Abstract
Xia–Gibbs syndrome (XGS) is a rare Mendelian disease typically caused by de novo stop-gain or frameshift mutations in the AT-hook DNA binding motif containing 1 (AHDC1) gene. Patients usually present in early infancy with hypotonia and developmental delay and later exhibit intellectual disability (ID). The overall presentation is variable, however, and the emerging clinical picture is still evolving. A detailed phenotypic analysis of 34 XGS individuals revealed five core phenotypes (delayed motor milestones, speech delay, low muscle tone, ID, and hypotonia) in more than 80% of individuals and an additional 12 features that occurred more variably. Seizures and scoliosis were more frequently associated with truncations that arise before the midpoint of the protein although the occurrence of most features could not be predicted by the mutation position. Transient expression of wild type and different patient truncated AHDC1 protein forms in human cell lines revealed abnormal patterns of nuclear localization including a diffuse distribution of a short truncated form and nucleolar aggregation in mid-protein truncated forms. Overall, both the occurrence of variable phenotypes and the different distribution of the expressed protein reflect the heterogeneity of this syndrome.
Keywords: AHDC1, AT-hook protein, phenotypic spectrum, protein truncation, Xia–Gibbs syndrome
1 |. INTRODUCTION
De novo autosomal dominant nonsense and frameshift mutations in the AT-hook DNA binding motif containing 1 (AHDC1) gene cause Xia–Gibbs syndrome (XGS; MIM #615829), a severe neurological disorder characterized by developmental anomalies and intellectual disability (Xia et al., 2014). The XGS phenotypic spectrum is variable and overlaps with other disorders, making it challenging to diagnose based on clinical presentation alone. All reported diagnoses have so far been established by DNA sequence-based approaches (Cardosodos-Santos et al., 2020; Cheng et al., 2019; Díaz-Ordoñez et al., 2019; García-Acero & Acosta, 2017; Gumus, 2020; He et al., 2020; Jiang et al., 2018; Mubungu et al., 2020; Murdock et al., 2019; Ritter et al., 2018; Xia et al., 2014; H. Yang et al., 2015; S. Yang et al., 2019). The AHDC1 gene contains one coding exon and pathogenic mutations are observed over most of its length. To date, more than 250 individuals with XGS are known worldwide. An XGS Registry, initiated in 2014 (Jiang et al., 2018), now has 70 registrants including 34 who have provided detailed clinical records and consented for further research activities.
Xia-Gibbs Syndrome symptoms appear in early childhood and usually include hypotonia, followed by speech delay and eventually, intellectual disability. Xia-Gibbs Syndrome individuals also display a varying series of clinical features that emerge at different ages, resulting in an overall complex clinical presentation. Etiological, demographic, or other genetic factors contributing to individual phenotypes are not known, nor is the relationship between these varying phenotypes and individual patient AHDC1 mutational changes or positions. A previous study of clinical records from 20 unrelated XGS individuals bearing 16 different mutations suggested that truncations toward the C-terminus of the protein were more likely to be associated with non-verbal progression (Jiang et al., 2018). The same study, however, identified three mutations that were each identical in more than one individual and those with matching mutations did not share fully matched phenotypes. Cardoso-dos-Santos et al. reported a single case in Brazil with a stop-gain mutation near the N-terminus of the protein with a severe phenotype and the suggestion that such “short” forms may be overall more severe (Cardoso-dos-Santos et al., 2020). Together these observations underscore the disease heterogeneity and that other factors, including genetic background, can influence outcome.
Little is known regarding how AHDC1 mutation gives rise to XGS. It is likely that AHDC1 has a normal function in nuclear processes, with a particular role in the brain. The protein contains AT-hook binding motifs that are regularly associated with DNA or RNA binding and immunohistochemistry analyses show AHDC1 protein expression in the nucleoli, nucleoplasm, and the whole nucleus (Thul et al., 2017; Uhlen et al., 2015). The Genotype-Tissue Expression and The Human Protein Atlas reveal variable AHDC1 expression patterns in all tissues but with elevated levels in the brain (GTEx Consortium, 2013; Uhlen et al., 2015).
A combination of detailed phenotypic characterization of additional XGS individuals and further molecular studies of the normal and mutant gene affords the opportunity to better understand the underlying XGS disease mechanism and to improve prognostic predictions to families. Here, we have analyzed 34 XGS individuals who provided detailed clinical notes with consent for research, including 14 who have newly joined the XGS Registry and 20 XGS Registry members who were reported previously (Jiang et al., 2018). In addition, we also generated a series of expressed proteins, to study the cellular localization of normal and mutant AHDC1 protein forms. The expression studies identified varying patterns of nuclear localization of different mutant forms, as well as the relative localization of the wild-type protein with the variant forms.
2 |. METHODS
2.1 |. Ethics, consent, and clinical data ascertainment
This study and ascertainment of patient clinical records were approved by the Baylor College of Medicine Institutional Review Board (H-39945). Patient records are stored in a Research Electronic Data Capture (RedCap) HIPAA compliant environment (Jiang et al., 2018). Parents or guardians provided informed consent to participate in the registry with their affected children. Participants in the registry were invited to provide genetic reports as well as clinical surveys for analysis.
2.2 |. Phenotypic analyses
Consistent phenotyping for all 34 individuals with XGS was carried out using clinical surveys administered through the referring physician or the individual’s guardians; 11 of the 20 individuals from the previous study had an in-person family meeting (Jiang et al., 2018). A total of 17 phenotypes with binary outcomes of yes or no were ascertained. The association between the occurrence of each phenotype with variant locations (as rank of the location through the gene) was calculated via a logistic regression model using sex, mutation type, and age as covariates. False discovery rate was calculated using the Benjamini–Hochberg procedure to adjust for multiple testing.
2.3 |. Generation of vectors
Wild type AHDC1 cloned into a Gateway-compatible vector (pENTR223.1) was obtained from the Human Orfeome Collaboration (ORFeome Collaboration, 2016). DNA oligonucleotides were designed for in vitro mutagenesis based on alleles observed in XGS individuals, using the New England Biolabs primer design tool. Primers were set to be compatible with the New England Biolabs Q5 Site-Directed Mutagenesis Kit (https://nebasechanger.neb.com). Larger constructs (missense and C-terminal mutants) required a higher fidelity polymerase (TaKaRa PrimeSTAR GXL DNA Polymerase) to be used in place of the Q5 polymerase provided. Mutants were screened using Zymo Research Plasmid Minipreps from single colonies on spectinomycin bacterial growth media per the manufacturer’s protocol. The mutation alleles and wild type were validated using Sanger sequencing and cloned into both pDEST-eGFP-N1 and pDEST-mCherry-N1 (Hong et al., 2010) using the Gateway LR Clonase II Enzyme mix. The constructs were then prepared using the QIAGEN Maxi preparation protocol, sequence confirmed again using Sanger sequencing, and utilized for transfection.
