Abstract
Changes in the gut microbiota during pathogen infection are often predicted to influence disease outcomes. However, studies exploring whether pathogens induce microbiota shifts have yielded inconsistent results. This suggests that variation in infection, rather than the presence of infection alone, might shape pathogen–microbiota relationships. For example, most hosts are coinfected with multiple pathogens simultaneously, and hosts vary in how long they are infected, which may amplify or diminish microbial shifts expected in response to a focal pathogen. We used a longitudinal anthelmintic treatment study of free-ranging African buffalo (Syncerus caffer) to examine whether (i) coinfection with bovine tuberculosis (Mycobacterium bovis, TB) and gastrointestinal nematodes, and (ii) the duration of TB infection, modified effects of single pathogens on the gut microbiota. By accounting for the interaction between TB and nematodes, we found that coinfection affected changes in microbial abundance associated with single infections. Furthermore, the duration of TB infection predicted more microbiota variation than the presence of TB. Importantly, coinfection and infection duration had nearly as much influence on microbial patterns as demographic and environmental factors commonly examined in microbiota research. These findings demonstrate that acknowledging infection heterogeneities may be crucial to understanding relationships between pathogens and the gut microbiota.
Subject terms: Microbiome, Infectious diseases
Introduction
Shifts in the mammalian gut microbiota often occur during pathogen infection and can influence disease outcomes [1, 2]. As such, there is growing interest in using infection-induced changes in the microbiota as biomarkers of disease status and progression [3–5]. However, observed changes in the gut microbiota during infection are rarely consistent, even with respect to single pathogens [6]. For example, the effects of human immunodeficiency virus (HIV) infection on gut microbial diversity vary dramatically [7]. In some cases, HIV-infected patients show higher gut microbial diversity than healthy controls [8], while in others, HIV patients show lower microbial diversity than healthy individuals [9]. Similarly, microbiota patterns vary across pathogens. For instance, Clostridium difficile infection is typically characterized by decreases in gut microbiota diversity [10], while Mycobacterium tuberculosis (Mtb) infection can increase gut microbial diversity [11, 12]. These inconsistencies were quantified in a recent meta-analysis comparing the gut microbiota of helminth-infected and helminth-uninfected hosts. Among non-rodent hosts there was no evidence of a systematic effect of helminth infection on microbial diversity, whereas microbial diversity generally decreased in the presence of helminth infection among rodent hosts [13]. Thus, understanding when and how pathogens affect the gut microbiota likely requires consideration of not only simple infection presence and absence, but also the context in which infection occurs.
Features of the infection process may themselves influence how the gut microbiota responds to infection [6]. First, although many studies investigate how single infections affect the microbiota, most humans and animals are concurrently infected with multiple pathogens simultaneously [14, 15]. Importantly, coinfection can dampen or amplify protective host immune responses [15, 16]. Host immunity is also strongly linked to changes in gut microbiota composition [1, 17], suggesting a mechanism by which coinfection could modify microbiota shifts in response to a focal infection. In support, a study of helminth-infected humans found that while a single-species helminth infection did not lead to changes in the gut microbiota, shifts in microbial abundance were apparent among those coinfected with multiple helminth species [18]. Second, hosts often vary in how long they have been infected with a pathogen, with the strength and function of the immune response to infection changing over time [19–21]. Accounting for these two key sources of infection heterogeneity might help better uncover how infections shape the gut microbiota.
In addition to differences in infection status, hosts also vary in a number of other traits such as diet and age that often emerge as strong predictors of gut microbiota patterns [22–25]. Consequently, infection-induced shifts in the gut microbiota can also be influenced by host heterogeneity [6, 26–29]. For example, gut microbial changes in astrovirus-infected wild bats depended on host age—infection was associated with a decrease in gut microbial diversity of young bats, but an increase in microbial diversity of adults [27]. Likewise, underlying variation in host environment can shape the microbiota and its response to infection. For example, helminth infection had different effects on the abundance of gut microbes in human patients from Liberia versus Indonesia [26]. Given the importance of host and environmental factors in shaping gut microbiota composition, considering demographic and environmental heterogeneity among hosts is likely crucial to understanding infection–microbiota relationships.
In this study, we examined the context-dependence of infection–microbiota relationships by testing the independent and combined effects of two infections on the gut microbiota of wild African buffalo (Syncerus caffer). African buffalo are host to a range of pathogens [30], including bovine tuberculosis (TB; causative agent Mycobacterium bovis, Mb) and gastrointestinal (GI) helminths (strongyle nematodes; Nematoda: Strongylidae). Interactions between these two infections are well-characterized in the buffalo system [31–33] and generate considerable variation in host immunity, which might feed back to shape the gut microbiota. In buffalo, coinfection with nematodes weakens protective immunity to Mb and exacerbates TB disease severity [33, 34]. Furthermore, in cattle, Mb infection causes chronic TB disease characterized by the upregulation of different immune components in early- versus late-stage infection [19]. Since human and rodent studies have not revealed consistent microbiota shifts during either TB [4] or helminth infections [6, 13], TB-helminth coinfection provides an ideal system to investigate if infection-induced shifts in the microbiota depend on coinfection or the time course of infection. To do this, we analyzed the gut microbiota of buffalo from a longitudinal study where TB incidence of anthelmintic-treated and control buffalo was monitored every 6 months [33]. First, we asked whether TB infection or anthelmintic treatment alone explained variation in the buffalo microbiota and if TB-nematode coinfection, represented by the interaction between TB status and anthelmintic treatment, modified these relationships. Second, we asked if effects of TB on the gut microbiota changed over the course of TB infection. For all analyses, we also accounted for the potentially confounding effects of variation in host demography and environment. By considering heterogeneities in infection status, demography, and environment, we characterized microbial shifts associated with TB and nematode infection and identified important drivers of variation in infection–microbiota relationships.
Materials and methods
Animal sampling
We captured and sampled wild female African buffalo (Syncerus caffer) in Kruger National Park, South Africa from 2008 to 2012 [33]. Prior to this study, buffalo were not subject to any veterinary interventions. At initial capture, animals were radio-collared and randomly assigned to a control or anthelmintic treatment group. At initial capture and all subsequent captures, treated animals received a slow-release fenbendazole bolus (Panacur, Intervet) placed in the rumen, while control animals received nothing [33]. Fenbendazole is effective against nematodes that comprise most of the buffalo GI helminth community, including Cooperia fuelleborni, Haemonchus placei, Haemonchus bedfordi, and Trichostrongylus sp. [35, 36], and our treatment regime significantly reduced both the occurrence (by 55%) and intensity (by 73%) of nematode infection in treated animals [33].
Animals were captured about every 180 days and at each capture we collected blood and fecal samples, assessed age and pregnancy status [37], and scored body condition on a 1–5 point scale, with higher scores indicating better condition [38]. Blood samples were used to test for TB infection using a whole-blood gamma interferon assay (BOVIGAM, Prionics) optimized for African buffalo [33]. Fecal samples were used to monitor host diet quality and gut microbiota composition. Diet quality was measured as fecal nitrogen content, which reliably tracks seasonal shifts in buffalo forage intake [39, 40]. Nitrogen analyses were performed using wet digestion followed by ammonia quantification on an Autoanalyzer [41]. A portion of each fecal sample was stored at −20 °C on the day of sample collection until DNA extraction for microbiota analysis. Microbial analyses were performed on 467 fecal samples from 149 individuals (age range: 29–192 months) sampled 1–6 times between June 2009 and December 2011.
Microbial DNA sequencing and bioinformatics
To characterize buffalo gut microbial communities, we used 16S rRNA gene sequencing. We prepared DNA libraries following standard Earth Microbiome Project protocols [42], with the exception that fecal DNA extractions were performed using the MO BIO PowerSoil DNA isolation kit with single PowerBead tubes rather than 96-well plates. The V4 region of the 16S rRNA gene was PCR-amplified using the 515 forward (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806 reverse (5′-GGACTACHVGGGTWTCTAAT-3′) primer pair [43], with Golay error-correcting barcodes (12 bp) included on the reverse primer for sample identification. We sequenced purified amplicons on an Illumina HiSeq 2000 (2 × 100-bp read length) at the University of Colorado, Boulder [44].
