Abstract
Several diagnostic tools have been developed for clinical and epidemiological assays. RT-PCR and antigen detection tests are more useful for diagnosis of acute disease, while antibody tests allow the estimation of exposure in the population. Currently, there is an urgent need for the development of diagnostic tests for COVID-19 that can be used for large-scale epidemiological sampling. Through a comprehensive strategy, potential 16 mer antigenic peptides suited for antibody-based SARS-CoV-2 diagnosis were identified. A systematic scan of the three structural proteins (S,N and M) and the non-structural proteins (ORFs) present in the SARS-CoV-2 virus was conducted through the combination of immunoinformatic methods, peptide SPOT synthesis and an immunoassay with cellulose-bound peptides (Pepscan). The Pepscan filter paper sheets with synthetic peptides were tested against pools of sera of COVID-19 patients. Antibody recognition showed a strong signal for peptides corresponding to the S, N and M proteins of SARS-CoV-2 virus, but not for the ORFs proteins. The peptides exhibiting higher signal intensity were found in the C-terminal region of the N protein. Several peptides of this region showed strong recognition with all three immunoglobulins in the pools of sera. The differential reactivity observed between the different immunoglobulin isotypes (IgA, IgM and IgG) within different regions of the S and N proteins, can be advantageous for ensuring accurate diagnosis of all infected patients, with different times of exposure to infection. Few peptides of the M protein showed antibody recognition and no recognition was observed for peptides of the ORFs proteins.
Keywords: SARS-CoV-2, COVID-19, Antigenic peptides, Synthetic peptide, SPOT technique, Pepscan
1. Introduction
Coronavirus disease (COVID-19) is the infectious disease associated with the novel coronavirus, named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (“Q&A on coronaviruses (COVID-19)”, n.d.). Since its emergence in December 2019 in Wuhan, China, the virus has infected around 100 million people and around 2 million deaths until January 2021 (“Home - Johns Hopkins Coronavirus Resource Center”, 2021). SARS-CoV-2 is the third highly pathogenic coronavirus to pass from an animal reservoir to infect humans in the 21st century: the emergence of recent outbreaks has proved that these viruses can mutate or recombine to become pathogenic and cross the species barriers. Since genetic changes are inevitable and a part of the evolutionary process, these viral outbreaks will keep on emerging (Rabaan et al., 2020). The high infectivity of SARS-CoV-2 and the worldwide spread of this ongoing outbreak highlight the urgent need for the development of effective diagnostic methods, therapeutics and vaccines.
SARS-CoV-2 is a spherical positive single-stranded RNA virus formed primarily of structural proteins currently known as spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins and a number of non-structural and accessory proteins, whose open reading frames (ORFs) localize between S and E genes, or between M and N genes. These non-structural proteins vary widely among different coronavirus species (Ashour et al., 2020; Yuan et al., 2005).
Several diagnostic tools have been developed for clinical and epidemiological use (Yüce et al., 2021). RT-PCR and antigen detection tests are more useful for diagnosis of acute disease, while antibody tests allow the estimation of exposure in the population. Mostly the viral Nucleoprotein (N) and the Spike (S) proteins, or part of them, have been used as antigens for antibody detection in infected or convalescent individuals.
Currently there is an urgent need for the development of serological tests for COVID-19 as diagnostic tools, playing a mayor role mitigating the coronavirus pandemic and a key step in returning to normality (Guglielmi, 2020).
The choice of antigen remains as the key component of the different immunodiagnostic tests (Noya et al., 2005). The identification of antigenic/immunogenic regions in antigenic proteins is a key step for the diagnosis of infectious diseases and for vaccine development. By contrast, the production and use of native antigens for diagnostic tests and vaccine assays is frequently hindered by variation in species and leads to high costs for purification, especially in vaccines production (Chen et al., 2011; Gomara and Haro, 2007; Van Regenmortel, 2006). In addition, some cross reactivity has been described in the immune diagnosis of SARS-CoV-2 infection (Latiano et al., 2020). The use of synthetic peptides may reduce great part of these problems. To date, there are few reports on the diagnostic value of protein-derived peptides of the SARS-CoV-2 virus using microarrays (Li et al., 2021; Musicò et al., 2021; Wang et al., 2020). Several peptides corresponding to the Spike (Li et al., 2021) and N (Musicò et al., 2021) proteins that exhibit potential diagnostic performance were identified for detecting SARS-CoV-2 IgG or IgM. The aim of this study is to evaluate the use of synthetic antigenic peptides for serological diagnostic tests in an effort to contribute to this important topic.
For this purpose, a systematic scan of the three structural proteins (S, N and M) and the non-structural proteins (ORFs) present in the SARS-CoV-2virus was conducted through computational methods to predict the potential antigenic regions (B-cell epitopes). Selected regions were synthesized using the SPOT technique and evaluated with the three most important classes of immunoglobulins (IgG, IgM and IgA) by Pepscan. The most antigenic peptides (spots) are the top candidates for the diagnosis of COVID-19.
2. Methods
2.1. Data source
The reference genome used throughout this study is from the severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1 (GenBank: MN908947.3). The protein sequences Spike protein (S) (QHD43416.1), nucleocapsid phosphoprotein (N) (QHR63298.1), membrane glycoprotein (M) (QHD43419.1) and the non-structural proteins ORF1ab (QHD43415), ORF3a (QHD43417.1), ORF6 (QHD43420.1), ORF7a (QHD43421), ORF8 (QHD43422.1) and ORF10 (QHI42199.1) were retrieved from NCBI GenBank database.
2.2. B-cell epitope prediction for SARS-CoV-2
Linear B cell epitope predictions were carried out on the spike glycoprotein (S), nucleocapsid phosphoprotein (N), membrane glycoprotein (M) and ORFs proteins (1ab, 3a. 6, 7a, 8 and 10) using several algorithms: Antheprot (stand-alone) (Deléage, 2012), SVMTrip (Yao et al., 2012), ABCPred (Saha and Raghava, 2006), Kolaskar&Tongaonkar Antigenicity (Kolaskar and Tongaonkar, 1990), Bebipred 2.0 (Jespersen et al., 2017) and Ellipro(Ponomarenko et al., 2008). The parameters are all set as default.
For prediction of discontinuous or conformational B-cell epitopes, coordinates of experimental structures of SARS-CoV-2 proteins were downloaded from the PDB. In case multiple structures were available, the structure with highest sequence coverage and highest resolution were selected. Conformational B-cell epitopes were predicted with Ellipro(Ponomarenko et al., 2008) using the 3D structures of the spike glycoprotein (S) (PDB 6VSB, 6M0J), the RNA binding domain of the N-terminal region (PDB: 6VYO) of the N protein. Where no PDB structure was available, high confidence I-Tasser (“Modeling of the SARS-COV-2 Genome using I-TASSER”, n.d.) models were used instead. Structural visualization was performed with Biovia Discovery Studio Visualizer 17.2.0 software.
In the case of spike protein (S) the crystallized structures were used to locate the 16 mer peptides and to adjust the epitopes. Additional characteristics like surface accessibility and hydrophobicity (GRAVY index < 0) were taken into account to obtain an initial peptide library. The library was enriched with peptides where some mutations were included (Cys → Ser). Seventy-five peptides were selected to be manually synthesized using the Spot technique.
