Abstract
Objectives
KRAS, NRAS, and BRAF mutations are commonly present in colorectal cancer (CRC). We estimated the frequency of KRAS, NRAS, and BRAF mutations and assessed their impact on survival and other clinical variables among Saudi patients.
Design
Retrospective cohort study design.
Settings
Oncology department of a tertiary hospital in Riyadh, Saudi Arabia. We gathered information from 2016 to 2018.
Participants
Cohort of 248 CRC patients to assess the demographic data, pathological tumour features, response to treatment modalities, disease progression, and metastasis.
Statistical analysis used
Correlation analysis using the chi-square test. Survival analysis using a Kaplan Meier method. Cox regression analysis to calculate the hazard ratios.
Results
Demographic data revealed that 84% of patients were diagnosed with CRC above the age of 50 years. Only 27% of patients presented with distant metastasis. KRAS mutations were the most prevalent (49.6%), followed by NRAS mutations (2%) and BRAF mutations (0.4%). Wild type tumours were found among 44.4% of patients. KRAS mutation showed no significant correlation with the site, type, pathological grade, and stage of the tumour.
The mean survival time was shorter among patients with KRAS mutations than among patients with wild type KRAS tumours (54.46 vs. 58.02 months). Adjusted analysis showed that the survival time was significantly affected by patients’ age at diagnosis (P = 0.04). Male patients had an increased risk of mortality by 77% (hazard ratio: 1.77).
Conclusions
Saudi CRC patients had a high frequency of KRAS mutations and a low frequency of BRAF mutations. The KRAS mutation status did not affect the patients’ survival.
Introduction
According to the Saudi Cancer Registry, colorectal cancer (CRC) is the most commonly diagnosed cancer among males and the third commonest among females [1]. Despite advancements made in CRC diagnosis and multimodal treatment, it remains the third commonest cause of cancer-related deaths worldwide; accounting for over half a million deaths annually [2,3]. CRC is a heterogeneous disease that arises from multiple cellular and genetic alterations. This can affect diagnosis and treatment. Efforts aimed at identifying molecular phenotypes for CRC are critical to provide more accurate prognoses, predict tumour response to treatment, and guide targeted therapies [2,4].
Advanced characterisation of the genetic alterations in CRC has suggested that the epidermal growth factor receptor (EGFR) and the downstream RAS–RAF–BRAF–MAPK pathways play a significant role in disease development [5]. RAS is a family of genes that encode for guanosine triphosphatases. The first identified RAS genes were HRAS, KRAS, and NRAS. RAS can subsequently activate other proteins that regulate transcription factors involved in cell cycle progression, apoptosis, differentiation, and proliferation [6].
KRAS mutations are found in approximately 30–50% of CRC lesions and comprise activating point mutations mostly in codons 12 and 13 and rarely in codon 61 [7,8]. Since KRAS is an important downstream component of the EGFR pathway, mutations in the coding gene result in a constitutively activated signalling cascade that acts independently from upstream signals. Prior studies of EGFR inhibitors (cetuximab and panitumumab) in metastatic CRC (mCRC) showed controversial results regarding therapeutic outcomes [9,10]. Responses to treatment were observed only in individuals with wild type KRAS tumours. Thus, KRAS mutations are now recognised as predictors of resistance to EGFR inhibitors [6]. However, the effect of KRAS on prognosis is still debatable, despite several investigations. Many studies have advocated that KRAS is a negative prognostic marker, while others have reported no prognostic significance [6,11].
Even among patients with wild type KRAS CRCs, responses to EGFR inhibitors were not observed among all cases, suggesting that other mutations downstream to EGFR might also lead to a lack of response to treatment [5]. NRAS mutations occur in approximately 3–5% of CRCs and are usually found in codon 60 [4]. Since NRAS mutations are rare and mainly investigated for their effect on resistance to therapy, their role as prognostic markers is not conclusively reported [4,12].
