Simulations and microscopy show that supercoiling enhances the dynamics of DNA by reducing threadings and entanglements.
Abstract
Ring polymers in dense solutions are among the most intriguing problems in polymer physics. Because of its natural occurrence in circular form, DNA has been extensively used as a proxy to study the fundamental physics of ring polymers in different topological states. Yet, torsionally constrained—such as supercoiled—topologies have been largely neglected so far. The applicability of existing theoretical models to dense supercoiled DNA is thus unknown. Here, we address this gap by coupling large-scale molecular dynamics simulations with differential dynamic microscopy of entangled supercoiled DNA plasmids. We find that, unexpectedly, larger supercoiling increases the size of entangled plasmids and concomitantly induces an enhancement in DNA mobility. These findings are reconciled as due to supercoiling-driven asymmetric and double-folded plasmid conformations that reduce interplasmid entanglements and threadings. Our results suggest a way to topologically tune DNA mobility via supercoiling, thus enabling topological control over the (micro)rheology of DNA-based complex fluids.
INTRODUCTION
The DNA not only is the central molecule of life but also is now increasingly used for biocompatible and responsive materials—such as DNA hydrogels (1) and origami (2)—with applications in medicine and nanotechnology (3). One feature that renders DNA a unique polymer is its ability to encode information, and this is now extensively leveraged to make complex structures (3, 4) and even self-replicating materials (5); another feature that distinguishes DNA from other synthetic polymers is its unique geometry, i.e., that of a (right-handed) helix with a well-defined pitch, which entails that DNA can display both bending and torsional stiffness (6). Unlike DNA’s information-encoding capabilities, its geometrical features are far less exploited to create synthetic materials. In fact, DNA is, at present, largely used to make up biopolymer complex fluids in its simplest geometrical forms, i.e., that of a linear or relaxed circular (torsionally unconstrained) molecule (7–9). Despite this, most naturally occurring DNA is under torsional and topological constraints, either because it is circular and non-nicked, as in bacteria (10), or because of the binding of proteins that restrict the relative rotation of base pairs, as in eukaryotes (11–13). The torsional stress stored in a closed DNA molecule cannot be mechanically relaxed (in the absence of topoisomerase proteins) but only rearranged or converted into bending to minimize the overall conformational free energy (14, 15). This entails that supercoiling—the linking deficit between sister DNA strands with respect to their relaxed state—can carry conformational information (16) that can affect the static and dynamic properties of DNA plasmids (14) and even regulate gene transcription (17). Here, we propose that supercoiling may also be leveraged to tune the dynamics of DNA plasmids in solution, thus potentially allowing fine control over the rheology of DNA-based complex fluids in a way that is orthogonal to varying DNA length (18), concentration (19), or architecture (7, 20). Last, entangled solutions of DNA plasmids are interesting not only because of their potential applications in bio- and nanotechnology but also because they enable us to study fundamental questions on the physics of ring polymers—one of the most active fields of soft matter research (21–29)—due to the extremely precise control over DNA lengths and topology (7–9) and access to sophisticated visualization techniques (30).
To characterize the effect of DNA supercoiling on the rheology of entangled solutions of plasmids, here, we perform large-scale molecular dynamics simulations of entangled DNA plasmids (Fig. 1, A to C), modeled as coarse-grained twistable chains (31). We find that while isolated DNA plasmids typically display a collapse with increasing levels of supercoiling [estimated via simulations (32) or gel electrophoresis (33)], here, we show that entangled DNA plasmids typically increase their average size with supercoiling. We further find that despite this swelling, larger supercoiling is accompanied by an enhanced mobility of the plasmids. This finding is counterintuitive and in marked contrast with standard polymer systems (34) in which larger polymer sizes correlate with slower diffusion. This speedup is also observed in differential dynamic microscopy (DDM) experiments performed on entangled plasmids with different supercoiling degrees. Last, we use sophisticated techniques involving minimal surface construction and primitive path analysis (PPA) to quantify the abundance of threadings and entanglements between plasmids in solution and find that larger supercoiling decreases both of these topological constraints, in turn explaining the enhanced mobility.
Fig. 1. Modeling supercoiled plasmids as twistable chains.
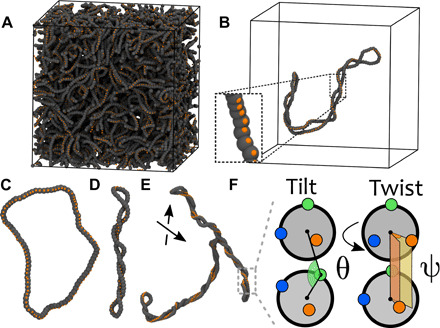
(A) Snapshot of simulation of entangled plasmids with length L = 200σb ≃ 1.47 kbp and σ = 0.04. (B) A single plasmid taken from (A), with an inset showing the patches in detail. (C to E) Snapshots of plasmids with (C) σ = 0, L = 100σb ≃ 750 bp, (D) σ = 0.06, L = 100σb ≃ 750 bp, and (E) σ = 0.06, L = 400σb ≃ 3 kbp. Backbone beads are shown in gray; one set of patches is shown in orange. The other patches are not shown for clarity. (F) Sketch of tilt θ and twist ψ between consecutive beads (another angle ψ is set between blue patches, not shown). The tilt angle θ is subject to a stiff potential with equilibrium θ0 = π to maintain the frame coplanar and aligned with the backbone.
We argue that our results will be key to enabling the design of complex fluids with rheology that can be precisely tuned using a combination of DNA length, concentration, topology, and supercoiling. Beyond providing blueprints for realizing the next generation of biomimetic DNA-based materials, our results can also shed light into the dynamics of DNA in vivo.
RESULTS
Computational model for DNA plasmids
DNA is represented as a twistable elastic chain (31) made of beads of size σb = 2.5 nm =7.35 base pairs (bp) connected by finitely extensible springs and interacting via a purely repulsive Lennard-Jones potential to avoid spontaneous chain crossing (see Fig. 1) (35). In addition to these potentials, a bending stiffness of lp = 50 nm (6) is set via a Kratky-Porod term and two torsional springs (dihedrals) constrain the relative rotation of consecutive beads, ψ, at a user-defined value ψ0. The torsional angle between consecutive beads ψ is determined by decorating each bead with three patches, which provides a reference frame running along the DNA backbone. We finally impose a stiff harmonic spring to constrain the tilt angle θ = π so to align the frame with the backbone, i.e., along its local tangent (see Fig. 1D). The simulations are performed at fixed monomer density (corresponding to a volume fraction ϕ = 4% and ϕ/ϕ* ≃ 16 with ϕ* = 0.26%) and by evolving the equation of motion for the beads coupled to a heat bath in LAMMPS (Large-scale Atomic/Molecular Massively Parallel Simulator) (see Materials and Methods) (36).
