Abstract
Introduction
We aimed to assess risk factors for multidrug-resistant Gram-negative bacilli (MDR-GNB) from a large amount of data retrieved from electronic health records (EHRs) and determine whether machine learning (ML) may be useful in assessing the risk of MDR-GNB infection at febrile neutropenia (FN) onset.
Methods
Retrospective study of almost 7 million pieces of structured data from all consecutive episodes of FN in hematological patients in a tertiary hospital in Barcelona (January 2008–December 2017). Conventional multivariate analysis and ML algorithms (random forest, gradient boosting machine, XGBoost, and GLM) were done.
Results
A total of 3235 episodes of FN in 349 patients were documented; MDR-GNB caused 180 (5.6%) infections in 132 patients. The most frequent MDR-GNBs were MDR-Pseudomonas aeruginosa (53%) and extended-spectrum beta-lactamase-producing Enterobacterales (46%). According to conventional logistic regression analysis, independent factors associated with MDR-GNB infection were age older than 45 years (OR 2.07; 95% CI 1.31–3.24), prior antibiotics (2.62; 1.39–4.92), first-ever FN in this hospitalization (2.94; 1.33–6.52), prior hospitalizations for FN (1.72; 1.02–2.89); at least 15 prior hospital visits (2.65; 1.31–5.33), high-risk hematological diseases (3.62; 1.12–11.67), and hospitalization in a room formerly occupied by patients with MDR-GNB isolation (1.69; 1.20–2.38). ML algorithms achieved the following AUC and F1 score for MDR-GNB prediction: random forest, 0.79–0.9711; GMB, 0.79–0.9705; XGBoost, 0.79–0.9670; and GLM, 0.78–0.9716.
Conclusion
Data generated in EHRs proved useful in assessing risk factors for MDR-GNB infections in patients with FN. The great number of analyzed variables allowed us to identify new factors related to MDR infection, as well as to train ML algorithms for infection predictions. This information may be used by clinicians to make better clinical decisions.
Keywords: Machine learning, Electronic health records, Multiresistance, Neutropenia
Key Summary Points
| Why carry out this study? |
| Hematological patients with febrile neutropenia presenting with multidrug-resistant Gram-negative bacilli (MDR-GNB) infections frequently receive inappropriate empirical antibiotic therapy which increases their morbidity and mortality. |
| Current studies aiming to identify patients at risk for MDR-GNB in this population use single predictive analysis focused on small sets of variables. |
| We hypothesized that machine learning using information stored in electronic health records could be useful to predict MDR-GNB in these patients. |
| What was learned from the study? |
| Clinical data stored directly in electronic health records can be used to identify risk factors for MDR-GNB infections in severe hematological patients at FN onset. |
| The high quantity of data allowed us to identify new risk factors for MDR infections. |
| Machine learning has proved useful for clinical predictors in MDR-GNB infections, thereby helping to provide personalized medical care. |
Digital Features
This article is published with digital features, including summary slide, to facilitate understanding of the article. To view digital features for this article go to 10.6084/m9.figshare.14248775.
Introduction
The increasing availability of data from daily clinical care electronic health records (EHRs) represents a major opportunity for progress in medicine. New statistical techniques, specifically machine learning (ML) approaches, can provide us with the ability to work with large amounts of data and provide optimal predictions in different scenarios [1–5]. However, there is very little information on the use of these techniques within the field of infectious diseases [6–9].
Our hypothesis was that the data directly retrieved from EHRs can be used to build practical tools to identify in real time which hematological patients with febrile neutropenia (FN) will have multidrug-resistant Gram-negative bacilli (MDR-GNB) infections. Identifying these patients is crucial, as patients with MDR-GNB frequently receive inadequate empirical antibiotic treatment [10–14], increasing their morbidity and mortality [11, 15–18]. Administration of broad-spectrum antibiotics to cover all potential microorganisms requires the use of 2–3 antibiotics; however, this can increase antibiotic pressure, as well as resistance selection, toxicity, and economic costs. Currently, few studies have identified risk factors for MDR-GNB infection in hematological patients with documented bloodstream infections. These studies were performed using simple predictive analytics and scoring systems focused on small sets of manual data entry, with a limited number of variables [11–13, 15]. There is a lack of current studies analyzing an entire population with FN.
We aimed to identify risk factors for MDR-GNB infections in hematologic patients at FN onset by performing analyses of a large amount of data obtained from EHRs through common statistical methods. Moreover, we trained ML algorithms to predict which patients will need broad antibiotic coverage for MDR-GNB infections. We also aimed to highlight differences offered by both mathematical approaches for general clinicians.
Methods
Setting, Study Population, Data Mining, and Study Design
This study was performed at the Hospital Clinic (Barcelona, Spain), a 700-bed university institution which provides care to a population of 500,000 inhabitants. For this study, we analyzed all consecutive episodes of FN occurring in hematological patients from January 2008 to December 2017. No major outbreaks occurred during this period.
