Abstract
The streptavidin-based enrichment of biotin-tagged molecules is a common methodology that is routinely used across multiple disciplines in biomedical research. Numerous and varied formats of immobilized streptavidin and related proteins are available, but predicting which product is most apt for a given application is complicated by the fact that there are numerous technical considerations and no universal reporting standards for describing the binding capacity of the beads. Here, we define criteria that should be considered when performing a fit-for-purpose evaluation of streptavidin beads. We also describe a colorimetric competitive displacement assay, the streptAVIdin binDing capacITY (AVIDITY) assay, a fast, easy, and inexpensive absorbance-based method to measure the binding capacity of streptavidin beads, which can be used to compare different products and evaluate variation among many of the same product. We expect that the fit-for-purpose criteria and the AVIDITY assay will benefit users across disciplines to make informed decisions regarding the most apt streptavidin bead products for their own experiments.
Keywords: binding capacity, streptavidin beads, proteomics, enrichment
Graphical Abstract

INTRODUCTION
The streptavidin-based enrichment of biotin-tagged molecules is a common methodology used in genomics, transcriptomics, and proteomics. In proteomics, streptavidin-based methods for the enrichment of specific proteins or classes of proteins include, but are not limited to, those used for the identification of protein–protein interactions (e.g., BioID and APEX1,2), newly synthesized proteins (e.g., BONCAT, PUNCH-P, and HILAQ3-5), post-translational modifications (e.g., farnesylation, O-GlcNAc, and nitrotyrosine6-8), and cell surface proteins (e.g., biotinylation of extracellular lysines or oligosaccharides and ligand receptor capture9-11). Given the popularity of streptavidin-based enrichment methods, there are currently a variety of commercially available products featuring streptavidin immobilized to beads. The form of streptavidin, the type of substrate it is bound to, and the size of the substrate differ among products. The protein may be avidin, streptavidin, or neutravidin, and the substrates include magnetic, paramagnetic, and superparamagnetic particles, sepharose, acrylamide, and agarose beads of various sizes (1–80 μm). Unfortunately, there are no universal reporting standards for describing the binding capacity of the beads (Table 1). Rather, product descriptions frequently do not specify the method used to determine the binding capacity and may reference the binding capacity for free biotin (nmol/mL, pmol/mg), biotinylated bovine serum albumin (reported as mg of biotinylated BSA per mL of resin), biotinylated peptide (nmol/mL, pmol/mg), biotinylated oligos (pmol/mL, pmol/mg), biotinylated antibody (mg/mL, μg/mg), or human biotinylated IgG (reported as μg or mg of human IgG per mg or mL of beads), which makes it challenging to accurately compare expectations among products. Binding capacities are commonly reported as a range or “greater than” a specified value instead of a discrete measurement that informs the binding capacity of a specific product lot. Further complicating interpretation of the binding capacity is that the concentration of beads within the transport solution may be undefined or described as the percent slurry, percent magnetite composition, or a defined unit (weight or number of beads) per unit volume. Finally, binding capacities reported using identical units cannot necessarily be directly compared among vendors because the size and substrate of the beads vary among products, adding an additional layer of complexity. Altogether, these variables, coupled to unknown or unreported binding capacities, pose challenges to obtaining and reporting results that are reproducible among experiments and laboratories.
Table 1.
Product Specifications for Various Streptavidin Bead Products Used in Proteomics Workflowsa
| vendor | description | typeb | catalog number | bead size (μm) |
content | binding capacity | method used to determine binding capacity |
|---|---|---|---|---|---|---|---|
| Cytiva (formerly GE Healthcare) | Sera-Mag streptavidin-coated magnetic beads | SPM | 30152105011150 | 1 | 40% magnetite content | 4500–5500 pmol per mg of particle | fluorescein-biotin assay |
| Cytiva (formerly GE Healthcare) | Sera-Mag SpeedBeads neutravidin-coated magnetic beads | SPM | 78152104011150 | 1 | not stated in datasheet | 3500–4500 pmol per mg of particle | fluorescein-biotin assay |
| GenScript | streptavidin MagBeads | SPM | L00424 | 40 | 25% slurry | typical binding capacity for 1 mL of settled beads free biotin: >60 nmol biotinylated peptide: ~10 nmol biotinylated antibody: ~1 mg biotinylated oligonucleotides: 50 nmol |
not stated in datasheet |
| Invitrogen | M-280 streptavidin | SPM | 11205D | 2.8 | 10 mg ((6 to 7) × 108) beads/mL | free biotin: 650–900 pmol/mg biotinylated peptide: ~200 pmol/mg biotinylated antibody: ~10 μg/mg biotinylated oligonucleotides: ~200 pmol/mg double-stranded DNA: ~10 μg/mg |
not stated in datasheet |
| Invitrogen | M-270 streptavidin | SPM | 65305 | 2.8 | 10 mg ((6 to 7) × 108) beads/mL | free biotin: 650–900 pmol/mg biotinylated peptide: ~200 pmol/mg biotinylated antibody: ~10 μg/mg biotinylated oligonucleotides: ~200 pmol/mg double-stranded DNA: ~10 μg/mg |
not stated in datasheet |
| Invitrogen | MyOne streptavidin C1 | SPM | 65001 | 1 | 10 mg ((7–10) × 109) beads/mL | free biotin: >2500 pmol/mg biotinylated peptide: ~400 pmol/mg biotinylated antibody: ~20 μg/mg biotinylated oligonucleotides: ~500 pmol/mg double-stranded DNA: ~20 μg/mg |
not stated in datasheet |
| Invitrogen | MyOne streptavidin T1 | SPM | 65601 | 1 | 10 mg ((7–10) × 109) beads/mL | free biotin: 1100–1700 pmol/mg (datasheet), 950–1500 pmoles free biotin (Web site) biotinylated peptide: ~400 pmol/mg biotinylated antibody: ~20 μg/mg biotinylated oligonucleotides: ~400 pmol/mg double-stranded DNA: ~20 μg/mg |
not stated in datasheet |
| Millipore Sigma | PureProteome streptavidin magnetic bead system | SPM | LSKMAGT02 | 10 | not stated in datasheet | >20 μg/mg settled beads (protein) | not stated in datasheet |
| New England Biolabs | streptavidin magnetic beads | M* | S1420S | 1 | not stated in datasheet | free biotin: >1000 pmol/mg biotinylated antibody: ≥30 μg/mg single-stranded biotinylated oligonucleotide: >500 pmol/mg (25 mer) |
not stated in datasheet |
| Pierce | streptavidin Plus UltraLink Resin | A | 53116 | 50–80 | 50% slurry | ≥4 mg of biotinylated BSA/mL resin | not stated in datasheet |
| ReSyn | MagReSyn streptavidin MAX | M* | MR-STM005 | 5–10 | not stated in datasheet | datasheet biotinylated IgG: ≥500 μg/mg biotinylated oligonucleotides: ≥12 000 pmol/mg (24 mer) web site biotinylated IgG: >5 mg/mL biotinylated oligonucleotides: >120 000 pmol/mL (25 mer) |
not stated in datasheet |
| Vector Laboratories | MagnaLINK streptavidin magnetic beads | SPM | M-1003–010 | 2.8 | 60% magnetite content | free biotin: ≥10 nmol/mg biotinylated IgG: ≥0.8 nmol/mg biotinylated oligonucleotides: ≥0.75 nmol/mg |
fluorescein-biotin assay |
This is not a comprehensive list of all available products.
