Summary
Ligand-gated ion channels mediate signal transduction at chemical synapses and transition between resting, open and desensitized states in response to neurotransmitter binding. Neurotransmitters that produce maximum open channel probabilities (Po) are full agonists whereas those that yield lower than maximum Po are partial agonists. Cys-loop receptors are an important class of neurotransmitter receptors yet a structure-based understanding of the mechanism of partial agonist action has proven elusive. Here we study the glycine receptor with the full agonist glycine and the partial agonists taurine and γ-amino butyric acid. We use electrophysiology to show how partial agonists populate agonist-bound, closed channel states, and cryo-EM reconstructions to illuminate the structures of intermediate, pre-open states, providing insights into previously unseen conformational states along the receptor reaction pathway. We further correlate agonist-induced conformational changes to Po across members of the receptor family, providing a hypothetical mechanism for partial and full agonist action at Cys-loop receptors.
Keywords: ligand-gated ion channels, partial agonists action, glycine receptor, gating mechanism, cryo-EM, SMA
ETOC:
Examination of the structure and electrophysiological response of the glycine receptor with full and partial agonists reveal agonist-bound closed states as well as open and desensitized states to illuminate conformational states along the receptor reaction pathway and structure-based insights into the mechanism of partial-agonist action.
Graphical Abstract

Introduction
Agonists can differ in their efficacy and produce different maximum responses on the same receptors (Colquhoun, 1998). “Partial agonists”, a term coined by Stephenson in 1956 (Stephenson, 1956), display submaximal efficacy. Early work on the muscle nicotinic acetylcholine receptor (nAChR) (Del Castillo and Katz, 1957), a Cys-loop or pentameric ligandgated channel (pLGIC), led to the hypothesis that efficacy is determined by the agonist’s ability to keep the channel open after it has bound. In pLGICs, agonist efficacy is reflected in the maximum open probability seen in single-channel recordings when the channel is fully occupied by the agonist yet is not desensitized. Neurotransmitters are often full agonists (Burzomato et al., 2004; Colquhoun and Sakmann, 1985), but partial agonists are of interest for therapeutics and include varenicline (Lam and Patel, 2007), a partial agonist of neuronal nAChR. In the auditory system, GABA, a partial agonist of glycine receptors (GlyRs), is co-released with glycine and both agonists act on the GlyR to sculpt the time course of postsynaptic responses (Lu et al., 2008). pLGICs are also important therapeutic targets and include the neuronal and muscle nAChR, GABAA receptors (GABAAR) and 5-HT3 receptors (5-HT3R) (Changeux and Edelstein, 1998).
GlyRs are an outstanding vehicle for investigating the mechanism of agonist efficacy. Studies by Katz and Thesleff outlined the canonical gating mechanism of pLGICs as the transition from closed/resting, open/activated and closed/desensitized states (Katz and Thesleff, 1957). Recent electrophysiological studies of GlyRs, however, showed that channel activation is not a simple isomerization from resting to open states, but rather involves a landscape of one or more pre-open intermediates (Auerbach, 2005; Burzomato et al., 2004; Lape et al., 2012). The presence of multiple agonist-bound closed states is a general feature of pLGICs, where the closed intermediate states have increased affinity for the agonist (Corradi et al., 2009; Jadey and Auerbach, 2012; Lape et al., 2008; Mukhtasimova et al., 2009). Fitting models that include activation intermediates to pLGIC single channel data has led to the proposal that the limited efficacy of partial agonists is due to their reduced ability to change the channel conformation to a short-lived pre-open intermediate (flipped/primed), rather than the reduced ability to open the receptor once the intermediate is reached (Corradi and Bouzat, 2014; Lape et al., 2008; Mukhtasimova et al., 2016).
Here we investigate the structural basis of partial agonist activation in GlyR, exploiting extensive functional characterization showing that taurine and GABA act as partial agonists (Fucile et al., 1999; Schmieden et al., 1992, 1993). While previous GlyR structures have shed light on the mechanism of full agonist, antagonist and modulator action (Du et al., 2015; Huang et al., 2015; Huang et al., 2017a; Huang et al., 2017b; Kumar et al., 2020), structures of the receptor bound to partial agonists are absent. Furthermore, the previously solved structures employed the receptor in detergent micelles as well as constructs lacking the M3/M4 cytoplasmic loop, factors that likely underlie the physiologically irrelevant ‘super open’ state of the ion channel pore and the anomalously high Po of the receptor constructs (Cerdan et al., 2018; Du et al., 2015; Gonzalez-Gutierrez et al., 2017). Recently, a cryo-EM structure of the glycine-bound, picrotoxin-blocked state of full length GlyR was elucidated (Kumar et al., 2020), where the receptor was reconstituted, together with soy bean lipids, into nanodiscs (Ritchie et al., 2009). This study provides insight into the full agonist-bound state of the receptor, but the blocking of the channel with picrotoxin yields an ion channel in a non conductive state.
To address the question of partial agonist action and to elucidate structures of the receptor in native, lipidic environment, we isolated the full length (FL) GlyR using the stryrene maleic acid polymer (SMA), enabling direct extraction of the receptor as a complex with endogenous lipids (Dorr et al., 2016; Knowles et al., 2009). We further reconstituted FL receptors into nanodiscs with brain lipids, and we then determined high resolution structures of the receptor bound to glycine, taurine or GABA in both the SMA and nanodisc environments. We explored both the SMA and nanodisc strategies because they both allow for the envelopment of the receptor transmembrane domain with native or native-like lipids (Dorr et al., 2016; Knowles et al., 2009; Ritchie et al., 2009), recognizing the sensitivity of membrane proteins (Autzen et al., 2019; Jamshad et al., 2011) and pLGICs to their membrane or membrane-mimic environment (Baenziger et al., 2015; Heidmann et al., 1980). These studies of GlyR bound to taurine or GABA reveal agonist-bound closed states as well as open and desensitized states, illuminating conformational states along the receptor reaction pathway and providing structure-based insights into the mechanism of partial-agonist action.
Results and Discussion
Glycine-bound complex populates multiple states
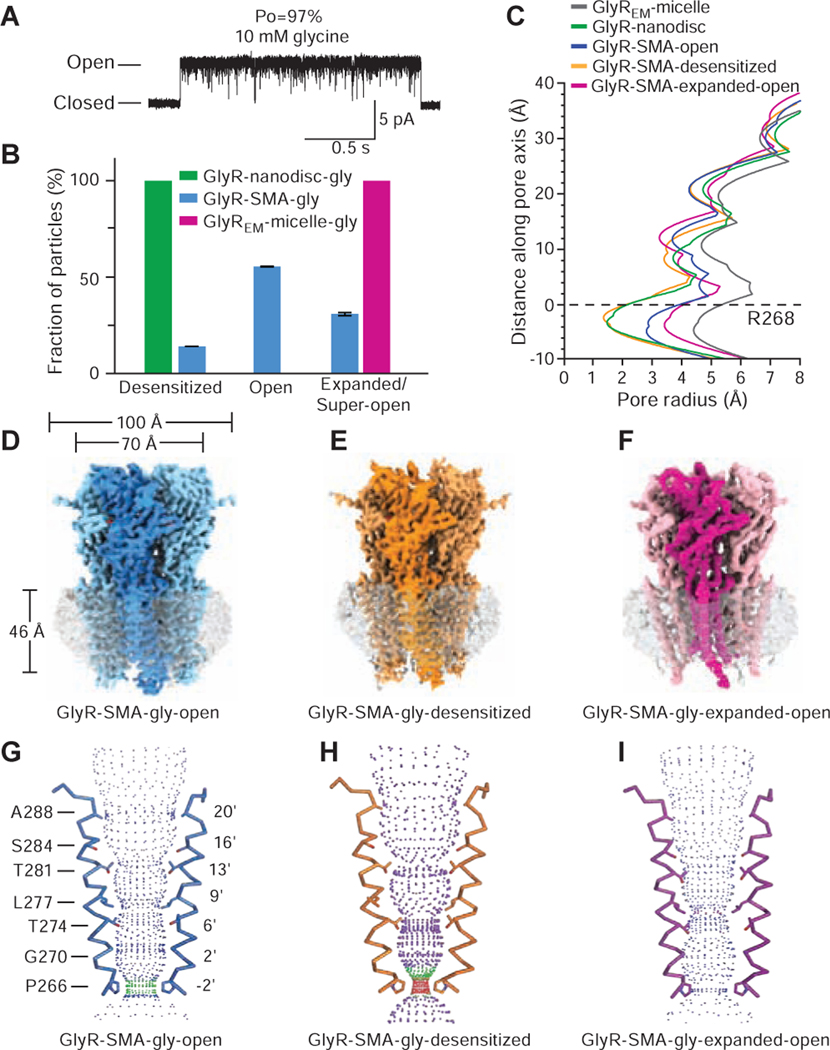
Glycine is a full agonist and single-channel analysis of clusters of receptor activity, excluding the long-lived desensitized states, reveals a maximum open probability (PO) of 97% (Figure 1A). Macroscopic current recordings elicited by saturating glycine show that the current decays to 57 ± 8% (n=8) of peak after 1s application because of desensitization (Figure S1A). Thus, under steady state conditions the glycine-bound receptor populates both open and desensitized states. To elucidate the structures of these states, we extracted the receptor using SMA or reconstituted the receptor into nanodiscs (see details in the STAR METHOD) and prepared cryo-EM grids in the presence of 10 mM glycine. Single particle cryo-EM studies of glycine-bound receptor in nanodiscs revealed a single closed desensitized or desensitized-like state (Figures S1C–S1D, S2, Table S1), which is at odds with electrophysiology data that shows the receptor populates both of open and desensitized states in the presence of glycine. We reasoned that either the lipids or perhaps the nanodisc complex itself shifts the conformational equilibrium of the receptor to the desensitized state and thus pursued studies of the receptor-SMA complex.
Figure 1. Single channel electrophysiology and cryo-EM analysis of the glycine complex.
A. Single-channel recording of GlyR openings in the presence of 10 mM glycine.
B. Fractions of GlyR particles in the desensitized, open and expanded/super-open states in nanodiscs, SMA, or detergent micelles. Error bars represent SEM, as described in the Methods.
C. Plots of pore radius for glycine-bound states. The Cα position of Arg 268 (M2 0’) is set to 0.
D-F. Cryo-EM density maps. The partially transparent surface represents lipid-SMA density.
G-I. Ion permeation pathways where M2 helices of two subunits are shown in ribbon and the side chains of pore-lining residues are shown in sticks, calculated by the program HOLE (radii coloring: red <1.8 Å, green 1.8–3.3 Å, and blue > 3.3 Å).
Single particle cryo-EM studies of the GlyR-SMA-gly complex revealed open (GlyR-SMA-gly-open), desensitized (GlyR-SMA-gly-desensitized) and expanded-open (GlyR-SMA-gly-expanded-open) states with overall resolutions of 2.9 Å, 3.1 Å and 4.0 Å, respectively (Figure 1B–1F, Figures S2 and Tables S2, Data S1). Importantly, these reconstructions have well resolved transmembrane domain (TMD) densities, especially for the M2 helices, allowing us to locate the 9’L and −2’P residues, thus informing the definition of functional state. In all of the structures the ion channel pores are open at 9’L but adopt three distinct conformations at the - 2’P position (Figure 1G–1I).
The three conformations at the −2’P position represent open, desensitized and expanded-open states. In the GlyR-SMA-gly-open state the constriction of the pore is ~5.6 Å, in agreement with the pore diameter ~5.3 Å estimated from functional studies and sufficient to allow permeation of partially hydrated chloride ions (Hille, 2001) and block by cyanotriphenylborate (Rundstrom et al., 1994) (Figure 1C, 1G). The GlyR-SMA-gly-desensitized state has a constriction at −2’P of 3 Å in diameter, indicative of a non-conducting state (Figure 1C, 1H). In the GlyR-SMA-gly-expanded-open state the pore has a minimum diameter of ~7 Å at −2’P, a dimension that is larger than that estimated by electrophysiological sizing experiments (Lynch, 2004) but smaller than that of the truncated receptor in detergent micelles (GlyREM-micelle-gly; PDB code: 3JAE) (Du et al., 2015) (Figure 1C, 1I, Figures S1E and S3A). To further assess the variable ‘open state’ pore diameters determined from structural studies (Du et al., 2015), we note that propionate, with an effective diameter of ~5.2 Å (Bormann et al., 1987), is one of the largest permeant organic anions. Isethionate, with an unhydrated diameter of ~6.2 Å (Sunesen et al., 2006), is an organic anion slightly larger than propionate and is impermeant in pLGIC anionic channels (Schwartz and Yu, 1995). Because the expanded-open state has a minimum diameter of 7 Å, and thus should be large enough to conduct isethionate, we suggest that the expanded-open state is too large to be the physiologically relevant open state. Thus, in addition to an expanded open state, the SMA-solubilized receptor yields conformations in physiologically relevant open and desensitized states, suggesting that SMA-extracted GlyR particles may provide insights into structure-based mechanisms of receptor function.