2.4 |. Localization studies
Polyethylenimine (PEI; #23966–2; Polysciences) transfection reagent was prepared using the manufacturer’s recommendation to a concentration of 1 μg/μl. HeLa or 293T cells (60k cells/well) were seeded on an ibidi 8-well μ-Slide at 12 h before transfection using Dulbecco’s modified Eagle’s medium (31053–028; Gibco™), 1× l-Glutamine (35050–061; Gibco™), and 10% fetal bovine serum (F2242; Sigma-Aldrich). Growth media was replaced before transfection and a 1 DNA:7 PEI (HeLa) or 1 DNA:4 PEI (293T) transfection mix was prepared in Opti-MEM (31985062; Gibco™). Otherwise, the transfection protocol described by Longo et al. (2013) was followed.
Cells were fixed with 4% paraformaldehyde (PFA) and stained with 4′,6-diamidino-2-phenylindole (DAPI) using the manufacturer’s protocol. Imaging was performed at the Integrated Microscopy Core at Baylor College of Medicine on a DV Live epifluorescence image restoration microscope (GE Healthcare). The system is equipped with an Olympus UPLS Apo ×100/1.40 N.A. objective and a 1.9k × 1.9k pco.edge sCMOS_5.5 camera with a 2048 × 2048 field of view (FOV). The filter sets used were: DAPI, fluorescein isothiocyanate, and A594. Whole wells were scanned to identify the presence of a consistent localization pattern with at least 100 cells examined in 15 different fields; at least three independent transfections per mutant were used to confirm a consistent localization pattern. A subset of cells was then imaged for further analyses to avoid photobleaching and to further validate the localization patterns. Z stacks (0.2 μm) covering selected cells (~8 μm) were acquired before applying a conservative restorative algorithm for quantitative image deconvolution using SoftWorx v7.0. Imaging was always performed with at least 3 fields per well at the same exposure time (across all wells) with a wild-type control and a negative control per batch. Images (34 z-plane images per cell on average) were analyzed with the default settings using ImageJ.
AHDC1 mid-protein truncations localized to the nucleoli in addition to forming foci in the nucleoplasm. This was better visualized with a counterstain to DAPI of the nucleoli. Specifically, transfections were repeated, and the cells were fixed with methanol to enable staining with SYTO™ RNASelect™ (S32703; Invitrogen) as per the manufacturer’s protocol; the SYTO™ RNASelect™ stain selectively binds RNA, giving a maximum fluorescence signal from the nucleoli. This was followed by DAPI staining which preferentially stains the nucleoplasm, using the manufacturer’s protocol. Imaging was performed on a Nikon A1R-s confocal laser scanning microscope equipped with a Nikon 60× Plan Apo/1.4 N.A. oil objective, four standard PMT detectors, and adjustable Galvano scanner. Laser lines used include 405, 488, and 561 nm. The whole well was then scanned to identify the presence of a consistent localization pattern with at least 100 cells examined in 15 different fields. A subset of cells was then imaged for further analyses to avoid photobleaching and to further validate the localization patterns. Single plane images were collected with an optimal pinhole diameter selected for the 488 channel (~38 μm) with a 2× line average applied. Files (12-bit grayscale) were saved and exported from the Nikon Elements software. Imaging was always performed with the same imaging parameters with at least 3 fields per well at the same exposure time (across all wells) with a wild-type control and a negative control per batch. Images were analyzed with the default settings using ImageJ.
2.5 |. Colocalization studies
Identical procedures to the localization studies using 4% PFA were followed (see above), except that mutant (enhanced green fluorescent protein [eGFP]) and wild-type (mCherry) plasmid constructs were mixed at 1:1 in Opti-MEM before transfection. Procedures for staining and image collection were as described in the previous section.
3 |. RESULTS
3.1 |. Patients and mutations
Thirty-four families with Xia–Gibbs individuals, from approximately 250 known worldwide, have joined the XGS Registry, consented for research, and completed systematic patient surveys (Table 1). These 34 individuals with complete records are the focus of this study and each have an XGS diagnosis based upon observation of de novo frameshift or stop-gain AHDC1 pathogenic variants, predicted to lead to truncation of protein synthesis (Figure 1). Among the 34 families who submitted complete records, 14 represent individuals who have not been previously reported, while 20 were described in Jiang et al. (2018). One of the 14 newly identified individuals has a mutation that has been previously described and 13 bear mutations that have not yet been reported. Records included mutation details, demographic features, clinical surveys, and detailed patient health histories, as previously described (Jiang et al., 2018). An additional 36 families who joined the XGS Registry have not yet completed all patient surveys or submitted clinical notes and were not included in this analysis.
TABLE 1.
