Abstract
The skin microbiome, especially the axillary microbiome, consists of odor-causing bacteria that decompose odorless sweat into malodor compounds, which contributes to the formation of body odor. Plant-derived products are a cheap source of bioactive compounds that are common ingredients in cosmetics. Microbial bioconversion of natural products is an ecofriendly and economical method for production of new or improved biologically active compounds. Therefore, in this study, we tested the potential of a Lactobacillus acidophilus KNU-02-mediated bioconverted product (BLC) of Lotus corniculatus seed to reduce axillary malodor and its effect on the associated axillary microbiota. A chemical profile analysis revealed that benzoic acid was the most abundant chemical compound in BLC, which increased following bioconversion. Moreover, BLC treatment was found to reduce the intensity of axillary malodor. We tested the axillary microbiome of 18 study participants, divided equally into BLC and placebo groups, and revealed through 16S rRNA gene sequencing that Staphylococcus, Corynebacterium, and Anaerococcus were the dominant taxa, and some of these taxa were significantly associated with axillary malodor. After one week of BLC treatment, the abundance of Corynebacterium and Anaerococcus, which are associated with well-known odor-related genes that produce volatile fatty acids, had significantly reduced. Likewise, the identified odor-related genes decreased after the application of BLC. BLC treatment enhanced the richness and network density of the axillary microbial community. The placebo group, on the other hand, showed no difference in the microbial richness, odor associated taxa, and predicted functional genes after a week. The results demonstrated that BLC has the potential to reduce the axillary malodor and the associated odor-causing bacteria, which makes BLC a viable deodorant material in cosmetic products.
Subject terms: Microbiology, Applied microbiology
Introduction
Human body odor generally creates an adverse impression in human-to-human interaction, causing psychological stress and negatively impacting social life in afflicted humans. Personality attributes, facial attractiveness, and health status are profoundly influenced by body odor1–3. The skin microbiome, particularly the armpit region, is covered by dense secretory glands, such as apocrine, eccrine, and sebaceous glands. These glands secret various nutrients and moisten skin folds such as the armpit, which provides habitable growth conditions for odor-causing bacteria4. The bacterial community in the armpit decomposes the odorless sweat into volatile odorous byproducts, such as volatile fatty acids (VFAs), 3-hydroxy hexanoic acid (3M3H), and 3-hydroxy-3-methylhexanoic acid (HMHA)5. Thus, the axillary microbiome plays a key role in the generation of malodor, making it a target for the inhibition of odor-producing bacteria as a means to substantially reduce body odor.
As body odor negatively affects personal life, numerous products that reduce malodor have been developed, including fragrances, antiperspirants, and deodorants, which mask body odor, suppress sweat secretion, and/or inhibit bacterial growth. However, these products carry their own risks upon exposure6–9. Masking body odor with a pleasant scent exacerbates odor-producing conditions due to a combination of fragrance and sweat, and exposure to certain ingredients in fragrances can cause allergic reactions6. In addition, aluminum salt is found in many antiperspirants to inhibit sweating, despite being a risk factor for Alzheimer’s disease and accelerating inflammatory reactions7,8. Deodorants with added fragrances can cause allergic reactions9. Therefore, it is necessary to find a substance that can reduce body odor without any adverse effects.
Natural ingredients, especially plant-derived products, have long been used in traditional medicine10,11 and are a cheap source of bioactive compounds that control antibiotic resistance12. Plant extracts have antioxidant, anti-inflammatory, and anticancer activities13. Phenolic compounds from plant extracts, which have antimicrobial and antiradical activities, have long been used as skin care ingredients14. The potential of phytochemicals to treat skin diseases has been previously documented15. Plant extracts, which are rich in piceatannol, are good skin care materials, as they moisturize dry skin16. Likewise, plant materials, which are also used as fragrance and deodorant substances for cosmetics, soaps, and perfumes, are also used to reduce odor, as they retain floral fragrances and absorb odor17. In addition, plants such as sage and tea leaves have been used as odor-reducing substances because of their antimicrobial activity18,19. Lotus corniculatus, which is an acidic-tolerant flowering plant containing condensed tannins20, has been found to show promising antimicrobial and anticancer properties21–23. Furthermore, L. corniculatus has been reported to possess substantial healing effects on skin wounds24. However, the effect of L. corniculatus extract on axillary odor and the associated microbiota has not been reported yet.
A bioconversion technique, which is the process of converting organic materials into usable products by biological agents, is an economically and ecologically important practical technology that is widely used to modify the structure of many types of biologically active products25. In addition, novel products can be isolated with new or different activities, such as antimicrobial and antioxidant properties25–27. In a previous study, probiotic bacterial bioconversion increased the antiradical activity of polyphenols in lotus seed epicarp28. Lactic acid bacteria (LAB) has been widely used for the bioconversion of natural products and in the food industry to ferment beverages29,30.
In this study, we bioconverted seeds of L. corniculatus using a LAB isolate, Lactobacillus acidophilus KNU-02. Next, we evaluated the effect of the bioconverted product of L. acidophilus KNU-02 (BLC) on the axillary microbiota structure and body odor. To validate the efficacy of BLC, we measured the odor intensity and collected armpit swab samples (for 16S rRNA gene sequencing) from 18 female Korean volunteers at baseline (0 week) and after one week of BLC treatment (1 week). Our study demonstrated that BLC reduced axillary malodor and inhibited odor-causing bacteria. This implies that BLC could be a potential source of ingredients for cosmetics.
Results
Bioconversion altered chemical profile of L. corniculatus seed extract
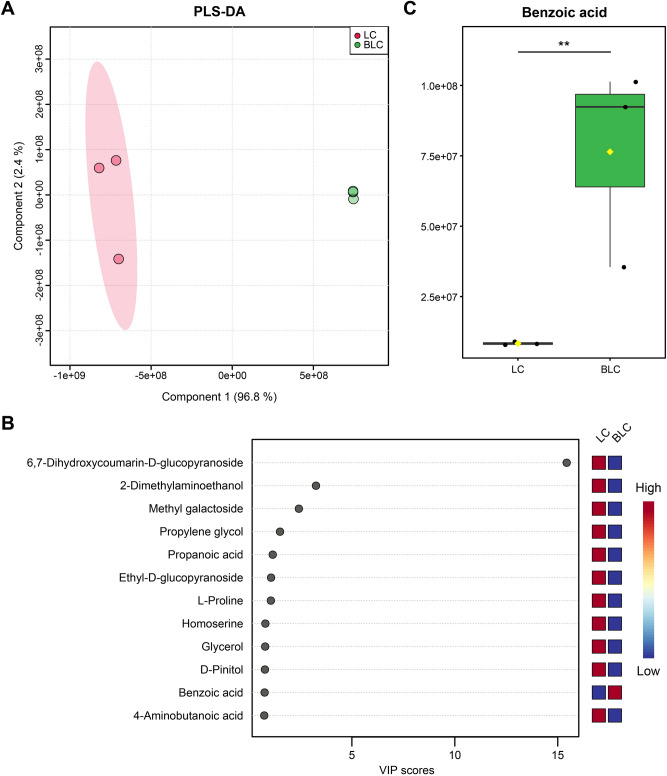
To identify changes in the chemical profile of the BLC seed extract, we analyzed the chemical compositions of BLC and the original seed extract using GCxGC/TOF–MS. The chemical profile of BLC showed that 12 major chemical compounds were more than 1% of the content. The predominant metabolites, each formed more than 3% of the profile, were benzoic acid, silanol, palmitic acid, propylene glycol, 4-hydroxybenzoic acid, and D-lactic acid (Table 1). Among the compounds, benzoic acid was the most abundant (17.9%) in BLC, followed by silanol and palmitic acid metabolites (14.6%). Note that benzoic acid was one of the least (< 1%) abundant in the original seed extract (Supplementary Table S1). The results of the partial least squares discriminant analysis (PLS-DA) revealed that the chemical composition of the BLC and original seed extract (LC) were distinct from each other. The total detected variation was 96.8% component1 and 2.4% component2 in PLS-DA (Fig. 1A). In addition, variable importance in projection (VIP) scores were used to identify discriminatory chemical compounds during the bioconversion process. 12 chemical compounds were identified as key features to discriminate BLC from LC (VIP > 0.7) (Fig. 1B, Supplementary Table S2). Among the key features that discriminate between the two extracts, only benzoic acid, a well-known cosmetic ingredient, was markedly increased (Fig. 1B,C) following the bioconversion process. Furthermore, the bioconversion process also resulted in a decline in the abundance of propionic acid (Fig. 1B), which can be detected by the strength of its pungent smell, implying the significance of this bioconversion technique.