RAF is a family of serine/threonine kinases that function as direct downstream targets of RAS and is composed of ARAF, BRAF, and CRAF. BRAF mutations are identified in approximately 5–20% of CRC lesions and are usually located at codon 600 [4]. Mostly, KRAS and BRAF mutations are mutually exclusive molecular events. A recent study conducted in Korea reported that Asian populations (including the Saudi Arabian population) had a lower incidence of BRAF mutations in mCRC [11]. However, present data are still insufficient to support this hypothesis. As with RAS mutations, BRAF mutations result in hyper-activation of the MAPK pathway. Therefore, BRAF mutations can be considered as a source of resistance to EGFR inhibitors [5]. However, current evidence is inadequate to conclusively evaluate their role as predictive markers [4,11].
In the past few years, considerable advances have been made in the elucidation of molecular alterations underlying mCRC. However, no definitive conclusions have been drawn regarding the role of KRAS, BRAF, and NRAS mutations as predictive and prognostic markers [2,11]. The present study aimed to estimate the prevalence of KRAS, NRAS, and BRAF mutations and highlight their prognostic value among Saudi patients. This can assist in offering a more accurate prognosis and personalising therapeutic approaches based on the presence or absence of biomarkers.
Methods
We conducted this retrospective cohort study at the oncology department of a tertiary hospital in Riyadh, Saudi Arabia. Since genetic testing started in 2016, we retrospectively reviewed all medical records of CRC patients who were admitted to the hospital between 2016 and 2018 and underwent surgery or were treated with chemotherapy, radiotherapy, or both (n = 248). There were no exclusion criteria. We used a non-probability consecutive sampling technique for all patients who met the inclusion criteria.
The research group members, using patients’ medical records from the Best Care system, collected the data from September 2018 to January 2019. Patients’ records were then reviewed until the 1st of January 2021 to estimate the survival time. We collected data regarding variables such as demographic characteristics (age and gender) and pathological tumour features (type, primary site, grade of differentiation, and staging). The overall mean survival time was defined as the mean of an interval between the date of diagnosis and death or last follow up.
DNA isolation and analysis of KRAS, BRAF, and NRAS mutations
For DNA isolation, 5–10 μm-thick sections from the patient formalin fixed paraffin-embedded tissue samples were used. Neoplastic cells then were lysed to extract genomic DNA and perform Realtime PCR amplification. Biocartis IdyllaTm KRAS mutations test was done to detect the presence of 21 KRAS mutations in exons 2,3, and 4. Biocartis IdyllaTm NRAS-BRAF mutations test was also performed to detect the presence of 18 mutations in NRAS exons 2,3, and 4 as well as 5 mutations in BRAF exon 15. The tumour mutational status and type of mutation were reported in the molecular testing reports provided by the pathology centre of our hospital.
Patient and public involvement
The study design was approved by King Abdullah’s International Medical Research Centre ethics review board. Patients and public were not involved in any stage of the research process. The need for informed consent was waived on account of the retrospective nature of the study. Confidentiality and anonymity were maintained throughout the study. No names or medical record numbers was taken from patients’ records. Only the research team had access to the data during the study and after completion. All data were retrieved from the Best Care system of King Abdulaziz Medical City in Riyadh from September 2018 to January 2019, and all were anonymized before accessing them. Patients sought treatment at different times between 2016 to 2018 depending on date of diagnosis.
Statistical analyses
We used frequencies and percentages to describe categorical data such as gender and mutation status. We described numerical data as the mean and standard deviation or the median and interquartile range, as appropriate. The frequencies of KRAS, NRAS, and BRAF mutations were estimated at a 95% confidence level. Due to the low number of patients with BRAF and NRAS mutated CRCs, these mutations were excluded from the survival analysis. We used the Chi-Squared test to assess the association between KRAS mutations and other clinical variables. A Kaplan Meier analysis was done to compare the mean survival time between patients with KRAS mutated and wild type CRCs. Hazard Ratios (HR) were calculated by the Cox regression analysis stratified by patients’ gender and age, tumour stage and mutational status. All statistical analyses were performed using SPSS version 22 (IBM Corporation, Armonk, NY, USA). Analysis items with a P-value of <0.05 were considered statistically significant.