For N and M proteins an initial library of overlapping peptides was built. Every peptide was 16 amino acid residues in length, offset by four and overlapped by eleven residues, obtaining a total of 135 peptides. Then, the sequences were reduced after elimination of transmembrane and highly hydrophobic peptides (GRAVY index > 0). Also, CBTope (Ansari and Raghava, 2010) was used to predict conformational B cell epitopes using primary sequence. Linear and conformational epitopes where inspected taking into account additional characteristics like Chou Fasman beta-turn prediction (Chou and Fasman, 2006), Emini Surface Accessibility Prediction (Emini et al., 1985), and Karplus&Schulz Flexibility Prediction (Karplus and Schulz, 1985). Additionally, IUPred2A (Mészáros et al., 2018) was used to identify disordered regions and RADAR (Heger and Holm, 2000) for repetitive regions. Then, consensus regions between the different predictors used were determined. A total of 73 peptides data set was obtained for these proteins.
For the ORF proteins (1ab, 3a, 6, 7a, 8 and 10) the same approach described for the N and M proteins was used. Consensus regions where determined and a final 58 peptides data set was obtained.
2.3. Solid phase peptide synthesis on cellulose paper (SPOTs array assay)
The procedure previously described was used with some modifications(Winkler et al., 2009): The selected peptides were synthesized in spots on filter paper Whatman® grade 540 as a solid support (rectangle 12 × 7 cm), functionalized with a solution of Fmoc-βAla-OH, (0.960 g, 3.0 mmoles), N,N′-diisopropylcarbodiimide (DIC) (560 μL, 3.6 mmoles) and N-methylimidazole (475 μL, 6.0 mmoles) in 25 mL of N-methylpirrolidine (NMP) overnight without agitation. Stock solutions of 0.33 M Fmoc-L-aminoacids and 0.5 M of 1-hydroxibenzotriazole (HOBt) were prepared in dry NMP. Aliquots of 100 μL of these solutions were activated by the addition 33 μL of a solution 1.1 M DIC in NMP to use during the whole day, after that the aliquots were discarded. The Fmoc group deprotection was achieved using a solution of 20% 4-methylpiperidine in NMP and the synthesis was monitored using a 0.02% bromophenol blue solution in ethanol. The final side chain deprotection was carried out in two steps, first by treatment with a cleavage solution I: 90% trifluoroacetic acid (TFA), 5% water 3%, triisopropylsilane (TIPS), 1% phenol and 1% dichlorometane (DCM) for 30 min; followed by a cleavage solution II: 50% TFA, 2% water, 3% TIS, 1% phenol and 44% DCM for 3 h. Both cleavages were performed without agitation. The reagents were from Iris Biotech and Merck Millipore, Germany.
2.4. Serum samples and ethical clearance
This retrospective study included sera from adult patients and healthy volunteers. Serum samples (n = 10) were obtained from COVID-19 patients confirmed by rapid test (Wondfo® and Avioq, Bio-tech) and/or RT-qPCR (Sansure®) and considered positive if reactive by two independent test. These sera were grouped in two pools of 5 sera each (pool 1 and pool 2). The days since the onset of the symptoms were collected from medical records. All patients were in the acute phase of infection and had a maximum of 30 days of evolution. Control non-SARS-CoV-2 samples (n = 5) anonymous stored residual serum samples collected in 2008 were used.
This research project was approved by the Independent Bioethics Committee for Research of the National Center for Bioethics (CIBI-CENABI, Venezuela). Serum samples were provided by patients who accepted to participate in this study and signed the Informed Consent.
2.5. Screening of SPOT membranes (Pepscan)
The dry paper with the different “spots” of peptides was blocked with phosphate buffer saline, pH 7.4 with 0.05% Tween 20 (PBS-Tween) and 5% non-fat dry milk and tested with two pools of five patients' sera with a final dilution of 1:200. Anti-Human Polyvalent Immunoglobulins (G,A,M), Anti-human IgG, anti-human IgA and anti-human IgM- horseradish-peroxidase (Sigma) were used at 1:30.000, 1:30.000, 1:3.000 and 1:10.000, respectively, as conjugates and revealed with a solution of SuperSignal™ West Pico PLUS. The chemiluminescence of the spots was measured using the ChemiDoc™ Imaging Systems equipment and the images obtained were processed with the ImageLab program (BioRad).
2.6. Regeneration of the membranes
The removal of bound proteins was carried out in two steps. In the first step the membranes were treated with the regeneration buffer I (solution 8 M of urea with 1% SDS, 3 times for 10 min). Then, they were treated with regeneration buffer II (solution of 100 mL of AcOH (99%), 500 mL of EtOH and 400 mL of water, 3 times for 10 min) both at room temperature. Finally, they were washed at least five times with EtOH and dried on air. At this point, the membranes were used immediately or stored at −80 °C until use. The regeneration was checked blocking the membranes and incubating with the corresponding enzyme-conjugate to ensure no spots remained. The regeneration process was carried out 3 times after the evaluation of antigenicity. In some cases, when it was not successful, the regeneration times using buffer I were increased from 1 h to 24 h.
3. Results and discussion
In the present study, our aim was the identification of potential antigenic peptides well suited for antibody-based SARS-CoV-2 diagnosis. First, we developed an immunoinformatic strategy to obtain a 16 mer peptides data set of 206 peptides of proteins S, N, M and ORFs.
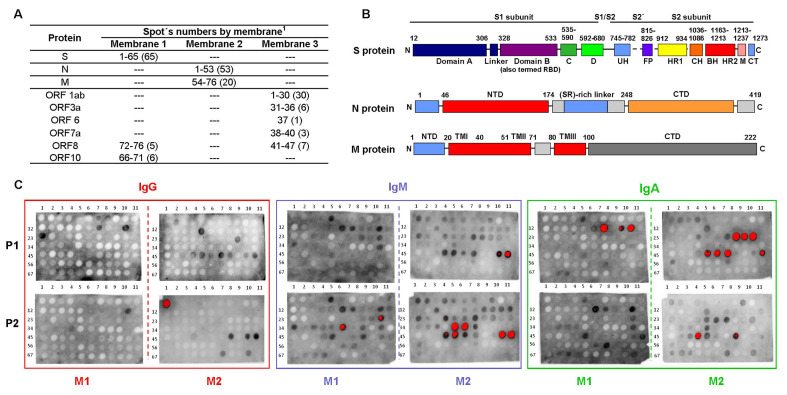
To experimentally determine the antigenicity of the selected peptides, the 206 sequences were synthesized on functionalized cellulose papers in a spot manner. Three membranes were prepared using the SPOT technique and identified as 1, 2 and 3. Each membrane contains 76 peptides (spots) covalently anchored to the cellulose paper, which was previously functionalized with the amino acid residue β-alanine. Additionally, a spacer was added using the same amino acid for C-terminus functionalization. Each membrane presents spots repeats as positive controls. All synthesized peptides were acetylated at the N-terminus to eliminate the non-physiological positive charge that would otherwise be present on the α-amino group of the N-terminal residue of an internal peptide sequence(Carter and Loomis-Price, 2004). Fig. 1A shows the distribution of the peptides (spots) of the proteins studied in each membrane.