Table 1.
The list of major compounds (> 1%) identified in bioconverted product by GCxGC/TOF–MS analysis.
| Compound | R.T. (s) | MW (g/mol) | Formula | Area (%) |
|---|---|---|---|---|
| Benzoic acid, 3,4-bis[(trimethylsilyl)oxy]-, trimethylsilyl ester (CAS) | 786.23 | 370.66 | C16H30O4Si3 | 17.90 |
| Silanol, trimethyl-, phosphate (3:1) (CAS) | 483.46 | 314.54 | C9H27O4PSi3 | 8.143 |
| Palmitic Acid, TMS derivative | 936.61 | 328.61 | C19H40O2Si | 6.532 |
| Propylene glycol, 2TMS derivative | 304.70 | 220.46 | C9H24O2Si2 | 5.045 |
| 4-Hydroxybenzoic acid, 2TMS derivative | 671.69 | 282.48 | C13H22O3Si2 | 3.517 |
| D-Lactic acid-DITMS | 346.62 | 234.44 | C9H22O3Si2 | 3.307 |
| Propanoic acid, 3-[(trimethylsilyl)oxy]-, trimethylsilyl ester (CAS) | 400.76 | 234.44 | C9H22O3Si2 | 2.853 |
| D-(-)-Ribofuranose, tetrakis(trimethylsilyl) ether (isomer 2) | 639.13 | 438.85 | C17H42O5Si4 | 1.449 |
| D-Psicose, pentakis(trimethylsilyl) ether, methyloxime (anti) | 828.64 | 570.10 | C22H55NO6Si5 | 1.265 |
| Maltol, TMS derivative | 494.95 | 198.29 | C9H14O3Si | 1.161 |
| D-Fucitol, 5TMS derivative | 764.81 | 527.08 | C21H54O5Si5 | 1.136 |
| 6,7-Dihydroxycoumarin-á-D-glucopyranoside, PENTA-TMS | 1470.8 | 700.00 | C30H56O9Si5 | 1.056 |
Figure 1.
Compositional analysis of bioconverted product (BLC) and the seed extract (LC). (A) PLS-DA score plot using all features. (B) PLS-DA variable importance in projection (VIP) score plot. The features were selected with a VIP score > 0.7 based on the corresponding PLS-DA model. (C) Box plot of normalized peak abundances for benzoic acid.
Effect of the bioconverted product on body odor intensity
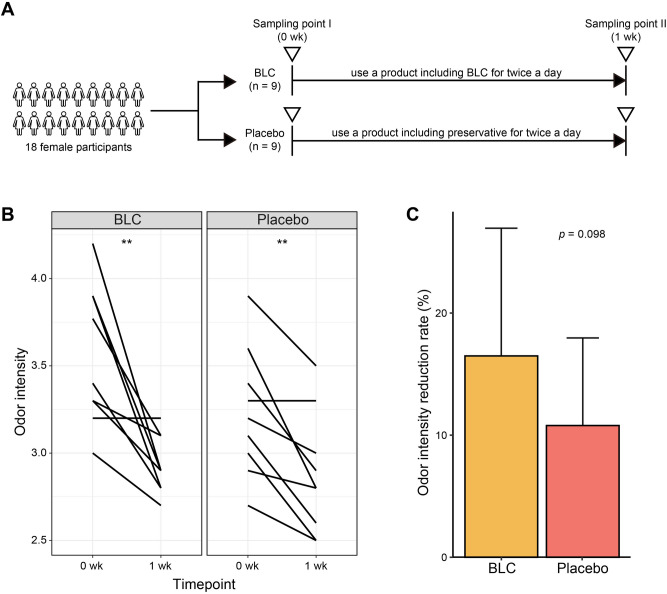
In this study, we determined the potential of BLC to reduce axillary malodor. We recruited nine participants for each treatment group (BLC and placebo groups). The two groups had no significant axillary odor intensity, age and gender differences at the baseline (0 week) (Supplementary Table S3). However, a week after treatment (1 week), odor intensity was significantly reduced in both groups; BLC had a significantly (p < 0.1) higher odor intensity decline than the placebo (Fig. 2B,C). This signifies the effectiveness of the BLC treatment in reducing body malodor.
Figure 2.
The study design and axillary odor intensity. (A) Pictorial representation of the study design. (B) The axillary odor intensity changes in each group. (C) Reduction rate of odor intensity in each group. A Wilcoxon matched-pairs test was conducted to compare the odor intensity between 0 and 1 week. The comparison of the odor intensity reduction rate between the two groups was evaluated with an unpaired t-test. A p value < 0.01 is indicated with asterisks (**).
Bioconverted product altered axillary microbiome
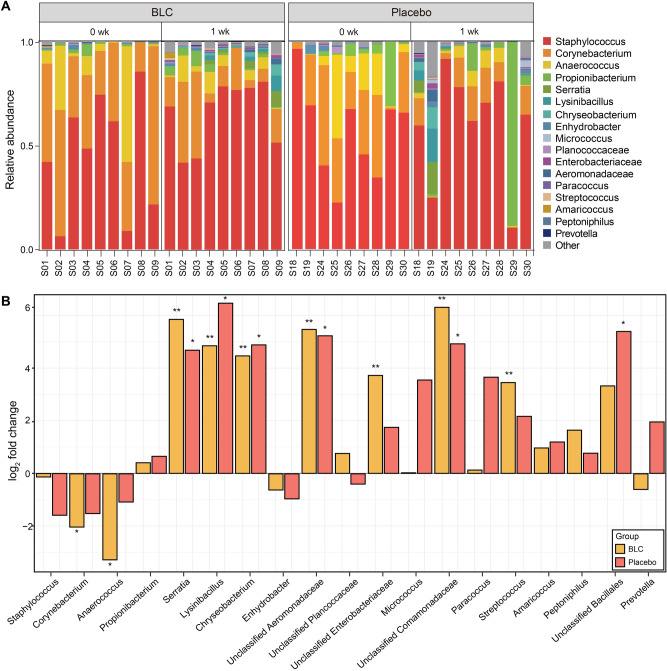
We also investigated the effect of BLC on the axillary microbiome of the study participants. Armpit swab samples were collected at 0 week and 1 week. Our results showed that Staphylococcus, Corynebacterium, Anaerococcus, and Propionibacterium were the predominant genera in the axillary microbiome of the two groups at 0 week and 1 week (Fig. 3A). The taxonomic differences between 0 and 1 week in BLC and placebo groups were compared, and the top 19 abundant taxa were included in the test. Nine and six taxa were found to be differentially abundant taxa at 1 week compared with that at 0 week in the BLC and placebo groups, respectively (Fig. 3B). Among the three dominant taxa, viz., Staphylococcus, Corynebacterium, and Anaerococcus, the latter two were significantly decreased (false discover rate [FDR] < 0.05) at 1 week following BLC treatment compared to those at 0 week. However, these two genera did not show significant differences from the placebo group after a week. Furthermore, seven taxa, including Streptococcus, Enterobacteriaceae, Chryseobacterium, Lysinibacillus, Aeromonadaceae, Serratia, and Comamonadaceae, were enriched at 1 week following BLC treatment. In the placebo at 1 week, six taxa, Aeromonadaceae, Bacillales, Chryseobacterium, Comamonadaceae, Lysinibacillus, and Serratia, were significantly (FDR > 0.05) increased compared with those at 0 week.
Figure 3.
Taxonomic changes in the axillary microbiome for bioconverted product (BLC) and placebo. (A) The relative abundance of the axillary microbiome at the genus level. (B) Log2-fold-change in the BLC (yellow) and placebo (red), which represents changes at 1 week after using applications in comparison with those of the baseline samples. False discover rate-adjusted values < 0.05 and 0.01 are indicated with asterisks (* and **, respectively).
