ABSTRACT
Chronic inflammation and gut microbiota dysbiosis are risk factors for colorectal cancer. In clinical practice, patients with inflammatory bowel disease (IBD) have a greatly increased risk of developing colitis-associated colorectal cancer (CAC). However, the underlying mechanism of the initiation of CAC remains unknown. Systematic analyses using an existing genome-wide association study (GWAS) and conditional deletion of Zfp90 (encoding zinc finger protein 90 homolog) in a CAC mouse model indicated that Zfp90 is a putative oncogene in CAC development.
Strikingly, depletion of the gut microbiota eliminated the tumorigenic effect of Zfp90 in the CAC mouse model. Moreover, fecal microbiota transplantation demonstrated that Zfp90 promoted CAC dependent on the gut microbiota. Analysis of 16s rDNA sequences in fecal specimens from the CAC mouse model allowed us to speculate that a Prevotella copri-defined microbiota might mediate the oncogenic role of Zfp90 in the development of CAC. Mechanistic studies revealed Zfp90 accelerated CAC development through the TLR4-PI3K-AKT-NF-κB pathway. Our findings revealed the crucial role of the Zfp90-microbiota-NF-κB axis in creating a tumor-promoting environment and suggested therapeutic targets for CAC prevention and treatment.
KEYWORDS: ZFP90, gut microbiota, colitis-associated colorectal cancer, gut barrier, prevotella copri
Introduction
Colorectal cancer (CRC) ranks as the third most common cancer and the second leading cause of cancer-related death worldwide.1,2 The development of CRC is influenced by multiple factors, which include genetic predisposition, dysbiosis of the gut microbiota, and environmental factors.3,4 Previous research showed that chronic inflammation could accelerate the development of CRC.5 In the clinical scenario, patients with inflammatory bowel disease (IBD) are predisposed to CRC, and colitis-associated colorectal cancer (CAC) has been recognized as a challenging complication in this population.6,7 A fundamental understanding of the evolutionary process underpinning carcinogenesis is vital to optimize the management of CAC risk in patients with IBD.
Genome-wide association study (GWAS) is an observational study of a genome-wide set of genetic variants in different individuals that allows the identification of disease susceptibility variants. Results from GWAS have been overlapped to identify potential pluripotent genes that are involved in diseases such as obesity and cardiovascular disease.8–10 GWAS and subsequent fine-mapping research have identified more than 50 genetic susceptibility loci for CRC11 and more than 240 susceptibility loci for IBD.12 Given the well-known association between IBD and CRC, exploring shared molecular pathways between the two diseases by taking advantage of existing GWAS might provide novel therapeutic targets for CAC prevention and treatment.13
Accumulating evidence shows that dysfunction of the gut barrier contributes to the development of both IBD and CRC.14,15 The monolayered gut epithelium, together with goblet cell-derived mucins and Paneth cell-derived antimicrobial peptides, forms a physical and biochemical barrier that segregates the intestinal content and the host immune system. Breach of intestinal barrier can lead to increased infiltration of molecules such as lipopolysaccharide (LPS), which then abnormally activates the host immune system, resulting in an inflammatory microenvironment. Thus, increased intestinal permeability is emerging as one of the hallmarks of CAC.15,16
The microbiota plays a major role in human health and diseases, including CRC and IBD. Many factors, including host genetics and external factors, such as diet, lifestyle, and medication, can affect the composition of the gut microbiota. Host genetics define the chemical and physical landscape in which the gut microbiota resides17 and genetic variations between individuals can result in significant differences in the gut microbiota.18,19 Many studies have found that the effects of certain genes on disease progression are partly mediated by modulating the intestinal microbiota.20–23 Additionally, as part of the gut barrier, the distribution and structure of tight junction proteins have been shown to be modified by the gut microbiota.24–26
In the present study, through overlapping findings from GWAS, we found that ZFP90 (encoding zinc finger protein 90 homolog) is associated with the development of CAC. We verified that deletion of Zfp90 in the mouse colonic epithelium reduced CAC formation. We showed that Zfp90ΔIEC mice (conditional deletion of Zfp90 in intestinal epithelial cells (IECs)) exhibited a better gut barrier function and milder inflammation upon azoxymethane and dextran sulfate sodium (AOM-DSS) treatment. Furthermore, we confirmed that the protective role of Zfp90 deletion in AOM-DSS-induced CAC is dependent on the gut microbiota.
Results
ZFP90 is associated with CRC and IBD and Zfp90ΔIEC mice are less susceptible to AOM-DSS-induced CAC
To identify a gene that might play a crucial role in the progression of CAC, we first searched HaploReg to explore candidate regulatory single nucleotide polymorphisms (SNPs) for CRC and IBD.27 Sixty-nine SNPs responsible for CRC and 386 SNPs responsible for IBD were identified. We then located the susceptibility loci of these SNPs using the UCSC Genome Browser.28 In total, 46 CRC susceptibility loci and 166 IBD susceptibility loci were identified. Among them, 16 loci were coincident (Figure 1a). To identify potential target genes regulated by the SNPs at these 16 loci, we performed electronic quantitative trait locus (eQTL) analyses in the Genotype-Tissue Expression (GTEx) portal and identified 13 candidate genes29 (Figure 1b). To seek genes that were differentially expressed in the process of AOM-DSS-induced CAC, the expression profile GSE44904 was obtained from the Gene Expression Omnibus (GEO) database.30,31 With the thresholds of adjusted P < .01 and |log-fold change (FC)| > 1, 918 genes were found to have significantly differential expression between AOM-DSS-induced CAC and the controls (Supplementary SFigure 1a). Next, we combined these differentially expressed genes with the 13 candidate genes mentioned above to discover the genes that are likely to play an important role in the initiation of CAC. After overlapping, ZFP90 was the only gene that appeared in both clusters (Figure 1c). Expression profile GSE44904 showed that Zfp90 expression increased during AOM-DSS treatment, indicating a potential oncogenic role of this gene (Supplementary SFigure 1b). Therefore, we chose Zfp90 as the candidate biological target gene and explored its underlying mechanisms in the development of CAC.
Figure 1.
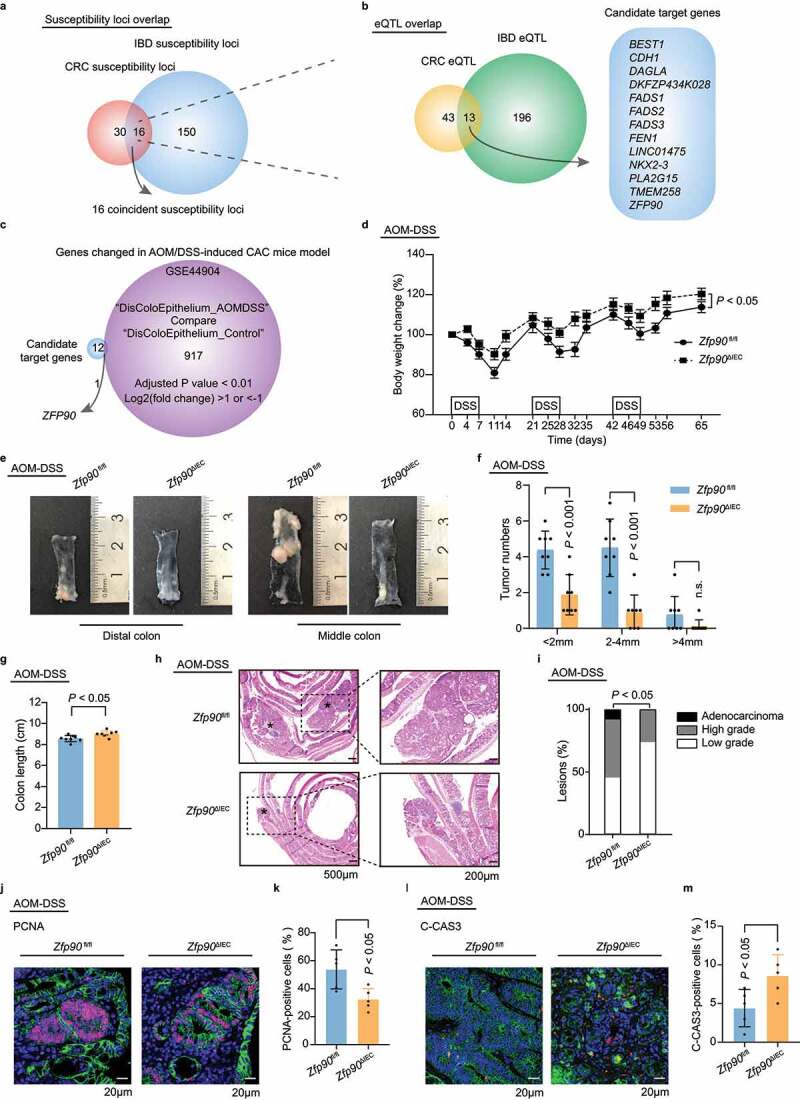
ZFP90 is associated with CRC and IBD and Zfp90ΔIEC mice are less susceptible to AOM-DSS-induced CAC a Venn diagram showing the chromosome loci accounting for CRC (46 loci), IBD (166 loci), and both (16 loci). b Venn diagram showing the genes regulated by CRC-related SNPs (56 genes), IBD-related SNPs (209 genes), and both (13 genes). c ZFP90 was identified after overlapping 13 candidate target genes with genes differentially expressed during AOM-DSS treatment (918 genes). d The body weight of Zfp90fl/fl and Zfp90ΔIEC mice was recorded throughout the experiment and was expressed as the ratio relative to the initial weight before DSS treatment (n = 8 per group). e Representative images of distal and middle colons in AOM-DSS-treated Zfp90fl/fl and Zfp90ΔIEC mice. f Tumor distribution in Zfp90fl/fl and Zfp90ΔIEC mice (n = 8 per group). g Colon length in Zfp90fl/fl and Zfp90ΔIEC mice at the end of the experiment (n = 8 per group). h Representative H&E staining of colon tumors from Zfp90fl/fl and Zfp90ΔIEC mice. * tumors. i Proportion of low-grade dysplasia, high-grade dysplasia, and adenocarcinoma in the colons. j Representative immunofluorescence staining of the PCNA protein in tumor tissues from Zfp90fl/fl and Zfp90ΔIEC mice. Sections were stained with DAPI (blue), CDH1 (green) and PCNA (red). k The percentage of PCNA-positive tumor cells was quantified (n = 5 per group). l Representative immunofluorescence staining of C-CAS3-positive cells in tumor tissues from Zfp90fl/fl and Zfp90ΔIEC mice. Sections were stained with DAPI (blue), CDH1 (green) and C-CAS3 (red). m The percentage of C-CAS3-positive tumor cells was quantified (n = 5 per group). Data with error bars represent the mean ± SD. Each panel is a representative experiment of at least three independent biological replicates. Two-way ANOVA (d, f), nonpaired two-tailed t-test (g, k, m), Fisher’s exact test (i)
To corroborate the role of intestinal ZFP90 in the development of CAC, Zfp90 was conditionally knocked out in the mouse IECs to generate IEC-specific Zfp90 deficient mice (Zfp90ΔIEC). The morphology of the colonic epithelium was similar between Zfp90fl/fl and Zfp90ΔIEC mice. (Supplementary SFigure 1c). Zfp90fl/fl and Zfp90ΔIEC mice were housed separately by genotypes after weaning. Mono-housed male Zfp90fl/fl and Zfp90ΔIEC mice were then treated with one dose of AOM followed by three cycles of feeding water containing DSS to induce a CAC model in a specific-pathogen-free (SPF) environment (Supplementary SFigure 1d). The Zfp90ΔIEC mice exhibited less weight loss than the Zfp90fl/fl mice during AOM-DSS treatment (Figure 1d). At the end of the experiment, the whole colon of each mouse was dissected for measurement. Compared with the Zfp90fl/fl mice, the Zfp90ΔIEC mice showed reduced numbers of colorectal tumors in the distal and middle region, their tumors were smaller (Figure 1(e, f) and Supplementary SFigure 1e), and milder colitis was characterized by a longer colon length (Figure 1g). In addition, the spleens were smaller in the AOM-DSS-treated Zfp90ΔIEC mice than in the Zfp90fl/fl mice, indicating alleviation of systemic inflammation in the Zfp90ΔIEC mice (Supplementary SFigure 1f). Although dysplasia was observed in both the Zfp90fl/fl and Zfp90ΔIEC mice, high-grade dysplasia was more common in the Zfp90fl/fl mice. Notably, adenocarcinoma invasion into the submucosa was only observed in the Zfp90fl/fl mice (Figure 1(h, i). Proliferative activities were assessed, and reduced proliferation and increased apoptosis were detected in the Zfp90ΔIEC mice, as shown by lower percentages of proliferating cell nuclear antigen (PCNA)-positive cells (Figure 1(j, k)) and higher numbers of cleaved caspase3 (C-CAS3)-positive cells (Figure 1(l, m)).