M = magnetic, SPM = superparamagnetic, A = agarose.
Unclear if magnetic or superparamagnetic.
While attempting to select a streptavidin bead product to use in a miniaturized protocol for cell surface proteomics, we encountered considerable variation in our mass spectrometry results that we eventually attributed to intralot variation in streptavidin bead binding capacity. Specifically, we found variations in the total number of proteins identified, including those specifically bound to beads. Whereas variation among products from different vendors is to be expected, the variation observed among different lots of the same product is a major concern. Our observations regarding intralot variation in bead performance are consistent with a recent study by St-Germain et al., which reported variations between two lots of a GE Healthcare product.12 In their study, levels of streptavidin shedding were found to inversely correlate with the quality of proteomics data, and they described a mass-spectrometry-based assay and an SDS-PAGE assay to evaluate the levels of streptavidin shedding to inform the selection of a higher quality product.12 Here, we echo the concerns raised in the study by St-Germain regarding the variation in bead performance among lots, but we did not find the same relationship between streptavidin shedding and performance. Rather, we found that performance is predicted based on the binding capacity for streptavidin, and here we describe a colorimetric competitive displacement assay, the streptAVIdin binDing capacITY (AVIDITY) assay, for assessing the binding capacity of streptavidin beads to ensure reproducibility and consistency of downstream applications. The AVIDITY assay is fast, inexpensive, and easy to perform, and the readout is based on an absorbance measurement that can be obtained using a standard spectrophotometer or plate reader. We expect that any laboratory will be able to perform this assay and that users across disciplines will find it helpful when evaluating the binding capacity of streptavidin beads to use in their own experiments. To promote its use, we provide a detailed standard operating procedure for the AVIDITY assay. We have found this protocol to be especially useful when negotiating with vendors to ensure that the products we purchase meet our own minimum standard for binding capacity.
MATERIALS AND METHODS
Unless otherwise noted, all chemicals were obtained from Sigma-Aldrich, St. Louis, MO. All experiments conducted for binding capacity measurements, including buffer compatibility tests and titration curves, were performed with manual pipetting and using a Neodymium magnetic rack of strength ≥ N38.
Categorization of Streptavidin Beads
Streptavidin beads were categorized according to (1) the rate at which they form a tight pellet when placed near a magnet, (2) whether cell pellets smear when aspirating supernatant, and (3) how easily bead pellets can be resuspended in binding or elution buffer, as this defines how the beads should be handled (Table 2).
Table 2.
Categorization of Streptavidin Bead Products Used in This Study Based on Their Behavior in Binding Buffer
| Group A | Group B | Group C | |
|---|---|---|---|
| Streptavidin beads | •GenScript streptavidin MagBeads •ReSyn MagReSyn streptavidin MAX beads |
•Cytiva Sera-Mag SpeedBeads neutravidin-coated magnetic beads | •New England Biolabs streptavidin magnetic beads •Invitrogen Dynabeads MyOne streptavidin C1 •Cytiva Sera-Mag streptavidin-coated magnetic beads |
| Pellet behavior on magnetic rack | Pellet fast | Slow, allow beads to settle for 30 s when placed on a magnetic rack | Slow, allow beads to settle for 30 s when placed on a magnetic rack |
| Pellet behavior during aspiration | Do not smear | Some smearing when supernatant is aspirated. Be cautious when aspirating supernatant | More smearing when supernatant is aspirated. Be cautious when aspirating supernatant |
| Resuspension behavior | Resuspend immediately | Triturate three to five times to fully resuspend pellet | Triturate five to ten times to fully resuspend pellet |
Development of the HABA and Biotin Titration Curves for the AVIDITY Assay
To determine the amount of 4-hydroxyazobenzene-2-carboxylic acid (HABA) and biotin to be used in the AVIDITY assay, 100 μL of GenScript streptavidin MagBeads (catalog no. L00424, lot C44261906), Cytiva Sera-Mag SpeedBeads neutravidin-coated magnetic beads (catalog no. 78152104011150; lot 17015347), and New England Biolabs (NEB) streptavidin magnetic beads (catalog no. S1420S; lot 10051761) were equilibrated three times with binding buffer (20 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH 7.5). A 10 mM solution of HABA (CAS number 1634-82-8, Thermo Scientific) was prepared in purified water (18 MΩ) with the addition of 0.2 mL of 1 N NaOH to completely dissolve HABA. A HABA titration curve was generated by successively adding 5–100 nmol HABA in binding buffer to streptavidin beads. After each successive addition of HABA, the absorbance was immediately measured at 350 nm (maximum absorbance for free HABA) using a Varioskan LUX multimode microplate reader with SkanIt 5.0 software (Thermo Fisher Scientific). A 4 mM solution of d-biotin was prepared in binding buffer. The maximum displacement level of d-biotin was determined by equilibrating GenScript streptavidin MagBeads (catalog no. L00424; lots C44261906 and C44242003), SpeedBeads neutravidin-coated magnetic beads (catalog no. 78152104011150; lot 17015347), Cytiva Sera-Mag streptavidin-coated magnetic beads (catalog no.30152105011150; lot 17013370), and Invitrogen Dynabeads MyOne streptavidin C1 (catalog no. 65001; lot 00866797) with binding buffer following the addition of 25 nmol HABA and cumulatively titrating 10–120 nmol d-biotin. Prior to and after the incubation with d-biotin, the absorbance was immediately measured at 350 nm. All measurements were performed in triplicate and graphed as the mean with standard deviation (SD) error bars (biotin titration) or with separate replicates (HABA titration). Graphs were generated using GraphPad Prism version 8.3.0 (GraphPad Software, San Diego, CA).