Raw forward sequence reads were uploaded to Qiita (v. 0.1.0 [45]) for demultiplexing and quality filtering according to default parameters [46]. Reads <100 bp in length were discarded, and Deblur (v. 1.0.4 [47]) was used to cluster putative error-free reads into sub-operational taxonomic units (sOTUs), which are sequence variants with up to single-nucleotide resolution. Deblurred data were then exported to Quantitative Insights into Microbial Ecology 2 (QIIME2, v. 2018-4 [48]) for further processing. A phylogenetic tree was constructed using the fragment insertion method (v. 2018.2.0 [49]). To focus our analyses on abundant taxa, we removed sOTUs that comprised <0.01% of the data set. To control for uneven sequencing depth across samples, the data were rarefied to 10,000 sOTUs per sample [50]. Alpha and beta diversity metrics were calculated in QIIME2. Taxonomy was assigned with a naïve Bayesian classifier trained against sequences in the Greengenes 13.8 99% database [51] trimmed to 100 bp. Detailed descriptions of sequence processing methods are in the Supplementary material.
Statistical analyses
Overall, we analyzed the effects of single TB and nematode infection, TB-nematode coinfection, and TB infection duration on the buffalo fecal microbiota by modeling four microbiota characteristics. (i) We measured microbial community structure using beta (between-sample) diversity metrics that are weighted by sOTU abundance: weighted UniFrac distance and Bray–Curtis dissimilarity. (ii) We measured microbial composition using unweighted beta diversity metrics that are based on sOTU presence-absence: unweighted UniFrac distance and the Jaccard index. (iii) We measured microbial diversity using alpha (within-sample) diversity metrics: sOTU richness, Faith’s phylogenetic diversity (PD) index, Pielou’s evenness, and Shannon’s diversity index. (iv) We measured microbial abundance by collapsing sOTU abundances at the genus level, resulting in 103 genera abundances. Furthermore, we incorporated a series of host traits and environmental variables as covariates into all of our analyses to account for the effects of host and environmental heterogeneity on gut microbial communities [23, 52–54], including: diet quality (fecal nitrogen content), age, year, body condition score, pregnancy status, and season (wet versus dry [55]).
Effects of single and coinfection on the gut microbiota
To evaluate effects of single and coinfection on microbial community structure and composition, we tested the effects of TB infection status, anthelmintic treatment, and their interaction on weighted and unweighted microbial beta diversity using permutational multivariate analysis of variance (PERMANOVA). Distance matrices of pairwise differences between samples were imported from QIIME2 and square-root transformed in vegan. Model predictors included host traits and environmental variables (fecal nitrogen content, age, year, body condition score, pregnancy status, and season), single infections (TB status and anthelmintic treatment), and coinfection (the interaction between TB status and anthelmintic treatment). Because the significance of model predictors is assessed sequentially, we ordered host and environmental covariates according to the number of microbial genera each affected in our analysis of microbial abundance, then tested for effects of infection. To account for repeated sampling of individuals, we restricted permutations within blocks specified by animal ID using permute (v. 0.9-4 [56]). PERMANOVAs were implemented in R with the adonis2 function in vegan (v. 2.5-2 [57]).
To evaluate effects of single and coinfection on microbial diversity, we used linear mixed models (LMMs) to test if TB status, anthelmintic treatment, or their interaction was associated with changes in microbial alpha diversity. Diversity values were normalized using Box–Cox transformations, then scaled without centering prior to analysis (Shapiro–Wilk’s test, W > 0.974). Model predictors included host traits and environmental variables (fecal nitrogen content, age, year, body condition score, season, pregnancy status), single infections (TB status and anthelmintic treatment), and coinfection (the interaction between TB status and anthelmintic treatment). To account for repeated sampling of individuals, animal ID was included as a random effect. LMMs were implemented in R using lme4 (v. 1.1–21 [58]) and lmerTest (v. 3.0-1 [59]). R2 values for LMMs were calculated using partR2 (v. 0.9.0 [60]). For models with a significant interaction effect, we used the difflsmeans function with Benjamini–Hochberg correction to identify significant differences in marginal means across the levels of interacting factors [59]. We were interested in four contrasts: (i) TB-negative, control vs. TB-negative, treated; (ii); TB-positive, control vs. TB-positive, treated; (iii) TB-negative, control vs. TB-positive, control; (iv) TB-negative, treated vs. TB-positive, treated.
To evaluate effects of single and coinfection on microbial abundance, we modeled genera abundance using a multivariate linear model (MLM) implemented in R with the manylm function in mvabund (v. 4.1.3 [61]). Since reference frames help account for the compositional nature of microbial abundance data [62], we transformed genera abundance counts into log-ratios prior to analysis. To do this, we added a pseudocount of 1 to rarefied counts, divided these counts by the abundance of a reference taxon (Ruminococcus), and then calculated the natural logarithm of each ratio. Sparsity of the data set was low (82.5% nonzero), suggesting that the addition of pseudocounts should not introduce bias. We chose Ruminoccocus as the reference taxon because it was found in all samples, improved the normality of log-ratio distributions and model residuals, and was insensitive to effects of infection in preliminary analyses (see Supplementary methods; Table S1). To account for repeated sampling of individuals, we used a restricted permutation approach. We generated a permutation matrix in which samples were permuted within blocks specified by animal ID using permute, then used the matrix to run the MLM with the “bootID” argument in mvabund [63]. The MLM fits separate linear models to each of the 102 log-ratios to test for associations between model predictors and microbial abundance at both the multivariate (global community) and univariate (individual taxa) levels. Model predictors included host traits and environmental variables (fecal nitrogen content, age, year, body condition score, season, pregnancy status), single infections (TB status and anthelmintic treatment), and coinfection (the interaction between TB status and anthelmintic treatment). Test statistics were estimated using likelihood ratio tests and p values were calculated using residual resampling. To account for correlations among multivariate response variables, we fit an unstructured correlation matrix to the abundance data using the cor.type = “R” argument in mvabund. Univariate tests were adjusted for multiple testing via a resampling-based Holm step-down procedure [63]. For the top ten taxa emerging with a significant interaction effect, we used individual LMMs and post hoc tests as above to identify significant differences across interacting factors.
Effects of infection duration on the gut microbiota
To evaluate the effect of TB infection duration, rather than TB status alone, on gut microbial community structure, composition, and diversity, we analyzed data from a subset of 14 buffalo that converted from TB-negative to TB-positive during the study period. For each animal, the last TB-negative sample collected prior to TB conversion was used to represent the baseline microbiota. This way, we could assess temporal patterns during infection relative to the most recent time point without infection. Across individuals, subsequent TB-positive samples were collected 78–539 days post-TB conversion, and categorized according to whether they were collected within 6, 12, or 18 months of the focal animal’s baseline TB-negative sample. To quantify TB duration effects on the microbiota during single TB infection and TB-nematode coinfection, we ran analyses separately in anthelmintic-treated and control buffalo. This resulted in a sample size of 18 for anthelmintic-treated animals (n0 = 7, n6 = 5, n12 = 3, n18 = 3) and 20 for control animals (n0 = 7, n6 = 4, n12 = 6, n18 = 3).
We tested for an effect of time since TB conversion (0, 6, 12, or 18 months) on microbial community structure and composition, measured as weighted and unweighted beta diversity, using PERMANOVAs. Model predictors included TB duration, as well as fecal nitrogen content, year, and season given the strong effects of these variables in analyses performed on the full data set (see Tables S2 and S3). Model permutations were restricted by animal ID. Finally, we tested for an effect of time since TB conversion on microbial alpha diversity using LMMs. Alpha diversity values were Box–Cox-transformed and scaled without centering for analysis. In these models, season was included as a key covariate (see Table S4) and animal ID was included as a random effect. Pairwise differences between time points were assessed using difflsmeans with Benjamini–Hochberg correction. Given the limited sample size for TB duration analyses, we could not calculate partial R2 values for the LMMs or account for all host and environmental variables simultaneously. For variables we did not include (age, body condition score), we ran a set of alternative models substituting in these variables as covariates.
