Abstract
Introduction
Clostridioides (Clostridium) difficile infection, the leading cause of healthcare-associated diarrhea, represents a significant burden on global healthcare systems. Despite being a global issue, information on C. difficile from a global perspective is lacking. The aim of this study is to model the global phylogeography of clinical C. difficile.
Methods
Using samples collected from the MODIFY I and II studies (NCT01241552, NCT01513239), we performed whole-genome sequencing of 1501 clinical isolates including 37 novel sequence types (STs), representing the largest worldwide collection to date.
Results
Our data showed ribotypes, multi-locus sequence typing clades, and whole-genome phylogeny were in good accordance. The clinical C. difficile genome was found to be more conserved than previously reported (61% core genes), and modest recombination rates of 1.4–5.0 were observed across clades. We observed a significant continent distribution preference among five C. difficile clades (Benjamini-Hochberg corrected Fisher’s exact test P < 0.01); moreover, weak association between geographic and genetic distance among ribotypes suggested sources beyond healthcare-related transmission. Markedly different trends of antibiotic susceptibility among lineages and regions were identified, and three novel mutations (in pyridoxamine 5′-phosphate oxidase family protein: Tyr130Ser, Tyr130Cys, and a promoter SNP) associated with metronidazole-reduced susceptibility were discovered on a nim-related gene and its promotor by genome-wide association study. Toxin gene polymorphisms were shown to vary within and between prevalent ribotypes, and novel severe mutations were found on the tcdC toxin regulator protein.
Conclusion
Our systematic characterization of a global set of clinical trial C. difficile isolates from infected individuals demonstrated the complexity of the genetic makeup of this pathogenic organism. The geographic variability of clades, variability in toxin genes, and mutations associated with antibiotic susceptibility indicate a highly complex interaction of C. difficile between host and environment. This dataset will provide a useful resource for validation of findings and future research of C. difficile.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40121-021-00426-6.
Keywords: Antibiotic susceptibility, CDI, Clostridioides (Clostridium) difficile, Evolution, Metronidazole, Moxifloxacin, Phylogeography, Rifaximin, Toxin, Vancomycin
Key Summary Points
| Why carry out this study? |
| Clostridioides (Clostridium) difficile infection poses a significant burden on global healthcare systems; however, a global perspective on the characteristics and distribution of C. difficile lineages is lacking |
| Using samples from the MODIFY I and II clinical trials, this study aimed to analyze the phylogeography, antibiotic susceptibility, and toxin polymorphisms of the largest whole-genome sequenced global collection of clinical trial CDI isolates documented to date |
| What was learned from the study? |
| This study found ribotypes, multi-locus sequence typing clades, and whole-genome phylogeny were in good accordance, and the clinical C. difficile genome was more conserved than previously reported (> 60% core genes) |
| Significant continent distribution preference was observed among five C. difficile clades, with markedly different trends of antibiotic susceptibility among lineages and regions, as well as three novel mutations discovered by GWAS, providing useful insight into this highly clinically relevant population |
| These results provide valuable insight into the spread and characteristics of epidemic C. difficile and allow better understanding of the distribution patterns of hypervirulent strains and antibiotic resistance. |
Digital Features
This article is published with digital features, including a summary slide, to facilitate understanding of the article. To view digital features for this article go to https://doi.org/10.6084/m9.figshare.14113565.
Introduction
Clostridioides (Clostridium) difficile infection (CDI) is the leading cause of healthcare-associated diarrhea, representing a significant burden upon global healthcare systems. Ribotyping and multi-locus sequence typing (MLST) have been the gold standard for the investigation of lineage prevalence and disease association. Determining phylogenetic relationships across geographic regions allows for the investigation of disease dissemination across landscapes. However, the correlation among ribotyping, MLST, and whole-genome single-nucleotide polymorphism (SNP) phylogeny has not been explored with an extensive dataset of cases collected prospectively across multiple continents.
Clostridioides (Clostridium) difficile (C. difficile) shares a paradoxical relationship with antibiotics: CDI is recognized as an antibiotic-associated infection caused by an imbalance in the gut microbiome; conversely, antibiotics are also the first-line treatment for CDI. A detailed review of the prevalence and known mechanisms of antibiotic susceptibility/resistance in C. difficile has been published [1]. Rates of resistance to the two first-line antibiotics, metronidazole (MET) and vancomycin (VAN), are reported to be low in the published literature [2]. Nonetheless, failure of treatment with MET has been increasingly reported in recent years [3]. The resistance mechanisms employed by C. difficile against both first-line antibiotics remain poorly understood [1].
Clinical manifestations of CDI are mainly caused by virulence factors Toxin A (TcdA) and Toxin B (TcdB), encoded and regulated by genes on the pathogenicity locus including the negative regulator Toxin C (TcdC), as well as the binary toxin cytolethal distending toxin (CDT), which may potentiate the toxicity of TcdA and TcdB and lead to more severe disease [4]. Many studies have attributed the hypervirulence of the C. difficile ribotype RT027 to its tcdB sequence heterogeneity, inactivity of tcdC alleles, and presence of CDT [5].
Information on the distribution of C. difficile lineages, ribotypes, geographic associations, resistance prevalence, and toxin polymorphisms from a global perspective is lacking. Using samples collected from the MODIFY I and II studies [6], the largest whole-genome sequenced global collection of clinical trial CDI isolates documented to date, our study aims to describe the characteristics of a representative clinical C. difficile population.
Methods
Sample Collection
The collection of the clinical C. difficile strains used in this study was previously described in detail by Wilcox et al. [6]. Briefly, 1501 CDI isolates were collected from two phase III, randomized, double-blind, placebo-controlled trials (MODIFY I [NCT01241552] and MODIFY II [NCT01513239]). No selection criteria were applied for the strains from the two trials that were analyzed in this study. This cohort represents all available strains cultured from baseline stool samples obtained from enrolled subjects [7].
DNA Extraction and Ribotyping
DNA was extracted from freshly isolated cultures generated from individual colonies, using standard large-scale DNA extraction with Qiagen Kits (QIAGEN, Venlo, The Netherlands). Ribotype analyses were conducted as previously described [8].
Whole-Genome Sequencing
Polymerase chain reaction-free index libraries, with an average insert size of 350 bp, were prepared for each isolate. Genome sequencing was performed using the Illumina® HiSeq™ 2000 platform to generate 100-bp paired-end (PE) or 90-bp PE reads (Illumina, Inc., San Diego, CA). Raw reads were filtered by SOAPnuke (https://github.com/BGI-flexlab/SOAPnuke) to remove sequencing adapters and low-quality reads, including those containing more than three unknown (denoted ‘N’) bases, those in which > 50% of the bases scored < Q20, and those with read length < 30 bp. After the quality-control process, high-quality data with an average of 686 Mb were generated, which has a coverage of 170-fold across the whole genome (~ 4 Mb).
Genome Assembly, Coding Sequence Prediction, and Core/Pangenome Identification
De novo genome assembly was performed using SPAdes v3.11.1 [9] (or SOAPdenovo2 [10] in toxin analysis for several isolates). Non-redundant coding sequence (CDS) prediction and annotation were performed using Prokka v1.13 [11]. Identification of core/pangenome was performed with Roary v3.12.0 [12] (default setting: minimum percentage identity for blastp is 95%, percentage of isolates a gene must be in to be core is 99%).