Application of AVIDITY Assay to Assess Biotin Binding Capacity among Bead Lots and Products
To assess the binding capacity of GenScript streptavidin MagBeads (catalog no. L00424; lots C44261906 and C44242003), SpeedBeads neutravidin-coated magnetic beads (catalog no. 78152104011150; lot 17015347), Cytiva Sera-Mag streptavidin-coated magnetic beads (catalog no. 30152105011150; lot 17013370), and Invitrogen Dynabeads MyOne streptavidin C1 (catalog no. 65001; lot 00945048), 100 μL of beads was equilibrated three times with binding buffer. Beads were incubated with 25 nmol HABA following the addition of 100 nmol d-biotin for 5 min. Prior to and after the incubation with d-biotin, the absorbance was measured at 350 nm.
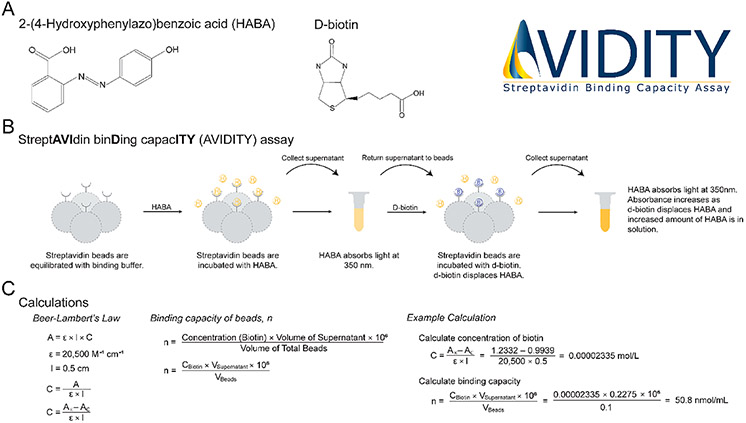
HABA is stoichiometrically displaced by d-biotin;13 therefore, the following formula can be used to determine the concentration of bound d-biotin, C (mol/L), , where AE is the experimental absorbance of free HABA measured after the incubation with d-biotin, AC is the control absorbance of free HABA before the incubation with d-biotin, ∈ is the molar extinction coefficient of HABA (ε = 20 500 M−1 cm−1),13 and l is the path length of the well. Knowing the concentration of bound d-biotin, the binding capacity, n, can be determined with the following formula, , where CBiotin is the concentration of d-biotin in mol/L, VSupernatant is volume of the supernatant in mL, VBeads is the volume of total beads (i.e., beads in the storage solution as pipetted from the storage container) in milliliters, and 106 is the conversion factor to obtain units of nmol/mL. Whereas the binding capacity can be reported as nmol/volume of “settled beads”, this requires the user to know the percent slurry, which is not always provided by the manufacturer (Table 1). Therefore, we report units of nmol/mL of total bead volume, as this only requires the user to record the volume of beads (i.e., beads in the storage solution as pipetted from the storage container) used for the assay. All measurements for binding capacities were performed in triplicate and graphed as the mean with SD error bars using GraphPad Prism.
Cell Culture
RPMI 1788 (human B lymphocytes (ATCC, CCL-156)) cells were cultured in Iscove’s modified Dulbecco’s medium (IMDM) with 100 U/mL penicillin, 100 μg/mL streptomycin, and 10% fetal bovine serum (FBS) and maintained in a humidifying atmosphere at 5% CO2 at 37 °C.
Cell Surface Capture
Cell surface capture (CSC), a chemoproteomic strategy for the identification of cell surface N-glycoproteins,9 was performed as we previously described in detail.14-17 To evaluate the correlation between the variation in the binding capacity and the proteomics results, we performed CSC on RPMI 1788 cells, starting with 1000 μg of peptide pre-enrichment, and used 100 μL of total bead volume each of the four different lots of GenScript streptavidin MagBeads catalog no. L00424 (lots C44251904, C44241809, C44261906, C44242003), Cytiva Sera-Mag SpeedBeads neutravidin-coated magnetic beads (catalog no. 78152104011150; lot 17015347), Cytiva Sera-Mag streptavidin-coated magnetic beads (catalog no. 30152105011150; lot 17013370), and Invitrogen Dynabeads MyOne streptavidin C1 (catalog no. 65001; lot 00945048) for the enrichment of N-glycopeptides, whereas all other steps were performed identically. Peptides were analyzed using an Orbitrap Fusion Lumos or Orbitrap Exploris 480 instrument (Thermo Fisher Scientific), and data were processed with ProteomeDiscoverer 2.4 (Thermo Fisher Scientific), implementing the Sequest18 and MSFragger19 search algorithms followed by Percolator20 for postsearch validation, as previously described.14-17 All methodological details for the MS analysis are described in the Supporting Information.
RESULTS AND DISCUSSION
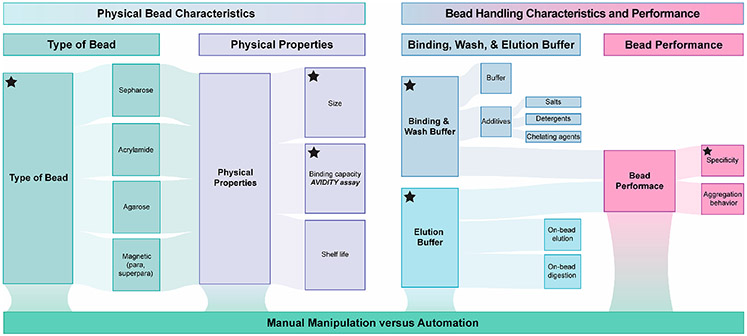
There are numerous technical aspects to consider when implementing streptavidin beads into a sample preparation workflow. Figure 1 summarizes these criteria, which we present as general considerations for evaluating the fit-for-purpose of streptavidin bead products and reporting key parameters to promote repeatability among studies. The general considerations can be divided into three main categories (1) physical bead characteristics, (2) bead handling characteristics and performance, and (3) manual manipulation versus automation.
Figure 1.
General considerations for evaluating the fit-for-purpose of streptavidin bead products and reporting key parameters to promote repeatability among studies. Key features to consider when implementing streptavidin beads into a sample preparation workflow are shown and include physical bead characteristics, bead handling characteristics and performance, and whether manual or automated sample processing is required. Technical details that should be included when reporting data from studies that implement streptavidin-bead-based enrichment are indicated with a black star.