herefore, we explored whether Zfp90 deletion had an impact on the intrinsic response to inflammatory stimuli. Mono-housed Zfp90fl/fl and Zfp90ΔIEC mice were given one period of DSS solution for 7 days and then transferred to autoclaved water for 5 days to induce an acute colitis model. Similarly, the Zfp90fl/fl mice exhibited more weight loss (Supplementary SFigure 1g) and a shorter colon length (Supplementary SFigure 1h) compared with those of the Zfp90ΔIEC mice under DSS-induced acute colitis.
According to our previous study,32 a SNP residing in the intron of CDH1 (encoding cadherin 1) could mediate a long-range regulation on ZFP90. Therefore, we could not exclude the possibility that the expression of CDH1 might also be regulated by ZFP90 because of their proximity, and such modulation might mediate the protective role of ZFP90 deletion, considering the importance of CDH1 in IBD and CRC.33,34 We thus tested the expression level of CDH1 in naïve and AOM-DSS-treated Zfp90fl/fl and Zfp90ΔIEC mice. However, no differences in the mRNA and protein levels of CDH1 were found in colon tissues of the Zfp90fl/fl and Zfp90ΔIEC mice (Supplementary SFigure 1i, j). This observation suggested that Cdh1 is not involved in the phenotypic differences between Zfp90fl/fl and Zfp90ΔIEC mice.
Zfp90 deletion in IECs improves the gut barrier and alleviates local inflammation during CAC development
Given the importance of barrier dysfunction in the development of IBD and CRC, we evaluated the gut permeability in mono-housed AOM-DSS-treated Zfp90fl/fl anfd Zfp90ΔIEC mice using fluorescein isothiocyanate (FITC)-dextran. The Zfp90fl/fl mice displayed higher gut permeability compared with the Zfp90ΔIEC mice under AOM-DSS treatment, as shown by more FITC-dextran leakage into the circulation (Figure 2a). We also assessed gut-derived LPS levels in the serum of AOM-DSS-treated Zfp90fl/fl and Zfp90ΔIEC mice. Consistently, the Zfp90fl/fl mice exhibited markedly increased LPS translocation compared with the Zfp90ΔIEC mice (Figure 2b). To determine whether gut microbes can traverse the impaired gut barrier in Zfp90fl/fl mice, we performed fluorescence in situ hybridization (FISH) using a bacterial 16S probe to visualize the bacteria-mucus-epithelium interface. More bacteria were found to be in direct contact with the epithelial cells, or even reached deep down into the crypts through the mucus in Zfp90fl/fl mice compared with those in the Zfp90ΔIEC mice (Figure 2c). Tight junctions, as part of the gut barrier composition, connect adjacent IECs and regulate gut permeability.35 We found that zona occludens 1 (ZO-1) was expressed at a lower level in both the non-neoplastic and neoplastic colon tissue of the Zfp90fl/fl mice treated with AOM-DSS compared with that in the Zfp90ΔIEC mice (Figure 2(d, e). Additionally, ZO-1 could be visualized with a tight honeycomb-like staining pattern in the non-neoplastic colon tissue of the Zfp90ΔIEC mice. Whereas disorganized structures were observed in the neoplastic colon tissue of Zfp90ΔIEC mice and the non-neoplastic colon tissue of Zfp90fl/fl mice. Notably, ZO-1 was almost invisible in the neoplastic colon tissue of Zfp90fl/fl mice (Figure 2f). Collectively, these results demonstrated that conditional deletion of Zfp90 in the intestines can alleviate the impaired gut barrier induced by AOM-DSS treatment.
Figure 2.
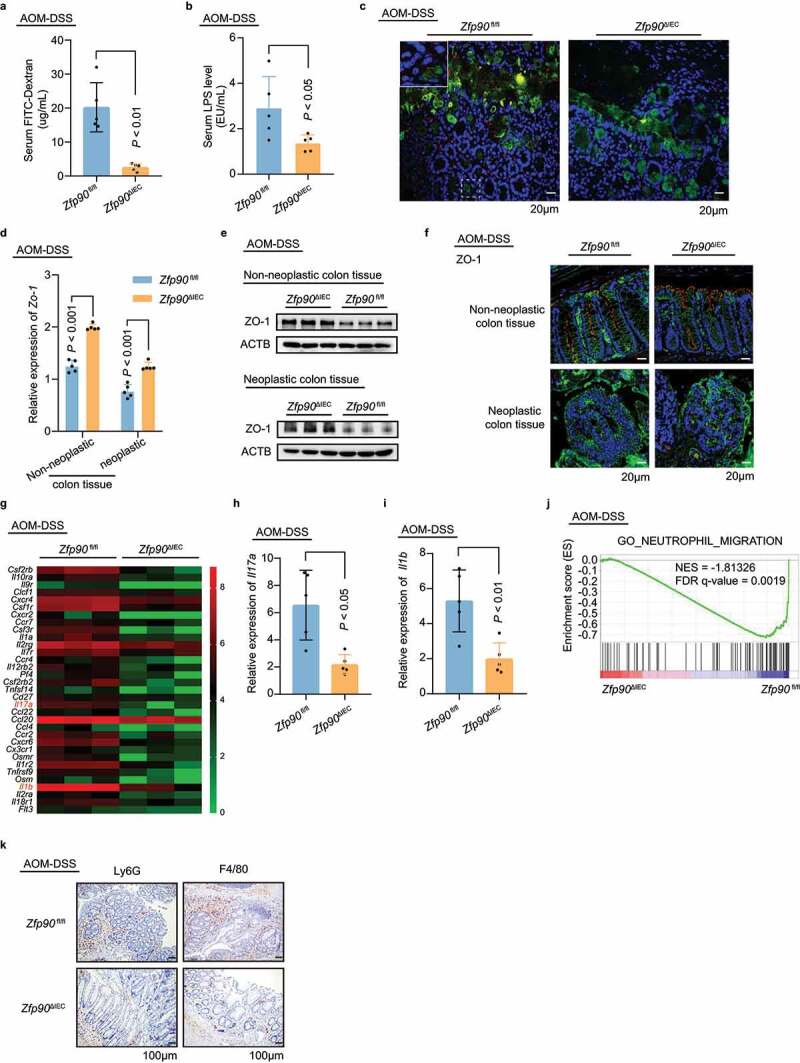
Zfp90 deletion in IECs improves the gut barrier and alleviates local inflammation during CAC development
a Intestinal permeability assessed by FITC-dextran in AOM-DSS-treated Zfp90fl/fl and Zfp90ΔIEC mice (n = 5 per group). b Serum levels of LPS in AOM-DSS-treated Zfp90fl/fl and Zfp90ΔIEC mice (n = 5 per group). c Representative immunofluorescence staining of the mucus UEA1 lectin and 16S bacterial probe in colon segments from AOM-DSS-treated Zfp90fl/fl and Zfp90ΔIEC mice. Inset: Magnified image of the area shown by the white dashed box, showing penetration of bacteria into the crypt region. Sections were stained with DAPI (blue), UEA1 lectin (green) and EUB338 universal bacterial FISH probe (red). d Real-time PCR was performed to determine the mRNA expression of Zo-1 in both neoplastic and non-neoplastic colon tissues from AOM-DSS-treated Zfp90fl/fl and Zfp90ΔIEC mice (n = 5 per group). e Representative data showing protein level of ZO-1 in both neoplastic and non-neoplastic colon tissues from AOM-DSS-treated Zfp90fl/fl and Zfp90ΔIEC mice. f Representative immunofluorescence staining of the ZO-1 protein in colon segments from AOM-DSS-treated Zfp90fl/fl and Zfp90ΔIEC mice. Sections were stained with DAPI (blue), CDH1 (green) and ZO-1 (red). g Heatmap representing the log2-normalized count of cytokine-cytokine receptor interaction pathway expression in the colonic epithelium from Zfp90fl/fl and Zfp90ΔIEC mice (n = 3 per group). h, i Real-time PCR was performed to determine the mRNA expression of Il17a and Il1b in the colonic epithelium from AOM-DSS-treated Zfp90fl/fl and Zfp90ΔIEC mice (n = 5 per group). j GSEA was used to identify the differential gene profiles between the colonic epithelium from AOM-DSS-treated Zfp90fl/fl and Zfp90ΔIEC mice. k Representative immunohistochemical staining for LY6G and F4/80 in colon segments from AOM-DSS-treated Zfp90fl/fl and Zfp90ΔIEC mice. Data with error bars represent the mean ± SD. Each panel is a representative experiment of at least three independent biological replicates. Mann–Whitney U test (a, h), nonpaired two-tailed t-test (b, d, i).
