Abstract
Type 1 diabetes (T1D) is an autoimmune disease that resulted from the severe destruction of the insulin-producing β cells in the pancreases of individuals with a genetic predisposition. Genome-wide studies have identified HLA and other risk genes associated with T1D susceptibility in humans. However, evidence obtained from the incomplete concordance of diabetes incidence among monozygotic twins suggests that environmental factors also play critical roles in T1D pathogenesis. Epigenetics is a rapidly growing field that serves as a bridge to link T1D risk genes and environmental exposures, thereby modulating the expression of critical genes relevant to T1D development beyond the changes of DNA sequences. Indeed, there is compelling evidence that epigenetic changes induced by environmental insults are implicated in T1D pathogenesis. Herein, we sought to summarize the recent progress in terms of epigenetic mechanisms in T1D initiation and progression, and discuss their potential as biomarkers and therapeutic targets in the T1D setting.
Keywords: Epigenetics, Type 1 diabetes, DNA methylation, Histone modifications, ncRNAs
Introduction
Type 1 diabetes (T1D) is a chronic autoimmune disease characterized by absolute insulin deficiency coupled with resultant hyperglycemia.[1] Both humoral and cellular immune responses are involved in the pathogenesis of T1D. After the early discoveries on humoral immunity in T1D, several T1D-associated autoantibodies have been identified, such as autoantibodies against insulin, glutamic acid decarboxylase (GAD), islet-associated protein-2 (IA-2), zinc transporter family member 8 (ZnT8), islet cell autoantigen 69,000 Da (ICA69), and chromogranin A (ChgA).[2] Autoantibodies can be detected months or even years before the presence of clinical symptoms, and therefore, they could serve as disease prediction markers in susceptible individuals. The pathological hallmark of T1D is insulitis, an inflammatory lesion of the islet accompanied by the destruction of pancreatic β cells.[3] The invading immune cells consist of various leukocytes, including natural killer cells, myeloid cells, as well as lymphoid cells such as CD4 and CD8 T cells, and B cells. During the early stage of insulitis, myeloid populations account for the majority of immunocytes, while lymphocytes gradually increase and become prominent as disease progression.[4] Importantly, the entry of regulatory T cells (Tregs) was less than that of conventional T cells (Tconv), which contributes to the imbalance of Tregs/Tconv in the inflamed islets.[5] The infiltrated immunocytes are generally observed within the islet parenchyma or in the islet periphery (peri-insulitis), leading to a dialogue with the insulin-producing β cells to commit the autoimmune attack.[6] Epidemiological studies have shown that at least 1 in 300 children in western countries develops T1D by age 18 years.[7] The disease requires lifelong exogenous insulin therapy coupled with the development of diverse complications such as cardiovascular diseases, peripheral neuropathies, nephropathy, retinopathy, and poor wound healing, thereby predisposing to a decline both in longevity and quality of life.[8]
The inherited genetic factors are a major component contributing to T1D pathogenesis. Currently, >60 susceptible loci of T1D have been identified by genetic studies. The HLA region, with its multiple genes and extreme polymorphism at those loci, is responsible for 40% to 50% of the genetic risk.[9] Moreover, multiple genetic variants have also been reported to be associated with T1D, including INS, CTLA4, PTPN22, IL2RA, IFIH1, CAPSL, IL7R, CLEC16A, and PTPN2. Similarly, we reported that the small ubiquitin-like modifier 4 (SUMO4) confers T1D susceptibility in Asian populations.[10] However, other than genetic factors environmental insults have also been noted contributing to T1D risk, as evidenced by that monozygotic twins display a discordance of T1D rate, especially for those manifested T1D onsets with age >15 years.[11] It is believed that epigenetic factors serve as a bridge to link risk genes and environmental insults, thereby regulating the expression of T1D susceptible genes involved in antigen presentation (such as HLA), tolerance homeostasis (such as FOXP3 and CTLA4), autoreactive T cell response (such as GAD65), and β-cell functions (such as INS).[12] Therefore, T1D susceptibility is determined by the interplay of multiple genes with environmental factors [Figure 1]. In this review, we seek to elucidate how epigenetic factors modulate T1D risk, and to discuss their potential as biomarkers to monitor T1D initiation and progression, or as therapeutic targets for intervention and treatment of T1D in clinical settings.
Figure 1.
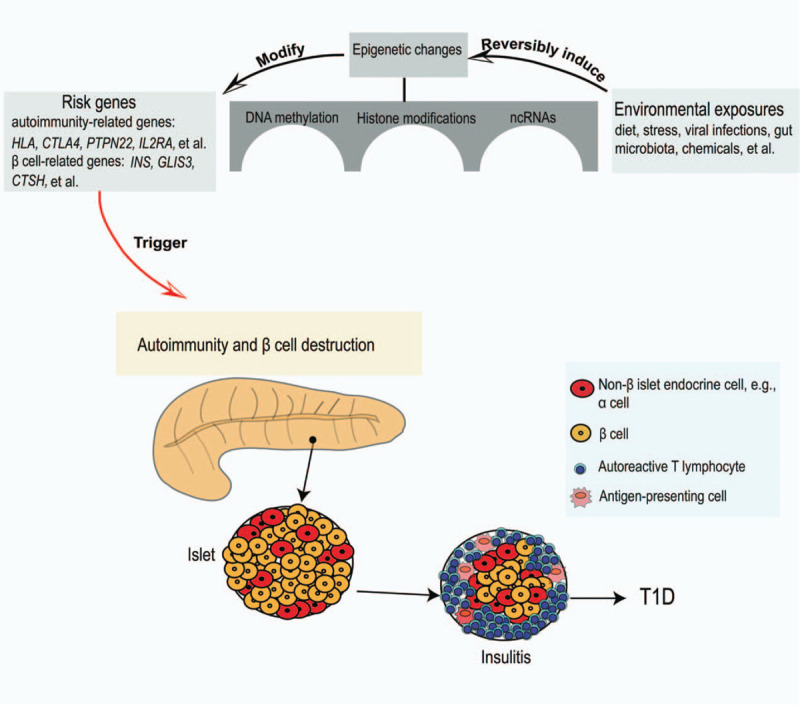
Environmental exposures induce epigenetic changes in genetically predisposed subjects to trigger the autoimmune destruction of the pancreatic β cells along with T1D onset. ncRNAs: Non-coding RNAs; T1D: Type 1 diabetes.
Environmental Factors Trigger Autoimmunity in T1D Setting
As mentioned above, many environmental factors, including viral infections, diet, gut microbiota, stress, and chemicals, are significantly associated with the development of autoimmune responses in T1D setting. For example, The environmental determinants of diabetes in the young (TEDDY), an ongoing observational cohort study, was designed to identify environmental exposures that may trigger islet autoimmunity and T1D in subjects at high genetic risk.[13] The TEDDY study provided a unique opportunity to shed light on the involvement of environmental factors in the etiology of T1D.
Viruses, especially enteroviruses such as coxsackieviruses (CVB) or echoviruses, have long been proposed as a potential environmental trigger for T1D in both animal and human studies.[14,15] However, the actual mechanisms by which viral infections may impact islet cell survival/function and contribute to T1D pathogenesis remains unclear. Different mechanisms have been proposed including direct cellular injury by cytolysis, molecular mimicry, phagocytosis of enterovirus-infected β cells, endoplasmic reticulum (ER) stress, increased intestinal permeability, loss of Tregs, and bystander activation,[16] which exacerbate dysfunction and loss of beta cells. Although the potential causative relationship between viral infections and T1D has been established in mice, epidemiological studies and conclusive trials to establish a causative link in humans are currently lacking. If the conclusive link is validated in humans, it would be feasible to develop an enterovirus vaccine to prevent T1D.
Dietary factors that have been evaluated include breastfeeding, the intake of cow's milk, vitamin D, early introduction of gluten, and food diversity during the first year of life.[17] There are conflicting conclusions for their role in the development of T1D, and some studies do not provide sufficient evidence to prove a causal effect. Nevertheless, most of the cohort studies showed that breastfeeding and vitamin D had protective effects, whereas cow's milk and early introduction of gluten were risk factors.[18] It is suggested that breastfeeding not only decreased intestinal permeability but also provided the protective effect against enteroviral infections mediated primarily by maternal antibodies in breast milk.[19,20] Conversely, early exposure to cow proteins increased intestinal permeability, making it accessible to potential T1D inciting exposures. Inflammation of the intestinal mucosa[21] and a dysregulated immune response to oral antigens[22] was also thought to mediate the predisposition caused by cow's milk. Intertwined with this concept is the effect of gut microbiota. Gut microbiota is important for nutrition metabolism, immunity, and neuroendocrine responses. Animal studies have clearly shown that the normal microbial consortium alleviated the progression of autoimmune diabetes.[23] In humans, there appears to be a trend of lower microbial diversity, as well as less abundance of lactate- and butyrate-producing bacteria to synthesize mucin and maintain gut integrity in T1D subjects.[24,25]
Unfortunately, despite extensive effort has been devoted to this field, current data obtained from past and ongoing studies are yet to draw a clear conclusion for the exact category of environmental exposures in T1D etiology. Therefore, more comprehensive and prospective studies with larger scale and carefully selected controls are needed to fully clarify those environmental risks associated with T1D pathoetiology.
