Abstract
Vascular invasion is a critical risk factor for hepatocellular carcinoma (HCC) recurrence and poor survival. The molecular drivers of vascular invasion in HCC are largely unknown. Deciphering the molecular landscape of invasive HCC will help identify novel therapeutic targets and noninvasive biomarkers.
To this end, we undertook this study to evaluate the genomic, transcriptomic, and proteomic profile of tumors with vascular invasion using the multi-platform cancer genome atlas (TCGA) data (n=373). In the TCGA liver hepatocellular carcinoma (LIHC) cohort, macrovascular invasion was present in 5% (n=17) of tumors and microvascular invasion in 25% (n=94) of tumors. Functional pathway analysis revealed that the MYC oncogene was a common upstream regulator of the mRNA, miRNA and proteomic changes in vascular invasion. We performed comparative proteomic analyses of invasive human HCC and MYC driven murine HCC and identified fibronectin to be proteomic biomarker of invasive HCC (mouse Fn1 p= 1.7 × 10−11; human FN1 p=1.5 × 10−4) conserved across the two species. Mechanistically, we show that FN1 promotes the migratory and invasive phenotype of HCC cancer cells. We demonstrate tissue overexpression of fibronectin in human HCC using a large independent cohort of human HCC tissue microarray (n=153; p<0.001). Lastly, we showed that plasma fibronectin levels were significantly elevated in patients with HCC (n=35, mean=307.7 μg/ml, SEM=35.9) when compared to cirrhosis (n=10, mean=41.8 μg/ml, SEM=13.3; p<0.0001).
Conclusion:
Our study evaluates the molecular landscape of tumors with vascular invasion, identifying distinct transcriptional, epigenetic and proteomic changes driven by the MYC oncogene. We show that MYC upregulates fibronectin expression which promotes HCC invasiveness. In addition, we identify fibronectin to be a promising non-invasive proteomic biomarker of vascular invasion in HCC.
INTRODUCTION
Hepatocellular carcinoma (HCC) is an aggressive malignancy with very high mortality (1). The prognosis for advanced HCC is dismal with 5-year survival around 15%. For patients diagnosed with HCC at an early stage, surgeries like resection and transplantation offer the promise of cure. But unfortunately recurrence after surgery is common, with the five-year recurrence rate reaching up to 50% post-resection and 10–20% post-transplant (2). Therefore, there is a critical need to understand the mechanisms that drive recurrence and identify those at high risk for recurrence to prevent the morbidity and mortality associated with it.
Vascular invasion, which is present in 25–50% of HCC (3–5), is a major risk factor for recurrence and it is associated with poor overall survival in HCC (6–8). The existing plasma biomarkers like alpha-fetoprotein (AFP) and des-carboxy prothrombin (DCP) have low sensitivity and are inadequate in determining tumor invasiveness (9). To further complicate the problem, HCC is a unique malignancy in which a diagnosis can be established with high level of confidence by its characteristic radiologic features, foregoing the need for tissue biopsy. Thus, unfortunately, histologic information about the presence of vascular invasion is not always available pre-operatively, and selection of surgical patients often rely solely on radiologic tests for tumor size characteristics. Hence, there clearly is a large unmet need for identification of biomarkers for the detection of invasive HCC to pre-operatively risk stratify patients and optimize organ allocation. Moreover, discerning the molecular drivers of vascular invasion in HCC may help identify novel therapeutic targets and lead to improved patient outcomes.
Previous studies have evaluated clinical risk factors and gene signatures for vascular invasion in HCC (10)(11), but predictive biomarkers are not part of current clinical practice yet. Although molecular mechanisms of HCC progression have been extensively studied (12,13), the specific driver genes and molecular pathways associated with vascular invasion remain poorly defined. Hence, we undertook this study to comprehensively analyze the genomic and proteomic landscape of HCC with vascular invasion. We harness the multi-platform genomic data from TCGA study (14) to determine major pathways which are dysregulated in tumors with vascular invasion in HCC, and identify fibronectin as a potential non-invasive biomarker for invasive HCC.
RESULTS
Vascular invasion is a prognostic factor associated with recurrence in HCC
In the TCGA liver hepatocellular carcinoma (LIHC) cohort (n=373), vascular invasion (VI) was present in 34.9% tumors (n=111)- macrovascular invasion in 5.8% (n=17) and microvascular invasion in 29.1% (n=94) (Figure 1a–1b). The prevalence of vascular invasion was higher with more advanced tumor stage (14% in Stage I, 64% in stage II and 53% in stage III)(p=0.03) (Figure 1c), more advanced tumor grade (15% in low grade, 46% in moderate/high grade) (p=0.03), and higher serum alpha fetoprotein (p=0.02) (Figure 1c). Recurrence rates were higher in tumors with either micro- (64%) or macrovascular invasion (65%) compared to those without vascular invasion (45%) (p<0.001, Figure 1d). The presence of macrovascular invasion was associated with worse overall survival when compared to those without vascular invasion (3 year 44% vs. 73%, p=0.001) (Figure 1e). The presence of either micro- or macrovascular invasion was associated with significantly worse recurrence free survival than those without vascular invasion (24% vs. 33% vs. 47%) (p=0.001) (Figure 1e). Thus, we demonstrate that the presence of vascular invasion in HCC is strongly associated with aggressive tumor behavior and poor clinical outcomes.
Figure 1. Vascular invasion is a prognostic factor associated with recurrence in hepatocellular Carcinoma (HCC).
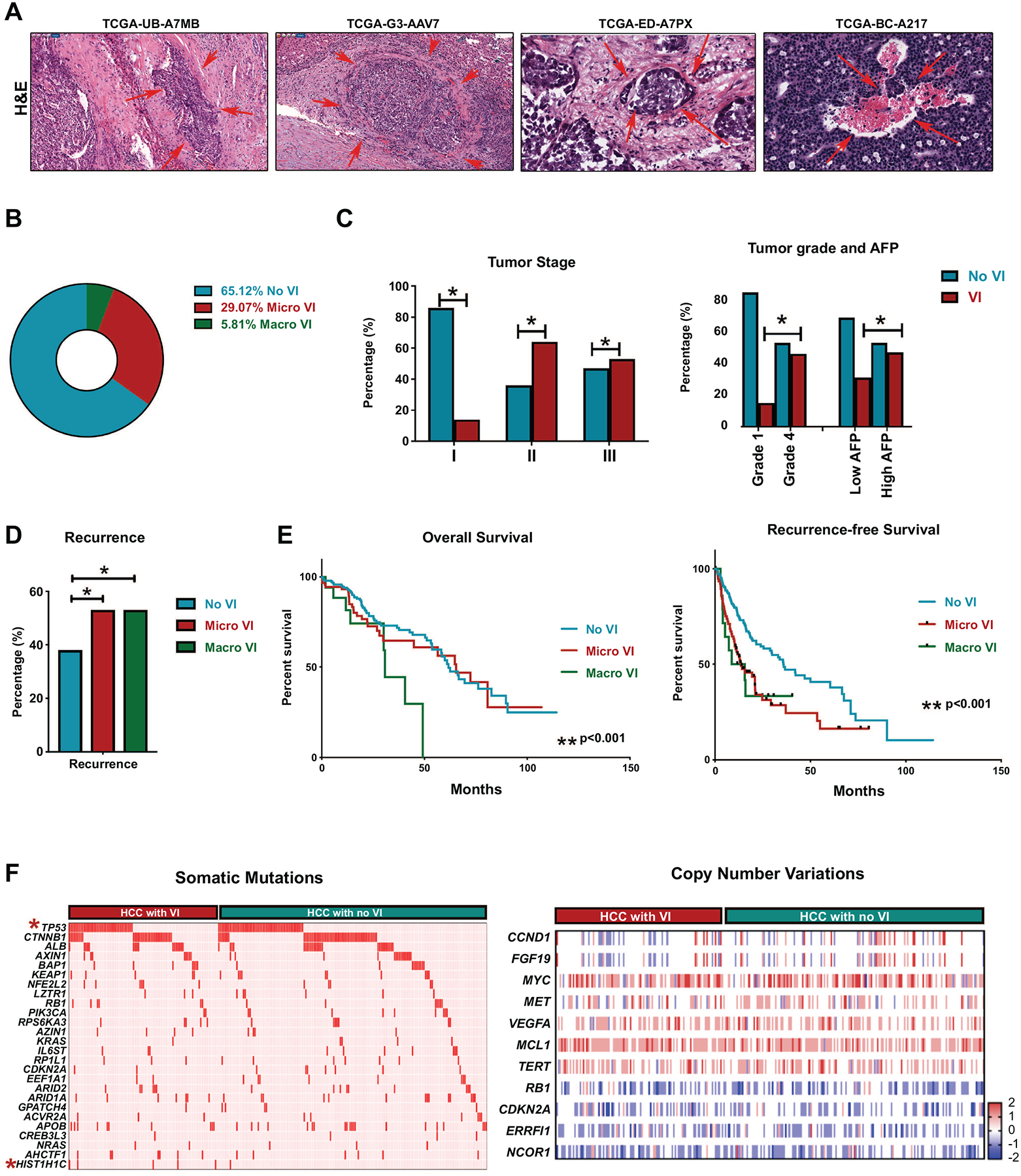
a. Representation H&E (10x) images from HCC tissue from TCGA dataset. Arrows denote the area of vascular invasion. Code represents the deidentified TCGA ID of the patient. b. Prevalence of vascular invasion in HCC TCGA cohort. c. Prevalence of vascular invasion in HCC stratified by tumor stage, tumor grade and AFP levels. d. Incidence of HCC recurrence stratified by micro or macrovascular invasion. e. Overall survival and recurrence free survival in patients stratified by presence of micro or macrovascular invasion. f. Incidence of major somatic mutations and major copy number variations stratified by presence of vascular invasion. Each row is a gene and each column is a patient. Star signifies statistically significant association.
To identify the genetic events that drive vascular invasion in human HCC we first compared the mutational profile of tumors with or without vascular invasion (Figure 1f). There was a higher incidence of TP53 mutation in tumors with vascular invasion (37.5%) than in those without (25%) (p=0.03). Mutation in HIST1H1C, a regulator of chromatin structure, was almost exclusively found in tumors with vascular invasion (4.8%) when compared to tumors without (0.5%) (p=0.01). The incidence of all other major somatic mutations was similar between the two groups (Supp Table 1). Also, we did not find any significant differences in major somatic copy number variations between tumors with or without vascular invasion (Figure 1f) (Supp Table 1). Thus, our data does not reveal a clear genotype-phenotype correlation in tumors with vascular invasion, prompting us to further study the transcriptomic and proteomic landscape.