The barrier function was also evaluated in the acute colitis model by testing FITC-dextran and LPS level in the serum of the Zfp90fl/fl and Zfp90ΔIEC mice (Supplementary SFigure 2a, b). Consistent with the CAC model, the Zfp90fl/fl mice showed more FITC-dextran leakage and increased LPS translocation than the Zfp90ΔIEC mice under DSS treatment. Moreover, the level of Zo-1 was decreased in colon tissue of the DSS-treated Zfp90fl/fl mice as compared with that in the Zfp90ΔIEC mice (Supplementary SFigure 2c).
The gut barriers in the AOM-DSS-treated Zfp90fl/fl and Zfp90ΔIEC mice showed different stabilities; therefore, we decided to determine the extent of colon inflammation in the two groups. RNA sequencing (RNA-seq) of the colon epithelium was conducted and the transcriptomes of the two groups were compared. Principal component analysis (PCA) showed that the transcripts were separately clustered according to genotype (Supplementary SFigure 2d). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed that the genes differentially expressed between the Zfp90fl/fl and Zfp90ΔIEC mice could be categorized into 27 pathways related to the immune system, signal transduction, and cancers (Supplementary SFigure 2e), such as the “Cytokine-cytokine receptor interaction”, “Inflammatory bowel disease (IBD)” and “NF-κB signaling pathway”. Gene expression in the “Cytokine-cytokine receptor interaction” pathway clearly showed milder colonic inflammation in the Zfp90ΔIEC mice compared with that in the Zfp90fl/fl mice (Figure 2g). The decreased expression of Il17a (encoding interleukin 17A (IL17A)) and Il1b (encoding interleukin 1 beta (IL1β)) in the colon epithelium of AOM-DSS-treated Zfp90ΔIEC mice were also confirmed using real-time PCR (Figure 2(h, i)). Gene set enrichment analysis (GSEA) revealed that the gene sets related to GO_NEUTROPHIL_MIGRATION were enriched in the Zfp90fl/fl mice (Figure 2j). Accordingly, our immunohistochemistry analysis of lymphocyte antigen 6 complex locus G6D (LY6G) showed higher abundance of neutrophil infiltration in colon segments from the Zfp90fl/fl mice (Figure 2k, Supplementary SFigure 2f). Additionally, increased infiltration of macrophages was also detected in lesions from the colons of the Zfp90fl/fl mice using F4/80 (also known as EGF module-containing mucin-like receptor) staining (Figure 2k, Supplementary SFigure 2g).
Taken together, these data revealed that the Zfp90ΔIEC mice possessed a relatively integral gut barrier upon AOM-DSS treatment, compared with the Zfp90fl/fl mice. A better gut barrier function reduced the translocation of small molecules such as LPS and even gut bacteria themselves, thus leading to an overall milder inflammatory response.
The gut microbiota is indispensable for the antitumor phenotype in Zfp90ΔIEC mice
Recent research indicated that gut microbiota is involved in the development of CAC.6 Therefore, we tried to determine whether the gut microbiota contributes to the discrepancy between Zfp90fl/fl and Zfp90ΔIEC mice.
To confirm the role of the gut microbiota in the tumorigenesis of Zfp90ΔIEC and Zfp90fl/fl mice, the gut microbiota was depleted in Zfp90fl/fl and Zfp90ΔIEC mice using an antibiotic cocktail for 2 weeks before AOM-DSS treatment. The efficacy of the microbiota depletion was confirmed by quantification of fecal 16S rRNA gene levels (Supplementary SFigure 3a). Depletion of the microbiota eliminated the differences in colon tumor number between the two groups (Figure 3(a, b)). Furthermore, cell proliferation and apoptosis, as shown by PCNA and C-CAS3 levels, were similar between the two groups after elimination of the microbiota (Figure 3(c-f)). We then evaluated the gut permeability in Zfp90fl/fl and Zfp90ΔIEC mice and found no differences in FITC-dextran leakage between the two groups, indicating a similar decline in gut barrier function after AOM-DSS treatment (Figure 3g). Accordingly, real-time PCR showed that the epithelial expression of Il17a and Il1b became similar between the Zfp90fl/fl and Zfp90ΔIEC mice after antibiotic treatment (Figure 3(h, i)). Immunohistochemistry image of LY6G and F4/80 showed similar abundance of neutrophil and macrophage infiltration in colon segments from Zfp90fl/fl and Zfp90ΔIEC mice (Figure 3j, Supplementary SFigure 3b).
Figure 3.
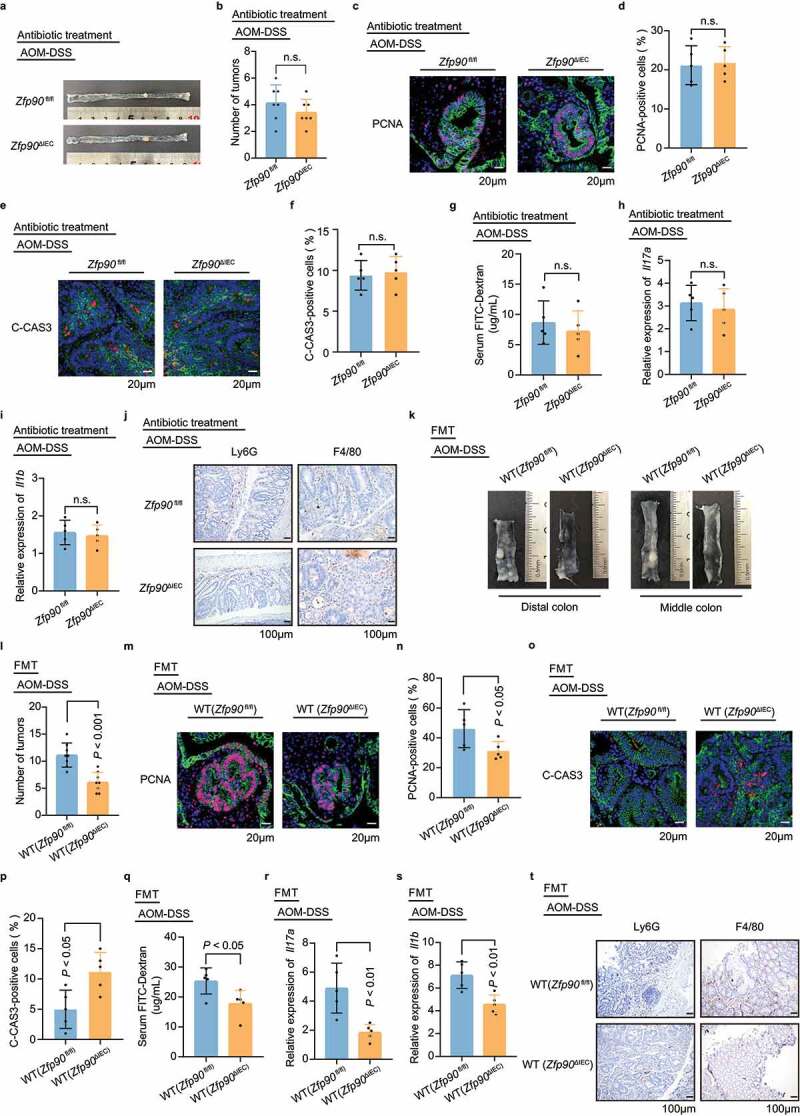
The gut microbiota is indispensable for the antitumor phenotype in Zfp90ΔIEC mice. a Representative images of colons in antibiotic-treated Zfp90fl/fl and Zfp90ΔIEC mice after AOM-DSS treatment. b Tumor numbers in antibiotic-treated Zfp90fl/fl and Zfp90ΔIEC mice (n = 7 per group). c Representative immunofluorescence staining of the PCNA protein in tumor tissues from antibiotic-treated Zfp90fl/fl and Zfp90ΔIEC mice after AOM-DSS treatment. Sections were stained with DAPI (blue), CDH1 (green) and PCNA (red). d The percentage of PCNA-positive tumor cells was quantified (n = 5 per group). e Representative immunofluorescence staining of C-CAS3 protein in tumor tissues from antibiotic-treated Zfp90fl/fl and Zfp90ΔIEC mice after AOM-DSS treatment. Sections were stained with DAPI (blue), CDH1 (green) and C-CAS3 (red). f The percentage of C-CAS3-positive tumor cells was quantified (n = 5 per group). g Intestinal permeability assessed by FITC-dextran in antibiotic-treated Zfp90fl/fl and Zfp90ΔIEC mice (n = 5 per group). h, i Real-time PCR was performed to determine the mRNA expression of Il17a and Il1b in the colonic epithelium from antibiotic-treated Zfp90fl/fl and Zfp90ΔIEC mice (n = 5 per group). j Representative immunohistochemical staining for LY6G and F4/80 in colon segments from antibiotic-treated Zfp90fl/fl and Zfp90ΔIEC mice with AOM-DSS treatment. k-t Gavage of WT mice with feces from Zfp90fl/fl and Zfp90ΔIEC mice (n = 5–8 per group). Representative images of distal and middle colons (k), number of tumors (l), representative immunofluorescence staining of PCNA (m), percentage of PCNA-positive tumor cells (n), representative immunofluorescence staining of C-CAS3 (o), percentage of C-CAS3-positive tumor cells (p), the serum FITC-Dextran level (q), mRNA expression of Il17a and Il1b in the colonic epithelium (r, s), and representative immunohistochemical staining for LY6G and F4/80 (t) from WT (Zfp90fl/fl mice) and WT (Zfp90ΔIEC mice) under AOM-DSS treatment. Sections were stained with DAPI (blue), CDH1 (green) and PCNA or C-CAS3 (red). Data with error bars represent the mean ± SD. Each panel is a representative experiment of at least three independent biological replicates. Nonpaired two-tailed t-test (b, d, f, g, h, i, l, n, p, r, s) and the Mann–Whitney U test (q) were used
We then sought to explore whether fecal microbiota transfer (FMT) could confer the antitumor ability of Zfp90ΔIEC mice to wild-type (WT) C57BL/6 J mice. Stools from mono-housed tumor-bearing Zfp90fl/fl and Zfp90ΔIEC mice were gavaged to antibiotic-treated WT mice in SPF background (Supplementary SFigure 3c). Gavage of antibiotic-treated mice with feces from the Zfp90ΔIEC mice led to a reduced number of colon tumors compared with those receiving feces from the Zfp90fl/fl mice under AOM-DSS treatment (Figure 3(k, l). Mice transplanted with fecal pellets from the Zfp90ΔIEC mice (WT (Zfp90ΔIEC mice)) also had lower proliferative activity and increased apoptosis compared with mice transplanted with fecal pellets from the Zfp90fl/fl mice (WT (Zfp90fl/fl mice)) (Figure 3(m-p)). Next, gut permeability was assessed between the two groups and a lower level of serum FITC-dextran was found in the WT (Zfp90ΔIEC) mice (Figure 3q). Real-time PCR showed lower expression of Il17a and Il1b in the WT (Zfp90ΔIEC) mice compared with that in WT (Zfp90fl/fl) mice (Figure 3(r, s)). Immunohistochemistry of LY6G and F4/80 showed higher levels of neutrophil and macrophage infiltration in colon segments from the WT (Zfp90fl/fl) mice than in those from the WT (Zfp90ΔIEC) mice (Figure 3t, Supplementary SFigure 3d).
