Abstract
Cellobiohydrolase I from Trichoderma reesei ( Tr Cel7A) is one of the best-studied cellulases, exhibiting high activity towards crystalline cellulose. Tryptophan residues at subsites -7 and -4 (Trp40 and Trp38 respectively) are located at the entrance and middle of the tunnel-like active site of Tr Cel7A, and are conserved among the GH family 7 cellobiohydrolases. Trp40 of Tr Cel7A is important for the recruitment of cellulose chain ends on the substrate surface, but the role of Trp38 is less clear. Comparison of the effects of W38A and W40A mutations on the binding energies of sugar units at the two subsites indicated that the contribution of Trp38 to the binding was greater than that of Trp40. In addition, the smooth gradient of binding energy was broken in W38A mutant. To clarify the importance of Trp38, the activities of Tr Cel7A WT and W38A towards crystalline cellulose and amorphous cellulose were compared. W38A was more active than WT towards amorphous cellulose, whereas its activity towards crystalline cellulose was only one-tenth of that of WT. To quantify the effect of mutation at subsite -4, we measured kinetic parameters of Tr Cel7A WT, W40A and W38A towards cello-oligosaccharides. All combinations of enzymes and substrates showed substrate inhibition, and comparison of the inhibition constants showed that the Trp38 residue increases the velocity of substrate intake ( k on for forming productive complex) from the minus side of the subsites. These results indicate a key role of Trp38 residue in processively loading the reducing-end of cellulose chain into the catalytic tunnel.
Keywords: cellobiohydrolase, steady-state kinetics, processive reaction, Trichoderma reesei
Abbreviations
GH, glycoside hydrolase; Tr Cel7A, cellobiohydrolase I from Trichoderma reesei ; WT, wild-type; C1 = glucose, C2 = cellobiose, C3 = cellotriose, C4 = cellotetraose, C5 = cellopentaose, C6 = cellohexaose; LPMOs, lytic polysaccharide monooxygenases, PASC, phosphoric acid swollen cellulose.
INTRODUCTION
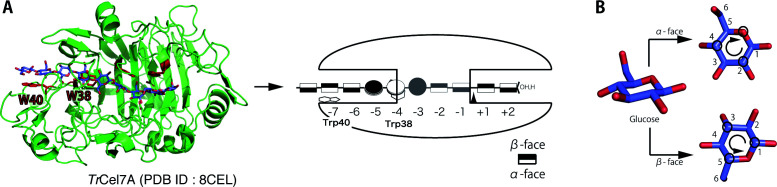
Cellobiohydrolases (CBHs) are the main components of enzyme cocktails employed for cellulosic biomass conversion to soluble sugars for the production of useful chemicals. CBHs can be divided into two main types according to their specificity towards the cellulose chain-end. Fungal CBH I hydrolyzes crystalline cellulose from the reducing-end and CBH II does so from the non-reducing-end. 1) The catalytic domains (CD) of these enzymes are classified into different families in the Carbohydrate-Active enZymes (CAZy) database according to the amino acid sequences and three-dimensional protein folds. The CDs of CBH I and CBH II are members of glycoside hydrolase (GH) family 7 and 6, respectively. 2) The best-studied CBH is CBH I from Trichoderma reesei ( Tr Cel7A), which has two domains, a cellulose-binding domain (CBD) classified in carbohydrate-binding module (CBM) family 1 and a GH family 7 CD, connected by a linker region. The unique feature of CD of GH family 7 cellobiohydrolases is a tunnel-like active-site structure, which consists of 9 subsites (−7 to +2); hydrolysis occurs between subsites −1 and +1, while subsites −4 to −2 are mostly covered by loop regions. 3) The Trp40 and Trp38 residues, located at subsites −7 and −4, respectively, are highly conserved in various GH family 7 CBHs, 4) and participate in formation of the entrance of the tunnel ( Fig.1 A ). The unique feature of Tr Cel7A is its processive mechanism of hydrolysis of the cellulose chain. We have observed the processive reaction of individual Tr Cel7A molecules on a crystalline cellulose surface in real-time by means of high-speed atomic force microscopy (HS-AFM), and discussed the importance of the tunnel-like structure. 5) 6) 7) We have also shown that the Trp40 residue at the tunnel entrance is important for recruiting a cellulose chain from the crystalline cellulose surface to initiate the processive hydrolysis reaction. 4) 5) As for Trp38, Kari et al. 8) compared the activity of Tr Cel7A W38A mutant with that of WT on Avicel, and concluded that the off-rate ( k off ) is higher and the processivity is lower for W38A. However, the reason why the Trp residue in subsite −4 is highly conserved in GH family 7 CBHs, and the difference between the roles of the tryptophan residues at −4 and −7 during crystalline cellulose hydrolysis remain to be fully established.
Fig. 1. (A) Positions of Trp40 and Trp38 residues in the active site of Tr Cel7A, (B) The faces of glucose rings.
Trp40 residue is located at the edge of the active-site tunnel (subsite −7) and Trp38 residue is located at the middle (subsite −4). The structure of Tr Cel7A (PDB ID: 8CEL) is illustrated, showing the recognized faces of glucose rings. The α-face and β-face are shown in white and black, respectively. The faces were named according to the rules for naming faces of ring compounds. 28)
Since the processive reaction is important for crystalline cellulose degradation, “processivity”, i.e. the amount of consecutive cuts of the cellulose chain by the enzyme before dissociation from the substrate, has been estimated by various methods. 9) The low solubility of cellulose, however, hinders determination of the actual processivity value, as it is difficult to determine the amount of active enzyme molecules in the reaction mixture. 10) On the other hand, analysis of CBH activity towards soluble cello-oligosaccharides is challenging due to the complexity of the overall reaction: 1) substrate chain can be loaded from both ends of the active-site tunnel, and possibly also through endo-type loading in the middle of the tunnel; 2) cleavage can occur at several different positions of the oligosaccharide chain, as well as in a processive manner, 3) both substrate and product inhibition can occur. Furthermore, 4) transglycosylation reactions are also possible, 11) and 5) the effect of anomeric conformer of reducing end was also suggested. 12)
Re-analysis of the kinetics of sucrose hydrolysis by invertase reported by Michaelis and Menten 13) by the application of global fitting (GF) with modern software provided new insights. 14) GF analysis can fit multiple equations at the same time with common parameters, and it is a powerful tool to analyze complex reaction models. Here, we have applied GF analysis to quantify the effects of W40A and W38A mutations on the kinetics of hydrolysis of soluble cello-oligosaccharides. Based on the results, we propose a possible role of the tryptophan 38 residue in promoting the processive reaction of Tr Cel7A.
RESULTS
Calculation of the interaction energy and the effect of tryptophan mutation on binding energy.