Results
Host traits and environmental variables influence gut microbial patterns
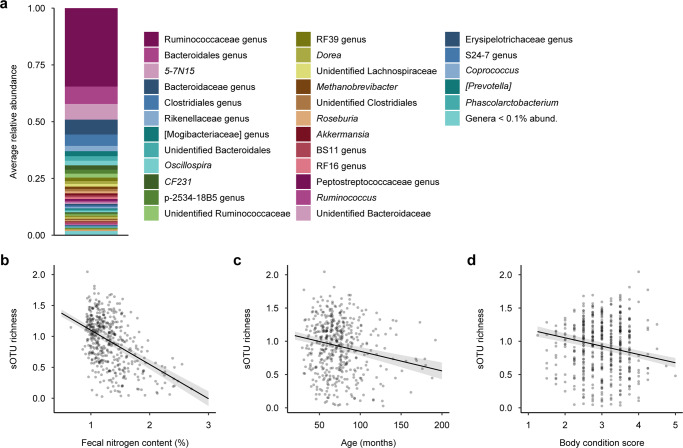
The fecal microbiota of African buffalo was dominated by Firmicutes (mean relative abundance: 58%) and Bacteroidetes (35%) at the phylum level (Fig. S1a), and Ruminococcaceae (39%), Bacteroidaceae (15%), and Lachnospiraceae (5%) at the family level (Fig. S1b). At the genus level, the most abundant taxa were an unidentified Ruminococcaceae genus (35%), an unidentified Bacteroidales genus (8%), and 5-7N15 (Bacteroidaceae; 7%). Genera common to ruminant intestinal microbiota, such as Oscillospira, Dorea, Methanobrevibacter, Roseburia, Akkermansia, and Ruminococcus were also present, each comprising ~1–2% of the total community (Fig. 1a).
Fig. 1. Characterization of the African buffalo gut microbiota.
a Genus-level composition profile representing average relative abundance values across all samples. b–d Gut microbial diversity, as measured by sub-operational taxonomic unit (sOTU) richness, was negatively associated with b fecal nitrogen content, a measure of intake diet quality, c age, and d body condition. Fitted lines constructed from a linear mixed model and shaded areas represent standard errors. Diversity values are Box–Cox-transformed and scaled.
Host traits and environmental variables had a range of effects on the gut microbiota. Effects on microbial community structure (weighted beta diversity) and composition (unweighted beta diversity) were generally weak to moderate. Fecal nitrogen content (R2 range = 0.027–0.030, p = 0.001), age (R2 = 0.007, p = 0.001), year (R2 range = 0.042–0.101, p = 0.001), and body condition score (R2 range = 0.006–0.007, p = 0.001) each explained ~0.6–10% of between-sample variation in microbial community structure, measured as weighted UniFrac distance and Bray–Curtis dissimilarity (Table S2). In addition, season explained 0.4% of variation in Bray–Curtis dissimilarity (R2 = 0.004, p = 0.001), while its effect on weighted UniFrac distance was marginal (R2 = 0.003, p = 0.073; Table S2); and pregnancy status explained 0.3% of variation in Bray–Curtis dissimilarity (R2 = 0.003, p = 0.029), but had no effect on weighted UniFrac distance (R2 = 0.002, p = 0.131; Table S2). In terms of microbial community composition, fecal nitrogen content (R2 range = 0.028–0.036, p = 0.001), age (R2 = 0.007, p = 0.001), year (R2 range = 0.021–0.024, p = 0.001), body condition score (R2 range = 0.009–0.010, p = 0.001), pregnancy status (R2 = 0.003, p ≤ 0.011), and season (R2 = 0.005, p = 0.001) each explained ~0.3–3.6% of the variance in unweighted UniFrac distance and the Jaccard index (Table S3).
For within-sample microbial diversity (alpha diversity), the effects of host traits and environmental variables ranged from weak to strong. Fecal nitrogen content (LMM: R2 range = 0.018–0.167, p ≤ 0.001; Fig. 1b), age (R2 range = 0.018–0.027, p ≤ 0.013; Fig. 1c), and body condition score (R2 range = 0.016–0.042, p ≤ 0.005; Fig. 1d) were all negatively correlated with gut microbial diversity, explaining ~1.6–16.7% of the variation across measures of alpha diversity (Table S4). Season affected microbial diversity as measured by sOTU richness and Faith’s PD index (R2 range = 0.011–0.019, p ≤ 0.003), but not Pielou’s evenness or Shannon’s diversity index (R2 range = 0.001–0.002, p ≥ 0.235; Table S4); and year affected microbial diversity as measured by Pielou’s evenness and Shannon’s diversity index (R2 range = 0.000–0.006, p ≤ 0.040), but not sOTU richness or Faith’s PD index (R2 range = 0.000–0.002, p ≥ 0.336; Table S4). In contrast, pregnancy status was not significantly associated with any measure of microbial diversity (R2 range = 0.000–0.001, p ≥ 0.446; Table S4).
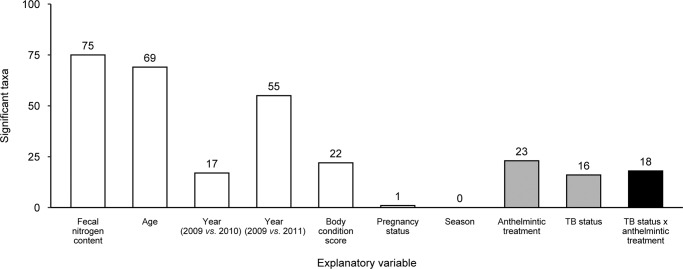
Last, the effects of host traits and environmental variables on microbial abundance were comparatively strong. At the community level, microbial abundance was significantly associated with fecal nitrogen content, age, year, body condition score, pregnancy status, and season (Table 1); and some of these variables predicted the abundance of a large fraction of individual microbial genera (Table S5 and Fig. 2). Fecal nitrogen content predicted the abundance of 74% of taxa (75 of 102; Fig. S2), age predicted the abundance of 68% of taxa (69 of 102; Fig. S3), year predicted the abundance of 17–54% of taxa (17–55 of 102; Fig. S4), and body condition score predicted the abundance of 22% of taxa (22 of 102; Fig. S5). In contrast, pregnancy status and season were poor predictors of microbial abundance, affecting one taxon and no taxa, respectively (Table S5).
Table 1.
Host traits and environmental variables, single infections, and coinfection shaped community-level patterns of microbial genera abundance in the African buffalo gut microbiota.
| LR value | p value | |
|---|---|---|
| Fecal nitrogen content | 497.8 | 0.001 |
| Age | 227.9 | 0.001 |
| Year (2009 vs. 2010) | 376.1 | 0.001 |
| Year (2009 vs. 2011) | 520.8 | 0.001 |
| Body condition score | 188.6 | 0.001 |
| Pregnancy status | 148.8 | 0.003 |
| Season | 280.4 | 0.001 |
| Anthelmintic treatment | 135.9 | 0.001 |
| TB status | 149.6 | 0.001 |
| TB status × anthelmintic treatment | 149.7 | 0.001 |
Multivariate output of a multivariate linear model. Significant values are shown in bold.
LR likelihood ratio statistic, TB bovine tuberculosis.
Fig. 2. Host traits and environmental variables, single infections, and coinfection shaped abundance patterns of individual microbial genera in the African buffalo gut microbiota.