Detailed genotypes, phenotypes, and demographic features of XGS individuals
| Patient ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
| Publication status | Not published | Jiang et al. (2018) | Not published | Not published | Not published | Not published | Jiang et al. (2018) | Jiang et al. (2018) | Not published |
| Mutation | |||||||||
| Nucleotide change | c.643dup | C.784C >T | c.979C>T | c,1122dup | c,1446del | c,1759C>T | c,1945del | C.2062C >T | C.2188G >T |
| Protein change | p.Ser215Lysfs*16 | p.Gln262* | p.Gln327* | p.Gly375Argfs*3 | p.Val483Tyrfs*16 | p.Arg587* | p.Ala649-Profs*83 | p.Arg688* | p.Glu730* |
| Age | 25 years | 9 years | 59 years | 5 years | 12 years | 15 years | 7 years | 24 years | 12 years |
| Sex | Male | Female | Male | Female | Male | Male | Male | Female | Male |
| Ethnicity | White | White | White | Latino or Hispanic | White | Black | White | White | White |
| Growth and feeding | |||||||||
| Height percentile (%) | 51.99 | 0.10 | 1.07 | 0.10 | 1.19 | 1.66 | 0.10 | 65.91 | |
| Scoliosis | Yes | No | Yes | No | No | No | No | Yes | Yes |
| Comprehensive skills and language | |||||||||
| M-CHAT score | 6 | 18 | 9 | 17 | 18 | 10 | 8 | 11 | 11 |
| Autism diagnosis | Yes | No | No | No | Yes | Yes | Yes | Yes | No |
| Current languagea | 3 | 3 | 3 | 0 | 3 | 0 | 0 | 2 | 0 |
| Age at first word | 12–18 months | 4 years | 3 years | N/A | 16 months | 1 year | No words | 5 years | No words |
| Age using two words together | 2 years | 5 years | 5 years | N/A | 30 months | Non-verbal | N/A | 6 years | N/A |
| Age at following command | Unknown | 5 years | 4 years | 3.5 years | 36 months | 9 years | 3 years, 9 months | 4 years | 7 years |
| Mobility | |||||||||
| Hypotonia diagnosis | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes |
| Independent walking | Yes | Yes | Walking with support | Yes | Yes | Yes | Yes | Yes | Not sitting independently |
| Age at independent walking | 2 years | 3 years | N/A | 2 years | 3 years | 4 years | 3 years, 5 months | 22 months | 3 years |
| Sleep/airway | |||||||||
| Sleep apnea | Yes | No | No | Yes | Yes | Yes | Yes | No | No |
| Using breathing support | No | No | No | No | No | No | No | No | No |
| Neurological | |||||||||
| MRI | Abnormal | Normal | Abnormal | Abnormal | Normal | Unknown | Normal | Abnormal | Abnormal |
| EEG | Abnormal | Normal | Normal | Abnormal | Abnormal | Abnorma | No | Normal | Abnormal |
| Seizure | Yes | No | Yes | No | Yes | Yes | No | Yes | Yes |
| Age at first seizure | 7 years | N/A | 12 years | N/A | 9 years | 9 years | N/A | 9 months | Unknown |
| Ataxia | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Vision | |||||||||
| Wearing glasses or contacts | Yes | Yes | No | Yes | No | No | Yes | Yes | No |
| Strabismus | Yes | Yes | No | Unknown | No | Yes | No | No | Unknown |
| Patient ID | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 |
| Publication status | Not published | Jiang et al. (2018) | Jiang et al. (2018) | Jiang et al. (2018) | Jiang et al. (2018) | Jiang et al. (2018) | Not published | Jiang et al. (2018) | Not published |
| Mutation | |||||||||
| Nucleotide change | C.2188G >T | c.2229del | c.2373_2374del | c.2373_2374del | c.2373_2374del | c.2373_2374del | c.2373_2374del | c.2415del | C.2473C >T |
| Protein change | p.Glu730* | p.Ser744Profs*188 | p.Cys791Trpfs*57 | p.Cys791Trpfs*57 | p.Cys791Trpfs*57 | p.Cys791Trpfs*57 | p.Cys791Trpfs*57 | p.Leu806Trpfs*126 | p.Gln825* |
| Age | 12 years | 16 years | 8 years | 15 years | 11 years | 24 years | 20 years | 9 years | 17 years |
| Sex | Male | Male | Female | Male | Male | Male | Female | Female | Female |
| Ethnicity | White | White | White | White | White | White | White | White | |
| Growth and feeding | |||||||||
| Height percentile (%) | 0.15 | 0.10 | 0.10 | 0.10 | 0.10 | 0.40 | 10.03 | 0.17 | 5.16 |
| Scoliosis | No | Yes | No | No | No | Yes | Yes | No | Yes |
| Comprehensive skills and language | |||||||||
| M-CHAT score | 11 | 7 | 7 | 15 | 15 | 13 | 7 | 17 | 9 |
| Autism diagnosis | No | No | Yes | No | No | No | No | No | Yes |
| Current languagea | 2 | 0 | N/A | 3 | 3 | 2 | 0 | 3 | 3 |
| Age at first word | 19 months | No words | 4 years | 12 months | 2 years | 5 years | N/A | 3 years | 4 years |
| Age using two words together | 24 months | N/A | N/A | 2 years | 2 years | 7 years | No use sentences | 3 years | 4–5 years |
| Age at following command | Does not follow command | 4 years | 2 years | 18 months | 4 years | 3 years | 4 years | 2 years | 5 years |
| Mobility | |||||||||
| Hypotonia diagnosis | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Independent walking | Yes | Yes | Yes | Yes | Yes | Walking with support | Yes | Yes | Yes |
| Age at independent walking | 18 months | 3 years | 3 years | 18 months | 4 years | N/A | 3 years | 2 years | 4.5 years |
| Sleep/airway | |||||||||
| Sleep apnea | No | Yes | Yes | Yes | Yes | Yes | No | No | |
| Using breathing support | No | No | Yes | Yes | No | Yes | No | No | |
| Neurological | |||||||||
| MRI | Abnormal | Normal | Abnormal | Abnormal | Abnormal | Abnormal | Abnormal | Normal | Abnormal |
| EEG | Abnormal | No | Normal | Abnormal | Yes | Abnormal | Abnormal | Normal | Normal |
| Seizure | Yes | No | No | Yes | Yes | Yes | Yes | Yes | Yes |
| Age at first seizure | 3 years | N/A | N/A | 3 years | 5 years | Unknown | 16 years | 2 years | 5 years |
| Ataxia | No | No | Yes | Yes | Yes | Yes | Yes | Yes | No |
| Vision | |||||||||
| Wearing glasses or contacts | Yes | No | Yes | No | No | No | Yes | No | Yes |
| Strabismus | No | No | Yes | No | No | Yes | Yes | No | No |