The user-defined angle ψ0 directly determines the thermodynamically preferred pitch of the twistable chains as p = 2π/ψ0, and in turn, this fixes the preferred linking number to Lk = M/p, where M is the number of beads in the plasmid. The twist is enforced by a harmonic potential with stiffness κt = 50σb = 125 nm comparable with the torsional persistence length of DNA (6). In this model, the degree of supercoiling is defined as σ ≡ Lk/M = 1/p. The twist is set by initializing the patchy polymer as a flat ribbon and by subsequently slowly increasing the stiffness of the potential associated with the twist degree of freedom. Ultimately, by imposing the angle ψ0, one can achieve the desired σ (which may be zero, if ψ0 = 0 or p = ∞). It should be noted that we will also consider nontorsionally constrained plasmids in which the torsional stiffness is set to κt = 0 mimicking nicked circular plasmids. We recall that for supercoiled circular DNA, the exchange of local torsion (twist Tw) into bending (writhe Wr) must obey the White-Fuller-Călugăreanu (WFC) (37) theorem, i.e., Lk = Tw + Wr, thus conserving the linking number Lk (and thus the supercoiling σ = Lk/M) between the two DNA single strands (Fig. 1, B to D). Notice that our polymer model is symmetric with respect to supercoiling; we will thus refer to σ without specifying its sign. Last, by simulating an ensemble of linear DNA molecules, we have computed the entanglement length for this model to be Me,linear = 54 ± 2 beads (about 400 bp) via standard PPA (see the Supplementary Materials).
Supercoiling increases the average size of DNA plasmids in entangled conditions
The conformational properties of polymers in solution are typically studied in terms of the gyration tensor
| (1) |
where denotes the coordinate α of the position of bead i. The (square) radius of gyration is then defined as the trace, . We find that the time and ensemble average of scales as , with metric exponents ν ≃ 3/5 for highly supercoiled plasmids (see Fig. 2A and fig. S1). Instead, relaxed chains display a short chain regime with ν ≃ 1/2 (M ≤ 200) and a crossover to smaller values of ν ≃ 0.35 for larger chains (M ≥ 400). These exponents suggests that relaxed plasmids in entangled solutions assume conformations similar to the ones of standard ring polymers (38), i.e., ν = 1/2 for small M/Me,linear ≲ 10 and ν ≃ 1/3 for large M/Me,linear ≳ 10 (note that for our longest plasmids M/Me,linear ≃ 16; hence, we capture the crossover to the compact regime). On the other hand, supercoiling-driven writhing induces stronger self-interactions that are no longer screened by the neighbors (see Fig. 1, B and C); in this case, we thus observe a larger metric exponent ν compatible with that of a self-avoiding walk. In the asymptotic limit M → ∞, we expect dense systems of supercoiled plasmids to fall into the universality class of ideal (annealed) branched polymers (39, 40), for which ν = 1/3. This is the same exponent expected for very long flexible ring polymers, although the precise folding structure will be different.
Fig. 2. Supercoiling increases plasmids size in entangled conditions.
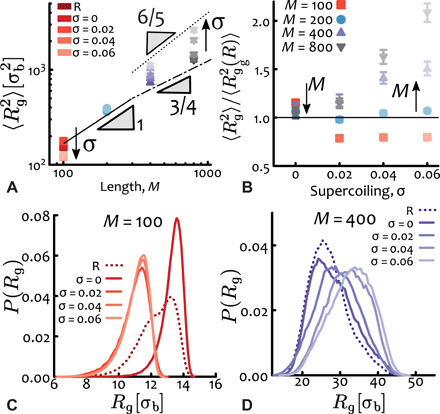
(A and B) Radius of gyrations Rg plotted against (A) contour length M and (B) supercoiling σ. Notice that for short lengths M = 100, increasing σ induces a collapse of the plasmids, whereas for longer lengths it drives swelling. The scaling of Rg as a function of plasmid length M is compatible with that of flexible rings [ν = 1/2 with crossover to ν ≃ 1/3 (38)] and that of self-avoiding walks (ν = 3/5) for relaxed and highly supercoiled plasmids, respectively. (C) The distribution of Rg for M = 100 is weakly bimodal, showing that plasmids can be in either an “open” or a “collapsed” state. Setting a supercoiling σ = 0 stabilizes the open state, whereas σ > 0 induces writhing and collapse. (D) For longer plasmids (M = 400), larger supercoiling σ broadens the distribution enlarges the average size. The unit of length is σb = 2.5 nm, and the entanglement length for linear counterparts is Me,linear = 54 beads.
The effect of supercoiling on the average size of plasmids can be better appreciated in Fig. 2B, where we show the (squared) radius of gyration rescaled by its value for relaxed plasmids and plotted against supercoiling. It is readily apparent that for long plasmids (e.g., M ≥ 400 ≃ 3 kb) the greater the supercoiling, the monotonically larger their typical size. We highlight that this behavior is highly counterintuitive, as one expects supercoiling to induce a compaction of plasmids, as indeed is found computationally in dilute conditions (32). At the same time, supercoiled plasmids travel faster than their relaxed counterparts in gel electrophoresis (33) because of their overall reduced size. Supercoiling is also often associated with the packaging of the bacterial genome (10, 41) and with organization into topological domains in eukaryotes (12, 13, 42). On the contrary, here, we observe a monotonic increase of Rg with supercoiling that is in marked contrast with the overall shrinking in dilute conditions (this shrinking is recapitulated by our model when simulated in dilute conditions; see fig. S1) (32).
We argue that this stark difference is due to interchain effects and the global topological invariance of the system. While supercoiled plasmids may want to reduce their overall size, they must also remain topologically unlinked from the neighbors. In turn, the competition between this global topological constraint and the torsional and bending rigidities appears to favor swelling of long molecules (L > 200σ ≃ 1.5 kbp) but still drives the collapse of short ones (Fig. 2B).
For the shortest plasmids considered here (M = 100 ≃ 730 bp), we observe an interesting exception to the behavior described above, whereby the typical size is nonmonotonic for increasing supercoiling levels. We attribute this peculiar behavior to a buckling transition (see below). More specifically, for σ = 0, we find that the conformations are typically larger than the relaxed ones, but they suddenly become more collapsed for σ > 0 (Fig. 2B). [Notice that with σ = 0, we mean plasmids that are intact and torsionally constrained to have linking number deficit equal to zero. These are different from relaxed (nicked) plasmids that are not torsionally constrained, as the latter do not need to obey the WFC theorem; we denote them with “R” throughout.]. We also examined the distributions of radius of gyration and noticed that relaxed short plasmids display a weakly bimodal distribution that is not found in larger plasmids (Fig. 2, C and D). This bimodal distribution reflects the fact that they can be found in two typical conformational states: either open (large Rg) or more collapsed (small Rg); imposing a certain supercoiling level appears to lock the molecules in one of the two states. Because the conformational space of non-nicked plasmids must satisfy the WFC topological conservation law, zero supercoiling (Lk = σ = 0) hinders the writhing of the plasmid because it would be energetically too costly for them to writhe multiple times with opposite sign to achieve a null global writhe, given their short length (L/lp = 5). This entails that short plasmids with σ = 0 are locked into open, not self-entangled, conformations. On the contrary, for σ > 0, the imposed writhing induces a conformational collapse, akin to a sharp buckling transition (43).
We note that the stable open state at σ = 0 for short plasmids is similar to the one computationally observed in dense solutions of semiflexible rings (44). These systems are expected to give rise to exotic columnar phases that would be thus intriguing to investigate in the context of dense solutions of short non-nicked plasmids with σ = 0.
We finally stress once more that the monotonic increase observed for long plasmids of their typical size with supercoiling is neither expected nor trivial and is in marked contrast with the overall shrinking behavior found in the literature for long dilute supercoiled plasmids (32). Because the monomer concentration is constant for all of the systems studied, and the critical overlap concentration scales as , one finds that c/c* increases with supercoiling. Thus, one would naïvely expect solutions of supercoiled plasmids to be effectively more entangled than their relaxed counterparts. As a consequence, we would also expect highly supercoiled long plasmids to display reduced mobility with respect to relaxed ones.