Canonical analysis of principal coordination (CAP) was based on the unweighted UniFrac distance metric. According to the CAP analysis, the BLC group showed a significantly different microbiota structure at 1 week compared with that at 0 week (p = 0.001), and there was a strong association with odor intensity (p = 0.003) (Fig. 4A). However, as expected, the placebo showed no significant difference between 0 and 1 week (p = 0.302) (Fig. 4B). In addition, there was no association between odor intensity and the axillary microbiome in the placebo (p = 0.073) (Fig. 4B). Furthermore, the placebo exhibited no changes between 0 and 1 week in all alpha diversity indices (Fig. 4C and Supplementary Fig. S1). On the contrary, BLC showed a significant increase of operational taxonomic unit (OTU) richness, as indicated by the Chao1 index (p = 0.014) (Fig. 4C), but not in the remaining alpha diversity indices (Supplementary Fig. S1).
Figure 4.
Shifts in the axillary microbiome of the bioconverted product (BLC) and placebo. Canonical analysis of the principal coordinates of the axillary microbiome in relation to the odor intensity and time point in the (A) BLC and (B) placebo groups. (C) The Chao1 index of each group. Paired t-test used to compare the Chao1 index. The color code and size of the canonical analysis of principal coordination analysis are the group and odor intensity, respectively.
We further explored the association between the top 4 genera and body odor using Spearman’s rank correlation coefficient (Table 2). In the correlation analysis, Corynebacterium (ρ = 0.422, p value = 0.010) and Anaerococcus (ρ = 0.416, p value = 0.012) were significantly positively correlated with odor intensity. Additionally, to adjust the effect of other taxa to one taxon, the association between the top 4 genera and odor intensity was further explored using multiple regression model. Among 4 genera, only Corynebacterium and Anaerococcus had significant association with odor intensity (Supplementary Figure S2). Collectively, our results showed that the BLC treatment reduced body odor by increasing the microbial diversity and inhibiting the growth of odor-causing bacteria, viz., Corynebacterium and Anaerococcus, in the axillary microbial community.
Table 2.
Correlation of the abundant 4 genera with odor intensity.
| Genus | Spearman’s rho | |
|---|---|---|
| ρ | p value | |
| Staphylococcus | − 0.183 | 0.285 |
| Corynebacterium | 0.422 | 0.010 |
| Anaerococcus | 0.416 | 0.012 |
| Propionibacterium | − 0.187 | 0.275 |
Network analysis
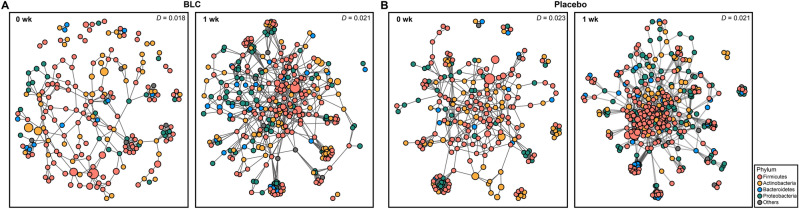
To investigate the interaction of OTUs in the axillary microbiome, OTU networks for each group (BLC and placebo) at 0 week and 1 week were generated by Sparse InversE Covariance estimation for Ecological Association Inference (SPIEC-EASI). The OTUs with < 0.01% relative abundance were not included in the analysis. Therefore, the network for the BLC treatment had 203 and 322 nodes at 0 week and 1wk, respectively. In the placebo, 228 and 362 nodes were obtained at 0 week and 1 week, respectively. The network density (D) of the axillary microbiome was calculated using the ratio of the number of edges for each time point in each group. The network analysis demonstrated that the D value was slightly higher following BLC treatment after 1 week (D = 0.021) than at to 0 week (D = 0.018) (Fig. 5A). On the other hand, the D value slightly decreased in the placebo after 1 week (D = 0.021) than at 0 week (D = 0.023) (Fig. 5B). This could be due to the reduced dominance of odor-causing taxa and the subsequent increase in the abundance of other bacterial taxa in the axillary microbiota after one week. Furthermore, the network topological coefficients were evaluated for each group (Supplementary Table S4). The network transitivity (T) value increased after one week compared to 0 week. The number of edge, degree, and node closeness centrality of the network were also increased after two treatments. The higher the degree, the higher the node dependence. Additionally, an increase of the node closeness centrality implies that the microbial network is the hub-based network which has high connection with other nodes. These results indicate that the BLC increased interdependence among bacteria in the community. Therefore, BLC makes the network more compact, complex and stable.
Figure 5.
Network analysis of the axillary microbiome at the bacterial operational taxonomic unit (OTU) level. (A) Network analyses of the bioconverted product treatment at 0 week and 1 week. (B) Network analyses of the placebo at 0 week and 1 week. Color markings indicate the phylum. Each node represents the OTUs, and the size of each node corresponds to the relative abundance. The D values, which represent the density, were calculated using the ratio of the number of edges.
Functional gene prediction using PICRUSt
To gain insight into the metabolic function of the microbial community in the armpit of the participants, functional gene prediction of the axillary microbiome from 16S rRNA gene sequencing was conducted using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt). Functional genes that were predicted to be related to the production of odorous compounds were analyzed. Among the odor-related genes, five genes that were predicted to be involved in odor production exhibited significant (FDR < 0.05) differential expression at 1 week of BLC treatment compared with that at 0 week (Fig. 6A). However, in the placebo, there were no significantly differentially expressed genes at 1 week compared with the baseline. After one week of BLC treatment, the two predicted genes that showed a significant decrease in abundance were found to encode long-chain acyl-CoA synthetase (FadD) and 3-hydroxybutyryl-CoA dehydrogenase (FadB). Long-chain acyl-CoA synthetase and 3-hydroxybutyryl-CoA dehydrogenase are essential enzymes for the production of VFAs by catabolizing lipids during β-oxidation cycle. In contrast, the remaining three genes that showed an increment in abundance were acyl-ACP dehydrogenase, acetyl-CoA acyltransferase, and Nα-acylglutamine aminoacylase. However, the relative abundance of these three genes in the axillary microbiome was less abundant, which might reduce their impact on body odor (Supplementary Table S5). Indeed, the total sum of the relative abundance of the predicted genes with odor-related functions decreased at 1 week of BLC treatment compared with that at 0 week (Supplementary Table S5). This implies that the BLC treatment significantly affected the odor-related functions of the axillary microbiota, as predicted.
Figure 6.
Functional gene prediction analysis of the axillary microbiome. (A) Log2-fold-change of odor-related genes in the bioconverted product (yellow) and placebo (red) group that represented changes at 1 week after using products in comparison with the baseline samples. False discover rate-adjusted values less than 0.05 and 0.01 are indicated with asterisks (* and **, respectively). (B) Correlation matrix showing a significant correlation (p < 0.05) between the most abundant taxa and odor-related genes. The color code and size of the circles are the ρ correlation coefficient.
Further correlation analysis was performed to determine which taxa had contributed more to the change in functional genes. Hence, we analyzed the correlation between the most abundant taxa and odor-related functional genes using Spearman’s rank correlation coefficient (Fig. 6B). Interestingly, at the genus level, Corynebacterium had a positive correlation with the four functional genes involved in odor production, namely 3-hydroxybutyryl-CoA dehydrogenase, pyruvate dehydrogenase, acetate kinase, and long-chain acyl-CoA synthetase. Moreover, Anaerococcus was also related to the four genes encoding 3-hydroxybutyryl-CoA dehydrogenase, acetate kinase, long-chain acyl-CoA synthetase and branched-chain amino acid (BCAA) aminotransferase. Anaerococcus had a positive correlation with all the above-mentioned genes except BCAA aminotransferase. Corynebacterium and Anaerococcus were positively associated with body-odor-related genes, such as 3-hydroxybutyryl-CoA dehydrogenase, pyruvate dehydrogenase, acetate kinase, and long-chain acyl-CoA synthetase. Furthermore, the contribution of each taxa to odor-related genes was also performed using metagenome_contributions.py command line in PICRUSt algorithm. Staphylococcus, Corynebacterium and Anaerococcus contributed the most to odor-related genes (Supplementary Figure S3). Therefore, the use of BLC plays a key role in reducing odor-related genes, primarily by inhibiting the growth of Corynebacterium and Anaerococcus in the axillary microbiome.