DNA Methylation in T1D Pathogenesis
Epigenetics refers to all mitotically and meiotically heritable changes in phenotype that are not caused by the alterations of the genetic code.[26] The types of epigenetic mechanisms generally consist of DNA methylation, post-translational modifications of histones, and non-coding RNAs (ncRNAs), which affect chromatin remodeling and accessibility of transcriptional machinery to the genome or determine gene expression post-transcriptionally [Figure 2].[27]
Figure 2.
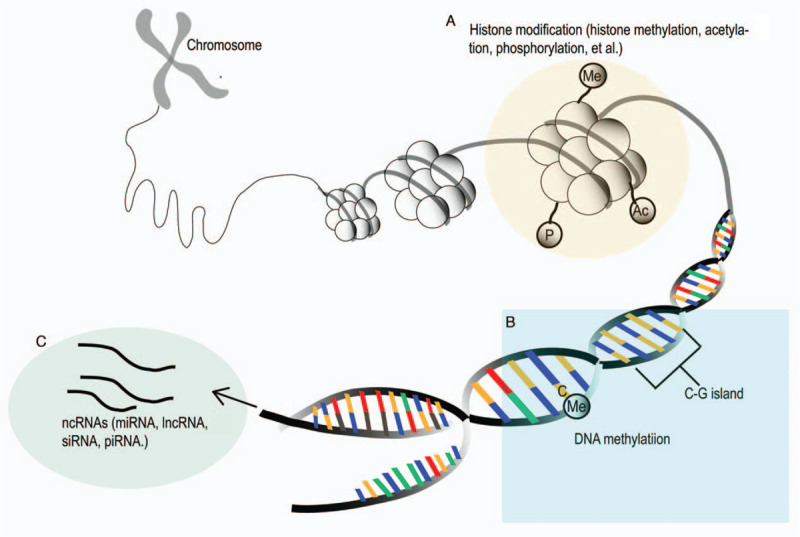
Schematic representation of main epigenetic mechanisms (Histone modifications [A], DNA methylation [B], and ncRNAs [C]) in the pathogenesis of type 1 diabetes. lncRNA: Long non-coding RNA; miRNA: MicroRNA; ncRNAs: Non-coding RNAs; piRNA: PIWI-interacting RNA; siRNA: Short-interfering RNA.
DNA methylation is a biochemical process in which a methyl group from the S-adenosyl-l-methionine (SAM) is transferred onto the C5 position of cytosines.[28] The process of DNA methylation is catalyzed by the DNA methyltransferases (DNMTs) such as DNMT1, DNMT3a, and DNMT3b. DNMT1 is involved in the maintenance of DNA methylation during cell replication, whereas DNMT3a and DNMT3b are de novo methyltransferases.[29] DNA methylation has been typically associated with transcriptional repression,[28] and in addition to transcriptional regulation, it is crucial for maintaining genome integrity.[27] Recently, dysregulated DNA methylation has been found in autoimmune diseases such as systemic lupus erythematosus (SLE), psoriasis, rheumatoid arthritis (RA), systemic sclerosis (SSc), and T1D.[30–34] However, DNA methylome could be affected by various factors, among which diets and environmental insults play an important role. There is increasing evidence that environmental factors, genetic variants, drugs, and microRNAs (miRNAs) impact the methylation levels of DNA to implicate in the pathogenesis of autoimmune disorders.[35]
Comparative analysis of DNA methylation patterns between monozygotic twin pairs who are discordant for T1D onset enables the characterization of T1D risks conferred by DNA methylome-encoded information, which highlights the role of epigenetic factors in T1D pathogenesis.[36] Indeed, genome-wide DNA methylation analysis of purified CD14+ monocytes (T1D-related effector cells) from monozygotic twin pairs with discordant T1D onset identified 132 T1D-methylation variable positions (T1D-MVPs), including the HLA class II gene HLA-DQB1, the strongest T1D susceptibility gene, and GAD2 which encodes the known autoantigen (GAD65) involved in T1D etiology.[37] As some of these T1D-MVPs are found in individuals before T1D diagnosis, they may occur very early during the course of autoimmune development that leads to overt T1D, which are unlikely caused by the antidiabetic therapies or long-term metabolic changes.[37] Follow-up studies also identified 88 CpG sites displaying significant methylation changes in 3 T1D-discordant monozygotic twin pairs, such as HLA, INS, IL-2RB, and CD226. Collectively, those findings further support that altered DNA methylation is implicated in the pathogenesis of T1D.
Other than HLA, the INS gene ranked second to be the high-risk gene for T1D susceptibility. Similarly, abnormal methylation changes have also been noted in INS. Seven CpG sites proximal to the transcription starting site (TSS) in the promoter of the INS gene have been characterized, and studies in T1D patients revealed a lower level of methylation at CpG-19, -135, and -234, but a higher methylation level at CpG-180 than that of controls.[38] Studies in NOD mice found that cytokines could induce methylation changes in the insulin DNA by enhancing the expression of methyltransferases,[39] which was confirmed by studies in human β cells in vitro.[39] Together, those data demonstrate that the changes of DNA methylome in response to immunological stressors may be a mechanism that affects insulin gene expression during the progression of T1D.[39]
It is noteworthy that altered DNA methylation also contributes to T1D risk by regulating the expression of autoimmunity-associated genes. For example, six CpG sites are located within the proximal promoter of the IL2RA gene, while studies in 252 T1D patients and 286 age-matched controls revealed that two CpG sites (−373 and −456) were characterized by the increased methylation levels in T1D patients when compared with that of controls, and more interestingly, DNA methylation levels at CpG-373 were correlated with 16 single-nucleotide polymorphisms (SNPs) located within the IL2RA locus, which is confirmed to be a T1D risk gene.[40] Similarly, we found that DNA hypermethylation impairs TLR9-induced Foxp3 expression in Tregs by attenuating IRF-7 binding activity in fulminant T1D patients.[34] In line with this observation, the FOXP3 promoter region was hypermethylated in CD4+ T cells originated from patients with latent autoimmune diabetes in adults (LADA).[41] Altogether, these findings highlight the importance of DNA methylation in the initiation and progression of autoimmune responses coupled with β-cell destruction during the course of T1D development, and more findings regarding DNA methylation in T1D pathogenesis are listed in Table 1.
Table 1.
Alterations of DNA methylation in T1D
| Genes | Cell types | Main findings | References |
| UNC13B | Whole blood | Involved in exocytosis, hypermethylation is associated with the risk of diabetic nephropathy in T1D | [96] |
| INS | Peripheral blood samples | DNA methylation near the INS gene is associated with INS genetic variation (rs689) and T1D | [97,98] |
| Human islets | Relevant to insulin secretion | ||
| IL-2RA | WBCs or tissue samples | CpGs (-373 and -456) showed increased methylation in T1D patients, associated with Tregs | [40] |
| Amylin DNA | β cells in the islet | Demethylated cfDNA may serve as a biomarker of β-cell death in T1D | [86] |
| FOXP3 | Tregs | FOXP3 promoter region was hypermethylated, FOXP3 expression was decreased, contributing to the pathogenesis of T1D | [34,41] |
| TNF | CD14+ cells | Hypermethylated in T1D, encodes protein TNF, a key inflammatory cytokine associated with T1D in animal models | [37] |
| TRAF6 | CD14+ cells | Hypermethylated in T1D, involved in NF-κB and MAPK kinase activation | [37] |
| CD6 | CD14+ cells | Hypermethylated in T1D, critical for T activation | [37] |
| HLA-DQB1 | CD14+ cells | Hypomethylated in T1D, carries the highest single genetic risk for T1D, involved in presenting peptides from extracellular proteins | [37] |
| NFKBIA | CD14+ cells | Hypomethylated in T1D, an important regulator of apoptosis and inflammatory immune responses | [37] |
| GAD2 | CD14+ cells | Hypomethylated in T1D, encodes GAD65, a major T1D autoantigen involved in disease etiology | [37] |
cfDNA: Circulating free amylin DNA; T1D: Type 1 diabetes; Tregs: Regulatory T cells; WBCs: Whole blood cells.
