Abstract
Humans are repeatedly exposed to variants of influenza virus throughout their lifetime. As a result, preexisting influenza-specific memory B cells can dominate the response after infection or vaccination. Memory B cells recalled by adulthood exposure are largely reactive to conserved viral epitopes present in childhood strains, posing unclear consequences on the ability of B cells to adapt to and neutralize newly emerged strains. We sought to investigate the impact of preexisting immunity on generation of protective antibody responses to conserved viral epitopes upon influenza virus infection and vaccination in humans. We accomplished this by characterizing monoclonal antibodies (mAbs) from plasmablasts, which are predominantly derived from preexisting memory B cells. We found that, whereas some influenza infection–induced mAbs bound conserved and neutralizing epitopes on the hemagglutinin (HA) stalk domain or neuraminidase, most of the mAbs elicited by infection targeted non-neutralizing epitopes on nucleoprotein and other unknown antigens. Furthermore, most infection-induced mAbs had equal or stronger affinity to childhood strains, indicating recall of memory B cells from childhood exposures. Vaccination-induced mAbs were similarly induced from past exposures and exhibited substantial breadth of viral binding, although, in contrast to infection-induced mAbs, they targeted neutralizing HA head epitopes. Last, cocktails of infection-induced mAbs displayed reduced protective ability in mice compared to vaccination-induced mAbs. These findings reveal that both preexisting immunity and exposure type shape protective antibody responses to conserved influenza virus epitopes in humans. Natural infection largely recalls cross-reactive memory B cells against non-neutralizing epitopes, whereas vaccination harnesses preexisting immunity to target protective HA epitopes.
INTRODUCTION
Influenza viruses are responsible for more than 5 million severe cases of respiratory tract infection and up to 650,000 deaths globally each year (1). Current influenza vaccine effectiveness is low (2), partly due to rapid antigenic evolution of the major surface glycoproteins of influenza virus: hemagglutinin (HA) and neuraminidase (NA) (3). Influenza vaccination platforms focus on eliciting protective antibody responses against HA, the most abundant and immunodominant viral glycoprotein (4). Antibodies against the HA head domain are potently neutralizing and inhibit receptor binding to sialic acid on epithelial cells. This is measured in vitro as the ability of an antibody to inhibit agglutination of red blood cells, referred to as hemagglutination inhibition (HAI) (5). Antibodies against HA are capable of limiting infection, and serum HAI titers have been used as the primary correlate of vaccine-induced protection against influenza for nearly 50 years (5, 6). However, the HA head is highly variable and easily mutates to evade host immunity (7-10), leading to seasonal epidemics and necessitating frequent reformulation of seasonal influenza vaccines.
In addition to rapid antigenic evolution, preexisting immunity profoundly affects protective immune responses upon exposure to novel influenza viruses or viruses that exhibit antigenic drift, the accumulation of mutations in viral surface glycoproteins (11-13). Original antigenic sin, also called imprinting, suggests that an individual’s first encountered strain takes a senior antigenic position in the memory B cell (MBC) repertoire (14-16). As a result, subsequent exposure to influenza viruses later in life leads to the recall of MBCs specific to epitopes present in strains from an individual’s childhood. Several reports have proposed beneficial and detrimental effects of imprinting on antibody responses to influenza viruses, depending on the context (17-19). It was recently suggested that the first HA group 1 or HA group 2 virus an individual was exposed to predicts protection against avian influenza viruses belonging to the same group and susceptibility to avian influenza viruses of the other group (11). Furthermore, the age distribution of seasonal influenza virus cases can be predicted by the likely subtype of first infection for each birth cohort (20, 21). The boosting of antibodies reactive to conserved protective or nonprotective viral epitopes may therefore play a major role in shaping influenza virus susceptibility or protective outcomes upon exposure.
To date, few studies have addressed the role of preexisting immunity in shaping the reactivity of antibodies elicited by natural influenza virus infection compared to vaccination at the single B cell level. Our current understanding of antibody immunodominance to influenza viruses in humans is largely limited to an understanding of vaccination-induced responses derived from serology studies, which fail to resolve the full spectrum of epitope targeting. It is well established that vaccination induces HA head–specific responses due to enrichment of HA during vaccine manufacturing (22, 23), but it is unclear how preexisting immunity shapes the targeting of conserved HA epitopes upon vaccination. Furthermore, past studies have suggested that viral antigens such as the HA stalk, NA, and internal nucleoprotein (NP) are frequently targeted upon infection, as these antigens are in high abundance (22, 24, 25). However, the frequencies by which conserved antigens are targeted by B cells upon natural infection remain to be determined. We hypothesized that plasmablasts that are induced early after natural infection would target conserved, yet less protective, viral epitopes on the HA stalk domain, NA, and NP rather than antigenically drifted epitopes on the HA head, as only conserved epitopes will be recognized by the preexisting MBC repertoire. By contrast, we hypothesized that vaccination may draw upon preexisting immunity to boost HA head–specific responses.
In this study, we characterized monoclonal antibodies (mAbs) cloned from plasmablasts induced early after influenza A virus infection and seasonal vaccination. Plasmablasts are a useful model as they are predominantly derived from the MBC compartment and typically express highly somatically mutated variable genes despite their rapid activation (5 to 14 days) after antigen exposure (12, 26). In addition, they are readily found to have clonal relationships with B cells activated by previously encountered influenza virus strains (12). By characterizing antigen reactivity, cross-reactivity toward historical and contemporary influenza virus strains, in vitro virus neutralization, and in vivo protective ability of human mAbs, we found key differences in how natural infection and seasonal vaccination rely on preexisting B cell memory to induce early protective antibody-mediated immunity. Our data demonstrate that preexisting immunity largely biases the early antibody response after infection toward conserved yet less protective viral epitopes, with only a small percentage of infection-induced mAbs targeting the HA head. Conversely, vaccination can draw upon preexisting immunity to boost cross-reactive and neutralizing responses against the HA head, likely resulting in superior protection when circulating strains are well matched to vaccine strains. This study emphasizes the necessity of understanding mechanisms of immune memory bias such as original antigenic sin in shaping protective antibody responses to influenza viruses, as harnessing or surmounting preexisting immunity is likely the most effective path toward universal vaccination.
RESULTS
A minority of antibodies induced early after influenza virus infection recognize the HA head
To examine how preexisting immunity shapes the antibody response after natural influenza virus infection and vaccination, we generated mAbs from single plasmablasts obtained from human participants after H1N1 or H3N2 virus infection (table S1) and from healthy individuals after seasonal vaccination (table S2). H1N1-infected individuals were recruited during the 2015–2016 flu season, and H3N2-infected individuals were recruited during the 2014–2015 flu season. Vaccinated individuals were studied after receipt of the northern hemisphere trivalent influenza vaccine in the 2010–2011 flu season or the quadrivalent influenza vaccine in the 2014–2015 flu season. The vaccine strains received by the vaccination cohort were distinct from infecting strains circulating during the time of sample isolation from the infection cohort (tables S1 and S2). Samples were collected at day 7 after vaccination and estimated days 7 to 11 after infection. All mAbs generated were initially screened for reactivity to influenza viruses (indicated in tables S1 and S2) and only included in the study if they exhibited specific binding.
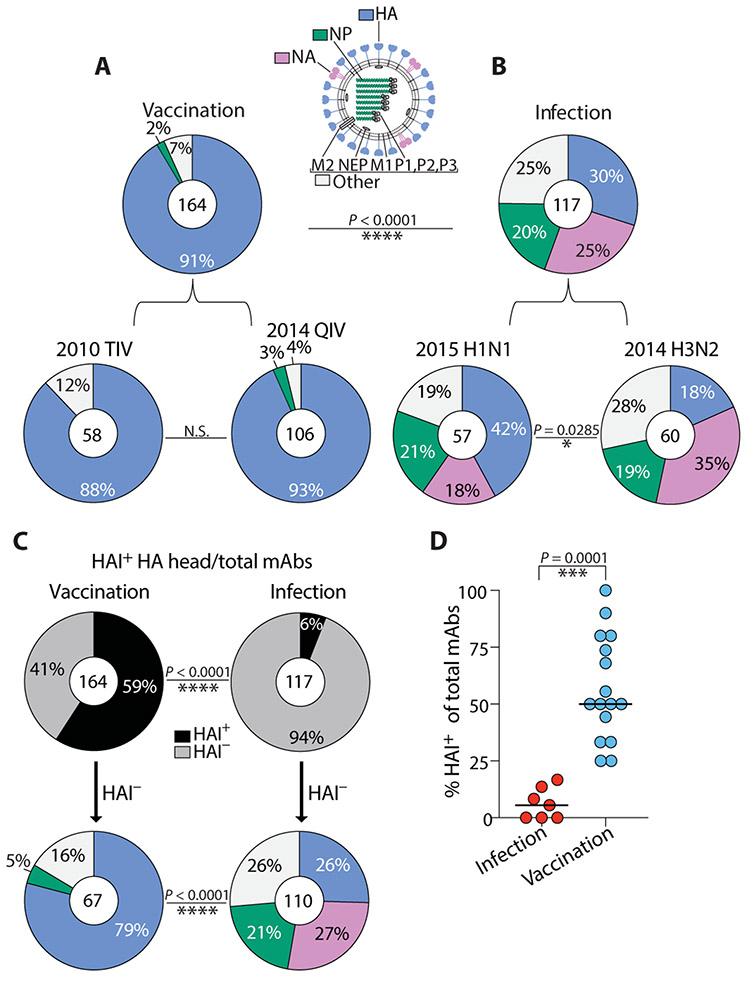
Whereas greater than 90% of mAbs induced by vaccination recognized HA by enzyme-linked immunosorbent assay (ELISA; Fig. 1A), only 30% of mAbs induced by natural infection with either H1N1 or H3N2 were HA reactive, including the HA head and stalk domain regions, with the majority recognizing other antigens such as NA and the highly conserved NP (P < 0.0001; Fig. 1B). Nearly half of all H1N1 infection–induced mAbs were HA reactive, although this was substantially less than the proportion of mAbs induced by seasonal vaccination (Fig. 1B, bottom left). Furthermore, H3N2 infection–induced mAbs mostly targeted NA and NP, whereas only 18% bound HA (Fig. 1B, bottom right). Because age-related factors likely affect antibody responses to conserved antigens (27-29), we next analyzed antigen reactivity in each individual within our infection cohort. Although there was variability in the proportion of antibodies targeting non-HA proteins per H3N2-infected individual, each individual mounted a non-HA–biased response overall, with roughly 80% of the response per individual directed toward non-HA antigens (fig. S1A). Similarly, for H1N1-infected individuals, a substantial fraction of all mAbs per person recognized non-HA antigens, although there was some degree of interindividual variability (fig. S1B). Accordingly, we found that only 6% of all mAbs induced by infection had HAI activity compared to 59% induced by vaccination (P < 0.0001; Fig. 1C), and the percentage of HAI+ mAbs per infected individual was, on average, only 5.5% (Fig. 1D).
Fig. 1. A minority of antibodies induced early after influenza virus infection recognize the HA head.
(A) Pie charts show binding of 2010–2011 trivalent and 2014–2015 quadrivalent vaccination-induced mAbs to a panel of hemagglutinin (HA), neuraminidase (NA), and nucleoprotein (NP) recombinant proteins by ELISA. (B) Pie charts show binding of 2015–2016 H1N1 and 2014–2015 H3N2 infection–induced mAbs to a panel of HA, NA, and NP recombinant proteins by ELISA. Recombinant proteins were chosen from the representative influenza A vaccine strains within each vaccine (table S2) or from viruses bearing resemblance to recently circulating strains during the year of mAb isolation from infected individuals (2014 H3N2: A/Switzerland/9715293/2013, A/Hong Kong/4801/2014; 2015 H1N1: A/California/7/2009, A/Michigan/45/2015). mAbs in the “Other” category bind virus, but not HA, NA, or NP, and likely bind other undetermined influenza virus antigens. (C) Pie charts demonstrate the percent of total isolated mAbs with HAI activity isolated from both cohorts (top) and the antigen reactivity of HAI− mAbs within each cohort (bottom panel). (D) The percentage of mAbs with HAI activity was compared in individuals from infected (n = 7) and vaccinated cohorts (n = 16). Numbers in the center of each pie chart indicate the number of mAbs tested. Statistical significance was determined by chi-square test (****P < 0.0001; *P = 0.0285) (A to C) and unpaired nonparametric Mann-Whitney test (***P = 0.0001) (D). Data are representative of two to three independent experiments performed in duplicate.
