Abstract
The gut microbiota has a critical role in the maintenance of immune homeostasis. Alterations in the intestinal microbiota and gut microbiota-derived metabolites have been recognized in many immune-related inflammatory disorders. These metabolites can be produced by gut microbiota from dietary components or by the host and can be modified by gut bacteria or synthesized de novo by gut bacteria. Gut microbiota-derived metabolites influence a plethora of immune cell responses, including T cells, B cells, dendritic cells, and macrophages. Some of these metabolites are involved in the pathogenesis of immune-related inflammatory diseases, such as inflammatory bowel diseases, diabetes, rheumatoid arthritis, and systemic lupus erythematosus. Here, we review the role of microbiota-derived metabolites in regulating the functions of different immune cells and the pathogenesis of chronic immune-related inflammatory diseases.
Keywords: gut microbiota, metabolites, T cells, B cells, autoimmune diseases
Subject terms: Mucosal immunology, Immunology
Introduction
The intestine harbors diverse and dynamic microbial communities, which have a critical role in human health and diseases. Many immune-related inflammatory disorders, such as inflammatory bowel diseases (IBDs),1,2 diabetes,3 rheumatoid arthritis (RA),4–6 and systemic lupus erythematosus (SLE),7,8 have been associated with altered gut microbiota. The crosstalk between gut microbiota and the immune system is intricate and is partially dependent on gut microbial metabolites.9 Both diet and gut microbiota species contribute to the production of various metabolites,10 which act locally in the intestine and remotely exert their diverse effects on other organs. It is well-recognized that microbial metabolites have important roles in regulating immune responses and chronic immune-related inflammatory diseases through different mechanisms, including activation of metabolic-specific receptors.11 More importantly, supplementation with some specific microbial metabolites,12–14 their receptor agonists,15,16 or a specific diet17 has been shown to regulate disease severity in mice and humans. For example, gut microbiota-derived short-chain fatty acids (SCFAs) have been the focus of intense research due to their profound effect on almost all types of immune cells and their potential for treating various chronic inflammatory diseases.18,19 In this review, we first introduce the key microbial metabolites that regulate immune responses, summarize the effect of these metabolites on immune cells, and discuss the relationship between microbial metabolites and chronic immune-mediated diseases. This review aims to provide a systematic understanding of microbiota-derived metabolites in immune cells and immune-related inflammatory diseases.
Gut microbiota regulation of immune responses
The host and gut microbiota have co-evolved to form a mutualistic relationship. The gut microbiota and host immunity interact in a complex, dynamic, and context-dependent manner. The role of intestinal microbiota in the development of the immune system and immune responses has been well established.20,21 Germ-free mice lack all microbial colonization, and therefore, their immune responses are “innocent” to the molecules of pathogens as well as beneficial gut microbiota. There are numerous immune deficits in germ-free mice, including impaired development of gut-associated lymphoid tissues (GALT) and systemic immune systems with decreased cellularity of GALT, mesenteric lymph nodes, and spleen.22,23 Both T and B cell responses are impaired in germ-free mice in response to antigen stimulation from challenge with foreign antigens or pathogens.21,24 Germ-free mice have fewer and smaller germinal centers within the spleen, fewer antibody-producing cells,21 and fewer IgG and IgM antibodies.24 Their T cells produce fewer cytokines in response to antigen challenge than conventionally colonized mice.24 Furthermore, the production of intestinal IgA, which is the most abundant antibody isotype in the host and provides the first line of immune protection at the mucosal surface,25,26 is nearly absent in germ-free mice.27 Reconstitution with gut microbiota normalizes the development of immune systems and immune responses,27,28 suggesting a crucial role of the microbiota in regulating the development and function of the immune system. Accumulating evidence indicates that not all gut bacteria function in the same way to regulate immune responses. Different gut bacteria differentially control the development of various components of the immune system. While segmented filamentous bacteria (SFB) promote Th17 cell development in the intestines, a cluster of Clostridia induces Treg cells.29,30 Functionally distinct Th17 cell populations are present in the intestines and are primarily determined by distinct bacteria. SFB-induced Th17 cells are homeostatic, while Citrobacter-induced Th17 cells are pro-inflammatory.31 Thus, it is essential to understand the molecular basis of such microbiota–host interactions, which could provide novel targets that shape immune function in treating various immune-related inflammatory diseases.
Gut microbiota-derived metabolites
The gut microbiota does not merely evade the immune system in the intestinal tract. In addition to well-recognized TLR ligands, including LPS, flagellins, and dsDNAs,32 which are beyond the scope of this review, gut microbiota-derived metabolites continuously regulate immune cells both locally and systemically. However, our understanding of these interactions is just beginning as most gut microbiota-derived metabolites are still unidentified, and many of their functions are yet to be defined.10 A relatively small but diverse range of gut microbiota-derived metabolites has been identified and functionally studied in the last decade. Depending on their origin and synthesis, these gut microbial metabolites can be broadly divided into three groups: (1) metabolites produced by gut bacteria from dietary components, (2) metabolites produced by the host and modified by gut bacteria, and (3) metabolites synthesized de novo by gut bacteria (Fig. 1 and Table 1).
Fig. 1.
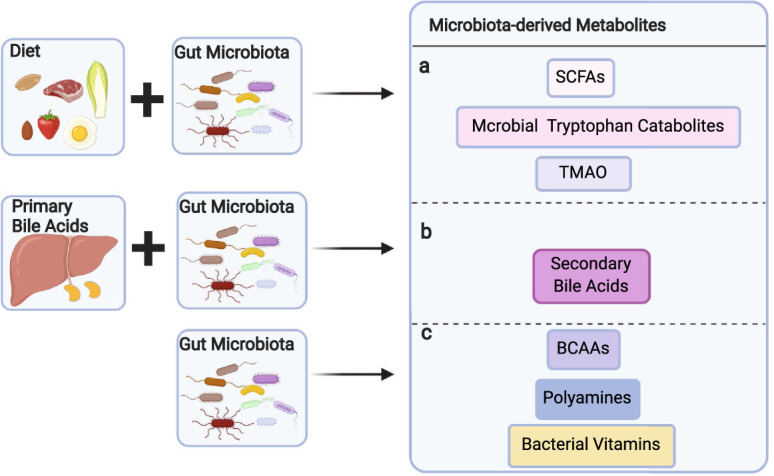
Production of gut microbiota-derived metabolites. Gut microbiota-derived metabolites can be produced by gut microbiota from dietary components (a), by the host and modified by gut bacteria (b), and synthesized de novo by gut bacteria (c). BCAAs branched-chain amino acids, SCFAs short-chain fatty acids, TMAO trimethylamine-N-oxide
Table 1.
The characteristics and functions of the major gut microbiota-derived metabolites
| Groups of microbial metabolites | Representatives of metabolites | Receptors | Functions | ||
|---|---|---|---|---|---|
| Metabolites | Examples | ||||
| Produced by gut bacteria from dietary components | SCFAs |
Acetate Propionate Butyrate |
GPR4142 GPR4343 GPR109a41 |
T cells |
Regulation of CD4+ T cell differentiation and cytokines production42,43,93–97,104,105,109,145,146 |
| B cell | Regulation of cell activation and antibody production122,123,126,129,130 | ||||
| DCs | Suppression of maturation and modulation of cytokines/chemokines production140,141,143,144 | ||||
| Macrophages | Enhancement of antimicrobial function and modulation of cytokines production135,150,151 | ||||
| Microbial tryptophan catabolites |
Indole IPA IAA |
PXR51 AhR52 |
T cell |
Induction of Treg cell but inhibition of Th17 development102,103,106 Induction of CD4+ CD8+ IELs119 |
|
| TMAO | TMAO |
TAARs59 PERK60 |
Macrophages | Induction of M1 polarization154 | |
| Produced by the host and modified by gut microbiota | Secondary bile acids |
DCA LCA |
FXR65 PXR67 VDR69 CAR70 TGR571 |
T cells |
Induction of Treg differentiation and RORγ+ Treg cell development, but suppression of Th17 differentiation99,100 Regulation of hepatic NKT cell accumulation114 |
| Macrophages |
Induction of Kupffer cells152 Inhibition of LPS-induced proinflammatory cytokines in macrophages71,153 |
||||
| Synthesized de novo by gut microbiota | BCAAs |
Leucine Isoleucine Valine |
unknown | T cells | Maintenance of Treg cells74 |
| Macrophages | Suppression of LPS-induced cytokines155 | ||||
| Polyamines |
Spermine Spermidine putrescine |
NMDA81 CaR82 |
T cells | Suppression of IFN-γ production107 | |
| DCs | Modulation cell activation and phenotypes148 | ||||
| Bacterial vitamins |
Ascorbate 6-FP 5-OP-RU |
MR1117,118 | T cells |
Suppression of CD4+ effector T cell88 |
|
BCAAs branched-chain amino acids, CaR calcium-sensing receptor, CAR constitutive androstane receptor, DCA deoxycholate, FXR farnesoid X receptor, GPR G protein-coupled receptor, IPA indolepropionic acid, IAA indoleacetic acid, LCA lithocholic acid, MR1 MHC-related molecule 1, NMDA N-methyl-d-aspartate receptors, PERK protein kinase R-like endoplasmic reticulum kinase, PXR pregnane X receptor, SCFAs short-chain fatty acids, TAARs trace amine-associated receptors, TGR5 G protein-coupled bile acid receptor 1, TMAO trimethylamine-N-oxide, 6-FP 6-FP 6-formylpterin, VDR vitamin D receptor, 5-OP-RU 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil.
Metabolites produced by gut bacteria from dietary components
Short-chain fatty acids
Undigested carbohydrates can be fermented to SCFAs by gut microbiota, which express carbohydrate-active enzymes. SCFAs, referring to fatty acids with fewer than six carbons, principally include acetate, propionate, and butyrate, which exist in a 3:1:1 ratio in the gut.33 SCFAs reach a concentration of ~100 mM in the intestinal lumen,34 depending on dietary fiber and the gut microbial composition. Acetate production is mainly formed from acetyl-CoA by enteric bacteria or by acetogens through the Wood–Ljungdahl pathway.35,36 The phylum Bacteroidetes primarily produces propionate through the succinate pathway or acrylate pathway by Lachnospiraceae and Negativicutes.37 Alternatively, propionate production can also occur through the propanediol pathway by Roseburia inulinivorans and Ruminococcus obeum.37 Butyrate is primarily produced by several species in the phylum Firmicutes, which express active CoA-transferase.38 After being absorbed in the colon, SCFAs are either utilized locally to provide colonocytes with energy or transported into the blood, where they are distributed to other organs for metabolism and consumption through the portal vein.34 SCFAs enter the cells in two primary ways: passive diffuse and carrier-mediated transportation via SMCT1 (SLC5A8) and MCT1 (SLC16A1).39 SCFAs regulate the function of different immune cells by inhibiting histone deacetylase (HDAC)40 and/or activating G protein-coupled receptors (GPRs),41–43 which include GPR41, GPR43, and GPR109a.