Supercoiling enhances DNA mobility
We study the dynamics of entangled plasmids at different levels of supercoiling by computing the time- and ensemble-averaged mean squared displacement (TAMSD) of the center of mass (CM) of the plasmids as g3(t) = 〈[rCM,i(t + t0) − rCM,i(t0)]2〉i, t0 (other gi quantities are reported in fig. S4). Curves for g3 are shown in Fig. 3 (A and B) for different values of plasmid supercoiling and length. At odds with the findings of the previous section, we find that higher values of σ yield faster mobility especially for longer plasmids.
Fig. 3. Supercoiling enhances plasmid mobility.
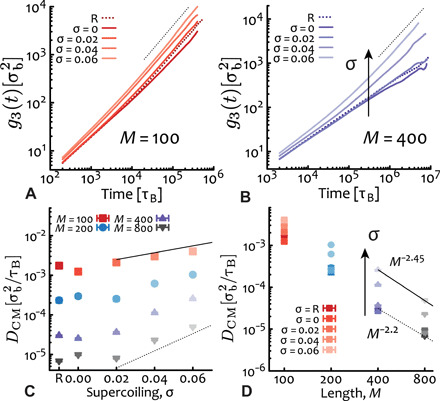
(A and B) Time-averaged mean squared displacement (TAMSD =g3) of the plasmids for (A) M = 100 ≃ 730 bp and (B) M = 400 ≃ 3 kbp. Dotted lines are linear functions of lag time as a guide to the eye. (C and D) Diffusion coefficient of the center of mass against (C) supercoiling σ and (D) length M. In (C), exponentials ~ exp (σ/0.05) (solid) and ∼ exp (σ/0.02) (dashed) are drawn as a guide to the eye (see below for a justification of exponential speedup). In (D), the best fits to the largest M for relaxed (nicked) and σ = 0.06 yield M−2.2 and M−2.45, respectively. Error bars are comparable to symbol size. R = “relaxed.”
The diffusion coefficient of the center of mass computed as allows us to more precisely quantify how the mobility of the plasmids changes as a function of length and supercoiling. We find that while DCM attains a plateau at small σ, at larger supercoiling it increases exponentially (see Fig. 3C), albeit more simulations are needed to confirm this conjecture (see below for an argument supporting the exponentially faster mobility). In addition, we find that the diffusion coefficient as a function of plasmid length scales as DCM ∼ M−2.2 and M−2.45 for relaxed and highly supercoiled large plasmids, and is compatible with the scaling of torsionally relaxed and flexible ring polymers (Fig. 3D) (22). The slightly stronger dependence on plasmid length for larger supercoiling suggests that these plasmids may effectively undergo a more traditional reptation-like relaxation and for which we expect D ∼ M−2.4 (22, 35). As we shall see below, this conjecture is confirmed by the fact that we find most of the plasmids to display two plectonemic tips and thus preferentially assume linear-like rather than branched structures (see also the Supplementary Materials).
We finally note that the solutions with M = 800 ≃ 6 kbp are not displaying a freely diffusive behavior despite the fact that we ran them for more than 107 Brownian times (see table S1); in turn, DCM is overestimated as its calculation assumes free diffusion. Despite this, values of DCM for M = 800 ≃ 6 kbp nicely follow the general trend of the other datasets (see Fig. 3, C and D).
DDM of DNA plasmids confirms simulations
To experimentally validate the prediction that supercoiling enhances the mobility of plasmids in dense solutions, we perform fluorescence microscopy experiments on 3 mg/ml solutions (corresponding to a volume fraction of 0.4%) made of 6-kb plasmids. We label 0.001% of the molecules in solution and use DDM to determine the diffusion coefficient from videos recorded on a custom fluorescence light-sheet microscope (Fig. 4A) (45). DDM, as compared to single-particle tracking, allows us to measure the dynamics of the diffusing molecules without having to resolve and track individual molecules over time—optimal for DNA of this size (Rg < 100 nm). To pinpoint the role of supercoiling, we compare a solution of plasmids extracted from Escherichia coli in the stationary phase against the same solution pretreated with topoisomerase I to relax the excess supercoiling (see Materials and Methods) (46).
Fig. 4. DDM of entangled plasmid DNA confirms the predictions from molecular dynamics simulations.
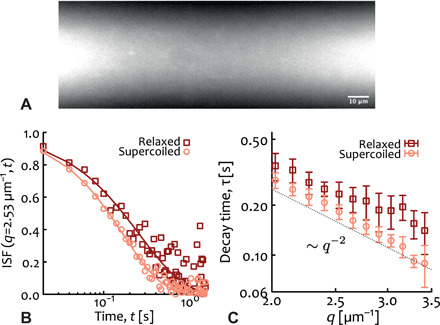
(A) Snapshot from light-sheet microscopy showing fluorescent 5.9-kbp DNA plasmids (comparable with M = 800 is the molecular dynamics simulations) at a concentration of 3 mg/ml concentration [c* ≃ 0.6 mg/ml (49) and c/c* ≃ 5]. (B) Intermediate scattering function (ISF) obtained from DDM measurements. (C) Scaling of the ISF decay time with wave vector, showing that it scales as q−2. The fitted diffusion coefficients are D = 0.34(1) μm2/s and D = 0.44(1) μm2/s for relaxed and supercoiled plasmids, respectively.
As one can notice (see Fig. 4B), the intermediate scattering function (ISF) shows a faster decay for supercoiled DNA compared to relaxed circular DNA, indicating faster dynamics. We fit each ISF with a stretched exponential f(q, t) = exp [ − (t/τ)γ] using γ ≃ 0.9 − 1 to determine the decay time τ as a function of q (Fig. 4C). As shown, the decay times are well fitted by a power law ∼q−2 that we use to extract the diffusion coefficients via the relation τ = (2Dq2)−1. The resulting diffusion coefficients are D = 0.34(1) μm2/s and D = 0.44(1) μm2/s for relaxed and supercoiled solutions, respectively.
We should note that while our choice of plasmid length allows us to purify them without introducing substantial nicks (∼80% are without nicks and thus supercoiled), determining their precise supercoiling level is not straightforward. In vivo, supercoiling for plasmids in the stationary phase of cell growth (the phase at which we extract our plasmids) is ∼2% (47, 48). Thus, these results suggest that increasing supercoiling in solutions of entangled plasmids speeds them up and are thus in qualitative agreement with the simulations.
We should mention that while the experiments are at lower volume fraction with respect to simulations (when considering bare DNA), the buffering condition effectively thickens the diameter of DNA (49), thus rendering the precise comparison of experimental and simulated volume fractions difficult. We also note that because of the small size of the plasmids, we are unable to accurately measure their size using single-molecule imaging. In turn, this renders the precise estimation of the overlap concentration also challenging [indirectly estimated to be about c* ≃ 0.6 mg/ml (18, 49)]. We are currently investigating alternative approaches, such as dynamic light scattering, so that in future work we can compare the intriguing predictions regarding the different sizes of supercoiled and relaxed circular DNA in dense solutions.
