z, Chen P, et al. Oncogene miR-154-5p regulates cellular function and acts as a molecular marker with poor prognosis in renal cell carcinoma. Life Sci. 2018;209:481–89. doi: 10.1016/j.lfs.2018.08.044. [DOI] [PubMed] [Google Scholar]
- 3.Heinemann FG, Tolkach Y, Deng M, et al. Serum miR-122-5p and miR-206 expression: Non-invasive prognostic biomarkers for renal cell carcinoma. Clin Epigenetics. 2018;10:11. doi: 10.1186/s13148-018-0444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan X, Li Z, Zhao L, et al. microRNA 572 functions as an oncogene and a potential biomarker for renal cell carcinoma prognosis. Oncol Rep. 2018;40(5):3092–101. doi: 10.3892/or.2018.6649. [DOI] [PubMed] [Google Scholar]
- 5.Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373(9669):1119–32. doi: 10.1016/S0140-6736(09)60229-4. [DOI] [PubMed] [Google Scholar]
- 6.Incorvaia L, Fanale D, Badalamenti G, et al. A “lymphocyte microRNA signature” as predictive biomarker of immunotherapy response and plasma PD-1/PD-L1 expression levels in patients with metastatic renal cell carcinoma: pointing towards epigenetic reprogramming. Cancers (Basel) 2020;12(11):3396. doi: 10.3390/cancers12113396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catto JW, Alcaraz A, Bjartell AS, et al. MicroRNA in prostate, bladder, and kidney cancer: A systematic review. Eur Urol. 2011;59(5):671–81. doi: 10.1016/j.eururo.2011.01.044. [DOI] [PubMed] [Google Scholar]
- 8.Naro Y, Ankenbruck N, Thomas M, et al. Small molecule inhibition of microRNA miR-21 rescues chemosensitivity of renal-cell carcinoma to topotecan. J Med Chem. 2018;61(14):5900–9. doi: 10.1021/acs.jmedchem.7b01891. [DOI] [PubMed] [Google Scholar]
- 9.Han J, LaVigne CA, Jones BT, et al. A ubiquitin ligase mediates target-directed microRNA decay independently of tailing and trimming. Science. 2020;370(6523):eabc9546. doi: 10.1126/science.abc9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10(10):704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsiakanikas P, Giaginis C, Kontos CK, et al. Current evidence and future perspectives. Expert Rev Mol Diagn. 2018;18(11):981–91. doi: 10.1080/14737159.2018.1539668. [DOI] [PubMed] [Google Scholar]
- 12.Yamada Y, Arai T, Kojima S, et al. Regulation of antitumor miR-144-5p targets oncogenes: Direct regulation of syndecan-3 and its clinical significance. Cancer Sci. 2018;109(9):2919–36. doi: 10.1111/cas.13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim EK, Choi EJ. Compromised MAPK signaling in human diseases: An update. Arch Toxicol. 2015;89(6):867–82. doi: 10.1007/s00204-015-1472-2. [DOI] [PubMed] [Google Scholar]
- 14.Song H, Rao Y, Zhang G, et al. MicroRNA-384 inhibits the growth and invasion of renal cell carcinoma cells by targeting astrocyte elevated gene 1. Oncol Res. 2018;26(3):457–66. doi: 10.3727/096504017X15035025554553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan Y-J, Wei L-L, Wu X-J, et al. MiR-106a-5p inhibits the cell migration and invasion of renal cell carcinoma through targeting PAK5. Cell Death Dis. 2017;8(10):e3155. doi: 10.1038/cddis.2017.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhai W, Ma J, Zhu R, et al. MiR-532-5p suppresses renal cancer cell proliferation by disrupting the ETS1-mediated positive feedback loop with the KRAS-NAP1L1/P-ERK axis. Br J Cancer. 2018;119(5):591–604. doi: 10.1038/s41416-018-0196-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin Z, Wei X, Jin N, et al. MiR-199a targeting ROCK1 to affect kidney cell proliferation, invasion and apoptosis. Artif Cells Nanomed Biotechnol. 2018;46(8):1920–25. doi: 10.1080/21691401.2017.1396224. [DOI] [PubMed] [Google Scholar]