When the interaction energies of a cellulose chain at the different subsites of Tr Cel7A WT were calculated, an increasing energy gradient from subsite −6 towards subsite −1 was found ( Fig. 2A ). On the other hand, a decreasing energy gradient was also found at the product side (at subsites +2 and +1) of the active-site tunnel; thus, the energy gradient was oppositely directed relative to the processive movement of the substrate. The interaction energy at subsite −7 was the highest among the all subsites. The interaction energy at subsite −7 is mostly due to electrostatic interactions via hydrogen bonding with Gln7 and Asn49. Individual mutations of the four tryptophans of the active-site tunnel of Tr Cel7A to alanine, i.e. , W40A (at −7), W38A (at −4), W367A (at −2), and W376A (at +1), affected the binding free energy as shown in Fig. 2B . The difference caused by the W38A mutation was higher than that caused by the W40A mutation, with the ∆∆G values for W38A and W40A being +5.89 and +2.89 kcal/mol, respectively. The ∆∆G values for W38A was about a half of those for W367A (+10.0 kcal/mol), but higher than ∆∆G values for W376A (+3.72 kcal/mol). Trp367 and Trp376 are known as important residues to form the stable Michaelis complex. These results suggest that the Trp38 residue has a substantial role in determining the affinity of Tr Cel7A for the cellulose chain.
Fig. 2. (A) A snap shot of trajectory and interaction energy at the subsites of Tr Cel7A WT, (B) Effects of tryptophan mutations on the binding free energy.
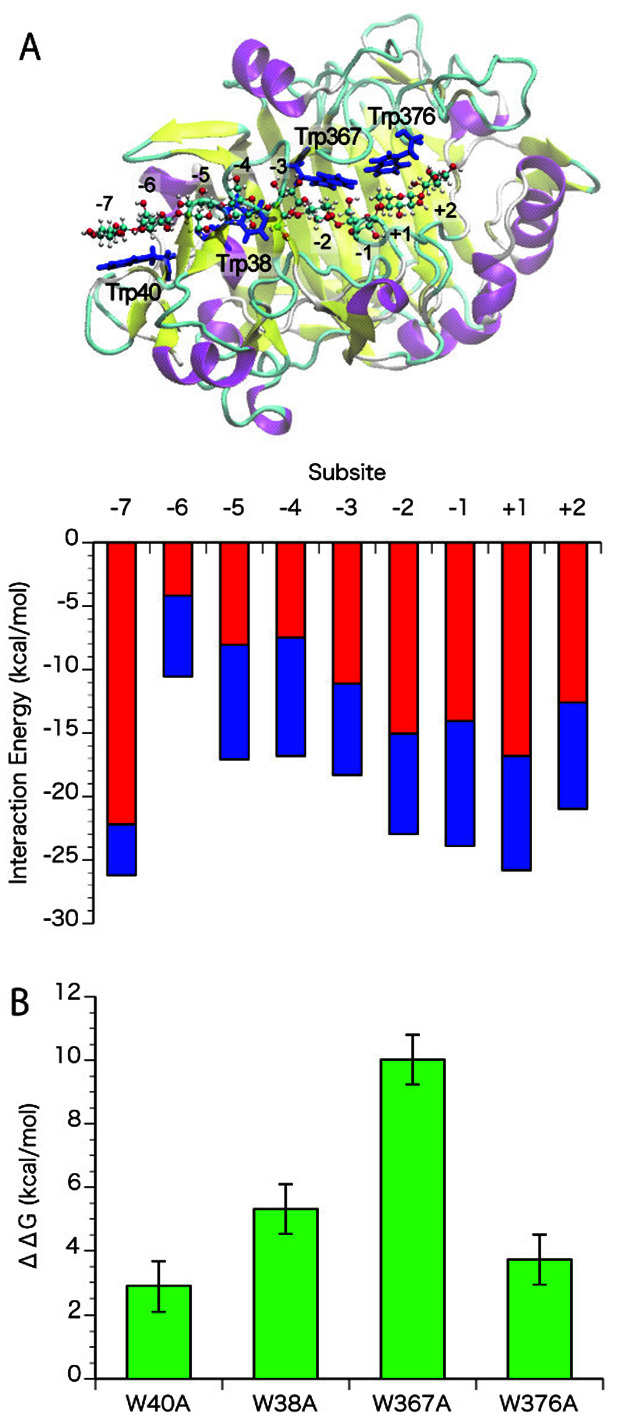
The interaction energy of cellononaose at the subsites in Tr Cel7A was calculated using the liganded structure (PDB ID: 8CEL). The electrostatic and the van der Waals interactions are shown in red and blue, respectively, in Fig. 2A. Standard errors for electrostatic and van der Waals interactions were about 0.4 and 0.1, respectively. The effects of mutation of each tryptophan (Trp40, 38, 367, and 376) to alanine were estimated by alanine-scanning, shown in green in Fig. 2B. Standard errors are shown as bars.
Comparison of cellulose degradation activity.
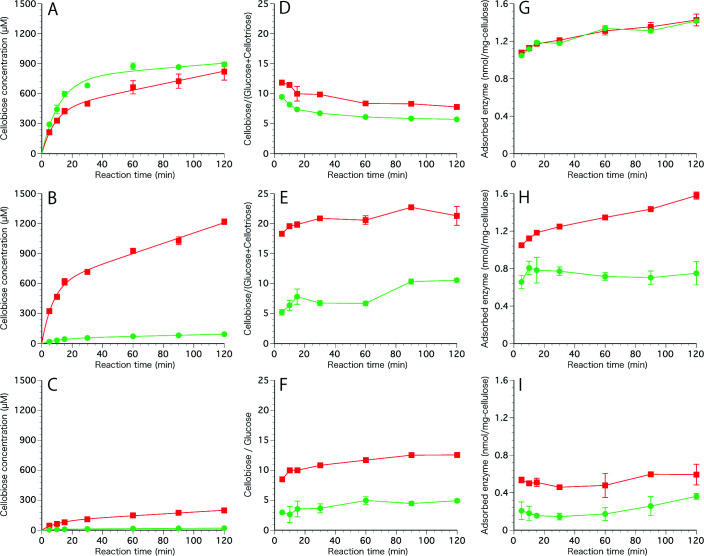
Tr Cel7A WT and W38A produced glucose (C1), cellobiose (C2) and cellotriose (C3) from phosphoric acid swollen cellulose (PASC) and crystalline cellulose III I , whereas they produced only C1 and C2 from crystalline cellulose I α . In all cases, cellobiose was the major soluble product. The time courses of C2 production by Tr Cel7A WT and W38A are shown in Fig. 3 . When PASC was used as a substrate, the velocity of C2 production by W38A was higher than that by WT ( Fig. 3A ). On the other hand, the activities of W38A towards crystalline celluloses were about 10 times lower than those of WT ( Fig. 3B and 3C ). The values of C2/(C1+C3), even/odds ratio, of WT and W38A of PASC were about 9 and 7 at 120 min, respectively ( Fig. 3D ), whereas the corresponding ratios of WT for cellulose I α and cellulose III I of WT were 10 and 20, respectively, being about 2-fold higher than those of W38A ( Fig. 3E and 3F ). The amount of W38A adsorbed on PASC was similar to that of WT ( Fig. 3G ), but the amount of W38A adsorbed on cellulose III I was about two-thirds of that of WT ( Fig. 3H ), while that of W38A on cellulose I α was almost half that of WT ( Fig. 3I ).