Supercoiling induces a buckling transition in short plasmids
The consequence of writhing on the plasmid conformations is not captured by Rg alone (50, 51). Instead, it is informative to study shape descriptors that can be computed via the eigenvalues of the gyration tensor RT (which we denote as a, b, and c, with a > b > c and ). Typical shape descriptors are the asphericity (50–52) , which quantifies the deviation from a perfectly spherical arrangement and the nature of asphericity quantified by either the prolateness (see fig. S2) or the anisotropy (shown in Fig. 5, A and B). These shape descriptors reveal that for M = 100 ≃ 730 bp and σ = 0, plasmids are stabilized in an open, highly symmetric, and oblate (M&M’s) state. Furthermore, they reveal that these short plasmids undergo a buckling transition to a closed, asymmetric, and prolate (rugby ball) shape for σ > 0. The sharp first-order–like buckling transition (see Fig. 5A and the Supplementary Materials) is weakened for larger contour lengths (see Fig. 5B), as self-writhing is energetically allowed even for σ = 0 (negative and positive self-crossings must cancel each other to satisfy the WFC conservation law). At the same time, both short and long plasmids display a general increase in asphericity, prolateness, and anisotropy with increasing supercoiling, strongly suggesting that the plasmids assume elongated and double-folded conformations (see fig. S2).
Fig. 5. Supercoiling induces buckling in short plasmids and reduces the threadable area.
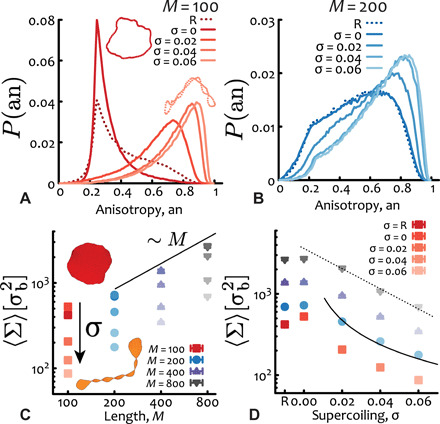
(A) The anisotropy shape descriptor (an, see text) for short plasmids M = 100 ≃ 730 bp displays a sharp buckling transition between an open and roughly symmetric state for σ = 0 and a collapsed and anisotropic one for σ > 0. In inset, two examples of conformations are shown. (B) For longer plasmids (M ≥ 200 ≃ 1.5 kbp), supercoiling shifts the anisotropy to larger values, indicating a smoother transition to more prolate conformations. (C) Scaling of the average minimal surface size 〈Σ〉 as a function of plasmid length (solid line shows the linear scaling). In inset, two examples of surfaces for M = 100 ≃ 730 bp are shown. (D) The size of the minimal surface area monotonically decreases with supercoiling (with the exception of short M ≤ 200 ≃ 1.5 kbp plasmids). The solid and dashed lines scale as 1/σ and e−σ/0.035, respectively, and are drawn as a guide to the eye. R = relaxed. The unit of length is σb = 2.5 nm. The error bars, typically smaller than the symbol size, represent the error of the mean area.
Supercoiling decreases the spanning minimal surface
It is natural to associate the open-oblate/closed-prolate conformations assumed by DNA plasmids to a larger/smaller (minimal) spanning area, respectively (53). The size of this area may be relevant for the dynamics because it could be “threaded” by neighboring plasmids, hence hindering the dynamics (24, 54, 55). To quantify this in more detail, we calculated the minimal surface (53) using the algorithm in (55, 56) for flexible ring polymers. We found that the minimal area grows approximately linearly with the plasmids’ contour, as expected for ν ≤ 1/2 (Fig. 5C) (55). We also observed that it overall decreased with supercoiling with the notable exception of short M ≤ 200 ≃ 1.5 kbp plasmids, for which there is a small increase for σ = 0 with respect to the relaxed case, again confirming the “topological locking” of open conformations (Fig. 2A).
A crude way to estimate the decrease in “threadable” area of a plasmid Σ is via recursive bisections of a perfect circle into several connected smaller circles joined at a vertex mimicking writhe-induced self-crossing. Each time a circle is split into two smaller ones, the new radii are R′ ≃ R/2 and thus n circles (with n − 1 self-crossings) have radii R′ = R/n, yielding an overall spanning surface Σ ≃ nπ(R/n)2 ∼ 1/n ∼ 1/σ. The same scaling of the threadable area is obtained if considers the supercoil as if wrapped around a cylinder of radius r (57) and projected in 2D; in this case, one would find that the enclosed area in each of the n superhelix turns is about rLee/(n − 1), where Lee is the end-to-end length of the plasmid. Given that r ∼ 1/σ (57) and that Lee is expected to be insensitive on σ for large supercoiling (see also Fig. 2B showing plateauing of Rg for M ≤ 400), one finds a total threadable area scaling as Σ ≃ nrLee/(n − 1) ∼ 1/σ. The fact that our data are instead more compatible with an exponential decrease of Σ as a function of supercoiling (Fig. 5D) suggests that the approximation of the supercoil wrapped around a cylinder may not be accurate. In fact, considering the large persistence length of DNA, it may be thermodynamically preferred to flatten and shrink the many inner openings at the expense of storing longer contour length at the fewer tips (see also snapshots in Fig. 1, B to E, and inset of Fig. 5C). These estimations are in good agreement with the scaling of the minimal surface, although we cannot rule out other functional forms (for instance, exponential; see Fig. 5D). [Note that the so-called magnetic moment and radius (58) give similar results, albeit different scaling (see fig. S3).]
Supercoiling reduces threadings
Motivated by the observation that the minimal surface—or “threadable area”—sharply decreases with supercoiling, we decided to quantify more precisely the number of threadings per plasmid for different levels of supercoiling. To this end, we identify a plasmid to be “passively threaded” by another when the minimal surface of the former is intersected by the contour of the latter (at least twice, as they are topologically unlinked) (Fig. 6A) (55). As shown in Fig. 6B, the average number of threadings per plasmid 〈nt〉 also appears to decrease exponentially with supercoiling and to mirror the behavior of the mean threadable area 〈Σ〉. [As for the minimal surface Σ, a notable exception to this general trend is the case of short plasmids (M = 100) for which we find that 〈nt〉 is statistically larger for σ = 0 than for relaxed plasmids because of the topological locking that we explained above.]
Fig. 6. Supercoiling reduces threadings and entanglements.
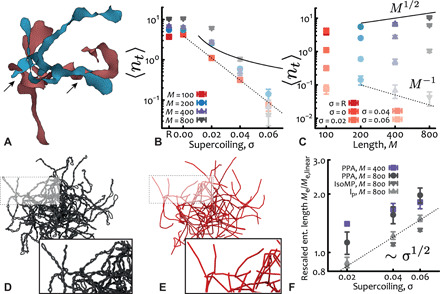
(A) Snapshot of two threading plasmids (relaxed, M = 800 ≃ 6 kbp) with minimal surfaces drawn and intersections highlighted by arrows. (B) Number of threadings per plasmid as a function of supercoiling (dashed = exponential, solid = 1/σ). (C) Number of threadings per plasmid as a function of DNA length (dashed = 1/M, solid = M1/2). (D and E) Snapshots of the PPA analysis run on a system with plasmids M = 800 ≃ 6 kbp and σ = 0.06. (F) The effective entanglement length increases with supercoiling as Me/Me,linear ∼ σα, with α ≃ 0.5 for both PPA and IsoMP methods. Note that Me,linear = 54 ± 2 (PPA) and Me,linear = 49 ± 2 (IsoMP). The effective persistence length lp/lp,linear also shows a scaling compatible with σ1/2 (lp,linear = 18 ± 1).
On the basis of these findings, we can also advance an argument as for why the diffusion coefficient of plasmids increases exponentially with supercoiling: Recent evidence suggests that the dynamics of ring polymers with threadings slow down exponentially with the number of threadings [e.g., entangled rings (55, 59, 60), melts of tadpole-shaped polymers (20, 61), or compressed long plasmids (62)]. We thus expect the dynamics of highly supercoiled (threading poor) plasmids to be exponentially faster than their relaxed (threading-rich) counterparts, as seen in Fig. 3C.