Preexisting immunity is expected to affect antibody responses to conserved viral epitopes, so we next assessed whether antibodies induced by influenza virus infection targeted typically more conserved and immunosubdominant viral epitopes. Of total HA-binding mAbs induced by vaccination, nearly two-thirds had HAI activity, reflective of their ability to target the HA head region (fig. S1C), compared to just 20% of HA-reactive infection-induced mAbs (P < 0.0001; fig. S1D). To determine mAbs binding epitopes on the stalk domain, we performed competition ELISAs with an anti-stalk mAb known to bind a well-characterized broadly neutralizing stalk epitope [mAb CR9114; (30)] as well as ELISA against a headless stalk construct (31) and chimeric HA constructs (cH5/1; cH7/3). Only 14% of vaccination-induced HA-reactive mAbs bound the HA stalk domain (fig. S1C), compared to 49% of all HA-reactive mAbs induced by infection (P < 0.0001; fig. S1D). Remarkably, 58% of all HA-reactive H1N1 infection–induced mAbs bound the stalk domain (fig. S1D, bottom left). Of all stalk domain–reactive antibodies induced by infection or vaccination, we did not see a difference in typical broadly neutralizing stalk epitopes bound (CR9114 competing) versus mAbs binding uncharacterized stalk epitopes (CR9114 non-competing, but positive for the headless stalk construct and chimeric HA constructs) (cH5/1; cH7/3) (fig. S1E). We also identified HA-reactive mAbs induced by infection and vaccination that did not confer HAI activity and were ruled out against binding stalk domain epitopes by our assays (fig. S1, C and D). These mAbs were categorized as HA+ mAbs binding to “unknown” epitopes, potentially binding undefined HAI− epitopes on the HA head or stalk. Together, these data reveal that the minority of infection-induced HA+ mAbs bind HAI+ head epitopes and instead tend to bind HAI− head and stalk epitopes.
Similar to HA, the head region of the influenza virus NA surface glycoprotein is susceptible to antigenic drift, although the active site and nearby epitopes are highly conserved (27). Because several infection-induced mAbs bound to NA, we asked whether NA-reactive mAbs bound to conserved sites. To address this, we used influenza NA inhibitor resistance detection (NA-STAR) and NA enzyme-linked lectin assays (ELLA), which can determine whether a mAb is binding conserved epitopes on the NA active site or regions within close proximity to it. In the NA-STAR assay, a small chemiluminescent substrate is bound directly to the NA enzymatic site unless an antibody binds the active site and blocks access of the substrate. For ELLA, the sialic acid substrate is present in glycans of the fetuin glycoprotein, and an antibody that binds to NA at or near the enzymatic site can sterically block access to the enzymatic site. We found that more than half of all infection-induced NA-reactive mAbs were NA-STAR+ or ELLA+ (fig. S1F). There was no difference between the percentages of combined NA-STAR+ and ELLA+ mAbs elicited by H1N1 and H3N2 infection, with 70% induced by H1N1 infection and more than half induced by H3N2 infection (fig. S1F, bottom). Overall, these data suggest that natural infection recalls preexisting MBCs reactive to more conserved influenza virus antigens, a hallmark of original antigenic sin.
Infection-induced antibodies are predominantly non-neutralizing
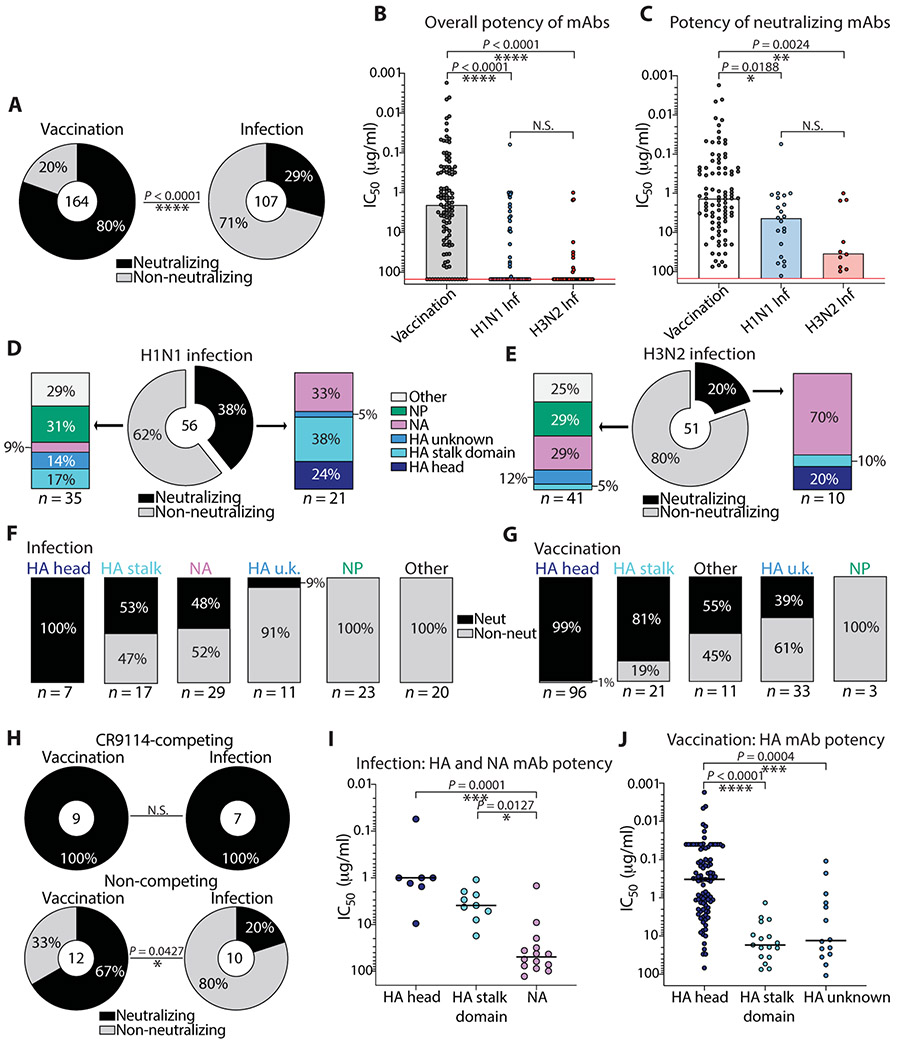
Only 6% of early infection–induced mAbs had HAI activity (Fig. 1C), which is the primary correlate of antibody-mediated protection against influenza. However, mAbs reactive to immunosubdominant and neutralizing regions such as the HA stalk and NA active site were also identified. These results led us to address whether these mAbs were neutralizing toward the inducing vaccinating and infecting strains. To do so, we determined the neutralization capacity of mAbs induced by infection or vaccination using an in vitro virus microneutralization assay or plaque reduction neutralization assay. Only 29% of infection-induced mAbs were neutralizing, relative to 80% of mAbs that were induced by seasonal vaccination (P < 0.0001; Fig. 2A). Moreover, we observed that the overall neutralization potency of all infection-induced mAbs was markedly reduced compared to vaccination-induced mAbs (P < 0.0001; Fig. 2B), and the potency was still reduced when non-neutralizing mAbs were not included in the analysis (P = 0.0188, H1N1; P = 0.0024, H3N2; Fig. 2C).
Fig. 2. Influenza virus infection–induced antibodies are predominantly non-neutralizing in vitro compared to vaccination-induced antibodies.
(A) Pie charts display percentages of all vaccination- and infection-induced mAbs with virus neutralization activity as assessed by microneutralization or plaque reduction assay using Madin-Darby canine kidney cell lines. (B) The overall potency of neutralizing and non-neutralizing vaccination-induced mAbs (n = 164) was compared to infection-induced mAbs (H1N1, n = 56; H3N2, n = 51), depicted as microneutralization IC50 values. Non-neutralizing mAbs are displayed on the red line above the highest test concentration at 150 μg/ml. N.S., not significant. (C) The potency of all neutralizing mAbs induced by the quadrivalent vaccine (n = 92) was compared to the potency of neutralizing infection–induced mAbs (H1N1, n = 21; H3N2, n = 10), depicted as microneutralization IC50 values. (D and E) Bar charts show the antigen reactivity of neutralizing and non-neutralizing H1N1 (D) and H3N2 (E) infection-induced mAbs. (F and G) Bar charts display the percent of total neutralizing and non-neutralizing mAbs induced by infection (F) and vaccination (G), subset by antigen reactivity. (H) Pie charts demonstrate the percentage of neutralizing and non-neutralizing stalk domain–reactive mAbs binding a broadly neutralizing stalk epitope, as determined by a CR9114 competition ELISA (top) or mAbs binding undefined stalk epitopes, determined by ELISA against a headless HA stalk construct and chimeric HA (bottom). (I and J) The potency of HA- and NA-reactive mAbs induced by infection (I) was compared to the potency of HA-reactive mAbs induced by vaccination (J). The numbers in the center of or below each chart indicate the number of mAbs tested. Statistical significance was determined by chi-square test (****P < 0.0001) (A), Fisher’s exact test (*P = 0.0427) (H), or unpaired nonparametric Kruskal-Wallis test with Dunn’s correction for multiple comparisons (B, C, I, and J). Data are representative of two independent experiments performed in duplicate.
We previously demonstrated that the distribution in antibody reactivity varied widely between H1N1 and H3N2 infections (P = 0.0285; Fig. 1B). We therefore analyzed the distribution in reactivity of neutralizing and non-neutralizing infection–induced mAbs. Of H1N1 infection–induced mAbs, only 38% were neutralizing, of which 67% bound HA and 33% bound NA (Fig. 2D and fig. S2A). Most of the stalk domain–reactive H1N1-induced mAbs were neutralizing, confirming the protective nature of mAbs against this region. Only 20% of all H3N2 infection–induced mAbs were neutralizing, and in sharp contrast to H1N1 infection, 70% of all H3N2 infection–induced neutralizing mAbs were NA reactive, with the minority recognizing HA (Fig. 2E and fig. S2B). All mAbs binding NP and other unknown antigens were non-neutralizing, with a substantial fraction of non-neutralizing HA- and NA-reactive mAbs detected in each group (Fig. 2, D and E).
We next addressed whether there were qualitative differences in the neutralization ability against distinct antigens between infection-induced and vaccination-induced mAbs. For both groups, most of the neutralizing mAbs targeted the HA head and stalk domain regions (Fig. 2, F and G). Unique to infection-induced mAbs, a substantial portion of NA-reactive mAbs were neutralizing (Fig. 2F). Most of the infection-induced mAbs targeting unknown regions of HA and other unknown antigens were non-neutralizing, in addition to several non-neutralizing NP-reactive mAbs isolated (Fig. 2, F and G). All mAbs isolated targeting the broadly neutralizing stalk epitope (CR9114 competing) were neutralizing, and several mAbs targeting other undefined stalk domain epitopes were also neutralizing (Fig. 2H). Last, we identified that HA head–reactive mAbs were typically more potent than HA stalk domain–reactive mAbs (Fig. 2, I and J, and fig. S2, C and D), and NA-reactive mAbs were less potent than HA head- and stalk domain–reactive mAbs (Fig. 2I). Together, these data demonstrate that most mAbs isolated from infected individuals targeted conserved yet non-neutralizing targets, although some neutralizing mAbs reactive to immunosubdominant epitopes of the HA stalk domain and NA were identified.