SNP Analysis
Whole-genome SNP calling: high-quality reads were mapped against reference genome CD196 (NC_013315) using a Burrows-Wheeler Aligner (BWA v0.6.2) [13]. SNPs were identified using SAMtools v0.1.19 [14]. An allele was only considered to be valid if align qual ≥ 100, depth ≥ 10, and alt rate ≥ 0.8 (found in ≥ 80% of reads); otherwise, it was treated as missing.
Repetitive region identification: repetitive regions in the reference genome sequence were identified using an in-house pipeline. Briefly, the reference genome was fragmented in silico into 20-bp reads using a sliding window approach; the reads generated were then mapped back to the reference, and reads mapped to multiple locations of the genome were identified. SNPs falling within these repetitive regions were excluded from all subsequent analyses (3,851,695/4,110,554 = 93.7%). The remaining SNPs were named whole-genome SNPs in our article.
Validation of the SNP pipeline: artificial variants were introduced to generate a pseudo-genome based on CD630 by INDELible v1.03 [15]. The pseudo-genome and CD630 were compared to extract the SNP and insertion/deletion (indel) sites (31,831 SNPs and 8524 indels). Repetitive regions were identified (3,923,283/4,290,244 = 91.4%) in the pseudo-genome using the pipeline described above. Realistic sequencing reads and reads with sequencing errors (based on the sequencing quality of a real sample) were generated by iMESSi [16], accounting for a depth from 20 to 200X of the genome separately. SNPs were called using the above pipeline. Valid accuracy rate was assessed by comparing these two SNP sets (the error rate of the average data size was ~ 0.06%).
Phylogenetic Analyses and Recombination Detection
Pseudo–whole-genome sequences were constructed for each isolate by replacing SNP positions in the CD196 reference genome with the consensus alleles identified by our SNP calling pipeline, with unknown sites being replaced by a gap character. These alignment sequences were adopted to construct the recombination-free maximum-likelihood (ML) phylogeny and detect the recombination rate (r/m ratio) within each clade by Gubbins [17] program. iTOL (https://itol.embl.de/) was used for tree visualization.
Genetic Diversity Analysis
SNP-level difference among isolates within a ribotype was calculated. Pairwise SNPs were counted based on defined bases only; missing data in either of two compared sites were ignored (for example, ‘–’ vs. ‘A’). The average number of nucleotide differences per site (nucleotide diversity, π) among ribotypes was calculated using DnaSP v5 [18].
MLST Analysis
Assembled contigs were aligned to a C. difficile MLST database (http://pubmlst.org/cdifficile), which organizes all allele sequences of seven C. difficile housekeeping genes to identify the sequence type and corresponding clade.
Geographical Epidemiology Analysis
The distribution pattern of the five clades across the six continents was tested by Fisher’s exact test with Monte Carlo simulation. The specific distribution preference of each clade was tested by Fisher’s exact test with the P value corrected using the Benjamini-Hochberg (BH) method. The prevalence patterns of clade 1 and clade 2 in the USA were fitted by polynomial curve (four states with only clade 1 were excluded). The genetic distance between any two isolates was calculated based on the whole-genome SNPs, using the function of dist.dna in the “ape” R package [19] with the substitute model TN93 adopted. The geographic distance between collection sites of any two isolates was calculated by the distCosine command in R, based on the longitude and latitude, which were generated by the geocode command ‘ggmap’ using region information for each isolate. The Mantel test from the R package “ade4” was used to determine correlations between the genetic and geographic distance between isolates, with a null distribution generated by 105 permutations, and the Kendall rank correlation coefficient (τ) was adopted according to the distribution of the two matrices and the sample size of each ribotype. P value was corrected by BH. Restriction of gene flow between geographic areas in each ribotype was tested based on the maximum-likelihood phylogenetic tree of each ribotype using the Slatkin-Maddison test [20] under 1000 permutations in the HyPhy software [21].
Antibiotic Susceptibility/Resistance Analysis
Antibiotic susceptibility testing was performed by R.M. Alden (Culver City, CA) using the agar dilution method approved in the CLSI M11-A8 standard [22]. The resistance or reduced susceptibility of an isolate to MOX, MET, and VAN was categorized according to EUCAST ECOFF (version 7.1) breakpoints (VAN 2 mg/l; MET 2 mg/l; MOX 4 mg/l; RIF 0.004 mg/l) [23]. Isolates with MIC ≥ breakpoint were considered as ‘resistant’ (for MOX and RIF) or as reduced susceptibility (for VAN and MET). Antibiotic resistance determinants were identified using Resistance Gene Identifier 4.0.3 of the Comprehensive Antibiotic Resistance Database (CARD 2.0.0) [24], supplemented with SNPs previously published in the literature [25]. Antibiotic preference of each clade or continent was tested by Fisher’s exact test with the P value corrected by BH.
Genome-Wide Association Study (GWAS)
The antibiotic-resistant GWAS was performed with treeWAS [26] based on a SNP matrix of 1474 isolates with MIC value (n.snps.sim = 5*ncol(snps), P = 1e−05) using a ML phylogenetic tree constructed by whole-genome SNP to test the association.
Toxin Gene Analysis
De novo assembled contigs were compared with reference genes (tcdB and tcdC of CD630 [RT012], which lacks binary toxin genes; cdtA and cdtB of CD196 [RT027], which has an incomplete tcdC gene), using BLAST, and CDS were extracted according to the alignment position for each sample. The presence of indels may prevent sequence alignment, which was addressed by connecting the truncated regions to identify start and end positions and deletion region. A single amino acid sequence (or nucleotide sequence for isolates with incomplete tcdC) was selected randomly from identical sequences as a variant type (VT) within each ribotype (i.e., VT was defined by any sequence variation). VT sequences were multiple-aligned by ClustalW [27] and then used to construct the corresponding maximum-likelihood gene tree. A specific non-synonymous variant of tcdB was displayed by MEGA7 [28]. Additionally, Geneious [29] was adopted to visualize the variants for tcdC. For cdtA and cdtB, the majority base in each site within a ribotype was extracted to construct one typical sequence for that ribotype based on the whole-genome SNPs, generating the gene consensus sequence for each ribotype.
Compliance with Ethics Guidelines
MODIFY I (NCT01241552) and MODIFY II (NCT01513239) were conducted in accordance with Good Clinical Practice guidelines and the provisions of the Declaration of Helsinki. The protocols and amendments were approved by the institutional review board or independent ethics committee at each study site (BGI IRB; IRB #13029). Written informed consent for specimen collection and use was provided by all participants before the trial began.
Results
Phylogeny of Global C. difficile Collection
We performed whole-genome sequencing (WGS) on 1501 C. difficile isolates from stools collected from participants enrolled in two phase III clinical trials (MODIFY I and MODIFY II) [6], which included both hospitalized and out-patients who had been diagnosed with CDI. Participants were enrolled between 2011 and 2015, from 322 sites in 30 countries/regions across six continents (Table S1 and Fig. S1).