Results
The present analysis included 248 CRC patients. Males represented a majority of patients compared to females (62% vs. 38%). The mean age at diagnosis was 63 ± 14 years with 84% of patients diagnosed above the age of 50 years. Approximately 45% of CRC patients had complete or partial response to treatment, and 27% had disease progression or metastasis.
The commonest primary tumour site was the rectum (27%), followed by the right colon (25%). Anorectal tumours were the least common and accounted for 0.4% of cases. Histologically, 99% of CRCs were adenocarcinomas in origin and 83% were moderately differentiated tumours. Advanced tumour stage (T stage) and metastasis stage (M stage) at the initial diagnosis had a negative impact on patients’ prognoses (P<0.001). Stage T3 (50%) and stage N1 (36%) were the most commonly reported stages. A large proportion of patients presented with stage M1 disease (28.2%).
KRAS mutations were most frequently detected among CRC patients (49.6%), followed by NRAS mutations (2%) and BRAF mutations (0.4%). Wild type CRCs were detected among 44.4% of patients (Figs 1 and 2). Compared to wild type KRAS CRCs, KRAS mutated tumours showed a trend toward female patients (56.3% vs. 43.7%), patients with right-sided tumours (66.7% vs. 33.3%), and a moderately differentiated histology (53.6% vs. 46.4%). However, these differences were not statistically significant (P = 0.404; P = 0.147; P = 0.436) (Table 1).
Fig 1. Distribution of mutation frequency of KRAS, NRAS and BRAF genes among 248 CRC patients stratified by gender.
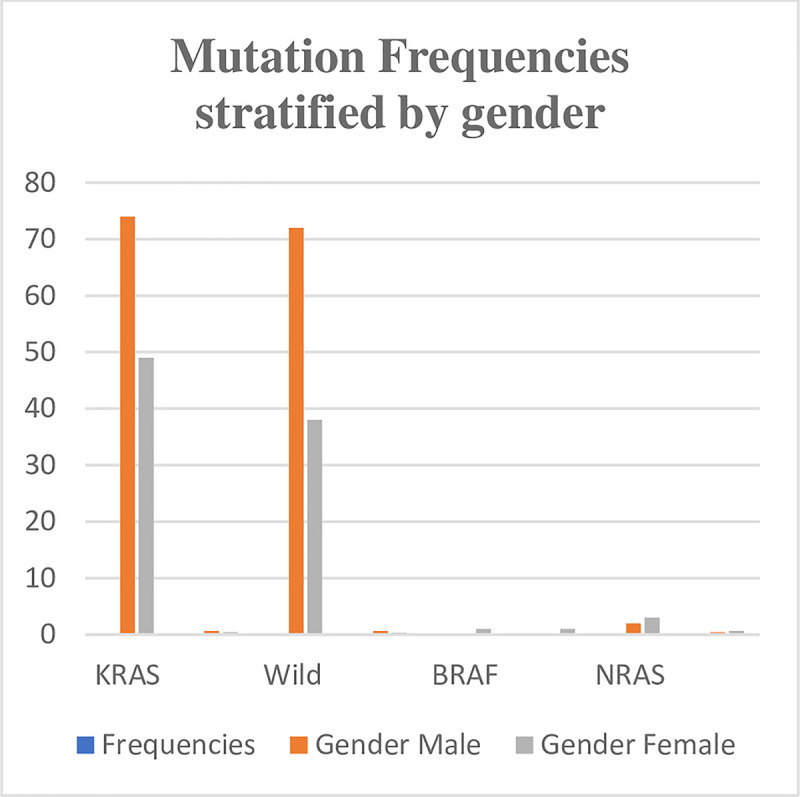
Fig 2. Distribution of mutation frequency of KRAS, NRAS and BRAF genes among 248 CRC patients stratified by primary tumor site.