Histone Modification in T1D Pathoetiology
The N-terminal tails of the histones are subject to an ever-growing list of posttranslational modifications such as methylation, acetylation, phosphorylation, deamination, ubiquitylation, ADP ribosylation, and sumoylation.[42] Among these biochemical modifications, histone methylation and acetylation are the most abundant and widely studied. Methylation of lysine residues within histones is regulated by the opposing action of two families of enzymes, the methyltransferases (KMTs) and the lysine demethylases (KDMs). Histone methylation represents either transcriptionally active or repressive marks, depending on the location of methylated lysine residues (eg, lysine 4, 9, 27, or 36) and the extent of methylation (eg, mono-, di-, or tri-methylation).[27] Histone acetylation is highly dynamic and regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs). Histone acetylation leads to gene activation, thus some HDAC inhibitors (HDACi) have been extensively synthesized or naturally isolated as an approach to increase the transcription of various genes, including those involved in immune disorders.[43–45]
A growing body of evidence now supports that histone modifications are relevant to autoimmune disorders and particularly to T1D development. Comparative analysis of genome-wide histone H3 lysine 9 dimethylation (H3K9me2) patterns in blood lymphocytes and monocytes between T1D patients and healthy controls indicated a significant increase in H3K9me2 in a subset of genes in lymphocytes, including CLTA4, a T1D susceptibility gene, which encodes a T cell receptor to regulate apoptosis.[46] Ingenuity pathway analysis characterized two high-scoring networks which comprise genes with manifestations of altered H3K9me2, and many of which are associated with autoimmune and inflammatory-related pathways, such as TGF-β, NF-κB, p38 MAPK, TLR, and IL-6.[46] Moreover, hyperglycemia also seems to impact chromatin structure by affecting histone methylation. For example, in models of transient hyperglycemia, the active transcriptional state of the NFκB-p65 gene is characterized by the enhanced H3K4 and reduced H3K9 methylation as a result of prior hyperglycemia.[47]
In addition to histone methylation, aberrant histone acetylation is also involved in T1D pathogenesis. Studies revealed that the expression of histone deacetylase genes is reduced in the CD4+ T cells from patients with new-onset T1D.[48] On the contrary, a concerted downregulation of histone H3 acetylation was found in CD4+ T lymphocytes from patients with LADA, a form of T1D that occurs in adults with a slower course of onset.[49] Marked variations in H3K9Ac levels at the upstream regions of the HLA-DRB1 and HLA-DQB1 genes were observed in T1D monocytes when compared with controls, implying that the promoter/enhancer architecture and chromatin status of key susceptible loci could be important determinants in their functional association to T1D susceptibility.[50]
In line with the above observations, FR901228, an HDACi can inhibit hyperglycemia and reverse diabetes in newly diagnosed diabetic NOD mice.[51] Consistently, treatment of NOD mice during the transition from prediabetic to the diabetic stage with the well-characterized histone deacetylase inhibitor, trichostatin A (TSA), effectively reduced the incidence of diabetes.[52] Mechanistically, protection against diabetes was accompanied by the histone hyperacetylation in the pancreas and spleen, and with the manifestation of higher frequency of splenic CD4+CD62L+ cells, but lower inflammatory infiltration in the islets coupled with the restoration of normoglycemia and glucose-induced insulin release by β cells.[52] On the contrary, TSA prevented the differentiation of human Tregs into IL-17-producing cells, suggesting that histone acetylation underlies Treg plasticity.[53] Furthermore, HDACs have been noted with different regulatory roles in mediating cytokine-induced β-cell apoptosis.[54] Collectively, all those data support that histone modifications play a vital role in T1D pathogenesis [Table 2].
Table 2.
Summary of studied histone modifications in T1D
| Genes | Modification alterations | Effects | References |
| HDAC | Decreased expression in human CD4+ T cell | Regulation of transcription | [48] |
| HLA-DRB1 and HLA-DQB1 | Increased H3K9 acetylation in monocytes | H3K9Ac status may be relevant to their regulation and transcriptional response toward external stimuli | [50] |
| H4 | Increased acetylated histone H4 in monocytes | Potentially reduces endothelial dysfunction, vascular remodeling, and atherothrombotic events | [99] |
| NFκB-p65 | Enhanced H3K4 and reduced H3K9 methylation | Involved in the inflammatory responses, plays a key role in regulating the immune response to infection | [47] |
| CTLA4 | Increased H3K9me2 methylation in T1D lymphocytes | Enhances T cell activation | [46] |
T1D: Type 1 diabetes.
Impact of ncRNAs on T1D Development
ncRNAs are functional RNA molecules that are transcribed from DNA but are not translated into proteins. Epigenetic-related ncRNAs include miRNA, short-interfering RNA (siRNA), PIWI-interacting RNA (piRNA), and long non-coding RNA (lncRNA).[55] miRNAs are an abundant class of approximately 22-nucleotide regulatory RNAs that are responsible for the regulation of about 60% of all mRNAs. miRNAs perform their functions by binding to the 3′-untranslated region (UTR) of target mRNA transcripts, leading to mRNA cleavage or repression of productive translation.[56] miRNAs are important epigenetic mechanisms involved in regulating cell differentiation, cell cycle, apoptosis, and immune responses, thereby influencing the pathogenesis of many human diseases.[57–60] lncRNAs represent a type of long transcribed RNA molecules that are longer than 200 nucleotides. Unlike miRNAs, lncRNAs localize both in the cytoplasm and nucleus and can either negatively or positively regulate gene expression by physically interacting with DNA (chromatin), RNA [mRNAs, miRNAs, circular RNAs (circRNAs)], and proteins.[61] lncRNAs are the rapidly expanding fields because their aberrant expression has been linked to a wide spectrum of disorders, and a few lncRNAs have been linked to the regulation of immune responses, for example, cytokine production, dendritic cell differentiation, and T cell development.[62–64]
Recent studies demonstrated evidence that miRNAs are implicated in autoimmune initiation, β-cell dysfunction, and apoptosis, which contribute to the development of T1D [Table 3]. For example, miR-326 is expressed at higher levels in the peripheral blood lymphocytes from T1D subjects, in which levels of this miRNA were highly correlated with ongoing islet autoimmunity and disease severity.[65] Similarly, miR-326 is highly correlated with disease severity in patients with multiple sclerosis and mice with experimental autoimmune encephalomyelitis (EAE). Mechanistically, miR-326 promotes Th17 differentiation by targeting erythroblastosis virus E26 oncogene homolog 1 (Ets-1) and its overexpression leads to higher severity of EAE, pointing to a more general role for miR-326 in autoimmunity.[66] miR-98, miR-23b, and miR-590-5p were found to be overexpressed in T1D patient-derived CD8+ T cells, and transfection of these miRNAs into primary T cells reduces FAS and TRAIL mRNA, and therefore, repression of pro-apoptotic pathways by these miRNAs contributes to the unrestricted expansion of diabetogenic cytotoxic T cells.[67] Tregs are critical regulators of autoimmune diseases, including T1D. The expression of miRNA-510 is significantly increased, both miRNA-342 and miRNA-191 expression are decreased in Tregs originated from T1D patients.[68] A recent study further revealed that miR142-3p is induced in islet autoimmunity and a miR142-3p/Tet2/Foxp3 axis impairs Treg differentiation and stability in models of T1D.[69]
Table 3.
miRNAs dysregulated in T1D
| miRNAs | Origins | Effects | Targets | References |
| miR-326 | Peripheral blood lymphocytes (+) | Associated with a state of ongoing autoimmunity | Ets-1, VDR | [65] |
| miR-21 | β cells (+) | NF-κB−miR-21−PDCD4 axis controls islet β-cell death | PDCD4 | [71,100] |
| PBMCs (−) | ||||
| miR-146 | Cytokine treated islets (+), NOD mice islets, MIN6 | Contribute to cytokine-mediated β-cell dysfunction | TRAF6, IRAK1 | [70,72,101] |
| miR-34a | VAMP2, BclII | |||
| miR-29a/b/c | Mcl1 | |||
| miR-101a | Neurod1 | |||
| miR-30b | Neurod1 | |||
| miR-375 | Islet-specific miRNA | A potential biomarker of β-cell death and predictor of diabetes | ? | [102] |
| Serum (+) | ||||
| miR-25 | Serum (+) | Negatively associated with residual β-cell function, positively associated with glycemic control (HbA1c) | Bim and Trail | [103] |
| miR-181a | Serum (+) | Mediates pancreatic β cells dysfunction | SMAD7 | [104] |
| miR-503 | β cells (+) | Knockdown of miR-503 enhances insulin secretion of pancreatic β cells, promotes cell proliferation, and protects cells from apoptosis | mTOR pathway | [105] |
| microRNA-16-5p, -17-5p, and -20a-5p | Plasma (+) | Novel diagnostic biomarkers for gestational diabetes mellitus | ? | [106] |
| miR-24, miR-26, miR-182, and miR-148 | β cells | Specific knockdown of miR-24, miR-26, miR-182, or miR-148 in cultured β cells or isolated primary islets downregulates insulin promoter activity and insulin mRNA levels | Bhlhe22, Sox6 | [107] |
| miR-33a | β cell | Influences insulin secretion | ABCA1 | [108] |
| miR-125a-5p | Tregs (+) | Increased expression of miR-125a-5p on Tregs results in reduced CCR2, thus limiting their migration and eventual function in the pancreas | CCR2 | [109] |
(+), increased; (−), decreased; ?, unknown function. Ets-1: Erythroblastosis virus E26 oncogene homolog 1; miRNAs: MicroRNAs; T1D: Type 1 diabetes; Tregs: Regulatory T cells; VDR: Vitamin D receptor.