Transcriptional landscape of hepatocellular carcinoma with vascular invasion
We analyzed the transcriptional profile of HCC to identify genes and pathways dysregulated in tumors with vascular invasion. We identified 102 genes which were differentially expressed in tumors with vascular invasion of which 92 genes were overexpressed and 10 genes were under-expressed (Figure 2a) (p<0.001, Fold change≥2, FDR<0.1) (Supp Table 2). Several genes like CDC20 (15), XPOT (16) and SIRT7 (17), which are part of this signature, have been previously implicated in the pathogenesis of HCC. Other genes like GTF2F2, ENGASE and RCCD1 have not been previously associated with HCC but have been implicated in other cancers. Upon comparison of the vascular invasion-related gene signature with multiple published prognostic gene signatures of HCC, we found enrichment of gene classes associated with poor outcome in HCC such as Hoshida subclass S1 (18), Boyault G3 subclass (19) and Woo liver cancer recurrence signature (20) (Figure 2b). Next, we performed upstream regulator pathway analysis to identify genetic drivers of the global transcriptional changes. We identified MYC (p=9.8 × 10−4), CREBZF (p=2.0 × 10−3), HOXD13 (4.2 × 10−3), ATF4 (5.0 × 10−3) and ZBTB17 (5.02 × 10−3) to be the most significant upstream regulators of the vascular invasion-related transcriptome (Figure 2c–2d). We further confirmed MYC is an important transcriptional regulator of this phenotype by demonstrating that the vascular invasion-related gene signature significantly overlapped with several other previously reported MYC gene signatures (Figure 2c).
Figure 2. Transcriptional landscape of hepatocellular carcinoma with vascular invasion.
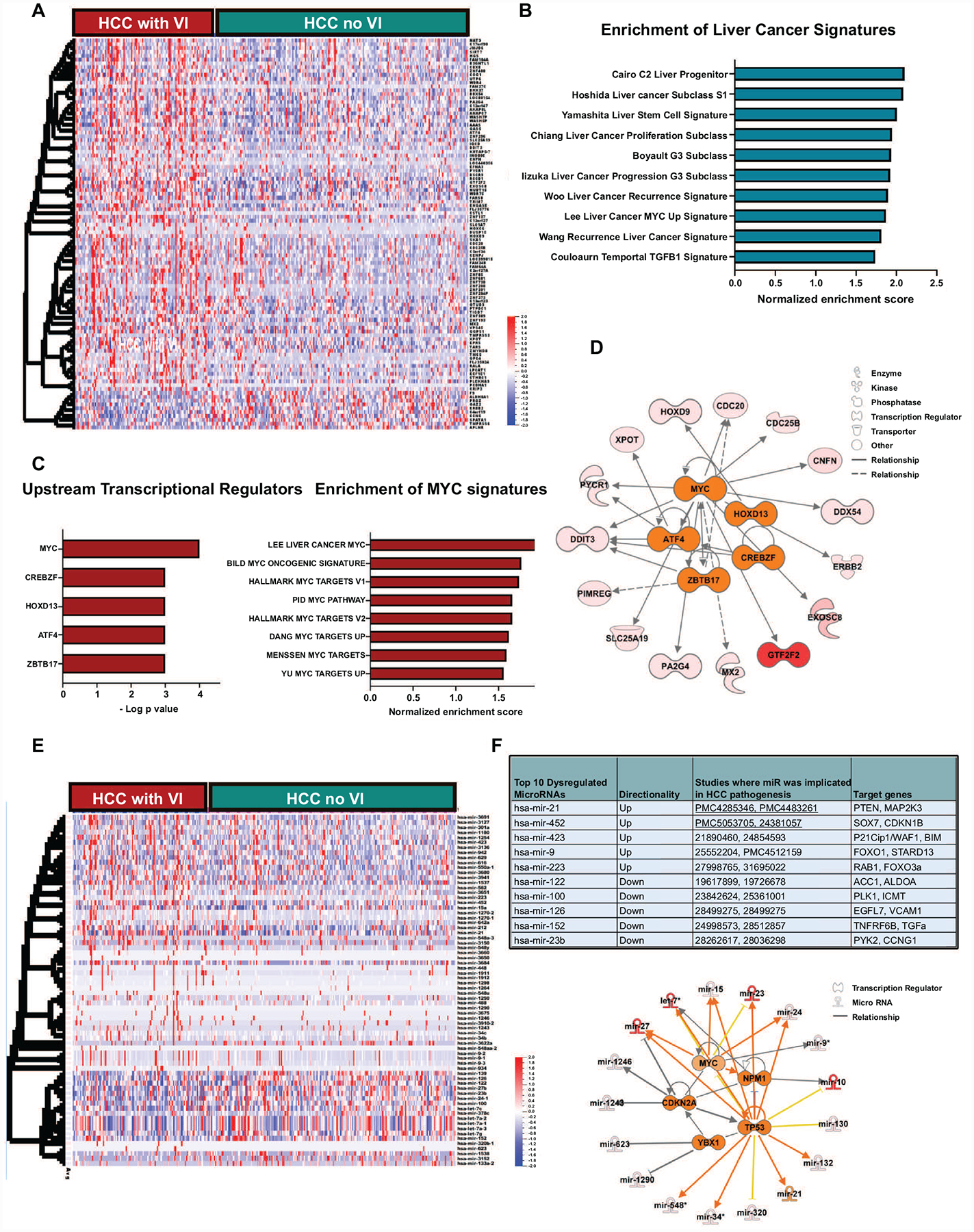
a. Heatmap shows the top differentially expressed genes between tumors with and without vascular invasion. b. Bar graph showing enrichment of major liver cancer signatures associated with poor prognosis in the tumors with vascular invasion. c. The top 5 upstream regulators of the transcriptional changes in tumors with vascular invasion.d. Bar graph showing enrichment of major MYC signatures associated in the tumors with vascular invasion. e. Network analysis displays the top transcriptional regulators and their associated network of genes. e. Heatmap shows the top differentially expressed microRNAs between tumors with and without vascular invasion. f. Top 10 microRNA differentially expressed between tumors with and without vascular invasion and analysis of their top transcriptional regulators and associated network of mRNAs.
We next identified 70 microRNAs which were differentially expressed in tumors with vascular invasion of which 23 were overexpressed and 47 were under-expressed (p<0.001, fold change≥2, FDR<0.1) (Figure 2e, Supp Table 2). The top 10 microRNAs and their target genes are shown in Figure 2f. Some of these microRNAs like mir-122, mir-21 and mir-100 have been previously implicated in HCC pathogenesis but not specifically in promoting vascular invasion (Figure 2f). Target genes of the upregulated microRNAs like PTEN, CDKN1B and CDKN1A are known tumor suppressors, while gene targets of the suppressed microRNAs, like TGFA, VCAM1 and ACC1, have known tumor promoting roles. These data thus demonstrate how the microRNAs can potentially enable the tumor to switch to a more invasive phenotype. We performed upstream regulator analysis and identified TP53 (p=1.4 × 10−10), MYC (3.4 × 10−5), CDKN2A (p=1.7 ×10−4), NPM1 (p=1.3 ×10−4), and YBX1 (p=9.7 ×10−4) to be the top five transcriptional regulators of the differentially expressed vascular invasion-related microRNAs (Figure 2f). Thus, our analysis of the transcriptional profile of tumors with vascular invasion reveals enrichment of genes and pathways associated with an aggressive phenotype and identifies, among others, the MYC oncogene as a common driver of vascular invasion in HCC.
Functional evaluation of the proteomic changes in HCC with vascular invasion
To understand the functional implications of the genomic and transcriptomic changes, we analyzed the proteomic landscape of tumors with vascular invasion given that proteins are ultimately the effectors of cellular function. We compared the expression of 219 proteins between tumors with and without vascular invasion and identified 87 proteins to be differentially expressed (Figure 3a) (n=130, p<0.001; FDR<0.1; Supp Table 3). The top 10 differentially expressed proteins in tumors with vascular invasion were noted to be involved in multiple cellular functions including cell adhesiveness, apoptosis and metabolism (Figure 3b). The top vascular invasion-related proteins like fibronectin, plasminogen activator inhibitor 1 and phospho PEA15 have been reported to promote cellular migration in other cancers, but not directly in vascular invasion in HCC (21–23).
Figure 3. Functional evaluation of the proteomic changes in HCC with vascular invasion.