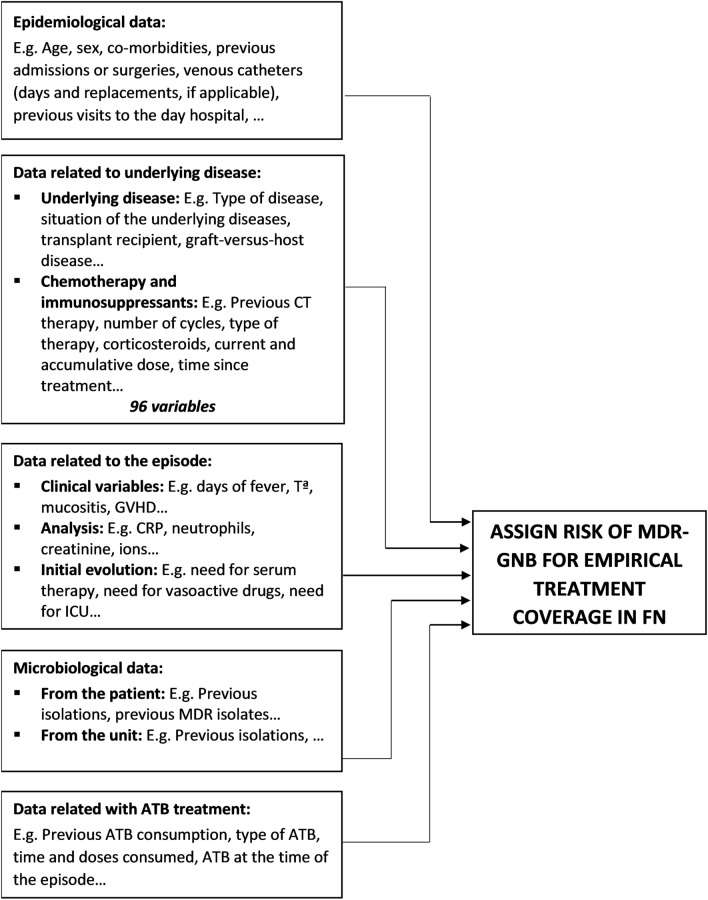
Our data mining approach was conducted as follows: (1) infectious diseases physicians listed data to create the study dataset. Patients’ medical history, physical examination, clinical and laboratory data, present and past results of microbiological tests from patients, and therapy, including current and prior antibiotic treatments, were selected. Figure 1 summarizes the most important variables selected for the dataset generation. Only structured data were used. (2) Computer scientists extracted a large set of data (6,768,767 pieces of data) from January 2008 to December 2017 directly from EHRs and worked on pre-processing data. (3) As it was the first time our department had used data from EHRs created from daily clinical practice, we manually performed a full data check of 100 patients. We achieved a perfect match between data obtained from EHRs and data reviewed. (4) A multidisciplinary team with experts across several scientific fields—clinical medicine, computer science, and statistics—worked on pre-processing data (selection, clearing, enrichment, and transformation of the database), as well as on subsequent statistical analyses. This study was performed in accordance with the Helsinki Declaration. The study was approved by the Ethics Committee Board of our institution (HCB/2018/0308) and followed privacy laws regarding active anonymity.
Fig. 1.
Main variables in dataset generation
Definitions
High-risk patients were defined as those with prolonged (more than 7 days’ duration) and profound neutropenia (less than 100 cells/mm3) and/or significant medical comorbid conditions, including hypotension or hyperlactacidemia, intensive care unit (ICU) requirement, pneumonia or hypoxemia, intravascular catheter infection, evidence of renal failure or hepatic insufficiency. Patients with FN were defined as those who had a temperature measurement greater than 38.0°C and an absolute neutrophil count of less than 500 cells/mm3. Separate episodes of FN were considered to be those whose febrile determination was preceded by more than 4 days of apyrexia. In accordance with hospital protocols, patients with expected neutropenia over 10 days received prophylaxis with a fluoroquinolone and an azole. Prior antibiotic therapy was explained as the usage of any antimicrobial agent prior to FN episode including antibiotic prophylaxis.
Following the current definitions [19], Gram-negative bacilli were considered to be MDR when these conditions were present: (1) extended-spectrum beta-lactamase (ESBL)-producing or AmpC-hyperproducing Enterobacterales, (2) MDR strains of non-fermenting GNB such as Pseudomonas aeruginosa, Acinetobacter baumannii, and Stenotrophomonas maltophilia. Non-fermenting GNB were defined as MDR strains when they were resistant to at least one antibiotic in three or more classes of antibiotics: carbapenems, ureidopenicillins, cephalosporins (ceftazidime and cefepime), monobactams, aminoglycosides, fluoroquinolones, fosfomycin, and colistin. Positive culture was considered related to FN event when collected during a time period of ± 24 h after FN onset. Empirical coverage for MDR-GNB was considered as needed in patients who have MDR-GNB or had had MDR-GNB infection within the prior 6 months.