More interestingly, analysis of miRNAs in cytokine-mediated β-cell cytotoxicity revealed that miR-21, miR-34a, and miR-146a could be novel players in β-cell failure elicited in vitro and in vivo by proinflammatory cytokines, notably during the development of peri-insulitis that precedes overt diabetes in NOD mice.[70] However, validation of these discoveries is necessary, as Ruan et al.[71] reported that c-Rel and p65 of the NF-κB family activate the mir21 gene promoter to increase miR-21 RNA levels in the pancreatic β cells, which in turn decreases PDCD4 to induce β-cell death through the Bax family of apoptotic proteins. Furthermore, altered expression of miR-29 family members contributes to cytokine-mediated β-cell dysfunction occurring during the initial phases of T1D, which is associated with diminished expression of the transcription factor Onecut2 coupled with the expression of Granuphilin, an inhibitor of β-cell exocytosis.[72] More recently, studies found that miR-142-3p, miR-142-5p, and miR-155 derived from exosomes released by effector T cells could be transferred into rodent and human pancreatic β cells, thereby triggering chemokine expression and apoptosis in β cells. As a result, the blockade of these miRNAs in recipient β cells prevents exosome-mediated apoptosis and decreases diabetes incidence in NOD mice.[73] These data suggest that the transfer of T cell exosomal-miRNAs could be a novel mechanism leading to T1D development.[73]
Other than miRNAs, lncRNAs have also emerged as important players in diverse biological processes, such as cell cycle progression, immune surveillance, genomic imprinting, and embryonic stem cell pluripotency.[74,75] As such, there is also feasible evidence that lncRNAs are implicated in T1D pathogenesis. Particularly, analysis of the pancreatic islets and β-cell transcriptome characterized >1000 lncRNAs in both humans and mice, most of which are islet-specific and dynamically regulated by glucose.[76] Studies further revealed that lncRNA mouse maternal expressed gene 3 (Meg3) may function as a novel regulator to maintain β-cell identity by affecting insulin production and cell apoptosis, and Meg3 in the islets was decreased in T1D (female NOD mice) and T2D (db/db mice) animals.[77] Additionally, GWAS identified that an SNP (rs941576: A>G) located in an intron of MEG3 is tightly linked to T1D.[78] Studies in a β-cell line, MIN6 cells, found that MIN6 cells express a large number of lncRNAs, and proinflammatory cytokine stimulation induced significant changes in many of those lncRNAs. Studies in four selected lncRNAs revealed that their expression increases along with the development of insulitis in NOD mice to promote β-cell apoptosis.[79] All lncRNAs potentially associated with T1D are listed in Table 4, which may provide an exciting opportunity to advance our understanding of T1D pathogenesis.
Table 4.
lncRNAs that have been implicated in T1D
| lncRNAs | Cell types | Effects | References |
| NONMMUT036704 | Pancreatic β-cell line (MIN6) | Upregulated after MIN6 cells exposed to the proinflammatory cytokines, may play an important role in the development of T1D through the regulation of NGAL | [110] |
| NONMMUT034373 | MIN6 | Upregulated after MIN6 cells exposed to the proinflammatory cytokines, sense overlap PD-L1 | [110,111] |
| IGF2-AS | β cells | Potentially promotes β-cell proliferation | [112,113] |
| MEG3 | β cells | A novel lncRNA regulator of insulin synthesis, secretion, and sensitivity | [77,114] |
| PVT1 | A variety of renal cell types such as mesangial cells | It can increase proliferation and inhibit apoptosis in multiple cell types, associated with ESRD attributed to T1D | [115,116] |
| HI-LNC25 | β cells | A β cell-specific lncRNA downregulates GLIS3 mRNA, thus exemplifies a gene regulatory function of islet lncRNAs | [76] |
| βlinc1 (HI-LNC15) | β cells | Necessary for the specification and function of insulin-producing β cells, deletion of βlinc1 results in defective islet development and disruption of glucose homeostasis in adult mice | [117] |
| MIN6 cells | |||
| PLUTO (HI-LNC71) | β cells | Affects local 3D chromatin structure and transcription of PDX1, thereby modulates the PDX1-dependent transcriptional program | [118] |
| EndoC-βH1 | |||
| Tunar (HI-LNC78) | β cells | A glucose-regulated islet transcript reduces insulin content and, consequently, impairs glucose-stimulated insulin secretion after knockdown of HILNC78 | [118] |
| EndoC-βH1 | |||
| MALAT1 | Endothelial cells | MALAT1 knockdown could obviously ameliorate diabetic retinopathy in vivo and regulate retinal endothelial cell proliferation, migration, and tube formation in vitro | [119,120] |
| TUG1 | β cells | Downregulation of lncRNA TUG1 expression affects apoptosis and insulin secretion in vitro and in vivo. lncRNA TUG1 may represent a factor that regulates the function of pancreatic β cells | [121] |
βlinc1: β-cell long intergenic non-coding RNA 1; ESRD: End-stage renal disease; IGF2-AS: Insulin-like growth factor 2 antisense RNA; lncRNAs: Long non-coding RNAs; MALAT1: Metastasis-associated lung adenocarcinoma transcript 1; NGAL: Neutrophil gelatinase-associated lipocalin; PD-L1: Programmed death-1 ligand-1; PVT1: Plasmacytoma variant translocation 1; T1D: Type 1 diabetes.
More recently, other types of ncRNAs, such as circRNAs, also receive increased attention in T1D pathogenesis. circRNAs are a class of covalently closed, single-stranded circular transcripts with no 5′ caps and 3′ poly (A) tails. They can regulate gene expression at transcriptional or post-transcriptional levels. Increasing evidence has shown that circRNAs exert an important effect on immune diseases, including multiple sclerosis,[80] SLE,[81] and T1D.[82,83] Recently, hsa_circ_0060450 was reported to release its target gene, SHP2, which suppresses the JAK-STAT signaling pathway triggered by type I interferon, thereby inhibiting macrophage-mediated inflammation in T1D.[83] Similarly, Arraystar human circRNA microarray characterized 93 differentially expressed circRNA in the peripheral blood between T1D patients and controls.[82] Among which, hsa_circ_0072697 may get involved in 50 circRNA–miRNA–mRNA signaling pathways associated with diabetes, such as hsa_circ_0072697–miR-15a–UBASH3A network, and five circRNAs (hsa_circ_0071224, hsa_circ_0002437, hsa_circ_0084429, hsa_circ_0072697, and hsa_circ_0000787) in T1D are considered to have the most coding potential.[82] Together, these results suggested a possible implication of circRNAs in T1D pathogenesis. However, experiments at the cell, tissue, and individual levels are necessary to reach a definite conclusion for their role in T1D pathoetiology.
Epigenetic Biomarkers and Therapeutic Targets in T1D Setting
Given the fact that epigenetic mechanisms regulate gene expressions, which in turn determine the dynamic molecular and cellular events during different stages of T1D, epigenetic factors could be sensitive biomarkers to monitor T1D progression and drug response. In addition, the reversible dynamic changes of epigenetic properties also support that some epigenetic genes could be viable targets for T1D prevention and treatment in clinical settings.
In general, circulating miRNAs are ideal biomarkers to monitor T1D initiation and progression as they do not degrade readily and can be detected non-invasively in body fluids. Indeed, some miRNAs with altered expressions are featured in a particular type of autoimmune disease. For example, miR-125b and miR-146a are specific for the SLE, miR-24 is featured in RA, while miR-130a and miR-181a are specific in T1D.[84] Furthermore, analysis of the miRNA expression profile revealed that miR-103a-3p, miR-155-5p, miR-200a-3p, and miR-210-3p are upregulated, while miR-146a-5p is downregulated in recently diagnosed T1D patients or in T1D patients with ≥5 years of diagnosis.[85] Additionally, miRNAs were shown to predict T1D-related complications. Similarly, methylated DNA has also been found to be potential biomarkers during T1D development. Specifically, circulating unmethylated insulin and amylin DNA are increased in recent-onset T1D patients, and detection of circulating β cell-derived DNA could be a viable approach to monitor β-cell destruction during the course of T1D development, and to assess β mass loss during T1D progression.[86,87] Although the above-mentioned epigenetic signatures are likely to be promising biomarkers for T1D prediction and diagnosis, their applications in clinical settings, however, are still awaiting further studies.