- 18.Li T, Sun X, Xu K. The suppressing role of miR-622 in renal cell carcinoma progression by down-regulation of CCL18/MAPK signal pathway. Cell Biosci. 2018;8(1):17. doi: 10.1186/s13578-018-0212-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Li Y, Guan B, Liu J, et al. MicroRNA-200b is downregulated and suppresses metastasis by targeting LAMA4 in renal cell carcinoma. EBioMedicine. 2019;44:439–51. doi: 10.1016/j.ebiom.2019.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie Y, Chen L, Gao Y, et al. miR-363 suppresses the proliferation, migration and invasion of clear cell renal cell carcinoma by downregulating S1PR1. Cancer Cell Int. 2020;20:227. doi: 10.1186/s12935-020-01313-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arai T, Okato A, Kojima S, et al. Regulation of spindle and kinetochore-associated protein 1 by antitumormiR-10a-5pin renal cell carcinoma. Cancer Sci. 2017;108(10):2088–101. doi: 10.1111/cas.13331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pal SK, Quinn DI. Differentiating mTOR inhibitors in renal cell carcinoma. Cancer Treat Rev. 2013;39(7):709–19. doi: 10.1016/j.ctrv.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okato A, Arai T, Yamada Y, et al. Dual strands of pre-miR-149 inhibit cancer cell migration and invasion through targeting FOXM1 in renal cell carcinoma. Int J Mol Sci. 2017;18(9):1969. doi: 10.3390/ijms18091969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao S, Wang Y, Lou Y, et al. MicroRNA 320a suppresses tumour cell proliferation and invasion of renal cancer cells by targeting FoxM1. Oncol Rep. 2018;40(4):1917–26. doi: 10.3892/or.2018.6597. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Li G, Chong T, Yang J, et al. Kinesin motor protein KIFC1 is a target protein of miR-338-3p and is associated with poor prognosis and progression of renal cell carcinoma. Oncol Res. 2018;27(1):125–37. doi: 10.3727/096504018X15213115046567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li G, Chong T, Xiang X, et al. Downregulation of microRNA-15a suppresses the proliferation and invasion of renal cell carcinoma via direct targeting of eIF4E. Oncol Rep. 2017;38(4):1995–2002. doi: 10.3892/or.2017.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei X, Yu L, Kong X. miR-488 inhibits cell growth and metastasis in renal cell carcinoma by targeting HMGN5. OncoTargets Ther. 2018;11:2205–16. doi: 10.2147/OTT.S156361. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Ding M, Lu X, Wang C, et al. The E2F1-miR-520/372/373-SPOP axis modulates progression of renal carcinoma. Cancer Res. 2018;78(24):6771–84. doi: 10.1158/0008-5472.CAN-18-1662. [DOI] [PubMed] [Google Scholar]
- 29.Dasgupta P, Kulkarni P, Majid S, et al. MicroRNA-203 inhibits long noncoding RNA HOTAIR and regulates tumorigenesis through epithelial-to-mesenchymal transition pathway in renal cell carcinoma. Mol Cancer Ther. 2018;17(5):1061–69. doi: 10.1158/1535-7163.MCT-17-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao H, Liu Z, Wang R, et al. miR-148a suppresses human renal cell carcinoma malignancy by targeting AKT2. Oncol Rep. 2017;37(1):147–54. doi: 10.3892/or.2016.5257. [DOI] [PubMed] [Google Scholar]
- 31.Kim E, Kim TG, Sung E-G, et al. miR-148a increases the sensitivity to cisplatin by targeting Rab14 in renal cancer cells. Int J Oncol. 2017;50(3):984–92. doi: 10.3892/ijo.2017.3851. [DOI] [PubMed] [Google Scholar]
- 32.Chen C, Xue S, Zhang J, et al. DNA-methylation-mediated repression of miR-766-3p promotes cell proliferation via targeting SF2 expression in renal cell carcinoma. Int J Cancer. 2017;141(9):1867–78. doi: 10.1002/ijc.30853. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Sun X. MicroRNA-375 inhibits the proliferation, migration and invasion of kidney cancer cells by triggering apoptosis and modulation of PDK1 expression. Environ Toxicol Pharmacol. 2018;62:227–33. doi: 10.1016/j.etap.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 34.Liu X, Zhong L, Li P, Zhao P. MicroRNA-100 enhances autophagy and suppresses migration and invasion of renal cell carcinoma cells via disruption of NOX4-dependent mTOR pathway. Clin Transl Sci. 2020 doi: 10.1111/cts.12798. [Online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goel HL, Mercurio AM. VEGF targets the tumour cell. Nat Rev Cancer. 