Fig. 1.
Protein of SARS-CoV-2 potential antigenic peptides tested by the Pepscan technique. (A) Peptides (spots) distribution of three structural and non-structural proteins of SARS-CoV-2. (B) Domain structure of the SARS-CoV-2 S, N and M proteins. (C) Peptide reactivity against pools of sera. The increasing reactivity of the peptides in the Pepscan spots, rises from gray to black and when it is very intense it turns red. In each block the upper pair shows the results for sera pool 1(P1) and the lower for pool 2 (P2). Reactivity (absent) to peptides included in Membrane 3 is not shown.
Membranes were exposed to two different pools (pool 1 and pool 2) of five COVID-19 patients´ sera that were highly reactive, moderately reactive and slightly reactive to Wondfo® y Avioq rapids test (Table 1 ). The serum samples were collected between March and April 2020 from adult volunteers during the acute phase of infection, seven men and three women.
Table 1.
Rapid test results Wondfo®, Avioq® Bio-Tech and RT-qPCR of COVID-19 patients' sera.
| Pool | Serum COVID-19 |
Rapid test |
RT-qPCR (Sansure®) |
||
|---|---|---|---|---|---|
| Wondfo® | Avioq® (Bio-tech) |
||||
| IgM | IgG | ||||
| 1 | 184 | + | + | ++ | nd |
| 179 | ++ | + | ++ | + | |
| 242 | ++ | +++ | +++ | + | |
| 178 | +++ | +++ | +++ | + | |
| 185 | +++ | +++ | +++ | nd | |
| 2 | 4 | + | − | − | + |
| 9 | + | − | − | + | |
| 161 | + | +++ | +++ | nd | |
| 170 | ++ | +++ | +++ | nd | |
| 194 | +++ | f+ | + | + | |
“+” positive result; number of “+” denotes relative increase in positive band intensity in the strip tests. “f+” stands for faint-positive. “−” negative result. “nd” not determined.
Membranes were evaluated for the three most important classes of immunoglobulins (IgG, IgA and IgM). A regeneration process was carried out and checked for each membrane before using different immunoglobulins, to ensure the removal of primary and secondary antibodies, and thus avoiding false positives during the assays.
When exposed to pools of positive COVID-19 sera, antibody recognition showed a strong signal for peptides corresponding to the S, N and M proteins of SARS-CoV-2 virus, but not for the ORFs proteins. Fig. 1C shows peptides reactivity against pools of sera (since no reactivity to ORFs peptides was observed, membrane 3 is not shown). This methodology was successfully used and allowed us to quickly identify antigens based on synthetic peptides. In general, higher IgA and IgM and reactivity was observed, compared to the IgG one. Regardless of the pool used, a higher number of peptides were recognized by IgM immunoglobulin, followed by IgA recognition. Few peptides (spots) were recognized by IgG, as expected. The serum samples used in the present study were collected from patients with COVID-19 at early stages of the viral infection (acute-phase) when IgM and IgA immunoglobulins are abundant, while IgG is the less abundant circulating antibody in the initial phase of infection. It has been reported that the level of IgG antibody reached the highest concentration on day 30, while the highest concentration of IgM antibody appeared on day 18 but then began to decline (Hueston et al., 2020; Qu et al., 2020). This might be one of the reasons for this differences in the isotypic responsiveness to the different peptides. The results obtained for each of the aforementioned proteins is described below.
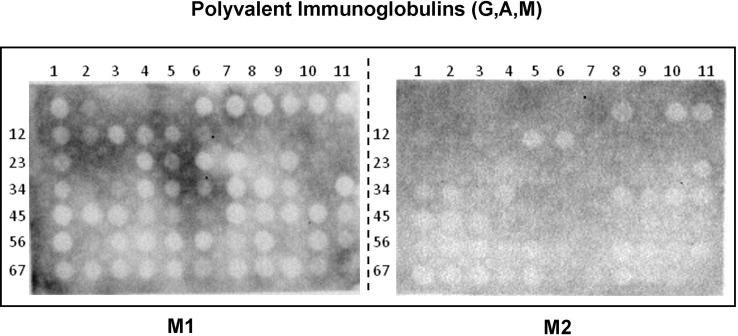
Membranes were also exposed to a pool of five non-COVID-19 patients´ sera, that were collected in 2008. For these assays a commercial mixture of total anti-immunoglobulins was used. No significant antibody recognition was observed Supplementary Fig. S1.
Supplementary Fig. S1.
Fig. S1 Protein of SARS-CoV-2 potential antigenic peptides tested by the Pepscan technique. The results for Membrane 1 and 2 when exposed to a sera pool of five non-COVID-19 patients are shown
3.1. Spike glycoprotein (S)
The spike glycoprotein (S) forms homotrimers protruding the viral surface and contains a S1 subunit, comprising the receptor binding domain (RBD) that mediates the binding to host cell ACE2 receptor, and the S2 subunit, which contains the fusion peptide and functions in membrane fusion. Spike protein has become the major target of antibodies responses, particularly the RBD domain(Barnes et al., 2020; Walls et al., 2020; Wrapp et al., 2020).
To select candidate diagnostic peptides, we performed predictions for linear B cell epitopes with Bepipred 2.0 and Ellipro, and for conformational epitopes with Ellipro and Discotope 2.0. To predict and map conformational B cell epitopes, we used the SARS-CoV-2 spike glycoprotein structures (PDB: 6VSB, 6MOJ). Several physicochemical characteristics, such as gravy index and hydrophobicity, were also considered.
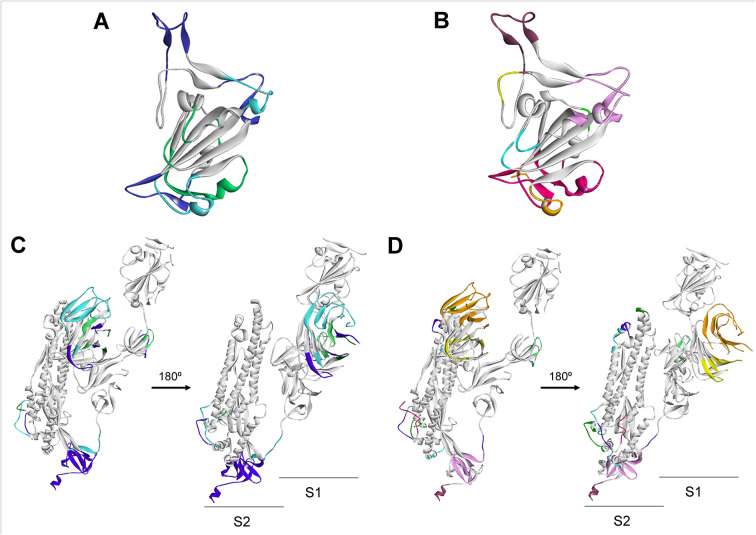
Then, the relevant amino acid positions were localized onto the model structures. The X-ray structure of the RBD domain of the S protein (PDB 6M0J) allowed us to map the epitopes and to determine the final 16 mer sequences for peptide synthesis (Fig. 2A, B). The x-ray structure of the S protein spike in the prefusion conformation (PDB 6VSB) was used to map the predicted epitopes and to determine the sequences of the peptides to be synthesized by the spot technique (Fig. 2C, D). Bepipred and Ellipro results when mapped on the 3D structures show the overlap between linear and conformational epitopes.