Bar plot illustrating the number of microbial taxa whose abundances were significantly influenced by host and environmental variables (open bars), single infections alone (gray bars), or bovine tuberculosis (TB)-nematode coinfection (black bar), as determined by univariate tests from a multivariate linear model.
Coinfection can modify the effects of single infections on gut microbial patterns
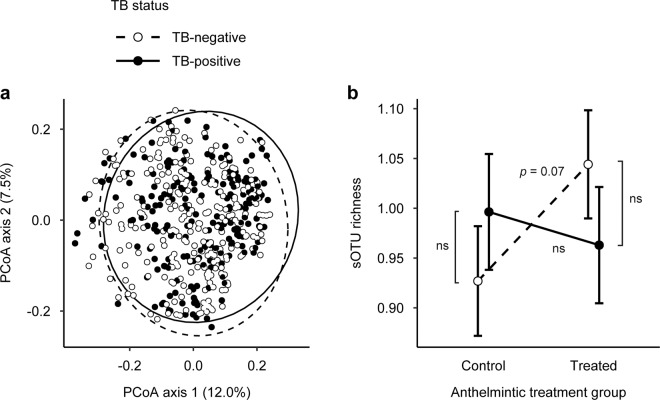
Compared to the effects of host traits and environmental variables, single infections had few effects on the gut microbiota, but some of these effects were modified by coinfection. After accounting for host and environment-related covariates, TB infection status explained 0.2% of the between-sample variation in microbial community structure as measured by Bray–Curtis dissimilarity (PERMANOVA: R2 = 0.002, p = 0.018; Table S2), while its effect on weighted UniFrac distance was similarly weak and marginal (R2 = 0.002, p = 0.054; Table S2). Anthelmintic treatment had no effect on community structure (R2 = 0.002, p ≥ 0.145; Table S2). As with microbial community structure, TB infection status showed weak effects on community composition, explaining 0.2% of observed variance in composition as measured by both unweighted UniFrac distance and the Jaccard index (R2 = 0.002, p ≤ 0.019; Table S3 and Fig. 3a). Again, anthelmintic treatment had no effect (R2 range = 0.002–0.003, p ≥ 0.613; Table S3). Furthermore, the interaction between TB status and anthelmintic treatment had no effect on either microbial community structure (R2 range = 0.002–0.003, p ≥ 0.185; Table S2) or composition (R2 = 0.003, p ≥ 0.274; Table S3).
Fig. 3. Single infections and coinfection had weak effects on African buffalo gut microbial community composition and diversity.
a Principle coordinate analysis (PCoA) of Jaccard indices revealed that bovine tuberculosis (TB) status predicted a small amount of variation in gut microbial community composition. Ellipses represent 95% confidence levels. b The interaction between TB status and anthelmintic treatment affected gut microbial diversity, as measured by sub-operational taxonomic unit (sOTU) richness. Post hoc tests revealed that differences in sOTU richness between anthelmintic-treated and control buffalo tended to be more prominent in TB-negative animals (p = 0.072) than in TB-positive animals (p = 0.559). Fitted points constructed from a linear mixed model and error bars represent standard errors. “ns” indicates nonsignificant contrasts from post hoc tests. Diversity values are Box–Cox-transformed and scaled.
On the other hand, accounting for coinfection helped reveal effects of single infections on gut microbial diversity. After controlling for host traits and environmental variables, neither TB status (LMM: R2 = 0.000, p ≥ 0.861), nor anthelmintic treatment (R2 = 0.000, p ≥ 0.280) alone were associated with microbial diversity (Table S4). However, the interaction between TB status and anthelmintic treatment had a weak effect on sOTU richness (R2 = 0.008, p = 0.041; Fig. 3b) and a marginal effect on Faith’s PD index (R2 = 0.007, p = 0.057; Table S4), but no effect on Pielou’s evenness or Shannon’s diversity index (R2 range = 0.000–0.001, p ≥ 0.655; Table S4). Post hoc tests revealed that anthelmintic treatment tended to increase sOTU richness in TB-negative buffalo (p = 0.072), but this effect disappeared in TB-positive buffalo (p = 0.559; Fig. 3b).
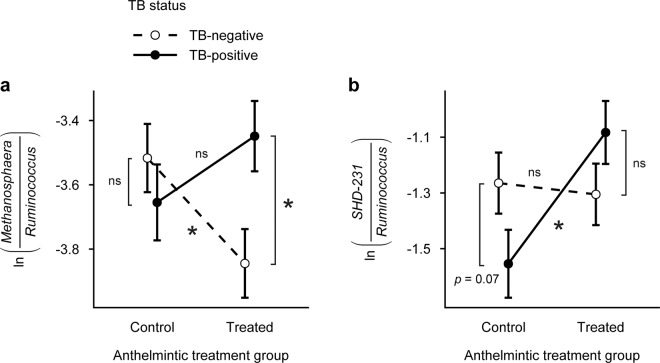
Coinfection most strongly influenced the effects of single infections on gut microbial abundance. Both TB status and anthelmintic treatment shaped community-level patterns of microbial abundance (Table 1). However, TB-nematode coinfection, as shown by the interaction between TB status and anthelmintic treatment, also had a significant effect (Table 1). In parallel, while anthelmintic treatment predicted the abundance of 23% of taxa (23 of 102) and TB status predicted the abundance of 16% of individual taxa (16 of 102), the interaction between TB status and anthelmintic treatment predicted the abundance of 18% of taxa (18 of 102; Fig. 2). Examining the ten taxa that emerged with the strongest model support for the TB-anthelmintic treatment interaction (Table S5 and Fig. 4, S6) revealed that, for two taxa, coinfection changed the magnitude of single infection effects on gut microbe abundance. For example, TB affected the abundance of Methanosphaera in anthelmintic-treated buffalo, but this effect disappeared in control buffalo (MLM: TB status × anthelmintic treatment, LR = 14.8, p = 0.001; Table S5 and Fig. 4a). In addition, anthelmintic treatment affected the abundance of SHD-231 in TB-positive buffalo, but this effect disappeared in TB-negative buffalo (MLM: TB status × anthelmintic treatment, LR = 12.8, p = 0.001; Table S5 and Fig. 4b).
Fig. 4. Coinfection influenced the effects of single infections on the predicted abundance of gut microbial genera in African buffalo.
The interaction between bovine tuberculosis (TB) status and anthelmintic treatment explained variation in the abundance of a Methanosphaera and b SHD-231. Post hoc tests revealed that coinfection changed the magnitude of relationships between single infections and genera abundance. Fitted points constructed from univariate tests of a multivariate linear model and error bars represent standard errors. Asterisks indicate significant contrasts among factor levels (p < 0.05) and “ns” indicates nonsignificant contrasts.
Duration of TB infection influences gut microbial patterns in coinfected animals
Accounting for another source of infection heterogeneity, the duration of infection, also unmasked effects of infection on the microbiota. Furthermore, these effects were more pronounced during coinfection. Although TB duration did not explain significant between-sample variation in microbial community structure (PERMANOVA: R2 range = 0.125–0.177, p ≥ 0.163; Table S6), it had a strong, context-dependent effect on microbial community composition. In control buffalo, TB duration explained ~17% of the variance in composition as measured by unweighted UniFrac distance (R2 = 0.174, p = 0.035) and the Jaccard index (R2 = 0.165, p = 0.030; Table S7 and Fig. 5a). In contrast, there was no effect of TB duration in anthelmintic-treated buffalo (PERMANOVA: R2 range = 0.162–0.165, p ≥ 0.564; Table S7 and Fig. 5b). Substituting age and body condition score as alternative covariates did not qualitatively change the observed effects of TB duration on microbial community structure (Table S8) or composition (Table S9).