Discussion
Microbial bioconversion of natural products has been used to produce new or improved products31,32. Such changes in the chemical constituents of bioconverted products are attributed to various chemical reactions, such as methylation, degradation, and glycosylation33–35. Several new bioactive compounds in bioconverted products are used for nutrition and health benefits, including anti-allergic, antioxidant, and anticancer effects32,36. Furthermore, the use of plant-based products in cosmetic markets is increasing33,37. This is due to the fact that plant-based skin care products are cheap and have huge potential to replace synthetic antimicrobials in skin care products38,39. These products help body’s cells to repair and reduce signs of skin aging. This has led to the growing of plant-based cosmetic brands and green-beauty market. Nevertheless, the untapped resource of plants for cosmetic purposes, however, has not been well exploited commercially40. Therefore, in this study, we determined the potential use of the BLC in reducing axillary odor. We also investigated the effects of BLC treatment on the associated axillary microbiome. At the genus level, the axillary microbiome of the participants mainly consisted of Staphylococcus, Corynebacterium, Anaerococcus, and Propionibacterium (Fig. 3A). Our study results support previous reports that established these four gram-positive bacteria as inhabitants of human skin41,42. Indeed, these genera are also the major taxa in the axillary region41–44. Staphylococcus and Corynebacterium are dominant taxa in moist body regions, such as the armpit and navel42,45. Previous studies demonstrated that Propionibacterium is particularly adapted to anaerobic or lipid-rich environments, such as sebaceous glands46,47.
We evaluated whether BLC could reduce axillary malodor and alter the axillary microbiome of the study participants. Indeed, the BLC treatment reduced body odor and shifted the axillary microbiome. Although body odor for the two groups decreased, the percentage of odor reduction following BLC treatment was higher than the placebo group (Fig. 2C). The decrease in body odor observed in a placebo group may be due to the placebo effect, which is commonly found in many clinical trials48,49. According to the previous studies, the ritual of taking medication50 and response expectancy51 is the key driving forces in the healing process as the placebo effect can modulate the nervous system-immune system communication52. In a separate study, the potential of plant-based extracts to reduce body odor was reported53. However, to the best of our knowledge, this is the first report showing the positive effects of a bioconverted product from L. corniculatus seed in reducing axillary malodor.
Such difference in odor reduction between the two groups can be explained by the observed difference in the axillary microbiome. Our study showed that the richness of the axillary microbiome increased after BLC treatment (Fig. 4C). Indeed, previous studies also showed that skin care products induce an increase in skin microbial richness54–56. The increase in richness may be caused by the colonization of non-skin commensal bacteria. Unlike the placebo, the relative abundance of Corynebacterium and Anaerococcus significantly decreased due to the BLC treatment (Fig. 3B). According to previous reports, skin commensal bacteria metabolize odorless sweat into volatile odorants57–59. Corynebacterium is one of the dominant genera that produces various odorants, including VFAs, HMHA, 3M2H, and 5α-androst-16-en-3α-ol5,60–62. Anaerococcus also plays a role in the production of odorants such as 3-hydroxy-3-methylhexanoic (3H3MH) acid and VFAs33,39,63. Moreover, Staphylococcus, Propionibacterium, and Micrococcus are also known to generate volatile odorants, viz., 3-methyl-3-sulfanylhexan-1-ol (3M3SH) and VFAs64–66. In the network analysis, BLC induced a more stable community for the axillary microbiome, as indicated by higher network density (D) and transitivity values (T) than the baseline (Fig. 5A and Supplementary Table S4). In previous studies, low network density and transitivity of the skin microbiome was correlated with dandruff skin and a fragile community, highlighting the need to maintain stable bacterial communities for good skin health67,68. Additionally, BLC induced a more complex and compact the axillary microbial community, as indicated by higher values of network topological coefficients including the node closeness centrality, edges, and degree (Supplementary Table S4). According to the previous study69, greater node closeness centrality and number of the degree are indicators of compact network with many hub-bacteria. Therefore, BLC induced more compact and stable axillary microbial network compared to placebo.
PICRUSt analysis also revealed that among the odor-associated functional groups5, the abundance of FadB and FadD significantly decreased in the BLC-treated group after a week. However, there was no significant decrease in the odor-related metabolic functional groups in the placebo group (Fig. 6A). PICRUSt analysis results are consistent with previous studies that demonstrated that FadD is a crucial membrane-associated functional gene that facilitates fatty acid degradation70. Likewise, FadB is also required for the degradation of short-chain fatty acids70. VFAs can be generated through β-oxidation cycle57,60. β-oxidation cycle is related to fatty acid catabolism, as it is the first cycle to break down long fatty acids into shorter fatty acyl-CoA chains60,71. In this study, FadB and FadD were positively correlated with Corynebacterium (Fig. 6B). Corynebacterium metabolites fatty acid into malodor compounds, such as VFAs, through the β-oxidation cycle60. In addition, Corynebacterium also exhibited a positive correlation with odor-associated functions, viz., pyruvate dehydrogenase and acetate kinase (Fig. 6B). Regarding properties of Corynebacterium, several studies reported that it is the primary cause of axillary malodor71. In addition, Anaerococcus was also positively related with L-lactate dehydrogenase, 3-hydroxybutyryl-CoA dehydrogenase, and acetate kinase (Fig. 6B). L-lactate dehydrogenase is related to the production of odorants, as it converts lactic acid into pyruvate66,72. Through oxidative degradation, pyruvate dehydrogenase converts pyruvate into acetic acid and sour odor66,73. Nevertheless, three genes significantly increased in the BLC (Fig. 6A). These genes have no association with abundant bacteria. Among the increased genes, acyl-ACP dehydrogenase (FadE) and acetyl-CoA acyltransferase (FadA) degrade fatty acid through the β-oxidation cycle70. Although these genes increased with BLC, they cannot metabolite substances when there is a decrease in FadD, which activates the long-chain fatty acid to transport into the inner membrane. Moreover, the abundance of Nα-acylglutamine aminoacylase is too low to significantly affect body odor production (Supplementary Table S5). Although previous studies presented a strong association of Staphylococcus with body odor74, we found that most genes had a negative correlation with Staphylococcus (Fig. 6B). Our findings are in agreement with the previous report that showed Anaerococcus is the key commensal microbiome75. Therefore, our study demonstrated that Corynebacterium and Anaerococcus contribute more than Staphylococcus to the body odor of our participants. Therefore, our results indicate that BLC has the potential to reduce body odor by altering the axillary microbiome.
The chemical composition of BLC was also profiled to determine the changes in the chemical profile of the seed extract after bioconversion. Benzoic acid was the predominant compound in BLC after four days of bioconversion (Table 1). Methyl galactoside was predominant and the abundance of benzoic acid was low (< 1%) in the original seed extract (Supplementary Table S1). In antibacterial activity experiment, BLC had antibacterial activity against Staphylococcus aureus, but LC had not (Supplementary Figure S4). The difference of antibacterial activity between BLC and LC may be due to benzoic acid. Indeed, benzoic acid and its derivatives have been previously reported to be nonspecific antibacterial agents that have been used as preservative agents76,77. Lipophilic acids, such as benzoic acid, inhibit the active uptake of some amino and oxo acids in Escherichia coli and Bacillus subtilis78. Furthermore, benzoic acid has been reported to possess several cosmetic properties that enhance skin health, like antioxidant and anti-inflammatory activity14. The relative composition of 4-hydroxybenzoic acid in the bioconverted seed extract was also abundant in the BLC (Table 1). 4-Hydroxybenzoic acid, which is a phenolic derivate of benzoic acid and also a paraben, is frequently used as a preservative in cosmetics and food industries, as it contains antibacterial and antifungal activities79,80. Likewise, palmitic acid, which is also abundant in the BLC, has antibacterial activity81. Therefore, such compounds with antibacterial properties in the BLC might have contributed to a reduction in the relative abundance of Corynebacterium and Anaerococcus in the axillary microbiome, thereby reducing the body odor. Palmitic acid has been used as a skin permeation enhancer82. Skin permeation enhancers interact with the constituents of a skin’s outermost layer and increase its permeability83. Compounds that have a skin permeability property might enhance the efficacy of the BLC as an axillary odor reducer. In addition, since the BLC contains compounds with various properties, such as antioxidant and anti-aging effects, the BLC could be used as a cosmetic ingredient with multiple beneficial properties for skin health. However, although the most commonly dominant odor-causing bacteria are only few in number5, it is worth considering race/ethnicity and gender variations in terms of the axillary microbiome43 and skin factors such as skin permeability, sweat secretion and skin hydration level84.