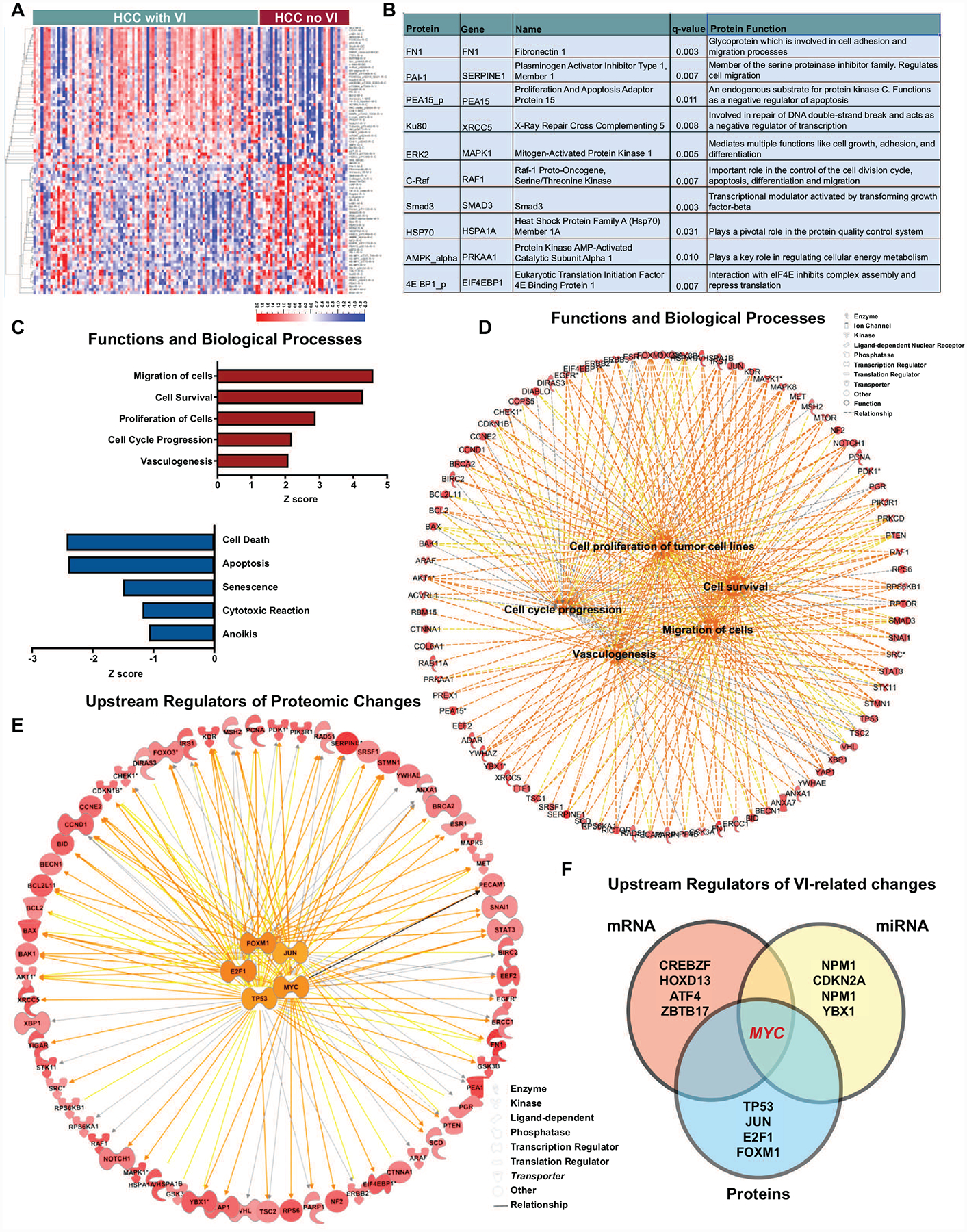
a. Heatmap shows the top differentially expressed proteins between tumors with and without vascular invasion. b. Top 10 proteins differentially expressed between tumors with and without vascular invasion. c. The top 5 upregulated and downregulated functions and biological processes in tumors with vascular invasion. d. Network analysis displays the top biological processes and their associated network of proteins. e. Network analysis displays the top transcriptional regulators and their associated network of proteins. f. Overlap of upstream regulators of vascular invasion-related mRNA, miRNA and proteomic changes.
Functional pathway analysis of the proteomic changes revealed that biological processes like cellular viability (p=6.4×10−39), cell migration (p=4.110−31), proliferation (p=1.5×10−49) and angiogenesis (p=3.5×10−14) were significantly upregulated in tumors with vascular invasion, while pathways involving apoptosis (3.1×10−54) and senescence (7.7×10−15) were significantly downregulated (Figure 3c–3d). We evaluated the upstream transcriptional regulators of the proteomic changes observed. The top 5 regulators were TP53 (p= 1.4×10−37), MYC (p=8.7×10−24), JUN (p=5.3×10−23), E2F1 (4.3×10−22) and FOXM1 (p=1.7×10−19) (Figure 3e). Meanwhile, tumor suppressor regulated pathways like RB1 pathway and VHL pathways were significantly downregulated in tumors with vascular invasion (Supp Fig 1). Thus, the functional analysis of the global proteomic changes associated with vascular invasion in HCC clearly portray features of an aggressive tumor phenotype.
Cross-species comparative proteomic profiling identifies fibronectin as a conserved biomarker of invasive HCC
Our data shows that MYC oncogene is a consistent upstream regulator of the vascular invasion-related mRNA, miRNA and proteomic changes in HCC (Figure 3f). This prompted us to further explore vascular invasion in HCC using the murine autochthonous transgenic model of MYC-driven HCC (LAP-tTA/tet-O-MYC) (Figure 4a). We found that vascular invasion was a highly prevalent feature of the murine MYC-HCC (75%, n=15/20 tumors) (Figure 4b). We evaluated the global proteomic changes associated with MYC-HCC using liquid chromatography–mass spectrometry (LC-MS). A total of 392 proteins were overexpressed and 360 proteins were under-expressed in HCC tumor tissue (n=3) when compared to control mouse liver (n=3) (P<0.05; FDR<0.05, fold change ±2.0) (Figure 4c). We confirmed that proteins like alpha fetoprotein (AFP), β-catenin and vimentin that are known to be elevated in human HCC were indeed higher in MYC-HCC, and proteins like carbamoyl-phosphate synthetase 1, acid phosphatase 1 and aldolase B which are suppressed in human HCC were lower in MYC-HCC (Figure 4d). The top upstream regulator of the proteomic changes was confirmed to be MYC (p=8.2 × 10−79) but other genetic drivers of human HCC like Trp53 (4.1 × 10−53) and β-catenin (4.2 × 10−23) were also found to be significant upstream regulators of the proteomic changes in MYC-HCC (Figure 4e). Biological functions like cancer cell migration (p=6.9 × 10−8) and invasiveness (1.4 × 10−7) were activated in murine MYC-HCC thus confirming these tumors have an invasive phenotype (Figure 4f). Overall, the above analysis demonstrates the robustness of the mouse model and its relevance to vascular invasion in human HCC.
Figure 4. Global proteomic profiling of murine MYC-driven HCC.
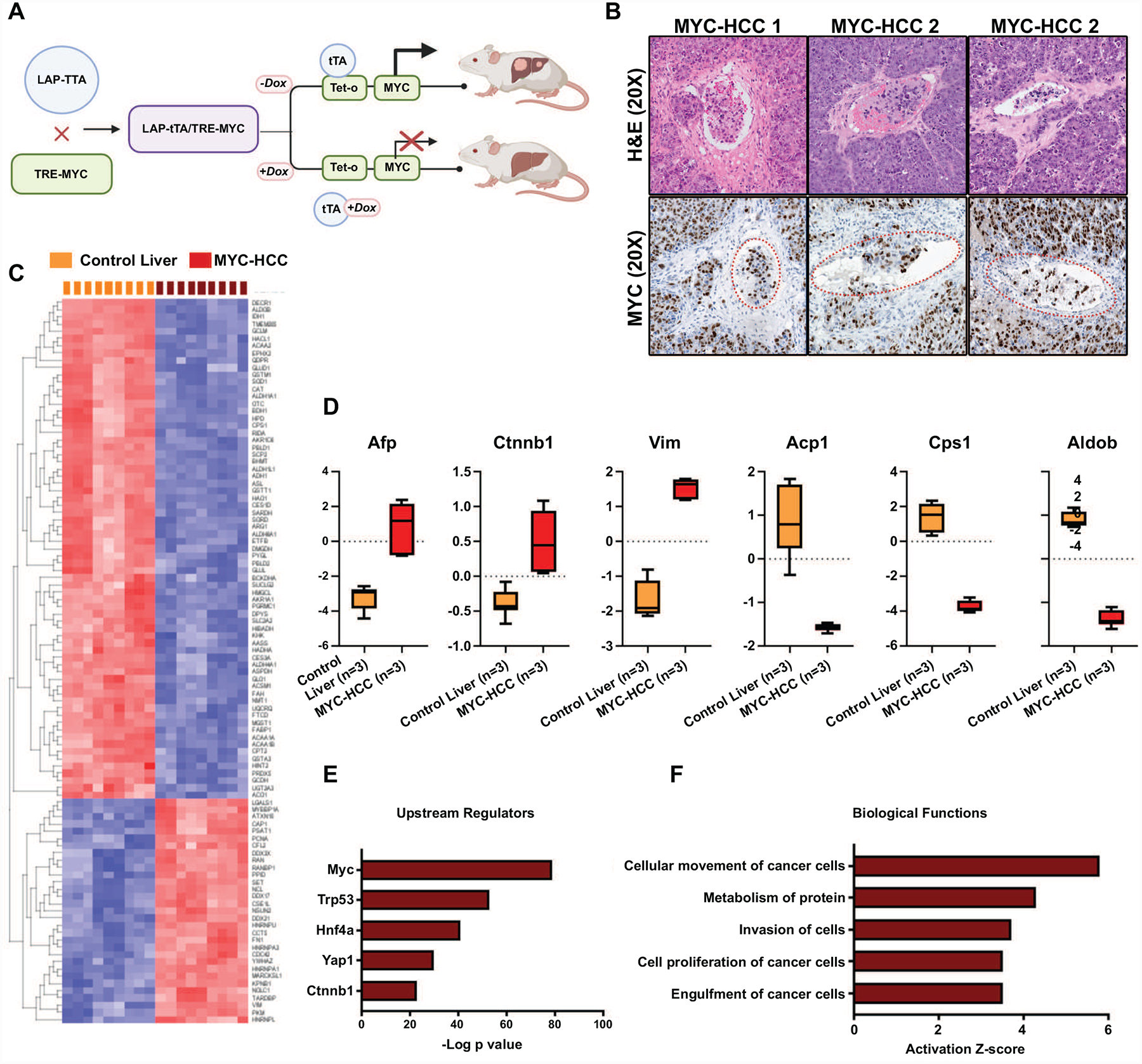
a. Schematic showing the mouse model of transgenic MYC-HCC. b. Representative H&E and IHC for MYC images from three MYC-HCC tumors demonstrating vascular invasion. c. Heatmap shows the top differentially expressed proteins between MYC-HCC and control liver tissue. d. Expression of known biomarkers of human HCC in murine MYC-driven HCC. e. Top upstream regulators of proteomic changes in MYC-HCC. f. Top biological processes that are dysregulated in the MYC-HCC.
We performed cross-species comparative analyses to identify conserved proteins that are enriched in both MYC-driven murine HCC (n=759) and human HCC with vascular invasion (n=87). We found seven proteins that overlapped between the two groups (Figure 5a–5b). Among them, fibronectin was the most highly overexpressed protein in both cohorts (mouse Fn1 p= 1.7 × 10−11; human FN1 p=1.5 × 10−4) (Figure 5c, Supp Fig 2). We further confirmed the overexpression of fibronectin in the murine MYC-driven HCC by immunoblotting (Figure 5d). Immunofluorescence staining of the primary tumors showed that fibronectin was observed to be overexpressed both in the cancer cells and also in the tumor stroma of MYC-HCC (Figure 5e). Moreover, stromal Fn1 expression was found predominantly in perivascular and peritumoral locations with 10% of cancer cells themselves expressing Fn1 on IHC staining of MYC-HCC (Figure 5f).
Figure 5. Fibronectin is a conserved biomarker of invasive HCC in cross-species analysis.