The reactivity of influenza virus infection–induced antibodies is biased by original antigenic sin
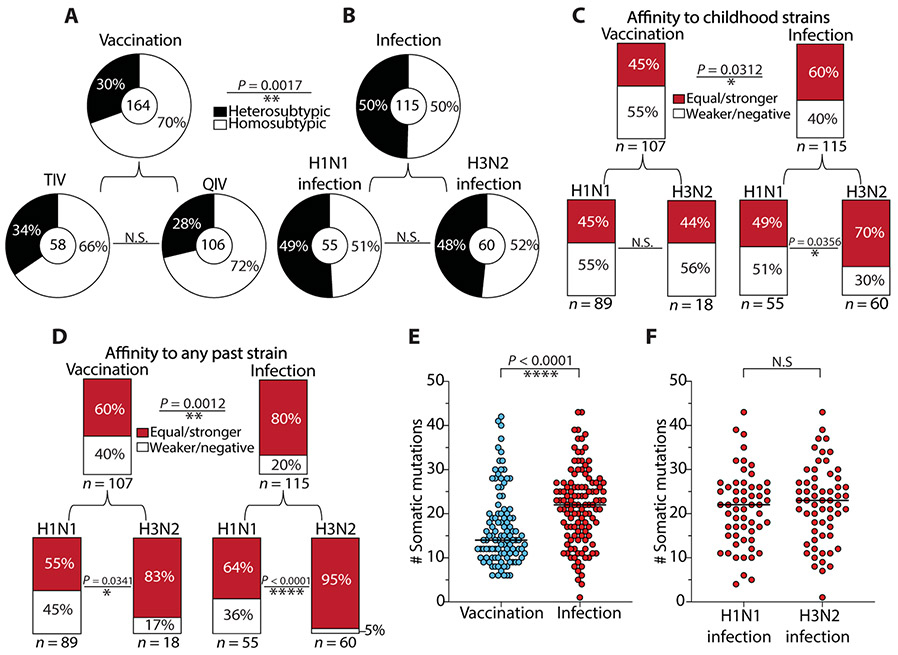
Most of the vaccination-induced mAbs bound antigenically drifted viral epitopes on the HA head. In contrast, most of the mAbs induced by infection bound to conserved epitopes, such as the HA stalk domain, NA active site, and NP (Fig. 1B and fig. S1). Accordingly, most of these mAbs were non-neutralizing (Fig. 2). To further investigate the role of original antigenic sin and previous immune history in shaping mAb reactivity, we assessed the cross-reactivity of mAbs induced by natural infection against heterosubtypic viral strains and past strains circulating during the lifetime of the individual. First, we tested each mAb against historical and contemporary H1N1 and H3N2 viral strains, which accounted for nearly 50 years of viral antigenic drift, to determine the degree of homosubtypic and heterosubtypic cross-reactivity within each infection and vaccination cohort. Consistent with the high prevalence of vaccination-induced mAbs that bound the HA head, most of the vaccination-induced mAbs were homosubtypic (Fig. 3A). Conversely, infection-induced mAbs displayed increased heterosubtypic cross-reactivity relative to vaccination-induced mAbs, regardless of the infecting subtype (p = 0.0017; Fig. 3B). As expected, and likely due to differences in age, immune histories, and other factors, we observed variation in heterosubtypic cross-reactivity among individual donors (fig. S3, A to C). Strikingly, 60% of all infection-induced mAbs displayed equal or stronger affinity for childhood viral strains than viral strains circulating at the time of infection (Fig. 3C and table S3), confirming a substantial role for original antigenic sin in shaping early antibody responses to natural infection. For vaccination-induced mAbs, this phenotype was also substantial, because 45% could bind with equal or stronger affinity to childhood strains as compared to viral strains circulating at the time of mAb isolation. Because responses to influenza viruses are thought to be shaped not only by childhood exposures but also by any past exposures (15), we analyzed the percentage of mAbs induced by infection or vaccination that had equal or stronger affinity toward any past strain relative to the inducing strains. We identified that 60% of vaccination-induced mAbs had equal or stronger reactivity toward past strains, compared to 80% for infection-induced mAbs (P = 0.0012; Fig. 3D). We observed a greater effect for original antigenic sin by past exposures on mAbs that were H3N2-reactive versus H1N1-reactive, patterns that were consistent among infected individuals [Fig. 3, C and D (bottom), and fig. S3, D and E]. Overall, we observed greater variation in both cross-reactivity and affinity toward past strains in vaccinated individuals (fig. S3, C and F).
Fig. 3. The cross-reactivity of influenza virus infection–induced antibodies is biased by original antigenic sin.
(A and B) Pie charts demonstrate the cross-reactivity of vaccination-induced mAbs (A) and infection-induced mAbs (B), which was inferred by ELISA binding to a panel of H1N1 and H3N2 viruses. Heterosubtypic cross-reactivity was defined on the basis of the ability of a mAb to bind to at least one or more strains opposite of the inducing subtype, such as an H1N1-induced mAb binding to one or more H3N2 strains. (C and D) Bar charts represent the percentage of infection-induced mAbs with equal or greater binding affinity to childhood strains (C) or any past strains (D) relative to contemporary strains circulating during the time of mAb isolation. Past strains in (D) include all available strains tested that were circulating before the year of the inducing strain for each cohort (H1N1 infection, n = 5; H3N2 infection, n = 6; H1N1-reactive vaccination, n = 5; H3N2-reactive vaccination, n = 3 strains analyzed). (E) The number of somatic mutations in the immunoglobulin heavy chain VH for 2014–2015 quadrivalent vaccine-induced mAbs (n = 106) was compared to H1N1 and H3N2 infection–induced mAbs combined (n = 117). (F) The number of somatic mutations in the immunoglobulin heavy chain VH for H1N1 infection–induced mAbs (n = 57) was compared to H3N2-infection induced mAbs (n = 60). The numbers in the center of or below each chart indicate the number of mAbs tested. Statistical significance was determined by chi-square or Fisher’s exact test (**P = 0.0017; *P = 0.0312; **P = 0.0012) (A to D) and unpaired nonparametric Mann-Whitney test (****P < 0.0001) (E and F; bars indicate median). Data are representative of two to three independent experiments performed in duplicate.
Because the cross-reactivity of infection-induced mAbs appeared to be more influenced by past exposures, we addressed whether these mAbs had increased immunoglobulin heavy chain variable region (VH) somatic mutations relative to vaccination-induced mAbs, as these antibodies were likely derived from MBCs that were continually recalled into germinal centers during the lifetime of the individual. Infection-induced mAbs had a median of 22 VH gene somatic mutations, whereas vaccination-induced mAbs had a median of 14 VH gene mutations (P < 0.0001; Fig. 3E). There was no difference in the number of somatic mutations between H1N1 and H3N2 infection–induced mAbs (Fig. 3F). Together, these data suggest that influenza virus infection recalls MBCs from long-past exposures, likely from childhood, supporting the original antigenic sin hypothesis. By contrast, vaccination may recall MBCs from more recent virus exposures, although the antibody response to vaccination is still largely biased by past exposures. mAbs derived from preexisting MBCs that bind with higher affinity to historical test strains relative to current test strains imply that those MBCs were originally generated upon exposure to a similar historical strain. However, recent exposures to divergent lineages similar to the historical test strain analyzed may also account for increased affinity.
Influenza virus antigen and subtype reactivities shape original antigenic sin–like antibody responses
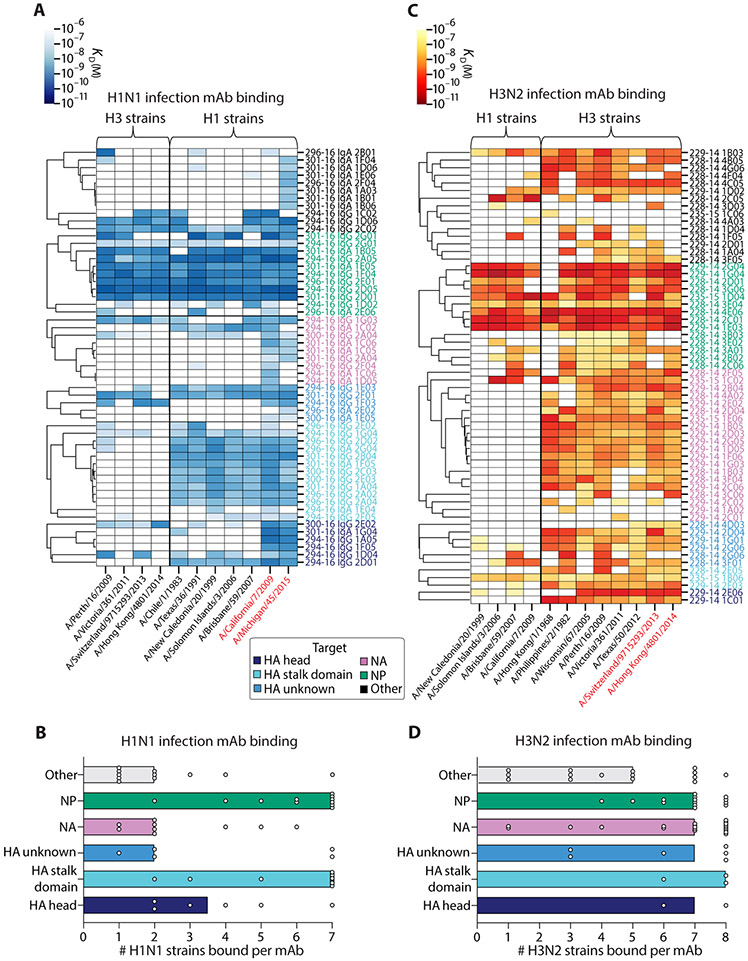
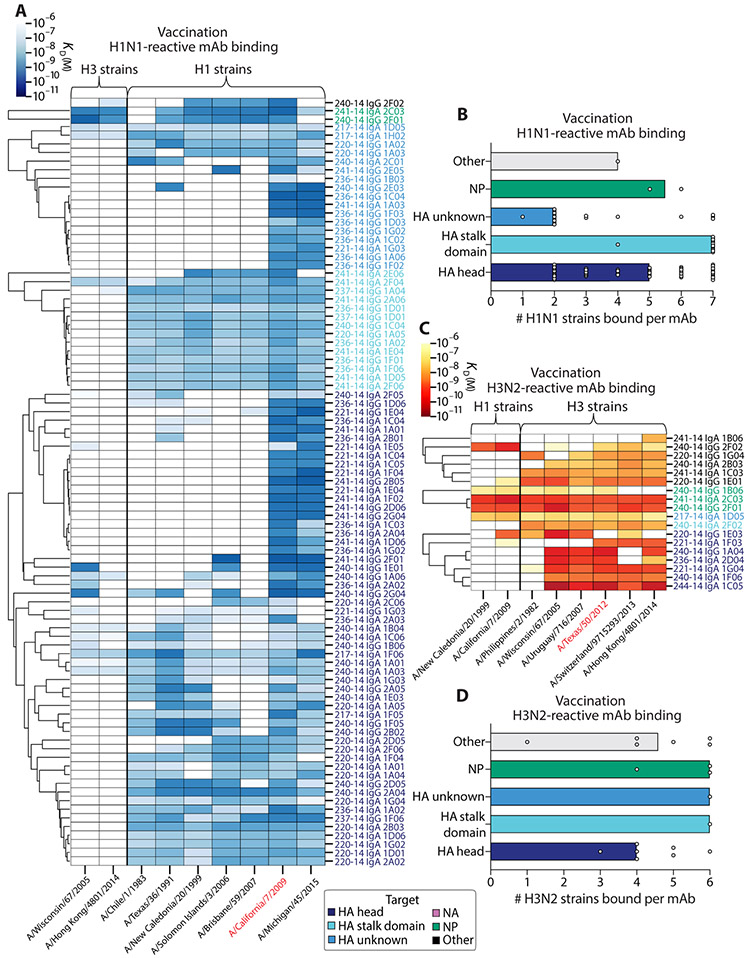
Reactivity toward distinct antigens is expected to shape mAb cross-reactivity. To determine how antigen targeting and subtype reactivity affect cross-reactivity of infection- and vaccination-induced mAbs, we clustered mAbs by antigen bound in heatmaps based on their affinity [ELISA dissociation constant (KD)] for several H1N1 and H3N2 virus strains. On the basis of patterns in reactivity, groups of mAbs predicted to bind similar epitopes could then be visualized on the basis of cluster formation. For H1N1 infection–induced mAbs, HA stalk domain– and NP-reactive mAbs displayed the greatest breadth of cross-reactivity to H1N1 strains, generally binding to all seven influenza H1N1 strains tested (Fig. 4, A and B, and fig. S3D). Most of the mAbs isolated from H1N1-infected individuals bound to the pandemic strains A/California/7/2009 and A/Michigan/45/2015, suggesting that those individuals were likely infected with an influenza virus variant most similar to these viral strains (Fig. 4A, right). Regardless of epitope reactivity, most mAbs cross-reacted with several H1N1 viral strains dating back to 1983 (A/Chile/1/1983). A distinct pattern in reactivity was seen for H3N2 infection–induced mAbs, which exhibited a marked degree of homosubtypic cross-reactivity, regardless of the epitope bound (Fig. 4, C and D, and fig. S3E). For both subtypes, heterosubtypic cross-reactivity was mainly observed for mAbs reactive to HA, NP, and other unknown antigens, whereas NA-reactive mAbs were largely homosubtypic. These results demonstrate both distinct and overlapping patterns in mAb breadth, dependent on the infecting subtype and viral epitopes targeted. Together, these results highlight the ability of H1N1 and H3N2 viruses to differentially induce antibodies to conserved viral epitopes.
