Abstract
Elevated CO2 stimulates plant growth and affects quantity and composition of root exudates, followed by response of its microbiome. Three scenarios representing nitrate fertilization regimes: limited (30 ppm), moderate (70 ppm) and excess nitrate (100 ppm) were compared under ambient and elevated CO2 (eCO2, 850 ppm) to elucidate their combined effects on root-surface-associated bacterial community abundance, structure and function. Wheat root-surface-associated microbiome structure and function, as well as soil and plant properties, were highly influenced by interactions between CO2 and nitrate levels. Relative abundance of total bacteria per plant increased at eCO2 under excess nitrate. Elevated CO2 significantly influenced the abundance of genes encoding enzymes, transporters and secretion systems. Proteobacteria, the largest taxonomic group in wheat roots (~ 75%), is the most influenced group by eCO2 under all nitrate levels. Rhizobiales, Burkholderiales and Pseudomonadales are responsible for most of these functional changes. A correlation was observed among the five gene-groups whose abundance was significantly changed (secretion systems, particularly type VI secretion system, biofilm formation, pyruvate, fructose and mannose metabolism). These changes in bacterial abundance and gene functions may be the result of alteration in root exudation at eCO2, leading to changes in bacteria colonization patterns and influencing their fitness and proliferation.
Subject terms: Microbiome, Microbial ecology, Metagenomics, Microbial ecology
Introduction
The atmospheric concentration of carbon dioxide (CO2) has been increasing since the industrial revolution due to fossil-fuel burning. With the rapid increase in industrialization, CO2 became a major greenhouse gas, and it is expected to increase from the current ambient level (aCO2) of ca. 400 ppm to 800–1000 ppm, or more, at the end of 21st century [1]. In addition to its many negative effects, elevated CO2 (eCO2) is known to improve the growth of agriculturally important plants [2], and promote production of root exudates [3], which can result in increased soil microbial biomass and subsequently, increased soil microbial nitrogen demand [4, 5]. The outcome of this may be enhanced competition in the root zone between soil microorganisms and plants for available nitrogen [6], leading to progressive nitrogen limitation for the plants [7, 8]. Indeed, plant nitrogen concentration generally decreases under eCO2 [9, 10], especially in C3 plants such as wheat [11]. Another possible explanation for this phenomenon is that eCO2 inhibits nitrate-N assimilation into proteins in the shoots of C3 plants [11–13]. Thus, with the predicted increase in atmospheric CO2 level, an increase in the supply of nitrogen will be needed to sustain wheat growth and satisfy soil microbial nitrogen demand [14].
Elevated CO2 affects the soil microbial community indirectly via plant metabolism and root secretions, particularly in C3 plants [15], an effect that is locally restricted to the apical root zone [6]. Not much is known about the effect of eCO2 on the structure, and especially function of the rhizosphere and root-surface-associated microbial communities. In fact, the studies published so far have focused on the structure of rhizosphere microbial communities and not on root-surface-associated communities [16–18], that are likely to be even more affected by eCO2 and increased nitrate supply [19]. Moreover, an increase in root exudates by eCO2 can stimulate microbial activity in the root system and thus increase the consumption of oxygen [20], creating anoxic niches along the root profile that can promote denitrification. Denitrification is essentially an anaerobic respiration pathway in which the oxidized form of nitrogen is used as a terminal electron acceptor in the absence of oxygen. It has been previously shown that with the addition of nitrogen supply, the activity and abundance of denitrifiers having nirS, nirK and nosZ genes are significantly affected in the wheat rhizosphere [21]. Moreover, with the addition of nitrogen, there are alterations in the quantity and composition of wheat exudates that may have a significant impact on root microbial community structure [22]. It was previously shown that the relative abundance of Proteobacteria, Actinobacteria and Bacteroidetes, which often dominate the roots [23, 24], is highly influenced by nitrogen supply [22] and CO2 level [25].
In this study, we have focused on the root-surface-associated bacterial community, which is the one in intimate interaction with the roots. It is hypothesized that they would be the first in responding to changes in the root system. Here we examined the combined effect of eCO2 and nitrate levels on the structure and function of the root-surface-associated (i.e., bacteria attached to root surface) bacterial community structure and functions based on metagenome extracted from the background of host-plant (wheat) genomic data. Three possible nitrate fertilization scenarios to sustain future wheat growth were studied: (1) limited (e.g., 30 ppm nitrate), moderate (e.g., 70 ppm nitrate) and excess nitrate (e.g., 100 ppm nitrate). We anticipated that changes in function of the root-surface-associated bacterial community over a period of weeks might react to and indicate the conditions developing in the root system, as a result of changing levels of atmospheric CO2 and nitrate supply. Hence, by observing changes in gene functions of the root-surface-associated microbiome, we sought to reveal possible conditions to which the roots will be exposed in the future, information that might enable accommodation of wheat growth to future environmental conditions.
Materials and methods
Greenhouse experiments and sampling
Wheat (Triticum turgidum cv. Negev) was cultivated in sandy loam soil (19% clay, 6% silt, 75% sand) classified as Calcic Haploxerept. The soil was obtained from intensive agriculture field located in Eshkol region, Israel (31.248,949, 34.379,872). Potatoes, wheat and peanuts were previously grown in this field. Initial soil parameters were: pH 8.78 ± 0.04, electrical conductivity 99 ± 1 (µS/m), NO3-N 0.22 ± 0.02 (mg/kg), NH4 0.30 ± 0.01 (mg/kg), P-PO4 0.09 ± 0.01(mg/kg), total soluble organic carbon 4.0 ± 0.04 (mg/kg) and total soluble nitrogen 0.70 ± 0.02 (mg/kg).
The plants were grown for 6 weeks (from December 2016 to February 2017) as described previously [25]. Briefly, 750 g of soil was distributed in a 700-mL plastic pot, with four seeds per pot. Those pots were able to sustain up to four wheat plants for six weeks under the experimental conditions. The wheat was grown in a greenhouse with two closed-system chambers at day/night temperatures of 25 °C/18 °C ± 1 °C, and with an automatically adjusted CO2-supply system (Emproco Ltd., Ashkelon, Israel). The photoperiod was 9 h and the daily light integral was 12.5 MJ/day. Wheat plants were grown in a sequence of three independent experimental cycles of 6 weeks each (five pots per treatment per cycle), with a 1-week shift between cycles. Plants were grown under either ambient (400 ppm) or elevated (850 ppm) atmospheric CO2 levels. Nutrient solution was prepared with 90% nitrogen supplied as nitrate and 10% supplied as ammonium using KNO3 and NH4NO3 to provide final concentrations of 30, 70 and 100 ppm nitrate [26]. Other macronutrients were supplied in each treatment at the following rate: P-15 ppm, K-150 ppm, Mg-24 ppm, Ca-120 ppm and S-40 ppm provide by NH4NO3, KNO3, CaCl2, KCl, MgCl2 and KH2PO4 salts. 40 ppm S and Ca were present in the tap water. Micronutrients were supplied at a rate of 1.3 ppm Fe, 0.7 ppm Mn, 0.3 ppm Zn, 0.05 ppm Cu, and 0.0375 ppm Mo using Korotin (Haifa Chemicals, Israel), a commercial micronutrient mix. Each pot was irrigated with 50 mL of the nutrient solution four times a week. The total amount of nitrogen in the 30 ppm nitrate treatment was 36 mg/pot (equivalent of ca. 73 kg N/ha), 70 ppm nitrate treatment was 84 mg/pot (equivalent of ca. 170 kg N/ha) and in the 100 ppm treatment, 120 mg/pot (equivalent of ca. 250 kg N/ha).