2013;13(12):871–82. doi: 10.1038/nrc3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang J, Wang X, Wen G, et al. miRNA-205-5p functions as a tumor suppressor by negatively regulating VEGFA and PI3K/Akt/mTOR signaling in renal carcinoma cells. Oncol Rep. 2019;42(5):1677–88. doi: 10.3892/or.2019.7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, Zheng D, Pan L, et al. Knockdown of TUG1 by shRNA inhibited renal cell carcinoma formation by miR-299-3p/VEGF axis in vitro and in vivo. Eur J Pharmacol. 2019;860:172536. doi: 10.1016/j.ejphar.2019.172536. [DOI] [PubMed] [Google Scholar]
- 38.Zhu J, Zhu DQ, Zhang Y, et al. MicroRNA-363 inhibits angiogenesis, proliferation, invasion, and migration of renal cell carcinoma via inactivation of the Janus tyrosine kinases 2-signal transducers and activators of transcription 3 axis by suppressing growth hormone receptor gene. J Cell Physiol. 2018;234(3):2581–92. doi: 10.1002/jcp.27020. [DOI] [PubMed] [Google Scholar]
- 39.Mu L, Guan B, Tian J, et al. MicroRNA-218 inhibits tumor angiogenesis of human renal cell carcinoma by targeting GAB2. Oncol Rep. 2020;44(5):1961–70. doi: 10.3892/or.2020.7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poole TJ, Finkelstein EB, Cox CM. The role of FGF and VEGF in angioblast induction and migration during vascular development. Dev Dyn. 2001;220(1):1–17. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1087>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 41.Zhang H, Ye Q, Du Z, et al. MiR-148b-3p inhibits renal carcinoma cell growth and pro-angiogenic phenotype of endothelial cell potentially by modulating FGF2. Biomed Pharmacother. 2018;107:359–67. doi: 10.1016/j.biopha.2018.07.054. [DOI] [PubMed] [Google Scholar]
- 42.Chen J, Li D. Telbivudine attenuates UUO-induced renal fibrosis via TGF-β/Smad and NF-κB signaling. Int Immunopharmacol. 2018;55:1–8. doi: 10.1016/j.intimp.2017.11.043. [DOI] [PubMed] [Google Scholar]
- 43.He Y, Liu J, Wang Y, et al. Role of miR-486-5p in regulating renal cell carcinoma cell proliferation and apoptosis via TGF-β-activated kinase 1. J Cell Biochem. 2019;120(3):2954–63. doi: 10.1002/jcb.26900. [DOI] [PubMed] [Google Scholar]
- 44.Bogusławska J, Rodzik K, Popławski P, et al. TGF-β1 targets a microRNA network that regulates cellular adhesion and migration in renal cancer. Cancer Lett. 2018;412:155–69. doi: 10.1016/j.canlet.2017.10.019. [DOI] [PubMed] [Google Scholar]
- 45.Liu S, Long J, Yuan B, et al. SUMO modification reverses inhibitory effects of smad nuclear interacting protein-1 in TGF-β responses. J Biol Chem. 2016;291(47):24418–30. doi: 10.1074/jbc.M116.755850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiao D, Wu M, Ji L, et al. MicroRNA-186 suppresses cell proliferation and metastasis through targeting sentrin-specific protease 1 in renal cell carcinoma. Oncol Res. 2018;26(2):249–59. doi: 10.3727/096504017X14953948675430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hanahan D, Weinberg RA. Hallmarks of cancer. The next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 48.Xiao W, Wang C, Chen K, et al. MiR-765 functions as a tumour suppressor and eliminates lipids in clear cell renal cell carcinoma by downregulating PLP2. EBioMedicine. 2020;51:102622. doi: 10.1016/j.ebiom.2019.102622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan DA, Sutphin PD, Nguyen P, et al. Targeting GLUT1 and the Warburg effect in renal cell carcinoma by chemical synthetic lethality. Sci Transl Med. 2011;3(94):94ra70. doi: 10.1126/scitranslmed.3002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, He Y, Bai H, et al. Phosphoinositide-dependent kinase 1-associated glycolysis is regulated by miR-409-3p in clear cell renal cell carcinoma. J Cell Biochem. 2019;120(1):126–34. doi: 10.1002/jcb.27152. [DOI] [PubMed] [Google Scholar]