Fig. 4. The degree of influenza virus infection–induced antibody cross-reactivity is influenced by antigen-reactivity and infecting subtype.
(A and B) The viral binding breadth of H1N1 infection–induced mAbs is represented by heatmap analysis displaying affinity (KD) for contemporary and historical H1N1 and H3N2 whole-virus strains (A) and bar graphs summarizing the number of homosubtypic H1N1 viral strains bound per H1N1 infection–induced mAb (n = 55), subset by antigen specificity (B; bars indicate median). (C and D) The viral binding breadth of H3N2 infection–induced mAbs is represented by heatmap analysis displaying affinity (KD) for contemporary and historical H3N2 and H1N1 whole viral strains (C) and bar graphs summarizing the number of homosubtypic H3N2 viral strains bound per H3N2 infection–induced mAb (n = 60), subset by antigen specificity (D; bars indicate median). Heatmap data are depicted as ELISA binding affinity (KD) values for each individual mAb tested against the respective viruses, and antigen reactivity of each mAb is indicated by the color coding in the legend. For both heatmaps, the strains colored in red text represent contemporary circulating strains during the time of mAb isolation. Data are representative of two to three independent experiments performed in duplicate.
The cross-reactivity of vaccination-induced mAbs was shaped by preexisting immunity, with H3N2-reactive mAbs characteristically having higher affinity toward past strains (Fig. 3, C and D). We therefore performed the same analyses with vaccination-induced mAbs to determine how antigen and subtype targeting affected cross-reactivity. Vaccination-induced H1N1-reactive mAbs displayed vast viral binding breadth to several strains, despite these mAbs predominantly targeting the antigenically variable HA head region (Fig. 5, A and B). Vaccination-induced H3N2-reactive mAbs were similarly cross-reactive and bound most of the test strains analyzed (Fig. 5, C and D). We observed similar degrees of homosubtypic breadth compared to infection-induced mAbs for H1N1- and H3N2-reactive mAbs targeting the HA head, HA stalk domain, NP, or other regions on HA, with H3N2-reactive vaccination-induced mAbs typically exhibiting greater breadth regardless of epitope targeted (Fig. 5, B and D). Together, our results point to inherent differences in the ability of H1N1 and H3N2 viruses to draw upon preexisting immunity and induce cross-reactive antibody responses, regardless of exposure type, with the degree of cross-reactivity dependent on antigens targeted. In addition, these results demonstrate the breadth of homosubtypic viral binding by HA head–reactive mAbs induced by seasonal vaccines, which are typically thought to elicit highly strain-specific responses.
Fig. 5. The degree of influenza virus vaccination–induced antibody cross-reactivity is influenced by antigen reactivity and vaccine strain reactivity.
(A and B) The viral binding breadth of H1N1-reactive quadrivalent vaccine-induced mAbs is represented by heatmap analysis displaying affinity (KD) to contemporary and historical H1N1 and H3N2 whole viral strains (A) and bar graphs summarizing the number of homosubtypic H1N1 viral strains bound per H1N1-reactive mAb (n = 90), subset by antigen specificity (B; bars indicate median). (C and D) The viral binding breadth of H3N2-reactive quadrivalent vaccine-induced mAbs is represented by heatmap analysis displaying affinity (KD) to contemporary and historical H3N2 and H1N1 whole viral strains (C) and bar graphs summarizing the number of homosubtypic H3N2 viral strains bound per H3N2-reactive mAb (n = 18), subset by antigen specificity (D; bars indicate median). Heatmap data are depicted as ELISA binding affinity (KD) values for each individual mAb tested against the respective viruses, and antigen reactivity of each mAb is indicated by the color coding in the legend. For both heatmaps, the strains colored in red text represent the vaccinating strains present in the vaccines at the time of mAb isolation. Data are representative of two to three independent experiments performed in duplicate.
Early infection–induced antibodies are less protective in mice compared to vaccination-induced antibodies
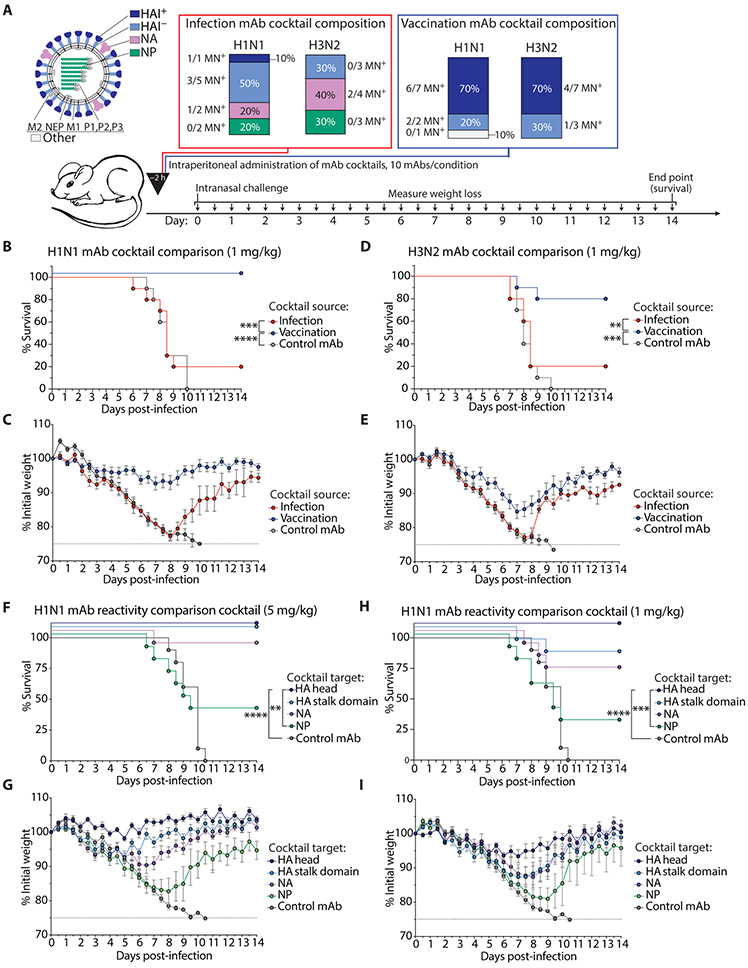
Most of the early cross-reactive infection-induced mAbs failed to neutralize virus in vitro, suggesting that they may have limited protective ability in vivo in comparison to HA head–reactive mAbs induced by vaccination. To evaluate this possibility, we prophylactically administered representative cocktails of mAbs covering the spectrum of reactivities induced by infection versus vaccination (Fig. 6A). The mAb cocktails were administered intraperitoneally at a dose titration of 5, 1, and 0.2 mg/kg to BALB/c mice, which were intranasally challenged 2 hours after mAb transfer with a lethal dose (10 LD50) of mouse-adapted A/Netherlands/602/2009 H1N1 or A/Philippines/2/1982 H3N2 influenza virus. We generated four separate cocktails each comprising 10 mAbs: an H1N1 infection–induced cocktail, an H3N2 infection–induced cocktail, and two cocktails of H1N1- and H3N2-reactive vaccination-induced mAbs (table S4); all of which were representative of typical binding and neutralization characteristics within each cohort.
Fig. 6. Influenza virus infection–induced antibodies are less protective in vivo than vaccination-induced antibodies.
(A) Bar charts display the composition of H1N1 and H3N2 infection– and vaccination-induced mAb cocktails. For the infection cocktails, all H1N1 mAbs were originally induced by H1N1 infection, and all H3N2 mAbs were originally induced by H3N2 infection. The vaccination cocktails were composed of either H1N1- or H3N2-reactive vaccination-induced mAbs. Each cocktail reflects the antigen reactivity and neutralization frequencies seen in our analyses. (B and C) Survival and weight loss curves display in vivo prophylactic protective ability of H1N1 infection– and vaccination-induced mAb cocktails administered intraperitoneally at 1 mg/kg to 6- to 8-week-old female BALB/c mice challenged with 10 LD50 mouse-adapted A/Netherlands/602/2009 H1N1 virus. (D and E) Survival and weight loss curves display in vivo prophylactic protective ability of H3N2 infection and vaccination-induced mAb cocktails administered intraperitoneally at 1 mg/kg to 6- to 8-week-old female BALB/c mice challenged with 10 LD50 mouse-adapted A/Philippines/2/1982 H3N2 virus. (F to I) Survival and weight loss curves display in vivo prophylactic protective ability of HA head–, HA stalk domain–, NA-, and NP-reactive mAb cocktails (5 mAbs per cocktail) administered intraperitoneally at 5 mg/kg (F and G) or 1 mg/kg (H and I) to 6- to 8-week-old female BALB/c mice challenged with 10 LD50 mouse-adapted A/Netherlands/602/2009 H1N1 virus. Data are representative of two independent experiments and depicted as survival (B, D, F, and H) and weight loss (C, E, G, and I) curves. Statistical significance for survival curves was determined using a Mantel-Cox log-rank test [(B) ****P < 0.0001 and ***P = 0.0003; (D) ***P = 0.0002 and **P = 0.0080; (F) ****P < 0.0001 and **P = 0.0047; (H) ****P < 0.0001 and ***P = 0.0004]. Weight loss is presented as means ± SEM (n = 9 to 10 mice per group).
H1N1-reactive vaccination-induced mAbs were potently protective against 10 LD50 of A/Netherlands/602/2009 virus at 1 mg/kg, whereas H1N1 infection–induced mAbs failed to provide protection, with mice displaying increased mortality and weight loss relative to mice receiving the vaccination cocktail (P = 0.0003, survival; Fig. 6, B and C). Whereas H3N2-reactive vaccination-induced mAbs could provide protection at 1 mg/kg against A/Philippines/2/1982 virus, H3N2 infection–induced mAbs provided substantially reduced protection when administered at the same dose (P = 0.0080, survival; Fig. 6, D and E). For both infection- and vaccination-induced mAb cocktails, 100% protection was provided at the highest dose of 5 mg/kg, and neither infection- nor vaccination-induced mAbs provided full protection at 0.2 mg/kg (fig. S4).