Fig. 2.
B Cell Immunodominant regions predicted for the SARS-CoV-2 Spike Glycoprotein. The corresponding B cell epitopes are shown in colors and mapped as predicted with Ellipro. Linear (A) and conformational (B) epitopes on the RBD domain. Linear (C) and conformational (D) epitopes on the monomer structure.
This analysis allowed us to select peptides that are clearly exposed. Three of the regions selected are located in the S1 subunit in the NTD and RBD domains, whereas others are in the S2 subunit.
Additionally, we used the SARS-CoV spike protein structure (PDB:6CRZ) to predict epitopes and to select the sequences to be synthesized (Data not shown).
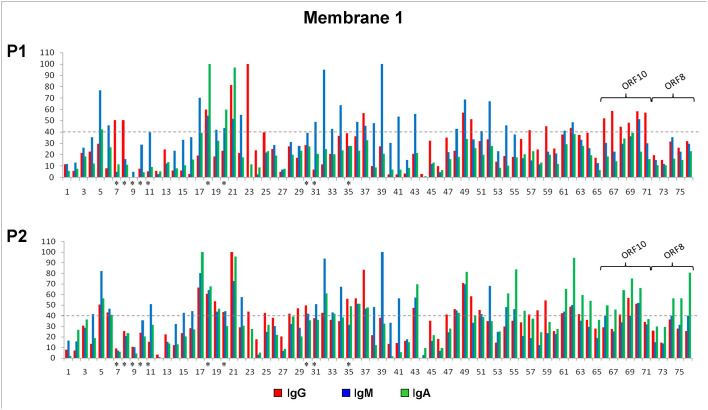
To determine the antigenicity of the selected peptides of the S protein, these were synthesized on functionalized cellulose papers in a spot manner (Fig. 1A) and exposed to two different pools of COVID-19 patients´ sera, as previously described. Antibody recognition was observed for several spots (Fig. 1C, M1 and Fig. 3 ). The sequences with higher signal intensity are localized in domains S1A (spot 5), RBD (spots 39, 41, 43, 49, 52, 55, 62; spot 62 was added as repetition of 55 for chemical control), and S1C (spots 17, 21, 22,23, 54, 61; spot 61 was added as repetition of 54 for chemical control), within the S1 subunit. At the S2 subunit, three sequences showed strong signal intensity (spot 32, 34 and 37). Other spots showed slight antibody recognition.
Fig. 3.
Antigenicity of the selected peptides of proteins Spike (S) ORF8 and ORF10 present in the SARS-CoV-2 virus. Volume tools were used to quantify the signal intensity of the Pepscan spots. The relative quantity is the adjusted volume normalized with the higher adjusted volume. The adjusted volume was established subtracting the smallest volume. The cutoff value is shown by the dotted line (denotes a positive result) and was based on the ability to see the spots on the membranes. Asterisks denote peptides of the SARS-CoV spike protein.
Serum antibody responses against the peptides (spots) with higher signal intensity can be characterized in more detail. Several peptides (spots 5, 17, 18, 20, 21, 22, 32, 34, 39, 41, 43, 49, 52, 54 and 62. Fig. 1C, M1 and Fig. 3) were strongly recognized by IgM and IgA immunoglobulins. Those are localized along the different antigenic regions at the S1 and S2 subunits and RBD domain. Peptides 5 and 17 (localized in domains S1A and S1C, respectively) showed strong IgM and IgA recognition with the two pools of sera. Peptides 43, 49 and 62, localized in the RBD domain, only showed a strong recognition with pool 2. Peptides 22, 34, 39, 41 and 52, showed recognition by the IgM immunoglobulin with both pool's of COVID-19 patients´ sera. These peptides are localized mainly in the RBD domain of the spike protein, with the exception of peptides 22 and 34, found in the S1C and S2 subunit, respectively. Among all evaluated peptides, IgG recognition was achieved by peptide 23, located at the S1C domain and peptide 37, located at de S2 domain. Spot 23 showed a strong signal when exposed to pool 1 of sera but a slight antibody recognition with pool 2. Only two peptides (spots 18 and 21, Fig. 1C, M1 and Fig. 3) were strongly recognized by all three IgG, IgM and IgA immunoglobulins. It is worth to mention that spots 18 and 20, corresponds to a selected sequence of the S protein of the SARS-CoV virus. Those peptides share 75% of sequence identity and are located in the same region when mapped in the structures of the corresponding proteins (PDB: 6VSB and 6CRZ, S proteins of SARS-CoV-2 and SARS-CoV, respectively. Data not shown).
Our analysis also considered several sequences of the SARS-CoV spike protein (Table 2 ). With the exception of spot 18 and 20, when those peptides were exposed to the two pools of COVID-19 patients´ sera, no significant antibody recognition was observed.
Table 2.
Synthetic peptides (spots) of S protein of SARS-CoV-2 and their antibody recognition by the pools of COVID-19 patients' sera.
| Spot (peptide) |
Region | IgG | IgM | IgA |
|---|---|---|---|---|
| 5 | S1A | −/+ | +++/+++ | +/++ |
| 17 | S1C | −/+++ | +++/+++ | −/+++ |
| 18* | S1C | ++/++ | +/++ | +++/++ |
| 20* | S1C | −/− | −/− | ++/− |
| 21 | S1C | +++/+++ | +/+++ | +++/+++ |
| 22 | S1C | −/− | ++/++ | −/− |
| 23 | S1C | +++/− | −/− | −/− |
| 32 | S2 | −/− | +++/+++ | −/++ |
| 34 | S2 | −/− | ++/++ | −/− |
| 37 | S2 | ++/+++ | −/− | −/− |
| 39 | RBD | −/− | +++/+++ | −/− |
| 41 | RBD | −/− | ++/++ | −/− |
| 43 | RBD | −/− | ++/++ | −/++ |
| 49 | RBD | ++/+++ | +++/+++ | −/+++ |
| 52 | RBD | −/− | +++/+++ | −/− |
| 54 | RBD | −/− | −/− | −/++ |
| 62 | RBD | −/− | −/− | −/+++ |
Results Pool 1/Pool 2. “+” denotes positive result; number of “+” signifies relative increase in positive spot intensity in the membrane; “−” denote negative result. Chemical control spots, 61 and 62, are repetition of peptides 54 and 55. ⁎ Peptides of SARS-CoV.
3.2. Nucleocapsid phosphoprotein (N)
Protein N (McBride et al., 2014) is frequently used structural protein for immunodiagnostic of SARS-CoV-2, it is produced at very high levels in the virus infected cells and is considered a good candidate for clinic diagnosis (Li et al., 2020). The N protein is a highly immunogenic, antigenic and abundantly expressed protein during infection, capable of inducing protective immune responses against SARS-CoV and SARS-CoV-2 (Kang et al., 2020).