We next investigated whether the gut microbiota also mediated the phenotypic variation in acute colitis model. Gut microbiota depletion was conducted for 2 weeks before the onset of DSS treatment. The severity of colitis was comparable between Zfp90fl/fl and Zfp90ΔIEC mice, as indicated by similar weight loss (Supplementary SFigure 3e) and colon length (Supplementary SFigure 3f). There were no differences in FITC-dextran leakage between the Zfp90fl/fl and Zfp90ΔIEC mice after 7 days of DSS treatment (Supplementary SFigure 3g). Furthermore, no significant differences in Zo-1 levels were detected between the colon tissues of the Zfp90fl/fl and Zfp90ΔIEC mice (Supplementary SFigure 3h).
Taken together, these results indicated that antitumor phenotype in the Zfp90ΔIEC mice is dependent on the gut microbiota and recipients transplanted with fecal microbiota from Zfp90ΔIEC mice could phenocopy their donors.
A Prevotella copri-defined microbiota underlies the mechanism of the deleterious effect of oncogene Zfp90
To explore the fecal bacterial composition in the Zfp90fl/fl and Zfp90ΔIEC mice, we performed 16S ribosomal DNA sequencing. PCA based on operational taxonomic unit (OTU) levels showed distinct gut microbiota compositions in the two groups under both naïve and AOM-DSS-treated conditions (Figure 4a and Supplementary SFigure 4a). Depletion of Zfp90 resulted in an increase in the community diversity and richness, as indicated by the Shannon and Sobs indices, respectively (Supplementary SFigure 4b, c), suggesting that the gut microbiota of the Zfp90ΔIEC mice had more species variation than that of the Zfp90fl/fl mice. KEGG analysis indicated that “lipopolysaccharide (LPS) biosynthesis proteins” was one of the key metabolic pathways downregulated in the Zfp90ΔIEC mice (Supplementary SFigure 4d). The different bacterial compositions between groups were further analyzed at the genus level. The abundance of Prevotellaceae_NK3B31_group and Prevotellaceae_UCG-001 was low in both the Zfp90fl/fl and Zfp90ΔIEC mice before AOM-DSS treatment, while the abundance increased significantly at the end of the treatment, especially in the Zfp90fl/fl mice (Figure 4b). Therefore, we speculated that the family Prevotellaceae could contribute to the AOM-DSS-induced CAC and might drive the different phenotypes of the two groups under these specific conditions. The family Prevotellaceae is composed of four genera, of which the genus Prevotella is known for its role in chronic inflammatory disease.36 The Prevotella genus encompasses more than 40 different culturable species, of which the Prevotella copri (P. copri) is rich among gut-commensal bacteria. Recent studies correlated higher abundance of P. copri with intestinal infection,37 insulin resistance,38 higher incidence of colon cancer,39 enhancement of rheumatoid arthritis,40 and metabolic syndrome.41 Therefore, we hypothesized that P. copri might be the driver microbe for CAC development in this context. To verify this hypothesis, real-time PCR was conducted in fecal pellets from untreated or AOM-DSS-treated Zfp90fl/fl and Zfp90ΔIEC mice. The abundance of P. copri in the fecal pellets from both mice increased significantly after AOM-DSS treatment and the abundance was markedly higher in the Zfp90fl/fl mice at the end of AOM-DSS treatment (Figure 4c).
Figure 4.
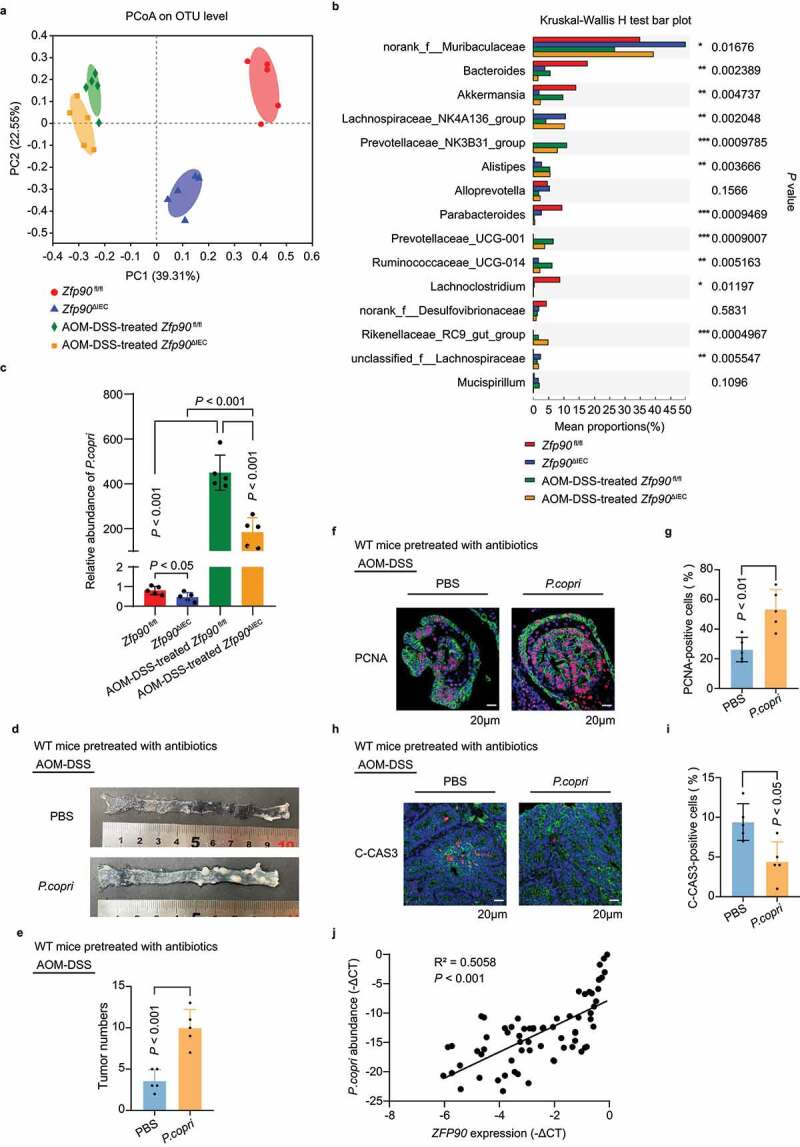
The gut microbiota is altered by intestine-specific deletion of oncogene Zfp90. a Principal component analysis plot based on bacterial 16S ribosomal DNA gene sequences of the fecal content from Zfp90fl/fl and Zfp90ΔIEC mice before and after AOM-DSS-treatment (n = 5 per group). b Differential abundances of bacteria in the fecal content of Zfp90fl/fl and Zfp90ΔIEC mice before and after AOM-DSS-treatment. c Real-time PCR was performed to determine the abundance of P. copri in the fecal content of Zfp90fl/fl and Zfp90ΔIEC mice before and after AOM-DSS-treatment. d Representative images of colons in PBS- and P. copri-treated WT mice after AOM-DSS treatment. e Tumor numbers in PBS- and P. copri-treated WT mice after AOM-DSS treatment. f Representative immunofluorescence staining of the PCNA protein in tumor tissues from PBS- and P. copri-treated WT mice after AOM-DSS treatment. Sections were stained with DAPI (blue), CDH1 (green) and PCNA (red). g The percentage of PCNA-positive tumor cells was quantified. h Representative immunofluorescence staining of C-CAS3 protein in tumor tissues from PBS- and P. copri-treated WT mice after AOM-DSS treatment. Sections were stained with DAPI (blue), CDH1 (green) and C-CAS3 (red). i The percentage of C-CAS3-positive tumor cells was quantified. j The correlation between P. copri abundance and ZFP90 expression in specimens from patients with CRC. Statistical analysis was performed using nonpaired two-tailed t-test (e, g, i), one-way ANOVA (c), and Wilcoxon’s rank-sum test (j)
To confirm the effect of P. copri on AOM-DSS-induced colorectal tumorigenesis, WT mice pretreated with antibiotics for 2 weeks were gavaged with 108 colony-forming units (cfu) of P. copri or phosphate-buffered saline (PBS) every other day during AOM-DSS treatment (Supplementary SFigure 4e). Eighty days after the induction of tumorigenesis, increased tumor number was observed in the mice colonized with P. copri (Figure 4(d, e)). Moreover, the mice colonized with P. copri exhibited higher proliferative activity and less apoptosis than the mice gavaged with PBS (Figure 4(f-i)). We then explored the role of P. copri in the acute colitis model. After 2 weeks of antibiotic treatment, the mice were first gavaged with P. copri for 1 week and then given DSS for 7 days, with the gavage of P. copri continuing throughout the experiment. More severe colitis was observed in the mice colonized with P. copri (Supplementary SFigure 4f, g). The gut barrier dysfunction was more severe in the mice colonized with P. copri, as indicated by more FITC-dextran leakage, more LPS translocation, and lower levels of Zo-1 (Supplementary SFigure 4h-j).
