Abstract
The genus Nitrospira is the most widespread group of nitrite-oxidizing bacteria and thrives in diverse natural and engineered ecosystems. Nitrospira marina Nb-295T was isolated from the ocean over 30 years ago; however, its genome has not yet been analyzed. Here, we investigated the metabolic potential of N. marina based on its complete genome sequence and performed physiological experiments to test genome-derived hypotheses. Our data confirm that N. marina benefits from additions of undefined organic carbon substrates, has adaptations to resist oxidative, osmotic, and UV light-induced stress and low dissolved pCO2, and requires exogenous vitamin B12. In addition, N. marina is able to grow chemoorganotrophically on formate, and is thus not an obligate chemolithoautotroph. We further investigated the proteomic response of N. marina to low (∼5.6 µM) O2 concentrations. The abundance of a potentially more efficient CO2-fixing pyruvate:ferredoxin oxidoreductase (POR) complex and a high-affinity cbb3-type terminal oxidase increased under O2 limitation, suggesting a role in sustaining nitrite oxidation-driven autotrophy. This putatively more O2-sensitive POR complex might be protected from oxidative damage by Cu/Zn-binding superoxide dismutase, which also increased in abundance under low O2 conditions. Furthermore, the upregulation of proteins involved in alternative energy metabolisms, including Group 3b [NiFe] hydrogenase and formate dehydrogenase, indicate a high metabolic versatility to survive conditions unfavorable for aerobic nitrite oxidation. In summary, the genome and proteome of the first marine Nitrospira isolate identifies adaptations to life in the oxic ocean and provides insights into the metabolic diversity and niche differentiation of NOB in marine environments.
Subject terms: Water microbiology, Microbial biooceanography, Marine microbiology, Bacterial genomics, Bacterial physiology
Introduction
Aerobic nitrite (NO2−) oxidation is the main biochemical nitrate (NO3−)-forming reaction, carried out during the second step of nitrification [1]. In marine ecosystems, nitrate is the dominant form of biologically available nitrogen, which is rapidly assimilated by phytoplankton in surface waters and accumulates in the deep sea [2].
Nitrite-oxidizing bacteria (NOB) are chemolithoautotrophic microorganisms found within four known bacterial phyla (Proteobacteria, Nitrospirae, Nitrospinae, and Chloroflexi) [3]. The genus Nitrospira, within the Nitrospirae phylum, is the most diverse NOB genus and consists of at least six phylogenetic sublineages [3]. Nitrospira are ubiquitously present in natural and engineered ecosystems, including oceans [4, 5], freshwater habitats [6], soils [7, 8], saline-alkaline lakes [9], hot springs [10], wastewater treatment plants [11–13], and aquaculture biofilters [14, 15]. In human-made ecosystems, Nitrospira is generally considered to be adapted to low NO2− concentrations [16]. In the open ocean, however, where NO2− concentrations are exceedingly low, NOB affiliated with the phylum Nitrospinae appear to be the dominant nitrite oxidizers [4], whereas Nitrospira are found in relatively high NO2− environments including sediments and deep sea hydrothermal vent plumes [17, 18]. Nitrospira also dominate over Nitrospinae-affiliated bacteria in some deep-sea trench environments [19, 20]. High NO2− concentrations are found coincident with low O2 concentrations in oxygen minimum zone (OMZ) waters [21, 22] in a feature known as the secondary nitrite maximum with concentrations reaching ~1–5 µM NO2− [23, 24]. Despite the O2-dependence of all known NOB, NO2− oxidation can still be detected at nanomolar O2 concentrations [25].
Some metabolic features appear to be common among Nitrospira, based on genomic analyses to date. Immunocytochemical analyses of a representative of Nitrospira sublineage I and metagenomic analyses of Nitrospira defluvii indicated a periplasmic orientation of nitrite oxidoreductase (NXR) [26, 27], the key enzyme for NO2− oxidation, and the presence of the O2-sensitive reductive tricarboxylic acid (rTCA) cycle for inorganic carbon fixation [26]. These results suggest that Nitrospira evolved from microaerophilic or anaerobic ancestors [26]. N. defluvii lacks genes for classical oxidative stress defense enzymes present in most aerobic organisms including catalase and superoxide dismutase (SOD), indicative of adaptations to low O2 environmental niches [26]. However, other Nitrospira species encode both types of enzymes [28, 29], suggesting different O2 tolerances within members of the Nitrospira genus. In addition to NO2− oxidation, Nitrospira exhibit a high metabolic versatility, growing aerobically on hydrogen [30] or anaerobically on organic acids while respiring nitrate [28]. Recently, the capability for the complete oxidation of ammonia to nitrate (comammox) was identified in representatives of sublineage II Nitrospira [14, 31], however, comammox Nitrospira appear to be absent in marine systems [31].
Nitrospira marina Nb-295T, the type species of the genus Nitrospira, was isolated from a water sample collected at a depth of 206 m from the Gulf of Maine in the Atlantic Ocean over 30 years ago [5]. It is the only Nitrospira species isolated from the oceanic water column and the environmental conditions favorable for Nitrospira in the ocean remain largely unexplored. Here, we investigated the metabolic potential of N. marina Nb-295T based on its complete genome sequence and compared the proteome signatures of cultures grown under atmospheric O2 tension and under low O2 conditions typically encountered in OMZs.
Materials and methods
Cultivation of N. marina Nb-295
A cryopreserved stock of N. marina Nb-295T was obtained from the culture collection of John B. Waterbury at the Woods Hole Oceanographic Institution. Strain Nb-295 was grown in 60 mL polycarbonate bottles (Nalgene) in 50 mL autotrophic mineral salts medium at pH 7.8 containing 2 mM NaNO2 (Table S1), and bottles were incubated at 25 °C in the dark without agitation. Mixotrophic growth was tested through the individual addition of the following organic carbon substrates to the culture medium of duplicate cultures (final concentrations): 150 mg L−1 yeast extract, 150 mg L−1 tryptone, 0.5 mM pyruvate, or 1 g L−1 glycerol. NO2− consumption was measured as previously described [32] and growth was monitored by flow cytometry (see Supplementary Methods). To test for chemoorganotrophic growth, the culture was transferred (2% inoculum) into NO2−-free medium containing either 1 mM formate or 1 mM pyruvate, and 100 µM ammonium chloride, which served as nitrogen source.
For incubations at different O2 concentrations, triplicate cultures of N. marina Nb-295 were grown at 22 °C in 400 mL of medium containing 70% natural seawater and 2 mM NaNO2 as described by Watson et al. [5] in 500 mL polycarbonate bottles (Nalgene) with a custom-made sparging rig. Bottles were constantly bubbled (flow rate: 15 mL min−1) with one of two sterile custom gas mixes containing either 0.5% or 20% oxygen, 300 ppm CO2, and a balance of high-purity N2. NO2− concentrations were measured as a proxy for growth as described above and 10 mL aliquot of each culture was fixed at the last time point (2% formaldehyde, 1 h, 4 °C) for cell enumeration on an epifluorescence microscope as previously described [33].
DNA extraction, genome sequencing, and annotation
High molecular weight genomic DNA was extracted from stationary phase cultures using a CTAB extraction protocol [34] and sequenced on the PacBio platform at the US Department of Energy Joint Genome Institute (JGI). 754,554 reads were produced, with 209,987 passing quality control. The assembly was conducted using HGAP (v 2.2.0.p1) with improvement with Quiver [35] resulting in a single contig.
Gene annotation was conducted using JGI’s Integrated Microbial Genomes and Microbiomes pipeline [36] and the MicroScope platform [37]. Manual curation included sequence similarity searches using BLASTP (e-value <1e−30) [38] against the Transporter Classification database [39] and protein domain searches using InterProScan (release 72.0) [40]. Signal peptides were identified with SignalP 5.0 [41] to determine if proteins were potentially addressed to the membrane and/or released to the periplasmic space. A list of manually curated annotations can be found in Data Set 1.
Phylogenomic analysis was performed using 120 concatenated phylogenetic marker genes from representatives of the phylum Nitrospirae/Nitrospirota as implemented in the Genome Taxonomy Database Toolkit (GTDB-tk) version 1.1.1 [42]. (see Supplementary Methods).
Protein extraction and proteome analyses
Cells for proteomic analysis were harvested when [NO2−] dropped to ~500 µM, corresponding to exponential growth of strain Nb-295. Each culture was mixed with an equal volume of a house-made fixative [43] similar to the commercially available solution RNALater (Thermo Fisher), and filtered by vacuum filtration onto 25 mm, 0.2 µm pore size Supor filters (Pall). 200 mL of fixed culture (equivalent to 100 mL of growth medium) were filtered for protein extraction and proteomic analyses. Filters were frozen at −80 °C until extraction. A detailed description of the procedures used for protein extraction and purification can be found in the Supplementary Methods. Global (untargeted) proteomes were analyzed on a Fusion Orbitrap mass spectrometer using one-dimensional nanospray separation and data-dependent acquisition based on Saito et al. [44] (see Supplementary Methods for detailed protocols). Select targeted quantitative proteomic assays using custom-made isotopically-labeled (15N) peptide standards were designed and samples were analyzed again by parallel reaction monitoring mass spectrometry using mass spectral information from the global proteome analyses as previously described [44] (also see Supplementary Methods). Tryptic peptides from ten proteins of interest, including nitrite oxidoreductase subunit alpha (NxrA), were targeted for absolute quantitation (see Data Set 2). Cellular NXR concentrations were calculated based on NxrA peptide concentrations using the average carbon content of N. marina cells (152 fg cell−1, Santoro et al., unpublished) and an estimated cellular protein:carbon ratio of 50% based on experimentally determined values [45]. NXR complex density on the cellular membrane was calculated using an estimated NXR complex size of 63 nm2 [27] and an estimated cell surface area of 2.45 µm2 (assuming a cylindrical shape with a length of 1.75 µm and a radius of 0.2 µm based on previously determined cell dimensions of N. marina [5]).