In our experience, the behavior of the beads (e.g., how they clump versus remain dispersed under various conditions, whether a minor population of beads is resistant to pelleting that leads to pellet smearing during the aspiration or adherence of beads to the side of the tube, and how well bead pellets resuspend in binding buffer or elution buffer) can vary among products (Tables 1 and 2). This behavior directly impacts our ability to manipulate them using a liquid handling workstation and affects how completely the beads can be collected using manual pipetting. We find that the bead behavior in the presence of standard binding and wash buffer solutions (e.g., PBS vs Tris-HCl) and elution buffer solutions can vary considerably among products and ultimately impacts the experimental workflow. For example, the degree to which the beads disperse and pellet in the presence of ammonium bicarbonate can be variable among products. This is an important consideration for workflows that include a trypsin, PNGase F, or other enzymatic digestion or elution steps. The volume of beads required for an experiment will affect the scale of the method. For example, when attempting to implement automated liquid handling devices or reduce the scale of the reaction volumes and tube sizes, bead volume may be a limiting factor due to physical constraints. Additionally, consistent with CRAPome studies,21 we have also found varying levels of nonspecific protein binding when comparing streptavidin beads made with different types of substrate despite using identical high-stringency washing conditions. Finally, we have found that bead performance can diminish over time in storage and recommend that users consider this when evaluating products to be used in experimental designs that take place over several months. On the basis of our experience, we strongly advocate that the user performs their own fit-for-purpose assessment to inform their experiments and, importantly, reports key parameters when publishing to promote repeatability among studies (Figure 1).
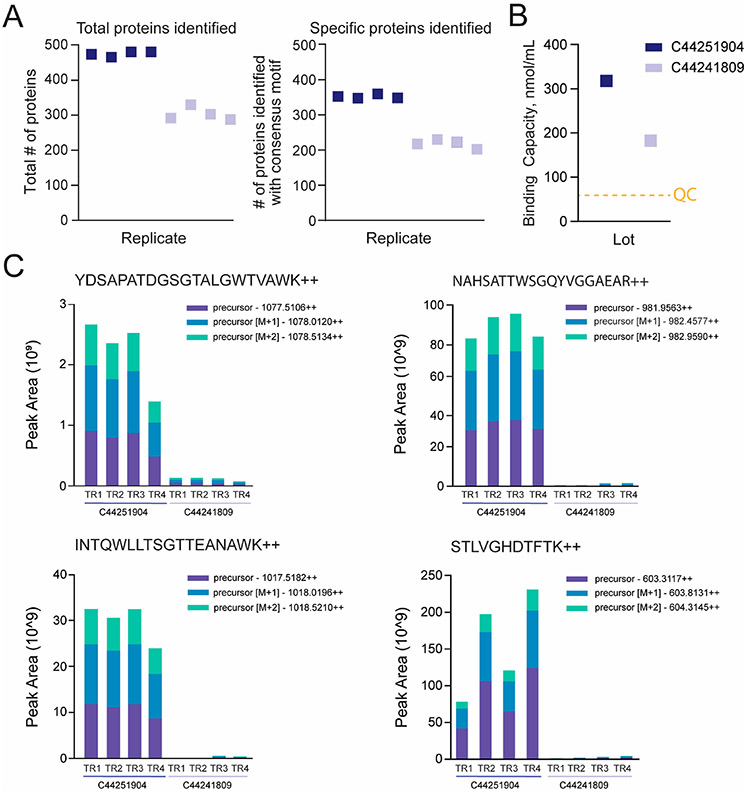
While testing beads for use in a miniaturized cell surface proteomics workflow, we found considerable differences in results between two different lots of GenScript streptavidin MagBeads. Whereas our observations are consistent with the St-Germain et al. study as far as finding differences in bead performance between lots of the same product, our results diverge with respect to the effect on the proteomics results and the underlying bead characteristic that we attribute to this variation. The St-Germain study found that whereas the total numbers of peptide spectrum matches and protein identifications were similar between lots, there was a difference in “bait” peptides and interactors identified. Namely, when a higher level of streptavidin peptides was “shed” from Streptavidin Sepharose High Performance beads (GE Healthcare; catalog no. GE17-5113-01) into the eluent, a lower number of high-confidence interactors (i.e., specific binders) was obtained from the BioID experiment, and data sets would cluster based on the lot used rather than by biological origin. In contrast, our assessment of GenScript beads found that the total number of protein identifications, including those specifically bound to the beads (i.e., cell surface N-glycoproteins containing consensus motif NxS/T/V/c, where x ≠ P) identified in CSC experiments, is not consistent between lots (Figure 2A). Upon consultation with the vendor, we learned that the binding capacities were different between the bead lots (314 versus 177.5 nmol/mL; Figure 2B). Ultimately, the lot with a higher binding capacity yielded higher numbers of total and specific proteins and a higher level of shed streptavidin peptides as compared with the lot with the lower binding capacity (Figure 2C).
Figure 2.
Intralot variation in binding capacity of streptavidin bead results in differences in number of proteins identified in a CSC experiment. (A) Total number of proteins and cell surface N-glycoproteins identified containing consensus motif (NxS/T/V/c, where x ≠ P) in CSC experiments (1000 μg total peptide) when using 100 μL each of two different lots of GenScript streptavidin MagBeads (C44251904 and C44241809). (B) Binding capacity as reported by GenScript. The orange dashed line highlights the minimum binding capacity necessary to pass quality control (60 nmol/mL). (C) Peak area replicate comparison view for precursors M, M+1, and M+2 for streptavidin peptides YDSAPATDGSGTALGWTVAWK, NAHSATTWSGQYVGGAEAR, INTGWLLTSGTTEANAWK, and STLVGHFTFTK (technical replicates = 4) detected in the CSC results from panel A.
The cause for the discrepancy between our observations and the St-Germain study is not yet clear, but it may be due to the fact that the BioID workflow uses streptavidin beads to enrich biotinylated proteins prior to trypsin digestion of the captured protein and streptavidin substrate, whereas CSC enriches at the peptide level (captures biotinylated glycopeptides and elutes deglycosylated peptides using PNGase F). There are also differences in the washing procedure used to remove nonspecific binders from the beads. (CSC uses 2% SDS in ultrapure water, 80 mM sodium phosphate, 2 M NaCl, 0.2% Tween20, 100 mM sodium carbonate, and 50 mM ammonium bicarbonate, whereas BioID uses 50 mM ammonium bicarbonate.)