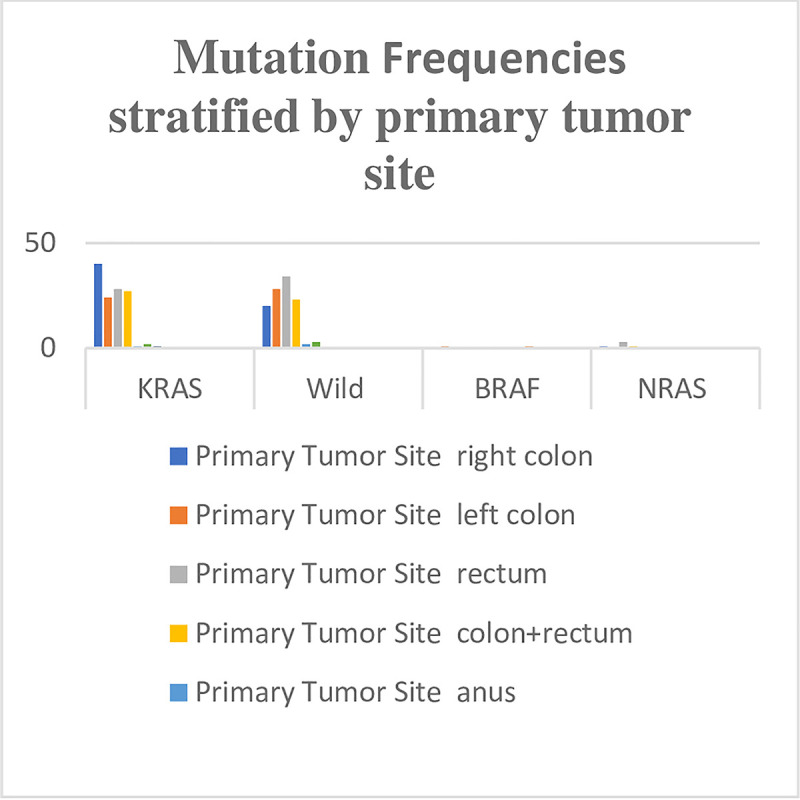
Table 1. Results of Chi-Squared tests of association between mutational status and demographic and clinicopathological characteristics.
| Variable | Category | Mutation status | P-value | |
|---|---|---|---|---|
| KRAS | Wild type | |||
| n (%) | n (%) | |||
| Gender | Male | 74 (50.7%) | 72 (49.3%) | 0.404 |
| Female | 49 (56.3%) | 38 (43.7%) | ||
| Age, years | <50 | 18 (46.2%) | 21 (53.8%) | 0.455 |
| 51–60 | 35 (54.7%) | 29 (45.3%) | ||
| 61–70 | 35 (60.3%) | 23 (39.7%) | ||
| ≥71 | 35 (48.6%) | 37 (51.4%) | ||
| T stage | T1 | 1 (33.3%) | 2 (66.7%) | 0.725 |
| T2 | 14 (45.2%) | 17 (54.8%) | ||
| T3 | 64 (53.3%) | 56 (46.7%) | ||
| T4 | 28 (52.8%) | 25 (47.2%) | ||
| Undocumented | 16 (61.5%) | 10 (38.5%) | ||
| N stage | N0 | 37 (54.4%) | 31 (45.6%) | 0.533 |
| N1 | 46 (52.9%) | 41 (47.1%) | ||
| N2 | 21 (44.7%) | 26 (55.3%) | ||
| X | 1 (33.3%) | 2 (66.7%) | ||
| Undocumented | 18 (64.3%) | 10 (35.7%) | ||
| M stage | M0 | 29 (43.9%) | 37 (56.1%) | 0.092 |
| M1 | 35 (53.8%) | 30 (46.2%) | ||
| MX | 42 (64.6%) | 23 (35.4%) | ||
| Undocumented | 17 (45.9%) | 20 (54.1%) | ||
| Location | Right colon | 40 (66.7%) | 20 (33.3%) | 0.147 |
| Left colon | 24 (46.2%) | 28 (53.8%) | ||
| Rectum | 28 (45.2%) | 34 (54.8%) | ||
| Colon and rectum | 27 (54%) | 23 (46%) | ||
| Anus | 1 (33.3%) | 2 (66.7%) | ||
| Small intestine | 2 (40%) | 3 (60%) | ||
| Anorectal | 1 (100%) | 0 (0%) | ||