Partial agonist bound states
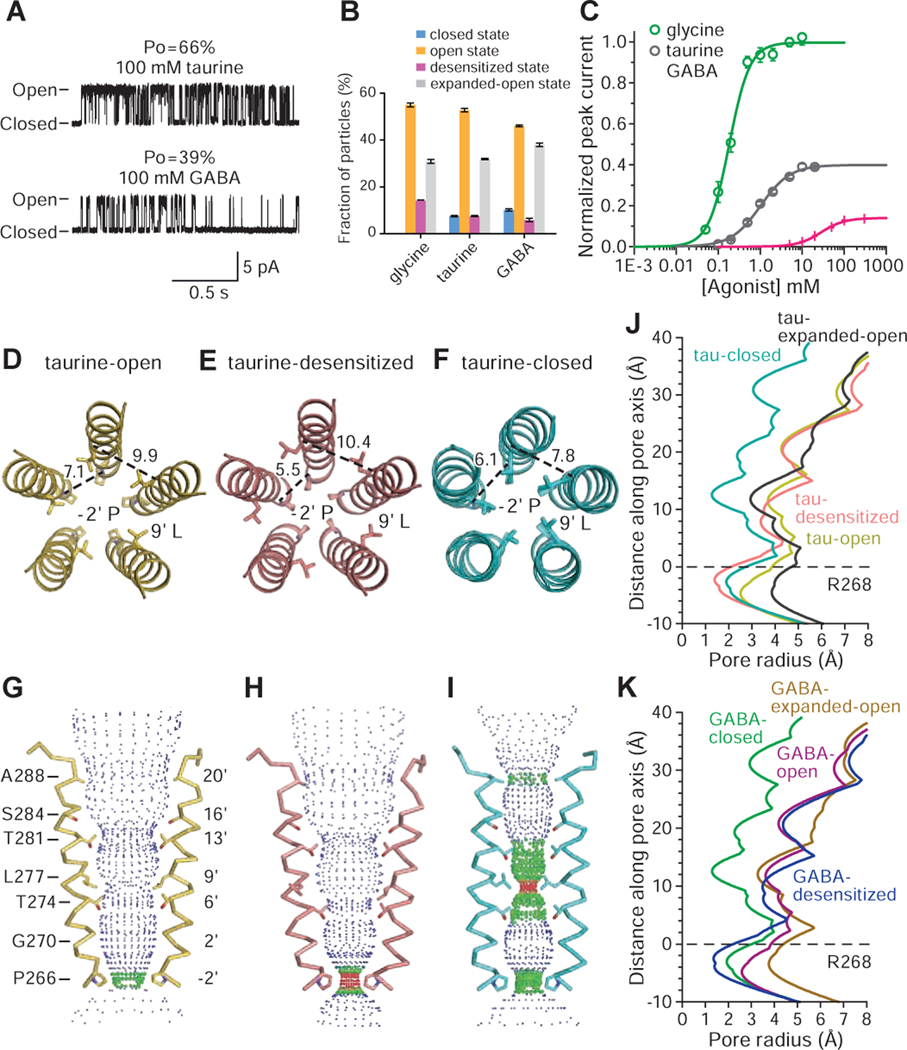
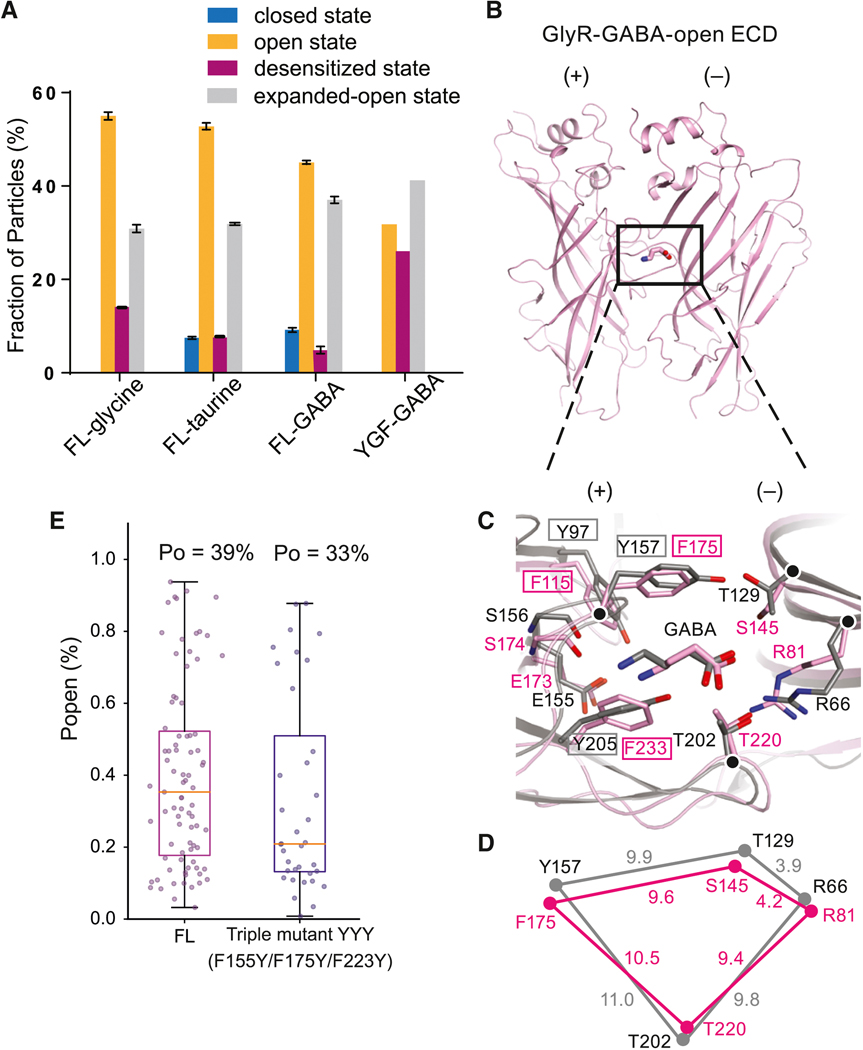
Single-channel recordings of GlyR in the presence of 100 mM taurine or GABA feature long-lived shut states while the channel is not desensitized, suggesting the presence of additional partial agonist-bound shut states (Figure 2A). Indeed, the maximum PO for taurine and GABA is 66 ± 3% and 39 ± 3%, respectively, substantially lower than that of glycine (98 ± 1%; 71, 83 and 83 clusters from 5–11 patches; mean ± SEM) (Figure 1A). The partial agonists are also less potent, with effective concentrations (EC50) for taurine and GABA of 1.05 ± 0.08 mM and 28.4 ± 0.9 mM, respectively (cf. glycine: 190 ± 20 μM; Figure 2C). In the subsequent structural studies, we applied 20 mM taurine and 40 mM GABA to the receptor prior to grid preparation for both the nanodisc and SMA data sets (Figures S4–S5, Methods S1–S2). The concentration of GABA employed for grid preparation was limited by its solubility and at higher concentrations GABA precipitated, rendering imaging the grids impossible.
Figure 2. Partial agonist complexes probed by electrophysiology and cryo-EM.
A. Representative cell-attached single-channel recordings of GlyR openings elicited by 100 mM taurine and 100 mM GABA, respectively.
B. Fractions of GlyR particles in the open, closed, desensitized and expanded-open states of the glycine, taurine and GABA complexes of the SMA-solubilized receptor, calculated by RELION and cisTEM as described in the Methods. Error bars represent SEM.
C. Whole cell, dose-response data for glycine, taurine and GABA. EC50 for glycine, taurine and GABA are 190 ± 20 μM, 1050 ± 80 μM and 28.4 ± 0.9 mM, respectively. Error bars represent SEM and n≥6 cells for all experiments. Curves are normalized to the glycine maximum current recorded in each cell.
D-F. Distances (in angstrom) and conformation of residues −2’P and 9’L in the taurine-bound, SMA-solubilized receptor in the open, desensitized and closed states.
G-I. Ion permeation pathway for taurine-bound states, calculated using HOLE and plotted as in Figure 1.
J-K. Pore radius as a function of distance along the pore axis for taurine-bound states (J) and GABA-bound states (K) of the SMA-solubilized receptor.
See also Figures S1, S3–S5, Tables S1–S3, S5, S7, Methods S1–S2 and Data S1.
Analysis of the partial agonist data sets reveals four 3D classes (Figures S4–S5, Tables S2–S3, Methods S2), with 3 of the taurine- or GABA-bound structures similar to the glycine-bound open, desensitized, and expanded-open states (Figure 2D–2K, Figure S3B–C). Importantly, we discovered a fourth, previously unseen, partial agonist-bound closed conformation of the receptor. There is clear density for the partial agonists in the neurotransmitter binding pocket (Figure S3E–H) and the M2 helices are oriented approximately perpendicular to the membrane, creating a constriction less than 3 Å in diameter at 9’L and rendering the channel impermeable to Cl- (Figure 2F, 2I, 2J, 2K). At the −2’P site, the pore has a diameter of 4 Å, intermediate between the open and desensitized states.
We hypothesize that the closed state, partial agonist-bound cryo-EM structures provide the visualization of the additional long-lived shut states that are seen in single-channel recordings with partial agonists (Figure 2A). These states are not seen with glycine (Figure 1A) because, by contrast, the glycine-bound closed conformation of the receptor is short-lived, as the receptor rapidly and nearly completely transitions to the brief intermediate “flipped” state (Lape et al., 2008), precluding its capture by single-particle cryo-EM. In comparison to glycine, taurine or GABA’s reduced ability to ‘flip’ GlyR, as well as their propensity to promote the receptor’s return from the flipped state to the closed state, allows us to capture agonist-bound closed states via cryo-EM. We suggest that the cryo-EM partial agonist-bound closed states most likely represent unflipped states. The particle fractions of the open or desensitized states are smaller for taurine as compared with glycine, and are smaller still for GABA. Conversely, the fraction of particles in the closed state is highest for the GABA dataset, lower for the taurine dataset, and absent in the glycine dataset (Figure 2B).
Neurotransmitter binding site
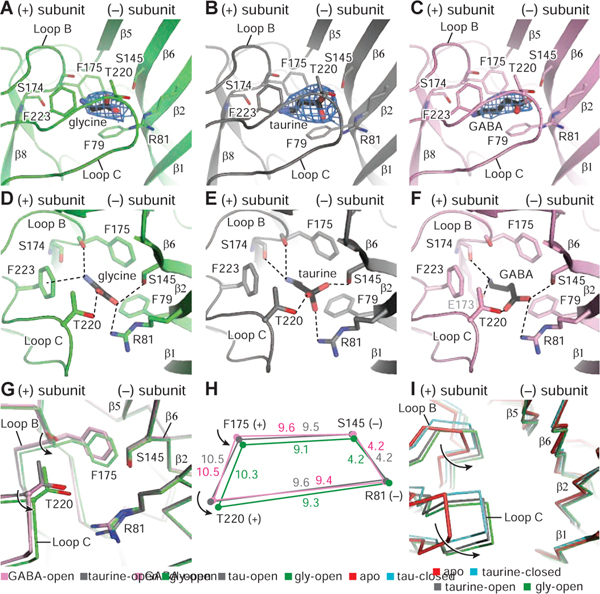
In all complexes, the densities for glycine, taurine and GABA are well defined, enabling positioning of the agonists in the neurotransmitter binding site (Figure 3A–3C). In the glycine complex, the agonist’s amino and carboxyl groups form multiple hydrogen bonding interactions and a cation-π contact (Figure 3D), consistent with the GlyR-α3 crystal structure (Huang et al., 2017b). While the orientations of taurine and GABA are similar to that of glycine, the interactions between the amino groups of the agonists and receptor residues are different (Figure 3E–3F) yet, as in the glycine complex, the sulfate group of taurine and the carboxylate group of GABA participate in multiple hydrogen bonds.
Figure 3. Neurotransmitter binding sites.
A-C. Illustration of the densities for glycine, taurine and GABA, contoured at 11 σ, 10 σ, and 8 σ, respectively. For the neurotransmitters, carbon and sulfate atoms are colored in black, whereas nitrogen and oxygen atoms are in blue and red, respectively.
D-F Glycine, taurine and GABA binding sites showing hydrogen bonds and cation-π interactions as dashed lines.
G. Illustration of conformational changes produced by the binding of glycine, taurine and GABA by superimposing the respective extracellular domains (ECDs).
H. Schematic diagram illustrating the changes in distances (angstrom) of key residues for the glycine, taurine and GABA-bound open states. The glycine, taurine and GABA structures are green, gray and pink, respectively.
I. Conformational changes of the binding pockets in the apo state, glycine-bound open, taurine-bound open and taurine-bound closed states by superimposing the main chain atoms of the ECDs.
See also Figures S8 and Table S6.
To identify the changes in the binding pocket upon agonist binding, we solved the structure of the apo/resting state in detergent micelles (apo-GlyREM; Figure S6, Table S4–S5). The apo-GlyREM structure has a pore constriction at 9’L, consistent with the apo-GluCl (Althoff et al., 2014), apo-GlyR in nanodisc (Kumar et al., 2020) and the taurine/GABA-bound GlyR closed states (Figure S1H). Comparison of the apo/resting state conformation of the agonist binding pocket with the open state/agonist-bound complexes shows that the binding pocket undergoes a ‘contraction’ where loop C transitions from an “uncapped” to “capped” configuration and loop B moves towards to the agonist (Figure 3I), as hypothesized for the muscle nicotinic acetylcholine receptor (Celie et al., 2004; Jadey and Auerbach, 2012; Mukhtasimova et al., 2009; Tripathy et al., 2019).
The extent of binding pocket contraction, however, is different between full and partial agonists, with the volume of the agonist binding pocket smallest in the glycine-bound structure (Hansen et al., 2005). Thr220 in loop C, Phe175 in loop B, Arg81 in β2, and Ser145 in β6 all coordinate taurine and GABA in a manner that is similar, yet slightly different than their coordination of glycine. In the glycine complex, Cα atoms of residues in loop C and in loop B ‘contract’ toward the agonist compared with the taurine-bound pocket (Figure 3G, 3H). We propose that these conformational changes are coupled to agonist-induced activation (Lynch, 2004), consistent with the notion that the agonist causes increased stabilization of the activated configuration (Changeux, 2012). Thus, in the full agonist-bound open state, loops B and C move closer to the ligand in comparison to partial agonists, resulting in a smaller binding pocket and a more efficient activation of the receptor (Figure 3H). Our structural data, together with agonist-dependent measurements on the nAChR, 5HT-3 and GABA receptor, show that full and partial agonist complexes of the open state of a pLGIC, within the confines of the agonist-binding site, are not identical and that full agonists yield a more ‘compact’ binding site in comparison to partial agonists. Indeed, the ligand volumes and distances spanning the binding site (DF175-S145 and DT220-R81) are correlated with agonist efficacy (Basak et al., 2018; Gharpure et al., 2019; Jayaram et al., 2012; Masiulis et al., 2019; Morales-Perez et al., 2016; Phulera et al., 2018; Polovinkin et al., 2018; Tripathy et al., 2019) (Table S6).
GABA acts on YGF mutant with a high Po
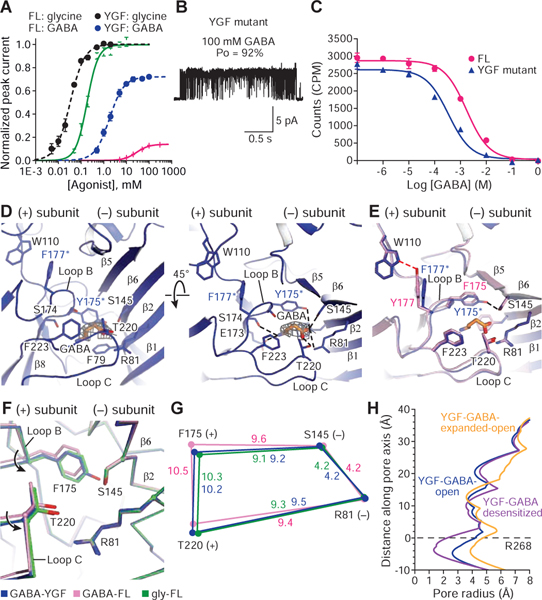
To test if the degree of contraction of the agonist binding pocket by partial agonists is correlated to agonist efficacy, we investigated the F175Y/Y177F variant (YGF) (Schmieden et al., 1993). Remarkably, on the YGF mutant GABA is an almost full agonist, with a Po of 92 ± 2% (n=18 clusters, Figure 4B) and is more potent in activating the channel or displacing strychnine (Figure 4A, 4C). Glycine remains a full agonist (Po of ~1.0; Figure 4A) and taurine has a Po of 95 ± 5% (n=26 clusters). We proceeded to solve the cryo-EM structure of the SMA-solubilized YGF mutant bound with GABA, obtaining high-resolution maps for the open, desensitized and expanded-open states (Figure 4H, Figures S3D, S6, Table S4). We did not observe any 3D classes associated with the closed state seen for partial agonists in FL GlyRs (Figure 5A).