More recently, compounds with the capability to block epigenetic changes are developed as potential new drugs (named epidrugs) against autoimmune disorders.[88] A typical example is the DNA demethylation agent, 5-Aza-2′-deoxycytidine (DAC), which has been widely used in cancer models and patients but now is found to exert a powerful beneficial effect on T1D.[89] Moreover, inhibition of KDM6 demethylases using a selective inhibitor, GSK-J4, protects β cells against autoimmune-induced dysfunction and apoptosis.[90] Currently, HDACi are the most widely studied epidrugs. HDACi are orally active, safe, and well-tolerated at low doses and some of them are currently being tested in clinical trials on cancer and inflammatory diseases.[91,92] As aforementioned, TSA, the well-characterized HDACi, protects mice against autoimmune diabetes.[52,93] Similarly, the clinically well-tolerated lysine deacetylase inhibitors (KDACi), vorinostat and givinostat, revert diabetes in NOD mice by attenuating autoimmune responses and protecting β cells from destruction.[94] In fact, one of the possible mechanisms for antidiabetic drug metformin is the deacetylation of non-histone and histone proteins, in which metformin activates SIRT-1, a protein deacetylase.[88] Similarly, SIRT-1 could also be an epigenetic target of fenofibrate (a drug against hyperlipidemia) and resveratrol (a red wine compound with antioxidant function). Administration of resveratrol not only prevented the reduction of SIRT-1 but also SIRT-2, SIRT-3, and SIRT-5 in the T1D rat heart. Therefore, SIRT-1 could be a viable target for the intervention of cardiac complications in T1D.[95] Together, these discoveries provided a rationale for clinical trials of the safety and efficacy of those epidrugs in the T1D setting.
Summary
T1D usually occurs in genetically susceptible individuals and is triggered by environmental factors. Therefore, it is crucial to understand the interaction between genetic susceptibility and environmental factors to define risk levels and to develop therapeutic strategies. Nowadays, the field of epigenetics has drawn extensive attention worldwide, as it serves as a bridge to link genetic factors and environmental triggers. Indeed, environmental insult-induced epigenetic changes modulate the expression of critical genes relevant to the initiation and progression of autoimmunity and β-cell destruction, and therefore, are implicated in T1D pathogenesis. Albeit exciting results have been achieved recently, forceful longitudinal follow-up studies should be carefully designed to exactly ascertain to what extent alterations in epigenetic factors influence islet autoimmunity and T1D progression. Unlike genetic hardwiring, epigenetic changes following environmental exposures are a reversible process, and as a result, those changes could be erased through specific mechanisms, which rendered them potential biomarkers to monitor T1D initiation and progression. More importantly, epigenetic changes encode somatically heritable information, which makes them good therapeutic candidates for the establishment of innovative therapies with epidrugs against T1D in clinical settings.
Funding
This study was supported by grants from the Ministry of Science and Technology (Nos. 2016YFC1305002 and 2017YFC1309603), the National Natural Science Foundation of China (Nos. 81530024, 91749207, 81920108009, 81770823, and 81670729), the NHC Drug Discovery Program (No. 2017ZX09304022-07), the Department of Science and Technology of Hubei State (No. 2017ACA096), the Integrated Innovative Team for Major Human Disease Programs of Tongji Medical College, Huazhong University of Science and Technology, and the Innovative Funding for Translational Research from Tongji Hospital.
Conflicts of Interest
None.
Footnotes
How to cite this article: Zhang J, Chen LM, Zou Y, Zhang S, Xiong F, Wang CY. Implication of epigenetic factors in the pathogenesis of type 1 diabetes. Chin Med J 2021;134:1031–1042. doi: 10.1097/CM9.0000000000001450
Jing Zhang and Long-Min Chen contributed equally to this work.
References
- 1.DiMeglio LA, Evans-Molina C, Oram RA. Type 1 diabetes. Lancet 2018; 391:2449–2462. doi: 10.1016/S0140-6736(18)31320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morran MP, Vonberg A, Khadra A, Pietropaolo M. Immunogenetics of type 1 diabetes mellitus. Mol Aspects Med 2015; 42:42–60. doi: 10.1016/j.mam.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J, Chen L, Wang F, Zou Y, Li J, Luo J, et al. Extracellular HMGB1 exacerbates autoimmune progression and recurrence of type 1 diabetes by impairing regulatory T cell stability. Diabetologia 2020; 63:987–1001. doi: 10.1007/s00125-020-05105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boldison J, Wong FS. Immune and pancreatic beta cell interactions in type 1 diabetes. Trends Endocrinol Metab 2016; 27:856–867. doi: 10.1016/j.tem.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Magnuson AM, Thurber GM, Kohler RH, Weissleder R, Mathis D, Benoist C. Population dynamics of islet-infiltrating cells in autoimmune diabetes. Proc Natl Acad Sci U S A 2015; 112:1511–1516. doi: 10.1073/pnas.1423769112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eizirik DL, Colli ML, Ortis F. The role of inflammation in insulitis and beta-cell loss in type 1 diabetes. Nat Rev Endocrinol 2009; 5:219–226. doi: 10.1038/nrendo.2009.21. [DOI] [PubMed] [Google Scholar]
- 7.Couzin-Frankel J. Mass screening weighed for type 1 diabetes risk. Science 2020; 368:353.doi: 10.1126/science.368.6489.353. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, Chen Z, Zhou Z, Yang P, Wang C-Y. Sumoylation modulates the susceptibility to type 1 diabetes. Adv Exp Med Biol 2017; 963:299–322. doi: 10.1007/978-3-319-50044-7_18. [DOI] [PubMed] [Google Scholar]
- 9.Noble JA, Erlich HA. Genetics of type 1 diabetes. Cold Spring Harb Perspect Med 2012; 2:a007732.doi: 10.1101/cshperspect.a007732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo D, Li M, Zhang Y, Yang P, Eckenrode S, Hopkins D, et al. A functional variant of SUMO4, a new I kappa B alpha modifier, is associated with type 1 diabetes. Nat Genet 2004; 36:837–841. doi: 10.1038/ng1391. [DOI] [PubMed] [Google Scholar]
- 11.Jerram ST, Dang MN, Leslie RD. The role of epigenetics in type 1 diabetes. Curr Diab Rep 2017; 17:89.doi: 10.1007/s11892-017-0916-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, Xie Z, Lu Q, Chang C, Zhou Z. Beyond genetics: what causes type 1 diabetes. Clin Rev Allergy Immunol 2017; 52:273–286. doi: 10.1007/s12016-016-8592-1. [DOI] [PubMed] [Google Scholar]
- 13.Elding Larsson H, Vehik K, Gesualdo P, Akolkar B, Hagopian W, Krischer J, et al. Children followed in the TEDDY study are diagnosed with type 1 diabetes at an early stage of disease. Pediatr Diabetes 2014; 15:118–126. doi: 10.1111/pedi.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coppieters KT, Wiberg A, Tracy SM, von Herrath MG. Immunology in the clinic review series: Focus on type 1 diabetes and viruses: The role of viruses in type 1 diabetes: a difficult dilemma. Clin Exp Immunol 2012; 168:5–11. doi: 10.1111/j.1365-2249.2011.04554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stene LC, Rewers M. Immunology in the clinic review series; focus on type 1 diabetes and viruses: The enterovirus link to type 1 diabetes: critical review of human studies. Clin Exp Immunol 2012; 168:12–23. doi: 10.1111/j.1365-2249.2011.04555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Precechtelova J, Borsanyiova M, Sarmirova S, Bopegamage S. Type I diabetes mellitus: genetic factors and presumptive enteroviral etiology or protection. J Pathog 2014; 2014:738512.doi: 10.1155/2014/738512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinonen MT, Moulder R, Lahesmaa R. New insights and biomarkers for type 1 diabetes: review for Scandinavian Journal of Immunology. Scand J Immunol 2015; 82:244–253. doi: 10.1111/sji.12338. [DOI] [PubMed] [Google Scholar]
- 18.Butalia S, Kaplan GG, Khokhar B, Rabi DM. Environmental risk factors and type 1 diabetes: Past, present, and future. Can J Diabetes 2016; 40:586–593. doi: 10.1016/j.jcjd.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Catassi C, Bonucci A, Coppa GV, Carlucci A, Giorgi PL. Intestinal permeability changes during the first month: effect of natural versus artificial feeding. J Pediatr Gastroenterol Nutr 1995; 21:383–386. doi: 10.1097/00005176-199511000-00003. [DOI] [PubMed] [Google Scholar]