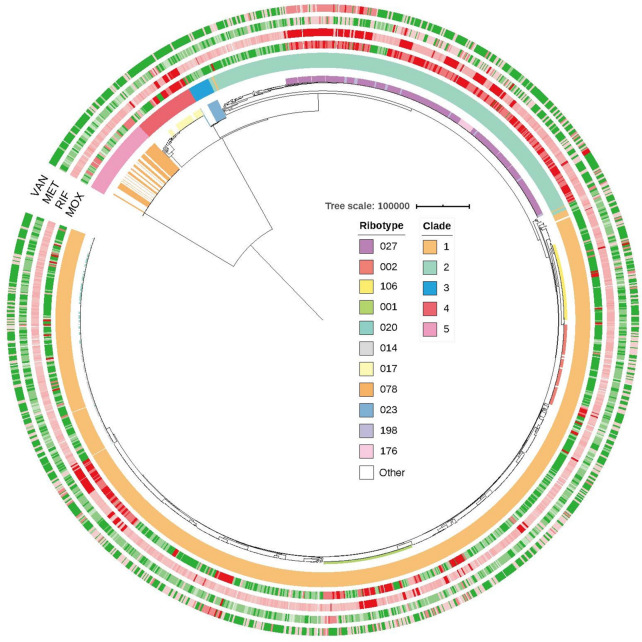
We compared consistency between the WGS-based, recombination-free, maximum-likelihood phylogeny and the two most commonly used molecular typing methods (MLST and ribotyping). Ribotyping analysis identified 164 distinct ribotypes, of which 32 ribotypes contained 10 or more isolates (termed major ribotypes hereafter). MLST analysis identified 129 sequence types (STs), including 29 novel gene alleles and 37 novel STs, which were submitted to the C. difficile MLST database (http://pubmlst.org/cdifficile). Eight isolates could not be identified using the MLST database (Table S2), such that a total of 1493 isolates (99.5%), belonging to 159 ribotypes, could be assigned to five predefined MLST clades. Among these ribotypes, 151 (containing 1467 isolates) were solely found in one of the MLST clades (Table S2). Generally, we observed high concordance among the WGS-based phylogenetic tree, ribotype, and MLST clade (Fig. 1). However, our results demonstrated a relatively poor correlation between ribotype and ST, with only 78 (49%) ribotypes corresponding to unique STs. The correspondence between ribotype and MLST is summarized in Table S2.
Fig. 1.
Maximum-likelihood phylogenetic tree with MIC information of four drugs. Circular presentation of the recombination-free ML phylogeny (midpoint rooted in order to display the structure clearly, five isolates were removed as outliers by Gubbins). MLST clades are labeled on the outer circle; predominant (top 1 for clade 3, 4, 5; top 3 for clade 2; top 5 for clade 1) ribotypes are highlighted in the inner circle. The MICs of MOX, RIF, MET, and VAN for each isolate are presented as heatmaps plotted on the phylogenic tree. The color key for the four outer circles indicates the drug sensitivity of isolates as per EUCAST guidelines: red, resistant or reduced susceptibility; green, sensitive. Branch lengths are indicative of the estimated substitution rate per variable site. CDI C. difficile infection, EUCAST European Committee on Antimicrobial Susceptibility Testing, MET metronidazole, MIC minimum inhibitory concentration, ML maximum-likelihood, MLST multi-locus sequence typing, MOX moxifloxacin, RIF rifaximin, VAN vancomycin
We next investigated the prevalence of ribotypes among the five MLST clades. Clade 1 possessed the greatest number of isolates (63.8%; n = 958) and ribotypes (65.9%; n = 108). The well-known non-toxigenic ribotypes RT009, RT010, and RT039 with toxigenic profile TcdAB– CDT– [30] belong to clade 1. Clade 2 was predominantly comprised of the hypervirulent ribotype RT027 (72.1%; n = 279), along with RT176 (5.4%; n = 21) and RT198 (3.6%; n = 14), which displayed an inseparable phylogenetic relationship with RT027. Clades 3 and 4 were smaller clades, dominated by toxigenic RT023 (65.0%; n = 13) and RT017 (57.9%; n = 33), respectively. Clade 5 was another hypervirulent clade, predominantly comprised of RT078 (62.0%; n = 44) and RT126 (26.8%; n = 19).
Genetic Diversity of the C. difficile Genome
To elucidate the genetic diversity of C. difficile, we carried out pangenome analysis on our collection. We identified a total of 29,921 CDSs, out of which 2317 CDSs were identified as core genes (defined as those present in ≥ 99% of all of the 1501 genomes), representing 61.1% of the average number of CDS per genome (3792 CDS), suggesting that clinical C. difficile isolates shared a conserved core genome.
To estimate the role played by homologous recombination, we calculated the rate of recombination (r) with respect to random point mutation (m) for each clade (r/m ratio). Clades 2 and 5, which include strains that have been shown to be hypervirulent, exhibited the lowest recombination rates at 1.8 and 1.4, respectively. Clades 3 and 4 showed similar recombination rates of 4.8 and 5.0, respectively, while the heterogeneous clade 1 had a slightly lower rate of 3.9. Overall, the C. difficile recombination rates reported here are modest.
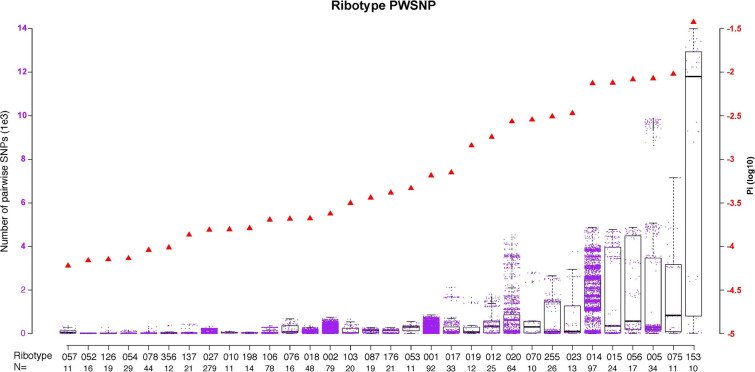
Next, we investigated the nucleotide diversity (π) among the 32 major ribotypes. The π for all major ribotypes correlated well with results from the median pairwise SNP (PWSNP) analysis (Pearson’s correlation coefficient 0.94) (Table S3). The number of PWSNPs within each ribotype varied markedly (Fig. 2), with the greatest genetic variation observed in RT153 (π = 0.037, median PWSNP = 11,792; Table S3).
Fig. 2.