Differential levels of expression between the two treatments (i.e., atmospheric O2 and low O2 conditions) were tested with the DESeq2 Bioconductor package (version 1.20.0) [46] in the R software environment (version 3.5.0) [47] using spectral counts as input data as previously described [48]. Proteins with a mean spectral count below 6 across all treatments were excluded from the analysis. In DESeq2, only proteins that increased in abundance under low O2 conditions were considered (‘altHyphothesis=greater’), P values were adjusted using the Benjamini-Hochberg method (“pAdjustmethod=BH”) and independent filtering was omitted (“independentFiltering=FALSE”). Changes in protein abundance (as determined by spectral counts) were considered statistically significant when adjusted P values were lower than or equal to 0.05 (see Data Set 3). While DESeq2 has a high precision and accuracy [49], it is more conservative than other methods on low-count transcripts/proteins [50]. Proteins of interest were visualized with the pheatmap package (version 1.0.12) [51] in the R software environment [47]. The normalized spectral abundance factor was calculated as proxy for relative protein abundances [52], and values were square-root transformed to improve visualization of low abundant proteins.
Results and discussion
Genome analysis
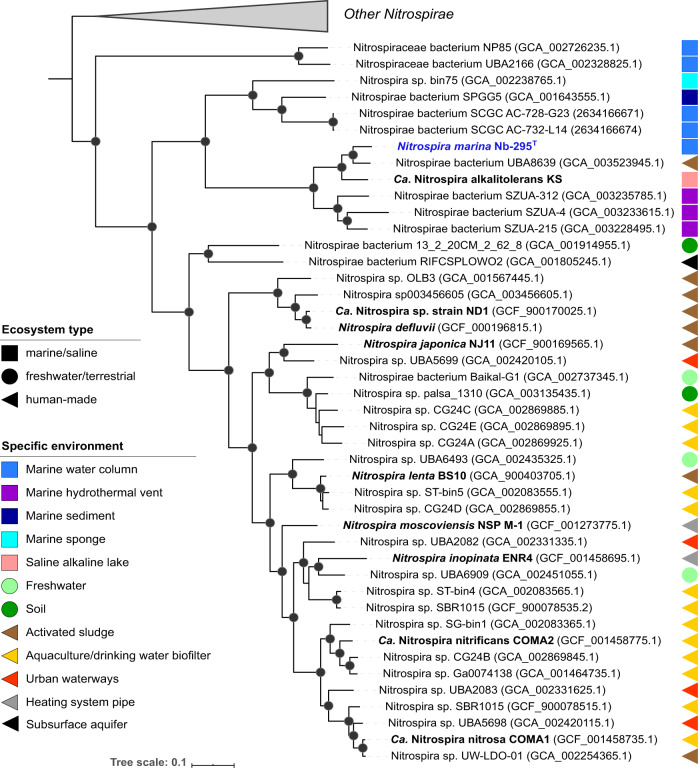
The genome of N. marina Nb-295 is a single element of 4,683,627 bp and contains 4272 coding DNA sequences (CDS) including one rRNA operon. A 5578 bp region of 99.7% identity on each end of the scaffold suggests circularization into a single chromosome. No plasmids or extra-chromosomal elements were identified. The G + C content is 50.04%, lower than in other Nitrospira species and the marine nitrite-oxidizers Nitrospina gracilis and Nitrococcus mobilis (Table S2). Phylogenomic analysis of available closed genomes, metagenome-assembled genomes (MAGs) and single-cell amplified genomes affiliated with the phylum Nitrospirae (see Supplementary Methods) placed N. marina Nb-295T within a cluster of genomes derived from marine and saline environments (Fig. 1). The most closely related Nitrospira MAG (UBA8639) was obtained from a laboratory-scale nitrification reactor; however, the reactor influent consisted of 33% untreated seawater [53], suggesting a marine origin of this MAG. The 16 S rRNA gene sequence of N. marina Nb-295T clustered together with environmental sequences derived from marine sediments and marine aquaculture biofilters (Fig. S1), and shared 99.1% and 97.9% sequence identity with the cultured lineage IV representatives Nitrospira sp. Ecomares 2.1 [15] and Ca. Nitrospira alkalitolerans [9], respectively.
Fig. 1. Maximum likelihood phylogenetic tree of representatives of the phylum Nitrospirae.
The multiple sequence alignment consisting of 120 concatenated phylogenetic marker genes contained 95 genomes and metagenome-assemble genomes (MAGs) from the Genome Taxonomy Database (GTDB) (Release 04-RS89, 19 June 2019), the genome of N. marina, the MAG of Ca. N. alkalitolerans and two open ocean single-cell amplified genomes (SAGs), AC-738-G23 and AC-732-L14. All MAGs and SAGs were estimated to be ≥50% complete with ≤5% contamination. Nodes with UFBoot support of at least 95% are indicated as black filled circles. Cultured representatives are shown in bold. The scale bar represents 0.1 substitutions per site.
Nitrogen metabolism and respiratory chain
The genome of N. marina encodes orthologs of all known proteins required for NO2− oxidation, including the putatively periplasm-oriented NXR complex. Three candidates each of genes for NxrA, NxrB, and NxrC, and two additional NxrC-like proteins were identified (Table S2, Data Set 1). The genes nxrA and nxrB, encoding the alpha and beta subunits of NXR, are colocalized in three clusters, whereas all nxrC candidate genes are localized separately from nxrAB, as previously described for Nitrospira moscoviensis [54]. The NxrA subunits share 87.3–88.9% amino acid identity, the NxrB subunits share 98.8–99.5% amino acid identity, and the putative NxrC subunits are less conserved, sharing between 33.7% and 86.9% amino acid identity.
Like all other analyzed Nitrospira genomes, N. marina encodes a putative copper-dependent NO-forming nitrite reductase (NirK), yet its function in Nitrospira and other NOB remains unknown. N. marina also encodes the ferredoxin-dependent nitrite reductase (NirA) for assimilatory nitrite reduction, which appears to be conserved in Ca. Nitrospira lenta and N. defluvii but absent in other Nitrospira [55]. In addition, N. marina encodes three high affinity ammonium transporters (Amt) enabling direct uptake of reduced N for assimilation, and cyanate lyase to hydrolyze cyanate to ammonium and CO2 (Data Set 1). In contrast to some Nitrospira species [13, 28, 31], N. marina does not encode a urea transporter or urease, which would catabolize urea to ammonia for N assimilation (Table S2).
Previously sequenced genomes of Nitrospira contain multiple copies of several complexes of the respiratory chain [26, 28, 29, 54]. N. marina encodes two paralogous copies of complex I, one of which contains a duplication of NADH:quinone oxidoreductase subunit M (NuoM) and lacks genes for NuoE, NuoF and NuoH (Data Set 1), which is a characteristic feature of Nitrospira genomes [54, 56]. Furthermore, the N. marina genome contains two copies of complex III, two cytochrome bd oxidases and seven putative cytochrome bd-like oxidases (Data Set 1), which show limited partial similarity to canonical cytochrome bd oxidase subunit I as described for N. moscoviensis [54]. One of these cytochrome bd-like oxidases (bd-like_6) contains putative heme b and copper binding sites potentially functioning as a novel terminal oxidase as previously proposed [26]. N. marina also encodes for a cbb3-type terminal oxidase, which usually exhibit high affinities for O2 [57]. This feature is shared with the closely related Ca. N. alkalitolerans [9] and with the more distantly related marine NOB Nitrospina gracilis [58], but absent in all other Nitrospira species sequenced thus far. In addition to the canonical H+-translocating F1F0-ATPase (complex V), N. marina also encodes a putative alternative Na+-translocating N-ATPase (Data Set 1), which potentially contributes to the maintenance of the membrane potential and the generation of a sodium motive force (SMF) as suggested for Ca. N. alkalitolerans [9]. Furthermore, a H+-translocating pyrophosphatase (H+-PPase) with homology to Leptospira/protozoan/plant-type enzymes [59] was identified. H+-PPases couple the translocation of H+ to the hydrolysis of the biosynthetic by-product pyrophosphate (PPi), which is suggested to be an adaptation to life under energy limitation [60]. The N. marina genome also contains an alternative complex III (ACIII) module, which shares similarity with that from sulfur-reducing Acidobacteria [61]. Like canonical complex III, ACIII also functions as a quinol oxidase transferring electrons to cytochrome c and contributes to energy conservation (Refojo et al., 2012). With the exception of the comammox bacterium Ca. Nitrospira nitrificans [14], no homologs of ACIII modules were identified in any other NOB genome.
N. marina encodes a putative hyb-like operon containing four subunits of a cytoplasmic Group 3b [NiFe] hydrogenase and six accessory proteins involved in hydrogenase assembly and maturation (Data Set 1). This type of hydrogenase appears to be conserved in the marine NOB Nitrospina gracilis 3/211 and Nitrococcus mobilis Nb-231 [58, 62], Ca. N. alkalitolerans [9] and in comammox Nitrospira [14, 31] (Table S2). In addition to catalyzing the reversible, NAD+-dependent oxidation of hydrogen, these so-called sulfhydrogenases have been reported to reduce elemental sulfur (So) or polysulfides to hydrogen sulfide (H2S) [63]. Furthermore, the N. marina genome encodes a putative periplasmic sulfite:cytochrome c oxidoreductase, which might couple sulfite (SO32−) oxidation to sulfate (SO42−) with the reduction of cytochrome c as previously suggested for Nitrospina gracilis [58]. Contrastingly, sulfide/quinone oxidoreductase, which is speculated to mediate sulfide oxidation in Nitrococcus [62], is lacking. Whether or not these enzymes are involved in energy conservation using H2S and SO32− as alternative substrates in NOB remains to be experimentally validated.
Central carbon metabolism
In agreement with other Nitrospira genomes [13, 26, 54, 55], N. marina encodes the complete gene repertoire for the rTCA cycle for carbon dioxide (CO2) fixation, including the key enzymes ATP-citrate lyase and 2-oxoglutarate/pyruvate:ferredoxin oxidoreductase (Data Set 1). In the ocean, inorganic carbon is predominately available in the form of bicarbonate (HCO3−) and to a much lesser extent as CO2. Five inorganic anion transporters (SulP family) with homology to BicA HCO3− uptake systems of the cyanobacterium Synechococcus [64] were identified in the N. marina genome (Data Set 1). Two of these putative BicA-like transporters are colocalized with genes encoding Na+/H+ antiporters (NhaB family), which could drive the uptake of HCO3− via Na+ extrusion under alkaline conditions as suggested for the cyanobacterium Aphanothece halophytica [65] and Ca. N. alkalitolerans [9]. N. marina also encodes one putative SulP-related bicarbonate transporter fused to a carbonic anhydrase and four genes encoding putative alpha, beta and gamma carbonic anhydrases (Data Set 1), which can convert the imported HCO3− to CO2 for inorganic carbon fixation via the rTCA cycle.