Intriguingly, in the case of short plasmids in which setting σ = 0 increases the threadable area and also the number of threadings, we also find a slower dynamics, in full agreement with our argument (see Figs. 3C and 6B).
Supercoiling reduces entanglements
The shape descriptors studied above suggest that long plasmids assume prolate double-folded conformations, but it remains unclear whether the conformations are simply plectonemic (linear-like) or more branched into comb, star, or tree-like structures (63). We thus computed the local absolute writhe along the contour length, W(s), from which the number and location of plectonemic tips can be identified as the local maxima of W(s) (see Materials and Methods and the Supplementary Materials) (64, 65). This calculation reveals that most of the conformations with σ ≥ 0.04 have two tips and so are mainly linear-like plectonemic conformations (see fig. S5). (For smaller supercoiling, it is difficult to unambiguously distinguish tips from other regions of large curvature.)
In light of this finding, another apparent controversy arises. Arguably, linear chains half the length as their ring counterparts are expected to diffuse slower than the rings due to reptation relaxation induced by ordinary entanglements (assuming that the entanglement length is the same for the two systems) (22); instead, we observe the opposite trend. To explain this result, we adapted the PPA (66) and isoconfigurational mean path (IsoMP) (67) methods to estimate the effective entanglement length of these systems (see Fig. 6, C and D, and fig. S10). For PPA, we determined an effective entanglement length Me by leveraging the fact that the tips of linear-like or branched conformations represent effective termini that can be pinned in space (see the Supplementary Materials for more details on PPA and IsoMP methods). (Note that the PPA method typically fails for standard flexible ring polymers because there are no well-defined ends to pin.)
We find that irrespective of the method chosen, the scaling of the effective entanglement length is compatible with Me ∼ σ1/2 (Fig. 6F), suggesting that the larger the supercoiling, the less entangled the plasmids. [The numerical difference of PPA and IsoMP is a known feature for topologically constrained ring polymers (67, 68) with Me,PPA/Me,IsoMP ≃ 3/2 and is in agreement with our findings for plasmids.] We argue that this effective reduction in entanglement [opposite to what one would naïvely expect considering or similar packing length arguments as , Lee being the end-to-end distance] is due to the fact that supercoiling (i) induces highly anisotropic conformations and (ii) increases the local concentration of intrachain beads (69). Because the superhelix radius of plasmids scales as r ∼ 1/σ (57) and most plasmids display only two tips (fig. S6), this entails that the intrachain density ρintra ∼ M/V ∼ σ2 (with V = πr2Lee the approximated cylindrical volume of the supercoil) grows with supercoiling.
Notably, we also find that the effective persistence length, computed as the decay length of the tangent-tangent correlation, , along the plasmid backbone (from tip to tip) scales as , in turn yielding or (Fig. 6F). This is compatible with the fact that our systems appear to be at the crossover between the semiflexible and stiff regimes, based on the values of density, stiffness, and chain diameter and as supported by the typical values of Me extracted from both PPA and IsoMP, which are of the order of the effective Kuhn length (Fig. 6F). In this crossover, both tube diameter and entanglement length scale linearly with the Kuhn length lk (70). The stiffening of the supercoiled plasmids can be naturally thought of as due to the self-writhing; in particular, one may argue that the shorter the contour length between self-crossings, i.e., the smaller 1/σ, the longer the effective persistence length displayed by the plasmids. A Flory-type estimate of the interaction free energy of n monomers Fint ∼ kTvn2/r3, per superhelix turn (n ∼ σ−1 and r ∼ σ−1), gives Fint ∼ σ. This can be viewed as if the excluded volume of the cylindrical Kuhn monomer v (composed of spherical beads in line) grew by a factor of σ. Because (71), the effective length Kuhn length .
Then, in the stiff regime, the plasmids behave as if they were rigid chains of diameter 2r ∼ 2/σ confined within narrow tubes with diameter a; in analogy with the classical Odijk problem (72), we can thus write . In turn, the value of the tube diameter can be obtained from the estimate that in each area element aMe spanned by the plasmid, there is about one transversal segment, i.e., ρsaMe ≃ 1, where ρs = ϕ/(2r)2 is the arc length density and ϕ is the polymer volume fraction (ϕ = r3M(b/r)/V in terms of supercoil turns with r ∼ b/σ). Combining these together (70), we expect Me ≃ lkϕ−2/5(d/lk)4/5 ∼ σ0.1 to be attained at very large values of σ, for which the stiff regime (a ≪ Me and lk ≪ Me) is justified.
Last, we note that for short plasmids, the PPA method cannot identify an entanglement length, confirming that these are very poorly entangled in the standard sense. Hence, their dynamics are mostly determined by threadings, which are abundant also in short plasmids (see Fig. 6, B and C).
DISCUSSION
In this work, we have studied the dynamics of entangled solutions of DNA plasmids to understand how supercoiling can be leveraged to tune the rheology of dense DNA solutions orthogonally to other traditional methods, such as varying length or concentration. We have found that, contrary to what is typically assumed, the size of long plasmids increases with supercoiling when in entangled solutions.
In dilute conditions, supercoiled plasmids are expected to fall into the universality class of interacting annealed branched polymers for which a metric exponent ν = 7/13 is expected asymptotically (39, 63). In the melt phase, the self-interactions are screened, and we thus expect supercoiled plasmids to behave as ideal annealed branched polymers or lattice animals for which ν = 1/4 (40); being unphysical in d = 3, we expect the size of very large supercoiled plasmids in the melt to scale with a metric exponent ν = 1/3. Although this is the same scaling expected for relaxed rings (23, 73), the folded structures are expected to be different. The supercoiling-driven swelling can still be achieved through a non-universal prefactor in front of a supercoiling independent universal scaling M1/3, for instance, because of an effectively larger persistence length (as we found in this work; Fig. 6F). The fact that we observe a metric exponent that depends on σ (Fig. 2A) thus suggests that our simulations are not in the asymptotic limit, and yet still in a regime that is experimentally interesting.
We find that the swelling of supercoiled plasmids is mirrored by an enhanced mobility. Our predictions are supported by experiments that show that the diffusion coefficient of entangled intact and supercoiled (σ ≃ 0.02) plasmids is larger than that of relaxed ones, i.e., with σ ≃ 0. We found that this enhanced mobility is due to severely asymmetric conformations that greatly reduce the threadable area and number of threadings. In parallel, entanglements are also reduced as supercoiling increases the effective entanglement length by increasing the local concentration of intra-chain contacts. We note that threadings are abundant also in short plasmids (Fig. 6B) that are poorly entangled in the standard sense; we observe that, in this case, threadings play a major role in determining the dynamics of short plasmids (notably for M = 100, the case with σ = 0 is slower and displays more threadings than the relaxed one). We have thus found that the unexpected enhanced diffusivity of entangled supercoiled DNA is due to a combination of reduced entanglements and, in particular, threadings.
We conjecture that beyond the range of lengths studied in this work (0.7 to 6 kbp), supercoiled plasmids in entangled solutions may display branched and annealed conformations (i.e., with nonfixed branching points), triggering the need of arm retraction or plectoneme diffusion/hopping relaxation mechanisms. These processes are notoriously slow, on the order of kbp2/s (74), and we thus predict a re-entrant slowing down of the diffusion of supercoiled plasmids. They ought to behave as quenched/annealed branched polymers on time scales shorter/longer than plectoneme diffusion, respectively. Ultimately, despite the expected onset of (exponentially) slowly diffusive “branched-polymer–like” regime for supercoiled plasmids, relaxed ones will still display many more threadings, which we argue will still (exponentially) slow down their dynamics also in the large length limit. Dissecting the contribution of these mechanisms will require longer simulations than currently possible.