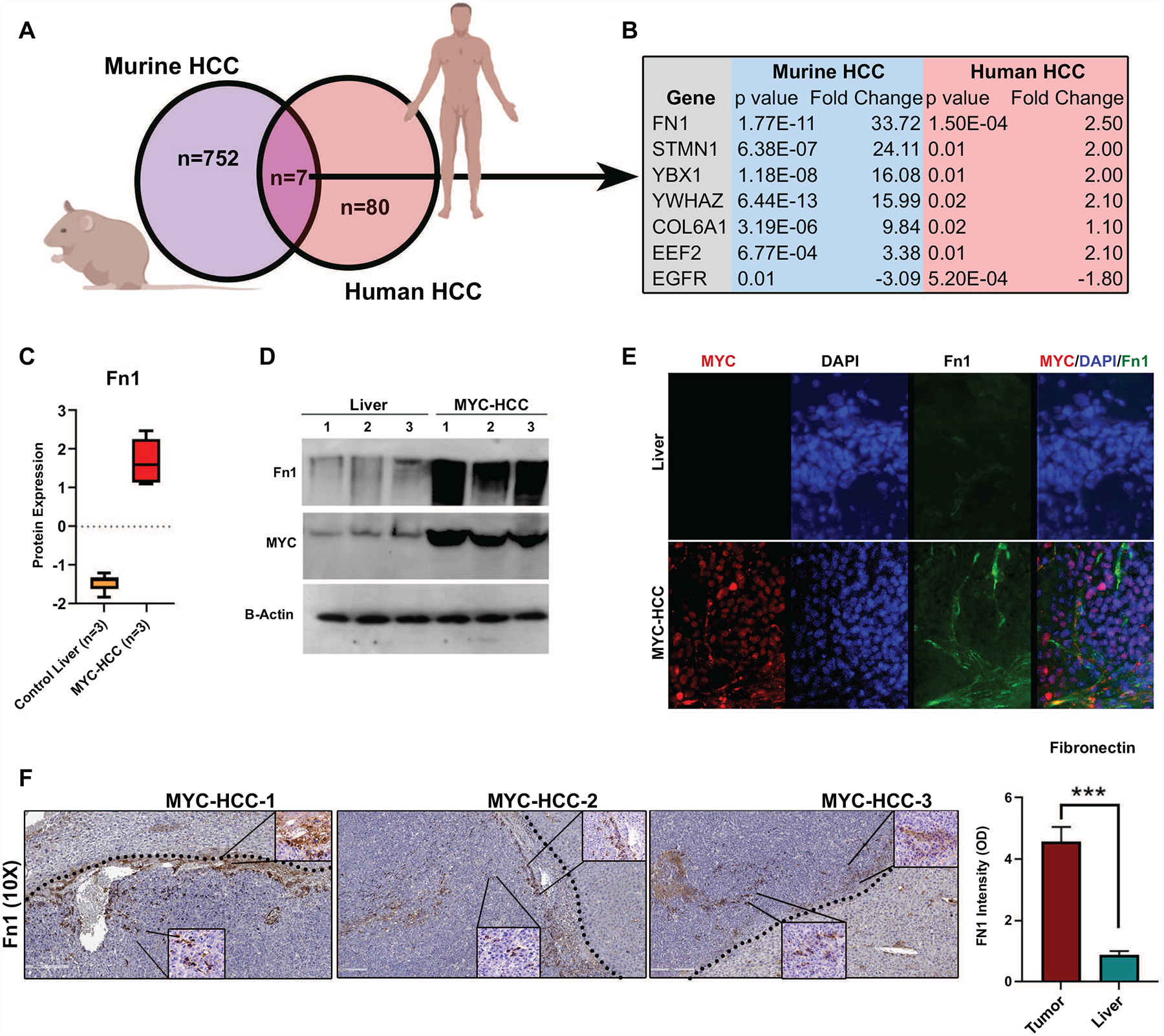
a. Cross-species comparative proteomics of MYC-HCC and human HCC with vascular invasion b. Common proteins overexpressed in murine MYC-HCC and human HCC with vascular invasion. c. Fibronectin expression in MYC-HCC based on LC/MS. d. Fibronectin expression in MYC-HCC based on immunoblotting. e. Fibronectin expression in MYC-HCC based on immunofluorescence. f. IHC for FN1 in MYC-HCC. The dotted line shows the tumor border with the surrounding liver. The bottom insert shows the FN1 cellular staining (60X) while the inserts in the top right demonstrate the stromal FN1 expression (60X).
We have shown that Fn1 is overexpressed in MYC-HCC. We found further mechanistic evidence that MYC promotes the transcription of fibronectin. First, a meta-analysis of Chip-seq experiments shows multiple instances of MYC binding upstream of the human FN1 gene in the Gene Transcription Regulation Database (GTRD) (24) (Supp Fig 3a). Second, motif finding analysis identified multiple MYC binding sites in the promoter region of FN1 in the data from Eukaryotic promoter database (EPD) (25) (Supp Fig 3b). Third, using the ENCODE database we identified FN1 to be a transcriptional target gene of MYC based on the presence of MYC binding site near the transcription start site of FN1 (26). Lastly, we have found that MYC levels positively and significantly correlated with FN1 levels in thirteen solid tumors in the TCGA pan-cancer database, including HCC (Supp Fig 3c). Taken together, these data show that MYC transcriptionally regulates the expression of FN1 in the cancer cells.
Fibronectin promotes human HCC cancer cell invasiveness and migration
We evaluated the functional role of fibronectin in human HCC. We analyzed data from the cancer dependency map data on genome wide CRISPRi and RNAi screens to evaluate if fibronectin knockout impacted HCC cell line survival or viability (27–29). Of the 20 HCC cell lines, none of them showed a low dependency score either on CRISPRi or RNAi screens, indicating that FN1 was not essential for HCC cancer cell viability or proliferation (Figure 6a). To evaluate the functional role of FN1 in HCC, we knocked down FN1 expression in two HCC cell lines SKHep1 and SNU182, both of which had neutral dependency scores, using dicer-substrate 27mer duplex siRNAs. We confirmed FN1 mRNA and protein knockdown using qPCR and immunofluorescence respectively (Figure 6b–6c, Supp Fig 4a). Since FN1 was found to be associated with vascular invasion in human HCC we evaluated if it played a role in cancer cell invasiveness using a basement membrane invasion assay. Cells with FN1 knockdown had decreased ability to invade through a layer of basement membrane extract in the Boyden chamber assay (mean percent invaded cells 18.3% vs. 64.5%; p<0.0001) (Figure 6d, Supp Fig 4b) compared to control cells. We also showed that SKHep1 cells with FN1 knockdown had decreased migratory capacity, in the wound healing assay, when compared to control cells (mean percent wound closure at 72 hours 40% vs. 92%; p=0.0004) (Figure 6e–6f). Thus, FN1 promotes the invasive and migratory capacity of HCC cancer cells.
Figure 6. Fibronectin promotes human HCC cancer cell invasiveness and migration.
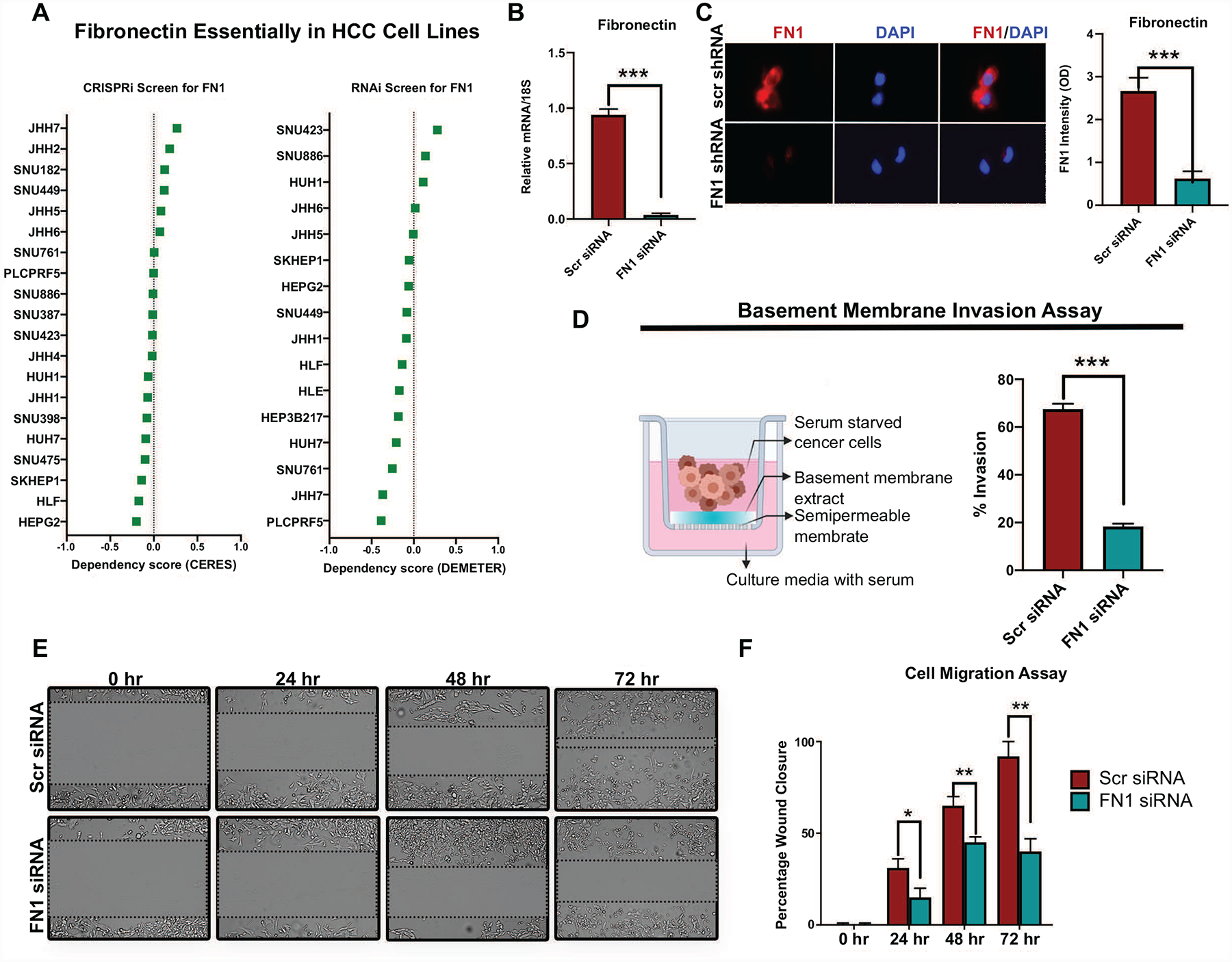
a. Fibronectin essentiality scores from genome-wide CRISPRi (CERES) and RNAi (DEMETER) screens in human HCC cell lines derived from the DepMap database. Score closer to −1 means the gene is likely to be essential for the survival or viability of the cell lines. All the HCC cell lines have a score greater than −0.5 in both screens. b. Demonstration of FN1 knockdown using qPCR of SKHep1 cells transfected with scr siRNA or FN1 siRNA. c. Immunofluorescence assay to confirm FN1 protein knockdown in SKHep1 cells transfected with scr siRNA or FN1 siRNA. d. Schematic of basement membrane invasion assay and quantification of invaded SKHep1 cells transfected with scr siRNA or FN1 siRNA. e-f. Wound healing assay and quantification of migrated SKHep1 cells transfected with scr siRNA or FN1 siRNA. **p<0.01; ***p<0.001.