Genetic diversity of all major ribotypes. Each purple dot represents a PWSNP (left y-axis) between two isolates within a ribotype. Each red triangle is the nucleotide diversity (π; right y-axis). The number of samples is provided below the corresponding ribotype. Ribotypes are arranged in order of ascending nucleotide diversity (π). PWSNP pairwise single nucleotide polymorphism
Geographical Epidemiology of C. difficile
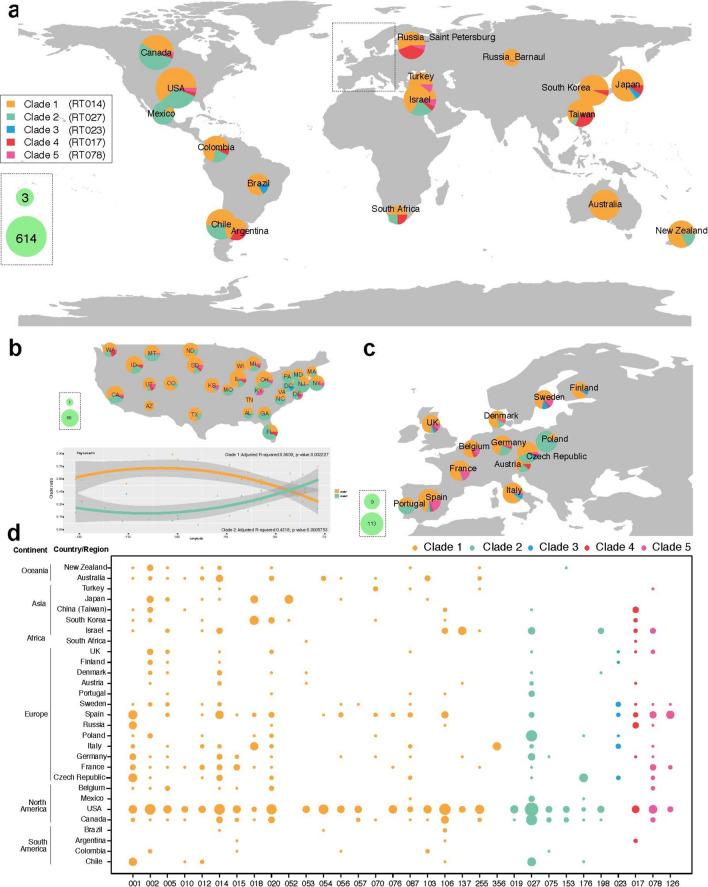
The isolates were collected globally to study C. difficile in the most affected and representative populations. We examined the distribution pattern of the five clades across the six continents and found a significant overall trend for clade distribution (Fisher’s exact test with Monte Carlo simulation, P = 1e−06). Furthermore, we explored the specific distribution preference of each clade by Fisher’s exact test with BH corrected P < 0.01 (Fig. 3a). While the most heterogeneous clade, clade 1, was identified in all sampled countries/regions, it was the most predominant clade found in Oceania (Oceania includes only Australia and New Zealand in our data) (odds ratio [OR] 9.35, 95% confidence interval [CI] 2.99–47.08), followed by Asia (OR 2.14, 95% CI 1.46–3.22). Hypervirulent clades 2 and 5, primarily represented by ribotypes RT027 and RT078, were more likely to be found in North America (OR 2.68, 95% CI 2.09–3.45) and Europe (OR 3.29, 95% CI 1.97–5.56), respectively. Interestingly, the prevalence of clade 1 and clade 2 in the USA (the region with the greatest number of isolates) showed regional divergence; for clade 2, prevalence was higher on the East Coast and West Coast compared with inland, and this observation was reversed for clade 1, which was more prevalent inland (Fig. 3b). Clade 3 was most likely to be found in Europe (OR 5.1, 95% CI 1.83–16.30) (Fig. 3c), and clade 4 was predominantly observed in Asia (OR 2.89, 95% CI 1.45–5.46).
Fig. 3.
Geographic distribution of ribotypes and clades. In panels a, b, and c, each pie chart represents a collection of isolates from a region (country/province/city). The size of the pie chart corresponds to the sample size collected. The proportion of MLST clades is presented in each pie chart; the most predominant ribotype in each clade is provided in parentheses in the color key. The panels show: a global prevalence data; b prevalence data for individual states in the USA; c prevalence data for individual European countries (except Russia). In panel d, the size of the circles corresponds to the number of isolates collected. MLST multi-locus sequence typing
To further understand the geographic distribution of isolates from a more refined perspective, we investigated the relationship between the genetic distance between isolates and the geographic distance between collection sites for each ribotype. Of the 32 major ribotypes analyzed, 9 showed a significant correlation between genetic and geographic distances (BH-corrected Mantel test, P < 0.01) (Table S4). The strongest significant correlation, calculated by Kendall rank correlation coefficient (τ), was detected with RT198 (τ = 0.63, P = 0.001), followed by RT176 (τ = 0.59, P = 0.001) and RT137 (τ = 0.40, P = 0.003). Analysis of gene flow restriction provided support for the above findings from a different angle: significantly fewer migration events were detected for these ribotypes (BH-corrected Slatkin-Maddison test P < 0.01) (Table S4). Although we did not see a correlation between genetic distance and geographic distance, we saw a clear restriction of gene flow across the globe. This suggests that a restriction of gene flow is driven by factors other than simple global distance.
Global Profile of Antibiotic Resistance in C. difficile
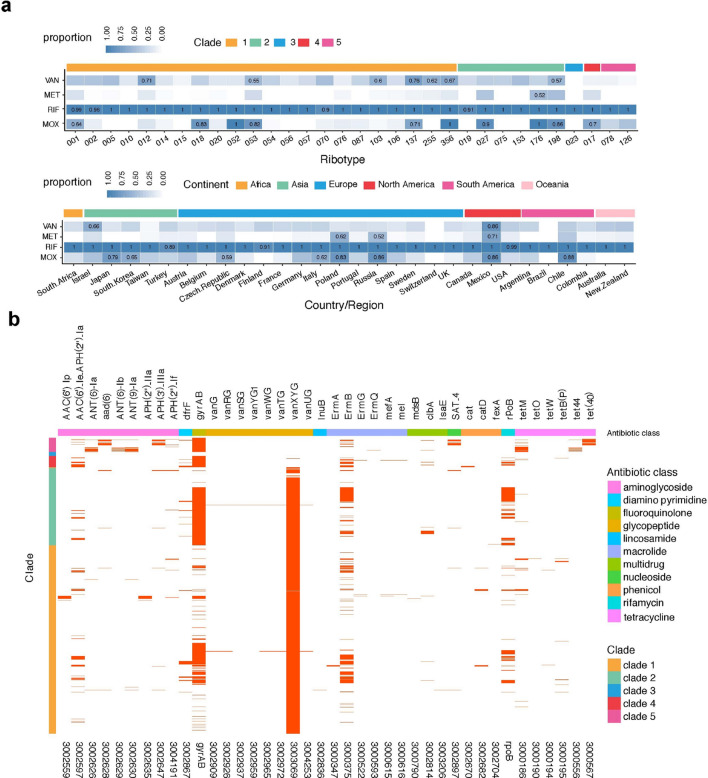
To elucidate the distribution of antibiotic susceptibility/resistance in C. difficile at a global level, we determined the in vitro minimum inhibitory concentrations (MICs) of four antibiotics (VAN, MET, moxifloxacin [MOX], rifaximin [RIF]) for the 1474 C. difficile isolates. Antibiotic susceptibility was determined according to European Committee on Antimicrobial Susceptibility Testing (EUCAST; version 7.1) epidemiological cut-off (ECOFF) breakpoints (VAN 2 mg/l; MET 2 mg/l; MOX 4 mg/l; RIF 0.004 mg/l). The overall reduced susceptibility/resistance rates were 32.8% for VAN, 14.5% for MET, 37.9% for MOX, and 99.5% for RIF. In total, 5.6% (83/1474) of isolates were determined to have reduced susceptibility/resistance to all four classes of antibiotics, out of which 77.1% (64/83) belonged to clade 2, having the highest mean MIC value among the population (Fig. 1). In total, 67% of RT027 strains showed reduced susceptibility or resistance to all four classes of antibiotic tested.