In summary, our results suggest a route for the topological tuning of the rheology of DNA-based complex fluids that uses supercoiling as a mean to control DNA mobility. We note that the fact that supercoiling regulates the number of threadings per plasmids can also be leveraged in polydisperse systems or in blends of linear and supercoiled DNA or other biopolymer composites, where threading of rings by the linear fraction is key to determine the stress relaxation of the fluids (20, 21, 61, 75).
In the future, it would be interesting to further investigate longer plasmids with selected or varying levels of supercoiling. Albeit experimentally difficult, this may be feasible using cesium chloride gradient separation techniques (76). Ultimately, understanding how DNA topology and supercoiling affect the dynamics and conformational properties of plasmids in entangled or crowded conditions may not only reveal novel pathways to finely tune the rheology of complex biopolymer fluids but also shed light on the role of supercoiling on chromosome dynamics in vivo (10, 77).
MATERIALS AND METHODS
Molecular dynamics
Each bead in our simulation is evolved through the Langevin equation , where ma and γa are the mass and the friction coefficient of bead a, and is its stochastic noise vector satisfying the fluctuation-dissipation theorem. U is the sum of the energy fields (see the Supplementary Materials). The simulations are performed in LAMMPS (36) with m = γ = kB = T = 1 and using a velocity-Verlet algorithm with integration time step Δt = 0.002 τB, where τB = γσ2/kBT ≃ 0.03 μs is the Brownian time (γ = 3πσηw, with ηw = 1 cP the viscosity of water).
Branching analysis
Following (64, 77), we compute the absolute writhe of a segment of a plasmid as with window l = 50 beads. This calculation yields a function W(s) whose maxima represent regions of high local writhe and can identify tips of plectonemes. In addition to being a local maximum, we require that W(s) > 0.35 to avoid false positives. See the Supplementary Materials for more details.
Primitive path analysis
Following (66), we fix certain polymer segments in space, turn intrachain repulsive interactions off, and keep interchain interactions on. We then run simulations at low temperature 0.01 to find a ground state. The resulting chain conformations (primitive paths) are made of straight segments connected by sharp kinks due to entanglements. The entanglement length is then given by , where ree is the mean endpoint distance, M is the number of monomers between the fixed points, and bpp is the mean bond length of the primitive path. We adapt the classical PPA for plasmids by fixing the tips of all detected plectonemes instead of the end points of linear chains (see the Supplementary Materials).
DNA preparation
Double-stranded 5.9-kbp DNA plasmids are replicated in E. coli, collected at the onset of stationary phase, before being extracted and purified using our previously described protocols (49). Following purification, the DNA solution is ∼80% supercoiled and ∼20% relaxed circular, as determined from gel electrophoresis (fig. S6). To produce concentrated solutions of relaxed circular DNA, topoisomerase I (New England Biolabs) is used to convert the DNA topology from supercoiled to relaxed circular (fig. S6) (79). Both supercoiled and relaxed circular DNA solutions are concentrated to 3 mg/ml using Eppendorf Vacufuge 5301.
Fluorescence imaging
To visualize DNA diffusion in concentrated solutions, supercoiled or relaxed circular DNA is labeled with YOYO-1 dye (Thermo Fisher Scientific) at a 4:1 bp:dye ratio and added at a concentration of 0.045 μg/ml to 3 mg/ml solutions of supercoiled or relaxed circular DNA described above. Glucose (0.9 mg/ml), glucose oxidase (0.86 mg/ml), and catalase (0.14 mg/ml) are added to inhibit photobleaching (46, 80). The DNA solutions are pipetted into capillary tubing that is index-matched to water and imaged using a custom-built light-sheet microscope with a 488-nm excitation laser, an excitation objective of 10× 0.25 numerical aperture (NA), an imaging objective of 20× 1.0 NA, and an Andor Zyla 4.2 CMOS camera. At least four sample videos are recorded at 50 frames per second for 2000 frames. The video dimensions are 256 pixels × 768 pixels, which are then analyzed by examining regions of interest (ROIs) of 256 pixels × 256 pixels (50 μm × 50 μm).
DDM analysis
We follow methods previously described to investigate DNA diffusion using DDM (45). Briefly, from each ROI, we obtain the image structure function or DDM matrix D(q, Δt), where q is the magnitude of the wave vector and Δt is the lag time. To extract the transport dynamics of the diffusing DNA molecules, we fit the structure functions to D(q, Δt) = A(q)[1 − f(q, Δt)] + B(q), where B is a measure of the camera noise, A depends on the optical properties of both the sample and microscope, and f(q, Δt) is the ISF. On the basis of our previous studies of microspheres and DNA diffusing in crowded environments, we fit the ISFs to stretched exponentials of the form f(q, Δt) = exp − (Δt/τ(q))γ(q), where τ is the characteristic decay time and γ is the stretching exponent, both of which depend on q (46).
For normal free diffusion, one expects ISFs described by a simple exponential, i.e., γ = 1, while our scattering functions are better fitted with stretching exponents between 0.9 and 1. Having extracted the decay times of density fluctuations τ over a range of spatial frequencies q, we fit the results to τ = (2Dq2)−1 to determine the diffusion coefficient, D, for the DNA plasmids.