Validation of fibronectin tissue expression in human HCC
Our exploratory cross-species proteomic analysis identifies fibronectin to be a conserved protein associated with invasive HCC. In the TCGA cohort, fibronectin protein expression in the tumor correlated not just with vascular invasion but also with more advanced tumor stage (p=0.01) and higher tumor grade (p=0.08) (Supp Fig 5a). Moreover, higher FN1 mRNA expression was associated with poor recurrence-free survival (high FN1 7.4 months vs low FN1 21.6 months; p=0.001; HR 2.4,) (Supp Fig 5b).
In order to further validate these findings, we employed an independent cohort of patients from whom paired samples of HCC and surrounding non-tumorous cirrhotic tissue (n=153) were available in a tissue microarray (TMA). We compared the expression of FN1 in HCC against expression in control-non-liver tissue (n=18) or peritumoral cirrhotic liver (n=153). FN1 expression was significantly higher in HCC tumor tissue compared to non-tumorous surrounding cirrhotic tissue (staining intensity score H=27.8 vs. H=17.5, p=0.0007) and also non-liver control tissue (staining intensity score H=27.8 vs. H=8.7, p=0.007) (Figure 7a–7b). The percentage of cells which were positive for FN1 expression in HCC was higher than in cirrhosis without HCC (25.3% vs. 17%, p=0.002) or control non-liver tissue (25.3% vs. 8.9%, p=0.01). Overall, 28% of the HCCs had low FN1 expression, and 72% had moderate-to-high FN1 expression. Additionally, we evaluated protein expression data for fibronectin from the Human Pathology Protein Atlas (HCPA) database of the public repository of liver cancer samples (n=12)(30). Consistent with our TMA findings, fibronectin was expressed in all HCC tumors and a majority (75%, n=9) had moderate-to-high expression of fibronectin with 25% having low expression (Supp Fig 6). These data confirm that fibronectin is significantly overexpressed in human HCC compared to non-cancerous cirrhosis tissue.
Figure 7. Validation of fibronectin tissue expression in human HCC.
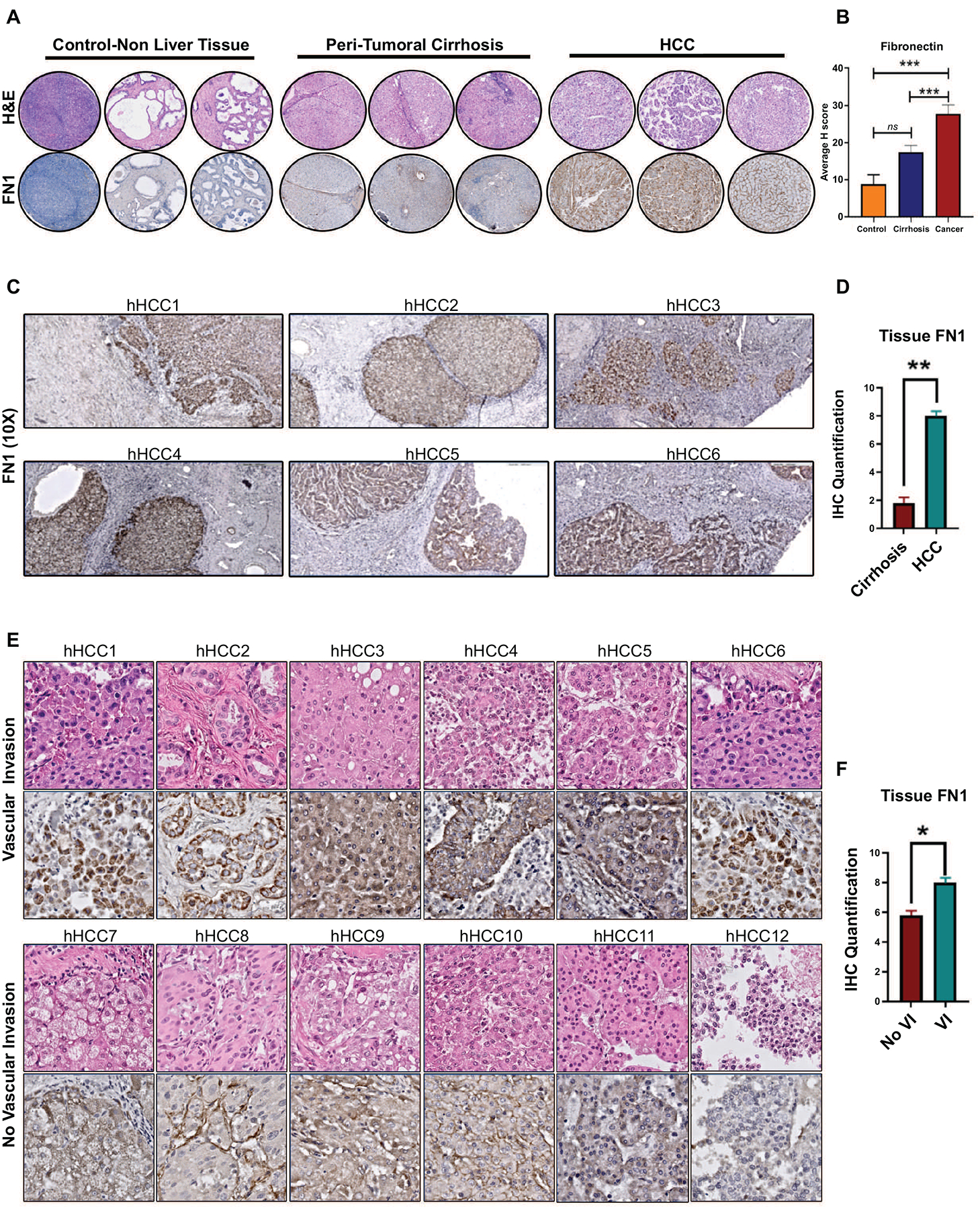
a. Immunohistochemistry for fibronectin in tissue microarray - representative H&E and IHC (10x) from non-liver tissue, peritumoral cirrhosis tissue and HCC tumor tissue. b. Quantification of Fn1 expression in the tissue microarray. c. Representative immunohistochemistry for FN1 in surrounding liver and human HCC tissues with vascular invasion. d. Quantification of Fn1 expression in the whole slide tissue. e. Representative immunohistochemistry for FN1 in human HCC whole slide tissues with (top panel) and without vascular invasion (bottom panel). f. Quantification of FN1 in tumors with and without vascular invasion. * p<0.05; **p<0.01; ***p<0.001.
To validate the association between FN1 expression and vascular invasion in HCC, we analyzed the expression of FN1 in a separate cohort of archived human HCC whole slide tissues (n=12) and compared tumors with known vascular invasion (n=6) and those without (n=6). The two groups were matched for primary tumor stage and etiology of cirrhosis. Consistent with the TMA findings, the HCC tissue showed higher levels of FN1 expression compared to the surrounding non-tumorous liver tissue (H score 8.0 vs. 1.8; p<0.001) (Figure 7c–7d). Further, we observed that FN1 expression was higher in tumors with microvascular invasion than in those without (H score 8.2 vs. 5.8; p=0.001) (Figure 7e–7f). Interestingly, we additionally found that fibronectin was consistently overexpressed in tumor emboli within the microvasculature of tumors with vascular invasion (Supp Fig 7). This data validates that tumor FN1 expression is associated with vascular invasion in human HCC.
Fibronectin as a non-invasive biomarker of advanced HCC
Lastly, we evaluated if fibronectin can serve as a non-invasive plasma biomarker of HCC. To do this, we performed a pilot case-control study by prospectively recruiting patients with HCC (n=35) and those with cirrhosis without HCC (n=10) who were matched for age, gender, ethnicity and etiology of cirrhosis (Figure 8a). Plasma fibronectin levels were significantly elevated in patients with HCC (mean=307.7 μg/ml, SEM=39.9) compared to non-HCC cirrhosis controls (mean=41.8 μg/ml, SEM=13.3) (p=0.0003) (Figure 8b). In contrast, plasma levels of PAI-1 and VEGFR2, two other proteins which were elevated in HCC tissue in the TCGA cohort, were not elevated in the plasma of patients with HCC-patients with cirrhosis-alone did not have a higher plasma PAI-1 (mean=4.1 μg/ml, SEM=7.1) or VEGFR2 (mean=13.20 μg/ml, SEM=3.81) compared to patients with HCC-PAI-1 (mean=4.3 μg/ml, SEM=4.2)(p=0.81) and VEGFR2 (mean=17.31 μg/ml, SEM=4.71(p=0.55) (Figure 8b). AFP, an established biomarker of HCC, was elevated but was not significantly different between patients with cirrhosis (mean=5.5 ng/ml, SEM=1.0) and HCC (mean=3446.9 μg/ml, SEM=2277.5) (p=0.196).
Figure 8. Evaluation of Fibronectin as a non-invasive biomarker of HCC.