Figure 4. YGF mutation renders GABA a high efficacy agonist.
A. Whole cell, dose response data for glycine and GABA for full length (FL) or the YGF mutant. EC50 for glycine at FL and YGF mutant are 190 ± 20 μM and 33 ± 3 μM, respectively. EC50 for GABA to FL and YGF mutant are 28.4 ± 0.9 mM and 1.05 ± 0.08 mM, respectively. Error bars represent SEM and n≥6 cells for all experiments. Responses are normalized to maximum responses to glycine in each cell.
B. Cell-attached single-channel recording showing openings of the YGF mutant produced by 100 mM GABA.
C. Competition ligand binding experiment using 3H-strychnine. Data are shown as means ± SEM (n=3). Ki of GABA to FL and YGF mutant are 1.41 ± 0.94 mM and 0.18 ± 0.07 mM, respectively. Kd of 3H-strychnine in the FL and YGF mutant are 220 ± 3 nM and 54 ± 4 nM, respectively.
D. GABA binding site in the YGF mutant viewed parallel to the membrane (left) or from the extracellular side of the membrane (right). GABA density is contoured at 0.013 σ. The possible hydrogen bonds and cation-π interactions are shown as dashed lines.
E. Superimposition of the binding pockets from the FL and YGF mutant in the presence of GABA to show the impact on the binding pocket of the swap between Y177 and F175.
F. Conformational changes in the binding pockets of the open states of GABA-bound YGF mutant and FL and glycine-bound FL shown by superposing the ECDs.
G. Schematic diagram illustrating changes in the distances in the binding pocket of the GABA-bound YGF mutant (blue) and FL (pink) as well as glycine-bound FL (green).
H. Plots for pore radius as a function of distance along the pore axis for GABA-bound YGF mutant in the open, desensitized and expanded-open states.
Figure 5. Particle distributions, binding pocket comparisons between GlyR and the α1β1γ2 GABA receptor-GABA complex and single channel open probabilities.
A. Fractions of GlyR particles in different states in the FL and YGF mutant, calculated by RELION and cisTEM. Error bars represent SEM.
B. Extracellular domain (ECD) of the open state of the GABA complex in SMA.
C. Superposition of the GABAAR (in grey; PDB code: 6DW1) and YGF mutant (in pink) structures highlights differences in the binding pockets. The Cα atoms of key residues are represented as spheres.
D. Schematic diagram illustrating the distances of the Cα atoms of R81, F175, S145 and T220 in the GlyR and corresponding residues in the GABAAR structure.
E. The open probabilities (Po) of the FL and the triple mutant (F115Y+F175Y+F223Y) elicited by 100 mM GABA. Error bars represent SEM and n≥6 cells for all experiments.
The increased binding affinity of GABA to the YGF mutant arises from additional interactions involving the carboxylate group of GABA, including a hydrogen bond with the hydroxyl oxygen of Tyr175 (Figure 4D). Superposition of the binding pockets of the FL and YGF mutant highlights other differences between interactions of receptor side chains. The mutation of Tyr177 to Phe disrupts the hydrogen bond formed with Trp110 through the hydroxyl oxygen of Tyr177, whereas the mutation of Phe175 to Tyr yields an additional hydrogen bond with Ser145 (Figure 4E). These interactions enable loop B to move closer to the (−) subunit, reducing the binding pocket volume similar to that of the FL glycine open complex (Figure 4F, 4G), thus supporting the notion that contraction of the agonist binding pocket is correlated to agonist efficacy.
To understand why GABA is almost a full agonist at the GABAAR but a partial agonist for GlyR, we also superimposed the binding pocket of GlyR with the canonical GABA binding site of the GABAAR (Mortensen et al., 2004; Phulera et al., 2018). Remarkably, most of the residues coordinating GABA in the two receptors are identical (Figure 5B–C). Moreover, the binding pocket of the GlyR open state has a smaller volume than GABA-bound GABAAR (Figure 5D), implying that the opening of GABAAR requires a smaller contraction of the binding pocket to fully transition the receptor to the open state. To explore if the simple substitution of the three tyrosine residues of GABAAR with the phenylalanine residues of GlyR enables GABA to act as a full agonist on the GlyR, we prepared the YYY mutant (F115Y/F175Y/F223Y) and found that GABA remained a weak partial agonist, with a similar Po in the YYY mutant as in the FL receptor (Figure 5C, E). Thus, the tuning of agonist efficacy requires precise changes in the pocket volume and in agonist-receptor interactions that go beyond which residues interact directly with the agonist (Mukhtasimova and Sine, 2018).
Partial agonists produce distinct ECD and ECD-TMD conformational changes
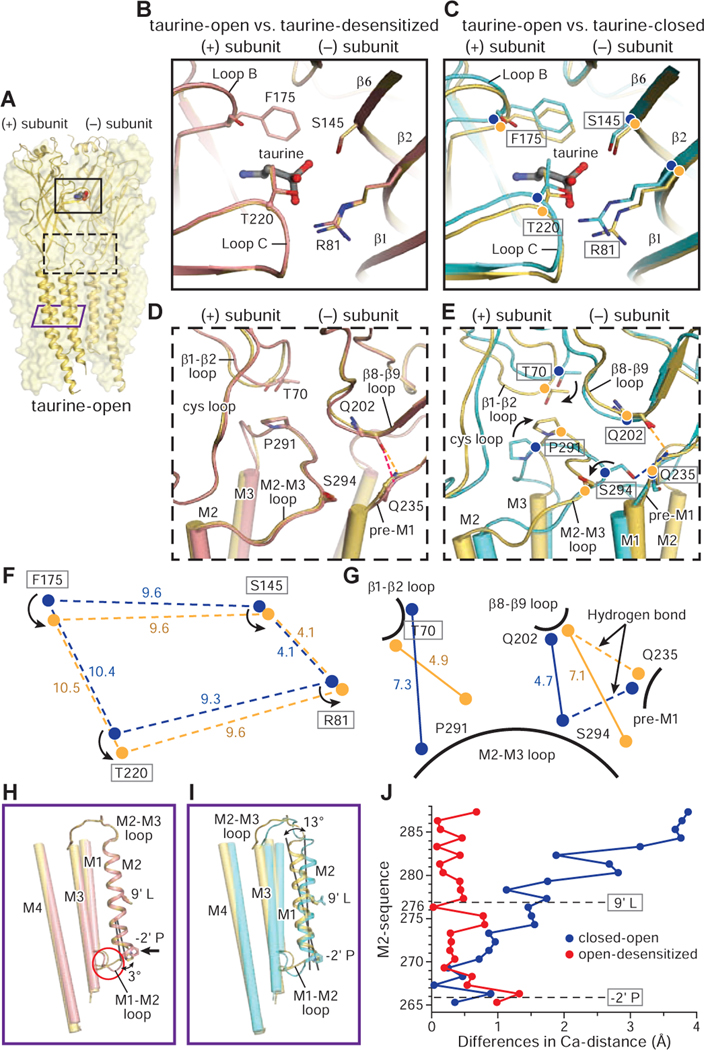
To understand the conformational changes associated with transition from the closed-to-open states, we used the well resolved, SMA-solubilized taurine-bound structures. Upon the closed-to-open transition, loop B and loop C move towards the agonist (Figure 6A, 6C, 6F) and residues in β6 and β2 shift away from the (+) subunit. Transduction of the conformational changes from the agonist binding pocket to the ECD-TMD interfaces include the repositioning of the β8-β9 loop by loop C and loop B, reshaping of β1-β2 loop by β2, reseating of the Cys-loop by β6 and loop B, and movement of pre-M1 by loop C (Figure 6E and Figure S1B). The M2-M3 loop acts as a “bridge”, forming contacts with the ECD and repositioning the M2 and M3 helices. From the taurine-closed to the taurine-open state, the M2-M3 loop moves away from the ion channel pore (Figure 6G). Pro291, positioned beneath Thr70, stabilizes the M2 and M3 helices in the taurine-open state, reminiscent of the position of Val45 and Pro268 in the GluCl in complex with ivermectin (Hibbs and Gouaux, 2011). Furthermore, in the taurine-closed state, the Ser294 to Gln235 hydrogen bond is ruptured during receptor activation, facilitating the outward movement of the M2 helix (Figure 6E, 6G). This action agrees with reports that the mutation of Ser294 to cysteine increases glycine EC50 by ~5 fold (Lynch et al., 2001).
Figure 6. Conformational changes between the open, desensitized and closed states.
A. Structure of the taurine-open state. Two subunits are shown in cartoon and the other three subunits are represented as partially transparent surfaces. Taurine is in sphere representation. The binding pocket in the ECD, the ECD-TMD interface and the TMD are indicated by a black solid, a black dashed and a purple solid rectangle, respectively.
B-G. Superposition of the (−) subunit illustrates the relative movements in the (+) subunit. Hydrogen bonds are shown in dashed lines. The taurine-open, taurine-desensitized and taurine-closed states are colored in yellow, salmon and cyan, respectively. (B. D) Comparison of the binding pocket (B) and ECD-TMD interfaces (D) between taurine-open and taurine-desensitized states. (C. E) Conformational changes in the binding pockets (C) and the ECD-TMD interfaces (F) between taurine-closed, taurine-open states. (F. G) Illustration of the changes in distances of the Cα atoms of key residues in the binding pockets (F) and ECD-TMD interfaces (G) between taurine-open and taurine-closed states. Cα atoms in the taurine-open and taurine-closed states are represented by yellow and blue balls, respectively.
H-I. Superimposition of a single subunit illuminates the changes in the TMD between taurine-closed, taurine-open and taurine-desensitized states. The M1, M3 and M4 helices are shown as cylinders while the M2 helix is shown in cartoon representation.
J. The plot of the differences in the position of Cα atoms from residues in the M2 helix derived from the taurine-open, taurine-desensitized and taurine-closed states. Two constriction sites are indicated by dash lines. From open to desensitized states, the major changes are concentrated at the lower half of the M2 helix with −2’P moving by 1.4 Å. By contrast, the M2 helix undergoes a larger conformational change upon transition from the closed to open states.
See also Figures S7–S8.
Comparison of the taurine-open to the taurine-desensitized state shows that the binding pockets and the ECD-TMD interfaces adopt the same conformation (Figure 6A, 6B, 6D). The transition from open to desensitized states thus involves local interactions in the ion permeation pathway (Auerbach and Akk, 1998; Plested, 2016), with desensitization proceeding from an uncoupling between the ECD and TMD (Zhang et al., 2013). In the ECD-TMD interfaces, a hydrogen bond between Gln235 and Gln202 stabilizes the receptor in the open and desensitized states (Figure 6D). Consistent with our results, ablation of this interaction, such as in the shaky startle mouse, reduces glycine potency (Janzen et al., 2017). While the overall conformational changes of the GABA and taurine structures are similar (Figure S7), in the taurine-closed state, the loop B and loop C are closer to the agonist, compared with the GABA-closed state (Figure S8F–S8H), providing structural insights into the differences between GABA and taurine in inducing the hypothetical flipped state of kinetic models, with the assumption that the taurine and GABA-bound closed states are not already flipped.
In comparison to pLGICs in closed/resting states (Basak et al., 2018; Du et al., 2015; Hilf and Dutzler, 2008; Huang et al., 2015; Miyazawa et al., 2003), the taurine-closed states illustrate how agonist binding promotes local rearrangements at the neurotransmitter binding site, prior to the overall changes associated with channel opening (Figure S8A–S8E, S8K–S8O). In the taurine-closed state, loop C is in a “capped” configuration as compared with the apo-GlyREM and apo 5-HT3AR structures, due to the binding of agonist (Figure S8B–C, S8L–M). By contrast, the ECD-TMD interfaces of the taurine-closed state and apo-GlyREM are similar, with identical conformations for M2-M3, β1-β2, β8-β9 and Cys loops (Figure S8D, S8N), showing that local movements of loop C alone cannot induce substantial conformational changes that affect the receptor ECD-TMD interfaces as well as the TMDs. The 5-HT3AR ECD-TMD interface shows substantial differences compared with the taurine/GABA-closed state (Figure S8E, S8O), indicating the contraction of the binding pocket around the agonist and the induced changes in the ECD-TMD interface are unique for each pLGIC.
Conformational changes of the ion channel pore
In the transition from the closed to the open states, the TMD of each subunit undergoes a counterclockwise rotation of ~8.6 °, expanding the pore and opening the ion channel, allowing for Cl- permeation (Figure 6A, 6I, 6J and Figure S8I). Within the M1-M4 helices, the M2 helices have a more pronounced movement, with a rotation of ~13°. During the transition from the open to the desensitized state, the major conformational changes are in the TMD, with each subunit undergoing a clockwise rotation by ~2° (Figure S8J). The lower half of the pore is occluded at - 2’P, moving by ~1.4 Å, in agreement with observations in the homomeric GlyR and GABAA receptor, and the α4β2 nicotinic receptor in the desensitized state (Kumar et al., 2020; Laverty et al., 2019; Masiulis et al., 2019; Walsh et al., 2018) (Figure 6A, 6H, 6J). In this process, the M2 helix exhibits a larger motion, with an inward rotation of ~3°. The M1-M2 loop undergoes a relatively large displacement during desensitization, in agreement with studies showing that mutation of the M1-M2 loop affects entry into desensitization (Breitinger et al., 2004; Gielen et al., 2015).