Microbial tryptophan catabolites
As an essential aromatic amino acid, tryptophan is supplied mostly from dietary protein. Tryptophan absorption primarily occurs in the small intestine for protein synthesis, whereas a variety of bacteria in the colon can directly degrade tryptophan into several metabolites, including indole, indole ethanol (IE), indolepropionic acid (IPA), indole lactic acid (ILA), indoleacetic acid (IAA), indolealdehyde (IAld), indoleacrylic acid (IA), skatole, and tryptamine.44 The types and levels of microbial tryptophan metabolites are primarily influenced by the gut microbiota, which possesses different catalytic enzymes for tryptophan metabolism. Tryptophan is converted into indole by numerous bacterial species (such as Escherichia coli45 and Proteus vulgaris46) using tryptophanase.47 Additionally, various other bacterial species produce several tryptophan metabolites through different metabolic pathways. For example, Clostridium sporogenes and Rominococcus gnavus can produce tryptamine by decarboxylation of tryptophan.48 Clostridium sporogenes and Peptostreptococcus spp. produce IA.49,50 Clostridium spp., such as Clostridium botulinum, Clostridium caloritolerans, and Clostridium sporogenes, can convert tryptophan into IPA.49 Gut microbiota-derived tryptophan catabolites regulate intestinal immune responses locally and systemically affect host physiology through the bloodstream, where they activate the pregnane X receptor (PXR)51 and/or aryl hydrocarbon receptor (AhR).52,53
Trimethylamine-N-oxide
Trimethylamine (TMA) is a gut microbiota metabolite derived from carnitine, choline, or choline-containing compounds in the diet.54 The formation of TMA depends on the balance and diversity of the gut microbiota. The sulfate-reducing bacteria Desulfovibrio alaskensis and Desulfovibrio desulfuricans, which possess the choline-utilization gene cluster (Cut), can convert choline to TMA in the gut,55 whereas the genera Acinetobacter and Serratia, encoding CntA and CntB, can generate TMA from l-carnitine.56,57 Furthermore, another gene pair, YeaW/YeaX, with homology to CntA/B, also contributes to the production of TMA.32 TMA can be absorbed in the intestine and then transferred to the liver, where it is oxidized to trimethylamine-N-oxide (TMAO) by flavin monooxygenases (FMOs).58 TMA, but not TMAO, selectively activates human trace amine-associated receptors (TAARs).59 It has been revealed recently that TMAO directly binds and activates protein kinase R-like endoplasmic reticulum kinase (PERK), a key sensor of intracellular stress.60
Metabolites produced by the host and modified by gut bacteria
Secondary bile acids
Primary bile acids, including cholate (CA) and chenodeoxycholate (CDCA), are synthesized by hepatocytes.61 After conjugation with glycine or taurine, which promotes the surfactant function of bile acids, the conjugated bile acids enter the intestinal lumen, where they promote digestion, transport, and absorption of nutrients.61,62 Most of the conjugated bile acids are absorbed in the small intestine and transported back to the liver. Approximately 5% of bile acids are converted into secondary bile acids by a limited number of microbiota species (e.g., Firmicutes species, notably Clostridium scindens) in the cecum and colon.63 Among secondary bile acids, deoxycholate (DCA) and lithocholic acid (LCA) are the two major types that are generated through 7a-dehydroxylation of CA and CDCA, respectively.63,64 In addition to directly affecting bacteria, bile acids activate cells by binding to nuclear hormone receptors, including farnesoid X receptor (FXR),65,66 PXR,67,68 vitamin D receptor (VDR),69 and constitutive androstane receptor (CAR),70 as well as a cell-surface receptor, G protein-coupled bile acid receptor 1 (GPBAR1, also called TGR5).71
Metabolites synthesized de novo by gut bacteria
Branched-chain amino acids
Among nine essential amino acids, leucine, isoleucine, and valine are branched-chain amino acids (BCAAs) having an aliphatic side chain with a branch.72 Although diet is the primary source of BCAAs in humans, BCAAs can also be synthesized by gut microbiota.73 Upon bacterial invasion, there is a markedly increased demand for synthesis substrates, including BCAAs, for immune responses and functions. In addition to enhancing protein synthesis and providing energy via catabolism, BCAAs regulate cell functions by activating the mTOR pathway.74
Polyamines
Polyamines, mainly spermine, spermidine, and putrescine, are widely distributed polycationic molecules in almost all living cells and are essential for biological functions.75 The polyamines in the intestinal lumen originate either from the diet or are synthesized de novo by host cells and intestinal microbiota.76 Dietary protein is the primary source of polyamines in the intestinal tract,77 and the majority of polyamine absorption occurs in the small intestine. The intestinal microbiota, such as Bacteroides spp. and Fusobacterium spp., are considered primary producers of polyamines in the lower parts of the intestine.78,79 In addition, diet can modulate microbial polyamine production. Specifically, dietary fiber increases SCFA production and lowers luminal pH, both of which activate intestinal bacteria to produce polyamines.80 Polyamines serve as ligands for various types of receptors, such as N-methyl-d-aspartate receptors (NMDA)81 and calcium-sensing receptor (CaR).82
Bacterial vitamins
Vitamins, essential nutrients for bacteria and host metabolism, are obtained from the diet or synthesized by gut microbiota. While dietary vitamins are absorbed in the small intestine,83 gut microbiota-derived vitamins are primarily utilized in the colon.84 For example, vitamin K2 (menaquinone) and vitamin B family members are the major vitamins produced by gut flora.85 Gut bacterial species, including Lactic acid bacteria and Bacteroides, produce vitamin K2.86,87 However, diet is the sole source of vitamin K1 (phylloquinone). Various gut bacteria produce vitamin B, including vitamin B1 (thiamine), B2 (riboflavin), B3 (nicotinic acid), B5 (pantothenic acid), B6 (pyridoxine), B7 (biotin), B9 (folates), and B12 (cobalamin). For instance, vitamin B2 is synthesized by several bacterial species in the phyla Bacteroidetes, Fusobacteria, Proteobacteria, and Firmicutes, while the synthesis of vitamin B12 occurs in the phylum Fusobacteria but rarely in Actinobacteria and Proteobacteria.84 It has also been recently found that gut microbiota can produce vitamin C (ascorbate).88
Gut microbiota-derived metabolite regulation of host immune responses
Accumulating evidence indicates that gut microbiota-derived metabolites regulate the development and function of multiple types of immune cells, including both adaptive and innate cells. Due to space limitations, we will mainly focus on their effects on major populations of immune cells, i.e., T cells, B cells, dendritic cells (DCs), and macrophages (Table 1 and Figs. 2, 3).
Fig. 2.
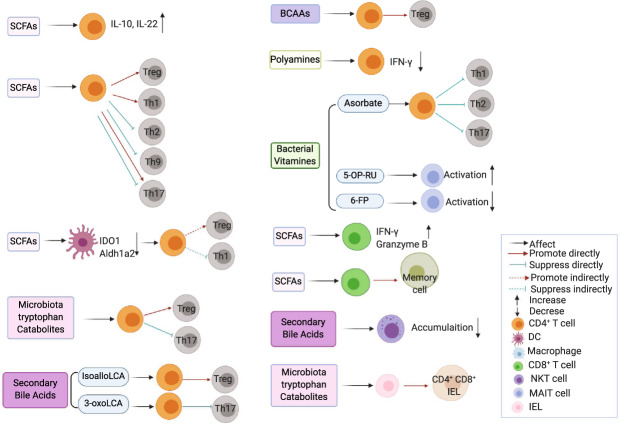
Effects of gut microbiota-derived metabolites on T cells. Microbiota-derived metabolites differentially modulate T cell functions. Aldh1a2 retinal dehydrogenase aldehyde dehydrogenase 1A2, BCAAs branched-chain amino acids, DC dendritic cell, IEL intraepithelial lymphocyte, IDO1 indoleamine 2,3-dioxygenase 1, LCA lithocholic acid, NKT natural killer T cells, MAIT mucosal-associated invariant T cell, SCFAs short-chain fatty acids, 5-OP-RU 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil, 6-FP 6-formylpterin
Fig. 3.
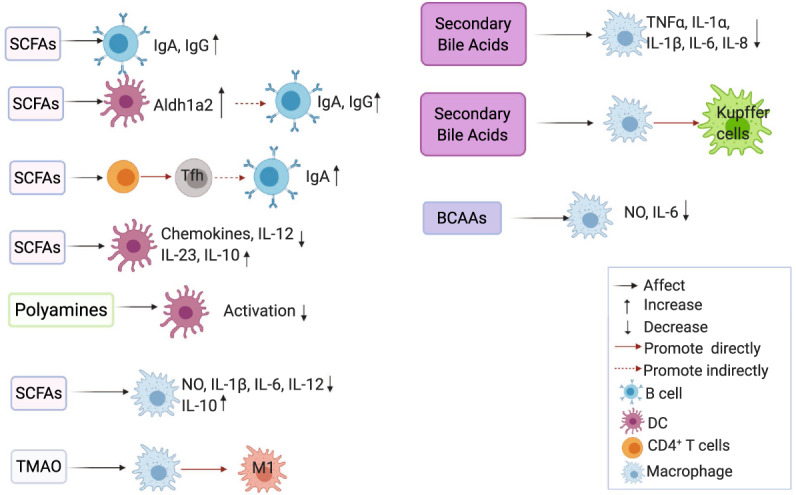
Effects of gut microbiota-derived metabolites on B cells, macrophages, and dendritic cells. Microbiota-derived metabolites regulate various types of immune cells. Aldh1a2 retinal dehydrogenase aldehyde dehydrogenase 1A2, BCAAs branched-chain amino acids, DC dendritic cell, DCA deoxycholate, SCFAs short-chain fatty acids, TMAO trimethylamine-N-oxide
T cells
T cells, the major mediators in regulating immune responses, are divided into two populations, TCRαβ and TCRγδ T cells.89 Among TCRαβ T cells, there are CD4+ T cells, CD8+ T cells, natural killer T cells (NKT cells), intraepithelial lymphocytes (IELs), and other innate TCRα-expressing T cells, i.e., mucosal-associated invariant T (MAIT) cells.90–92 In this section, we will summarize the effects of gut microbiota-derived metabolites on T cells (Table 1 and Fig. 2).