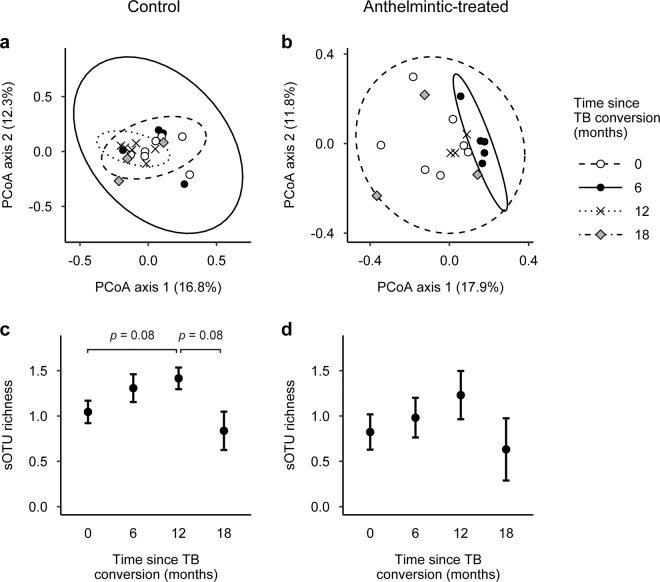
Fig. 5. The duration of bovine tuberculosis (TB) infection affected African buffalo gut microbial community composition and diversity during TB-nematode coinfection.
a, b Principle coordinate analysis (PCoA) of Jaccard indices revealed that TB duration explained significant variation in microbial community composition of control buffalo, but not anthelmintic-treated buffalo. Ellipses represent 95% confidence levels. c, d There was also a marginal association between TB duration and gut microbial diversity in control buffalo. Post hoc tests revealed that sub-operational taxonomic unit (sOTU) richness tended to increase between 0- and 12-months after TB conversion (p = 0.084) and decrease between 12- and 18-months after TB conversion (p = 0.084). However, TB duration was not associated with gut microbial diversity in anthelmintic-treated buffalo. Fitted points constructed from linear mixed models and error bars represent standard errors. Diversity values are Box–Cox-transformed and scaled.
The effect of TB duration on microbial diversity was also more pronounced during coinfection. In control buffalo, there was a marginal association between TB duration and sOTU richness (LMM: F = 3.688, p = 0.051; Fig. 5c) and Faith’s PD index (F = 2.886, p = 0.087), but not Pielou’s evenness or Shannon’s diversity index (F range = 0.347–1.249, p ≥ 0.340; Table S10). Post hoc comparisons revealed that sOTU richness tended to increase from 0- to 12-months post-TB conversion (p = 0.084) and decrease from 12- to 18-months post-TB conversion (p = 0.084; Table S11 and Fig. 5c). In contrast, there was no association between TB duration and microbial diversity in anthelmintic-treated buffalo (LMM: F range = 0.672–1.511, p ≥ 0.286; Table S10 and Fig. 5d). Again, substituting age and body condition score as alternative covariates did not qualitatively change the observed effects of TB duration on microbial diversity (Table S12).
Discussion
Changes in the gut microbiota are thought to influence outcomes of host pathogen infection, but our understanding of infection–microbiota relationships is challenged by conflicting results among studies. By studying the gut microbiota of a wild mammal and using a combination of anthelmintic treatment and natural acquisition of TB to compare microbiota patterns between single nematode infection (TB-negative, control), single TB infection (TB-positive, treated), and TB-nematode coinfection (TB-positive, control), we found that infection-associated microbiota shifts depended on both the presence of other pathogens and the duration of infection. Moreover, by taking advantage of natural heterogeneities in host traits, we showed that some effects of infection were nearly as important as demographic and environmental factors that are well-known drivers of gut microbial patterns. These results suggest that considering infection heterogeneity is crucial to understanding how pathogens shape the gut microbiota.
One of our key findings was that pathogen interactions can alter the effects of single infections on gut microbial patterns. Specifically, coinfection diminished some pathogen-associated shifts in microbial diversity and abundance. For example, nematode-associated decreases in sOTU richness tended to be weakened by TB, which might be explained by immune-mediated interactions between TB and nematodes. Nematodes suppress Th1 immunity in buffalo [33], and reduced Th1 immunity has been associated with reduced gut microbiota diversity in mice [64, 65]. Thus, the upregulation of Th1 immunity in response to TB infection may increase microbial diversity, explaining the tendency for buffalo with TB-nematode coinfection (TB-positive, control) to have similar levels of microbial diversity as buffalo with single TB infection (TB-positive, treated). Similarly, immune dynamics might explain why TB-associated differences in the abundance of a specific microbial genus, Methanosphaera, were dampened during coinfection. Methanosphaera stimulates Th17 and proinflammatory cell responses [66, 67], which contribute to TB defense [16]. However, helminth infections have been shown to decrease the abundance of methanogens [68]. Thus, while Methanosphaera may increase during TB infection alongside anti-TB immune responses, nematodes may counteract this effect during coinfection. Intriguingly, since nematodes might modulate host immunity by altering the gut microbiota [69–71], future work should explore whether nematode-associated decreases in Methanosphaera weaken anti-TB immunity during coinfection. Nonetheless, these results suggest that immune-mediated interactions between pathogens can mask microbiota changes induced by single infections.
Unlike the pattern we observed for Methanosphaera, coinfection amplified the effects of single pathogens on another microbial taxon, SHD-231. In this case, nonimmune-mediated interactions between pathogens may explain effects of coinfection on the microbiota. We found that helminth-associated differences in the abundance of SHD-231 were only observed in the presence of concurrent TB infection. Specifically, in TB-positive buffalo, control individuals had a lower abundance of SHD-231 than treated individuals. Additive effects of two species on the abundance of a third are a common outcome of species interactions [72, 73], including pathogen interactions [74, 75]. One common mechanism explaining such additive interactions is resource competition [76]. Both TB and nematode infections are frequently associated with changes in host nutrition [77, 78], and in combination, the two infections reduce buffalo body condition [31]. Both of these effects could have implications for substrate utilization by gut microbes [52, 79, 80]. Since SHD-231 abundance has been linked to dietary changes in wild primates [81] and domestic ruminants [82, 83], we speculate that TB-nematode coinfection may exacerbate single pathogen–microbe relationships via impacts on host nutrition and microbial resource availability. However, more research is required to fully understand the mechanisms by which pathogen interactions shape the gut microbiota.
It is important to note that we examined the effects of nematode infection on the gut microbiota by giving some buffalo an anthelmintic drug and this treatment itself may have affected the microbiota. However, the drug we used, fenbendazole, and related drugs, have not been found to have significant effects on the gut microbiota [84–86]. Thus, the effects of anthelmintic treatment we observed likely represent the effects of nematode removal rather than effects of the drug [85], although it is possible that the physical placement of the long-lasting fenbendazole bolus in the rumen of hosts also disturbed the microbiota. In addition, while our treatment effectively reduced nematode burdens compared to control buffalo, the incomplete clearance of nematodes in some treated individuals may have dampened the effects of treatment on gut microbes. Nonetheless, our repeated finding that the microbiota was affected by interactions between TB and anthelmintic treatment supports our conclusion that microbiota responses to infection can differ in the presence of other pathogens.
In addition to coinfection, accounting for the duration of infection also clarified how pathogens affect the microbiota. We found that TB duration explained more variation in microbial composition and diversity than TB status alone. This difference suggests that pathogen effects on the microbiota can change over time. For example, microbiota composition changed over the course of TB infection in coinfected, but not singly-infected, animals. In addition, there was a tendency for microbial diversity to increase early in TB infection (first 12 months of infection), then decrease later in infection (12- and 18-months post-infection). In Mtb-infected humans, changes in microbial diversity also varied with stage of TB infection [12]. Likewise, Mtb-infected mice showed similar trends in microbial diversity over time, with diversity decreasing shortly after Mtb challenge, then gradually increasing throughout infection, and decreasing prior to host death [87]. Interestingly, these diversity patterns appear to mirror changes in Th1 and Th2 immune responses [87–89]. Since Mb-infected cattle shift from Th1- to Th2-dominated responses as infection progresses [19], the diversity patterns we observed might also reflect immune dynamics. This immune mechanism is also consistent with our finding that temporal microbiota patterns were more pronounced in coinfected buffalo, which show reduced Th1 immune function and accelerated TB mortality [33]. Therefore, our results again point to a role for infection heterogeneities, especially those linked to immune variation, in shaping the gut microbiota.