Microbiological Methods
Our center follows international guideline recommendations to collect and incubate cultures [20]. Blood samples were treated using the BACTEC 9240 system or Bactec FX system (Becton–Dickinson Microbiology Systems), with an incubation period of 5 days. Isolates were recognized by standard techniques. Antimicrobial susceptibility testing was performed by using a microdilution system (Microscan WalkAway Dade Behring, West Sacramento, CA or Phoenix system, Becton Dickinson, Franklin Lakes, NJ) or the Etest method (AB Biodisk, Solna, Sweden/bioMérieux, Marcy l’Etoile, France). Current CLSI (from 2008 to 2010) and EUCAST breakpoints (from 2010 to 2018) were employed to describe susceptibility or resistance to such antimicrobial agents; intermediate susceptibility was perceived as resistance. ESBL were detected by minimum inhibitory concentration (MIC) results and double-disk synergy test using disks containing cefotaxime, ceftazidime, and cefepime that are applied to plates next to a disk with clavulanic acid.
Statistical Analysis, Model Development, and Validation
Descriptive analysis of the entire cohort was provided. Categorical variables were detailed as counts and percentages, whereas continuous variables were described as either means and standard deviations (SD) or medians and interquartile ranges (IQRs). For independent variables, we chose parameters that showed predictive value using a univariate analysis (age older than 45 years, autologous stem cell transplant, prior antibiotic treatment, first-ever episode of FN in this hospitalization, more than three FN episodes in this hospitalization, more than 90 days since a past episode of febrile neutropenia, prior hospitalizations for FN, more than 15 prior hospital visits, ICU admission, breakthrough bacteremia, high-risk hematological diseases, prior positive culture, prior MDR, more than 14 days with neutropenia, and hospitalization in a room formerly occupied within the last 3 months by a patient with MDR-GNB isolation (same pathogen, same resistance pattern)). A logistic regression model with step-forward procedure in the overall cohort of patients was performed to identify independent factors related to the need for empirical MDR-GNB coverage, and significance (p) was set at the value of 0.05. The goodness of fit of the multivariate models was assessed by the Hosmer–Lemeshow test. The accuracy of the rule was assessed by the area under receiver operating characteristic (ROC) curve (AUC). These analyses were performed using the SPSS software (version 23.0; SPSS, Inc., Chicago, IL).
In the second part of the study, ML algorithms were used to predict which patients will need empirical coverage for MDR-GNB. We started by performing a descriptive analysis of the data to ensure data quality of each of the available variables. Coherence of the obtained results was checked. Correlation between different variables was also analyzed. Patients with positive microbiology were a minority among the total number of patients available, and the rules to classify a patient as positive were exclusively in adherence to definitions. The list of patients, together with the variables used to perform the classification and the resulting target variable, was provided. It was validated by doctors on a case-by-case basis. Some numerical variables needed to be within a certain range (provided by the team at Hospital Clinic). A similar approach was taken for observations with incoherent data. This step was also guided by doctors. Observations with missing data in categorical variables were either classified into a “missing” or “blank” category or dropped. Missing data in numerical variables such as results from blood tests was usually substituted by the mean of the valid interval. Class imbalances were managed in the way most suitable to the selected model. Variable importance was measured by calculating the increase in the model’s prediction error after permuting the features. A variable is considered to be important if shuffling its values increases the model error, and unimportant if the permutation leaves the model error unchanged. Decision tree algorithms were used [4]. We trained four models typically used for classification problems: (1) random forest—an ensemble method that uses decision trees as base models, and are good for capturing complex data structures; (2) gradient boosting machine (GBM)—an ensemble method that sequentially fits new models to improve the estimate on the response variable; (3) XGBoost is another tree boosting implementation which uses a clever penalization of individual trees, as well as Newton boosting; (4) and a logistic regression, using R and with the dataset methodology used for ML techniques. The study cohort was divided into training and test sets. Each model was trained using the training set. We then used these trained models to predict the response variable for the episodes in the test set. This separation is standard procedure used to be able to assess ML model performance. We followed a 70–30 time split, meaning 70% of the episodes were in the training set and 30% were reserved for the test. We followed a time split for building these two data sets instead of a random split for two reasons: (i) time consistency; (ii) stress the capability of these algorithms behaving in a real-life scenario. Test accuracy was measured by the F1 score, which considers both the precision (number of correct positive results divided by the number of all positive results) and the recall (number of correct positive results divided by the number of all relevant samples) of the test. ML analyses were done by using the R language and environment for statistical computing (Version 3.5.1-07/2018). The ML models mentioned here come from the following R packages: (1) glmnet 2.0-16; (2) XGBoost 0.71.2; (3) Random Forest 4.6-14; and (4) gbm 2.1.4.