Soil and plant analyses
At the end of the 6th week of growth, 15 pots (5 pots per cycle) from each treatment were sampled for soil, shoots and roots, and the following parameters were measured: soil nitrate and ammonia content, soil EC and soil pH, shoot and root dry biomass, nitrogen concentration and content in shoot and roots. Soil properties and relevant methods were as described previously [25]. Briefly, soil EC and pH were determined in a solution of 1:5 air dry sieved soil:distilled water (w/v). Nitrate and ammonium concentrations were determined using an autoanalyzer (Lachat Instruments, Milwaukee, WI or Gallery Plus, Thermo Fisher Scientific, Waltham, MA, USA). Sampled shoots and roots were dried at 60 °C for 48 h, ground and weighed to obtain dry biomass. Total nitrogen concentration was determined using an autoanalyzer (Lachat Instruments or Gallery Plus) following digestion with sulfuric acid and peroxide [27].
Root DNA extraction for sequencing and qPCR
At the end of the 6th week of wheat growth, pots were randomly selected for DNA extraction. To obtain the root-surface-associated microbiome, wheat roots were collected in triplicate from each of the three cycles and were vortexed three time with 85% saline solution, until no visible soil particles were attached to the roots. Total DNA was extracted from 0.4 g of complete root system, using the Exgene Soil DNA mini isolation kit (GeneAll, Seoul, Korea) according to the manufacturer’s instructions.
Generation of qPCR plasmid standards
Plasmids containing the 16S rRNA gene were generated as described previously [28, 29]. Each PCR amplification product was ligated into pGEM-T Easy Vector (Promega, Madison, WI, USA) and plasmids were transformed into BioSuper Escherichia coli DH5α competent cells (Bio-Lab, Jerusalem, Israel). Circular plasmid DNAs were used as the standards to create calibration curves at 10-fold dilutions for gene quantification by real-time qPCR.
Assessment of gene copy numbers by qPCR
Copy numbers of the total bacterial community (16S rRNA gene) and translation elongation factor 1 (TEF, a plant housekeeping gene) were assessed using selected primers (Table S1) in roots of 6-week-old wheat plants with the StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Triplicates from whole genomic DNA were diluted to 6 ng/µL and 1 µL was used in a 20-µL final reaction volume together with 50 µM forward and reverse primers and 10 µL 1X FAST MasterMix (Thermo Fisher Scientific). Three biological and three technical replicates were analyzed for each root DNA sample. Reaction efficiency was monitored in each run by means of an internal standard curve (constructed plasmids) using duplicates of 10-fold dilutions of standards ranging from 108–102 copies per reaction. Efficiency was 89–98% for all target genes and runs, and R2 values were greater than 0.99. Copy numbers of the target genes were calculated based on the relative calibration curve of the plasmid copy numbers. All data analyses were conducted using StepOne software v2.3 (Applied Biosystems).
Shotgun sequencing
Root DNA was extracted from each of the biological triplicates, in each of the three cycles. For sequencing, the DNA of the triplicates was combined, resulting in three biological replicates per treatment (one from each batch) and 18 samples altogether. Shotgun metagenome libraries were prepared using the Celero DNA-Seq library preparation kit (NuGen, Takara Bio, USA) with enzymatic shearing, according to the manufacturer’s instructions. All libraries were then pooled in equal volumes and size selection (350–400 bp fragments) was performed using a Blue PippinPrep instrument (Sage Scientific). The libraries were then sequenced using an Illumina MiniSeq instrument employing a mid-output kit. Based on the number of reads per sample, the samples were repooled with varying volumes, and size selection was performed again using the same size range. The final size-selected pool was sequenced on an Illumina NovaSeq instrument with an S4 flow cell, employing 2 × 150 base reads. Library preparation and pooling were performed at the University of Illinois at Chicago Sequencing Core (UICSQC), and sequencing was performed by Novogene Corporation (Chula Vista, CA, USA).
In total, we obtained 310 Gb of information, with 30–44 million sequences per root sample. These sequence data were submitted to the Sequence Read Archive (SRA) of the NCBI databases under accession numbers SUB6631533 and SUB8385777, BioProject: PRJNA592741.
All reads were subjected to quality control using FastQC v0.11.3 [30] and barcode trimming using Trimmomatics v0.32 [31]. Reads were mapped to the whole wheat metagenome using Bowtie2 v2.3.5.1 [32], and mapped reads were filtered out from each sample. Then, short Illumina reads from triplicates of each nitrate treatment (30, 70 and 100 ppm) were assembled using SPADES v3.13.0 [33] into longer contigs, to create three wheat root microbiome catalogs for each treatment separately. The 30 ppm nitrate catalog had 677,271 contigs with N50 of 964 bp, 70 ppm nitrate catalog had 644,394 contigs with N50 of 971 bp, and the 100 ppm catalog had 677,271 contigs with N50 of 964 bp. Those three catalogs were combined and Prodigal v2.6.2 [34] was used for protein-coding gene prediction. To create a non-redundant set of genes, we used CD-HIT-EST software v4.8.1 [35] with a similarity threshold of 95%. Those genes were used as the root gene catalog, which included 35 million partial genes. This gene catalog was searched against the non-redundant NCBI protein database using DIAMOND sensitive algorithm v0.9.24.125 [36] to assign taxonomic and functional annotations. Results were then uploaded to MEGAN Ultimate edition software v6.15.2 [37]. The LCA (lowest common ancestor) algorithm was applied (parameters used with minimum bit-score of 70, minimum support of 5% and 30% top threshold) to compute the assignment of genes to specific taxa. For functional annotation, the Kyoto Encyclopedia of Genes and Genomes (KEGG) database [38] was used. Following annotation, to generate taxonomic and functional count tables, each library was mapped to the gene catalog with Trinity mapping software v2.8.4 [39], with Bowtie2-modified parameters (--no-unal --gbar 99999999 -k 250 --dpad 0 --mp 1,1 --np 1 --score-min L,0,−0.9 -L 20 -i S,1,0.50).
