Abstract
Tumor cells release nucleic acid-containing proinflammatory complexes, termed nucleic acid-containing damage-associated molecular patterns (NA DAMPs), passively upon death and actively during stress. NA DAMPs activate pattern recognition receptors on cells in the tumor microenvironment leading to prolonged and intensified inflammation that potentiates metastasis. No strategy exists to control endogenous or therapy-induced inflammation in cancer patients. We discovered that the generation 3.0 polyamidoamine dendrimer (PAMAM-G3) scavenges NA DAMPs and mitigates their proinflammatory effects. In this study, we tested if the nucleic acid scavenger (NAS) PAMAM-G3 reduces lung metastasis in murine models of breast cancer. Our data indicate that PAMAM-G3 treatment decreases cell-free DNA levels and reduces lung metastasis in the experimental intravenous tumor-injection model and the postsurgical tumor-resection model of 4T1 breast cancer. Reduction in lung metastasis is associated with reduction in inflammatory immune cell subsets and proinflammatory cytokine levels in the tumor and the periphery. This study is the first example of NAS-mediated inhibition of metastasis to the lung. The study results provide a strong rationale for inclusion of NAS therapy in women with breast cancer undergoing standard-of-care surgery.
Keywords: breast cancer lung metastasis, inflammation, inflammatory immune cells, nucleic acid scavenger, PAMAM-G3
Graphical abstract
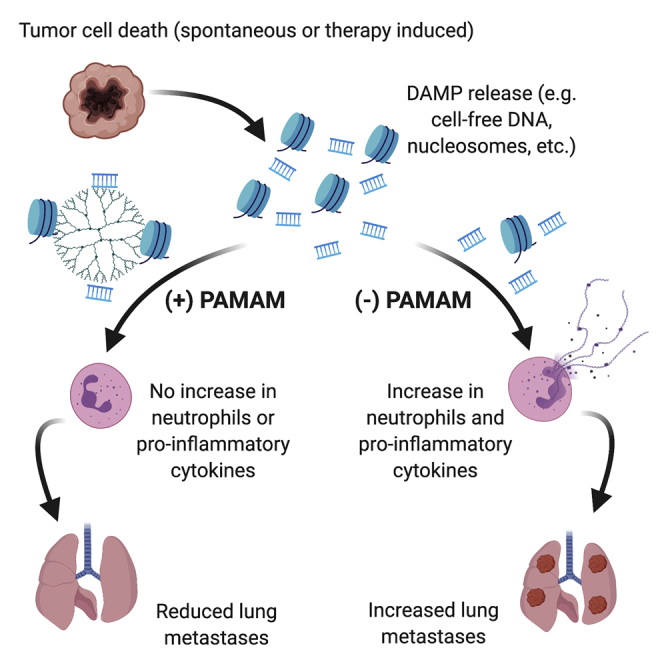
Tumor cell death releases nucleic acids that activate TLR, causing systemic inflammation, thereby enabling tumor metastasis. Nair and colleagues demonstrate that controlling cancer-induced inflammation with the nucleic acid scavenger PAMAM-G3 reduces lung metastasis in murine breast cancer models. PAMAM-G3 treatment reduces inflammatory immune cells and proinflammatory cytokines in the tumor and the periphery.
Introduction
Nucleic acid molecules were thought to be largely immunologically inert until Krieg1 discovered that unmethylated CpG motifs in plasmid DNA-based vaccines were critical components of such vaccines. Subsequently, it was discovered that the innate immune system employs pattern recognition receptors (PRRs) to recognize various molecular patterns associated with harmful pathogens and damaged cells and to initiate inflammatory responses. These signals alert the immune system of an infection or tissue damage and are called pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs).2,3 Bacterial- and viral-derived DNA and RNA molecules activate several transmembrane and cytoplasmic PRRs, including Toll-like receptors (TLRs), nucleotide-binding oligomerization domain-like receptors (NLRs), C-type lectin receptors (CLRs), and RIG-I like receptors (RLRs).4, 5, 6, 7, 8, 9, 10, 11 Given their potent immunostimulatory and proinflammatory effects, inappropriate activation of PRRs is associated with a broad range of inflammatory diseases, including cancer.9,11,12 It has become increasingly apparent that nucleic acid-sensing TLRs and PRRs play a central role in numerous inflammatory disorders, presumably because dead and dying cells release nucleic acids and nucleic acid-containing complexes into extracellular space and activate PRR-expressing cells.9,11,12 Thus, nucleic acid-sensing PRRs have become attractive therapeutic targets for treatment of many diseases, from cancer to autoimmune disease.
When excessive DAMPs are released from stressed/dying tumor cells, inflammation becomes prolonged, intensified, and detrimental, a process that is accelerated with standard-of-care cytotoxic chemotherapy and surgical therapies.13, 14, 15, 16, 17, 18, 19, 20, 21, 22 DAMPs released from tumor cells include cell-free (cf) nucleic acids23,24 (cfDNA, cfRNA, and cf microRNA [miRNA]), referred to as nucleic acid-containing DAMPs (NA DAMPs), and associated proteins, such as nucleosomes and high-mobility group box-1 (HMGB-1).25, 26, 27 We have observed that cancer patients have higher levels of cfDNA in blood when compared to healthy controls, and cfDNA levels increase with disease burden and following chemoradiation therapy and surgery.28 NA DAMPs have also been implicated in tumor progression and metastasis. We demonstrated that circulating cfDNA-containing DAMPs from cancer patients activate PRRs (e.g., TLR4 and -9), whereas serum from healthy donors does not.28 Recent studies in breast, lung, and pancreatic cancers have shown that the presence of cfDNA in blood results in tumor cells that are more invasive and aggressive in vitro.26,29, 30, 31 Although the mechanism(s) by which NA DAMPs mediate tumor progression and metastasis are not fully elucidated, a likely explanation is the recognition of such complexes by and subsequent activation of innate immune receptors on cells in the tumor microenvironment. Unfortunately, the complex network and redundancy of PRRs and their ability to sense a variety of structurally different nucleic acid ligands have made it challenging to develop effective inhibitors that can ameliorate the multimodal proinflammatory effects of RNA/DNA and NA DAMPs.
We have pioneered an innovative method to ameliorate the downstream proinflammatory effects of cfDNA, cfRNA, and associated protein complexes using nucleic acid-binding polymers, such as the generation 3.0 polyamidoamine dendrimer (PAMAM-G3). We have previously demonstrated that PAMAM-G3 can specifically block nucleic acid-mediated activation of TLRs in vitro32 and can serve as a therapeutic anti-inflammatory agent in mouse models of systemic lupus erythematosus33 without engendering nonspecific immune suppression.34 In Naqvi et al.,28 we reported that PAMAM-G3 inhibits nucleic acid-mediated activation of TLRs on and invasion of human pancreatic tumor cells in vitro and dramatically reduces liver metastasis in a pancreatic cancer model in mice. Given that PAMAM-G3 is cleared by the liver, the ability to control metastasis may be restricted to tumors that metastasize to the liver. In the present study, we therefore examined metastasis to lungs and investigated if reducing inflammation with the nucleic acid scavenger (NAS) PAMAM-G3 reduces lung metastasis in two murine models of breast cancer: an experimental intravenous tumor-injection model and an orthotopic surgical tumor-resection model. We also examined changes in innate and adaptive immune cell profiles in metastatic lung tumors and periphery in mice treated with PAMAM-G3.