The results in Figure 2 suggest that the biotin binding capacity could be a useful predictor of bead performance in a proteomics workflow. However, as evidenced in Table 1, there is no universal reporting format for binding capacity among vendors. We therefore sought to implement a binding capacity assay that could be used routinely for intralot comparisons and to directly assess the binding capacity of different products in a way that avoids reliance on vendor product information. We first tested an assay that uses biotin 4-nitrophenyl ester (BNPE) but encountered several challenges when performing this assay. In brief, to avoid the autohydrolysis activity of streptavidin at alkaline pH,22,23 we tested acidic pH but were unsuccessful in obtaining reliable results. Whereas the fluorescein-biotin assay is used by several vendors (Table 1), we decided against this assay because it was reported that beads are incubated with a fluorescein-biotin standard for 60 min (Vector Laboratories user manual version 10.30.2012; MagnaLink streptavidin magnetic beads catalog no. M-1003); therefore, we opted for an assay that was faster and less expensive. We turned to the HABA reaction because it is the basis of several commercially available assays to determine the biotinylation levels of labeled antibodies and other proteins and has been applied in previous studies where the absorbance of the HABA–avidin or HABA–streptavidin complexes was measured at 500 nm.13
The AVIDITY assay described here is based on the competitive displacement of HABA from streptavidin by biotin, resulting in a spectroscopic change due to HABA converting from the bound to the unbound state (Figure 3A,B). HABA is a colorimetric compound that binds avidin and streptavidin at the same binding sites as biotin. HABA is a versatile reagent for biochemical assays because it binds over a wide range of pH and salt concentrations. Because the affinity of biotin for streptavidin is high (dissociation constant, Kd = 10−14 moL/L) compared with HABA, biotin displaces HABA, leading to an increase in the unbound HABA in solution. Subsequently, upon the displacement of HABA, the absorbance of free HABA in solution can be measured at 350 nm (Figure 3B).13 Unlike previously described methods for measuring HABA–avidin or HABA–streptavidin complexes, in the AVIDITY assay developed here, the absorbance of free HABA is measured, which can be used to infer the amount of biotin bound to a given volume of streptavidin beads (Figure 3C).
Figure 3.
Overview of the AVIDITY assay. (A) Molecular structures of key reagents. (B) Overview of the AVIDITY assay workflow. (C) Key calculations used to determine the binding capacity from the AVIDITY assay measurement. AVIDITY logo: Copyright 2020 Rebekah L. Gundry.
In these studies, a HABA titration curve was generated for GenScript streptavidin MagBeads (lot C44261906), Cytiva SpeedBeads neutravidin-coated magnetic beads (lot 17015347), and NEB streptavidin magnetic beads (catalog no. S1420S; lot 10051761). We found the linear range of absorbance for HABA at 350 nm for lot C44261906 to be between 0.2078 ± 0.0182 (5 nmol HABA) and 3.6738 ± 0.0531 (85 nmol) with an R2 value of 0.9994, that for lot 17015347 to be between 0.2242 ± 0.0106 (5 nmol HABA) and 3.7298 ± 0.0653 (70 nmol) with an R2 value of 0.9999, and that for lot 10051761 to be between 0.3589 ± 0.0248 (5 nmol HABA) and 2.8829 ± 0.0120 (50 nmol) with an R2 value of 0.9997 (Figure 4A). The addition of ≥100, 85, and 70 nmol HABA was beyond the limit of detection for GenScript streptavidin MagBeads, Cytiva SpeedBeads neutravidin-coated magnetic beads, and NEB streptavidin magnetic beads, respectively. From this experiment, we determined a baseline absorbance of free HABA between 0.9 and 1.5 to be suitable for all products assessed in this study because this range corresponds to the lower-middle portion of the graph and represents the most accurate and sensitive values when determining the amount of free HABA (Figure 4B). A titration curve of d-biotin allowed us to determine the change in absorbance at maximum displacement of HABA by d-biotin from 100 μL of GenScript streptavidin MagBeads (lots C44261906 and C44242003) and Cytiva Sera-Mag SpeedBeads neutravidin-coated magnetic beads (lot 17015347) (Figure 4C). From these experiments, we determined that 100 nmol d-biotin would be most suitable to perform the assessment of binding capacities, as all beads showed maximum displacement at 100 nmol d-biotin.
Figure 4.
AVIDITY assay is compatible with a range of streptavidin bead products. (A) HABA titration curves for GenScript streptavidin MagBeads (lot C44261906), Cytiva Sera-Mag Magnetic neutravidin-coated particles (lot 17015347), and New England Biolabs streptavidin magnetic beads (lot 10051761). (B) Absorbances of free HABA of five different bead types at 25 nmol of HABA are shown as means with SD error bars (n = 3). An absorbance range of 0.9 to 1.5 was suitable for all beads tested. (C) Biotin titration curves for two different lots of GenScript streptavidin MagBeads (lots C44261906 and C44242003) and one lot of Cytiva Sera-Mag SpeedBeads neutravidin-coated magnetic beads (lot 17015347). Changes in absorbance (Δ absorbance) as determined by subtracting the control absorbance reading (beads incubated with HABA) from the experimental absorbance reading (beads incubated with HABA and d-biotin) are shown as means with SD error bars (n = 3).
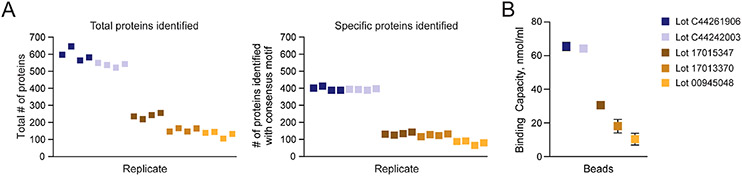
To test the ability of the AVIDITY assay to evaluate differences in binding capacity that are predictive of proteomics results, we compared CSC results from two lots of GenScript streptavidin MagBeads (lots C44261906 and C44242003), Cytiva Sera-Mag SpeedBeads neutravidin-coated magnetic beads (lot 17015347), Cytiva Sera-Mag streptavidin-coated magnetic beads (lot 17013370), and Invitrogen Dynabeads MyOne streptavidin C1 (lot 00945048). The total number of protein identifications, including those specifically bound to the beads (i.e., cell surface N-glycoproteins containing consensus motif NxS/T/V/c, where x ≠ P) in a CSC experiment starting with 1000 μg of total peptide ranged from 105 to 647 and 65 to 412, respectively (Figure 5A). The GenScript beads with the highest binding capacity as measured by the AVIDITY assay yielded the highest number of protein identifications (Figure 5B). Although we were not able to successfully generate a biotin titration curve due to the extensive smearing of the Cytiva Sera-Mag streptavidin-coated magnetic beads (lot 17013370) and Invitrogen Dynabeads MyOne streptavidin C1 (lot 00945048) beads when using the amount of biotin that worked well for GenScript streptavidin MagBeads and Cytiva Sera-Mag SpeedBeads neutravidin-coated magnetic beads, the trend in binding capacities as measured by the AVIDITY assay for the two different streptavidin bead products from Group C is consistent with CSC results. The beads with the lowest measured binding capacity yielded the lowest number of cell surface proteins (Figure 5A).