Results
Demographics and Epidemiology
A total of 3235 FN episodes in 349 hematological patients were documented. Median age was 57 (IQR 44–67) years and 1841 (56.9%) were male. Most patients had acute leukemia (1221, 38%) and stem cell transplantation (914, 28%). Table 1 summarizes the main demographic and clinical characteristics of the patients.
Table 1.
Main demographic and clinical characteristics of the patients
| Episodes N = 3235 (%) |
|
|---|---|
| Demographics | |
| Male sex | 1841 (56.9) |
| Age, median (IQR) years | 57 (44–67) |
| Older than 45 years | 2378 (73.5) |
| Baseline disease | |
| Acute leukemia | 1221 (38) |
| Hematopoietic stem cell transplant | 914 (28) |
| Main clinical conditions | |
| Prior antibiotic consumption | 2086 (64.5) |
| First-ever episode of FN | 1542 (47.7) |
| First FN in this hospitalization | 2540 (78.5) |
| Median days from hospital admission to FN episode (IQR) | 9 (1–17) |
| More than 15 prior visits to hospital | 255 (7.9) |
| Severe mucositis | 591 (18.3) |
| FN in ICU | 319 (9.9) |
A total of 395 (12.2%) episodes have confirmed infection by cultures, primarily bacteremia (245; 7.6%). MDR-GNB accounted for 180 (5.6%) episodes in 132 patients. The most frequent MDR-GNB were MDR-P. aeruginosa, 96 episodes (53%) and ESBL Enterobacterales, 84 episodes (46%). In total, 295 (9.1%) were patients considered in need of empirical coverage for MDR-GNB.
Independent Factors Associated with Need for MDR-GNB Coverage by Conventional Logistic Regression Model
Independent factors in the logistic regression model associated with the need for MDR-GNB coverage among patients with FN using all dataset were age older than 45 years (OR 2.07; 95% CI 1.31–3.24), prior antibiotic treatment (OR 2.62; 95% CI 1.39–4.92), first FN in this hospitalization (OR 2.94; 95% CI 1.33–6.52), prior hospitalizations for FN (OR 1.72; 95% CI 1.02–2.89), more than 15 prior hospital visits (OR 2.65; 95% CI 1.31–5.33), high-risk hematological diseases (OR 3.62; 95% CI 1.12–11.67), and hospitalization in a room formerly occupied within the last 3 months by a patient with GNB-MDR isolation (OR 1.69; 95% CI 1.20–2.38). The goodness of fit of the multivariate model was assessed by the Hosmer–Lemeshow test (0.76). The discriminatory power of the model, as evaluated by the area under the ROC curve, was 0.849 (95% 0.814–0.871), demonstrating a robust ability to identify factors related to the need for MDR-GNB coverage among patients with FN.
Prediction of Need for MDR-GNB Coverage by Machine Learning Models
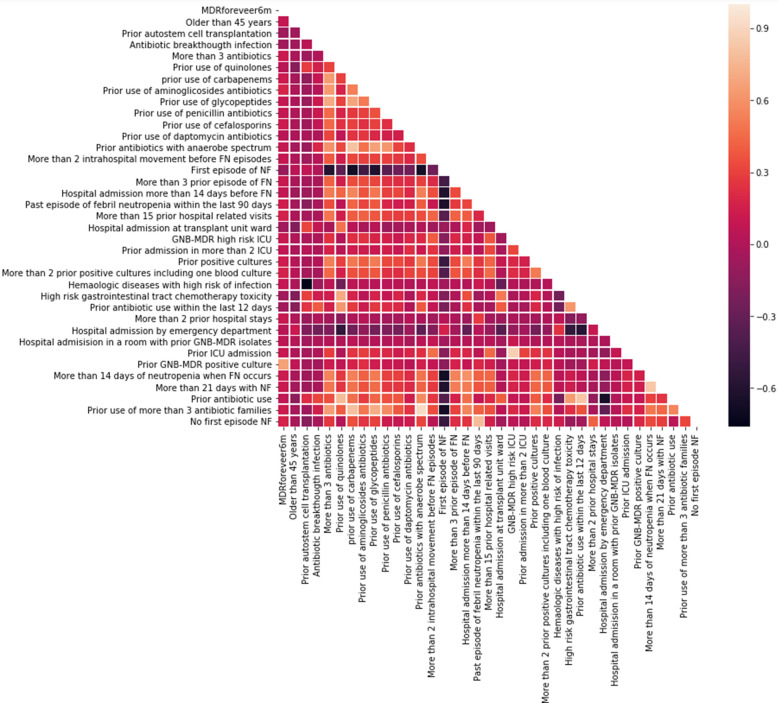
Figure 2 shows the correlation among main variables in the dataset. The correlation between the target variable of having MDR and that capturing whether patient had MDR before was positive and important (correlation 0.67). As mentioned before, the whole data was randomly split into two different datasets: 70% to train (2262 episodes) and 30% to test (973 episodes). Based on the training set, a prediction model to select the need for MDR-GNB antibiotic coverage was developed.