Host demographic and environmental factors are common drivers of variation in the gut microbiota [22–25], and our observations support this pattern. We found that diet quality (fecal nitrogen content), age, and body condition were associated with all attributes of the buffalo gut microbiota we quantified. Surprisingly, though, some effects of single and coinfection on the gut microbiota were of comparable importance to these demographic and environmental variables. For example, single and coinfections affected nearly as many microbial genera (16–23%) as body condition and year (17–22%). Moreover, during coinfection, TB duration explained more variation (~17%) in microbiota composition than diet quality (~8–9%) or year (~12.5%). A similar pattern was described in a study of Trichostrongylus-infected rabbits, where infection appeared to have equal or larger effects on gut microbial diversity as nutritional constraints [29].
It is now recognized that pathogen dynamics cannot be fully understood by considering only single pathogens or time points of infection in isolation [90–92]. Yet, this perspective is rarely extended to the study of how pathogens affect the microbiota [6, 14]. Our findings emphasize that both coinfection and infection duration can alter effects of single pathogens on gut microbial patterns. In addition, these nuances in infection status had nearly as much influence on the microbiota as well-studied demographic and environmental factors. As such, acknowledging the heterogeneities that exist among real-world hosts may be key to improving our understanding of the gut microbiota and its relationship to infectious diseases across host–pathogen systems.
Supplementary information
Acknowledgements
This work was supported by the National Science Foundation (Grant #s: DEB-1102493, DGE-1545433), the Earth Microbiome Project, and an ARCS Foundation Award. Animal protocols for this study were approved by the University of Georgia (UGA) and Oregon State University (OSU) Institutional Animal Care and Use Committees (UGA AUP A2010 10-190-Y3-A5; OSU AUP 3822 and 4325). We thank South African National Parks (SANParks) for permission to conduct this study in Kruger; M. Hofmeyr, P. Buss, and the entire SANParks Veterinary Wildlife Services Department for assistance with animal captures and project logistics; and R. Spaan, J. Spaan, K. Thompson, B. Beechler, S. Budischak, and K. Lane-deGraaf for work on animal captures and sample processing. We also thank D. Warton, J. Morton, and JP Schmidt for advice on statistical analyses, and members of the Ezenwa Lab for thoughtful comments on drafts of the manuscript.
Data availability
Sequence data and metadata are available through the Earth Microbiome Project data portal (qiita.microbio.me/emp) under study ID 1736, file 30386.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-020-00855-0) contains supplementary material, which is available to authorized users.
References
- 1.Libertucci J, Young VB. The role of the microbiota in infectious diseases. Nat Microbiol. 2019;4:35–45. doi: 10.1038/s41564-018-0278-4. [DOI] [PubMed] [Google Scholar]
- 2.Leung JM, Graham AL, Knowles SCL. Parasite-microbiota interactions with the vertebrate gut: Synthesis through an ecological lens. Front Microbiol. 2018;9:1–20. doi: 10.3389/fmicb.2018.00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hols P, Ledesma-García L, Gabant P, Mignolet J. Mobilization of microbiota commensals and their bacteriocins for therapeutics. Trends Microbiol. 2019;27:690–702. doi: 10.1016/j.tim.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Eribo OA, du Plessis N, Ozturk M, Guler R, Walzl G, Chegou NN. The gut microbiome in tuberculosis susceptibility and treatment response: guilty or not guilty? Cell Mol Life Sci. 2020;77:1497–509. doi: 10.1007/s00018-019-03370-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lv L-X, Jiang H-Y, Yan R, Li L. Interactions between gut microbiota and hosts and their role in infectious diseases. Infect Microbes Dis. 2019;1:3–9. [Google Scholar]
- 6.Cortes A, Peachey LE, Jenkins TP, Scotti R, Cantacessi C. Helminths and microbes within the vertebrate gut—not all studies are created equal. Parasitology. 2019;146:1–8. doi: 10.1017/S003118201900088X. [DOI] [PubMed] [Google Scholar]
- 7.Tuddenham SA, Koay WLA, Zhao N, White JR, Ghanem KG, Sears CL. The impact of human immunodeficiency virus infection on gut microbiota α-diversity: an individual-level meta-analysis. Clin Infect Dis. 2020;70:615–27. doi: 10.1093/cid/ciz258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lozupone CA, Li M, Campbell TB, Flores SC, Linderman D, Gebert MJ, et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe. 2013;14:329–39. doi: 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McHardy IH, Li X, Tong M, Ruegger P, Jacobs J, Borneman J, et al. HIV Infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome. 2013;1:1. doi: 10.1186/2049-2618-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, et al. Decreased diversity of the fecal microbiome in recurrent clostridium difficile-associated diarrhea. J Infect Dis. 2008;197:435–8. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 11.Maji A, Misra R, Dhakan DB, Gupta V, Mahato NK, Saxena R, et al. Gut microbiome contributes to impairment of immunity in pulmonary tuberculosis patients by alteration of butyrate and propionate producers. Environ Microbiol. 2018;20:402–19. doi: 10.1111/1462-2920.14015. [DOI] [PubMed] [Google Scholar]
- 12.Luo M, Liu Y, Wu P, Luo DX, Sun Q, Zheng H, et al. Alternation of gut microbiota in patients with pulmonary tuberculosis. Front Physiol. 2017;8:822. doi: 10.3389/fphys.2017.00822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scotti R, Southern S, Boinett C, Jenkins TP, Cortés A, Cantacessi C. MICHELINdb: a web-based tool for mining of helminth-microbiota interaction datasets, and a meta-analysis of current research. Microbiome. 2020;8:10. doi: 10.1186/s40168-019-0782-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kreisinger J, Bastien G, Hauffe HC, Marchesi J, Perkins SE. Interactions between multiple helminths and the gut microbiota in wild rodents. Philos Trans R Soc B Biol Sci. 2015;370:20140295. doi: 10.1098/rstb.2014.0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ezenwa VO. Helminth-microparasite co-infection in wildlife: lessons from ruminants, rodents and rabbits. Parasite Immunol. 2016;38:527–34. doi: 10.1111/pim.12348. [DOI] [PubMed] [Google Scholar]
- 16.Babu S, Nutman TB. Helminth-tuberculosis co-infection: an immunologic perspective. Trends Immunol. 2016;37:597–607. doi: 10.1016/j.it.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341–52. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper P, Walker AW, Reyes J, Chico M, Salter SJ, Vaca M, et al. Patent human infections with the whipworm, trichuris trichiura, are not associated with alterations in the faecal microbiota. PLoS ONE. 2013;8:e76573. doi: 10.1371/journal.pone.0076573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welsh MD, Cunningham RT, Corbett DM, Girvin RM, McNair J, Skuce RA, et al. Influence of pathological progression on the balance between cellular and humoral immune responses in bovine tuberculosis. Immunology. 2005;114:101–11. doi: 10.1111/j.1365-2567.2004.02003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann M, Pantazis N, Martin GE, Hickling S, Hurst J, Meyerowitz J, et al. Exhaustion of activated CD8 T cells predicts disease progression in primary HIV-1 infection. PLoS Pathog. 2016;12:1–19. doi: 10.1371/journal.ppat.1005661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy L, Nalpas N, Stear M, Cattadori IM. Explaining patterns of infection in free-living populations using laboratory immune experiments. Parasite Immunol. 2011;33:287–302. doi: 10.1111/j.1365-3024.2011.01281.x. [DOI] [PubMed] [Google Scholar]
- 22.Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–5. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 23.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Z, Knight R. Dietary effects on human gut microbiome diversity. Br J Nutr. 2015;113:S1–5. doi: 10.1017/S0007114514004127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graf D, Di Cagno R, Fåk F, Flint HJ, Nyman M, Saarela M, et al. Contribution of diet to the composition of the human gut microbiota. Micro Ecol Heal Dis. 2015;26:1–11. doi: 10.3402/mehd.v26.26164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosa BA, Supali T, Gankpala L, Djuardi Y, Sartono E, Zhou Y, et al. Differential human gut microbiome assemblages during soil-transmitted helminth infections in Indonesia and Liberia. Microbiome. 2018;6:1–19. doi: 10.1186/s40168-018-0416-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wasimuddin Brändel SD, Tschapka M, Page R, Rasche A, Corman VM, et al. Astrovirus infections induce age-dependent dysbiosis in gut microbiomes of bats. ISME J. 2018;12:2883–93. doi: 10.1038/s41396-018-0239-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arrazuria R, Elguezabal N, Juste RA, Derakhshani H, Khafipour E. Mycobacterium avium subspecies paratuberculosis infection modifies gut microbiota under different dietary conditions in a rabbit model. Front Microbiol. 2016;7:1–14. doi: 10.3389/fmicb.2016.00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cattadori IM, Sebastian A, Hao H, Katani R, Albert I, Eilertson KE, et al. Impact of helminth infections and nutritional constraints on the small intestine microbiota. PLoS ONE. 2016;11:1–23. doi: 10.1371/journal.pone.0159770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ezenwa VO, Jolles AE, Beechler BR, Budischak SA, Gorsich EE. The causes and consequences of parasite interactions: African buffalo as a case study. In: Wilson K, Fenton A, Tompkins D (eds). Wildlife Disease Ecology: Linking Theory to Data and Application. 1st edn. (Cambridge University Press, Cambridge, 2019). pp 129–60.