We then sought to determine the correlation between the expression of ZFP90 and the abundance of P. copri in clinical patients. There was a lack of CAC specimens; therefore, we verified the relationship in patients with CRC, because chronic inflammation is frequently observed in this population.16 Remarkably, a positive correlation was detected between the abundance of P. copri and the expression of ZFP90 in 81 patients with CRC (Figure 4j).
Taken together, our results indicated that P. copri plays an important role in the development of CAC, and that the characteristics caused by a P. copri-defined microbiota, including increased LPS biosynthesis and a compromised gut barrier function, might partly account for the susceptibility of Zfp90fl/fl mice to colitis and AOM-DSS-induced CAC.
Decreased TLR4-dependent PI3K/AKT/NF-κB signaling in the colon tissues of Zfp90ΔIEC mice
Researchers have found that antigen presenting cells (APCs) in the Prevotella-rich mucosa showed a profile similar to those activated by LPS.42 Considering the increased abundance of P. copri and the enrichment of the “LPS biosynthesis proteins” pathway in Zfp90fl/fl mice, we speculated that a P. copri-defined microbiota might have a more potent ability to produce LPS. Accompanied by a compromised gut barrier, the elevated production of LPS might then translocate and play a vital role in the development of CAC. Previous studies demonstrated that LPS could bind with Toll-like receptor 4 (TLR4) at the surface of intestinal cells and triggered signal transductions and activated the expression of genes encoding inflammatory effectors, such as nuclear factor κB (NF-κB).43,44
Based on these results, we first detected the expression of TLR4 in the Zfp90ΔIEC and Zfp90fl/fl mice using real-time PCR, western blot, and immunofluorescence. However, the expression of TLR4 in the colon tissue was not significantly different between mono-housed Zfp90ΔIEC and Zfp90fl/fl mice during AOM-DSS treatment (Figure 5(a-c)). These data revealed that deletion of Zfp90 in the colon epithelium did not change the expression of TLR4.
Figure 5.
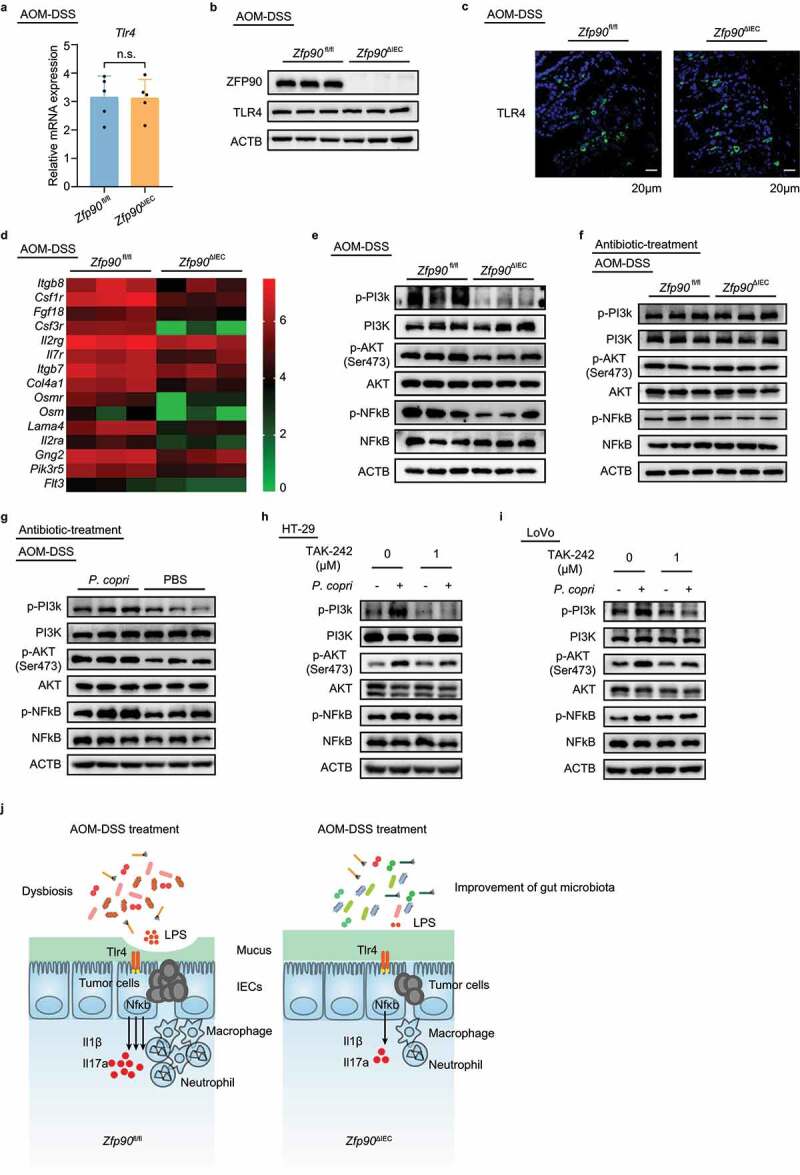
Decreased TLR4-dependent PI3K/AKT/NF-κB signaling in colon tissue of Zfp90ΔIEC mice
a Real-time PCR was performed to determine the mRNA expression of Tlr4 in AOM-DSS-treated Zfp90fl/fl and Zfp90ΔIEC mice (n = 5 per group). b Representative data showing protein level of TLR4 in AOM-DSS-treated Zfp90fl/fl and Zfp90ΔIEC mice. c Representative immunofluorescence staining of the TLR4 protein in tumor tissues from AOM-DSS-treated Zfp90fl/fl and Zfp90ΔIEC mice. Sections were stained with DAPI (blue) and TLR4 (green). d Heatmap representing log2-normalized count of the PI3K-AKT pathway expression on the colonic epithelium from Zfp90fl/fl and Zfp90ΔIEC mice (n = 3 per group). e Representative data showing the activity of the PI3K-AKT-NFκB pathway in AOM-DSS-treated Zfp90fl/fl and Zfp90ΔIEC mice. f Representative data showing the activity of the PI3K-AKT-NFκB pathway in antibiotic-treated Zfp90fl/fl and Zfp90ΔIEC mice after AOM-DSS treatment. g Representative data showing the activity of the PI3K-AKT-NFκB pathway in PBS- and P. copri-treated WT mice after AOM-DSS treatment. h, i HT-29 (h) and LoVo (i) cell lines were pretreated with TLR4 inhibitor (TAK-242) 30 minutes before co-culturing with P. copri. Representative data showing the effect of TLR4 inhibitor on the activation of the PI3K-AKT-NFκB pathway. j Model for the P. copri-defined microbiota-mediated colorectal tumorigenesis. Data with error bars represent the mean ± SD. Each panel is a representative experiment of at least three independent biological replicates. Statistical analysis was performed using nonpaired two-tailed t-test.
To determine whether the deletion of Zfp90 in the colon epithelium had an impact on the activation of TLR4, we assessed the activity of its downstream signaling pathway. It has been established that phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K) -protein kinase B (AKT)-NF-κB is one of primary signaling transduction pathways following the activation of TLR4.45,46 In accordance with this, our RNA-seq results revealed that the “PI3K-Akt signaling pathway” was downregulated in the Zfp90ΔIEC mice (Supplementary SFigure 2e, Figure 5d). The inhibitory function of Zfp90 deletion on the activity of the PI3K-Akt signaling pathway was confirmed by western blotting (Figure 5e). Whereas the mRNA expression levels of Pi3k, Akt, and Nfkb showed no differences between the Zfp90fl/fl and Zfp90ΔIEC mice (Supplementary Figure 5a-c). To further verify that the gut microbiota mediated the activation of PI3K/AKT/NF-κB signaling, we detected the activity of this pathway in antibiotic-treated Zfp90fl/fl and Zfp90ΔIEC mice. At the end of AOM-DSS treatment, no significant differences were found in the activity of the PI3K-AKT signaling pathway between antibiotic-treated Zfp90fl/fl and Zfp90ΔIEC mice (Figure 5f). Moreover, stronger activation of PI3K/AKT/NF-κB signaling was observed in antibiotic-treated WT mice colonized with P. copri (Figure 5g).
To further verify that the activation of PI3K/AKT/NF-κB signaling was TLR4-dependent, HT-29 cells and LoVo cells were first treated with a TLR4 inhibitor (TAK-242) and then co-cultured with P. copri. Western blotting showed that P. copri-mediated PI3K-AKT-NF-κB activation was largely abrogated by the TLR4 inhibitor (Figure 5(h, i)), corroborating the role of TLR4 in this process.
Taken together, these data showed that a P. copri-defined microbiota might participate in the development of CAC by activating TLR4 and the PI3K-AKT-NF-κB signaling pathway (Figure 5j).
Discussion
ZFP90 is a zinc-finger protein containing a KRAB box. Dysregulation of ZFP90 has been proven to be associated with several diseases including obesity,47 cardiac dysfunction,48 and mental retardation.49 In addition, we previously showed that ZFP90 is a target gene of CRC susceptibility locus 16q22.132. Using an AOM-induced sporadic CRC mouse model and clinical specimens from patients with CRC, we validated its oncogenic role in the CRC. However, the underlying mechanisms of ZFP90 in the development of CAC warrant further investigation.
In this study, through overlapping findings from GWAS and the GEO database, we found that ZFP90 might be a key gene that regulates the pathogenesis of CAC. We observed that conditional knock-out of Zfp90 in IECs could protect mice from AOM-DSS-induced CAC.