Results
NAS PAMAM-G3 reduces lung metastasis in an intravenous lung metastasis mouse model of breast cancer
In this study, we investigated the ability of PAMAM-G3 to block lung metastasis in a syngeneic immunocompetent murine model of breast cancer. In this model, intravenously injected tumor cells seed the lung and serve as a surrogate for lung metastasis. The model is especially aggressive, as cells colonize in the lung, and macroscopic metastatic lesions develop in approximately 7 days. Female BALB/c mice were intravenously injected with 4T1 breast cancer cells expressing luciferase (luc), as described in Materials and methods.
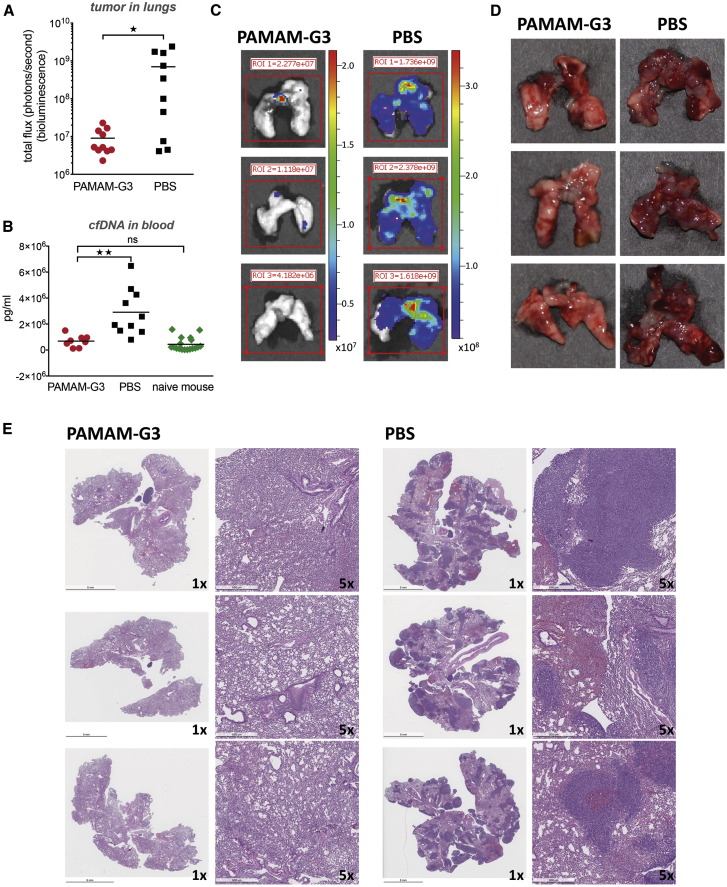
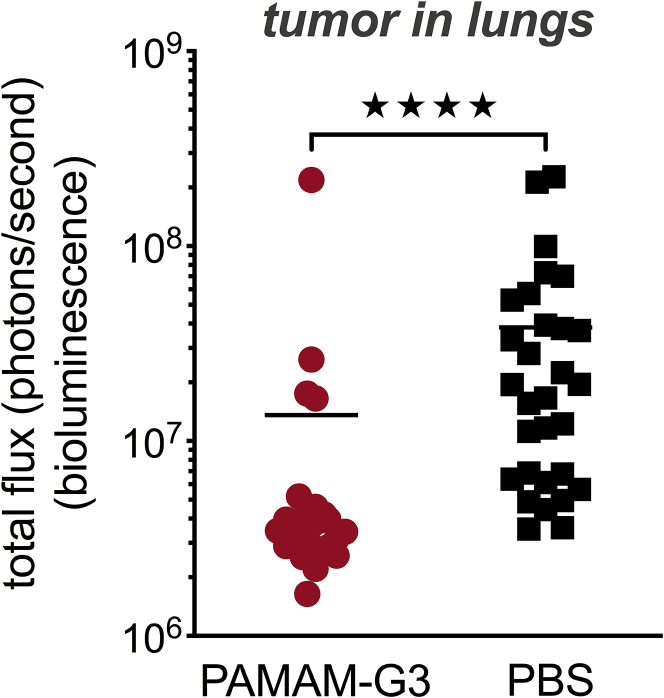
As shown in Figure 1A, PAMAM-G3 treatment resulted in a significant reduction in metastatic tumor burden in the lungs (p < 0.05), as measured by tumor cell bioluminescence in lungs. This reduction in metastatic tumor burden in the lungs correlated with a significant reduction in levels of plasma cfDNA (Figure 1B). Figure 1C illustrates 3 representative examples of bioluminescence in lungs from mice treated with PAMAM-G3 or PBS. Figure 1D illustrates 3 representative photographs of harvested lungs demonstrating a significant tumor burden in PBS-treated mice as compared to PAMAM-G3-treated mice. Figure 1E illustrates histopathology of harvested lungs using hematoxylin and eosin (H&E) staining and indicates the presence of multiple tumor foci throughout the lungs of PBS-treated mice, which was not observed in PAMAM-G3-treated mice. The data in Figure 1 indicate that PAMAM-G3 effectively reduces metastasis to the lungs in the 4T1 intravenous experimental lung metastasis mouse model of breast cancer. This reduction in tumor burden correlates with diminished cfDNA DAMP levels.
Figure 1.
PAMAM-G3 reduces lung metastasis in the intravenous experimental lung metastasis mouse model of breast cancer
Female BALB/c mice were injected intravenously with 104 4T1-luciferase cells (day 0). Mice were injected intraperitoneally with PAMAM-G3 at 20 mg/kg or PBS prior to tumor injection on days −4 and −2. Following tumor injection on day 0, they were treated with either PAMAM-G3 or PBS on days 2, 6, 9, and 13. Luciferase imaging was used to monitor tumor growth and lung metastasis. Lungs were harvested posteuthanasia and imaged to record bioluminescence ex vivo. (A) Mice (n = 10) were sacrificed 15−17 days post-tumor injection, and lung tumor burden was assessed using bioluminescence. ∗p < 0.05, unpaired t test (Mann-Whitney test). (B) Blood was obtained from mice (n = 8−10) at the time of study termination, and circulating levels of cfDNA were assessed in plasma using a PicoGreen assay. Naive mice (nontumor bearing; n = 18) were used as controls. Circulating cfDNA levels are expressed as picogram per milliliter. ∗∗p < 0.01; ns, not significant; unpaired t test (Kruskal-Wallis test). (C) Representative images of bioluminescent lungs obtained using PerkinElmer In Vivo Imaging System. Luciferase expression was assessed for all tumor-bearing mice treated with PAMAM-G3 or PBS. (D) Representative photographs of harvested lungs demonstrating tumor burden in PAMAM-G3- or PBS-treated mice. (E) Gross histopathology of harvested lungs was determined using H&E staining. Images represent 1× or 5× magnification.