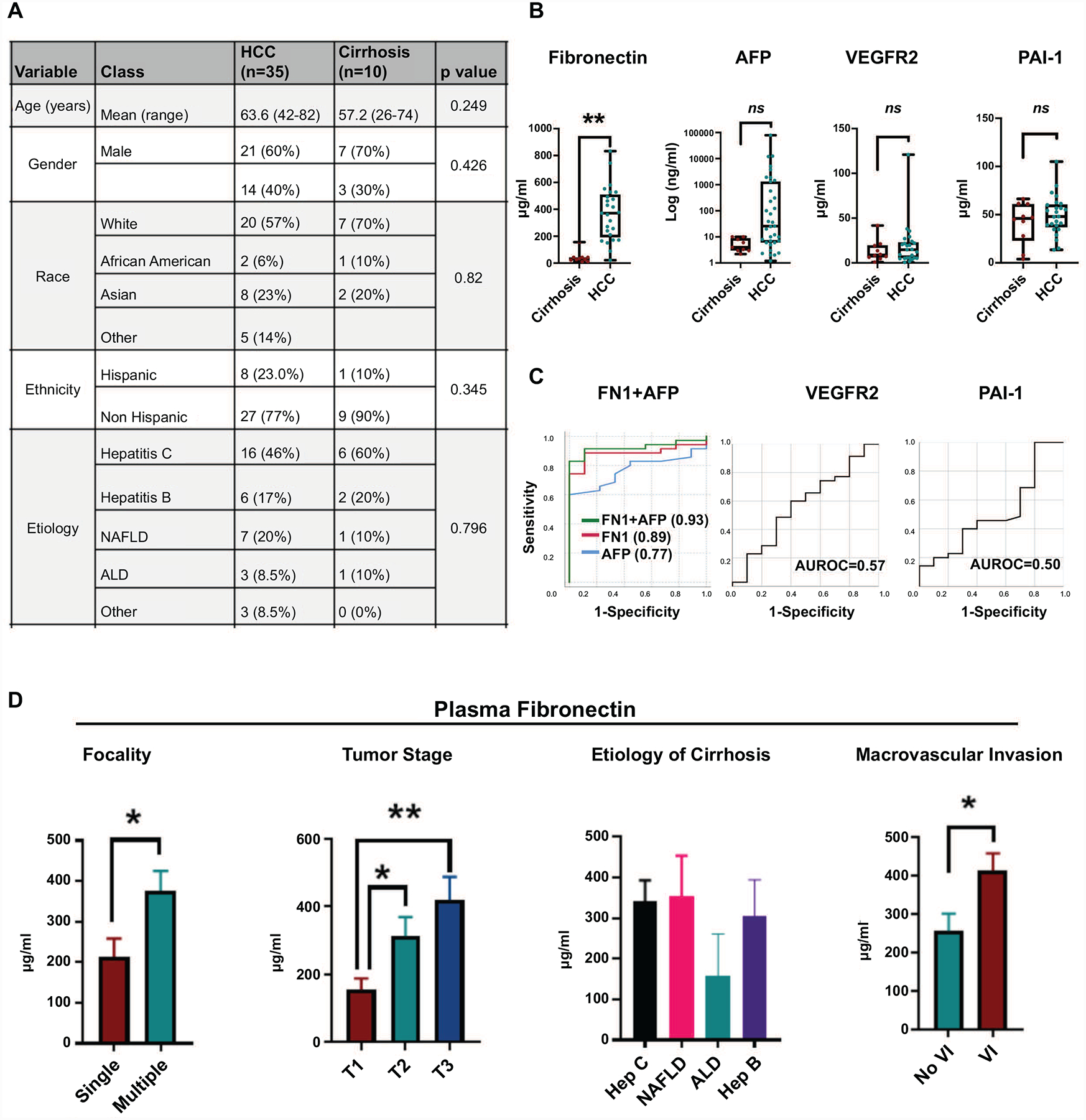
a. Clinical data for the case-control study to assess plasma biomarkers of vascular invasion. b. Plasma levels of FN1, AFP, VEGFR2 and PAI-1 in patients with HCC compared to cirrhosis-alone. c. ROC curves for the diagnosis of HCC of FN1, AFP, combined FN1+AFP, VEGFR2 and PAI-1. d. Correlation of plasma fibronectin levels with HCC tumor focality, tumor stage, etiology of cirrhosis and vascular invasion. * p<0.05; **p<0.01; ***p<0.001.
The sensitivity of FN1 in diagnosing HCC was 92% and specificity 88% with FN1 cutoff of 100 μg/ml. When the cut-off was increased to 160 μg/ml, the sensitivity fell to 88% and specificity increased to 100%. AFP had a sensitivity of 88% and a specificity of 40% with a cut-off value of 4ng/ml. Combining AFP and FN1 (combined AUC 0.93 (0.84–1.00)) improved AUC than either FN1 (AUC 0.89 (0.79–0.99)) or AFP alone (AUC 0.77 (0.63–0.90)) (Figure 8c). The AUC for PAI-1 0.56 (95% CI 0.35–0.78; p=0.535) and VEGFR2 0.57 (95% CI 0.35–0.78; p=0.596) were poor (Figure 8c). FN1 levels were statistically higher in patients with multifocal HCC (mean 377 vs. 214 μg/ml) (p=0.02) and in advanced stage of HCC (mean T1 (n=9)=154 vs T2 (n=14) =312, vs T3 (n=11)=419 μg/ml) (p<0.05) (Figure 8d). FN1 levels did not vary based on the etiology of liver disease (Figure 8d). Lastly, consistent with tissue fibronectin expression, plasma FN1 was higher in patients whose tumors had vascular invasion (mean 414 μg/ml) vs those who did not (mean 258 μg/ml) (p=0.04) (Figure 8d). In contrast, serum alpha fetoprotein (AFP) levels were not predictive or tumor stage (p=0.482) or vascular invasion (0.415). Thus, fibronectin is a promising non-invasive plasma proteomic biomarker of invasive HCC.
DISCUSSION
The presence of vascular invasion portends aggressive tumor behavior and poor clinical outcome in HCC. Our study evaluates the proteogenomic topography of tumors with vascular invasion to elucidate the key molecular drivers of this phenotype. The functional pathway analysis of the transcriptomic and proteomic changes reveals upregulation of multiple pathways and processes which promote invasiveness, thus demonstrating the coordinated impact of these changes. We found the MYC oncogene was a consistent upstream regulator of the significant changes in the vascular invasion-related mRNAs, miRNA and proteins. We performed a cross-species comprehensive proteomic analysis comparing the proteomic changes in MYC-driven murine HCC and human HCC with vascular invasion and identified fibronectin as a biomarker conserved across both species. We demonstrate that fibronectin mechanistically promotes HCC cancer cell migration and invasiveness. Moreover, we validate the role of fibronectin as a tissue and plasma biomarker of vascular invasion in HCC using two independent clinical cohorts. Thus, we perform a multi-platform functional analysis of HCC, identifying MYC as a major genetic driver of vascular invasion and fibronectin as a potential biomarker.
Our functional analysis of the molecular landscape of HCC reveals pathways involved in cellular migration, invasion and angiogenesis to be enriched in tumors with vascular invasion, while anti-tumor pathways involving apoptosis and senescence were suppressed. These global genetic and epigenetic changes thus appear to promote vascular invasion in a coordinated fashion. Previous studies have generally focused on individual genes or proteins associated with vascular invasion, but the precise causal drivers of this phenotype are yet to be established. We performed upstream regulatory pathway analysis of the mRNA, miRNA and proteomic changes to identify the driver mechanisms. The transcription factor MYC was consistently found to be an upstream regulator of the changes in all three platforms- mRNA, miRNA and proteomics. The MYC oncogene is frequently genetically amplified or transcriptionally overexpressed in HCC (14,31). MYC has been shown to promote angiogenesis and invasiveness in other cancers (32,33) but has not been shown to play a causal role in vascular invasion in HCC. We did not find that MYC amplifications or MYC mRNA expression levels directly predicted vascular invasion. But the vascular invasion-related gene signature strongly overlapped with multiple MYC signatures suggesting that the downstream transcriptional effects of MYC drive vascular invasion. Consistent with this finding, our analysis of the MYC-driven murine HCC demonstrated high prevalence of vascular invasion. In addition, we present evidence that MYC upregulates the transcription of fibronectin which in turn promotes cancer cell migration and invasiveness thus providing a mechanistic link between MYC, fibronectin and invasive HCC. There has been significant recent progress in developing direct or indirect anti MYC therapeutics, which in the future can potentially be used to target tumors with vascular invasion (34–37).
We adopted a cross-species comparative approach to identify proteins predictive of vascular invasion which were conserved across species, and hence more likely to have translational relevance. This approach allowed us to determine fibronectin as a promising predictive biomarker of vascular invasion in HCC. Transcriptome based analysis of tissues with vascular invasion has previously identified multiple predictive gene signatures (10,38). But difficulty in obtaining tissue samples for validation, lack of tractable assays and inter-test variability are challenges in translating these microarray- or RNAseq based gene signatures to the bedside. Conversely, proteins have been successfully used both for early detection and risk stratification in multiple cancers. Fibronectin, a glycoprotein which modulates interactions with the extracellular matrix, has been shown to be overexpressed in HCC tissue (39,40) and also identified as a potential plasma biomarker of HCC in an Asian cohort of predominantly hepatitis B patients (41,42). We here show that fibronectin has the potential to serve as a non-invasive biomarker of vascular invasion, especially in combination with AFP. We validated the overexpression of fibronectin in independent cohorts of human HCC tissue and also in the plasma of patients with HCC. Further evaluation of FN1 as a biomarker of vascular invasion will require a large, rigorous prospective study, which we plan to pursue next. Fibronectin-targeting MRI contrast agents have been developed and used successfully to detect breast cancer micro metastases (43), and can potentially be used as an imaging biomarker of vascular invasion in HCC.
In conclusion, the current study highlights the distinct molecular profile of tumors with vascular invasion, thus advancing our understanding of this clinically relevant, aggressive subtype of HCC. We identify the MYC oncogene as a major driver of this phenotype and propose that fibronectin is a promising predictor of invasive HCC.
MATERIALS AND METHODS
TCGA Data
In the TCGA study, surgical resection specimens were collected from patients with HCC who had not received prior treatment. Institutional review boards (IRB) at each tissue source site (TSS) approved submission to TCGA. Clinical and histopathological data including data on vascular invasion were submitted from each TSS after detailed pathology review of the entire tumor specimen. The TCGA biospecimen core subjected each tissue to independent pathology review to confirm the diagnosis of HCC. Complete details of the pathology review are available in the TCGA manuscript on HCC (14)The liver cancer (LIHC) clinical data, processed genomic data, level 3 normalized RNAseq data, level 3 microRNA data and level 3 reverse phase protein array (RPPA) data were downloaded from GDC data portal https://portal.gdc.cancer.gov/ (44).