CD4+ T cells
The role of microbiota-derived metabolites in regulating CD4+ T cells is an area of intensive investigation. Given that Treg cells are essential for limiting immune responses, various types of metabolites have been investigated for their effects on Treg cells. SCFAs have been found to promote colonic Treg differentiation by inhibiting HDAC expression and activity in a GPR43-dependent manner93 or by enhancing histone acetylation in the conserved noncoding sequence region (CNS) 1 of the Foxp3 locus in a GPR43-independent manner.94,95 However, SCFAs did not induce Treg cells under Th1 and Th17 polarization conditions with a high anti-CD3 activation level, whereas Foxp3 expression was upregulated under low T cell activation conditions.96 Furthermore, butyrate failed to generate Treg cells in the absence of TGFβ.97 Therefore, SCFAs affect Treg cells in a context-dependent manner. Secondary bile acids are known to affect innate immunity,16,71,98 but their effects on adaptive immune responses have been poorly investigated until recently. It has been shown that isoalloLCA, an LCA derivative, promoted Treg differentiation via histone modification of CNS3 of the Foxp3 locus and production of mitochondrial reactive oxygen species.99 Furthermore, dominant intestinal bile acids along with their potent secondary bile acids induced RORγ+ Treg cells, which are critical in maintaining intestinal immune homeostasis, in a VDR-dependent manner.100 AhR, a transcription factor widely expressed in immune cells, has a crucial role in regulating immune responses, including CD4+ T cells.101 AhR ligands, including bacterial tryptophan catabolites, modulate CD4+ T cell responses. It has been reported that indoles and indole derivatives promoted Treg differentiation both in vitro and in vivo in an AhR-dependent manner.102,103 Amino acids are essential for activated T cells with high metabolic demands. The maintenance of Treg cells was regulated by BCAAs, which activated mTOR through the transporter SLC3A2.74
Effector CD4+ T cells have an essential role in modulating immune-related inflammatory diseases. SCFAs were found to promote naïve CD4+ T cell differentiation into Th1 cells through activation of the mTOR–S6K pathway, which was dependent on the inhibition of HDAC activity but not GPR41 and GPR43.96,104 However, the effects of SCFAs on Th17 cells are context-dependent. Although SCFAs have been reported to induce Th17 differentiation,96 butyrate has also been shown to inhibit T cell IL-17 expression through induction of T-bet, a key transcription factor for Th1 cells,97 and by inhibiting the expression of the transcription factors retinoid-related orphan receptor (Ror)α and Rorγt.104 In addition, SCFAs suppressed Th2 and Th9 differentiation.97,105 It has been shown that an LCA derivative, 3-oxoLCA, suppressed Th17 cell differentiation by directly binding to Rorγt.99 Indoles and indole derivatives, which are bacterial tryptophan catabolites, were found to suppress Th17 development through the AhR pathway.102,103,106 Spermine, one type of polyamine, suppressed Th1 cytokine IFN-γ secretion by spleen cells,107 suggesting that it might regulate T cell responses. Ascorbate, a novel microbial metabolite of bacterial vitamins, was reported to suppress T effector cells and inhibit T cell activation.88
IL-10 produced by effector T cells is an important self-regulatory mechanism for maintaining immune homeostasis.108 It has been reported that SCFAs promoted IL-10 production in differentiated Th1 cells in a GPR43-dependent manner,43 while SCFA induction of IL-10 during the differentiation of Th1 and Th17 cells was dependent on inhibiting HDAC activity.96,104,109 IL-22, a member of the IL-10 family, is critical to host protection against intestinal inflammation. CD4+ T cells are considered major sources of IL-22 during chronic intestinal inflammation. We recently identified SCFAs as important metabolites in inducing IL-22 in CD4+ T cells through the GPR41 pathway and in inhibiting HDAC activity.42
CD8+ T cells
The investigation into how gut microbiota-derived metabolites affect CD8+ T cells, which are indispensable in mediating immune defenses against intracellular pathogens and tumor surveillance, is still in the early stage. It has been demonstrated that SCFAs regulate CD8+ T cell functions.110 Systemic acetate levels were increased following bacterial infection, and upregulated acetate production facilitated the rapid recall responses of memory CD8+ T cells by promoting glycolysis.110 A recent study reported that butyrate enhanced memory CD8+ T cell responses, which were dependent on both GPR41 and GPR43.111 In addition, GPR41 is critical in SCFA-mediated expansion of CD8+ T cell functionality.112 Conversely, butyrate suppressed IL-17 but promoted IFN-γ and granzyme B in CD8+ T cells, which was due to the inhibition of HDAC activity but not by activating GPR41 and GPR43.113 Collectively, SCFAs and their receptors are essential for modulating the functions of CD8+ T cells.
Other T cells
There are only a few reports on how gut microbiota-derived metabolites regulate NKT cells, which can recognize a diverse repertoire of lipids presented by CD1d and participate in the regulation of immune responses. A recent study demonstrated that primary bile acids and secondary bile acids regulated hepatic NKT cell accumulation in opposing ways in liver tumors,114 which was attributed to hepatic CXCR6 expression induced by primary bile acids but suppressed by secondary bile acids. Further investigation is needed to clarify the underlying mechanisms as well as whether bile acids affect NKT cells in other conditions.
Accumulating evidence indicates that gut microbiota-derived metabolites regulate MAIT cells, a subset of T cells restricted by MHC-related molecule 1 (MR1). MAIT cells can respond to diverse microbiota in an MR1-dependent manner.115,116 The pyrimidine-based intermediate 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil (5-OP-RU), along with the vitamin B2 riboflavin biosynthetic pathway, are the most potent agonists for MAIT cells.117 The vitamin B9 metabolite 6-formylpterin (6-FP) inhibits MAIT cell activation by competitively binding to MR1,118 indicating that specific vitamin metabolites have important roles in activating MAIT cells.
IELs, which reside within the intestinal epithelium, are involved in immune protection against invading pathogens and microbiota. There is scant literature on how gut microbiota-derived metabolites regulate IELs. An elegant recent study revealed that the bacterial species Lactobacillus reuteri produced indole derivatives of dietary tryptophan to induce CD4+ CD8αα+ double-positive intraepithelial lymphocytes from interepithelial CD4+ T cells in an AhR-dependent manner,119 which underscores the delicate interplay between gut microbiota and the diet in the maintenance of intestinal homeostasis.
B cells
Among the microbial metabolites, SCFAs and microbial tryptophan catabolites have been shown to regulate B cell activation and antibody responses (Table 1 and Fig. 3).
B cell activation and antibody production depend on energy and metabolites from glycolysis and oxidative phosphorylation.120,121 SCFAs directly enhanced cellular acetyl-CoA levels in B cells, leading to higher mitochondrial energy production, fatty acid synthesis, and mTOR-mediated glycolysis, which contributed to plasma cell differentiation and antibody production.122 Additionally, SCFAs regulated the expression of genes (i.e., Aicda, Xbp1, and Prdm1) associated with B cell differentiation via HDAC inhibition.122 However, it has also been reported that SCFAs at lower doses directly impacted B cell-intrinsic functions to moderately enhance antibody production, while SCFAs at higher doses inhibited antibody production through regulation of AID and Blimp1 expression, class switching response (CSR), somatic hypermutation, and plasma cell differentiation,123 suggesting that SCFAs at different doses differentially affect B cell responses. Intestinal IgA undergoes CSR and affinity maturation in germinal centers (GCs) with the help of T follicular helper cells and follicular DCs.124,125 SCFAs can also regulate B cell IgA production through interactions with T cells and DCs. Dietary fiber promoted Tfh cell differentiation, which further promoted GC formation and IgA CSR.122,126 Retinoic acid (RA), converted from a vitamin A metabolite by retinal dehydrogenase aldehyde dehydrogenase 1A2 (Aldh1A2) in DCs, maintained the development of B cells and intestinal IgA production.127,128 SCFAs induced the expression of Aldh1A2 in DCs to promote intestinal IgA responses126,129 and mucosal adjuvant activity of cholera toxin in a GPR43-dependent manner.130 Given that IL-10 is a key cytokine for B cell differentiation and antibody maturation,131,132 SCFAs also indirectly affected B cell functions through the induction of IL-10 in CD4+ T cells,43,133 DCs,134,135 and macrophages.134 Collectively, SCFAs regulate B cell activation and antibody production by directly acting on B cells or indirectly acting on other cell types.
There are a few reports on the role of microbiota-derived tryptophan in B cells. Considering that microbial tryptophan catabolites (i.e., indole and its derivatives) can bind to AhR, which profoundly regulates B cell development,136 differentiation,137 cytokine production, and regulatory activity,138 microbial tryptophan catabolites may act on B cells through AhR signaling. Interestingly, butyrate administration increased tryptophan-metabolizing bacteria in mice, promoting IL-10-producing regulatory B cells in an AhR-dependent manner,139 indicating the complex network between microbial metabolites and the regulation of B cell function. However, it is still unknown whether and how microbiota-derived tryptophan directly regulates B cell IL-10 production as well as antibody production.
Dendritic cells
DCs, one of the major professional antigen-presenting cells, have a critical role in the interaction between innate and adaptive immune responses. Accumulating evidence demonstrates the role of gut microbiota-derived metabolites in regulating DC functions (Table 1 and Fig. 3).
SCFAs suppressed the maturation of DCs from mouse bone marrow stem cells140–142 and their generation from human monocytes.143 Mechanistically, SCFAs inhibited HDAC activity in DCs via the Na+-coupled monocarboxylate transporter Slc5a8, thereby inhibiting the transcription factors associated with the maturation of DCs, namely, PU.1 and RelB.140 In addition, SCFAs modulated the cytokines and chemokines secreted by DCs. It has been shown that SCFAs suppressed IL-12 production,143 whereas they upregulated IL-23 and IL-10 production in DCs.141,143 Furthermore, SCFAs decreased DC production of chemokines, i.e., CCL3, CCL4, CCL5, CXCL9, CXCL10, and CXCL11.144 SCFAs also indirectly regulate T cell functions through modulation of DCs. SCFAs impaired the capacity of DCs to induce CD4+ and CD8+ T cell proliferation,142,143 which was likely due to upregulated IL-10 in DCs.143 Butyrate-treated DCs promoted Treg differentiation but inhibited Th1 differentiation by inducing DC expression of the immunosuppressive enzymes indoleamine 2,3-dioxygenase 1 (IDO1) and Aldh1a2 in a Slc5a8-dependent manner.145 In addition, lung DCs purified from propionate-treated mice were ineffective at driving Th2 cell responses.146 All these studies indicate that SCFA-treated DCs promote Treg cell function but inhibit effector T cell responses. DCs exposed to SCFAs are also involved in regulating humoral immune responses.129,130 Our study showed that acetate induced B cell IgA responses in the presence of DCs, which was dependent on GPR43.129 This effect was due to acetate induction of Aldh1a2 expression in DCs.129 We further found that the administration of SCFAs facilitated cholera toxin mucosal adjuvanticity through DCs in a GPR43-dependent manner.130 Therefore, SCFAs not only directly affect DC function but also regulate other adaptive cell functions through modulation of DCs.