PAMAM-G3-mediated reduction in breast cancer metastasis results in changes in the local and systemic immune landscape
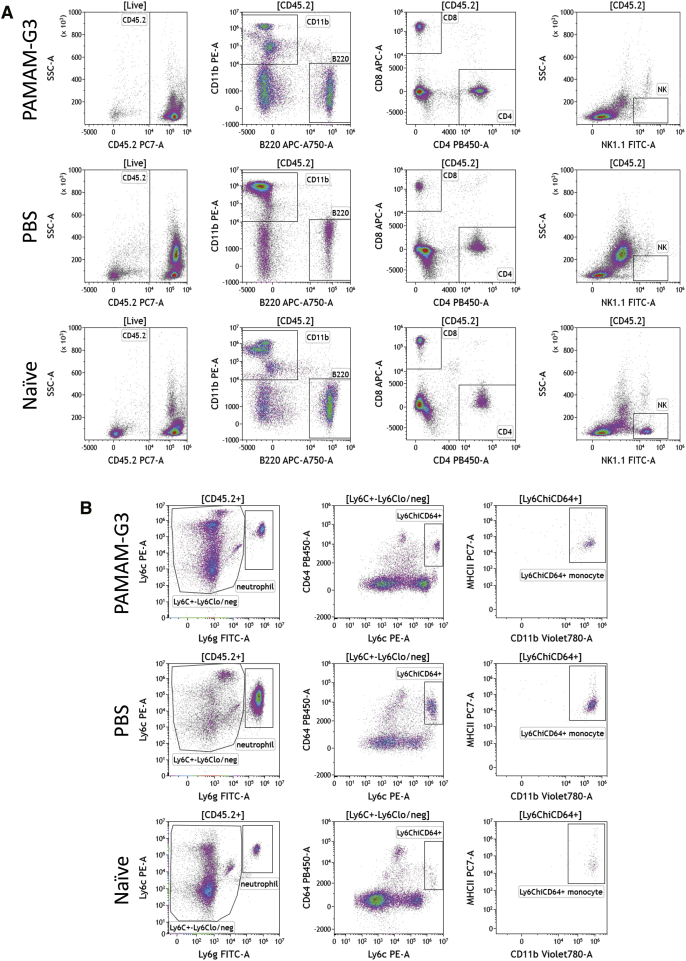
In Figure 1, we demonstrated that PAMAM-G3 treatment prevents lung metastasis and reduces the levels of the DAMP, cfDNA, in the 4T1 experimental intravenous metastasis model of breast cancer. Recognition of DAMPs/PAMPs by PRRs on immune cells, especially innate immune cells, induces their activation, which can lead to inflammation. Given that DAMPs activate immune cells, we investigated if systemic PAMAM-G3 treatment and corresponding reduction in systemic cfDNA levels are associated with changes in the local (tumor/lung) and systemic (spleen) innate and adaptive immune cell profiles of these mice. Figure 2 illustrates the gating strategy used to identify the following: adaptive immune cells (CD4 and CD8 T cells and B cells) and natural killer (NK) cells (Figure 2A) and innate/myeloid inflammatory immune cells (neutrophils [Ly6G+Ly6C+]) and Ly6ChiCD64+major histocompatibility complex class II (MHC-II)+ inflammatory monocytes (i-monocytes)35,36 in lungs (Figure 2B). In some studies, Ly6ChiCD64+MHC-II+ i-monocytes are termed monocyte-derived cells37 or metastasis-associated macrophage precursor cells.38 Similar analysis was conducted to analyze immune subsets in spleen (Figure S1).
Figure 2.
Gating strategy to examine immune cell landscape in tumor/lungs of tumor-bearing mice treated with PAMAM-G3 or PBS
The intravenous experimental lung metastasis 4T1 mouse model of breast cancer described in Figure 1 was used for this analysis. Lungs were collected from PAMAM-G3- or PBS-treated tumor-bearing mice on day 15 and processed for immune cell analysis via flow cytometry. Lungs from naive nontumor-bearing mice were used as controls. One representative mouse from each group is shown. (A) Hematopoietic cells in lungs were identified via CD45.2 cell surface expression. Immune cell populations were identified based on expression of CD11b (innate/myeloid), B220 (B cells), CD4 and CD8 T cells, and NK1.1 (NK cells). (B) Neutrophils within the CD45.2 gate were further identified as Ly6C+Ly6G+. Ly6C+Ly6Clo/neg cells that did not express Ly6G were analyzed for expression of Ly6C and CD64. Ly6ChiCD64+ monocytes were examined for expression of MHC class II (MHC-II) and were identified as inflammatory monocytes (also known as monocyte-derived cells).
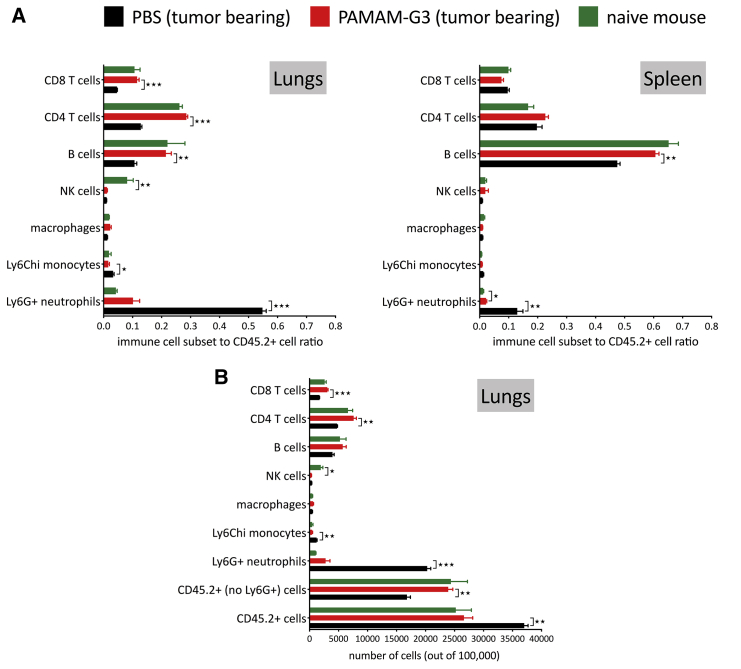
Significant changes in immune cell populations (as identified in Figures 2 and S1) were noted in PAMAM-G3-treated animals and are presented as a ratio of immune cell subsets within the CD45+ immune cell compartment in the lungs and spleen (Figure 3A). Lungs and spleen from naive mice were used to understand the relevance of the changes observed in lungs and spleen from 4T1 tumor-bearing mice treated with either PAMAM-G3 or PBS. Analysis of innate/myeloid immune cells revealed changes in two inflammatory cell subsets: neutrophils and Ly6ChiCD64+MHC-II+ i-monocytes. We did not observe any changes in the macrophages (CD45+CD11b+Ly6G−Ly6C− class II ± CD11c+F4/80+) in lungs or spleen of PAMAM-G3- or PBS-treated mice. Clearly, the most striking finding from this analysis was the observed decrease in Ly6G+ neutrophils in lungs obtained from PAMAM-G3-treated mice compared to PBS-treated mice. Neutrophils are the primary responders to sites of insult/injury and signify an inflammatory response; elimination of neutrophils following inflammation (also called resolution of inflammation) promotes transition from innate immunity to adaptive immunity.39, 40, 41 To further appreciate the changes in immune cell subsets in lungs, we looked at a number of cells in 100,000 acquired events (Figure 3B). PBS-treated lungs had a significantly higher number of CD45.2+ immune cells as compared to PAMAM-G3-treated lungs. Notably, a considerable fraction of these immune cells was neutrophils. Next, we compared the number of CD45.2+ immune cells, excluding the Ly6G+ neutrophil population (CD45.2+ [no Ly6G+] cells). We observed that the number of this subset of immune cells in PBS-treated lungs was in fact significantly lower than CD45.2+ (no Ly6G+) cells in PAMAM-G3-treated lungs (Figure 3B), even though the total number of CD45.2+ cells in the lungs was higher. Of note, the number of CD45.2+ immune cells in lungs was similar in PAMAM-G3-treated lungs and naive nontumor-bearing mouse lungs (Figures 3A, left, and 3B). Importantly, we observed that reduction in neutrophils was evident locally in the metastatic lung tumor and systemically in the spleen of PAMAM-G3-treated mice (Figure 3A, right).
Figure 3.