Data analyses
Significance of interactions between CO2 and nitrate levels on soil and plant parameters was calculated using two-way ANOVA the least-squares method, in JMP 14 Pro software (SAS Institute Inc., Cary, NC, USA). Differences between soil and plant parameters as influenced by interactions between CO2 and nitrate levels was calculated using Student’s t test in JMP 14 Pro software and statistical significance was set at P < 0.05 The abundance of genes measured using qPCR, was calculated using Student’s t test and statistical significance was set at P < 0.05. All sequencing data analyses were performed using R statistical software. An nMDS (non-metric multidimensional scaling) ordination plot was constructed using R package VEGAN v.2.5-5 [40]. The data matrix was transformed using normalized count transformation, and the batch effect of three experimental cycles was removed using the DESeq2 v.1.22.2 package [41] limma::removeBatchEffect function. For community structure, ordination was generated using the Bray–Curtis dissimilarity matrix, and for community function of KEGG orthologous (KO) groups, ordination was generated using the Euclidean dissimilarity matrix. Permutational multivariate analysis was used to compute the variance between bacterial community structure or function and experimental parameters (CO2, nitrate and cycle), using the Adonis function in the Vegan R package [42]. For comparison of taxonomic changes and functional traits, differential abundance was estimated using DESeq2 [41] and was considered significant when the difference in abundance between genes had a FDR-adjusted P value < 0.05. For comparison of changes in taxonomic and functional traits, the bacterial read counts table was binned into KO groups, based on DIAMOND-MEGAN annotation. The links between gene functions (denitrification and type VI secretion system (T6SS)) and wheat root microbial community structure at the order level, represented as networks, were constructed using Cytoscape v.3.7.2 software [43].
Results
Wheat was grown for 6 weeks in a greenhouse under 400 ppm CO2 (aCO2) or 850 ppm CO2 (eCO2), and three nitrate levels representing three possible wheat-growth-sustaining fertilization scenarios: limited (30 ppm), moderate (70 ppm) and excess (100 ppm). Soil, shoots and roots were sampled at the end of week 6. This single time point was chosen for two reasons. First, in a previous study, the difference in plant nitrogen and soil soluble nitrate contents, as influenced by CO2 level, was evident at the end of week 6 [25], suggesting a measurable effect on the root-surface-associated microbiome. Moreover, threefold accumulation of root-surface-associated bacteria was observed at this time point compared to all previous weeks [25]. Secondly, this time point is just before spike formation, which is known to alter root exudates, and thus may alter the root-associated microbial community [44]. The impact of spike formation on plant parameters and subsequent effects on the microbiome may therefore interfere with measuring the effects of CO2 and nitrate on the plant microbiome.
Influence of eCO2 and nitrate levels on soil and wheat parameters
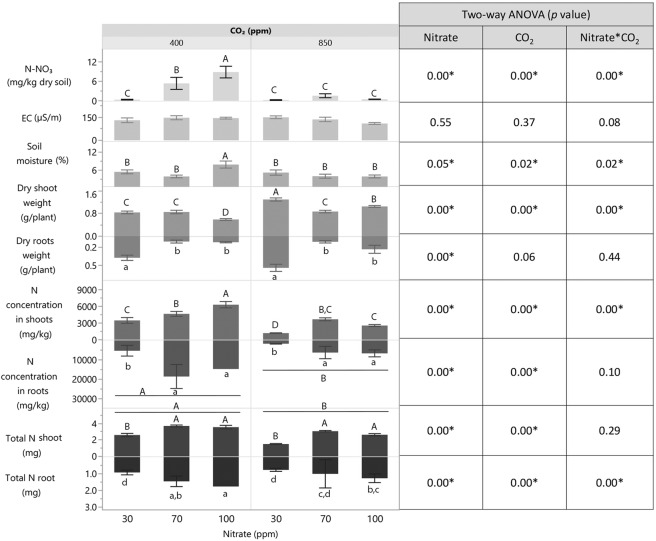
The changes in soil parameters (soluble nitrate content, electrical conductivity [EC] and moisture) and plant properties (shoot and root dry weight, nitrogen concentration and content) effected by nitrate and CO2 levels at the end of the 6-week period are presented in Fig. 1. Two-way ANOVA of interactions between CO2 and nitrate levels on soil and plant parameters was calculated. Interestingly, most of the soil and plant parameters were influenced by interactions between CO2 and nitrate levels (marked with an asterisk in Fig. 1). As expected, under aCO2, soil nitrate content was positively correlated with nitrate supply. However, under eCO2, soil nitrate content was negligible at all nitrate concentrations and was comparable to that under aCO2 and limited nitrate supply (30 ppm). Under eCO2, dry shoot weight increased under limited and excess nitrate, supporting the notion of plant biomass accumulation with increasing CO2 level. Moreover, under eCO2 vs. aCO2, plant nitrogen concentration (shoots and roots combined) and shoot total nitrogen content decreased. This indicated decreased plant nitrate accumulation with increasing CO2 level.
Fig. 1. Soil and plant properties as influenced by nitrate and CO2 levels in 6-weeks-old wheat.
Each bar represents the average of replicates with standard error (exact number of replicates can be found in Table S6). Different uppercase and lowercase letters indicate significant difference (P ≤ 0.05) by Student’s t test. Two-way ANOVA p values are provided in table on the right. NO-N3 nitrate nitrogen.
Influence of eCO2 and nitrate levels on the abundance of root-surface-associated bacteria
The effect of CO2 and nitrate levels on the abundance of total bacteria on the roots of 6-week-old wheat plants was measured in two ways: (1) quantitative PCR (qPCR) and (2) shotgun sequencing. For the qPCR measurement, the abundance of total bacteria on the plant roots was estimated as copy numbers of the 16S rRNA gene, and calculated in three ways: (i) per plant housekeeping gene TEF, (ii) per 1 g dry root, and (iii) per plant, i.e., per whole root dry weight (Fig. 2). For the shotgun sequencing, the relative abundance of total bacteria per plant was calculated by dividing the total number of bacterial genes by the total number of wheat genes. All four calculations showed a similar trend: with excess (100 ppm) nitrate supply, but not 30 or 70 ppm, there was a significant increase in the relative abundance of total bacteria per plant with increasing CO2 level (Fig. 2). This increased proliferation of root-surface-associated bacteria may result from improved plant status, and increased availability of root exudates and a change in their composition with rising CO2 level, combined with the excess nitrate supply.
Fig. 2. Effect of CO2 and nitrate levels on abundance of total bacteria per plant in roots of 6-weeks-old wheat.