Partial agonist gating mechanism
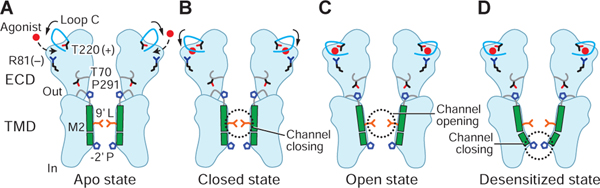
We have captured partial agonist-bound closed states, which we hypothesize are poised between the resting and the open/desensitized states, thus enabling visualization of intermediate or preopen-shut states of the ion channel, in accord with partial agonist-bound, long-lived shut states in the single-channel recordings (Figure 2A). Binding of a partial agonist induces a contraction in the binding pocket, yet to a lesser extent than the full agonist (Figure 7A–C). These ECD changes promote the receptor transition from closed-to-open, in which the TMD undergoes an outward rotation and expands the channel pore at the −2’P and 9’L positions (Figure 7C). Sustained binding of agonist promotes a further transition into the desensitized state, involving an inward rotation of the TMD and occlusion of the pore at −2’P (Figure 7D). Because of the structural diversity of ligands and the complexity of functional behavior of each pLGIC, the precise details of the contraction of the binding site may be specific to each subgroup of the superfamily. Nevertheless, we hypothesize that the agonist-bound closed, open and desensitized states link efficacy to the contraction of the agonist binding site, thus providing a speculative structural proposal for partial agonist gating of GlyR and of pLGICs as a whole.
Figure 7. GlyR gating mechanism.
The ECD changes promote the transition from closed to the open state (A-C), while sustained binding of partial agonist promotes a further transition into the desensitized state (C-D).
STAR★METHODS
RESOURCE AVAILABLITY
Lead Contact
Further information and requests for materials should be directed to the Lead Contact, Eric Gouaux (gouauxe@ohsu.edu).
Materials Availability
We are glad to share the associated plasmids in this work upon completion of a Material/Data Transfer Agreement for non-commercial usage. This study did not generate new unique reagents.
Data and Code Availability
The data that support the findings of this study are available from the corresponding author upon request. The following Protein Data Bank (PDB) and Electron Microscopy Data Bank (EMDB) accession codes are provided: 6PM6 and 20389 (GlyR-gly-open-SMA), 6PM5 and 20388 (GlyR-gly-desensitized-SMA), 6PM4 and 20386 (GlyR-gly-expanded-open-SMA), 6PM3 and 20385 (GlyR-taurine-closed-SMA), 6PM2 and 20384 (GlyR-taurine-open-SMA), 6PM1 and 20383 (GlyR-taurine-desensitized-SMA) 6PM0 and 20382 (GlyR-taurine-expanded-open-SMA), 6PLZ and 20381 (GlyR-GABA-closed-SMA), 6PLY and EMD-20380 (GlyR-GABA-open-SMA), 6PLX and 20379 (GlyR-GABA-desensitized-SMA), 6PLW and 20378 (GlyR-GABA-expanded-open-SMA), 6PLO and 20370 (YGF-GABA-open-SMA), 6PLP and 20371(YGF-GABA-desensitized-SMA), 6PLQ and 20372 (YGF-GABA-expanded-open-SMA), 6PXD and 20518 (apo-GlyREM), 6PLR and 20373 (GlyR-gly-desensitized-nanodisc), 6PLT and 20375 (GlyR-taurine-desensitized-nanodisc), 6PLS and 20374 (GlyR-GABA-closed-nanodisc), 6PLV and 20377 (GlyR-GABA-desensitized-nanodisc), and 6PLU and 20376 (GlyR-GABA-closed-nanodisc).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines-SF9 cells (CRL-1573) were cultured in sf9–900 III SFM (ThermolFisher Scientific) at 27 °C. These cells were used for expression of Baculovirus and receptor. They are female in origin. Human embryonic kidney 293A cells (HEK293A; ATCC, Teddington, UK, Cat. No. PTA-4488, RRID:CVCL0045) were used for transient expression of GlyRs. HEK293A cells were cultured in 25 cm2 vented culture flasks containing 5 ml of Dulbecco’s modified Eagle’s medium (DMEM; Gibco Thermo Fisher, Loughborough, UK, Cat. No. 41966–029) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Gibco Cat. No. 10500–064), 2 mM glutamine (Invitrogen) and 100 units/ml penicillin/ 100 microg/ml streptomycin (Gibco Cat. No. 15140–122). For expression, cells were plated on poly-L-lysine–coated glass coverslips (Sigma-Aldrich and VWR, respectively) in 35-mm culture dishes (Scientific Laboratory Supplies) containing 2 ml of DMEM, and then transfected via the calcium phosphate- precipitation method with pcDNA3.1 plasmids coding for GlyRs. Cells were washed 5–16 hrs later and electrophysiological experiments were performed 1–2 days after transfection.
METHOD DETAILS
Protein purification
The cDNA encoding the full-length zebrafish α1 glycine receptor (GlyR, NP_571477), with the following modifications, was cloned into the pFastBac1 vector for baculovirus expression in Sf9 insect cells (Du et al., 2015). We used two GlyR constructs for the studies described here. For all of the experiments, other than the apo state cryo-EM studies, we used the full length GlyR modified only by the addition of a thrombin site (Leu-Val-Pro-Arg-Gly-Ser) and an octa-histidine tag at the carboxy terminus. We refer to this construct as the full length (FL) GlyR. For structure determination of the apo state, we employed the previously described GlyREM construct in which the M3-M4 loop was replaced by an Ala-Gly-Thr tripeptide (Du et al., 2015). The YGF mutant, where residues Phe175 and Tyr177 were swapped, was generated from FL using site-directed mutagenesis.
Following transduction of insect cells with baculovirus and cell culture at 27°C for 72 hours, cells of the FL receptor and apo-GlyREM were collected and resuspended in a buffer composed of 20 mM Tris pH 8.0 and 150 mM NaCl (TBS) in the presence of 0.8 μM aprotinin, 2 μg/ml leupeptin, 2 mM pepstain A and 1 mM phenylmethylsulfonyl fluoride and disrupted by sonication. Membranes were isolated by centrifugation for 1 h at 200,000 g. For the FL receptor, membrane fraction was collected and solubilized in 20 mM n-dodecyl-β-D-maltopyranoside (C12M) and 2.5 mM cholesteryl hemisuccinate Tris salt (CHS) for 2 hours with slow stirring at 4°C. After ultracentrifugation to clarify the solution, the supernatant was collected and incubated with Talon resin for 1 hour and washed with TBS buffer supplemented with 1 mM C12M, 0.25 mM CHS and 35 mM imidazole. The receptor was eluted from the affinity resin using a buffer containing 250 mM imidazole. Fractions were pooled togethe3r and applied to a size-exclusion chromatography (SEC) Superose 6 10/300 GL column for further purification using a TBS buffer supplemented with 1 mM C12M and 0.25 mM CHS. The peak fractions were collected and concentrated to ~16 μM receptor concentration for reconstitution into lipid nanodiscs studies.
For the apo-GlyREM, to remove unbound and bound glycine to enable determination of the apo-GlyREM structure, all the buffers were prepared using HPLC water (Sigma) and extensive dialysis was carried out as follows. The pelleted membrane with a volume of 30 ml was homogenized and incubated with 100 nM strychnine. The strychnine saturated membranes were transferred to an 8–10 kDa molecular weight cut off cellulose ester dialysis tubing (Spectra/PorBiotech) and dialyzed in 200-fold volume of glycine-free buffer containing 150 mM NaCl, 20 mM Tris 8.0 (TBS buffer in HPLC water) in the presence of 1 mM PMSF, 0.8 μM aprotinin, 2 μg/ml leupeptin, and 2 mM pepstatin A. Buffer exchange was carried out twice a day over the course of five days at 4°C, resulting in 10 changes and an estimated dilution of the original buffer by 1020 fold. The purification strategy was the same as that for the FL receptor. The peak fractions after SEC were used for cryo-EM studies.
Nanodisc reconstitution
To incorporate the receptor in lipid nanodiscs (Denisov and Sligar, 2016), brain total lipid extract (BTL, Avanti) dissolved in chloroform was dried down using a rotary evaporator and resuspended in a TBS buffer supplemented with 20 mM C12M and 2.5 mM CHS to a final lipid concentration of 10 mM. Because we estimate that the diameter of the GlyR transmembrane domain is ~80 Å, we used the MSP1E3D1 scaffold protein which, in turn, yields nanodiscs with an outer diameter of ~12 nm. MSP1E3D1 was purified through a metal chelate affinity column (Ni-NTA) and extensive dialysis against TBS buffer (Alvarez et al., 2010). FL receptor was mixed with MSP1E3D1 and BTL in a final ratio of 1: 7.5: 200 (molar ratios) and incubated for 40 min by gentle agitation at 4°C. Detergent was removed by adding Bio-Beads (SM2, Bio-Rad) to a final concentration of 150 mg/ml. After incubation for 2 hours, the Bio-Beads were replaced with fresh beads. The mixture was incubated overnight at 4°C. After separation from Bio-beads, the protein was loaded onto a Superose 6 10/300 GL column equilibrated with TBS buffer. Peak fractions corresponding to reconstituted GlyR were collected for cryo-EM analysis and scintillation-proximity assay (SPA) (Nelson, 1987).
We reconstituted the detergent purified receptor into nanodiscs in the presence of saturating solutions of glycine, taurine or GABA. For the full agonist (glycine), the cryo-EM data set yielded a reconstruction (GlyR-nanodisc-gly) with a single 3D class at 3.2 Å resolution (Figures S1C–S1D, S5 and S7, Table S1). The receptor features are well resolved and there is clear density for lipids and the membrane scaffold protein (MSP). Within the receptor transmembrane domain (TMD), at the 9’L and −2’P constriction sites on the M2 helix, the pore has a radius of 4 Å and 1.5 Å, respectively, suggesting that the 9’L gate is ‘open’ and the −2’P gate is closed, consistent with the receptor occupying a desensitized-like conformation (Figure 1C, Figure S1C–S1D).
For each partial agonist taurine and GABA, three-dimensional classification of the nanodisc cryo-EM data sets, reveals 2 distinct classes, respectively, with overall resolutions of 2.8 to 3.3 Å and local resolutions as high as 2.5 Å in the extracellular domain (ECD) (Figures S4–S5, Tables S1 and S5, Methods S1). One of the two structures is reminiscent of the glycine-bound desensitized state. Another one is partial agonist-bound closed state with M2 helices are approximately perpendicular to the membrane, similar as the closed state observed in the SMA data sets (Figure S1F–S1H). We did not observe the open and super-open states as in the nanodisc data sets. Because we did not observe any open states in the nanodisc-derived cryo-EM data sets, we focused our attention on the SMA-isolated receptor.
Protein purification via styrene maleic acid (SMA) copolymer solubilization
The SMA copolymer XIRAN 30010 (~2.3:1 molar ratio of stryrene: maleic acid) was purchased from Polyscope as an aqueous solution of 20% (w/v) SMA. The purification procedures in the context of the SMA copolymer are similar to those previously described except for the following differences. Following collection of the membrane fraction by centrifugation, the membranes were resuspended in TBS buffer and XIRAN 30010 was added to a final concentration of 0.5% with slow stirring at 4°C for 1 h. After ultracentrifugation for 1 hr at 200,000 g, the supernatant was collected and incubated with NTA metal ion affinity resin for 5–6 hours. The remaining purification steps were carried out as described above with the caveat that no additional SMA or detergents were added throughout the remaining purification steps. Following SEC chromatography, the FL/YGF mutant-SMA complex was concentrated to 2 mg/ml for cryo-EM analysis.
Determination of glycine contamination
Potential glycine contamination in 50 mg/ml stocks of taurine or GABA was analyzed using liquid chromatography tandem mass spectrometry (LC-MS/MS) with electrospray ionization in positive mode. MRM (multiple reaction monitoring) transitions monitored specific product ion fragment from glycine. The reported major fragments were 75.9/48 and 75.9/30.2. Standards spiked with 0.1 μg/ml to 50 μg/ml glycine in the presence of a constant concentration of 50 mg/ml of GABA or taurine were prepared as a control to test assay sensitivity. The glycine contents in the 50 mg/ml stocks of taurine or GABA were below the lower limit of detection, 0.1 μg/ml. These experiments allowed us to estimate that the extent of glycine contamination was less than 1.33 μM of glycine in 0.4 M of taurine or 0.49 M of GABA.
Ligand binding assay
Competition ligand binding experiments were carried out using the SPA (Nelson, 1987) and a solution containing 40 nM FL receptor or YGF mutant reconstituted into the SMA, 1 mg/ml Cu-Ysi beads, 0.1% bovine serum albumin, TBS buffer and 100 nM 3H-labelled strychnine (1:9 3H: 1H). The stocks of cold competitors - glycine, taurine and GABA – were prepared in TBS at final concentrations of 0.1M, 0.5M and 1M respectively, followed by serial dilution. To estimate the extent of non-specific binding, 250 mM imidazole was used for background subtraction. The binding reactions reached equilibration after an overnight (<12 hr) incubation and the scintillation counts from the beads were read on a Micro Beta TriLux 1450 LSC & Luminescence Counter (Perkin Elmer). Ki values were determined with the Cheng–Prusoff equation in GraphPad Prism.
Cryo-EM sample preparation and data acquisition
To prepare the GlyR complexes with different agonists, the FL receptor complexes in nanodisc or SMA were mixed with 10 mM glycine, 20 mM taurine or 40 mM GABA immediately before the samples were placed onto the grids. Apo-GlyREM sample was placed onto the grid directly. Preparation of the YGF mutant complexes with 40 mM GABA followed the same procedures as with the FL type receptor. We estimate that the time elapsed between mixing of receptor with agonist and plunge freezing of the grids was around 10 s. In addition, the extent of glycine contamination in the taurine and GABA chemicals was estimated as less than 1.33 μM in 0.4 M of taurine or 0.49 M of GABA, as described above. To minimize preferred orientation, the nanodisc-reconstituted receptor solutions were supplemented with 60 μM perfluoro octyl maltoside (FOM) prior to grid preparation. A 2.5 μL sample of receptor was applied to glow-discharged Quantifoil R1.2/1.3 or 2/2 200 mesh gold holey carbon grids which were then blotted for 2.5 s under 100% humidity at 12 °C. The grids were then flash-frozen in liquid ethane, maintained at melting temperature, using a FEI Mark IV cryo-plunge instrument.
Cryo-EM datasets for the receptor-nanodisc complexes were collected on a Talos Arctica microscope (FEI) operated at 200 kV. Images were acquired on a K2 Summit direct-detector (Gatan) using super-resolution mode at a magnification of x45,000, corresponding to a pixel size of 0.455 Å. The defocus range was set from −0.8 μm to −2.2 μm. Each micrograph was recorded over 100 frames at a dose rate of ~5.49 e−/pixel/s and a total exposure time of 10 s, resulting in a total dose rate of 61 e−/Å2.