- 31.Ezenwa VO, Etienne RS, Luikart G, Beja-Pereira A, Jolles AE. Hidden consequences of living in a wormy world: nematode‐induced immune suppression facilitates tuberculosis invasion in African buffalo. Am Nat. 2010;176:613–24. doi: 10.1086/656496. [DOI] [PubMed] [Google Scholar]
- 32.Jolles AE, Ezenwa VO, Etienne RS, Turner WC, Olff H. Interactions between macroparasites and microparasites drive infection patterns in free-ranging African buffalo. Ecology. 2008;89:2239–50. doi: 10.1890/07-0995.1. [DOI] [PubMed] [Google Scholar]
- 33.Ezenwa VO, Jolles AE. Opposite effects of anthelmintic treatment on microbial infection at individual versus population scales. Science. 2015;347:175–7. doi: 10.1126/science.1261714. [DOI] [PubMed] [Google Scholar]
- 34.Ezenwa VO, Budischak SA, Buss P, Seguel M, Luikart G, Jolles AE, et al. Natural resistance to worms exacerbates bovine tuberculosis severity independently of worm coinfection. bioRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 35.Budischak SA, Hoberg EP, Abrams ART, Jolles AE, Ezenwa VO. A combined parasitological molecular approach for noninvasive characterization of parasitic nematode communities in wild hosts. Mol Ecol Resour. 2015;15:1112–9. doi: 10.1111/1755-0998.12382. [DOI] [PubMed] [Google Scholar]
- 36.Williams JC. Efficacy of albendazole, levamisole and fenbendazole against gastrointestinal nematodes of cattle, with emphasis on inhibited early fourth stage Ostertagia ostertagi larvae. Vet Parasitol. 1991;40:59–71. doi: 10.1016/0304-4017(91)90083-8. [DOI] [PubMed] [Google Scholar]
- 37.Jolles AE, Cooper DV, Levin SA. Hidden effects of chronic tuberculosis in African buffalo. Ecology. 2005;86:2358–64. [Google Scholar]
- 38.Ezenwa VO, Jolles AE, O’Brien MP. A reliable body condition scoring technique for estimating condition in African buffalo. Afr J Ecol. 2009;47:476–81. [Google Scholar]
- 39.Ryan SJ, Cross PC, Winnie J, Hay C, Bowers J, Getz WM. The utility of normalized difference vegetation index for predicting African buffalo forage quality. J Wildl Manag. 2012;76:1499–508. [Google Scholar]
- 40.Budischak SA, O’Neal D, Jolles AE, Ezenwa VO. Differential host responses to parasitism shape divergent fitness costs of infection. Funct Ecol. 2018;32:324–33. [Google Scholar]
- 41.Coetsee C, February EC, Bond WJ. Nitrogen availability is not affected by frequent fire in a South African savanna. J Trop Ecol. 2008;24:647–54. [Google Scholar]
- 42.Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature. 2017;551:457–63. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci. 2011;108:4516–22. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–4. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gonzalez A, Navas-Molina JA, Kosciolek T, McDonald D, Vázquez-Baeza Y, Ackermann G, et al. Qiita: rapid, web-enabled microbiome meta-analysis. Nat Methods. 2018;15:796–8. doi: 10.1038/s41592-018-0141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods. 2013;10:57–9. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amir A, McDonald D, Navas-Molina JA, Kopylova E, Morton JT, Zech Xu Z, et al. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems. 2017;2:e00191–16. doi: 10.1128/mSystems.00191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–7. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Janssen S, McDonald D, Gonzalez A, Navas-Molina JA, Jiang L, Xu ZZ, et al. Phylogenetic placement of exact amplicon sequences improves associations with clinical information. mSystems. 2018;3:e00021–18. doi: 10.1128/mSystems.00021-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A, et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 2017;5:27. doi: 10.1186/s40168-017-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–8. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sonnenburg JL, Bäckhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535:56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Viney M. The gut microbiota of wild rodents: challenges and opportunities. Lab Anim. 2019;53:252–8. doi: 10.1177/0023677218787538. [DOI] [PubMed] [Google Scholar]
- 54.Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Kling Bäckhed H, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150:470–80. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beechler BR, Jolles AE, Budischak SA, Corstjens PLAM, Ezenwa VO, Smith M, et al. Host immunity, nutrition and coinfection alter longitudinal infection patterns of schistosomes in a free ranging African buffalo population. PLoS Negl Trop Dis. 2017;11:1–24. doi: 10.1371/journal.pntd.0006122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simpson GL. permute: Functions for generating restricted permutations of data. R package version 0.9-4. https://cran.r-project.org/package=permute.
- 57.Oksanen JF, Blanchet G, Friendly M, Kindt R, Legendre P, McGlinn D, et al. vegan: Community ecology package. R package version 2.5-2. https://cran.r-project.org/package=vegan.
- 58.Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67:1–48. [Google Scholar]
- 59.Kuznetsova A, Brockhoff PB, Christensen RHB. lmerTest Package: tests in linear mixed effects models. J Stat Softw. 2017;82:1–26. [Google Scholar]
- 60.Stoffel M, Nakagawa S, Schielzeth H. partR2: Partitioning R2 in GLMMs. R package version 0.9.0. 2020. [DOI] [PMC free article] [PubMed]
- 61.Wang Y, Naumann U, Eddelbuettel D, Wilshire J, Warton D. mvabund: Statistical methods for analysing multivariate abundance data. R package version 4.1.3. https://cran.r-project.org/package=mvabund.