| Prognosis | Response to treatment | 53 (49.5%) | 54 (50.5%) | 0.509 |
| No response to treatment | 33 (52.4%) | 30 (47.6%) | ||
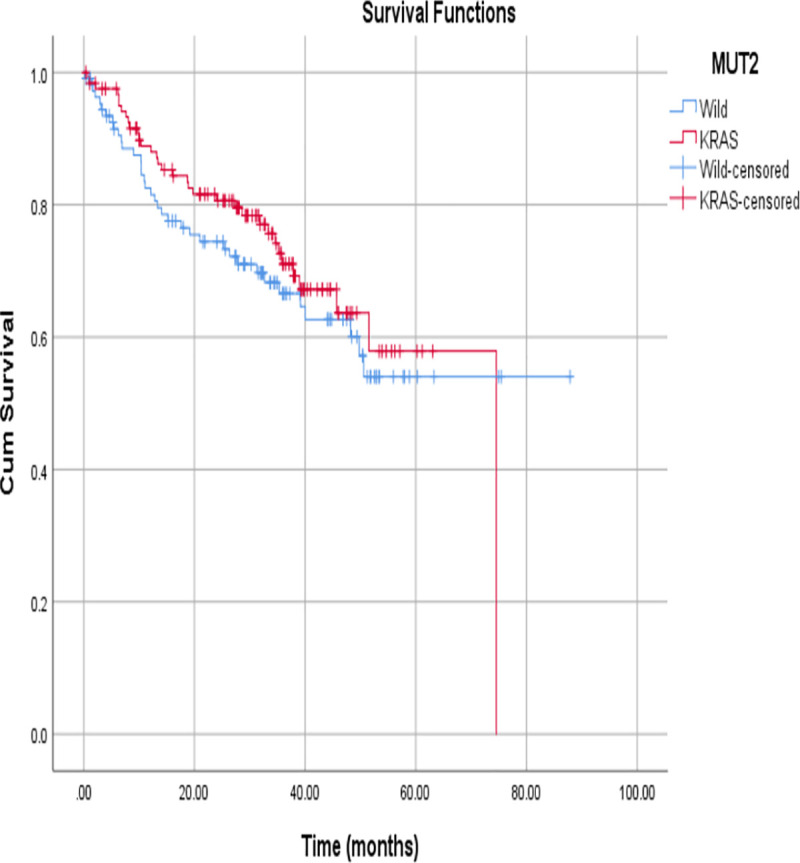
The mortality rate was estimated to be 31.9%. The overall mean survival time was 58.22 months. The mean survival time was shorter among patients with KRAS mutations than among patients with wild type KRAS tumours (54.46 vs. 58.02 months), but the difference was not statistically significant (P = 0.444) (Fig 3). The survival time was better among females compared to males (62 vs. 50.75 months), but this was not statistically significant (P = 0.518).
Fig 3. A Kaplan Meier analysis of KRAS mutated tumours vs wild-type tumours.
Adjusted analysis showed that the survival time was significantly affected by patients’ age at diagnosis (P = 0.04). Patients who are younger than 60 years had a lower risk of mortality. Compared to those aged under 50 years or over 60 years, patients aged between 51–60 years had the lowest risk of mortality. Moreover, presence of metastasis significantly increased the risk of death (hazard ratio: 3.81, P = <0.001). The difference between males and females for the hazard of mortality approached the borderline of significance (P = 0.08). Male patients had an increased risk of mortality by 77% (hazard ratio: 1.77). There was no significant difference between KRAS mutated tumours and wild type KRAS tumours for the hazard of mortality (hazard ratio:1.68, P = 0.09) (Table 2).