Fig. 3. Progress curves of hydrolysis and adsorption of Tr Cel7A WT and W38A mutant on amorphous and crystalline cellulose.
1.0 µM Tr Cel7A WT (red box) and W38A (green circle) were incubated with 0.1 % PASC (A), cellulose III I (B) and crystalline cellulose I α (C) in 50 mM sodium acetate buffer pH 5.0 at 30 °C. The ratios of produced cellobiose / glucose or (glucose + cellotriose) from PASC (D), crystalline cellulose III I (E) and cellulose I α (F) are shown in the middle. The amounts of adsorbed enzyme on PASC (G), crystalline cellulose III I (H) and cellulose I α (I) are shown on the right side. Each reaction was repeated three times.
Time course of cello-oligosaccharide hydrolysis.
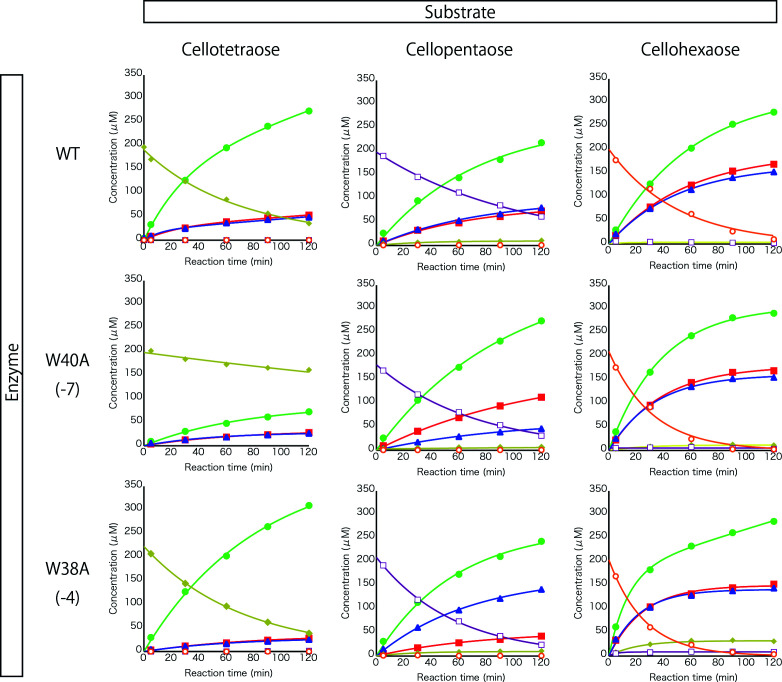
Time courses of substrate and product concentrations were monitored for each enzyme ( Tr Cel7A WT, W38A, and W40A). As shown in Fig. 4 , when cellotetraose (C4) was used as a substrate, Tr Cel7A WT and W38A showed similar activity, but the activity of W40A was clearly lower. On the other hand, in the case of cellopentaose (C5) hydrolysis, W40A showed drastically increased velocity. W38A showed similar hydrolytic velocity to W40A, while WT was slowest. In cellohexaose (C6) hydrolysis, WT was slowest, whereas the velocities of W38A and W40A were similar. Focusing on the products, the biggest difference is in the ratio between C1 and C3 derived from C5. WT produced equal amounts of C1 and C3, whereas W40A produced twice as much C1 as C3. On the other hand, glucose production by W38A was one-third of that of C3. Another difference is in the formation of products with longer DP from C5 and C6. Only a small amount of C4 was detected in the reaction mixture of WT and W40A, while W38A produced more C4 from C5. In addition, W38A produced C4 and C5 from C6, but little C4 or C5 was produced by WT or W40A.
Fig. 4. Time courses of hydrolysis of cello-oligosaccharides (cellotetraose to cellohexaose) by Tr Cel7A WT and the two tryptophan mutants.
Reactions were conducted in 50 mM sodium acetate pH 5.0 at 30 °C. Initial concentration of substrates was 200 µM and enzyme concentrations were 0.1 µM. Filled square: glucose, filled circle: cellobiose, triangle: cellotriose, diamond: cellotetraose, open square: cellopentaose and open circle: cellohexaose.
Measurements of initial velocity for different concentrations of cello-oligosaccharides.
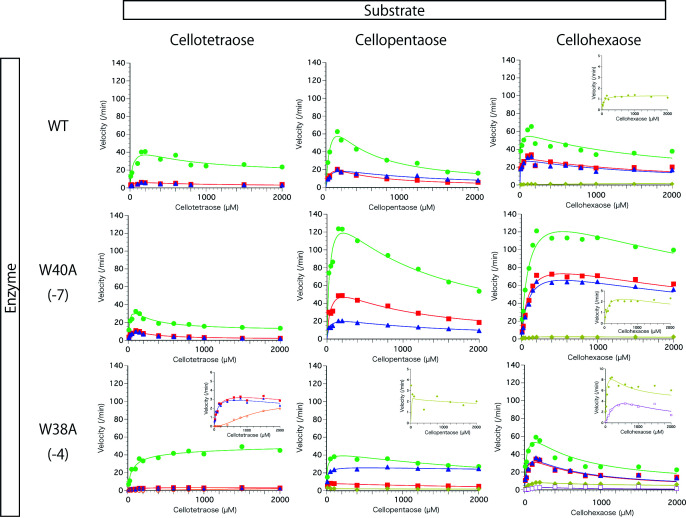
Plots of initial velocities of cello-oligosaccharide production from different concentrations of C4, C5, and C6 are shown in Fig. 5 . Major products were C2, C1, and C3 under all conditions examined. All of the enzyme and substrate combinations showed clear substrate inhibition, except for C4 hydrolysis by W38A. The production velocities of C2 from C4 by all enzymes remained relatively high even at high substrate concentration. C3 production by W38A from C5 did not show substrate inhibition. These results indicate that small amounts of products can be generated when two molecules of the substrate are bound in the active-site tunnel. Furthermore, W38A produced C6 at high C4 concentration, indicating that trans-glycosylation occurs during C4 hydrolysis.
Fig. 5. Plots of initial velocity at each concentration of cello-oligosaccharides and fitted curves.