- 62.Morton JT, Marotz C, Washburne A, Silverman J, Zaramela LS, Edlund A, et al. Establishing microbial composition measurement standards with reference frames. Nat Commun. 2019;10:2719. doi: 10.1038/s41467-019-10656-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y, Naumann U, Wright ST, Warton DI. mvabund—An R package for model-based analysis of multivariate abundance data. Methods Ecol Evol. 2012;3:471–4. [Google Scholar]
- 64.Leung JM, Budischak SA, Chung The H, Hansen C, Bowcutt R, Neill R, et al. Rapid environmental effects on gut nematode susceptibility in rewilded mice. PLoS Biol. 2018;16:1–28. doi: 10.1371/journal.pbio.2004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qian LJ, Kang SM, Xie JL, Huang L, Wen Q, Fan YY, et al. Early-life gut microbial colonization shapes Th1/Th2 balance in asthma model in BALB/c mice. BMC Microbiol. 2017;17:1–8. doi: 10.1186/s12866-017-1044-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bang C, Weidenbach K, Gutsmann T, Heine H, Schmitz RA. The intestinal archaea Methanosphaera stadtmanae and Methanobrevibacter smithii activate human dendritic cells. PLoS ONE. 2014;9:1–9. doi: 10.1371/journal.pone.0099411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bernatchez E, Gold MJ, Langlois A, Blais-Lecours P, Boucher M, Duchaine C, et al. Methanosphaera stadtmanae induces a type IV hypersensitivity response in a mouse model of airway inflammation. Physiol Rep. 2017;5:1–13. doi: 10.14814/phy2.13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peachey LE, Molena RA, Jenkins TP, Di Cesare A, Traversa D, Hodgkinson JE, et al. The relationships between faecal egg counts and gut microbial composition in UK Thoroughbreds infected by cyathostomins. Int J Parasitol. 2018;48:403–12. doi: 10.1016/j.ijpara.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Su C, Su L, Li Y, Chang J, Zhang W, Walker WA, et al. Helminth-induced alterations of the gut microbiota exacerbate bacterial colitis. Mucosal Immunol. 2018;11:144–57. doi: 10.1038/mi.2017.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reynolds LA, Finlay BB, Maizels RM. Cohabitation in the intestine: interactions among helminth parasites, bacterial microbiota, and host immunity. J Immunol. 2015;195:4059–66. doi: 10.4049/jimmunol.1501432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martin I, Kaisar MMM, Wiria AE, Hamid F, Djuardi Y, Sartono E, et al. The effect of gut microbiome composition on human immune responses: an exploration of interference by helminth infections. Front Genet. 2019;10:1028. doi: 10.3389/fgene.2019.01028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Preston DL, Henderson JS, Johnson PTJ. Community ecology of invasions: direct and indirect effects of multiple invasive species on aquatic communities. Ecology. 2012;93:1254–61. doi: 10.1890/11-1821.1. [DOI] [PubMed] [Google Scholar]
- 73.Sih A, Englund G, Wooster D. Emergent impacts of multiple predators on prey. Trends Ecol Evol. 1998;13:350–5. doi: 10.1016/s0169-5347(98)01437-2. [DOI] [PubMed] [Google Scholar]
- 74.Telfer S, Lambin X, Birtles R, Beldomenico P, Burthe S, Paterson S, et al. Species interactions in a parasite community drive infection risk in a wildlife population. Science. 2010;330:243–7. doi: 10.1126/science.1190333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Johnson PTJ, Hoverman JT. Parasite diversity and coinfection determine pathogen infection success and host fitness. Proc Natl Acad Sci USA. 2012;109:9006–11. doi: 10.1073/pnas.1201790109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Griffiths EC, Pedersen AB, Fenton A, Petchey OL. Analysis of a summary network of co-infection in humans reveals that parasites interact most via shared resources. Proc R Soc B Biol Sci. 2014;281:20132286. doi: 10.1098/rspb.2013.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kant S, Gupta H, Ahluwalia S. Significance of nutrition in pulmonary tuberculosis. Crit Rev Food Sci Nutr. 2015;55:955–63. doi: 10.1080/10408398.2012.679500. [DOI] [PubMed] [Google Scholar]
- 78.Coop RL, Kyriazakis I. Influence of host nutrition on the development and consequences of nematode parasitism in ruminants. Trends Parasitol. 2001;17:325–30. doi: 10.1016/s1471-4922(01)01900-6. [DOI] [PubMed] [Google Scholar]
- 79.Hu Y, Feng Y, Wu J, Liu F, Zhang Z, Hao Y, et al. The gut microbiome signatures discriminate healthy from pulmonary tuberculosis patients. Front Cell Infect Microbiol. 2019;9:1–8. doi: 10.3389/fcimb.2019.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li RW, Wu S, Li W, Navarro K, Couch RD, Hill D, et al. Alterations in the porcine colon microbiota induced by the gastrointestinal nematode Trichuris suis. Infect Immun. 2012;80:2150–7. doi: 10.1128/IAI.00141-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hicks AL, Lee KJ, Couto-Rodriguez M, Patel J, Sinha R, Guo C, et al. Gut microbiomes of wild great apes fluctuate seasonally in response to diet. Nat Commun. 2018;9:1786. doi: 10.1038/s41467-018-04204-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bi Y, Zeng S, Zhang R, Diao Q, Tu Y. Effects of dietary energy levels on rumen bacterial community composition in Holstein heifers under the same forage to concentrate ratio condition. BMC Microbiol. 2018;18:1–11. doi: 10.1186/s12866-018-1213-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Omoniyi LA, Jewell KA, Isah OA, Neumann AP, Onwuka CFI, Onagbesan OM, et al. An analysis of the ruminal bacterial microbiota in West African Dwarf sheep fed grass- and tree-based diets. J Appl Microbiol. 2014;116:1094–105. doi: 10.1111/jam.12450. [DOI] [PubMed] [Google Scholar]
- 84.Fujishiro MA, Lidbury JA, Pilla R, Steiner JM, Lappin MR, Suchodolski JS. Evaluation of the effects of anthelmintic administration on the fecal microbiome of healthy dogs with and without subclinical Giardia spp. and Cryptosporidium canis infections. PLoS ONE. 2020;15:1–17. doi: 10.1371/journal.pone.0228145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Korte SW, Franklin CL, Dorfmeyer RA, Ericsson AC. Effects of fenbendazole-impregnated feed and topical moxidectin during quarantine on the gut microbiota of C57BL/6 mice. J Am Assoc Lab Anim Sci. 2018;57:229–35. [PMC free article] [PubMed] [Google Scholar]
- 86.Martin I, Martin I, Djuardi Y, Supali T, Sartono E, Yazdanbakhsh M, et al. Dynamic changes in human-gut microbiome in relation to a placebo-controlled anthelminthic trial in Indonesia. PLoS Negl Trop Dis. 2018;12:e0006620. doi: 10.1371/journal.pntd.0006620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Winglee K, Eloe-Fadrosh E, Gupta S, Guo H, Fraser C, Bishai W. Aerosol mycobacterium tuberculosis infection causes rapid loss of diversity in gut microbiota. PLoS ONE. 2014;9:1–9. doi: 10.1371/journal.pone.0097048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chackerian AA, Alt JM, Perera TV, Dascher CC, Behar SM. Dissemination of mycobacterium tuberculosis is influenced by host factors and precedes the initiation of T-cell immunity. Infect Immun. 2002;70:4501–9. doi: 10.1128/IAI.70.8.4501-4509.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hernández-Pando R, Orozcoe H, Sampieri A, Pavón L, Velasquillo C, Larriva-Sahd J, et al. Correlation between the kinetics of Th1, Th2 cells and pathology in a murine model of experimental pulmonary tuberculosis. Immunology. 1996;89:26–33. [PMC free article] [PubMed] [Google Scholar]
- 90.Pedersen AB, Antonovics J. Anthelmintic treatment alters the parasite community in a wild mouse host. Biol Lett. 2013;9:20130205. doi: 10.1098/rsbl.2013.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pedersen AB, Fenton A. Emphasizing the ecology in parasite community ecology. Trends Ecol Evol. 2007;22:133–9. doi: 10.1016/j.tree.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 92.Thakar J, Pathak AK, Murphy L, Albert R, Cattadori IM. Network model of immune responses reveals key effectors to single and co-infection dynamics by a respiratory bacterium and a gastrointestinal helminth. PLoS Comput Biol. 2012;8:e1002345. doi: 10.1371/journal.pcbi.1002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data and metadata are available through the Earth Microbiome Project data portal (qiita.microbio.me/emp) under study ID 1736, file 30386.