Taurochenodeoxycholic acid (TCDCA), a taurine-conjugated form of the primary bile acid CDCA, induced monocytes to differentiate towards an IL-12 hypoproducing DC phenotype through TGR5 but not the FXR pathway.147 In addition, FXR activation inhibited DC differentiation from monocytes and suppressed DC production of TNFα.16 Given that secondary bile acids can also activate TGR5 and FXR, it can be speculated that these bacterial metabolites may also regulate DC functions.
Polyamine also regulates DC functions. The polyamine compound deoxyspergualin (DSG) suppresses shock protein-induced activation of DCs.148 Putrescine, a polyamine molecule, affects the phenotype of DCs and reduces the capacity of DCs to activate T cells.149 However, the underlying mechanisms are still unclear.
Macrophages
Macrophages can ingest and kill pathogens, produce proinflammatory and anti-inflammatory cytokines, and present antigens to T cells and are thus indispensable in maintaining immune responses. Increasing studies have indicated that gut microbiota-derived metabolites regulate macrophage function (Table 1 and Fig. 3).
SCFAs suppressed proinflammatory cytokines through inhibition of HDAC activity135 but promoted anti-inflammatory IL-10 expression in macrophages.150 In addition, butyrate inhibited HDAC3 to reduce the activation of mTOR and glycolysis, which contributed to the enhanced antimicrobial function of macrophages,151 indicating that SCFAs promote the anti-inflammatory function of macrophages against infection and inflammation.
Bile acids also regulate macrophage functions. DCA, a secondary bile acid, induced Kupffer cells, which are specialized macrophages located in the liver, to generate reactive oxygen species.152 Both primary and secondary bile acids inhibited LPS induction of proinflammatory cytokines in macrophages in a TGR5-dependent manner.71,153 BAR501, a TGR5 agonist, promoted the M1 shift to M2 macrophages.15 Foam cells, macrophages containing low-density lipoproteins (LDL), are involved in the pathogenesis of atherogenesis. INT-777, a TGR5-specific semisynthetic bile acid, inhibited macrophage foam cell formation by reducing the uptake of LDL.98
In addition, TMAO has been shown to induce M1 polarization,154 and BCAAs were able to suppress LPS-induced NO and IL-6 and reduce the damage caused by H2O2 in macrophages in vitro.155 It is unclear whether they also function in the regulation of macrophages in vivo, which will be crucial in dissecting their roles in macrophages in the real world.
Gut microbiota-derived metabolite regulation of immune-related inflammatory diseases
Many chronic inflammatory diseases are immune-related disorders caused by dysregulated immune responses to autoantigens or environmental antigens. It has been established that the gut microbiota regulates the pathogenesis of various immune-related inflammatory diseases. In this section, we will discuss the effects of gut microbiota-derived metabolites on the regulation of the pathogenesis of these immune-related chronic inflammatory diseases, including IBD, diabetes, RA, and SLE (Fig. 4).
Fig. 4.
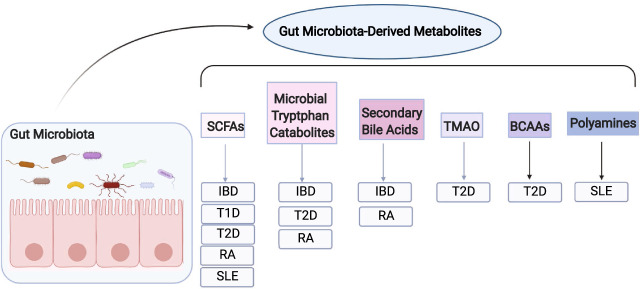
Gut microbiota-derived metabolites and immune-related inflammatory diseases. Certain gut microbiota-derived metabolites regulate the pathogenesis of various immune-related chronic inflammatory diseases. SCFAs short-chain fatty acids, TMAO trimethylamine-N-oxide, BCAAs branched-chain amino acids, IBD inflammatory bowel diseases, T1D type-1 diabetes, T2D type-2 diabetes, RA rheumatoid arthritis, SLE systemic lupus erythematosus
Inflammatory bowel diseases
IBD, which mainly includes Crohn’s disease and ulcerative colitis, is a group of chronic inflammatory intestine disorders. In addition to genetic factors, diet and intestinal microbiota have been implicated in the development of IBD. Fecal metabolomic studies demonstrated that bacterial metabolites were altered in patients with IBD, including decreased SCFAs,156–158 secondary bile acids,158 vitamin B,158 TMA,157 BCAAs,157 and increased primary bile acids158–160 and polyamines.160 Furthermore, microbiota-derived metabolites, mainly SCFAs, bacterial tryptophan catabolites, and bile acids, have been found to regulate the pathogenesis of IBD.43,100,161
SCFAs have been reported to regulate intestinal inflammation in different experimental colitis models. Butyrate attenuated colitis severity in mice with transferred CD4+ CD45RBhi T cells,94 which is a T cell-dependent chronic colitis model. We and others further showed the beneficial effect of butyrate on T cell-induced colitis.43,96,104 However, it is still controversial whether butyrate attenuates or worsens intestinal inflammation in dextran sulfate sodium (DSS)-induced colitis, an acute colitis model that causes epithelial injury, compromises barrier integrity, and subsequently induces inflammation. Maslowski et al.162 and our group43 found that administration of acetate or butyrate in drinking water attenuated DSS-induced colitis.43,162 However, it has also been shown that butyrate did not affect colitis induced by DSS.135 These differences might be attributed to the concentration of DSS used, the butyrate dose, the route by which butyrate was given, and the gut microbiota of the mice housed in different animal facilities. Indeed, the route of butyrate treatment has been shown to be crucial for its effect on colitis.141 Butyrate given orally in drinking water exacerbated colitis, whereas butyrate given systemically by intraperitoneal injection decreased the severity of colitis in the DSS-induced model.141 Interestingly, GPR43, the receptor of SCFAs, was decreased in CD patients,163 indicating that GPR43 might be involved in the protection of IBD. In agreement with this argument, GPR43−/− mice and GPR109a−/− mice were more susceptible to colitis.15,134 Several studies have reported that SCFA enemas demonstrated positive results in treating distal UC.13,164,165 However, some other studies showed no significant beneficial effect of SCFA enemas on colitis severity compared with placebo in IBD patients.166,167 Therefore, many questions on the effects and mechanisms of SCFAs on intestinal immune responses, especially in patients with IBD, require further investigation. More efforts are warranted to examine whether SCFAs only work among specific groups of IBD patients, as well as to examine the doses and route of treatment of SCFAs for IBD patients.
The association of tryptophan metabolites with IBD has been reported recently. Fecal tryptophan and a bacterial tryptophan metabolite, IAA, were decreased, while kynurenine, a host-derived tryptophan metabolite, was increased in both CD and UC patients.168 Serum IPA, another bacterial tryptophan metabolite, was also reduced in patients with active UC.169 Furthermore, serum tryptophan levels were associated with IBD.161 These studies suggest that the dysregulation of tryptophan might be associated with IBD.161 The expression of AhR, the receptor of bacterial tryptophan metabolites, in intestinal tissue was increased in patients with IBD.170 Furthermore, reduced AhR activity was found in feces from IBD patients.168 In experimental colitis, AhR−/− mice developed more severe intestinal inflammation induced by DSS53 or Citrobacter rodentium,171 whereas the administration of an AhR agonist attenuated intestinal inflammation induced by adoptive T cell transfer or chemicals.170 Thus, the development of indole- or AhR-based therapies could be a potential approach for treating IBD.
Although the effects of primary bile acids and secondary bile acids on immune cells are not identical, a recent study showed that primary and secondary bile acid mixtures protected mice against DSS-induced colitis.100 Bile acid receptors, including FXR, TGR5, and VDR, have been reported to regulate experimental colitis. INT-747, an FXR agonist, reduced chemical-induced colitis,16 while FXR−/− mice developed more severe intestinal inflammation.172 Furthermore, activation of TGR5 by BAR501 was protective in a trinitrobenzene sulfonic acid (TNBS)-induced model,15 which is a T cell-mediated colitis model. Additionally, bile acids failed to protect VDR−/− mice from colitis development. Overall, these studies suggest that bile acids and their receptors are potentially involved in the pathogenesis of IBD, although their clinical effects remain to be defined.
Diabetes
Diabetes refers to a group of metabolic disorders with sustained high blood glucose caused by defects in insulin secretion, insulin efficiency, or both, which are mainly divided into two categories: type-1 diabetes and type-2 diabetes. In addition to the traditional major risk factors, i.e., genetics, age, sex, and lifestyle, the gut microbiota and their metabolites have been implicated in the development of diabetes.3,173–175
Type-1 diabetes
Type-1 diabetes is characterized by the destruction of insulin-producing pancreatic β cells, which is mediated by T cells. Studies on the relationship between gut microbiota-derived metabolites and type-1 diabetes are still in the early stage. It has been shown recently that treatment with a combined acetate- and butyrate-yielding diet protected against type-1 diabetes in nonobese diabetic mice,176 demonstrating that the benefit of a high dietary fiber diet in type-1 diabetes might be attributed to SCFAs.
Type-2 diabetes
Type-2 diabetes is characterized by obesity and insulin resistance, which reduces the ability of cells to absorb and utilize glucose. Interestingly, the abundance of butyrate-producing bacteria was decreased in patients with type-2 diabetes.3 Treatment with SCFAs increased insulin sensitivity in rats177,178 and protected against diet-induced obesity and insulin resistance in mice.179 Additionally, GPR43−/− mice exhibited obesity on a regular diet and reduced insulin sensitivity on a high-fat diet,180 suggesting that SCFAs may be protective against the development of type-2 diabetes. However, whether SCFAs affect patients with type-2 diabetes remains to be investigated.
TMAO was increased in diabetic mice181 and patients with type-2 diabetes.182–184 Additionally, the TMAO-producing enzyme FMO3 was negatively correlated with insulin sensitivity in humans.182 Dietary TMAO impaired glucose tolerance and increased high-fat-induced insulin resistance,14 which could be mediated by TMAO directly binding to PERK, the endoplasmic reticulum stress kinase, which induces FoxO1 to drive metabolic diseases.60 Interestingly, administration of 3,3′-diindolylmethane, which reduced TMAO, benefited insulin-resistant mice,60 suggesting a potential therapeutic target for treating type-2 diabetes.