The enzymes and substrates were incubated in 20 mM sodium acetate pH 5.0 at 30 °C. Filled square: glucose, filled circle: cellobiose, triangle: cellotriose, diamond: cellotetraose, open square: cellopentaose and open circle: cellohexaose.
Construction of reaction models and global curve fitting.
From the results of hydrolysis of cello-oligosaccharides, we constructed a hydrolysis model and corresponding equations. Cello-oligosaccharides have two faces, because the glucose ring has an α-face and a β-face ( Fig. 1B ). Facial recognition is important, because Tr Cel7A can hydrolyze the chain only when the glycoside bond is correctly oriented towards the catalytic residues. For example, C4 can be hydrolyzed to two C2 molecules or a C1 and C3 pair, depending on the initial substrate recognition. Thus, C4 ( S ) and enzyme ( E ) can form two different enzyme-substrate complexes ( ES and ES ’), and they will produce different products, as shown in the model in Supplementary Method 1(see J. Appl. Glycosci. Web site). Substrate inhibition should be considered, because production velocities were decreased at high substrate concentration. Substrate inhibition means that the enzyme cannot hydrolyze the substrate if two substrate molecules are bound to the enzyme at the same time. Tr Cel7A has at least seven subsites on the minus side (−7 to −1) and two subsites on the plus side (+1 and +2). Therefore, a substrate cannot be attacked by the catalytic residues if another substrate molecule is already occupying subsites close to the catalytic center; these states are called ESS and ES ’ S . Additionally, the reaction model should include production of C2 at high substrate concentration, because the experimental data indicated that the production velocity of C2 is tolerant to increase of substrate concentration. Thus, the production of C2 from ESS and ES ’ S complexes should be considered; for example, Tr Cel7A can produce C2 even if one substrate is bound on −7 to −4. The last issue is C6 production by W38A. This reaction is a trans-glycosylation reaction. There are two possible ways to form C6 from C4. One is the connection of C2 and C4, and the other is the hydrolysis of cello-octaose produced by the connection of two C4 molecules. These two models are different, but they give the same equations for fitting. The effect of anomer conformations at reducing end is not included in models, but it can be discussed from obtained parameters.
The principles of the models for C5 (Supplementary Method 2; see J. Appl. Glycosci. Web site) and C6 hydrolysis (Supplementary Method 3; see J. Appl. Glycosci. Web site) were almost the same as in the model for C4. They also contain two types of the enzyme-substrate complex ( ES and ES ’) and substrate inhibition by two substrates binding at the same time ( ESS and ES ’ S ). In addition, the production of C2 and C3 at high concentrations of substrate was added to the model for C5, and the production of C2 and C4 at high concentrations of C6 was added to account for the tolerance of the reaction velocity to high concentrations of substrate. Production of C2 and C3 at high concentration of C5 has already been reported. 15) In the case of C5, however, the products generated from ES and ES ’ are the same even if the positions of the cleaved bonds are different. Therefore, turnovers of C2 and C3 pair production from ES ( k cat1 ) and ES ’ ( k cat5 ) cannot be distinguished. Also, k cat3 (from ES ) and k cat7 (from ES ’) cannot be distinguished, because both reactions produce a pair of C1 and C4. The relationship between k cat2 and k cat6 (production of C1 and two C2) is similar. The main difference from the model for C4 is the effect of processive reaction. Tr Cel7A can cut the substrate into C2 units, and thus C5 should be hydrolyzed to C1 and two C2 molecules in a processive manner, while C6 should be hydrolyzed into three C2 molecules or a mixture of C1, C2 and C3. Additionally, possible failure of the second hydrolysis was included in the models. For the C6 hydrolytic model, one further reaction, which affords C3 without production of glucose, was added because although W38A produces C5 and C1 from C6, the velocities of C3 and C1 production were almost the same at all substrate concentrations. One possible explanation is the production of two C3 molecules from C6.
The parameters obtained by the global fittings are shown in Tables 1 , 2 , and 3 . Some parameters were set as 0 when the corresponding product was not detected or when the value converged to less than 0. The value of K m is considered as an indicator of the affinity between enzyme and substrate for a simple Michaelis-Menten reaction (although this assumes that enzyme-substrate complex formation is much faster than the reaction). In the reaction of Tr Cel7A with cello-oligosaccharides, Tr Cel7A has many subsites, and in addition the substrates have two faces. Further, if the affinity between enzyme and substrate is very high, a second substrate will bind to the empty subsites to form a multiple substrate-bound state. Therefore, it is difficult to precisely understand the meaning of these K m values. On the other hand, K m2 , constant for the substrate inhibition, is more clear-cut, because the influence of the two-substrate-bound state, which inhibits the hydrolysis reaction, is limited. In the case of C4 hydrolysis, K m2 for W40A was the smallest, whereas K m2 for WT and W38A was the smallest for C5 and C6 hydrolysis, respectively. The k cat values also showed variation: for example, the velocity of C4 production from C5 ( k cat3 + k cat7 ) by W38A was 3.06 ± 1.12 min -1 , whereas those of WT and W40A were assumed to be 0. This indicates that C5 was hydrolyzed into one C1 and two C2s by WT and W40A, but C4 and C1 were produced by W38A. Additionally, k cat6 for C5 production from C6 by WT and W40A were both 0, whereas that of W38A was 4.30 ± 2.04 min -1 . This result also indicates that WT and W40A hydrolyze C6 to C1, C2 and C3 processively, whereas W38A releases C1 and C5. Moreover, the velocity of C2 and C3 production from double-bound C6s ( k cat3 + k cat7 ) could be estimated, whereas those of WT and W40A were assumed to be 0. This tendency is explained by the difference of K m2 , because the K m2 values of WT and W40A were 2,060 ± 250 and 4,290 ± 490 µM, respectively, which are too high for two substrate molecules to enter the active site under the conditions of this analysis. The product ratio of C4 + C2 ( k cat2 ) and C2 + C2 + C2 ( k cat1 ) from C6 with ES complex of WT were 1:9 and more biased to C2 production than expected value C4:C2 = 3:7 from anomeric conformation ratio α:β = 3:7. 12) The effect of anomer conformations at reducing end for oligo-saccharides hydrolysis should be smaller than the effect ofinteraction with Trp38, because this C4:C2 production ratio of W40A was similar to WT, but that of W38A was drastically changed (C4:C2 = 1:1).
Table 1.