| Patient ID | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 |
| Publication status | Jiang et al. (2018) | Jiang et al. (2018) | Jiang et al. (2018) | Jiang et al. (2018) | Jiang et al. (2018) | Not published | Jiang et al. (2018) | Jiang et al. (2018) | Jiang et al. (2018) |
| Mutation | |||||||||
| Nucleotide change | c.2520del | c.2644C>T | c.2691del | c.2773C>T | c.2773C>T | c.2849del | c.2898del | C.2908C >T | c.2908C>T |
| Protein change | p.Arg841Alafs*91 | p.Gln882* | p.Val898Trpfs*34 | p.Arg925* | p.Arg925* | p.Pro950Argfs*192 | p.Tyr967Thrfs*175 | p.Gln970* | p.Gln970* |
| Age | 12 years | 13 years | 7 years | 6 years | 9 years | 10 years | 11 years | 9 years | 13 years |
| Sex | Male | Male | Female | Male | Female | Male | Female | Female | Female |
| Ethnicity | White | White | White | White | White | White | Asian | White | White |
| Growth and feeding | |||||||||
| Height percentile (%) | 0.36 | 0.10 | 0.10 | 0.10 | 0.10 | 38.97 | 0.10 | 0.10 | |
| Scoliosis | No | Yes | No | No | No | No | No | No | No |
| Comprehensive skills and language | |||||||||
| M-CHAT score | 6 | 12 | 17 | 12 | 11 | 9 | 13 | 18 | 8 |
| Autism diagnosis | No | Yes | No | No | No | Yes | No | No | No |
| Current languagea | 0 | 0 | 1 | 0 | 0 | 2 | 1 | 3 | 3 |
| Age at first word | No words | No words | 2.5 years | 2.5 years | 2.5–3 years | 3 years | Unknown | 2.5 years | 24 months |
| Age using two words together | N/A | N/A | 3.5 years | N/A | No two words | 3–4 years | 6 years | 3.5 years | 3 years |
| Age at following command | N/A | 8 years | 2 years | 23 months | 3 years | 4 years | 6 years | 3 years | 6 years |
| Mobility | |||||||||
| Hypotonia diagnosis | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes |
| Independent walking | Not walking but sitting independently | Yes | Yes | Walking with support | Yes | Yes | Yes | Yes | Yes |
| Age at independent walking | N/A | 3 years | 2.5 years | N/A | 28 months | 3 years | 4 years | 2.5 years | 2 years |
| Sleep/airway | |||||||||
| Sleep apnea | No | No | No | Yes | No | No | Yes | No | No |
| Using breathing support | No | No | No | No | No | No | No | No | No |
| Neurological | |||||||||
| MRI | Normal | Normal | Abnormal | Normal | Abnormal | Normal | Normal | Normal | Normal |
| EEG | Normal | Normal | Normal | Normal | No | Abnormal | No | Normal | Normal |
| Seizure | No | No | No | No | No | Yes | No | No | No |
| Age at first seizure | N/A | N/A | N/A | N/A | N/A | 5 years | N/A | N/A | N/A |
| Ataxia | Yes | No | Yes | No | Yes | Yes | No | Yes | Yes |
| Vision | |||||||||
| Wearing glasses or contacts | Yes | No | No | No | Yes | No | No | No | No |
| Strabismus | No | Yes | Yes | Yes | Yes | No | No | Yes | No |
| Patient ID | 28 | 29 | 30 | 31 | 32 | 33 | 34 | ||
| Publication status | Not published | Not published | Not published | Jiang et al. (2018) | Jiang et al. (2018) | Jiang et al. (2018) | Not published | ||
| Mutation | |||||||||
| Nucleotide change | C.2932C >T | c.3204C>G | C.3466C >T | c.3773C>G | c.3809del | C.3989C >A | c.4438del | ||
| Protein change | p.Gln978* | p.Tyrl068* | p.Glnll56* | p.Serl258* | p.Glnl270Argfs*75 | p.Serl330* | p.Glul480Lysfs*67 | ||
| Age | 2 years | 3 years | 3 years | 4 years | 15 years | 20 years | 33 years | ||
| Sex | Female | Female | Male | Male | Male | Male | Female | ||
| Ethnicity | White Hispanic | White | White | White | Asian | White | White | ||
| Growth and feeding | |||||||||
| Height percentile (%) | 11.90 | 0.10 | 0.10 | 0.10 | 0.10 | 0.10 | 71.90 | ||
| Scoliosis | No | No | No | No | No | No | No | ||
| Comprehensive skills and language | |||||||||
| M-CHAT score | 11 | 0 | 4 | 6 | 13 | 9 | 15 | ||
| Autism diagnosis | No | No | No | No | No | Yes | No | ||
| Current languagea | 1 | 0 | 0 | 0 | 0 | 0 | 1 | ||
| Age at first word | 15 months | N/A | No words | No words | No words | No words, just noises | 6 years | ||
| Age using two words together | N/A | N/A | N/A | No words | N/A | No words | Unknown | ||
| N/A | N/A | 8 years | 5 years | 3 years | |||||
| Age at following command | Does not follow command | Does not follow command | |||||||
| Mobility | |||||||||
| Hypotonia diagnosis | Yes | Yes | Yes | Yes | Yes | Yes | Yes | ||
| Independent walking | Walking with support | Not walking but sitting independently | Yes | Walking with support | Yes | Yes | Yes | ||
| Age at independent walking | N/A | 18 months | N/A | 6 years | 19 months | 2 years, 2 months | |||
| Sleep/airway | |||||||||
| Sleep apnea | No | Yes | No | No | No | No | No | ||
| Using breathing support | No | No | No | No | No | No | No | ||
| Neurological | |||||||||
| MRI | No | Abnormal | Normal | Abnormal | Yes | Normal | Abnormal | ||
| EEG | No | Normal | Normal | Normal | Unknown | No | Abnormal | ||
| Seizure | No | No | No | No | No | No | Yes | ||
| Age at first seizure | N/A | N/A | N/A | N/A | N/A | N/A | 6 years | ||
| Ataxia | Yes | No | Yes | No | Yes | No | No | ||
| Vision | |||||||||
| Wearing glasses or contacts | Yes | Yes | No | No | No | No | Yes | ||
| Strabismus | Yes | Yes | No | No | No | No | Yes | ||
Note: Mutations are classified as N-terminal, mid-protein, and C-terminal based on which one-third of the protein they occurred in, respectively.
Abbreviations: EEG, electroencephalography; M-CHAT, Modified Checklist for Autism in Toddlers; MRI, magnetic resonance imaging; XGS, Xia–Gibbs syndrome.
Current language: 0, no words; 1, <50 words, 2, no sentence but >50 words; 3, full sentence >200 words.
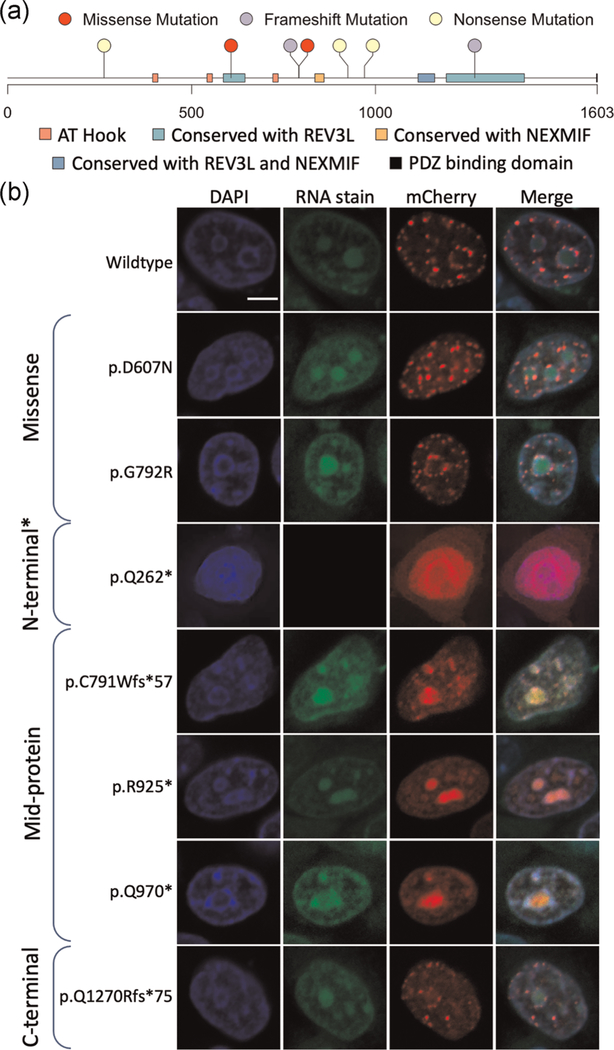
FIGURE 1.