Potential B cell epitopes of the N protein were selected from a library of 82 linear peptides of overlapping sequences of 16 residues. These predicted peptides were then reduced to 71 by eliminating highly hydrophobic ones. To select the most antigenic peptides from the library, a collection of linear B-cell epitope predictors that utilize different algorithms were used (propensity scales, neural networks, among others). Once identified, as a consensus between the different predictors, these regions were located in the library helping us to eliminate those with less antigenic capacity. The final sequences were manually synthesized using the SPOT technique (Fig. 1C, M2).
The N protein have three distinct and highly conserved domains: an N-terminal RNA-binding domain (NTD), a C-terminal dimerization domain (CTD) which are separated by an intrinsically disordered central Ser/Arg (SR)-rich linker (Kang et al., 2020). To date, the 3D structures of the NTD and CTD domains of the SARS-CoV-2 nucleocapsid phosphoprotein are available. To complement our epitope prediction strategy, the available experimental and modeled structures were downloaded from the PDB (6VYO) and I-Tasser server (code QHD43423) and used for prediction of discontinuous or conformational B-cell epitopes (Fig. 4 ).
Fig. 4.
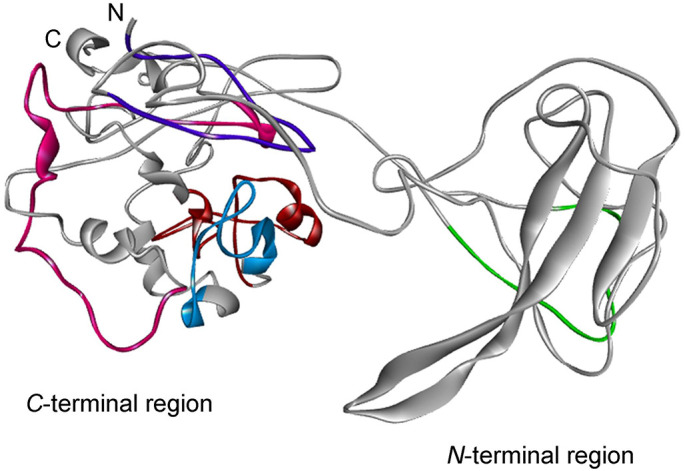
B Cell Immunodominant regions predicted for the Nucleocapsid phosphoprotein (N) of SARS-CoV-2. The corresponding B cell epitopes are shown in colors and mapped as predicted as a consensus of the methods used.
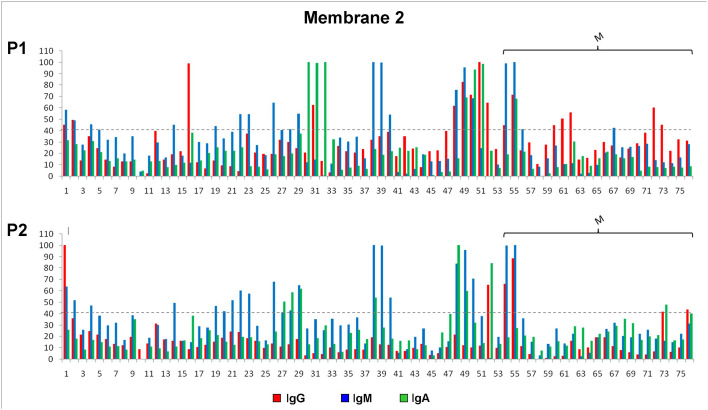
The immunoreactivity of the selected peptides of the N protein was evaluated against two different pools of COVID-19 patients´ sera through the Pepscan methodology (Fig. 1C, M2 and Fig. 5 ). The sequences with higher signal intensity are localized in three antigenic regions. The sequences with higher signal intensity are localized in the C-terminal (spots 48 to 52), SR-rich linker (spot 26) and CDT regions (spots 30–32, 38–39).
Fig. 5.
Antigenicity of the selected peptides of Nucleocapsid phosphoprotein (N) and Membrane glycoprotein (M) present in the SARS-CoV-2 virus. Volume tools were used to quantify the signal intensity of the Pepscan spots. The relative quantity is the adjusted volume normalized with the higher adjusted volume. The adjusted volume was established subtracting the smallest volume. The cutoff value is shown by the dotted line (denotes a positive result) and was based on the ability to see the spots on the membranes.
The sequences with higher signal intensity are localized in the C-terminal region of the N protein, represented by several peptides (spots 48 to 52, Fig. 1C, M2 and Fig. 5). These spots showed antibody recognition with different signal intensities when exposed with the two pool of sera. Nevertheless, peptide 52 showed anti-IgG positive strong recognition only with the pool 2 of sera, IgA recognition with the two pool of sera but no IgM recognition (Fig. 5 and Table 3 ). Peptides of this region were highly hydrophilic (GRAVY index of −2.2) when compared with the other peptides synthesized in membrane 2.
Table 3.
Synthetic peptides (spots) of structural N protein of SARS-CoV-2 and their antibody recognition by the pools of COVID-19 patients' sera.
| Spot (peptide) |
Region | IgG | IgM | IgA |
|---|---|---|---|---|
| 1 | N-terminal | −/+++ | ++/++ | +/+ |
| 2 | N-terminal | +/f+ | +/++ | ++/f+ |
| 16 | NTD | +++/− | f+/− | ++/+ |
| 26 | SR-rich linker | −/− | +/+++ | ++/f+ |
| 30 | CTD | −/− | ++/f + | +++/− |
| 31 | CDT | ++/− | +/+ | +++/f+ |
| 32 | CDT | −/− | +/f+ | +++/+ |
| 38 | CDT | −/− | +/+++ | −/++ |
| 39 | CDT | −/− | −/+++ | −/f+ |
| 48 | C-terminal | +/+ | +++/+++ | f+/+++ |
| 49 | C-terminal | ++/− | +++/+++ | +++/++ |
| 50 | C-terminal | +/− | +/+++ | +++/f + |
| 51 | C-terminal | ++/− | ++/+ | +++/− |
| 52 | C-terminal | +/++ | −/− | f+/+++ |
Results Pool 1/Pool 2. “+” denotes positive result; number of “+” signifies relative increase in positive spot intensity in the membrane; “f+” stands for faint-positive. “−” denote negative result.
Other regions outside the C-terminal fragment of the N protein exhibited strong IgG, IgM and IgA recognition (Fig. 5 and Table 3). One of these regions are localized in the SR-rich linker and CDT regions (spots 26, 30–32, 38–39, Figure1C, M2), and showed different immunoreactive response with the two pools of sera and with the all three IgG, IgM and IgA immunoglobulins (Fig. 5). In Table 3 it can be clearly observed there is no IgG recognition for any of the peptides, regardless of the pool evaluated, except for peptide 31 that showed IgG recognition. However, these peptides were strongly recognized by IgM immunoglobulin with the two pool of sera. These regions are less hydrophilic that the C-terminal fragment of the N protein, according to the GRAVY index analysis and could support the relationship between antigenic response and hydrophilicity (data not shown). Antibody recognition was also observed for several spots at the N-terminal region (spots 1, 2 and 16), where no specific recognition pattern was observed.