Intestinal barrier breach and subsequent inflammation have been proven to contribute to chronic inflammatory and malignant colorectal diseases;50 therefore, we compared the gut permeability in the AOM-DSS-treated Zfp90fl/fl mice with that in the AOM-DSS-treated Zfp90ΔIEC mice. As expected, the Zfp90fl/fl mice displayed significantly higher intestinal permeability. Furthermore, our RNA-seq and PCR results confirmed higher inflammatory responses in the Zfp90fl/fl mice after AOM-DSS treatment. Taken together, our results suggested that the increased gut permeability in the Zfp90fl/fl mice might contribute to the exaggerated inflammation and subsequent AOM-DSS-induced carcinogenesis.
Accumulating evidence demonstrates that the human gut microbiota is involved in various pathological conditions, including CRC. We therefore investigated whether the gut microbiota actively mediates the protective role of Zfp90 deletion in AOM-DSS-induced CAC. We treated Zfp90fl/fl and Zfp90ΔIEC mice with antibiotics before the initiation of AOM-DSS treatment. After depletion of the microbiota, we failed to find differences between the two groups in terms of tumor development, gut permeability, and inflammation. We then employed FMT, and found that mice transplanted with the fecal microbiota from Zfp90fl/fl and Zfp90ΔIEC mice phenocopied their donors in terms of tumor development, gut permeability, and inflammation. Furthermore, our high-throughput sequencing results indicated that the microbiota compositions were different between AOM-DSS-treated Zfp90ΔIEC and Zfp90fl/fl mice. Specifically, we detected a higher abundance of P. copri in the Zfp90fl/fl mice. It has been reported that a P. copri-defined microbiota was associated with exacerbated colitis in mice.37,51 Accordingly, our functional analysis demonstrated that “lipopolysaccharide (LPS) biosynthesis proteins” was one of the key metabolic pathways that was upregulated in the microbiota of the Zfp90fl/fl mice. Considering that the gut barrier was greatly compromised in the Zfp90fl/fl mice during AOM-DSS treatment, we therefore speculated that the increased LPS production from the intestinal flora could subsequently translocate and induce inflammation by activating TLR45. As such, a P. copri-defined microbiota would exacerbate inflammation and contribute to the subsequent development of AOM-DSS-induced CAC.
To further corroborate the enhanced activation of TLR4 by LPS, the expression of TLR4 and activity of its downstream signaling pathway were assessed. Although the expression of TLR4 was not significantly different between Zfp90fl/fl and Zfp90ΔIEC mice, PI3K-AKT-NF-κB signaling pathway activation was strongly decreased in the Zfp90ΔIEC mice. PI3K-AKT-NF-κB is one of primary signaling transduction pathways responsible for the synthesis of pro-inflammatory mediators following the activation of TLR4.52,53 Our in vitro experiments further demonstrated that pretreatment with a TLR4 inhibitor could greatly reduce P. copri-induced activation of PI3K-AKT-NF-κB signaling. Thus, these results validated that TLR4 activation was increased by the P. copri-defined microbiota, which then contributed to the development of inflammatory and malignant pathologies.
In conclusion, our work demonstrated the oncogenic role of Zfp90 in AOM-DSS-induced CAC and this relationship is, at least partly, mediated by the gut microbiota. The P. copri-defined microbiota in Zfp90fl/fl mice promotes the production and translocation of LPS upon a compromised gut barrier, which contributes to the subsequent inflammatory and carcinogenic processes by activating the TLR4-dependent PI3K-AKT-NF-κB signaling pathway. This study highlighted that manipulation of the gut microbiota might represent a potential strategy to prevent and treat CAC.
Methods
Study population and clinical specimens
Patients with CRC were recruited from Renji Hospital affiliated to Shanghai Jiao Tong University School of Medicine. All the patients with CRC were of Han Chinese ethnicity. Written informed consents were obtained from all participants in this study. All the research was carried out in accordance with the provisions of the Helsinki Declaration of 1975. There were 81 snap frozen, colorectal cancerous tissues. DNA and RNA were extracted from the snap frozen tissues using an AllPrep DNA/RNA Mini Kit (QIAGEN, Hilden, Germany) under the manufacturer’s guidelines to detect ZFP90 expression and P. copri abundance.
Generation of Zfp90ΔIEC Mice
Zfp90ΔIEC mice have been described previously.32 Briefly, to generate the Zfp90fl/+,Villin-cre/+ mice, exon 4 and the 3′ UTR were flanked by loxP sites using C57BL/6 J ES cells with a 2 kb fragment upstream and downstream as the 5′ and 3′ arms. Then, the Zfp90fl/+mice were crossed to C57BL/6 J Villin-cre mice to generate Zfp90fl/+,Villin-cre/+ mice. Zfp90fl/+,Villin-cre/+ mice were each intercrossed to generate homozygous knockout mice (Zfp90ΔIEC) and littermates (Zfp90fl/fl). Mice were genotyped via PCR and Sanger sequencing using genomic DNA obtained from mouse tails.
All mice were bred in an SPF environment. Mouse experiments were conducted in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals. The study procedures were approved by the Institutional Animal Care and Use Committee of Renji Hospital, Shanghai Jiao Tong University School of Medicine.
Mouse model
To construct the CAC model,54 Zfp90fl/fl and Zfp90ΔIEC mice at 8 weeks old were injected intraperitoneally with AOM (10 mg/kg, Sigma-Aldrich, St. Louis, MO, USA). After 5 days, 2.5% DSS was added to the drinking water for 7 consecutive days, followed by 14 days of regular water. Three cycles of DSS treatment were used. The mice were sacrificed on day 80 after the induction of tumorigenesis. All colon tissues, tumor tissues, feces, serum, and spleens were collected on day 80. To deplete the gut microbiota, drinking water supplemented with an antibiotic cocktail (0.5 g/L of ampicillin, neomycin, and metronidazole and 0.25 g/L of vancomycin) was given to the mice for 2 weeks before AOM-DSS treatment and the procedure was repeated once in the middle of the experiment. For the gut microbiota transfer experiment, fresh stool samples from tumor-bearing Zfp90fl/fl and Zfp90ΔIEC mice were collected, suspended in 30% glycerin/PBS, aliquoted, and stored at – 80 °C. Fecal microbiota was then gavaged into the antibiotic-treated mice three times a week. Colons were removed and were flushed with cold PBS after the mice were sacrificed. The colon length, tumor number, and tumor size were measured for each mouse. The colons of different experimental batches were used for histopathological analysis or epithelium cell isolation. Spleens were also separated from each mouse.
Isolation of colonic epithelium cells
Colonic epithelium cells were isolated as described previously, with some modifications.55 Colons were collected from mice and cleared by holding with forceps and flushing with a syringe filled with ice-cold PBS. Next, the colons were opened longitudinally and cut into 1–2 cm pieces, followed by several PBS washes. The pieces were then incubated with 2.5 mM ethylenediaminetetraacetic acid (EDTA) and 1 mM Dithiothreitol (DTT) in Hank’s balanced salt solution (HBSS) for 20 min at 37 °C under slow rotation. Supernatants were then passed through a 100 μm cell strainer. Colonic epithelium cells were collected by centrifugation at 800 × g and were used for the following analysis.
Immunohistochemistry (IHC)
For immunohistochemistry, 4-μm-thick paraffin colon tissue slides were subjected to heat-induced antigen retrieval, followed by peroxidase blocking treatment to remove endogenous hydrogen peroxide. After blocking with 10% sheep serum in PBS, the slides were incubated with anti-LY6G (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and anti-F4/80 (Santa Cruz Biotechnology) antibodies in a humidified chamber at 4°C overnight. Then, the slides were incubated with horseradish peroxidase (HRP)-labeled secondary antibodies at room temperature for 30 minutes and then stained with 3,3’-Diaminobenzidine (DAB) substrate solution.
Immunofluorescence staining and confocal microscopy
For immunofluorescence, 4-μm-thick paraffin colon tissue slides were subjected to heat-induced antigen retrieval, followed by sodium borohydride treatment to remove tissue autofluorescence. After blocking with 5% bovine serum albumin in PBS, the slides were incubated with anti-C-CAS3 (Cell Signaling Technology (CST), Danvers, MA, USA) and anti-PCNA (Santa Cruz Biotechnology) antibodies in a humidified chamber at 4°C overnight. Then, the slides were incubated with fluorophore-labeled secondary antibodies at room temperature for 1 hour. After rinsing with PBS, the slides were mounted using antifade mounting medium with 4′,6-diamidino-2-phenylindole (DAPI), and the fluorescence was detected under a laser-scanning confocal microscope.
Fluorescence in situ hybridization (FISH)
FISH was performed as described previously.56 Briefly, colon tissues with fecal pellets were fixed in methanol-carnoy fixative (60% methanol, 30% chloroform, and 10% glacial acetic acid). After fixation, tissues were washed with the following solutions: twice in methanol, twice in ethanol, and twice in xylene, and then embedded in paraffin. Paraffin sections of 6 μm were subjected to mucin 2 staining with fluorescein-labeled Anti-Ulex Europaeus-I (UEA-I) lectin, and bacteria were detected using the universal bacteria FISH probe, 5′-Cy3-labeled EUB338I. After rinsing with washing buffer, the slides were mounted with DAPI and imaged under a laser-scanning confocal microscope.
FITC–dextran intestinal permeability assay
Colonic permeability was assessed by oral gavage of 4 kDa FITC-labeled dextran (Sigma-Aldrich). Mice were fasted for 3 hours before the assay and then administered with 150 μL FITC-dextran (80 mg/mL). After 4 hours, 100 μL of blood was collected and centrifuged at 10000 × g for 10 minutes to acquire serum. The serum was diluted 1:4 in water and 100 μL/well was added to 96-well plates. The FITC-labeled dextran levels were then measured by fluorometry with 485 excitation/528 emission.
Analysis of LPS levels in serum
The serum LPS levels were quantified using a simple and sensitive amebocyte lysate assay (Pierce Chromogenic Endotoxin Quant Kit, Pierce Biotechnology, Rockford, IL, USA). The experiment was conducted according to the manufacturer’s instructions. Briefly, serum samples were diluted 50-fold and heat-shocked at 70 °C for 15 minutes. Then, the Amebocyte Lysate Reagent, the pre-warmed Chromogenic Substrate Solution, and the Stop Solution were added sequentially added to the serum sample. The optical density (OD) was read at 405 nm immediately after assay completion.