Figure 5.
Binding capacity of streptavidin beads as measured by the AVIDITY assay is predictive of proteomics results. (A) Total numbers of proteins and cell surface N-glycoproteins identified containing the consensus motif (NxS/T/V/c, where x ≠ P) in CSC experiments (1000 μg of total peptide) when using 100 μL each of two different lots of GenScript streptavidin MagBeads and one lot each for Cytiva Sera-Mag SpeedBeads neutravidin-coated magnetic beads (lot 17015347), Cytiva Sera-Mag streptavidin-coated magnetic beads (lot 17013370), and Invitrogen Dynabeads MyOne streptavidin C1 (lot 00945048) are shown. (B) Binding capacity as determined by the AVIDITY assay using 25 nmol HABA and 100 nmol d-biotin shown as means with SD error bars (n = 3).
Notably, whereas we were able to reduce the amount of bead smearing for Group C beads by including SDS, Tween20, or 2 M NaCl in the binding buffer, these compounds interfere with the absorbance reading. Therefore, alternative binding buffers that improve the bead behavior yet do not interfere with the absorbance measurement would be required to generate titration curves for the AVIDITY assay for Group C beads. Despite the challenges with the Group C beads, these results demonstrate that the AVIDITY assay works well among beads that are different sizes and composed of different substrates and is capable of detecting differences in binding capacities among products that are predictive of proteomics results. To promote its use, a detailed standard operating procedure, including tips for adaptation to beads not used in the current study, is provided in the Supporting Information. Included in this protocol is the recommendation to use a Neodymium magnetic rack of strength ≥ N38 or an alternative magnet of similar strength, as this greatly enhances the “tightness” of the pellet to avoid bead loss during the aspiration of the supernatant.
Altogether, the AVIDITY assay is sensitive enough to detect differences in binding capacities that are predictive of proteomics results, at least for this peptide-centric workflow. Importantly, these data are based on using 100 μL of slurry volume, as that is optimal for our current automated sample handling workflow. Because the size and concentration of the beads within the transport solution vary among products, it is possible that for beads with the lower apparent binding capacity (based on 100 μL of total slurry), using a greater slurry volume would yield results similar to those for 100 μL of the GenScript beads. However, users should consider the cost of using larger amounts of beads and whether the experimental format can support a larger volume.
The AVIDITY assay described here is inexpensive and easy to use. Given the small volumes and concentrations of HABA and d-biotin required to achieve maximum displacement, the format of our AVIDITY assay is particularly well suited for microfuge tube and multiwell plate formats, and it can be applied to a variety of products. Compared with the streptavidin shedding readout proposed by St-Germain et al., the AVIDITY assay provides a discrete value that can be reported in publications to inform other researchers. However, there are several limitations of the current study and alternative approaches to consider. First, we have validated the AVIDITY assay for one neutravidin product (Cytiva Sera-Mag SpeedBeads neutravidin-coated magnetic beads), but the application of the assay for the assessment of avidin products would require further development and validation. Second, whether differences in the binding capacity measured by the AVIDITY assay are equally predictive for proteomic workflows that capture intact proteins is yet to be tested.
Third, we have not directly compared the AVIDITY assay to all alternative strategies. Alternative strategies include BNPE, biotinylated alkaline phosphatase assay, and the fluorescein-biotin assay.24-26 As previously described, we were unsuccessful in obtaining reliable data for the BNPE assay. Whereas the fluorescein-biotin assay is expected to be more sensitive, the sensitivity of the AVIDITY assay is sufficient to detect differences in the binding capacity of beads that are predictive of the performance of our proteomics experiments (Figure 5) and it is less expensive and requires fewer experimental data points than the fluorescein-biotin assay. Furthermore, it is questionable whether the fluorescein-biotin binding assay is predictive for proteomics workflows. The AVIDITY assay showed a higher binding capacity for Cytiva Sera-Mag SpeedBeads neutravidin-coated magnetic beads compared with streptavidin-coated magnetic beads (30.8 versus 18.3 nmol/mL, Figure 5), which is consistent with CSC results. However, Cytivia reported the binding capacity, as measured by the fluorescein-biotin binding assay, for neutravidin beads to be lower than that for streptavidin beads, 3462 versus 4920 pmol/mg, respectively. The reason for this discrepancy is not obvious, and details of how the measurements were performed are not provided by the vendor. Therefore, any underlying reasons why the measurement from the vendor does not predict performance in a proteomics experiment are unclear.
Fourth, although 100 nmol d-biotin used was suitable for all beads tested here, a user may find the solubility of d-biotin to be too low to prepare a stock concentration that achieves the maximum displacement for other beads. In this case, it is possible to use the biotin (long arm) NHS (Vector Laboratories, catalog no. SP-1210) because it can be prepared in higher concentrations than d-biotin. Fifth, the AVIDITY assay, like other binding capacity assays, cannot provide an assessment of binding specificity. It is possible that some beads may be inherently prone to more nonspecific binding than others, which can have important implications for interpreting the results of proteomic workflows. Finally, the AVIDITY assay relies on interactions of a surrogate molecule with streptavidin, which may not perfectly represent the physiochemical properties of the biomolecules captured in an affinity enrichment experiment (e.g., complex mixture of biotinylated proteins or peptides). Therefore, it may not be possible to precisely quantify the true binding capacity of streptavidin beads for the biological molecule of interest. However, because the AVIDITY assay is capable of detecting differences in the binding capacity that correlate to differences in the proteomics results, it should be useful for the routine comparison of streptavidin beads. Finally, whereas adsorptive capacity may be the most appropriate term to describe the measurement performed here, all streptavidin bead vendors use the term binding capacity in product descriptions; therefore, we adopted this term throughout this manuscript to avoid confusion.