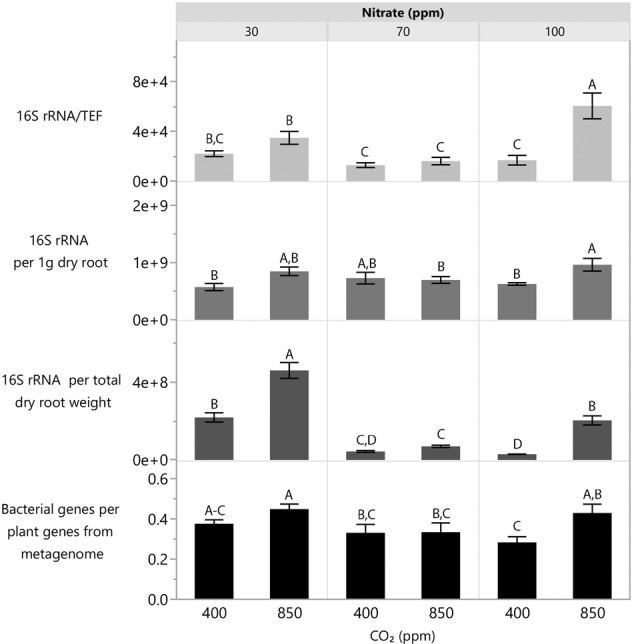
Gray bars represent measurement using qPCR, where each bar represents the average of nine replicates (three biological replicates, each composed of three technical ones) with standard error. Black bars represent measurement using shotgun sequencing, where each bar represents the average of three replicates (each replicate contains a mix of three technical replicates) with standard error. Different uppercase letters indicate significant difference (P ≤ 0.05) by Student’s t test.
Influence of eCO2 and nitrate levels on structure of root-surface-associated bacterial community
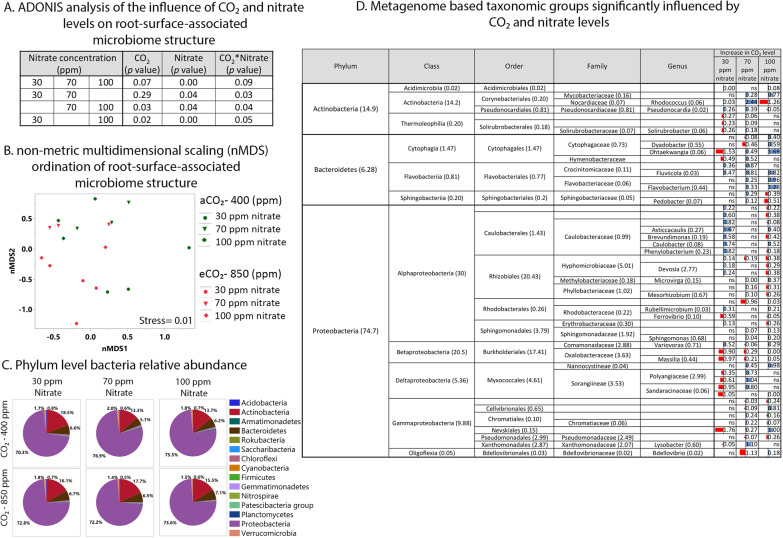
After filtering out the plant host genes from those of the root-surface-associated microbiome, around 84% of the genes were related to Bacteria, 0.56% to Eukaryota, 0.06% to Archaea, and 14% could not be assigned to any known taxonomic group. The Eukaryota, Archaea, and unassigned genes were filtered out from the root-surface-associated microbiome and the bacterial genes were further analyzed. Bacterial community structure on the roots of 6-week-old wheat plants was significantly influenced by nitrate level (calculated using the Adonis function in the Vegan R package; P < 0.006), and less so by CO2 level (Adonis; P < 0.07) (Fig. 3a, b). A list of all taxonomic groups whose abundance significantly changed as a function of CO2 and nitrate levels is provided in Table S2. The most dominant phylum in all samples was Proteobacteria (70–77% relative abundance), followed by Actinobacteria (13–18%) and Bacteroidetes (5–7%) (Fig. 3c). Pairwise comparisons between supplied nitrate concentrations showed that bacterial community structure was influenced by the interaction between CO2 and nitrate levels (Fig. 3a). The increase in abundance of almost all taxonomic groups originated from the three most abundant phyla (Proteobacteria, Actinobacteria and Bacteroidetes) was depended on the CO2–nitrate combination (Fig. 3d). Abundance of Actinobacteria and Bacteroidetes taxonomic groups was most significantly influenced by CO2 level at 70 ppm nitrate, whereas abundance of Proteobacteria taxonomic groups was most significantly influenced by CO2 level at 100 ppm nitrate.
Fig. 3. Combined influence of CO2 and nitrate levels on 6-weeks-old wheat root-surface-associated bacterial community structure.
a ADONIS analysis of the effect of CO2 and different combinations of nitrate on bacterial community structure. b nMDS ordination plot showing clustering patterns of root-surface-associated bacterial community structure as influenced by all nitrate and CO2 levels. Data matrix was transformed using normalized count transformation and batch effect was removed using DESeq2 package, and then ordination was generated using Bray–Curtis dissimilarity. c Bacterial community structure at the phylum level as influenced by nitrate and CO2 level. Changes in microbiome gene abundance were calculated using DESeq2 with cutoff <0.05 of FDR-adjusted P value. d Significantly changed groups with increase in CO2 level at three nitrate levels at all taxonomic levels. Numbers in parentheses indicate relative abundance of this specific taxa out of whole phylogeny assigned from metagenome. Blue indicates positive and red indicates negative log2 fold change. n = 3.