3.3. Membrane glycoprotein (M)
To date, there are very few reports about the antigenic properties of the transmembrane M glycoprotein and its importance in the diagnosis of COVID-19. Both SARS-CoV-2 and SARS-CoV are closely related, belonging to the β-coronavirus genus, and the M protein function is expected to be similar.
The M protein (also known as E1 membrane glycoprotein or matrix protein) is one of major membrane proteins of coronaviruses, together with the S (spike) and the E (envelope) proteins. The M protein contains three domains: a short N terminal ectodomain, a triple-spanning transmembrane domain, and a C-terminal endodomain (Armstrong et al., 1984). It has been postulated that the M protein is related to viral infectivity through binding to the viral S protein and the host surface receptor(s), thus promoting membrane fusion. It appears to be involved in the antigenicity demonstrated by the virus-induced immune responses of the host (Hu et al., 2003).
For diagnostic peptide selection, a similar strategy that the one applied to protein N, was followed. To date, the crystal structure of the SARS-CoV-2 M protein have not been reported. To map the predicted epitopes and to design the peptides finally synthesized, the homology model from the I-Tasser server was used (code QHD43419) (“Modeling of the SARS-COV-2 Genome using I-TASSER”, n.d.) (Fig. 6 ).
Fig. 6.

B Cell Immunodominant regions predicted for the SARS-CoV-2 Membrane protein (M). The corresponding B cell epitopes are shown in colors and mapped as predicted.
The antigenicity of the selected peptides of the M protein was evaluated against the two pools of COVID-19 patients´ sera (pool 1 and 2) (Fig. 1C, M2). Strong antibody recognition, by the three IgG, IgM and IgA immunoglobulins, was observed for peptides 54 and 55, located at the N-terminal domain. Spot 55 showed the higher signal intensity (Fig. 1C, M2, Table 4 and Fig. 5). Differences between these peptides are at the first and the last amino acid residues (1 and 16 aa), showing how modifications at peptides ends could significantly modify the antigenic response. A difference was observed in the antigenic response between the two different pool of sera. Spots 54 and 55 showed strong IgM recognition when exposed to both pool of sera, whereas their IgG and IgA recognition was variable (Fig. 1C, M2 and Fig. 5). Peptide 54 showed slight IgG and IgA recognition with pool 1 and no IgA recognition with pool 2 of sera. Peptide 55 showed slight IgA recognition with pool 1 when compared with other immunoglobulins.
Table 4.
Synthetic peptides (spots) of SARS-CoV-2 M protein and their antibody recognition by the pools of COVID-19 patients' sera.
| Spot (peptide) |
Region | IgG | IgM | IgA |
|---|---|---|---|---|
| 54 | NTD | ++/+++ | +++/+++ | −/− |
| 55 | NTD | +++/+++ | +++/+++ | +++/− |
| 56 | CTD | −/− | +/− | −/− |
| 62 | CTD | +/− | −/− | −/− |
| 73 | CTD | ++/− | −/− | −/++ |
| 76 | CTD | −/++ | −/− | −/+ |
Results Pool 1/Pool 2. “+” denotes positive result; number of “+” signifies relative increase in positive spot intensity in the membrane; “−” denote negative result.
Other sequences (spots 56, 62, 73 y 76) of the C-terminal region of protein M, showed weak to intermediate antibody recognition and not all the immunoglobulins were recognized (Table 4).
It is worth to mention that pool 1 contained highly reactive of COVID-19 patients´ sera (2 of 5), reactive (2 of 5) and one low reactive (1 of 5). On the other hand, pool 2 consisted of low reactive sera (3 of 5), one high reactive and one moderately reactive. The results obtained using pool 1 showed that the immunoglobulins (IgA, IgM and IgG) recognition by the different spots were similar. When the peptides were exposed to pool 2, IgA mild recognition was observed for a greater number of spots (Fig. 1C, M2 and Fig. 5).
None of the evaluated peptides were able to discriminate between the acute or chronic infection. Nevertheless, to make this statement, additional evaluation is required with individual COVID-19 patients's sera. The heterogeneous recognition of the peptides by different immunoglobulins of the patients whose appearance is determined by the time of evolution of the infection, indicates that before the development of a diagnostic kit, polyvalent conjugates capable of capturing the three main immunoglobulins should be used.
3.4. ORF proteins
Although there are few reports on the usefulness of ORF proteins in diagnosis, the immunoinformatic analysis identified some possible antigenic regions that were evaluated by the Pepscan method. ORF3b and ORF8 are the least identical proteins to SARS-CoV and homologous proteins do not exist in other sarbecoviruses, and very little is known about their function and expression in SARS-CoV-2(Fuk-Woo Chan et al., 2020).
Most of the peptides evaluated from these proteins were weakly recognized, when compared with the structural proteins S, N and M. Of all the non-structural proteins tested, only peptides derived from the ORF8 and ORF10 proteins were recognized by any of the different immunoglobulins (Fig. 1C, M1, Table 5 and Fig. 3).
Table 5.
Synthetic peptides (spots) of SARS-CoV-2 ROF10 and ORF8 proteins and their antibody recognition by the pools of COVID-19 patients' sera.
| Spot (peptide) |
IgG | IgM | IgA |
|---|---|---|---|
| ORF10 | |||
| 66 | ++/− | −/+ | −/++ |
| 69 | ++/+++ | −/++ | +/+++ |
| 70 | +++/++ | ++/++ | −/+++ |
| 71 | +++/− | −/− | −/− |
| ORF8 | |||
| 74 | −/− | −/+ | −/+++ |
| 75 | −/− | −/− | −/+++ |
| 76 | −/− | −/− | −/+++ |
Results Pool 1/Pool 2. “+” denotes positive result; number of “+” signifies relative increase in positive spot intensity in the membrane; “−” denote negative result.
Several peptides of the ORF10 protein were evaluated, 66, 67, 68, 69, 70 and 71 (spot 69 and 70 were added as repetition of 68 and 67, respectively, for chemical control). In Table 5 it can be clearly observed that the selected peptides, localized in the C-terminal region of the ORF10 protein, showed recognition by the IgG immunoglobulin only with pool 1 of COVID-19 patients´ sera and that peptides 66, 99 and 70, showed recognition by the IgA immunoglobulin but only with pool 2 of sera. One peptide (spot 70) was recognized by the IgM immunoglobulin with the two pools of sera.
The ORF8 protein is a small protein with only 121 amino acid residues, represented by peptides (spots 72 to 76) of the N-terminal region. A recent work show that the combinational use of ORF3b and ORF8 tests alone could be sufficient to detect individuals exposed to COVID-19 at any time point of infection (Hachim et al., 2020). However, in our case, at least with the evaluated peptides, only a moderate IgA recognition was observed with pool 2 of COVID-19 patients´ sera.