- 20.Sadeharju K, Knip M, Virtanen SM, Savilahti E, Tauriainen S, Koskela P, et al. Maternal antibodies in breast milk protect the child from enterovirus infections. Pediatrics 2007; 119:941–946. doi: 10.1542/peds.2006-0780. [DOI] [PubMed] [Google Scholar]
- 21.Westerholm-Ormio M, Vaarala O, Pihkala P, Ilonen J, Savilahti E. Immunologic activity in the small intestinal mucosa of pediatric patients with type 1 diabetes. Diabetes 2003; 52:2287–2295. doi: 10.2337/diabetes.52.9.2287. [DOI] [PubMed] [Google Scholar]
- 22.Luopajarvi K, Savilahti E, Virtanen SM, Ilonen J, Knip M, Akerblom HK, et al. Enhanced levels of cow's milk antibodies in infancy in children who develop type 1 diabetes later in childhood. Pediatr Diabetes 2008; 9:434–441. doi: 10.1111/j.1399-5448.2008.00413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 2008; 455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One 2011; 6:e25792.doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Goffau MC, Luopajarvi K, Knip M, Ilonen J, Ruohtula T, Harkonen T, et al. Fecal microbiota composition differs between children with beta-cell autoimmunity and those without. Diabetes 2013; 62:1238–1244. doi: 10.2337/db12-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerna M. Epigenetic regulation in etiology of type 1 diabetes mellitus. Int J Mol Sci 2019; 21:36.doi: 10.3390/ijms21010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fodor A, Cozma A, Karnieli E. TBC update: personalized epigenetic management of diabetes. Per Med 2017; 14:531–549. doi: 10.2217/pme-2017-0043. [DOI] [PubMed] [Google Scholar]
- 28.Schubeler D. Function and information content of DNA methylation. Nature 2015; 517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 29.Jurkowska RZ, Jeltsch A. Mechanisms and biological roles of DNA methyltransferases and DNA methylation: From past achievements to future challenges. Adv Exp Med Biol 2016; 945:1–17. doi: 10.1007/978-3-319-43624-1_1. [DOI] [PubMed] [Google Scholar]
- 30.Zhao M, Wang J, Liao W, Li D, Li M, Wu H, et al. Increased 5-hydroxymethylcytosine in CD4 (+) T cells in systemic lupus erythematosus. J Autoimmun 2016; 69:64–73. doi: 10.1016/j.jaut.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 31.Zhang P, Zhao M, Liang G, Yin G, Huang D, Su F, et al. Whole-genome DNA methylation in skin lesions from patients with psoriasis vulgaris. J Autoimmun 2013; 41:17–24. doi: 10.1016/j.jaut.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 32.Nakano K, Whitaker JW, Boyle DL, Wang W, Firestein GS. DNA methylome signature in rheumatoid arthritis. Ann Rheum Dis 2013; 72:110–117. doi: 10.1136/annrheumdis-2012-201526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang YY, Shu Y, Xiao YF, Wang Q, Kanekura T, Li YP, et al. Hypomethylation and overexpression of ITGAL (CD11a) in CD4 (+) T cells in systemic sclerosis. Clin Epigenetics 2014; 6:25.doi: 10.1186/1868-7083-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z, Zheng Y, Hou C, Yang L, Li X, Lin J, et al. DNA methylation impairs TLR9 induced Foxp3 expression by attenuating IRF-7 binding activity in fulminant type 1 diabetes. J Autoimmun 2013; 41:50–59. doi: 10.1016/j.jaut.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 35.Mazzone R, Zwergel C, Artico M, Taurone S, Ralli M, Greco A, et al. The emerging role of epigenetics in human autoimmune disorders. Clin Epigenetics 2019; 11:34.doi: 10.1186/s13148-019-0632-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang H, Pollin TI. Epigenetics variation and pathogenesis in diabetes. Curr Diab Rep 2018; 18:121.doi: 10.1007/s11892-018-1091-4. [DOI] [PubMed] [Google Scholar]
- 37.Rakyan VK, Beyan H, Down TA, Hawa MI, Maslau S, Aden D, et al. Identification of type 1 diabetes-associated DNA methylation variable positions that precede disease diagnosis. PLoS Genet 2011; 7:e1002300.doi: 10.1371/journal.pgen.1002300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fradin D, Le Fur S, Mille C, Naoui N, Groves C, Zelenika D, et al. Association of the CpG methylation pattern of the proximal insulin gene promoter with type 1 diabetes. PLoS One 2012; 7:e36278.doi: 10.1371/journal.pone.0036278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rui J, Deng S, Lebastchi J, Clark PL, Usmani-Brown S, Herold KC. Methylation of insulin DNA in response to proinflammatory cytokines during the progression of autoimmune diabetes in NOD mice. Diabetologia 2016; 59:1021–1029. doi: 10.1007/s00125-016-3897-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belot M-P, Fradin D, Mai N, Le Fur S, Zelenika D, Kerr-Conte J, et al. CpG methylation changes within the IL2RA promoter in type 1 diabetes of childhood onset. PLoS One 2013; 8:e68093.doi: 10.1371/journal.pone.0068093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng C, Zhou Z, Yang L, Lin J, Huang G, Li X, et al. Fulminant type 1 diabetes mellitus exhibits distinct clinical and autoimmunity features from classical type 1 diabetes mellitus in Chinese. Diabetes Metab Res Rev 2011; 27:70–78. doi: 10.1002/dmrr.1148. [DOI] [PubMed] [Google Scholar]
- 42.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell 2012; 48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zwergel C, Valente S, Jacob C, Mai A. Emerging approaches for histone deacetylase inhibitor drug discovery. Expert Opin Drug Discov 2015; 10:599–613. doi: 10.1517/17460441.2015.1038236. [DOI] [PubMed] [Google Scholar]
- 44.Chistiakov DA, Orekhov AN, Bobryshev YV. Treatment of cardiovascular pathology with epigenetically active agents: Focus on natural and synthetic inhibitors of DNA methylation and histone deacetylation. Int J Cardiol 2017; 227:66–82. doi: 10.1016/j.ijcard.2016.11.204. [DOI] [PubMed] [Google Scholar]
- 45.Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov 2014; 13:673–691. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- 46.Miao F, Smith DD, Zhang L, Min A, Feng W, Natarajan R. Lymphocytes from patients with type 1 diabetes display a distinct profile of chromatin histone H3 lysine 9 dimethylation: An epigenetic study in diabetes. Diabetes 2008; 57:3189–3198. doi: 10.2337/db08-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brasacchio D, Okabe J, Tikellis C, Balcerczyk A, George P, Baker EK, et al. Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes 2009; 58:1229–1236. doi: 10.2337/db08-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orban T, Kis J, Szereday L, Engelmann P, Farkas K, Jalahej H, et al. Reduced CD4+ T-cell-specific gene expression in human type 1 diabetes mellitus. J Autoimmun 2007; 28:177–187. doi: 10.1016/j.jaut.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 49.Liu X-Y, Xu J-F. Reduced histone H3 acetylation in CD4 (+) T lymphocytes: Potential mechanism of latent autoimmune diabetes in adults. Dis Markers 2015; 2015:285125.doi: 10.1155/2015/285125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miao F, Chen Z, Zhang L, Liu Z, Wu X, Yuan YC, et al. Profiles of epigenetic histone post-translational modifications at type 1 diabetes susceptible genes. J Biol Chem 2012; 287:16335–16345. doi: 10.1074/jbc.M111.330373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skov S, Rieneck K, Bovin LF, Skak K, Tomra S, Michelsen BK, et al. Histone deacetylase inhibitors: A new class of immunosuppressors targeting a novel signal pathway essential for CD154 expression. Blood 2003; 101:1430–1438. doi: 10.1182/blood-2002-07-2073. [DOI] [PubMed] [Google Scholar]
- 52.Patel T, Patel V, Singh R, Jayaraman S. Chromatin remodeling resets the immune system to protect against autoimmune diabetes in mice. Immunol Cell Biol 2011; 89:640–649. doi: 10.1038/icb.2010.144. [DOI] [PubMed] [Google Scholar]
- 53.Koenen HJPM, Smeets RL, Vink PM, van Rijssen E, Boots AMH, Joosten I. Human CD25highFoxp3pos regulatory T cells differentiate into IL-17-producing cells. Blood 2008; 112:2340–2352. doi: 10.1182/blood-2008-01-133967. [DOI] [PubMed] [Google Scholar]
- 54.Lundh M, Christensen DP, Rasmussen DN, Mascagni P, Dinarello CA, Billestrup N, et al. Lysine deacetylases are produced in pancreatic beta cells and are differentially regulated by proinflammatory cytokines. Diabetologia 2010; 53:2569–2578. doi: 10.1007/s00125-010-1892-8. [DOI] [PubMed] [Google Scholar]