Kinetic parameters of Tr Cel7A wild-type and mutants for cellotetraose.
| kcat1 (min -1 ) | kcat2 + k cat5 (min -1 µM -1 ) | kcat3 (min -1 µM -1 ) | kcat4 (min -1 ) | Km (µM) | Km2 (µM) | |
|---|---|---|---|---|---|---|
| WT | 25.2 ± 2.5 | 0.00720 ± 0.00110 | ✢ | 8.51 ± 1.72 | 41.1 ± 6.9 | 783 ± 432 |
| W40A | 25.9 ± 3.1 | 0.0282 ± 0.0103 | ✢ | 18.1 ± 2.8 | 37.5 ± 8.7 | 170 ± 48 |
| W38A | 21.6 ± 1.1 | 0.00267 ± 0.00068 | 0.00124 ± 0.00073 | 3.30 ± 0.65 | 56.4 ± 7.8 | 11800 ± 3900 |
✢ The value was set at 0, because the amount of product was below the detection limit (0.1 µM). The values are mean ± fitting error.
Table 2.
Kinetic parameters of Tr Cel7A wild-type and mutants for cellopentaose.
| kcat1 + k cat5 (min -1 ) | kcat2 + k cat6 (min -1 ) | kcat3 + k cat7 (min -1 ) | kcat4 + k cat8 (min -1 µM -1 ) | Km (µM) | Km2 (µM) | |
|---|---|---|---|---|---|---|
| WT | 41.7 ± 5.2 | 48.0 ± 7.1 | - § | 0.0305 ± 0.0125 | 102 ± 21 | 190 ± 44 |
| W40A | 30.2 ± 3.0 | 68.5 ± 4.1 | - § | - § | 44.7 ± 6.8 | 1020 ± 130 |
| W38A | 29.0 ± 1.4 | 8.85 ± 0.89 | 3.06 ± 1.12 | 0.0277 ± 0.0077 | 18.0 ± 2.5 | 778 ± 182 |
§ The value was set at 0, because it was estimated to be lower than 0 during fitting. The values are mean ± fitting error.
Table 3.
Kinetic parameters of Tr Cel7A wild-type and mutants for cellohexaose.
| kcat1 (min -1 ) | kcat2 (min -1 ) | kcat3 + k cat7 (min -1 µM -1 ) | kcat4 (min -1 ) | kcat5 (min -1 ) | kcat6 (min -1 ) | Km (µM) | Km2 (µM) | |
|---|---|---|---|---|---|---|---|---|
| WT | 9.56 ± 0.71 | 1.38 ± 1.24 | - § | ✢ | 30.1 ± 1.2 | - § | 6.45 ± 1.37 | 2060 ± 250 |
| W40A | 20.9 ± 1.2 | 2.93 ± 2.03 | - § | ✢ | 87.9 ± 3.1 | - § | 84.1 ± 7.8 | 4290 ± 490 |
| W38A | 8.54 ± 1.48 | 10.2 ± 2.3 | 0.00761 ± 0.00379 | 4.30 ± 2.04 | 41.4 ± 4.2 | 3.14 ± 1.76 | 34.2 ± 6.4 | 548 ± 90 |
§ The value was set at 0, because it was estimated to be lower than 0 during fitting. ✢ The value was set at 0, because the amount of product was below the detection limit (0.1 µM). The values are mean ± fitting error.
The hydrolytic pattern and velocities at 200 µM substrate were calculated from the kinetic parameters (Fig. S1; see J. Appl. Glycosci. Web site). The relationships of calculated velocities are consistent with the experimental data of cello-oligosaccharides hydrolysis shown in Fig. 4 . For example, the velocities of C4 hydrolysis by W40A and C5 hydrolysis by WT are the slowest among the three, possibly because their K m2 values are close to 200 µM. The models we created in this study also explain well the product ratios of C1 and C3 from C5 for the three enzymes.
DISCUSSION
The three-dimensional structure of Tr Cel7A was solved more than 25 years ago. 16) The enzyme has a tunnel-like active-site architecture lined by hydrogen bond donors, together with four tryptophan residues. The importance of the tunnel-like active site for the degradation of crystalline cellulose by CBHs has been addressed and compared with the case of endoglucanases, which have more cleft-like active sites. However, the importance of the tryptophan residues in the active-site tunnel of CBHs has not been fully established. One of the ways to quantify the effect of CBH mutations is to examine their effects on the hydrolysis of soluble substrates. However, the kinetics of Tr Cel7A with soluble cello-oligosaccharides has not been reported yet, although the catalytic reaction of Tr Cel7A is well known and products can be easily measured, e.g., by HPLC. The biggest barrier has been the complexity of the reactions, which make curve-fitting difficult. However, recent advances in computing power allow better analysis of complex enzyme reactions. 17) A good example is the re-analysis of the original data for this enzyme, simultaneously considering the hydrolysis of sucrose and the effects of product inhibition, by means of global fitting. In the present study, we used this approach to examine the role of the Trp38 residue in the hydrolysis of crystalline cellulose.
PASC hydrolysis rate by W38A was higher than that by WT, even though the activities towards carboxyl methyl cellulose are the same (Fig. S2; see J. Appl. Glycosci. Web site). J. Kari et al. reported that Tr Cel7A W38A has higher dissociation rate than WT. 8) Our calculation of the interaction energy of cellononaose at the subsites of Tr Cel7A WT also revealed a relatively large contribution (−5.31 kcal/mol) from the Trp38 residue at subsite −4. The higher hydrolysis activity of W38A to PASC indicates that the rate-limiting step of amorphous cellulose is dissociation from the chain. On the other hand, the calculation results showed a gradient of interaction energy from subsite −6 to −1 of WT was broken by the W38A mutation. This result suggests an important role of Trp38 residue in the processive reaction. Indeed, the even/odds ratio of W38A on crystalline celluloses was about a half that of WT, further suggesting the significance of Trp38 for processive reaction as reported by R. Kont et al . 18) . Moreover, when we compared product formation from crystalline cellulose I α and III I by WT and W38A, W38A was about one-tenth as effective as WT. Since the amounts of W38A adsorbed on crystalline cellulose I α and III I were about half and two-thirds of those of WT, respectively, the specific activities of adsorbed WT on crystalline celluloses were more than 5 times higher than those of W38A. These results suggest that the Trp38 residue increases the productivity of binding molecule for the degradation of crystalline cellulose.