- 51.Qu F, Ye J, Pan X, et al. MicroRNA-497-5p down-regulation increases PD-L1 expression in clear cell renal cell carcinoma. J Drug Target. 2019;27(1):67–74. doi: 10.1080/1061186X.2018.1479755. [DOI] [PubMed] [Google Scholar]
- 52.Wang W, Zhao E, Yu Y, et al. MiR-216a exerts tumor-suppressing functions in renal cell carcinoma by targeting TLR4. Am J Cancer Res. 2018;8(3):476. [PMC free article] [PubMed] [Google Scholar]
- 53.Paolino M, Choidas A, Wallner S, et al. The E3 ligase Cbl-b and TAM receptors regulate cancer metastasis via natural killer cells. Nature. 2014;507(7493):508–12. doi: 10.1038/nature12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ding M, Sun X, Zhong J, et al. Decreased miR-200a-3p is a key regulator of renal carcinoma growth and migration by directly targeting CBL. J Cell Biochem. 2018;119(12):9974–85. doi: 10.1002/jcb.27326. [DOI] [PubMed] [Google Scholar]
- 55.Chao CC. Mechanisms of p53 degradation. Clin Chim Acta. 2015;438:139–47. doi: 10.1016/j.cca.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 56.Kulkarni P, Dasgupta P, Bhat NS, et al. Elevated miR-182-5p associates with renal cancer cell mitotic arrest through diminished MALAT-1 expression. Mol Cancer Res. 2018;16(11):1750–60. doi: 10.1158/1541-7786.MCR-17-0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Francis R, Guo H, Streutker C, et al. Gastrointestinal transcription factors drive lineage-specific developmental programs in organ specification and cancer. Sci Adv. 2019;5(12):eaax8898. doi: 10.1126/sciadv.aax8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang K, Jin W, Jin P, et al. miR-211-5p suppresses metastatic behavior by targeting SNAI1 in renal cancer. Mol Cancer Res. 2017;15(4):448–56. doi: 10.1158/1541-7786.MCR-16-0288. [DOI] [PubMed] [Google Scholar]
- 59.Chen J, Zhong Y, Li L. miR-124 and miR-203 synergistically inactivate EMT pathway via coregulation of ZEB2 in clear cell renal cell carcinoma (ccRCC) J Transl Med. 2020;18(1):69. doi: 10.1186/s12967-020-02242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamada Y, Nohata N, Uchida A, et al. Replisome genes regulation by anti-tumor miR-101-5p in clear cell renal cell carcinoma. Cancer Sci. 2020;111(4):1392–406. doi: 10.1111/cas.14327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bai J, Yeh S, Qiu X, et al. TR4 nuclear receptor promotes clear cell renal cell carcinoma (ccRCC) vasculogenic mimicry (VM) formation and metastasis via altering the miR490-3p/vimentin signals. Oncogene. 2018;37(44):5901. doi: 10.1038/s41388-018-0269-1. [DOI] [PubMed] [Google Scholar]
- 62.Wang M, Sun Y, Xu J, et al. Preclinical studies using miR-32-5p to suppress clear cell renal cell carcinoma metastasis via altering the miR-32-5p/TR4/HGF/Met signaling. Int J Cancer. 2018;143(1):100–12. doi: 10.1002/ijc.31289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ding X, Yang DR, Lee SO, et al. TR4 nuclear receptor promotes prostate cancer metastasis via upregulation of CCL2/CCR2 signaling. Int J Cancer. 2015;136(4):955–64. doi: 10.1002/ijc.29049. [DOI] [PubMed] [Google Scholar]
- 64.Hua H, Kong Q, Zhang H, et al. Targeting mTOR for cancer therapy. J Hematol Oncol. 2019;12(1):71. doi: 10.1186/s13045-019-0754-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang C, Uemura M, Tomiyama E, et al. MicroRNA-92b-3p is a prognostic oncomiR that targets TSC1 in clear cell renal cell carcinoma. Cancer Sci. 2020;111(4):1146–55. doi: 10.1111/cas.14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Patergnani S, Guzzo S, Mangolini A, et al. The induction of AMPK-dependent autophagy leads to P53 degradation and affects cell growth and migration in kidney cancer cells. Exp Cell Res. 2020;395(1):112190. doi: 10.1016/j.yexcr.2020.112190. [DOI] [PubMed] [Google Scholar]