4. Conclusions
In this study, we use a comprehensive strategy to identify potential 16 mer antigenic peptides suited for antibody-based SARS-CoV-2 diagnosis, in order to be used with different immunological techniques. A systematic scan of the three structural proteins (S, N and M) and the non-structural proteins (ORFs) present in the SARS-CoV-2 virus was conducted through the combination of immunoinformatic methods, peptide SPOT synthesis and immunoassay by cellulose-bound peptides (Pepscan). The membranes with synthetic peptides were tested against two pools of sera from COVID-19 acute infected patients. A strong antibody recognition was observed for peptides corresponding to the S, N and M proteins of SARS-CoV-2 virus, but not to the ORFs proteins. IgM and IgA immunoglobulins highly recognized several peptides of the S protein. Those are localized along the different antigenic regions at the S1 and S2 subunits and RBD domain. The sequences with higher signal intensity were localized in the C-terminal region of the N protein. Sequences of the SR-rich linker and CDT regions showed different immunoreactivity, but all of them were strongly recognized by IgM immunoglobulin with the two pools of sera. Three sequences of the N-terminal region also showed antibody recognition but with no specific pattern. Strong antibody recognition, by the three IgG, IgM and IgA immunoglobulins, was also observed for two peptides of the N-terminal domain of the M protein.
The differential reactivity observed between the immunoglobulin isotypes within different regions of the S and N proteins, can be exploited for ensuring accurate diagnosis of all infected patients, with different times of exposure to infection. However, a possible limitation of this study was the use of serum from individuals with a recent COVID-19 episode. This might reduce the strength of reactivity against the tested peptides. On the other hand, this allowed the selection of peptides that are recognized by IgA and IgM at the earliest stages of infection, without excluding their possible importance once the acute phase is over, when detected by IgG. Our results are extremely encouraging and our overall strategy led us to reduce a data set of 200 peptides of proteins S, N, M and ORFs of SARS-CoV-2 virus to a reduced number of antigenic 16 mer peptides, in total 33. These antigenic peptides are on their way in a larger scale by solid-phase synthesis and will be tested against a higher number of positive for SARS-CoV-2 patients' sera with different times of evolution by the Multiple Antigen Blot Assay (MABA), by ELISA and by an immunochromatographic rapid test. Likewise, each of the selected peptides will be synthesized and evaluated in different formats (monomers, dimers, trimers, MAPS, etc.) creating chimeras with greater antigenic properties. These results show the potential of synthetic peptides as one of the strategies for the production of reagents of great diagnostic value, due to their low production cost, stability and high yield.
The following is the supplementary data related to this article.
Declaration of Competing Interest
The authors declare no conflict of interests.
Acknowledgments
This work is dedicated to three researchers who made possible the development of a protein chemistry laboratory in the Biohelminthiasis Section of the Institute of Tropical Medicine: Manuel Elkin Patarroyo, who opened the doors of the Institute of Immunology of Colombia for the training of our personnel and motivated us to look at to chemistry for the main tool to produce antigens; Fanny Guzman, the researcher who taught us the different strategies for the production of synthetic peptides and quality control, and Johan Hoebeke, who taught us the power of bioinformatics in the analysis of protein structure and the use of predictive algorithms.
This work was possible thanks to the support of three enthusiastic advisors, who believed in the capabilities of our team, Carlota Perez, Nydia Ruiz and Pablo Liendo, who managed to obtain most of the funds through the donation of a benefactor, Mr. James Anderson. Likewise, there was additional financial support from a project of the Academy of Physical, Mathematical and Natural Sciences (ACFIMAN) of Venezuela “COVID-19 in Venezuela: Modeling epidemiological scenarios and validation of immunodiagnostic techniques to improve the capacity of relevant stakeholders to mitigate its impact on the population” (INT 2021/VEC C1901), financed by the British Embassy in Caracas. Finally, additional support was secured through the project “2020 to Present. COVID Rapid Response. University of Glasgow. GCRF Grant, Engagement Network, Small Grant Funds. Co-PI”. We also appreciate the donation of the rapid tests for COVID made by Gabriela Redondo, from LANA. Finally, the authors of this work wish to thank the comments of the referees, since they served to improve the content of this publication.
References
- Ansari H.R., Raghava G.P. Identification of conformational B-cell epitopes in an antigen from its primary sequence. Immunome Res. 2010;6:6. doi: 10.1186/1745-7580-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong J., Niemann H., Smeekens S., Rottier P., Warren G. Sequence and topology of a model intracellular membrane protein, E1 glycoprotein, from a coronavirus. Nature. 1984;308:751–752. doi: 10.1038/308751a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashour H.M., Elkhatib W.F., Rahman M.M., Elshabrawy H.A. Insights into the recent 2019 novel coronavirus (Sars-coV-2) in light of past human coronavirus outbreaks. Pathogens. 2020 doi: 10.3390/pathogens9030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., West A.P., Huey K.E., Robbiani D.F., Nussenzweig M.C., Correspondence P.J.B., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., Lorenzi J.C.C., Finkin S., Hä Gglö T., Hurley A., Millard K.G., Weisblum Y., Schmidt F., Hatziioannou T., Bieniasz P.D., Caskey M., Bjorkman P.J. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies ll structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182:1–15. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter J.M., Loomis-Price L. B cell epitope mapping using synthetic peptides. Curr. Protoc. Immunol. 2004;60:9.4.1–9.4.23. doi: 10.1002/0471142735.im0904s60. [DOI] [PubMed] [Google Scholar]
- Chen P., Rayner S., Hu K.H. Advances of bioinformatics tools applied in virus epitopes prediction. Virol. Sin. 2011;26:1–7. doi: 10.1007/s12250-011-3159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P.Y., Fasman G.D. Advances in Enzymology and Related Areas of Molecular Biology. Wiley Blackwell; 2006. Prediction of the secondary structure of proteins from their amino acid sequence; pp. 45–148. [DOI] [PubMed] [Google Scholar]
- Deléage G. An interactive 3D viewer of molecules compatible with the suite of ANTHEPROT programs. J. Biophys. Chem. 2012;03:35–38. doi: 10.4236/jbpc.2012.31004. [DOI] [Google Scholar]
- Emini E.A., Hughes J.V., Perlow D.S., Boger J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J. Virol. 1985;55:836–839. doi: 10.1128/jvi.55.3.836-839.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuk-Woo Chan J., Kok K.-H., Zhu Z., Chu H., Kai-Wang To, K, Yuan S., Yuen K.-Y. 2020. Genomic Characterization of the 2019 Novel Human-Pathogenic Coronavirus Isolated from a Patient with Atypical Pneumonia after Visiting Wuhan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomara M., Haro I. Synthetic peptides for the immunodiagnosis of human diseases. Curr. Med. Chem. 2007;14:531–546. doi: 10.2174/092986707780059698. [DOI] [PubMed] [Google Scholar]
- Guglielmi G. The explosion of new coronavirus tests that could help to end the pandemic. Nature. 2020;583:506–509. doi: 10.1038/d41586-020-02140-8. [DOI] [PubMed] [Google Scholar]
- Hachim A., Kavian N., Cohen C.A., Chin A.W.H., Chu D.K.W., Mok C.K.P., Tsang O.T.Y., Yeung Y.C., Perera R.A.P.M., Poon L.L.M., Peiris J.S.M., Valkenburg S.A. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat. Immunol. 2020;21:1293–1301. doi: 10.1038/s41590-020-0773-7. [DOI] [PubMed] [Google Scholar]
- Heger A., Holm L. Rapid automatic detection and alignment of repeats in protein sequences. Proteins Struct. Funct. Bioinforma. 2000;41:224–237. doi: 10.1002/1097-0134(20001101)41:2<224::AID-PROT70>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Home - Johns Hopkins Coronavirus Resource Center [WWW Document], n.d. URL https://coronavirus.jhu.edu/ (accessed 1.19.21). 2021.