We studied the prevalence of reduced susceptibility/resistance to each of the four antibiotics with EUCAST ECOFF breakpoints among MLST clades and continents. Reduced susceptibility to VAN was significantly less likely among the samples from clade 4 (OR 0.23, 95% CI 0.08–0.55) and clade 5 (OR 0.15, 95% CI 0.05–0.37) as well as the samples from Europe (OR 0.61, 95% CI 0.48–0.79). The three ribotypes with the highest antibiotic susceptibility/resistance rates are RT137, 012, and 356 (0.76, 0.71, and 0.67, respectively). The reduced susceptibility/resistance to both MET and MOX was significantly more likely in clade 2 (OR 9.07, 95% CI 6.52–12.72 and OR 8.15, 95% CI 6.21–10.75 respectively); MOX resistance was significantly more likely to be observed among the samples from Europe (OR 1.58, 95% CI 1.25–1.98) and Asia (OR 2.67, 95% CI 1.91–3.75) than among those from other continents, but was not observed in the samples collected in Oceania. For MET, only RT176 has a high resistant rate (defined as > 0.5), and the high resistant regions are Mexico, Russia, and Poland. For MOX, RT052, 356, and 176 are 100% resistant, and the three regions with the highest resistance rates are Chile, Mexico, and Russia. Totally, the ribotypes which have a high resistance rate in > 3 drugs are 053, 137, 356, 176, and 198, and the regions with high resistance rates are Poland, Russia, and Mexico (Fig. 4a). Given the high prevalence of RIF resistance in our collection, no association with specific clades or locations was found.
Fig. 4.
Prevalence of resistance based on in-vitro data (MIC) and in silico prediction (CARD). a Reduced antibiotic susceptibility/resistance rate across ribotypes, clades, and geographic region. Heatmap of proportions of antibiotic-reduced susceptibility/resistant samples for VAN, MET, RIF, and MOX are plotted from top to bottom against ribotypes, color-coded according to clade or continent of sampling; values are shown for proportions > 0.5. b The presence of resistance determinants, identified using the Comprehensive Antibiotic Resistance Database and literature reports, is indicated with a red line. The resistance determinants are indicated by gene name (top) and Antibiotic Resistance Ontology accession number (bottom), and the corresponding antibiotic class and clade are indicated by the color-coded X and Y axes, respectively. The heatmaps represent the proportions of antibiotic-reduced susceptibility/resistant samples; values are shown for proportions > 0.5. EUCAST European Committee on Antimicrobial Susceptibility Testing, MET metronidazole, MIC minimum inhibitory concentration, MOX moxifloxacin, RIF rifaximin, VAN vancomycin
We next sought to understand to what level the antibiotic susceptibility phenotype can be explained by known genetic determinants and predict the resistance to other drugs. We first identified the reduced susceptibility/resistance determinants in our genome collection using the Comprehensive Antibiotic Resistance Database (CARD 2.0.0). However, the predictive power with CARD annotation for C. difficile is poor for the MOX and RIF resistance: no isolate has resistance based on the CARD reports (0% sensitivity and 100% specificity). The van operons have been recognized as the genetic basis of acquired VAN resistance in Enterococci [31], but the dominant prevalence of the vanXY gene in our collection indicates that such a mechanism cannot explain the observed VAN reduced-susceptibility phenotypes. Due to the poor performance of CARD in predicting MOX and RIF resistance, we manually annotated resistance determinants reported in the literature [25] (rpoB and gyrAB mutations). This manual annotation largely improved the predictive power for MOX (sensitivity = 96.95%, specificity = 87.3%) and RIF resistance (sensitivity = 97.54%, specificity = 98.58%) under the previous study breakpoint of 32 mg/l [32] (Table S5). Based on the combined results of CARD and our curated annotation, clade 2 was noticeably enriched with reported determinants of fluoroquinolone (MOX; gyrA), RIF (rpoB), and macrolide (CLI; ermB) resistance (Fig. 4b). Strains from clade 5, particularly RT126, carried a broader variety of aminoglycoside and tetracycline resistance genes and a higher prevalence of the nucleoside resistance compared with the rest of the population (Fig. S2).
Because there are no known genetic determinants for C. difficile resistance to vancomycin and metronidazole, we performed a GWAS to identify potential novel markers of resistance. To test the performance of the GWAS pipeline used, we first conducted the analysis on MOX, the mechanism of resistance to which is well defined, and were able to identify significant signals for the SNPs Thr82Ile on gyrA and Asp426Asn on gyrB, which are both well-known markers of MOX resistance. Among potential markers, three novel SNPs demonstrated strong association with MET reduced susceptibility: two mutations identified in CD196–1331, Tyr130Ser, and Tyr130Cys, and a SNP located in the promoter region of CD196–1331. CD196–1331 encodes a pyridoxamine 5′-phosphate oxidase family protein (POFP), an oxidoreductase putatively involved in the transfer of electrons during the synthesis of pyridoxal 5′-phosphate (vitamin B6).
Polymorphism of Toxin Genes
To elucidate the genetic basis of toxin-mediated hypervirulence in C. difficile, we analyzed the polymorphism of four toxin-associated genes, tcdB, tcdC, cdtA, and cdtB, across major ribotypes.
In total, 62 VTs of the tcdB gene were identified. VT diversity varied across different ribotypes, the ribotypes in clades 3, 4, and 5 having only one tcdB VT, whereas some ribotypes in clade 2 (RT027, 075, and 153) and clade 1 (50% of ribotypes) had 2–7 VTs. There was an evident clade-associated pattern for tcdB variation (Fig. S3A); clade 5 is distant from the other clades based on WGS or MLST data, but the tcdB gene was not divergent with other clades which suggests relative conservation of tcdB. More interestingly, though whole-gene phylogeny is clade-associated, the diversity of receptor-binding domain sequences (CROP domain) showed a different pattern: RT017 (clade 4) has similar variation to clade 1 (Fig. S3B). Besides, the VT019.1–0042 (12/12) clade 2 is worthy of attention, as the variant pattern could obviously be divided into two regions; the first half (about 1000 residues) was similar to VT017.1–0067 (33/33) (clade 4) with the same single amino acid insertion (at p310), while the latter half was consistent with other VTs in clade 2 isolates.
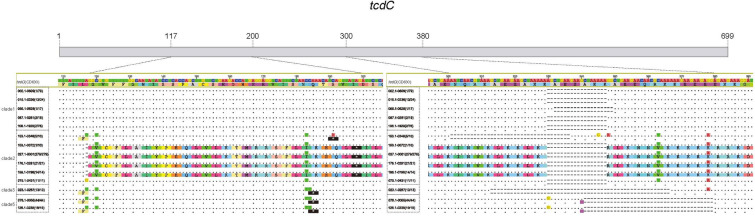
We identified 48 VTs for tcdC in the 32 major ribotypes with 14 VTs containing severe variants (stop-gain mutations or deletions), which generate a truncated TcdC protein (Fig. 5). Among these severe variants, we found the well-documented VT for RT027 [33, 34], RT078 [33, 35], and RT023 [36] and also identified novel variants. The nonsense mutation (C191A) and 36-base pair (bp) deletion have been reported without ribotype information [37]; here, we identified that 50% of RT153 isolates possess this tcdC VT. In addition, a 39-bp deletion has been reported in RT126 [35]; however, we also found a nonsense mutation at position 184 (C184T) for this ribotype, which is the same as RT078. Furthermore, Persson et al. reported that the △117 deletion was 100% specific for RT027 [33], whereas our data indicated that this single bp deletion was also present in one isolate of RT153 and in all isolates of RT176 and RT198. Also, an 18-bp deletion was observed in some isolates in clade 1 and clade 2, which may also produce non-functional proteins (Fig. 5).
Fig. 5.