BCAAs and the bacterial tryptophan metabolite IPA have also been associated with type-2 diabetes. BCAA levels were associated with insulin resistance,185–187 which has been considered a predictive biomarker of the development of type-2 diabetes. BCAAs aggravated obesity-associated insulin resistance,188 and depletion of BCAAs in diets improved insulin sensitivity.189 Furthermore, IPA was negatively associated with the incidence of diabetes.190 Thus, BCAAs could promote whereas IPA could inhibit the pathogenesis of type-2 diabetes.
Rheumatoid arthritis
RA is an autoimmune disease that systemically affects joints and is caused by both genetic and environmental factors.191 The link between gut microbiota, gut microbiota-derived metabolites, and RA has been established in recent decades.139,162,192,193 A recent study demonstrated that SCFAs were positively associated with RA at the genetic, functional, and phenotypic levels.192 Furthermore, butyrate in stool samples was decreased in RA patients.139 Treatment with SCFAs attenuated disease severity in various RA mouse models, including collagen-induced arthritis (CIA),193 K/BxN serum-transfer arthritis,162 and antigen-induced arthritis.139 Consistently, GPR43−/− mice developed more severe arthritis,162 whereas an HDAC inhibitor reduced the severity of collagen-induced arthritis.194 Overall, SCFAs and their receptors have a critical role in regulating the pathogenesis and progression of RA, and supplementation with SCFAs could benefit RA patients.
Abnormal bile metabolism has long been recognized in RA patients.195 However, whether bile acids regulate the development of RA is still not completely understood. LCA, a secondary bile acid, exhibited anti-inflammatory effects in collagen-induced arthritis,196 suggesting a potential role of bile acids in treating RA. The correlation between bacterial tryptophan catabolites and RA, however, remains unclear. A recent report demonstrated that indole-3-carbinol (I3C), a dietary indole derivative, reduced adjuvant-induced arthritis,197 indicating that gut bacterial-derived indole and indole derivatives might be beneficial for RA treatment.
Systemic lupus erythematosus
SLE is an autoimmune disorder that occurs when the immune system mistakenly attacks the host’s own tissues and organs. Many SLE patients have altered intestinal microbiota,7,8 and the role of gut microbiota in the pathogenesis of SLE has also been implicated.198,199 Furthermore, altered production of SCFAs was related to altered intestinal microbiota in patients with SLE.200 It has been shown that both resistant starch and SCFAs suppress the development of TLR7-dependent systemic autoimmunity exacerbated by a Lactobacillus strain.199 The benefit of SCFAs on murine SLE was also demonstrated.123 However, it is still unknown whether SCFAs will benefit SLE patients, and more basic research and clinical investigations are needed to determine the potential role of SCFAs in treating SLE patients.
Given that polyamine synthesis reduced S-adenosylmethionine (SAM), a methyl donor, polyamines might have a role in autoimmune diseases, including SLE. Polyamine profiles in serum are different in SLE patients, with decreased levels of cadaverine and increased levels of N1-acetylcadaverine, spermidine, N1-acetylspermidine, and spermine.201 It will be interesting to determine whether serum polyamine profiles could be biomarkers for the diagnosis of SLE in patients. Administration of difluoromethylornithine, an inhibitor of the first enzyme of polyamine synthesis, ornithine decarboxylase (ODC), reduced disease severity in a murine model of SLE.202 Thus, targeting polyamine synthesis could be a potential therapeutic approach in treating SLE.
Concluding remarks
The relationship between microbiota and host immune responses is complicated, and their interactions occur not only in the intestine but also elsewhere in the body. Gut microbiota-derived metabolites differentially regulate immune responses and the pathogenesis of various immune-related inflammatory diseases. Among these microbial metabolites, SCFAs and secondary bile acids in the regulation of immune cell responses have been relatively extensively investigated, but their potential clinical uses in the treatment of these immune-related chronic inflammatory diseases remain unclear. Regarding the other microbial metabolites mentioned in this review, it is necessary to investigate their effects on different immune cells and their potential roles in various immune-related chronic inflammatory diseases. As the effects of most gut microbiota-derived metabolites on immune cells are largely context-dependent, it is unknown whether they function differently under homeostatic and inflammatory conditions. Because the environments of mucosal and systemic tissues are dramatically different, it is also unclear whether they function differently at the mucosal surface and systemic tissues. Therefore, more studies are required under various conditions. As most gut microbiota-derived metabolites have not been identified and the functions of known metabolites are still not fully understood, more efforts are warranted to determine the microbial metabolite pathways that are important in human diseases. Understanding the interplay between gut microbiota-derived metabolites, the immune system, and diseases will help develop future therapeutics to treat various inflammatory diseases.
Acknowledgements
This work was supported by NIH grants DK105585, DK112436, DK125011, AI150210, and DK124132; the University of Texas System STARs award (Y.C.); and supported by the James W. McLaughlin Fellowship Fund, UTMB (W.Y.). We appreciate Dr. Sherry Haller of The University of Texas Medical Branch for proofreading the manuscript. All images were created with BioRender.com.
Competing interests
The authors declare no competing interests.
References
- 1.Halfvarson J, et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat. Microbiol. 2017;2:17004. doi: 10.1038/nmicrobiol.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willing BP, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–1854.e1841. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 3.Qin J, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 4.Vaahtovuo J, Munukka E, Korkeamaki M, Luukkainen R, Toivanen P. Fecal microbiota in early rheumatoid arthritis. J. Rheumatol. 2008;35:1500–1505. [PubMed] [Google Scholar]
- 5.Scher JU, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 2015;21:895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 7.Luo XM, et al. Gut microbiota in human systemic lupus erythematosus and a mouse model of lupus. Appl. Environ. Microbiol. 2018;84:e02288–17. doi: 10.1128/AEM.02288-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hevia A, et al. Intestinal dysbiosis associated with systemic lupus erythematosus. mBio. 2014;5:e01548-14. doi: 10.1128/mBio.01548-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shapiro H, Thaiss CA, Levy M, Elinav E. The cross talk between microbiota and the immune system: metabolites take center stage. Curr. Opin. Immunol. 2014;30:54–62. doi: 10.1016/j.coi.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016;16:341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim CH. Immune regulation by microbiome metabolites. Immunology. 2018;154:220–229. doi: 10.1111/imm.12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiminez JA, Uwiera TC, Abbott DW, Uwiera RRE, Inglis GD. Butyrate supplementation at high concentrations alters enteric bacterial communities and reduces intestinal inflammation in mice infected with Citrobacter rodentium. mSphere. 2017;2:e00243-17. doi: 10.1128/mSphere.00243-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vernia P, et al. Short-chain fatty acid topical treatment in distal ulcerative colitis. Aliment. Pharmacol. Therap. 1995;9:309–313. doi: 10.1111/j.1365-2036.1995.tb00386.x. [DOI] [PubMed] [Google Scholar]
- 14.Gao X, et al. Dietary trimethylamine N-oxide exacerbates impaired glucose tolerance in mice fed a high fat diet. J. Biosci. Bioeng. 2014;118:476–481. doi: 10.1016/j.jbiosc.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Biagioli M, et al. The bile acid receptor GPBAR1 regulates the M1/M2 phenotype of intestinal macrophages and activation of GPBAR1 rescues mice from murine colitis. J. Immunol. 2017;199:718–733. doi: 10.4049/jimmunol.1700183. [DOI] [PubMed] [Google Scholar]
- 16.Gadaleta RM, et al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut. 2011;60:463–472. doi: 10.1136/gut.2010.212159. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Banares F, et al. Randomized clinical trial of Plantago ovata seeds (dietary fiber) as compared with mesalamine in maintaining remission in ulcerative colitis. Spanish Group for the Study of Crohn’s Disease and Ulcerative Colitis (GETECCU) Am. J. Gastroenterol. 1999;94:427–433. doi: 10.1111/j.1572-0241.1999.872_a.x. [DOI] [PubMed] [Google Scholar]
- 18.Sun M, Wu W, Liu Z, Cong Y. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J. Gastroenterol. 2017;52:1–8. doi: 10.1007/s00535-016-1242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bilotta AJ, Cong Y. Gut microbiota metabolite regulation of host defenses at mucosal surfaces: implication in precision medicine. Precis. Clin. Med. 2019;2:110–119. doi: 10.1093/pcmedi/pbz008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noverr MC, Huffnagle GB. Does the microbiota regulate immune responses outside the gut? Trends Microbiol. 2004;12:562–568. doi: 10.1016/j.tim.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 21.Bauer H, Horowitz RE, Levenson SM, Popper H. The response of the lymphatic tissue to the microbial flora. Studies on germfree mice. Am. J. Pathol. 1963;42:471–483. [PMC free article] [PubMed] [Google Scholar]
- 22.Cebra JJ. Influences of microbiota on intestinal immune system development. Am. J. Clin. Nutr. 1999;69:1046S–1051S. doi: 10.1093/ajcn/69.5.1046s. [DOI] [PubMed] [Google Scholar]
- 23.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 24.Ohwaki M, Yasutake N, Yasui H, Ogura R. A comparative study on the humoral immune responses in germ-free and conventional mice. Immunology. 1977;32:43–48. [PMC free article] [PubMed] [Google Scholar]
- 25.Macpherson AJ, McCoy KD, Johansen FE, Brandtzaeg P. The immune geography of IgA induction and function. Mucosal Immunol. 2008;1:11–22. doi: 10.1038/mi.2007.6. [DOI] [PubMed] [Google Scholar]
- 26.Mestecky J, Russell MW, Elson CO. Perspectives on mucosal vaccines: is mucosal tolerance a barrier? J. Immunol. 2007;179:5633–5638. doi: 10.4049/jimmunol.179.9.5633. [DOI] [PubMed] [Google Scholar]
- 27.Benveniste J, Lespinats G, Salomon J. Serum and secretory IgA in axenic and holoxenic mice. J. Immunol. 1971;107:1656–1662. [PubMed] [Google Scholar]