Cell lines
Human CRC cell lines, HT-29 and LoVo were used in the study. All CRC cell lines were cultured according to ATCC culture methods. HT-29 was cultured in McCoy’s 5a Medium (GIBCO, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (GIBCO, Carlsbad, CA). LoVo was cultured in F-12 K Medium (GIBCO, Carlsbad, CA) supplemented with 10% FBS. These two cell lines were cultured at 37°C with 5% CO2.
Western Blot and Chemical Reagents
Western blot was performed using standard protocols. Cell extracts were collected and quantified using BCA Protein Assay Kit (Thermo Scientific, Rockford, U.S.A.). 40 μg of protein was electrophoresed through 7.5% SDS polyacrylamide gels and were then transferred to PVDF membranes (Bio-Rad, Hercules, CA). The membranes were blocked in 5% BSA for one hour and then incubated with primary antibodies at 4°C overnight. Secondary antibodies were labeled with HRP (KangChen, China) and the signals were detected using Pierce ECL Western Blotting Substrate (Thermo Scientific, Rockford, U.S.A.) by ChemiDoc Imaging System (BIO-RAD, U.S.A.). A β-actin antibody was used as a control for whole-cell lysates.
16S ribosomal DNA gene sequencing
Fecal genomic DNA was extracted using QIAamp PowerFecal DNA Kit (QIAGEN). The final DNA concentration and purity were determined using a NanoDrop 2000 UV-vis spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA), and the DNA quality was checked using 1% agarose gel electrophoresis. The V3–V4 hypervariable regions of the bacteria 16S rRNA gene were amplified using primers 338 F (5′- ACTCCTACGGGAGGCAGCAG-3′) and 806 R (5′-GGACTACHVGGGTWTCTAAT-3′) in a thermocycler PCR system (GeneAmp 9700, Applied Biosystems, Foster City, CA, USA). The PCR reactions were conducted using the following program: 3 min of denaturation at 95°C; 27 cycles of 30 s at 95°C, 30 s for annealing at 55°C, and 45 s for elongation at 72°C; and a final extension at 72°C for 10 min. The PCR reactions were performed in triplicate in a 20 μL mixture containing 4 μL of 5 × FastPfu Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu Polymerase, and 10 ng of template DNA. The resulting PCR products were extracted from a 2% agarose gel and further purified using an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) and quantified using a QuantiFluor™-ST (Promega, Madison, WI, USA) according to the manufacturer’s protocol.
The purified amplicons were pooled in equimolar amounts and paired-end sequenced (2 × 300 bp) on an Illumina MiSeq platform (Illumina, San Diego, CA, USA) according to the standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). The raw reads were deposited in the NCBI Sequence Read Archive (SRA) database (Accession Number: SRP277482).
Raw fastq files were quality-filtered by Trimmomatic and merged by FLASH with the following criteria: (i) The reads were truncated at any site receiving an average quality score < 20 over a 50 bp sliding window. (ii) Sequences whose overlap was longer than 10 bp were merged according to their overlap with a mismatch of no more than 2 bp. (iii) Sequences of each sample were separated according to barcodes (exactly matching) and Primers (allowing two nucleotide mismatches), and reads containing ambiguous bases were removed.
Operational taxonomic units (OTUs) were clustered with a 97% similarity cutoff using UPARSE (version 7.1 http://drive5.com/uparse/) with a novel ‘greedy’ algorithm that performs chimera filtering and OTU clustering simultaneously. The taxonomy of each 16S rRNA gene sequence was analyzed using the RDP Classifier algorithm (http://rdp.cme.msu.edu/) against the Silva (SSU123) 16S rRNA database, using a confidence threshold of 70%.
Fecal DNA extraction and P. copri quantification
Fecal genomic DNA was extracted using a QIAamp DNA Mini Kit (QIAGEN) according to the manufacturer’s instructions. Real-time PCR was performed to detect the P. copri level using 20 ng of genomic DNA on an Applied Biosystems 7900 quantitative PCR system. P. copri quantitation was measured relative to the 16s rDNA gene.
RNA Extraction and Quantitative Real-time Reverse Transcription PCR
Total RNA was extracted from mouse colonic epithelium cells using the TRIzol reagent and from human colorectal tissues using AllPrep DNA/RNA Mini Kit (QIAGEN) following the manufacturer’s guidelines. Total RNA (1 μg) was reverse transcribed using the PrimeScript RT Reagent Kit (Perfect Real Time; Takara, Shiga, Japan) to detect relative mRNAs. Quantitative real-time PCR was performed in triplicate on an Applied Biosystems 7900 quantitative PCR system as described above. The Ct values obtained from different samples were compared using the 2−ΔCt method. ACTB (beta actin) served as the internal reference gene.
RNA-seq
Samples were sequenced in an Illumina HiSeq 3000 instrument for 2 × 150-bp paired-end sequencing. Reads were mapped to the human genome (hg19) using TopHat v2.0.117 (http://tophat.cbcb.umd.edu) with the default options with a TopHat transcript index built from Ensembl_GRCh37.57 Transcript expression was estimated with an improved version of Cuffdiff (http://cufflinks.cbcb.umd.edu). Cuffdiff was run with the default options against the UCSC iGenomes GTF file from Illumina (http://cufflinks.cbcb.umd.edu/igenomes.html). The workflow used to analyze the data was described in detail in Trapnell et al.58 To identify a gene or transcript as differentially expression, Cuffdiff2 tests the observed log-fold-change in its expression against the null hypothesis of no change (i.e., the true log-fold-change is zero). Clustering of gene expression profiles was achieved using the csDendro function from CummeRbund (http://compbio.mit.edu/cummeRbund/). The RNA sequence data has been deposited in NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and is accessible through GEO Series accession number GSE156241 (reviewer access token: sfwpqmoqpzchraz)
Statistical Analysis
Statistical analyses were carried out using the program R (www.r-project.org). Data from at least three independent experiments performed in multiple replicates are presented as the means ± SD. Error bars in the scatterplots and the bar graphs represent the SD. Data was examined to determine whether they were normally distributed using the One-Sample Kolmogorov-Smirnov test. If the data was normally distributed, comparisons of measurement data between two groups were performed using an independent sample t test. If the results showed a significant difference, when the data presented a skewed distribution, comparisons were performed using a nonparametric test. Measurement data between two groups was compared using nonparametric Mann–Whitney U test or Wilcoxon’s rank-sum test. Spearman correlation analysis was performed to determine the correlation between two variables. Fisher’s exact test was used to analyze the pathological grade between two groups. One-way ANOVA was performed to test the difference in P. copri numbers in the fecal content between the mice. Two-way ANOVA was used to analyze the body weight and tumor distribution between two groups.
Supplementary Material
Acknowledgments
This project was supported in part by grants from the National Natural Science
Foundation of China (81421001, 81530072, 81830081, and 31970718), the Shanghai Municipal Health Commission, Collaborative Innovation Cluster Project (2019CXJQ02), and the Innovative research team of high-level local universities in Shanghai (SSMU-ZLCX20180200). We thank all the patients who participated in this study.
Funding Statement
This work was supported by the National Natural Science Foundation of China [81421001, 81530072, 81830081,31970718].
Disclosure statement
The authors declare no potential conflicts of interest.
AUTHOR Contributions
Conceptualization: J-X.H., C-Y.Y., and J-Y.F.; Investigation: J-X.H., Z-H.T., Y. Q., C-Y.Y., J.L., Z-R.K., S.L., Y.X., J.H., H.C., and Y-X.C.; Writing-Original Draft: J-X.H., Z-H.T.; Statistical analyses: J-X.H. and Z-H.T.; Writing-Reviewing & Editing: J-Y.F.; Funding Acquisition: J-Y.F.; Supervision: J-Y.F.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Siegel RL, Miller KD, Jemal A.. Cancer statistics, 2019. CA: A Cancer Journal for Clinicians. 2019;69(1):7–20. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Peters U, Bien S, Zubair N. Genetic architecture of colorectal cancer. Gut. 2015;64(10):1623–1636. doi: 10.1136/gutjnl-2013-306705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Keefe SJ. Diet, microorganisms and their metabolites, and colon cancer. Nature Reviews. Gastroenterology & Hepatology. 2016;13(691–706): doi: 10.1038/nrgastro.2016.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clevers H. At the crossroads of inflammation and cancer. Cell. 2004;118(6):671–674. doi: 10.1016/j.cell.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Choi C-HR, Bakir IA, Hart AL, Graham TA. Clonal evolution of colorectal cancer in IBD. Nature Reviews. Gastroenterology & Hepatology. 2017;14(218–229): doi: 10.1038/nrgastro.2017.1. [DOI] [PubMed] [Google Scholar]
- 7.Olén O, Erichsen R, Sachs MC, Pedersen L, Halfvarson J, Askling J, et al. Colorectal cancer in ulcerative colitis: a Scandinavian population-based cohort study. Lancet 2020;395:123–31. [DOI] [PubMed] [Google Scholar]
- 8.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, et al. An atlas of genetic correlations across human diseases and traits. Nature genetics 2015;47:1236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rankinen T, Sarzynski MA, Ghosh S, Bouchard C. Are there genetic paths common to obesity, cardiovascular disease outcomes, and cardiovascular risk factors? Circ Res. 2015;116:909–922. doi: 10.1161/CIRCRESAHA.116.302888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amare AT, Schubert KO, Klingler-Hoffmann M, Cohen-Woods S, Baune BT. The genetic overlap between mood disorders and cardiometabolic diseases: a systematic review of genome wide and candidate gene studies. Transl Psychiatry. 2017;7(1):e1007–e1007. doi: 10.1038/tp.2016.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu Y, Kweon -S-S, Tanikawa C, Jia W-H, Xiang Y-B, Cai Q, Zeng C, Schmit SL, Shin A, Matsuo K, et al. Large-scale genome-wide association study of East Asians identifies loci associated with risk for colorectal cancer. Gastroenterology. 2019;156(5):1455–1466. doi: 10.1053/j.gastro.2018.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furey TS, Sethupathy P, Sheikh SZ. Redefining the IBDs using genome-scale molecular phenotyping. Nature Reviews Gastroenterology & Hepatology. 2019;16(5):296–311. doi: 10.1038/s41575-019-0118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Consortium UIG, Barrett JC, Lee JC, Lees CW, Prescott NJ, Anderson CA, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nature genetics 2009; 41:1330–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14(3):141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 15.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448(7152):427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 16.Lasry A, Zinger A, Ben-Neriah Y. Inflammatory networks underlying colorectal cancer. Nat Immunol. 2016;17(3):230–240. doi: 10.1038/ni.3384. [DOI] [PubMed] [Google Scholar]