Variant types of tcdC. VTs with a stop-gain mutation or deletion in the tcdC gene. Each VT has two sequences; the upper sequence is the nucleotide sequence, and the lower sequence is the corresponding amino acid sequence. “-” indicates a deletion; “.” indicates presence of the same base or residue as the reference gene; color indicates bases or residues that differ from the reference gene; “*” with black background indicates a stop codon. VT variant type
Our data also demonstrated three types of structure for the binary genes: RT010 (clade 1) and RT017 (clade 4) lack cdtA and cdtB. Other ribotypes in clade 1 have incomplete cdtA and cdtB genes, lacking about 850 bp from the 3′ end of cdtA and about 1000 bp from the 5′ end of cdtB. All isolates in clades 2, 3, and 5 possessed complete cdtA and cdtB genes. The maximum-likelihood trees of cdtA and cdtB were congruent within the clade (Fig. S4). All VTs for tcdB, tcdC, cdtA and cdtB were summarized in Table S6.
Discussion
In this study, we performed WGS of 1501 CDI isolates collected in two global phase III clinical trials. Our results demonstrated a concordant relationship among ribotypes, MLST clades, and the high-resolution phylogeny inferred by whole-genome SNP analysis. This finding was consistent with a previous report that ribotyping has high discriminative power to distinguish clonal relationships [38], and the relatively poor correlation between ribotype and ST is a comprehensive supplement to a previous study on a smaller dataset [39].
Our study generated the largest clinical-based collection of C. difficile genomes documented to date; the number of pangenome CDS identified was much greater than the previously reported estimate of 9640 CDSs [40]. Although conservation of the core genome of C. difficile was previously reported to be as low as 16% in studies using strains isolated from humans and animals [41, 42], our results showed a much more conserved genome content, with an average of 61% of core genome shared across our collection. Similarly, the number of core genes (2317) identified in clinical samples collected over 3 years is more than that of a recent study (1322) that analyzed samples across a longer time span and diverse host sources [43]. Therefore, it appears that C. difficile strains collected from clinical CDI cases share a much closer genetic relationship than those collected from different hosts, although our samples come from a large geographic span.
Given that recombination has been reported to play an important role in determining C. difficile genomic diversity [44], we further investigated the rate of recombination in our global collection. Our average recombination rates (r/m ratio), ranging from 1.4–5.0 across the five clades, were higher than previously reported. One previous study reported a C. difficile r/m ratio estimate of 0.2 using MLST genes [45]; given the use of only housekeeping genes, this value was perhaps understandably lower than that reported in the current study. Another study, using WGS data, reported a C. difficile r/m ratio of 0.63–1.13 under 95% CI [44]. Indeed, in our study, recombination rates appeared to vary by clade, with the hypervirulent clades 2 and 5 having lower r/m ratios than other clades. Nonetheless, the recombination rates of C. difficile reported here, and by others, are modest when compared with those of other bacterial species [45].
A significantly uneven distribution of clades was found across the globe. Clade 1, a heterogeneous group with a notable absence of hypervirulent ribotypes, was found in every sampling site and was the predominant clade in Oceania. Clades 2 and 5, predominantly represented by the hypervirulent ribotypes RT027 and RT078, had a stronger association with North America and Europe, respectively, which is in line with where these hypervirulent ribotypes were first reported [46, 47]. A previous study that focused on the isolates in the USA showed that the prevalence of RT027 (clade 2) was highest in the Northeast and the South, while RT014/020 (clade 1) was more common in the West, which is partially consistent with our data [48]. Clade 5 was the most genetically distanced from the other clades, possibly because of the well-established livestock origin of its main ribotypes [47]; hence, it is possible that the C. difficile ribotypes comprising this clade may be subject to different selective pressures than those in other clades. Clade 3 was mostly comprised of the toxigenic RT023 ribotype and was predominantly collected from Europe, both in our study and as reported by others [49]. Clade 4 included the hypervirulent RT017 ribotype and was significantly more likely to be found in Asia in our dataset. RT017 has previously been reported to have caused widespread CDI outbreaks in Europe, North America, and particularly Asia [42]. Although the distribution of clades was significantly correlated with continents, the correlation between genetic distance and geographic distance between strains was not strong. In our correlation analysis, stratified by ribotypes, a significant correlation between genetic and geographic distance was demonstrated in only nine ribotypes (28%), of which one was RT027, a well-reported hospital-associated ribotype. Notably, in a pan-European surveillance study, reduced ribotype diversity in individual countries was associated with increased antimicrobial resistance. Resistance to multiple antibiotics was most evident in certain epidemic ribotypes, such as RT027 and RT001, but was also notable in RT017, RT012, emerging RT198 (exclusive to Hungary), and RT356 (exclusive to Italy) [50]. Our data suggest the dissemination of closely related strains across global regions, over long distances; possible explanations for this include the formation of spores to facilitate the remote transmission of C. difficile without genetic changes, foodborne transmission, or that gene flow is strong between C. difficile strains.
Clade 5 exhibited a considerably different resistance-gene pattern to other clades based on in silico annotation, with a broad variety of resistance-conferring alleles, likely because of selective pressure from a range of antibiotics, given that the predominant ribotype in this clade, RT078, is also the predominant strain in livestock entering the human food chain [47]. The accessory genome of RT078 is indistinguishable between animal and human isolates [51], and given it has a broad geographic distribution, high prevalence of resistance determinants, and bidirectional exchange between livestock and humans, the currently rising prevalence of RT078 is a potential threat, and continued global surveillance is important. Fluoroquinolone resistance in RT027, the most predominant ribotype in clade 2, is well documented, and elevated MICs of other antibiotics have also been reported for RT027 isolates [1]. Our results showed that a significant proportion (67%) of RT027 of C. difficile strains demonstrated resistance or reduced susceptibility to all four classes of antibiotics tested, including the first-line antibiotics for CDI treatment, VAN and MET. The high rates of reduced susceptibility/resistance of RT027 may have conferred an advantage in terms of its selective success and expansion. Three novel SNPs in CD196–1331 were significantly associated with MET resistance: one at the promoter region and two at amino acid position 130. CD196–1331 encodes a POFP, which has recently been shown to be distantly related to nim genes (nimA–nimK) encoding 5-nitroimidazole reductase enzymes responsible for reduced MET susceptibility in Bacteroides spp. [52]. CD196–1331 shared 47.5% similarity with the nimE gene, the expression of which has been identified in uncultured bacteria from human stool samples [52], placing the presence of the nimE gene within the habitat of C. difficile. Functional and expressional analyses are needed to confirm the potential role of CD196–1331 in MET resistance in C. difficile.
Different ribotypes could share identical toxin gene VTs (RT078 and RT126), and, conversely, multiple toxin gene VTs could occur within a ribotype. The specific gene character of ribotype 19 raised the possibility that some genetic exchange events may have occurred between ribotype 017 and 109. The diversity in the CROP domain of tcdB is partially associated with clades; ribotype 017 (clade 4) is indiscrete with clade 1. Among all ribotypes, 153 was of special interest because the divergence of its toxin gene VTs was higher than that of other ribotypes. Totally, our analysis of toxin VTs (tcdB, tcdC, cdtAB) demonstrated the diversity of the genetic reservoir for these genes.
Despite the aforementioned insight into the global population characteristics of C. difficile, our study has some limitations. Although the isolates were collected globally, the trials were not designed for the purpose of a balanced global sampling of C. difficile, but to investigate the safety and efficacy of monoclonal antibodies on preventing CDI recurrence in countries/regions where the disease is prevalent, so the isolates were not perfectly randomly sampled to present a representation of global epidemiology. We also expect further studies to establish WGS-based antibiotic susceptibility/resistance prediction models for C. difficile and corresponding experiment to validate the potential resistant determinates. In addition, the role of well-known toxin profiles (TcdB+, CDT+, incomplete TcdC) are partially associated with hypervirulent ribotypes, so other virulence targets should be investigated based on clinical severity phenotypes.