- 28.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 29.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Omenetti S, et al. The intestine harbors functionally distinct homeostatic tissue-resident and inflammatory Th17 cells. Immunity. 2019;51:77–89.e76. doi: 10.1016/j.immuni.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salzman NH. Microbiota-immune system interaction: an uneasy alliance. Curr. Opin. Microbiol. 2011;14:99–105. doi: 10.1016/j.mib.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandes J, Su W, Rahat-Rozenbloom S, Wolever TM, Comelli EM. Adiposity, gut microbiota and faecal short chain fatty acids are linked in adult humans. Nutr. Diabetes. 2014;4:e121. doi: 10.1038/nutd.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ragsdale SW, Pierce E. Acetogenesis and the Wood-Ljungdahl pathway of CO(2) fixation. Biochim. Biophys. Acta. 2008;1784:1873–1898. doi: 10.1016/j.bbapap.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 37.Reichardt N, et al. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 2014;8:1323–1335. doi: 10.1038/ismej.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Louis P, Young P, Holtrop G, Flint HJ. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ. Microbiol. 2010;12:304–314. doi: 10.1111/j.1462-2920.2009.02066.x. [DOI] [PubMed] [Google Scholar]
- 39.Sivaprakasam S, Bhutia YD, Yang S, Ganapathy V. Short-chain fatty acid transporters: role in colonic homeostasis. Compr. Physiol. 2017;8:299–314. doi: 10.1002/cphy.c170014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davie JR. Inhibition of histone deacetylase activity by butyrate. J. Nutr. 2003;133:2485S–2493S. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- 41.Thangaraju M, et al. GPR109A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. 2009;69:2826–2832. doi: 10.1158/0008-5472.CAN-08-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang W, et al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat. Commun. 2020;11:4457. doi: 10.1038/s41467-020-18262-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun M, et al. Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nat. Commun. 2018;9:3555. doi: 10.1038/s41467-018-05901-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roager HM, Licht TR. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018;9:3294. doi: 10.1038/s41467-018-05470-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith T. A modification of the method for determining the production of indol by bacteria. J. Exp. Med. 1897;2:543–547. doi: 10.1084/jem.2.5.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hickman FW, Steigerwalt AG, Farmer JJ, 3rd, Brenner DJ. Identification of Proteus penneri sp. nov., formerly known as Proteus vulgaris indole negative or as Proteus vulgaris biogroup 1. J. Clin. Microbiol. 1982;15:1097–1102. doi: 10.1128/jcm.15.6.1097-1102.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee JH, Lee J. Indole as an intercellular signal in microbial communities. FEMS Microbiol. Rev. 2010;34:426–444. doi: 10.1111/j.1574-6976.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- 48.Williams BB, et al. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe. 2014;16:495–503. doi: 10.1016/j.chom.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dodd D, et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature. 2017;551:648–652. doi: 10.1038/nature24661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wlodarska M, et al. Indoleacrylic acid produced by commensal Peptostreptococcus species suppresses inflammation. Cell Host Microbe. 2017;22:25–37 e26. doi: 10.1016/j.chom.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Venkatesh M, et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity. 2014;41:296–310. doi: 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hubbard TD, et al. Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci. Rep. 2015;5:12689. doi: 10.1038/srep12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zelante T, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 54.Zeisel SH, Warrier M. Trimethylamine N-oxide, the microbiome, and heart and kidney disease. Annu. Rev. Nutr. 2017;37:157–181. doi: 10.1146/annurev-nutr-071816-064732. [DOI] [PubMed] [Google Scholar]
- 55.Craciun S, Balskus EP. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc. Natl Acad. Sci. USA. 2012;109:21307–21312. doi: 10.1073/pnas.1215689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu Y, et al. Carnitine metabolism to trimethylamine by an unusual Rieske-type oxygenase from human microbiota. Proc. Natl Acad. Sci. USA. 2014;111:4268–4273. doi: 10.1073/pnas.1316569111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koeth RA, et al. gamma-Butyrobetaine is a proatherogenic intermediate in gut microbial metabolism of L-carnitine to TMAO. Cell Metab. 2014;20:799–812. doi: 10.1016/j.cmet.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bennett BJ, et al. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab. 2013;17:49–60. doi: 10.1016/j.cmet.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wallrabenstein I, et al. Human trace amine-associated receptor TAAR5 can be activated by trimethylamine. PLoS ONE. 2013;8:e54950. doi: 10.1371/journal.pone.0054950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen S, et al. Trimethylamine N-oxide binds and activates PERK to promote metabolic dysfunction. Cell Metab. 2019;30:1141–1151.e1145. doi: 10.1016/j.cmet.2019.08.021. [DOI] [PubMed] [Google Scholar]
- 61.Long SL, Gahan CGM, Joyce SA. Interactions between gut bacteria and bile in health and disease. Mol. Asp. Med. 2017;56:54–65. doi: 10.1016/j.mam.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 62.Hofmann AF. Biliary secretion and excretion in health and disease: current concepts. Ann. Hepatol. 2007;6:15–27. [PubMed] [Google Scholar]
- 63.Stellwag EJ, Hylemon PB. 7alpha-Dehydroxylation of cholic acid and chenodeoxycholic acid by Clostridium leptum. J. lipid Res. 1979;20:325–333. [PubMed] [Google Scholar]
- 64.Gerard P. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens. 2013;3:14–24. doi: 10.3390/pathogens3010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Forman BM, et al. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell. 1995;81:687–693. doi: 10.1016/0092-8674(95)90530-8. [DOI] [PubMed] [Google Scholar]
- 66.Wang H, Chen J, Hollister K, Sowers LC, Forman BM. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- 67.Bertilsson G, et al. Identification of a human nuclear receptor defines a new signaling pathway for CYP3A induction. Proc. Natl Acad. Sci. USA. 1998;95:12208–12213. doi: 10.1073/pnas.95.21.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xie W, et al. An essential role for nuclear receptors SXR/PXR in detoxification of cholestatic bile acids. Proc. Natl Acad. Sci. USA. 2001;98:3375–3380. doi: 10.1073/pnas.051014398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Makishima M, et al. Vitamin D receptor as an intestinal bile acid sensor. Science. 2002;296:1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- 70.Zhang J, Huang W, Qatanani M, Evans RM, Moore DD. The constitutive androstane receptor and pregnane X receptor function coordinately to prevent bile acid-induced hepatotoxicity. J. Biol. Chem. 2004;279:49517–49522. doi: 10.1074/jbc.M409041200. [DOI] [PubMed] [Google Scholar]
- 71.Kawamata Y, et al. A G protein-coupled receptor responsive to bile acids. J. Biol. Chem. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- 72.Harper AE, Miller RH, Block KP. Branched-chain amino acid metabolism. Annu. Rev. Nutr. 1984;4:409–454. doi: 10.1146/annurev.nu.04.070184.002205. [DOI] [PubMed] [Google Scholar]
- 73.Amorim Franco TM, Blanchard JS. Bacterial branched-chain amino acid biosynthesis: structures, mechanisms, and drugability. Biochemistry. 2017;56:5849–5865. doi: 10.1021/acs.biochem.7b00849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ikeda K, et al. Slc3a2 mediates branched-chain amino-acid-dependent maintenance of regulatory T cells. Cell Rep. 2017;21:1824–1838. doi: 10.1016/j.celrep.2017.10.082. [DOI] [PubMed] [Google Scholar]
- 75.Sanchez-Jimenez F, Medina MA, Villalobos-Rueda L, Urdiales JL. Polyamines in mammalian pathophysiology. Cell. Mol. life Sci. 2019;76:3987–4008. doi: 10.1007/s00018-019-03196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Di Martino ML, et al. Polyamines: emerging players in bacteria-host interactions. Int. J. Med. Microbiol. 2013;303:484–491. doi: 10.1016/j.ijmm.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 77.Milovic V. Polyamines in the gut lumen: bioavailability and biodistribution. Eur. J. Gastroenterol. Hepatol. 2001;13:1021–1025. doi: 10.1097/00042737-200109000-00004. [DOI] [PubMed] [Google Scholar]
- 78.Matsumoto M, Benno Y. The relationship between microbiota and polyamine concentration in the human intestine: a pilot study. Microbiol. Immunol. 2007;51:25–35. doi: 10.1111/j.1348-0421.2007.tb03887.x. [DOI] [PubMed] [Google Scholar]
- 79.Noack J, Dongowski G, Hartmann L, Blaut M. The human gut bacteria Bacteroides thetaiotaomicron and Fusobacterium varium produce putrescine and spermidine in cecum of pectin-fed gnotobiotic rats. J. Nutr. 2000;130:1225–1231. doi: 10.1093/jn/130.5.1225. [DOI] [PubMed] [Google Scholar]
- 80.Noack J, Kleessen B, Proll J, Dongowski G, Blaut M. Dietary guar gum and pectin stimulate intestinal microbial polyamine synthesis in rats. J. Nutr. 1998;128:1385–1391. doi: 10.1093/jn/128.8.1385. [DOI] [PubMed] [Google Scholar]
- 81.Zhang L, et al. Spermine potentiation of recombinant N-methyl-D-aspartate receptors is affected by subunit composition. Proc. Natl Acad. Sci. USA. 1994;91:10883–10887. doi: 10.1073/pnas.91.23.10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hofer AM, Brown EM. Extracellular calcium sensing and signalling. Nat. Rev. Mol. Cell Biol. 2003;4:530–538. doi: 10.1038/nrm1154. [DOI] [PubMed] [Google Scholar]
- 83.Said HM. Intestinal absorption of water-soluble vitamins in health and disease. Biochem. J. 2011;437:357–372. doi: 10.1042/BJ20110326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Magnusdottir S, Ravcheev D, de Crecy-Lagard V, Thiele I. Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front. Genet. 2015;6:148. doi: 10.3389/fgene.2015.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hill MJ. Intestinal flora and endogenous vitamin synthesis. Eur. J. Cancer Prev. 1997;6:S43–S45. doi: 10.1097/00008469-199703001-00009. [DOI] [PubMed] [Google Scholar]
- 86.Morishita T, Tamura N, Makino T, Kudo S. Production of menaquinones by lactic acid bacteria. J. Dairy Sci. 1999;82:1897–1903. doi: 10.3168/jds.S0022-0302(99)75424-X. [DOI] [PubMed] [Google Scholar]
- 87.Ramotar K, Conly JM, Chubb H, Louie TJ. Production of menaquinones by intestinal anaerobes. J. Infect. Dis. 1984;150:213–218. doi: 10.1093/infdis/150.2.213. [DOI] [PubMed] [Google Scholar]
- 88.Chang YL, et al. A screen of Crohn’s disease-associated microbial metabolites identifies ascorbate as a novel metabolic inhibitor of activated human T cells. Mucosal Immunol. 2019;12:457–467. doi: 10.1038/s41385-018-0022-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Heinonen KM, Perreault C. Development and functional properties of thymic and extrathymic T lymphocytes. Crit. Rev. Immunol. 2008;28:441–466. doi: 10.1615/critrevimmunol.v28.i5.40. [DOI] [PubMed] [Google Scholar]