- 67.Kuei CH, Lin HY, Lee HH, et al. IMPA2 downregulation enhances mTORC1 activity and restrains autophagy initiation in metastatic clear cell renal cell carcinoma. J Clin Med. 2020;9(4):956. doi: 10.3390/jcm9040956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin Y-F, Chou J-L, Chang J-S, et al. Dysregulation of the miR-25-IMPA2 axis promotes metastatic progression in clear cell renal cell carcinoma. EBioMedicine. 2019;45:220–230. doi: 10.1016/j.ebiom.2019.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bogusławska J, Popławski P, Alseekh S, et al. MicroRNA-mediated metabolic reprograming in renal cancer. Cancers. 2019;11(12):1825. doi: 10.3390/cancers11121825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guo H, German P, Bai S, et al. The PI3K/AKT pathway and renal cell carcinoma. J Genet Genomics. 2015;42(7):343–53. doi: 10.1016/j.jgg.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pan Y, Hu J, Ma J, et al. MiR-193a-3p and miR-224 mediate renal cell carcinoma progression by targeting alpha-2,3-sialyltransferase IV and the phosphatidylinositol 3 kinase/Akt pathway. Mol Carcinog. 2018;57(8):1067–77. doi: 10.1002/mc.22826. [DOI] [PubMed] [Google Scholar]
- 72.Jing Z-F, Bi J-B, et al. mir-19 promotes the proliferation of clear cell renal cell carcinoma by targeting the FrK-PTen axis. Onco Targets Ther. 2019;12:2713. doi: 10.2147/OTT.S199238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nie W, Ni D, Ma X, et al. [Corrigendum] miR 122 promotes proliferation and invasion of clear cell renal cell carcinoma by suppressing Forkhead box O3. Int J Oncol. 2019;54(4):1496. doi: 10.3892/ijo.2019.4694. [Erratum for: Int J Oncol. 2019 Feb;54(2): 559–571] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cheng YC, Chen PH, Chiang HY, et al. Candidate tumor suppressor B-cell translocation gene 3 impedes neoplastic progression by suppression of AKT. Cell Death Dis. 2015;6(1):e1584. doi: 10.1038/cddis.2014.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu L, Liu S, Duan Q, et al. MicroRNA-142-5p promotes cell growth and migration in renal cell carcinoma by targeting BTG3. Am J Transl Res. 2017;9(5):2394–402. [PMC free article] [PubMed] [Google Scholar]
- 76.Attisano L, Labbé E. TGFbeta and Wnt pathway cross-talk. Cancer Metastasis Rev. 2004;23(1–2):53–61. doi: 10.1023/a:1025811012690. [DOI] [PubMed] [Google Scholar]
- 77.Zhai W, Li S, Zhang J, et al. Sunitinib-suppressed miR-452-5p facilitates renal cancer cell invasion and metastasis through modulating SMAD4/SMAD7 signals. Mol Cancer. 2018;17(1):157. doi: 10.1186/s12943-018-0906-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.García de Vinuesa A, Abdelilah-Seyfried S, Knaus P, et al. BMP signaling in vascular biology and dysfunction. Cytokine Growth Factor Rev. 2016;27:65–79. doi: 10.1016/j.cytogfr.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 79.Yoshino H, Yonezawa T, Yonemori M, et al. Downregulation of microRNA-1274a induces cell apoptosis through regulation of BMPR1B in clear cell renal cell carcinoma. Oncol Rep. 2018;39(1):173–81. doi: 10.3892/or.2017.6098. [DOI] [PubMed] [Google Scholar]
- 80.Hirata H, Hinoda Y, Nakajima K, et al. Wnt antagonist DKK1 acts as a tumor suppressor gene that induces apoptosis and inhibits proliferation in human renal cell carcinoma. Int J Cancer. 2011;128(8):1793–803. doi: 10.1002/ijc.25507. [DOI] [PubMed] [Google Scholar]
- 81.Ueno K, Hirata H, Majid S, et al. Wnt antagonist DICKKOPF-3 (Dkk-3) induces apoptosis in human renal cell carcinoma. Mol Carcinog. 2011;50(6):449–57. doi: 10.1002/mc.20729. [DOI] [PubMed] [Google Scholar]