Conclusions
Using samples from participants with clinically diagnosed CDI from across the globe, this analysis improved understanding of genomic, geographic, and clinical features of CDI infection in a clinically relevant epidemic C. difficile population and will serve as a valuable resource for future studies. These findings indicated that within these epidemic populations, the core genome was more highly conserved than previously reported, with modest recombination rates. Genetic variation within the entire genome was most recapitulated by the MLST data. Moreover, material gene transfer (i.e., recombination) was a negligible signal in the genome. Highly distinct geographic prevalence of clade may be related to local selection pressures such as use of certain antibiotics. Antibiotic susceptibility rates are variable among the ribotype/clade and geographic location, which may result from localized selection pressures as suggested by the inverse correlation observed between ribotype diversity and antibiotic resistance in European surveillance samples [50]. The genetics of antibiotic resistance clearly resides on the genomic DNA, as resistance has been shown to be a co-variate with clade designation. The novel potential resistant determinants of MET susceptibility and the novel severe mutations on tcdC need further validation to understand any implications. All these results provide valuable insight into the spread and characteristics of epidemic C. difficile to potentially support future investigation. Better understanding of the spread of hypervirulent strains and antibiotic resistance may aid development of more effective treatment plans, drug development, and ultimately a better outcome for patients.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgments
Funding
This research was funded by the National Science and Technology Major Project of the Ministry of Science and Technology of China (grant number 2017ZX10303406), Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA, the National Natural Science Foundation of China (grant number 31601073), and the National Key Research and Development Program of China (grant number 2018YFC1200501). The Rapid Service Fee was funded by Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Editorial and Other Assistance
Editorial support was provided by Paul O’Neill, PhD, of CMC AFFINITY, a division of McCann Health Medical Communications Ltd, Glasgow, UK, in accordance with Good Publication Practice (GPP3) guidelines. This assistance was funded by Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA. We sincerely thank Karsten Kristiansen (Professor of Molecular Biology, Department of Biology, University of Copenhagen) for helpful discussion and the support provided by China National GeneBank. We also acknowledge Xiaoyun Li for her previous analysis on the project.
Disclosures
David C. Nickle was an employee of Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA, at the time the study was conducted and may own stock and/or stock options. He is currently affiliated with University of Washington, Seattle, WA, USA and Gossamer Bio, San Diego, CA, USA. Zhen Zeng, Lan Chen, Judong Shen, Mary Beth Dorr, and Peter M. Shaw are employees of Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA (MSD), and may own stock and/or stock options in Merck & Co., Inc., Kenilworth, NJ, USA. Rebecca L. Blanchard was an employee of Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA, at the time the study was conducted and may own stock and/or stock options. She is currently affiliated with CRISPR Therapeutics, Cambridge, MA, USA. Mark H. Wilcox has received consulting fees from Abbott Laboratories, Actelion, AiCuris, Antabio, Astellas, AstraZeneca, Bayer, Biomèrieux, Cambimune, Cerexa, Da Volterra, Ferring, Menarini, Meridian, Motif Biosciences, MSD, Nabriva, Paratek, Pfizer, Phico Therapeutics, Qiagen, Roche, Seres, Spero, Surface Skins, Sanofi-Pasteur, Seres, Summit, Synthetic Biologics, The European Tissue Symposium, The Medicines Company, Valneva, and Vaxxilon; lecture fees from Abbott, Alere, Allergan, Astellas, AstraZeneca, MSD, Nabriva, Pfizer, Roche, and Seres; and grant support from Abbott, Actelion, Astellas, Biomèrieux, Cubist, Da Volterra, Merck, MicroPharm, Morphochem AG, Motif Biosciences, Nabriva, Paratek, Pfizer, Sanofi-Pasteur, Seres, Summit, and The European Tissue Symposium. Hailong Zhao, Pierra Y.T. Law, Ye Peng, Jie Meng, Ziqing Deng, Huanzi Zhong, Xun Xu, Shida Zhu, and Junhua Li have nothing to disclose.
Compliance with Ethics Guidelines
MODIFY I (NCT01241552) and MODIFY II (NCT01513239) were conducted in accordance with Good Clinical Practice guidelines and the provisions of the Declaration of Helsinki. The protocols and amendments were approved by the institutional review board or independent ethics committee at each study site (BGI IRB; IRB #13029). Written informed consent for specimen collection and use was provided by all participants before the trial began.
Data Availability
The datasets generated during and/or analysed during the current study are available in the CNGB Nucleotide Sequence Archive repository, https://db.cngb.org/cnsa (accession number CNP0000099). To make our data accessible, user-friendly, and interactive, we developed the C. difficile Pathogen Database (http://dev.dc.org/pathodb/), which not only allows deposition of the genome sequence data and corresponding phenotype data for downloading but also automatically performs phylogenetic and toxin analysis for a given user’s data.
Footnotes
Hailong Zhou, David C. Nickle, Zhen Zeng contributed equally to the work.
Contributor Information
Peter M. Shaw, Email: peter_shaw3@merck.com
Junhua Li, Email: lijunhua@genomics.cn.
References
- 1.Baines SD, Wilcox MH. Antimicrobial resistance and reduced susceptibility in Clostridium difficile: potential consequences for induction, treatment, and recurrence of C. difficile infection. Antibiotics (Basel) 2015;4:267–298. doi: 10.3390/antibiotics4030267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banawas SS. Clostridium difficile infections: a global overview of drug sensitivity and resistance mechanisms. Biomed Res Int. 2018;2018:8414257. doi: 10.1155/2018/8414257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leffler DA, Lamont JT. Clostridium difficile infection. N Engl J Med. 2015;372:1539–1548. doi: 10.1056/NEJMra1403772. [DOI] [PubMed] [Google Scholar]
- 4.Gonçalves C, Decré D, Barbut F, Burghoffer B, Petit JC. Prevalence and characterization of a binary toxin (actin-specific ADP-ribosyltransferase) from Clostridium difficile. J Clin Microbiol. 2004;42:1933–1939. doi: 10.1128/JCM.42.5.1933-1939.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denève C, Janoir C, Poilane I, Fantinato C, Collignon A. New trends in Clostridium difficile virulence and pathogenesis. Int J Antimicrob Agents. 2009;33(Suppl 1):S24–S28. doi: 10.1016/S0924-8579(09)70012-3. [DOI] [PubMed] [Google Scholar]
- 6.Wilcox MH, Gerding DN, Poxton IR, et al. Bezlotoxumab for prevention of recurrent Clostridium difficile infection. N Engl J Med. 2017;376:305–317. doi: 10.1056/NEJMoa1602615. [DOI] [PubMed] [Google Scholar]
- 7.Johnson S, Citron DM, Gerding DN, et al. Efficacy of bezlotoxumab in trial participants infected with Clostridioides difficile strain BI associated with poor outcomes. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1035. [DOI] [PubMed] [Google Scholar]
- 8.Fawley WN, Freeman J, Smith C, et al. Use of highly discriminatory fingerprinting to analyze clusters of Clostridium difficile infection cases due to epidemic ribotype 027 strains. J Clin Microbiol. 2008;46:954–960. doi: 10.1128/JCM.01764-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo R, Liu B, Xie Y, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 12.Page AJ, Cummins CA, Hunt M, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stabler RA, He M, Dawson L, et al. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 2009;10:R102. doi: 10.1186/gb-2009-10-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fletcher W, Yang Z. INDELible: a flexible simulator of biological sequence evolution. Mol Biol Evol. 2009;26:1879–1888. doi: 10.1093/molbev/msp098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mende DR, Waller AS, Sunagawa S, et al. Assessment of metagenomic assembly using simulated next generation sequencing data. PLoS ONE. 2012;7:e31386. doi: 10.1371/journal.pone.0031386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Croucher NJ, Page AJ, Connor TR, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 19.Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 20.Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. 2.75 ed2011.