Differential expression and functional pathway analysis
Heatmaps were generated to show a visually interpretable overview of the mRNA, miRNA and proteomic expression profile of HCC stratified by the presence of vascular invasion (Qlucore Omics Explorer v. 2.2). Differential expression analysis was performed using Qlucore Omics Explorer. Variance filtering was used to reduce the noise, and the projection score to set the filtering threshold. The ‘mean = 0, var = 1’ setting was used to scale the data. QIAGEN’s Ingenuity® Pathway Analysis (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity) software was used for pathway analysis and upstream regulatory analysis of differentially expressed genes, microRNAs and proteins. The differential gene expression was compared against a proprietary gene interaction network database to estimate the probability that a given pathway is differentially enriched. Upstream regulator analysis in IPA was used to predict the upstream transcriptional regulators from the dataset based on the literature and compiled in the Ingenuity® Knowledge Base. The significance (p-value) and activation score (z-score) was computed by analyzing overlap between the differentially expressed genes in human HCC with microvascular invasion and known targets regulated by the transcriptional regulator. Graphic representations of the data were prepared using the IPA Canonical Pathways Molecular Activity Predictor tool and GraphPad Prism 7 software.
For survival analysis of TCGA data, we derived the tumor stratification based on FN1 mRNA expression from cBioportal (https://www.cbioportal.org/) to identify the tumors expressing fibronectin two times the relative expression (z-score) when compared to its expression distribution in all tumors.
Transgenic Mice
Liver specific LAP-tTa/tet-o-MYC transgenic lines have previously been described (45). Mice received 0.1 mg/ml of doxycycline (Sigma) in drinking water until 4 weeks of age and then were taken off doxycycline. Age-matched control mice were of the same genotype but they remained on doxycycline continuously since birth. All procedures and housing of animals were in accordance with Stanford’s APLAC protocols.
Liquid Chromatography – Mass Spectrometry Analysis (LC/MS)
Frozen MYC-HCC and liver tissue samples were used for the assay. Three technical replicate LC-MS/MS runs were performed on each sample. Sample preparation and proteomic quantification methods were based on standard pipelines as described before (46).The resulting MS raw files were searched using Byonic 2.11.0 software (Protein Metrics) with a Swiss-Prot database referencing mouse proteome (2017; 17,191 entries).
Tissue Microarray analysis
De-identified tissue microarray slides were obtained from the Mayo Clinic Hepatobiliary SPORE. This project was supported by Award Number P50 CA210964 (Mayo Clinic Hepatobiliary SPORE) from the National Cancer Institute. The slides contained HCC core tissue and matched non-tumorous surrounding tissue from a total of 153 patients with HCC and 18 control samples from non-liver tissues. Tissue microarray slides were digitally scanned with the 20x objective (0.24μm/pixel resolution) by a P1000 Pannoramic scanner (3DHistech Corporation, Budapest Hungary). Quantitative digital image analysis was performed in HALO (Indica Labs, New Mexico USA). Tissue microarray cores were automatically recognized with the TMA add-on module. Blank cores, cores with <5% intact tissue, or cores with no morphologically recognizable tumor were excluded from analysis. H-scores were calculated with the brightfield area quantification that binned di-amino benzidine (DAB) positive signals based on optical density for each core.
Immunohistochemistry
Paraffin embedded tumor sections were deparaffinized by successive incubations in xylene, graded washes in ethanol, and PBS. Epitope unmasking was performed by steaming in 0.01 mol/L citrate buffer (pH 6.0) for 45 minutes. Paraffin embedded sections were immunostained with fibronectin (1:100, R&D MAB1918) or MYC (1:150, Epitomics EP121) overnight at 4°C. The tissue was washed with PBS and incubated with biotinylated anti-rabbit or anti-mouse for 30 minutes at room temperature (1:300 Vectastain ABC kit, Vector Labs). Sections were developed using 3,3’-Diaminobenzidine (DAB), counterstained with hematoxylin, and mounted with permount. Images were scanned using a Philips Digital scanner.
FN1 knockdown with Migration and Invasion Assays
Two human HCC cell lines SK-Hep1 and SNU182 were purchased from ATCC (American Type Culture Collection, Manassas, VA, USA) and maintained in complete DMEM media (Gibco, USA) with 10% FBS in a 37C incubator. Scr siRNA (SR30004) and three unique FN1 siRNA duplexes (SR320193) were purchased from Origene, Inc. Trilencer-27 fluorescent labeled transfection control (SR30002) was first used to determine optimal transfection efficiency for each cell line. The siRNA duplex #1 was confirmed to have the highest efficacy of FN1 knockdown among the three unique duplexes. We used qPCR and immunofluorescence assays to confirm FN1 mRNA and protein knockdown respectively.
Cell invasion across a basement membrane was assessed using a 24 well transwell chamber with 8 μm filter inserts (Abcam ab235882). We coated the cell insert with a basement membrane extract (BME) and seeded 2×105 serum starved SK-Hep1 or SNU182 transfected with scr siRNA or FN1 siRNA in the top chamber. Complete media is added to the bottom chamber. Cells invaded the basement membrane matrix and then migrated through a semipermeable membrane over 48 hours. The percent cell invasion can be analyzed directly in a plate reader at 530/590 nm.
Cell migration was assessed using a wound healing assay. We seeded 1×106 SK-Hep1 cells transfected either with scr siRNA or FN1 siRNA per well in a 6-well plate and cultured overnight in culture medium. Thereafter, a scratch (wound) was introduced in the confluent cell layer using a yellow tip. Pictures of a defined wound spot were made with a Leica DM16000 microscope at t = 0, 24, 48 and 72 hr. The area of the wound in the microscopic pictures was measured using Image J software (National Institutes of Health, MD). The percentage wound healing was calculated relative to the total wound area at t = 0 hr. of the same wound spot.
Clinical study
This study was approved by the institutional review board (IRB) of Stanford University (IRB Number: 28374), and all patients provided informed consent before being enrolled in this study. We prospectively collected blood from patients with HCC or cirrhosis alone. Blood samples were obtained at the time of diagnosis of HCC before treatment initiation. Plasma was separated, aliquoted and stored at −80C. Clinical data was gathered from their medical records. Plasma fibronectin, PAI-1 and VEGFR2 were measured using respective Quantikine ELISA kits from R&D Systems. The ELISA assay for fibronectin recognizes two large human fibronectin fragments amino acid (aa) 32–1908 and aa 607–1265 and has high specificity. After samples and standards had been appropriately diluted, 50 uL of the appropriate standard or sample was added to each well in duplicates with some amount of Assay Diluent. After the appropriate time, each well was then washed with Wash Buffer solution and then the appropriate conjugate was added to each well. The solution was then washed again, and a substrate solution was added. After which, a stop solution was added and the plate was read on a plate reader.
Statistics
Differences between groups were analyzed using Student’s t-test or one-way analysis of variance (ANOVA). Chi square test was used to compare categorical variables. Kaplan Meier analysis with Log Rank test was performed for survival analysis. A P value of less than 0.05 was considered to be significant. All graphs are presented as the mean +/− SEM. Analyses were performed with Prism, version 7 (GraphPad Software, San Diego, CA).
Supplementary Material
Acknowledgements
RD- National Institutes of Health (NIH) grant CA222676 from the National Cancer Institute (NCI), American College of Gastroenterology Junior Faculty Career Development Grant.
DF- National Institutes of Health (NIH) grant CA208735, NIH R01 CA184384, NIH U01 CA188383 from the National Cancer Institute (NCI).
Pauline Chu- Tissue processing. Ivanshi Ahuja- Image analysis.
Mayo Clinic HB SPORE- The content is solely the responsibility of the authors of this manuscript and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Footnotes
No conflicts of interest.