Fig. 2.
Correlation matrix—full dataset (heatmap, generated with Seaborn library)
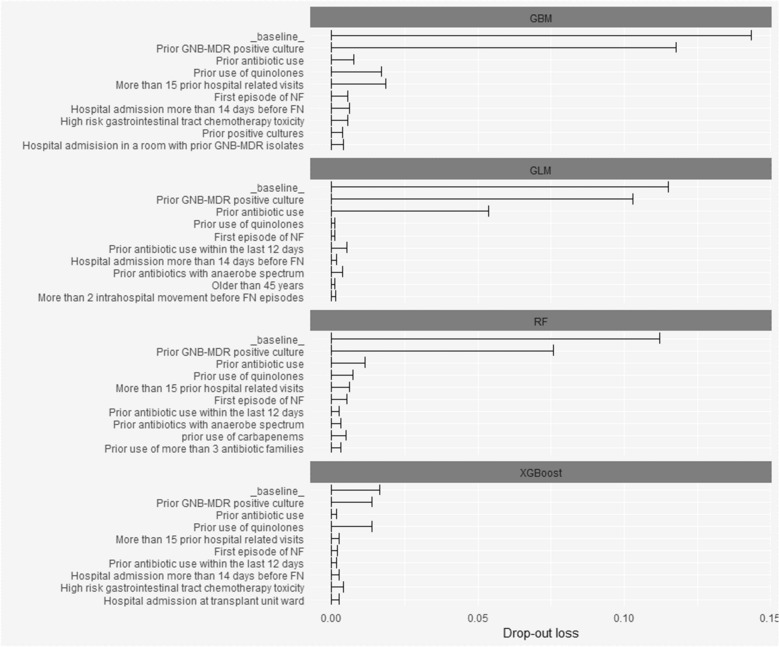
Figure 3 details plots showing the global varying importance of the many variables for different models. Among them, “prior GNB-MDR positive culture” is the most influential variable in the pool of potential predictors.
Fig. 3.
Variable importance plots for the four models (GBM, GLM, RF, XGBoost using data in the training set)
Table 2 shows the results of different models in the test set according to varying metrics, always applying the standard rule that the probability should be higher than 50%, so that the episode is to be labelled as the most “probable” category. Provided that MDR episodes are a small sample in the dataset, an F1 score accuracy metric could be a better comparison tool. With this metric in mind, there is no significant difference in the results obtained from the four models. As we established a cutoff of a 50% probability, the models had high specificity, high negative predictive value, and fair sensitivity.
Table 2.
Metrics of ML models to predict the need for MDR-GNB coverage in patients with FN in the test set
| Models | AUC | F1 score | Sensitivity | Specificity | Negative predictive value | Positive predictive value |
|---|---|---|---|---|---|---|
| GBM | 0.7872 | 0.9705 | 0.4583 | 0.9988 | 0.9438 | 0.9778 |
| XGBoost | 0.7945 | 0.9670 | 0.4895 | 0.9886 | 0.9464 | 0.8246 |
| Random forest | 0.7896 | 0.9711 | 0.4583 | 1.00 | 0.9439 | 1.0 |
| GLM | 0.7827 | 0.9716 | 0.4687 | 1.00 | 0.9449 | 1.0 |
Discussion
This study is innovative in several sections of its approach. The study was originated from a large amount of data obtained from daily clinical practice, contrasting with the common practice of using specific datasets constructed for research. This approach allowed us to evaluate risk factors usually difficult to assess, as well as demonstrate associations among such factors like hospital epidemiology and the risk of MDR-GNB infection. Importantly, the use of data from EHRs can allow for the creation of a real-time prediction tool. Another significant novelty is that prediction of which patients will need coverage for MDR-GNB infections was performed at FN onset, and not when clinicians received microbiological confirmations, as done in many prior studies. Consequently, our study provides a clinical recommendation based on data obtained at the moment when the clinician must make a decision regarding antibiotics. Finally, our study demonstrates that ML can be used to train data from some episodes and predict new episodes, namely which high-risk hematological neutropenic patients will need broad empirical antibiotic coverage for MDR-GNB infection when the patient has a fever.