In addition to the rTCA cycle, N. marina encodes all required genes for the oxidative TCA cycle for pyruvate oxidation via acetyl-CoA, complete gluconeogenesis and glycolysis pathways, and the oxidative and non-oxidative branches of the pentose phosphate pathway (Data Set 1), which are common features of all sequenced Nitrospira genomes [9, 26, 28, 29, 55]. Furthermore, biosynthetic pathways for all amino acids except methionine were identified in the N. marina genome. Although N. marina encodes a vitamin B12-dependent methionine synthase (MetH) (Data Set 1), it appears to lack additional enzymes of known methionine biosynthesis pathways, a trait shared by all other sequenced Nitrospira species and Nitrospina gracilis [9, 26, 28, 29, 55, 58]. However, as N. marina can grow in artificial seawater medium without added methionine, we hypothesize that an alternative unknown pathway for the early steps of methionine biosynthesis functions in Nitrospira and Nitrospina. The N. marina genome contains genes for the biosynthesis and degradation of the storage compounds glycogen and polyphosphate (Data Set 1). In contrast to other Nitrospira and the marine NOB N. gracilis and N. mobilis that encode a glgA-type glycogen synthase, N. marina encodes alpha-maltose-1-phosphate synthase (glgM) and alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase (glgE) for the synthesis of glycogen via alpha-maltose-1-phosphate.
Use of organic carbon substrates
N. marina has been reported to be an obligate chemolithotroph that grows best in medium supplemented with low concentrations of organic compounds including pyruvate, glycerol, yeast extract and peptone [5]. Thus, we investigated the genomic basis for this observation and conducted additional physiological experiments.
In addition to its complete glycolysis pathway and oxidative TCA cycle, a putative carbohydrate degradation operon was identified, consisting of a sugar ABC transporter module, beta-glucosidase, a putatively secreted glycoside hydrolase (GH15) and a carbohydrate-binding protein (Data Set 1). N. marina also encodes two putative carbohydrate-selective porins (OprB), a sugar:sodium symporter (SSS family), a putative galactonate/glucarate transporter (MFS superfamily) and a putative carboxylate transporter (DASS family). The genomic repertoire for the catabolic degradation and assimilation of peptides and amino acids, including transporter proteins for di- and oligopeptides (ABC and POT/PTR families), multiple amino acid:cation symporters (SSS, DAACS and AGCS families), amino acid/polyamine transporters (APC superfamily) and multiple putatively secreted peptidases are present (Data Set 1).
In agreement with Watson et al. [5], NO2− oxidation activity was enhanced when undefined organic compound mixtures such as tryptone and yeast extract were added to the culture medium (Fig. 2). Interestingly, growth was greatly stimulated by tryptone (Fig. 2), while amendment with defined organic carbon compounds had no effect on NO2− oxidation or growth (Table S3). Parallel incubations with ammonium as an N source did not increase activity or growth (Table S3), suggesting that the stimulating effect of tryptone, and to a lesser extent of yeast extract, most likely can be attributed to direct amino acid assimilation and does not only reflect the eliminated energy demand for assimilatory NO2− reduction. No growth on yeast extract and tryptone was observed in the absence of NO2− (Fig. 3), corroborating their use as source of amino acids rather than for energy conservation.
Fig. 2. The effect of undefined organic carbon substrates on nitrite oxidation and growth of N. marina Nb-295T.
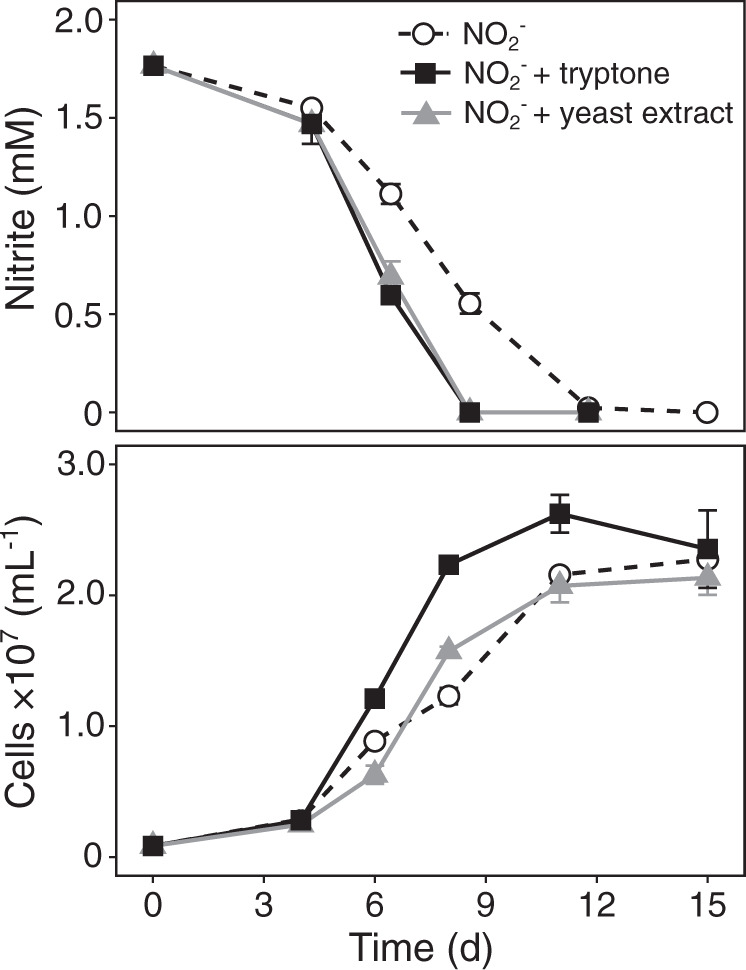
Error bars represent the range of measurements from duplicate cultures.
Fig. 3. Growth of N. marina Nb-295T on formate (1 mM), pyruvate (1 mM) and yeast extract and tryptone (150 mg L−1 each) in the absence of nitrite.
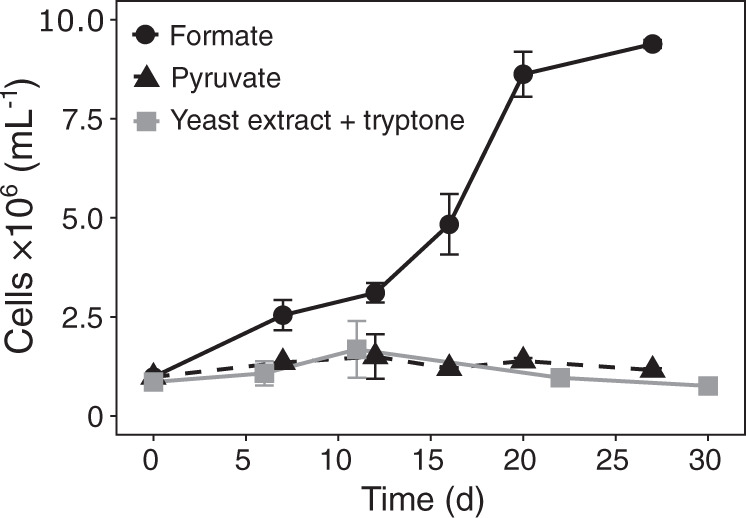
Error bars represent the range of measurements from duplicate cultures.
In addition to undefined organic carbon substrates, defined organic compounds such as glycerol and pyruvate have been reported to enhance the growth of N. marina and N. defluvii, respectively [5, 12]. Formate has also been shown to serve as an electron donor and carbon source for some lineage I and II Nitrospira [11, 28]. N. marina encodes a putative formate dehydrogenase (FdhA) (Data Set 1), which is divergent from those found in N. moscoviensis and N. defluvii (∼24 and 27% amino acid identity, respectively), but shares a relatively high sequence similarity (∼48% amino acid identity) to the functionally characterized formate dehydrogenase Fdh4 from Methylobacterium extorquens [66]. N. marina was able to grow chemoorganotrophically on 1 mM formate in the absence of NO2− (Fig. 3) and is thus not an obligate chemolithotrophic organism. However, the use of formate instead of NO2− as electron donor resulted in slower growth rates, as previously also observed in N. moscoviensis [28]. In contrast to earlier reports [5], additions of 1 g L−1 glycerol did not stimulate NO2− oxidation activity or growth (Table S3). Furthermore, pyruvate could neither be used as alternative energy source (Fig. 3), nor did it stimulate metabolic activity in the presence of NO2− (Table S3).
Protection against oxidative, osmotic, and UV light-induced stress
The formation of reactive oxygen species is prevalent in oxic environments and oxidative stress defense is an important component of the stress response in marine organisms [67]. N. marina encodes multiple enzymes to reduce oxidative stress, including a cytoplasmic Mn/Fe-binding SOD, a periplasmic Cu/Zn-binding SOD, two heme-containing catalases, and various peroxiredoxins (Data Set 1). In contrast, Nitrospina gracilis and marine ammonia-oxidizing archaea lack catalase [48, 58], suggesting that N. marina is less susceptible to oxidative stress compared to other marine nitrifiers. In addition to its plethora of oxidative stress defense-related proteins, N. marina also encodes two putative photolyases—enzymes known to be involved in the repair of UV light-induced DNA damage [68]—suggesting that it is well adapted to conditions characteristic for euphotic environments.