Acknowledgments
We would like to acknowledge the networking support by the “European Topology Interdisciplinary Action” (EUTOPIA) CA17139. Funding: This project has received funding from the European Union’s Horizon 2020 programme under grant agreement no. 731019 (EUSMI). D.M. acknowledges the computing time provided on the supercomputer JURECA at Jülich Supercomputing Centre and the support by the Leverhulme Trust (ECF-2019-088), the Royal Society, and ERC (StG TAP 947918). J.S. acknowledges the support from the Austrian Science Fund (FWF) through Lise-Meitner Fellowship No. M2470-N28. J.S. is grateful for the computational time at Vienna Scientific Cluster. R.R.-A. is supported by the Air Force Office of Scientific Research (grant no. AFOSR-FA9550-17-1-0249). Sample codes can be found at git.ecdf.ed.ac.uk/dmichiel/supercoiledplasmids. Author contributions: D.M. conceived the project. D.M. and J.S. performed and analyzed simulations. J.G. and R.R.-A. performed experiments. All authors wrote the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/20/eabf9260/DC1
REFERENCES AND NOTES
- 1.Um S. H., Lee J. B., Park N., Kwon S. Y., Umbach C. C., Luo D., Enzyme-catalysed assembly of DNA hydrogel. Nat. Mater. 5, 797–801 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Rothemund P. W. K., Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Seeman N. C., Sleiman H. F., DNA nanotechnology: Building big with DNA bricks. Nat. Rev. Mater. 3, 17092 (2018). [Google Scholar]
- 4.Mao C., Sun W., Seeman N. C., Assembly of Borromean rings from DNA. Nature 386, 137–138 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Leunissen M. E., Dreyfus R., Sha R., Wang T., Seeman N. C., Pine D. J., Chaikin P. M., Towards self-replicating materials of DNA-functionalized colloids. Soft Matter 5, 2422 (2009). [Google Scholar]
- 6.A. Bates, A. Maxwell, DNA Topology (Oxford Univ. Press, 2005). [Google Scholar]
- 7.Robertson R. M., Smith D. E., Strong effects of molecular topology on diffusion of entangled DNA molecules. Proc. Natl. Acad. Sci. U.S.A. 104, 4824–4827 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teixeira R. E., Dambal A. K., Richter D. H., Shaqfeh E. S. G., Chu S., The Individualistic dynamics of entangled DNA in solution volume 40, number 7, April 3, 2007, pp 2461−2476. Macromolecules 40, 3514 (2007). [Google Scholar]
- 9.Fitzpatrick R., Michieletto D., Peddireddy K. R., Hauer C., Kyrillos C., Gurmessa B. J., Robertson-Anderson R. M., Synergistic interactions between DNA and actin trigger emergent viscoelastic behavior. Phys. Rev. Lett. 121, 257801 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Wu F., Japaridze A., Zheng X., Wiktor J., Kerssemakers J. W. J., Dekker C., Direct imaging of the circular chromosome in a live bacterium. Nat. Commun. 10, 2194 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Racko D., Benedetti F., Goundaroulis D., Stasiak A., Chromatin loop extrusion and chromatin unknotting. Polymers 10, 1126 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benedetti F., Dorier J., Burnier Y., Stasiak A., Models that include supercoiling of topological domains reproduce several known features of interphase chromosomes. Nucleic Acids Res. 42, 2848–2855 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naughton C., Avlonitis N., Corless S., Prendergast J. G., Mati I. K., Eijk P. P., Cockroft S. L., Bradley M., Ylstra B., Gilbert N., Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat. Struct. Mol. Biol. 20, 387–395 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irobalieva R. N., Fogg J. M., Catanese D. J. Jr., Sutthibutpong T., Chen M., Barker A. K., Ludtke S. J., Harris S. A., Schmid M. F., Chiu W., Zechiedrich L., Structural diversity of supercoiled DNA. Nat. Commun. 6, 8440 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fosado Y. A. G., Michieletto D., Marenduzzo D., Dynamical scaling and phase coexistence in topologically constrained DNA melting. Phys. Rev. Lett. 119, 118002 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Sutthibutpong T., Matek C., Benham C., Slade G. G., Noy A., Laughton C., Doye J. P. K., Louis A. A., Harris S. A., Long-range correlations in the mechanics of small DNA circles under topological stress revealed by multi-scale simulation. Nucleic Acids Res. 44, 9121–9130 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding Y., Manzo C., Fulcrand G., Leng F., Dunlap D., Finzi L., DNA supercoiling: A regulatory signal for the λ repressor. Proc. Natl. Acad. Sci. U.S.A. 111, 15402–15407 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laib S., Robertson R. M., Smith D. E., Preparation and characterization of a set of linear DNA molecules for polymer physics and rheology studies. Macromolecules 39, 4115–4119 (2006). [Google Scholar]
- 19.Zhu X., Kundukad B., Van Der Maarel J. R., Viscoelasticity of entangled lambda-phage DNA solutions. J. Chem. Phys. 129, 185103 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Rosa A., Smrek J., Turner M. S., Michieletto D., Threading-induced dynamical transition in tadpole-shaped polymers. ACS Macro Lett. 9, 743–748 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapnistos M., Lang M., Vlassopoulos D., Pyckhout-Hintzen W., Richter D., Cho D., Chang T., Rubinstein M., Unexpected power-law stress relaxation of entangled ring polymers. Nat. Mater. 7, 997–1002 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halverson J. D., Lee W. B., Grest G. S., Grosberg A. Y., Kremer K., Molecular dynamics simulation study of nonconcatenated ring polymers in a melt. II. Dynamics. J. Chem. Phys. 134, 204905 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Rosa A., Everaers R., Ring polymers in the melt state: The physics of crumpling. Phys. Rev. Lett. 112, 118302 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Michieletto D., Turner M. S., A topologically driven glass in ring polymers. Proc. Natl. Acad. Sci. U.S.A. 113, 5195–5200 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krajina B. A., Zhu A., Heilshorn S. C., Spakowitz A. J., Active DNA olympic hydrogels driven by topoisomerase activity. Phys. Rev. Lett. 121, 148001 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Wu Q., Rauscher P. M., Lang X., Wojtecki R. J., de Pablo J. J., Hore M. J. A., Rowan S. J., Poly[n]catenanes: Synthesis of molecular interlocked chains. Science 358, 1434–1439 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Rauscher P. M., Schweizer K. S., Rowan S. J., De Pablo J. J., Thermodynamics and structure of Poly[n]catenane melts. Macromolecules 53, 3390–3408 (2020). [Google Scholar]
- 28.Mei B., Dell Z. E., Schweizer K. S., Microscopic theory of long-time center-of-mass self-diffusion and anomalous transport in ring polymer liquids. Macromolecules 53, 10431–10445 (2020). [Google Scholar]
- 29.Gómez L. R., García N. A., Pöschel T., Packing structure of semiflexible rings. Proc. Natl. Acad. Sci. U.S.A. 117, 3382–3387 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abadi M., Serag M. F., Habuchi S., Entangled polymer dynamics beyond reptation. Nat. Commun. 9, 5098 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brackley C. A., Morozov A. N., Marenduzzo D., Models for twistable elastic polymers in Brownian dynamics, and their implementation for LAMMPS. J. Chem. Phys. 140, 135103 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Krajina B. A., Spakowitz A. J., Large-scale conformational transitions in supercoiled DNA revealed by coarse-grained simulation. Biophys. J. 111, 1339–1349 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cebrián J., Chen T., Amendola M., van Steensel B., Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 3112, e168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.M. Doi, S. Edwards, The Theory of Polymer Dynamics (Oxford Univ. Press, 1988). [Google Scholar]
- 35.Kremer K., Grest G. S., Dynamics of entangled linear polymer melts: A molecular-dynamics simulation. J. Chem. Phys. 92, 5057–5086 (1990). [Google Scholar]
- 36.Plimpton S., Fast Parallel algorithms for short-range molecular dynamics. J. Comp. Phys. 117, 1–19 (1995). [Google Scholar]
- 37.Dennis M. R., Hannay J. H., Geometry of Călugăreanu’s theorem. Proc. R. Soc. A 461, 3245–3254 (2005). [Google Scholar]
- 38.Halverson J. D., Lee W. B., Grest G. S., Grosberg A. Y., Kremer K., Molecular dynamics simulation study of nonconcatenated ring polymers in a melt. I. Statics. J. Chem. Phys. 134, 204904 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Gutin A. M., Grosberg A. Y., Shakhnovich E. I., Microphase separation in randomly branched polymers. Macromolecules 26, 3598–3600 (1993). [Google Scholar]