In conclusion, to the best of our knowledge, the present study reports for the first time the potential of a microbial bioconverted product of BLC to reduce axillary malodor and alter the associated microbiota. The bioconversion process resulted in the predominance of benzoic acid, which is known to possess several cosmetic application properties that enhance skin health. BLC was found to alter the axillary microbiota and thereby the axillary odor. The BLC treatment was effective in reducing the abundance of the most dominant and odor-associated taxa, Corynebacterium and Anaerococcus. The BLC treatment also increased the richness and slightly enhanced the network density of the axillary microbial community. Thus, the BLC contains cheap bioactive compounds and could be a potential source of ingredients for cosmetics. Additionally, as Corynebacterium and Anaerococcus are most common odor-causing bacteria across race/ethnicity and gender group5,43, BLC could be an ingredient for universal skin-care products. However, further research with a large number of participants is warranted to ensure the potential of the BLC for future applications in cosmetic products.
Methods
Subject recruitment and sample collection
Total 18 Korean women, without chronic skin diseases, were recruited for this study. The participants were divided into two groups, the BLC and the placebo groups. Participants were assigned divided to BLC and placebo groups. Additionally, the treatment assignment was randomized by double-blind test without informing to the experimenters and participants about the distinction between BLC and placebo. Participants of each group were asked to apply their respective treatments twice a day for one week, and they were asked to stop taking showers or using any other products, such as lotion, deodorant, or antiperspirant, on their armpits for at least 12 h prior to the axillary microbiota sample collection (Fig. 2A). Sampling was conducted in a controlled room at 20–24 °C, 45–55% RH. During the 30 min prior to sampling, the participants stayed still without moving their armpit. For the sample collection, a 50 cm2 area from the armpit of each subject was swabbed with sterile cotton-tipped swabs pre-moistened with a preservative, which contained 0.1% Tween 20, 0.15 M sodium chloride, and 0.1% agar. The sampling area was swabbed for at least 3 min. Each swab was placed into a tube containing a preservative, immediately frozen, and stored at -80 °C prior to DNA extraction. The odor intensity of the axillae of the participants was assessed at 0 week and 1 week by using an odor meter (Shinyei OMX-ADM, Japan) before axillary microbial collection. In addition, all participants were required to answer a questionnaire related to body odor at 0 week and 1 week. This study was approved by the Institutional Review Boards (IRBs) of the Global Medical Research Center (IRB: GIRB-19O01-AK). Both verbal and written informed consent was obtained from all participants. We confirm that all methods used in the study were performed in accordance with the relevant guidelines and regulations.
DNA extraction and sequencing
Axillary microbial genomic DNA was extracted from swab samples using the DNeasy PowerSoil Pro Kit (Qiagen, Germany) with a modified method to obtain additional genomic DNA. The preservative solution was transferred to a 1.5 mL microcentrifuge tube and centrifuged at 15,000g for 2 min. The pellet was suspended in a CD1 solution that was included in the extraction kit and was transferred to a PowerBead Pro tube with each swab head. After cell lysis of each sample, genomic DNA was extracted by following the manufacturer’s protocols. To prepare a skin microbiome library for sequencing, the 16S rRNA gene was amplified at V4 to V5 hypervariable region using a 515 F (5′-barcode-CGCTCTTCCGATCTGTGNCAGCMGCCGCGGTRA-3′) forward primer attached to 5′ Illumina adapter and 907 R (5′-barcode-GTGCTCTTCCGATCCGYCWATTYHTTTRAGTTT-3′) indexed reverse primer. Sequencing of all samples was performed with an Illumina MiSeq using a MiSeq Reagent Kit v3 (300 bp paired-end reads) (Illumina, Inc.).
Sequencing data processing and network analysis
Quality check of the raw sequence data was performed using a Quantitative Insights into Microbial Ecology (QIIME) pipeline (version 1.9.1)85. The total reads were grouped into OTUs using the Greengenes database86 with a sequence identity threshold of 97%. All samples were rarefied to a depth of 2,720 reads. Furthermore, a prediction of the metabolic functions in the axillary microbiome was performed using the PICRUSt algorithm87. For a network analysis of the axillary microbiome, we used SPIEC-EASI88. OTUs < 0.01% were filtered for network analysis. We constructed the network of the axillary microbiome using the neighborhood algorithm and the Stability Approach to Regularization Selection method with a minimum lambda threshold of 0.1. This analysis was performed using the SPIEC-EASI R package (version 1.1.0). The network topological statistic coefficients were calculated using igraph R package (version 1.2.6). These features includes nodes (number of OTUs), edges (number of potential links between associated nodes), density (ratio of the number of nodes and the number of possible edges), degree (mean number of connected edges per node), transitivity (the probability that the adjacent nodes of a node are connected), and node closeness centrality (the number of steps required to access other nodes from one node).
Bioconversion of L. corniculatus seeds using L. acidophilus KNU-02
Lotus corniculatus seeds were grinded and extracted with 70% ethanol at the ratio of 1:9. The filtered sample extract was dried using a SPD1010 SpeedVac System (Thermo Fisher Scientific, Waltham, MA, USA) at 45 °C for 2 h. A LAB isolate, L. acidophilus KNU-02, was cultivated in MRS (de Man, Rogosa, Sharpe) broth (Difco, Sparks, MD, USA) for 2 d at 37 °C in a shaking incubator at 180 rpm. The activated bacterial cells were obtained by centrifugation and washed with phosphate-buffered saline (pH 7.4). The bacterial cells were suspended (1.0 × 108 cells/mL) in a phosphate-buffered saline mixed with 100 mg/mL of the L. corniculatus seed extract (100 mg/mL) in the ratio of 19:1 (v/v) and incubated for 4 d at 37 °C with shaking at 180 rpm. After centrifugation of the mixture, 1 volume of ethyl acetate with 0.1% acetic acid was added to the supernatant. The resulting solution was mixed thoroughly and placed for 5 min. The upper-layered solution was collected and dried using the SPD1010 SpeedVac System (Thermo Fisher Scientific, Waltham, MA, USA) at 45 °C for 2 h. The product was prepared with 3% 1,2-hexanediol, 0.16% tromethamine, 0.05% ethylhexylglycerin, 20% cabomer, and 0.1% extract powder (v/v).
Compositional analysis
To investigate the compounds in the BLC and LC, a compositional analysis was performed by using GCxGC/TOF–MS. The compositional analysis was conducted using three replicates of each extract. Then, 100 mg of the dried extracts was transferred to a 2 mL centrifuge tube with 1 mL isopropanol–acetonitrile aqueous solution (3:2:2, v/v/v) added to the tube. The mixtures were vortexed for 10 min, followed by centrifugation at 8,000 g for 8 min. The supernatant was filtered with a 0.22 μm polyvinylidene fluoride filter, and 100 μL filtered supernatant was dried using a nitrogen evaporator. For the derivatization reaction, 100 μL methoxyamine hydrochloride in pyridine (20 mg/mL) was added to the dried extract and incubated at 30 °C for 90 min. Then, for silylation, 50 μL N-methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA) was added and incubated at 37 °C for 30 min. GCxGC-TOFMS, an Agilent 7890B Gas Chromatograph coupled with a LECO Pegasus 4D time-of-flight mass spectrometer (TOFMS) (Leco, St. Joseph, MI, USA), was used for the compositional analysis of the BLC. A column was used with an Rtx-5MS (Restek, Bellefonte, PA) with a length of 30 m, an internal diameter of 0.25 mm, and film thickness of 0.25 μm. The carrier gas was helium with a flow rate of 1.5 mL/min. A split ratio for the injector was set to 20:1, with a total injection volume of 1 μL. The front inlet, ion source, and transfer temperature were kept at 250 °C, 200 °C, and 260 °C, respectively. The oven temperature was maintained at 80 °C for 2 min, and the sample was injected. The temperature was increased at a rate of 15 °C/min to 200 °C. Then, the temperature was further increased by 5 °C/min to reach 300 °C and held for 3 min to complete the run. The MS was set at -70 eV with 1600 V of detector voltage. The mass scan range was m/z 35–650, and the acquisition rate was 15 Hz.