- 55.Huang Z, Zhou J-K, Peng Y, He W, Huang C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol Cancer 2020; 19:77.doi: 10.1186/s12943-020-01188-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen C-Z, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004; 303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 57.Pattarayan D, Thimmulappa RK, Ravikumar V, Rajasekaran S. Diagnostic potential of extracellular microRNA in respiratory diseases. Clin Rev Allergy Immunol 2018; 54:480–492. doi: 10.1007/s12016-016-8589-9. [DOI] [PubMed] [Google Scholar]
- 58.Meroni PL, Penatti AE. Epigenetics and systemic lupus erythematosus: Unmet needs. Clin Rev Allergy Immunol 2016; 50:367–376. doi: 10.1007/s12016-015-8497-4. [DOI] [PubMed] [Google Scholar]
- 59.Renauer P, Coit P, Sawalha AH. Epigenetics and vasculitis: a comprehensive review. Clin Rev Allergy Immunol 2016; 50:357–366. doi: 10.1007/s12016-015-8495-6. [DOI] [PubMed] [Google Scholar]
- 60.Bao Y, Cao X. Epigenetic control of B cell development and B-cell-related immune disorders. Clin Rev Allergy Immunol 2016; 50:301–311. doi: 10.1007/s12016-015-8494-7. [DOI] [PubMed] [Google Scholar]
- 61.Jathar S, Kumar V, Srivastava J, Tripathi V. Technological developments in lncRNA biology. Adv Exp Med Biol 2017; 1008:283–323. doi: 10.1007/978-981-10-5203-3_10. [DOI] [PubMed] [Google Scholar]
- 62.Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, et al. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A 2014; 111:1002–1007. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 2014; 344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 64.Hu G, Tang Q, Sharma S, Yu F, Escobar TM, Muljo SA, et al. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol 2013; 14:1190–1198. doi: 10.1038/ni.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sebastiani G, Grieco FA, Spagnuolo I, Galleri L, Cataldo D, Dotta F. Increased expression of microRNA miR-326 in type 1 diabetic patients with ongoing islet autoimmunity. Diabetes Metab Res Rev 2011; 27:862–866. doi: 10.1002/dmrr.1262. [DOI] [PubMed] [Google Scholar]
- 66.Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol 2009; 10:1252–1259. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- 67.de Jong VM, van der Slik AR, Laban S, van’t Slot R, Koeleman BPC, Zaldumbide A, et al. Survival of autoreactive T lymphocytes by microRNA-mediated regulation of apoptosis through TRAIL and Fas in type 1 diabetes. Genes Immun 2016; 17:342–348. doi: 10.1038/gene.2016.29. [DOI] [PubMed] [Google Scholar]
- 68.Hezova R, Slaby O, Faltejskova P, Mikulkova Z, Buresova I, Raja KR, et al. MicroRNA-342, microRNA-191 and microRNA-510 are differentially expressed in T regulatory cells of type 1 diabetic patients. Cell Immunol 2010; 260:70–74. doi: 10.1016/j.cellimm.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 69.Scherm MG, Serr I, Zahm AM, Schug J, Bellusci S, Manfredini R, et al. miRNA142-3p targets Tet2 and impairs Treg differentiation and stability in models of type 1 diabetes. Nat Commun 2019; 10:5697.doi: 10.1038/s41467-019-13587-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roggli E, Britan A, Gattesco S, Lin-Marq N, Abderrahmani A, Meda P, et al. Involvement of microRNAs in the cytotoxic effects exerted by proinflammatory cytokines on pancreatic beta-cells. Diabetes 2010; 59:978–986. doi: 10.2337/db09-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ruan Q, Wang T, Kameswaran V, Wei Q, Johnson DS, Matschinsky F, et al. The microRNA-21-PDCD4 axis prevents type 1 diabetes by blocking pancreatic beta cell death. Proc Natl Acad Sci U S A 2011; 108:12030–12035. doi: 10.1073/pnas.1101450108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roggli E, Gattesco S, Caille D, Briet C, Boitard C, Meda P, et al. Changes in microRNA expression contribute to pancreatic beta-cell dysfunction in prediabetic NOD mice. Diabetes 2012; 61:1742–1751. doi: 10.2337/db11-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guay C, Kruit JK, Rome S, Menoud V, Mulder NL, Jurdzinski A, et al. Lymphocyte-derived exosomal microRNAs promote pancreatic beta cell death and may contribute to type 1 diabetes development. Cell Metab 2019; 29:348–361. e6. doi: 10.1016/j.cmet.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 74.Santoro F, Mayer D, Klement RM, Warczok KE, Stukalov A, Barlow DP, et al. Imprinted Igf2r silencing depends on continuous Airn lncRNA expression and is not restricted to a developmental window. Development 2013; 140:1184–1195. doi: 10.1242/dev.088849. [DOI] [PubMed] [Google Scholar]
- 75.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 2009; 106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moran I, Akerman I, van de Bunt M, Xie R, Benazra M, Nammo T, et al. Human beta cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab 2012; 16:435–448. doi: 10.1016/j.cmet.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.You LH, Wang N, Yin DD, Wang LT, Jin FY, Zhu YN, et al. Downregulation of long noncoding RNA Meg3 affects insulin synthesis and secretion in mouse pancreatic beta cells. J Cell Physiol 2016; 231:852–862. doi: 10.1002/jcp.25175. [DOI] [PubMed] [Google Scholar]
- 78.Bradfield JP, Qu H-Q, Wang K, Zhang H, Sleiman PM, Kim CE, et al. A genome-wide meta-analysis of six type 1 diabetes cohorts identifies multiple associated loci. PLoS Genet 2011; 7:e1002293.doi: 10.1371/journal.pgen.1002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Motterle A, Gattesco S, Caille D, Meda P, Regazzi R. Involvement of long non-coding RNAs in beta cell failure at the onset of type 1 diabetes in NOD mice. Diabetologia 2015; 58:1827–1835. doi: 10.1007/s00125-015-3641-5. [DOI] [PubMed] [Google Scholar]
- 80.Iparraguirre L, Munoz-Culla M, Prada-Luengo I, Castillo-Trivino T, Olascoaga J, Otaegui D. Circular RNA profiling reveals that circular RNAs from ANXA2 can be used as new biomarkers for multiple sclerosis. Hum Mol Genet 2017; 26:3564–3572. doi: 10.1093/hmg/ddx243. [DOI] [PubMed] [Google Scholar]
- 81.Li L-J, Huang Q, Pan H-F, Ye D-Q. Circular RNAs and systemic lupus erythematosus. Exp Cell Res 2016; 346:248–254. doi: 10.1016/j.yexcr.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 82.Luo S, Deng M, Xie Z, Li X, Huang G, Zhou Z. Circulating circular RNAs profiles associated with type 1 diabetes. Diabetes Metab Res Rev 2020; e3394.doi: 10.1002/dmrr.3394. [DOI] [PubMed] [Google Scholar]
- 83.Yang L, Han X, Zhang C, Sun C, Huang S, Xiao W, et al. Hsa_circ_0060450 negatively regulates type I interferon-induced inflammation by serving as miR-199a-5p sponge in type 1 diabetes mellitus. Front Immunol 2020; 11:576903.doi: 10.3389/fimmu.2020.576903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang L, Wu H, Zhao M, Lu Q. Identifying the differentially expressed microRNAs in autoimmunity: a systemic review and meta-analysis. Autoimmunity 2020; 53:122–136. doi: 10.1080/08916934.2019.1710135. [DOI] [PubMed] [Google Scholar]
- 85.Assmann TS, Recamonde-Mendoza M, Punales M, Tschiedel B, Canani LH, Crispim D. MicroRNA expression profile in plasma from type 1 diabetic patients: Case-control study and bioinformatic analysis. Diabetes Res Clin Pract 2018; 141:35–46. doi: 10.1016/j.diabres.2018.03.044. [DOI] [PubMed] [Google Scholar]
- 86.Olsen JA, Kenna LA, Spelios MG, Hessner MJ, Akirav EM. Circulating differentially methylated amylin DNA as a biomarker of beta-cell loss in type 1 diabetes. PLoS One 2016; 11:e0152662.doi: 10.1371/journal.pone.0152662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Akirav EM, Lebastchi J, Galvan EM, Henegariu O, Akirav M, Ablamunits V, et al. Detection of beta cell death in diabetes using differentially methylated circulating DNA. Proc Natl Acad Sci U S A 2011; 108:19018–19023. doi: 10.1073/pnas.1111008108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zullo A, Sommese L, Nicoletti G, Donatelli F, Mancini FP, Napoli C. Epigenetics and type 1 diabetes: Mechanisms and translational applications. Transl Res 2017; 185:85–93. doi: 10.1016/j.trsl.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 89.Zheng Q, Xu Y, Liu Y, Zhang B, Li X, Guo F, et al. Induction of Foxp3 demethylation increases regulatory CD4+CD25+ T cells and prevents the occurrence of diabetes in mice. J Mol Med (Berl) 2009; 87:1191–1205. doi: 10.1007/s00109-009-0530-8. [DOI] [PubMed] [Google Scholar]