Our study was based on data at FN onset. There is a lack of current information of antibiotic resistance rates in the overall population with FN. More studies report the percentage of MDR-GNB among patients with documented infection and our data is concordant with these papers [10, 11, 21–23]. However, patients with documented infection account for a small subset of patients among hematological patients with FN; clinicians must decide antibiotic treatment at FN onset, and not when documented infection is confirmed. In our study, we found that infections caused by MDR-GNB are uncommon among the entire population with FN. Consequently, a personalized antibiotic approach in patients with FN can be an important measure to save the use of antibiotics, when not necessary.
Our study agrees with prior studies that describe some factors related to the need for MDR-GNB coverage: older age, prior antibiotic treatment, some specific hematological diseases, or previous episodes of FN [10, 11, 13, 24, 25]. Additionally, the possibility to comprehensively analyze non-common variables via our approach has allowed us to document the relationship between multiresistance and factors such as more than 15 prior hospital visits, first febrile episode recorded during current hospital admission, or hospitalization in a room formerly occupied within the last 3 months by patients with MDR-GNB isolation. These factors are closely related to the likelihood of colonization by multiresistant bacteria due to changes in microbiota caused by treatments—mainly antibiotics—as well as contact with hospital environments where MDR-GNB colonize inert surfaces.
We employed an ML approach to predict which patients would necessitate coverage for MDR-GNB. The main difference between common medicine statistics and ML is that the ML approach extends beyond the comprehension of causal relationships, focusing on a potential set of variables and algorithms to predict an event [26, 27]. Logistic regression is one approach that pertains to ML, given that its ability to identify risk factors also helps to predict when an event can happen. ML techniques cannot easily express the reasoning behind the assignment. For this reason, clinicians may have a “black box” feeling concerning ML predictive models, and results of ML algorithms might be difficult to introduce in the clinical decision-making process [28]. In our study, we demonstrate that factors used by different ML techniques to perform algorithms are very similar to those used by our conventional logistic regressions. One of the main strengths of the ML approach is that their predictions will be always done on the basis of input variables. Within the setting of MDR prediction, geographical differences in resistance rates and patterns are important. Thus, following our approach, input data will always be its own data center. Prediction of function and output is useful in the area explored as well. In our study, predictive accuracy of ML algorithms is good, but not optimal yet. Factors such as including a higher number of patients, integrating more data, working on the learning process of ML models, or the integration of different models may result in more precise predictions. The disparity found between sensitivity and the high predictive values is perhaps related to the lower number of MDR events. Likewise, our metrics are calculated applying a rule that probability must be higher than 50%, so that the episode is labelled as an event. Different calibrations of this parameter can provide varying values on sensitivity, specificity, and predictive values.
This study has several limitations. First, our study provides predictions validated in a test dataset. All data were obtained from hospital EHRs. Some outpatient data might be missing. However, algorithm prospective validation will be needed. Second, as we previously commented, ML algorithms are typically more opaque than classic statistical models: their predictions might be difficult for physicians to understand. Closing the gap between computer algorithm results and medical clinical understanding will prove to be a challenge for the future. Our study does shed some light, though, in that results obtained from ML are not very different than those obtained by usual regression models. Third, the study was conducted in a single center, with its particular epidemiology. If ML algorithms are applied to a different population, ML will use data from the receiving center. It is unknown what type of impact the new hospital epidemiology will have on the sensitivity and specificity of the algorithm. Moreover, computing power and infrastructure necessary to real-time models are not available everywhere, and patients could be admitted to different healthcare areas, making medical backgrounds misleading. Finally, the percentage of patients with MDR-GNB was very low, jeopardizing the sensitivity of the mathematical approaches.
Conclusion
This is the first study that demonstrates that clinical data stored directly in EHRs can be used to identify risk factors for MDR-GNB infections in severe hematological patients at FN onset. ML approach has proved useful for clinical predictors in MDR-GNB infections and helps pave the way for personalized medical care.
Acknowledgements
Funding
This study has been co-funded by a research grant from the Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III [FIS PI18/01061] and a European Regional Development Fund (EDRD). CG-V has received the INTENSIFICACIÓ Grant—a grant supported by the Catalan Health Agency [PERIS (Pla estratègic de recerca i innovació en salut – “Strategic Plan for Research and Innovation in HealthCare”)]. PP-A has received a pre-doctoral grant supported by the Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III [CM18/00132]. No funding bodies had any role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No Rapid Service Fee was received by the journal for the publication of this article.
Medical Writing, Editorial, and Other Assistance
Anthony Armenta, an independent English corrector, provided English language/syntax corrections to the manuscript. His assistance was funded with private research resources from the Department of Infectious Diseases of the Hospital Clínic de Barcelona.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Authorship Contributions
CG-V and PP-A: Literature search, study design, data collection, data analysis, data interpretation, writing, and decision to submit. MAO: Data mining and extraction. GB and JL: pre-processing data (selection, clearing, enrichment and transformation of database), and statistical analyses. All authors: writing-review and editing. CG-V, PP-A, CC, JAM, MC, JAM, JM and AS: Data interpretation, writing and decision to submit. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted.