Distribution of mutations and phenotypes in the Xia–Gibbs registry among patients with a frameshift or nonsense variant. AT-hook domains, PDZ binding domain, and REV3L/NEXMIF (KIAA2022) conservation regions are shown. Each column in the heatmap represents a patient ranked by the position of the variant carried by the patient. Only frameshift and nonsense variants were included in the analysis. Mutations were classified as N-terminal, mid-protein, and C-terminal based on which one-third of the protein they occurred in, respectively. Each row represents a clinical phenotype from patient surveys with rows labeled in red representing phenotypes occurring in more than 80% of individuals. The phenotypes were ranked by p values of logistic regression tests for associations between the binary outcome and positions of all the variants
3.2 |. Patient phenotypes are heterogeneous
Analysis of the phenotypes of the 34 XGS individuals showed features that were each present in more than 80% of XGS individuals in this study (>27/34). These features were designated as the “core defining phenotypes” and included delayed motor milestones (94.1%), speech delay (94.1%), low muscle tone (88.2%), intellectual disability (88.2%), and hypotonia (88.2%). Other features that were not shared by at least 80% of individuals but were reported in clinical records of at least two individuals, were designated “secondary phenotypes” (Figures 1 and S1).
We tested the association of each of the patient phenotypes with mutation type and location, as well as with demographic features (Figure 1). While we observed no association between disease phenotypes and sex, age, or ethnicity (p > .05), we identified nominal associations between the position of the variants along the length of the protein, with the occurrences of seizures (p = .021) and scoliosis (p = .033). Thus, those truncating mutations that mapped to the N-terminal to mid-protein positions, were more likely to be associated with these phenotypes. Conversely, the mutations closer to the C-terminus were less likely to be associated with seizures or scoliosis. This trend was not previously observed in the study of 20 XGS individuals (Jiang et al., 2018).
Among the 34 individuals reported here, five bore the identical recurrent frameshift mutation (p.Cys791TrpfsTer57). While these individuals overall shared a majority of features, they did not have the exact same phenotypes. Ten of the 17 scored phenotypes were shared among at least four of these five patients with the identical genotype (Table 1). As this mutation is located proximal to the midpoint of the protein, it is within the group anticipated to be associated with seizures and scoliosis. Four of the five individuals had seizures and the remaining individual was the youngest of the five (8 years old) and may yet develop this clinical manifestation as the earliest age of other reported seizures is 9 years old. Similarly, 2/5 patients have scoliosis and are above the age of 20 while 3/5 are 8, 11, and 15 years old with the earliest reported age for scoliosis being 12 years old. Thus, the phenotypes associated with this recurrent mutation maintain the association between proximal mutations, seizures, and scoliosis, but the individuals also show heterogeneity consistent with the influence of age, genetic background, and environmental effects.
3.3 |. AHDC1 protein domains and cellular localization
To investigate the cellular localization of AHDC1, we transiently expressed AHDC1 wild-type and mutant forms as fluorescently labeled fusion proteins. We selected five different patient de novo variants across the length of the protein representing N-terminal truncating, mid-protein truncating, and C-terminal truncating mutations. The truncations progressively interrupted known regions of homology or functional motifs, including either: (i) the C-terminal REV3L conserved region; (ii) the C-terminal REV3L and the mid protein NEXMIF (KIAA2022) conserved regions, or (iii) all of these regions and all three known AT-hook binding motifs. Clones bearing missense mutations were also generated (Figure 2a). All mutations were expressed and imaged in HeLa cell lines and a subset was examined in the 293T cell line (Figure S2).
FIGURE 2.
AHDC1 mutations tested in this study. (a) Series of cloned variants along the length of the AHDC1 protein representing patients with Xia–Gibbs syndrome or an unconfirmed diagnosis. AT-hook domains, PDZ binding domain, and REV3L/NEXMIF (KIAA2022) conservation regions are shown. (b) Representative z-plane images of the nucleus in the transiently expressing HeLa cells with DNA stained with 4′,6-diamidino-2-phenylindole (DAPI) (first column), Syto RNASelect-stained nucleoli (second column), wild-type, missense, or truncated alleles (third column), and a merged image (fourth column). Scale bar = 5 μm. *p.Gln262Ter was not amenable to methanol fixation due to mutant short-form diffuse expression and it is alternatively shown with paraformaldehyde fixation without the nucleolar stain (see Section 2)
As anticipated from previous reports (Thul et al., 2017), we observed that wild-type AHDC1 was expressed at distinct foci that surrounded the nucleoli and the inner perimeter of the nucleus. There were also distinct foci distributed throughout the nucleoplasm (Figure 2b and Table S1). These foci coincide with regions marked by DAPI staining and suggest the normal association of AHDC1 with heterochromatin-rich sites. The variant forms with missense mutations showed a similar localization pattern to the wild-type with distinct foci in the nucleoplasm (Figure 2b and Table S1).
Expression of the different truncated AHDC1 protein forms yielded patterns that were distinguishable from that of the wild type. An N-terminal truncation (p.Gln262Ter) resulted in a diffuse expression pattern with the absence of foci observed in wild-type AHDC1 (Figure 2b and Table S1). Expression of mid-AHDC1 truncation alleles (p.Cys791TrpfsTer57, p.Arg925Ter, p.Gln970Ter) resulted in a more dramatic shift in the observed localization patterns, with a much stronger signal from within the nucleoli. The boundaries of the suborganelle were clearly discernible. These mid-AHDC1 truncation patterns appeared less like that from the DAPI patterns and suggested aggregation of the protein within the nucleolus as a hallmark feature (Figure 2 and Table S1). C-terminal truncation of AHDC1 (Q1270Rfs*75) resulted in a more similar expression pattern to that observed from the wild-type and missense mutants, with protein localizing to distinct foci in the nucleoplasm with some occurrences at the nucleolar boundary.
A subset of these experiments studying mutant expression was repeated in 293T cells, demonstrating identical patterns to those described above for the HeLa cell line, for the wild-type, missense and mid-length truncated forms (see Figure S2). Overall, the in situ studies supported a pattern of diffuse expression of the shortest truncated AHDC1 form, with an observable accumulation of the mid-protein truncations in the nucleolus and a return to a pattern more similar to the wild type from the C-terminal truncation form. The patterns were, in general, consistent with the normal forms aggregating in regions presumed to be rich in heterochromatin, while the most dramatic differences were from the more substantial truncations that deleted regions with putative functional roles, including the AHDC1 AT-Hook domains and conserved REV3L and NEXMIF homologies.