Variation in the binding capacity of streptavidin beads has several important implications for proteomics. First, beyond the obvious challenges to reproducibility among experiments within a laboratory where multiple different product lots are used, reproducibility among laboratories is also a concern. In cases where new ultrasensitive methods are described in the literature, laboratories attempting to replicate the methods would benefit from knowing the binding capacity of the product used in the original study so that they can manage expectations and troubleshoot more effectively. Thus, routine reporting of the binding capacity in addition to the assay used to determine the binding capacity, the volume of beads, and the amount of biotin substrate (i.e., d-biotin, biotinylated biomolecules) used to perform the assay would be tremendously valuable in moving the field forward toward more reproducible studies, and as such, we include this as a key reporting metric in Figure 1.
In the course of investigating the variation in bead performance, our discussions with multiple vendors revealed that whereas the minimum binding capacity threshold is provided in product information, the actual binding capacity specific to each lot may not be provided unless requested by the consumer. As we show in Figure 2 and Table 1, GenScript provided the minimum binding capacity in their product description and provided lot-specific data upon request. Despite both lots passing the vendor’s quality control testing, we found considerable variation in actual binding capacity among lots. In our experience, the lower limit of binding capacity required (i.e., the minimum binding capacity that the product must meet before it can be sold) is far below the actual capacity such that merely meeting this threshold during quality-control testing does not ensure lot-to-lot reproducibility and comparability in terms of proteomics results (Figure 2). Although the causes of this variation are not known, we learned that many vendors of streptavidin bead products do not produce the streptavidin protein in house and therefore may not have full control over the quality of the streptavidin protein. For these reasons, we have adopted a strategy whereby we request the vendor to produce a product with a stated minimum binding capacity. Only once our in-house assessment verifies that the lot meets our requirements do we proceed with purchasing the reagent. This strategy has resulted in the acquisition of two lots with similar binding capacities for our most recent studies (lots C44261906 and C44242003, Figure 5).
It is not our goal in this study to provide specific recommendations regarding the minimum binding capacity or the maximum amount of variation in binding capacity that is allowable. That will be best determined by the experimenter and will be application-dependent. We also do not speculate regarding the mathematical relationship between the binding capacity and the proteomics results. Whereas we do see that a higher binding capacity yields more protein identifications for our proteomics workflow, the exact relationship is likely to be application-dependent. Importantly, we do not endorse any specific vendor, nor did we test all of the available products. The data in Figures 4 and 5 provide evidence that the AVIDITY assay is compatible with a range of product types. Because each of these products has been successfully used in published studies, we emphasize the importance of evaluating the considerations described in Figure 1 and the Supporting Information when performing a fit-for-purpose assessment and selecting the most apt product for a particular experimental workflow. In the case of differences between products shown in Figure 5, it remains possible that equivalent numbers of proteins could be identified among all bead types if the volume of beads was increased for those with lower binding capacity. However, as previously mentioned, balancing yield with reagent costs and total reaction volumes is a driving force when deciding on the product to use.
CONCLUSIONS
We provide guidance for selecting streptavidin beads and describe an inexpensive and fast assay for measuring binding capacity. Our study also shows the problem of intralot variation in streptavidin bead binding capacity. Whereas our study included a comparison of GenScript streptavidin MagBeads lots, the St-Germain study compared lots from GE Healthcare, suggesting that the issue of intralot variability is not vendor-specific. To address this problem, we developed an assay to directly compare binding capacities among lots of the same product and to predict differences in proteomics performance among different products. Finally, we applied the assay to multiple different vendor products to demonstrate that it works across different superparamagnetic bead formats. In the future, new assays capable of simultaneously assessing the binding specificity and the binding capacity in a way that is reflective of how the beads will perform under conditions of typical biological heterogeneity will be of tremendous value, as this would further promote the reproducibility of downstream applications. Until such methods are developed, we hope that the AVIDITY assay will promote standardizing such assessments in the proteomics community and be used as leverage to advocate for higher quality and more reproducible products.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (R01-HL134010 and R01-HL126785 to R.L.G.; TL1TR001437 and UL1TR001436 to L.B.L.), American Heart Association (20PRE35200049 to L.B.L.), and Juvenile Diabetes Research Foundation (2-SRA-2019-829-S-B to R.L.G.). L.B.L. is a member of the MCW-MSTP, which is partially supported by a T32 grant from NIGMS, GM080202. Funding sources were not involved in the study design, data collection, interpretation, analysis, or publication. We appreciate valuable discussions and input from past and present Gundry lab members, especially Dr. Matthew Waas.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.0c00772.
Figure S1. Biotin titration curves for Cytiva Sera-Mag streptavidin-coated magnetic beads and Invitrogen Dynabeads MyOne streptavidin C1. Table S1. Mass spectrometry acquisition settings. Table S2. Peptide search and postsearch validation parameters. Supporting methods. General considerations for assessing streptavidin bead fit-for-purpose. Standard operating procedure: AVIDITY assay (PDF)
The authors declare no competing financial interest.
The Skyline file to enable the extraction of streptavidin peptides is available at https://panoramaweb.org/XNdVsp.url.
Contributor Information
Linda Berg Luecke, Department of Biochemistry, Medical College of Wisconsin, Milwaukee, Wisconsin 53226, United States; CardiOmics Program, Center for Heart and Vascular Research; Division of Cardiovascular Medicine; and Department of Cellular and Integrative Physiology, University of Nebraska Medical Center, Omaha, Nebraska 68198, United States.
Rebekah L. Gundry, CardiOmics Program, Center for Heart and Vascular Research; Division of Cardiovascular Medicine; and Department of Cellular and Integrative Physiology, University of Nebraska Medical Center, Omaha, Nebraska 68198, United States.