List of Investigators
ID-INNOVATION study group: L. Morata, A. Urbano, T. Baumann, M. Suarez-Lledó, A. Costa, S. Orozco Martin, L. Cozma, K. Ritter, A. Barreiro Carrillo, M. Rivero, A. Suarez, D. Vidal, X. Pastor. Our group is recognized by the AGAUR (Project 2017SGR1432) of the Catalan Health Agency.
Disclosures
Carolina Garcia-Vidal has received honoraria for talks on behalf of Gilead Science, Merck Sharp and Dohme, Pfizer, Jannsen, Novartis, Lilly and a grant support from Gilead Science and Merck Sharp and Dohme. Pedro Puerta-Alcalde has received honoraria for talks on behalf of Gilead Science and Pfizer and a personal grant of the Instituto de Salud Carlos III (CM18/00132). GB and JL are employees of the firm Accenture Advance Analytics. However, the firm itself played no role in the design of the study or the decision to submit. Alex Soriano has received honoraria for talks on behalf of Merck Sharp and Dohme, Pfizer, Novartis, Angellini, and a grant support from Pfizer. Jaime Lagunas has received honoraria for talks on behalf of Merck Sharp and Dohme, Pfizer, Novartis and Angellini. Celia Cardozo, Miquel A Orellana, Gaston Besanson, Francesc Marco, Ana Del Río, Jose A Martínez, Mariana Chumbita, Nicole Garcia-Pouton, Josep Mensa, Montserrat Rovira and Jordi Esteve have nothing to disclose.
Compliance with Ethics Guidelines
This study was performed in accordance with the Helsinki Declaration. The study was approved by the Ethics Committee Board of our institution (HCB/2018/0308) and followed privacy laws regarding active anonymity.
Data Availability
The datasets generated during and analyzed during the current study are available from the corresponding author on reasonable request.
Footnotes
Carolina Garcia-Vidal and Pedro Puerta-Alcalde contributed equally to this manuscript.
Contributor Information
Carolina Garcia-Vidal, Email: cgarciav@clinic.cat.
Pedro Puerta-Alcalde, Email: pedro.puerta84@gmail.com.
References
- 1.Shouval R, Labopin M, Bondi O, et al. Prediction of allogeneic hematopoietic stem-cell transplantation mortality 100 days after transplantation using a machine learning algorithm: a European Group for blood and marrow transplantation acute leukemia working party retrospective data mining study. J Clin Oncol. 2015;33:3144–3151. [DOI] [PubMed]
- 2.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. [DOI] [PMC free article] [PubMed]
- 3.Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402. [DOI] [PubMed]
- 4.Schwalbe N, Wahl B. Artificial intelligence and the future of global health. Lancet. 2020;1579–1586. [DOI] [PMC free article] [PubMed]
- 5.Garcia-Vidal C, Sanjuan G, Puerta-Alcalde P, Moreno-García E, Soriano A. Artificial intelligence to support clinical decision-making processes. EBioMedicine. 2019;46:27–29. doi: 10.1016/j.ebiom.2019.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mani S, Ozdas A, Aliferis C, et al. Medical decision support using machine learning for early detection of late-onset neonatal sepsis. J Am Med Inform Assoc. 2014;21:326–336. [DOI] [PMC free article] [PubMed]
- 7.Wiens J, Campbell WN, Franklin ES, Guttag JV, Horvitz E. Learning data-driven patient risk str.jpegication models for Clostridium difficile. Open Forum Infect Dis. 2014;1:ofu045. [DOI] [PMC free article] [PubMed]
- 8.Taylor RA, Pare JR, Venkatesh AK, et al. Prediction of in-hospital mortality in emergency department patients with sepsis: a local big data-driven, machine learning approach. Jones A, editor. Acad Emerg Med. 2016;23:269–278. [DOI] [PMC free article] [PubMed]
- 9.Komorowski M, Celi LA, Badawi O, Gordon AC, Faisal AA. The Artificial Intelligence Clinician learns optimal treatment strategies for sepsis in intensive care. Nat Med. 2018;24:1716–1720. [DOI] [PubMed]
- 10.Gudiol C, Calatayud L, Garcia-Vidal C, et al. Bacteraemia due to extended-spectrum β-lactamase-producing Escherichia coli (ESBL-EC) in cancer patients: clinical features, risk factors, molecular epidemiology and outcome. J Antimicrob Chemother. 2009;65:333–341. doi: 10.1093/jac/dkp411. [DOI] [PubMed] [Google Scholar]
- 11.Averbuch D, Tridello G, Hoek J, et al. Antimicrobial resistance in Gram-negative rods causing bacteremia in hematopoietic stem cell transplant recipients: intercontinental prospective study of the Infectious Diseases Working Party of the European Bone Marrow Transplantation Group. Clin Infect Dis. 2017;65:1819–1828. doi: 10.1093/cid/cix646. [DOI] [PubMed] [Google Scholar]