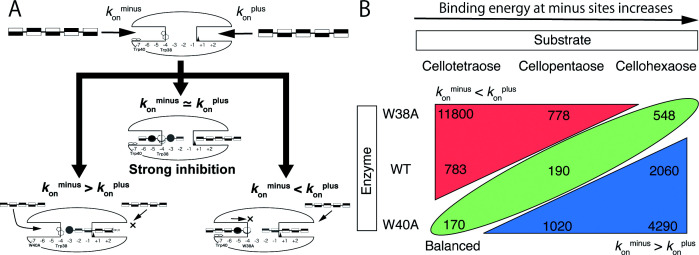
In order to understand the importance of Trp38, we focused on K m2 , which is a key parameter for understanding the complicated reaction of Tr Cel7A with soluble substrates. It is typically an indicator of substrate inhibition and is also written as K s . Substrate inhibition occurs when two or more substrates form a complex with the enzyme in such a way that catalysis is blocked. Initially, before any product is formed, two substrate molecules compete for binding to the catalytic position, approaching from the minus side (tunnel entrance) and the plus side (tunnel exit) as shown in Fig. 6A . The substrate coming from minus side cannot be hydrolyzed if the plus side is occupied by another substrate, and vice versa . In other words, substrate inhibition in Tr Cel7A involves competition of two substrate molecules for one catalytic center, just as in competitive inhibition. Therefore, K m2 will be small if the rates of substrate intake from each side ( k on minus and k on plus , rate constants for the substrate intake to active site from minus and plus subsites, respectively) are similar. However, if the rate from one side is higher than that of the other side, K m2 will be high, and the inhibition will be relatively low. Therefore, K m2 represents an indicator of the balance between k on minus and k on plus . By making a matrix of K m2 values among enzymes and substrate lengths, we can visualize the relationship, as shown in Fig. 6B . The values of K m2 for W40A, WT and W38A were smallest with C4, C5, and C6, respectively, and the relationship is consistent with the two-substrate competition model. Three enzymes have similar k on plus values, because W38A and W40A mutations have less effect on k on plus (and k on endo , rate constant for direct binding of substrate to active site) and the catalytic event and cellotetraose is enough long to occupy the plus side. At the minus side, these two mutations affect to the binding energy gradient, and values of k on minus are different among the three enzymes. Furthermore, the energy gradient getting stronger with longer substrate, because minus side is longer than the substrate lengths. Therefore, the combinations of three enzymes (WT, W38A, W40A) and three substrates (C4, C5, C6) make a matrix of different balances between k on minus and k on plus . Focusing on cellotetraose hydrolysis, k cat1 and k cat4 of W40A were the largest among the three enzymes, and those of W38A were the smallest. This result means that k on minus of W40A is the largest, and that of W38A is the smallest. This relationship is reasonable, because W40A mutation makes the binding energy gradient smoother than that of WT and W38A mutation break that gradient. Furthermore, W38A produced cellotetraose from cellopentaose ( k cat3 + k cat7 ), and cellotetraose and cellopentaose from cellohexaose ( k cat2 and k cat6 respectively). These results clearly show that the W38A mutant is less processive than WT or W40A.
Fig. 6. (A) Scheme of the example relationship between k on plus and k on minus for cellopentaose hydrolysis, (B) Matrix of K m2 values.
The values of K m2 were extracted from the Tables 1–3.
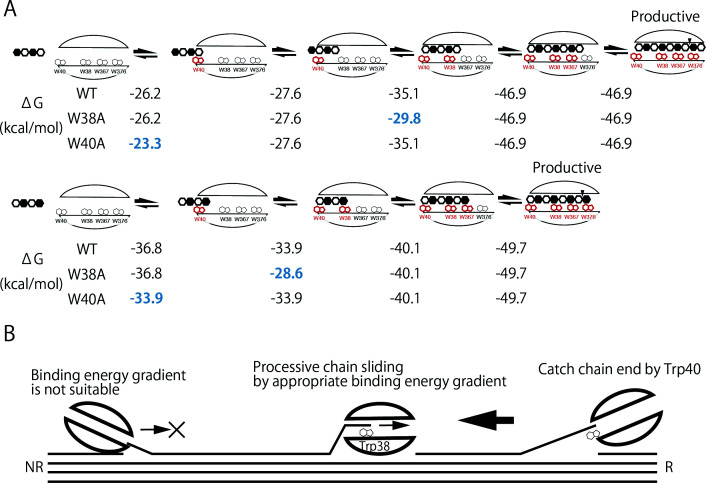
The differences of binding energy (ΔG) of the three enzymes to the cellulose molecular chain are illustrated in Fig. 7A . W38A is clearly disadvantageous for cellulose chain intake from the minus side, because W38A mutation causes unfavorable changes in binding free energy. The complexes spontaneously change to the states with lower ΔG, thus W38A mutation slows down the chain sliding at the middle of tunnel when compared to WT as observed in oligo-saccharides hydrolysis. This finding indicates that Trp38 is necessary to pull the substrate chain efficiently into the catalytic site from the crystal surface after initiation by Trp40, as shown in Fig. 7B . This hypothesis can explain why the specific activity of W38A on crystalline cellulose is much lower than that of WT, because strong binding energies are required to release cellobiose units from the crystal surface and to step forward along the chain. Without Trp38, many CBH molecules may just bind to a substrate chain end (using Trp40) and remain waiting for intake deeper to the active-site tunnel. On the other hand, these results indicate that W38A mutant may be a better enzyme than WT for applications in combination with assisting enzymes, such as endoglucanases and/or LPMOs, which can produce chain ends on a crystalline surface. This is because the effect of low k on minus will be smaller and will be overcome by the benefit of high k off (easy detachment from the crystal surface after stacking).
Fig. 7. (A) Differences of binding free energies during cellulose hydrolysis by Tr Cel7A WT and mutants, (B) Scheme of cellulose hydrolysis by Tr Cel7A WT.
Differences of binding free energies was calculated from Fig. 2. Interacting tryptophan residues are shown in red, and unfavorable changes of free energy compared with WT are shown in blue.
In this paper, we have shown that, besides Trp40 at the entrance of the tunnel (at subsite −7), Trp38 deeper in the active-site tunnel (at subsite −4 of Tr Cel7A) is important for degradation of crystalline cellulose. By applying global fitting analysis, we were able to explain the courses of the reactions of Tr Cel7A with different lengths of cello-oligosaccharides and to elucidate the contribution of Trp38 to substrate intake from the tunnel entrance (minus side). Our results indicate that the Trp38 residue of Tr Cel7A contributes to make a smooth energy gradient of the subsites, promoting processive introduction of a cellulose chain from the crystalline cellulose surface into the active-site tunnel.
EXPERIMENTAL
Materials.
Cellooligosaccharides, i.e. , cellobiose (C2), cellotriose (C3), cellotetraose (C4), cellopentaose (C5), and cellohexaose (C6), were purchased from Seikagaku Corporation (Tokyo, Japan), glucose was from Wako Pure Chemical Industries, Ltd., Osaka, Japan, and p -nitrophenyl-β-D-lactoside ( p NPL) from Sigma-Aldrich Corp., Saint Louis, MO, USA.
Enzyme production and purification.