Statistical analysis
All microbial analysis in this study was conducted within R software (version 4.0.2; http://www.r-project.org/)89. CAP allows a constrained ordination based on any distances or dissimilarity90. This method maximizes the separation of samples by continuous or factorial variables. CAP of the axillary microbiome was performed with an unweighted UniFrac distance matrix. We performed ANOVA to analyze the association between the microbiota and odor intensity using CAP analysis. This analysis was performed using the phyloseq (version 1.32.0) and vegan (version 2.5-6)91,92 R packages. A Wilcoxon matched-pairs test and a paired t-test were performed to compare 0 week with 1 week. The comparison of the baseline information for the BLC and placebo groups was performed using an unpaired t-test. A correlation analysis was performed using Spearman’s rank correlation coefficient with the Hmisc R package (version 4.4-1). A fold-change analysis of taxa and predicted genes was conducted using the DESeq2 R package (version 1.28.1). The p value in the fold-change analysis was calculated using multiple tests through the FDR (Benjamini-Hochberg). To adjust the effect of other taxa to one taxon, the multivariable regression model was used for investigating the association between taxa and odor intensity. This analysis was performed using MaAsLin2 R package (version 1.4.0). PLS-DA is a multiple linear regression technique to find the direction of maximum covariance between a data set and the class membership93. A VIP score is calculated as a weighted sum of the squared correlations between the PLS-DA components and the original variable. Therefore, PLS-DA was performed using a MetaboAnalyst for a compositional analysis, and the comparison of discriminant feature between two extracts was conducted using two-tailed student’s t-test94.
Supplementary Information
Acknowledgements
The sample collection for axillary microbiome analyses was performed by the Global Medical Research Center (GMRC). This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through Agricultural Microbiome R&D Program funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (918010043SB010).
Author contributions
M.-J.K., D.-G.L. and J.-H.S designed the research study; M.-J.K., H.W.J., M.-C.K., Y.G.J., Y.-J.P., J.-H.S., H.J.K., H.J.K., D.-G.L., and S.K., performed the research; M.-J.K., S.B., and J.-H.S. analyzed the data; M.-J.K. prepared all the figures; M.-J.K., S.B. and J.-H.S wrote the main manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-89606-5.
References
- 1.Sorokowska A, Sorokowski P, Havlícek J. Body odor based personality judgments: The effect of fragranced cosmetics. Front. Psychol. 2016;7:1–8. doi: 10.3389/fpsyg.2016.00530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Demattè ML, Österbauer R, Spence C. Olfactory cues modulate facial attractiveness. Chem. Senses. 2007;32:603–610. doi: 10.1093/chemse/bjm030. [DOI] [PubMed] [Google Scholar]
- 3.Moshkin M, et al. Scent recognition of infected status in humans. J. Sex. Med. 2012;9:3211–3218. doi: 10.1111/j.1743-6109.2011.02562.x. [DOI] [PubMed] [Google Scholar]
- 4.Wilke K, Martin A, Terstegen L, Biel SS. A short history of sweat gland biology. Int. J. Cosmet. Sci. 2007;29:169–179. doi: 10.1111/j.1467-2494.2007.00387.x. [DOI] [PubMed] [Google Scholar]
- 5.Fredrich E, Barzantny H, Brune I, Tauch A. Daily battle against body odor: Towards the activity of the axillary microbiota. Trends Microbiol. 2013;21:305–312. doi: 10.1016/j.tim.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Heisterberg MV, et al. Deodorants are the leading cause of allergic contact dermatitis to fragrance ingredients. Contact Dermatitis. 2011;64:258–264. doi: 10.1111/j.1600-0536.2011.01889.x. [DOI] [PubMed] [Google Scholar]
- 7.Graves AB, et al. The association between aluminum-containing products and Alzheimer’s disease. J. Clin. Epidemiol. 1990;43:35–44. doi: 10.1016/0895-4356(90)90053-R. [DOI] [PubMed] [Google Scholar]
- 8.Campbell A, Yang EY, Tsai-Turton M, Bondy SC. Pro-inflammatory effects of aluminum in human glioblastoma cells. Brain Res. 2002;933:60–65. doi: 10.1016/S0006-8993(02)02305-3. [DOI] [PubMed] [Google Scholar]
- 9.Zirwas J, Moennich J. Antiperspirant and deodorant allergy: diagnosis and management. J. Clin. Contact Dermatol. Case. 2008;1:38–43. [PMC free article] [PubMed] [Google Scholar]
- 10.Savoia D. Plant-derived antimicrobial compounds: Alternatives to antibiotics. Future Microbiol. 2012 doi: 10.2217/fmb.12.68. [DOI] [PubMed] [Google Scholar]
- 11.Ribeiro AS, Estanqueiro M, Oliveira MB, Lobo JMS. Main benefits and applicability of plant extracts in skin care products. Cosmetics. 2015 doi: 10.3390/cosmetics2020048. [DOI] [Google Scholar]
- 12.Subramani R, Narayanasamy M, Feussner KD. Plant-derived antimicrobials to fight against multi-drug-resistant human pathogens. 3 Biotech. 2017 doi: 10.1007/s13205-017-0848-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alkhalaf MI, Alansari WS, Ahmed E, Elhalwagy MEA. Anti-oxidant, anti-inflammatory and anti-cancer activities of avocado (Persea americana) fruit and seed extract. J. King Saud Univ. Sci. 2019;31:1358–1362. doi: 10.1016/j.jksus.2018.10.010. [DOI] [Google Scholar]
- 14.Velioglu YS, Mazza G, Gao L, Oomah BD. Antioxidant activity and total phenolics in selected fruits, vegetables, and grain products. J. Agric. Food Chem. 1998;46:4113–4117. doi: 10.1021/jf9801973. [DOI] [Google Scholar]
- 15.Asong JA, et al. Antimicrobial activity, antioxidant potential, cytotoxicity and phytochemical profiling of four plants locally used against skin diseases. Plants. 2019 doi: 10.3390/plants8090350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiroko M, Minoru M, Yoshikazu Y, Masahiko S. Effect of passion fruit seed extract rich in piceatannol on the skin of women: A randomized, placebo-controlled, double-blind trial. J. Nutr. Sci. Vitaminol. 2018;64:75–80. doi: 10.3177/jnsv.64.75. [DOI] [PubMed] [Google Scholar]
- 17.González-Minero FJ, Bravo-Díaz L. The use of plants in skin-care products, cosmetics and fragrances: Past and present. Cosmetics. 2019 doi: 10.3390/cosmetics5030050. [DOI] [Google Scholar]
- 18.Shahtalebi MA, et al. Deodorant effects of a sage extract stick: Antibacterial activity and sensory evaluation of axillary deodorancy. J. Res. Med. Sci. 2013;18:833. [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi T, Aso Y, Kasai W, Kondo T. Synergetic deodorant effect and antibacterial activity of composite paper containing waste tea leaves. J. Wood Sci. 2011 doi: 10.1007/s10086-010-1171-9. [DOI] [Google Scholar]
- 20.Ramírez-Restrepo CA, Barry TN, López-Villalobos N, Kemp PD, McNabb WC. Use of Lotus corniculatus containing condensed tannins to increase lamb and wool production under commercial dryland farming conditions without the use of anthelmintics. Anim. Feed Sci. Technol. 2004;117:85–105. doi: 10.1016/j.anifeedsci.2004.05.005. [DOI] [Google Scholar]
- 21.Girardi FA, et al. Phytochemical profile and antimicrobial properties of Lotus spp. (Fabaceae) An. Acad. Bras. Cienc. 2014;86:1295–1302. doi: 10.1590/0001-3765201420130220. [DOI] [PubMed] [Google Scholar]