The limited protective ability of infection-induced mAbs suggested that the specificities recalled by infection in this cohort were subpar for protection compared to vaccination-induced mAbs, which largely targeted the HA head. We therefore tested the protective ability of mAbs targeting distinct epitopes by prophylactically administering cocktails of H1N1-reactive mAbs targeting the HA head, HA stalk domain, NA, or NP (table S5) into BALB/c mice intraperitoneally, which were subsequently intranasally challenged with 10 LD50 of mouse-adapted A/Netherlands/602/2009 H1N1. Of note, HA stalk domain–, NA-, and NP-reactive mAbs were derived from infected individuals, whereas HA head–reactive mAbs were induced by both infection and vaccination. At 5 mg/kg, we saw clear resolution of the different potencies of these cocktails. Mice receiving the HA head–reactive mAb cocktail lost the least weight, followed by the HA stalk domain mAb cocktail, the NA mAb cocktail, which provided intermediate protection, and, last, the NP-reactive mAb cocktail, which provided the least protection (Fig. 6, F and G). At 1 mg/kg, the protective ability of the HA stalk domain–reactive mAb cocktail decreased to the same degree as the NA-reactive mAb cocktail, and mice in these groups lost substantially more weight relative to the HA head–reactive mAb cocktail (Fig. 6, H and I). At both 5 and 1 mg/kg, the NP-reactive mAbs provided minimal protection in vivo.
DISCUSSION
It is well appreciated that protective antibody responses to influenza in adults are biased by past exposures, explained by the concept of original antigenic sin. However, it remains unclear how preexisting immunity shapes the immunodominance hierarchy among influenza virus proteins targeted early after natural infection and seasonal vaccination in humans and how such hierarchies correlate with protection. In this study, we revealed that preexisting immunity biased the early antibody response to infection toward conserved yet less protective viral epitopes in an original antigenic sin-like fashion. Most of the B cells activated as plasmablasts in our infected cohort targeted the HA stalk domain, NA, NP, and other unknown viral antigens, and the majority exhibited equal or stronger affinity to past strains relative to inducing strains. The minority of mAbs bound potently neutralizing HA stalk and NA epitopes, reflecting the immunosubdominance of these regions. Conversely, we identified that seasonal influenza vaccines induced early antibodies targeting highly cross-reactive and protective HA head epitopes, highlighting the value of influenza vaccination and the potential of universal vaccine strategies that draw upon preexisting immunity to elicit protective antibody responses to conserved epitopes.
Whereas the landscape of protective specificities was diverse between infection and vaccination, we observed inherent differences in the ability of H1N1 and H3N2 viruses to induce cross-reactive antibody responses, regardless of exposure type. Reactivity to childhood and past viral strains was especially prominent for H3N2-reactive mAbs whether they were induced by vaccination or infection, which may result from the rapid evolutionary rate of the H3N2 HA (27, 32). As the HA head of H3N2 viruses rapidly mutates, repeated exposures to antigenically drifted H3N2 viruses could further bias the antibody response toward more conserved epitopes on HA or other viral antigens, as these are the only epitopes that will be recognized by the preexisting MBC repertoire upon exposure. We observed increased targeting of NA, NP, and other unknown viral antigens in response to H3N2 infection, whereas H1N1 infection was more biased toward generating HA-reactive responses. Furthermore, we observed that vaccination generally induced a greater magnitude of H1N1-reactive plasmablasts relative to H3N2, consistent with findings from a recent study that suggested that these differences may account for differential vaccine effectiveness against H1N1 and H3N2 viruses (33). In the future, it will be important to investigate the role of preexisting immunity in shaping antibody responses to H1N1 versus H3N2 viruses, which are largely contrasting in the severity of cases they inflict and in their rates of antigenic evolution.
Through this study, we uncovered a role for preexisting immunity in shaping broadly reactive and neutralizing anti-HA head responses to vaccination. It is currently appreciated that influenza vaccination provides effective immunity to vaccine strains; however, immune pressure drives antigenic drift of the major head epitopes of circulating strains, limiting the effectiveness of seasonal vaccines (7-10, 34, 35). The extent to which seasonal vaccines elicit cross-reactive protective responses to the HA head remains unclear, and we hypothesized that preexisting immunity to HA may bias the induction of highly cross-reactive HA head–specific responses. Most of the HA head–reactive mAbs induced by vaccination exhibited broad binding to current, past, and drifted strains circulating years after mAb isolation, with the minority being strain specific. As the overwhelming majority of these mAbs were neutralizing and potently protective in vivo, these data suggest that vaccination can harness preexisting immunity to induce more potently protective antibody responses than currently appreciated. Moving forward, it will be important to investigate mechanisms by which cross-reactive responses can be preferentially induced by vaccination in a durable fashion to develop vaccines that effectively elicit long-lived protective responses at the population level.
In contrast to vaccination, infection induced a higher frequency of antibodies targeting conserved influenza virus epitopes on the HA stalk domain, NA, and NP, supported by previous serology studies (24). Whereas numerous studies have shown that antibodies reactive to the HA stalk and NA can provide potent viral neutralization and protection (22, 36-39), about half of all HA stalk domain– and NA-reactive infection-induced mAbs isolated in this study were non-neutralizing, potentially reflecting an unappreciated role for original antigenic sin in inducing antibodies targeting conserved yet less protective epitopes on these antigens. Conversely, more than 80% of stalk domain-binding antibodies induced by vaccination targeted neutralizing epitopes, highlighting the ability of influenza virus vaccination to induce protective stalk antibody responses. We identified several HA stalk domain–specific mAbs that did not compete with CR9114, a mAb specific for a well-characterized neutralizing stalk epitope (30), but did bind a headless HA stalk construct (31), signifying that these antibodies bind stalk epitopes yet to be characterized.
Consistent with the reduced neutralization potential of infection-induced mAbs, we identified that mixed-epitope cocktails of H1N1 and H3N2 infection–induced mAbs were less protective in vivo compared to vaccination-induced mAbs targeting the HA head. To date, few studies have directly compared the protective ability of mAbs targeting distinct conserved influenza virus epitopes on the HA stalk domain, NA, and NP relative to the HA head in vivo (40). Previous studies have shown that antibodies against the HA stalk are less potent than antibodies against the HA head in vitro but have the potential to confer robust protection in vivo, consistent with our findings (36). It has also been shown that NA-reactive mAbs are protective in vivo (22, 41), although the NA-reactive mAbs from our cohort were less prophylactically protective relative to HA head domain– and stalk domain–reactive mAbs. Whereas NA-inhibiting antibodies reduce influenza illness and transmission, past studies have debated their role as an independent correlate of protection from initial infection, likely due to their inability to directly inhibit viral entry (42-46). All of the NP-reactive mAbs isolated in this study were non-neutralizing in vitro and provided limited protection in vivo, although some studies have suggested that anti-NP mAbs may play a role in protection through antibody-dependent cellular cytotoxicity or complement-dependent cytotoxicity (47). In summary, our in vivo data highlight differences in how preexisting B cells reactive to distinct influenza epitopes provide protection against infection.
Inherent differences in exposure route as well as the nature of inactivated virus vaccines versus live viral infections are likely to create differences in antibody epitope targeting. The greater abundance of non-HA antigens during natural infection compared to vaccination may bias MBC recall to conserved yet less protective non-HA head epitopes, as MBCs targeting these regions are likely at an early competitive advantage. Moreover, memory CD4 T cells recognizing H1N1 and H3N2 viruses largely target NP- and NA-derived peptides, suggesting that CD4 T cell help could influence MBC recall toward these epitopes (48). The antibody response to vaccination characterized in the elderly is similarly biased toward non-HA head epitopes, which may be due to the lack of new naïve B cells and adaptability of MBCs to new influenza viruses encountered with age, increased reliance on CD4 T cell help for B cell activation, and the competitive advantage of MBCs targeting conserved epitopes (27).
Our study was limited in that we were only capable of characterizing the early plasmablast response to infection in symptomatic patients from a single time point. Previous studies have shown that influenza virus infection can induce broad and durable antibody responses, which may be due to sustained antigen availability and priming of B cell and T cell responses (25, 49). In addition, it has been shown that infection induces MBCs with broad reactivity toward both the HA head and stalk, with evidence of adaptation to the infecting strain (36, 50). The induction of long-lived MBCs targeting conserved viral epitopes after natural infection could provide superior protection against future infections relative to the short-lived immunity induced by vaccination. Although we were able to show that our collection of infection-induced mAbs provided sub-optimal protection in mice, it is unclear how antibody targeting of these epitopes affords protection in humans. Past studies have shown that non-HAI antibodies can serve as a correlate of protection against infection in humans, and natural infection is capable of providing long-lived protection from reinfection with the same subtype (43, 45, 46, 51). Future studies should assess the kinetics of epitope targeting across multiple time points at both the cellular and serological level to better determine correlates of protection. In addition, household cohort studies, controlled human infections, or vaccine efficacy studies will be required to determine whether preexisting immune biases toward particular viral epitopes directly affect susceptibility or protection in humans.
To understand the formation of bias within the adaptive immune repertoire, it is critical that we study the response to natural influenza virus infection versus vaccination in cohorts of different ages, particularly in naïve infants and children. In our study, we were only able to assess the antibody response in a limited number of adults, but future pediatric studies assessing mechanisms of imprinting will be key for evaluating which B cell specificities are recruited by primary infection. A pressing question in the field of influenza vaccine development is whether priming naïve children by initial infection or vaccination leads to better B cell–mediated immunity. It will be important to determine the extent to which children are capable of mounting antibody responses to conserved protective viral epitopes, and how age and degree of viral exposure correlates with antibody adaptability. On the opposite end, it is important to study whether and how high-dose and adjuvanted influenza vaccination of elderly individuals can overcome immunosenescence and promote adaption to new influenza viruses.
The precise epitopes that should be targeted for increased vaccine effectiveness is an area of active investigation. Our data confirm the well-accepted notion that vaccine-induced antibodies against the HA head domain are potently protective, but the age-old problem remains: Constant immune pressure drives antigenic drift of the HA head, limiting the effectiveness of seasonal vaccines when circulating strains are not well matched (7-10, 34, 35). Moreover, it remains to be determined whether cross-reactive antibody responses to HA elicited by vaccination can provide potent and durable protection against antigenically drifted strains in humans. Although not as potently protective as antibodies against the HA head, antibodies against conserved HA stalk and NA epitopes are associated with protection against influenza virus infection (46, 52-54), and several vaccine platforms targeting these epitopes are in preclinical and clinical trials (31, 55-57). The best vaccination platform to achieve both viral binding breadth and potent protection may be one that drives naïve and MBC maturation against antigenically drifted HA head epitopes but can also induce potent antibodies against conserved HA and NA epitopes to protect against antigenically drifted and novel influenza viruses. Further studies on B cell immunodominance patterns in response to such vaccine platforms are necessary to understand whether antibodies can be induced against both conserved and nonconserved protective epitopes. In addition, future research is warranted to address differences in the potency of antibodies targeting distinct epitopes on HA and NA, as not all conserved viral epitopes will be equally protective to target. In summary, our results point to inherent differences in the ability of influenza virus infection and vaccination to induce early protective antibody responses, largely dependent on preexisting immunity to distinct conserved epitopes and the influenza A virus subtype encountered.