For the FL receptor complexes in SMA, all the datasets were collected on a 300 kV FEI Titan Krios microscope equipped with an energy filter (Gatan Image Filter) set to 20 eV. Images were acquired on a K2 Summit direct-detector positioned after the energy filter using super-resolution mode at 165,000 magnification and a pixel size of 0.412 Å. Images were collected using ‘multishot’ methods driven by SerialEM (Mastronarde, 2005) with seven shots per hole, utilizing 2/2 200 mesh grids and a defocus ranging from −1.0 μm to −2.0 μm. Each micrograph was recorded over 40 frames with a total dose of 42 e−/Å2. For the YGF mutant in complex with GABA, the dataset was collected on the same Titan Krios microscope as the FL, but images were acquired on a K3 BioQuantum direct-detector using super-resolution mode at 135,000 magnification with a pixel size of 0.65 Å. Each micrograph was recorded over 60 frames at a dose rate of ~18.76 e−/pixel/s and a total exposure time of 1.5 s, resulting in total dose rate of 62 e−/Å2.
The data for apo-GlyREM was collected on a 300 kV FEI Titan Krios at the cryo-EM facility at HHMI’s Janelia Research Campus. This microscope is equipped with a CETCOR Image Corrector for spherical aberration correction and an energy filter. A 20eV energy slit centered on the zero-loss peak, and a Cs of 0.01 mm were used during data collection. All micrographs were recorded on a Gatan K2 Summit direct electron detector operated in super-resolution counting mode with a pixel size of 1.04 Å. The dose-fractionated images were recorded using the automated acquisition program SerialEM (Mastronarde, 2005). Each micrograph was recorded over 30 frames with a total exposure time of 6 s, resulting in a total dose rate of 54 e−/Å2.
Image processing
Super-resolution image stacks were 2×2 binned in the Fourier space and motion corrected by Motion Cor2 (Zheng et al., 2016). The contrast transfer function (CTF) parameters were estimated by Gctf (Zhang, 2016) and particles were picked by DoG-picker (Voss et al., 2009) for all the datasets except where indicated.
We first analyzed the taurine-bound GlyR-nanodisc complex data, culling through the total micrographs and retaining 1,264 micrographs that had Thon rings extending to at least 4 Å. Approximately 669,253 particles were picked from these micrographs and subjected to 2D classification in RELION 2.1 (Scheres, 2012). After several rounds of classification, 2D classes with clearly defined features of the glycine receptor contained 244,309 particles. This particle stack was then employed for subsequent analysis. An initial model generated from cryoSPARC (Punjani et al., 2017) was then used for 3D classification and refinement. A soft mask with extending binary map of 4 pixels and a soft-edge of 6 pixels and C5 symmetry were applied during all the following steps of classification and refinement. Two of the 3D classes (class 1 and class 3) showed reasonable secondary structure features and were subjected to 3D refinement, yielding two different maps belonging to what we define as the desensitized and closed states, respectively. To reduce heterogeneity among these two classes and improve the density in the transmembrane regions, class 1 and class 3 were re-classified into three classes. For class 1, this resulted in an additional class 1a (45,300 particles) and class 1b (79,557 particles) with the same transmembrane helix orientation and density. We therefore combined these classes and carried out 3D refinement, which yielded a map that we identify as the taurine-bound desensitized state. For class 3, the resulting class 3b contained only 15,292 particles, albeit with a better density in the transmembrane region. This class was selected for the 3D refinement, yielding a map that we identify as the taurine-bound closed state (Figure S4, Methods S1).
A similar procedure was used to process data associated with the GABA-bound receptor in nanodiscs. Briefly, 493,873 particles were auto-picked from 1,109 micrographs and were subjected to 2D and then 3D classification. Two different classes containing 39,926 and 38,383 particles were selected for 3D reconstruction and 3D auto-refinement respectively, giving rise to one map that we identify as the GABA-bound desensitized state and a second map that we believe represents the GABA-bound receptor in a closed state (Figure S5).
The single particle cryo-EM data for the glycine-bound receptor in nanodiscs was processed using similar procedures. To do this, 472,910 particles were auto-picked from 1,231 micrographs and subjected to 2D and 3D classification. Notably, the 3D classes shared the same overall shape and transmembrane helix orientation, suggesting the preponderance of one conformational state in this glycine-bound data set. Three classes (80,121 particles) with good secondary structure features were combined and used for 3D auto-refinement, yielding a map that we define as the glycine-bound desensitized state (Figure S2).
For the glycine bound receptor in SMA, a total of 5309 micrographs were retained following the same criteria as above. A total of 1,578k particles were picked by DoG-picker. RELION 3.0 (Zivanov et al., 2018) was used for 2D classification, 3D auto-refinement and 3D classification. 4× binned particles were extracted followed by one round of 2D classification by limiting the resolution to 8 Å. After this round of 2D classification, only the classes with clear receptor features and clean background were selected. 804k ‘good’ particles were selected and 2× binned particles were re-extracted followed by one round of 3D refinement using a filtered map and an initial model derived from the previous structure determination of GlyR (PDB code: 3jaf). Additional 3D classification was performed using the refined 2× binned particles by applying C1 or C5 symmetry with alignment with the angular sampling interval of 15° for 25 iterations. The 3D classifications with C1 symmetry yielded similar classes as with C5 symmetry, albeit with slightly weaker TM density, thus justifying use of C5 symmetry. The 172k particles belonging to the dominant class 6 were selected and unbinned particles were re-extracted and re-centered followed by another round of 3D refinement. To facilitate the further 3D classification, a mask which includes as much as GlyR density while excluding as much as SMA density as possible was designed. Using the refined unbinned particle stack along with the mask, 6 classes were generated during the 3D classification without alignment until convergence, after 170 iterations. Based on the differences of the M2 helices in these classes, 84,978, 155,456 and 39,586 particles belonging to the putative open state (class 6a), the desensitized state (class 6b), and the ‘expanded-open’ state (class 6c), respectively, were re-extracted for 3D auto-refinement (Figure S2).
Similar procedures were performed for the taurine-bound SMA data set. Here, 5,004 movies yielded 1,124k particles that were automatically selected using template based particle picking in RELION 3.0 (Zivanov et al., 2018). 4× binned particles were extracted followed by one round of 2D classification. 543k ‘good’ particles were retained and unbinned particles were extracted. After 3D classification, four different classes containing 17,873, 17,271, 122,322 and 74,365 particles were isolated and were used in 3D auto-refinement, giving rise to the density maps of the taurine-bound closed, taurine-bound desensitized, taurine-bound open and taurine-bound expanded-open states, respectively (Figure S4, Methods S2). For the GABA-bound complex in SMA, a total of 2,062k particles were extracted from 5,200 micrographs and were subjected to 2D and 3D classification, and then the classes without obvious receptor features were dumped. Four different classes containing 121,470, 150,199, 20,845 and 31,667 particles were selected for 3D reconstruction and 3D auto-refinement respectively, giving rise to four maps that we identify as expanded-open, open, desensitized and closed states, respectively (Figure S5). For the GABA-bound YGF mutant complex in SMA, a total of 1,800k particles extracted from 9,300 micrographs were subjected to 2D and 3D classification. The resulted 407k particles produced two classes containing 32,386, 27,496 and 42,097 particles that we identified as open, desensitized and expanded-open states, respectively (Figure S6).
To further explore the particle fractions in all of the SMA data sets of the FL GlyR complexes, we also used cisTEM (Grant et al., 2018; Zhang et al., 2019) to perform the 3D classification following the procedure and using the parameters as described below. We first created the particle stacks after 2D classification by RELION 3.0 (Scheres, 2016; Zivanov et al., 2018) and then imported these particle stacks into cisTEM. A subsequent auto 3D refinement was performed in cisTEM. Based on the cisTEM refined stacks, we then created a new stack by specifying 15 3D classes. A focused refinement without alignment targeting the TMD was performed with the specified center of 131,131,145 (in Å) located at the TMD, a radius of 36 Å and high resolution limit of 5 Å for 30 iterations. Shown in Table S7 are the results of 3D classification using RELION 3.0 and cisTEM.
For the apo-GlyREM image processing, approximately 1,000 particles were manually picked for an initial reference-free 2D classification. Seven to eight representative 2D class averages were selected as templates for automated particle picking using RELION 2.1 (Scheres, 2012) or GAUTOMATCH (www.mrc-lmb.cam.ac.uk/kzhang/Gautomatch/). The auto-picked particles were visually checked and false positives were removed, giving rise to 204,323 total particles. The particles were further cleaned-up by 2D classification using RELION 2.1 (Scheres, 2012). The reconstruction of the published gly-EM complex (EMD-6345) was low-pass filtered to 60 Å and used as an initial model. No symmetry was applied during 3D classification in RELION 2.1 (Scheres, 2012). Classes with characteristic features of pLGICs were then selected. Particles belonging to the chosen classes, with a number of 115,864, were used for 3D auto-refinement (Figure S6).
To improve the density in the transmembrane region, all the refined maps from RELION 2.1/3.0 (Scheres, 2012; Zivanov et al., 2018) were further refined using auto refinement in cisTEM (Grant et al., 2018), yielding well-defined maps. The final reconstructions for the receptor in the nanodiscs were subjected to post-processing in RELION 2.1 ( Scheres, 2012 ), and for the reconstructions of the receptor in SMA the maps were sharpened by Localscale (Jakobi et al., 2017). Local resolution was determined by RELION 3.0 (Zivanov et al., 2018). The reported resolutions in the Table S1 and Table S2 were estimated by gold standard FSC 0.143 criteria (Chen et al., 2013).
Model building
The model building commenced with rigid body fitting of an initial model derived from the prior GlyR structures to each density map followed by real space refinement in Coot (Emsley and Cowtan, 2004) and by using Phenix (Afonine et al., 2018). To build all of the desensitized protein structures, the taurine-bound desensitized state structure in nanodisc (taurine_desensitized_nanodisc) was first built and then was used as the initial structure for the other desensitized state complexes. We began by fitting the glycine/ivermectin-bound protein structure (PDB code: 3jaf), excluding the ligands, into the taurine-bound desensitized state map using UCSF Chimera (Pettersen et al., 2004). The resulting structure was manually adjusted in Coot, guided by well resolved side chain densities. Amino acid residues missing from the previously determined glycine/ivermectin protein structure were added, including H327 at the end of the M3 helix and E390, M391 and R392at the beginning of the M4 helix. Prominent tube-shaped electron density surrounding the receptor transmembrane domain (TMD) likely arises from ordered lipid molecules and thus alkane chains with appropriate lengths were placed in the density features. The structure was further refined using Phenix in the context of restraints for stereochemistry of the alkane chains and carbohydrate groups. After Phenix refinement, the map to model cross correlation (cc) value between the final model and the map was 0.83. To build the other desensitized state structures, the above determined structure taurine_desensitized_nanodisc was fitted to the density maps using UCSF chimera. After rigid-body fitting of the taurine_desensitized_nanodisc model, taurine was replaced with the appropriate ligands, alkane chains were either replaced with appropriate lengths in these nanodisc structures or removed in the SMA structures, and the models were then real-space refined in Coot and Phenix. Specifically, two amino acids - V388 and E399 - were added at the beginning of the M4 helix in these protein structures of the desensitized state in SMA.
The procedures to build the open/expanded-open state protein structures were similar to the model building of the desensitized protein structures. The glycine-bound open state protein structure in SMA (glycine_open_SMA) was achieved by fitting the glycine-bound desensitized protein structure in SMA (glycine_desensitized_SMA) to its corresponding map using UCSF Chimera. Coot was then used to manually adjust the glycine_desensitized_SMA structure, mainly focusing on the TMD, followed by Phenix refinement, yielding a map to model cc of 0.80. To build the taurine-bound and GABA-bound open/expanded-open state protein structures derived from the receptor-SMA complex, the glycine_open_SMA structure was simply fit to the corresponding maps using UCSF Chimera, following by fitting of the appropriate ligands to the density in the neurotransmitter binding pocket. Phenix real space refinement was next performed to refine the protein structures within their corresponding density maps.
To build the GABA-bound YGF mutant open, desensitized and expanded-open states, the two amino acids 175F and 177Y were firstly mutated into 175Y and 177F by Coot, respectively, within their corresponding FL GABA-bound structures. After Phenix refinement, the resulted map to model cc values were 0.80, 0.79 and 0.72 for YGF mutant open, desensitized and expanded-open states, respectively.
For model building of the taurine-bound closed state in nanodisc (taurine_closed_nanodisc), we found that the previously determined strychnine-bound structure (PDB code: 3jad) fit the map well and thus it was used as an initial model. The structure was fit to the taurine-bound closed state map using UCSF Chimera. Chain A of the superimposed model was further optimized by real space refinement using Coot, including fitting of a taurine molecule to the density in the neurotransmitter binding pocket, adding Q326 and H327 at the end of the M3 and E390, M391 and R392 at the beginning of M4 helices, manually building the M2-M3 loop and adding lipid-derived alkane chains surrounding TMD. The model generated in Coot was further refined against the density map by Phenix. The cc between the taurine_closed_nanodisc and its corresponding map was 0.81. To build the other state protein structures either in nanodisc or SMA, the Phenix refined taurine_closed_nanodisc coordinates were used as the initial structure. Coot was used to change the ligands and remove the alkane chains in SMA structures or replace with appropriate chain lengths in nanodisc structures based on the density features, followed by Phenix refinement. All of the final models have good stereochemistry as evaluated by molprobity (Chen et al., 2010) (Tables S1–S4). All the figures were prepared by Pymol (Schrödinger, LLC) and UCSF chimera (Pettersen et al., 2004). Pore radii were calculated using program HOLE (Smart et al., 1996).
Patch-clamp and single-channel recording
Heterologous expression in human embryonic kidney cells.
Full-length zebrafish α1 GlyR was subcloned into the pcDNA3.1 for expression in the human embryonic kidney 293A cells (HEK293A). Point mutations were made using site-directed mutagenesis. HEK293A cells were transiently transfected using the calcium phosphate-DNA precipitation method. The DNA mix consisted of pcDNA3.1 plasmids with inserts coding for the enhanced Green Fluorescence Protein (eGFP; GenBank accession number U55763), and GlyR (FL or mutant). Plasmid without an open reading frame (“empty”) was added to the transfection mix to optimize the expression level (Groot-Kormelink et al., 2002). The final mixture of cDNA contained 2% α1, 18% eEGFP and 80% empty pcDNA3.1 plasmid. Cells were washed 5–16 hrs later and electrophysiological experiments were performed 1–2 days after transfection.
Patch-clamp recordings and analysis.
Macroscopic and single-channel currents were recorded from transfected HEK293A cells in the whole-cell, outside-out and cell-attached patch-clamp configurations at 20ºC. Patch electrodes were pulled from thick-walled borosilicate glass capillary tubes (GC150F-7.5; Harvard Apparatus) on a Flaming/Brown puller (Model-P97, Sutter Instruments) and were fire-polished to give a final resistance of 3–5 MΩ for whole-cell recordings and 5–15 MΩ for cell-attached or outside-out recordings (when filled with the appropriate internal solution).