REFERENCES
- (1).Roux KJ; Kim DI; Raida M; Burke B A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol 2012, 196 (6), 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Rhee H-W; Zou P; Udeshi ND; Martell JD; Mootha VK; Carr SA; Ting AY Proteomic Mapping of Mitochondria in Living Cells via Spatially Restricted Enzymatic Tagging. Science 2013, 339 (6125), 1328–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Dieterich DC; Link AJ; Graumann J; Tirrell DA; Schuman EM Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc. Natl. Acad. Sci. U. S. A 2006, 103 (25), 9482–9487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ma Y; McClatchy DB; Barkallah S; Wood WW; Yates JR Quantitative analysis of newly synthesized proteins. Nat. Protoc 2018, 13 (8), 1744–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Aviner R; Geiger T; Elroy-Stein O PUNCH-P for global translatome profiling: Methodology, insights and comparison to other techniques. Translation (Austin) 2013, 1 (2), No. e27516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Kho Y; Kim SC; Jiang C; Barma D; Kwon SW; Cheng J; Jaunbergs J; Weinbaum C; Tamanoi F; Falck J; Zhao Y A tagging-via-substrate technology for detection and proteomics of farnesylated proteins. Proc. Natl. Acad. Sci. U. S. A 2004, 101 (34), 12479–12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Sprung R; Nandi A; Chen Y; Kim SC; Barma D; Falck JR; Zhao Y Tagging-via-Substrate Strategy for Probing O-GlcNAc Modified Proteins. J. Proteome Res. American Chemical Society 2005, 4 (3), 950–957. [DOI] [PubMed] [Google Scholar]
- (8).Abello N; Barroso B; Kerstjens HAM; Postma DS; Bischoff R Chemical labeling and enrichment of nitrotyrosine-containing peptides. Talanta 2010, 80 (4), 1503–1512. [DOI] [PubMed] [Google Scholar]
- (9).Wollscheid B; Bausch-Fluck D; Henderson C; O’Brien R; Bibel M; Schiess R; Aebersold R; Watts JD Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat. Biotechnol 2009, 27 (4), 378–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Frei AP; Moest H; Novy K; Wollscheid B Ligand-based receptor identification on living cells and tissues using TRICEPS. Nat. Protoc 2013, 8 (7), 1321–1336. [DOI] [PubMed] [Google Scholar]
- (11).Sabarth N; Lamer S; Zimny-Arndt U; Jungblut PR; Meyer TF; Bumann D Identification of Surface Proteins of Helicobacter pylori by Selective Biotinylation, Affinity Purification, and Two-dimensional Gel Electrophoresis. J. Biol. Chem 2002, 277 (31), 27896–27902. [DOI] [PubMed] [Google Scholar]
- (12).St-Germain J; Samavarchi Tehrani P; Wong C; Larsen B; Gingras A-C; Raught B Variability in streptavidin-sepharose matrix quality can significantly affect proximity-dependent biotinylation (BioID) data. J. Proteome Res 2020, 19, 3554. [DOI] [PubMed] [Google Scholar]
- (13).Green NM Methods Enzymol. 2020, 418–424. [Google Scholar]
- (14).Haverland NA; Waas M; Ntai I; Keppel T; Gundry RL; Kelleher NL Cell Surface Proteomics of N-Linked Glycoproteins for Typing of Human Lymphocytes. Proteomics 2017, 17 (19), 1770141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Boheler KR; Bhattacharya S; Kropp EM; Chuppa S; Riordon DR; Bausch-Fluck D; Burridge PW; Wu JC; Wersto RP; Chan GCF; Rao S; Wollscheid B; Gundry RL A human pluripotent stem cell surface N-glycoproteome resource reveals markers, extracellular epitopes, and drug targets. Stem Cell Rep. 2014, 3 (1), 185–2C03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Mallanna SK; Waas M; Duncan SA; Gundry RL N-glycoprotein surfaceome of human induced pluripotent stem cell derived hepatic endoderm. Proteomics 2017, 17 (5), 1600397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Fujinaka CM; Waas M; Gundry RL Mass Spectrometry-Based Identification of Extracellular Domains of Cell Surface N-Glycoproteins: Defining the Accessible Surfaceome for Immunophenotyping Stem Cells and Their Derivatives. Methods Mol. Biol 2018, 1722, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Eng JK; McCormack AL; Yates JR An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom 1994, 5 (11), 976–989. [DOI] [PubMed] [Google Scholar]
- (19).Kong AT; Leprevost FV; Avtonomov DM; Mellacheruvu D; Nesvizhskii AI MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry–based proteomics. Nat. Methods 2017, 14 (5), 513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Käll L; Canterbury JD; Weston J; Noble WS; MacCoss MJ Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 2007, 4 (11), 923–925. [DOI] [PubMed] [Google Scholar]
- (21).Mellacheruvu D; Wright Z; Couzens AL; Lambert J-P; St-Denis NA; Li T; Miteva YV; Hauri S; Sardiu ME; Low TY; Halim VA; Bagshaw RD; Hubner NC; Al-Hakim A; Bouchard A; Faubert D; Fermin D; Dunham WH; Goudreault M; Lin Z-Y; Badillo BG; Pawson T; Durocher D; Coulombe B; Aebersold R; Superti-Furga G; Colinge J; Heck AJR; Choi H; Gstaiger M; Mohammed S; Cristea IM; Bennett KL; Washburn MP; Raught B; Ewing RM; Gingras A-C; Nesvizhskii AI The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 2013, 10 (8), 730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Eisenberg-Domovich Y; Pazy Y; Nir O; Raboy B; Bayer EA; Wilchek M; Livnah O Structural elements responsible for conversion of streptavidin to a pseudoenzyme. PNAS. Proc. Natl. Acad. Sci. U. S. A 2004, 101 (16), 5916–5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Pazy Y; Raboy B; Matto M; Bayer EA; Wilchek M; Livnah O Structure-based Rational Design of Streptavidin Mutants with Pseudo-catalytic Activity. J. Biol. Chem 2003, 278 (9), 7131–7134. [DOI] [PubMed] [Google Scholar]
- (24).Kada G; Falk H; Gruber HJ Accurate measurement of avidin and streptavidin in crude biofluids with a new, optimized biotin-fluorescein conjugate. Biochim. Biophys. Acta, Gen. Subj 1999, 1427 (1), 33–43. [DOI] [PubMed] [Google Scholar]
- (25).Kada G; Kaiser K; Falk H; Gruber HJ Rapid estimation of avidin and streptavidin by fluorescence quenching or fluorescence polarization. Biochim. Biophys. Acta, Gen. Subj 1999, 1427 (1), 44–48. [DOI] [PubMed] [Google Scholar]
- (26).Dorgan L; Magnotti R; Hou J; Engle T; Ruley K; Shull B Methods to determine biotin-binding capacity of streptavidin-coated magnetic particles. J. Magn. Magn. Mater 1999, 194, 69. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.