Changes in immune cell landscape in tumor/lungs and spleen of tumor-bearing mice treated with PAMAM-G3 or PBS
The intravenous experimental lung metastasis 4T1 mouse model of breast cancer described in Figure 1 was used for this analysis. Immune cell subsets in lungs and spleen were identified using the gating strategy described in Figure 2. Lungs and spleen from naive nontumor-bearing mice were used as controls. Data represent an average of 3 mice per group ± standard error of mean (SEM). (A) The ratio of immune cell subsets to total CD45.2+ immune cells in lungs (left) and spleen (right) is presented in the figure. (B) Number of immune cells (CD45.2+ cells) and immune cell subsets in lungs in 100,000 cellular events acquired for each sample. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, unpaired t test.
The significant increase in neutrophil number in lungs from PBS-treated, cancer-bearing mice had a major impact on the ratio of adaptive immune cells to total immune cells in lungs (Figure 3A, left). Lungs from PAMAM-G3-treated, 4T1 tumor-bearing mice had higher proportions of CD8 and CD4 T cells and B cells as compared to lungs from PBS-treated 4T1 tumor-bearing mice. Moreover, ratios of CD8 and CD4 T cells and B cells to total immune cells in lungs from PAMAM-G3-treated, 4T1 tumor-bearing mice were similar to those observed in naive mouse lungs that were not injected with 4T1 tumor cells. Interestingly however, lungs from 4T1 tumor-bearing mice have significantly reduced NK cells regardless of treatment (PBS or PAMAM-G3) as compared to lungs from naive mice not challenged with 4T1 breast cancer cells. Thus, intravenous injection of 4T1 tumor cells in mice alters the repertoire of immune cells both locally in the metastatic lung tumor and systemically in the spleen; however, PAMAM-G3 treatment tempers such changes.
In Naqvi et al.,28 we reported that PAMAM-G3 administration (6 injections, 20 mg/kg) in naive nontumor-bearing mice does not cause hepatic toxicity, renal toxicity, or changes in blood cell counts that are indicative of systemic toxicity. We measured the enzymes aspartate aminotransferase and alanine aminotransferase enzymes in plasma as a marker of liver function and plasma creatinine and blood urea nitrogen as a marker of kidney function. To determine how PAMAM-G3 treatment impacts immune cell subsets in nontumor-bearing mice, naive mice with no tumors were injected with PAMAM-G3 (20 mg/kg) or PBS, 6 times at 3-day interval. 1 day after the final injection, we harvested lungs and spleen and examined immune cell subsets using flow cytometry. As shown in Figure S2, we observed no changes in immune cell subsets in lungs from PAMAM-G3- versus PBS-treated naive mice with no tumors. Similarly, we did not observe differences in immune cell subsets in the spleen, with the exception of a modest reduction of CD4 T cells in the spleens of PAMAM-G3-treated mice.
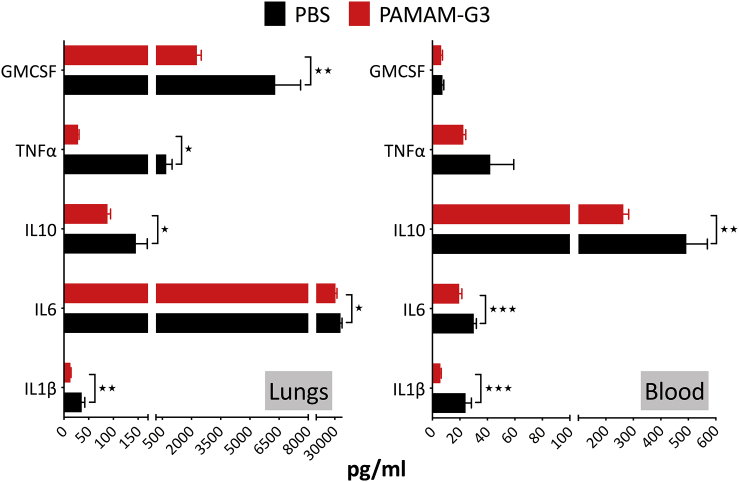
PAMAM-G3-mediated reduction in lung metastasis is associated with a decrease in proinflammatory cytokines in metastatic lung tumor and blood
The larger metastatic burden observed in PBS-treated 4T1 tumor-bearing mice is associated with an elevation in the number of neutrophils and Ly6ChiCD64+MHC-II+ i-monocytes in tumor-bearing lungs (Figure 3A, left). PAMAM-G3 treatment results in a reduction in the number of neutrophils and Ly6ChiCD64+MHC-II+ i-monocytes. Given that inflammation and mobilization of inflammatory innate myeloid cells, such as neutrophils, are associated with an increase in proinflammatory cytokines, we examined proinflammatory cytokine levels both locally (in tumor/lung) and systemically (in blood) in the 4T1 intravenous metastasis model of breast cancer. Data in Figure 4 indicate that lungs from PBS-treated, 4T1 tumor-bearing mice have elevated levels of the proinflammatory cytokines interleukin (IL)-1β, IL-6, IL-10, tumor necrosis factor (TNF)-α, and GMCSF, when compared to 4T1 tumor-bearing mice treated with PAMAM-G3. This reduction in proinflammatory cytokines is consistent with a reduction in levels of cfDNA DAMPs and inflammatory cells, such as neutrophils and Ly6ChiCD64+MHC-II+ i-monocytes, in PAMAM-G3-treated mice. Along with the local response in lungs, we observed a reduction in circulating levels of proinflammatory cytokines IL-1β, IL-6, and IL-10 in blood from 4T1 tumor-bearing mice treated with PAMAM-G3 as compared to mice treated with PBS.
Figure 4.
Proinflammatory cytokine levels in tumor/lungs and blood of tumor-bearing mice treated with PAMAM-G3 or PBS
The intravenous experimental lung metastasis 4T1 mouse model of breast cancer described in Figure 1 was used for this analysis. Lungs and blood were collected from PAMAM-G3- or PBS-treated animals on day 15. Plasma and processed lung supernatant (described in Materials and methods) were used for analysis. Analysis was conducted using the BioLegend LEGENDplex bead-based multiplex assay. Data represent an average of 5 mice per group ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, unpaired t test.
To determine how PAMAM-G3 treatment impacts circulating levels of proinflammatory cytokines in nontumor-bearing mice, naive mice with no tumors were injected with PAMAM-G3 or PBS, 6 times at a 3-day interval. 1 day after the final injection, we harvested blood and examined cytokine levels in plasma. As shown in Figure S3, there were no differences in circulating levels of the proinflammatory cytokines IL-1β, IL-6, IL-10, and GM-CSF in blood from PAMAM-G3- versus PBS-treated naive mice with no tumors. TNF-α levels in blood were higher in PAMAM-G3-treated naive mice with no tumors, which is in contrast to what we observed in Figure 4, where PAMAM-G3-treated, tumor-bearing mice had lower levels of TNF-α.
Collectively, these results indicate that the PAMAM-G3-mediated reduction in breast cancer metastasis to the lung is accompanied by a reduction in cfDNA DAMPs in circulation, a decrease in neutrophils and i-monocytes in lungs, a decrease in neutrophils in spleen, as well as a reduction in local and circulating levels of proinflammatory cytokines.
NAS PAMAM-G3 reduces lung metastasis in a postsurgical-resection metastasis model of breast cancer
In Figure 1, we determined that PAMAM-G3 treatment reduces lung metastasis in the 4T1 intravenous lung metastasis mouse model of breast cancer. However, clinically, breast cancer metastasis is most often diagnosed after the patient has undergone surgical resection of the primary tumor. Therefore, we established a clinically relevant mouse model that closely resembles standard-of-care for breast cancer patients. As shown in Figure 5, treatment with PAMAM-G3 significantly reduced lung metastasis, as compared to PBS treatment (p < 0.0001), following surgical resection of a primary breast cancer. These data indicate that NAS PAMAM-G3 treatment significantly reduces lung metastasis in a clinically relevant, postsurgical breast cancer-resection mouse.