- 22.Dalmarco J, Dalmarco E, Koelzer J, Pizzolatti M, Fröde T. Isolation and identification of bioactive compounds responsible for the anti-bacterial efficacy of Lotus corniculatus var So gabriel. Int. J. Green Pharm. 2010;4:108–114. doi: 10.4103/0973-8258.63886. [DOI] [Google Scholar]
- 23.Barone CD, et al. In vitro screening of 51 birdsfoot trefoil (Lotus corniculatus L.; Fabaceae ) strains for anti-parasitic effects against Haemonchus contortus. Parasitology. 2019 doi: 10.1017/S0031182018002214. [DOI] [PubMed] [Google Scholar]
- 24.Yerlikaya S, et al. Investigation of chemical profile, biological properties of Lotus corniculatus L. extracts and their apoptotic-autophagic effects on breast cancer cells. J. Pharm. Biomed. Anal. 2019 doi: 10.1016/j.jpba.2019.05.068. [DOI] [PubMed] [Google Scholar]
- 25.Bianchini LF, et al. Microbial biotransformation to obtain new antifungals. Front. Microbiol. 2015 doi: 10.3389/fmicb.2015.01433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhatti HN, et al. Biotransformation of monoterpenoids and their antimicrobial activities. Phytomedicine. 2014 doi: 10.1016/j.phymed.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 27.Li S, et al. Increasing antioxidant activity of procyanidin extracts from the pericarp of Litchi chinensis processing waste by two probiotic bacteria bioconversions. J. Agric. Food Chem. 2013 doi: 10.1021/jf305213e. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Hui X, Ibrahim SA, Huang W. Increasing antiradical activity of polyphenols from lotus seed epicarp by probiotic bacteria bioconversion. Molecules. 2018;23:1–12. doi: 10.3390/molecules23102667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linares DM, et al. Lactic acid bacteria and bifidobacteria with potential to design natural biofunctional health-promoting dairy foods. Front. Microbiol. 2017 doi: 10.3389/fmicb.2017.00846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee NK, Paik HD. Bioconversion using lactic acid bacteria: Ginsenosides, gaba, and phenolic compounds. J. Microbiol. Biotechnol. 2017 doi: 10.4014/jmb.1612.12005. [DOI] [PubMed] [Google Scholar]
- 31.Mihreteab M, Stubblefield BA, Gilbert ES. Microbial bioconversion of thermally depolymerized polypropylene by Yarrowia lipolytica for fatty acid production. Appl. Microbiol. Biotechnol. 2019 doi: 10.1007/s00253-019-09999-2. [DOI] [PubMed] [Google Scholar]
- 32.Ling TJ, et al. New triterpenoids and other constituents from a special microbial-fermented tea—Fuzhuan brick tea. J. Agric. Food Chem. 2010 doi: 10.1021/jf9043524. [DOI] [PubMed] [Google Scholar]
- 33.Zhu M, et al. Microbial bioconversion of the chemical components in dark tea. Food Chem. 2020 doi: 10.1016/j.foodchem.2019.126043. [DOI] [PubMed] [Google Scholar]
- 34.Li Z, et al. Revealing the influence of microbiota on the quality of Pu-erh tea during fermentation process by shotgun metagenomic and metabolomic analysis. Food Microbiol. 2018 doi: 10.1016/j.fm.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Zhu YF, et al. Changes of major tea polyphenols and production of four new B-ring fission metabolites of catechins from post-fermented Jing-Wei Fu brick tea. Food Chem. 2015 doi: 10.1016/j.foodchem.2014.08.075. [DOI] [PubMed] [Google Scholar]
- 36.Abari AH, Tayebi M. Bioconversion of genistein to orobol by Bacillus subtilis Spore displayed tyrosinase and monitoring the anticancer effects of orobol on MCF-7 breast cancer cells. Biotechnol. Bioprocess Eng. 2019 doi: 10.1007/s12257-019-0067-9. [DOI] [Google Scholar]
- 37.Pham JV, et al. A review of the microbial production of bioactive natural products and biologics. Front. Microbiol. 2019 doi: 10.3389/fmicb.2019.01404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amaral LFB, Moriel P, Foglio MA, Mazzola PG. Caryocar brasiliense supercritical CO2 extract possesses antimicrobial and antioxidant properties useful for personal care products. BMC Complement. Altern. Med. 2014 doi: 10.1186/1472-6882-14-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barbulova A, Colucci G, Apone F. New trends in cosmetics: By-products of plant origin and their potential use as cosmetic active ingredients. Cosmetics. 2015 doi: 10.3390/cosmetics2020082. [DOI] [Google Scholar]
- 40.Suroowan, S. & Mahomoodally, M. F. Alternative antimicrobials from natural products against dermatological infections. in The Microbiology of Skin, Soft Tissue, Bone and Joint Infections (2017). doi:10.1016/b978-0-12-811079-9.00012-4.
- 41.Perez GIP, et al. Body site is a more determinant factor than human population diversity in the healthy skin microbiome. PLoS ONE. 2016;11:1–15. doi: 10.1371/journal.pone.0151990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grice EA, et al. Topographical and temporal diversity of the human skin microbiome. Science (80-.) 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li M, et al. The influence of age, gender and race/ethnicity on the composition of the human axillary microbiome. Int. J. Cosmet. Sci. 2019;41:371–377. doi: 10.1111/ics.12549. [DOI] [PubMed] [Google Scholar]
- 44.Troccaz M, et al. Mapping axillary microbiota responsible for body odours using a culture-independent approach. Microbiome. 2015 doi: 10.1186/s40168-014-0064-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kong HH, Segre JA. Skin microbiome: Looking back to move forward. J. Investig. Dermatol. 2012 doi: 10.1038/jid.2011.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bek-Thomsen M, Lomholt HB, Kilian M. Acne is not associated with yet-uncultured bacteria. J. Clin. Microbiol. 2008;46:3355–3360. doi: 10.1128/JCM.00799-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeeuwen PLJM, et al. Microbiome dynamics of human epidermis following skin barrier disruption. Genome Biol. 2012 doi: 10.1186/gb-2012-13-11-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corsi N, Colloca L. Placebo and nocebo effects: The advantage of measuring expectations and psychological factors. Front. Psychol. 2017 doi: 10.3389/fpsyg.2017.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Colloca L, Barsky AJ. Placebo and nocebo effects. N. Engl. J. Med. 2020 doi: 10.1056/nejmra1907805. [DOI] [PubMed] [Google Scholar]
- 50.Kam-Hansen S, et al. Altered placebo and drug labeling changes the outcome of episodic migraine attacks. Sci. Transl. Med. 2014 doi: 10.1126/scitranslmed.3006175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller FG, Wendler D, Swartzman LC. Deception in research on the placebo effect. PLoS Med. 2005 doi: 10.1371/journal.pmed.0020262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vits S, Schedlowski M. Learned placebo effects in the immune system. Zeitschrift fur Psychologie/J. Psychol. 2014 doi: 10.1027/2151-2604/a000184. [DOI] [Google Scholar]
- 53.Lali Mathew A, Saji L, Eliza Joseph L, Abraham E. Natural deodorants and antiperspirants. Int. J. Novel Trends Pharm. Sci. 2017;7:49–53. [Google Scholar]
- 54.Urban J, et al. The effect of habitual and experimental antiperspirant and deodorant product use on the armpit microbiome. PeerJ. 2016;2016:1–20. doi: 10.7717/peerj.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Callewaert C, Hutapea P, Van de Wiele T, Boon N. Deodorants and antiperspirants affect the axillary bacterial community. Arch. Dermatol. Res. 2014;306:701–710. doi: 10.1007/s00403-014-1487-1. [DOI] [PubMed] [Google Scholar]