Marine microorganisms counteract the external osmotic stress from high salt concentrations by accumulating a variety of organic solutes (=osmolytes) in the cytoplasm, which can either be synthesized de-novo or transported into the cell from the surrounding environment [69]. In addition to select amino acids that can serve as compatible solutes (e.g., proline and glutamate) [70], biosynthesis pathways for the osmolytes glycine betaine and trehalose were identified in the N. marina genome (see Supplementary Results and Discussion). The production and concomitant release of osmolytes (i.e., via diffusion, excretion, predation or cell lysis) could potentially fuel heterotrophic metabolism in the ocean [71] representing a link between chemolithoautotrophic production and heterotrophic consumption of DOM as recently suggested for ammonia-oxidizing archaea [72].
Vitamin B12 auxotrophy
B vitamins are important biochemical co-factors required for cellular metabolism and their concentrations are depleted to near zero across large areas of the global ocean [73]. N. marina encodes complete biosynthetic pathways for the B vitamins thiamin (B1), riboflavin (B2), pantothenate (B5), pyridoxine (B6), biotin (B7) and tetrahydrofolate (B9). However, an incomplete vitamin B12 biosynthesis pathway was identified (Fig. S2), lacking genes for multiple precorrin conversion reactions that ultimately lead to the biosynthesis of the molecule’s corrin ring [74]. Since N. marina only encodes the cobalamin-dependent versions of ribonucleotide reductase, methionine synthase and methylmalonyl-CoA mutase, it must rely on the supply of vitamin B12 or its precursors by other members of the microbial community. Indeed, nitrite consumption by N. marina ceased after repeated transfers into an artificial seawater mineral medium without the addition of vitamin B12 (Fig. 4), and nitrite oxidation activity was restored after adding vitamin B12 to B12-deplete cultures (data not shown). Interestingly, the N. marina genome contains genes for multiple reactions that convert the precursor cobyrinate/hydrogenobyrinate to cobalamin, and encodes all genes required for cobalamin salvage from cobinamide (Fig. S2, Data Set 1). Hence, in contrast to many other bacteria that lack the complete vitamin B12 biosynthesis pathway [75], N. marina appears to obtain its B12 from salvage of multiple intermediates from the environment. Vitamin B12 auxotrophy has recently also been observed for several marine Nitrospinae strains [76]. In contrast, Park et al. [77] suggested that the two novel marine NOB strains MSP and DJ, one of which is closely related to N. marina, might be able to synthesize cobalamin. However, given that these cultures were not axenic and that genes involved in precorrin conversion reactions absent in N. marina (Fig. S2) are also missing from their genomes, it is likely that both strains depend on the exogenous supply of vitamin B12 as well.
Fig. 4. Nitrite consumption by N. marina Nb-295T in artificial seawater mineral medium with and without addition of vitamin B12 (Table S1).
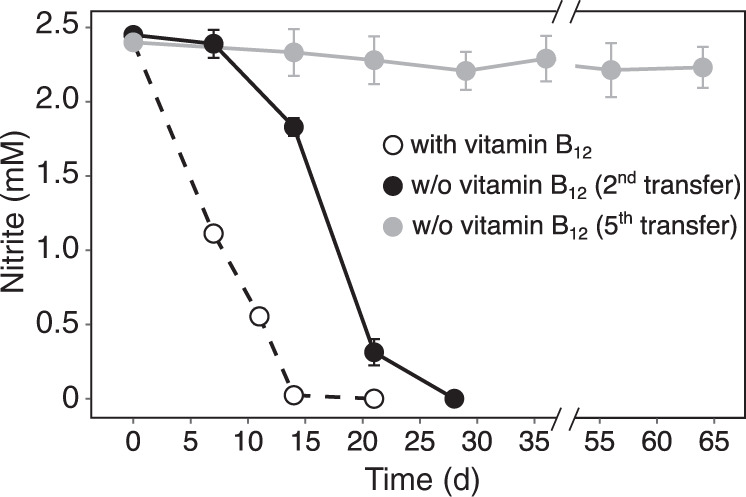
Cultures of strain Nb-295T were repeatedly transferred into medium without vitamin B12 using 2% inocula. Error bars represent the range of measurements from duplicate cultures.
When N. marina was grown in natural seawater, cellular concentrations of ribonucleotide reductase were approximately 39-fold higher than for the cyanobacterium Prochlorococcus [78] (Table 1) despite only having a ~two-fold greater cell volume, consistent with the importance of B12 nutrition to N. marina. Moreover, this salvage acquisition mode is interesting in the context of dissolved cobalt speciation, which is complexed by strong organic ligands hypothesized to include B12 precursors or degradation products [79]. In the Northwest Atlantic near where N. marina was isolated, organic cobalt complexes are abundant, comprising about half the dissolved cobalt inventory [80].
Table 1.
Targeted quantitative analyses of selected proteins using isotopically-labeled (15N) peptide standards.
| Target proteina | 15N-Peptide sequence | Peptide concentration (copies cell −1) | |
|---|---|---|---|
| atm. O2 | low O2 | ||
| Nitrite oxidoreductase, alpha subunit 1 | LVVITPEYNPTAYR | 845 (52) | 1924 (275) |
| DYAFPDFANSYSGK | 241 (40) | 451 (91) | |
| HPFWEETNESKPQWTR | 673 (39) | 1571 (388) | |
| Nitrite oxidoreductase, alpha subunit 2 | VVVITPEYNPTAQR | 1,154 (27) | 851 (187) |
| Nitrite oxidoreductase, alpha subunit 3 | DYQFPDFTSTYSGK | 1,761 (159) | 1,261 (242) |
| SGIDPALTGTHR | 4,519 (516) | 3,400 (526) | |
| IAVITPEYNPTAYR | 4,860 (496) | 3,741 (710) | |
| Nitrite oxidoreductase, alpha subunit (all) | GWKPSDPYYK | 11,467 (1,957) | 9,564 (1,083) |
| AIALDTGYQSNFR | 13,996 (962) | 13,116 (2,934) | |
| Pyruvate:ferredoxin oxidoreductase, | TPSFFTGSEVIK | 2,026 (192) | 1,550 (248) |
| alpha subunit | EAIAILEEEGIR | 1,362 (80) | 1,054 (170) |
| EVSATVPNNER | 2,453 (169) | 1,736 (215) | |
| Ribonucleotide reductase | TGESPYQTIPFSHR | 300 (46) | 198 (30) |
| EAAVPEPYIHR | 705 (132) | 472 (77) | |
| IINQSLPPALR | 750 (88) | 546 (68) | |
Nitrite oxidoreductase peptide targets were designed to match a specific NxrA copy (1, 2, or 3), or to match all three as indicated in the target protein column. Values represent the mean of triplicate measurements and standard deviations are shown in brackets.
aA complete list of quantified proteins can be found in Data Set 2.
Metabolic response to low oxygen concentrations
Given the presence of multiple signatures of microaerophilic adaptation and metabolic diversity of nitrite oxidizers [26, 28, 58, 62] and their occurrence in low oxygen environments [25, 81, 82], we sought to further investigate potential adaptations of Nitrospira marina Nb-295T to low O2 conditions. N. marina was grown at O2 concentrations characteristic for the upper ocean (∼200 µM) and at O2-limiting conditions (∼5.6 µM O2) found in environments with elevated NO2− concentrations such as OMZ or sediments [83, 84].
When grown under atmospheric O2 concentration, N. marina oxidized 1.5 mM NO2− within 12 days, whereas under O2-limiting conditions depletion of 1.5 mM substrate took 27 days (Fig. 5). This result is in agreement with the initial description by Watson et al. [5], who reported partial, though unquantified, inhibition at low O2 partial pressure. Cell abundances at the final timepoint (after 1.5 mM NO2− was oxidized) were comparable for both treatments (Fig. S3), indicating that the reduced NO2− oxidation rate during O2-limiting conditions ultimately resulted in similar cell yields.
Fig. 5. Nitrite consumption by N. marina Nb-295T grown under atmospheric (filled circles) and low O2 conditions (open circles).
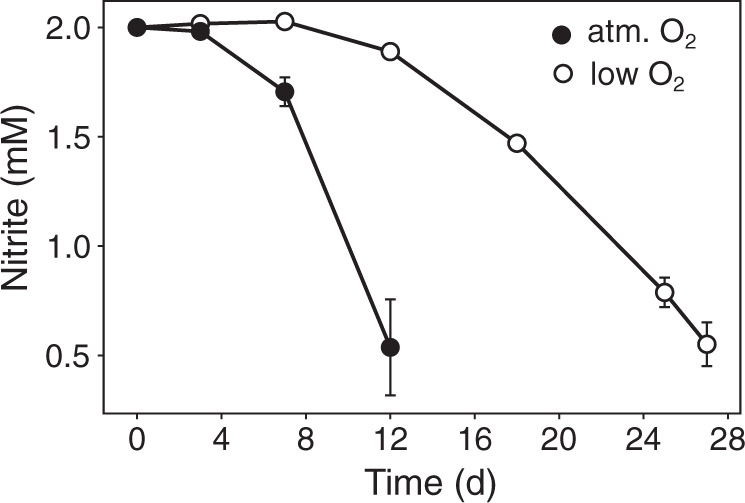
Cells for proteome analysis were harvested after 12 and 27 days, respectively. Error bars represent standard deviations from measurements of triplicate cultures.