- 90.Wan YY. Multi-tasking of helper T cells. Immunology. 2010;130:166–171. doi: 10.1111/j.1365-2567.2010.03289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rossjohn J, Pellicci DG, Patel O, Gapin L, Godfrey DI. Recognition of CD1d-restricted antigens by natural killer T cells. Nat. Rev. Immunol. 2012;12:845–857. doi: 10.1038/nri3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Koay HF, Godfrey DI, Pellicci DG. Development of mucosal-associated invariant T cells. Immunol. cell Biol. 2018;96:598–606. doi: 10.1111/imcb.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 95.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Park J, et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 2015;8:80–93. doi: 10.1038/mi.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kespohl M, et al. The microbial metabolite butyrate induces expression of Th1-associated factors in CD4(+) T cells. Front. Immunol. 2017;8:1036. doi: 10.3389/fimmu.2017.01036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pols TW, et al. TGR5 activation inhibits atherosclerosis by reducing macrophage inflammation and lipid loading. Cell Metab. 2011;14:747–757. doi: 10.1016/j.cmet.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hang S, et al. Author Correction: Bile acid metabolites control TH17 and Treg cell differentiation. Nature. 2020;579:E7. doi: 10.1038/s41586-020-2030-5. [DOI] [PubMed] [Google Scholar]
- 100.Song X, et al. Microbial bile acid metabolites modulate gut RORgamma(+) regulatory T cell homeostasis. Nature. 2020;577:410–415. doi: 10.1038/s41586-019-1865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wheeler MA, Rothhammer V, Quintana FJ. Control of immune-mediated pathology via the aryl hydrocarbon receptor. J. Biol. Chem. 2017;292:12383–12389. doi: 10.1074/jbc.R116.767723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Singh NP, et al. Dietary indoles suppress delayed-type hypersensitivity by inducing a switch from proinflammatory Th17 cells to anti-inflammatory regulatory T cells through regulation of microRNA. J. Immunol. 2016;196:1108–1122. doi: 10.4049/jimmunol.1501727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rouse M, Singh NP, Nagarkatti PS, Nagarkatti M. Indoles mitigate the development of experimental autoimmune encephalomyelitis by induction of reciprocal differentiation of regulatory T cells and Th17 cells. Br. J. Pharmacol. 2013;169:1305–1321. doi: 10.1111/bph.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen L, et al. Microbiota metabolite butyrate differentially regulates Th1 and Th17 cells’ differentiation and function in induction of colitis. Inflamm. Bowel Dis. 2019;25:1450–1461. doi: 10.1093/ibd/izz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vieira RS, et al. Butyrate attenuates lung inflammation by negatively modulating Th9 cells. Front. Immunol. 2019;10:67. doi: 10.3389/fimmu.2019.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wilck N, et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature. 2017;551:585–589. doi: 10.1038/nature24628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hasko G, et al. Spermine differentially regulates the production of interleukin-12 p40 and interleukin-10 and suppresses the release of the T helper 1 cytokine interferon-gamma. Shock. 2000;14:144–149. doi: 10.1097/00024382-200014020-00012. [DOI] [PubMed] [Google Scholar]
- 108.Jankovic D, Kugler DG, Sher A. IL-10 production by CD4+ effector T cells: a mechanism for self-regulation. Mucosal Immunol. 2010;3:239–246. doi: 10.1038/mi.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Luu M, et al. The short-chain fatty acid pentanoate suppresses autoimmunity by modulating the metabolic-epigenetic crosstalk in lymphocytes. Nat. Commun. 2019;10:760. doi: 10.1038/s41467-019-08711-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Balmer ML, et al. Memory CD8(+) T cells require increased concentrations of acetate induced by stress for optimal function. Immunity. 2016;44:1312–1324. doi: 10.1016/j.immuni.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 111.Bachem A, et al. Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8(+) T cells. Immunity. 2019;51:285–297 e285. doi: 10.1016/j.immuni.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 112.Trompette A, et al. Dietary fiber confers protection against flu by shaping Ly6c(−) patrolling monocyte hematopoiesis and CD8(+) T cell metabolism. Immunity. 2018;48:992–1005 e1008. doi: 10.1016/j.immuni.2018.04.022. [DOI] [PubMed] [Google Scholar]
- 113.Luu M, et al. Regulation of the effector function of CD8(+) T cells by gut microbiota-derived metabolite butyrate. Sci. Rep. 2018;8:14430. doi: 10.1038/s41598-018-32860-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ma C, et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science. 2018;360:eaan5931. doi: 10.1126/science.aan5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Le Bourhis L, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat. Immunol. 2010;11:701–708. doi: 10.1038/ni.1890. [DOI] [PubMed] [Google Scholar]
- 116.Gold MC, et al. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. 2010;8:e1000407. doi: 10.1371/journal.pbio.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Corbett AJ, et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 2014;509:361–365. doi: 10.1038/nature13160. [DOI] [PubMed] [Google Scholar]
- 118.Kjer-Nielsen L, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012;491:717–723. doi: 10.1038/nature11605. [DOI] [PubMed] [Google Scholar]
- 119.Cervantes-Barragan L, et al. Lactobacillus reuteri induces gut intraepithelial CD4(+)CD8alphaalpha(+) T cells. Science. 2017;357:806–810. doi: 10.1126/science.aah5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jellusova J. Metabolic control of B cell immune responses. Curr. Opin. Immunol. 2020;63:21–28. doi: 10.1016/j.coi.2019.11.002. [DOI] [PubMed] [Google Scholar]
- 121.Blair D, Dufort FJ, Chiles TC. Protein kinase Cβ is critical for the metabolic switch to glycolysis following B-cell antigen receptor engagement. Biochem. J. 2012;448:165–169. doi: 10.1042/BJ20121225. [DOI] [PubMed] [Google Scholar]
- 122.Kim M, Qie Y, Park J, Kim Chang H. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe. 2016;20:202–214. doi: 10.1016/j.chom.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sanchez HN, et al. B cell-intrinsic epigenetic modulation of antibody responses by dietary fiber-derived short-chain fatty acids. Nat. Commun. 2020;11:60. doi: 10.1038/s41467-019-13603-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lycke NY, Bemark M. The role of Peyer’s patches in synchronizing gut IgA responses. Front. Immunol. 2012;3:329. doi: 10.3389/fimmu.2012.00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Reboldi A, et al. IgA production requires B cell interaction with subepithelial dendritic cells in Peyer’s patches. Science. 2016;352:aaf4822. doi: 10.1126/science.aaf4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tan J, et al. Dietary fiber and bacterial SCFA enhance oral tolerance and protect against food allergy through diverse cellular pathways. Cell Rep. 2016;15:2809–2824. doi: 10.1016/j.celrep.2016.05.047. [DOI] [PubMed] [Google Scholar]
- 127.Feng T, Cong Y, Qin H, Benveniste EN, Elson CO. Generation of mucosal dendritic cells from bone marrow reveals a critical role of retinoic acid. J. Immunol. 2010;185:5915–5925. doi: 10.4049/jimmunol.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mora JR, et al. Generation of gut-homing IgA-secreting B cells by intestinal dendritic cells. Science. 2006;314:1157–1160. doi: 10.1126/science.1132742. [DOI] [PubMed] [Google Scholar]
- 129.Wu W, et al. Microbiota metabolite short-chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal Immunol. 2017;10:946–956. doi: 10.1038/mi.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang W, et al. Microbiota metabolite short-chain fatty acids facilitate mucosal adjuvant activity of cholera toxin through GPR43. J. Immunol. 2019;203:282–292. doi: 10.4049/jimmunol.1801068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Laidlaw BJ, et al. Interleukin-10 from CD4+ follicular regulatory T cells promotes the germinal center response. Sci. Immunol. 2017;2:eaan4767. doi: 10.1126/sciimmunol.aan4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Itoh K, Hirohata S. The role of IL-10 in human B cell activation, proliferation, and differentiation. J. Immunol. 1995;154:4341–4350. [PubMed] [Google Scholar]
- 133.Park J, et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR–S6K pathway. Mucosal Immunol. 2015;8:80–93. doi: 10.1038/mi.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Singh N, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–139. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl Acad. Sci. USA. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Li J, et al. Aryl hydrocarbon receptor activation suppresses EBF1 and PAX5 and impairs human B lymphopoiesis. J. Immunol. 2017;199:3504–3515. doi: 10.4049/jimmunol.1700289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Vaidyanathan B, et al. The aryl hydrocarbon receptor controls cell-fate decisions in B cells. J. Exp. Med. 2017;214:197–208. doi: 10.1084/jem.20160789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Piper CJM, et al. Aryl hydrocarbon receptor contributes to the transcriptional program of IL-10-producing regulatory B cells. Cell Rep. 2019;29:1878–1892.e1877. doi: 10.1016/j.celrep.2019.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rosser EC, et al. Microbiota-derived metabolites suppress arthritis by amplifying aryl-hydrocarbon receptor activation in regulatory B cells. Cell Metab. 2020;31:837–851 e810. doi: 10.1016/j.cmet.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Singh N, et al. Blockade of dendritic cell development by bacterial fermentation products butyrate and propionate through a transporter (Slc5a8)-dependent inhibition of histone deacetylases. J. Biol. Chem. 2010;285:27601–27608. doi: 10.1074/jbc.M110.102947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Berndt BE, et al. Butyrate increases IL-23 production by stimulated dendritic cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2012;303:G1384–G1392. doi: 10.1152/ajpgi.00540.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Andrade-Oliveira V, et al. Gut bacteria products prevent AKI induced by ischemia-reperfusion. J. Am. Soc. Nephrol. 2015;26:1877–1888. doi: 10.1681/ASN.2014030288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liu L, et al. Butyrate interferes with the differentiation and function of human monocyte-derived dendritic cells. Cell. Immunol. 2012;277:66–73. doi: 10.1016/j.cellimm.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 144.Nastasi C, et al. The effect of short-chain fatty acids on human monocyte-derived dendritic cells. Sci. Rep. 2015;5:16148. doi: 10.1038/srep16148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gurav A, Sivaprakasam S, Bhutia YD, Boettger T, Singh N. Ganapathy V. Slc5a8, a Na+-coupled high-affinity transporter for short-chain fatty acids, is a conditional tumour suppressor in colon that protects against colitis and colon cancer under low-fibre dietary conditions. Biochem. J. 2015;469:267–278. doi: 10.1042/BJ20150242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Trompette A, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 147.Ichikawa R, et al. Bile acids induce monocyte differentiation toward interleukin-12 hypo-producing dendritic cells via a TGR5-dependent pathway. Immunology. 2012;136:153–162. doi: 10.1111/j.1365-2567.2012.03554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sugawara A, et al. Polyamine compound deoxyspergualin inhibits heat shock protein-induced activation of immature dendritic cells. Cell Stress Chaperones. 2009;14:133–139. doi: 10.1007/s12192-008-0064-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gervais A, et al. Dendritic cells are defective in breast cancer patients: a potential role for polyamine in this immunodeficiency. Breast Cancer Res. 2005;7:R326–R335. doi: 10.1186/bcr1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Liu T, et al. Short-chain fatty acids suppress lipopolysaccharide-induced production of nitric oxide and proinflammatory cytokines through inhibition of NF-kappaB pathway in RAW264.7 cells. Inflammation. 2012;35:1676–1684. doi: 10.1007/s10753-012-9484-z. [DOI] [PubMed] [Google Scholar]