Table 2. Results of Cox regression analysis of the hazard ratios stratified patients’ gender and age, tumour stage and mutational status.
| Variable | Category | P-value | HR | 95% CI for HR | |
|---|---|---|---|---|---|
| Lower | Upper | ||||
| Gender | Male | 0.081 | 1.77 | 0.93 | 3.36 |
| Female* | 1.00 | ||||
| Age | = < 50 | 0.042 | 0.41 | 0.18 | 0.97 |
| 51–60 | 0.002 | 0.26 | 0.11 | 0.61 | |
| 61–70 | 0.091 | 0.54 | 0.26 | 1.10 | |
| = 71+ * | 1.00 | ||||
| Stage T | T2 | 0.851 | 1.09 | 0.44 | 2.70 |
| T3 | 0.238 | 0.68 | 0.35 | 1.30 | |
| T4* | 1.00 | ||||
| Stage N | N0 | 0.839 | 0.91 | 0.369 | 2.248 |
| N1 | 0.417 | 0.737 | 0.352 | 1.541 | |
| N2* | 1.00 | ||||
| Stage M | M0 | 0.256 | 0.61 | 0.26 | 1.44 |
| M1 | 0 | 3.81 | 1.83 | 7.93 | |
| Mx* | 1.00 | ||||
| Mutation | Wild | 0.099 | 1.68 | 0.91 | 3.11 |
| KRAS* | 1.00 | ||||
Discussion
KRAS mutations were found in 49% of CRCs. This proportion was higher than those reported in western countries and the Asian region (35–40%) [13,14]. However, it is in accordance with the mutation rates reported in Iraq, Spain, and Slovenia (46.2–48.0%) [15,16]. On the contrary, BRAF mutations were observed in only 0.4% of CRCs. To the best of our knowledge, this is the lowest proportion reported in the literature. The mutation rates reported in western countries ranged from 10–15% and are estimated to be 4% in the Asian region. NRAS mutations were evident in 2% of CRCs, which is similar to global estimates (3–5%) [13,14].
In this study, the age of the patient did not significantly affect the mutation status. Instead, CRCs among female patients were likelier to harbour KRAS mutations. Approximately 56% of female and 50% of male patients had KRAS mutations. This finding corresponds to the results of other studies conducted in the US, Indonesia, and China [14,17,18]. The reasons behind this phenomenon are not yet known. We found no significant difference in the tumour stage at diagnosis between mutant and wild type CRCs. Moreover, the type of tumour and grade of differentiation was unaffected by the mutation status. Both findings are in accordance with those of similar studies [14,19]. Right colon tumours were likelier to harbour KRAS mutations than left colon tumours; similar findings were observed in previous studies [18,19]. This could explain the relationship between right colon tumours and poor prognosis.
The survival times among patients with KRAS mutant and wild type CRCs were estimated to be 54 months and 58 months, respectively. Although this difference is not statistically significant, it is clinically significant.
The survival rate among CRC patients was estimated to be 68%. According to the Saudi Cancer Registry, the 5-year survival rate among CRC patients from 1994 to 1999 was 44% [20]. We believe that this significant difference is linked to an improvement in diagnostic and treatment modalities. Moreover, our data were extracted from a tertiary hospital with better patient care and multidisciplinary teams. By contrast, the Saudi Cancer Registry included all CRC patients from different cities in multiple facilities.
CRC was more prevalent among male patients (61.7%) than among female patients (38.3%), consistent with the results of other studies conducted worldwide [2,3]. According to a study published in the Annals of Saudi Medicine in 2015, Saudi males had higher rates of CRC than did males in both developed and developing countries, while females had similar rates to those detected in developed countries [21]. This disparity between the sexes is not well understood; however, it could be linked to increased exposure to risk factors such as smoking and red meat consumption [22].
The median age at diagnosis of colon cancer in the United States (US) is 68 years among males and 72 years among females [20]. In the present study, the median ages for the development of CRC were 63 years and 61 years among males and females, respectively. The younger median age at which Saudi patients are diagnosed compared to patients in the US is possibly linked to a younger Saudi population. It is estimated that 71.1% of the Saudi population is younger than 45 years [20]. This observation highlights the importance of screening Saudi patients earlier than at 50 years, which is considered the standard age for CRC screening in the US.