Influence of eCO2 and nitrate levels on functions of root-surface-associated bacterial community
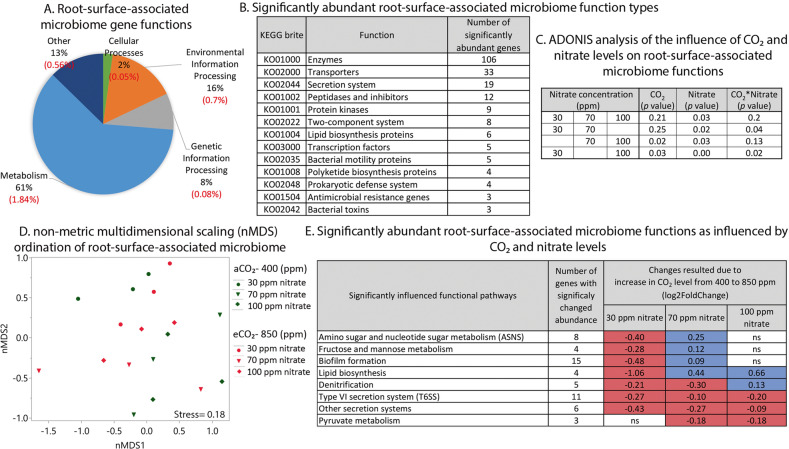
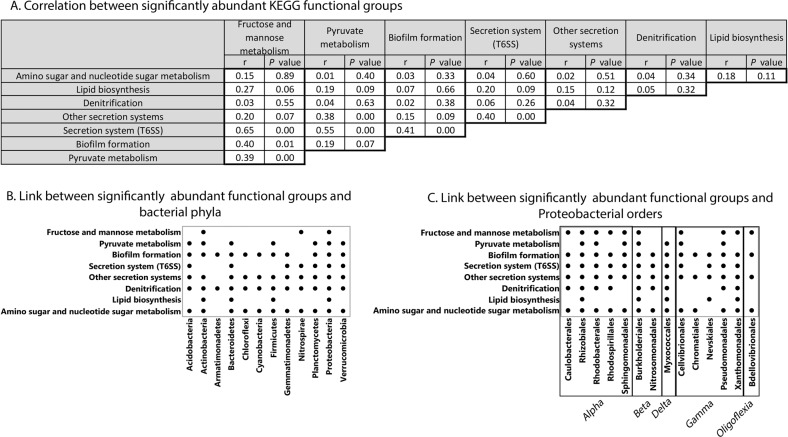
To reveal changes in root microbiome functions under different CO2 and nitrate levels, changes in microbiome gene abundance were calculated using DESeq2 with cutoff <0.2 for FDR-adjusted P value. More than half of the functional genes in the wheat root community metagenome were found to be involved in metabolic processes, 16% in environmental information processing, and 8% in genetic information processing (Fig. 4a). Around 3% of the functional genes of the root-surface-associated bacterial community were influenced by CO2 and nitrate levels. Metabolism-related KOs were the most strongly influenced group, with 1.8% out of the total 3% influenced genes. Less than 1% of the influenced genes was involved in environmental information processing and around 0.5% in other unknown processes. Half of the functional genes influenced by CO2 and nitrate levels encoded enzymes, 16% encoded transporters and around 10% were involved in the secretion system (Fig. 4b). A list of gene functions, divided into known pathways, whose abundance significantly changed with CO2 and nitrate levels, is given in Table S3. Functional genes were significantly influenced by the three nitrate levels (Adonis; P < 0.035) and not by CO2 level (Adonis; P < 0.21) (Fig. 4c, d). Pairwise comparisons between different combinations of supplied nitrate levels revealed that root-surface-associated bacterial community gene functions, similar to their community structures, were influenced by the interaction between CO2 and nitrate levels (Fig. 4c). Eight distinct functional gene groups were significantly influenced by the CO2–nitrate interaction (Fig. 4e): genes related to denitrification, three groups related to carbon metabolism (amino sugar and nucleotide sugar [ASNS], fructose and mannose, and pyruvate), groups related to biofilm formation, secretion systems, and in particular the type VI secretion system (T6SS), and lipid biosynthesis-related genes. Under eCO2 with limited (30 ppm) nitrate, the abundance of genes in seven groups (ASNS metabolism, fructose and mannose metabolism, biofilm formation, secretion systems, T6SS, lipid biosynthesis and denitrification) decreased (Fig. 4e). However, at 70 ppm nitrate, gene abundance in half of the groups increased with CO2 level (ASNS metabolism, fructose and mannose metabolism, biofilm formation and lipid biosynthesis) and that in the other half decreased (denitrification, pyruvate metabolism, T6SS, and other secretion systems). In the presence of excess (100 ppm) nitrate, gene abundance increased with CO2 level in two groups (denitrification and lipid biosynthesis), decreased in three groups (pyruvate metabolism, T6SS, and other secretion systems) and did not change in the other groups. Mantel test showed that five out of the eight significantly abundant root-surface-associated bacterial functional groups were correlated with one another under both CO2 levels and the three nitrate concentrations (Fig. 5a): fructose and mannose metabolism, pyruvate metabolism, biofilm formation, T6SS, and other secretion systems.
Fig. 4. Changes in root microbiome functions (KEGG) of 6-weeks-old wheat as influenced by nitrate and CO2 levels.
Changes in microbiome gene abundance were calculated using DESeq2 with cutoff < 0.2 of FDR-adjusted P value. a Pie chart of all functional genes in wheat root microbiome. Significantly changed pathways are marked in red. In parentheses is the percentage of that pathway out of whole metagenome which was significantly changed as a function of nitrate and CO2 levels. b Significantly abundant root-surface-associated microbiome function types. c ADONIS analysis of summarized effect of CO2 and different combinations of nitrate supply on bacterial community functional genes. d nMDS ordination plot showing clustering patterns of root-surface-associated microbiome functional genes as influenced by nitrate and CO2 levels. Data matrix was transformed using normalized count transformation and batch effect removal with the DESeq2 package, and then ordination was generated using Euclidean dissimilarity, n = 3. e Significantly abundant root-surface-associated microbiome functional gene pathways with increase in CO2 level. Blue indicates positive and red indicates negative log2 fold change in gene abundance between 850 and 400 ppm CO2. ns not significant.
Fig. 5. Correlation between significantly abundant root-surface-associated bacterial functional groups and association to their respective taxonomy.
a Correlation between significantly abundant root-surface-associated bacterial functional groups. Pearson correlation (r) was calculated using Mantel test and P is the significance level. Link between significantly abundant root-surface-associated bacterial functional groups and bacterial phyla (b) and proteobacterial order level (c).