- 151.Schulthess J, et al. The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity. 2019;50:432–445.e437. doi: 10.1016/j.immuni.2018.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ljubuncic P, Fuhrman B, Oiknine J, Aviram M, Bomzon A. Effect of deoxycholic acid and ursodeoxycholic acid on lipid peroxidation in cultured macrophages. Gut. 1996;39:475–478. doi: 10.1136/gut.39.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Haselow K, et al. Bile acids PKA-dependently induce a switch of the IL-10/IL-12 ratio and reduce proinflammatory capability of human macrophages. J. Leukoc. Biol. 2013;94:1253–1264. doi: 10.1189/jlb.0812396. [DOI] [PubMed] [Google Scholar]
- 154.Wu K, et al. The gut microbial metabolite trimethylamine N-oxide aggravates GVHD by inducing M1 macrophage polarization in mice. Blood. 2020;136:501–515. doi: 10.1182/blood.2019003990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lee JH, et al. Anti-inflammatory and anti-genotoxic activity of branched chain amino acids (BCAA) in lipopolysaccharide (LPS) stimulated RAW 264.7 macrophages. Food Sci. Biotechnol. 2017;26:1371–1377. doi: 10.1007/s10068-017-0165-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bjerrum JT, et al. Metabonomics of human fecal extracts characterize ulcerative colitis, Crohn’s disease and healthy individuals. Metabolomics. 2015;11:122–133. doi: 10.1007/s11306-014-0677-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Marchesi JR, et al. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J. Proteome Res. 2007;6:546–551. doi: 10.1021/pr060470d. [DOI] [PubMed] [Google Scholar]
- 158.Lloyd-Price J, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Franzosa EA, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat. Microbiol. 2019;4:293–305. doi: 10.1038/s41564-018-0306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Le Gall G, et al. Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J. Proteome Res. 2011;10:4208–4218. doi: 10.1021/pr2003598. [DOI] [PubMed] [Google Scholar]
- 161.Nikolaus S, et al. Increased tryptophan metabolism is associated with activity of inflammatory bowel diseases. Gastroenterology. 2017;153:1504–1516 e1502. doi: 10.1053/j.gastro.2017.08.028. [DOI] [PubMed] [Google Scholar]
- 162.Maslowski KM, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Agus A, et al. Western diet induces a shift in microbiota composition enhancing susceptibility to adherent-invasive E. coli infection and intestinal inflammation. Sci. Rep. 2016;6:19032. doi: 10.1038/srep19032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Patz J, Jacobsohn WZ, Gottschalk-Sabag S, Zeides S, Braverman DZ. Treatment of refractory distal ulcerative colitis with short chain fatty acid enemas. Am. J. Gastroenterol. 1996;91:731–734. [PubMed] [Google Scholar]
- 165.Breuer RI, et al. Rectal irrigation with short-chain fatty acids for distal ulcerative colitis. Preliminary report. Digestive Dis. Sci. 1991;36:185–187. doi: 10.1007/BF01300754. [DOI] [PubMed] [Google Scholar]
- 166.Steinhart AH, Hiruki T, Brzezinski A, Baker JP. Treatment of left-sided ulcerative colitis with butyrate enemas: a controlled trial. Aliment. Pharmacol. Therap. 1996;10:729–736. doi: 10.1046/j.1365-2036.1996.d01-509.x. [DOI] [PubMed] [Google Scholar]
- 167.Breuer RI, et al. Short chain fatty acid rectal irrigation for left-sided ulcerative colitis: a randomised, placebo controlled trial. Gut. 1997;40:485–491. doi: 10.1136/gut.40.4.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lamas B, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016;22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Alexeev EE, et al. Microbiota-derived indole metabolites promote human and murine intestinal homeostasis through regulation of interleukin-10 receptor. Am. J. Pathol. 2018;188:1183–1194. doi: 10.1016/j.ajpath.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Monteleone I, et al. Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology. 2011;141:237–248. doi: 10.1053/j.gastro.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 171.Qiu J, et al. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36:92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S. The bile acid receptor FXR is a modulator of intestinal innate immunity. J. Immunol. 2009;183:6251–6261. doi: 10.4049/jimmunol.0803978. [DOI] [PubMed] [Google Scholar]
- 173.Velmurugan G, Ramprasath T, Gilles M, Swaminathan K, Ramasamy S. Gut microbiota, endocrine-disrupting chemicals, and the diabetes epidemic. Trends Endocrinol. Metab. 2017;28:612–625. doi: 10.1016/j.tem.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 174.Boerner BP, Sarvetnick NE. Type 1 diabetes: role of intestinal microbiome in humans and mice. Ann. N. Y. Acad. Sci. 2011;1243:103–118. doi: 10.1111/j.1749-6632.2011.06340.x. [DOI] [PubMed] [Google Scholar]
- 175.Karlsson FH, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 176.Marino E, et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat. Immunol. 2017;18:552–562. doi: 10.1038/ni.3713. [DOI] [PubMed] [Google Scholar]
- 177.De Vadder F, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156:84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 178.Lin HV, et al. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS ONE. 2012;7:e35240. doi: 10.1371/journal.pone.0035240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gao Z, et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009;58:1509–1517. doi: 10.2337/db08-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kimura I, et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat. Commun. 2013;4:1829. doi: 10.1038/ncomms2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Dambrova M, et al. Diabetes is associated with higher trimethylamine N-oxide plasma levels. Exp. Clin. Endocrinol. Diabetes. 2016;124:251–256. doi: 10.1055/s-0035-1569330. [DOI] [PubMed] [Google Scholar]
- 182.Schugar RC, et al. The TMAO-producing enzyme flavin-containing monooxygenase 3 regulates obesity and the beiging of white adipose tissue. Cell Rep. 2017;19:2451–2461. doi: 10.1016/j.celrep.2017.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Shan Z, et al. Association between microbiota-dependent metabolite trimethylamine-N-oxide and type 2 diabetes. Am. J. Clin. Nutr. 2017;106:888–894. doi: 10.3945/ajcn.117.157107. [DOI] [PubMed] [Google Scholar]
- 184.Tang WH, et al. Increased trimethylamine N-oxide portends high mortality risk independent of glycemic control in patients with type 2 diabetes mellitus. Clin. Chem. 2017;63:297–306. doi: 10.1373/clinchem.2016.263640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Felig P, Wahren J, Sherwin R, Palaiologos G. Amino acid and protein metabolism in diabetes mellitus. Arch. Intern. Med. 1977;137:507–513. [PubMed] [Google Scholar]
- 186.Nakamura H, et al. Plasma amino acid profiles are associated with insulin, C-peptide and adiponectin levels in type 2 diabetic patients. Nutr. Diabetes. 2014;4:e133. doi: 10.1038/nutd.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Batch BC, et al. Branched chain amino acids are novel biomarkers for discrimination of metabolic wellness. Metab. Clin. Exp. 2013;62:961–969. doi: 10.1016/j.metabol.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Newgard CB, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Xiao F, et al. Effects of individual branched-chain amino acids deprivation on insulin sensitivity and glucose metabolism in mice. Metab. Clin. Exp. 2014;63:841–850. doi: 10.1016/j.metabol.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 190.Tuomainen M, et al. Associations of serum indolepropionic acid, a gut microbiota metabolite, with type 2 diabetes and low-grade inflammation in high-risk individuals. Nutr. Diabetes. 2018;8:35. doi: 10.1038/s41387-018-0046-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Deane KD, et al. Genetic and environmental risk factors for rheumatoid arthritis. Best Pract. Res. Clin. Rheumatol. 2017;31:3–18. doi: 10.1016/j.berh.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wang Q, Xu R. Data-driven multiple-level analysis of gut-microbiome-immune-joint interactions in rheumatoid arthritis. BMC Genomics. 2019;20:124. doi: 10.1186/s12864-019-5510-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Mizuno M, Noto D, Kaga N, Chiba A, Miyake S. The dual role of short fatty acid chains in the pathogenesis of autoimmune disease models. PLoS ONE. 2017;12:e0173032. doi: 10.1371/journal.pone.0173032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Saouaf SJ, et al. Deacetylase inhibition increases regulatory T cell function and decreases incidence and severity of collagen-induced arthritis. Exp. Mol. Pathol. 2009;87:99–104. doi: 10.1016/j.yexmp.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Bruusgaard A, Andersen RB. Abnormal bile acid metabolism in rheumatoid arthritis. Preliminary communication. Dan. Med. Bull. 1976;23:95–98. [PubMed] [Google Scholar]
- 196.Li ZY, Zhou JJ, Luo CL, Zhang LM. Activation of TGR5 alleviates inflammation in rheumatoid arthritis peripheral blood mononuclear cells and in mice with collagen IIinduced arthritis. Mol. Med. Rep. 2019;20:4540–4550. doi: 10.3892/mmr.2019.10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hasan H, Ismail H, El-Orfali Y, Khawaja G. Therapeutic benefits of Indole-3-Carbinol in adjuvant-induced arthritis and its protective effect against methotrexate induced-hepatic toxicity. BMC Complem. Alternat. Med. 2018;18:337. doi: 10.1186/s12906-018-2408-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Mu Q, et al. Control of lupus nephritis by changes of gut microbiota. Microbiome. 2017;5:73. doi: 10.1186/s40168-017-0300-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Zegarra-Ruiz DF, et al. A diet-sensitive commensal Lactobacillus strain mediates TLR7-dependent systemic autoimmunity. Cell Host microbe. 2019;25:113–127.e116. doi: 10.1016/j.chom.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Rodriguez-Carrio J, et al. Intestinal dysbiosis is associated with altered short-chain fatty acids and serum-free fatty acids in systemic lupus erythematosus. Front. Immunol. 2017;8:23. doi: 10.3389/fimmu.2017.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kim HA, et al. Polyamine patterns in plasma of patients with systemic lupus erythematosus and fever. Lupus. 2018;27:930–938. doi: 10.1177/0961203317751860. [DOI] [PubMed] [Google Scholar]
- 202.Gunnia UB, Amenta PS, Seibold JR, Thomas TJ. Successful treatment of lupus nephritis in MRL-lpr/lpr mice by inhibiting ornithine decarboxylase. Kidney Int. 1991;39:882–890. doi: 10.1038/ki.1991.111. [DOI] [PubMed] [Google Scholar]