In the present study, the proportion of Saudi patients presenting with distant metastases was approximately 27%, which corresponds to other results reported locally. According to the Saudi Cancer Registry, the percentage of patient presented with distant metastasis was 29.2% and 28.4% in 2008 and in 2010 respectively [20]. In the US, about 20% of patients have tumour metastasis. Moreover, the proportion of patients presenting with localised CRCs in the present study was approximately 15%, compared to 39% in the US [17]. This indicates that proportionally more Saudi patients present at an advanced stage. This may be a result of CRC screening programmes in the US, which are lacking in Saudi Arabia.
Limitations
The genetic testing started three years ago in our hospital, which affected the total number of patients with known mutation status. Instead of 447 patients, which was the recommended sample size, only 248 patients were included in this study. Moreover, this study was conducted in a single tertiary hospital. Such limitations might affect both the overall distribution of variables, and the identified associations as well as the generalizability of the study results. Since only one patient had BRAF mutation, and five patients had NRAS mutation, this study could not assess the impact of BRAF and NRAS mutations on patients’ survival.
Conclusions
Saudi patients had a higher frequency of KRAS mutations and a lower frequency of BRAF mutations when compared to Asian and western countries. KRAS mutation status did not impact CRC patients’ survival. Further local studies with a larger number of patients are necessary to determine the frequency of KRAS, NRAS and BRAF mutations and to truly understand their impact on prognosis and survival.
Acknowledgments
We would like to thank Dr.Saeed Alturki for his kind help and support, and the research unit in King Saud bin Abdulaziz University for Health Sciences for their continuous support.
Data Availability
• As per KAIMRC ethics committee policy, the data of this study are not approved to be released publicly, and the data supporting the findings of this study will be available upon reasonable request. Data requests can be sent to the corresponding author or King Abdullah International Medical Research Center, mailing address P.O. Box 3660, Riyadh 11481, Mail Code 1515 (KAIMRC), Tel:(+966) 4294444, Email: KAIMRC@NGHA.MED.SA.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Cancer Registry reports[internet]. Riyadh: Saudi Cancer Registry; 2017Sep [cited 2018 Feb22]. Available from: https://www.nhic.gov.sa/eServices/Documents/2014.pdf.
- 2.Kocarnik JM, Shiovitz S, Phipps AI. Molecular phenotypes of colorectal cancer and potential clinical applications. Gastroenterol Rep. 2015. November;3(4):269–76. 10.1093/gastro/gov046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer [Internet]. Geneva: World Health Organization; 2018 Feb [Cited 2018 Feb24]. Available from: http://www.who.int/mediacentre/factsheets/fs297/en/.
- 4.4.Lech G, Słotwiński R, Słodkowski M, et al. Colorectal cancer tumour markers and biomarkers: Recent therapeutic advances. World J Gastroenterol. 2016. February 7;22(5):1745–55. 10.3748/wjg.v22.i5.1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao B, Wang L, Qiu H, Zhang M, Sun L, Peng P et al. Mechanisms of resistance to anti-EGFR therapy in colorectal cancer. Oncotarget. 2016;8(3):3980–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prenen H, Tejpar S, Van Cutsem E. New strategies for treatment of KRAS mutant metastatic colorectal cancer. Clin Cancer Res. 2010. June 1;16(11):2921–6. 10.1158/1078-0432.CCR-09-2029 [DOI] [PubMed] [Google Scholar]
- 7.De Roock W, Piessevaux H, De Schutter J, Janssens M, De Hertogh G, Personeni N et al. KRAS wild-type state predicts survival and is associated to early radiological response in metastatic colorectal cancer treated with cetuximab. Ann Oncol. 2008;19(3):508–515. 10.1093/annonc/mdm496 [DOI] [PubMed] [Google Scholar]