REFERENCES
- 1.Global Burden of Disease Liver Cancer Collaboration, Akinyemiju T, Abera S, Ahmed M, Alam N, Alemayohu MA, et al. The Burden of Primary Liver Cancer and Underlying Etiologies From 1990 to 2015 at the Global, Regional, and National Level: Results From the Global Burden of Disease Study 2015. JAMA Oncol [Internet]. 2017;3:1683–1691. Available from: 10.1001/jamaoncol.2017.3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dhir M, Melin AA, Douaiher J, Lin C, Zhen WK, Hussain SM, et al. A Review and Update of Treatment Options and Controversies in the Management of Hepatocellular Carcinoma. Ann. Surg. [Internet] 2016;263:1112–1125. Available from: 10.1097/SLA.0000000000001556 [DOI] [PubMed] [Google Scholar]
- 3.Lee Y-H, Hsu C-Y, Huang Y-H, Hsia C-Y, Chiou Y-Y, Su C-W, et al. Vascular invasion in hepatocellular carcinoma: prevalence, determinants and prognostic impact. J. Clin. Gastroenterol. [Internet] 2014;48:734–741. Available from: 10.1097/MCG.0b013e3182a8a254 [DOI] [PubMed] [Google Scholar]
- 4.Kurokawa T, Yamazaki S, Mitsuka Y, Moriguchi M, Sugitani M, Takayama T. Prediction of vascular invasion in hepatocellular carcinoma by next-generation des-r-carboxy prothrombin. Br. J. Cancer [Internet] 2016;114:53–58. Available from: 10.1038/bjc.2015.423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gouw ASH, Balabaud C, Kusano H, Todo S, Ichida T, Kojiro M. Markers for microvascular invasion in hepatocellular carcinoma: where do we stand? Liver Transpl. [Internet] 2011;17 Suppl 2:S72–80. Available from: 10.1002/lt.22368 [DOI] [PubMed] [Google Scholar]
- 6.Pommergaard H-C, Rostved AA, Adam R, Thygesen LC, Salizzoni M, Gómez Bravo MA, et al. Vascular invasion and survival after liver transplantation for hepatocellular carcinoma: a study from the European Liver Transplant Registry. HPB [Internet]. 2018;20:768–775. Available from: 10.1016/j.hpb.2018.03.002 [DOI] [PubMed] [Google Scholar]
- 7.Hsieh C-H, Wei C-K, Yin W-Y, Chang C-M, Tsai S-J, Wang L-Y, et al. Vascular invasion affects survival in early hepatocellular carcinoma. Mol Clin Oncol [Internet]. 2015;3:252–256. Available from: 10.3892/mco.2014.420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roayaie S, Blume IN, Thung SN, Guido M, Fiel M-I, Hiotis S, et al. A system of classifying microvascular invasion to predict outcome after resection in patients with hepatocellular carcinoma. Gastroenterology [Internet]. 2009;137:850–855. Available from: 10.1053/j.gastro.2009.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JU, Cox IJ, Taylor-Robinson SD. The Quest for Relevant Hepatocellular Carcinoma Biomarkers. Cell Mol Gastroenterol Hepatol [Internet]. 2017;4:283–284. Available from: 10.1016/j.jcmgh.2017.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mínguez B, Hoshida Y, Villanueva A, Toffanin S, Cabellos L, Thung S, et al. Gene-expression signature of vascular invasion in hepatocellular carcinoma. J. Hepatol. [Internet] 2011;55:1325–1331. Available from: 10.1016/j.jhep.2011.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Sun L, Gao F, Yang X, Li Y, Zhang Q, et al. A new scoring model predicting macroscopic vascular invasion of early-intermediate hepatocellular carcinoma. Medicine [Internet]. 2018;97:e13536. Available from: 10.1097/MD.0000000000013536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang Y, Han Q-J, Zhang J. Hepatocellular carcinoma: Mechanisms of progression and immunotherapy [Internet]. World Journal of Gastroenterology. 2019;25:3151–3167. Available from: 10.3748/wjg.v25.i25.3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nault J-C, Cheng A-L, Sangro B, Llovet JM. Milestones in the pathogenesis and management of primary liver cancer. J. Hepatol. [Internet] 2020;72:209–214. Available from: 10.1016/j.jhep.2019.11.006 [DOI] [PubMed] [Google Scholar]
- 14.Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell [Internet]. 2017;169:1327–1341.e23. Available from: 10.1016/j.cell.2017.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Gao J-Z, Du J-L, Huang Z-X, Wei L-X. Increased CDC20 expression is associated with development and progression of hepatocellular carcinoma. Int. J. Oncol. [Internet] 2014;45:1547–1555. Available from: 10.3892/ijo.2014.2559 [DOI] [PubMed] [Google Scholar]
- 16.Lin J, Hou Y, Huang S, Wang Z, Sun C, Wang Z, et al. Exportin-T promotes tumor proliferation and invasion in hepatocellular carcinoma. Mol. Carcinog. [Internet] 2019;58:293–304. Available from: 10.1002/mc.22928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao J, Wozniak A, Adams A, Cox J, Vittal A, Voss J, et al. SIRT7 regulates hepatocellular carcinoma response to therapy by altering the p53-dependent cell death pathway. J. Exp. Clin. Cancer Res. [Internet] 2019;38:252. Available from: 10.1186/s13046-019-1246-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoshida Y, Nijman SMB, Kobayashi M, Chan JA, Brunet J-P, Chiang DY, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. [Internet] 2009;69:7385–7392. Available from: 10.1158/0008-5472.CAN-09-1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyault S, Rickman DS, de Reyniès A, Balabaud C, Rebouissou S, Jeannot E, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology [Internet]. 2007;45:42–52. Available from: 10.1002/hep.21467 [DOI] [PubMed] [Google Scholar]
- 20.Woo HG, Lee J-H, Yoon J-H, Kim CY, Lee H-S, Jang JJ, et al. Identification of a cholangiocarcinoma-like gene expression trait in hepatocellular carcinoma. Cancer Res. [Internet] 2010;70:3034–3041. Available from: 10.1158/0008-5472.CAN-09-2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kenny HA, Chiang C-Y, White EA, Schryver EM, Habis M, Romero IL, et al. Mesothelial cells promote early ovarian cancer metastasis through fibronectin secretion. J. Clin. Invest. [Internet] 2014;124:4614–4628. Available from: 10.1172/JCI74778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein RM, Bernstein D, Higgins SP, Higgins CE, Higgins PJ. SERPINE1 expression discriminates site-specific metastasis in human melanoma. Exp. Dermatol. [Internet] 2012;21:551–554. Available from: 10.1111/j.1600-0625.2012.01523.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quintavalle C, Hindupur SK, Quagliata L, Pallante P, Nigro C, Condorelli G, et al. Phosphoprotein enriched in diabetes (PED/PEA15) promotes migration in hepatocellular carcinoma and confers resistance to sorafenib. Cell Death Dis. [Internet] 2017;8:e3138. Available from: 10.1038/cddis.2017.512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yevshin I, Sharipov R, Kolmykov S, Kondrakhin Y, Kolpakov F. GTRD: a database on gene transcription regulation—2019 update [Internet]. Nucleic Acids Research. 2019;47:D100–D105. Available from: 10.1093/nar/gky1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dreos R, Ambrosini G, Groux R, Cavin Périer R, Bucher P. The eukaryotic promoter database in its 30th year: focus on non-vertebrate organisms. Nucleic Acids Res. [Internet] 2017;45:D51–D55. Available from: 10.1093/nar/gkw1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rouillard AD, Gundersen GW, Fernandez NF, Wang Z, Monteiro CD, McDermott MG, et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins [Internet]. Database. 2016;2016:baw100. Available from: 10.1093/database/baw100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Website [Internet]. [cited 2020 Sep 1];Available from: DepMap, Broad (2020): DepMap 20Q2 Public. figshare. Dataset. 10.6084/m9.figshare.12280541.v4. [DOI]
- 28.McFarland JM, Ho ZV, Kugener G, Dempster JM, Montgomery PG, Bryan JG, et al. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat. Commun. [Internet] 2018;9:4610. Available from: 10.1038/s41467-018-06916-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matteson R DepMap: Dependency Mapping of Applications Using Operating System Events [Internet]. Available from: 10.15368/theses.2010.180 [DOI]
- 30.Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, et al. A pathology atlas of the human cancer transcriptome. Science [Internet]. 2017;357. Available from: 10.1126/science.aan2507 [DOI] [PubMed] [Google Scholar]
- 31.Fujimoto A, Furuta M, Totoki Y, Tsunoda T, Kato M, Shiraishi Y, et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat. Genet. [Internet] 2016;48:500–509. Available from: 10.1038/ng.3547 [DOI] [PubMed] [Google Scholar]
- 32.Baudino TA, McKay C, Pendeville-Samain H, Nilsson JA, Maclean KH, White EL, et al. c-Myc is essential for vasculogenesis and angiogenesis during development and tumor progression. Genes Dev. [Internet] 2002;16:2530–2543. Available from: 10.1101/gad.1024602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Udager AM, Ishikawa MK, Lucas DR, McHugh JB, Patel RM. MYC immunohistochemistry in angiosarcoma and atypical vascular lesions: practical considerations based on a single institutional experience. Pathology [Internet]. 2016;48:697–704. Available from: 10.1016/j.pathol.2016.08.007 [DOI] [PubMed] [Google Scholar]
- 34.Han H, Jain AD, Truica MI, Izquierdo-Ferrer J, Anker JF, Lysy B, et al. Small-Molecule MYC Inhibitors Suppress Tumor Growth and Enhance Immunotherapy. Cancer Cell [Internet]. 2019;36:483–497.e15. Available from: 10.1016/j.ccell.2019.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dhanasekaran R, Gabay-Ryan M, Baylot V, Lai I, Mosley A, Huang X, et al. Anti-miR-17 therapy delays tumorigenesis in MYC-driven hepatocellular carcinoma (HCC). Oncotarget [Internet]. 2018;9:5517–5528. Available from: 10.18632/oncotarget.22342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Struntz NB, Chen A, Deutzmann A, Wilson RM, Stefan E, Evans HL, et al. Stabilization of the Max Homodimer with a Small Molecule Attenuates Myc-Driven Transcription. Cell Chem Biol [Internet]. 2019;26:711–723.e14. Available from: 10.1016/j.chembiol.2019.02.009 [DOI] [PubMed] [Google Scholar]
- 37.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell [Internet]. 2011;146:904–917. Available from: 10.1016/j.cell.2011.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ho M-C, Lin J-J, Chen C-N, Chen C-C, Lee H, Yang C-Y, et al. A gene expression profile for vascular invasion can predict the recurrence after resection of hepatocellular carcinoma: a microarray approach. Ann. Surg. Oncol. [Internet] 2006;13:1474–1484. Available from: 10.1245/s10434-006-9057-1 [DOI] [PubMed] [Google Scholar]
- 39.Jagirdar J, Ishak KG, Colombo M, Brambilla C, Paronetto F. Fibronectin patterns in hepatocellular carcinoma and its clinical significance. Cancer [Internet]. 1985;56:1643–1648. Available from: 3.0.co;2-o”> [DOI] [PubMed] [Google Scholar]
- 40.Torbenson M, Wang J, Choti M, Ashfaq R, Maitra A, Wilentz RE, et al. Hepatocellular carcinomas show abnormal expression of fibronectin protein. Mod. Pathol. [Internet] 2002;15:826–830. Available from: 10.1097/01.MP.0000024257.83046.7C [DOI] [PubMed] [Google Scholar]
- 41.Kim H, Park J, Kim Y, Sohn A, Yeo I, Jong Yu S, et al. Serum fibronectin distinguishes the early stages of hepatocellular carcinoma. Sci. Rep. [Internet] 2017;7:9449. Available from: 10.1038/s41598-017-09691-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin H, Tan Z, Wu J, Zhu J, Shedden KA, Marrero J, et al. Mass-Selected Site-Specific Core-Fucosylation of Serum Proteins in Hepatocellular Carcinoma. J. Proteome Res. [Internet] 2015;14:4876–4884. Available from: 10.1021/acs.jproteome.5b00718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Z, Qutaish M, Han Z, Schur RM, Liu Y, Wilson DL, et al. MRI detection of breast cancer micrometastases with a fibronectin-targeting contrast agent. Nat. Commun. [Internet] 2015;6:7984. Available from: 10.1038/ncomms8984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, et al. Toward a Shared Vision for Cancer Genomic Data [Internet]. New England Journal of Medicine. 2016;375:1109–1112. Available from: 10.1056/nejmp1607591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shachaf CM, Kopelman AM, Arvanitis C, Karlsson Å, Beer S, Mandl S, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer [Internet]. Nature. 2004;431:1112–1117. Available from: 10.1038/nature03043 [DOI] [PubMed] [Google Scholar]
- 46.Going CC, Tailor D, Kumar V, Birk AM, Pandrala M, Rice MA, et al. Quantitative Proteomic Profiling Reveals Key Pathways in the Anticancer Action of Methoxychalcone Derivatives in Triple Negative Breast Cancer. J. Proteome Res. [Internet] 2018;17:3574–3585. Available from: 10.1021/acs.jproteome.8b00636 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