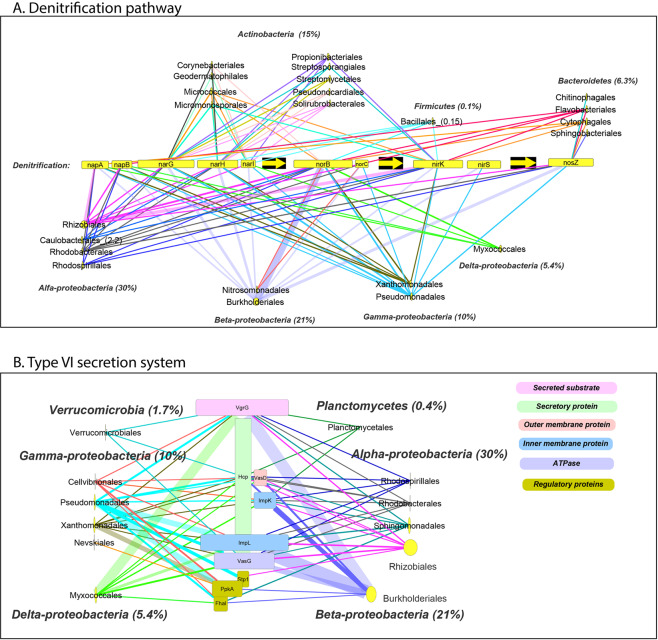
Link between taxonomy and significantly changed functions
Bacterial taxa responsible for the eight functional groups described above are presented in Fig. 5b, c. Proteobacteria (as well as its two orders Rhizobiales and Burkholderiales) was the only phylum in which genes from all eight functional groups changed in abundance. To further study the link between taxonomy and these functional groups, two were selected (denitrification and T6SS) and all of their functional genes (significantly changed and others) were associated to their respective taxonomy (Fig. 6a, b). Genes involved in denitrification originated from seven distinct bacterial groups, belonging mostly to the Actinobacteria, Bacteroidetes and Proteobacteria (especially Alphaproteobacteria) (Fig. 6a). The only bacterial groups that had all functional denitrifying genes were Burkholderiales and Pseudomonadales, while Rhizobiales lacked nirS. When calculating the absolute amount of all genes necessary for denitrification per relative abundance of specific orders, Burkholderiales and Pseudomonadales had comparable numbers of genes (45 and 51, respectively), whereas Rhizobiales (the most abundant order) had only 12 genes (Table S4). In addition, a distinct taxonomic separation was observed for each denitrifying gene. For example, Actinobacteria had only nitrate reductase genes related to nar but not nap. Another gene, nosZ, originated predominantly from Burkholderiales and partly from Pseudomonadales, Rhizobiales and Rhodobacterales. To calculate the absolute amount of genes in a specific group, gene copy number (i.e., number of reads from a specific gene per sample normalized with DESeq2 to total number of reads per treatment) was divided by the relative abundance of the orders. Rhodobacterales had an absolute amount of 31 nosZ genes compared to 6, 5 and 0.3 in Burkholderiales, Pseudomonadales and Rhizobiales, respectively (Table S4).
Fig. 6. The link between selected root microbiome functions and their associated taxonomy.
Link between order level root microbiome and genes of denitrification (a) and type VI secretion system (T6SS) (b). Relative abundance of each taxon under aCO2 and 100 ppm nitrate is represented by the size of the yellow node, and relative abundance is indicated in brackets near each taxon. Line width represents amount of gene. Width of gene rectangle indicates relative abundance of this gene in metagenome.
Genes involved in T6SS originated from six distinct bacterial orders, with most reads per gene belonging to the Proteobacteria (alpha, beta, gamma and delta) and negligible amounts in the phyla Verrucomicrobia and Planctomycetes (Fig. 6b). The Proteobacteria with the most T6SS reads were Rhizobiales, followed by Burkholderiales, Myxococcales and Pseudomonadales; their number of T6SS reads was consistent with their relative abundance in the roots. Looking at the absolute amount of all genes required for the construction of T6SS per relative abundance of each group, Pseudomonadales had the highest number of genes (121), followed by Myxococcales (58), Burkholderiales (34) and Rhizobiales (1.2) (Table S5). Interestingly, the groups with higher relative abundance possessed lower absolute amounts of T6SS genes. Moreover, Pseudomonadales, with very small relative abundance of 3%, was the only order with all of the genes necessary to construct a complete T6SS.
Discussion
Agricultural crops require nitrogen for their growth and development, especially at elevated CO2 levels, when plant growth is enhanced [35]. In this study, three scenarios representing nitrate fertilization regimes were compared under ambient and elevated CO2 levels: limited (30 ppm), moderate (70 ppm) and excess (100 ppm) nitrate.
The interaction between CO2 and nitrate levels significantly influenced soil parameters and plant development, as well as root-surface-associated bacterial abundance and community structure, and the abundance of bacterial genes with different functions. Limited nitrate supply resulted in negligible nitrogen concentrations in the soil at both CO2 levels. Under eCO2, nitrogen concentration and total content in the plants decreased, while shoot biomass increased (Fig. 1). Moderate nitrate supply under eCO2 resulted in a decrease in soil nitrate and plant total nitrogen, with no difference in biomass. Excess nitrate under eCO2 resulted in decreased soil nitrate and plant nitrogen with increased shoot biomass. The decrease in plant nitrogen could be because 90% of the nitrogen was supplied as nitrate and only 10% as ammonium, and it has been suggested that eCO2 inhibits nitrate-N assimilation into proteins in wheat [11–13]. Thus, with nitrogen mainly supplied as nitrate, its concentration generally decreases in plants under eCO2 [9, 10]. Indeed, in our study, under eCO2, nitrate fertilization did not result in nitrogen accumulation in the plants. Nevertheless, its content in the soil decreased, indicating an alternative nitrate-consumption process—probably denitrification. Indeed, the abundance of denitrification-associated functional genes in the metagenome of the root-surface-associated bacterial community increased significantly under eCO2 and excess nitrate (Fig. 4e).
Previous studies have shown that under eCO2, increased carbon availability in the soil originated from plants, lead to bacterial proliferation and activity [45, 46]. In this study, we used several methods to directly quantify the abundance of root-surface-associated bacteria. The relative abundance of total bacteria per plant increased under eCO2 with excess nitrate, but not with limited or moderate nitrate (Fig. 2). In addition, eCO2 significantly influenced the abundance of genes encoding enzymes (with changes in 3% of all enzymes in the metagenome), transporters (changes in 4% of total transporters) and the secretion system (changes in 8% of all secretion system genes). These alterations in bacterial abundance and in some of their functional genes may be the result of increased root exudates under eCO2, enabling bacteria to respond to the environmental shifts and proliferate.
In agreement with previous reports [22, 24, 47], the wheat root microbial community consisted mostly of Proteobacteria, Actinobacteria and Bacteroidetes (Fig. 3c). Proteobacteria, the largest taxonomic group in the wheat root-surface-associated bacterial metagenome (~ 75%), was also the group that was most influenced by eCO2 at all three nitrate levels (Fig. 3d). Moreover, Proteobacteria is the only phylum that contained genes from all of the functional gene groups affected by the CO2–nitrate interaction (Fig. 5a). At the order level, Rhizobiales was the dominant group, followed by Burkholderiales, Myxococcales, Sphingomonadales and Pseudomonadales. The relative abundance of those groups is in agreement with a recent study in which several wheat genotypes were grown under field conditions in different soil types [22]: not only was Rhizobiales the dominant group, but its functions seemed to play an important role in the wheat root microbiome. In another study, removing Rhizobiales-related functions from wheat roots in silico had the highest impact on the functional network of a root microbial community [48], whereas removing Burkholderiales from the network had a smaller impact, and removing Pseudomonadales had almost no impact at all [48]. In the current study, Rhizobiales and Burkholderiales were the only two orders of Proteobacteria to have all of the significantly changed functional genes, while Pseudomonadales lacked the significantly changed lipid biosynthesis genes (Fig. 5b). Moreover, despite the dominance of Rhizobiales compared to Burkholderiales and Pseudomonadales, they had less (in terms of gene number per relative abundance of the group) of the genes required for both denitrification and construction of T6SS (Tables S4 and S5). This suggests that Burkholderiales and Pseudomonadales play a major role in the functional variations occurring in the root microbiome as a result of eCO2 and nitrate level (the eight functional groups affected by these parameters’ interaction). While denitrification in the root microbial community is well understood, the function and role of T6SS in providing fitness and colonization advantages to root-surface-associated bacteria have only recently been investigated [49–51]. In the current study, a strong correlation was observed between five functional gene groups whose abundance was significantly changed (T6SS and other secretion systems, biofilm formation, pyruvate metabolism, and fructose and mannose metabolism) (Fig. 5a). A recent study identified acetic, succinic and malic acids as organic acids secreted by wheat roots, with succinic acid being the major organic acid exudate and its secreted concentration changing as a function of nitrogen supply [22]. Those three organic acids are linked to pyruvate metabolism (https://www.genome.jp/kegg/kegg2.html). This may support the notion that variations in root exudates resulting from eCO2 and nitrate levels affect colonization patterns and community assembly via interactions among root-surface-associated bacteria.