- 82.Chen Z-y, Du Y, Wang L, et al. MiR-543 promotes cell proliferation and metastasis of renal cell carcinoma by targeting Dickkopf 1 through the Wnt/β-catenin signaling pathway. J Cancer. 2018;9(20):3660. doi: 10.7150/jca.27124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yan L, Ding B, Liu H, et al. Inhibition of SMYD2 suppresses tumor progression by down-regulating microRNA-125b and attenuates multi-drug resistance in renal cell carcinoma. Theranostics. 2019;9(26):8377. doi: 10.7150/thno.37628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Meng G, Li G, Yang X, et al. Inhibition of miR146b-5p suppresses CT-guided renal cell carcinoma by targeting TRAF6. J Cell Biochem. 2018 doi: 10.1002/jcb.27566. [Online ahead of prin.] [DOI] [PubMed] [Google Scholar]
- 85.Masilamani AP, Ferrarese R, Kling E, et al. KLF6 depletion promotes NF-κB signaling in glioblastoma. Oncogene. 2017;36(25):3562–75. doi: 10.1038/onc.2016.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang F, Ma J, Tang Q, et al. MicroRNA-543 promotes the proliferation and invasion of clear cell renal cell carcinoma cells by targeting Kruppel-like factor 6. Biomed Pharmacother. 2018;97:616–23. doi: 10.1016/j.biopha.2017.10.136. [DOI] [PubMed] [Google Scholar]
- 87.Lee NJ, Oh JH, Ban JO, et al. 4-O-methylhonokiol, a PPARγ agonist, inhibits prostate tumour growth: p21-mediated suppression of NF-κB activity. Br J Pharmacol. 2013;168(5):1133–45. doi: 10.1111/j.1476-5381.2012.02235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Geng J, Ito Y, Shi L, et al. Regulation of RIPK1 activation by TAK1-mediated phosphorylation dictates apoptosis and necroptosis. Nat Commun. 2017;8(1):359. doi: 10.1038/s41467-017-00406-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao C, Zhou Y, Ran Q, et al. MicroRNA-381-3p functions as a dual suppressor of apoptosis and necroptosis and promotes proliferation of renal cancer cells. Front Cell Dev Biol. 2020;8:290. doi: 10.3389/fcell.2020.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xiao W, Wang X, Wang T, et al. MiR-223-3p promotes cell proliferation and metastasis by downregulating SLC4A4 in clear cell renal cell carcinoma. Aging (Albany NY) 2019;11(2):615. doi: 10.18632/aging.101763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jingushi K, Kashiwagi Y, Ueda Y, et al. High miR-122 expression promotes malignant phenotypes in ccRCC by targeting occludin. Int J Oncol. 2017;51(1):289–97. doi: 10.3892/ijo.2017.4016. [DOI] [PubMed] [Google Scholar]
- 92.Li X, Xin S, Yang D, et al. Down-regulation of PDCD4 expression is an independent predictor of poor prognosis in human renal cell carcinoma patients. J Cancer Res Clin Oncol. 2012;138(3):529–35. doi: 10.1007/s00432-011-1121-y. [DOI] [PubMed] [Google Scholar]
- 93.Fan B, Jin Y, Zhang H, et al. MicroRNA 21 contributes to renal cell carcinoma cell invasiveness and angiogenesis via the PDCD4/c Jun (AP 1) signalling pathway. Int J Oncol. 2020;56(1):178–92. doi: 10.3892/ijo.2019.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu Z, Lu Y, Xiao Y, et al. Upregulation of miR-21 expression is a valuable predicator of advanced clinicopathological features and poor prognosis in patients with renal cell carcinoma through the p53/p21-cyclin E2-Bax/caspase-3 signaling pathway. Oncol Rep. 2017;37(3):1437–44. doi: 10.3892/or.2017.5402. [DOI] [PubMed] [Google Scholar]
- 95.Lennon AM, Buchanan AH, Kinde I, et al. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science. 2020;369(6499):eabb9601. doi: 10.1126/science.abb9601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kurahashi R, Kadomatsu T, Baba M, et al. MicroRNA-204-5p: A novel candidate urinary biomarker of Xp11. 2 translocation renal cell carcinoma. Cancer Sci. 2019;110(6):1897. doi: 10.1111/cas.14026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dias F, Teixeira AL, Nogueira I, et al. Extracellular vesicles enriched in hsa-miR-301a-3p and hsa-miR-1293 dynamics in clear cell renal cell carcinoma patients: Potential biomarkers of metastatic disease. Cancers (Basel) 2020;12(6):1450. doi: 10.3390/cancers12061450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xiao CT, Lai WJ, Zhu WA, et al. MicroRNA derived from circulating exosomes as noninvasive biomarkers for diagnosing renal cell carcinoma. Onco Targets Ther. 2020;13:10765–74. doi: 10.2147/OTT.S271606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang L, Yang G, Zhao D, et al. CD103-positive CSC exosome promotes EMT of clear cell renal cell carcinoma: role of remote MiR-19b-3p. Mol Cancer. 