MATERIALS AND METHODS
Study design
We recruited human participants about 7 to 11 days after onset of influenza virus infection (2015–2016 H1N1 and 2014–2015 H3N2) or 7 days after vaccination (2010–2011 trivalent influenza vaccine and 2014–2015 quadrivalent influenza vaccine) to assess the role of immune history in shaping antibody targeting of conserved viral epitopes. We generated mAbs from single cell–sorted plasmablasts, which peak in expansion between days 5 and 14 and largely derive from the preexisting MBC pool. Plasmablasts were not antigen bait-sorted, and there is no obvious bias in our mAb generation protocol toward particular influenza virus antigens. Therefore, the sampling of mAbs per individual is expected to be representative of the overall plasmablast specificities elicited. We generated and characterized mAbs from 7 infected individuals and 18 vaccinated individuals across multiple influenza seasons. Because vaccinated individuals from controlled seasonal vaccine trials were readily accessible to our group, we included more vaccinated individuals than infected individuals in our study. We characterized all influenza-reactive mAbs that could be isolated from individual participants within the limitations of the labor-intensive process of generating mAbs. We comprehensively characterized antigen specificity and viral cross-reactivity of all mAbs by ELISA, neutralization potency by microneutralization or plaque assay, and in vivo protective ability in a mouse model to identify how preexisting immunity shaped early antibody responses to natural influenza virus infection versus vaccination. Because these groups were vaccinated or infected in different years with the vaccinating and infecting strains distinct for each year, this study was unblinded and not randomized.
All studies were performed with the approval of the University of Rochester and the University of Chicago institutional review boards. Informed consent was obtained after the research applications, and possible consequences of the studies were disclosed to study participants. Clinical information is detailed in tables S1 and S2. Polymerase chain reaction (PCR)–confirmed influenza virus–infected individuals were recruited and only included in the study if they did not have coinfections and were not being treated with immunosuppressive therapies.
All experiments were done in accordance with the University of Chicago Institutional Animal Care and Use Committee and in adherence to the NIH Guide for the Care and Use of Laboratory Animals. Six- to 8-week-old female BALB/c mice were used for these studies, as the virus dose was titrated in mice of this same age and sex. Nine to 10 mice were used per group, and a power analysis was used to determine the number of mice per experiment. All mice from independent experiments were included in data analysis until the point of euthanasia, which occurred upon 25% weight loss from the initial starting weight or upon completion of the experiment (14 days). Mice were provided standard diet chow and water and were housed in the ABSL-2 (animal biosafety level-2) facility within the Carlson Animal Research Facility at the University of Chicago.
Cell culture
Human embryo kidney (HEK) 293T cells (Thermo Fisher Scientific) were maintained at 37°C with 5% CO2 in advanced Dulbecco’s modified Eagle’s medium (DMEM; Gibco) with 2% ultralow immunoglobulin G (IgG) fetal bovine serum (FBS; Gibco), 1% l-glutamine (Gibco), and 1% antibiotic-antimycotic (Gibco). Madin-Darby canine kidney (MDCK) cells (American Type Culture Collection or London) were maintained in culture at 37°C with 5% CO2 in DMEM (Gibco) supplemented with 10% FBS (Gibco), 1% l-glutamine (Gibco), and 1% penicillin-streptomycin (Gibco).
Viruses and recombinant proteins
Influenza viruses used in all assays were grown in-house in specific pathogen–free eggs, harvested, purified, and titered. Recombinant HA and NA proteins derived from A/Michigan/45/2015 (H1N1), A/California/7/2009 (H1N1), A/Hong Kong/4801/2014 (H3N2), A/Switzerland/9715293/2013 (H3N2), A/Texas/50/2012 (H3N2), A/Perth/16/2009 (H3N2), and A/Philippines/2/1982 (H3N2) were obtained from Biodefense and Emerging Infections Research Resources Repository or provided as gifts from the Krammer laboratory at Icahn School of Medicine at Mount Sinai. A/Texas/36/1991 and A/Chile/1/1983 H1N1 viruses were provided by the Hensley laboratory at The University of Pennsylvania. Stabilized trimeric headless HA protein was provided by L. Coughlan at Icahn School of Medicine at Mount Sinai. Recombinant NP proteins derived from A/Puerto Rico/8/1934 (H1N1) and A/Aichi/2/1968 (H3N2) were obtained from Sino Biological.
Monoclonal antibodies
mAbs were generated as previously described (26, 58, 59). Peripheral blood was obtained from each individual 7 days after vaccination or about 7 to 11 days after onset of infection. Lymphocytes were isolated and enriched for B cells using RosetteSep human B cell negative selection enrichment cocktail (STEMCELL Technologies). Enriched B cells were stained for 30 min with anti-human CD3 fluorescein isothiocyanate (FITC) (clone 7D6; Invitrogen; 1:50 dilution), anti-human CD19 Pacific Blue (clone H1B19; BioLegend; 1:100 dilution), anti-human CD27 phycoerythrin (PE) (clone O323; BioLegend; 1:100 dilution), and anti-human CD38 AF647 (clone HIT2; BioLegend; 1:200 dilution) in 500 μl of 1× PBS supplemented with 0.2% bovine serum albumin (BSA) (Sigma-Aldrich). Plasmablasts (CD3−CD19+CD27hiCD38hi) were single cell–sorted into 96-well plates, and immunoglobulin heavy and light chain genes were amplified by reverse transcription PCR. Briefly, the first PCR of a two-step nested PCR was performed on amplified complementary DNA templates from individual plasmablasts. Three separate PCRs were carried out to amplify heavy, kappa, and lambda chain genes using DreamTaq Green PCR 2× MasterMix (Thermo Fisher Scientific). A second nested PCR was then performed using the first PCR product as a template. Second PCR products were then sequenced, and the first PCR product was used to perform a cloning PCR for cloning heavy and light chain genes into human IgG1 expression vectors. The first PCR, second PCR, and cloning PCR primers for heavy, kappa, and lambda chain genes were obtained from IDT Technologies and detailed in table 1 of a previously published protocol from our group (58). Expression vectors for heavy and light chain pairs corresponding to individual mAbs were then cotransfected into HEK293T cells (Thermo Fisher Scientific). Secreted mAbs were purified from the supernatant using protein A agarose beads (Thermo Fisher Scientific).
Enzyme-linked immunosorbent assay
High-protein binding microtiter plates (Costar) were coated with eight hemagglutination units (HAU) of virus in carbonate buffer per well or with recombinant HA, NA, or NP, or headless HA stalk construct at 1 μg/ml in phosphate-buffered saline (PBS) overnight at 4°C. Plates were washed the next morning with PBS + 0.05% Tween 20 and blocked with PBS containing 20% FBS for 1 hour at 37°C. Antibodies were then serially diluted 1:3 starting at 10 μg/ml and incubated for 1 hour at 37°C. Horseradish peroxidase (HRP)–conjugated goat anti-human IgG antibody diluted 1:1000 (Jackson ImmunoResearch) was used to detect binding of mAbs, and plates were subsequently developed with Super AquaBlue ELISA substrate (eBioscience). Absorbance was measured at 405 nm on a microplate spectrophotometer (Bio-Rad). To standardize the assays, control antibodies with known binding characteristics were included on each plate and the plates were developed when the absorbance of the control reached 3.0 OD405 (optical density at 405 nm) units. To determine mAbs that bound the HA head, HAI assays were performed. To determine which mAbs bound the HA stalk domain, competition ELISAs were carried out using the known stalk-binding mAb CR9114 as a competitor mAb (30) or by performing ELISAs using chimeric HA (cH5/1; cH7/3) and a headless HA stalk construct derived from the stalk of A/Brisbane/59/2007 (31). All viruses used in virus-specific ELISAs for the study are shown in Figs. 4 and 5. All experiments were performed in duplicate two to three times.
NA enzyme-linked lectin assay
ELLAs were performed as previously described (60). Briefly, flat-bottom 96-well plates (Thermo Fisher Scientific) were coated with 100 μl of fetuin (Sigma-Aldrich) at 25 μg/ml and incubated at 4°C overnight. mAbs were serially diluted twofold at a starting concentration of 300 μg/ml in Dulbecco’s PBS containing CaCl2 (0.133 g/liter) and MgCl2 (0.1 g/liter) with 0.05% Tween 20 and 1% BSA (DPBSTBSA) and then incubated in fetuin-coated plates with an equal volume of the desired antigen dilution in DPBSTBSA. Plates were sealed and incubated for 20 hours at 37°C. Plates were then washed six times with PBS–0.05% Tween 20, and 100 ml per well of HRP-conjugated peanut agglutinin lectin (Sigma-Aldrich) in DPBSTBSA was added at room temperature (RT) for 2 hours in the dark. Plates were washed six more times and subsequently developed with Super AquaBlue ELISA substrate (eBioscience). Absorbance was read at 405 nm on a microplate spectrophotometer (Bio-Rad). Data were analyzed using Prism software, and the 50% inhibitory concentration (IC50) was determined as the concentration at which 50% of NA activity was inhibited compared to negative control (PBS). All experiments were performed in duplicate two times.
NA-STAR assay
NA-STAR assays were performed with the Resistance Detection Kit according to the manufacturer’s instructions (Applied Biosystems). Briefly, 25-μl test mAbs (starting concentration of 100 μg/ml) were prepared in serial twofold dilutions in NA-STAR assay buffer, mixed with 25 μl of 4× EC50 (median effective concentration) of virus, and incubated at 37°C for 20 min. After adding 10 μl of 1000× diluted NA-STAR substrate, the plates were incubated at RT for 30 min. The reaction was stopped by adding 60 μl of NA-STAR accelerator. Chemiluminescence was determined by using a DTX 880 plate reader (Beckman Coulter). IC50 values were determined using the Prism software. All experiments were performed in duplicate two times.
HAI assay
Viruses were diluted to 8 HAU/50 μl in PBS. Twenty-five microliters was combined with an equal volume of mAb serially diluted 1:3 in PBS in duplicate and subsequently incubated at RT for 45 min. Fifty microliters of 0.5% Turkey red blood cells (Lampire Biological) was added to each well and incubated for 1 hour at RT. Minimum effective concentrations were then calculated on the basis of the final dilution of mAb for which HAI was observed. All experiments were performed in duplicate two to three times.
Microneutralization assay
Microneutralization assay for mAb characterization was carried out as previously described (22, 61). MDCK cells were maintained in DMEM supplemented with 10% FBS, 1% l-glutamine, and 1% penicillin-streptomycin at 37°C with 5% CO2. On the day before the experiment, confluent MDCK cells in a 96-well format were washed twice with sterile PBS and incubated in minimal essential medium (MEM) supplemented with tosyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (1 μg/ml). Serial twofold dilutions (starting concentration of 128 μg/ml) of mAb were mixed with an equal volume of 100 50% tissue culture infectious doses (TCID50) of virus and incubated for 1 hour at 37°C. The mixture was removed, and cells were cultured for 20 hours at 37°C with 1× MEM supplemented with TPCK-treated trypsin (1 μg/ml) and appropriate mAb concentration. Cells were washed twice with PBS, fixed with 80% ice-cold acetone at −20°C for at least 1 hour, washed three times with PBS, blocked for 30 min with 3% BSA in PBS (BSA-PBS), and then treated for 30 min with 2% H2O2. An anti-NP-biotinylated antibody (1:1000) in 3% BSA-PBS was incubated for 1 hour at RT. The plates were developed with Super AquaBlue ELISA substrate at 405 nm. The signal from uninfected wells was averaged to represent 100% inhibition. The signal from infected wells without mAb was averaged to represent 0% inhibition. Duplication wells were used to calculate the mean and SD of neutralization, and IC50 was determined by a sigmoidal dose-response curve. The inhibition ratio (%) was calculated as below: [(OD positive control – OD sample)/(OD positive control – OD negative control)] * 100. The IC50 was determined using Prism software (GraphPad). All experiments were performed in duplicate two to three times.