W40A, W38A mutants of Tr Cel7A were produced by T. reesei , into which genes encoding the mutant enzymes had been introduced as described in a previous report. 19) Tr Cel7A wild-type (WT) was purified from Celluclast® 1.5L (Novozymes A/S, Bagsvaerd, Denmark). The crude enzyme was desalted on a 50 mL gel filtration column (TOYOPEARL® HW-40S: TOSOH Corporation, Tokyo, Japan), which was equilibrated with 20 mM potassium phosphate buffer pH 7.0. The protein concentration of each fraction was measured with a Protein Assay kit (Bio-Rad Laboratories Inc., Hercules, CA, USA), and the fractions containing protein were mixed and injected to a 150 mL anion exchange column (TOYOPEARL® DEAE-650S), which was equilibrated with the same buffer. Proteins were eluted with a linear gradient of 300 mM potassium chloride. After measurement of protein concentration, the hydrolytic activity was measured, and the purity of each fraction was estimated by SDS-PAGE. The fractions containing enzyme of approximately 60 kDa, which showed activity towards p NPL, were collected and mixed with an equal volume of 2.0 M ammonium sulfate solution. The enzyme solution was injected into a 70 mL hydrophobic interaction column (TOYOPEARL® Phenyl-650S), which was equilibrated with 20 mM sodium acetate buffer pH 5.0 containing 1.0 M ammonium sulfate. The enzyme was eluted with a linear reverse-gradient of ammonium sulfate. The protein concentration, activity towards p NPL and purity of each fraction were analysed by the same methods as above, and the enzyme was collected. The buffer solution of the enzyme was changed to 20 mM sodium acetate buffer pH 5.0 by ultra-filtration, and the enzyme solution was filtered through a 0.1 μm PVDF membrane (Ultrafree Centrifugal Filters: Merck-Millipore, Germany). Enzyme concentration was calculated from the absorbance at 280 nm. Molar absorbance coefficient of Tr Cel7A WT was 83,370 M -1 •cm -1 . The tryptophan mutant enzymes were purified similarly from culture mediums, but the value of molar absorbance coefficient was in each case was taken to be 77,680 M -1 •cm -1 .
Comparison of cellulose degradation activities.
1.0 µM Tr Cel7A WT and W38A were incubated with 0.1 % phosphoric acid swollen cellulose (PASC), crystalline cellulose I α and crystalline cellulose III I in 50 mM sodium acetate buffer pH 5.0 at 30 ºC. After 5, 10, 15, 30, 60, 90, and 120 min incubation, the reaction mixtures were filtered through a 0.22 µm PVDF membrane, and the concentrations of soluble products were measured by HPLC (series 2000, JASCO Corporation, Japan). The products were separated on a Shodex NH2P-4E column (Showa Denko K.K., Tokyo, Japan) with a linear gradient of water and acetonitrile. 20) The amounts of glucose and cellooligosaccharides were quantified with a Corona CAD detector (Thermo Fisher Scientific Inc., Waltham, MA, USA), based on a standard curve for each oligosaccharide. Plots of cellobiose concentration were fitted by the equation:
where a , b , and c are constants. Cellulosic substrates were prepared as described in a previous report. 7) The amounts of free enzymes in reaction mixtures were quantified by HPLC with a DEAE-5PW column (TOSOH Corporation) and photodiode array detector (JASCO Corporation). The amount of remaining cellulose was estimated from the amounts of products. Processivity of enzymes were compared by the ratio of even/odd length of products. 12)
Time course of cello-oligosaccharide hydrolysis.
200 μM cello-oligosaccharides (C4 to C6) were incubated with 0.1 μM enzyme in 50 mM sodium acetate buffer pH 5.0 at 30 ºC. After 5, 30, 60, 90, and 120 min incubation, the reaction mixture was subjected to HPLC. The plots of substrate concentration vs time were fitted to single exponential decay:
wher a and b are constants. Additionally plots of products were fitted to single or double exponential equations:
or
where a , b , c and d are constants.
Determination of kinetic parameters of cello-oligosaccharides.
Various concentrations of cello-oligosaccharides (C4 to C6) were incubated with the enzymes in 20 mM sodium acetate buffer pH 5.0 for 30 min at 30 ºC. 0.0125 μM enzyme was incubated with 10 μM and 25 μM substrate, and 0.025 μM enzyme was incubated with 50 to 100 μM substrate. 150 μM or more of each substrate was hydrolyzed with 0.05 μM enzyme. The methods of separation and quantification of products were as described above. All plots were fitted by the global fitting program packaged in Igor Pro 6 (WaveMetrics, Portland, OR, USA) under the restrictions calculated from the reaction models. The values of individual parameters were shared among the equations during fitting. The equations used are shown in Supplementary material. Details of the calculation of parameters are also available as Supplementary data.
Calculation of the interaction energy and the effects of tryptophan mutations of Tr Cel7A.
Molecular dynamics (MD) simulations were first conducted for structure sampling as described previously, 4) then the MM/PBSA method was applied to the obtained trajectories as follows. The initial coordinates of the Tr Cel7A catalytic domain with cellononaose were taken from Protein Data Bank (PDB) structure 8CEL (3). The protonation states at pH 7.0 were determined using the PDB2PQR server 21) (except for Glu212 as a nucleophile). The systems were first solvated by the 3D-RISM method 22) (including crystal water), then fully solvated with an explicit solvent box, and counterions were added to neutralize the system. All the MD calculations were performed using the AMBER 12 package. 23) The AMBER ff99SB force field was used for proteins, the general AMBER force field (GAFF) with the RESP partial charges 24) based on HF/6-31G(d) level calculation (Gaussian09 25) ) for the N-terminal pyroglutamic acid, the GLYCAM_06h force field for cellononaose, and the TIP3P model for water molecules. The systems were energetically minimized for 5,000 steps, and equilibrated for 2.0 ns with gradually reducing restraints. Then, two 30-ns production runs were performed with different initial velocities. The temperature and pressure were controlled at 298 K and at 1 bar using the Berendsen rescaling method, 26) and the long-range electrostatic forces were calculated using the particle mesh Ewald method 27) . Finally, using the last 10 ns trajectories, the interaction energy of cellulose chain at the subsites (−7 to +2) of Tr Cel7A were calculated by the MM/PBSA method, and the effects of mutation of each tryptophan (Trp40, Trp38, Trp367, and Trp376) to alanine on the binding free energy were estimated using the alanine-scanning module implemented in the MM-PBSA method.
CONFLICTS OF INTEREST
The authors declare no conflict of interests.
Acknowledgments
We thank Riitta Nurmi (VTT) for skilful technical assistance in Trichoderma transformations. The authors are grateful for Grants-in-Aid for Scientific Research (B) (18H02252 and 19H03013 to K.I. and 19H03094 to A. N.) from the Japan Society for the Promotion of Science (JSPS), a Grant-in-Aid for Innovative Areas from the Japanese Ministry of Education, Culture, Sports, and Technology (MEXT) (No. 18H05494 to K.I.) and Advanced Technology Institute Research Grants 2015 (RG2709 to A.N.) and Leading Initiative for Excellent Young Researchers program (A.N.). In addition, K.I. thanks Business Finland (BF, formerly the Finnish Funding Agency for Innovation (TEKES)) for support via the Finland Distinguished Professor (FiDiPro) Program “Advanced approaches for enzymatic biomass utilization and modification (BioAD)”.