- 90.Backe MB, Andersson JL, Bacos K, Christensen DP, Hansen JB, Dorosz JJ, et al. Lysine demethylase inhibition protects pancreatic beta cells from apoptosis and improves beta-cell function. Mol Cell Endocrinol 2018; 460:47–56. doi: 10.1016/j.mce.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 91.Vojinovic J, Damjanov N. HDAC inhibition in rheumatoid arthritis and juvenile idiopathic arthritis. Mol Med 2011; 17:397–403. doi: 10.2119/molmed.2011.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Smith KT, Workman JL. Histone deacetylase inhibitors: anticancer compounds. Int J Biochem Cell Biol 2009; 41:21–25. doi: 10.1016/j.biocel.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 93.Jayaraman S, Patel A, Jayaraman A, Patel V, Holterman M, Prabhakar B. Transcriptome analysis of epigenetically modulated genome indicates signature genes in manifestation of type 1 diabetes and its prevention in NOD mice. PLoS One 2013; 8:e55074.doi: 10.1371/journal.pone.0055074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Christensen DP, Gysemans C, Lundh M, Dahllof MS, Noesgaard D, Schmidt SF, et al. Lysine deacetylase inhibition prevents diabetes by chromatin-independent immunoregulation and beta-cell protection. Proc Natl Acad Sci U S A 2014; 111:1055–1059. doi: 10.1073/pnas.1320850111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bagul PK, Dinda AK, Banerjee SK. Effect of resveratrol on sirtuins expression and cardiac complications in diabetes. Biochem Biophys Res Commun 2015; 468:221–227. doi: 10.1016/j.bbrc.2015.10.126. [DOI] [PubMed] [Google Scholar]
- 96.Bell CG, Teschendorff AE, Rakyan VK, Maxwell AP, Beck S, Savage DA. Genome-wide DNA methylation analysis for diabetic nephropathy in type 1 diabetes mellitus. BMC Med Genomics 2010; 3:33.doi: 10.1186/1755-8794-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Carry PM, Vanderlinden LA, Johnson RK, Dong F, Steck AK, Frohnert BI, et al. DNA methylation near the INS gene is associated with INS genetic variation (rs689) and type 1 diabetes in the diabetes autoimmunity study in the young. Pediatr Diabetes 2020; 21:597–605. doi: 10.1111/pedi.12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Olsson AH, Volkov P, Bacos K, Dayeh T, Hall E, Nilsson EA, et al. Genome-wide associations between genetic and epigenetic variation influence mRNA expression and insulin secretion in human pancreatic islets. PLoS Genet 2014; 10:e1004735.doi: 10.1371/journal.pgen.1004735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen SSH, Jenkins AJ, Majewski H. Elevated plasma prostaglandins and acetylated histone in monocytes in type 1 diabetes patients. Diabet Med 2009; 26:182–186. doi: 10.1111/j.1464-5491.2008.02658.x. [DOI] [PubMed] [Google Scholar]
- 100.Salas-Perez F, Codner E, Valencia E, Pizarro C, Carrasco E, Perez-Bravo F. MicroRNAs miR-21a and miR-93 are down regulated in peripheral blood mononuclear cells (PBMCs) from patients with type 1 diabetes. Immunobiology 2013; 218:733–737. doi: 10.1016/j.imbio.2012.08.276. [DOI] [PubMed] [Google Scholar]
- 101.Zheng Y, Wang Z, Tu Y, Shen H, Dai Z, Lin J, et al. miR-101a and miR-30b contribute to inflammatory cytokine-mediated beta-cell dysfunction. Lab Invest 2015; 95:1387–1397. doi: 10.1038/labinvest.2015.112. [DOI] [PubMed] [Google Scholar]
- 102.Erener S, Mojibian M, Fox JK, Denroche HC, Kieffer TJ. Circulating miR-375 as a biomarker of beta-cell death and diabetes in mice. Endocrinology 2013; 154:603–608. doi: 10.1210/en.2012-1744. [DOI] [PubMed] [Google Scholar]
- 103.Nielsen LB, Wang C, Sorensen K, Bang-Berthelsen CH, Hansen L, Andersen ML, et al. Circulating levels of microRNA from children with newly diagnosed type 1 diabetes and healthy controls: Evidence that miR-25 associates to residual beta-cell function and glycaemic control during disease progression. Exp Diabetes Res 2012; 2012:896362.doi: 10.1155/2012/896362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nabih ES, Andrawes NG. The association between circulating levels of miRNA-181a and pancreatic beta cells dysfunction via SMAD7 in type 1 diabetic children and adolescents. J Clin Lab Anal 2016; 30:727–731. doi: 10.1002/jcla.21928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xu K, Bian D, Hao L, Huang F, Xu M, Qin J, et al. MicroRNA-503 contribute to pancreatic beta cell dysfunction by targeting the mTOR pathway in gestational diabetes mellitus. EXCLI J 2017; 16:1177–1187. doi: 10.17179/excli2017-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cao YL, Jia YJ, Xing BH, Shi DD, Dong XJ. Plasma microRNA-16-5p, -17-5p and -20a-5p: Novel diagnostic biomarkers for gestational diabetes mellitus. J Obstet Gynaecol Res 2017; 43:974–981. doi: 10.1111/jog.13317. [DOI] [PubMed] [Google Scholar]
- 107.Melkman-Zehavi T, Oren R, Kredo-Russo S, Shapira T, Mandelbaum AD, Rivkin N, et al. miRNAs control insulin content in pancreatic beta-cells via downregulation of transcriptional repressors. EMBO J 2011; 30:835–845. doi: 10.1038/emboj.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wijesekara N, Zhang L-H, Kang MH, Abraham T, Bhattacharjee A, Warnock GL, et al. miR-33a modulates ABCA1 expression, cholesterol accumulation, and insulin secretion in pancreatic islets. Diabetes 2012; 61:653–658. doi: 10.2337/db11-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sebastiani G, Ventriglia G, Stabilini A, Socci C, Morsiani C, Laurenzi A, et al. Regulatory T-cells from pancreatic lymphnodes of patients with type-1 diabetes express increased levels of microRNA miR-125a-5p that limits CCR2 expression. Sci Rep 2017; 7:6897.doi: 10.1038/s41598-017-07172-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sun C, Xue L, Zhu Z, Zhang F, Yang R, Yuan X, et al. Insights from lncRNAs profiling of MIN6 beta cells undergoing inflammation. Mediators Inflamm 2016; 2016:9275106.doi: 10.1155/2016/9275106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang CJ, Chou FC, Chu CH, Wu JC, Lin SH, Chang DM, et al. Protective role of programmed death 1 ligand 1 (PD-L1) in nonobese diabetic mice: the paradox in transgenic models. Diabetes 2008; 57:1861–1869. doi: 10.2337/db07-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.He X, Ou C, Xiao Y, Han Q, Li H, Zhou S. lncRNAs: Key players and novel insights into diabetes mellitus. Oncotarget 2017; 8:71325–71341. doi: 10.18632/oncotarget.19921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mutskov V, Felsenfeld G. The human insulin gene is part of a large open chromatin domain specific for human islets. Proc Natl Acad Sci U S A 2009; 106:17419–17424. doi: 10.1073/pnas.0909288106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Leti F, DiStefano JK. Long noncoding RNAs as diagnostic and therapeutic targets in type 2 diabetes and related complications. Genes (Basel) 2017; 8:207.doi: 10.3390/genes8080207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Millis MP, Bowen D, Kingsley C, Watanabe RM, Wolford JK. Variants in the plasmacytoma variant translocation gene (PVT1) are associated with end-stage renal disease attributed to type 1 diabetes. Diabetes 2007; 56:3027–3032. doi: 10.2337/db07-0675. [DOI] [PubMed] [Google Scholar]
- 116.Alvarez ML, DiStefano JK. Functional characterization of the plasmacytoma variant translocation 1 gene (PVT1) in diabetic nephropathy. PLoS One 2011; 6:e18671.doi: 10.1371/journal.pone.0018671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Arnes L, Akerman I, Balderes DA, Ferrer J, Sussel L. βlinc1 encodes a long noncoding RNA that regulates islet beta-cell formation and function. Genes Dev 2016; 30:502–507. doi: 10.1101/gad.273821.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Akerman I, Tu Z, Beucher A, Rolando DMY, Sauty-Colace C, Benazra M, et al. Human pancreatic beta cell lncRNAs control cell-specific regulatory networks. Cell Metab 2017; 25:400–411. doi: 10.1016/j.cmet.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yan B, Tao ZF, Li XM, Zhang H, Yao J, Jiang Q. Aberrant expression of long noncoding RNAs in early diabetic retinopathy. Invest Ophthalmol Vis Sci 2014; 55:941–951. doi: 10.1167/iovs.13-13221. [DOI] [PubMed] [Google Scholar]
- 120.Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li YJ, et al. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis 2014; 5:e1506.doi: 10.1038/cddis.2014.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yin DD, Zhang EB, You LH, Wang N, Wang LT, Jin FY, et al. Downregulation of lncRNA TUG1 affects apoptosis and insulin secretion in mouse pancreatic beta cells. Cell Physiol Biochem 2015; 35:1892–1904. doi: 10.1159/000373999. [DOI] [PubMed] [Google Scholar]