Figure 5.
PAMAM-G3 reduces lung metastasis in a postsurgical-resection lung metastasis mouse model of breast cancer
Female BALB/c mice were injected with 4T1-luciferase cells (8 × 105) orthotopically in the left abdominal mammary fat pad #4 on day 0. Mice were weighed and treated according to weight with either PAMAM-G3 (20 mg/kg) or PBS intraperitoneally on days 0, 3, 10, 13, 16, and 20. Tumors were surgically resected on day 7 post-tumor implantation. Luciferase imaging was used to monitor tumor growth and lung metastasis. Mice were euthanized 22−24 days post-tumor injection. Lungs were harvested posteuthanasia and imaged to record bioluminescence ex vivo. ∗∗∗∗p < 0.0001, unpaired t test (Mann-Whitney test); n = 26 in PAMAM-G3 group; n = 30 in PBS group.
Discussion
In this study, we tested if controlling systemic inflammation with PAMAM-G3 reduces breast cancer metastasis to the lung in mice. Our data indicate the following: (1) the control of inflammation by decreasing NA DAMPs (e.g., cfDNA) using the NAS PAMAM-G3results in reduced lung metastasis in the experimental intravenous tumor-injection model and the postsurgical tumor-resection model of 4T1 breast cancer (Figures 1 and 5), and (2) the NAS PAMAM-G3 significantly reduces inflammatory immune cell subsets and proinflammatory cytokine levels in the tumor and the periphery (Figures 2, 3, 4, and S1). Collectively, these results highlight the potential utility of the NAS approach to treat cancer-associated chronic inflammation and thereby inhibit cancer metastasis. In addition, these studies provide the first example of NAS-mediated inhibition of metastasis to the lung, complement our prior studies examining the ability of this approach to limit pancreatic cancer metastasis to the liver,28 and indicate that the scavenger-based anti-metastasis approach is generalizable to other solid tumors that metastasize to various organs and tissues.
Although the mechanism(s) by which NA DAMPs mediate tumor progression and metastasis are not fully elucidated, a likely explanation is the recognition of such DAMP complexes by innate immune receptors. For example, cfDNA is thought to exert its downstream effects via stimulation of TLR9 via genomic DNA or mtDNA (mitochondrial DNA) containing unmethylated CpG motifs. mtDNA evolutionarily resembles bacterial DNA and retains many of its proinflammatory properties.26,42 RNA-sensing TLR3 and TLR7 have also been shown to play critical roles in inflammation and potentially, in cancer progression.25,26,43,44 In addition, cfDNA-associated proteins can activate TLR2 and -4 and TLR-associated receptors, such as the receptor for advanced glycation end products (RAGE). Nucleosomes and HMGB-1 have been shown to ligate TLR4 and RAGE, both independently and complexed to DNA, leading to nuclear factor κB (NF-κB) activation in tumor cells and immune cells. Palha De Sousa et al.45 have shown that 4T1 tumor cells express TLR2, -3, -4, -5, and -9 with TLR2 and TLR4 expressed at the greatest levels. Thus, cfDNA and associated proteins can have diverse and potent effects on tumor progression and metastasis via TLR and RAGE signaling axes.27,46, 47, 48, 49, 50
Cancer development and progression are associated with immune suppression and loss of immune homeostasis. Recognition of DAMPs/PAMPs by PRRs on immune cells, especially innate immune cells, elicits inflammation. An inflammatory response prompts innate and adaptive immune cell infiltration and is designed to re-establish immune homeostasis. In cancer, this same inflammatory response can be co-opted by the tumor cells to enable tumor growth and progression by prolonging inflammation and simultaneously modulating immune cell function to promote cancer growth. This pathological response was corroborated in the 4T1 experimental lung metastasis model. Data in Figures 2, 3, 4, and S1 indicate that 4T1 tumor growth is associated with higher number of inflammatory cells (neutrophils and i-monocytes) in the metastatic lung tumors as well as in the spleen. This increase in inflammatory cells is also associated with higher levels of cancer-promoting proinflammatory cytokines in the tumor environment and in the periphery (blood). As the data indicate, PAMAM-G3 treatment restored the balance between the innate immune cells and the adaptive T/B cells in the tumor and the spleen. We saw a significant decrease in the numbers of neutrophils in the tumor and periphery in PAMAM-G3-treated mice. Neutrophils are a key component of the acute inflammatory response, which is required for initiation of an immune response. The subsequent elimination of neutrophils and resolution of inflammation promote the transition from innate immunity to adaptive immunity.39, 40, 41 However, in cancer, this response goes awry, and neutrophils are often reprogrammed to become tumor-associated neutrophils (or granulocytic myeloid-derived suppressor cells), which help promote cancer growth. Similarly, i-monocytes or monocyte-derived cells that infiltrate tumors can differentiate into monocytic myeloid-derived suppressor cells.
In the clinical setting, breast cancer metastasis is most often diagnosed after the patient has undergone surgical resection of the primary tumor. It is known that DAMPs are released from stressed/dying tumor cells, and DAMP release is accelerated with standard-of-care cytotoxic chemotherapy and surgical therapies.13, 14, 15, 16, 17, 18, 19, 20, 21, 22 Therefore, we tested the utility of PAMAM-G3 in a clinically relevant mouse model in which cancer metastasis was monitored after surgical resection of the primary tumor, closely resembling standard-of-care for breast cancer patients. In this model, we observed a significant reduction in lung metastasis in mice treated with PAMAM-G3 (Figure 5). These data provide guidance on how to incorporate NA DAMP-scavenging agents into standard-of-care treatment of women with breast cancer to attempt to limit metastatic spread of their cancer. Inflammation blocking with NAS might be particularly useful during surgical intervention, as surgery has been shown to increase the levels of circulating DAMPs,28 which might promote metastasis.13, 14, 15, 16 Importantly, this treatment approach should be generalizable to any type of breast cancer. Whereas targeted therapies are available for receptor-positive cancers, patients with triple-negative breast cancer (TNBC) have limited options. Although TNBC accounts for roughly 10% of all breast cancers, it is more common in younger patients and has a worse prognosis due to high rates of local recurrence and distant metastasis.51 The study results provide a strong rationale for inclusion of inflammation-blocking agents such as NAS in cancer therapy and facilitates the development of a novel class of anti-cancer agents to mitigate counterproductive inflammation in women with breast cancer.
Materials and methods
Ethics statement
All in vivo experiments were conducted in accordance with protocols approved by Duke University Institutional Animal Care and Use Committee (IACUC).
Cell lines
The 4T1-luciferase murine mammary carcinoma cell line stably expressing firefly luciferase gene was generously provided by Mark Dewhirst, PhD, at Duke University Medical Center. Cells were passaged for less than 5 generations before use in animal experiments. Cell lines were tested and found to be free of mycoplasma and mouse pathogens. The 4T1 cell line is considered a TNBC model (ER-PR-HER2-). 4T1-luciferase cells were grown in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% fetal bovine serum (FBS), 1 mM sodium pyruvate, and 4 mM glutamine, in a 37°C humidified 5% CO2 incubator.
Reagents
PAMAM-G3 was purchased from Sigma-Aldrich.