3.4 |. Wild-type and mutant AHDC1 colocalization
Mutations in AHDC1 in XGS individuals are heterozygous and as the message contains a single coding exon, the mutant RNA is not expected to undergo nonsense-mediated decay (Lindeboom et al., 2016). It is not known if the mature protein is monomeric or has multiple subunits. To examine interactions between transiently expressed AHDC1 wild-type and mutant protein forms, we coexpressed equimolar amounts of AHDC1 fusion proteins, tagged with mCherry (wild type) or eGFP (mutant), respectively. As expected, wild-type controls (AHDC1|eGFP and AHDC1|mCherry) colocalized in distinct foci in the nucleoplasm (Figure 3 and Table S2). This pattern is consistent with the distinct foci representing multimeric forms and/or large protein aggregations. Missense mutations (p.Asp607Asn and p.Gly792Arg) also colocalized with wild-type AHDC1 in distinct foci in the nucleoplasm. Thus, we did not observe a difference in colocalization of the products of missense alleles, compared with wild type.
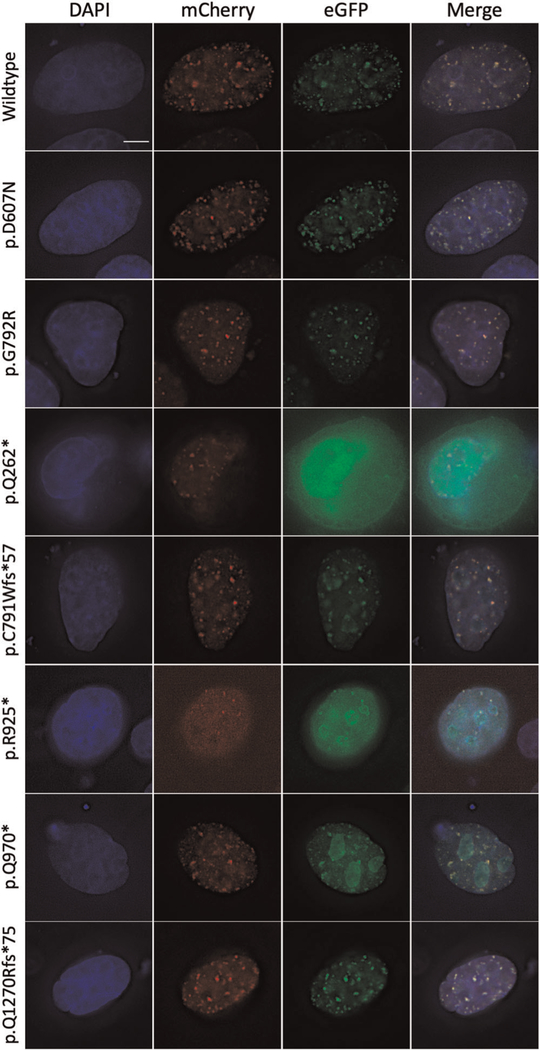
FIGURE 3.
Representative z-plane images of cells with transient expression and colocalization of the wild-type and mutated AHDC1 allelic series in the HeLa cell line. The respective images show the 4′,6-diamidino-2-phenylindole (DAPI)-stained nucleus in the first column, wild-type AHDC1|mCherry in the second column, mutated allele tagged with enhanced green fluorescent protein in the third column, and a merge of all channels in the last column. Scale bar = 5 μm
The N-terminal truncation (p.Gln262Ter) was diffusely expressed, extending beyond the nucleus, consistent with the patterns observed with transfection of this form, alone (see above). The expression of this truncation variant did not appear to perturb the localization pattern of the wild-type protein when the labels were reversed.
Mid-protein truncations (p.Cys791TrpfsTer57, p.Arg925Ter, p.Gln970Ter) colocalized with the wild type in the nucleoplasm, suggesting that either form was competent to aggregate in these observed foci. More significantly, while the mutant form demonstrated a similar aggregation within the nucleolus to that observed above, the labeled wild-type form did not show any propensity to aggregate in the nucleolus in the presence of the unlabeled mid-protein truncated forms. Thus, it appeared that the mutant protein alone was demonstrating unusual localization, including aggregation in the nucleolus, and was not influencing the patterns from the accompanying wild type.
The C-terminal truncation mutant (Q1270Rfs*75) colocalized with the wild-type protein in the nucleoplasm in distinct foci, where it did not show a distinct localization pattern or density, from the wild type. The nucleolus did not show the intense aggregation of the mid-protein truncated forms, also representing a pattern that was much more similar to wild type (Figure 3 and Table S1).
Overall, the position of the different AHDC1 truncating mutations did not seem to have an effect on wild-type AHDC1 localization. More strikingly, the position of truncating mutations affected the general distribution of the mutant AHDC1, with the “short form” diffusely expressed, the mid form most associated with nucleolar aggregation and the C-terminus truncation more consistent with the wild-type distribution pattern.
4 |. DISCUSSION
XGS individuals exhibit considerable phenotypic heterogeneity. The detailed analysis of the 34 individuals described here, identifies five “core” phenotypes of delayed motor milestones, speech delay, low muscle tone, intellectual disability, and hypotonia that are associated with more than 80% (>27/34) of XGS individuals in this study. These findings are consistent with previous reports of XGS. Additional phenotypes occur more variably, however, ranging from few instances to more than half of the XGS individuals in this study. In addition, individuals harboring the same mutation (e.g., p.Cys791TrpfsTer57) showed differences in their phenotypic spectrum and although this in part is explained by age-dependent expressivity effects, it underscores the overall phenotypic heterogeneity of the disorder and explains much of the difficulty of diagnosis without DNA sequence data.
Examination of individual phenotypic features, demographics, and mutation position revealed no associations for 15 of 17 XGS features, including the five core phenotypes. Seizures and scoliosis were, however, each more likely in individuals with truncating mutations near the N-terminus of the protein. The association was modest, but statistically significant and warrants further study including broad phenotypic surveys to record the occurrence of these features in larger XGS populations. Ideally, future surveys will be comprehensive and will also detect any additional features that are infrequent and not yet reported in the syndrome.
Further understanding of the XGS disease mechanism demands detailed molecular studies of AHDC1 in model systems, which has been challenging to implement. The protein is difficult to isolate, antibodies display limited sensitivity and recombinant mice do not show either an obvious phenotype or ability to breed (data not shown). To further investigate the molecular mechanisms underlying the role of AHDC1 in XGS, we, therefore, generated in vitro allelic variants based on the known patterns of mutation in XGS individuals. The chosen mutations truncated the AHDC1 protein and progressively removed the downstream AT-hook binding domains and regions of known homologies. This series of variants was tested in an assay based upon transient transfection and in situ fluorescent imaging.