- 12.Gudiol C, Tubau F, Calatayud L, et al. Bacteraemia due to multidrug-resistant Gram-negative bacilli in cancer patients: risk factors, antibiotic therapy and outcomes. J Antimicrob Chemother. 2011;66:657–663. [DOI] [PubMed]
- 13.Oliveira AL, de Souza M, Carvalho-Dias VMH, et al. Epidemiology of bacteremia and factors associated with multi-drug-resistant gram-negative bacteremia in hematopoietic stem cell transplant recipients. Bone Marrow Transplant. 2007;39:775–781. doi: 10.1038/sj.bmt.1705677. [DOI] [PubMed] [Google Scholar]
- 14.Weisser M, Theilacker C, Tschudin Sutter S, et al. Secular trends of bloodstream infections during neutropenia in 15 181 haematopoietic stem cell transplants: 13-year results from a European multicentre surveillance study (ONKO-KISS). Clin Microbiol Infect. 2017;23:854–859. [DOI] [PubMed]
- 15.Garcia-Vidal C, Cardozo-Espinola C, Puerta-Alcalde P, et al. Risk factors for mortality in patients with acute leukemia and bloodstream infections in the era of multiresistance. PLoS One. 2018;13:1–12. [DOI] [PMC free article] [PubMed]
- 16.Lodise TP, Patel N, Kwa A, et al. Predictors of 30-day mortality among patients with Pseudomonas aeruginosa bloodstream infections: impact of delayed appropriate antibiotic selection. Antimicrob Agents Chemother. 2007;51:3510–3515. doi: 10.1128/AAC.00338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Girmenia C, Rossolini GM, Piciocchi A, et al. Infections by carbapenem-resistant Klebsiella pneumoniae in SCT recipients: a nationwide retrospective survey from Italy. Bone Marrow Transplant. 2015;50:282–288. [DOI] [PubMed]
- 18.Trecarichi EM, Pagano L, Martino B, et al. Bloodstream infections caused by Klebsiella pneumoniae in onco-hematological patients: clinical impact of carbapenem resistance in a multicentre prospective survey. Am J Hematol. 2016;91:1076–1081. [DOI] [PubMed]
- 19.Magiorakos A-P, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. [DOI] [PubMed]
- 20.Baron EJ, Miller JM, Weinstein MP, et al. A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2013 recommendations by the Infectious Diseases Society of America (IDSA) and the American Society for Microbiology (ASM). Clin Infect Dis. 2013;57:e22–121. [DOI] [PMC free article] [PubMed]
- 21.Marty FM, Ostrosky-Zeichner L, Cornely OA, et al. Isavuconazole treatment for mucormycosis: a single-arm open-label trial and case-control analysis. Lancet Infect Dis. 2016;16:828–837. [DOI] [PubMed]
- 22.Vehreschild MJGT, Hamprecht A, Peterson L, et al. A multicentre cohort study on colonization and infection with ESBL-producing Enterobacteriaceae in high-risk patients with haematological malignancies. J Antimicrob Chemother. 2014;69:3387–3392. [DOI] [PubMed]
- 23.Trecarichi EM, Pagano L, Candoni A, et al. Current epidemiology and antimicrobial resistance data for bacterial bloodstream infections in patients with hematologic malignancies: an Italian multicentre prospective survey. Clin Microbiol Infect. 2015;21:337–343. doi: 10.1016/j.cmi.2014.11.022. [DOI] [PubMed] [Google Scholar]
- 24.Viasus D, Puerta-Alcalde P, Cardozo C, et al. Predictors of multidrug-resistant Pseudomonas aeruginosa in neutropenic patients with bloodstream infection. Clin Microbiol Infect. 2020;26:345–350. doi: 10.1016/j.cmi.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Tofas P, Samarkos M, Piperaki E-T, et al. Pseudomonas aeruginosa bacteraemia in patients with hematologic malignancies: risk factors, treatment and outcome. Diagn Microbiol Infect Dis. 2017;88:335–341. [DOI] [PubMed]
- 26.Deo RC. Machine learning in medicine. Circulation. 2015;132:1920–1930. [DOI] [PMC free article] [PubMed]
- 27.Wiens J, Shenoy ES. Machine learning for healthcare: on the verge of a major shift in healthcare epidemiology. Clin Infect Dis. 2018;66:149–153. [DOI] [PMC free article] [PubMed]
- 28.Shah ND, Steyerberg EW, Kent DM. Big data and predictive analytics: recalibrating expectations. JAMA. 2018;320:27–28. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and analyzed during the current study are available from the corresponding author on reasonable request.