- 17.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, et al. Evolution of mammals and their gut microbes. Science (New York, N.Y.). 2008;320(5883):1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Consortium HMP. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(207–214): doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodrich JK, Davenport E, Beaumont M, Jackson M, Knight R, Ober C, Spector T, Bell J, Clark A, Ley R, et al. Genetic determinants of the gut microbiome in UK twins. Cell Host & Microbe. 2016;19(5):731–743. doi: 10.1016/j.chom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang T, Fan C, Yao A, Xu X, Zheng G, You Y, Jiang C, Zhao X, Hou Y, Hung M-C, et al. The adaptor protein CARD9 protects against colon cancer by restricting mycobiota-mediated expansion of myeloid-derived suppressor cells. Immunity. 2018;49(3):504–514.e504. doi: 10.1016/j.immuni.2018.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plantamura E, Dzutsev A, Chamaillard M, Djebali S, Moudombi L, Boucinha L, et al. MAVS deficiency induces gut dysbiotic microbiota conferring a proallergic phenotype. Proceedings of the National Academy of Sciences of the United States of America 2018;115:10404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Tinoco R, Elmén L, Segota I, Xian Y, Fujita Y, et al. Gut microbiota dependent anti-tumor immunity restricts melanoma growth in Rnf5 mice. Nature communications 2019;10:1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamas B, Richard ML, Leducq V, Pham H-P, Michel M-L, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad JM, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nature Medicine. 2016;22(6):598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cani PD, Jordan BF. Gut microbiota-mediated inflammation in obesity: a link with gastrointestinal cancer. Nature Reviews Gastroenterology & Hepatology. 2018;15(11):671–682. doi: 10.1038/s41575-018-0025-6. [DOI] [PubMed] [Google Scholar]
- 25.Cani PD, Possemiers S, Van De Wiele T, Guiot Y, Everard A, Rottier O, Geurts L, Naslain D, Neyrinck A, Lambert DM, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58(8):1091–1103. doi: 10.1136/gut.2008.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slowicka K, Petta I, Blancke G, Hoste E, Dumas E, Sze M, et al. Zeb2 drives invasive and microbiota-dependent colon carcinoma. Nature Cancer 2020;1:620–34. [DOI] [PubMed] [Google Scholar]
- 27.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(D1):D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler AD. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ardlie KG, Deluca DS, Segre AV, Sullivan TJ, Young TR, Gelfand ET, Trowbridge CA, Maller JB, Tukiainen T, Lek M; Human genomics . The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science (New York, N.Y.). 2015;348(6235):648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Y, Li X, Yang M, Zhao Q, Liu X, Wang G, Lu X, Wu Q, Wu J, Yang Y, et al. Colitis-accelerated colorectal cancer and metabolic dysregulation in a mouse model. Carcinogenesis. 2013;34(8):1861–1869. doi: 10.1093/carcin/bgt135. [DOI] [PubMed] [Google Scholar]
- 31.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(Database issue):D991–5. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu C-Y, Han J-X, Zhang J, Jiang P, Shen C, Guo F, Tang J, Yan T, Tian X, Zhu X, et al. A 16q22.1 variant confers susceptibility to colorectal cancer as a distal regulator of ZFP90. Oncogene. 2020;39(6):1347–1360. doi: 10.1038/s41388-019-1055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lees CW, Barrett JC, Parkes M, Satsangi SJ. New IBD genetics: common pathways with other diseases. Gut. 2011;60(12):1739–1753. doi: 10.1136/gut.2009.199679. [DOI] [PubMed] [Google Scholar]
- 34.Wheeler JM, Kim HC, Efstathiou JA, Ilyas M, Mortensen NJ, Bodmer WF. Hypermethylation of the promoter region of the E-cadherin gene (CDH1) in sporadic and ulcerative colitis associated colorectal cancer. Gut. 2001;48:367–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su L, Shen L, Clayburgh DR, Nalle SC, Sullivan EA, Meddings JB, Abraham C, Turner JR. Targeted epithelial tight junction dysfunction causes immune activation and contributes to development of experimental colitis. Gastroenterology. 2009;136(2):551–563. doi: 10.1053/j.gastro.2008.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larsen JM. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology. 2017;151(4):363–374. doi: 10.1111/imm.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rolhion N, Chassaing B, Nahori M-A, De Bodt J, Moura A, Lecuit M, Dussurget O, Bérard M, Marzorati M, Fehlner-Peach H, et al. A listeria monocytogenes bacteriocin can target the commensal prevotella copri and modulate intestinal infection. Cell Host & Microbe. 2019;26(5):691–701.e5. doi: 10.1016/j.chom.2019.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BAH, Forslund K, Hildebrand F, Prifti E, Falony G, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535(7612):376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 39.Bellone M, Brevi A, Huber S. Microbiota-propelled T helper 17 cells in inflammatory diseases and cancer. Microbiol Mol Biol Rev. 2020;84(2): doi: 10.1128/MMBR.00064-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alpizar-Rodriguez D, Lesker TR, Gronow A, Gilbert B, Raemy E, Lamacchia C, Gabay C, Finckh A, Strowig T. Prevotella copri in individuals at risk for rheumatoid arthritis. Ann Rheum Dis. 2019;78(5):590–593. doi: 10.1136/annrheumdis-2018-214514. [DOI] [PubMed] [Google Scholar]
- 41.Leite AZ, Rodrigues NDC, Gonzaga MI, Paiolo JCC, De Souza CA, Stefanutto NAV, Omori WP, Pinheiro DG, Brisotti JL, Matheucci Junior E, et al. Detection of increased plasma interleukin-6 levels and prevalence of prevotella copri and bacteroides vulgatus in the feces of type 2 diabetes patients. Frontiers in Immunology. 2017;8(1107): doi: 10.3389/fimmu.2017.01107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gosmann C, Anahtar MN, Handley SA, Farcasanu M, Abu-Ali G, Bowman BA, Padavattan N, Desai C, Droit L, Moodley A, et al. Lactobacillus-deficient cervicovaginal bacterial communities are associated with increased HIV acquisition in young South African women. Immunity. 2017;46(1):29–37. doi: 10.1016/j.immuni.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boulangé CL, Neves AL, Chilloux J, Nicholson JK, Dumas M-E. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Medicine. 2016;8(1): doi: 10.1186/s13073-016-0303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saad MJA, Santos A, Prada PO. Linking gut microbiota and inflammation to obesity and insulin resistance. Physiology (Bethesda). 2016;31(283–293): doi: 10.1152/physiol.00041.2015. [DOI] [PubMed] [Google Scholar]
- 45.Jiang K, Guo S, Yang C, Yang J, Chen Y, Shaukat A, Zhao G, Wu H, Deng G. Barbaloin protects against lipopolysaccharide (LPS)-induced acute lung injury by inhibiting the ROS-mediated PI3K/AKT/NF-κB pathway. International Immunopharmacology. 2018;64(140):140–150. doi: 10.1016/j.intimp.2018.08.023. [DOI] [PubMed] [Google Scholar]
- 46.Piao X, Liu B, Sui X, Li S, Niu W, Zhang Q, Shi X, Cai S, Fan Y. Picroside II improves severe acute pancreatitis-induced intestinal barrier injury by inactivating oxidative and inflammatory TLR4-dependent PI3K/AKT/NF-κB signaling and improving gut microbiota. Oxidative Medicine and Cellular Longevity. 2020;2020(3589497):1–12. doi: 10.1155/2020/3589497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, GuhaThakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nature Genetics. 2005;37(7):710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hata L, Murakami M, Kuwahara K, Nakagawa Y, Kinoshita H, Usami S, Yasuno S, Fujiwara M, Kuwabara Y, Minami T, et al. Zinc-finger protein 90 negatively regulates neuron-restrictive silencer factor-mediated transcriptional repression of fetal cardiac genes. Journal of Molecular and Cellular Cardiology. 2011;50(6):972–981. doi: 10.1016/j.yjmcc.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 49.Palka Bayard de Volo C, Alfonsi M, Gatta V, Novelli A, Bernardini L, Fantasia D, Antonucci I, Angelucci D, Zori R, Stuppia L, et al. 16q22.1 microdeletion detected by array-CGH in a family with mental retardation and lobular breast cancer. Gene. 2012;498(2):328–331. doi: 10.1016/j.gene.2012.01.028. [DOI] [PubMed] [Google Scholar]
- 50.Grivennikov SI Inflammation and colorectal cancer: colitis-associated neoplasia. Seminars in immunopathology 35, 229–244, doi: 10.1007/s00281-012-0352-6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EG, Abramson SB, et al. Expansion of intestinal prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2(e01202): doi: 10.7554/eLife.01202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Velloso LA, Folli F, Saad MJ. TLR4 at the crossroads of nutrients, gut microbiota, and metabolic inflammation. Endocrine Reviews. 2015;36(245–271): doi: 10.1210/er.2014-1100. [DOI] [PubMed] [Google Scholar]
- 53.Tilg H, Zmora N, Adolph TE, Elinav E. The intestinal microbiota fuelling metabolic inflammation. Nature Reviews. Immunology. 2019. doi: 10.1038/s41577-019-0198-4. [DOI] [PubMed] [Google Scholar]
- 54.Neufert C, Becker C, Neurath MF. An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nature Protocols. 2007;2(1998–2004): doi: 10.1038/nprot.2007.279. [DOI] [PubMed] [Google Scholar]
- 55.Weigmann B, Tubbe I, Seidel D, Nicolaev A, Becker C, Neurath MF. Isolation and subsequent analysis of murine lamina propria mononuclear cells from colonic tissue. Nature protocols. 2007;2:2307–11. [DOI] [PubMed] [Google Scholar]
- 56.Johansson ME, Hansson GC. Preservation of mucus in histological sections, immunostaining of mucins in fixed tissue, and localization of bacteria with FISH. Methods in Molecular Biology (Clifton, N.J.). 2012;842(229–235): doi: 10.1007/978-1-61779-513-8_13. [DOI] [PubMed] [Google Scholar]
- 57.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics (Oxford, England). 2009;25(1105–1111): doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7:562–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