General proteomic response and upregulation of gene clusters
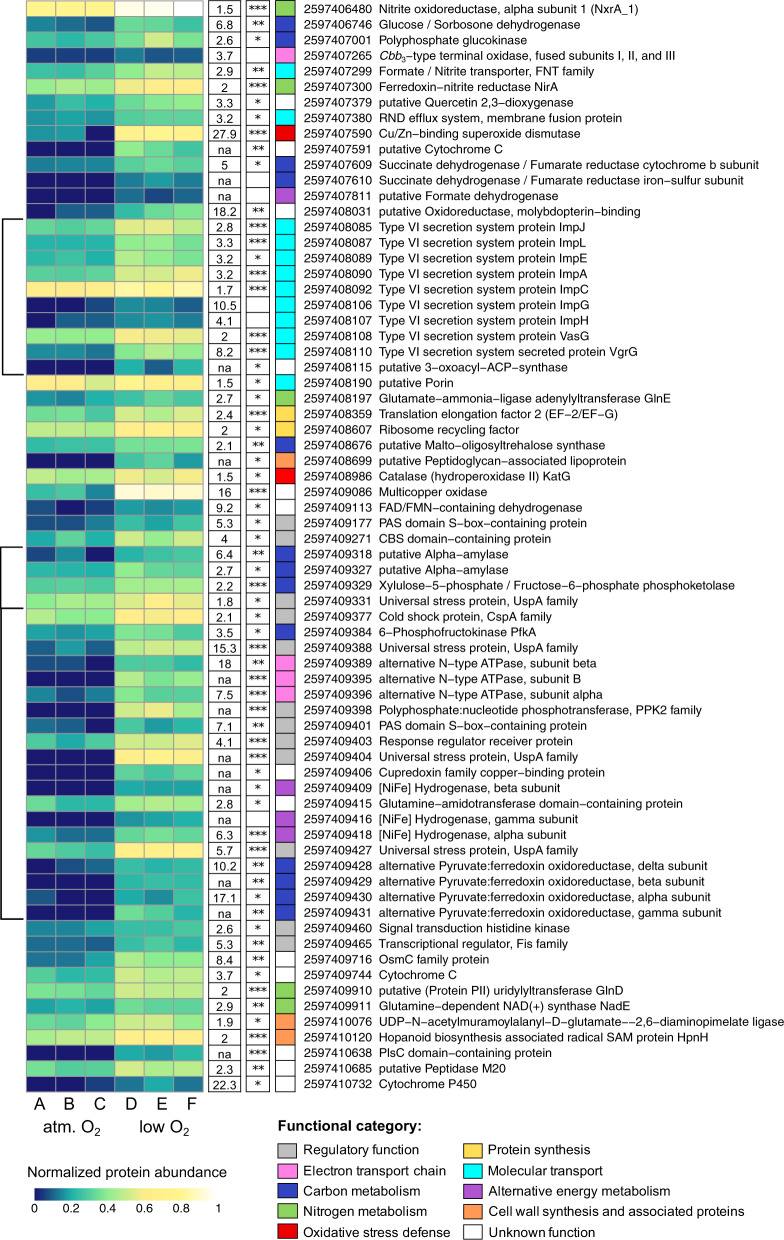
Cultures grown under both O2 treatments were harvested for proteomic analysis when [NO2−] dropped to ~500 µM, corresponding to exponential growth of strain Nb-295 (see “Material and Methods”). A total of 2031 and 2046 proteins were identified by liquid chromatography-tandem mass spectrometry in the atmospheric and low O2 treatments, respectively, accounting for 48.1 and 48.5% (49.7% combined from a total of 175,653 peptides) of the predicted protein coding sequences in the N. marina genome. As previously reported for N. marina and Nitrococcus mobilis, proteins exhibiting the highest abundances were associated with NO2− oxidation [44] (Data Set 1). NXR made up on average 4% of all peptide spectral counts and cellular NXR concentrations were ~13,500 copies cell−1 as determined by targeted quantitative proteomic analyses (Table 1), covering an estimated 35% of the membrane surface (see “Material and Methods”). All three NxrA copies were detected in the proteome as determined by detection of unique peptides of each, and NxrA_3 appeared to be more abundant compared to NxrA_1 and NxrA_2 (Table 1). Under low O2 conditions, NxrA_1 increased in abundance (Table 1, Fig. 6) indicating different metabolic or regulatory roles of the highly similar subunits.
Fig. 6. Heat map of N. marina Nb-295T proteins that were more abundant under low O2 concentrations compared to atmospheric O2 concentration.
Relative protein abundance values were square-root transformed and hypothetical proteins were excluded to improve readability. The complete set of untransformed values can be found in Data Set 3. Fold-changes and significance values (adj. P value ≤ 0.001, ***; ≤0.01, **; ≤0.05, *) are shown in white boxes next to the corresponding protein. Select low abundant proteins of interest with high fold-changes were included despite being not statistically significant (see “Material and Methods”). Fold-changes of proteins that were not detected under atmospheric O2 conditions are omitted to avoid dividing by zero (not available, na). Functional categories of depicted proteins are indicated by different colors. Gene clusters are indicated by black brackets.
Proteins involved in CO2 fixation, DNA replication, electron transport and central carbon metabolism were also highly abundant under both conditions, indicating that N. marina retained its central metabolism during O2 deficiency (Data Set 1). Although proteomic spectral counts remained constant for the majority of proteins during both treatments, spectral counts of 93 proteins significantly increased (adjusted P value ≤ 0.05) during growth at low O2 concentrations (Fig. 6, Data Set 3). These results are supported by the targeted quantitative proteomic analyses (Fig. S4, Data Set 2), suggesting that spectral counts are a good proxy for changes in absolute protein abundance in our dataset.
Multiple universal stress proteins (UspA superfamily) were among the proteins that showed the highest increase in abundance under low O2 conditions compared to the control treatment (Fig. 6, Data Set 3). UspA proteins have versatile regulatory and protective functions to enable survival under diverse external stresses [85], and are induced during growth inhibition [86] and oxygen starvation [87]. In N. marina, all four upregulated UspA proteins are located upstream or downstream of operons containing genes that also increased in abundance under low O2 conditions (Fig. 6), suggesting a regulatory role of UspA-related proteins upon O2 limitation. These upregulated gene clusters include proteins involved in electron transport, carbon metabolism, and alternative energy metabolism (Fig. 6).
In addition to putative UspA-regulated gene clusters, a gene cluster containing type VI secretion system (T6SS)-related proteins exhibited higher abundances under low O2 conditions (Fig. 6). The T6SS is typically involved in the secretion of effectors required for pathogenesis, bacterial competition, biofilm formation, and cell communication (e.g., quorum sensing) [88, 89]. While quorum sensing has recently been shown for diverse NOB including N. moscoviensis [90], no LuxI autoinducer synthases and/or LuxR signal receptor homologs were identified in the N. marina genome.
Induction of a putative O2-sensitive 2-oxoacid:ferredoxin oxidoreductase complex
The key enzymes of the rTCA cycle, pyruvate:ferredoxin oxidoreductase (POR) and 2-oxoglutarate:ferredoxin oxidoreductase (OGOR), are typically highly O2 sensitive because they contain easily oxidized iron-sulfur clusters [91]. In Hydrogenobacter thermophilus, five-subunit O2-tolerant forms of POR and OGOR mainly support aerobic growth, while a O2-sensitive two-subunit form is used under anaerobic conditions [92]. N. marina encodes three 2-oxoacid:ferredoxin oxidoreductase gene clusters that could exhibit POR or OGOR activity (Data Set 1). Two of these gene clusters consist of five CDS that exhibit a high sequence similarity to the O2-tolerant five-subunit POR/OGOR of H. thermophilus [93, 94], as previously described for N. defluvii [26]. Both complexes were highly abundant in N. marina proteomes from atmospheric and low O2 treatments (Data Set 1, Table 1) confirming their important role in central carbon metabolism. The third cluster contains alpha, beta, and gamma subunits of a putative POR with homology to the functionally characterized four-subunit PORs of the anaerobic thermophiles Pyrococcus and Thermotoga [95] and is absent in all other Nitrospira with the exception of Ca. N. alkalitolerans [9]. A protein with a 4Fe-4S binding domain was identified in the same operon, potentially representing the missing delta subunit of the POR complex (Data Set 1). This putative four-subunit POR was among the proteins that showed the highest increase in abundance under low O2 conditions in N. marina (Fig. 6). The O2-tolerant POR/OGOR isoforms were reported to have a >5-times lower specific activity [92] and might therefore constitute a substantial part of the cellular soluble protein content in H. thermophilus [96]. Hence, it is tempting to speculate that N. marina increases the expression of a more efficient (i.e., higher specific activity), O2-sensitive four-subunit POR under O2 limitation. While oxidative stress typically decreases under low O2 conditions, it might still be high enough to damage O2-sensitive enzymes. In N. marina, the abundance of a periplasmic Cu/Zn-binding SOD and a cytoplasmic catalase (KatG) increased under O2-limited conditions (Fig. 6), while the abundances of other oxidative stress defense-related proteins remained constant (Data Set 1 and 3). SOD has been shown to be efficient in protecting POR activity from oxidative damage in Entamoeba histolytica [97], and was among the proteins with the highest increase in abundance under low O2 conditions in N. marina (fold-change: 27.9), suggesting a role in POR protection.
Expression of a high O2-affinity cbb3-type terminal oxidase
The majority of proteins related to electron transport showed similar abundance levels at atmospheric and limiting O2 concentrations (Data Set 1 and 3). Despite its overall low abundance, a putative high-affinity cytochrome cbb3-type terminal oxidase was 3.6-times more abundant under low O2 concentrations compared to atmospheric O2 tension (Fig. 6). The cbb3-type terminal oxidase of Bradyrhizobium japonicum was reported to have a Km value of 7 nmol L−1 O2 [98] and NO2− oxidation rates have been detected at O2 concentrations in the low nanomolar range (5–33 nmol L−1 O2) [25]. This suggests that the cbb3-type terminal oxidase might enable N. marina to continue aerobic respiration at low O2 concentrations, albeit at lower NO2− oxidation rates (Fig. 5). Low-affinity terminal oxidases are typically more efficient in energy conservation [99], suggesting that N. marina benefits from the presence of a terminal oxidase with lower O2 affinity in well-oxygenated environments. While N. gracilis encodes a highly similar high affinity cbb3-type terminal oxidase to N. marina [58], this enzyme is lacking in other Nitrospinae, including those identified in OMZs and sediments [76, 82, 100]. Interestingly, these genomes also lack the putative terminal oxidase proposed for Nitrospira [26]. Still, in an OMZ where Nitrospinae bacteria were the only detected NOB, NO2− oxidation rates already approached saturation at ∼1 μmol L−1 O2 [25], indicating that Nitrospinae bacteria might be better adapted to low O2 concentrations compared to N. marina, but the enzyme conferring high O2 affinity in Nitrospinae remains to be identified.