References
- 1).Arai M., Sakamoto R., and Murao S.: Different action by two avicelases from Aspergillus aculeatus . Agric. Biol. Chem. Tokyo,, 53, 1411–1412 (1989). [Google Scholar]
- 2).Henrissat B.: A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J., 280, 309–316 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Divne C., Stahlberg J., Teeri T.T., and Jones T.A.: High-resolution crystal structures reveal how a cellulose chain is bound in the 50 Å long tunnel of cellobiohydrolase I from Trichoderma reesei . J. Mol. Biol., 275, 309–325 (1998). [DOI] [PubMed] [Google Scholar]
- 4).Nakamura A., Tsukada T., Auer S., Furuta T., Wada M., Koivula A., Igarashi K., and Samejima M.: the tryptophan residue at the active site tunnel entrance of Trichoderma reesei cellobiohydrolase Cel7A is important for initiation of degradation of crystalline cellulose. J. Biol. Chem., 288, 13503–13510 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Igarashi K., Koivula A., Wada M., Kimura S., Penttila M., and Samejima M.: High speed atomic force microscopy visualizes processive movement of Trichoderma reesei cellobiohydrolase I on crystalline cellulose. J. Biol. Chem., 284, 36186–36190 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Igarashi K., Uchihashi T., Koivula A., Wada M., Kimura S., Okamoto T., Penttila M., Ando T., and Samejima M.: Traffic jams reduce hydrolytic efficiency of cellulase on cellulose surface. Science, 333, 1279–1282 (2011). [DOI] [PubMed] [Google Scholar]
- 7).Nakamura A., Watanabe H., Ishida T., Uchihashi T., Wada M., Ando T., Igarashi K., and Samejima M.: Trade-off between processivity and hydrolytic velocity of cellobiohydrolases at the surface of crystalline cellulose. J. Biol. Chem., 136, 4584–4592 (2014). [DOI] [PubMed] [Google Scholar]
- 8).Kari J., Olsen J., Borch K., Cruys-Bagger N., Jensen K., and Westh P.: Kinetics of cellobiohydrolase (Cel7A) variants with lowered substrate affinity. J. Biol. Chem., 289, 32459–32468 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Horn S.J., Sørlie M., Vårum K.M., Valjamae P., and Eijsink V.G.H. (2012) Measuring processivity. Methods Enzymol., 510, 69–95. [DOI] [PubMed] [Google Scholar]
- 10).Kurasin M. and Valjamae P.: Processivity of cellobiohydrolases is limited by the substrate. J. Biol. Chem., 286, 169–177 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Harjunpaa V., Helin J., Koivula A., Siika-aho M., and Drakenberg T.: A comparative study of two retaining enzymes of Trichoderma reesei transglycosylation of oligosaccharides catalysed by the cellobiohydrolase I, Cel7A, and the β-mannanase, Man5A. FEBS Lett., 443, 149–153 (1999). [DOI] [PubMed] [Google Scholar]
- 12).Kari J., Kont R., Borch K., Buskov S., Olsen J.P., Cruyz-Bagger N., Valjamae P., and Westh P.: Anomeric selectivity and product profile of a processive cellulase. Biochemistry, 56, 167–178 (2017). [DOI] [PubMed] [Google Scholar]
- 13).Michaelis L. and Menten M.L.: Die Kinetik der Invertinwirkung. Biochem. Z., 49, 333–369 (1913). [Google Scholar]
- 14).Johnson K.A. and Goody R.S.: The original Michaelis constant: Translation of the 1913 Michaelis-Menten paper. Biochemistry, 50(39), 8264–8269 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Vrs̆anská M. and Biely P.: The cellobiohydrolase I from Trichoderma reesei QM 9414: action on cello-oligosaccharides. Carbohydr. Res., 227, 19–27 (1992). [Google Scholar]
- 16).Divne C., Stahlberg J., Reinikainen T., Ruohonen L., Pettersson G., Knowles J.K., Teeri T.T., and Jones T.A.: The three-dimensional crystal structure of the catalytic core of cellobiohydrolase I from Trichoderma reesei. Science, 265, 524–528 (1994). [DOI] [PubMed] [Google Scholar]
- 17).Johnson K.A.: A century of enzyme kinetic analysis, 1913 to 2013. FEBS Lett., 587, 2753–2766 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18).Kont R., Kari J., Borch K., Westh P., and Valjamae P.: Inter-domain synergism is required for efficient feeding of cellulose chain into active site of cellobiohydrolase Cel7A. J. Biol. Chem., 291, 26013–26023 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Nakamura A., Tsukada T., Auer S., Furuta T., Wada M., Koivula A., Igarashi K., and Samejima M.: The tryptophan residue at the active site tunnel entrance of Trichoderma reesei cellobiohydrolase Cel7A is important for initiation of degradation of crystalline cellulose. J. Biol. Chem., 288, 13503–13510 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).Igarashi K., Ishida T., Hori C., and Samejima M.: Characterization of an endoglucanase belonging to a new subfamily of glycoside hydrolase family 45 of the basidiomycete Phanerochaete chrysosporium . Appl. Environ. Microbiol., 74, 5628–5634 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Dolinsky T.J., Nielsen J.E., McCammon J.A., and Baker N.A.: PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res., 32, W665–W667 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).Sindhikara D.J., Yoshida N., and Hirata F.: Placevent: An algorithm for prediction of explicit solvent atom distribution—Application to HIV‐1 protease and F‐ATP synthase. J. Comp. Chem., 33, 1536–1543 (2012). [DOI] [PubMed] [Google Scholar]
- 23).Case A.D., Darden AT., Cheatham III E.T.. et al.: AMBER 12University of California, San Francisco: (2012). [Google Scholar]
- 24).Bayly C.I., Cieplak P., Cornell W., and Kollman P.A.: A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem., 97, 10269–10280 (1993). [Google Scholar]
- 25).Frisch J.M., Trucks W.G., Schlegel B.H.. et al.: Gaussian 09. Gaussian, Inc., Wallingford, CT, USA: (2009). [Google Scholar]
- 26).Berendsen H.J C., Postma J.P.M., van Gunsteren W.F., DiNola A., and Haak J.R.: Molecular dynamics with coupling to an external bath. J. Chem. Phys., 81, 3684–3690 (1984). [Google Scholar]
- 27).Darden T., York D., and Pedersen L.: Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys., 98, 10089 (1993). [Google Scholar]
- 28).Rose I.A., Hanson K.R., Wilkinson K.D., and Wimmer M.J.: A suggestion for naming faces of ring compounds. Proc. Natl. Acad. Sci. USA, 77, 2439 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]