- 40.Lubensky T. C., Isaacson J., Statistics of lattice animals and dilute branched polymers. Phys. Rev. A 20, 2130–2146 (1979). [Google Scholar]
- 41.Sinden R. R., Pettijohn D. E., Chromosomes in living Escherichia coli cells are segregated into domains of supercoiling. Proc. Natl. Acad. Sci. U.S.A. 78, 224–228 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benedetti F., Racko D., Dorier J., Burnier Y., Stasiak A., Transcription-induced supercoiling explains formation of self-interacting chromatin domains in S. pombe. Nucleic Acids Res. 45, 9850–9859 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ott K., Martini L., Lipfert J., Gerland U., Dynamics of the buckling transition in double-stranded DNA and RNA. Biophys. J. 118, 1690–1701 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernabei M., Bacova P., Moreno A. J., Narros A., Likos C. N., Fluids of semiflexible ring polymers: Effective potentials and clustering. Soft Matter 9, 1287–1300 (2013). [Google Scholar]
- 45.Cerbino R., Trappe V., Differential dynamic microscopy: Probing wave vector dependent dynamics with a microscope. Phys. Rev. Lett. 100, 188102 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Wulstein D. M., Regan K. E., Garamella J., McGorty R. J., Robertson-Anderson R. M., Topology-dependent anomalous dynamics of ring and linear DNA are sensitive to cytoskeleton crosslinking. Sci. Adv. 5, eaay5912 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balke V. L., Gralla J. D., Changes in the linking number of supercoiled DNA accompany growth transitions in Escherichia coli. J. Bacteriol. 169, 4499–4506 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Z., Dröge P., Long-range effects in a supercoiled DNA domain generated by transcription in vitro. J. Mol. Biol. 271, 499–510 (1997). [DOI] [PubMed] [Google Scholar]
- 49.Robertson R. M., Laib S., Smith D. E., Diffusion of isolated DNA molecules: Dependence on length and topology. Proc. Natl. Acad. Sci. U.S.A. 103, 7310–7314 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rawdon E. J., Kern J. C., Piatek M., Plunkett P., Stasiak A., Millett K. C., Effect of knotting on the shape of polymers. Macromolecules 41, 8281–8287 (2008). [Google Scholar]
- 51.Benedetti F., Japaridze A., Dorier J., Racko D., Kwapich R., Burnier Y., Dietler G., Stasiak A., Effects of physiological self-crowding of DNA on shape and biological properties of DNA molecules with various levels of supercoiling. Nucleic Acids Res. 43, 2390–2399 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosa A., Orlandini E., Tubiana L., Micheletti C., Structure and dynamics of ring polymers: Entanglement effects because of solution density and ring topology. Macromolecules 44, 8668–8680 (2011). [Google Scholar]
- 53.Lang M., Ring conformations in bidisperse blends of ring polymers. Macromolecules 46, 1158–1166 (2013). [Google Scholar]
- 54.Michieletto D., Nahali N., Rosa A., Glassiness and heterogeneous dynamics in dense solutions of ring polymers. Phys. Rev. Lett. 119, 197801 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Smrek J., Grosberg A. Y., Minimal surfaces on unconcatenated polymer rings in melt. ACS Macro Lett. 5, 750–754 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Smrek J., Kremer K., Rosa A., Threading of unconcatenated ring polymers at high concentrations: Double-folded vs time-equilibrated structures. ACS Macro Lett. 8, 155–160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boles T. C., White J. H., Cozzarelli N. R., Structure of plectonemically supercoiled DNA. J. Mol. Biol. 213, 931–951 (1990). [DOI] [PubMed] [Google Scholar]
- 58.Schram R. D., Rosa A., Everaers R., Local loop opening in untangled ring polymer melts: A detailed “Feynman test” of models for the large scale structure. Soft Matter 15, 2418–2429 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Michieletto D., Marenduzzo D., Orlandini E., Alexander G. P., Turner M. S., Dynamics of self-threading ring polymers in a gel. Soft Matter 10, 5936–5944 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Smrek J., Chubak I., Likos C. N., Kremer K., Active topological glass. Nat. Commun. 11, 26 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doi Y., Takano A., Takahashi Y., Matsushita Y., Melt rheology of tadpole-shaped polystyrenes. Macromolecules 48, 8667–8674 (2015). [Google Scholar]
- 62.Soh B. W., Klotz A. R., Robertson-Anderson R. M., Doyle P. S., Long-lived self-entanglements in ring polymers. Phys. Rev. Lett. 123, 048002 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Everaers R., Grosberg A. Y., Rubinstein M., Rosa A., Flory theory of randomly branched polymers. Soft Matter 13, 1223–1234 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Michieletto D., On the tree-like structure of rings in dense solutions. Soft Matter 12, 9485–9500 (2016). [DOI] [PubMed] [Google Scholar]
- 65.Vologodskii A. V., Levene S. D., Klenin K. V., Frank-Kamenetskii M., Cozzarelli N. R., Conformational and thermodynamic properties of supercoiled DNA. J. Mol. Biol. 227, 1224–1243 (1992). [DOI] [PubMed] [Google Scholar]
- 66.Everaers R., Sukumaran S. K., Grest G. S., Svaneborg C., Sivasubramanian A., Kremer K., Rheology and microscopic topology of entangled polymeric liquids. Science 303, 823–826 (2004). [DOI] [PubMed] [Google Scholar]
- 67.Bisbee W., Qin J., Milner S. T., Finding the tube with isoconfigurational averaging. Macromolecules 44, 8972–8980 (2011). [Google Scholar]
- 68.Hoy R. S., Kröger M., Unified analytic expressions for the entanglement length, tube diameter, and plateau modulus of polymer melts. Phys. Rev. Lett. 124, 147801 (2020). [DOI] [PubMed] [Google Scholar]
- 69.A. V. Vologodskii, N. R. Cozzarelli, Conformational and thermodynamic properties of supercoiled DNA. Annu. Rev. Biophys. 1224–1243 (1994); http://www.annualreviews.org/doi/pdf/10.1146/annurev.bb.23.060194.003141. [DOI] [PubMed]
- 70.Milner S. T., Unified entanglement scaling for flexible, semiflexible, and stiff polymer melts and solutions. Macromolecules 53, 1314–1325 (2020). [Google Scholar]
- 71.Rubinstein M., Colby H. R., Polymer Physics 2003 (Oxford Univ. Press, 1995). [Google Scholar]
- 72.Odijk T., The statistics and dynamics of confined or entangled stiff polymers. Macromolecules 16, 1340–1344 (1983). [Google Scholar]
- 73.Grosberg A., Rabin Y., Havlin S., Neer A., Crumpled globule model of the three-dimensional structure of DNA. Europhys. Lett. 23, 373–378 (1993). [Google Scholar]
- 74.van Loenhout M. T., de Grunt M. V., Dekker C., Dynamics of DNA supercoils. Science 338, 94–97 (2012). [DOI] [PubMed] [Google Scholar]
- 75.Parisi D., Ahn J., Chang T., Vlassopoulos D., Rubinstein M., Stress relaxation in symmetric ring-linear polymer blends at low ring fractions. Macromolecules 53, 1685–1693 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Clewell D. B., Helinski D. R., Supercoiled circular DNA-protein complex in Escherichia coli: Purification and induced conversion to an open circular DNA form. Proc. Natl. Acad. Sci. U.S.A. 62, 1159–1166 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Japaridze A., Gogou C., Kerssemakers J. W., Nguyen H. M., Dekker C., Direct observation of independently moving replisomes in Escherichia coli. Nat. Commun. 11, 3109 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Klenin K., Langowski J., Computation of writhe in modeling of supercoiled DNA. Biopolymers 54, 307–317 (2000). [DOI] [PubMed] [Google Scholar]
- 79.Chapman C., Shanbhag S., Smith D. E., Robertson-Anderson R. M., Complex effects of molecular topology on diffusion in entangled biopolymer blends. Soft Matter 8, 9177–9182 (2012). [Google Scholar]
- 80.Garamella J., Regan K., Aguirre G., McGorty R. J., Robertson-Anderson R. M., Anomalous and heterogeneous DNA transport in biomimetic cytoskeleton networks. Soft Matter 16, 6344–6353 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ge T., Panyukov S., Rubinstein M., Self-similar conformations and dynamics in entangled melts and solutions of nonconcatenated ring polymers. Macromolecules 49, 708–722 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Michieletto D., Sakaue T., Dynamical entanglement and cooperative dynamics in entangled solutions of ring and linear polymers. ACS Macro Lett. 10, 129–134 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/20/eabf9260/DC1