Increase in abundance of proteins involved in alternative energy metabolism
The abundance of proteins putatively involved in alternative energy metabolisms increased under low O2 concentrations. These included Group 3b [NiFe] hydrogenase and formate dehydrogenase (Fig. 6). Alternative electron donors such as H2 and formate are common reaction products at oxic/anoxic interfaces [101]. N. moscoviensis can couple H2 and formate oxidation to NO3− reduction to remain active under anoxia [28, 30] (whereas no net growth was observed for the former [30]), however, it is unlikely that H2 or formate were present at the culture conditions in this study. While abundances of hydrogenase and formate dehydrogenase increased under low O2 conditions, they were overall still comparably low (Fig. 6), suggesting that their expression might be upregulated when conditions become more unfavorable for aerobic NO2– oxidation. Curiously, multiple subunits of the Na+-translocating N-type ATPase exhibited higher abundances at low O2 concentration (Fig. 6). Na+-translocating ATPases are suggested to be ancient enzymes that were later replaced by energetically more favorable H+-translocating ATPases [102]. While only few obligate anaerobes with very tight energy budgets (which cannot cover the losses caused by proton leaks) primarily use Na+ energetics, many organisms retained Na+ pumps and utilize them under energetically less favorable conditions such as anaerobiosis [102]. Hence, expression of a Na+-translocating ATPase suggests an adaptation of N. marina to overcome periods of starvation when energetically favorable electron donors or acceptors are short in supply.
Conclusions
Although the vast majority of the ocean is well oxygenated, oxygen-depleted zones exist within the oceanic water column and in marine sediments [83, 84] with consequences for microbial adaptation and evolution. Our results show that, in contrast to Nitrospinae-dominated NOB populations in low oxygen waters [25], NO2− oxidation activity of Nitrospira marina was reduced when grown at ∼5.6 µM O2, suggesting different O2 adaptations among different marine NOB. We confirm that N. marina benefits from the addition of undefined organic carbon substrates, which were shown to be inhibitory for Nitrospina gracilis [103], potentially further contributing to ecological niche partitioning within marine NOB. Our results indicate that N. marina is highly metabolically versatile, which might enable it to survive under unfavorable conditions with fluctuating levels of electron donors and acceptors. Hence, while Nitrospinae bacteria are the dominant nitrite oxidizers in oligotrophic oceanic regions and OMZs, Nitrospira might be better adapted to well-oxygenated high productivity regions including coastal systems, deep-sea trenches and hydrothermal vents. Finally, our results also indicate several ways that NOB may interact with other members of the marine microbial community—through the supply of organic carbon-containing osmolytes and their requirement for exogenous vitamin B12.
Supplementary information
Acknowledgements
We thank John B. Waterbury and Frederica Valois for providing the culture of Nitrospira marina Nb-295T and for continued advice about cultivation. The N. marina genome was sequenced as part of US Department of Energy Joint Genome Institute Community Sequencing Project 1337 to CLD, AES, and MAS in collaboration with the user community. We thank Claus Pelikan for bioinformatic assistance. This research was supported by a Simons Foundation Early Career Investigator in Marine Microbiology and Evolution Award (345889) and US National Science Foundation (NSF) award OCE-1924512 to AES. Proteomics analysis was supported by NSF awards OCE-1924554 and OCE-1850719, and NIH award R01GM135709 to MAS. BB was supported by the Austrian Science Fund (FWF) Project Number: J4426-B (“The influence of nitrifiers on the oceanic carbon cycle”), SL by the Netherlands Organization for Scientific Research (NWO) grant 016.Vidi.189.050, and CLD by NSF award OCE-125999.
Data availability
The genome of Nitrospira marina Nb-295T is available in the JGI IMG/M repository under genome ID number 2596583682. Manually curated protein annotations are available in Data Set 1, targeted quantitative proteomic analyses results are available in Data Set 2, global proteomic spectral counts and differential expression analysis results are available in Data Set 3. Raw mass spectra are available in PRIDE as project number PXD021606. Data is archived at Biological and Chemical Oceanography Data Management office (BCO-DMO) under project 806565.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Barbara Bayer, Email: bbayer@ucsb.edu.
Alyson E. Santoro, Email: asantoro@ucsb.edu
Supplementary information
The online version of this article (10.1038/s41396-020-00828-3) contains supplementary material, which is available to authorized users.
References
- 1.Kuypers MMM, Marchant HK, Kartal B. The microbial nitrogen-cycling network. Nat Rev Microbiol. 2018;16:263–76. doi: 10.1038/nrmicro.2018.9. [DOI] [PubMed] [Google Scholar]
- 2.Gruber N. The marine nitrogen cycle: Overview and challenges. In: G Capone, DA Bronk, MR Mulholland, EJ Carpenter (eds). Nitrogen in the marine environment, 2nd ed. Academic Press: New York, NY, 2008, pp. 1–50.
- 3.Daims H, Lücker S, Wagner M. A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol. 2016;24:699–712. doi: 10.1016/j.tim.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pachiadaki MG, Sintes E, Bergauer K, Brown JM, Record NR, Swan BK, et al. Major role of nitrite-oxidizing bacteria in dark ocean carbon fixation. Science. 2017;358:1046–51. doi: 10.1126/science.aan8260. [DOI] [PubMed] [Google Scholar]
- 5.Watson SW, Bock E, Valois FW, Waterbury JB, Schlosser U. Nitrospira marina gen. nov. sp. nov.: a chemolithotrophic nitrite-oxidizing bacterium. Arch Microbiol. 1986;144:1–7. doi: 10.1007/BF00454947. [DOI] [Google Scholar]
- 6.Altmann D, Stief P, Amann R, De Beer D, Schramm A. In situ distribution and activity of nitrifying bacteria in freshwater sediment. Environ Microbiol. 2003;5:798–803. doi: 10.1046/j.1469-2920.2003.00469.x. [DOI] [PubMed] [Google Scholar]
- 7.Freitag TE, Chang L, Clegg CD, Prosser JI. Influence of inorganic nitrogen management regime on the diversity of nitrite-oxidizing bacteria in agricultural grassland soils. Appl Environ Microbiol. 2005;71:8323–34. doi: 10.1128/AEM.71.12.8323-8334.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pester M, Maixner F, Berry D, Rattei T, Koch H, Lücker S, et al. NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environ Microbiol. 2014;16:3055–71. doi: 10.1111/1462-2920.12300. [DOI] [PubMed] [Google Scholar]
- 9.Daebeler A, Kitzinger K, Koch H, Herbold CW, Steinfeder M, Schwarz J, et al. Exploring the upper pH limits of nitrite oxidation: diversity, ecophysiology, and adaptive traits of haloalkalitolerant Nitrospira. ISME J. 2020. 10.1038/s41396-020-0724-1. [DOI] [PMC free article] [PubMed]
- 10.Lebedeva EV, Alawi M, Fiencke C, Namsaraev B, Bock E, Spieck E. Moderately thermophilic nitrifying bacteria from a hot spring of the Baikal rift zone. FEMS Microbiol Ecol. 2005;54:297–306. doi: 10.1016/j.femsec.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 11.Gruber-Dorninger C, Pester M, Kitzinger K, Savio DF, Loy A, Rattei T, et al. Functionally relevant diversity of closely related Nitrospira in activated sludge. ISME J. 2015;9:643–55. doi: 10.1038/ismej.2014.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spieck E, Hartwig C, McCormack I, Maixner F, Wagner M, Lipski A, et al. Selective enrichment and molecular characterization of a previously uncultured Nitrospira-like bacterium from activated sludge. Environ Microbiol. 2006;8:405–15. doi: 10.1111/j.1462-2920.2005.00905.x. [DOI] [PubMed] [Google Scholar]
- 13.Ushiki N, Fujitani H, Aoi Y, Tsuneda S. Isolation of Nitrospira belonging to sublineage II from a wastewater treatment plant. Microbes Environ. 2013;28:346–53. doi: 10.1264/jsme2.ME13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Kessel MAHJ, Speth DR, Albertsen M, Nielsen PH, Op Den Camp HJM, Kartal B, et al. Complete nitrification by a single microorganism. Nature. 2015;528:555–9. doi: 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keuter S, Kruse M, Lipski A, Spieck E. Relevance of Nitrospira for nitrite oxidation in a marine recirculation aquaculture system and physiological features of a Nitrospira marina-like isolate. Environ Microbiol. 2011;13:2536–47. doi: 10.1111/j.1462-2920.2011.02525.x. [DOI] [PubMed] [Google Scholar]
- 16.Nowka B, Off S, Daims H, Spieck E. Improved isolation strategies allowed the phenotypic differentiation of two Nitrospira strains from widespread phylogenetic lineages. FEMS Microbiol Ecol. 2015;91:fiu031. doi: 10.1093/femsec/fiu031. [DOI] [PubMed] [Google Scholar]
- 17.Hongxiang X, Min W, Xiaogu W, Junyi Y, Chunsheng W. Bacterial diversity in deep-sea sediment from northeastern Pacific Ocean. Acta Ecol Sin. 2008;28:479–85. doi: 10.1016/S1872-2032(08)60026-8. [DOI] [Google Scholar]
- 18.Baker BJ, Sheik CS, Taylor CA, Jain S, Bhasi A, Cavalcoli JD, et al. Community transcriptomic assembly reveals microbes that contribute to deep-sea carbon and nitrogen cycling. ISME J. 2013;7:1962–73. doi: 10.1038/ismej.2013.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nunoura T, Takaki Y, Hirai M, Shimamura S, Makabe A, Koide O, et al. Hadal biosphere: Insight into the microbial ecosystem in the deepest ocean on Earth. Proc Natl Acad Sci USA. 2015;112:E1230–6. doi: 10.1073/pnas.1421816112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiraoka S, Hirai M, Matsui Y, Makabe A, Minegishi H, Tsuda M, et al. Microbial community and geochemical analyses of trans-trench sediments for understanding the roles of hadal environments. ISME J. 2020;14:740–56. doi: 10.1038/s41396-019-0564-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lam P, Kuypers MMM. Microbial nitrogen cycling processes in oxygen minimum zones. Ann Rev Mar Sci. 2011;3:317–45. doi: 10.1146/annurev-marine-120709-142814. [DOI] [PubMed] [Google Scholar]
- 22.Codispoti LA, Friederich GE, Packard TT, Glover HE, Kelly PJ, Spinrad RW, et al. High nitrite levels off northern Peru: a signal of instability in the marine denitrification rate. Science. 1986;233:1200–2. doi: 10.1126/science.233.4769.1200. [DOI] [PubMed] [Google Scholar]