The in situ data demonstrated characteristic patterns of distribution and colocalization with the wild-type protein, differing between the varying length mutant forms. Short forms were diffusely distributed, and most strikingly, the mid-length protein form displayed a pattern of aggregation of accumulation in the nucleolus. Long forms were indistinguishable from wild type in this assay. The effect was reproduced for two different mid-length mutations in parallel experiments in the 293T cell line.
Colocalization of the mutant and wild-type forms was also studied in the transient transfection assay. The wild-type protein appears to normally aggregate in foci throughout the nucleoplasm. None of the mutant forms perturbed the pattern of distribution of the wild-type protein although the mid-length and longer forms co-aggregated with the wild type in the nucleoplasm. The accumulation of the mid-length forms in the nucleolus did not appear to involve an unusual distribution of the wild-type form.
Two of the variants studied here were identical to AHDC1 missense mutations observed in children with XGS-like symptoms and may be pathogenic (Gumus, 2020). In the assay used here, these missense variant proteins behaved essentially the same as the wild type and the in situ images did not support that these changes could result in abnormal AHDC1 localization. In the absence of more complete knowledge of normal AHDC1 function, the failure of missense mutations to manifest altered protein localization in this transient expression assay, does not, however, suggest that they are benign.
Overall, these expression data show that different truncating mutations can result in observable differences in subcellular AHDC1 protein localization patterns. The transient expression assay that was used can result in overexpression of the tagged protein, limiting the direct physiological relevance of the observations (Gibson et al., 2013). Nevertheless, the different mutations that were tested here demonstrate different effects in this model system. We observed that “short” forms of the protein, truncated near the N-terminus were distributed diffusely. This loss of localization is consistent with the removal of the AT-hook domains and comparable to that observed when the protein MECP2 has disrupted AT-hook domains (Lyst et al., 2016; Vermeulen et al., 2010). In contrast to the diffuse patterns from the “short form,” we observed that mid-protein truncations maintained the ability to form foci in the nucleoplasm and resulted in partial mislocalization of mutant AHDC1 protein to the nucleolus. The nucleolus is a site of active transcription that is critical for ribosomal RNA synthesis, as well as the site of assembly of mature ribosomes. These mid-protein forms have disrupted or lost C-terminal REV3L/NEXMIF conserved domains but maintain AT-hook binding domains that are closer to the N-terminus. The C-terminal truncated forms did not show a change in localization of foci, relative to the wild type. Together these data show that different lengths of AHDC1 truncation can result in different patterns of protein localization.
An ongoing question in this disorder is whether the primary disease mechanism is one of haploinsufficiency or else potentially a dominant-negative mechanism. Some elements of this study support the notion of the dominant-negative disease model, as the N-terminal and mid-protein truncated forms can exhibit protein mislocalization without involving the aggregation of wild-type protein in foci within the nucleoplasm. Further resolution of this issue may come from other ongoing studies of patients with large chromosomal deletions, that involve AHDC1 along with other genes in the region, or from the identification of additional cellular phenotypes and other molecular reagents.
Overall, this study illustrates the heterogeneity of both the XGS phenotypes and the subcellular localization patterns of different AHDC1 mutant forms in the nucleus, in a transient expression assay. A subset of 17 scored XGS phenotypic features, seizures, and scoliosis, are associated more frequently with N-terminal mutations that lead to the shortest protein forms. These short protein forms also have a characteristic diffuse distribution in the nucleus, and it is tempting to speculate that there is a mechanistic connection between the cellular and the high-level phenotypes. While such phenotypic heterogeneity is more often explained as resulting from different genetic backgrounds or environmental effects, it is also possible that common factors related to the mutational damage to AHDC1 contribute to each (Chow et al., 2019; Geschwind & Levitt, 2007).
Supplementary Material
ACKNOWLEDGMENTS
This study would have not been possible without the contributions of family members participating in the AHDC1 Xia–Gibbs Registry and the coordination efforts of Yunyun Jiang at the Human Genome Sequencing Center. This study was funded in part by the NHGRI awards to Richard A. Gibbs (HG008898) and James R. Lupski (NHGRI/NHLBI UM1 HG006542) and by private donation. He Li was partially supported by an award from the Xia-Gibbs Society. Varuna Chander was supported by the training fellowship from the NLM Training Program in Biomedical Informatics & Data Science (T15LM007093). Jennifer E. Posey was supported by NHGRI K08 HG008986. Imaging for this project was supported by the Integrated Microscopy Core at Baylor College of Medicine with funding from NIH (DK56338 and CA125123), CPRIT (RP150578 and RP170719), the Dan L. Duncan Comprehensive Cancer Center, and the John S. Dunn Gulf Coast Consortium for Chemical Genomics.
Funding information National Human Genome Research Institute, Grant/Award Numbers: HG008898, HG008986; National Human Genome Research Institute and National Heart, Lung, and Blood Institute, Grant/Award Number: HG006542; NIH, Grant/Award Numbers: CA125123, DK56338; National Library of Medicine, Grant/Award Number: T15LM007093; Cancer Prevention and Research Institute of Texas, Grant/Award Numbers: RP150578, RP170719
Footnotes
CONFLICT OF INTERESTS
James R. Lupski has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, and is a coinventor on multiple US and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis and clinical exome sequencing offered in the Baylor Genetics Laboratory (http://baylorgenetics.com).
SUPPORTING INFORMATION
Additional Supporting Information may be found online in the supporting information tab for this article.
DATA AVAILABILITY STATEMENT
The pathogenic variants reported here are deposited in the ClinVar database (ClinVar accessions: SCV001480339-SCV001480365). Additional data that support the findings of this study are available upon request from the corresponding author, subject to privacy or ethical restrictions.