- 21.Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 22.The Clinical and Laboratory Standards Institute (CLSI). M11-A8: methods for antimicrobial susceptibility testing of anaerobic bacteria; approved standard. 8th ed. Wayne, PA, USA; 2012.
- 23.The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 9.02019 June 03, 2019. https://www.eucast.org/mic_distributions_and_ecoffs/.
- 24.Jia B, Raphenya AR, Alcock B, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spigaglia P. Recent advances in the understanding of antibiotic resistance in Clostridium difficile infection. Ther Adv Infect Dis. 2016;3:23–42. doi: 10.1177/2049936115622891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collins C, Didelot X. A phylogenetic method to perform genome-wide association studies in microbes that accounts for population structure and recombination. PLoS Comput Biol. 2018;14:e1005958. doi: 10.1371/journal.pcbi.1005958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drummond AJ, Ashton B, Buxton S, et al. Geneious v5.4. Auckland, New Zealand.
- 30.Romano V, Pasquale V, Krovacek K, et al. Toxigenic Clostridium difficile PCR ribotypes from wastewater treatment plants in southern Switzerland. Appl Environ Microbiol. 2012;78:6643–6646. doi: 10.1128/AEM.01379-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chancey ST, Zahner D, Stephens DS. Acquired inducible antimicrobial resistance in Gram-positive bacteria. Future Microbiol. 2012;7:959–978. doi: 10.2217/fmb.12.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Connor JR, Galang MA, Sambol SP, et al. Rifampin and rifaximin resistance in clinical isolates of Clostridium difficile. Antimicrob Agents Chemother. 2008;52:2813–2817. doi: 10.1128/AAC.00342-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Persson S, Jensen JN, Olsen KEP. Multiplex PCR method for detection of Clostridium difficile tcdA, tcdB, cdtA, and cdtB and internal in-frame deletion of tcdC. J Clin Microbiol. 2011;49:4299–4300. doi: 10.1128/JCM.05161-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuijper EJ, Coignard B, Tull P. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin Microbiol Infect. 2006;12(Suppl 6):2–18. doi: 10.1111/j.1469-0691.2006.01580.x. [DOI] [PubMed] [Google Scholar]
- 35.Wu YC, Lee JJ, Tsai BY, et al. Potentially hypervirulent Clostridium difficile PCR ribotype 078 lineage isolates in pigs and possible implications for humans in Taiwan. Int J Med Microbiol. 2016;306:115–122. doi: 10.1016/j.ijmm.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 36.Persson S, Torpdahl M, Olsen KEP. New multiplex PCR method for the detection of Clostridium difficile toxin A (tcdA) and toxin B (tcdB) and the binary toxin (cdtA/cdtB) genes applied to a Danish strain collection. Clin Microbiol Infect. 2008;14:1057–1064. doi: 10.1111/j.1469-0691.2008.02092.x. [DOI] [PubMed] [Google Scholar]
- 37.Curry SR, Marsh JW, Muto CA, et al. tcdC genotypes associated with severe TcdC truncation in an epidemic clone and other strains of Clostridium difficile. J Clin Microbiol. 2007;45:215–221. doi: 10.1128/JCM.01599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurka H, Ehrenreich A, Ludwig W, et al. Sequence similarity of Clostridium difficile strains by analysis of conserved genes and genome content is reflected by their ribotype affiliation. PLoS ONE. 2014;9:e86535. doi: 10.1371/journal.pone.0086535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dingle KE, Griffiths D, Didelot X, et al. Clinical Clostridium difficile: clonality and pathogenicity locus diversity. PLoS ONE. 2011;6:e19993. doi: 10.1371/journal.pone.0019993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scaria J, Ponnala L, Janvilisri T, et al. Analysis of ultra low genome conservation in Clostridium difficile. PLoS ONE. 2010;5:e15147. doi: 10.1371/journal.pone.0015147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janvilisri T, Scaria J, Thompson AD, et al. Microarray identification of Clostridium difficile core components and divergent regions associated with host origin. J Bacteriol. 2009;191:3881–3891. doi: 10.1128/JB.00222-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knight DR, Elliott B, Chang BJ, Perkins TT, Riley TV. Diversity and evolution in the genome of Clostridium difficile. Clin Microbiol Rev. 2015;28:721–741. doi: 10.1128/CMR.00127-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar N, Browne HP, Viciani E, et al. Adaptation of host transmission cycle during Clostridium difficile speciation. Nat Genet. 2019;51:1315–1320. doi: 10.1038/s41588-019-0478-8. [DOI] [PubMed] [Google Scholar]
- 44.He M, Sebaihia M, Lawley TD, et al. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc Natl Acad Sci USA. 2010;107:7527–7532. doi: 10.1073/pnas.0914322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vos M, Didelot X. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 2009;3:199–208. doi: 10.1038/ismej.2008.93. [DOI] [PubMed] [Google Scholar]
- 46.He M, Miyajima F, Roberts P, et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet. 2013;45:109–113. doi: 10.1038/ng.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goorhuis A, Bakker D, Corver J, et al. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin Infect Dis. 2008;47:1162–1170. doi: 10.1086/592257. [DOI] [PubMed] [Google Scholar]
- 48.Tickler IA, Goering RV, Whitmore JD, et al. Strain types and antimicrobial resistance patterns of Clostridium difficile isolates from the United States, 2011 to 2013. Antimicrob Agents Chemother. 2014;58:4214–4218. doi: 10.1128/AAC.02775-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bauer MP, Notermans DW, van Benthem BH, et al. Clostridium difficile infection in Europe: a hospital-based survey. Lancet. 2011;377:63–73. doi: 10.1016/S0140-6736(10)61266-4. [DOI] [PubMed] [Google Scholar]
- 50.Freeman J, Vernon J, Pilling S, et al. The ClosER study: results from a three-year pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes, 2011–2014. Clin Microbiol Infect. 2018;24:724–731. doi: 10.1016/j.cmi.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 51.Knetsch CW, Kumar N, Forster SC, et al. Zoonotic transfer of Clostridium difficile harboring antimicrobial resistance between farm animals and humans. J Clin Microbiol. 2018;56:e01384. doi: 10.1128/JCM.01384-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alauzet C, Aujoulat F, Lozniewski A, Marchandin H. A sequence database analysis of 5-nitroimidazole reductase and related proteins to expand knowledge on enzymes responsible for metronidazole inactivation. Anaerobe. 2019;55:29–34. doi: 10.1016/j.anaerobe.2018.10.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available in the CNGB Nucleotide Sequence Archive repository, https://db.cngb.org/cnsa (accession number CNP0000099). To make our data accessible, user-friendly, and interactive, we developed the C. difficile Pathogen Database (http://dev.dc.org/pathodb/), which not only allows deposition of the genome sequence data and corresponding phenotype data for downloading but also automatically performs phylogenetic and toxin analysis for a given user’s data.