Mouse models of breast cancer
Intravenous experimental lung metastasis mouse model of breast cancer
Female BALB/c mice (Jackson Laboratory) were injected intravenously in the tail vein with 104 4T1-luciferase murine tumor cells (day 0). Mice were weighed and treated according to weight with either PAMAM-G3 (20 mg/kg) or PBS via intraperitoneal injection. Mice were injected with PAMAM-G3 or PBS on days −5/−4 and −2 prior to tumor injection on day 0. Following tumor injection on day 0, they were treated with PAMAM-G3 or PBS on days 2, 6, 9, and 12/13. Mice were monitored throughout the course of the study with the PerkinElmer IVIS (In Vivo Imaging System) Lumina XRMS100, starting on days 3−4 post-tumor injection. Imaging was performed three times per week. Mice were euthanized after observing a detectable signal in lungs that remained constant or increased in three consecutive luciferase imaging sessions. This method is chosen to prevent unnecessary suffering to mice due to tumor burden in the lungs and possible respiratory distress. All mice were euthanized when 50% of the mice in the control/PBS-treated group succumbed to tumor burden. In general, all mice were euthanized around days 15−17. Mice were euthanized using approved protocols, and lungs were harvested, photographed, weighed, and imaged to record bioluminescence ex vivo. In addition, lungs and spleens were collected and saved for flow cytometry and histology. Cardiac puncture was performed to collect blood immediately after CO2 euthanasia for analysis of blood cell counts, cfDNA, and cytokines/chemokines.
Naive, nontumor-bearing, female, age-matched BALB/c mice were used as controls. Naive mice were untreated or treated with either PAMAM-G3 (20 mg/kg) or PBS (6 injections, 3 days apart). Lungs, spleen, and blood were collected and analyzed in a similar manner as experimental mice.
Postsurgical-resection lung metastasis mouse model of breast cancer
Female BALB/c mice (Jackson Laboratory) were injected with 4T1-luciferase cells (8 × 105) orthotopically in the left abdominal mammary fat pad #4 on day 0. Mice were weighed and treated according to weight with either PAMAM-G3 (20 mg/kg) or PBS via intraperitoneal injection on days 0, 3, 10, 13, 16, and 20. Tumors were surgically resected on day 7 post-tumor implantation (tumors are 5−7 mm diameter on day 7). Mice were subcutaneously injected with a nonsteroidal anti-inflammatory drug, 15 min prior to surgery. Resections were performed under a general inhalation anesthetic (Isoflurane). Once mice were completely anesthetized, the surgery site was shaved and sterilized with 70% alcohol, followed by 2% chlorhexidine solution. Incisions were made to the right of the tumor using an 8-mm surgical blade, and tumors were removed using sterile forceps and scissors by gently splaying the tumor away from the dermis and intraperitoneal wall. Once the tumor was removed, the dermis was closed using wound clips. A local anesthetic was then applied to the surgical site. Mice were closely monitored until they fully recovered. Another subcutaneous injection of a nonsteroidal anti-inflammatory was administered at 24 h post-surgery. Surgery wound clips were removed 7 days after surgery. Mice were monitored throughout the course of the study with the PerkinElmer IVIS Lumina XRMS100 starting 7 days post-tumor resection. Imaging was performed three times per week.
Mice were euthanized after observing a detectable signal in lungs that remained constant or increased in three consecutive luciferase imaging sessions. This method is chosen to prevent unnecessary suffering to mice due to tumor burden in the lungs and possible respiratory distress. Mice were also euthanized if we observed tumor regrowth at the primary/orthotopic site that reached 2,000 mm3. All mice were euthanized when 50% of the mice in the control/PBS-treated group succumbed to tumor burden. In general, all mice were euthanized, and the study was terminated around days 22−24. Mice were euthanized using approved protocols, and lungs were harvested, photographed, and imaged to record bioluminescence ex vivo.
Luciferase imaging to monitor tumor metastasis
Mice were intraperitoneally injected with 200 μL (150 mg/kg) of VivoGlo Luciferin (Promega). 15 min following the injection, mice were anesthetized (Isoflurane) and imaged with the PerkinElmer IVIS Lumina XRMS100. After imaging, mouse recovery was closely monitored. On the last day of study, mice were intraperitoneally injected with 200 μL (150 mg/kg) of VivoGlo Luciferin. After live imaging, mice were immediately euthanized, and lungs were harvested from each mouse for ex vivo quantification of bioluminescence.
Living Image software was used to record IVIS data. The parameters used to acquire IVIS data (lens aperture size [f/stop] and exposure time) were consistent for the duration of the study. The ROI (region-of-interest) area was identical for all images. Bioluminescence for the ROI was defined automatically. Bioluminescence is expressed as total flux (photons/second).
Flow cytometry
Mouse lung tissue was dissociated by enzymatic degradation using the Lung Dissociation Kit enzymes and protocol (Miltenyi Biotec), followed by mechanical dissociation using the gentleMACS Dissociator (Miltenyi Biotec). Once fully dissociated, lung cells were strained through a 70-μm cell strainer to obtain a single cell suspension. Spleens were placed in a 70-μm cell strainer, minced using scissors, and gently and repeatedly pressed through the strainer with a 5-mL syringe plug to obtain a single cell suspension. Lung and spleen single cell suspensions were subsequently analyzed by flow cytometry. Cells were incubated with anti-mouse-CD16/CD32 Fc blocking antibody for 15 min. Cells were then stained with the following antibody panels (BioLegend) for 20 min: (1) live/dead, Ly6G-FITC, Ly6C-PE, MHC-II-PC7, CD11c-APC, CD45.2-APC-Cy7, CD64-BV421, CD24-BV510, and CD11b-BV785; (2) live/dead, Ly6G-FITC, Ly6C-PE, F4/80-PC7, CD11c-APC, CD45.2-APC-Cy7, CD11b-BV421, and CD103-BV510; and (3) live/dead, NK1.1-FITC, CD11b-PE, CD45.2-PC7, CD8-APC, B220-APC-Cy7, and CD4-BV421. Stained cells were immediately acquired using a 13-color CytoFLEX flow cytometer (Beckman Coulter). Data were analyzed using Kaluza Software (Beckman Coulter).
Cytokine arrays
The BioLegend LEGENDplex Mouse Anti-Virus Response Panel is a bead-based multiplex assay that allows quantification of 13 mouse cytokines, including interferons (IFNs; α, β, γ), ILs (1β, 6, 10, 12), chemokines (CCL2 [MCP-1], CCL5 [RANTES], CXCL1 [KC], CXCL10 [IP-10]), TNF-α, and GM-CSF. Blood was obtained from experimental mice, and plasma was used to analyze circulating levels of inflammatory cytokines and chemokines using LEGENDplex. For lung supernatants, lung tissue was minced using an 8-mm surgical blade in 10% FBS/RPMI and incubated for 18 h prior to analysis. Supernatants were collected at the end of incubation and analyzed using LEGENDplex. Samples were acquired using a CytoFLEX flow cytometer, and data were analyzed using BioLegend’s Bio-Bits cloud-based software platform.
Lung immunopathology
Lungs were obtained from experimental mice and fixed in 10% formalin for 24 h, followed by 70% ethanol. They were then processed and sectioned for subsequent H&E staining. All processing and staining services were performed by Histowiz (Brooklyn, NY, USA).
Circulating cfDNA assay
Total DNA was obtained from plasma using the QIAamp DNA Blood Mini Kit (QIAGEN). Once purified, cfDNA concentration was determined using the Quant-iT PicoGreen assay (Invitrogen).
Statistical analysis
Statistical analyses were performed using Prism 6 (GraphPad Software), as specified in figure legends. Error bars are ± standard error of the mean (SEM). Statistical significance indicates p <0.05.