In this study, we demonstrate that interactions between CO2 and nitrate levels affect plant development, as well as root-surface-associated bacterial community structure and functions that probably serve for root colonization and fitness. Analyzing the connections between phylogeny and gene functions revealed that Rhizobiales, Burkholderiales and Pseudomonadales are responsible for most of the significant functional changes in the root microbiome. To the best of our knowledge, this is the first metagenomics study to shed light on alterations in soil and wheat properties and root-surface-associated bacteria as a result of combined atmospheric CO2 levels and nitrate fertilization. Further understanding the response of the root microbiome to changing environmental conditions may help sustain future wheat cultivation by improving fertilization to compensate for nitrate loss by denitrification. Furthermore, we believe that such studies should be carried out on other agriculturally important crops to improve yield and support growing populations, even under eCO2.
Supplementary information
Acknowledgements
This research was supported by the Israel Science Foundation (Research Project 1281/13). We acknowledge the computational help of Matan Cohen and Moshe Halpern for helping in the greenhouse experiment maintenance.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-020-00831-8) contains supplementary material, which is available to authorized users.
References
- 1.Plattner G-K. IPCC Climate change 2014. Synthesis report. Versión inglés. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. 2014.
- 2.Ainsworth EA, Long SP. What have we learned from 15 years of free-air CO2 enrichment (FACE)? A meta-analytic review of the responses of photosynthesis, canopy properties and plant production to rising CO2. N. Phytol. 2004;165:351–72. doi: 10.1111/j.1469-8137.2004.01224.x. [DOI] [PubMed] [Google Scholar]
- 3.Paterson E, Hall JMM, Rattray EASAS, Griffiths BSS, Ritz K, Killham K. Effect of elevated CO2 on rhizosphere carbon flow and soil microbial processes. Glob Chang Biol. 1997;3:363–77. doi: 10.1046/j.1365-2486.1997.t01-1-00088.x. [DOI] [Google Scholar]
- 4.de Graaff MA, van Groenigen KJ, Six J, Hungate B, van Kessel C. Interactions between plant growth and soil nutrient cycling under elevated CO2: a meta-analysis. Glob Chang Biol. 2006;12:2077–91. doi: 10.1111/j.1365-2486.2006.01240.x. [DOI] [Google Scholar]
- 5.de Graaff MA, Six J, van Kessel C. Elevated CO2 increases nitrogen rhizodeposition and microbial immobilization of root-derived nitrogen. N. Phytol. 2007;173:778–86. doi: 10.1111/j.1469-8137.2006.01974.x. [DOI] [PubMed] [Google Scholar]
- 6.Haase S, Philippot L, Neumann G, Marhan S, Kandeler E. Local response of bacterial densities and enzyme activities to elevated atmospheric CO2 and different N supply in the rhizosphere of Phaseolus vulgaris L. Soil Biol Biochem. 2008;40:1225–34. doi: 10.1016/j.soilbio.2007.12.025. [DOI] [Google Scholar]
- 7.Luo Y, Hui D, Zhang D. Elevated CO2 stimulates net accumulations of carbon and nitrogen in land ecosystems: a meta-analysis. Ecology. 2006;87:53–63. doi: 10.1890/04-1724. [DOI] [PubMed] [Google Scholar]
- 8.Feng Z, Rütting T, Pleijel H, Wallin G, Reich PB, Kammann CI, et al. Constraints to nitrogen acquisition of terrestrial plants under elevated CO2. Glob Chang Biol. 2015;21:3152–68. doi: 10.1111/gcb.12938. [DOI] [PubMed] [Google Scholar]
- 9.Cotrufo MF, Ineson P, Scott A. Elevated CO2 reduces the nitrogen concentration of plant tissues. Glob Chang Biol. 1998;4:43–54. doi: 10.1046/j.1365-2486.1998.00101.x. [DOI] [Google Scholar]
- 10.Long SP, Ainsworth EA, Rogers A, Ort DR. Rising atmospheric carbon dioxide: plants FACE the Future. Annu Rev Plant Biol. 2004;55:591–628. doi: 10.1146/annurev.arplant.55.031903.141610. [DOI] [PubMed] [Google Scholar]
- 11.Bloom AJ, Smart DR, Nguyen DT, Searles PS. Nitrogen assimilation and growth of wheat under elevated carbon dioxide. Proc Natl Acad Sci. 2002;99:1730–5. doi: 10.1073/pnas.022627299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.J. Bloom A, Burger M, A. Kimball B, J. Pinter P., Jr Nitrate assimilation is inhibited by elevated CO2 in field-grown wheat. Nat Clim Chang. 2014;4:477–80. doi: 10.1038/nclimate2183. [DOI] [Google Scholar]
- 13.Asensio JSR, Rachmilevitch S, Bloom AJ. Responses of Arabidopsis and wheat to rising CO2 depend on nitrogen source and nighttime CO2 levels. Plant Physiol. 2015;168:156–63. doi: 10.1104/pp.15.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li P, Hao X, Aryal M, Thompson M, Seneweera S. Elevated carbon dioxide and nitrogen supply affect photosynthesis and nitrogen partitioning of two wheat varieties. J Plant Nutr. 2019;42:1290–300. doi: 10.1080/01904167.2019.1616758. [DOI] [Google Scholar]
- 15.Drigo B, Kowalchuk GA, Van Veen JA. Climate change goes underground: effects of elevated atmospheric CO2 on microbial community structure and activities in the rhizosphere. Biol Fertil Soils. 2008;44:667–79. doi: 10.1007/s00374-008-0277-3. [DOI] [Google Scholar]
- 16.He Z, Xu M, Deng Y, Kang S, Kellogg L, Wu L, et al. Metagenomic analysis reveals a marked divergence in the structure of belowground microbial communities at elevated CO2. Ecol Lett. 2010;13:564–75. doi: 10.1111/j.1461-0248.2010.01453.x. [DOI] [PubMed] [Google Scholar]
- 17.Drigo B, Kowalchuk GA, Knapp BA, Pijl AS, Boschker HTS, van Veen JA. Impacts of 3 years of elevated atmospheric CO2 on rhizosphere carbon flow and microbial community dynamics. Glob Chang Biol. 2013;19:621–36. doi: 10.1111/gcb.12045. [DOI] [PubMed] [Google Scholar]