Cells were bathed in an extracellular solution consisting of (in mM): 20 Na D-gluconate, 112.7 NaCl, 2 KCl, 2 CaCl2, 1.2 MgCl2, 10 HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid), 40 glucose (all Sigma) and 10 tetraethylammonium chloride (TEACl) (Alfa Aesar); the pH adjusted to 7.4 with NaOH. For the cell-attached recordings, pipettes were filled with extracellular solution containing the required concentration of agonist. To record macroscopic currents, the pipette was filled with a 30 mM chloride intracellular solution, containing (in mM): 101.1 K gluconate, 1 ethylene glycol tetraacetic acid (EGTA), 1 CaCl2, 1 MgCl2, 10 HEPES, 2 MgATP, 40 sucrose, 6 KCl (all Sigma) and 20 TEACl, the pH adjusted to 7.4 with NaOH. All solutions were prepared in bi-distilled water or, for cell-attached single channel recordings, in high-performance liquid chromatography grade (HPLC) water (VWR Chemicals), and filtered through a 0.2 μm cellulose nitrate membrane filter (Whatman) before use. Glycine, taurine or GABA-containing solutions were prepared by diluting a 1 M glycine, 1 M taurine or 1 M GABA stock in extracellular solution.
Taurine and GABA were purified by 2x recrystallization from aqueous ethanol and subsequently tested for glycine contamination by an HPLC assay: samples were resuspended in 10 μl 50% EtOH, reacted for 30 min with 90% EtOH:triethylamine:phenylisothiocyanate (7:2:1), evaporated to dryness at room temperature and redissolved in 100 μl 5% acetonitrile in 0.1 M ammonium acetate. A 150×4.6 mm Hypersil C18 column was used and detection was monitored at 254 nm. The purified taurine and GABA contained no detectable glycine, which in this assay is less than 1 part in 10,000.
Whole-cell recording.
Whole-cell currents were recorded from isolated transfected cells held at a holding potential of −40 mV (corrected for the junction potential of 11 mV) with an Axopatch 200B (Molecular Devices) and filtered at 5 kHz by the amplifier’s low pass 4-pole Bessel filter. Recordings were digitized with a Digidata 1440A (Molecular Devices) at a sampling frequency of 20 kHz and acquired to PC using Clampex 10.2 software (Molecular Devices) for offline analysis.
Agonist solutions were applied to the cells for approximately 1–2 s by a U-tube application system (Krishtal and Pidoplichko, 1980), with a 10–90% exchange time <1 ms (as tested by application of 50% diluted extracellular solution to the open tip pipette). The saturating agonist concentration was applied every third or fourth application to check response stability throughout the recording. Only cells where the test response rundown was less than 30% during the experiment were accepted for further analysis. No correction for rundown was applied. Different agonist concentrations were applied in random order to obtain concentration-response curves. The peak current amplitudes for each glycine application were measured using Clampfit 10.2 software (Molecular Devices). The concentration-response data for each cell were then fitted with the Hill equation using the CVFIT programme (DCPROGS; https://github.com/DCPROGS/CVFIT) to estimate EC50 (agonist concentration required to elicit 50% maximal response), nH (Hill-slope) and Imax (maximum peak current).
Fast agonist application.
Macroscopic currents evoked in outside-out patches by fast agonist application pulses were recorded at a pipette holding potential of −100 mV (−111 mV when corrected for a junction potential of 11 mV). The internal solution was the same as the one for whole cell recordings and contained 30 mM chloride. Agonist, dissolved in extracellular solution, was applied to outside-out patches with a theta tube (Hilgenberg GmbH) cut to a final diameter of ≈ 150 μm at the tip. The tube was driven by a piezo stepper PZ-150M (Burleigh Instruments Inc). The exchange time was measured by the application of diluted bath solution (e.g., 30:70 bath solution: water) before the experiment (to optimize the electrode position) and after the rupture of the patch. The rise and decay times for these open-tip currents were measured using Clampfit 10.2 as the times from 20 to 80% of the peak response. Patches in which the open tip response had a 20–80% exchange time slower than 200 μs were rejected from further analysis.
In order to study the kinetics of macroscopic currents, 10–20 responses were recorded in response to pulses of agonist applied at intervals of at least 10 s and averaged. Only experiments in which the rundown between the first and last three responses was < 30% were included in the analysis. The time course of the macroscopic currents was characterized by fitting the rise time between 20 and 100% and the decay time from 80 to 20% of the peak response with one or more exponentials.
Single-channel recording.
Low-noise single-channel currents were recorded in the cell-attached configuration at a pipette holding potential of +100 mV with an Axopatch 200B and filtered at 10 kHz using the amplifier’s low pass 4-pole Bessel filter. The data was digitized with a Digidata 1322A (Molecular Devices) at a sampling frequency of 100 kHz and acquired to PC using Clampex 10.2 for offline analysis.
Single channel current amplitude and cluster PO measurements.
In order to compare single-channel current amplitudes and cluster open probability, single-channel recordings were filtered offline using the Clampfit 10.2 low-pass Gaussian filter with a final cut-off of 5 kHz and resampled at 50 kHz. At high agonist concentration, channel openings occurred in clusters delimited by long closed intervals, likely to be desensitized. These clusters are likely to originate from the activity of a single ion channel molecule and were used for PO measurements. Clusters longer than 100 ms with more than 10 openings were selected for analysis. The gap between clusters was at least 100 ms or 300 ms (for GABA measurements). Channel activity in the selected clusters was idealized using the half-amplitude threshold method implemented in Clampfit 10.2 and open probability was calculated as the ratio of cluster open time over total cluster length. The amplitude of single channel currents was measured in Clampfit 10.2 as the average of all detected opening amplitudes inside a cluster. Electrophysiological data are reported as means ± SEM.
QUANTIFICATION AND STATISTICAL ANALYSIS
For the competitive inhibition radioligand-binding experiment, specific counts with standard deviations were counted by subtracting background nonspecific counts from triplicate experiments. Ki values were obtained from the IC50 values using the Cheng-Prusoff equation by fitting data to a standard single site competition equation. All the values are included in the analysis.
Supplementary Material
Data 1 Representative densities of the glycine or taurine complex maps. Related to Figures1 and 2.
The structures are shown in stick and maps are in mesh. For each panel, the M1 and M2 helices together with the M1-M2 loop starting from Y239 to S286 are isolated, contoured at 6.5 σ to 8 σ. In the nanodisc maps, a density feature of a putative lipid molecule has been modeled as an alkane chain. The M2-M3 loop from A288 to Y295 is illustrated, contoured at 6.5 σ to 8 σ. The β2 strand of the ECD from residue D73 to W84 is isolated, contoured at 7.5 σ to 8 σ. The maps associated with the nanodisc and SMA reconstructions were sharpened by RELION and Localscale, respectively.
Methods S1 Data processing flow chart for the taurine-bound GlyR complex in nanodiscs. Related to Figure 2.
Methods S2 Data processing flow chart for the taurine-bound GlyR isolated using SMA. Related to Figure 2.
Figure S1 Cryo-EM studies on the GlyR reconstituted into nanodisc. Related to Figures 1 and 2.
A. Macroscopic responses of GlyR to the rapid application of 10 mM glycine, 100 mM taurine and 100 mM GABA to the same outside-out patch. In this patch the agonist current left at the end of the 1-s pulse was 17% for glycine (cf. average of 57±8%, range 17–89%, 8 patches).
B. A single subunit of the glycine-bound open state of GlyR viewed parallel to the membrane with secondary structure elements and key loops labeled.
C. Cryo-EM reconstruction map of GlyR complexed with glycine in the nanodisc environment, viewed parallel to the membrane (GlyR-nanodisc-gly). One subunit is highlighted in green. The nanodisc density is indicated with a partially transparent blue surface.
D-E. Ion permeation pathways for GlyR-nanodisc-gly (green) and GlyREM-micelle-gly (grey, PDB code: 3JAE). The M2 helices of two subunits are shown in ribbon and the side chains of pore-lining residues are shown in sticks representation. As calculated by the program HOLE, different colors define different radii: red <1.8 Å, green 1.8–3.3 Å, and blue > 3.3 Å.
F-G. Conformational differences in the M2 helices between the desensitized and closed states of GlyR complexed with taurine (F) and GABA (G) in nanodiscs. The residues at two constrictions, 9’L and −2’P, are shown as sticks.
H. Plots of pore radius as a function of distance along the pore axis for desensitized and closed states of GlyR complexed with taurine and GABA in nanodiscs as well as the apo-GlyREM structure determined in a detergent micelle.
Figure S2 3D reconstructions for glycine-bound states in nanodiscs (green box) and SMA (blue box). Related to Figure 1.
A. SEC trace for GlyR in the nanodisc and SDS-PAGE analysis of peak fractions.
B. A typical cryo-EM micrograph for the glycine-bound GlyR - nanodisc complex.
C. Selected 2D class averages.
D. Local resolution estimate of the unsharpened map.
E. FSC plots before (unmasked) and after (masked) RELION post processing, and between the model and the final map.
F. Particle angular distribution.
G. SEC trace for glycine-bound GlyR in SMA and SDS-PAGE analysis of the peak fraction.
H. Typical cryo-EM micrograph.
I. Selected 2D class averages.
J. M and P. Local resolution estimation of unsharpened open, desensitized and expanded-open maps, respectively.
K. N and Q. FSC curves before (unmasked) and after (masked) RELION post processing, and between the model and the final maps for open, desensitized and expanded-open states, respectively.
l. O and R. Particle angular distributions for open, desensitized and expanded-open maps, respectively.
Table S1 | Statistics for 3D reconstruction and model refinement in the nanodisc. Related to Figures 1 and 2.
Figure S3 Representative cryo-EM densities of the M2 helices in the expanded-open states and of the partial agonists in the closed states. Related to Figures 1–2.
A-D. Cryo-EM densities and models for the pore lining M2 helices in the expanded-open states of GlyR or YGF mutant complexes solved using receptor isolated in SMA, showing the protruding densities at the intracellular end of M2 helices. The origin of these extra densities is unknown because of the low resolution, thus making them difficult to model. The constriction site at the −2’P position is indicated.
E-H. GABA densities in the nanodisc (E) and SMA (F) complexes are contoured at 3 σ and 4 σ, respectively. Taurine densities in the nanodisc (G) and SMA (H) complexes are contoured at 4 σ and 0.02 σ, respectively.
Figure S4 3D reconstructions for taurine-bound states in nanodiscs (green box) and SMA (blue box). Related to Figure 2.
A. A typical cryo-EM micrograph of the taurine-bound nanodisc complex. Scale bar represents the size of 30 nm.
B. Selected 2D class averages.
C. and E. Local resolution maps for unsharpened taurine bound desensitized and closed states, respectively.
D. and F. FSC curves before (unmasked) and after (masked) RELION post processing, and between the model and the final map.
G. A typical cryo-EM micrograph of the taurine-bound SMA complex. Scale bar represents the size of 30 nm.
H. Selected 2D class averages.
I. K. M and O. Local resolution maps for unsharpened open, desensitized, closed and expanded-open states, respectively.
J. L. N and P. FSC curves before (unmasked) and after (masked) RELION post processing, and between the model and the final map for open, desensitized, closed and expanded-open maps, respectively.
Figure S5 3D reconstructions for GABA-bound states in the nanodisc (green box) and SMA (blue box). Related to Figure 2.
A. A typical cryo-EM micrograph for GABA-bound GlyR in nanodiscs. Scale bar represents the size of 30 nm.
B. Selected 2D class averages.
C and E. Unsharpened local resolution maps for the GABA-bound desensitized and closed states, respectively.
D. and F. FSC curves before (unmasked) and after (masked) RELION post processing, and between the model and the final map for GABA-bound GlyR at desensitized and closed states, respectively.
G. A typical cryo-EM micrograph for taurine-bound GlyR in SMA. Scale bar represents the size of 100 nm.
H. Selected 2D class averages.
I. K. M and O. Local resolution maps for unsharpened open, desensitized, closed and expanded-open, respectively.
J. L. N and P. FSC curves before (unmasked) and after (masked) RELION post processing, and between the model and the final map for open, desensitized, closed and expanded-open, respectively.
Figure S6 3D reconstructions for the GABA-bound states of the YGF mutant in SMA and the apo state of GlyREM in detergent micelles. Related to Figures 3–4.
A. A typical cryo-EM micrograph. Scale bar represents the size of 100 nm.
B. Selected 2D class averages.
C. E and G. Local resolution maps for unsharpened open, desensitized and expanded-open states.
D. F and H. FSC curves before (unmasked) and after (masked) RELION post processing, and between the model and the final map for open, desensitized and expanded-open states, respectively.
J. A typical cryo-EM micrograph. Scale bar represents the size of 20 nm.
K. Selected 2D class averages.
L. M. Local resolution map and FSC plot for the apo-GlyREM reconstruction.
Figure S7 Conformational changes between the open, desensitized and closed states of GABA-bound GlyR in SMA. Related to Figure 6.
A. Reference orientation of the GABA-open state. The binding pocket in the ECD, the ECD-TMD interface and TMD are indicated by a black solid, a black dashed and a purple solid rectangle.
B-C. Conformational changes in the TMD between the GABA-closed, GABA-open and GABA-desensitized states. The centers of mass (COM) of the four TM helices from one subunit are indicated. The rotation angle of the COM relative to the receptor center is labeled. The GABA-open, GABA-desensitized and GABA-closed states are colored in gray, violet and green, respectively.
D-I. Superposition of the (−) subunit to visualize the relative movements in the (+) subunit. Hydrogen bonds are shown in dashed lines. The Cα atoms of key residues in the GABA-open and GABA-closed state are highlighted by gray and green spheres, respectively. (D. G) Comparison of the binding pocket (D) and ECD-TMD interfaces (G) between GABA-open and GABA-desensitized states. (E. H) Conformational changes in the binding pockets (E) and the ECD-TMD interfaces (H) between GABA-closed, GABA-open states. (F. I) Schematic diagrams illustrating the changes in distances of the Cα atoms of key residues in the binding pockets (F) and ECD-TMD interfaces (I) between GABA-open and GABA-closed states.
Figure S8 Conformational changes in the ECD, ECD-TMD and TMD in the partial agonists bound closed states. Related to Figures 3 and 6.
A-E. Superposition of two adjacent subunits from the closed state in complex with taurine in SMA (taurine-closed) with the resting state of the GlyR (apo-GlyREM) and 5-HT3A receptor (5-HT3AR) (PDB code: 6BE1) to show the differences in the binding pocket (black box) and ECD-TMD interfaces (red box). The taurine-closed, apo-GlyREM and 5-HT3AR are colored in cyan, red and purple, respectively. Taurine is shown in sticks.