Plaque reduction assay
Plaque assays were performed using the MDCK London cell line, as previously described (36), with the exception that cells were incubated with the agar overlay for 48 hours. Plaques were counted, and the final mAb concentration that reduced the number of plaques to 50% was determined using GraphPad Prism software. The assay was performed in duplicate two to three times.
In vivo challenge experiments
mAbs were passively transferred into 6- to 8-week-old female BALB/c mice by intraperitoneal injection of mAb cocktail (0.2, 1, and 5 mg/kg). Negative control mice received 5 mg/kg of the anthrax-specific mAb 003-15D03 as an isotype control. Two hours after mAb injection, mice were anesthetized with isoflurane and intranasally challenged with 10 LD50 of mouse-adapted A/Netherlands/602/2009 H1N1 virus diluted in 20 μl of sterile 1× PBS to test protective ability of H1N1-binding mAbs or A/Philippines/2/1982 H3N2 virus to test protective ability of H3N2-binding mAbs. As a readout, survival and weight loss were monitored twice a day for 2 weeks.
Clustering of mAbs
mAbs were clustered on the basis of their affinity for a panel of test viruses using a hierarchical clustering approach. First, each mAb was defined as a vector whose elements were the binding affinity (ELISA KD) of that mAb against each of the H1N1 and H3N2 viruses in our test panel. Then, the Euclidean distance between each mAb was calculated. The unweighted pair group method with arithmetic mean (UPGMA) was applied to the distance matrix to obtain a hierarchical clustering of all mAbs, which was visualized as a dendrogram (Figs. 4, A and C, and 5, A and C). Clustering was performed separately for groups of mAbs belonging to the depicted epitope reactivity categories.
Statistical analysis
All statistical analyses were performed using Prism software (GraphPad version 8.0). Sample sizes (n) for the number of mAbs are indicated directly in the figures or in the corresponding figure legends, and sample sizes (n) for animals, number of biological replicates for experiments, and specific tests for statistical significance used are indicated in the corresponding figure legends. P values less than or equal to 0.05 were considered significant: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Supplementary Material
Fig. S1. Reactivity of influenza virus infection–induced antibodies from individual participants and conserved epitope reactivity.
Fig. S2. Neutralization potency of influenza virus infection– and vaccination-induced antibodies stratified by subtype reactivity.
Fig. S3. Antibody cross-reactivity and affinity toward past strains by individual participants.
Fig. S4. Prophylactic protection of influenza virus infection– and vaccination-induced antibodies at 0.2 and 5 mg/kg.
Table S1. Participant characteristics for influenza virus infection cohorts.
Table S2. Participant characteristics for influenza virus vaccination cohorts.
Table S3. Reference childhood strains used to characterize infection-induced mAbs.
Table S4. mAbs used for the infection- and vaccination-induced antibody cocktails.
Table S5. mAbs used in the antigen comparison cocktails.
Raw data.
Acknowledgments:
We thank S. E. Hensley and S. Zost for providing influenza viruses and recombinant HA proteins. We are thankful to all individuals who participated in this study. We thank K. Gostic and M. Knight for providing thoughtful discussion related to the manuscript. Last, we thank S. F. Andrews for the initial sample collection for the 2010–2011 trivalent influenza vaccine cohort.
Funding:
This project was funded, in part, by the National Institute of Allergy and Infectious Disease; NIH grant numbers U19AI082724 (P.C.W.), U19AI109946 (P.C.W.), and U19AI057266 (P.C.W.); NIAID Centers of Excellence for Influenza Research and Surveillance (CEIRS) grant numbers HHSN272201400005C (P.C.W.) and HHSN272201400008C (F.K. and L.C.); and University of Chicago Committee on Immunology T32-AI007090 (H.L.D.) and T32-HL007605 (J.J.G.). S.C. and P.A. were supported by NIH Young Investigator Award DP2AI117921 (S.C.) and NIH F32AI145177 (P.A.).
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data associated with this study are present in the paper or the Supplementary Materials. All antibody sequences have been deposited to GenBank and are available under the following accession numbers: MW067375-MW067608 and MW079531-MW079816.
REFERENCES AND NOTES
- 1.Iuliano AD, Roguski KM, Chang HH, Muscatello DJ, Palekar R, Tempia S, Cohen C, Gran JM, Schanzer D, Cowling BJ, Wu P, Kyncl J, Ang LW, Park M, Redlberger-Fritz M, Yu H, Espenhain L, Krishnan A, Emukule G, van Asten L, Pereira da Silva S, Aungkulanon S, Buchholz U, Widdowson M-A, Bresee JS; Global Seasonal Influenza-associated Mortality Collaborator Network, Estimates of global seasonal influenza-associated respiratory mortality: A modelling study. Lancet 391, 1285–1300 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dawood FS, Chung JR, Kim SS, Zimmerman RK, Nowalk MP, Jackson ML, Jackson LA, Monto AS, Martin ET, Belongia EA, McLean HQ, Gaglani M, Dunnigan K, Foust A, Sessions W, DaSilva J, Le S, Stark T, Kondor RJ, Barnes JR, Wentworth DE, Brammer L, Fry AM, Patel MM, Flannery B, Interim estimates of 2019-20 seasonal influenza vaccine effectiveness—United States, February 2020. MMWR Morb. Mortal. Wkly Rep 69, 177–182 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Webster RG, Laver WG, Air GM, Schild GC, Molecular mechanisms of variation in influenza viruses. Nature 296, 115–121 (1982). [DOI] [PubMed] [Google Scholar]
- 4.Krammer F, Palese P, Advances in the development of influenza virus vaccines. Nat. Rev. Drug Discov 14, 167–182 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Hobson D, Curry RL, Beare AS, Ward-Gardner A, The role of serum haemagglutination-inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. J. Hyg. (Lond.) 70, 767–777 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohmit SE, Petrie JG, Cross RT, Johnson E, Monto AS, Influenza hemagglutination-inhibition antibody titer as a correlate of vaccine-induced protection. J. Infect. Dis 204, 1879–1885 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Doud MB, Lee JM, Bloom JD, How single mutations affect viral escape from broad and narrow antibodies to H1 influenza hemagglutinin. Nat. Commun 9, 1386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heaton NS, Sachs D, Chen C-J, Hai R, Palese P, Genome-wide mutagenesis of influenza virus reveals unique plasticity of the hemagglutinin and NS1 proteins. Proc. Natl. Acad. Sci. U.S.A 110, 20248–20253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doud MB, Bloom JD, Accurate measurement of the effects of all amino-acid mutations on influenza hemagglutinin. Viruses 8, 155 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirkpatrick E, Qiu X, Wilson PC, Bahl J, Krammer F, The influenza virus hemagglutinin head evolves faster than the stalk domain. Sci. Rep 8, 10432 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gostic KM, Ambrose M, Worobey M, Lloyd-Smith JO, Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science 354, 722–726 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andrews SF, Huang Y, Kaur K, Popova LI, Ho IY, Pauli NT, Henry Dunand CJ, Taylor WM, Lim S, Huang M, Qu X, Lee J-H, Salgado-Ferrer M, Krammer F, Palese P, Wrammert J, Ahmed R, Wilson PC, Immune history profoundly affects broadly protective B cell responses to influenza. Sci. Transl. Med 7, 316ra192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fonville JM, Wilks SH, James SL, Fox A, Ventresca M, Aban M, Xue L, Jones TC, Le NMH, Pham QT, Tran ND, Wong Y, Mosterin A, Katzelnick LC, Labonte D, Le TT, van der Net G, Skepner E, Russell CA, Kaplan TD, Rimmelzwaan GF, Masurel N, de Jong JC, Palache A, Beyer WEP, Le QM, Nguyen TH, Wertheim HFL, Hurt AC, Osterhaus ADME, Barr IG, Fouchier RAM, Horby PW, Smith DJ, Antibody landscapes after influenza virus infection or vaccination. Science 346, 996–1000 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guthmiller JJ, Wilson PC, Harnessing immune history to combat influenza viruses. Curr. Opin. Immunol 53, 187–195 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Henry C, Palm A-KE, Krammer F, Wilson PC, From original antigenic sin to the universal influenza virus vaccine. Trends Immunol. 39, 70–79 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davenport FM, Hennessy AV, Francis T Jr., Epidemiologic and immunologic significance of age distribution of antibody to antigenic variants of influenza virus. J. Exp. Med 98, 641–656 (1953). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skountzou I, Koutsonanos DG, Kim JH, Powers R, Satyabhama L, Masseoud F, Weldon WC, Del Pilar Martin M, Mittler RS, Compans R, Jacob J, Immunity to pre-1950 H1N1 influenza viruses confers cross-protection against the pandemic swine-origin 2009 A (H1N1) influenza virus. J. Immunol 185, 1642–1649 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linderman SL, Chambers BS, Zost SJ, Parkhouse K, Li Y, Herrmann C, Ellebedy AH, Carter DM, Andrews SF, Zheng N-Y, Huang M, Huang Y, Strauss D, Shaz BH, Hodinka RL, Reyes-Terán G, Ross TM, Wilson PC, Ahmed R, Bloom JD, Hensley SE, Potential antigenic explanation for atypical H1N1 infections among middle-aged adults during the 2013–2014 influenza season. Proc. Natl. Acad. Sci. U.S.A 111, 15798–15803 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linderman SL, Hensley SE, Antibodies with 'original antigenic sin' properties are valuable components of secondary immune responses to influenza viruses. PLOS Pathog. 12, e1005806 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arevalo P, McLean HQ, Belongia EA, Cobey S, Earliest infections predict the age distribution of seasonal influenza A cases. eLife 9, e50060 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gostic KM, Bridge R, Brady S, Viboud C, Worobey M, Lloyd-Smith JO, Childhood immune imprinting to influenza A shapes birth year-specific risk during seasonal H1N1 and H3N2 epidemics. PLOS Pathog. 15, e1008109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y-Q, Wohlbold TJ, Zheng N-Y, Huang M, Huang Y, Neu KE, Lee J, Wan H, Rojas KT, Kirkpatrick E, Henry C, Palm A-KE, Stamper CT, Lan LY-L, Topham DJ, Treanor J, Wrammert J, Ahmed R, Eichelberger MC, Georgiou G, Krammer F, Wilson PC, Influenza infection in humans induces broadly cross-reactive and protective neuraminidase-reactive antibodies. Cell 173, 417–429.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henry C, Palm A-KE, Utset HA, Huang M, Ho IY, Zheng N-Y, Fitzgerald T, Neu KE, Chen Y-Q, Krammer F, Treanor JJ, Sant AJ, Topham DJ, Wilson PC, Monoclonal antibody responses after recombinant HA vaccine versus subunit inactivated influenza virus vaccine: A comparative study. J. Virol 93, e01150–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He X-S, Holmes TH, Sanyal M, Albrecht RA, García-Sastre A, Dekker CL, Davis MM, Greenberg HB, Distinct patterns of B-cell activation and priming by natural influenza virus infection versus inactivated influenza vaccination. J. Infect. Dis 211, 1051–1059 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Margine I, Hai R, Albrecht RA, Obermoser G, Harrod AC, Banchereau J, Palucka K, Garcia-Sastre A, Palese P, Treanor JJ, Krammer F, H3N2 influenza virus infection induces broadly reactive hemagglutinin stalk antibodies in humans and mice. J. Virol 87, 4728–4737 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng N-Y, Mays I, Garman L, Helms C, James J, Air GM, Capra JD, Ahmed R, Wilson PC, Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature 453, 667–671 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henry C, Zheng N-Y, Huang M, Cabanov A, Rojas KT, Kaur K, Andrews SF, Palm A-KE, Chen Y-Q, Li Y, Hoskova K, Utset HA, Vieira MC, Wrammert J, Ahmed R, Holden-Wiltse J, Topham DJ, Treanor JJ, Ertl HC, Schmader KE, Cobey S, Krammer F, Hensley SE, Greenberg H, He X-S, Wilson PC, Influenza virus vaccination elicits poorly adapted B cell responses in elderly individuals. Cell Host Microbe 25, 357–366.