REFERENCES
- Cardoso-dos-Santos AC, Silva TO, Faccini AS, Kowalski TW, Bertoli-Avella A, Morales Saute JA, Schuler-Faccini L, & de Oliveira Poswar F (2020). Novel AHDC1 gene mutation in a Brazilian individual: Implications of Xia-Gibbs syndrome. Molecular Syndromology, 11(1), 24–29. 10.1159/000505843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Tang F, Hu X, Li H, Li M, Fu Y, Yan L, Li Z, Gou P, Su N, Gong C, He W, Xiang R, Bu D, & Shen Y (2019). Two Chinese Xia-Gibbs syndrome patients with partial growth hormone deficiency. Molecular Genetics & Genomic Medicine, 7(4), e00596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J, Jensen M, Amini H, Hormozdiari F, Penn O, Shifman S, Girirajan S, & Hormozdiari F (2019). Dissecting the genetic basis of comorbid epilepsy phenotypes in neurodevelopmental disorders. Genome Medicine, 11(1), 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Ordoñez L, Ramirez-Montaño D, Candelo E, Cruz S, & Pachajoa H (2019). Syndromic intellectual disability caused by a novel truncating variant in AHDC1: A case report. Iranian Journal of Medical Sciences, 44(3), 257–261. [PMC free article] [PubMed] [Google Scholar]
- García-Acero M, & Acosta J (2017). Whole-exome sequencing identifies a de novo mutation in a Colombian patient with Xia-Gibbs syndrome. Molecular Syndromology, 8(6), 308–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, & Levitt P (2007). Autism spectrum disorders: Developmental disconnection syndromes. Current Opinion in Neurobiology, 17(1), 103–111. [DOI] [PubMed] [Google Scholar]
- Gibson TJ, Seiler M, & Veitia RA (2013). The transience of transient overexpression. Nature Methods, 10(8), 715–721. [DOI] [PubMed] [Google Scholar]
- GTEx Consortium. (2013). The Genotype-Tissue Expression (GTEx) project. Nature Genetics, 45(6), 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumus E (2020). Extending the phenotype of Xia-Gibbs syndrome in a two-year-old patient with craniosynostosis with a novel de novo AHDC1 missense mutation. European Journal of Medical Genetics, 63(1), 103637. [DOI] [PubMed] [Google Scholar]
- He P, Yang Y, Zhen L, & Li D-Z (2020). Recurrent hypoplasia of corpus callosum as a prenatal phenotype of Xia-Gibbs syndrome caused by maternal germline mosaicism of an AHDC1 variant. European Journal of Obstetrics, Gynecology, and Reproductive Biology, 244, 208–210. [DOI] [PubMed] [Google Scholar]
- Hong T-T, Smyth JW, Gao D, Chu KY, Vogan JM, Fong TS, Jensen BC, Colecraft HM, & Shaw RM (2010). BIN1 localizes the L-type calcium channel to cardiac T-tubules. PLOS Biology, 8(2), e1000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Wangler MF, McGuire AL, Lupski JR, Posey JE, Khayat MM, Murdock DR, Sanchez-Pulido L, Ponting CP, Xia F, Hunter JV, Meng Q, Murugan M, & Gibbs RA (2018). The phenotypic spectrum of Xia-Gibbs syndrome. American Journal of Medical Genetics, Part A, 176(6), 1315–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom RGH, Supek F, & Lehner B (2016). The rules and impact of nonsense-mediated mRNA decay in human cancers. Nature Genetics, 48(10), 1112–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo PA, Kavran JM, Kim M-S, & Leahy DJ (2013). Transient mammalian cell transfection with polyethylenimine (PEI). Methods in Enzymology, 529, 227–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyst MJ, Connelly J, Merusi C, & Bird A (2016). Sequence-specific DNA binding by AT-hook motifs in MeCP2. FEBS Letters, 590(17), 2927–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mubungu G, Makay P, Boujemla B, Yanda S, Posey JE, Lupski JR, Bours V, Lukusa P, Devriendt K, & Lumaka A (2020). Clinical presentation and evolution of Xia-Gibbs syndrome due to p. Gly375ArgfsTer3 variant in a patient from DR Congo (Central Africa). American Journal of Medical Genetics, Part A, 182, 1572–1575. 10.1002/ajmg.a.62049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdock DR, Jiang Y, Wangler M, Khayat MM, Sabo A, Juusola J, McWalter K, Schatz KS, Gunay-Aygun M, & Gibbs RA (2019). Xia-Gibbs syndrome in adulthood: A case report with insight into the natural history of the condition. Cold Spring Harbor Molecular Case Studies, 5(3), a003608. 10.1101/mcs.a003608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ORFeome Collaboration. (2016). The ORFeome Collaboration: A genome-scale human ORF-clone resource. Nature Methods, 13(3), 191–192. [DOI] [PubMed] [Google Scholar]
- Ritter AL, McDougall C, Skraban C, Medne L, Bedoukian EC, Asher SB, Balciuniene J, Campbell CD, Baker SW, Denenberg EH, Mazzola S, Fiordaliso SK, Krantz ID, Kaplan P, Ierardi-Curto L, Santani AB, Zackai EH, & Izumi K (2018). Variable clinical manifestations of Xia-Gibbs syndrome: Findings of consecutively identified cases at a single children’s hospital. American Journal of Medical Genetics, Part A, 176(9), 1890–1896. [DOI] [PubMed] [Google Scholar]
- Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Björk L, Breckels LM, Bäckström A, Danielsson F, Fagerberg L, Fall J, Gatto L, Gnann C, Hober S, Hjelmare M, Johansson F, … Lundberg E (2017). A subcellular map of the human proteome. Science, 356(6340), eaal3321. 10.1126/science.aal3321 [DOI] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA-K, Odeberg J, Djureinovic D, Takanen JO, Hober S, … Ponten F (2015). Proteomics. Tissue-based map of the human proteome. Science, 347(6220), 1260419. [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, & Mann M (2010). Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell, 142(6), 967–980. [DOI] [PubMed] [Google Scholar]
- Xia F, Bainbridge MN, Tan TY, Wangler MF, Scheuerle AE, Zackai EH, Harr MH, Sutton VR, Nalam RL, Zhu W, Nash M, Ryan MM, Yaplito-Lee J, Hunter JV, Deardorff MA, Penney SJ, Beaudet AL, Plon SE, Boerwinkle EA, … Gibbs RA (2014). De novo truncating mutations in AHDC1 in individuals with syndromic expressive language delay, hypotonia, and sleep apnea. American Journal of Human Genetics, 94(5), 784–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Douglas G, Monaghan KG, Retterer K, Cho MT, Escobar LF, Tucker ME, Stoler J, Rodan LH, Stein D, Marks W, Enns GM, Platt J, Cox R, Wheeler PG, Crain C, Calhoun A, Tryon R, Richard G, … Chung WK (2015). De novo truncating variants in the AHDC1 gene encoding the AT-hook DNA-binding motif-containing protein 1 are associated with intellectual disability and developmental delay. Cold Spring Harbor Molecular Case Studies, 1(1), a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Li K, Zhu M-M, Yuan X-D, Jiao X-L, Yang Y-Y, Li J, Li L, Zhang H-N, Du Y-H, Wei Y-X, & Qin Y-W (2019). Rare mutations in AHDC1 in patients with obstructive sleep apnea. BioMed Research International, 2019, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The pathogenic variants reported here are deposited in the ClinVar database (ClinVar accessions: SCV001480339-SCV001480365). Additional data that support the findings of this study are available upon request from the corresponding author, subject to privacy or ethical restrictions.