F-H. Superposition of the two adjacent subunits of the taurine-closed state with the GABA-closed state to illustrate the changes in the ECD and ECD-TMD interfaces. Cα atoms of key residues are represented as colored circles. The schematic diagram illustrates the changes in distances (angstrom) of key residues for the taurine-closed and GABA-closed states. The taurine-closed and GABA-closed are cyan and green, respectively.
I-J. Conformational changes in the TMD between the taurine-closed, taurine-open and taurine-desensitized states. The centers of mass (COM) of the four TM helices from one subunit are indicated. The rotation angle of the COM relative to the receptor center is labeled. The taurine-closed, taurine-open and taurine-desensitized states are in cyan, yellow and salmon, respectively.
K. Two subunits derived from the GABA-bound SMA closed state, highlighting the neurotransmitter binding site (solid box) and the ECD-TMD interface (dashed box).
(L-O). Superposition of two adjacent subunits from the closed state of GlyR in complex with GABA in SMA (GABA-closed) with the resting state of GlyR (apo-GlyREM) (L, N) and 5-HT3A receptor (5-HT3AR) (PDB code: 6BE1) (M, O) to show the differences in the binding pocket and ECD-TMD interfaces. The GABA-closed, apo-GlyREM and 5-HT3AR are colored in green, red and purple, respectively.
Highlights:
Single channel and single particle cryo-EM studies inform partial agonist mechanism
Partial agonists populate agonist bound, previously unseen shut channel states
These pre-open bound states vary in degree of closure of the agonist binding site
Apo, partial and full agonist-bound states illuminate ion channel reaction pathway
Acknowledgements
We thank L. Vaskalis for assistance with figures, F. Jalali-Yazdi for discussion and Polyscope for the SMA. Electron microscopy was performed at OHSU at the Multiscale Microscopy Core. This work was supported by MRC grant MR/R009074/1 to L.G.S. and NIH grant R01 GM100400 to E.G.. E.G. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, and Adams PD (2018). Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althoff T, Hibbs RE, Banerjee S, and Gouaux E. (2014). X-ray structures of GluCl in apo states reveal a gating mechanism of Cys-loop receptors. Nature 512, 333–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez FJ, Orelle C, and Davidson AL (2010). Functional reconstitution of an ABC transporter in nanodiscs for use in electron paramagnetic resonance spectroscopy. J Am Chem Soc 132, 9513–9515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach A. (2005). Gating of acetylcholine receptor channels: brownian motion across a broad transition state. Proc Natl Acad Sci U S A 102, 1408–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach A, and Akk G. (1998). Desensitization of mouse nicotinic acetylcholine receptor channels. A two-gate mechanism. J Gen Physiol 112, 181–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autzen HE, Julius D, and Cheng Y. (2019). Membrane mimetic systems in CryoEM: keeping membrane proteins in their native environment. Curr Opin Struct Biol 58, 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baenziger JE, Henault CM, Therien JP, and Sun J. (2015). Nicotinic acetylcholine receptor-lipid interactions: Mechanistic insight and biological function. Biochim Biophys Acta 1848, 1806–1817. [DOI] [PubMed] [Google Scholar]
- Basak S, Gicheru Y, Samanta A, Molugu SK, Huang W, Fuente M, Hughes T, Taylor DJ, Nieman MT, Moiseenkova-Bell V, et al. (2018). Cryo-EM structure of 5-HT3A receptor in its resting conformation. Nat Commun 9, 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bormann J, Hamill OP, and Sakmann B. (1987). Mechanism of anion permeation through channels gated by glycine and gamma-aminobutyric acid in mouse cultured spinal neurones. J Physiol 385, 243–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitinger HG, Lanig H, Vohwinkel C, Grewer C, Breitinger U, Clark T, and Becker CM (2004). Molecular dynamics simulation links conformation of a pore-flanking region to hyperekplexia-related dysfunction of the inhibitory glycine receptor. Chem Biol 11, 1339–1350. [DOI] [PubMed] [Google Scholar]
- Burzomato V, Beato M, Groot-Kormelink PJ, Colquhoun D, and Sivilotti LG (2004). Single-channel behavior of heteromeric alpha1beta glycine receptors: an attempt to detect a conformational change before the channel opens. J Neurosci 24, 10924–10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celie PH, van Rossum-Fikkert SE, van Dijk WJ, Brejc K, Smit AB, and Sixma TK (2004). Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron 41, 907–914. [DOI] [PubMed] [Google Scholar]
- Cerdan AH, Martin NE, and Cecchini M. (2018). An Ion-Permeable State of the Glycine Receptor Captured by Molecular Dynamics. Structure 26, 1555–1562 e1554. [DOI] [PubMed] [Google Scholar]
- Changeux JP (2012). Allostery and the Monod-Wyman-Changeux model after 50 years. Annu Rev Biophys 41, 103–133. [DOI] [PubMed] [Google Scholar]
- Changeux JP, and Edelstein SJ (1998). Allosteric receptors after 30 years. Neuron 21, 959980. [DOI] [PubMed] [Google Scholar]
- Chen S, McMullan G, Faruqi AR, Murshudov GN, Short JM, Scheres SH, and Henderson R. (2013). High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 136, 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun D. (1998). Binding, gating, affinity and efficacy: the interpretation of structure-activity relationships for agonists and of the effects of mutating receptors. Br J Pharmacol 125, 924–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun D, and Sakmann B. (1985). Fast events in single-channel currents activated by acetylcholine and its analogues at the frog muscle end-plate. J Physiol 369, 501–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradi J, and Bouzat C. (2014). Unraveling mechanisms underlying partial agonism in 5-HT3A receptors. J Neurosci 34, 16865–16876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradi J, Gumilar F, and Bouzat C. (2009). Single-channel kinetic analysis for activation and desensitization of homomeric 5-HT(3)A receptors. Biophys J 97, 1335–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Castillo J, and Katz B. (1957). Interaction at end-plate receptors betwen different choline derivatives. Proc Roy Soc B 146, 369–381. [DOI] [PubMed] [Google Scholar]
- Denisov IG, and Sligar SG (2016). Nanodiscs for structural and functional studies of membrane proteins. Nat Struct Mol Biol 23, 481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorr JM, Scheidelaar S, Koorengevel MC, Dominguez JJ, Schafer M, van Walree CA, and Killian JA (2016). The styrene-maleic acid copolymer: a versatile tool in membrane research. Eur Biophys J 45, 3–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Lu W, Wu S, Cheng Y, and Gouaux E. (2015). Glycine receptor mechanism elucidated by electron cryo-microscopy. Nature 526, 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Fucile S, de Saint Jan D, David-Watine B, Korn H, and Bregestovski P. (1999). Comparison of glycine and GABA actions on the zebrafish homomeric glycine receptor. J Physiol 517 ( Pt 2), 369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharpure A, Teng J, Zhuang Y, Noviello CM, Walsh RM Jr., Cabuco R, Howard RJ, Zaveri NT, Lindahl E, and Hibbs RE (2019). Agonist Selectivity and Ion Permeation in the alpha3beta4 Ganglionic Nicotinic Receptor. Neuron 104, 501–511 e506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielen M, Thomas P, and Smart TG (2015). The desensitization gate of inhibitory Cys-loop receptors. Nat Commun 6, 6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Gutierrez G, Wang Y, Cymes GD, Tajkhorshid E, and Grosman C. (2017). Chasing the open-state structure of pentameric ligand-gated ion channels. J Gen Physiol 149, 1119–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant T, Rohou A, and Grigorieff N. (2018). cisTEM, user-friendly software for single-particle image processing. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot-Kormelink PJ, Beato M, Finotti C, Harvey RJ, and Sivilotti LG (2002). Achieving optimal expression for single channel recording: a plasmid ratio approach to the expression of alpha 1 glycine receptors in HEK293 cells. J Neurosci Methods 113, 207–214. [DOI] [PubMed] [Google Scholar]
- Hansen SB, Sulzenbacher G, Huxford T, Marchot P, Taylor P, and Bourne Y. (2005). Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. EMBO J 24, 3635–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidmann T, Sobel A, Popot JL, and Changeux JP (1980). Reconstitution of a functional acetylcholine receptor. Conservation of the conformational and allosteric transitions and recovery of the permeability response; role of lipids. Eur J Biochem 110, 35–55. [DOI] [PubMed] [Google Scholar]
- Hibbs RE, and Gouaux E. (2011). Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature 474, 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilf RJ, and Dutzler R. (2008). X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature 452, 375–379. [DOI] [PubMed] [Google Scholar]
- Hille B. (2001). Ion channels of excitable membranes, 3rd edn (Sunderland, MA: Sinauer Associates, Inc.). [Google Scholar]
- Huang X, Chen H, Michelsen K, Schneider S, and Shaffer PL (2015). Crystal structure of human glycine receptor-alpha3 bound to antagonist strychnine. Nature 526, 277–280. [DOI] [PubMed] [Google Scholar]
- Huang X, Chen H, and Shaffer PL (2017a). Crystal Structures of Human GlyRalpha3 Bound to Ivermectin. Structure 25, 945–950 e942. [DOI] [PubMed] [Google Scholar]
- Huang X, Shaffer PL, Ayube S, Bregman H, Chen H, Lehto SG, Luther JA, Matson DJ, McDonough SI, Michelsen K, et al. (2017b). Crystal structures of human glycine receptor alpha3 bound to a novel class of analgesic potentiators. Nat Struct Mol Biol 24, 108–113. [DOI] [PubMed] [Google Scholar]
- Jadey S, and Auerbach A. (2012). An integrated catch-and-hold mechanism activates nicotinic acetylcholine receptors. J Gen Physiol 140, 17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobi AJ, Wilmanns M, and Sachse C. (2017). Model-based local density sharpening of cryo-EM maps. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamshad M, Lin YP, Knowles TJ, Parslow RA, Harris C, Wheatley M, Poyner DR, Bill RM, Thomas OR, Overduin M, et al. (2011). Surfactant-free purification of membrane proteins with intact native membrane environment. Biochem Soc Trans 39, 813–818. [DOI] [PubMed] [Google Scholar]
- Janzen D, Schaefer N, Delto C, Schindelin H, and Villmann C. (2017). The GlyR Extracellular beta8-beta9 Loop - A Functional Determinant of Agonist Potency. Front Mol Neurosci 10, 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaram B, Singh T, Mukherjee G, Mathur A, Shekhar S, and Shekhar V. (2012). Sanjeevini: a freely accessible web-server for target directed lead molecule discovery. BMC Bioinformatics 13 Suppl 17, S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz B, and Thesleff S. (1957). A study of the ‘desensitization’ produced by acetylcholine at the motor end-plate. J Physiol 138, 63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles TJ, Finka R, Smith C, Lin YP, Dafforn T, and Overduin M. (2009). Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J Am Chem Soc 131, 7484–7485. [DOI] [PubMed] [Google Scholar]
- Krishtal OA, and Pidoplichko VI (1980). A receptor for protons in the nerve cell membrane. Neuroscience 5, 2325–2327. [DOI] [PubMed] [Google Scholar]
- Kumar A, Basak S, Rao S, Gicheru Y, Mayer ML, Sansom MSP, and Chakrapani S. (2020). Mechanisms of activation and desensitization of full-length glycine receptor in lipid nanodiscs. Nat Commun 11, 3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S, and Patel PN (2007). Varenicline: a selective alpha4beta2 nicotinic acetylcholine receptor partial agonist approved for smoking cessation. Cardiol Rev 15, 154–161. [DOI] [PubMed] [Google Scholar]
- Lape R, Colquhoun D, and Sivilotti LG (2008). On the nature of partial agonism in the nicotinic receptor superfamily. Nature 454, 722–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lape R, Plested AJ, Moroni M, Colquhoun D, and Sivilotti LG (2012). The alpha1K276E startle disease mutation reveals multiple intermediate states in the gating of glycine receptors. J Neurosci 32, 1336–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laverty D, Desai R, Uchanski T, Masiulis S, Stec WJ, Malinauskas T, Zivanov J, Pardon E, Steyaert J, Miller KW, et al. (2019). Cryo-EM structure of the human alpha1beta3gamma2 GABAA receptor in a lipid bilayer. Nature 565, 516–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Rubio ME, and Trussell LO (2008). Glycinergic transmission shaped by the corelease of GABA in a mammalian auditory synapse. Neuron 57, 524–535. [DOI] [PubMed] [Google Scholar]
- Lynch JW (2004). Molecular structure and function of the glycine receptor chloride channel. Physiol Rev 84, 1051–1095. [DOI] [PubMed] [Google Scholar]
- Lynch JW, Han NL, Haddrill J, Pierce KD, and Schofield PR (2001). The surface accessibility of the glycine receptor M2-M3 loop is increased in the channel open state. J Neurosci 21, 2589–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiulis S, Desai R, Uchanski T, Martin IS, Laverty D, Karia D, Malinauskas T, Zivanov J, Pardon E, Kotecha A, et al. (2019). Author Correction: GABAA receptor signalling mechanisms revealed by structural pharmacology. Nature 566, E8. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51. [DOI] [PubMed] [Google Scholar]
- Miyazawa A, Fujiyoshi Y, and Unwin N. (2003). Structure and gating mechanism of the acetylcholine receptor pore. Nature 423, 949–955. [DOI] [PubMed] [Google Scholar]
- Morales-Perez CL, Noviello CM, and Hibbs RE (2016). X-ray structure of the human alpha4beta2 nicotinic receptor. Nature 538, 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen M, Kristiansen U, Ebert B, Frolund B, Krogsgaard-Larsen P, and Smart TG (2004). Activation of single heteromeric GABA(A) receptor ion channels by full and partial agonists. J Physiol 557, 389–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtasimova N, daCosta CJ, and Sine SM (2016). Improved resolution of single channel dwell times reveals mechanisms of binding, priming, and gating in muscle AChR. J Gen Physiol 148, 43–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtasimova N, Lee WY, Wang HL, and Sine SM (2009). Detection and trapping of intermediate states priming nicotinic receptor channel opening. Nature 459, 451–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtasimova N, and Sine SM (2018). Full and partial agonists evoke distinct structural changes in opening the muscle acetylcholine receptor channel. J Gen Physiol 150, 713–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson N. (1987). A novel method for the detection of receptors and membrane proteins by scintillation proximity radioassay. Anal Biochem 165, 287–293. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Phulera S, Zhu H, Yu J, Claxton DP, Yoder N, Yoshioka C, and Gouaux E. (2018). Cryo-EM structure of the benzodiazepine-sensitive alpha1beta1gamma2S tri-heteromeric GABAA receptor in complex with GABA. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plested AJ (2016). Structural mechanisms of activation and desensitization in neurotransmitter-gated ion channels. Nat Struct Mol Biol 23, 494–502. [DOI] [PubMed] [Google Scholar]
- Polovinkin L, Hassaine G, Perot J, Neumann E, Jensen AA, Lefebvre SN, Corringer PJ, Neyton J, Chipot C, Dehez F, et al. (2018). Conformational transitions of the serotonin 5-HT3 receptor. Nature 563, 275–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, and Sligar SG (2009). Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol 464, 211–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundstrom N, Schmieden V, Betz H, Bormann J, and Langosch D. (1994). Cyanotriphenylborate: subtype-specific blocker of glycine receptor chloride channels. Proc Natl Acad Sci U S A 91, 8950–8954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH (2016). Processing of Structurally Heterogeneous Cryo-EM Data in RELION. Methods Enzymol 579, 125–157. [DOI] [PubMed] [Google Scholar]
- Schmieden V, Kuhse J, and Betz H. (1992). Agonist pharmacology of neonatal and adult glycine receptor alpha subunits: identification of amino acid residues involved in taurine activation. EMBO J 11, 2025–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieden V, Kuhse J, and Betz H. (1993). Mutation of glycine receptor subunit creates beta-alanine receptor responsive to GABA. Science 262, 256–258. [DOI] [PubMed] [Google Scholar]
- Schwartz RD, and Yu X. (1995). Optical imaging of intracellular chloride in living brain slices. J Neurosci Methods 62, 185–192. [DOI] [PubMed] [Google Scholar]
- Smart OS, Neduvelil JG, Wang X, Wallace BA, and Sansom MS (1996). HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph 14, 354–360, 376. [DOI] [PubMed] [Google Scholar]
- Stephenson RP (1956). A modification of receptor theory. Brit J Pharmacol 11, 379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunesen M, de Carvalho LP, Dufresne V, Grailhe R, Savatier-Duclert N, Gibor G, Peretz A, Attali B, Changeux JP, and Paas Y. (2006). Mechanism of Cl- selection by a glutamate-gated chloride (GluCl) receptor revealed through mutations in the selectivity filter. J Biol Chem 281, 14875–14881. [DOI] [PubMed] [Google Scholar]
- Tripathy S, Zheng W, and Auerbach A. (2019). A single molecular distance predicts agonist binding energy in nicotinic receptors. J Gen Physiol 151, 452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss NR, Yoshioka CK, Radermacher M, Potter CS, and Carragher B. (2009). DoG picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol 166, 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh RM Jr., Roh SH, Gharpure A, Morales-Perez CL, Teng J, and Hibbs RE (2018). Structural principles of distinct assemblies of the human alpha4beta2 nicotinic receptor. Nature 557, 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Cantara W, Jeon Y, Musier-Forsyth K, Grigorieff N, and Lyumkis D. (2019). Analysis of discrete local variability and structural covariance in macromolecular assemblies using Cryo-EM and focused classification. Ultramicroscopy 203, 170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Xue F, Liu Y, Yang H, and Wang X. (2013). The structural mechanism of the Cys-loop receptor desensitization. Mol Neurobiol 48, 97–108. [DOI] [PubMed] [Google Scholar]
- Zhang K. (2016). Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Palowcak E, Armache J-P, Cheng Y, and Agard D. (2016). Anisotropic correction of beam-induced motion for improved single-particle electron cryo-microscopy. bioRxiv doi: 10.1101/061960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data 1 Representative densities of the glycine or taurine complex maps. Related to Figures1 and 2.