- 23.Buchwald C, Santoro AE, Stanley RHR, Casciotti KL. Nitrogen cycling in the secondary nitrite maximum of the eastern tropical North Pacific off Costa Rica. Glob Biogeochem Cycles. 2015;29:2061–81. doi: 10.1002/2015GB005187. [DOI] [Google Scholar]
- 24.Lam P, Jensen MM, Kock A, Lettmann KA, Plancherel Y, Lavik G, et al. Origin and fate of the secondary nitrite maximum in the Arabian Sea. Biogeosciences. 2011;8:1565–77. doi: 10.5194/bg-8-1565-2011. [DOI] [Google Scholar]
- 25.Bristow LA, Dalsgaard T, Tiano L, Mills DB, Bertagnolli AD, Wright JJ, et al. Ammonium and nitrite oxidation at nanomolar oxygen concentrations in oxygen minimum zone waters. Proc Natl Acad Sci USA. 2016;113:10601–6. doi: 10.1073/pnas.1600359113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lücker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B, et al. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci USA. 2010;107:13479–84. doi: 10.1073/pnas.1003860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spieck E, Ehrich S, Aamand J, Bock E. Isolation and immunocytochemical location of the nitrite-oxidizing system in Nitrospira moscoviensis. Arch Microbiol. 1998;169:225–30. doi: 10.1007/s002030050565. [DOI] [PubMed] [Google Scholar]
- 28.Koch H, Lücker S, Albertsen M, Kitzinger K, Herbold C, Spieck E, et al. Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc Natl Acad Sci USA. 2015;112:11371–6. doi: 10.1073/pnas.1506533112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ushiki N, Fujitani H, Shimada Y, Morohoshi T, Sekiguchi Y, Tsuneda S. Genomic analysis of two phylogenetically distinct Nitrospira species reveals their genomic plasticity and functional diversity. Front Microbiol. 2018;8:2637. doi: 10.3389/fmicb.2017.02637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koch H, Galushko A, Albertsen M, Schintlmeister A, Gruber-Dorninger C, Lücker S, et al. Growth of nitrite-oxidizing bacteria by aerobic hydrogen oxidation. Science. 2014;345:1052–4. doi: 10.1126/science.1256985. [DOI] [PubMed] [Google Scholar]
- 31.Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, et al. Complete nitrification by Nitrospira bacteria. Nature. 2015;528:504–9. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strickland JDH, Parsons TR A practical handbook of seawater analysis, 2nd ed. 1972. Fish. Res. Bd. Can., Bull. No. 167
- 33.Porter KG, Feig YS. The use of DAPI for identifying aquatic microflora. Limnol Oceanogr. 1980;25:943–8. doi: 10.4319/lo.1980.25.5.0943. [DOI] [Google Scholar]
- 34.Wilson K. Preparation of genomic DNA from bacteria. Curr Protoc Mol Biol. 2001;56:2.4.1–5. doi: 10.1002/0471142727.mb0204s56. [DOI] [PubMed] [Google Scholar]
- 35.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–9. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 36.Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Grechkin Y, et al. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res. 2012;40:D115–22. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vallenet D, Calteau A, Dubois M, Amours P, Bazin A, Beuvin M, et al. MicroScope: an integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucleic Acids Res. 2020;48:D579–89. doi: 10.1093/nar/gkz926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 39.Saier MH. TCDB: the transporter classification database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–6. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, et al. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–20. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 2019. 10.1038/s41587-019-0036-z. [DOI] [PubMed]
- 42.Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2019;36:1925–7. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malmstrom R RNAlater Recipe. protocols.io 2015. 10.17504/protocols.io.c56y9d.
- 44.Saito MA, Mcilvin MR, Moran DM, Santoro AE, Dupont CL, Rafter PA, et al. Abundant nitrite-oxidizing metalloenzymes in the mesopelagic zone of the tropical Pacific Ocean. Nat Geosci. 2020;13:355–62. doi: 10.1038/s41561-020-0565-6. [DOI] [Google Scholar]
- 45.Simon M, Azam F. Protein content and protein synthesis rates of planktonic marine bacteria. Mar Ecol Prog Ser. 1989;51:201–13. doi: 10.3354/meps051201. [DOI] [Google Scholar]
- 46.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2013. http://www.R-project.org.
- 48.Bayer B, Pelikan C, Bittner MJ, Reinthaler T, Könneke M, Herndl GJ, et al. Proteomic response of three marine ammonia-oxidizing archaea to hydrogen peroxide and their metabolic interactions with a heterotrophic alphaproteobacterium. mSystems. 2019;4:e00181–19. doi: 10.1128/mSystems.00181-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langley SR, Mayr M. Comparative analysis of statistical methods used for detecting differential expression in label-free mass spectrometry proteomics. J Proteom. 2015;129:83–92. doi: 10.1016/j.jprot.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 50.Raithel S, Johnson L, Galliart M, Brown S, Shelton J, Herndon N, et al. Inferential considerations for low-count RNA-seq transcripts: a case study on the dominant prairie grass Andropogon gerardii. BMC Genomics. 2016;17:140. doi: 10.1186/s12864-016-2442-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kolde R pheatmap: Pretty Heatmaps. R package version 1.0.8. 2015. http://CRAN.R-project.org/package_pheatmap.
- 52.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res. 2006;5:2339–47. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 53.Ye L, Shao MF, Zhang T, Tong AHY, Lok S. Analysis of the bacterial community in a laboratory-scale nitrification reactor and a wastewater treatment plant by 454-pyrosequencing. Water Res. 2011;45:4390–8. doi: 10.1016/j.watres.2011.05.028. [DOI] [PubMed] [Google Scholar]
- 54.Mundinger AB, Lawson CE, Jetten MSM, Koch H, Lücker S. Cultivation and transcriptional analysis of a canonical Nitrospira under stable growth conditions. Front Microbiol. 2019;10:1325. doi: 10.3389/fmicb.2019.01325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sakoula D, Nowka B, Spieck E, Daims H, Lücker S. The draft genome sequence of “Nitrospira lenta” strain BS10, a nitrite oxidizing bacterium isolated from activated sludge. Stand Genom Sci. 2018;13:32. doi: 10.1186/s40793-018-0338-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chadwick GL, Hemp J, Fischer WW, Orphan VJ. Convergent evolution of unusual complex I homologs with increased proton pumping capacity: energetic and ecological implications. ISME J. 2018;12:2668–80. doi: 10.1038/s41396-018-0210-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cosseau C, Batut J. Genomics of the ccoNOQP-encoded cbb3 oxidase complex in bacteria. Arch Microbiol. 2004;181:89–96. doi: 10.1007/s00203-003-0641-5. [DOI] [PubMed] [Google Scholar]
- 58.Lücker S, Nowka B, Rattei T, Spieck E, Daims H. The genome of Nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer. Front Microbiol. 2013;4:27. doi: 10.3389/fmicb.2013.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luoto HH, Belogurov GA, Baykov AA, Lahti R, Malinen AM. Na+-translocating membrane pyrophosphatases are widespread in the microbial world and evolutionarily precede H+-translocating pyrophosphatases. J Biol Chem. 2011;286:21633–42. doi: 10.1074/jbc.M111.244483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luoto HH, Baykov AA, Lahti R, Malinen AM. Membrane-integral pyrophosphatase subfamily capable of translocating both Na+ and H+ Proc Natl Acad Sci USA. 2013;110:1255–60. doi: 10.1073/pnas.1217816110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hausmann B, Pelikan C, Herbold CW, Köstlbacher S, Albertsen M, Eichorst SA, et al. Peatland Acidobacteria with a dissimilatory sulfur metabolism. ISME J. 2018;12:1729–42. doi: 10.1038/s41396-018-0077-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Füssel J, Lücker S, Yilmaz P, Nowka B, van Kessel MAHJ, Bourceau P, et al. Adaptability as the key to success for the ubiquitous marine nitrite oxidizer Nitrococcus. Sci Adv. 2017;3:2–10. doi: 10.1126/sciadv.1700807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma K, Weiss R, Adams MWW. Characterization of hydrogenase II from the hyperthermophilic archaeon Pyrococcus furiosus and assessment of its role in sulfur reduction. J Bacteriol. 2000;182:1864–71. doi: 10.1128/JB.182.7.1864-1871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Price GD, Woodger FJ, Badger MR, Howitt SM, Tucker L. Identification of a SulP-type bicarbonate transporter in marine cyanobacteria. Proc Natl Acad Sci USA. 2004;101:18228–33. doi: 10.1073/pnas.0405211101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fukaya F, Promden W, Hibino T, Tanaka Y, Nakamura T, Takabe T. An Mrp-like cluster in the halotolerant cyanobacterium Aphanothece halophytica functions as a Na+/H+ antiporter. Appl Environ Microbiol. 2009;75:6626–9. doi: 10.1128/AEM.01387-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chistoserdova L, Crowther GJ, Vorholt JA, Skovran E, Portais JC, Lidstrom ME. Identification of a fourth formate dehydrogenase in Methylobacterium extorquens AM1 and confirmation of the essential role of formate oxidation in methylotrophy. J Bacteriol. 2007;189:9076–81. doi: 10.1128/JB.01229-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lesser MP. Oxidative stress in marine environments: biochemistry and physiological ecology. Annu Rev Physiol. 2006;68:253–78. doi: 10.1146/annurev.physiol.68.040104.110001. [DOI] [PubMed] [Google Scholar]
- 68.Heelis PF, Kim ST, Okamura T, Sancar A. The photo-repair of pyrimidine dimers by DNA photolyase and model systems. J Photochem Photobio B. 1993;17:219–28. doi: 10.1016/1011-1344(93)80019-6. [DOI] [PubMed] [Google Scholar]
- 69.Roberts M. Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Syst. 2005;1:5. doi: 10.1186/1746-1448-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Csonka L. Prokaryotic osmoregulation: genetics and physiology. Annu Rev Microbiol. 1991;45:569–606. doi: 10.1146/annurev.mi.45.100191.003033. [DOI] [PubMed] [Google Scholar]
- 71.Boysen AK, Carlson LT, Durham BP, Groussman RD, Aylward FO, Heal KR, et al. Diel Oscillations of Particulate Metabolites Reflect Synchronized Microbial Activity in the North Pacific Subtropical Gyre. bioRxiv 2020. 10.1101/2020.05.09.086173.