- 8.Vogelstein B, Fearon E, Hamilton S, Kern S, Preisinger A, Leppert M et al. Genetic Alterations during Colorectal-Tumor Development. Engl J Med. 1988;319(9):525–532. 10.1056/NEJM198809013190901 [DOI] [PubMed] [Google Scholar]
- 9.Tanaka M, Omura K, Watanabe Y, Oda Y, Nakanishi I. Prognostic factors of colorectal cancer: K-ras mutation, overexpression of the p53 protein, and cell proliferative activity. J Surg Oncol. 1994;57(1):57. 10.1002/jso.2930570115 [DOI] [PubMed] [Google Scholar]
- 10.Dix B, Robbins P, Soong R, Jenner D, House A, Iacopetta B. The common molecular genetic alterations in Dukes’ B and C colorectal carcinomas are not short-term prognostic indicators of survival. IJC. 1994;59(6):747–751. [DOI] [PubMed] [Google Scholar]
- 11.Won DD, Lee JI, Lee IK, et al. The prognostic significance of KRAS and BRAF mutation status in Korean colorectal cancer patients. BMC Cancer. 2017. June 5;17(1):403. 10.1186/s12885-017-3381-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irahara N, Baba Y, Nosho K, et al. NRAS mutations are rare in colorectal cancer. Diagn Mol pathol. 2010. September;19(3):157–63. 10.1097/PDM.0b013e3181c93fd1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010. August;11(8):753–62. 10.1016/S1470-2045(10)70130-3 [DOI] [PubMed] [Google Scholar]
- 14.Guo F, Gong H, Zhao H, et al. Mutation status and prognostic values of KRAS, NRAS, BRAF and PIK3CA in 353 Chinese colorectal cancer patients. Sci Rep.2018. April 17;8:6076. 10.1038/s41598-018-24306-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bisht S, Ahmad F, Sawaimoon S, et al. Molecular spectrum of KRAS, BRAF, and PIK3CA gene mutation: determination of frequency, distribution pattern in Indian colorectal carcinoma. Med Oncol. 2014. September;31(9):124. 10.1007/s12032-014-0124-3 [DOI] [PubMed] [Google Scholar]
- 16.Malhotra P, Anwar M, Nanda N, et al. Alterations in K-ras, APC and p53-multiple genetic pathway in colorectal cancer among Indians. Tumour Biol. 2013. June;34(3):1901–11. 10.1007/s13277-013-0734-y [DOI] [PubMed] [Google Scholar]
- 17.Cancer of the colon and Rectum–Cancer Stat Facts [Internet]. National Cancer Institute, Surveillance, Epidemiology and End-Results Program; 2020. [cited 20 Jan 2020]. Available from: https://seer.cancer.gov/statfacts/html/colorect.html. [Google Scholar]
- 18.Jauhri M, Bhatnagar A, Gupta S, et al. Prevalence and coexistence of KRAS, BRAF, PIK3CA, NRAS, TP53, and APC mutations in Indian colorectal cancer patients: Next-generation sequencing-based cohort study. Tumour Biol. 2017. February;39(2):1010428317692265. 10.1177/1010428317692265 [DOI] [PubMed] [Google Scholar]
- 19.Xie M, Li J, Cai Z, et al. Impact of primary colorectal Cancer location on the KRAS status and its prognostic value. BMC Gastroenterol. 2019. March 27;19:46. 10.1186/s12876-019-0965-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alsanea N, Abduljabbar A, Alhomoud S, et al. Colorectal cancer in Saudi Arabia: incidence, survival, demographics and implications for national policies. Ann Saudi Med. 2015. June;35(3):196–202. 10.5144/0256-4947.2015.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bazarbashi S, Al Eid H, Minguet J. Cancer Incidence in Saudi Arabia: 2012 Data from the Saudi Cancer Registry. Asian Pac J Cancer Prev. 2017; 18(9): 2437–2444. 10.22034/APJCP.2017.18.9.2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy G, Devesa SS, Cross AJ, et al. Sex disparities in colorectal cancer incidence by anatomic subsite, race and age. Int J Cancer. 2011. April 1;128(7):1668–75. 10.1002/ijc.25481 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
• As per KAIMRC ethics committee policy, the data of this study are not approved to be released publicly, and the data supporting the findings of this study will be available upon reasonable request. Data requests can be sent to the corresponding author or King Abdullah International Medical Research Center, mailing address P.O. Box 3660, Riyadh 11481, Mail Code 1515 (KAIMRC), Tel:(+966) 4294444, Email: KAIMRC@NGHA.MED.SA.