The structures are shown in stick and maps are in mesh. For each panel, the M1 and M2 helices together with the M1-M2 loop starting from Y239 to S286 are isolated, contoured at 6.5 σ to 8 σ. In the nanodisc maps, a density feature of a putative lipid molecule has been modeled as an alkane chain. The M2-M3 loop from A288 to Y295 is illustrated, contoured at 6.5 σ to 8 σ. The β2 strand of the ECD from residue D73 to W84 is isolated, contoured at 7.5 σ to 8 σ. The maps associated with the nanodisc and SMA reconstructions were sharpened by RELION and Localscale, respectively.
Methods S1 Data processing flow chart for the taurine-bound GlyR complex in nanodiscs. Related to Figure 2.
Methods S2 Data processing flow chart for the taurine-bound GlyR isolated using SMA. Related to Figure 2.
Figure S1 Cryo-EM studies on the GlyR reconstituted into nanodisc. Related to Figures 1 and 2.
A. Macroscopic responses of GlyR to the rapid application of 10 mM glycine, 100 mM taurine and 100 mM GABA to the same outside-out patch. In this patch the agonist current left at the end of the 1-s pulse was 17% for glycine (cf. average of 57±8%, range 17–89%, 8 patches).
B. A single subunit of the glycine-bound open state of GlyR viewed parallel to the membrane with secondary structure elements and key loops labeled.
C. Cryo-EM reconstruction map of GlyR complexed with glycine in the nanodisc environment, viewed parallel to the membrane (GlyR-nanodisc-gly). One subunit is highlighted in green. The nanodisc density is indicated with a partially transparent blue surface.
D-E. Ion permeation pathways for GlyR-nanodisc-gly (green) and GlyREM-micelle-gly (grey, PDB code: 3JAE). The M2 helices of two subunits are shown in ribbon and the side chains of pore-lining residues are shown in sticks representation. As calculated by the program HOLE, different colors define different radii: red <1.8 Å, green 1.8–3.3 Å, and blue > 3.3 Å.
F-G. Conformational differences in the M2 helices between the desensitized and closed states of GlyR complexed with taurine (F) and GABA (G) in nanodiscs. The residues at two constrictions, 9’L and −2’P, are shown as sticks.
H. Plots of pore radius as a function of distance along the pore axis for desensitized and closed states of GlyR complexed with taurine and GABA in nanodiscs as well as the apo-GlyREM structure determined in a detergent micelle.
Figure S2 3D reconstructions for glycine-bound states in nanodiscs (green box) and SMA (blue box). Related to Figure 1.
A. SEC trace for GlyR in the nanodisc and SDS-PAGE analysis of peak fractions.
B. A typical cryo-EM micrograph for the glycine-bound GlyR - nanodisc complex.
C. Selected 2D class averages.
D. Local resolution estimate of the unsharpened map.
E. FSC plots before (unmasked) and after (masked) RELION post processing, and between the model and the final map.
F. Particle angular distribution.
G. SEC trace for glycine-bound GlyR in SMA and SDS-PAGE analysis of the peak fraction.
H. Typical cryo-EM micrograph.
I. Selected 2D class averages.
J. M and P. Local resolution estimation of unsharpened open, desensitized and expanded-open maps, respectively.
K. N and Q. FSC curves before (unmasked) and after (masked) RELION post processing, and between the model and the final maps for open, desensitized and expanded-open states, respectively.
l. O and R. Particle angular distributions for open, desensitized and expanded-open maps, respectively.
Table S1 | Statistics for 3D reconstruction and model refinement in the nanodisc. Related to Figures 1 and 2.
Figure S3 Representative cryo-EM densities of the M2 helices in the expanded-open states and of the partial agonists in the closed states. Related to Figures 1–2.
A-D. Cryo-EM densities and models for the pore lining M2 helices in the expanded-open states of GlyR or YGF mutant complexes solved using receptor isolated in SMA, showing the protruding densities at the intracellular end of M2 helices. The origin of these extra densities is unknown because of the low resolution, thus making them difficult to model. The constriction site at the −2’P position is indicated.
E-H. GABA densities in the nanodisc (E) and SMA (F) complexes are contoured at 3 σ and 4 σ, respectively. Taurine densities in the nanodisc (G) and SMA (H) complexes are contoured at 4 σ and 0.02 σ, respectively.
Figure S4 3D reconstructions for taurine-bound states in nanodiscs (green box) and SMA (blue box). Related to Figure 2.
A. A typical cryo-EM micrograph of the taurine-bound nanodisc complex. Scale bar represents the size of 30 nm.
B. Selected 2D class averages.
C. and E. Local resolution maps for unsharpened taurine bound desensitized and closed states, respectively.
D. and F. FSC curves before (unmasked) and after (masked) RELION post processing, and between the model and the final map.
G. A typical cryo-EM micrograph of the taurine-bound SMA complex. Scale bar represents the size of 30 nm.
H. Selected 2D class averages.
I. K. M and O. Local resolution maps for unsharpened open, desensitized, closed and expanded-open states, respectively.
J. L. N and P. FSC curves before (unmasked) and after (masked) RELION post processing, and between the model and the final map for open, desensitized, closed and expanded-open maps, respectively.
Figure S5 3D reconstructions for GABA-bound states in the nanodisc (green box) and SMA (blue box). Related to Figure 2.
A. A typical cryo-EM micrograph for GABA-bound GlyR in nanodiscs. Scale bar represents the size of 30 nm.
B. Selected 2D class averages.
C and E. Unsharpened local resolution maps for the GABA-bound desensitized and closed states, respectively.
D. and F. FSC curves before (unmasked) and after (masked) RELION post processing, and between the model and the final map for GABA-bound GlyR at desensitized and closed states, respectively.
G. A typical cryo-EM micrograph for taurine-bound GlyR in SMA. Scale bar represents the size of 100 nm.
H. Selected 2D class averages.
I. K. M and O. Local resolution maps for unsharpened open, desensitized, closed and expanded-open, respectively.
J. L. N and P. FSC curves before (unmasked) and after (masked) RELION post processing, and between the model and the final map for open, desensitized, closed and expanded-open, respectively.
Figure S6 3D reconstructions for the GABA-bound states of the YGF mutant in SMA and the apo state of GlyREM in detergent micelles. Related to Figures 3–4.
A. A typical cryo-EM micrograph. Scale bar represents the size of 100 nm.
B. Selected 2D class averages.
C. E and G. Local resolution maps for unsharpened open, desensitized and expanded-open states.
D. F and H. FSC curves before (unmasked) and after (masked) RELION post processing, and between the model and the final map for open, desensitized and expanded-open states, respectively.
J. A typical cryo-EM micrograph. Scale bar represents the size of 20 nm.
K. Selected 2D class averages.
L. M. Local resolution map and FSC plot for the apo-GlyREM reconstruction.
Figure S7 Conformational changes between the open, desensitized and closed states of GABA-bound GlyR in SMA. Related to Figure 6.
A. Reference orientation of the GABA-open state. The binding pocket in the ECD, the ECD-TMD interface and TMD are indicated by a black solid, a black dashed and a purple solid rectangle.
B-C. Conformational changes in the TMD between the GABA-closed, GABA-open and GABA-desensitized states. The centers of mass (COM) of the four TM helices from one subunit are indicated. The rotation angle of the COM relative to the receptor center is labeled. The GABA-open, GABA-desensitized and GABA-closed states are colored in gray, violet and green, respectively.
D-I. Superposition of the (−) subunit to visualize the relative movements in the (+) subunit. Hydrogen bonds are shown in dashed lines. The Cα atoms of key residues in the GABA-open and GABA-closed state are highlighted by gray and green spheres, respectively. (D. G) Comparison of the binding pocket (D) and ECD-TMD interfaces (G) between GABA-open and GABA-desensitized states. (E. H) Conformational changes in the binding pockets (E) and the ECD-TMD interfaces (H) between GABA-closed, GABA-open states. (F. I) Schematic diagrams illustrating the changes in distances of the Cα atoms of key residues in the binding pockets (F) and ECD-TMD interfaces (I) between GABA-open and GABA-closed states.
Figure S8 Conformational changes in the ECD, ECD-TMD and TMD in the partial agonists bound closed states. Related to Figures 3 and 6.
A-E. Superposition of two adjacent subunits from the closed state in complex with taurine in SMA (taurine-closed) with the resting state of the GlyR (apo-GlyREM) and 5-HT3A receptor (5-HT3AR) (PDB code: 6BE1) to show the differences in the binding pocket (black box) and ECD-TMD interfaces (red box). The taurine-closed, apo-GlyREM and 5-HT3AR are colored in cyan, red and purple, respectively. Taurine is shown in sticks.
F-H. Superposition of the two adjacent subunits of the taurine-closed state with the GABA-closed state to illustrate the changes in the ECD and ECD-TMD interfaces. Cα atoms of key residues are represented as colored circles. The schematic diagram illustrates the changes in distances (angstrom) of key residues for the taurine-closed and GABA-closed states. The taurine-closed and GABA-closed are cyan and green, respectively.
I-J. Conformational changes in the TMD between the taurine-closed, taurine-open and taurine-desensitized states. The centers of mass (COM) of the four TM helices from one subunit are indicated. The rotation angle of the COM relative to the receptor center is labeled. The taurine-closed, taurine-open and taurine-desensitized states are in cyan, yellow and salmon, respectively.
K. Two subunits derived from the GABA-bound SMA closed state, highlighting the neurotransmitter binding site (solid box) and the ECD-TMD interface (dashed box).
(L-O). Superposition of two adjacent subunits from the closed state of GlyR in complex with GABA in SMA (GABA-closed) with the resting state of GlyR (apo-GlyREM) (L, N) and 5-HT3A receptor (5-HT3AR) (PDB code: 6BE1) (M, O) to show the differences in the binding pocket and ECD-TMD interfaces. The GABA-closed, apo-GlyREM and 5-HT3AR are colored in green, red and purple, respectively.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request. The following Protein Data Bank (PDB) and Electron Microscopy Data Bank (EMDB) accession codes are provided: 6PM6 and 20389 (GlyR-gly-open-SMA), 6PM5 and 20388 (GlyR-gly-desensitized-SMA), 6PM4 and 20386 (GlyR-gly-expanded-open-SMA), 6PM3 and 20385 (GlyR-taurine-closed-SMA), 6PM2 and 20384 (GlyR-taurine-open-SMA), 6PM1 and 20383 (GlyR-taurine-desensitized-SMA) 6PM0 and 20382 (GlyR-taurine-expanded-open-SMA), 6PLZ and 20381 (GlyR-GABA-closed-SMA), 6PLY and EMD-20380 (GlyR-GABA-open-SMA), 6PLX and 20379 (GlyR-GABA-desensitized-SMA), 6PLW and 20378 (GlyR-GABA-expanded-open-SMA), 6PLO and 20370 (YGF-GABA-open-SMA), 6PLP and 20371(YGF-GABA-desensitized-SMA), 6PLQ and 20372 (YGF-GABA-expanded-open-SMA), 6PXD and 20518 (apo-GlyREM), 6PLR and 20373 (GlyR-gly-desensitized-nanodisc), 6PLT and 20375 (GlyR-taurine-desensitized-nanodisc), 6PLS and 20374 (GlyR-GABA-closed-nanodisc), 6PLV and 20377 (GlyR-GABA-desensitized-nanodisc), and 6PLU and 20376 (GlyR-GABA-closed-nanodisc).