Acknowledgments
We are grateful to Rachel Rempel, PhD, and Ibtehaj Naqvi, MD, PhD, for reviewing the manuscript and providing insightful comments and Ibtehaj Naqvi, MD, PhD, for the graphical abstract. This study was funded by the Department of Defense Breast Cancer Research Program awards W81XWH-16-1-0512 (to S.K.N.) and W81XWH-16-1-0513 (to B.S.), National Cancer Institute Advanced Immunobiology Training Program for Surgeons T32AI141342-01 award from 2019 to 2020 (to K.L.), and National Cancer Institute Translation Research in Surgical Oncology T32CA093245-12 award from 2017 to 2019 (to K.L.).
Author contributions
Study conceptualization and resources, S.K.N. and B.S.; study design and data analysis, S.K.N., E.K.H., and V.F.; study implementation, E.K.H., V.F., D.B., and K.L.; manuscript writing − original draft, S.K.N.; manuscript editing & review, E.K.H., V.F., K.L., D.B., and B.S.
Declaration of interests
Duke University has submitted a patent application on the use of nucleic acid scavengers to limit cancer progression, and B.S. is named as an inventor on this patent. All other authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2020.12.026.
Supplemental information
References
- 1.Krieg A.M. CpG motifs in bacterial DNA and their immune effects. Annu. Rev. Immunol. 2002;20:709–760. doi: 10.1146/annurev.immunol.20.100301.064842. [DOI] [PubMed] [Google Scholar]
- 2.Kono H., Rock K.L. How dying cells alert the immune system to danger. Nat. Rev. Immunol. 2008;8:279–289. doi: 10.1038/nri2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medzhitov R., Janeway C.A., Jr. Innate immunity: impact on the adaptive immune response. Curr. Opin. Immunol. 1997;9:4–9. doi: 10.1016/s0952-7915(97)80152-5. [DOI] [PubMed] [Google Scholar]
- 4.Palm N.W., Medzhitov R. Pattern recognition receptors and control of adaptive immunity. Immunol. Rev. 2009;227:221–233. doi: 10.1111/j.1600-065X.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- 5.Leulier F., Lemaitre B. Toll-like receptors--taking an evolutionary approach. Nat. Rev. Genet. 2008;9:165–178. doi: 10.1038/nrg2303. [DOI] [PubMed] [Google Scholar]
- 6.Kawai T., Akira S. TLR signaling. Semin. Immunol. 2007;19:24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Takeda K., Akira S. Toll-like receptors in innate immunity. Int. Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 8.Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 9.Marshak-Rothstein A., Rifkin I.R. Immunologically active autoantigens: the role of toll-like receptors in the development of chronic inflammatory disease. Annu. Rev. Immunol. 2007;25:419–441. doi: 10.1146/annurev.immunol.22.012703.104514. [DOI] [PubMed] [Google Scholar]
- 10.O’Neill L.A. Primer: Toll-like receptor signaling pathways--what do rheumatologists need to know? Nat. Clin. Pract. Rheumatol. 2008;4:319–327. doi: 10.1038/ncprheum0802. [DOI] [PubMed] [Google Scholar]
- 11.Chen K., Huang J., Gong W., Iribarren P., Dunlop N.M., Wang J.M. Toll-like receptors in inflammation, infection and cancer. Int. Immunopharmacol. 2007;7:1271–1285. doi: 10.1016/j.intimp.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 12.Deane J.A., Bolland S. Nucleic acid-sensing TLRs as modifiers of autoimmunity. J. Immunol. 2006;177:6573–6578. doi: 10.4049/jimmunol.177.10.6573. [DOI] [PubMed] [Google Scholar]
- 13.Al-Sahaf O., Wang J.H., Browne T.J., Cotter T.G., Redmond H.P. Surgical injury enhances the expression of genes that mediate breast cancer metastasis to the lung. Ann. Surg. 2010;252:1037–1043. doi: 10.1097/SLA.0b013e3181efc635. [DOI] [PubMed] [Google Scholar]
- 14.Minn A.J., Gupta G.P., Siegel P.M., Bos P.D., Shu W., Giri D.D., Viale A., Olshen A.B., Gerald W.L., Massagué J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Retsky M., Demicheli R., Hrushesky W.J., Forget P., De Kock M., Gukas I., Rogers R.A., Baum M., Sukhatme V., Vaidya J.S. Reduction of breast cancer relapses with perioperative non-steroidal anti-inflammatory drugs: new findings and a review. Curr. Med. Chem. 2013;20:4163–4176. doi: 10.2174/09298673113209990250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neeman E., Zmora O., Ben-Eliyahu S. A new approach to reducing postsurgical cancer recurrence: perioperative targeting of catecholamines and prostaglandins. Clin. Cancer Res. 2012;18:4895–4902. doi: 10.1158/1078-0432.CCR-12-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosseini H., Obradović M.M.S., Hoffmann M., Harper K.L., Sosa M.S., Werner-Klein M., Nanduri L.K., Werno C., Ehrl C., Maneck M. Early dissemination seeds metastasis in breast cancer. Nature. 2016;540:552–558. doi: 10.1038/nature20785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harper K.L., Sosa M.S., Entenberg D., Hosseini H., Cheung J.F., Nobre R., Avivar-Valderas A., Nagi C., Girnius N., Davis R.J. Mechanism of early dissemination and metastasis in Her2+ mammary cancer. Nature. 2016;540:588–592. doi: 10.1038/nature20609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park C.G., Hartl C.A., Schmid D., Carmona E.M., Kim H.J., Goldberg M.S. Extended release of perioperative immunotherapy prevents tumor recurrence and eliminates metastases. Sci. Transl. Med. 2018;10:eaar1916. doi: 10.1126/scitranslmed.aar1916. [DOI] [PubMed] [Google Scholar]
- 20.Revesz L. Effect of tumour cells killed by x-rays upon the growth of admixed viable cells. Nature. 1956;178:1391–1392. doi: 10.1038/1781391a0. [DOI] [PubMed] [Google Scholar]
- 21.Huang Q., Li F., Liu X., Li W., Shi W., Liu F.F., O’Sullivan B., He Z., Peng Y., Tan A.C. Caspase 3-mediated stimulation of tumor cell repopulation during cancer radiotherapy. Nat. Med. 2011;17:860–866. doi: 10.1038/nm.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sulciner M.L., Serhan C.N., Gilligan M.M., Mudge D.K., Chang J., Gartung A., Lehner K.A., Bielenberg D.R., Schmidt B., Dalli J. Resolvins suppress tumor growth and enhance cancer therapy. J. Exp. Med. 2018;215:115–140. doi: 10.1084/jem.20170681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hawes M.C., Wen F., Elquza E. Extracellular DNA: A Bridge to Cancer. Cancer Res. 2015;75:4260–4264. doi: 10.1158/0008-5472.CAN-15-1546. [DOI] [PubMed] [Google Scholar]
- 24.Schwarzenbach H., Hoon D.S., Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 25.Pandey S., Singh S., Anang V., Bhatt A.N., Natarajan K., Dwarakanath B.S. Pattern Recognition Receptors in Cancer Progression and Metastasis. Cancer Growth Metastasis. 2015;8:25–34. doi: 10.4137/CGM.S24314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhatelia K., Singh K., Singh R. TLRs: linking inflammation and breast cancer. Cell. Signal. 2014;26:2350–2357. doi: 10.1016/j.cellsig.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 27.Wittwer C., Boeck S., Heinemann V., Haas M., Stieber P., Nagel D., Holdenrieder S. Circulating nucleosomes and immunogenic cell death markers HMGB1, sRAGE and DNAse in patients with advanced pancreatic cancer undergoing chemotherapy. Int. J. Cancer. 2013;133:2619–2630. doi: 10.1002/ijc.28294. [DOI] [PubMed] [Google Scholar]