- 56.Bouslimani A, et al. The impact of skin care products on skin chemistry and microbiome dynamics. BMC Biol. 2019;17:1–20. doi: 10.1186/s12915-019-0660-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.James AG, Austin CJ, Cox DS, Taylor D, Calvert R. Microbiological and biochemical origins of human axillary odour. FEMS Microbiol. Ecol. 2013;83:527–540. doi: 10.1111/1574-6941.12054. [DOI] [PubMed] [Google Scholar]
- 58.Okamoto H, Koizumi S, Shimizu H, Cho O, Sugita T. Characterization of the axillary microbiota of Japanese male subjects with spicy and milky odor types by pyrosequencing. Biocontrol Sci. 2018 doi: 10.4265/bio.23.1. [DOI] [PubMed] [Google Scholar]
- 59.Taylor D, et al. Characterization of the microflora of the human axilla. Int. J. Cosmet. Sci. 2003 doi: 10.1046/j.1467-2494.2003.00181.x. [DOI] [PubMed] [Google Scholar]
- 60.James AG, Casey J, Hyliands D, Mycock G. Fatty acid metabolism by cutaneous bacteria and its role in axillary malodour. World J. Microbiol. Biotechnol. 2004;20:787–793. doi: 10.1007/s11274-004-5843-8. [DOI] [Google Scholar]
- 61.Emter R, Natsch A. The sequential action of a dipeptidase and a β-lyase is required for the release of the human body odorant 3-methyl-3-sulfanylhexan-1-ol from a secreted Cys-Gly-(S) conjugate by Corynebacteria. J. Biol. Chem. 2008 doi: 10.1074/jbc.M800730200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Starkenmann C, Mayenzet F, Brauchli R, Troccaz M. 5α-Androst-16-en-3α-ol β-D-glucuronide, precursor of 5α-androst-16-en-3α-ol in human sweat. Chem. Biodivers. 2013 doi: 10.1002/cbdv.201300286. [DOI] [PubMed] [Google Scholar]
- 63.Fujii T, Shinozaki J, Kajiura T, Iwasaki K, Fudou R. A newly discovered Anaerococcus strain responsible for axillary odor and a new axillary odor inhibitor, pentagalloyl glucose. FEMS Microbiol. Ecol. 2014;89:198–207. doi: 10.1111/1574-6941.12347. [DOI] [PubMed] [Google Scholar]
- 64.Troccaz M, Starkenmann C, Niclass Y, Van De Waal M, Clark AJ. 3-methyl-3-sulfanylhexan-1-ol as a major descriptor for the human axilla-sweat odour profile. Chem. Biodivers. 2004 doi: 10.1002/cbdv.200490077. [DOI] [PubMed] [Google Scholar]
- 65.Leyden JJ, McGinley KJ, Holzle E, Labows JN, Kligman AM. The microbiology of the human axilla and its relationship to axillary odor. J. Investig. Dermatol. 1981 doi: 10.1111/1523-1747.ep12494624. [DOI] [PubMed] [Google Scholar]
- 66.James AG, Hyliands D, Johnston H. Generation of volatile fatty acids by axillary bacteria. Int. J. Cosmet. Sci. 2004 doi: 10.1111/j.1467-2494.2004.00214.x. [DOI] [PubMed] [Google Scholar]
- 67.Park T, et al. Collapse of human scalp microbiome network in dandruff and seborrhoeic dermatitis. Exp. Dermatol. 2017 doi: 10.1111/exd.13293. [DOI] [PubMed] [Google Scholar]
- 68.Kim HJ, et al. Fragile skin microbiomes in megacities are assembled by a predominantly niche-based process. Sci. Adv. 2018 doi: 10.1126/sciadv.1701581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Centrality and Hubs. in Fundamentals of Brain Network Analysis (2016). 10.1016/b978-0-12-407908-3.00005-4.
- 70.Jimenez-Diaz, L., Caballero, A. & Segura, A. Pathways for the degradation of fatty acids in bacteria. in Aerobic Utilization of Hydrocarbons, Oils and Lipids (2017). 10.1007/978-3-319-39782-5_42-1.
- 71.James AG, Austin CJ, Cox DS, Taylor D, Calvert R. Microbiological and biochemical origins of human axillary odour. FEMS Microbiol. Ecol. 2013 doi: 10.1111/1574-6941.12054. [DOI] [PubMed] [Google Scholar]
- 72.Moat AG, Foster JW, Spector MP. Microbial Physiology. Microb. Physiol. 2002 doi: 10.1002/0471223867. [DOI] [Google Scholar]
- 73.Lam TH, et al. Understanding the microbial basis of body odor in pre-pubescent children and teenagers. Microbiome. 2018 doi: 10.1186/s40168-018-0588-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Callewaert C, et al. Characterization of Staphylococcus and Corynebacterium clusters in the human axillary region. PLoS ONE. 2013 doi: 10.1371/journal.pone.0070538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.James AG. The axillary microbiome and its relationship with underarm odor. Skin Microbiome Handb. 2020 doi: 10.1002/9781119593058.ch5. [DOI] [Google Scholar]
- 76.Park ES, et al. Antimicrobial activity of phenol and benzoic acid derivatives. Int. Biodeterior. Biodegrad. 2001 doi: 10.1016/S0964-8305(01)00058-0. [DOI] [Google Scholar]
- 77.del Olmo A, Calzada J, Nuñez M. Benzoic acid and its derivatives as naturally occurring compounds in foods and as additives: Uses, exposure, and controversy. Crit. Rev. Food Sci. Nutr. 2017 doi: 10.1080/10408398.2015.1087964. [DOI] [PubMed] [Google Scholar]
- 78.Shannon K. Understanding antibacterial action and resistance. J. Hosp. Infect. 1996 doi: 10.1016/s0195-6701(96)90077-8. [DOI] [Google Scholar]
- 79.Kim HW, Seok YS, Rhee MS. Synergistic staphylocidal interaction of benzoic acid derivatives (benzoic acid, 4-hydroxybenzoic acid and β-resorcylic acid) and capric acid: Mechanism and verification study using artificial skin. J. Antimicrob. Chemother. 2020 doi: 10.1093/jac/dkz494. [DOI] [PubMed] [Google Scholar]
- 80.Soni MG, Burdock GA, Taylor SL, Greenberg NA. Safety assessment of propyl paraben: A review of the published literature. Food Chem. Toxicol. 2001 doi: 10.1016/S0278-6915(00)00162-9. [DOI] [PubMed] [Google Scholar]
- 81.Mazzutti S, et al. Supercritical fluid extraction of Agaricus brasiliensis: Antioxidant and antimicrobial activities. J. Supercrit. Fluids. 2012 doi: 10.1016/j.supflu.2012.06.010. [DOI] [Google Scholar]
- 82.Kim MJ, et al. Skin permeation enhancement of diclofenac by fatty acids. Drug Deliv. 2008 doi: 10.1080/10717540802006898. [DOI] [PubMed] [Google Scholar]
- 83.Kováčik A, Kopečná M, Vávrová K. Permeation enhancers in transdermal drug delivery: Benefits and limitations. Expert Opin. Drug Deliv. 2020 doi: 10.1080/17425247.2020.1713087. [DOI] [PubMed] [Google Scholar]
- 84.Lee HJ, et al. Effects of cosmetics on the skin microbiome of facial cheeks with different hydration levels. Microbiologyopen. 2018;7:1–14. doi: 10.1002/mbo3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Caporaso JG, et al. correspondEnce QIIME allows analysis of high-throughput community sequencing data Intensity normalization improves color calling in SOLiD sequencing. Nat. Publ. Gr. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Desantis TZ, et al. Greengenes, a Chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Langille MGI, et al. A n a ly s i s Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kurtz ZD, et al. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput. Biol. 2015;11:1–25. doi: 10.1371/journal.pcbi.1004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2020. [Google Scholar]
- 90.Anderson MJ, Willis TJ. Canonical analysis of principal coordinates: A useful method of constrained ordination for ecology. Ecology. 2003 doi: 10.1890/0012-9658(2003)084[0511:CAOPCA]2.0.CO;2. [DOI] [Google Scholar]
- 91.McMurdie PJ, Holmes S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Oksanen, J. et al. The vegan package. Community Ecol. Packag. 190 (2008).
- 93.Ballabio D, Consonni V. Classification tools in chemistry. Part 1: Linear models. PLS-DA. Anal. Methods. 2013;5:3790. doi: 10.1039/c3ay40582f. [DOI] [Google Scholar]
- 94.Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat. Protoc. 2011 doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.