- Hu Y., Wen J., Tang L., Zhang H., Zhang X., Li Y., Wang Jing, Han Y., Li G., Shi J., Tian X., Jiang F., Zhao X., Wang Jun, Liu S., Zeng C., Wang Jian, Yang H. The M protein of SARS-CoV: basic structural and immunological properties. Genom. Proteom. Bioinforma. Beijing Genomics Inst. 2003;1:118–130. doi: 10.1016/S1672-0229(03)01016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueston L., Kok J., Guibone A., McDonald D., Hone G., Goodwin J., Carter I., Basile K., Sandaradura I., Maddocks S., Sintchenko V., Gilroy N., Chen S., Dwyer D.E., O’Sullivan M.V.N. The antibody response to SARS-CoV-2 infection. Open Forum Infect. Dis. 2020;7 doi: 10.1093/ofid/ofaa387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen M.C., Peters B., Nielsen M., Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45 doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Ziliang, Zhou Zhechong, Chen Q., Yan Y., Zhang C., Shan H., Chen S. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B. 2020 doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus P.A., Schulz G.E. Prediction of chain flexibility in proteins - A tool for the selection of peptide antigens. Naturwissenschaften. 1985;72:212–213. doi: 10.1007/BF01195768. [DOI] [Google Scholar]
- Kolaskar A.S., Tongaonkar P.C. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990;276:172–174. doi: 10.1016/0014-5793(90)80535-Q. [DOI] [PubMed] [Google Scholar]
- Latiano A., Tavano F., Panza A., Palmieri O., Niro G.A., Andriulli N., Latiano T., Corritore G., Gioffreda D., Gentile A., Fontana R., Guerra M., Biscaglia G., Bossa F., Carella M., Miscio G., di Mauro L. False positive results of IgM/IgG antibodies against antigen of the SARS-CoV-2 in sera stored before the 2020 Endemia in Italy. Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.12.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Jin R., Peng Y., Wang C., Ren W., Lv F., Gong S., Fang F., Wang Q., Li J., Shen T., Sun H., Zhou L., Cui Y., Song H., Sun L. Generation of antibodies against COVID-19 virus for development of diagnostic tools. medRxiv. 2020 doi: 10.1101/2020.02.20.20025999. 2020.02.20.20025999. [DOI] [Google Scholar]
- Li Y., Lai D. Yun, Lei Q., Xu Z. Wei, Wang F., Hou H., Chen L., Wu J., Ren Y., Ma M. Liang, Zhang B., Chen H., Yu C., Xue J. Biao, Zheng Y. Xiao, Wang X. Ning, Jiang H. Wei, Zhang H. Nan, Qi H., Guo S. Juan, Zhang Y., Lin X., Yao Z., Pang P., Shi D., Wang W., Yang X., Zhou J., Sheng H., Sun Z., Shan H., Fan X., Tao S. Ce. Systematic evaluation of IgG responses to SARS-CoV-2 spike protein-derived peptides for monitoring COVID-19 patients. Cell. Mol. Immunol. 2021;18:621–631. doi: 10.1038/s41423-020-00612-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014 doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modeling of the SARS-COV-2 Genome using I-TASSER [WWW Document], n.d. URL https://zhanglab.ccmb.med.umich.edu/COVID-19/ (accessed 7.28.20).
- Mészáros B., Erdős G., Dosztányi Z. IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018;46(W1):329–337. doi: 10.1093/nar/gky384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musicò A., Frigerio R., Mussida A., Barzon L., Sinigaglia A., Riccetti S., Gobbi F., Piubelli C., Bergamaschi G., Chiari M., Gori A., Cretich M. SARS-CoV-2 epitope mapping on microarrays highlights strong immune-response to n protein region. Vaccines. 2021;9:1–11. doi: 10.3390/vaccines9010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noya O., Patarroyo M., Guzman F., de Noya B. Immunodiagnosis of parasitic diseases with synthetic peptides. Curr. Protein Pept. Sci. 2005;4:299–308. doi: 10.2174/1389203033487153. [DOI] [PubMed] [Google Scholar]
- Ponomarenko J., Bui H.H., Li W., Fusseder N., Bourne P.E., Sette A., Peters B. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics. 2008:9. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Q&A on coronaviruses (COVID-19) [WWW Document], n.d. URL https://www.who.int/emergencies/diseases/novel-coronavirus-2019/question-and-answers-hub/q-a-detail/q-a-coronaviruses (accessed 8.26.20).
- Qu J., Wu C., Li Xiaoyong, Zhang G., Jiang Z., Li Xiaohe, Zhu Q., Liu L. Profile of immunoglobulin G and IgM antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2020;71:2255–2258. doi: 10.1093/cid/ciaa489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabaan A.A., Al-Ahmed S.H., Haque S., Sah R., Tiwari R., Singh Malik Y., Dhama K., Iqbal Yatoo M., Katterine Bonilla-Aldana D., Rodriguez-Morales A.J. SARS-CoV-2, SARS-CoV, and MERS-CoV: a comparative overview. Infez Med. 2020;28:174–184. [PubMed] [Google Scholar]
- Saha S., Raghava G.P.S. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins Struct. Funct. Genet. 2006;65:40–48. doi: 10.1002/prot.21078. [DOI] [PubMed] [Google Scholar]
- Van Regenmortel M.H.V. Immunoinformatics may lead to a reappraisal of the nature of B cell epitopes and of the feasibility of synthetic peptide vaccines. J. Mol. Recognit. 2006;19:183–187. doi: 10.1002/jmr.768. [DOI] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181 doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wu X., Zhang X., Hou X., Liang T., Wang D., Teng F., Dai J., Duan H., Guo S., Li Y., Yu X. 2020. SARS-CoV-2 Proteome Microarray for Mapping COVID-19 Antibody Interactions at Amino Acid Resolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler D.F.H., Hilpert K., Brandt O., Hancock R.E.W. Synthesis of peptide arrays using SPOT-technology and the CelluSpots-method. Methods Mol. Biol. 2009;570:157–174. doi: 10.1007/978-1-60327-394-7_5. [DOI] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (80-) 2020;367:1260–1263. doi: 10.1126/science.aax0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao B., Zhang L., Liang S., Zhang C. SVMTriP: A method to predict antigenic epitopes using support vector machine to integrate tri-peptide similarity and propensity. PLoS One. 2012;7 doi: 10.1371/journal.pone.0045152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Li J., Shan Y., Yang Z., Zhao Z., Chen B., Yao Z., Dong B., Wang S., Chen J., Cong Y. Subcellular localization and membrane association of SARS-CoV 3a protein. Virus Res. 2005;109:191–202. doi: 10.1016/j.virusres.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yüce M., Filiztekin E., Özkaya K.G. COVID-19 diagnosis —A review of current methods. Biosens. Bioelectron. 2021;172:112752. doi: 10.1016/j.bios.2020.112752. [DOI] [PMC free article] [PubMed] [Google Scholar]