2019;18(1):86. doi: 10.1186/s12943-019-0997-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Molina AM, Lin X, Korytowsky B, et al. Sunitinib objective response in metastatic renal cell carcinoma: analysis of 1059 patients treated on clinical trials. Eur J Cancer. 2014;50(2):351–58. doi: 10.1016/j.ejca.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 101.Kovacova J, Juracek J, Poprach A, et al. Mir-376b-3p is associated with long-term response to sunitinib in metastatic renal cell carcinoma patients. Cancer Genomics Proteomics. 2019;16(5):353–59. doi: 10.21873/cgp.20140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ralla B, Busch J, Flörcken A, et al. miR-9-5p in nephrectomy specimens is a potential predictor of primary resistance to first-line treatment with tyrosine kinase inhibitors in patients with metastatic renal cell carcinoma. Cancers. 2018;10(9):321. doi: 10.3390/cancers10090321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Incorvaia L, Fanale D, Badalamenti G, et al. Baseline plasma levels of soluble PD-1, PD-L1, and BTN3A1 predict response to nivolumab treatment in patients with metastatic renal cell carcinoma: A step toward a biomarker for therapeutic decisions. Oncoimmunology. 2020;9(1):1832348. doi: 10.1080/2162402X.2020.1832348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lawrie CH, Armesto M, Fernandez-Mercado M, et al. Noncoding RNA expression and targeted next-generation sequencing distinguish tubulocystic renal cell carcinoma (TC-RCC) from other renal neoplasms. J Mol Diagn. 2018;20(1):34–45. doi: 10.1016/j.jmoldx.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 105.Luo W, Wang L, Luo MH, et al. hsa-mir-3199-2 and hsa-mir-1293 as novel prognostic biomarkers of papillary renal cell carcinoma by Cox ratio risk regression model screening. J Cell Biochem. 2017;118(10):3488–94. doi: 10.1002/jcb.26008. [DOI] [PubMed] [Google Scholar]
- 106.Lokeshwar SD, Talukder A, Yates TJ, et al. A potential three-MicroRNA prognostic signature. Cancer Epidemiol Biomarkers Prev. 2018;27(4):464–72. doi: 10.1158/1055-9965.EPI-17-0700. [DOI] [PubMed] [Google Scholar]
- 107.Kowalik CG, Palmer DA, Sullivan TB, et al. Profiling microRNA from nephrectomy and biopsy specimens: Predictors of progression and survival in clear cell renal cell carcinoma. BJU Int. 2017;120(3):428–40. doi: 10.1111/bju.13886. [DOI] [PubMed] [Google Scholar]
- 108.Luo Y, Chen L, Wang G, et al. Identification of a three-miRNA signature as a novel potential prognostic biomarker in patients with clear cell renal cell carcinoma. J Cell Biochem. 2019;120(8):13751–64. doi: 10.1002/jcb.28648. [DOI] [PubMed] [Google Scholar]
- 109.Liu Y, Zheng X, Yu Q, et al. Epigenetic activation of the drug transporter OCT2 sensitizes renal cell carcinoma to oxaliplatin. Sci Transl Med. 2016;8(348):348ra97. doi: 10.1126/scitranslmed.aaf3124. [DOI] [PubMed] [Google Scholar]
- 110.Chen L, Chen L, Qin Z, et al. Upregulation of miR-489-3p and miR-630 inhibits oxaliplatin uptake in renal cell carcinoma by targeting OCT2. Acta Pharm Sin B. 2019;9(5):1008–20. doi: 10.1016/j.apsb.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim E, Jang J-H, Sung E-G, et al. MiR-1208 increases the sensitivity to cisplatin by targeting TBCK in renal cancer cells. Int J Mol Sci. 2019;20(14):3540. doi: 10.3390/ijms20143540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Osako Y, Yoshino H, Sakaguchi T, et al. Potential tumor-suppressive role of microRNA-99a-3p in sunitinib-resistant renal cell carcinoma cells through the regulation of RRM2. Int J Oncol. 2019;54(5):1759–70. doi: 10.3892/ijo.2019.4736. [DOI] [PubMed] [Google Scholar]