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nachbagauer R, Choi A, Izikson R, Cox MM, Palese P, Krammer F, Age dependence and isotype specificity of influenza virus hemagglutinin stalk-reactive antibodies in humans. MBio 7, e01996–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajendran M, Nachbagauer R, Ermler ME, Bunduc P, Amanat F, Izikson R, Cox M, Palese P, Eichelberger M, Krammer F, Analysis of anti-influenza virus neuraminidase antibodies in children, adults, and the elderly by ELISA and enzyme inhibition: Evidence for original antigenic sin. MBio 8, e02281–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, van der Vlugt R, Lamrani M, Korse HJWM, Geelen E, Sahin Ö, Sieuwerts M, Brakenhoff JPJ, Vogels R, Li OTW, Poon LLM, Peiris M, Koudstaal W, Ward AB, Wilson IA, Goudsmit J, Friesen RHE, Highly conserved protective epitopes on influenza B viruses. Science 337, 1343–1348 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Impagliazzo A, Milder F, Kuipers H, Wagner MV, Zhu X, Hoffman RMB, van Meersbergen R, Huizingh J, Wanningen P, Verspuij J, de Man M, Ding Z, Apetri A, Kükrer B, Sneekes-Vriese E, Tomkiewicz D, Laursen NS, Lee PS, Zakrzewska A, Dekking L, Tolboom J, Tettero L, van Meerten S, Yu W, Koudstaal W, Goudsmit J, Ward AB, Meijberg W, Wilson IA, Radošević K, A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 349, 1301–1306 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Ferguson NM, Galvani AP, Bush RM, Ecological and immunological determinants of influenza evolution. Nature 422, 428–433 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Abreu RB, Kirchenbaum GA, Clutter EF, Sautto GA, Ross TM, Preexisting subtype immunodominance shapes memory B cell recall response to influenza vaccination. JCI Insight 5, e132155 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hensley SE, Das SR, Bailey AL, Schmidt LM, Hickman HD, Jayaraman A, Viswanathan K, Raman R, Sasisekharan R, Bennink JR, Yewdell JW, Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science 326, 734–736 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerdil C, The annual production cycle for influenza vaccine. Vaccine 21, 1776–1779 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Wrammert J, Koutsonanos D, Li G-M, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, Mehta A, Razavi B, Del Rio C, Zheng N-Y, Lee J-H, Huang M, Ali Z, Kaur K, Andrews S, Amara RR, Wang Y, Das SR, O'Donnell CD, Yewdell JW, Subbarao K, Marasco WA, Mulligan MJ, Compans R, Ahmed R, Wilson PC, Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J. Exp. Med 208, 181–193 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacobsen H, Rajendran M, Choi A, Sjursen H, Brokstad KA, Cox RJ, Palese P, Krammer F, Nachbagauer R, Influenza virus hemagglutinin stalk-specific antibodies in human serum are a surrogate marker for in vivo protection in a serum transfer mouse challenge model. MBio 8, e01463–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ekiert DC, Bhabha G, Elsliger M-A, Friesen RHE, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA, Antibody recognition of a highly conserved influenza virus epitope. Science 324, 246–251 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ekiert DC, Friesen RHE, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJWM, Brandenburg B, Vogels R, Brakenhoff JPJ, Kompier R, Koldijk MH, Cornelissen LAHM, Poon LLM, Peiris M, Koudstaal W, Wilson IA, Goudsmit J, A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333, 843–850 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wohlbold TJ, Chromikova V, Tan GS, Meade P, Amanat F, Comella P, Hirsh A, Krammer F, Hemagglutinin stalk- and neuraminidase-specific monoclonal antibodies protect against lethal H10N8 influenza virus infection in mice. J. Virol 90, 851–861 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stadlbauer D, Zhu X, McMahon M, Turner JS, Wohlbold TJ, Schmitz AJ, Strohmeier S, Yu W, Nachbagauer R, Mudd PA, Wilson IA, Ellebedy AH, Krammer F, Broadly protective human antibodies that target the active site of influenza virus neuraminidase. Science 366, 499–504 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maier HE, Nachbagauer R, Kuan G, Ng S, Lopez R, Sanchez N, Stadlbauer D, Gresh L, Schiller A, Rajabhathor A, Ojeda S, Guglia AF, Amanat F, Balmaseda A, Krammer F, Gordon A, Pre-existing anti-neuraminidase antibodies are associated with shortened duration of influenza A(H1N1)pdm virus shedding and illness in naturally infected adults. Clin. Infect. Dis 70, 2290–2297 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng S, Nachbagauer R, Balmaseda A, Stadlbauer D, Ojeda S, Patel M, Rajabhathor A, Lopez R, Guglia AF, Sanchez N, Amanat F, Gresh L, Kuan G, Krammer F, Gordon A, Novel correlates of protection against pandemic H1N1 influenza A virus infection. Nat. Med 25, 962–967 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Couch RB, Atmar RL, Franco LM, Quarles JM, Wells J, Arden N, Nino D, Belmont JW, Antibody correlates and predictors of immunity to naturally occurring influenza in humans and the importance of antibody to the neuraminidase. J. Infect. Dis 207, 974–981 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monto AS, Petrie JG, Cross RT, Johnson E, Liu M, Zhong W, Levine M, Katz JM, Ohmit SE, Antibody to influenza virus neuraminidase: An independent correlate of protection. J. Infect. Dis 212, 1191–1199 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Memoli MJ, Shaw PA, Han A, Czajkowski L, Reed S, Athota R, Bristol T, Fargis S, Risos K, Powers JH, Davey RT Jr., Taubenberger JK, Evaluation of antihemagglutinin and antineuraminidase antibodies as correlates of protection in an influenza A/H1N1 virus healthy human challenge model. MBio 7, e00417–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujimoto Y, Tomioka Y, Takakuwa H, Uechi G-I, Yabuta T, Ozaki K, Suyama H, Yamamoto S, Morimatsu M, Mai LQ, Yamashiro T, Ito T, Otsuki K, Ono E, Cross-protective potential of anti-nucleoprotein human monoclonal antibodies against lethal influenza A virus infection. J. Gen. Virol 97, 2104–2116 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Richards KA, Treanor JJ, Nayak JL, Sant AJ, Overarching immunodominance patterns and substantial diversity in specificity and functionality in the circulating human influenza A and B virus-specific CD4+ T-cell repertoire. J. Infect. Dis 218, 1169–1174 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nachbagauer R, Choi A, Hirsh A, Margine I, Iida S, Barrera A, Ferres M, Albrecht RA, García-Sastre A, Bouvier NM, Ito K, Medina RA, Palese P, Krammer F, Defining the antibody cross-reactome directed against the influenza virus surface glycoproteins. Nat. Immunol 18, 464–473 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tesini BL, Kanagaiah P, Wang J, Hahn M, Halliley JL, Chaves FA, Nguyen PQT, Nogales A, DeDiego ML, Anderson CS, Ellebedy AH, Strohmeier S, Krammer F, Yang H, Bandyopadhyay S, Ahmed R, Treanor JJ, Martinez-Sobrido L, Golding H, Khurana S, Zand MS, Topham DJ, Sangster MY, Broad hemagglutinin-specific memory B cell expansion by seasonal influenza virus infection reflects early-life imprinting and adaptation to the infecting virus. J. Virol 93, e00169–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ranjeva S, Subramanian R, Fang VJ, Leung GM, Ip DKM, Perera RAPM, Peiris JSM, Cowling BJ, Cobey S, Age-specific differences in the dynamics of protective immunity to influenza. Nat. Commun 10, 1660 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee J, Paparoditis P, Horton AP, Frühwirth A, McDaniel JR, Jung J, Boutz DR, Hussein DA, Tanno Y, Pappas L, Ippolito GC, Corti D, Lanzavecchia A, Georgiou G, Persistent antibody clonotypes dominate the serum response to influenza over multiple years and repeated vaccinations. Cell Host Microbe 25, 367–376.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murphy BR, Kasel JA, Chanock RM, Association of serum anti-neuraminidase antibody with resistance to influenza in man. N. Engl. J. Med 286, 1329–1332 (1972). [DOI] [PubMed] [Google Scholar]
- 54.He W, Mullarkey CE, Duty JA, Moran TM, Palese P, Miller MS, Broadly neutralizing anti-influenza virus antibodies: Enhancement of neutralizing potency in polyclonal mixtures and IgA backbones. J. Virol 89, 3610–3618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krammer F, Pica N, Hai R, Margine I, Palese P, Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J. Virol 87, 6542–6550 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wohlbold TJ, Nachbagauer R, Xu H, Tan GS, Hirsh A, Brokstad KA, Cox RJ, Palese P, Krammer F, Vaccination with adjuvanted recombinant neuraminidase induces broad heterologous, but not heterosubtypic, cross-protection against influenza virus infection in mice. MBio 6, e02556 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bernstein DI, Guptill J, Naficy A, Nachbagauer R, Berlanda-Scorza F, Feser J, Wilson PC, Solórzano A, Van der Wielen M, Walter EB, Albrecht RA, Buschle KN, Chen Y-Q, Claeys C, Dickey M, Dugan HL, Ermler ME, Freeman D, Gao M, Gast C, Guthmiller JJ, Hai R, Henry C, Lan LY-L, McNeal M, Palm A-KE, Shaw DG, Stamper CT, Sun W, Sutton V, Tepora ME, Wahid R, Wenzel H, Wohlbold TJ, Innis BL, García-Sastre A, Palese P, Krammer F, Immunogenicity of chimeric haemagglutinin-based, universal influenza virus vaccine candidates: Interim results of a randomised, placebo-controlled, phase 1 clinical trial. Lancet Infect. Dis 20, 80–91 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guthmiller JJ, Dugan HL, Neu KE, Lan LY-L, Wilson PC, An efficient method to generate monoclonal antibodies from human B cells. Methods Mol. Biol 1904, 109–145 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Smith K, Garman L, Wrammert J, Zheng N-Y, Capra JD, Ahmed R, Wilson PC, Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat. Protoc 4, 372–384 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Westgeest KB, Bestebroer TM, Spronken MIJ, Gao J, Couzens L, Osterhaus ADME, Eichelberger M, Fouchier RAM, de Graaf M, Optimization of an enzyme-linked lectin assay suitable for rapid antigenic characterization of the neuraminidase of human influenza A(H3N2) viruses. J. Virol. Methods 217, 55–63 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Henry Dunand CJ, Leon PE, Kaur K, Tan GS, Zheng N-Y, Andrews S, Huang M, Qu X, Huang Y, Salgado-Ferrer M, Ho IY, Taylor W, Hai R, Wrammert J, Ahmed R, García-Sastre A, Palese P, Krammer F, Wilson PC, Preexisting human antibodies neutralize recently emerged H7N9 influenza strains. J. Clin. Invest 125, 1255–1268 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Reactivity of influenza virus infection–induced antibodies from individual participants and conserved epitope reactivity.
Fig. S2. Neutralization potency of influenza virus infection– and vaccination-induced antibodies stratified by subtype reactivity.
Fig. S3. Antibody cross-reactivity and affinity toward past strains by individual participants.
Fig. S4. Prophylactic protection of influenza virus infection– and vaccination-induced antibodies at 0.2 and 5 mg/kg.
Table S1. Participant characteristics for influenza virus infection cohorts.
Table S2. Participant characteristics for influenza virus vaccination cohorts.
Table S3. Reference childhood strains used to characterize infection-induced mAbs.
Table S4. mAbs used for the infection- and vaccination-induced antibody cocktails.
Table S5. mAbs used in the antigen comparison cocktails.
Raw data.