- 72.Bayer B, Hansman RL, Bittner MJ, Noriega‐Ortega BE, Niggemann J, Dittmar T, et al. Ammonia‐oxidizing archaea release a suite of organic compounds potentially fueling prokaryotic heterotrophy in the ocean. Environ Microbiol. 2019;21:4062–75. doi: 10.1111/1462-2920.14755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sañudo-Wilhelmy SA, Gómez-Consarnau L, Suffridge C, Webb EA. The role of B vitamins in marine biogeochemistry. Ann Rev Mar Sci. 2014;6:339–67. doi: 10.1146/annurev-marine-120710-100912. [DOI] [PubMed] [Google Scholar]
- 74.Graham RM, Deery E, Warren MJ. Vitamin B12: biosynthesis of the corrin ring. Tetrapyrroles: birth, life and death. New York, NY: Springer; 2009. pp. 286–99. [Google Scholar]
- 75.Shelton AN, Seth EC, Mok KC, Han AW, Jackson SN, Haft DR, et al. Uneven distribution of cobamide biosynthesis and dependence in bacteria predicted by comparative genomics. ISME J. 2019;13:789–804. doi: 10.1038/s41396-018-0304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mueller AJ, Jung M-Y, Strachan CR, Herbold CW, Kirkegaard RH, Wagner M, et al. Genomic and kinetic analysis of novel Nitrospinae enriched by cell sorting. ISME J. 2020. 10.1038/s41396-020-00809-6. [DOI] [PMC free article] [PubMed]
- 77.Park S-J, Andrei A-Ş, Bulzu P-A, Kavagutti VS, Ghai R, Mosier AC Expanded diversity and metabolic versatility of marine nitrite-oxidizing bacteria revealed by cultivation- and genomics-based approaches. Appl Environ Microbiol 2020. 10.1128/AEM.016667-20. [DOI] [PMC free article] [PubMed]
- 78.Hawco NJ, McIlvin MM, Bundy RM, Tagliabue A, Goepfert TJ, Moran DM, et al. Minimal cobalt metabolism in the marine cyanobacterium Prochlorococcus. Proc Natl Acad Sci USA. 2020;12. 10.1073/pnas.2001393117. [DOI] [PMC free article] [PubMed]
- 79.Saito MA, Rocap G, Moffett JW. Production of cobalt binding ligands in a Synechococcus feature at the Costa Rica upwelling dome. Limnol Oceanogr. 2005;50:279–90. doi: 10.4319/lo.2005.50.1.0279. [DOI] [Google Scholar]
- 80.Noble AE, Ohnemus DC, Hawco NJ, Lam PJ, Saito MA. Coastal sources, sinks and strong organic complexation of dissolved cobalt within the US North Atlantic GEOTRACES transect GA03. Biogeosciences. 2017;14:2715–39. doi: 10.5194/bg-14-2715-2017. [DOI] [Google Scholar]
- 81.Füssel J, Lam P, Lavik G, Jensen MM, Holtappels M, Günter M, et al. Nitrite oxidation in the Namibian oxygen minimum zone. ISME J. 2012;6:1200–9. doi: 10.1038/ismej.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun X, Kop LFM, Lau MCY, Frank J, Jayakumar A, Lücker S, et al. Uncultured Nitrospina-like species are major nitrite oxidizing bacteria in oxygen minimum zones. ISME J. 2019;13:2391–402. doi: 10.1038/s41396-019-0443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.D’Hondt S, Inagaki F, Zarikian CA, Abrams LJ, Dubois N, Engelhardt T, et al. Presence of oxygen and aerobic communities from sea floor to basement in deep-sea sediments. Nat Geosci. 2015;8:299–304. doi: 10.1038/ngeo2387. [DOI] [Google Scholar]
- 84.Karstensen J, Stramma L, Visbeck M. Oxygen minimum zones in the eastern tropical Atlantic and Pacific oceans. Prog Oceanogr. 2008;77:331–50. doi: 10.1016/j.pocean.2007.05.009. [DOI] [Google Scholar]
- 85.Vollmer AC, Bark SJ Twenty-five years of investigating the universal stress protein: function, structure, and applications. advances in applied microbiology, 1st ed. 2018. Elsevier Inc., pp 1–36. [DOI] [PubMed]
- 86.Nyström T, Neidhardt FC. Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol Microbiol. 1994;11:537–44. doi: 10.1111/j.1365-2958.1994.tb00334.x. [DOI] [PubMed] [Google Scholar]
- 87.O’Toole R, Smeulders MJ, Blokpoel MC, Kay EJ, Lougheed K, Williams HD. A two-component regulator of universal stress protein expression and adaptation to oxygen starvation in Mycobacterium smegmatis. J Bacteriol. 2003;185:1543–54. doi: 10.1128/JB.185.5.1543-1554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Coulthurst S. The Type VI secretion system: a versatile bacterial weapon. Microbiology. 2019;165:503–15. doi: 10.1099/mic.0.000789. [DOI] [PubMed] [Google Scholar]
- 89.Gallique M, Bouteiller M, Merieau A. The type VI secretion system: a dynamic system for bacterial communication? Front Microbiol. 2017;8:1454. doi: 10.3389/fmicb.2017.01454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mellbye BL, Spieck E, Bottomley PJ, Sayavedra-Soto LA. Acyl-homoserine lactone production in nitrifying bacteria of the genera Nitrosospira, Nitrobacter, and Nitrospira identified via a survey of putative quorum-sensing genes. Appl Environ Microbiol. 2017;83:e01540–17. doi: 10.1128/AEM.01540-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Erb TJ. Carboxylases in natural and synthetic microbial pathways. Appl Environ Microbiol. 2011;77:8466–77. doi: 10.1128/AEM.05702-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yamamoto M, Arai H, Ishii M, Igarashi Y. Role of two 2-oxoglutarate:ferredoxin oxidoreductases in Hydrogenobacter thermophilus under aerobic and anaerobic conditions. FEMS Microbiol Lett. 2006;263:189–93. doi: 10.1111/j.1574-6968.2006.00415.x. [DOI] [PubMed] [Google Scholar]
- 93.Ikeda T, Ochiai T, Morita S, Nishiyama A, Yamada E, Arai H, et al. Anabolic five subunit-type pyruvate:ferredoxin oxidoreductase from Hydrogenobacter thermophilus TK-6. Biochem Biophys Res Commun. 2006;340:76–82. doi: 10.1016/j.bbrc.2005.11.155. [DOI] [PubMed] [Google Scholar]
- 94.Yun NR, Yamamoto M, Arai H, Ishii M, Igarashi Y. A novel five-subunit-type 2-oxoglutalate:ferredoxin oxidoreductases from Hydrogenobacter thermophilus TK-6. Biochem Biophys Res Commun. 2002;292:280–6. doi: 10.1006/bbrc.2002.6651. [DOI] [PubMed] [Google Scholar]
- 95.Kletzin A, Adams MWW. Molecular and phylogenetic characterization of pyruvate and 2-ketoisovalerate ferredoxin oxidoreductases from Pyrococcus furiosus and pyruvate ferredoxin oxidoreductase from Thermotoga maritima. J Bacteriol. 1996;178:248–57. doi: 10.1128/jb.178.1.248-257.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Berg IA. Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol. 2011;77:1925–36. doi: 10.1128/AEM.02473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pineda E, Encalada R, Rodríguez-Zavala JS, Olivos-García A, Moreno-Sánchez R, Saavedra E. Pyruvate:ferredoxin oxidoreductase and bifunctional aldehyde-alcohol dehydrogenase are essential for energy metabolism under oxidative stress in Entamoeba histolytica. FEBS J. 2010;277:3382–95. doi: 10.1111/j.1742-4658.2010.07743.x. [DOI] [PubMed] [Google Scholar]
- 98.Preisig O, Zufferey R, Thöny-Meyer L, Appleby CA, Hennecke H. A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J Bacteriol. 1996;178:1532–8. doi: 10.1128/jb.178.6.1532-1538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Han H, Hemp J, Pace LA, Ouyang H, Ganesan K, Roh JH, et al. Adaptation of aerobic respiration to low O2 environments. Proc Natl Acad Sci USA. 2011;108:14109–14. doi: 10.1073/pnas.1018958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ngugi DK, Blom J, Stepanauskas R, Stingl U. Diversification and niche adaptations of Nitrospina-like bacteria in the polyextreme interfaces of Red Sea brines. ISME J. 2016;10:1383–99. doi: 10.1038/ismej.2015.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shao MF, Zhang T, Fang HHP. Sulfur-driven autotrophic denitrification: diversity, biochemistry, and engineering applications. Appl Microbiol Biotechnol. 2010;88:1027–42. doi: 10.1007/s00253-010-2847-1. [DOI] [PubMed] [Google Scholar]
- 102.Mulkidjanian AY, Dibrov P, Galperin MY. The past and present of the sodium energetics: May the sodium-motive force be with you. Biochim Biophys Acta. 2008;1777:985–92. doi: 10.1016/j.bbabio.2008.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Watson SW, Waterbury JB. Characteristics of two marine nitrite oxidizing bacteria, Nitrospina gracilis nov. gen. nov. sp. and Nitrococcus mobilis nov. gen. Arch Mikrobiol. 1971;77:203–30. doi: 10.1007/BF00408114. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome of Nitrospira marina Nb-295T is available in the JGI IMG/M repository under genome ID number 2596583682. Manually curated protein annotations are available in Data Set 1, targeted quantitative proteomic analyses results are available in Data Set 2, global proteomic spectral counts and differential expression analysis results are available in Data Set 3. Raw mass spectra are available in PRIDE as project number PXD021606. Data is archived at Biological and Chemical Oceanography Data Management office (BCO-DMO) under project 806565.