- 113.Liu W, Chen H, Wong N, et al. Pseudohypoxia induced by miR-126 deactivation promotes migration and therapeutic resistance in renal cell carcinoma. Cancer Lett. 2017;394:65–75. doi: 10.1016/j.canlet.2017.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu S, Wang Y, Li W, et al. miR-221-5p acts as an oncogene and predicts worse survival in patients of renal cell cancer. Biomed Pharmacother. 2019;119:109406. doi: 10.1016/j.biopha.2019.109406. [DOI] [PubMed] [Google Scholar]
- 115.Pan X, Quan J, Li Z, et al. miR-566 functions as an oncogene and a potential biomarker for prognosis in renal cell carcinoma. Biomed Pharmacoth. 2018;102:718–27. doi: 10.1016/j.biopha.2018.03.072. [DOI] [PubMed] [Google Scholar]
- 116.Zhou L, Pan X, Li Z, et al. Oncogenic miR-663a is associated with cellular function and poor prognosis in renal cell carcinoma. Biomed Pharmacother. 2018;105:1155–63. doi: 10.1016/j.biopha.2018.05.082. [DOI] [PubMed] [Google Scholar]
- 117.Quan J, Pan X, Li Y, et al. MiR-23a-3p acts as an oncogene and potential prognostic biomarker by targeting PNRC2 in RCC. Biomed Pharmacother. 2019;110:656–66. doi: 10.1016/j.biopha.2018.11.065. [DOI] [PubMed] [Google Scholar]
- 118.Bhat NS, Colden M, Dar AA, et al. MicroRNA-720 regulates E-cadherin-αE-catenin complex and promotes renal cell carcinoma. Mol Cancer Ther. 2017;16(12):2840–48. doi: 10.1158/1535-7163.MCT-17-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pan X, Zhao L, Quan J, et al. MiR-378a-5p acts as a tumor suppressor in renal cell carcinoma and is associated with the good prognosis of patients. Am J Transl Res. 2019;11(4):2207. [PMC free article] [PubMed] [Google Scholar]
- 120.Li Y, Quan J, Chen F, et al. MiR-31-5p acts as a tumor suppressor in renal cell carcinoma by targeting cyclin-dependent kinase 1 (CDK1) Biomed Pharmacother. 2019;111:517–26. doi: 10.1016/j.biopha.2018.12.102. [DOI] [PubMed] [Google Scholar]
- 121.Yamada Y, Arai T, Sugawara S, et al. Impact of novel oncogenic pathways regulated by antitumor miR-451a in renal cell carcinoma. Cancer Sci. 2018;109(4):1239–53. doi: 10.1111/cas.13526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hou P, Li H, Yong H, et al. PinX1 represses renal cancer angiogenesis via the mir-125a-3p/VEGF signaling pathway. Angiogenesis. 2019;22(4):507–19. doi: 10.1007/s10456-019-09675-z. [DOI] [PubMed] [Google Scholar]
- 123.Zhang J, Ye Y, Chang DW, et al. Global and targeted miRNA expression profiling in clear cell renal cell carcinoma tissues potentially links miR-155-5p and miR-210-3p to both tumorigenesis and recurrence. Am J Pathol. 2018;188(11):2487–96. doi: 10.1016/j.ajpath.2018.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chan JJ, Tay Y. Noncoding RNA: RNA regulatory networks in cancer. Int J Mol Sci. 2018;19(5):1310. doi: 10.3390/ijms19051310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhao J, Liu C, Li Y, et al. Thermophoretic detection of exosomal microRNAs by nanoflares. J Am Chem Soc. 2020;142(11):4996–5001. doi: 10.1021/jacs.9b13960. [DOI] [PubMed] [Google Scholar]
- 126.Hashimoto K, Inada M, Ito K. Multiplex real-time loop-mediated isothermal amplification using an electrochemical DNA chip consisting of a single liquid-flow channel. Anal Chem. 2019;91(5):3227–32. doi: 10.1021/acs.analchem.8b05284. [DOI] [PubMed] [Google Scholar]
- 127.Conde J, Oliva N, Atilano M, et al. Self-assembled RNA-triple-helix hydrogel scaffold for microRNA modulation in the tumour microenvironment. Nat Mater. 2016;15(3):353–63. doi: 10.1038/nmat4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Costales MG, Aikawa H, Li Y, et al. Small-molecule targeted recruitment of a nuclease to cleave an oncogenic RNA in a mouse model of metastatic cancer. Proc Natl Acad Sci USA. 2020;117(5):2406–11. doi: 10.1073/pnas.1914286117. [DOI] [PMC free article] [PubMed] [Google Scholar]