- 28.Naqvi I., Gunaratne R., McDade J.E., Moreno A., Rempel R.E., Rouse D.C., Herrera S.G., Pisetsky D.S., Lee J., White R.R., Sullenger B.A. Polymer-Mediated Inhibition of Pro-invasive Nucleic Acid DAMPs and Microvesicles Limits Pancreatic Cancer Metastasis. Mol. Ther. 2018;26:1020–1031. doi: 10.1016/j.ymthe.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merrell M.A., Ilvesaro J.M., Lehtonen N., Sorsa T., Gehrs B., Rosenthal E., Chen D., Shackley B., Harris K.W., Selander K.S. Toll-like receptor 9 agonists promote cellular invasion by increasing matrix metalloproteinase activity. Mol. Cancer Res. 2006;4:437–447. doi: 10.1158/1541-7786.MCR-06-0007. [DOI] [PubMed] [Google Scholar]
- 30.Ren T., Wen Z.K., Liu Z.M., Liang Y.J., Guo Z.L., Xu L. Functional expression of TLR9 is associated to the metastatic potential of human lung cancer cell: functional active role of TLR9 on tumor metastasis. Cancer Biol. Ther. 2007;6:1704–1709. doi: 10.4161/cbt.6.11.4826. [DOI] [PubMed] [Google Scholar]
- 31.Wen F., Shen A., Choi A., Gerner E.W., Shi J. Extracellular DNA in pancreatic cancer promotes cell invasion and metastasis. Cancer Res. 2013;73:4256–4266. doi: 10.1158/0008-5472.CAN-12-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J., Sohn J.W., Zhang Y., Leong K.W., Pisetsky D., Sullenger B.A. Nucleic acid-binding polymers as anti-inflammatory agents. Proc. Natl. Acad. Sci. USA. 2011;108:14055–14060. doi: 10.1073/pnas.1105777108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holl E.K., Shumansky K.L., Borst L.B., Burnette A.D., Sample C.J., Ramsburg E.A., Sullenger B.A. Scavenging nucleic acid debris to combat autoimmunity and infectious disease. Proc. Natl. Acad. Sci. USA. 2016;113:9728–9733. doi: 10.1073/pnas.1607011113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holl E.K., Shumansky K.L., Pitoc G., Ramsburg E., Sullenger B.A. Nucleic acid scavenging polymers inhibit extracellular DNA-mediated innate immune activation without inhibiting anti-viral responses. PLoS ONE. 2013;8:e69413. doi: 10.1371/journal.pone.0069413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai S.M., Sheng J., Gupta P., Renia L., Duan K., Zolezzi F., Karjalainen K., Newell E.W., Ruedl C. Organ-Specific Fate, Recruitment, and Refilling Dynamics of Tissue-Resident Macrophages during Blood-Stage Malaria. Cell Rep. 2018;25:3099–3109.e3. doi: 10.1016/j.celrep.2018.11.059. [DOI] [PubMed] [Google Scholar]
- 36.Chen Z., Feng X., Herting C.J., Garcia V.A., Nie K., Pong W.W., Rasmussen R., Dwivedi B., Seby S., Wolf S.A. Cellular and Molecular Identity of Tumor-Associated Macrophages in Glioblastoma. Cancer Res. 2017;77:2266–2278. doi: 10.1158/0008-5472.CAN-16-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salmon H., Idoyaga J., Rahman A., Leboeuf M., Remark R., Jordan S., Casanova-Acebes M., Khudoynazarova M., Agudo J., Tung N. Expansion and Activation of CD103(+) Dendritic Cell Progenitors at the Tumor Site Enhances Tumor Responses to Therapeutic PD-L1 and BRAF Inhibition. Immunity. 2016;44:924–938. doi: 10.1016/j.immuni.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kitamura T., Doughty-Shenton D., Cassetta L., Fragkogianni S., Brownlie D., Kato Y., Carragher N., Pollard J.W. Monocytes Differentiate to Immune Suppressive Precursors of Metastasis-Associated Macrophages in Mouse Models of Metastatic Breast Cancer. Front. Immunol. 2018;8:2004. doi: 10.3389/fimmu.2017.02004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greenlee-Wacker M.C. Clearance of apoptotic neutrophils and resolution of inflammation. Immunol. Rev. 2016;273:357–370. doi: 10.1111/imr.12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones H.R., Robb C.T., Perretti M., Rossi A.G. The role of neutrophils in inflammation resolution. Semin. Immunol. 2016;28:137–145. doi: 10.1016/j.smim.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 41.Geering B., Stoeckle C., Conus S., Simon H.U. Living and dying for inflammation: neutrophils, eosinophils, basophils. Trends Immunol. 2013;34:398–409. doi: 10.1016/j.it.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 42.Rakoff-Nahoum S., Medzhitov R. Toll-like receptors and cancer. Nat. Rev. Cancer. 2009;9:57–63. doi: 10.1038/nrc2541. [DOI] [PubMed] [Google Scholar]
- 43.Mehmeti M., Allaoui R., Bergenfelz C., Saal L.H., Ethier S.P., Johansson M.E., Jirström K., Leandersson K. Expression of functional toll like receptor 4 in estrogen receptor/progesterone receptor-negative breast cancer. Breast Cancer Res. 2015;17:130. doi: 10.1186/s13058-015-0640-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.González-Reyes S., Marín L., González L., González L.O., del Casar J.M., Lamelas M.L., González-Quintana J.M., Vizoso F.J. Study of TLR3, TLR4 and TLR9 in breast carcinomas and their association with metastasis. BMC Cancer. 2010;10:665. doi: 10.1186/1471-2407-10-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palha De Sousa C., Blum C.M., Sgroe E.P., Crespo A.M., Kurt R.A. Murine mammary carcinoma cells and CD11c(+) dendritic cells elicit distinct responses to lipopolysaccharide and exhibit differential expression of genes required for TLR4 signaling. Cell. Immunol. 2010;266:67–75. doi: 10.1016/j.cellimm.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Todorova J., Pasheva E. High mobility group B1 protein interacts with its receptor RAGE in tumor cells but not in normal tissues. Oncol. Lett. 2012;3:214–218. doi: 10.3892/ol.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tafani M., Schito L., Pellegrini L., Villanova L., Marfe G., Anwar T., Rosa R., Indelicato M., Fini M., Pucci B., Russo M.A. Hypoxia-increased RAGE and P2X7R expression regulates tumor cell invasion through phosphorylation of Erk1/2 and Akt and nuclear translocation of NF-kappaB. Carcinogenesis. 2011;32:1167–1175. doi: 10.1093/carcin/bgr101. [DOI] [PubMed] [Google Scholar]
- 48.Kostova N., Zlateva S., Ugrinova I., Pasheva E. The expression of HMGB1 protein and its receptor RAGE in human malignant tumors. Mol. Cell. Biochem. 2010;337:251–258. doi: 10.1007/s11010-009-0305-0. [DOI] [PubMed] [Google Scholar]
- 49.Kang R., Tang D., Schapiro N.E., Loux T., Livesey K.M., Billiar T.R., Wang H., Van Houten B., Lotze M.T., Zeh H.J. The HMGB1/RAGE inflammatory pathway promotes pancreatic tumor growth by regulating mitochondrial bioenergetics. Oncogene. 2014;33:567–577. doi: 10.1038/onc.2012.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan W., Chang Y., Liang X., Cardinal J.S., Huang H., Thorne S.H., Monga S.P., Geller D.A., Lotze M.T., Tsung A. High-mobility group box 1 activates caspase-1 and promotes hepatocellular carcinoma invasiveness and metastases. Hepatology. 2012;55:1863–1875. doi: 10.1002/hep.25572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Foulkes W.D., Smith I.E., Reis-Filho J.S. Triple-negative breast cancer. N. Engl. J. Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.