- 18.Gschwendtner S, Leberecht M, Engel M, Kublik S, Dannenmann M, Polle A, et al. Effects of elevated atmospheric CO2 on microbial community structure at the plant-soil interface of young beech trees (Fagus sylvatica L.) grown at two sites with contrasting climatic conditions. Micro Ecol. 2015;69:867–78. doi: 10.1007/s00248-014-0527-x. [DOI] [PubMed] [Google Scholar]
- 19.Benlloch-Gonzalez M, Bochicchio R, Berger J, Bramley H, Palta JA. High temperature reduces the positive effect of elevated CO2 on wheat root system growth. F Crop Res. 2014;165:71–9. doi: 10.1016/j.fcr.2014.04.008. [DOI] [Google Scholar]
- 20.Luo J, White RE, Roger Ball P, Tillman RW. Measuring denitrification activity in soils under pasture: optimizing conditions for the short-term denitrification enzyme assay and effects of soil storage on denitrification activity. Soil Biol Biochem. 1996;28:409–17. doi: 10.1016/0038-0717(95)00151-4. [DOI] [Google Scholar]
- 21.Hamonts K, Clough TJ, Stewart A, Clinton PW, Richardson AE, Wakelin SA, et al. Effect of nitrogen and waterlogging on denitrifier gene abundance, community structure and activity in the rhizosphere of wheat. FEMS Microbiol Ecol. 2013;83:568–84. doi: 10.1111/1574-6941.12015. [DOI] [PubMed] [Google Scholar]
- 22.Chen S, Waghmode TR, Sun R, Kuramae EE, Hu C, Liu B. Root-associated microbiomes of wheat under the combined effect of plant development and nitrogen fertilization. Microbiome. 2019;7:136. doi: 10.1186/s40168-019-0750-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donn S, Kirkegaard JA, Perera G, Richardson AE, Watt M. Evolution of bacterial communities in the wheat crop rhizosphere. Environ Microbiol. 2015;17:610–21. doi: 10.1111/1462-2920.12452. [DOI] [PubMed] [Google Scholar]
- 24.Mahoney AK, Yin C, Hulbert SH. Community structure, species variation, and potential functions of rhizosphere-associated bacteria of different winter wheat (Triticum aestivum) cultivars. Front Plant Sci. 2017;8:1–14. doi: 10.3389/fpls.2017.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Usyskin-Tonne A, Hadar Y, Yermiyahu U, Minz D. Elevated CO2 has a significant impact on denitrifying bacterial community in wheat roots. Soil Biol Biochem. 2020;142:107697. doi: 10.1016/j.soilbio.2019.107697. [DOI] [Google Scholar]
- 26.Halpern M, Bar-Tal A, Lugassi N, Egbaria A, Granot D, Yermiyahu U. The role of nitrogen in photosynthetic acclimation to elevated [CO2] in tomatoes. Plant Soil. 2019;434:397–411. doi: 10.1007/s11104-018-3857-5. [DOI] [Google Scholar]
- 27.Snell FD, Snell CT. Colorimetric Methods of Analysis. 3rd edn. New York: D. Van Nostran Co Inc.; 1949. [Google Scholar]
- 28.Usyskin-Tonne A, Hadar Y, Minz D. Altering N2O emissions by manipulating wheat root bacterial community. Sci Rep. 2019;9:7613. doi: 10.1038/s41598-019-44124-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rocha J, Cacace D, Kampouris I, Guilloteau H, Jäger T, Marano RBM, et al. Inter-laboratory calibration of quantitative analyses of antibiotic resistance genes. J Environ Chem Eng. 2020;8:102214. doi: 10.1016/j.jece.2018.02.022. [DOI] [Google Scholar]
- 30.Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 31.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–2. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2014;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 37.Huson DH, Beier S, Flade I, Górska A, El-Hadidi M, Mitra S, et al. MEGAN community edition - interactive exploration and analysis of large-scale microbiome sequencing data. PLOS Comput Biol. 2016;12:e1004957. doi: 10.1371/journal.pcbi.1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci. 2003;14:927–30. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 41.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- 43.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pausch J, Kuzyakov Y. Carbon input by roots into the soil: quantification of rhizodeposition from root to ecosystem scale. Glob Chang Biol. 2018;24:1–12. doi: 10.1111/gcb.13850. [DOI] [PubMed] [Google Scholar]
- 45.Zak DR, Pregitzer KS, King JS, Holmes WE. Elevated atmospheric CO2, fine roots and the response of soil microorganisms: a review and hypothesis. N. Phytol. 2000;147:201–22. doi: 10.1046/j.1469-8137.2000.00687.x. [DOI] [Google Scholar]
- 46.Hu S, Tu C, Chen X, Gruver JB. Progressive N limitation of plant response to elevated CO2: a microbiological perspective. Plant Soil. 2006;289:47–58. doi: 10.1007/s11104-006-9093-4. [DOI] [Google Scholar]
- 47.Ofek-Lalzar M, Sela N, Goldman-Voronov M, Green SJ, Hadar Y, Minz D. Niche and host-associated functional signatures of the root surface microbiome. Nat Commun. 2014;5:1–9. doi: 10.1038/ncomms5950. [DOI] [PubMed] [Google Scholar]
- 48.Ofaim S, Ofek-Lalzar M, Sela N, Jinag J, Kashi Y, Minz D. Analysis of microbial functions in the rhizosphere using a metabolic-network based framework for metagenomics interpretation. Front Microbiol. 2017;8:1606. doi: 10.3389/fmicb.2017.01606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bernal P, Llamas MA, Filloux A, Type VI. secretion systems in plant-associated bacteria. Environ Microbiol. 2018;20:1–15. doi: 10.1111/1462-2920.13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coulthurst S. The Type VI secretion system: a versatile bacterial weapon. Microbiology. 2019;165:503–15. doi: 10.1099/mic.0.000789. [DOI] [PubMed] [Google Scholar]
- 51.Kim N, Kim JJ, Kim I, Mannaa M, Park J, Kim J, et al. Type VI secretion systems of plant‐pathogenic Burkholderia glumae BGR1 play a functionally distinct role in interspecies interactions and virulence. Mol Plant Pathol. 2020;21:1055–69. doi: 10.1111/mpp.12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.